
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
TECHNOLOGY AND CODE article
Front. Earth Sci. , 24 September 2020
Sec. Biogeoscience
Volume 8 - 2020 | https://doi.org/10.3389/feart.2020.585087
The flow of energy and elements between the geosphere and biosphere can be traced through changing redox chemistry of Earth’s surface. Deep-time trends in the mineral record, including mineral age and elemental composition, reveal a dynamic history of changing redox states and chemical speciation. We present a user-friendly exploratory network analysis platform called dragon (Deep-time Redox Analysis of the Geobiology Ontology Network) to facilitate investigation of the expanding redox chemical network preserved in the mineral record throughout Earth’s history and beyond. Given a user-indicated focal element or set of focal elements, dragon constructs interactive bipartite networks of minerals and their constituent elements over a specified range in geologic-time using information from the Mineral Evolution Database (https://rruff.info/evolution/). Written in the open-source language R as a Shiny application, dragon launches a browser-based dashboard to explore mineral evolution in deep-time. We demonstrate dragon’s utility through examining the mineral chemistry of lithium over deep-time. dragon is freely available from CRAN under a GPL-3 License, with source code and documentation hosted at https://github.com/sjspielman/dragon.
Major events in Earth history, such as the formation of continents (Cawood et al., 2018), enhanced chemical weathering (Satkoski et al., 2016), and atmospheric oxygenation (Farquhar et al., 2000) have dramatically influenced the chemistry and redox state of Earth’s atmosphere, oceans, and crust. Changes in Earth’s chemistry are accompanied by shifts in reduction/oxidation potential (also known as redox potential), which measures the propensity of a chemical species to gain negatively-charged electrons and thereby be reduced. Earth surface redox conditions govern the flow of electrons among chemical species in aqueous systems and directly influenced microbial metabolic pathways and the chemistry of potential metal cofactors preserved in minerals during the Hadean and Archean Eons (Jelen et al., 2016; Morrison et al., 2018). A thorough understanding of Earth’s redox evolution and its specific impacts on planetary surface chemistry is crucial for identifying the driving forces behind planetary evolution and habitability.
Minerals comprise an abundant source of geochemical evidence for reconstructing Earth’s redox history (Golden et al., 2013; Liu et al., 2016). A mineral’s elemental composition implicitly records information about the chemical speciation and redox state of the surrounding environment, as well as the bioavailability of life’s critical elements, at the time of mineral formation (Hazen et al., 2008).
The Mineral Evolution Database [https://rruff.info/evolution/ (Golden et al., 2019), last accessed 8/24/20] is a comprehensive resource for studying mineralogy in geologic time. It contains the chemical formulas, element redox states, and age data (including youngest- and oldest-known ages) for documented mineral occurrences of 5,603 known minerals, nearly 5,000 of which have associated locality pairings, found throughout Earth’s history and from extraterrestrial sources such as asteroids, meteorites, and pre-solar sources. However, analyzing and visualizing this extensive data resource remains a considerable challenge due to limited cyberinfrastructure resources and methodologies.
Network analysis has emerged as a useful tool for investigating mineralogical systems, allowing researchers to investigate the evolution of Earth’s surface over spatial and temporal scales (Morrison et al., 2017; Hazen et al., 2019; Hystad et al., 2019). In particular, bipartite networks, which feature two distinct types of nodes, are well-suited for mineral-chemistry analysis as they allow for precise examination of the associations between minerals and their constituent elements. Recent research has successfully employed this approach to interrogate relationships between minerals and inherent properties of their constituent elements for the redox evolution of both cobalt (Co) (Moore et al., 2018) and vanadium (V) (Moore et al., 2020). As such, data-driven network analysis represents an emerging and promising avenue for discoveries in Earth sciences.
Here, we introduce a user-friendly platform to facilitate network-based exploratory analysis of Earth’s mineral-chemistry network over geologic time scales called dragon (Deep-time Redox Analysis of the Geobiology Ontology Network). We demonstrate how dragon can be used to reveal trends in the evolving redox history of Earth’s surface and crust. These trends can in turn be used to generate testable hypotheses about factors that impacted geochemical cycling and the evolution of metabolic electron transfer.
