- 1Key Laboratory of Shaanxi Province for Craniofacial Precision Medicine Research, College of Stomatology, Xi'an Jiaotong University, Xi'an, China
- 2The State Key Laboratory Breeding Base of Basic Science of Stomatology (Hubei-MOST) and Key Laboratory of Oral Biomedicine Ministry of Education, School and Hospital of Stomatology, Wuhan University, Wuhan, China
- 3Department of Periodontology, College of Stomatology, Xi'an Jiaotong University, Xi'an, China
Stem cells derived from dental tissues (DSCs) exhibit multipotent regenerative potential in pioneering tissue engineering regimens. The multipotency of DSCs is critically regulated by an intricate range of factors, of which the epigenetic influence is considered vital. To gain a better understanding of how epigenetic alterations are involved in the DSC fate determination, the present review overviews the current knowledge relating to DSC epigenetic modifications, paying special attention to the landscape of epigenetic modifying agents as well as the related signaling pathways in DSC regulation. In addition, insights into the future opportunities of epigenetic targeted therapies mediated by DSCs are discussed to hold promise for the novel therapeutic interventions in future translational medicine.
Introduction
Dental stem cells (DSCs), a subgroup of mesenchymal stem cells (MSCs), are isolated mainly from dental pulp or periodontium-associated tissues. Current identified DSCs are dental pulp stem cells (DPSCs) (1, 2), stem cells from human exfoliated deciduous teeth (SHED) (3), periodontal ligament stem cells (PDLSCs) (4), dental follicle precursor cells (DFPCs) (5), stem cells from apical papilla (SCAP) (6), gingival-derived MSCs (GMSCs) (7), and alveolar bone marrow-derived MSCs (ABMSCs) (8, 9). In vitro identification demonstrated that DSCs positively expressed MSC-related markers, including CD13, CD29, CD44, CD59, CD73, CD90, CD105, CD146, and STRO-1; while negatively expressed hematopoietic markers, such as CD14, CD31, CD34, and CD45 (10–12). The key pluripotency markers (Oct4, Sox2, and Nanog) that are involved in maintaining the undifferentiated state were also expressed in DSCs with different levels (13). More importantly, a recent study revealed that Oct4 was only expressed in the nuclei of DPSCs, suggesting a better potential of DPSCs to differentiate into multi-lineage tissues (14). DSCs are easily accessible with no ethical controversies. Compared with other MSCs, DSCs presented better proliferative properties and odontogenic differentiation abilities, making them attractive candidates in the future application of tissue regeneration (15, 16).
It is generally believed that MSCs prefer to present regenerative potential corresponding to their origin in ectopic (17). In this regard, DSC-mediated oral and craniofacial regenerative therapies have lead a way in decades. Wang et al. revealed that the PDLSC/PRF/ABMSC sheet composites successfully regenerated periodontal ligament- and bone-like structures in nude mice (18). When SCAP and PDLSCs were co-transplanted into the alveolar bone of mini-pigs, biological roots were formed (19). Expressing neural and angiogenic markers, such as neurofilament, nestin, vascular endothelial growth factor (VEGF), and angiogenin-1, SCAP presented great neurogenic and angiogenic potential (6, 20, 21). More importantly, clinical trials in recent years have provided more convincing evidence that the implanted DPSCs might induce functional dental pulp (22). Application of DPSCs in deep intrabony defects significantly improved clinical and radiographic parameters of periodontal regeneration 1 year after implantation (23). Nevertheless, despite the current encouragements, to improve the regenerative efficiency of DSCs, the underlying mechanisms that modulate DSC function are needed to be critically explored.
Emerging evidence has indicated that DSC fate is of great importance in directing their functions (11, 24). Potentiated to osteogenesis, adipogenesis, chondrogenesis, neurogenesis, and angiogenesis, DSCs may facilitate multiple tissue regeneration (25). To take full advantage of DSCs, key regulators that affect DSC fate should be critically discussed ahead of their applications. Various factors were reported to be involved in determining cell fate, of which epigenetic regulation has been considered as a vital cue (26). Without gene editing, epigenetic regulation plays a stable and heritable role in DSC function (27, 28).
Epigenetic modifications are generally orchestrated by DNA methylation, histone modifications, RNA modifications, and non-coding RNAs (29). DNA methylation, one of the most well-studied epigenetic modifications, is catalyzed by DNA methyltransferases (DNMTs) (24). In CpG islands, the C5 position of cytosine can be methylated into 5-methylcytosine [5-mC] by DNMTs, such as DNMT1, DNMT3A, and DNMT3B (24). DNA methylation can also be reversed by enzymes of the ten-eleven translocation (TET) family (e.g., TET1, TET2, and TET3) (30).
Histone methyltransferases (HMTs) result in chromatin compaction or relaxation (31). Histone 3 (H3) is the most extensively modified histone. Histone 3 lysine 9 (H3K9) and H3K27 are associated with specific and dynamic repression of transcriptional regulation. By contrast, H3K4, H3K36, and H3K79 facilitate target gene transcriptional activation (32). Histone lysine demethylases (KDMs), removing methyl groups from histones, mainly include the lysine-specific demethylase 1 (LSD1) and Jumonji C (JMJC) families (33). Histone acetyltransferases (HATs) result in a more open chromatin structure for transcription factor binding that induces gene transcription (34). However, the effects of HATs can be counteracted by histone deacetylases (HDACs), leading to chromatin compaction and transcriptional repression (35).
Internal mRNA modifications, such as N6-methyladenosine (m6A), N1-methyladenosine (m1A), 5-mC, and 5-hydroxymethylcytosine (5-hmC), are critical in the maintenance of mRNA stability (36). As the most prevalent internal RNA modification, m6A modification is catalyzed by methyltransferases and reversed by demethylases (29). Abnormal regulation that leads to an imbalance in m6A levels might result in incompetent osteogenic differentiation of bone marrow MSCs (BMMSCs) (37) and impaired odontogenic differentiation of human DPSCs (38). Besides, micro-RNAs (miRNAs) (39), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs) (40) are critical components in epigenetic regulation mediated by non-coding RNAs. Dysregulation of these non-coding RNAs was found to have relevance with not only DSC fate but also their potential to regulate tissue regeneration.
To better understand how these epigenetic alterations are involved in DSC fate, the present review critically overviews the epigenetic modifiers in the regulation of DSCs and the relevant key pathways, providing a theoretical basis for the future translation of DSCs in regenerative medicine. Insights into the future perspectives are also provided for the identification of novel epigenetic targeted regenerative therapies mediated by DSCs.
