- 1Dental Research Institute, Faculty of Dentistry, University of Toronto, Toronto, ON, Canada
- 2Endodontics Department, Faculty of Dentistry, Ain Shams University, Cairo, Egypt
- 3School of Graduate Studies, University of Toronto, Toronto, ON, Canada
- 4Department of Dentistry, Mount Sinai Health System, Mount Sinai Hospital, Toronto, ON, Canada
Apical periodontitis is an inflammatory reaction of the periradicular tissues as a consequence of multispecies microbial communities organized as biofilms within the root canal system. Periradicular tissue changes at the molecular level initiate and orchestrate the inflammatory process and precede the presentation of clinical symptoms. Inflammatory mediators have been studied at either the proteomic, metabolomic, or transcriptomic levels. Analysis at the protein level is the most common approach used to identify and quantify analytes from diseased periradicular tissues during root canal treatment, since it is more representative of definitive and active periradicular inflammatory mediator than its transcript expression level. In disease, proteins expressed in an altered manner could be utilized as biomarkers. Biomarker proteins in periradicular tissues have been qualitatively and quantitatively assessed using antibodies (immunoassays and immunostaining) or mass spectrometry-based approaches. Herein, we aim to provide a comprehensive understanding of biomarker proteins identified in clinical studies investigating periradicular lesions and pulp tissue associated with apical periodontitis using proteomics. The high throughput mass spectrometry-based proteomics has the potential to improve the current methods of monitoring inflammation while distinguishing between progressive, stable, and healing lesions for the identification of new diagnostic and therapeutic targets. This method would provide more objective tools to (a) discover biomarkers related to biological processes for better clinical case selection, and (b) determine tissue response to novel therapeutic interventions for more predictable outcomes in endodontic treatment.
Introduction
Apical periodontitis is an inflammation occurring as a consequence of endodontic infections caused by multispecies microbial communities organized as biofilms within the root canal system (1). Once inflammation is initiated, molecular changes orchestrate the inflammatory process and precede the presentation of clinical symptoms. From a clinical perspective, the ability to distinguish between a progressive lesion and a stable lesion that is undergoing regression and healing is critical to determine the appropriate treatment approach (2). Increased pathogenicity of the intracanal microbiota and complex host response results in increased inflammatory and oxidative damage (3, 4). Several studies have reported changes in the expression of proteins, metalloproteinases, and inflammatory cytokines related to endodontic clinical symptoms (5–8). The degree of periradicular bone destruction has been linked to the concentrations of inflammatory mediators present within periradicular lesions (9). Thus, studying the levels of inflammatory mediators and protein expression profiles within periradicular lesions would shed new light on the disease process.
A biomarker is defined as a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention (10). Evaluation of the biomarkers associated with apical periodontitis is crucial for understanding the pathogenesis, diagnosis and development of therapeutic strategies (11). The inflammatory profile of periradicular lesions is highly dynamic with various inflammatory cells, their mediators, and effector molecules being involved in the disease process (12). Hence, defining a single or few biomarkers for apical periodontitis is a challenging task due to the complex interplay of several parameters which affect the state and progression of the disease.
Periradicular inflammatory markers have been studied at either the proteomic, metabolomic, or transcriptomic level (13). Transcriptomic-based assay techniques utilize transcribed mRNA sequences as biomarkers whereas proteomics identifies the actual secreted protein/metabolite (14). A strong correlation is anticipated when mRNA is translated into its respective protein, and consequently, both can be used to quantify the presence of a specific mediator. However, in human cells, a weak correlation between concentrations of protein and its respective mRNA expression has been observed (14), which could be attributed to various post-transcriptional modifications or translational mechanisms. During highly dynamic phases of cell activity, such as cellular differentiation or cellular stress response to specific microenvironmental stimuli, post-transcriptional processes may lead to higher deviations from an ideal correlation (15). In this case, proteomic analysis which involves studying the whole set of proteins expressed by cell/tissue/organism in a particular environment during a specific stage of the cell cycle would provide a better idea of the actually produced proteins.
Studying molecular biomarkers at the protein level is crucial since proteins are the direct biofunctional effector molecules in living organisms (16). Proteins biosynthesis is critical for cell and tissue signaling, development, homeostasis as well as structural stability (17). In dentistry, proteomics-based methods have been used to study various oral tissues/samples such as saliva (18–20), acquired pellicle (21), gingival crevicular fluid (22), dentin (23, 24), pulp tissue (25), and dental stem cells (26, 27). Currently, proteomic analysis of the tissues represents a powerful and unbiased approach to identify important differentially expressed proteins within diseased tissue in comparison to their normal counterparts (28). Proteins could also serve as prognostic markers and therapeutic targets, for example, antioxidant enzymes and heat shock protein 27 (29, 30). Therefore, researchers have conducted proteomics-based studies to investigate oral diseases including caries (31), gingivitis (32), periodontitis (33), pulp and periradicular diseases (34), as well as oral cancer (35).
Proteomics has been applied in endodontics to identify the bacterial proteins associated with apical periodontitis, for a better understanding of the pathogenicity and virulence factors of bacterial community (36–38). In addition to the analysis of bacterial proteins, studying proteomics provide relevant data on the host's response in specific clinical conditions, allowing a broad insight into the host response in homeostasis and in apical periodontitis, through qualitative and quantitative analysis of protein expression (39). Within this context, herein we focus on reviewing endodontic literature investigating human protein expression in different pathological conditions and healing of periradicular tissue using proteomics-based approach, and how these tools can improve knowledge in the diagnosis, intervention, and prognosis of endodontic treatment.
Protein Detection Approaches
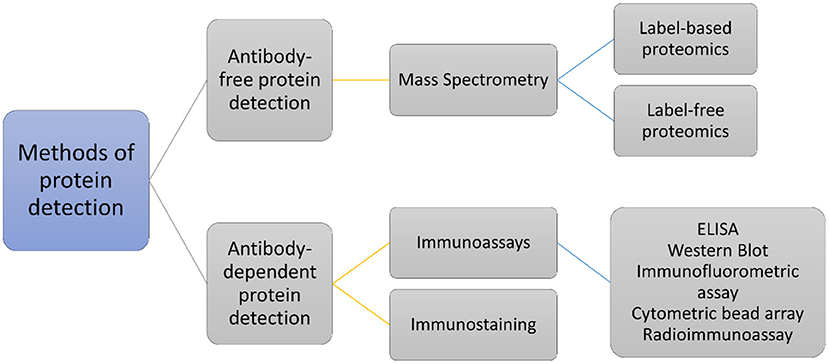
Biomarkers for different pathological states and healing have been investigated at the protein level using antibody-based assays and antibody-free protein detection using mass spectrometry (MS)-based techniques (or proteomics) (Figure 1) (5, 7, 13, 37–40). Specific techniques and protocols using antibodies for the detection of particular proteins, or their modified forms, have been applied to assess various inflammatory mediators in periradicular lesions (7, 41). The antibody-based techniques, such as western blot, immunohistochemistry assay, or enzyme-linked immunosorbent assay (ELISA), are very sensitive; however, they have limitations such as cross-reactivity, long development time, and high variability (42). MS-based proteomics has rapidly become the analytical method of choice for the identification and characterization of proteins. Quantification of proteins in MS-based proteomics can be carried out using label-based or label-free strategies (43). Label-based quantitation methods utilize stable-isotope labels which are incorporated within the peptides, introducing a known mass difference within the experimental conditions. In contrast, label-free proteomics quantitates both relative and absolute protein quantity by directly utilizing a peptide's intensity or inferring quantity indirectly from spectral counting of peptides (44). MS-based antibody-free protein detection methods offer multiple advantages such as the ability to determine the sequence of a protein or peptide, providing higher throughput than antibody-based assays, accurate quantification, high level of specificity, ability to discriminate between isoforms while avoiding interference caused by cross-reactivity of antibodies, while identifying and quantifying proteins for which no antibody exists (45–47).
Technological advances in MS-based proteomics have enabled the large-scale detection and quantitation of proteins, including measurement of properties such as their abundance, isoform expression, turnover rate, subcellular localization, post-translational modifications, and interactions (48). Protein expression profiling is defined as identifying the proteins expressed in a particular tissue, under specific conditions and at a particular time, typically compared to protein expression in reference samples (49). Analysis of differentially expressed proteins allows a better understanding of the overall physiologic profile of biological systems (cells and tissues) under a given condition (50). Different proteomics-based strategies are summarized in Figure 2. The bottom-up approach involves proteolysis of proteins into short peptides (8–30 amino acids) using a proteolytic enzyme (mostly trypsin due to its efficiency and specificity of cleavage), and MS/MS fragmentation to determine the peptide sequences (51). In top-down technique, the intact protein is introduced into the mass spectrometer to measure its intact and fragment ions masses (52). This approach enables 100% sequence coverage and full characterization of proteoforms, the specific molecular form of the protein resulting from combinations of genetic variation, alternative splicing, and post-translational modifications (53).