Written in the R open-source language (R Core Team, 2019) using the shiny package (Chang et al., 2020), dragon is an interactive browser-based application that allows users of any disciplinary background to explore, manipulate, visualize, and statistically analyze mineral-chemistry networks at a user-selected time range within the 4.7 billion year history of the mineral record of Earth and extraterrestrial sources (i.e., meteorites, asteroids, pre-solar sources). Specifically, given a focal element or set of focal elements and a specified time range, dragon constructs a bipartite network, where the two node classes represent elements and minerals, using information from the Mineral Evolution Database (Golden et al., 2019). Within this framework, edges connect minerals to their constituent elements, which in turn are connected to all minerals where the element is found (Figure 1A). Core dragon functions are performed using several R packages including igraph for network construction and evaluation (Csardi and Nepusz, 2006), core tidyverse libraries for data management and manipulation (Wickham et al., 2019), and visNetwork for rendering of the interactive network (Almende et al., 2019).
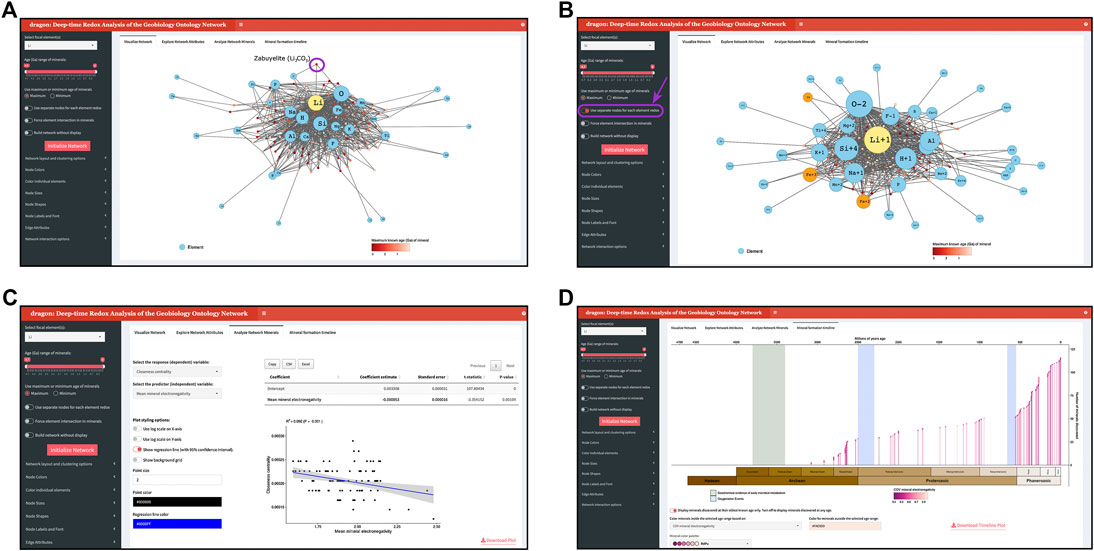
FIGURE 1. Screenshots of dragon views of the lithium (Li) mineral-chemistry network. Larger labeled nodes represent elements in the network, and smaller unlabeled nodes represent minerals in the network. (A) Li mineral-chemistry network for all known Li-containing minerals. Element nodes are shown in light blue (with Li in yellow for emphasis), and mineral nodes are colored based on their maximum known age. (B) Li mineral-chemistry network for all known Li-containing minerals, where element nodes are separated based on their redox state. For example, there are now three different nodes for iron (Fe) (colored orange) for each of the Fe redox states present in the network’s minerals: Fe2+, Fe3+, and Fe [representing unknown redox state(s)]. (C) Example of a linear model analysis examining the relationship between mean mineral electronegativity and node closeness centrality for the Li mineral-chemistry network shown in panel (A). (D) Mineral formation timeline for all Li-containing minerals, where each line and point represents the oldest-known age of each mineral in the network.
dragon is organized into four main tabs: 1) “Visualize Network” for network construction and dynamic visualization, 2) “Explore Network Attributes” for examining various properties of each node in the constructed network, 3) “Analyze Network Minerals” for analyzing attributes of mineral nodes in the constructed network, and 4) “Mineral formation timeline” for visualizing the scope of mineral formation in the context of major oxygenation events in Earth’s history and geochemical evidence for early Archean microbial metabolic pathways.
