
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Dent. Med. , 14 October 2021
Sec. Regenerative Dentistry
Volume 2 - 2021 | https://doi.org/10.3389/fdmed.2021.747068
This article is part of the Research Topic Epigenetic Regulation of Stem Cells Derived from Craniofacial Tissues View all 8 articles
 Xiao Han1
Xiao Han1 Zhipeng Fan1,2*
Zhipeng Fan1,2*Mesenchymal stem cells (MSCs) are a kind of pluripotent stem cell with the potential of self-renewal and multidirectional differentiation. They can be obtained from a variety of tissues and can differentiate into a variety of cell types under different induction conditions, including osteoblasts. Because of this osteogenic property, MSCs have attracted much attention in the treatment of bone metabolism-related diseases. MicroRNAs (miRNAs), as an epigenetic factor, are thought to play an important regulatory role in the process of osteogenic differentiation of MSCs. In recent years, increasingly evidence shows that miRNAs imbalance is involved in the regulation of osteoporosis and fracture. In this review, miRNAs involved in osteogenic differentiation and their mechanisms for regulating the expression of target genes are reviewed. In addition, we also discuss the potential clinical applications and possible directions of this field in the future.
Mesenchymal stem cells (MSCs), derived from the mesoderm at the early stage of embryonic development, are a kind of adult stem cells with self-renewal and multidirectional differentiation potential. They can differentiate into different cell types under different induction conditions, such as adipocytes, osteoblasts, nerve cells and chondrocytes. Friedenstein et al. first discovered in bone marrow a group of adherent cells capable of forming a single clone, which could differentiate into cells of all kinds of sources, such as adipocytes, osteoblasts and chondrocytes, in vitro or in vivo (1). Therefore, these cells are called MSCs. In addition to bone marrow tissue, researchers have successively isolated and cultured MSCs from the dental apical papilla (SCAPs), dental pulp stem cells (DPSCs) and periodontal ligament stem cells (PDLSCs) (2–4). The differentiation of MSCs into osteoblasts is crucial for maintaining bone remodeling. Studies indicated that MSCs have low immunogenicity and can secrete vast chemokines and migrate to injured site during tissue injury and inflammation, thus exerting the functions of inflammation regulation and tissue repair (5).
Osteoblasts can secrete organic bone matrix, promote mineralization of bone matrix, and maintain bone homeostasis (6). The dysplasia or dysfunction of osteoblasts can result in the destruction of bone microstructure and defect of bone formation, leading to bone metabolic diseases such as osteoarthritis and osteoporosis. However, differentiation of MSCs into osteoblasts is the main pathway of osteoblastic formation and plays a crucial role in bone formation and functional maintenance (7). Numerous studies have shown that microRNAs, as endogenous post-transcriptional regulators, play a key biological role in regulating proliferation and differentiation of stem cells (8, 9). In this paper, the role and mechanism of miRNAs in the osteogenic differentiation of MSCs were summarized, and the regulatory function of miRNAs in this process was recognized, so as to have a deeper understanding of bone metabolic diseases related to this process.
In 1993, Lee (10) et al. found a gene that does not encode protein: LIN-4 when conducting genetic screening for the early development of C. elegans. LIN-4 does not encode a protein, but produces a small pair of RNAs. Later, it was recognized that Lin-4 is a member of a rich class of small regulatory RNAs, called microRNAs, and that it controls the timing of nematode embryo development. MicroRNA is a kind of small non-coding single-stranded RNA about 21–23 nucleotides, which is widely present in eukaryotes and acts on the 3 'UTR of target genes, inhibit the translation of target gene RNA (mRNA), or make it degrade to play an important regulatory role in stem cell renewal, differentiation, cell cycle and cell apoptosis and other biological processes. MicroRNAs undergo a series of changes before maturation. The classic microRNA synthesis pathway is that first, microRNAs are transcribed in the nucleus under the action of RNA polymerase (Pol) into cap-structured primary transcripts, namely pri-microRNAs. In the presence of a microprocessing complex consisting of endonuclease Drosha and cofactor double stranded RNA binding protein DGCR8, pri-miRNAs are further processed to form hairpin precursor microRNAs (pre-miRNAs) of about 70 nucleotides. After the pre-miRNA is transported to the cytoplasm by Exportin5, Doubly-stranded microRNAs with about 22 nucleotides are formed by clipping by Dicer endoribonuclease. Doubly-stranded miRNAs have unstable 5 'ends and bind with miRNA-induced silencing complex to form miRNA-RISC, while the other strand is hydrolyzed under the action of hydrolase (11, 12). The miRNA-RISC complex recognizes the target mRNA through the principle of base complementary pairing, mainly binds to the 3 'UTR of the target mRNA, and directs the silencing complex to avoid the production of proteide by inhibiting the translation or degradation of mRNA according to the different degree of complementarity. When miRNA is fully complementary to the target gene, the target gene is completely degraded, which mainly occurs in plants (13). When miRNA is not completely complementary to the target gene, the transcription of the target gene will be inhibited to varying degrees, which mainly occurs in animals (14). In addition, miRNAs are specific in time and space as well as fine-tuning. Through these characteristics, miRNAs regulate nearly 30% of genes (15), making them play a key role in physiological and pathological processes such as cell proliferation, apoptosis, differentiation, growth and development, and tumor.
With the continuous exploration of osteogenic differentiation of MSCs in recent years, studies indicated that the osteogenic differentiation of MSCs is a complex process jointly completed under the expression of multiple signaling pathways and multiple genes, and confirmed that miRNA is closely related to multiple osteogenic signaling pathways and multiple osteogenic genes (16, 17). Therefore, this paper summarizes the current research results of miRNA, multiple osteogenic signaling pathways and multiple osteogenic related genes.
Osteogenic differentiation of MSCs is regulated by osteogenic transcription factors, such as Runx2, OSX, DLX5 (18), etc. Among which Runx2 and OSX are two important regulatory factors. More and more studies have shown that miRNAs regulate the osteogenic differentiation of MSCs by directly targeting osteogenic transcription factors. This paper focuses on the summary of these two regulatory factors (Figure 1).