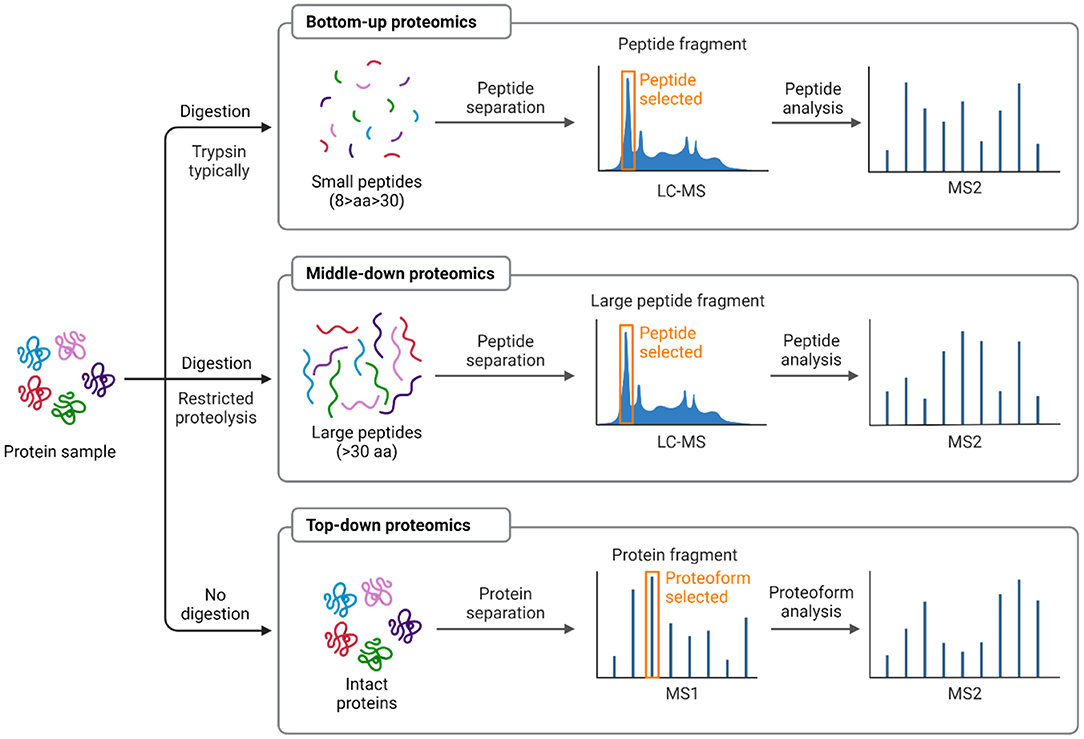
Figure 2. Schematic representing proteomic-based strategies: bottom-up vs. top-down vs. middle-down. In bottom-up approach, a protein is typically digested with an enzyme (i.e., trypsin) into many small peptides either in gel or in solution. The recovered peptides will be detected by mass spectrometer and a specific peptide can be isolated and fragmented by MS/MS to identify the protein from the database. Whereas in top-down method, the whole protein is analyzed directly in the mass spectrometer without digestion. The middle-down strategy analyzes larger peptides resulting from limited digestion or more selective proteases. One or more protein or peptide fractionation techniques can be applied prior to MS analysis and database searching. Modified and adapted from (16, 51). Schematic created with BioRender.com.
Several techniques have been applied to extract and separate proteins including the 2 commonly used methods, 2-dimensional electrophoresis (2-DE) and liquid chromatography (LC) (54). In 2-DE, separation of proteins is based on two principles; first, isoelectric points, then; two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to separate proteins according to the molecular weights of these proteins (55). Whereas in LC, a solution containing protein is introduced to flow through a column containing various substances under the action of external force (56). The different interactions between different proteins and chromatographic column materials, result in differences in the retention time of various proteins in the chromatographic column which in turn allows the separation and purification of different proteins according to their retention time. For data interpretation, dedicated search engines that rely on a “protein sequence database” are explored to compare experimental MS/MS spectra to theoretical peptide fragmentation patterns and identify the best match to interpret acquired data (51). Bioinformatics tools applied to analyze the identified proteins would facilitate the mechanistic understanding of the involved biological processes (57). The next two sections review the proteomics-based studies investigating apical periodontitis in endodontic literature. According to the sampled tissue site, the protein expression profiles of the periradicular lesion itself and the pulp tissue associated with periradicular lesions are discussed.
Proteomic Profile of Periradicular Lesions
Proteomic analysis of tissues involved in apical periodontitis represents a powerful strategy to detect differentially expressed proteins with significant role in the biological pathways involved in lesion progression or resolution. Clinical studies in endodontic literature investigating the proteomic profile of pulp and/or periradicular lesions in different conditions, using MS (2, 37–39, 58–61), are summarized in Supplementary Table 1. The main findings of studies assessing protein expression in periradicular lesions are presented in Figure 3. The identification of human proteins expressed in endodontic infections was addressed for the first time by Provenzano et al. in samples collected from teeth with asymptomatic apical periodontitis and acute apical abscesses, describing the host response and potential biomarkers in the activity of endodontic diseases (37). Subsequent studies qualitatively analyzed samples from post-treatment apical periodontitis and acute apical abscesses (38, 39, 58). Proteins were identified more frequently in samples from periapical abscesses compared to samples from asymptomatic teeth (37), due to the acute nature of host response in abscesses and the associated high killing rate of host cells and bacteria which would generate more proteins, including cytoplasmic proteins (37). Irrigating the canals with chlorohexidine reduced the number of identified proteins substantially in post-treatment samples, whereas NaOCl irrigation increased the number of proteins, which could be attributed to the tissue dissolving ability of NaOCl and its high destructive effects on cells, releasing cytoplasmic proteins to the environment (37). Identified proteins were mostly involved in cellular processes and metabolism followed by proteins related to the immune system. Proteins were involved in tissue destruction and different mechanisms against infection as well as protection against tissue damage. These included immune-related proteins such as, immunoglobulins, alarmins, defensins, complement proteins, heat shock proteins, protease inhibitors, and circulatory proteins, as well as cytoskeletal proteins.
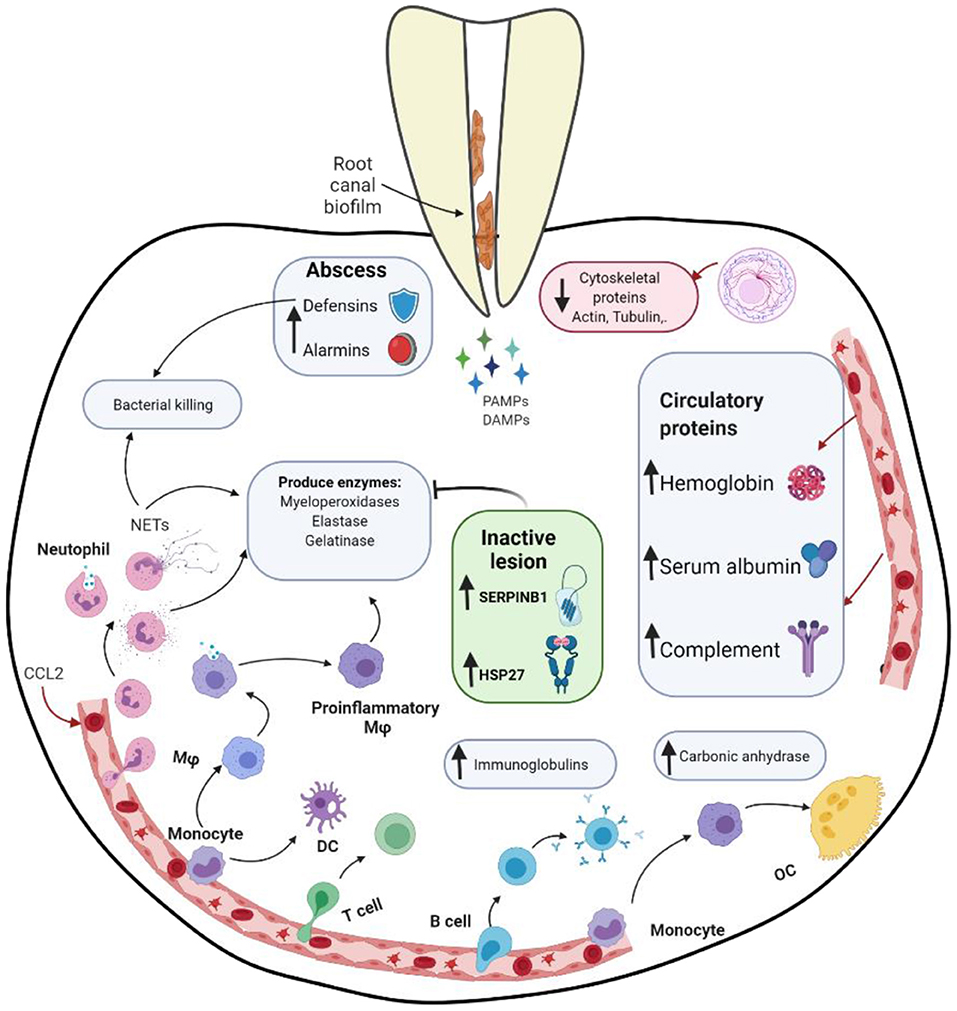
Figure 3. Schematic representing different proteins expression and interactions in apical periodontitis. DAMPs, danger associated molecular patterns; DC, dendritic cell; Mφ, macrophage; NET, neutrophil extracellular traps; OC, osteoclast; PAMPs, pathogen associated molecular patterns; SERPINB1, Serpin Family B Member 1. Schematic created with BioRender.com.