Upon launching dragon, users select their desired network focal element(s) and an age range for minerals to include in the network. dragon will include all minerals that contain the element(s) whose oldest, or youngest if the user prefers, known age, based on mineral discovery information provided by MED, is within the selected age range. If multiple focal elements are selected, dragon will, by default, construct a network containing all minerals in the selected age range which contain at least one of the focal elements. By instead activating the feature “Force element intersection in minerals,” dragon will construct a network featuring only those minerals which contain all focal elements. dragon additionally supports two modes to render element nodes: The default mode will create a single node for each element, and the redox mode (“Use separate nodes for each element redox”) will create a separate element node for each element redox state that exists in the network (Figure 1B). Finally, for extremely large networks (e.g., the full network containing all elements and minerals) which may experience prohibitive rendering times, dragon offers the option to construct the network for analysis without the interactive display (“Build network without display”).
After setting these baseline options, clicking the “Initialize Network” button will trigger dragon to construct and render the specified bipartite network for dynamic interactive visualization, exploration, and analysis. Users can further modify stylistic components of the network display. First, users can specify a network layout from several options for deterministic, dynamic physics, and force-directed algorithms, with an option to set the random seed for stochastic network layout algorithms to ensure reproducibility. By default, dragon will use the force-directed Fruchterman–Reingold algorithm (Fruchterman and Reingold, 1991) to set initial node positions. Node position can be further customized through clicking on and dragging nodes to their desired location. Users can additionally select the algorithm that performs network community clustering [performed by igraph (Csardi and Nepusz, 2006)] from either the (default) Louvain (Blondel et al., 2008) or Leading Eigenvector (Newman, 2006) community detection methods. Network appearance, including node color, shape, and size as well as edge color and weight, can either be set according to user preference (i.e., set all element nodes to be a specific color or size), or according to dozens of node-specific attributes (Table 1). When coloring nodes or edges by a given attribute, users can choose a color scheme from a set of colorblind-friendly ColorBrewer palettes (Neuwirth, 2014). We emphasize that, given dragon’s dynamic and interactive nature, network edge lengths do not carry specific meaning.
Users can directly obtain information and attributes about nodes in the network. Hovering over any given node will reveal various key attributes, e.g., number of known localities for mineral nodes or Pauling electronegativity (Pauling, 1932) for element nodes. When a given node is clicked, a table in the dashboard box below the interactive network entitled “Examine individual nodes” will reveal all of the given nodes’ first-degree connections. Users can also directly select nodes for more in-depth attribute examination using an associated dropdown menu in the “Examine individual nodes” box, and the resulting table can be exported in either CSV or Excel format.
dragon’s second tab “Explore Network Attributes” contains tables of all element and mineral node attributes (Table 1) calculated and consumed by dragon, as well as the number of element and mineral nodes, the number of edges, and the modularity of the network as determined by community detection. These tables can be also exported for external use in either CSV or Excel format.
dragon’s third tab “Analyze Network Minerals” allows users to analyze properties of minerals in the current network by constructing linear models for a given response and predictor variable (Figure 1C). For example, among minerals in the currently rendered network, users can examine the strength of the relationship between the maximum known age of each mineral and the number of localities at which each has been recovered. Notably, this tab includes the option to assess whether certain mineral properties statistically differ across community clusters, using both an ANOVA and a post-hoc Tukey test to directly compare network clusters to one another. To ensure robust statistical interpretation, dragon will check that all such comparisons contain sufficient amounts of data and adhere to modeling assumptions such as equal variance among groups. That said, users must take care to perform and interpret analysis with their own scientific goals in mind. While this feature enables construction of linear models, it does not transform data or assess any other assumptions of linear models before analyzing the data.