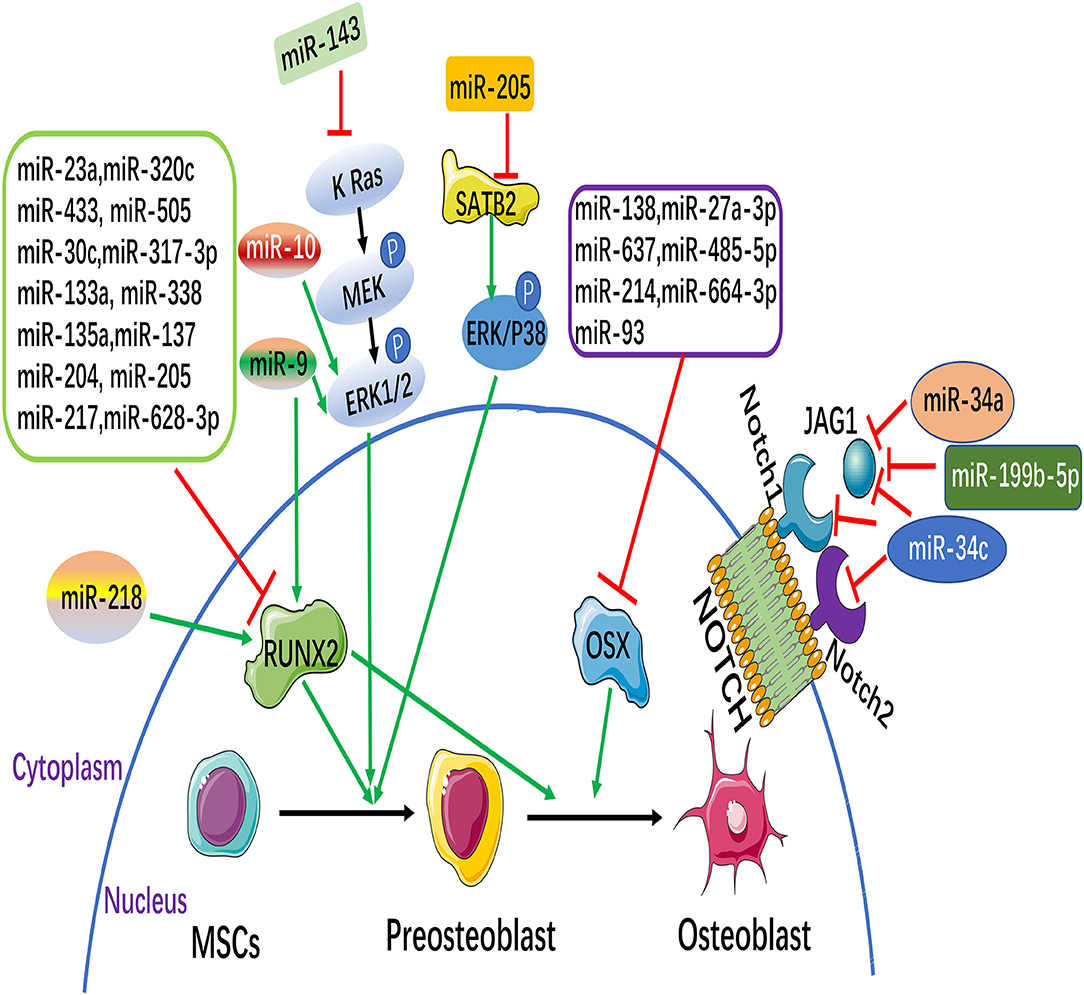
Figure 1. MicroRNAs (miRNAs) and signaling pathways involved in the osteogenesis of mesenchymal stem cells (MSCs). MiRNAs can inhibit or promote osteoblast differentiation through the direct targeting of their target genes, some of which are osteoblastic markers [such as runt-related transcription factor 2 (Runx2) and osterix (Osx)]. Notch and MAPK signaling pathways participate in regulating the osteoblastic differentiation of MSCs. (The green arrow: promotion, Red symbol: inhibition).
Runx2, a member of the Runx family of transcription factors, is a key transcription factor specific to osteoblasts and is essential for osteoblastic differentiation. Runx2 is regulated by multiple cytokines and multiple signaling pathways in osteogenic differentiation of MSCs, among which miRNAs can directly target Runx2 to regulate osteogenic differentiation of MSCs. Recently, 11 miRNAs that directly target Runx2 (miR-23a, miR-30c, miR-34c, miR-133a, miR-135a, miR-137, miR-204, miR-205, miR-217, miR-218, and miR-338) were highly expressed in bone mesenchymal stem cells (BMSCs), and further study showed that 11 miRNAs (except miR-218) inhibited osteogenic differentiation of BMSCs by directly targeting Runx2 (19). Studies found that the miR-320 family (miR-320a, 320b, 320c, 320d, and 320e) was up-regulated during the adipose differentiation of MSCs. Further, miR-320c can promote adipogenic differentiation and inhibit osteogenic differentiation of MSCs. It has been proved that Runx2 is a direct target gene of the miR-320 family by dual luciferase report assay. Studies indicated that the miR-320 family regulates the differentiation process of MSCs by directly targeting the 3'UTR of Runx2 (20). Kim et al. found that the level of miR-433 was down-regulated in the osteoblast cell line C3H10T1/2 of MSCs during osteoblast differentiation. Over-expression miR-433 reduced the expression of Runx2. Further studies demonstrated that Runx2 was a direct target gene of miR-433. Therefore, miR-433 suppressed the differentiation of osteoblasts by directly targeting the 3'UTR of Runx2 (21). Recent studies have found that miR-505 is considered as a new regulator of Runx2. miR-505 was down regulated during osteoblast differentiation in MC3T3-E1 cells. Furthermore, over-expression miR-505 inhibits osteoblast differentiation by directly targeting to Runx2 (22).
Currently, miRNAs have been used in other bone diseases or altered osteogenic differentiation. Recent studies have shown that miR-137-3p is involved in avascular necrosis of the femoral head by directly binding to the 3 'UTR of Runx2. In addition, it also targets CXCL12, which encodes SDF-1 α, an angiogenic factor. Further studies show that silencing miR-137-3p promotes osteogenesis and angiogenesis by targeting Runx2 and CXCL12 (23). Recently, study has shown that miR-628-3p expression is up-regulated in patients with atrophic nonunion and may inhibit osteoblastic differentiation by inhibiting its target gene Runx2.Atrophic nonunion is a serious complication of fracture. In vitro experiments, overexpression of miR-628-3p resulted in reduced osteoblastic differentiation in human osteoblast-like cells (MG63) (24). Recently, it has been found that RSVL can promote osteogenesis of BMSCs and inhibit adipogenesis in a dose-dependent manner. Further, miR-320c and its target gene Runx2 were identified during the process of osteoblastic differentiation of BMSCs treated with RSVL. Over-expression miR-320c suppressed osteoblastic differentiation of BMSCs by targeting Runx2 in RSVL, while down-regulation of miR-320c accelerated osteoblastic differentiation of BMSCs. In conclusion, miR-320c can be used as a new target to regulate the biological function of BMSCs with RSVL (25).