Immune-related proteins have been consistently detected in apical periodontitis. A study conducted by Alfenas et al. analyzed the exoproteome, a set of extracellular proteins including those that are secreted via specific pathways. In this investigation fluids (pus) sampled from acute periapical abscesses were analyzed for host proteins (39). Multiple proteins associated with the immune defense against bacteria were found in pus samples, indicating an active protective response against invading bacteria. Several proteins related to polymorphonuclear neutrophils (PMNs), macrophages and their mediators were identified. PMNs play an important role in the innate immune response to bacterial infection and in abscess formation. PMNs kill bacteria intracellularly after engulfing them or extracellularly upon activation and further cell lysis by forming neutrophil extracellular traps (NETs). NETs are large complexes of DNA molecules associated with nucleic and cytoplasmic proteins, including histones, elastase, myeloperoxidase (MPO), and lactoferrin (62); all of which were detected in sampled tissues included in the study (acute periapical abscess) (39). NETs play a significant role in the innate immune response against a wide range of gram-positive and gram-negative bacteria via degradation of their virulence factors and killing (63). Further, NETs contain up to 20 different inflammatory mediators and effector molecules, such as cathepsin G (CTSG) and elastase. CTSG is one of the key proteolytic effector molecules responsible for direct tissue degradation and pro-inflammatory signaling. In addition to PMNs involvement in microbial killing, they release factors, such as MPO, elastase, and gelatinase, which may cause damage to the adjacent tissues and contribute to the abscess pathogenesis (64). These factors were also detected in this study. Similarly, a study that analyzed the correlation between brain abscess volume and extracellular protein levels, demonstrated that MPO, aminopeptidase N, azurocidin, lactotransferrin, cathelicidin, CTSG, neutrophil collagenase, and resistin, which could be related to neutrophils or macrophages, were among proteins with correlation coefficients > 0.5 (65).
Other immunity-related proteins including protease inhibitors have been identified in both asymptomatic apical periodontitis and abscess samples (37). SERPINB1 (Serpin Family B Member 1 or leukocyte elastase inhibitor), a potent inhibitor of neutrophil serine proteases (elastase and CTSG), was expressed in higher levels in apical periodontitis, with significantly more up-regulation in stable lesions than in progressive lesions (2). These findings were validated using quantitative PCR. SERPINB1 plays a vital role in tissue protection against neutrophil-induced damage, maintaining PMNs survival by antagonizing intracellular CTSG activity (66). SERPINB1 was exclusively expressed in the epithelium and in infiltrating PMNs present in the granulation tissue, suggesting a protective role in inhibiting the cathepsin G-mediated apoptosis of neutrophils and epithelial cells to limit the damage derived from the diffuse and uncontrolled secretion of neutrophil elastase and CTSG (2). Moreover, SERPINB1 was negatively correlated with multiple biomarkers of acute inflammation including CXCR1, matrix metalloproteinase (MMP)-8, MPO, and CTSG (2). The strongest negative correlation was noted for CTSG, indicative of a direct regulatory link between the SERPINB1 expression and the inhibition of the CTSG expression (67). The up-regulation of SERPINB1 might regulate and reduce the destructive potential of NET release, inhibiting the direct degradation of the tissue and helping to stabilize the lesion (68). Overexpression of another serpin family member (SERPINE1) has been previously reported in inactive apical periodontitis (69). SERPINE1 is involved in the acute phase response to infections, inflammatory reactions, and protection of the tissues against proteolytic enzymes (70). Additionally, it is involved in essential healing processes; promotes adhesion, cell migration, and collagen deposition (69). This further reinforces the evidence for the role of this protein family in the pathogenesis as well as successful repair of periradicular disease.
Alarmins are multifunctional immune-activating protein/peptides that act as danger signals and together with pathogen-associated molecular patterns (PAMPs), are considered damage-associated molecular patterns (DAMPs) (71). They are involved in the defense response to bacterial infection as well as lipopolysaccharide, chemokines, and chronic inflammation. They are expressed by PMNs, macrophages, and dendritic cells and released to the extracellular milieu after infection or tissue damage (72). They interact with receptors expressed by host defense cells to stimulate the initiation of innate and adaptive immune responses, triggering inflammation, which allow them to be used as markers of destructive tissue processes (73). S100 proteins (alarmins) have been detected in cases of asymptomatic primary apical periodontitis (37), post-treatment apical periodontitis (38), and for the first time in abscesses (39). S100 proteins exhibit antimicrobial properties through the process of metal limitation (74) and have been reported as potential candidate biomarkers for diagnosis and a predictor of therapeutic responses to inflammation-associated diseases (75).
Heat shock proteins (HSPs), also referred to as molecular chaperones, are known to play a protective role for cells under stress conditions, including infection and inflammation (76). HSPs have been identified in acute periradicular abscesses (39). Interactions between host cells and microbial factors in periradicular lesions result in substantial stress of the involved host cells, which cause damage to cellular proteins and adverse effects on cellular metabolism and lead to the induction of HSPs production. HSPs can stabilize new proteins to fold correctly and help refold proteins damaged by the stress (76). They have been suggested to participate in the pathogenesis of apical periodontitis and influence the prognosis of root canal treatment (77). HSP27 has been found to be 3.79-fold up-regulated in apical periodontitis with significantly higher expression in inactive lesions, implicating a potential role for this molecule in regulating lesion progression (41). HSP27 can modulate neutrophil chemotaxis and activity (78), which could account for the local immune regulation of apical periodontitis. It has an important role in protecting the cells during thermal, oxidative, and chemical stress providing antioxidant and anti-apoptotic functions (30). Additionally, HSP27 suppresses cytokine expression, inhibits neutrophil infiltration, and promotes cell proliferation contributing to the acceleration of wound healing (79).
Defensins are low-molecular-weight cysteine-rich cation proteins with antimicrobial activity. They are primarily involved in innate immunity against pathogens and have been identified in pus sampled from periapical abscesses (37, 39). They are released by neutrophils and macrophages into the extracellular environment during the infection process. Defensins have broad activity spectrum; kill gram-positive and gram-negative bacteria, mycobacteria, fungi, and viruses (80). These antimicrobial peptides may promote the expression of proinflammatory cytokines (81). Another group of proteins involved in the host antibacterial response; neutrophil gelatinase-associated lipocalin, lactoferrin, and transferrin, were also identified in periapical abscesses (39). They exert their antibacterial action through sequestration of iron, which is essential for the growth and virulence of many pathogens, at the site of infection (82). Iron-sequestrating proteins not only deprive bacteria of this essential factor but also some of them, transferrin and lactoferrin, have been suggested to suppress the formation of bacterial biofilms (83).
Circulatory proteins, such as hemoglobin and albumin, as well as components of the complement system, were abundant in samples collected from abscesses, as anticipated since blood components were involved in purulent exudates (39). The complement system is a network of interacting soluble and cell surface-associated molecules that play a key role in both innate and adaptive immune responses to infection (84). It is involved in chemotaxis, opsonization, and lysis of pathogens via formation of a membrane attack complex. The expression of these proteins has been reported to be elevated during periodontal inflammation (85). Complement (C3) has been suggested as a therapeutic target in periodontitis, its inhibition would block the generation of downstream effector molecules regardless of the initiation mechanism of complement activation (86).
Cytoskeleton proteins have been shown to be downregulated in apical periodontitis, which might reflect the tissue destruction resulting from the chronic inflammatory process. Actin, a protein involved in the cytoskeleton structure, participates in the generation of forces and cell adhesion, stabilizes the cell, and determines the shape of the plasma membrane, cell migration, and membrane trafficking (87). Actin cytoskeletal regulation has also been demonstrated to impact the function of immune cells including mast cells, dendritic cells, PMNs, and macrophages (88). Structural proteins were quantitatively more expressed in asymptomatic compared to symptomatic apical periodontitis, including 8 types of actins and 2 types of profilins (61), which might suggest higher cytoskeletal structure injury in symptomatic lesions.
Proteins involved in the adaptive immune response were detected in abscess and post-treatment apical periodontitis such as immunoglobulins and proteins related to T-cell and B-cell activation (38, 39). Immunoglobulins play important roles in the defense against pathogens, including neutralization and clearance of virulence factors, opsonization, and activation of the classical pathway of the complement system (89). Immunoglobulins (mainly IgG) specific against many bacterial species associated with endodontic infections have been previously reported in apical periodontitis lesions (90). Antigen-antibody immune complexes have been reported to be three-fold more abundant in patients with apical abscesses compared to healthy subjects (91). Receptors involved in the regulation of T cells, such as CTLA4 (Cytotoxic T-lymphocyte protein 4), and T-LAK cell originated protein kinase (Lymphokine-activated killer T-cell originated protein kinase), detected in abscess are involved in the activation of lymphoid cells (37).
