- 1Department of Orofacial Sciences, University of California, San Francisco, San Francisco, CA, United States
- 2Departments of Pediatrics, Anatomy/Neurobiology, Neurology, University of California, Irvine, Irvine, CA, United States
- 3Department of Stomatology, Faculty of Dental Medicine, Université de Montréal, Montreal, QC, Canada
In a systemic effort to survive environmental stress, organ systems fluctuate and adapt to overcome external pressures. The evolutionary drive back toward homeostasis makes it difficult to determine if an organism experienced a toxic exposure to stress, especially in early prenatal and neonatal periods of development. Previous studies indicate that primary human teeth may provide historical records of experiences related to stressors during that early time window. To assess the molecular effects of early-life adversity on enamel formation, we used a limited bedding and nesting (LBN) mouse model of early-life adversity (ELA) to assess changes in the enamel organ gene expression and enamel matrix mineralization. On average, postnatal day 12 (P12) ELA mice weighed significantly less than the controls. When adjusted for animal weight, ELA molar enamel volume was reduced as compared with the controls, and the relative mineral density of molar enamel was significantly increased. There were no obvious changes in enamel matrix crystal morphology or structure in ELA as compared with the control mouse enamel. RNAseq showed extracellular matrix organization to be the most significantly affected GO and reactome pathways, whereas butanote metabolism was the most significantly altered KEGG pathway. Transcripts expressing the enamel matrix proteins amelogenin (Amelx) and enamelin (Enam) were among the top 4 most differentially expressed genes. When evaluating molecular mechanisms for the changes in gene expression in ELA enamel organs, we found significantly increased expression of Dlx3, while transcripts for clock genes Per1 and Nrd1 were downregulated. These findings support the possibility that the developing enamel organ is sensitive to the pressures of early-life adversity and produces molecular and structural biomarkers reflecting these challenges.
Introduction
The exposome, which includes environmental stress affecting the hypothalamus–pituitary–adrenal (HPA) axis, affects gene transcription and organ phenotype (1, 2). Evidence from our work in characterizing phenotypes of human primary teeth suggests that exposure to early-life adversity alters the tooth phenotype (3–5). Adverse early-life experiences can affect physiology, including immune function and cognitive and emotional development, and can influence the risk of developing stress-related psychopathology (6, 7). Therefore, biomarkers, such as those reflected in altered phenotypes in tooth enamel, could be useful screening tools for identifying children at risk of early-life stress-related diseases. However, further studies are needed to understand the mechanisms by which alterations in the HPA axis, such as those resulting from early-life adversity, affect tooth formation.
To study how early-life adversity (ELA) affects tooth enamel formation, we used the limited bedding and nesting mouse model (LBN) developed in the Baram lab (8–10). In this model, pup stress is evoked via fragmented maternal care, generated by reducing the amount of nesting material available to the dam beginning at P2 through the end of the study period at P12.
In mice, the incisor continuously erupts so that all stages of enamel development are present over the length of the incisor, while molars are rooted teeth that develop through sequential stages of development similar to human teeth. In molars, enamel formation begins at P2 with the secretion of enamel matrix proteins, including amelogenin, enamelin, and ameloblastin. Following the secretion of matrix proteins, the protein matrix is hydrolyzed first by MMP-20, followed by further hydrolysis with KLK4 in the maturation stage. As the protein is removed, it is replaced by minerals to form the highly mineralized mature enamel matrix. In mouse molars, the maturation stage begins from about P8 and is complete by P15 (11). The coincident timing of mouse molar tooth enamel matrix protein secretion and maturation to the timing of early-life stress in the LBN model for ELA makes this ELA model ideal to assess the effects of stress of early life on tooth enamel formation.
Methods
Limited Bedding and Nesting Early-Life Adversity (ELA) Mouse Protocol
Animals
Dams were Crh-IRES-Cre +/+ (12), and they were paired with Ai14 tdTomato (13) males, both on a C57Bl6 background. The resulting offspring were Crh-IRES-Cre, Ai14 tdTomato, as previously described (14). Animals were housed in a 12-h light cycle and provided ad libitum food and water. All experiments were carried out in accordance with the University of California, Irvine Institutional Animal Care and Use Committee at the University of California-Irvine and were consistent with Federal guidelines.
Early-life adversity was imposed on neonatal mice using simulated poverty by limiting nesting and bedding materials in cages between P2 and P12 (9, 10). For the ELA group, a plastic-coated mesh platform was placed ~2.5 cm above the floor of a standard cage. Cobb bedding was reduced to cover the cage floor sparsely, and one-half of a single nestlet was provided for nesting material on the platform. Control dams and litters resided in standard cages containing ample cobb bedding and one whole nestlet for nesting. Control and experimental cages were undisturbed during P2–P12, housed in temperature-controlled rooms (22°C).