OSX is a downstream gene of Runx2, which is specifically expressed in osteoblasts and is a key gene for the pre-differentiation of osteoblasts into mature osteoblasts. Studies found that in mice with OSX knockout, embryonic cartilage development was normal, but there was no mature differentiation and bone formation of osteocytes. The study demonstrates the need for OSX in bone formation (26). Studies have shown that miR-138 inhibited osteogenic differentiation of MSCs in vitro, and in vivo overexpression of miR-138 reduced ectopic bone formation by 85%. On the contrary, knockdown of miR-138 increased bone formation by 60% in vivo. Further studies showed that miR-138 inhibited phosphorylation of ERK1/2 and thus OSX phosphorylation, showing that it inhibited osteogenic differentiation of MSCs (27). Studies have reported that miRNAs such as miR-637, miR-93, and miR-214 directly regulate OSX, inhibit its translation and transcription, and inhibit the osteogenic differentiation of MSCs (28–30).
Recently, studies have indicated that the level of miR-485-5p is up-regulated in osteoporosis patients, and which is down-regulated in the process of osteogenic differentiation. Further luciferase detection confirms that 3'UTR region of OSX is directly targeted to miR-485-5p. The upregulation of miR-485-5p resulted in decreased OSX expression, which could inhibit the expression levels of cell viability and osteogenesis markers. This study revealed the miR-485-5p can promote osteoporosis, indicating that miR-485-5p may be a potential therapeutic strategy for osteoporosis (31). MiR-27a-3p is a key inhibitor of osteogenesis. Luciferase reporting analysis suggested that miR-27a-3p controlled the mRNA level of OSX through targeting the 3'UTR region of OSX, and miR-27a-3p inhibited the differentiation of preosteoblasts by decreasing OSX expression. The expression level of miR-27a-3p in patients with osteoporosis is reduced, and miR-27a-3p weakens the potential of osteogenic differentiation in vitro, therefore, miR-27a-3p may be used as a therapeutic target and diagnostic biomarker for osteoporosis (32). It was found that osteoblast miR-664-3p transgenic mice reduced bone mass by inhibiting osteoblastic function. Further luciferase detection confirms that Smad4 and OSX are direct targets of miR-664-3p. In addition, subperiosteal injection of miR-664-3p antagomir protects against bone loss induced by oophorectomy. In conclusion, miR-664-3 may serve as a new diagnostic and therapeutic target for osteoporosis patients (33).
Wnt signaling pathways are mainly divided into the classical Wnt/β-catenin signaling pathway and the non-classical Wnt signaling pathway. The participation of Wnt signaling pathway in osteogenic differentiation is mainly due to the activation of the classical Wnt/β-catenin signaling pathway (34) (Table 1). On the classical Wnt/β-catenin signaling pathway, Wnt proteins pass through the domain of Frizzled receptor or LDL receptor-related protein 5/6, through a series of cytoplasmic protein interactions, β-catenin accumulates in the cytoplasm, and enters the nucleus, where it is combined with intraconuclear T cytokine 3(TCF3)/Lymphoid enhancer factor 1(LEF1) forms a complex that initiates transcription of downstream target genes. Long et al. showed that miR-139-5p bound to the 3'UTR of CTNNB1 and FZD4 in BMSCs by regulating the Wnt/β-catenin signaling pathway to negatively regulate osteogenic differentiation BMSCs (35). β-catenin is a multifunctional protein encoded by CTNNB1. Wnt protein binds to FZD4 on the cell membrane of BMSCs, activates the accumulation of β-catenin into the nucleus, and initiates the transcriptional activity of Runx2, which can promote osteogenic differentiation. Cao et al. found that knockdown miR-214 can inhibit the Wnt/β-catenin signaling pathway by inhibiting the expression of β-catenin, thereby promoting the osteogenic differentiation of PDLSCs (36). Osteogenic differentiation is promoted by suppressing Glycogen synthase kinase-3 activation of Wnt signaling pathway (52). Numerous studies have found that GSK-3β is the target gene of miR-346, miR-199b-5p and miR-26a. miR-346, miR-199b-5p and miR-26a target to the 3'UTR region of GSK-3β, which promoted osteogenic differentiation of BMSCs by reducing the expression level of GSK-3β and activating the Wnt signaling pathway (37–39).
Some studies have found that, Secreted Human DickkopF-related Protein 1(DKK1), Kremen2 and Secreted Frizzled Related Protein 2 (SFRP2), is an inhibitor of the Wnt signaling pathway. Kapinas et al. found that DKK1, Kremen2 and SFRP2 are the target genes of miR-29a, and when miR-29a is overexpressed, the expression levels of DKK1, Kremen2 and SFRP2 are reduced, which initiates the Wnt signaling pathway and stimulates osteoblastic differentiation of osteoblasts (40). In addition, miR-9, miR-433-3p, miR-335-5p and miR-217 promote osteoblast differentiation of murine and human MSCs, binding to the 3'UTR region of DKK1 (41–44). It has been found that miR-218 can effectively promote the osteogenic differentiation of human adipose derived mesenchymal stem cells (hADSCs) by directly targeting SFRP2 and DKK2 and activating Wnt signaling pathway Zhang et al. (45). During osteogenic differentiation of human fetal osteoblasts (hFOBs), the expression of miR-27a was up-regulated and targeted to the extracellular antagonist SFRP1 of the Wnt signaling pathway. Therefore, miR-27a positively regulates the Wnt signaling pathway by down-regulating the expression level of SFRP1 (47).
In recent years, some Wnt ligands, including Wnt3a, Wnt5a, Wnt6, and Wnt10a, have been found to be involved in initiating the Wnt/β-catenin cascade and stimulating osteoblastic differentiation. In the process of osteoblast differentiation, miR-16-2-3p expression is observably reduced in BMSCs (48). Further studies showed that miR-16-2-3p blocks the Wnt signaling pathway and thereby inhibits BMSCs osteoblast differentiation by targeting the 3'UTR region of Wnt5a (48). In addition, Wnt5a could also be used as a target of miR-1297 in BMSCs. miR-1297 is highly expressed in the serum of patients with osteoporosis, but it is significantly decreased in the process of osteogenic differentiation induced by BMSCs. Therefore, miR-1297 may be involved in the progress of osteoporosis by combining with Wnt5a (46). Zhang et al. found that miR-9-5p can inhibit osteogenesis and promote adipogenesis by targeting Wnt3a in BMSCs, thus promoting the occurrence and progress of osteoporosis (49). Recently, it has been found that miR-381 can target to the 3'UTR regions of Wnt5a and FZD3 in BMSCs, inhibit the mRNA expression of Wnt5a and FZD3, and thus suppress the osteogenic differentiation of BMSCs (50). Fang et al. found that overexpression of miR-378 suppressed the osteogenesis of MSCs, while antimir-378 promoted its osteogenic differentiation, thus confirming that miR-378 could bind to the 3'UTR of Wnt6 and Wnt10a and inhibited osteogenesis by blocking Wnt/β-catenin signaling (51).