Comparing symptomatic to asymptomatic apical periodontitis, proteins related to bone remodeling activity were exclusive to the symptomatic cases, including carbonic anhydrase 2, carbonic anhydrase 12, and protein-tyrosine kinase 2-beta. On the other hand, carbonic anhydrase 1, demonstrated higher expression in symptomatic apical periodontitis (61). They inhibit the activity and differentiation of osteoprogenitor cells while stimulating the differentiation and augmentation of resorptive activity of osteoclasts (92). Conversely, proteins that promote the differentiation of osteoblasts and organization of the extracellular matrix, specifically collagen alpha-1 (I) chain and collagen alpha-2 (I) chain, were exclusively detected in the asymptomatic apical periodontitis (61). Osteoclastic activity in periapical lesions has been correlated with the expression of semaphorin 7A (Sema7a) (93), a membrane-associated glycophosphatidylinositol-linked protein, which is involved in the regulation of bone homeostasis, showing high expression during osteoclastic fusion (94). Sema7a plays a critical role during the effector phase of inflammatory immune responses; stimulates pro-inflammatory cytokines, activates monocytes, modulates T-cell function, and regulates chemokine expression, macrophage recruitment, as well as leukocyte trafficking (95–98). Upregulation of Sema7a in symptomatic apical periodontitis and its colocalization with MMP-1 and MMP-3 in vascular vessels and extracellular matrix indicate its involvement in the tissue destruction and infiltration of immune cells in periapical lesions (99).
Proteomic Profile of Pulp Tissue Associated With Periradicular Lesions
Dental pulp is a highly vascularized and richly innervated connective tissue. It has relatively low compliance since it is enclosed in hard dentin tissue (100). Exposure of pulp tissue to non-microbial (traumatic, mechanical, or chemical) or microbial insults, triggers an inflammatory process that varies according to the stimulant type and intensity. As pulp disease progresses, innate effector cells and cells of adaptive immune response alter the physiology of the dental pulp attempting to remove the invading agents. The immune cells recruited within the pulp contribute to the release of soluble molecules or mediators aiming to control bacterial infection. However, they can cause significant collateral host tissue damage. When irritation persists, pulp inflammation will progress leading eventually to pulp necrosis. In the process, bacterial colonization of the root canal system and subsequent neuro-vascular and immunological response would trigger the development of periradicular pathosis (101, 102).
Degradative enzymes, such as matrixins or MMPs necessary for the immune cell migration through the tissue matrix, cause degradative damage and the increased levels of reactive oxygen species (ROS) utilized by immune cells for antimicrobial action (103). ROS, including superoxide anions, hydrogen peroxide, and hydroxyl radicals, can subsequently induce cytokine release by activating the key inflammatory intracellular signaling pathways regulated by the p38 MAPK and NF-κB proteins in several immune and tissue structural cell types (104). ROS could cause major metabolic, transcriptomic, and proteomic changes and greatly challenge the cellular control over a healthy proteome (i.e., proteastasis) (105).
A quantitative approach has been recently applied to describe the proteomic changes in the progression of pulp pathogenesis. This approach allows a more accurate interpretation of the host inflammatory response by comparing the protein expression of normal, inflamed, and necrotic pulp tissues (59, 61). The main findings of studies assessing protein expression in inflamed pulp and necrotic pulp associated with apical periodontitis are presented in Figure 4. Necrotic pulp tissue associated with apical periodontitis has been found to possess the highest number of identified proteins compared to other pulp conditions (inflamed and healthy pulps) (60). Proteins were mainly found in the cytosol and were related to pulp response to stimuli, multicellular organismal process, immune response functions, and stress processes (60). Proteins exclusively detected in necrotic pulp tissue associated with symptomatic apical periodontitis were mainly related to the host's response to viral infection, oxidative stress and proteolytic enzymes, whereas proteins involved in the acute phase of inflammation were more frequently identified in comparison to asymptomatic lesions (61).
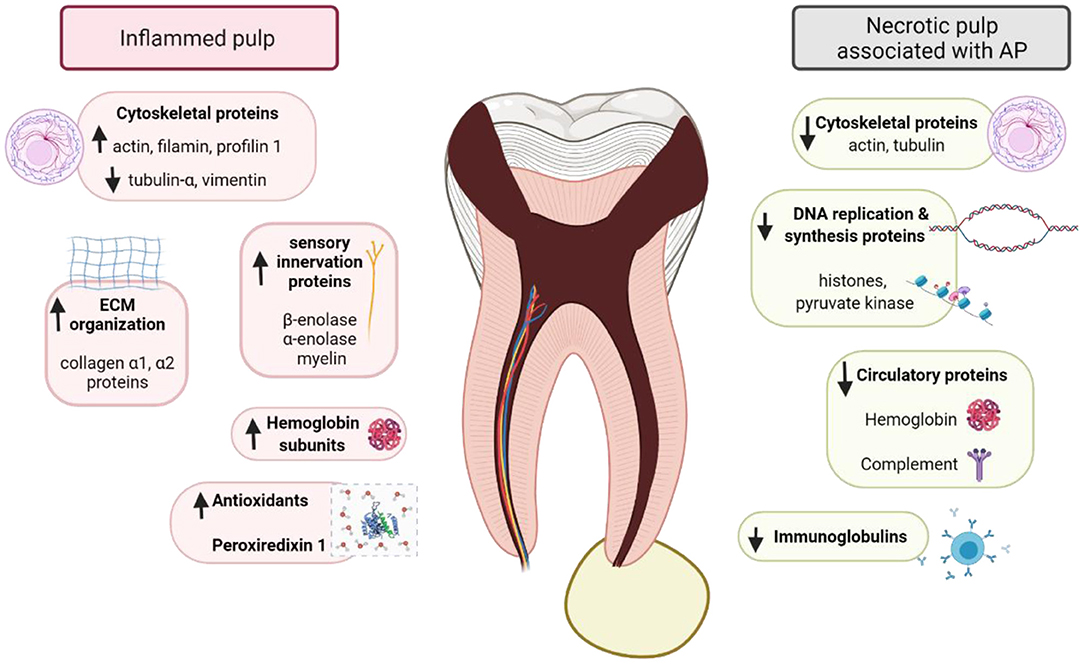
Figure 4. Schematic representing different proteins expression and interactions in inflamed pulp and necrotic pulp associated with apical periodontitis (AP). Schematic created with BioRender.com.
Necrotic pulp associated with chronic apical periodontitis have demonstrated high level of immunity-related proteins, such as immunoglobulins and protease inhibitors, involved in antigen presentation, defense cell activation, and stress response (59), suggesting that host cells react to root canal system infections (37). Immunoglobulins may be related to the immune response as they participate in reducing the diffusion of antigens adherent to the dentinal tubules (106). Protease inhibitors such alpha-2-macroglobulin and SERPINE1 were up-regulated in the necrotic pulp associated with symptomatic apical periodontitis, indicating their significant role in the pulp inflammatory process (107). Alpha-2-macroglobulin (α2M) protects the body against bacterial endotoxins, regulates apoptosis, and inhibits the generation of hydrogen peroxide. It provides a superior action to SERPINE1 in inhibiting proteases, being effective in inactivating all protease groups (59, 61). The increase in its concentration has been reported to be directly related to the severity of inflammatory responses (107). Its high levels in symptomatic cases might be attributed to the increased demand for protease inactivation during the exacerbated host response (61). Further, α2M has been suggested as a biomarker for the diagnosis and prognosis of various diseases (108). Similarly, characterization of the proteome of root canals in teeth with post-treatment apical periodontitis demonstrated several proteins related to immune system process. Histone-lysine Nmethyltransferase 2A, HLA-C protein (fragment), gremlin-1, tumor necrosis factor receptor superfamily member 8, immunoglobulin heavy constant gamma 1, proteoglycan 4, and proteins S100 are some examples (58).
Comparing necrotic and inflamed pulp tissues revealed that larger number of proteins related to stress response were identified in necrotic pulp indicating an ongoing host response to fight invading microbes (109, 110). Stress-related proteins could be due to cell death, besides regulation of proliferation and autophagy (111). However, proteins related to DNA replication and synthesis (histone, pyruvate kinase), extracellular matrix organization (collagen α), cytoskeleton organization (actin cytoplasmic, cofilin, profilin, tubulin-α), cell death regulation (14-3-3 3 protein ζΔ), and sensory innervation (β-enolase) were detected in reduced abundance compared to inflamed pulp (60). Actin, showed down-regulation of 4 of its isoforms indicating that the degradation process of the pulp tissue leads to the destruction of cytoskeleton and rupture of actin microfilaments. Necrotic pulp tissue also showed down-regulation of hemoglobin, when compared with inflamed pulp. This is due to the changes in the microcirculation that led to reduced pulp blood flow, followed by hypoxia and tissue necrosis (112). Contrarily, serum albumin and albumin, which constitute fluids and exudates that infiltrate the apical and lateral foramen of the root canal, were among the up-regulated proteins in the necrotic pulp.