MicroCT Imaging and Analysis of P12 Mouse Mandibles
Mandible Collection
On postnatal day 12 (P12), mandibles were collected from 3 separate litters of control (N = 9) and 3 separate litters of ELA (N = 10) mice. The mandibles were fixed in 4% PFA for 24 h, and a total of 5 male and 4 female control and 6 male and 4 female ELA mice were selected for microCT imaging.
MicroCT Scanning
One hemimandible from each mouse was scanned by micro-computed tomography (microCT) using a Scanco Medical μCT50 at the UCSF Core Center of Muscoloskeletal Biology and Medicine under the Skeletal Biology core. Specimens were scanned at 10.0 μm resolution with 500 ms integration time within a field of view of 15.2 mm (energy parameters of 55 kVP, 109 μA, 6 W, 0.5 mm AI filter). Reconstructions were generated using Scanco Medical's integrated μCT Evaluation Program V6.5-3 and converted into DICOM files for post-process analysis.
Image Analysis
DICOM files were uploaded into Amira software (ThermoFisher, Version 2020.3.1). A non-local means filter [spatial StdDev = 5, intensity StdDev = 0.2, search window [px] = 9, local neighborhood [px] = 3] and an unsharp masking [interpretation = 3D, edge size [px] = 6, edge contrast = 0.5, brightness threshold = 0] image processing filter were applied to increase the contrast between the mineralized enamel matrix and the surrounding dentin. An enamel masking threshold of 8,500–18,000 Hounsfield units (HU) was applied to the segment mineralizing enamel matrix. Relative enamel mineral density was calculated by averaging the grayscale values in Hounsfield units (HU) of all the voxels within this segment. A 3D volume smoothing (px size = 3) was applied to exclude partial volume components. The volume of the mineralizing matrix was calculated by simple voxel counting of the labeled enamel material. Relative enamel density and volume of all the samples were compared relative to body weight by student t-tests.
Enamel Matrix Crystal Structure
To examine the enamel matrix structure, hemimandibles were fixed by immersion in 4% PFA for 24 h and then stored in PBS at 4°C. They were then post-fixed with a 50:50 mixture of 1.5% aqueous potassium ferrocyanide and 1% aqueous osmium tetroxide, dehydrated in a 30–100% graded ethanol series, and processed for embedding in LR White resin (Electron Microscopy Sciences). The polymerized resin blocks were sectioned perpendicular to the hemimandible with an IsoMet low-speed saw (Buehler, Lake Bluff, Il) and then polished using a polisher PowerPro 5000 (Buehler). The samples were imaged using a Hitachi Regulus 8220 scanning electron microscope (SEM) operated at 1 kV using the low-angle backscattered detector (LA-BSE) for imaging.
RNAseq Pathway Analysis
Mandibles of 4 P12 male controls (bodyweight = 5.9 ± 0.4 gm) and 5 P12 male ELA (bodyweight = 5.30 ± 0.4 g) mouse pups were placed in an RNAlater stabilization solution (Invitrogen) and then transferred to phosphate-buffered saline. Enamel organs were removed from the first molars, and mRNA was extracted and purified using a Direct-zol RNA MiniPrep kit (Zymo Research). RNA quality and quantity were assessed using a NanoDrop spectrophotometer and sent to Novogene Corporation Inc. (Sacramento, CA) for RNA sequencing and analysis.
Gene expression was quantified and normalized, and the differential gene expression was assessed using DESeq2 at a p-value of < 0.05 (15), followed by an assessment of FDR values (false discovery rate) (16). Pathway enrichment analyses included gene ontology (GO), KEGG, which integrates genomic, chemical, and systemic functional information (17), and reactome pathway analyses.
qPCR Amplification
P12 enamel organs from control male mice from 2 separate control litters (N = 4) and male mice from 2 separate ELA litters (N = 5) were collected for qPCR transcript analysis. The expressions of enamel matrix genes, Amelx and Enam, and maturation stage enamel matrix proteinase Klk4, of clock genes, Nr1d1 and Per2 (18, 19), and of Dlx3 (20) and Igfbp2, Igfbp3 (21), as well as Hsd11b2, were amplified from ELA and control enamel organs following conversion of mRNA to cDNA using SuperScript IV VILO Master Mix (Invitrogen). Relative mRNA expression was quantified by qPCR with PowerUp SYBR Green Master Mix (Applied Biosystems) using primer sets generated by Elim Biopharmaceuticals, Hayward, CA, with Rpl19 used as the reference gene (Table 1). The relative expression of target genes was analyzed using the ΔΔCt method (22). Significant differences in expression were determined by an independent student t-test using fold-change levels.
Results
Animal Weight Was Associated With the Density and Volume of the Mineralizing Enamel Matrix
Early-life adversity mice collected for further dissection and analyses (4.97 ± 0.8 g, N = 18; 6 l) weighed on average 20% less than their control counterparts (6.0 ± 0.5 g, N = 26; 7 l). Density, as measured by relative intensity, and volume of the mineralized segment of the enamel matrix were negatively correlated with body weight (Figure 1). These data show that body weight influences mineralized enamel volume and density.