TGF-β belongs to a group of TGF-β superfamily that regulate cell growth and differentiation, including Activins, Inhibin, transforming growth factors (TGF-βs) and bone morphogenetic protein (BMP) (53) (Table 2). TGF- β Signal promotes the activation and proliferation of pre osteoblasts and makes osteoprogenitor cells differentiate into immature osteoblasts. In the later stage, especially in the process of matrix mineralization, it inhibits osteoblast maturation, mineralization and transformation to osteoblasts (65–67). Studies have shown that after the overexpression of miR-181a in C2C12 and MC3T3, the activity of alkaline phosphatase and calcium-salt nodules stained with Alizarin red, a marker of late osteogenesis, are increased. Further studies have found that, G-protein signal transduction regulator 4(RGS4) and transcriptional regulator 6 (GATA6) are TGF-β signaling molecules, and miR-181a can bind to the 3'UTR of RGS4 and GATA6, indicating that miR-181a accelerates osteoblastic differentiation of C2C12 and MC3T3 cells by down-regulating the expression of RGS4 and GATA6 and blocking the TGF-β signaling pathway (54).
SMAD (mothers against decapentaplegic) is a key transcription factor in the TGF-β signaling pathway during osteogenic differentiation. Some miRNAs target TGF-β signal-induced SMAD 2/3, BMP signal-induced smad1/5 and TGF-β/BMP signal-induced co-induced SMAD4 affect osteogenic differentiation. In TGF-β signal-induced SMADs, miR-10b promotes osteogenesis in vivo and enhances bone formation by targeting SMAD2 in ADSCs, and its expression is associated with OPN and Runx2 in clinical samples from patients with osteoporosis (55). Pais et al. proved that Smad3 may be a target of miR-140.MiR-140 promotes osteogenic differentiation of BMSCs by binding to Smad3 and blocking the TGF-β signaling pathway (56). MiR-708 is highly expressed in MSCs of patients with hormone-induced femoral osteonecrosis (ONFH), while Smad3 is low expressed. miR-708 binds to 3'UTR of Smad3, which can inhibit osteogenic and adipogenic differentiation of MSCs in patients with ONFH, suggesting that miR-708 may be a novel therapeutic biomarker to prevent and treat ONFH (57). Recently, Studies showed that miR-221-5p inhibited osteogenic differentiation of myeloma osteopathic mesenchymal stem cells (MBD-MSCs) by binding to Smad3 affecting osteogenic differentiation and negatively regulated PI3K/ Akt /mTOR signaling pathway (58). Overexpression of miR-222-3p decreased expression of Smad5 and Runx2, while knockout increased Smad5 and Runx2 expression in BMSCs. Further, miR-222-3p suppressed osteogenic differentiation by directly targeting Smad5 and Runx2 Yan et al. (59). Recently, the study found that miR-203-3p targeted the 3'UTR of Smad1 and inhibited osteogenesis in vivo and in vitro in diabetic rats, indicating that miR-203-3p may be a therapeutic target for diabetic bone and may improve osteoporosis and fracture healing (60). Huang et al. found that miR-144-3p suppressed osteoblastic differentiation of C3H mouse (C3H10T1/2) embryonic cells by directly targeting Smad4 in TGF-β/BMP signal-induced common Smads (61). In addition, the expression of miR-146a was down-regulated in BMP2-induced rat adipose-derived mesenchymal stem cells (ADSCs). Overexpression of miR-146a significantly inhibited osteogenic differentiation of ADSCs, and it was further found that Smad4 was a direct target of miR-146a (62).
BMPs is a member of the TGF-β superfamily, in which BMP-2 is the most significant bone growth factor promoting osteogenic differentiation of MSCs. MiR-320a has been reported to inhibit osteogenic differentiation of BMSCs by targeting Homeobox A10 (HOXA10). HOXA10 is a direct target gene of miR-320, and the expression of HOXA10 will be inhibited after binding with miR-320a.HOXA10 can induce BMP2 expression gene to activate Runx2, inhibit Runx2 transcription through the BMP2/ RUNX2 signaling pathway, and inhibit osteogenic differentiation (63). In the process of osteogenesis, BMP-2 regulates the transcription of osteogenic genes by activating Smads signal transduction, thus playing its role in osteogenesis (68). In this study, it was found that knockdown of miR-106b-5p and miR-17-5p promoted bone differentiation of C2C12 and MC3T3-E1 cells. Further study revealed that knockdown of miR-106b-5p and miR-17-5p activated the BMP-2 signaling pathway.BMP-2 activates SMAD5 signal transduction protein, indicating that knockdown of miR-106b-5p and miR-17-5p can promote osteogenic differentiation of C2C12 and MC3T3-E1 cells by activating the BMP-2/ SMAD5 signaling pathway (64).
Mitogen-activated protein kinase (MAPK) belongs to the class of intracellular serine/threonine protein kinases, and is an major transmitter of signal transduction from cell surface to nucleus. The MAPK signaling pathway is activated by growth factors that play an important role in the process of osteogenic differentiation (Figure 1). It has been reported that Extracelluar signal-regulated kinase (ERK) c-Jun N-terminal kinase (JNK) is an Extracelluar signal-regulated kinase.JNK) and p38 are involved in MAPK signaling pathway mediated osteogenic differentiation of BMSCs (69). It has been shown that knockdown of miR-205 can promote osteogenic differentiation of BMSCs.MiR-205 negatively regulates the expression of Special AT-rich sequence-binding protein 2(SATB2), an osteogenic transcription factor. Overexpression of SATB2 activates Runx2. In conclusion, it is confirmed that down-regulation of miR-205 can promote the expression of STAB2 and RUNX2, thus activating the ERK/ P38 signaling pathway to promote the osteogenic differentiation of BMSCs (70). It was found that overexpression of miR-9 promoted the osteogenic differentiation of BMSCs by up-regulating the expression of Runx2, while miR-10 negatively regulated the osteogenic differentiation of BMSCs by negatively regulating the expression of Runx2. Further investigation found that the expression changes of miR-9 and miR-10 can cause the phosphorylation of ERK and activate the ERK signaling pathway to further regulate the osteogenic differentiation of BMSCs (71). Recently, it has been found that miR-143 is significantly down-regulated during osteogenic differentiation of ADSCs. Overexpression of miR-143 blocked the extracellular signal-regulated kinase 1/2 (ERK1/2) pathway to inhibit osteoblastic differentiation, while knockdown of miR-143 enhanced the activation of the ERK1/2 pathway to promote osteoblastic differentiation. Bioinformatics analysis further revealed that miR-143 had a KRas was the target and directly bound to its mRNA 3'UTR. In conclusion, miR-143 inhibits osteogenic differentiation of ADSCs by the K Ras/MEK/ERK pathway, further revealing its molecular mechanism (72).