Finally, dragon’s fourth tab “Mineral formation timeline” provides a chronological view of mineral formation scaffolded along geologic eras, including periods of tectonic assembly, oxygenation events in Earth’s history (Farquhar et al., 2000; Scott et al., 2008; Och and Shields-Zhou, 2012; Sahoo et al., 2012; Luo et al., 2016; Warke et al., 2020), and early geochemical evidence for microbial metabolisms (Moore et al., 2017) (Figure 1D).
dragon is released as an R package, structured with the robust golem framework for building and managing robust Shiny applications. Maintained on GitHub with Travis-CI (https://travis-ci.org/), source code and full documentation are available from https://github.com/sjspielman/dragon. dragon can either be run locally or can be accessed freely on a dedicated server whose current link is available from the dragon GitHub site. Locally, dragon can be run on any standard operating system (MacOS, Windows, and Linux) where R and associated dependencies have been installed. The dragon library is freely available for download from CRAN, the central repository for open-source R packages.
We present an example of performing network analysis with dragon by exploring the evolution of minerals containing lithium (Li) over deep time (Figure 1). Unlike other elements with complex redox chemistry, such as Fe or S, Li has straightforward redox chemistry and relatively smaller associated networks. We emphasize that dragon is designed to analyze complex and larger mineral-chemistry networks with complex redox changes, as associated with oxygenation events for example (Scott et al., 2008; Och and Shields-Zhou, 2012; Sahoo et al., 2012; Warke et al., 2020), but we focus on the simpler Li network here to clearly showcase dragon’s functionality.
The alkali metal Li is the third lightest chemical element with just three protons in its nucleus and is predicted to have been one of the three elements synthesized in the big bang (Boesgaard and Steigman, 1985). Figure 1A highlights the bipartite nature of dragon networks within the full lithium network: Edges connect the mineral node zabuyelite (Li2CO3) to element nodes Li, C, and O. Here, element nodes are sized according to network degree centrality, and minerals are colored by their maximum known age. Figure 1B displays the same network as in Figure 1A but with the setting “Use separate nodes for each element redox” turned on. While lithium has only a single redox state in the network (Li1+), many other elements that form minerals with Li have different redox states in different mineral species, e.g., there are now three separates nodes representing iron (Fe2+, Fe3+, and Fe for unknown redox states).
Figure 1C demonstrates a linear modeling analysis performed on minerals in this full Li network, revealing a significant, negative relationship between mineral closeness centrality (a measure of a given node’s average inverse distance from other network nodes) and mean mineral electronegativity, calculated as the average Pauling scale electronegativity (Pauling, 1932) for all elements in a given mineral. In other words, minerals with higher mean electronegativities tend to be less central in this network. We emphasize that results from dragon’s linear models must be interpreted with caution on a case-by-case basis as it applies to a given scientific question. Finally, Figure 1D depicts the “Mineral Formation Timeline” view, where minerals have been colored according to their mean electronegativity. By default, this tab displays each mineral in the network at its oldest-discovered age.
Notably, the lithium network shown in Figures 1A,B is constructed from all known Li-minerals. One of dragon’s key features is the ability to construct networks that consider only minerals formed within a specified time range, thereby allowing for exploration of changing redox trends in mineral formation over deep-time. In Figure 2, we show the Li mineral-chemistry network, with element nodes separated by redox state, across three different points in time: all Li minerals dated to ≥2.5 Ga (billions of years ago; Figures 2A,B), all Li minerals dated to ≥1.5 Ga (Figures 2C,D), and finally all known Li minerals at present day (Figures 2E,F). We particularly emphasize how lithium forms minerals with iron (Fe) and manganese (Mn), two elements crucial to metabolic processes which exist at a range of redox states. We find that, at ≥2.5 Ga, Fe2+ does not form Li minerals with Mn at any redox state (Figure 2A), but Fe3+ forms Li minerals with Mn2+ (Figure 2B). Moving forward in time to the Li mineral-chemistry network at ≥1.5 Ga, we find that Fe2+ now indeed forms Li minerals with Mn2+, and Fe3+ has expanded to also form Li minerals with Mn3+ in addition to Mn2+. Finally, in the full Li network at 0 Ga, Fe2+ forms Li minerals with Mn2+ and Fe3+, whereas only Fe3+ forms Li minerals with Mn2+ and Mn3+. Moreover, a node Mn also exists in the Li network, but it does not form Li minerals with either Fe or other Mn redox states. The observed redox associations between Li, Fe and Mn provide an example that can be used to further investigate recently described mineral evolution redox trends of Mn and other elements (Hazen et al., 2019).