Epigenetic Modulations of DSCS
DNA Methylation
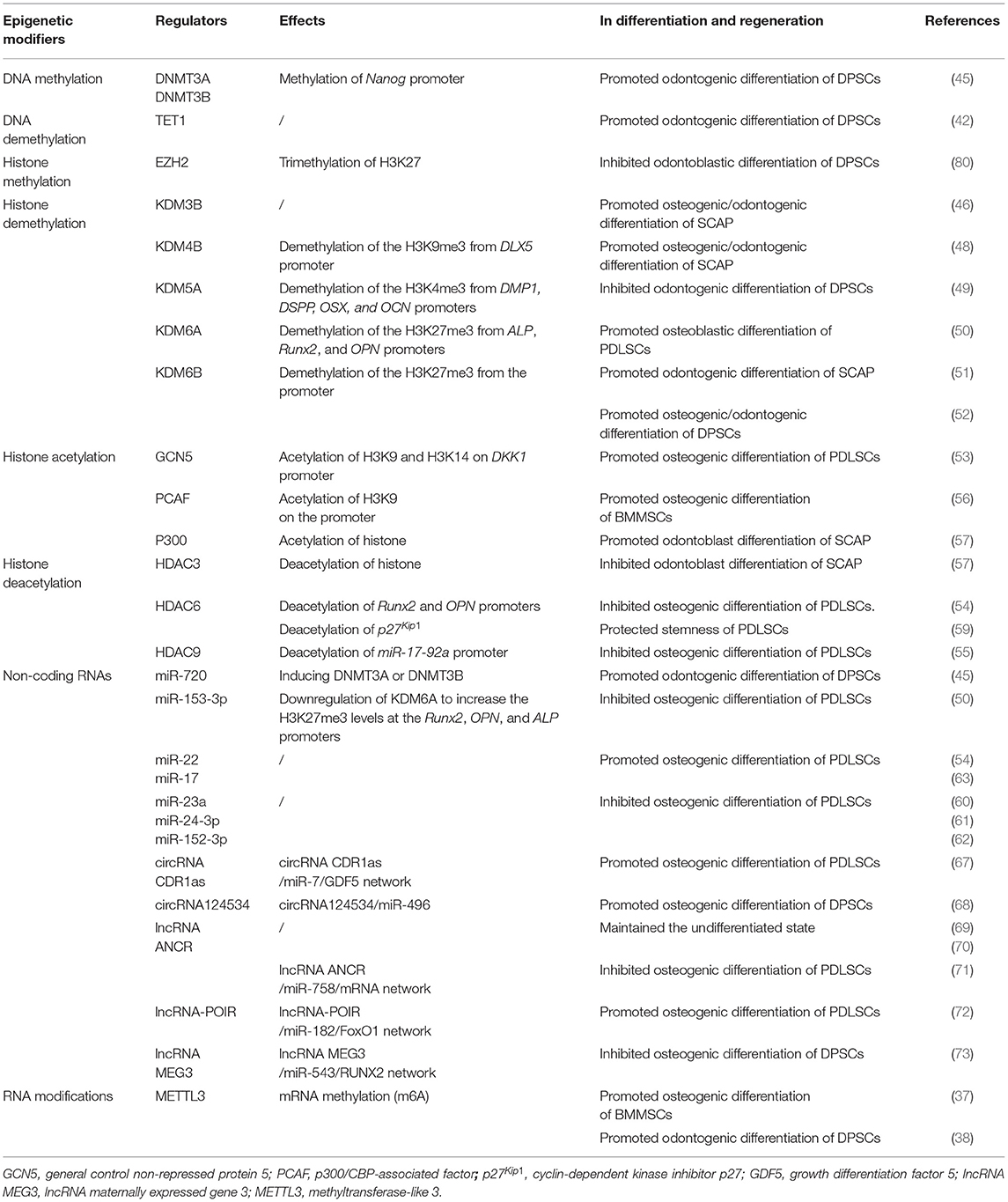
It has been proved that DNA methylation profiles are associated with the osteogenic potential of DSCs. Using the HumanMethylation450 Beadchip, Ai et al. indicated that DPSCs, DFPCs, and PDLSCs showed highly similar DNA methylation patterns that were related to bone formation. Compared with DPSCs and DFPCs, PDLSCs had higher transcription levels of osteogenic-related factors indicating a better osteogenic capacity in vitro and in vivo (41). Besides the regulation of osteogenic differentiation, DNA methylation is also critical in modulating the odontogenic differentiation of DSCs. TET1 knockdown suppressed the proliferation and odontogenic differentiation of DPSCs, suggesting TET1-dependent DNA demethylation in dental tissue regeneration (42, 43). When DPSCs were treated with lipopolysaccharide, DNMT1 was downregulated, while the transcription of inflammatory cytokines was increased (44). This study provided a new rationale to reveal the molecular mechanisms of the inflamed dental pulp and was of instructive significance in revealing the effect of DNMT1 on the differentiation capacity of DPSCs in inflammatory conditions. It has also been indicated that by inducing DNMT3A/DNMT3B, the odontogenic differentiation of DPSCs was promoted, providing a potential target for DPSC mediated odontogenic regeneration (45). Taken together, DNA methylation is vital to the regulation of osteogenic and odontoblastic differentiation of DSCs, contributing to the regeneration of bone-related defects.
Histone Methylation
In addition to DNA methylation, histone modifications are also responsible for DSC fate, of which KDMs are of great importance. KDM3B enhanced the osteo-/odontogenic differentiation potential of SCAP (46). KDM4B removed H3K9me3 by binding with DLX promoters, leading to target gene activation (47). Yang et al. indicated that KDM4B and DLX5 regulated each other via a positive feedback loop in SCAP. DLX5 enhanced the alkaline phosphatase (ALP) activity and the expression of DSPP, DMP1, OPN, and OSX, inducing osteogenic differentiation. In vivo studies on nude mice further demonstrated that DLX5 promoted osteo-/dentinogenesis via upregulating KDM4B in SCAP (48). KDM5A inhibited the odontogenic differentiation potentiality of DPSCs by removing H3K4me3 from DMP1, DSPP, OSX, and OCN promoters (49). KDM6A promoted osteoblastic differentiation of PDLSCs by decreasing H3K27me3 on promoters of ALP, Runx2, and OPN, suggesting a potential intervention for periodontal repair and regeneration (50). KDM6B was dynamically expressed in the bell stage of human tooth germs. KDM6B knockdown induced H3K27me3, which repressed the transcriptional activity of Wnt5A and ultimately suppressed odontogenic differentiation of SCAP (51). In addition, inhibition of KDM6B also resulted in a marked decrease of mineralization-related genes (ALP, BMP2, BMP4, DLX2, OCN, and OPN) during odontogenic/osteogenic differentiation of DPSCs (52).
Histone Acetylation
HATs and HDACs are also involved in DSC fate decisions. The acetyltransferase GCN5 regulated DKK1 expression by acetylation of H3K9 and H3K14 at its promoter region. Inhibiting the Wnt/β-catenin pathway through DKK1, GCN5 increased the osteogenic differentiation of PDLSCs. In vivo injection of aspirin that targeted GCN5 would rescue periodontitis of rats by promoting osteogenic differentiation of PDLSCs (53). By suppressing HDAC6, miR-22 would promote osteogenic differentiation of PDLSCs (54). Similarly, HDAC9 and miR-17 formed a loop of mutual inhibition during osteogenic differentiation of PDLSCs (55). Zhang et al. also revealed that PCAF knockdown resulted in critical repression of the osteogenic differentiation of BMMSCs by reducing H3K9 acetylation of BMP signaling (56), suggesting different osteogenic regulation of DSCs and other MSCs. Tao et al. indicated that the coordinated expression of p300 and HDAC3 was critical for odontoblast differentiation of SCAP, providing new hints for restorative dentistry (57). It was also reported that the expression levels of SIRT7 and HDAC6 were decreased in stem cell aging models, whereas SIRT7 overexpression rescued the miR-152-induced senescence phenotype of DPSCs (58). Li et al. further suggested that HDAC6 played an important role in PDLSC aging (59). The aforementioned literature provided candidate targets to improve the functional and therapeutic potential of DSCs.
Non-coding RNAs
It has been demonstrated that several miRNAs participated in the regulation of osteogenic differentiation in DSCs, such as miR-23a (60), miR-24-3p (61), miR-152-3p (62), miR-17 (63), and miR-22 (54). miR-23a acted as a negative regulator during osteogenesis of PDLSCs from patients with periodontitis by inhibiting the phosphorylation of Smad1/5/ 9, key transcription factors of the BMP pathway (60). In addition, miR-24-3p and miR-152-3p suppressed the osteogenic differentiation of PDLSCs (61, 62). On the contrary, miR-22 and miR-17 promoted the osteogenic differentiation of PDLSCs (54, 63). The above findings demonstrated miRNAs' different effects on periodontal and alveolar bone regeneration.