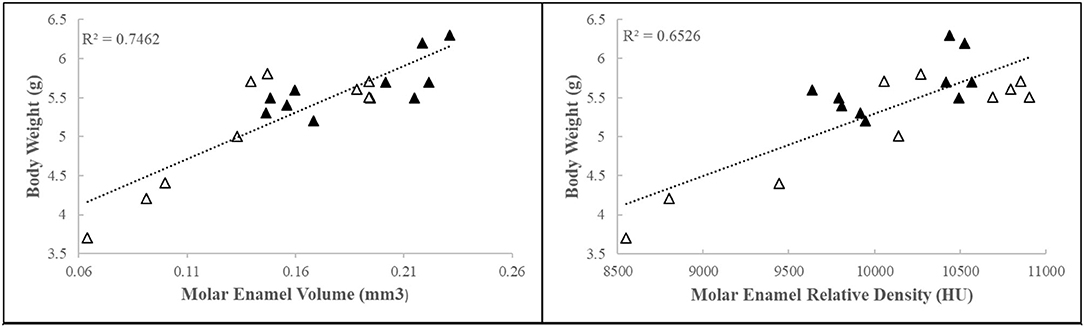
Figure 1. First molar relative enamel mineral density and enamel volume is associated with animal weight. Black triangles = control, white triangles = ELA.
When we adjusted for body weight, we found that normalized enamel volume and density were similar in male and female mice (data not shown). However, the relative volume of mineralized enamel in ELA was significantly less than that of the controls, and the relative density of ELA enamel was significantly increased (Figure 2). Given the large effect of body weight, we checked for normal distribution of weights within the groups using the Shapiro–Wilk test, and while the CTL group passed normality test (W=0.89, p=0.21), the ELA mice did not (W=0.83, p=0.04). After excluding the lowest weight values below 4.5 gm, the ELA mice passed the normality test (W=0.83, p=0.075). Therefore, to minimize the effects of weight, these lowest-weight ELA mice were excluded from the remaining analyses.
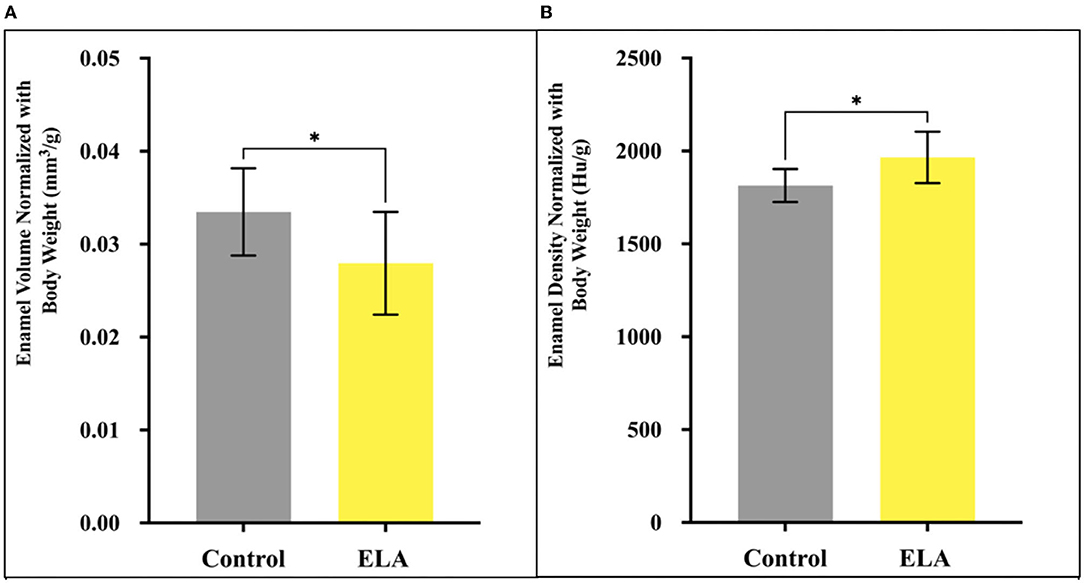
Figure 2. Mineralized enamel volume and relative density in molars from P12 mice, normalized for body weight. (A) Enamel volume was significantly less in ELA (n = 10) as compared to controls (n = 10). (B) Enamel density was significantly higher in ELA mice as compared to controls *p < 0.05. Error bars: SD.
SEM Imaging Revealed Normal Enamel Morphology and Crystal Structure
Backscattered SEM images of enamel from both P12 control mice (Figures 3A,B) and ELA mice (Figures 3C,D) showed no difference in the structural organization of the enamel layer and appearance of the enamel crystals.