Challenges of Proteomics-Based Studies in Pulp and Periradicular Diseases
High-throughput proteomic methods are powerful screening tools that provide opportunity to understand the protein function and interaction networks (113). In spite of their advantages, it is important to consider some of the technical challenges associated with sampling, processing, and analysis of proteomics data in pulp as well as periradicular tissues. Human clinical samples that could be obtained from a small environment like the root canal or the periapical exudate may not provide sufficient biomass for processing in proteomics assays. Thus, in multiple proteomics-based studies, root canal samples had to be pooled to be properly analyzed (37, 60). Pooling of samples in proteomics has been reported as a valid and potentially valuable procedure, however some issues related to the experimental design should be taken into consideration. The issues of pooling approach include: the protein expression of some proteins in individual samples does not match their expression in the pool; the biological variance between pools is reduced compared with that between individual samples resulting in potential reduction in the statistical power; and proteins evident in individual samples may not always be detectable in pooled samples due to dilution effect (114, 115). The number of proteins identified could be affected by multiple factors; some proteins expression levels may not be enough for detection, while highly abundant peptides may mask the detection of low-abundance peptides (16). Detection of less abundant proteins could be unfeasible, because, unlike DNA, proteins cannot be amplified (116). Additionally, the principle of parsimony adopted by the software used in some studies reduces the occurrence of redundant sequences (37). Different protein normalization methods could be applied to account for variations during proteomics workflows (117).
Analysis of the exoproteome also presents some limitations. For instance, in the study conducted by Alfenas et al. several cytoplasmic proteins were detected, although its objective was to evaluate the exoproteome (39). This might be attributed to rupture of cells during the inflammatory process in response to infection and cells that recently died and released their protein content in the environment (especially in abscesses and in samples taken after treatment), which are still considered as part of the exoproteome if these proteins are stable and remain in abundance. Remnants of necrotic pulp tissue in the canal could also interfere with the detection of exoproteome per se. A few proteins of bacterial subcellular origin were also identified while no intentional attempt was made to lyse the bacterial cells present in the sample (37). This could be due to autolyzed cells or cell lysis caused by vortexing with glass beads, which was performed to dislocate bacteria from the paper points following sample collection. Further, sample collection and processing including cryopulverization could result in lysis of some cells, while taking samples with paper points canals might not provide a good representative sample of the root canal system (118). Cryogenic grinding generates a more representative sample, yet it is only feasible for specimens obtained by tooth extraction or periradicular surgery (38).
Inferences about host responses should be interpreted with caution (3), especially in investigations that did not include healthy controls of root apexes and periradicular tissues. Further, apical periodontitis is a complex and dynamic disease and at a given time, different stages of disease process (pathological changes) can be observed throughout the pulp. The coronal part could be necrotic due to bacterial invasion, while the radicular pulp might remain non-inflamed, thus proteins related to both necrotic and inflamed pulp could be detected simultaneously. Likewise, components of both innate and adaptive immune responses have been found to coexist in abscesses because they often occur as acute exacerbation of previously chronic apical periodontitis lesions (38). Proteomic profiling of different pulp and periradicular conditions revealed differential expression of proteins that are considered constitutively produced and ideally should be unchanged such as beta-actin, glyceraldehyde-3 phosphate dehydrogenase, macroglobulin, actin cytoplasmic, cofilin, tubulin-α and -β (59, 60). Therefore, validation experiments using quantitative methods such as real time-PCR and western blotting analysis should avoid considering such genes/proteins as reference when analyzing samples related to pulp or periradicular diseases (59). Additionally, many proteins could be found only in one of validation assays, possibly due to different aliquots used in each identification method, thus influencing the detection of proteins in low abundance.
Numerous important proteins have been identified through proteomic analyses of periradicular tissues, nevertheless, the diversity of proteins involved in periradicular abscesses may still have been underestimated and further studies are required for validation. Applying rigid filters, such as the 1% false discovery rate, and filtering of data generated from Proteome Discoverer may have contributed to a limited but more reliable protein identification (119, 120). Quantitative assessment of identified proteins was only carried out in 2 studies (59, 61), while only one attempt has been made to correlate the size of periradicular lesions or clinical signs/symptoms to the expression levels of detected proteins in which no significant correlations were found (58). This might be attributed to the low number of samples and also because only one endodontic disease condition (post-treatment endodontic disease) was investigated (58). Studying the correlation between the expression levels of key proteins and clinical signs/symptoms could give an insight about what determines the extent of tissue damage in apical periodontitis. Hence, more studies are warranted to expand the protein database in different endodontic conditions.
Conclusions and Future Perspectives
Identification of biomarkers in host reaction to invading pathogens and subsequent tissue injury is an asset to comprehend the pathogenesis of periradicular diseases. Proteomic-based techniques provide relevant data on host tissue response in various pathological conditions, allowing a broad view of the host's physiology, and the involved biological processes through qualitative and quantitative protein expression profiles. Recently, more studies have been conducted to evaluate the host proteins expressed in response to endodontic infection to identify the biological networks and signaling pathways involved in disease resolution or progression. The cell-specific proteins detected in samples obtained from different pulp and periradicular lesions evidenced the range of cells that take part in the response to endodontic infection in which components of both innate and adaptive immune responses function together. Proteins associated with tissue destruction coexist with those maintaining homeostasis, as mechanisms of protection in apical periodontitis.
Qualitative analysis using bottom-up approach represents the most frequent analytical proteomic technique in endodontic studies (38, 39, 58). However, quantitative analysis would enable more insight on the potential disease/healing biomarkers and help in setting the threshold of differential protein expression in comparison to normal tissues. Quantitative data would also allow correlation analysis to be conducted between immunological and microbiological markers. Performing validation assays, which is essential to verify the differentially expressed proteins obtained from proteomics analysis, is lacking in most proteomics-based clinical studies in endodontics. Future studies could include more validation experiments using antibody-based targeted assays, or antibody-free methods in order to confirm the proteomics findings. Recent rapid advances in MS-based proteomics technologies allow the usage of powerful targeted MS-based methods such as selected reaction monitoring (SRM) and parallel reaction monitoring (PRM) in the verification process (121).
Comprehensive knowledge of protein biomarkers and expression profiles in different endodontic conditions would allow clinicians to inform prognosis and provide treatment with more predictable outcomes. It could also provide researchers with more objective tools to investigate the biological processes involved in periradicular disease and their response to novel therapeutic interventions, which has the potential to help in developing therapeutics targeting certain cells/mediators customized to the patient's lesion condition. Specific protein expression profiles of different sampling sites should be taken into consideration while designing diagnostic tools based on proteomic markers. Establishing predictor markers to monitor patient response to treatment would be another interesting venue for further investigations.