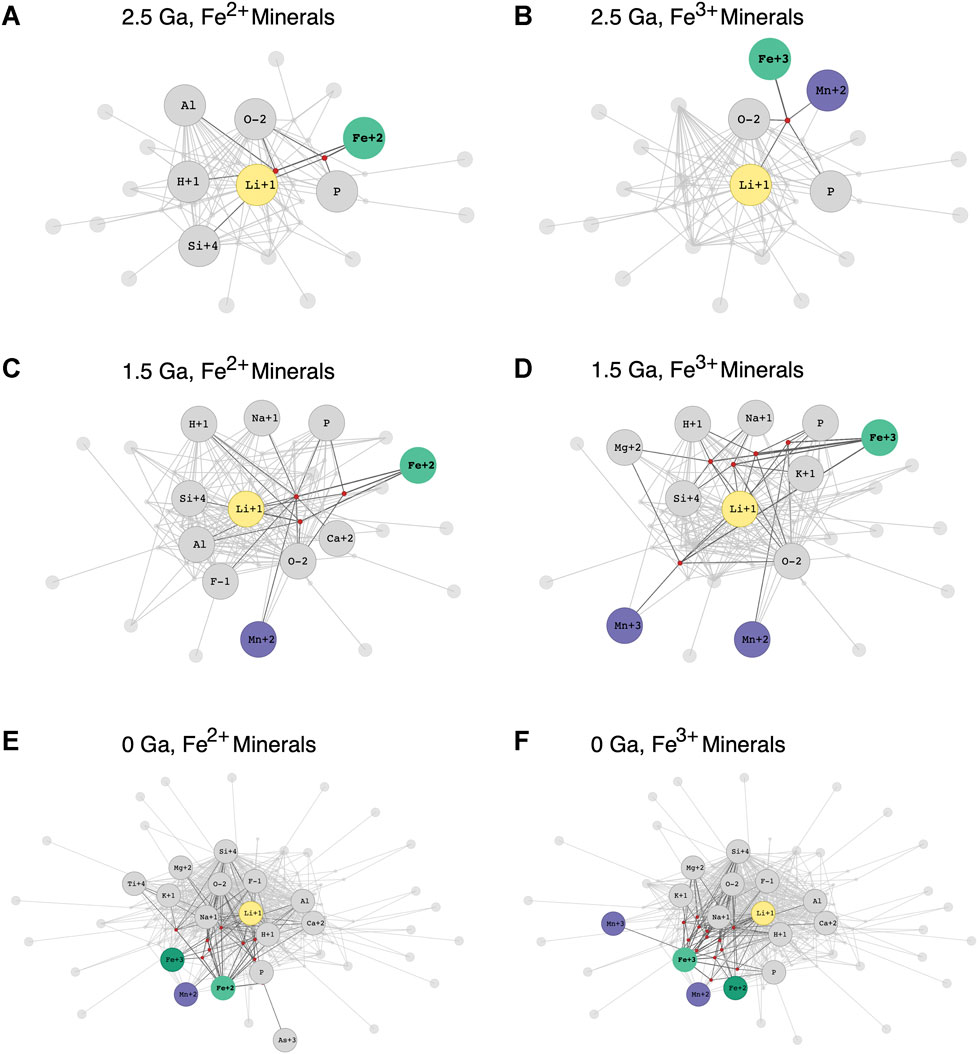
FIGURE 2. Lithium mineral-chemistry network with element nodes separated by redox states over time. In all panels, Li is highlighted in yellow, and Iron (Fe) and manganese (Mn) nodes at different redox states are shown in green and purple, respectively. All other element nodes are shown in gray, all mineral nodes that contain the highlighted elements are shown in red, and minerals that do not contain the highlighted elements are shown in gray. In each network image, only second-degree connections from the emphasized Fe node (Fe2+ in panels A, C, and E and Fe3+ in panels B, D, and F) are labeled. (A) The Li mineral-chemistry network for all minerals with oldest-known formation dates ≥2.5 Ga, with Li-minerals containing Fe2+ emphasized. (B) The Li mineral-chemistry network for all minerals with oldest-known formation dates ≥2.5 Ga, with Li-minerals containing Fe3+ emphasized. (C) The Li mineral-chemistry network for all minerals with oldest-known formation dates ≥1.5 Ga, with Li-minerals containing Fe2+ emphasized. (D) The Li mineral-chemistry network for all minerals with oldest-known formation dates ≥1.5 Ga, with Li-minerals containing Fe3+ emphasized. (E) The full Li mineral-chemistry network, with Li-minerals containing Fe2+ emphasized. (F) The full Li mineral-chemistry network, with Li-minerals containing Fe3+ emphasized.