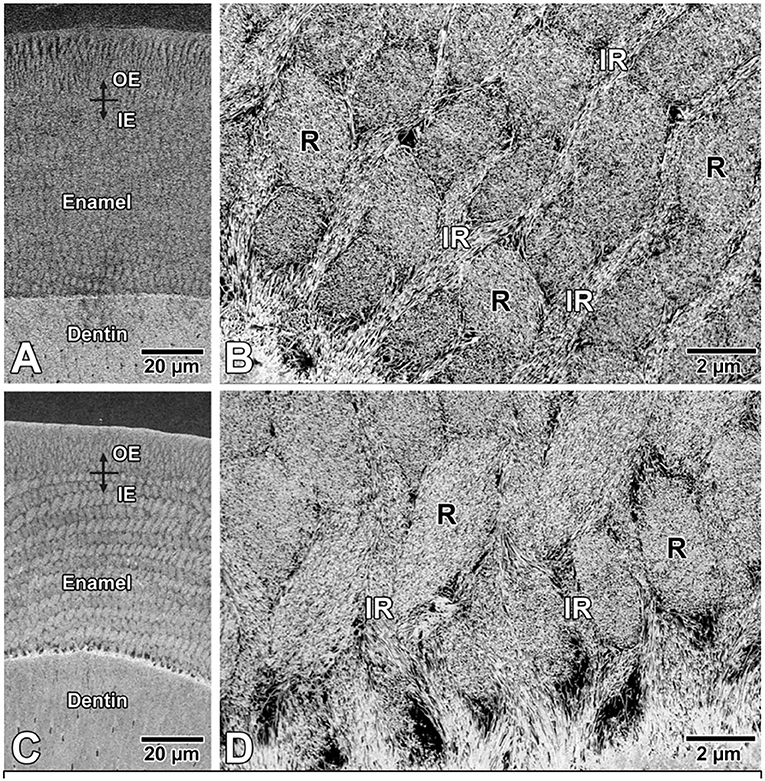
Figure 3. Backscattered electron SEM images of enamel in (A,B) control and (C,D) ELA mice. Qualitatively, there are no differences in the structural organization of rod (R)-interrod (IR) enamel and (B,D) also no apparent difference in appearance of crystal profiles. IE, inner enamel; OE, outer enamel.
RNAseq Seq Analysis Showed Significant Differences in Gene Expression in Enamel Organs From ELA as Compared With Control Mice
RNAseq of P12 enamel organs from control and ELA mice showed 437 genes uniquely expressed in ELA and 345 genes uniquely expressed in control enamel organs (GEO#GSE199982; Figure 4).
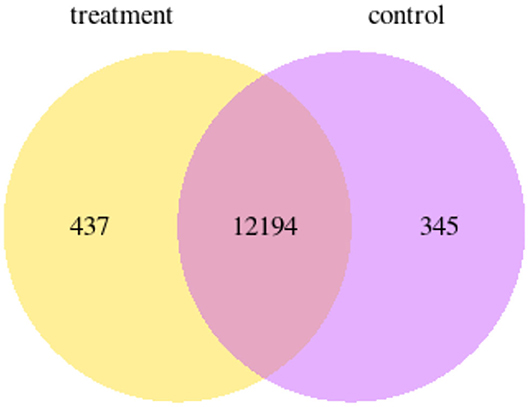
Figure 4. Venn diagram showing the number of genes that are uniquely expressed within each group; yellow = ELA mice, purple = weight-matched controls.
There were 81 differentially expressed genes (DEGs) upregulated in the first molar enamel organs of the ELA p12 mouse and 62 downregulated genes as compared with the controls (p adjusted < 0.05). Secretory enamel matrix proteins, including amelogenin, enamelin, and ameloblastin, were among the most highly upregulated genes in ELA mice. However, there were no differences in the relative expression of the maturation stage matrix proteins, odam and amelotin. Genes for metalloproteinases, including MMP20, found primarily in the secretory stage, and KLK4, found primarily in the maturation stage, were also not differentially expressed in ELA as compared with control enamel organs.
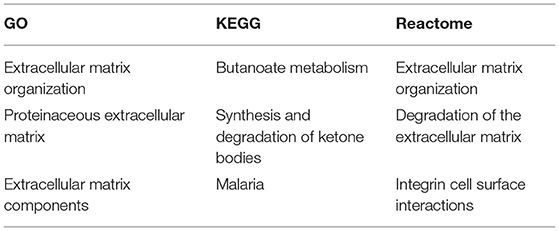
Go pathway analysis showed extracellular matrix organization and structure as the most significantly ELA-altered pathways. KEGG pathways that were most significantly altered by ELA were butanoate metabolism and synthesis and degradation of ketone bodies. The most significantly altered reactome pathway was extracellular matrix organization (see Table 2 and Supplementary Figures 1–3).
qPCR Analysis Showed Significant Differences in the Expression of Genes Associated With Amelogenin Expression in ELA and Control P12 Molars
PCR amplification and analysis showed a significant increase in the expression of amelogenin and enamelin in ELA as compared with control mice. Clock genes Nr1d1 and Per2, which are associated with increased amelogenin expression (18, 19), were, however, downregulated in ELA enamel organs. Of Igfb2, which is associated with IGF-related amelogenin expression (21), was upregulated, and Dlx3, a transcription factor associated with amelogenin expression (20), was significantly increased. There were no differences in the expression of Klk4 and Hsd11b2 gene expression (Figure 5).