Author Contributions
HH contributed to conception, design, literature search, drafted, and critically reviewed the manuscript. AK contributed to conception, design, and interpretation and critically revised the manuscript. Both authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by a research grant from the Natural Sciences and Engineering Research Council of Canada: Discovery Grant—AK (RGPIN-2020-05844) and the University of Toronto.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fdmed.2022.814603/full#supplementary-material
References
1. Siqueira JF, Rôças IN. Diversity of endodontic microbiota revisited. J Dent Res. (2009) 88:969–81. doi: 10.1177/0022034509346549
2. Cavalla F, Biguetti C, Jain S, Johnson C, Letra A, Garlet GP, et al. Proteomic profiling and differential messenger RNA expression correlate HSP27 and Serpin Family B member 1 to apical periodontitis outcomes. J Endod. (2017) 43:1486–93. doi: 10.1016/j.joen.2017.03.014
3. Hernández-Ríos P, Pussinen PJ, Vernal R, Hernández M. Oxidative stress in the local and systemic events of apical periodontitis. Front Physiol. (2017) 8:869. doi: 10.3389/fphys.2017.00869
4. Vengerfeldt V, Mändar R, Saag M, Piir A, Kullisaar T. Oxidative stress in patients with endodontic pathologies. J Pain Res. (2017) 10:2031–40. doi: 10.2147/JPR.S141366
5. Martinho FC, Nascimento GG, Leite FR, Gomes AP, Freitas LF, Camões IC. Clinical influence of different intracanal medications on Th1-type and Th2-type cytokine responses in apical periodontitis. J Endod. (2015) 41:169–75. doi: 10.1016/j.joen.2014.09.028
6. Fernández A, Cárdenas AM, Astorga J, Veloso P, Alvarado A, Merino P, et al. Expression of Toll-like receptors 2 and 4 and its association with matrix metalloproteinases in symptomatic and asymptomatic apical periodontitis. Clin Oral Investig. (2019) 23:4205–12. doi: 10.1007/s00784-019-02861-9
7. Martinho FC, Teixeira FF, Cardoso FG, Ferreira NS, Nascimento GG, Carvalho CA, et al. Clinical investigation of matrix metalloproteinases, tissue inhibitors of matrix metalloproteinases, and matrix metalloproteinase/tissue inhibitors of matrix metalloproteinase complexes and their networks in apical periodontitis. J Endod. (2016) 42:1082–8. doi: 10.1016/j.joen.2016.04.001
8. Gomes BPFA, Herrera DR. Etiologic role of root canal infection in apical periodontitis and its relationship with clinical symptomatology. Braz Oral Res. (2018) 32(suppl 1):e69. doi: 10.1590/1807-3107bor-2018.vol32.0069
9. Kawashima N, Stashenko P. Expression of bone-resorptive and regulatory cytokines in murine periapical inflammation. Arch Oral Biol. (1999) 44:55–66. doi: 10.1016/S0003-9969(98)00094-6
10. Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. (2001) 69:89–95. doi: 10.1067/mcp.2001.113989
11. Braz-Silva PH, Bergamini ML, Mardegan AP, De Rosa CS, Hasseus B, Jonasson P. Inflammatory profile of chronic apical periodontitis: a literature review. Acta Odontol Scand. (2019) 77:173–80. doi: 10.1080/00016357.2018.1521005
12. Graunaite I, Lodiene G, Maciulskiene V. Pathogenesis of apical periodontitis: a literature review. J Oral Maxillofac Res. (2012) 2:e1. doi: 10.5037/jomr.2011.2401
13. Virdee SS, Butt K, Grant M, Camilleri J, Cooper PR, Tomson PL. A systematic review of methods used to sample and analyse periradicular tissue fluid during root canal treatment. Int Endod J. (2019) 52:1108–27. doi: 10.1111/iej.13104
14. Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. (2012) 13:227–32. doi: 10.1038/nrg3185
15. Liu Y, Beyer A, Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. (2016) 165:535–50. doi: 10.1016/j.cell.2016.03.014
16. Zhang Y, Fonslow BR, Shan B, Baek M-C, Yates JR. Protein analysis by shotgun/bottom-up proteomics. Chem Rev. (2013) 113:2343–94. doi: 10.1021/cr3003533
17. Buszczak M, Signer RAJ, Morrison SJ. Cellular differences in protein synthesis regulate tissue homeostasis. Cell. (2014) 159:242–51. doi: 10.1016/j.cell.2014.09.016
18. Siqueira WL, Dawes C. The salivary proteome: challenges and perspectives. Proteomics Clin Appl. (2011) 5:575–9. doi: 10.1002/prca.201100046
19. Zhang A, Sun H, Wang P, Wang X. Salivary proteomics in biomedical research. Clin Chim Acta. (2013) 415:261–5. doi: 10.1016/j.cca.2012.11.001
20. Wang K, Wang X, Zheng S, Niu Y, Zheng W, Qin X, et al. iTRAQ-based quantitative analysis of age-specific variations in salivary proteome of caries-susceptible individuals. J Transl Med. (2018) 16:293. doi: 10.1186/s12967-018-1669-2
21. PelÁ VT, Ventura TMO, Buzalaf MAR. Optimizing the formation of the acquired enamel pellicle in vitro for proteomic analysis. J Appl Oral Sci. (2020) 28:e20200189. doi: 10.1590/1678-7757-2020-0189
22. Tsuchida S, Satoh M, Takiwaki M, Nomura F. Current status of proteomic technologies for discovering and identifying gingival crevicular fluid biomarkers for periodontal disease. Int J Mol Sci. (2018) 20:86. doi: 10.3390/ijms20010086
23. Park ES, Cho HS, Kwon TG, Jang SN, Lee SH, An CH, et al. Proteomics analysis of human dentin reveals distinct protein expression profiles. J Proteome Res. (2009) 8:1338–46. doi: 10.1021/pr801065s
24. Jágr M, Eckhardt A, Pataridis S, Mikšík I. Comprehensive proteomic analysis of human dentin. Eur J Oral Sci. (2012) 120:259–68. doi: 10.1111/j.1600-0722.2012.00977.x
25. Eckhardt A, Jágr M, Pataridis S, Mikšík I. Proteomic analysis of human tooth pulp: proteomics of human tooth. J Endod. (2014) 4:1961–6. doi: 10.1016/j.joen.2014.07.001
26. Wang H, Ma D, Zhang X, Xu S, Ning T, Wu B. Comparative proteomic profiling of human dental pulp stem cells and periodontal ligament stem cells under in vitro osteogenic induction. Arch Oral Biol. (2018) 89:9–19. doi: 10.1016/j.archoralbio.2018.01.015
27. Akpinar G, Kasap M, Aksoy A, Duruksu G, Gacar G, Karaoz E. Phenotypic and proteomic characteristics of human dental pulp derived mesenchymal stem cells from a natal, an exfoliated deciduous, and an impacted third molar tooth. Stem Cells Int. (2014) 2014:457059. doi: 10.1155/2014/457059
28. Arentz G, Weiland F, Oehler MK, Hoffmann P. State of the art of 2D DIGE. Proteomics Clin Appl. (2015) 9:277–88. doi: 10.1002/prca.201400119
29. Forman HJ, Zhang H. Targeting oxidative stress in disease: promise and limitations of antioxidant therapy. Nat Rev Drug Discov. (2021) 20:689–709. doi: 10.1038/s41573-021-00233-1
30. Vidyasagar A, Wilson NA, Djamali A. Heat shock protein 27 (HSP27): biomarker of disease and therapeutic target. Fibrogenesis Tissue Repair. (2012) 5:7–. doi: 10.1186/1755-1536-5-7
31. Jágr M, Eckhardt A, Pataridis S, Foltán R, Myšák J, Mikšík I. Proteomic analysis of human tooth pulp proteomes—comparison of caries-resistant and caries-susceptible persons. J Proteomics. (2016) 145:127–36. doi: 10.1016/j.jprot.2016.04.022
32. Huynh AH, Veith PD, McGregor NR, Adams GG, Chen D, Reynolds EC, et al. Gingival crevicular fluid proteomes in health, gingivitis and chronic periodontitis. J Periodontal Res. (2015) 50:637–49. doi: 10.1111/jre.12244
33. Tsuchida S. Proteome analysis of molecular events in oral pathogenesis and virus: a review with a particular focus on periodontitis. Int J Mol Sci. (2020) 21:5184. doi: 10.3390/ijms21155184
34. Bao K, Li X, Poveda L, Qi W, Selevsek N, Gumus P, et al. Proteome and microbiome mapping of human gingival tissue in health and disease. Front Cell Infect Microbiol. (2020) 10:588155. doi: 10.3389/fcimb.2020.588155
35. Carnielli CM, Macedo CCS, De Rossi T, Granato DC, Rivera C, Domingues RR, et al. Combining discovery and targeted proteomics reveals a prognostic signature in oral cancer. Nat Commun. (2018) 9:3598. doi: 10.1038/s41467-018-05696-2
36. Nandakumar R, Madayiputhiya N, Fouad AF. Proteomic analysis of endodontic infections by liquid chromatography-tandem mass spectrometry. Oral Microbiol Immunol. (2009) 24:347–52. doi: 10.1111/j.1399-302X.2009.00520.x
37. Provenzano JC, Siqueira JF, Rôças IN, Domingues RR, Paes Leme AF, Silva MR. Metaproteome analysis of endodontic infections in association with different clinical conditions. PLoS One. (2013) 8:e76108. doi: 10.1371/journal.pone.0076108
38. Provenzano JC, Antunes HS, Alves FR, Rôças IN, Alves WS, Silva MR, et al. Host-bacterial interactions in post-treatment apical periodontitis: a metaproteome analysis. J Endod. (2016) 42:880–5. doi: 10.1016/j.joen.2016.02.013
39. Alfenas CF, Mendes TAO, Ramos HJO, Bruckner FP, Antunes HS, Rôças IN, et al. Human exoproteome in acute apical abscesses. J Endod. (2017) 43:1479–85. doi: 10.1016/j.joen.2017.04.019
40. Alptekin NO, Ari H, Haliloglu S, Alptekin T, Serpek B, Ataoglu T. The effect of endodontic therapy on periapical exudate neutrophil elastase and prostaglandin-E2 levels. J Endod. (2005) 31:791–5. doi: 10.1097/01.don.0000158010.43884.59
41. Goodman SC, Letra A, Dorn S, Araujo-Pires AC, Vieira AE, Chaves de Souza L, et al. Expression of heat shock proteins in periapical granulomas. J Endod. (2014) 40:830–6. doi: 10.1016/j.joen.2013.10.021
42. Zhang S, Jian W. Recent advances in absolute quantification of peptides and proteins using LC-MS. Rev Anal Chem. (2014) 33:31–47. doi: 10.1515/revac-2013-0019
43. Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem. (2007) 389:1017–31. doi: 10.1007/s00216-007-1486-6
44. Bantscheff M, Lemeer S, Savitski MM, Kuster B. Quantitative mass spectrometry in proteomics: critical review update from 2007 to the present. Anal Bioanal Chem. (2012) 404:939–65. doi: 10.1007/s00216-012-6203-4
45. Viodé A, Fournier C, Camuzat A, Fenaille F, Bank NB, Latouche M, et al. New antibody-free mass spectrometry-based quantification reveals that C9ORF72 long protein isoform is reduced in the frontal cortex of hexanucleotide-repeat expansion carriers. Front Neurosci. (2018) 12:589. doi: 10.3389/fnins.2018.00589
46. Jedrychowski MP, Wrann CD, Paulo JA, Gerber KK, Szpyt J, Robinson MM, et al. Detection and quantitation of circulating human irisin by tandem mass spectrometry. Cell Metab. (2015) 22:734–40. doi: 10.1016/j.cmet.2015.08.001
47. McAlister GC, Huttlin EL, Haas W, Ting L, Jedrychowski MP, Rogers JC, et al. Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal Chem. (2012) 84:7469–78. doi: 10.1021/ac301572t
48. Larance M, Lamond AI. Multidimensional proteomics for cell biology. Nat Rev Mol Cell Biol. (2015) 16:269–80. doi: 10.1038/nrm3970
49. Bradley BP, Kalampanayil B, O'Neill MC. Protein expression profiling. Methods Mol Biol. (2009) 519:455–68. doi: 10.1007/978-1-59745-281-6_30
50. Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. (2003) 422:198–207. doi: 10.1038/nature01511
51. El Kennani S, Crespo M, Govin J, Pflieger D. Proteomic Analysis of histone variants and their ptms: strategies and pitfalls. Proteomes. (2018) 6:29. doi: 10.3390/proteomes6030029
52. Catherman AD, Skinner OS, Kelleher NL. Top down proteomics: facts and perspectives. Biochem Biophys Res Commun. (2014) 445:683–93. doi: 10.1016/j.bbrc.2014.02.041
53. Smith LM, Kelleher NL, Consortium for Top Down P. Proteoform: a single term describing protein complexity. Nat Methods. (2013) 10:186–7. doi: 10.1038/nmeth.2369
54. Liu S, Li Z, Yu B, Wang S, Shen Y, Cong H. Recent advances on protein separation and purification methods. Adv Colloid Interface Sci. (2020) 284:102254. doi: 10.1016/j.cis.2020.102254
55. Marín-García J, Goldenthal MJ, Moe GW. “CHAPTER 2—Molecular and biochemical methodology in the post-genomic era”. In: Marín-García J, editor. Post-Genomic Cardiology. Burlington: Academic Press (2007). p. 11–25.