Recent studies indicated that Notch signaling pathway is bound up with the osteogenic differentiation of MSCs (Figure 1). Notch signaling pathway is mainly composed of Notch receptor, Notch ligand and DNA protein binding CSL3 part (73). So far it has been found that there are 4 homologous Notch receptors and five homologous ligand in mammals. Some Notch target genes, effector molecules, have been identified, including Hes1-6(Hairy enhancement of Split-1) NF-κB(Nuclear Factor κB) Cyclin D1C - myc, etc., (74, 75). It was found that over-expression miR-34a suppressed osteogenic differentiation of MSCs in vitro Jagged1, a key transcription factor in Notch signaling pathway, was further confirmed as the direct target gene of miR-34a by bioinformatics prediction of target genes. In an in vivo preclinical model of bone, overexpression of miR-34a in MSCs resulted in a 60% reduction in ectopic bone formation, while knockdown of miR-34a resulted in 200% in vivo bone formation. In conclusion, the study indicated that miR-34a decreased Jagged1 expression and inhibited Notch signaling pathway by targeting Jagged1, and played a negative role in the regulation of osteogenic differentiation of MSCs and osteogenesis in vivo (76). Studies found that the increase of mouse osteoblast-specific miR-34c can lead to age-dependent osteoporosis with the growth of age in mice. In osteoblasts, miR-34c directly acts on multiple effector molecules of the Notch signaling pathway, including Notch1, Notch2, and Jagged1, to regulate osteoclast differentiation in an involuntary manner. The study revealed that miR-34c plays a part in bone homeostasis by regulating multiple targets in osteoblasts and osteoclasts in vivo (77).
Studies have shown that the expression level of miR-199b-5p is down-regulated in the process of osteoblastic differentiation of Ligamentum flavum cells (LF), and the overexpression of miR-199b-5p inhibits osteoblastic differentiation. In addition, the expression of JAG1 was upregulated in LF cells during osteogenic differentiation, and knocking down JAG1 inhibited Notch signaling and osteogenic differentiation. Further, dual luciferase reporting analysis indicated that miR-199b-5p targeted the 3'UTR region of JAG1, and miR-199b-5p down-regulated both JAG1 and Notch. These results suggest that miR-199b-5p may play an inhibitory role in osteoblastic differentiation of LF cells by binding to JAG1 and affecting Notch signaling pathway (78).
In summary, during the process of osteogenic differentiation of MSCs, a variety of miRNAs directly influence the osteogenic differentiation of MSCs by regulating their target genes, or they affect the expression levels of upstream or downstream regulatory factors of each signaling pathway.
Several studies have reported that circulating miRNAs have been analyzed and validated in serum or plasma and whole blood as prognostic or diagnostic markers for differentiating osteoporosis (OP) patients from non-OP subjects. Serum miRNA miR-140-3p and miR-23b-3p can be considered as potential biomarkers for OP and OP fractures in postmenopausal women (79). miR-338 clusters are elevated in postmenopausal women with OP, and in vivo and in vitro studies suggest that RUNX2 and SOX4 may be two key functional targets of miR-338 by mediating their regulation during bone formation (80). Abnormal expression of miR-135b-5p in bone tissue of OP patients suggests that miRNA is involved in the occurrence and development of OP (81). Interestingly, TAmiRNA, a European miRNA diagnostics company, recently commercialized a product called Osteo-miR panel, which uses a set of 19 miRNA to identify the first fracture risk of postmenopausal osteoporosis and type 2 diabetes women (82).
Although non-viral vectors or small RNAs are safer and non-toxic, the transfection efficiency is lower. However, many studies have shown that biomaterials can be used as miRNA-based scaffolders to improve transfection efficiency (83). Some of these materials have already shown preclinical success in treating bone defects. Several researchers have demonstrated the efficacy of periosteal injection miRNA therapy at fracture sites in rodent models. This results in accelerated repair, reduced callus width, osteogenic marker genes and increased callus bone density (84, 85).
Bone metabolic diseases are common clinical diseases at present. With the increase of age, the incidence of bone metabolic diseases is also increasing, troubling all mankind. Therefore, the use of bone regeneration to treat bone metabolic diseases is the focus of our current research. The development of this influential feature of miRNA is a new field of bone therapy. However, most of the studies on miRNAs and bone formation are obtained through in vitro studies, and further exploration and in vivo studies are needed before they are applied in clinic. It is important to note that there may be multiple targets for a single miRNA, so it is necessary to understand its mechanism and target before using miRNA as treatment to avoid the suppression of genetically important genes. The process of bone regeneration is regulated by a series of growth factors, among which miRNAs can regulate the osteogenic differentiation of MSCs by regulating several osteogenic signaling pathways and osteogenic transcription factors. However, the precise mechanism of the regulation remains to be further clarified. With the continuous development of tissue engineering technology and the deepening of research on miRNAs, it is expected to reveal the pathogenesis of a variety of bone metabolic diseases, and provide a theoretical basis for the treatment of bone metabolic diseases, in order to benefit the clinic.
XH: design and conception, manuscript writing, and final approval of the manuscript. ZF: design and conception, manuscript revising, financial support, and final approval of the manuscript. Both authors have read and approved the final version of the manuscript.