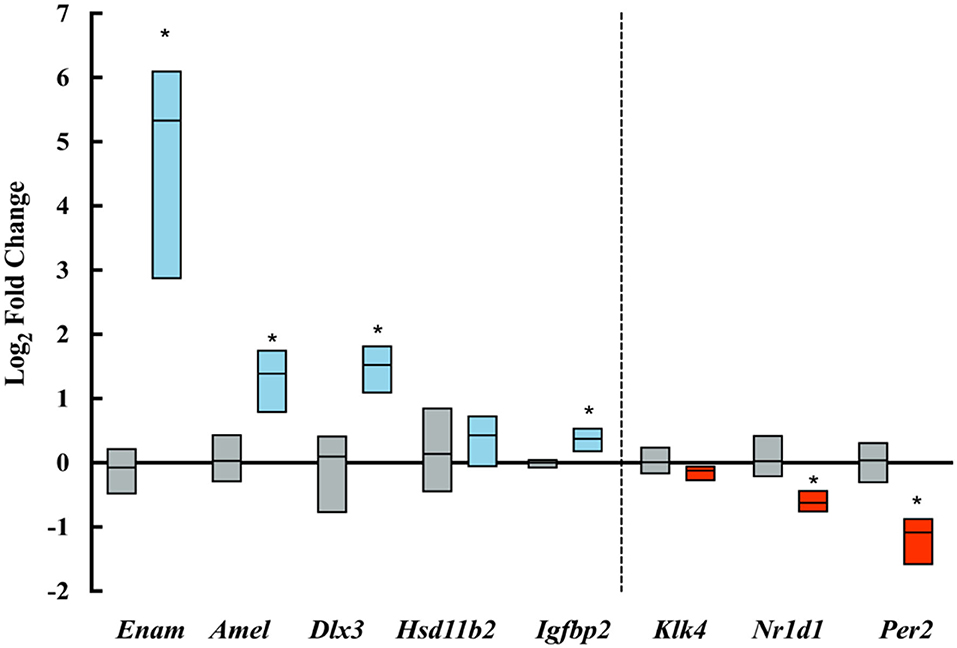
Figure 5. Log-fold changes of transcripts amplified by qPCR. Blue boxes show upregulated transcripts and red boxes show downregulated transcripts expressed in ELA enamel organs as compared to controls (gray boxes). *p < = 0.05.
Discussion
Our results show that in mice, the experience of early-life adversity alters tooth enamel formation. Overall, we found a negative association between body weight and the relative density and volume of the mineralized enamel. This indicates that factors that influence overall growth, as reflected in body weight, also influence enamel mineralization. The bodyweight of ELA mice is negatively associated with plasma corticosterone levels (23), and intracellular corticosterone concentration can be regulated by corticosteroid 11-beta-dehydrogenase isozyme 2, expressed by Hsd11b2. However, we found low expression of Hsd11b2 in the enamel organ, and PCR amplification showed no significant differences in Hsd11b2 expression in ELA as compared with weight-matched controls. This suggests that cellular corticosterone in the enamel organ does not have a major role in altering enamel matrix mineralization, but rather, in ELA mice, these changes are associated with systemic effects resulting from the dysregulation of the hypothalamus pituitary adrenal axis (HPA).
A cellular effect of ELA that may affect amelogenesis is suggested by the enriched KEGG pathway, butyrate metabolism. Butyrate metabolism describes the metabolic fate of short-chain fatty acids or short-chain alcohols that are typically produced by intestinal fermentation. Our findings of alterations in the KEGG butyrate metabolism pathway in enamel organs of ELA mice are consistent with studies that have shown LBN models of ELA rats to have increased intestinal permeability, decreased microbial alpha diversity, and reduced butyrate-producing microbes (24). Butyrate metabolism is associated with the production of ketone bodies (25), a pathway that was also affected in ELA enamel organs.
What was unexpected to us was the significant increase in relative density of the mineralized enamel matrix in the weight-normalized ELA mice as compared with controls. This relative increase in mineral density in ELA mouse enamel is consistent with our findings in human primary mandibular incisors. Enamel density in primary mandibular incisors is positively associated with internalizing symptoms (5), and internalizing symptoms in humans are associated with early-life adversity (26). These studies of ELA mice, therefore, support the possibility that early-life adversity, related to disrupted maternal care (23), can be reflected in increased enamel mineralization. SEM analysis did not show obvious differences in the structure of enamel crystals in the mineralizing enamel matrix of ELA molars as compared to controls, though qualitative SEM studies to assess possible changes related to the timing of enamel mineralization will require additional studies.