56. Coskun O. Separation techniques: chromatography. North Clin Istanb. (2016) 3:156–60. doi: 10.14744/nci.2016.32757
57. Chen C, Hou J, Tanner JJ, Cheng J. Bioinformatics methods for mass spectrometry-based proteomics data analysis. Int J Mol Sci. (2020) 21:2873. doi: 10.3390/ijms21082873
58. Francisco PA, Delboni MG, Lima AR, Xiao Y, Siqueira WL, Gomes BPFA. Proteomic profile of root canal contents in teeth with post-treatment endodontic disease. Int Endod J. (2019) 52:451–60. doi: 10.1111/iej.13021
59. Loureiro C, Buzalaf MAR, Pessan JP, Moraes FRNd, Pelá VT, Ventura TMO, et al. Comparative analysis of the proteomic profile of the dental pulp in different conditions. A pilot study. Braz Dent J. (2020) 31:319–36. doi: 10.1590/0103-6440202003167
60. Silva PAO, Lima SMF, Freire MS, Murad AM, Franco OL, Rezende TMB. Proteomic analysis of human dental pulp in different clinical diagnosis. Clin Oral Investig. (2020) 25:3285–95. doi: 10.1007/s00784-020-03660-3
61. Loureiro C, Buzalaf MAR, Moraes FRN, Ventura TMO, Pelá VT, Pessan JP, et al. Quantitative proteomic analysis in symptomatic and asymptomatic apical periodontitis. Int Endod J. (2021) 54:834–47. doi: 10.1111/iej.13480
62. Lögters T, Margraf S, Altrichter J, Cinatl J, Mitzner S, Windolf J, et al. The clinical value of neutrophil extracellular traps. Med Microbiol Immunol. (2009) 198:211–19. doi: 10.1007/s00430-009-0121-x
63. Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, et al. Neutrophil extracellular traps kill bacteria. Science. (2004) 303:1532–5. doi: 10.1126/science.1092385
64. Klebanoff SJ. Myeloperoxidase: friend and foe. J Leukoc Biol. (2005) 77:598–625. doi: 10.1189/jlb.1204697
65. Hassel B, De Souza GA, Stensland ME, Ivanovic J, Voie Ø, Dahlberg D. The proteome of pus from human brain abscesses: host-derived neurotoxic proteins and the cell-type diversity of CNS pus. J Neurosurg. (2018) 129:829–37. doi: 10.3171/2017.4.JNS17284
66. Baumann M, Pham CTN, Benarafa C. SerpinB1 is critical for neutrophil survival through cell-autonomous inhibition of cathepsin G. Blood. (2013) 121:3900–7. doi: 10.1182/blood-2012-09-455022
67. Henry CM, Sullivan GP, Clancy DM, Afonina IS, Kulms D, Martin SJ. Neutrophil-derived proteases escalate inflammation through activation of IL-36 family cytokines. Cell Rep. (2016) 14:708–22. doi: 10.1016/j.celrep.2015.12.072
68. Yang H, Biermann MH, Brauner JM, Liu Y, Zhao Y, Herrmann M. New insights into neutrophil extracellular traps: Mechanisms of formation and role in inflammation. Front Immunol. (2016) 7:302. doi: 10.3389/fimmu.2016.00302
69. Garlet GP, Horwat R, Ray HL, Garlet TP, Silveira EM, Campanelli AP, et al. Expression analysis of wound healing genes in human periapical granulomas of progressive and stable nature. J Endod. (2012) 38:185–90. doi: 10.1016/j.joen.2011.09.011
70. Carrell RW, Jeppsson JO, Laurell CB, Brennan SO, Owen MC, Vaughan L, et al. Structure and variation of human alpha 1-antitrypsin. Nature. (1982) 298:329–34. doi: 10.1038/298329a0
71. Bianchi ME. DAMPs, PAMPs and alarmins: all we need to know about danger. J Leukoc Biol. (2007) 81:1–5. doi: 10.1189/jlb.0306164
72. Yang D, Han Z, Oppenheim JJ. Alarmins and immunity. Immunol Rev. (2017) 280:41–56. doi: 10.1111/imr.12577
73. Boteanu RM, Suica VI, Uyy E, Ivan L, Dima SO, Popescu I, et al. Alarmins in chronic non-communicable diseases: atherosclerosis, diabetes and cancer. J Proteomics. (2017) 153:21–9. doi: 10.1016/j.jprot.2016.11.006
74. Zackular JP, Chazin WJ, Skaar EP. Nutritional immunity: S100 proteins at the host-pathogen interface. J Biol Chem. (2015) 290:18991–8. doi: 10.1074/jbc.R115.645085
75. Wang S, Song R, Wang Z, Jing Z, Wang S, Ma J. S100A8/A9 in inflammation. Front Immunol. (2018) 9:1298. doi: 10.3389/fimmu.2018.01298
76. De Maio A. Heat shock proteins: facts, thoughts, and dreams. Shock. (1999) 11:1–12. doi: 10.1097/00024382-199901000-00001
77. Maheshwari K, Silva RM, Guajardo-Morales L, Garlet GP, Vieira AR, Letra A. Heat shock 70 protein genes and genetic susceptibility to apical periodontitis. J Endod. (2016) 42:1467–71. doi: 10.1016/j.joen.2016.07.010
78. Jog NR, Jala VR, Ward RA, Rane MJ, Haribabu B, McLeish KR. Heat shock protein 27 regulates neutrophil chemotaxis and exocytosis through two independent mechanisms. J Immunol. (2007) 178:2421. doi: 10.4049/jimmunol.178.4.2421
79. Crowe J, Aubareda A, McNamee K, Przybycien PM, Lu X, Williams RO, et al. Heat shock protein B1-deficient mice display impaired wound healing. PLoS One. (2013) 8:e77383. doi: 10.1371/journal.pone.0077383
80. Jarczak J, Kościuczuk EM, Lisowski P, Strzałkowska N, Józwik A, Horbańczuk J, et al. Defensins: Natural component of human innate immunity. Hum Immunol. (2013) 74:1069–79. doi: 10.1016/j.humimm.2013.05.008
81. Menendez A, Brett Finlay B. Defensins in the immunology of bacterial infections. Curr Opin Immunol. (2007) 19:385–91. doi: 10.1016/j.coi.2007.06.008
82. Ganz T. Iron in innate immunity: starve the invaders. Curr Opin Immunol. (2009) 21:63–7. doi: 10.1016/j.coi.2009.01.011
83. Weinberg ED. Suppression of bacterial biofilm formation by iron limitation. Med Hypotheses. (2004) 63:863–5. doi: 10.1016/j.mehy.2004.04.010
84. Rus H, Cudrici C, Niculescu F. The role of the complement system in innate immunity. Immunol Res. (2005) 33:103–12. doi: 10.1385/IR:33:2:103
85. Gonçalves LDR, Soares MR, Nogueira FCS, Garcia C, Camisasca DR, Domont G, et al. Comparative proteomic analysis of whole saliva from chronic periodontitis patients. J Proteomics. (2010) 73:1334–41. doi: 10.1016/j.jprot.2010.02.018
86. Hajishengallis G, Maekawa T, Abe T, Hajishengallis E, Lambris JD. Complement involvement in periodontitis: molecular mechanisms and rational therapeutic approaches. Adv Exp Med Biol. (2015) 865:57–74. doi: 10.1007/978-3-319-18603-0_4
87. Navarro-Garcia F, Serapio-Palacios A, Ugalde-Silva P, Tapia-Pastrana G, Chavez-Dueñas L. Actin cytoskeleton manipulation by effector proteins secreted by diarrheagenic Escherichia coli pathotypes. Biomed Res Int. (2013) 2013:374395. doi: 10.1155/2013/374395
88. Kopecki Z, Ludwig RJ, Cowin AJ. Cytoskeletal regulation of inflammation and its impact on skin blistering disease epidermolysis bullosa acquisita. Int J Mol Sci. (2016) 17:1116. doi: 10.3390/ijms17071116
89. Schroeder HW, Cavacini L. Structure and function of immunoglobulins. J Allergy Clin Immunol. (2010) 125:S41–S52. doi: 10.1016/j.jaci.2009.09.046
90. Baumgartner JC, Falkler WA. Reactivity of IgG from explant cultures of periapical lesions with implicated microorganisms. J Endod. (1991) 17:207–12. doi: 10.1016/S0099-2399(06)81922-9