The expression profiles of circRNAs and lncRNAs have revealed an extremely complex regulatory network (64). Specific circRNAs are involved in the osteoblastic differentiation of DSCs (65, 66). For instance, circRNA CDR1as inhibited miR-7 and alleviated the negative regulatory effect of miR-7 on osteoblastic differentiation (67). CircRNA124534 promoted the osteogenic differentiation of DPSCs by inhibiting miR-496, suggesting the functions and underlying mechanisms of circRNAs in the osteogenic differentiation (68). Anti-differentiation ncRNA (ANCR), a subclass of lncRNAs, can maintain the undifferentiated stem cell state (69, 70). Furthermore, lncRNA–miRNA–mRNA networks are also critical in osteogenesis. For instance, overexpression of ANCR downregulated miR-758, inhibiting the osteogenic differentiation of PDLSCs by reducing ALP, RUNX2, and OSX (71). The osteogenesis impairment-related lncRNA (lncRNA-POIR) competitively inhibited miR-182 in PDLSCs, leading to de-repression of its target gene FoxO1. FoxO1 increased bone formation of PDLSCs by competing for β-catenin with TCF-4 (a transcriptional cofactor) and inhibiting the canonical Wnt pathway (72). LncRNA MEG3 inhibited osteogenic differentiation of DPSCs via miR-543/RUNX2 regulatory network, which might contribute to the precise regulation of DPSC differentiation (73).
Key Pathways Regulated by Epigenetic Mechanisms
Wnt Signaling Pathway
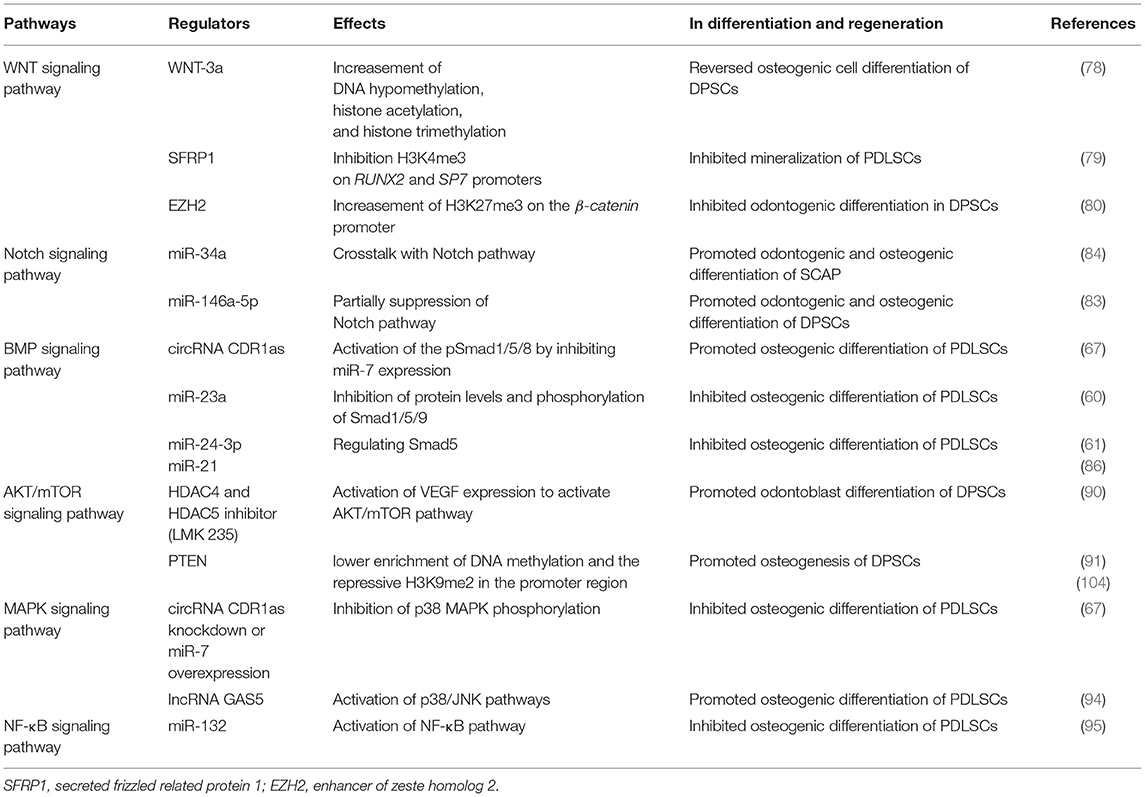
Wnt ligands bind to the Wnt receptors on the cell surfaces or induce β-catenin release to bind TCF in the nucleus to regulate downstream signaling (74). It has been proved that the Wnt signaling pathway is involved in the regulation of DSC stemness (75) and multipotency (76, 77). Through mass spectroscopy, Uribe-Etxebarria et al. revealed that activation of Wnt signaling induced epigenetic remodeling in DPSCs, mainly by inducing DNA demethylation, histone acetylation, and histone methylation (78). Moreover, SFRP1 (a Wnt antagonist) inhibited the mineralization of PDLSCs through H3K4me3-mediated regulation, maintaining the nonmineralized state of PDLSCs (79). EZH2 depletion activated the Wnt/β-catenin pathway led to the promotion of odontogenic differentiation in DPSCs (80).
Notch Signaling Pathway
Notch receptors and ligands are highly conserved type I transmembrane proteins. Once activated, Notch would release the Notch intracellular domain (NICD), allowing its consequent translocation to the nucleus and activating the expression of Notch targets (81). When nuclear NICD is absent, a repressor complex CSL/RBP-Jk will form in the nucleus by recruiting ubiquitous corepressors and HATs, repressing Notch targets (82). Activation of Notch enhanced the stemness and potency of DPSCs, providing a novel approach in DPSC related therapeutics (75). Notch signaling regulated by non-coding RNAs is also critical in DSC fate decisions. miR-146a-5p/Notch signaling played an important role in supporting odontogenic and osteogenic differentiation of STRO-1+ DPSCs, indicating that the application of DSCs may be facilitated by epigenetic modifications in regenerative medicine and tissue engineering (83). miR-34a interacted with Notch signaling and promoted both odontogenic and osteogenic differentiation of SCAP (84).
BMP Signaling Pathway
Among the variety of signaling and transcription factors involved in osteogenesis, BMP/Smads signaling is considered to play a central role in controlling osteogenic differentiation. Exploration of all known factors affecting osteogenic differentiation and their interactions is of major importance in the field of DSC-mediated regenerative medicine. In the canonical BMP pathway, activated BMP phosphorylates the transcription factors Smad1/5/8. The phosphorylated Smads (pSmad1/5/8) form a heterodimeric complex with Smad4, which is then translocated into the nucleus to activate the expression of downstream osteogenic genes (85). At the epigenetic level, circRNA CDR1as activated pSmad1/5/8 by inhibiting the miR-7 expression and further promoted the osteogenesis of PDLSCs (67). Downregulation of miR-24-3p or miR-21 promoted osteogenic differentiation of PDLSCs by targeting Smad5 (61, 86). Under long-term BMP stimulations, DFPCs significantly increased ALP activity and mineralization (87).
AKT/mTOR Signaling Pathway
Phosphatidylinositide 3 kinases (PI3Ks), serine/threonine kinase AKT (also known as protein kinase B), and mTOR (also known as mechanistic TOR) are considered as core components of the PI3K/AKT/mTOR signaling cascade, participating in the regulation of DSC proliferation and differentiation. Under physiological conditions, the receptor tyrosine kinase (RTK) is activated by growth factors or cytokines and then phosphorylates the tyrosine residue to recruit PI3Ks (88, 89). It has been confirmed that the expression of vascular endothelial growth factor (VEGF) is repressed by HDACs, especially HDAC4. The VEGF/AKT/mTOR pathway promoted odontoblast differentiation of DPSCs after treatment with LMK235, a specific inhibitor of HDAC4 and HDAC5 (90). Additionally, the phosphatase and tensin homolog (PTEN) exhibited higher enrichment of DNA methylation and the repressive H3K9me2 in the promoter region of BMMSCs compared to DPSCs. PTEN suppression activated the AKT pathway to promote adipogenesis and inhibit osteogenesis of DPSCs, suggesting a differed differentiation potential of DPSCs and BMMSCs (91).