We found that mRNA transcript for the enamel matrix proteins, amelogenin and enamelin, were highly upregulated in ELA enamel organs as compared with controls. To explore possible mechanisms by which ELA alters amelogenin expression, we evaluated several candidate pathways. C/EBPα, a transcription factor for amelogenin in ELA and controls, showed no evidence of differential expression by RNAseq (data not shown). Clock genes, Nr1d1 and Per2, which have been positively associated with amelogenin expression (18), were downregulated in ELA mice. Thrombospondin 2, a member of the multifunctional family of glycoproteins, has been shown to increase amelogenin expression (27); however, our RNA seq data showed downregulation of thrombospondin 2 in ELA mouse enamel organs. Consistent with previous studies showing that insulin-like growth factor binding proteins (Igfbp2 and Igfbp3) can regulate the expression pattern of both Amelx (21) and Enam (28), RNAseq and qPCR data showed a small but significant upregulation in Igfbp2 in ELA as compared with the control enamel organ.
Dlx3, which in vitro has been shown to upregulate expression of the enamel matrix protein genes Amelx, Enam, Klk4, and Odam (29), was the most highly upregulated of the candidate genes. However, while our RNAseq analysis showed the upregulation of amelogenin and enamelin expression in ELA enamel organs, we found no changes in KLK4 and Odam expression. Duverger et al. (30) reported that in vivo DLX3 loss of function has no effect on the expression of the major enamel matrix proteins (including Amelx and Enam) and proteinases (including KLK4); however, the expression of ion transporters and carbonic anhydrase is affected. Our RNAseq analysis showed a significant upregulation of carbonic anhydrase transcripts Car6, Car3, and Car12, which in maturation stage enamel function to synthesize bicarbonate, which is then transported to the mineralizing enamel matrix to neutralize protons produced by the formation of hydroxyapatite (31, 32). This neutralization of the mineralizing enamel matrix then allows the continued growth of hydroxyapatite crystals. It may be that the relative increase in enamel mineral in weight-matched ELA mice is related to a Dlx3-mediated increase in bicarbonate synthesis to allow more rapid growth of hydroxyapatite crystals.
In the brain, Dlx isoforms induce the synthesis of glutamic acid decarboxylase (GAD1 and GAD2) (33), which increases GABA synthesis. Though Dlx3 has not been associated with the regulation of GABA synthesis, RNA seq data showed upregulated genes for enzymes involved in the breakdown of GABA (Abat and Aldh5A1) in ELA mice, suggesting the possibility that GABA-related pathways are involved in changes in gene expression in enamel organs of the ELA mouse model. Dlx3 transcription is mediated through Wnt signaling, which has a critical role in amelogenesis (34) and hippocampal neurogenesis (35). These findings suggest the importance of further studies to characterize the effects of early-life adversity on Wnt signaling and its effects on enamel maturation.
Taken together, our findings show the dysregulation of multiple genes in the enamel organ of mice exposed to early-life adversity through limited bedding and nesting and disrupted maternal behaviors. It is not clear why amelogenin and enamelin expression are upregulated in enamel organs from ELA mice, or how increased expression of these genes influences enamel matrix mineralization relative to body weight in the ELA mouse model. However, enhanced enamel mineralization relative to body weight in ELA mice may be specifically associated with increased Dlx3 expression to drive the upregulation of carbonic anhydrase synthesis, resulting in more rapid mineralization of hydroxyapatite crystals.
Increased relative mineral density in molars of mice exposed to early-life adversity is also consistent with the association of early-life adversity with accelerated biological aging (36, 37). Therefore, it may be that the enamel from human primary teeth, which mineralize in early life, can provide biomarkers to identify individuals with risk factors associated with early-life stress. Taken together, these studies support the concept that the developing tooth enamel organ is a useful model to explore cellular mechanisms related to changes in the HPA axis during development.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, GSE199982.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee at UC Irvine and The Institutional Animal Care and Use Committee at UCSF.
Author Contributions
IS, YN, YZ, and PD contributed to the conception and design of the study. IS, AP, AS, AN, YN, and RS completed experiments and provided data. IS, YN, and RS performed the statistical analysis. IS and PD wrote the draft of the manuscript. All authors contributed to manuscript revision and read and approved the submitted version.