This work was supported by the National Sciences Foundation of China (81625005 to ZF), CAMS Innovation Fund for Medical Sciences (2019-I2M-5-031 to ZF), and the Program for Hundred-Thousand- Ten Thousand Talents in Beijing (2018A16 to ZF).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Friedenstein AJ, Piatetzky-Shapiro II, Petrakova KV. Osteogenesis in transplants of bone marrow cells. J Embryol Exp Morphol. (1996) 16:381–90. doi: 10.1242/dev.16.3.381
2. Na S, Zhang H, Huang F, et al. Regeneration of dental pulp/dentine complex with a three-dimensional and scaffold-free stem-cell sheet-derived pellet. J Tissue Eng Regen Med. (2016) 10:261–70. doi: 10.1002/term.1686
3. Huang GT, Yamaza T, Shea LD, et al. Stem/progenitor cell-mediated de novo regeneration of dental pulp with newly deposited continuous layer of dentin in an invivomodel. TissueEngPartA. (2010) 16:605–15. doi: 10.1089/ten.tea.2009.0518
4. Yu S, Long J, Yu J, et al. Analysis of differentiation potentials and gene expression profiles of mesenchymal stem cells derived from periodontal ligament and Wharton's jelly of the umbilical cord. Cells Tissues Organs. (2013) 197:209–23. doi: 10.1159/000343740
5. Rustad KC, Gurtner GC. Mesenchymal stem cells home to sites of injury and inflammation. Adv Wound Care. (2012) 1:147–52. doi: 10.1089/wound.2011.0314
6. Marie PJ. Osteoblast dysfunctions in bone diseases: from cellular and molecular mechanisms to therapeutic strategies. Cell Mol Life Sci. (2015) 72:1347–61. doi: 10.1007/s00018-014-1801-2
7. Huang C, Geng J, Jiang S. MicroRNAs in regulation of osteogenic differentiation of mesenchymal stem cells. Cell Tissue Res. (2017) 368:229–38. doi: 10.1007/s00441-016-2462-2
8. Buhagiar A, Borg J, Ayers D. Overview of current microRNA biomarker signatures as potential diagnostic tools for leukaemic conditions. Noncoding RNA Res. (2020) 5:22–26. doi: 10.1016/j.ncrna.2020.02.001
9. Ferguson SW, Wang J, Lee CJ, et al. The microRNA regulatory landscape of MSC-derived exosomes: a systems view. Sci Rep. (2018) 8:1419. doi: 10.1038/s41598-018-19581-x
10. Lee RC, Feinbaum RL, Ambros V. The C elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. (1993) 75:843–54. doi: 10.1016/0092-8674(93)90529-Y
11. Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. (2009) 136:642–55. doi: 10.1016/j.cell.2009.01.035
12. Zeng Y, Cullen BR. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. (2004) 32:4776–85. doi: 10.1093/nar/gkh824
13. Chen X. MicroRNA biogenesis and function in plants. FEBS Lett. (2005) 579:5923–31. doi: 10.1016/j.febslet.2005.07.071
14. Smalheiser NR, Torvik VI. Complications in mammalian microRNA target prediction. Methods Mol Biol. (2006) 342:115–27. doi: 10.1385/1-59745-123-1:115
15. Lewis BP, Shih IH, Jones-Rhoades MW, et al.prediction of mammalian microRNA targets. Cell. (2003) 115:787–98. doi: 10.1016/S0092-8674(03)01018-3
16. Hata A, Kang H. Functions of the bone morphogenetic protein signaling pathway through microRNAs (review). Int J Mol Med. (2015) 35:563–8. doi: 10.3892/ijmm.2015.2060
17. Arfat Y, Xiao WZ, Ahmad M, et al. Role of microRNAs in osteoblasts differentiation and bone disorders. Curr Med Chem. (2015) 22:748–58. doi: 10.2174/0929867321999141106121227
18. Yang H, Fan J, Cao Y, Gao R, Fan Z. Distal-less homeobox 5 promotes the osteo-/dentinogenic differentiation potential of stem cells from apical papilla by activating histone demethylase KDM4B through a positive feedback mechanism. Exp Cell Res. (2019) 374:221–30. doi: 10.1016/j.yexcr.2018.11.027
19. Zhang Y, Xie RL, Croce CM, et al. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci U S A. (2011) 108:9863–8. doi: 10.1073/pnas.1018493108
20. Hamam D, Ali D, Vishnubalaji R, et al. microRNA-320/RUNX2 axis regulates adipocytic differentiation of human mesenchymal (skeletal) stem cells. Cell Death Dis. (2014). 5:e1499. doi: 10.1038/cddis.2014.462
21. Kim EJ, Kang IH, Lee JW, Jang WG, Koh JT. MiR-433 mediates ERRγ-suppressed osteoblast differentiation via direct targeting to Runx2 mRNA in C3H10T1/2 cells. Life Sci. (2013) 92:562–8. doi: 10.1016/j.lfs.2013.01.015
22. Li W, Chen Z, Cai C, Li G, Wang X, Shi Z. MicroRNA-505 is involved in the regulation of osteogenic differentiation of MC3T3-E1 cells partially by targeting RUNX2. J Orthop Surg Res. (2020). 15:143. doi: 10.1186/s13018-020-01645-2
23. Kong L, Zuo R, Wang M, et al. Silencing microRNA-137-3p, which targets RUNX2 and CXCL12 prevents steroid-induced osteonecrosis of the femoral head by facilitating osteogenesis and angiogenesis. Int J Biol Sci. (2020) 16:655–70. doi: 10.7150/ijbs.38713
24. Chen H, Ji X, She F, Gao Y, Tang P. miR-628-3p regulates osteoblast differentiation by targeting RUNX2: Possible role in atrophic non-union. Int J Mol Med. (2017) 39:279–86. doi: 10.3892/ijmm.2016.2839
25. Zou J, Du J, Tu H, et al. Resveratrol benefits the lineage commitment of bone marrow mesenchymal stem cells into osteoblasts via miR-320c by targeting Runx2. J Tissue Eng Regen Med. (2021) 15:347–60. doi: 10.1002/term.3176
26. Nakashima K, Zhou X, Kunkel G, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. (2002) 108:17–29. doi: 10.1016/S0092-8674(01)00622-5
27. Eskildsen T, Taipaleenmäki H, Stenvang J, et al. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc Natl Acad Sci U S A. (2011) 108:6139–44. doi: 10.1073/pnas.1016758108
28. Zhang JF, Fu WM, He ML, et al. MiR-637 maintains the balance between adipocytes and osteoblasts by directly targeting Osterix. Mol Biol Cell. (2011) 22:3955–61. doi: 10.1091/mbc.e11-04-0356
29. Yang L, Cheng P, Chen C, et al. miR-93/Sp7 function loop mediates osteoblast mineralization. J Bone Miner Res. (2012) 27:1598–606. doi: 10.1002/jbmr.1621
30. Shi K, Lu J, Zhao Y, et al. MicroRNA-214 suppresses osteogenic differentiation of C2C12 myoblast cells by targeting Osterix. Bone. (2013) 55:487–94. doi: 10.1016/j.bone.2013.04.002
31. Zhang SY, Gao F, Peng CG, Zheng CJ, Wu MF. miR-485-5p promotes osteoporosis via targeting Osterix. Eur Rev Med Pharmacol Sci. (2018) 22:4792–9.