MAPK Signaling Pathway
The MAPK pathways consist of extracellular signal-related kinases (ERK1/2), C-Jun N-terminal kinase (JNK), p38 MAPK, and ERK5 (92, 93). Decreased phosphorylation of MAPK mediated by circRNA CDR1as knockdown would lead to the inhibited osteogenic differentiation of PDLSCs (67). While lncRNA GAS5 enhanced the osteogenic differentiation of PDLSCs via GDF5 and p38/JNK signaling pathway (94). These findings provided the theoretical basis for understanding the osteogenesis mechanism in PDLSCs.
NF-κB Signaling Pathway
NF-κB is a transcription factor that is ubiquitous in the cytoplasm. It has pivotal roles in the regulation of inflammatory response and osteogenic differentiation of DSCs (95–97). NF-κB stays in an inactive state when it binds to the inhibitory protein IκB. Once activated by the activators, NF-κB dissociates from IκB and translocates into the nucleus to bind with the target genes and enhance their expression (98). Martins et al. revealed that NF-κB was involved in the epigenetic regulation of oral epithelial cells, suggesting a potential mechanism of their roles in DSCs (99). Activating NF-κB signaling by miR-132, the osteogenic ability of PDLSCs was inhibited, suggesting a potential role of NF-κB in future periodontal related therapy (95).
Future Perspectives
Knowing the promising roles of epigenetic regulation in DSC fate (Figure 1, Table 1), new insights into future opportunities would provide promise for the identification of novel targets for DSC-mediated regenerative therapies. DSCs may partially lose their potential during long-term cell culture. In this context, how to ensure stemness of DSCs in normal culture conditions is critical to address. Diomede et al. revealed that 5-Aza (a DNMT inhibitor) induced the direct conversion of GMSCs into embryonic lineages. When treated with 5-Aza for 48 h, GMSCs were organized as round 3D structures and expressed markers related to three germ layers (100). Above findings suggested a possible application of epigenetic regulation in future translational medicine. It has further been demonstrated that 5-Aza treatment is responsible for GMSC dedifferentiation into embryonic lineages other than neural precursor cells after prolonged expansion (101, 102), suggesting the potential role of 5-Aza in DSC stemness maintaining for future application.
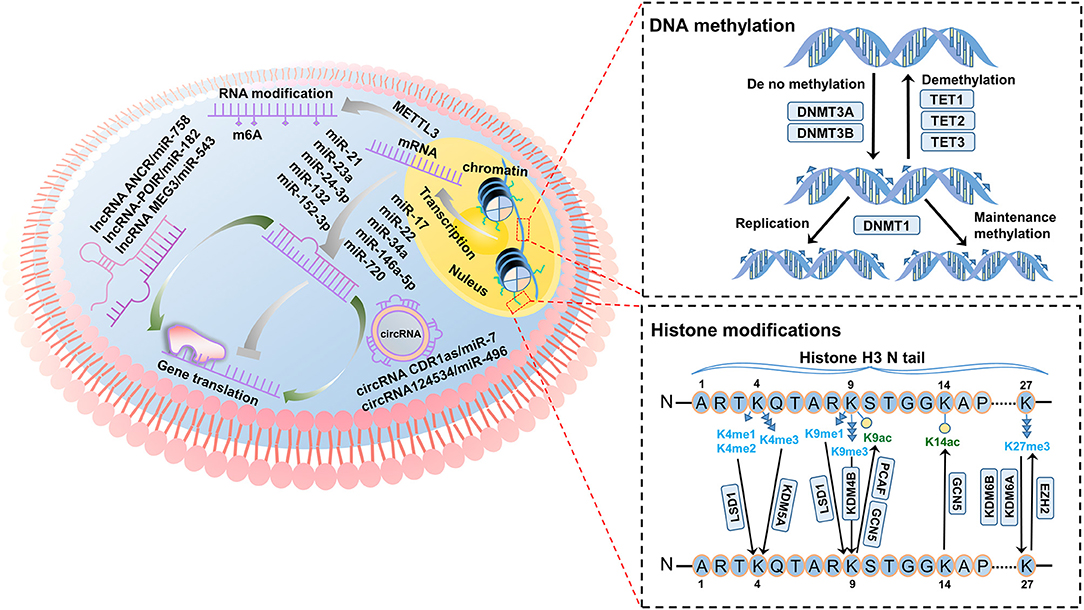
Figure 1. Overview of the principal epigenetic modifiers in the regulation of dental stem cells (DSCs). DNA methylation: DNMT3A and DNMT3B modify unmethylated double strands of DNA in DSCs, while TETs remove methyl groups from DNA. DNMT1 is responsible for the maintenance of methylation levels of DSCs. Histone modifications: KDMs remove methyl groups from lysine mediated by LSD1, KDM4B, KDM5A, KDM6A, and KDM6B in DSCs. Histone acetylation (K9ac and K14ac) is regulated by HATs (PCAF and GCN5) in DSCs. Non-coding RNAs: In the epigenetic regulation of DSC fate, non-coding RNAs are of great importance. miR-17, miR-22, miR-34a, miR-146a-5p, and miR-720 promote DSC differentiation, while miR-21, miR-23a, miR-24-3p, miR-132 and miR-152-3p inhibit DSC differentiation. LncRNAs and circRNAs act as competing endogenous RNAs for miRNAs, constituting a complex regulatory network. LncRNA-POIR, circRNA124534, and circRNA CDR1as promote DSC differentiation. LncRNA MEG3 and ANCR inhibit DSC differentiation.
The key to regenerative medicine lies also in the differentiation of stem cells into specific tissues. Inhibition of DNMTs with RG108 increased the level of transcription factor Klf4, enhancing the efficiency of odontoblastic differentiation of DSCs, thus presenting great prospects in future application (103). DPSCs exhibited higher osteogenic as well as lower adipogenic potential compared with BMMSCs. As revealed by Shen et al. the PTEN promoter of BMMSCs presented higher levels of DNA methylation mediated by increased DNMT3B and enrichment of the repressive H3K9me2 (91). Altering epigenetics of PTEN that is responsible for inhibiting adipogenesis and promoting osteogenesis (104), we may control the lineage commitment of DSCs and facilitate their future translation. In this regard, histone deacetylase inhibitors (HDACi) have been critically studied. As a potent HDACi, trichostatin A (TSA) promoted odontoblast differentiation of DPSCs at certain concentrations (105). While at higher concentrations, TSA significantly accelerated mineralization of DPSCs by promoting DMP1 and DSPP expression (106). In rat models of periodontitis, TSA treatment resulted in increased alveolar bone volume and decreased inflammatory infiltration levels, suggesting HDACi as potential candidates for the treatment of periodontal disease (107).
Conclusions
Epigenetic modifications participate in the determination of DSC fate by regulating various critical signal pathways (Figure 2, Table 2). Summarizing the current state of knowledge regarding epigenetic cues would substantially promote the clinical research of DSCs to a new level. Moreover, deciphering the epigenetic code of DSCs would provide potential targets for DSC-mediated regenerative therapies, facilitating DSC applications from bench to bedside.