Funding
Funding for these studies was provided by the National Institutes of Health Grants MH73136, P50 MH96889 (TZB), R01DE027971 (PD), F30DE029399 (IS), and T32DE007306.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the author and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Dr. Tallie Z. Baram for her enthusiastic support and constructive discussions for these studies.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fdmed.2022.894753/full#supplementary-material
References
1. Gao P. The exposome in the era of one health. Environ Sci Technol. (2021) 55:2790–9. doi: 10.1021/acs.est.0c07033
2. Wright ML, Starkweather AR, York TP. Mechanisms of the maternal exposome and implications for health outcomes. ANS Adv Nurs Sci. (2016) 39:E17–30. doi: 10.1097/ANS.0000000000000110
3. Boyce WT, Den Besten PK, Stamperdahl J, Zhan L, Jiang Y, Adler NE, et al. Social inequalities in childhood dental caries: the convergent roles of stress, bacteria and disadvantage. Soc Sci Med. (2010) 71:1644–52. doi: 10.1016/j.socscimed.2010.07.045
4. Davis KA, Mountain RV, Pickett OR, Den Besten PK, Bidlack FB, Dunn EC. Teeth as potential new tools to measure early-life adversity and subsequent mental health risk: an interdisciplinary review and conceptual model. Biol Psychiatry. (2020) 87:502–13. doi: 10.1016/j.biopsych.2019.09.030
5. Dunn EC, Mountain RV, Davis KA, Shaffer I, Smith ADAC, Roubinov DS, et al. Association between measures derived from children's primary exfoliated teeth and psychopathology symptoms: results from a community-based study. Front Dental Med. (2022) 3:803364. doi: 10.3389/fdmed.2022.803364
6. Boyce WT, Adams S, Tschann JM, Cohen F, Wara D, Gunnar MR. Adrenocortical and behavioral predictors of immune responses to starting school. Pediatr Res. (1995) 38:1009–17. doi: 10.1203/00006450-199512000-00030
7. Krugers HJ, Arp JM, Xiong H, Kanatsou S, Lesuis SL, Korosi A, et al. Early life adversity: lasting consequences for emotional learning. Neurobiol Stress. (2016) 6:14–21. doi: 10.1016/j.ynstr.2016.11.005
8. Gilles EE, Schultz L, Baram TZ. Abnormal corticosterone regulation in an immature rat model of continuous chronic stress. Pediatr Neurol. (1996) 15:114–9. doi: 10.1016/0887-8994(96)00153-1
9. Rice CJ, Sandman CA, Lenjavi MR, Baram TZ. A novel mouse model for acute and long-lasting consequences of early life stress. Endocrinology. (2008) 149:4892–900. doi: 10.1210/en.2008-0633
10. Molet J, Maras PM, Avishai-Eliner S, Baram TZ. Naturalistic rodent models of chronic early-life stress. Dev Psychobiol. (2014) 56:1675–88. doi: 10.1002/dev.21230
11. Lungová V, Radlanski RJ, Tucker AS, Renz H, Míšek I, Matalová E. Tooth-bone morphogenesis during postnatal stages of mouse first molar development. J Anatomy. (2011) 218:699–716. doi: 10.1111/j.1469-7580.2011.01367.x
12. Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, et al. A resource of cre driver lines for genetic targeting of gabaergic neurons in cerebral cortex. Neuron. (2011) 71:995–1013. doi: 10.1016/j.neuron.2011.07.026
13. Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, et al. A robust and high-throughput cre reporting and characterization system for the whole mouse brain. Nature neuroscience. (2010) 13:133–40. doi: 10.1038/nn.2467
14. Short AK, Thai CW, Chen Y, Kamei N, Pham AL, Birnie MT, et al. Single-cell transcriptional changes in hypothalamic corticotropin-releasing factor expressing neurons after early-life adversity inform enduring alterations in vulnerabilities to stress. Biol Psychiatry Global Open Sci. (In press). doi: 10.1016/j.bpsgos.2021.12.006
15. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for Rna-Seq data with Deseq2. Genome Biol. (2014) 15:550. doi: 10.1186/s13059-014-0550-8
16. Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of Rna-Seq data. Genome Biol. (2010) 11:R25. doi: 10.1186/gb-2010-11-3-r25
17. Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. Kegg for linking genomes to life and the environment. Nucleic Acids Res. (2008) 36:D480–4. doi: 10.1093/nar/gkm882
18. Athanassiou-Papaefthymiou M, Kim D, Harbron L, Papagerakis S, Schnell S, Harada H, et al. Molecular and circadian controls of ameloblasts. Eur J Oral Sci. (2011) 119(Suppl. 1):35–40. doi: 10.1111/j.1600-0722.2011.00918.x
19. Babajko S, de La Dure-Molla M, Jedeon K, Berdal A. Msx2 in ameloblast cell fate and activity. Front Physiol. (2015) 5:510. doi: 10.3389/fphys.2014.00510
20. Lézot F, Descroix V, Mesbah M, Hotton D, Blin C, Papagerakis P, et al. Cross-talk between Msx/Dlx homeobox genes and vitamin d during tooth mineralization. Connect Tissue Res. (2002) 43:509–14. doi: 10.1080/03008200290000583