32. Xu Y, Li D, Zhu Z, et al. miR-27a-3p negatively regulates osteogenic differentiation of MC3T3-E1 preosteoblasts by targeting osterix. Mol Med Rep. (2020) 22:1717–26. doi: 10.3892/mmr.2020.11246
33. Xu Y, Jin Y, Hong F, et al. MiR-664-3p suppresses osteoblast differentiation and impairs bone formation via targeting Smad4 and Osterix. J Cell Mol Med. (2021) 25:5025–37. doi: 10.1111/jcmm.16451
34. Teufel S, Hartmann C. Wnt-signaling in skeletal development. Curr Top Dev Biol. (2019) 133:235–79. doi: 10.1016/bs.ctdb.2018.11.010
35. Long H, Sun B, Cheng L, et al. miR-139-5p Represses BMSC osteogenesis via targeting Wnt/β-Catenin signaling pathway. DNA Cell Biol. (2017) 36:715–24. doi: 10.1089/dna.2017.3657
36. Cao F, Zhan J, Chen X, Zhang K, Lai R, Feng Z. miR-214 promotes periodontal ligament stem cell osteoblastic differentiation by modulating Wnt/β-catenin signaling. Mol Med Rep. (2017) 16:9301–8. doi: 10.3892/mmr.2017.7821
37. Wang Q, Cai J, Cai XH, Chen L. miR-346 regulates osteogenic differentiation of human bone marrow-derived mesenchymal stem cells by targeting the Wnt/β-catenin pathway. PLoS One. (2013). 8:e72266. doi: 10.1371/journal.pone.0072266
38. Su X, Liao L, Shuai Y, et al. MiR-26a functions oppositely in osteogenic differentiation of BMSCs and ADSCs depending on distinct activation and roles of Wnt and BMP signaling pathway. Cell Death Dis. (2015). 6:e1851. doi: 10.1038/cddis.2015.221
39. Zhao R, Li Y, Lin Z, et al. miR-199b-5p modulates BMSC osteogenesis via suppressing GSK-3β/β-catenin signaling pathway. Biochem Biophys Res Commun. (2016) 477:749–54. doi: 10.1016/j.bbrc.2016.06.130
40. Kapinas K, Kessler C, Ricks T, Gronowicz G, Delany AM. miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J Biol Chem. (2010) 285:25221–31. doi: 10.1074/jbc.M110.116137
41. Liu X, Xu H, Kou J, Wang Q, Zheng X, Yu T. MiR-9 promotes osteoblast differentiation of mesenchymal stem cells by inhibiting DKK1 gene expression. Mol Biol Rep. (2016) 43:939–46. doi: 10.1007/s11033-016-4030-y
42. Zhang J, Tu Q, Bonewald LF, et al. Effects of miR-335-5p in modulating osteogenic differentiation by specifically downregulating Wnt antagonist DKK1. J Bone Miner Res. (2011) 26:1953–63. doi: 10.1002/jbmr.377
43. Tang X, Lin J, Wang G, Lu J. MicroRNA-433-3p promotes osteoblast differentiation through targeting DKK1 expression. PLoS ONE. (2017). 12:e0179860. doi: 10.1371/journal.pone.0179860
44. Dai Z, Jin Y, Zheng J, et al. MiR-217 promotes cell proliferation and osteogenic differentiation of BMSCs by targeting DKK1 in steroid-associated osteonecrosis. Biomed Pharmacother. (2019) 109:1112–9. doi: 10.1016/j.biopha.2018.10.166
45. Zhang WB, Zhong WJ, Wang LA. signal-amplification circuit between miR-218 and Wnt/β-catenin signal promotes human adipose tissue-derived stem cells osteogenic differentiation. Bone. (2014) 58:59–66. doi: 10.1016/j.bone.2013.09.015
46. Guo D, Li Q, Lv Q, Wei Q, Cao S, Gu J. MiR-27a targets sFRP1 in hFOB cells to regulate proliferation, apoptosis and differentiation. PLoS ONE. (2014). 9:e91354. doi: 10.1371/journal.pone.0091354
47. Zhang HG, Wang XB, Zhao H, Zhou CN. MicroRNA-9-5p promotes osteoporosis development through inhibiting osteogenesis and promoting adipogenesis via targeting Wnt3a. Eur Rev Med Pharmacol Sci. (2019) 23:456–63.
48. Duan L, Zhao H, Xiong Y, Tang X, Yang Y, Hu Z, et al. miR-16-2* Interferes with WNT5A to Regulate Osteogenesis of Mesenchymal Stem Cells. Cell Physiol Biochem. (2018) 51:1087–102. doi: 10.1159/000495489
49. Wang Q, Wang CH, Meng Y. microRNA-1297 promotes the progression of osteoporosis through regulation of osteogenesis of bone marrow mesenchymal stem cells by targeting WNT5A. Eur Rev Med Pharmacol Sci. (2019) 23:4541–50.
50. Long H, Zhu Y, Lin Z, et al. miR-381 modulates human bone mesenchymal stromal cells (BMSCs) osteogenesis via suppressing Wnt signaling pathway during atrophic nonunion development. Cell Death Dis. (2019) 10:470. doi: 10.1038/s41419-019-1693-z
51. Feng L, Zhang JF, Shi L, et al. MicroRNA-378 suppressed osteogenesis of mscs and impaired bone formation via inactivating Wnt/β-Catenin signaling. Mol Ther Nucleic Acids. (2020) 21:1017–28. doi: 10.1016/j.omtn.2020.07.018
52. Yan DY, Tang J, Chen L, et al. Imperatorin promotes osteogenesis and suppresses osteoclast by activating AKT/GSK3 β/β-catenin pathways. J Cell Mol Med. (2020) 24:2330–41. doi: 10.1111/jcmm.14915
53. Augello A, De Bari C. The regulation of differentiation in mesenchymal stem cells. Hum Gene Ther. (2010) 21:1226–38. doi: 10.1089/hum.2010.173
54. Bhushan R, Grünhagen J, Becker J, Robinson PN, Ott CE, Knaus P. miR-181a promotes osteoblastic differentiation through repression of TGF-β signaling molecules. Int J Biochem Cell Biol. (2013) 45:696–705. doi: 10.1016/j.biocel.2012.12.008
55. Li H, Fan J, Fan L, et al. MiRNA-10b Reciprocally stimulates osteogenesis and inhibits adipogenesis partly through the TGF-β/SMAD2 signaling pathway. Aging Dis. (2018). 9:1058–73. doi: 10.14336/AD.2018.0214
56. Pais H, Nicolas FE, Soond SM, et al. Analyzing mRNA expression identifies Smad3 as a microRNA-140 target regulated only at protein level. RNA. (2010) 16:489–94. doi: 10.1261/rna.1701210
57. Hao C, Yang S, Xu W, et al. MiR-708 promotes steroid-induced osteonecrosis of femoral head, suppresses osteogenic differentiation by targeting SMAD3. Sci Rep. (2016) 6:22599. doi: 10.1038/srep22599
58. Fan FY, Deng R, Lai SH, et al. Inhibition of microRNA-221-5p induces osteogenic differentiation by directly targeting smad3 in myeloma bone disease mesenchymal stem cells. Oncol Lett. (2019) 18:6536–44. doi: 10.3892/ol.2019.10992
59. Yan J, Guo D, Yang S, Sun H, Wu B, Zhou D. Inhibition of miR-222-3p activity promoted osteogenic differentiation of hBMSCs by regulating Smad5-RUNX2 signal axis. Biochem Biophys Res Commun. (2016) 470:498–503. doi: 10.1016/j.bbrc.2016.01.133