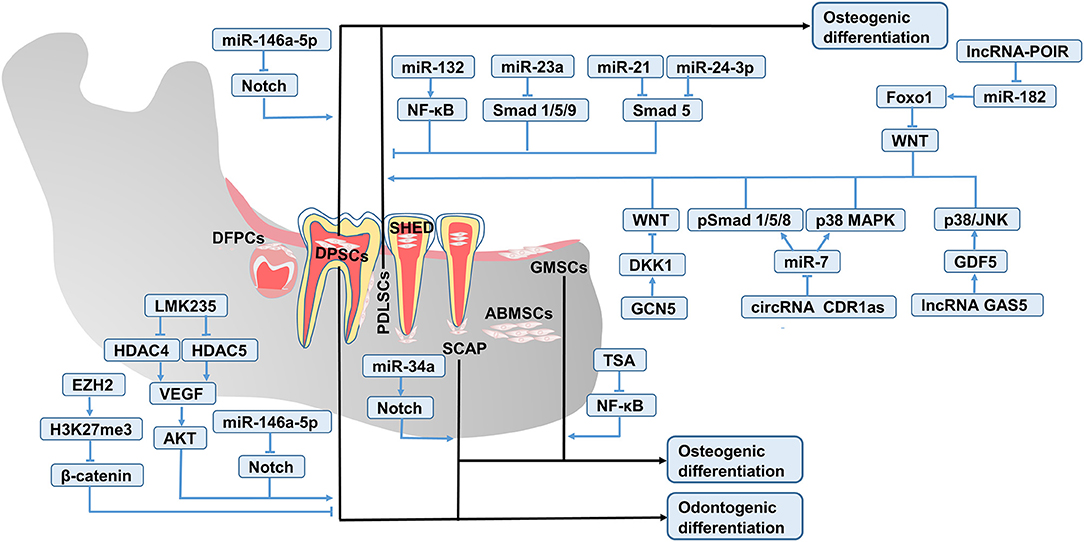
Figure 2. Key pathways involved in the epigenetic regulation of DSC fate. The odontogenic and osteogenic differentiation of DSCs is mediated by the interaction of critical signal pathways such as Wnt, Notch, BMP, AKT/mTOR, MAPK, and NF-κB.
Author Contributions
AL and YL contributed to the conception of the study. YL, XZ, and MS wrote the manuscript. DP and AL revised the manuscript. All authors approved the final manuscript.
Funding
Current research was funded by China Postdoctoral Innovation Science Foundation (BX20200254), National Natural Science Foundation of China (Nos. 82071078, 81870798, and 82170927), the Key Industrial Innovation Chain Project in Social Development Domain of Shaanxi Key Research and Development Program (No. 2020ZDLSF02-04), and China Postdoctoral Science Foundation (2021M692474).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Kobayashi T, Torii D, Iwata T, Izumi Y, Nasu M, Tsutsui TW. Characterization of proliferation, differentiation potential, and gene expression among clonal cultures of human dental pulp cells. Hum Cell. (2020) 33:490–501. doi: 10.1007/s13577-020-00327-9
2. Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. P Natl Acad Sci USA. (2000) 97:13625–30. doi: 10.1073/pnas.240309797
3. Miura M, Gronthos S, Zhao M, Bai L, Fisher LW, Robey PG, et al. SHED: Stem cells from human exfoliated deciduous teeth. P Natl Acad Sci USA. (2003) 100:5807–12. doi: 10.1073/pnas.0937635100
4. Seo BM, Miura M, Gronthos S, Bartold PM, Batouli S, Brahim J, et al. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet. (2004) 364:149–55. doi: 10.1016/S0140-6736(04)16627-0
5. Morsczeck C, Götz W, Schierholz J, Zeilhofer F, Kühn U, Möhl C, et al. Isolation of precursor cells (PCs) from human dental follicle of wisdom teeth. Matrix Biol. (2005) 24:155–65. doi: 10.1016/j.matbio.2004.12.004
6. Nada OA, Backly R. Stem cells from the apical papilla (SCAP) as a tool for endogenous tissue regeneration. Front Bioeng Biotech. (2018) 6:103. doi: 10.3389/fbioe.2018.00103
7. Jin SH, Lee JE, Yun JH, Kim I, Ko Y, Park JB. Isolation and characterization of human mesenchymal stem cells from gingival connective tissue. J Periodontal Res. (2015) 50:461–7. doi: 10.1111/jre.12228
8. Pekovits K, Kröpfl JK, Stelzer I, Payer M, Hutter H, Dohr G. Human mesenchymal progenitor cells derived from alveolar bone and human bone marrow stromal cells: A comparative study. Histochem Cell Biol. (2013) 140:611–21. doi: 10.1007/s00418-013-1140-7
9. Matsubara T, Suardita K, Ishii M, Sugiyama M, Igarashi A, Oda R, et al. Alveolar bone marrow as a cell source for regenerative medicine: Differences between alveolar and iliac bone marrow stromal cells. J Bone Miner Res. (2010) 20:399–409. doi: 10.1359/JBMR.041117
10. Chen J, Zheng X, Rao N, Huang Y, Liu J, Li Y, et al. Key markers and epigenetic modifications of dental-derived mesenchymal stromal cells. Stem Cells Int. (2021) 2021:5521715. doi: 10.1155/2021/5521715
11. Yan X, Yan F, Mohammed HAG, Liu O. Maxillofacial-derived mesenchymal stem cells: Characteristics and progress in tissue regeneration. Stem Cells Int. (2021) 2021:5516521. doi: 10.1155/2021/5516521
12. Gardin C, Ricci S, Ferroni L. Dental stem cells (DSCs): Classification and properties. Springer International Publishing. (2016) 1–25. doi: 10.1007/978-3-319-33299-4_1
13. Hu L, Zhao B, Gao Z, Xu J, Wang S. Regeneration characteristics of different dental derived stem cell sheets. J Oral Rehabil. (2019) 1–7. doi: 10.1111/joor.12839
14. Kotova A, Lobov A, Dombrovskaya J, Sannikova V, Ryumina N, Klausen P, et al. Comparative analysis of dental pulp and periodontal stem cells: Differences in morphology, functionality, osteogenic differentiation and proteome. Biomedicines. (2021) 9:1606. doi: 10.3390/biomedicines9111606
15. Ansari S, Seagroves JT, Chen C, Shah K, Moshaverinia A. Dental and orofacial mesenchymal stem cells in craniofacial regeneration: the prosthodontist's point of view. J Prosthet Dent. (2017) 118:455. doi: 10.1016/j.prosdent.2016.11.021
16. Son YB, Kang YH, Lee HJ, Jang SJ, Bharti D, Lee SL, et al. Evaluation of odonto/osteogenic differentiation potential from different regions derived dental tissue stem cells and effect of 17β-estradiol on efficiency. BMC Oral Health. (2021) 21:1–14. doi: 10.1186/s12903-020-01366-2
17. Wang L, Shen H, Zheng W, Tang L, Yang Z, Gao Y, et al. Characterization of stem cells from alveolar periodontal ligament. Tissue Eng. (2011) 17:1015–26. doi: 10.1089/ten.tea.2010.0140
18. Wang ZS, Feng ZH, Wu GF, Bai SZ, Dong Y, Chen FM, et al. The use of platelet-rich fibrin combined with periodontal ligament and jaw bone mesenchymal stem cell sheets for periodontal tissue engineering. Sci Rep. (2016) 6:28126. doi: 10.1038/srep28126
19. Sonoyama W, Liu Y, Fang D, Yamaza T, Seo B, Zhang C, et al. Mesenchymal stem cell-mediated functional tooth regeneration in swine. PLoS ONE. (2006) 1:e79. doi: 10.1371/journal.pone.0000079
20. Bakopoulou A, Leyhausen G, Volk J, Koidis P, Geurtsen W. Comparative characterization of STRO-1neg/CD146pos and STRO-1pos/CD146pos apical papilla stem cells enriched with flow cytometry. Arch Oral Biol. (2013) 58:1556–68. doi: 10.1016/j.archoralbio.2013.06.018
21. Almeida JD, Chen P, Henry MA, Diogenes A. Stem cells of the apical papilla regulate trigeminal neurite outgrowth and targeting through a BDNF-dependent mechanism. Tissue Eng. (2015) 20:3089–100. doi: 10.1089/ten.tea.2013.0347
22. Xuan K, Li B, Guo H, Sun W, Kou X, He X, et al. Deciduous autologous tooth stem cells regenerate dental pulp after implantation into injured teeth. Sci Transl Med. (2018) 10:3227. doi: 10.1126/scitranslmed.aaf3227
23. Ferrarotti F, Romano F, Gamba MN, Quirico A, Giraudi M, Audagna M, et al. Human intrabony defect regeneration with micrografts containing dental pulp stem cells: A randomized controlled clinical trial. J Clin Periodontol. (2018) 45:841–50. doi: 10.1111/jcpe.12931
24. Rodas-Junco BA, Michel CC, Rojas-Herrera RA, Clelia D, Nic-Can GI. Stem cells from dental pulp: What epigenetics can do with your tooth. Front Physiol. (2017) 8:999. doi: 10.3389/fphys.2017.00999