dragon provides a user-friendly browser-based tool for exploration of bipartite mineral-chemistry networks over geologic time scales, with a particular focus on tracking trends in mineral speciation associated with evolving element redox states. dragon provides exceptional flexibility for users to visualize chemical and geological characteristics embedded in mineral-chemistry networks and perform associated statistical analyses. All network nodes and associated metadata can be directly exported to flat CSV or Excel files, and the network itself can additionally be exported as a publication-ready figure, or to a plain text file in a format supported by the R igraph package (Csardi and Nepusz, 2006), such as DOT or LGL (Adai et al., 2004).
dragon additionally maintains a data cache of the most recent information from MED. Upon launch, dragon will always check (provided there is an internet connection) whether the current MED cache is up to date. If dragon’s cached MED data has been superseded by a new release of MED data, dragon will issue a prompt to the user with the option to download the most recent MED data for use in the current dragon session.
dragon requires only ∼800 MB of RAM for the most complex mineral-chemistry networks and leverages asynchronous processing, performed with the future (Bengtsson, 2020) and promises (Cheng, 2020) R libraries, for time-consuming operations to ensure scalability. That said, due to limitations with the visNetwork library, itself an R wrapper for the vis.js JavaScript library, it may take a prohibitively long time to render the interactive visualization for extremely large networks. For example, the interactive full mineral-chemistry network of all known minerals (which contains, as of 7/10/20, 4,786 mineral nodes, 74 element nodes, and 22,797 edges) can take several minutes to render, and users may experience lags when manipulating nodes and edges. To ameliorate this issue, dragon offers the option “Build network without display.” When this option is activated, dragon will still construct a network for full analysis and exploration, but it will not render an interactive visualization.
Users should bear in mind several additional limitations when using dragon. First, while the Mineral Evolution Database provides highly reliable information about mineral formation and age, the mineral record itself is biased toward more recently-formed minerals due to geologic processes. As such, most mineral-chemistry networks will tend to expand dramatically from roughly 500 million years ago to present, and this expansion is not necessarily due to shifts in Earth surface conditions. Instead, this expansion may be an artifact driven by the preservation of younger crustal materials rather than specific evidence of expanded mineral chemistries. Second, while dragon does analyze data using linear models through the “Analyze Network Minerals” tab, it is the users’ responsibility to interpret and apply the modeling results. For example, dragon will neither transform any data before linear model analysis nor check for linearity (in the case of a numeric predictor variable), so users should take care to ensure modeling assumptions are met for any given analysis.
dragon provides a new useful tool for the Earth Sciences and geobiology communities to apply state-of-the-art network analyses to exploration of redox trends found in Earth’s mineral record. We further recommend that, when citing dragon, users also reference the Mineral Evolution Database (https://rruff.info/evolution/;Golden et al., 2019) which maintains all mineral record data that dragon consumes.
All code and associated data used by dragon is freely available from the GitHub repository https://github.com/sjspielman/dragon. MED data used by and cached within dragon is publicly available from https://rruff.info/evolution/.
EM initially conceptualized the work presented here. SS wrote all code and is the active maintainer for dragon. EM and SS wrote the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by start-up funds from Rowan University to EM and SS as well as NSF EAR2020520 to EM and SS.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Joshua Golden at the University of Arizona for his assistance in working with the Mineral Evolution Database and for his input in the development of dragon. We also thank Robert Downs at the University of Arizona and Nathan Yee at Rutgers University for their input in the development of dragon.