60. Tang Y, Zheng L, Zhou J, et al. miR-203-3p participates in the suppression of diabetes-associated osteogenesis in the jaw bone through targeting Smad1. Int J Mol Med. (2018) 41:1595–607. doi: 10.3892/ijmm.2018.3373
61. Huang C, Geng J, Wei X, Zhang R, Jiang S. MiR-144-3p regulates osteogenic differentiation and proliferation of murine mesenchymal stem cells by specifically targeting Smad4. FEBS Lett. (2016) 590:795–807. doi: 10.1002/1873-3468.12112
62. Xie Q, Wei W, Ruan J, et al. Effects of miR-146a on the osteogenesis of adipose-derived mesenchymal stem cells and bone regeneration. Sci Rep. (2017) 7:42840. doi: 10.1038/srep42840
63. Huang J, Meng Y, Liu Y, et al. MicroRNA-320a regulates the osteogenic differentiation of human bone marrow-derived mesenchymal stem cells by targeting HOXA10. Cell Physiol Biochem. (2016) 38:40–8. doi: 10.1159/000438607
64. Fang T, Wu Q, Zhou L, Mu S, Fu Q. miR-106b-5p and miR-17-5p suppress osteogenic differentiation by targeting Smad5 and inhibit bone formation. Exp Cell Res. (2016) 347:74–82. doi: 10.1016/j.yexcr.2016.07.010
65. Wu M, Chen G, Li YP. TGF-β and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. (2016) 4:16009. doi: 10.1038/boneres.2016.9
66. Rahman MS, Akhtar N, Jamil HM, Banik RS, Asaduzzaman SM. TGF-β/BMP signaling and other molecular events: regulation of osteoblastogenesis and bone formation. Bone Res. (2015) 3:15005. doi: 10.1038/boneres.2015.5
67. Aslani S, Abhari A, Sakhinia E, Sanajou D, Rajabi H, Rahimzadeh S. Interplay between microRNAs and Wnt, transforming growth factor-β, and bone morphogenic protein signaling pathways promote osteoblastic differentiation of mesenchymal stem cells. J Cell Physiol. (2019) 234:8082–93. doi: 10.1002/jcp.27582
68. Liu T, Gao Y, Sakamoto K, et al. BMP-2 promotes differentiation of osteoblasts and chondroblasts in Runx2-deficient cell lines. J Cell Physiol. (2007) 211:728–35. doi: 10.1002/jcp.20988
69. Robinson MJ, Cobb MH. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. (1997) 9:180–6. doi: 10.1016/S0955-0674(97)80061-0
70. Hu N, Feng C, Jiang Y, Miao Q, Liu H. Regulative effect of Mir-205 on osteogenic differentiation of bone mesenchymal stem cells (BMSCs): possible role of SATB2/Runx2 and ERK/MAPK pathway. Int J Mol Sci. (2015) 16:10491–506. doi: 10.3390/ijms160510491
71. Luo H, Gao H, Liu F, Qiu B. Regulation of Runx2 by microRNA-9 and microRNA-10 modulates the osteogenic differentiation of mesenchymal stem cells. Int J Mol Med. (2017) 39:1046–52. doi: 10.3892/ijmm.2017.2918
72. Zhang Y, Zhou K, Wu L, Gu H, Huang Z, Xu J. Downregulation of microRNA-143 promotes osteogenic differentiation of human adipose-derived mesenchymal stem cells through the k-Ras/MEK/ERK signaling pathway. Int J Mol Med. (2020) 46:965–76. doi: 10.3892/ijmm.2020.4651
73. Liu W, Singh SR, Hou SX JAK-STAT. is restrained by Notch to control cell proliferation of the Drosophila intestinal stem cells. J Cell Biochem. (2010) 109:992–9. doi: 10.1002/jcb.22482
74. Ba K, Yang X, Wu L, et al. Jagged-1-mediated activation of notch signalling induces adipogenesis of adipose-derived stem cells. Cell Prolif. (2012) 45:538–44. doi: 10.1111/j.1365-2184.2012.00850.x
75. Chu Q, Liu L, Wang W. Overexpression of hCLP46 enhances Notch activation and regulates cell proliferation in a cell type-dependent manner. Cell Prolif. (2013) 46:254–62. doi: 10.1111/cpr.12037
76. Chen L, Holmstrøm K, Qiu W, et al. MicroRNA-34a inhibits osteoblast differentiation and in vivo bone formation of human stromal stem cells. Stem Cells. (2014) 32:902–12. doi: 10.1002/stem.1615
77. Bae Y, Yang T, Zeng HC, et al. miRNA-34c regulates Notch signaling during bone development. Hum Mol Genet. (2012) 21:2991–3000. doi: 10.1093/hmg/dds129
78. Qu X, Chen Z, Fan D, et al. MiR-199b-5p inhibits osteogenic differentiation in ligamentum flavum cells by targeting JAG1 and modulating the Notch signalling pathway. J Cell Mol Med. (2017) 21:1159–70. doi: 10.1111/jcmm.13047
79. Ramírez-Salazar EG, Carrillo-Patiño S, Hidalgo-Bravo A, et al. Serum miRNAs miR-140-3p and miR-23b-3p as potential biomarkers for osteoporosis and osteoporotic fracture in postmenopausal Mexican-Mestizo women. Gene. (2018) 679:19–27. doi: 10.1016/j.gene.2018.08.074
80. Lin C, Yu S, Jin R, et al. Circulating miR-338 Cluster activities on osteoblast differentiation: Potential Diagnostic and Therapeutic Targets for Postmenopausal Osteoporosis. Theranostics. (2019) 9:3780–97. doi: 10.7150/thno.34493
81. Chen B, Yang W, Zhao H, et al. Abnormal expression of miR-135b-5p in bone tissue of patients with osteoporosis and its role and mechanism in osteoporosis progression. Exp Ther Med. (2020) 19:1042–50. doi: 10.3892/etm.2019.8278
82. Ladang A, Beaudart C, Locquet M, Reginster JY, Bruyère O, Cavalier E. Evaluation of a panel of MicroRNAs that predicts fragility fracture risk: a pilot study. Calcif Tissue Int. (2020) 106:239–47. doi: 10.1007/s00223-019-00628-8
83. Hadjiargyrou M, Komatsu DE. The therapeutic potential of MicroRNAs as orthobiologics for skeletal fractures. J Bone Miner Res. (2019) 34:797–809. doi: 10.1002/jbmr.3708
84. Tu M, Tang J, He H, Cheng P, Chen C. MiR-142-5p promotes bone repair by maintaining osteoblast activity. J Bone Miner Metab. (2017) 35:255–64. doi: 10.1007/s00774-016-0757-8
Keywords: MSCs, miRNAs, osteogenesis differentiation, signaling pathway, osteogenic transcription factors
Citation: Han X and Fan Z (2021) MicroRNAs Regulation in Osteogenic Differentiation of Mesenchymal Stem Cells. Front. Dent. Med. 2:747068. doi: 10.3389/fdmed.2021.747068
Received: 25 July 2021; Accepted: 17 September 2021;
Published: 14 October 2021.
Edited by:
Rachel Jane Waddington, Cardiff University, United KingdomReviewed by:
John Colombo, University of Nevada, Las Vegas, United StatesCopyright © 2021 Han and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhipeng Fan, enBmYW5AY2NtdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.