25. Li B, Ouchi T, Cao Y, Zhao Z, Men Y. Dental-Derived Mesenchymal Stem Cells: State of the Art. Front Cell Dev Biol. (2021) 9:654559. doi: 10.3389/fcell.2021.654559
26. Sharma S, Bhonde R. Genetic and epigenetic stability of stem cells: Epigenetic modifiers modulate the fate of mesenchymal stem cells. Genomics. (2020) 112:3615–23. doi: 10.1016/j.ygeno.2020.04.022
27. McGuire AL, Gabriel S, Tishkoff SA, Wonkam A, Chakravarti A, Furlong EEM, et al. The road ahead in genetics and genomics. Nat Rev Genet. (2020) 21:581–96. doi: 10.1038/s41576-020-0272-6
28. Broeders M, Herrero-Hernandez P, Ernst M, Ploeg A, Pijnappel W. Sharpening the molecular scissors: Advances in gene-editing technology. iScience. (2019) 3:100789. doi: 10.1016/j.isci.2019.100789
29. Sui BD, Zheng CX, Li M, Jin Y, Hu CH. Epigenetic regulation of mesenchymal stem cell homeostasis. Trends Cell Biol. (2020) 30:97–116. doi: 10.1016/j.tcb.2019.11.006
30. Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine. Science. (2011) 333:1300–3. doi: 10.1126/science.1210597
31. Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. (2011) 21:381–95. doi: 10.1038/cr.2011.22
32. Francis M, Gopinathan G, Foyle D, Fallah P, Gonzalez M, Luan X, et al. Histone methylation: Achilles heel and powerful mediator of periodontal homeostasis. J Dent Res. (2020) 99:1332–40. doi: 10.1177/0022034520932491
33. Lan F, Nottke AC, Shi Y. Mechanisms involved in the regulation of histone lysine demethylases. Curr Opin Cell Biol. (2008) 20:316–25. doi: 10.1016/j.ceb.2008.03.004
34. Galvani A, Thiriet C. Nucleosome dancing at the tempo of histone tail acetylation. Genes (Basel). (2015) 6:607–21. doi: 10.3390/genes6030607
35. Meier K, Brehm A. Chromatin regulation: How complex does it get? Epigenetics. (2014) 9:1485–95. doi: 10.4161/15592294.2014.971580
36. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. (2017) 169:1187–200. doi: 10.1016/j.cell.2017.05.045
37. Wu Y, Xie L, Wang M, Xiong Q, Guo Y, Liang Y, et al. Mettl3-mediated m(6)A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat Commun. (2018) 9:1–12. doi: 10.1038/s41467-018-06898-4
38. Sheng R, Wang Y, Wu Y, Wang J, Zhang S, Li Q, et al. METTL3-mediated m6A mRNA methylation modulates tooth root formation via affecting NFIC translation. J Bone Miner Res. (2021) 36:412–23. doi: 10.1002/jbmr.4180
39. Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. (2015) 16:421–33. doi: 10.1038/nrg3965
40. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. (2014) 505:344–52. doi: 10.1038/nature12986
41. Ai T, Zhang J, Wang X, Zheng X, Qin X, Zhang Q, et al. DNA methylation profile is associated with the osteogenic potential of three distinct human odontogenic stem cells. Signal Transduct Tar. (2018) 3:1. doi: 10.1038/s41392-017-0001-6
42. Rao LJ, Yi BC, Li QM, Xu Q. TET1 knockdown inhibits the odontogenic differentiation potential of human dental pulp cells. Int J Oral Sci. (2016) 8:110–6. doi: 10.1038/ijos.2016.4
43. Li Q, Yi B, Feng Z, Meng R, Tian C, Xu Q. FAM20C could be targeted by TET1 to promote odontoblastic differentiation potential of human dental pulp cells. Cell Prolif. (2018) 51:e12426. doi: 10.1111/cpr.12426
44. Cai L, Zhan M, Li Q, Li D, Xu Q. DNA methyltransferase DNMT1 inhibits lipopolysaccharide-induced inflammatory response in human dental pulp cells involving the methylation changes of IL-6 and TRAF6. Mol Med Rep. (2020) 21:959–68. doi: 10.3892/mmr.2019.10860
45. Hara ES, Ono M, Eguchi T, Kubota S, Pham HT, Sonoyama W, et al. miRNA-720 controls stem cell phenotype, proliferation and differentiation of human dental pulp cells. PLoS ONE. (2013) 8:e.83545. doi: 10.1371/journal.pone.0083545
46. Zhang C, Han X, Liang Y, Liu H, Fan Z, Zhang J. The histone demethylase KDM3B promotes osteo-/odontogenic differentiation, cell proliferation, and migration potential of stem cells from the apical papilla. Stem Cells Int. (2020) 2020:8881021. doi: 10.1155/2020/8881021
47. Ye L, Fan Z, Yu B, Chang J, Al Hezaimi K, Zhou X, et al. Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell. (2012) 11:50–61. doi: 10.1016/j.stem.2012.04.009
48. Yang H, Fan J, Cao Y, Gao R, Fan Z. Distal-less homeobox 5 promotes the osteo-/dentinogenic differentiation potential of stem cells from apical papilla by activating histone demethylase KDM4B through a positive feedback mechanism. Exp Cell Res. (2019) 374:221–30. doi: 10.1016/j.yexcr.2018.11.027
49. Li QM, Li JL, Feng ZH, Lin HC, Xu Q. Effect of histone demethylase KDM5A on the odontogenic differentiation of human dental pulp cells. Bioengineered. (2020) 11:449–62. doi: 10.1080/21655979.2020.1743536
50. Jiang H, Jia P. MiR-153-3p inhibits osteogenic differentiation of periodontal ligament stem cells through KDM6A-induced demethylation of H3K27me3. J Periodontal Res. (2021) 56:379–87. doi: 10.1111/jre.12830
51. Zhou Y, Zheng L, Li F, Wan M, Fan Y, Zhou X, et al. Bivalent histone codes on WNT5A during odontogenic differentiation. J Dent Res. (2018) 97:99–107. doi: 10.1177/0022034517728910
52. Hoang M, Kim JJ, Kim Y, Tong E, Trammell B, Liu Y, et al. Alcohol-induced suppression of KDM6B dysregulates the mineralization potential in dental pulp stem cells. Stem Cell Res. (2016) 17:111–21. doi: 10.1016/j.scr.2016.05.021
53. Li B, Sun J, Dong Z, Xue P, He X, Liao L, et al. GCN5 modulates osteogenic differentiation of periodontal ligament stem cells through DKK1 acetylation in inflammatory microenvironment. Sci Rep. (2016) 6:26542. doi: 10.1038/srep26542
54. Yan GQ, Wang X, Yang F, Yang ML, Zhang GR, Wang GK, et al. MicroRNA-22 promoted osteogenic differentiation of human periodontal ligament stem cells by targeting HDAC6. J Cell Biochem. (2017) 118:1653–8. doi: 10.1002/jcb.25931
55. Li L, Liu W, Wang H, Yang Q, Zhang L, Jin F, et al. Mutual inhibition between HDAC9 and miR-17 regulates osteogenesis of human periodontal ligament stem cells in inflammatory conditions. Cell Death Dis. (2018) 9:480. doi: 10.1038/s41419-018-0480-6