Adai, A. T., Date, S. V., , S., W., and Wieland, E. M. (2004). Lgl: creating a map of protein function with an algorithm for visualizing very large biological networks. J. Mol. Biol. 340, 179–190. doi:10.1016/j.jmb.2004.04.047
Almende, B. V., Thieurmel, B., and Robert, T. (2019). visNetwork: network visualization using ‘vis.js’ library. R package version 2.0.9.
Bengtsson, H. (2020). Future: unified parallel and distributed processing in R for everyone. R package version 1.18.0.
Blondel, V. D., Guillaume, J.-L., Lambiotte, R., and Lefebvre, E. (2008). Fast unfolding of communities in large networks. J. Stat. Mech. 2008, P10008. doi:10.1088/1742-5468/2008/10/P10008
Boesgaard, A. M., and Steigman, G. (1985). Big bang nucleosynthesis: theories and observations. Annu. Rev. Astron. Astrophys. 23, 319–378. doi:10.1146/annurev.aa.23.090185.001535
Cawood, P. A., Hawkesworth, C. J., Pisarevsky, S. A., Dhuime, B., Capitanio, F. A., and Nebel, O. (2018). Geological archive of the onset of plate tectonics. Phil. Trans. R. Soc. A. 376, 20170405. doi:10.1098/rsta.2017.0405
Chang, W., Cheng, J., Allaire, J., Xie, Y., and McPherson, J. (2020). Shiny: web application framework for R. R package version 1.5.0.
Cheng, J. (2020). Promises: abstractions for promise-based asynchronous programming. R package version 1.1.1. https://CRAN.R-project.org/package=promises.
Csardi, G., and Nepusz, T. (2006). The igraph software package for complex network research, InterJournal, Complex Systems 1695. https://igraph.org
Farquhar, J., Bao, H., and Thiemens, M. (2000). Atmospheric influence of earth’s earliest sulfur cycle. Science 289, 756–758. doi:10.1126/science.289.5480.756
Fruchterman, T. M. J., and Reingold, E. M. (1991). Graph drawing by force-directed placement. Softw Pract. Exper. 21, 1129–1164. doi:10.1002/spe.4380211102
Golden, J., Downs, R., Hazen, R, Pires, A., and Ralph, J. (2019). Mineral evolution database: data-driven age assignment, how does a mineral get an age?. doi:10.1130/abs/2019AM-334056
Golden, J., McMillan, M., Downs, R., Hystad, G., Goldstein, I., Stein, H. J., et al. (2013). Rhenium variations in molybdenite (MoS2): evidence for progressive subsurface oxidation. Earth Planet. Sci. Lett. 366, 1–5. doi:10.1016/j.epsl.2013.01.034
Hazen, R. M., Downs, R. T., Eleish, A., Fox, P., Gagné, O. C., Golden, J. J., et al. (2019). Data-driven discovery in mineralogy: recent advances in data resources, analysis, and visualization. Engineering 5, 397–405. doi:10.1016/j.eng.2019.03.006
Hazen, R. M., Papineau, D., Bleeker, W., Downs, R. T., Ferry, J. M., McCoy, T. J., et al. (2008). Mineral evolution. Am. Mineral. 93, 1693–1720. doi:10.2138/am.2008.2955
Hystad, G., Morrison, S. M., and Hazen, R. M. (2019). Statistical analysis of mineral evolution and mineral ecology: the current state and a vision for the future. Appl. Comput. Geosci. 1, 100005. doi:10.1016/j.acags.2019.100005
Jelen, B. I., Giovannelli, D., and Falkowski, P. G. (2016). The role of microbial electron transfer in the coevolution of the biosphere and geosphere. Annu. Rev. Microbiol. 70, 45–62. doi:10.1146/annurev-micro-102215-095521
Liu, X.-M., Kah, L. C., Knoll, A. H., Cui, H., Kaufman, A. J., Shahar, A., et al. (2016). Tracing earth’s O2 evolution using zn/fe ratios in marine carbonates. Geochem. Persp. Lett. 2, 24–34. doi:10.7185/geochemlet.1603
Luo, G., Ono, S., Beukes, N. J., Wang, D. T., Xie, S., and Summons, R. E. (2016). Rapid oxygenation of earth’s atmosphere 2.33 billion years ago. Sci. Adv. 2, e1600134. doi:10.1126/sciadv.1600134