21. Takahashi K, Yamane A, Bringas P, Caton J, Slavkin HC, Zeichner-David M. Induction of amelogenin and ameloblastin by insulin and insulin-like growth factors (Igf-I and Igf-Ii) during embryonic mouse tooth development in vitro. Connect Tissue Res. (1998) 38:269–78. doi: 10.3109/03008209809017047
22. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative Pcr and the 2(-Delta Delta C(T)) method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
23. Walker CD, Bath KG, Joels M, Korosi A, Larauche M, Lucassen PJ, et al. Chronic early life stress induced by limited bedding and nesting (Lbn) material in rodents: critical considerations of methodology, outcomes and translational potential. Stress. (2017) 20:421–48. doi: 10.1080/10253890.2017.1343296
24. Moussaoui N, Jacobs JP, Larauche M, Biraud M, Million M, Mayer E, et al. Chronic early-life stress in rat pups alters basal corticosterone, intestinal permeability, and fecal microbiota at weaning: influence of sex. J Neurogastroenterol Motil. (2017) 23:135–43. doi: 10.5056/jnm16105
25. Stilling RM, van de Wouw M, Clarke G, Stanton C, Dinan TG, Cryan JF. The neuropharmacology of butyrate: the bread and butter of the microbiota-gut-brain axis? Neurochem Int. (2016) 99:110–32. doi: 10.1016/j.neuint.2016.06.011
26. Jensen SKG, Dickie EW, Schwartz DH, Evans CJ, Dumontheil I, Paus T, et al. Effect of early adversity and childhood internalizing symptoms on brain structure in young men. JAMA Pediatr. (2015) 169:938–46. doi: 10.1001/jamapediatrics.2015.1486
27. Huang Z, Newcomb CJ, Lei Y, Zhou Y, Bornstein P, Amendt BA, et al. Bioactive nanofibers enable the identification of thrombospondin 2 as a key player in enamel regeneration. Biomaterials. (2015) 61:216–28. doi: 10.1016/j.biomaterials.2015.05.035
28. Catón J, Bringas P Jr, Zeichner-David M. Igfs increase enamel formation by inducing expression of enamel mineralizing specific genes. Arch Oral Biol. (2005) 50:123–9. doi: 10.1016/j.archoralbio.2004.11.012
29. Zhang Z, Tian H, Lv P, Wang W, Jia Z, Wang S, et al. Transcriptional factor Dlx3 promotes the gene expression of enamel matrix proteins during amelogenesis. PLoS ONE. (2015) 10:e0121288. doi: 10.1371/journal.pone.0121288
30. Duverger O, Ohara T, Bible PW, Zah A, Morasso MI. Dlx3-dependent regulation of ion transporters and carbonic anhydrases is crucial for enamel mineralization. J Bone Miner Res. (2017) 32:641–53. doi: 10.1002/jbmr.3022
31. Lacruz RS, Smith CE, Moffatt P, Chang EH, Bromage TG, Bringas P, et al. Requirements for ion and solute transport, and pH regulation during enamel maturation. J Cell Physiol. (2012) 227:1776–85. doi: 10.1002/jcp.22911
32. Yin K, Paine ML. Bicarbonate transport during enamel maturation. Calcified Tissue Int. (2017) 101:457–64. doi: 10.1007/s00223-017-0311-2
33. Le TN, Zhou Q-P, Cobos I, Zhang S, Zagozewski J, Japoni S, et al. Gabaergic interneuron differentiation in the basal forebrain is mediated through direct regulation of glutamic acid decarboxylase isoforms by Dlx homeobox transcription factors. J Neurosci. (2017) 37:8816–29. doi: 10.1523/JNEUROSCI.2125-16.2017
34. Fan L, Deng S, Sui X, Liu M, Cheng S, Wang Y, et al. Constitutive activation of B-catenin in ameloblasts leads to incisor enamel hypomineralization. J Mol Histol. (2018) 49:499–507. doi: 10.1007/s10735-018-9788-x
35. Arredondo SB, Valenzuela-Bezanilla D, Mardones MD, Varela-Nallar L. Role of Wnt signaling in adult hippocampal neurogenesis in health and disease. Front Cell Dev Biol. (2020) 8. doi: 10.3389/fcell.2020.00860
36. Sun Y, Fang J, Wan Y, Su P, Tao F. Association of early-life adversity with measures of accelerated biological aging among children in China. JAMA Netw Open. (2020) 3:e2013588. doi: 10.1001/jamanetworkopen.2020.13588
Keywords: enamel, early-life adversity (ELA), mineralization, ameloblasts, RNAseq, limited bedding and nesting (LBN)
Citation: Shaffer IC, Nakano Y, Pham A, Short A, Nanci A, Zhang Y, Shemirani R and Den Besten PK (2022) Effects of Early-Life Adversity on Tooth Enamel Formation. Front. Dent. Med. 3:894753. doi: 10.3389/fdmed.2022.894753
Received: 12 March 2022; Accepted: 27 April 2022;
Published: 15 June 2022.
Edited by:
Olivier Duverger, National Institute of Dental and Craniofacial Research (NIH), United StatesReviewed by:
Michel Goldberg, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceMarianna Bei, Harvard Medical School, United States
Copyright © 2022 Shaffer, Nakano, Pham, Short, Nanci, Zhang, Shemirani and Den Besten. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pamela K. Den Besten, cGFtZWxhLmRlbmJlc3RlbiYjeDAwMDQwO3Vjc2YuZWR1
 Ida C. Shaffer
Ida C. Shaffer Yukiko Nakano
Yukiko Nakano Aidan Pham2
Aidan Pham2 Pamela K. Den Besten
Pamela K. Den Besten