56. Zhang P, Liu YS, Jin CY, Zhang M, Lv LW, Zhang X, et al. Histone H3K9 acetyltransferase PCAF is essential for osteogenic differentiation through bone morphogenetic protein signaling and may be involved in osteoporosis. Stem Cells. (2016) 34:2332–41. doi: 10.1002/stem.2424
57. Tao H, Li Q, Lin Y, Zuo H, Liu H. Coordinated expression of p300 and HDAC3 upregulates histone acetylation during dentinogenesis. J Cell Biochem. (2019) 121:2478–88. doi: 10.1002/jcb.29470
58. Gu S, Ran S, Liu B, Liang J. miR-152 induces human dental pulp stem cell senescence by inhibiting SIRT7 expression. FEBS Lett. (2016) 590:1123–31. doi: 10.1002/1873-3468.12138
59. Li Q, Ma Y, Zhu Y, Zhang T, Zhou Y. Declined expression of histone deacetylase 6 contributes to periodontal ligament stem cell aging. J Periodontol. (2017) 88:e12–23. doi: 10.1902/jop.2016.160338
60. Zhang Y, Li S, Yuan S, Zhang H, Liu J. MicroRNA-23a inhibits osteogenesis of periodontal mesenchymal stem cells by targeting bone morphogenetic protein signaling. Arch Oral Biol. (2019) 102:93–100. doi: 10.1016/j.archoralbio.2019.04.001
61. Li Z, Sun Y, Cao S, Zhang J, Wei J. Downregulation of miR-24-3p promotes osteogenic differentiation of human periodontal ligament stem cells by targeting SMAD family member 5. J Cell Physiol. (2019) 234:7411–9. doi: 10.1002/jcp.27499
62. Wu D, Ma L. Downregulating microRNA-152-3p promotes the viability and osteogenic differentiation of periodontal ligament stem cells via targeting integrin alpha 5. Arch Oral Biol. (2020) 120:104930. doi: 10.1016/j.archoralbio.2020.104930
63. Liu Y, Liu W, Hu C, Xue Z, Wang G, Ding B, et al. MiR-17 modulates osteogenic differentiation through a coherent feed-forward loop in mesenchymal stem cells isolated from periodontal ligaments of patients with periodontitis. Stem Cells. (2011) 29:1804–16. doi: 10.1002/stem.728
64. Gu X, Li M, Jin Y, Liu D, Wei F. Identification and integrated analysis of differentially expressed lncRNAs and circRNAs reveal the potential ceRNA networks during PDLSC osteogenic differentiation. BMC Genomic Data. (2017) 18:100. doi: 10.1186/s12863-017-0569-4
65. Zheng Y, Li X, Huang Y, Jia L, Li W. The circular RNA landscape of periodontal ligament stem cells during osteogenesis. J Periodontol. (2017) 88:906–14. doi: 10.1902/jop.2017.170078
66. Li Z, Li N, Ge X, Pan Y, Yu J. Differential circular RNA expression profiling during osteogenic differentiation of stem cells from apical papilla. Epigenomics. (2019) 11:1057–73. doi: 10.2217/epi-2018-0184
67. Li X, Zheng Y, Zheng Y, Huang Y, Zhang Y, Jia L, et al. Circular RNA CDR1as regulates osteoblastic differentiation of periodontal ligament stem cells via the miR-7/GDF5/SMAD and p38 MAPK signaling pathway. Stem Cell Res Ther. (2018) 9:232. doi: 10.1186/s13287-018-0976-0
68. Ji F, Pan J, Shen Z, Yang Z, Wang J, Bai X, et al. The circular RNA circRNA124534 promotes osteogenic differentiation of human dental pulp stem cells through modulation of the miR-496/β-catenin pathway. Front Cell Dev Biol. (2020) 8:230. doi: 10.3389/fcell.2020.00230
69. Qian J, Chen X, Jiang W, Wang W, Ni L. The regulatory effects of long noncoding RNA-ANCR on dental tissue-derived stem cells. Stem Cells Int. (2016) 2016:1–12. doi: 10.1155/2016/3146805
70. Jia Q, Jiang W, Ni L. Down-regulated non-coding RNA (lncRNA-ANCR) promotes osteogenic differentiation of periodontal ligament stem cells. Arch Oral Biol. (2015) 60:234–41. doi: 10.1016/j.archoralbio.2014.10.007
71. Peng W, Deng W, Zhang J, Pei G, Rong Q, Zhu S. Long noncoding RNA ANCR suppresses bone formation of periodontal ligament stem cells via sponging miRNA-758. Biochem Bioph Res Co. (2018) 503:815–21. doi: 10.1016/j.bbrc.2018.06.081
72. Wang L, Wu F, Song Y, Li X, Wu Q, Duan Y, et al. Long noncoding RNA related to periodontitis interacts with miR-182 to upregulate osteogenic differentiation in periodontal mesenchymal stem cells of periodontitis patients. Cell Death Dis. (2016) 7:e2327. doi: 10.1038/cddis.2016.125
73. Zhao LD, Xu WC, Cui J, Liang YC, Cheng WQ, Xin BC, et al. Long non-coding RNA maternally expressed gene 3 inhibits osteogenic differentiation of human dental pulp stem cells via microRNA-543/smad ubiquitin regulatory factor 1/runt-related transcription factor 2 axis. Arch Oral Biol. (2020) 118:104838. doi: 10.1016/j.archoralbio.2020.104838
74. Clevers H, Loh KM, Nusse R. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. (2014) 346:54–54. doi: 10.1126/science.1248012
75. Uribe-Etxebarria V, Luzuriaga J, García-Gallastegui P, Agliano A, Ibarretxe G. Notch/Wnt cross-signalling regulates stemness of dental pulp stem cells through expression of neural crest and core pluripotency factors. Eur Cells Mater. (2017) 34:249–70. doi: 10.22203/eCM.v034a16
76. Lu X, Chen X, Xing J, Lian M, Huang D, Lu Y, et al. miR-140-5p regulates the odontoblastic differentiation of dental pulp stem cells via the Wnt1/β-catenin signaling pathway. Stem Cell Res Ther. (2019) 10:226. doi: 10.1186/s13287-019-1344-4
77. Chen L, Song Z, Huang S, Wang R, Qin W, Guo J, et al. lncRNA DANCR suppresses odontoblast-like differentiation of human dental pulp cells by inhibiting wnt/β-catenin pathway. Cell Tissue Res. (2016) 364:309–18. doi: 10.1007/s00441-015-2333-2
78. Uribe-Etxebarria V, Garcia-Gallastegui P, Perez-Garrastachu M, Casado-Andres M, Irastorza I, Unda F, et al. Wnt-3a induces epigenetic remodeling in human dental pulp stem cells. Cells. (2020) 9:652. doi: 10.3390/cells9030652
79. Gopinathan G, Foyle D, Luan X, Diekwisch TGH. The Wnt antagonist SFRP1: A key regulator of periodontal mineral homeostasis. Stem Cells Dev. (2019) 28:1004–14. doi: 10.1089/scd.2019.0124
80. Li B, Yu F, Wu F, Hui T, A P, Liao X, et al. EZH2 impairs human dental pulp cell mineralization via the Wnt/β-Catenin pathway. J Dent Res. (2018) 7:571–9. doi: 10.1177/0022034517746987
81. Bray S, Bernard F. Notch targets and their regulation. Curr Top Dev Biol. (2010) 92:253–75. doi: 10.1016/S0070-2153(10)92008-5
82. Perdigoto CN, Bardin AJ. Sending the right signal: Notch and stem cells. Biochim Biophys Acta. (2013) 1830:2307–22. doi: 10.1016/j.bbagen.2012.08.009