Moore, E. K., Hao, J., Prabhu, A., Zhong, H., Jelen, B. I., Meyer, M., et al. (2018). Geological and chemical factors that impacted the biological utilization of cobalt in the archean eon. J. Geophys. Res. Biogeosci. 123, 743–759. doi:10.1002/2017JG004067
Moore, E. K., Hao, J., Spielman, S. J., and Yee, N. (2020). The evolving redox chemistry and bioavailability of vanadium in deep time. Geobiology 18, 127–138. doi:10.1111/gbi.12375
Moore, E. K., Jelen, B. I., Giovannelli, D., Raanan, H., and Falkowski, P. G. (2017). Metal availability and the expanding network of microbial metabolisms in the archaean eon. Nat. Geosci. 10, 629–636. doi:10.1038/ngeo3006
Morrison, S. M., Liu, C., Eleish, A., Prabhu, A., Li, C., Ralph, J., et al. (2017). Network analysis of mineralogical systems. Am. Mineral. 102, 1588–1596. doi:10.2138/am-2017-6104CCBYNCND
Morrison, S. M., Runyon, S., and Hazen, R. (2018). The paleomineralogy of the hadean eon revisited. Life 8, 64. doi:10.3390/life8040064
Neuwirth, E. (2014). RColorBrewer: ColorBrewer palettes.R package version 1.1-2. https://CRAN.R-project.org/package=RColorBrewer.
Newman, M. E. J. (2006). Finding community structure in networks using the eigenvectors of matrices. Phys. Rev. E 74, 036104. doi:10.1103/PhysRevE.74.036104
Och, L. M., and Shields-Zhou, G. A. (2012). The neoproterozoic oxygenation event: environmental perturbations and biogeochemical cycling. Earth-Sci. Rev. 110, 26–57. doi:10.1016/j.earscirev.2011.09.004
Pauling, L. (1932). The nature of the chemical bond. IV. The energy of single bonds and the relative electronegativity of atoms. J. Am. Chem. Soc. 54, 3570–3582. doi:10.1021/ja01348a011
Pearson, R. G. (1963). Hard and soft acids and bases. J. Am. Chem. Soc. 85, 3533–3539. doi:10.1021/ja00905a001
R Core Team. (2019). R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing.
Sahoo, S. K., Planavsky, N. J., Kendall, B., Wang, X., Shi, X., Scott, C., et al. (2012). Ocean oxygenation in the wake of the marinoan glaciation. Nature 489, 546–549. doi:10.1038/nature11445
Satkoski, A. M., Lowe, D. R., Beard, B. L., Coleman, M. L., and Johnson, C. M. (2016). A high continental weathering flux into paleoarchean seawater revealed by strontium isotope analysis of 3.26 ga barite. Earth Planet. Sci. Lett. 454, 28–35. doi:10.1016/j.epsl.2016.08.032
Scott, C., Lyons, T. W., Bekker, A., Shen, Y., Poulton, S. W., Chu, X., et al. (2008). Tracing the stepwise oxygenation of the proterozoic ocean. Nature 452, 456–459. doi:10.1038/nature06811
Warke, M. R., Di Rocco, T., Zerkle, A. L., Lepland, A., Prave, A. R., Martin, A. P., et al. (2020). The great oxidation event preceded a paleoproterozoic "snowball Earth". Proc. Natl. Acad. Sci. USA 117, 13314–13320. doi:10.1073/pnas.2003090117
Keywords: dragon, redox, network, mineral chemistry, mineral evolution, geobiology
Citation: Spielman SJ and Moore EK (2020) dragon: A New Tool for Exploring Redox Evolution Preserved in the Mineral Record. Front. Earth Sci. 8:585087. doi: 10.3389/feart.2020.585087
Received: 19 July 2020; Accepted: 31 August 2020;
Published: 24 September 2020.
Edited by:
Alexandra V. Turchyn, University of Cambridge, United KingdomReviewed by:
Malcolm S. W. Hodgskiss, Stanford University, United StatesCopyright © 2020 Spielman and Moore. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephanie J. Spielman, c3BpZWxtYW5Acm93YW4uZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.