91. Kettering JD, Torabinejad M. Concentrations of immune complexes, IgG, IgM, IgE, and C3 in patients with acute apical abscesses. J Endod. (1984) 10:417–21. doi: 10.1016/S0099-2399(84)80262-9
92. Katagiri M, Ogasawara T, Hoshi K, Chikazu D, Kimoto A, Noguchi M, et al. Suppression of adjuvant-induced arthritic bone destruction by cyclooxygenase-2 selective agents with and without inhibitory potency against carbonic anhydrase II. J Bone Miner Res. (2006) 21:219–27. doi: 10.1359/JBMR.051025
93. He M, Bian Z. Expression of hypoxia-induced semaphorin 7A correlates with the severity of inflammation and osteoclastogenesis in experimentally induced periapical lesions. Arch Oral Biol. (2017) 75:114–9. doi: 10.1016/j.archoralbio.2016.10.032
94. Delorme G, Saltel F, Bonnelye E, Jurdic P, Machuca-Gayet I. Expression and function of semaphorin 7A in bone cells. Biol Cell. (2005) 97:589–97. doi: 10.1042/BC20040103
95. Kamata M, Tada Y, Uratsuji H, Kawashima T, Asano Y, Sugaya M, et al. Semaphorin 7A on keratinocytes induces interleukin-8 production by monocytes. J Dermatol Sci. (2011) 62:176–82. doi: 10.1016/j.jdermsci.2011.02.004
96. Morote-Garcia JC, Napiwotzky D, Köhler D, Rosenberger P. Endothelial Semaphorin 7A promotes neutrophil migration during hypoxia. Proc Natl Acad Sci U S A. (2012) 109:14146–51. doi: 10.1073/pnas.1202165109
97. Holmes S, Downs AM, Fosberry A, Hayes PD, Michalovich D, Murdoch P, et al. Sema7A is a potent monocyte stimulator. Scand J Immunol. (2002) 56:270–5. doi: 10.1046/j.1365-3083.2002.01129.x
98. Suzuki K, Okuno T, Yamamoto M, Pasterkamp RJ, Takegahara N, Takamatsu H, et al. Semaphorin 7A initiates T-cell-mediated inflammatory responses through alpha1beta1 integrin. Nature. (2007) 446:680–4. doi: 10.1038/nature05652
99. Song Y, Wang L, Li J, Yang F, Gao Y, Song D, et al. The expression of Semaphorin 7A in human periapical lesions. J Endod. (2021) 47:1631–9. doi: 10.1016/j.joen.2021.06.005
100. Kawashima N, Okiji T. Odontoblasts: Specialized hard-tissue-forming cells in the dentin-pulp complex. Congenital Anomalies. (2016) 56:144–53. doi: 10.1111/cga.12169
101. Siqueira JF, Rôças IN. Community as the unit of pathogenicity: an emerging concept as to the microbial pathogenesis of apical periodontitis. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. (2009) 107:870–8. doi: 10.1016/j.tripleo.2009.01.044
102. Jacinto RC, Gomes BP, Ferraz CC, Zaia AA, Filho FJ. Microbiological analysis of infected root canals from symptomatic and asymptomatic teeth with periapical periodontitis and the antimicrobial susceptibility of some isolated anaerobic bacteria. Oral Microbiol Immunol. (2003) 18:285–92. doi: 10.1034/j.1399-302X.2003.00078.x
103. Löffek S, Schilling O, Franzke CW. Biological role of matrix metalloproteinases: a critical balance. Eur Respir J. (2011) 38:191. doi: 10.1183/09031936.00146510
104. Simon S, Smith AJ, Berdal A, Lumley PJ, Cooper PR. The MAP kinase pathway is involved in odontoblast stimulation via p38 phosphorylation. J Endod. (2010) 36:256–9. doi: 10.1016/j.joen.2009.09.019
105. Reichmann D, Voth W, Jakob U. Maintaining a healthy proteome during oxidative stress. Mol Cell. (2018) 69:203–13. doi: 10.1016/j.molcel.2017.12.021
106. Hahn CL, Best AM. The pulpal origin of immunoglobulins in dentin beneath caries: an immunohistochemical study. J Endod. (2006) 32:178–82. doi: 10.1016/j.joen.2005.08.016
107. McClanahan SB, Turner DW, Kaminski EJ, Osetek EM, Heuer MA. Natural modifiers of the inflammatory process in the human dental pulp. J Endod. (1991) 17:589–93. doi: 10.1016/S0099-2399(06)81830-3
108. Rehman AA, Ahsan H, Khan FH. α-2-Macroglobulin: a physiological guardian. J Cell Physiol. (2013) 228:1665–75. doi: 10.1002/jcp.24266
109. Siqueira JF, Rôças IN. Bacterial pathogenesis and mediators in apical periodontitis. Braz Dent J. (2007) 18:267–80. doi: 10.1590/S0103-64402007000400001
110. Márton IJ, Kiss C. Protective and destructive immune reactions in apical periodontitis. Oral Microbiol Immunol. (2000) 15:139–50. doi: 10.1034/j.1399-302x.2000.150301.x
111. Asea A. Stress proteins and initiation of immune response: chaperokine activity of hsp72. Exerc Immunol Rev. (2005) 11:34–45.
112. Abbott PV, Yu C. A clinical classification of the status of the pulp and the root canal system. Aust Dent J. (2007) 52(1 Suppl):S17–31. doi: 10.1111/j.1834-7819.2007.tb00522.x
113. Pääkkönen V, Tjäderhane L. High-throughput gene and protein expression analysis in pulp biologic research: review. J Endod. (2010) 36:179–89. doi: 10.1016/j.joen.2009.10.016
114. Diz AP, Truebano M, Skibinski DOF. The consequences of sample pooling in proteomics: an empirical study. Electrophoresis. (2009) 30:2967–75. doi: 10.1002/elps.200900210
115. Zolg W. The proteomic search for diagnostic biomarkers: lost in translation?. Mol Cell Proteom. (2006) 5:1720–6. doi: 10.1074/mcp.R600001-MCP200
116. Cho WC. Proteomics technologies and challenges. Genom Proteom Bioinform. (2007) 5:77–85. doi: 10.1016/S1672-0229(07)60018-7
117. Gokce E, Shuford CM, Franck WL, Dean RA, Muddiman DC. Evaluation of normalization methods on GeLC-MS/MS label-free spectral counting data to correct for variation during proteomic workflows. J Am Soc Mass Spectrom. (2011) 22:2199–208. doi: 10.1007/s13361-011-0237-2
118. Alves FR, Siqueira JF, Carmo FL, Santos AL, Peixoto RS, Rôças IN, et al. Bacterial community profiling of cryogenically ground samples from the apical and coronal root segments of teeth with apical periodontitis. J Endod. (2009) 35:486–92. doi: 10.1016/j.joen.2008.12.022
119. Gupta N, Pevzner PA. False discovery rates of protein identifications: a strike against the two-peptide rule. J Proteome Res. (2009) 8:4173–81. doi: 10.1021/pr9004794
120. Searle BC. Scaffold: a bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics. (2010) 10:1265–9. doi: 10.1002/pmic.200900437
Keywords: apical periodontitis, inflammatory mediators, biomarkers, proteomics, mass spectrometry
Citation: Hussein H and Kishen A (2022) Application of Proteomics in Apical Periodontitis. Front. Dent. Med. 3:814603. doi: 10.3389/fdmed.2022.814603
Received: 13 November 2021; Accepted: 20 January 2022;
Published: 10 February 2022.
Edited by:
Frederico C. Martinho, University of Maryland, United StatesReviewed by:
Rogerio Jacinto, São Paulo State University, BrazilOleh Andrukhov, University Dental Clinic Vienna, Austria
Copyright © 2022 Hussein and Kishen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anil Kishen, YW5pbC5raXNoZW5AZGVudGlzdHJ5LnV0b3JvbnRvLmNh
 Hebatullah Hussein
Hebatullah Hussein Anil Kishen
Anil Kishen