83. Qiu Z, Lin S, Hu X, Zeng J, Xiao T, Ke Z, et al. Involvement of miR-146a-5p/neurogenic locus notch homolog protein 1 in the proliferation and differentiation of STRO-1(+) human dental pulp stem cells. Eur J Oral Sci. (2019) 127:294–303. doi: 10.1111/eos.12624
84. Sun F, Wan M, Xu X, Gao B, Zhou Y, Sun J, et al. Crosstalk between miR-34a and Notch signaling promotes differentiation in apical papilla stem cells (SCAPs). J Dent Res. (2014) 93:589–95. doi: 10.1177/0022034514531146
85. Lowery JW, Rosen V. The BMP pathway and its inhibitors in the skeleton. Physiol Rev. (2018) 98:2431–52. doi: 10.1152/physrev.00028.2017
86. Wei F, Yang S, Guo Q, Zhang X, Ren D, Lv T, et al. MicroRNA-21 regulates osteogenic differentiation of periodontal ligament stem cells by targeting Smad5. Sci Rep. (2017) 7:1–12. doi: 10.1038/s41598-017-16720-8
87. Kémoun P, Laurencin-Dalicieux S, Rue J, Farges JC, Gennero I, Conte-Auriol F, et al. Human dental follicle cells acquire cementoblast features under stimulation by BMP-2/-7 and enamel matrix derivatives (EMD) in vitro. Cell Tissue Res. (2007) 329:283–94. doi: 10.1007/s00441-007-0397-3
88. Hoxhaj G, Manning BD. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat Rev Cancer. (2020) 20:74–88. doi: 10.1038/s41568-019-0216-7
89. Jason SL, Wei C. Proliferation, survival and metabolism: the role of PI3K/AKT/ mTOR signalling in pluripotency and cell fate determination. Development. (2016) 143:3050–60. doi: 10.1242/dev.137075
90. Liu Z, Chen T, Han Q, Chen M, You J, Fang F, et al. HDAC inhibitor LMK235 promotes the odontoblast differentiation of dental pulp cells. Mol Med Rep. (2018) 17:1445–52. doi: 10.3892/mmr.2017.8055
91. Shen WC, Lai YC, Li LH, Liao K, Lai HC, Kao SY, et al. Methylation and PTEN activation in dental pulp mesenchymal stem cells promotes osteogenesis and reduces oncogenesis. Nat Commun. (2019) 10:2226. doi: 10.1038/s41467-019-10197-x
92. Yue J, Lopez JM. Understanding MAPK signaling pathways in apoptosis. Int J Mol Sci. (2020) 21:2346. doi: 10.3390/ijms21072346
93. Kholodenko BN, Birtwistle MR. Four-dimensional dynamics of MAPK information processing systems. Wires Syst Biol Med. (2010) 1:28–44. doi: 10.1002/wsbm.16
94. Yang Q, Han Y, Liu P, Huang Y, Li X, Jia L, et al. Long noncoding RNA GAS5 promotes osteogenic differentiation of human periodontal ligament stem cells by regulating GDF5 and p38/JNK signaling pathway. Front Pharmacol. (2020) 11:701. doi: 10.3389/fphar.2020.00701
95. Xu Y, Ren C, Zhao X, Wang W, Zhang N. microRNA-132 inhibits osteogenic differentiation of periodontal ligament stem cells via GDF5 and the NF-κB signaling pathway. Pathol Res Pract. (2019) 215:152722. doi: 10.1016/j.prp.2019.152722
96. Li J, Yan M, Wang Z, Jing S, Li Y, Liu G, et al. Effects of canonical NF-κB signaling pathway on the proliferation and odonto/osteogenic differentiation of human stem cells from apical papilla. Biomed Res Int. (2014) 2014:319651. doi: 10.1155/2014/319651
97. Diomede F, Thangavelu SR, Merciaro I, D'Orazio M, Bramanti P, Mazzon E, et al. Porphyromonas gingivalis lipopolysaccharide stimulation in human periodontal ligament stem cells: Role of epigenetic modifications to the inflammation. Eur J Histochem. (2017) 61:2826. doi: 10.4081/ejh.2017.2826
98. Hayden MS, Ghosh S. Shared principles in NF-κB signaling. Cell. (2008) 132:344–62. doi: 10.1016/j.cell.2008.01.020
99. Martins MD, Jiao Y, Larsson L, Almeida LO, Garaicoa-Pazmino C, Le JM, et al. Epigenetic modifications of histones in periodontal disease. J Dent Res. (2015) 95:215–22. doi: 10.1177/0022034515611876
100. Diomede F, Zini N, Pizzicannella J, Merciaro I, Pizzicannella G, D'Orazio M, et al. 5-Aza exposure improves reprogramming process through embryoid body formation in human gingival stem cells. Front Genet. (2018) 9:419. doi: 10.3389/fgene.2018.00419
101. Rajan TS, Scionti D, Diomede F, Piattelli A, Bramanti P, Mazzon E, et al. Prolonged expansion induces spontaneous neural progenitor differentiation from human gingiva-derived mesenchymal stem cells. Cell Reprogram. (2017) 19:389–401. doi: 10.1089/cell.2017.0012
102. Diomede F, Rajan TS, Gatta V, D'Aurora M, Merciaro I, Marchisio M, et al. Stemness maintenance properties in human oral stem cells after long-term passage. Stem Cells Int. (2017) 2017:5651287. doi: 10.1155/2017/5651287
103. Sun Z, Yu S, Chen S, Liu H, Chen Z. SP1 regulates KLF4 via SP1 binding motif governed by DNA methylation during odontoblastic differentiation of human dental pulp cells. J Cell Biochem. (2019) 120:14688–99. doi: 10.1002/jcb.28730
104. Dangaria SJ, Ito Y, Luan X, Diekwisch T. Differentiation of neural-crest-derived intermediate pluripotent progenitors into committed periodontal populations involves unique molecular signature changes, cohort shifts, and epigenetic modifications. Stem Cells Dev. (2011) 20:39–52. doi: 10.1089/scd.2010.0180
105. Jin H, Park JY, Choi H, Choung PH. HDAC inhibitor trichostatin A promotes proliferation and odontoblast differentiation of human dental pulp stem cells. Tissue Eng. (2013) 19:613–24. doi: 10.1089/ten.tea.2012.0163
106. Duncan HF, Smith AJ, Fleming GJP, Cooper PR. Histone deacetylase inhibitors epigenetically promote reparative events in primary dental pulp cells. Exp Cell Res. (2013) 319:1534–43. doi: 10.1016/j.yexcr.2013.02.022
Keywords: epigenetic regulation, stem cells derived from dental tissues (DSCs), cell differentiation, signaling pathways, regenerative therapies
Citation: Li Y, Zhao X, Sun M, Pei D and Li A (2022) Deciphering the Epigenetic Code of Stem Cells Derived From Dental Tissues. Front. Dent. Med. 2:807046. doi: 10.3389/fdmed.2021.807046
Received: 01 November 2021; Accepted: 20 December 2021;
Published: 12 January 2022.
Edited by:
Xiaoshan Wu, Central South University, ChinaReviewed by:
Dawei Liu, Peking University School and Hospital of Stomatology, ChinaJinhua Yu, Nanjing Medical University, China
Copyright © 2022 Li, Zhao, Sun, Pei and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dandan Pei, cGVpZGFuZGFuMTk4NkAxMjYuY29t; Ang Li, ZHJsaWFuZ0BtYWlsLnhqdHUuZWR1LmNu
†These authors have contributed equally to this work
 Ye Li
Ye Li Xitong Zhao
Xitong Zhao Meng Sun3
Meng Sun3 Ang Li
Ang Li