
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Dent. Med., 14 April 2021
Sec. Regenerative Dentistry
Volume 2 - 2021 | https://doi.org/10.3389/fdmed.2021.651334
This article is part of the Research TopicMolecular Pathways and Novel Therapeutic Targets in Regenerative DentistryView all 7 articles
This study aimed to elucidate the role of the Sonic hedgehog (Shh)–Patched (Ptch)–Gli signaling pathway in maintaining dental epithelial and pulp stem/progenitor cells and regulating the function of odontoblasts. Doxycycline (dox)-inducible histone 2B (H2B)–green fluorescent protein (GFP) transgenic mice ingested dox at prenatal embryonic days 14.5 or 15.5 and their offspring were collected from postnatal day 1 (P1) to week 3 (P3W). Immunohistochemistry for Gli1, Ptch1, and Ptch2 and in situ hybridization for Shh and Ptch1 were conducted. Mandibular incisors of postnatal day 2 H2B-GFP transgenic and wild-type mice were cultivated in a nutrient medium with Shh antibody for 4 days and subsequently processed for immunohistochemistry for Sox2. In molars, dense H2B-GFP-label-retaining cells (H2B-GFP-LRCs) were densely distributed throughout the dental pulp during P1 to postnatal week 2 (P2W) and decreased in number by postnatal P3W, whereas the number of dense H2B-GFP-LRCs in the subodontoblastic layer increased in number at P2W. Gli1+ and Pthc1+ cells were distributed throughout the enamel organ and dental pulp, including the odontoblast and subodontoblastic layers. Shh mRNA was expressed in the inner enamel epithelium and shifted into odontoblasts after dentin deposition. Ptch1 mRNA was expressed in the inner enamel epithelium and cuspal pulpal tissue on P1 and decreased in intensity from postnatal week 1 to P3W. In incisors, the apical bud contained H2B-GFP-LRCs, Gli1+ cells, and Ptch1+ cells. The addition of Shh antibody to explants induced a decrease in the number of Sox2+ cells due to the increase in apoptotic cells in the apical bud. Thus, the Shh–Ptch–Gli signaling pathway plays a role in maintaining quiescent adult stem cells and regulating the function of odontoblasts.
Committed multipotent stem cells, or adult stem cells, exist in renewal tissues such as the epidermis, intestinal crypt, and bone marrow and are crucial for physiological tissue renewal and regeneration after injury (1). Current stem cell biology suggests that quiescent (removed from the cell cycle) and active (present in the cell cycle) subpopulations of stem cells may coexist within renewal tissues (2). Epithelial stem cells (ESCs) in the epidermis and mesenchymal stem cells (MSCs) in the dermis play crucial roles in the process of skin regeneration following injury (3). In contrast, dental epithelial tissue, or enamel, never regenerates after tooth injury, and exogenous injury to the dentin induces scar formation, referred to as tertiary dentin via pulpal irritation (4). A weak injury activates preexisting odontoblasts, resulting in the production of reactionary dentin, whereas a heavy injury elicits degeneration of injured odontoblasts followed by replacement with new odontoblast-like cells that contribute to reparative dentin (4). Dental pulp stem/progenitor cells play a crucial role in the differentiation of odontoblast-like cells in the process of pulpal healing (5). It has been difficult to identify the location of these cells in the mature pulp using the 5-bromo-2′-deoxyuridine (BrdU) pulse-chase method, because the pulp tissue rarely proliferates after tooth eruption. The prenatal BrdU labeling method, in which BrdU is injected into the pregnant mother to label proliferative cells in the prenatal stages, enables us to identify long-term label-retaining cells (LRCs) in the perivascular niche (5, 6). These LRCs are considered to be quiescent stem cells that actively proliferate following exogenous stimuli (5, 7, 8) and play an important role in pulpal homeostasis and the reparative process. These cells maintain their regenerative capability throughout life and are capable of producing odontoblast-like cells to repair injured dentin. Dental MSCs share characteristics similar to bone marrow-derived MSCs in terms of proliferative capacity, cell markers, and differentiation capability into plural lineages, such as osteoblasts, chondrocytes, and adipocytes (9). Sonic hedgehog (Shh) signaling plays crucial roles in the development of numerous organs, and Gli1+ cells are co-localized with LRCs during molar tooth development (10, 11). Even BrdU labeling in the stem cells becomes sparse if these LRCs continuously provide transit-amplifying cells (5, 7, 8). In contrast, long-term LRCs labeled by BrdU are regarded as quiescent stem cells (10). However, the localization of stem/progenitor cells in the dental pulp, the differences in their differentiation capacity in different cell populations, and the mechanisms maintaining their niches are not fully understood.
Rodent incisors peculiarly evolved as continuously growing teeth. The homeostasis of this tooth type is maintained by the balance between continuous cell proliferation at the apical end and attrition of the incisal edge (12, 13). The adult stem cells that provide transit-amplifying cells exist in the apical end of rodent incisors (14), and the epithelial bulge, including the stem cell niche, shows the human head-like appearance referred to as an apical bud or cervical loop (12, 13). Sox2+ apical bud cells provide cells of all lineages in the enamel organ (15). The LRCs in the apical bud that have been identified by prenatal BrdU labeling are regarded as quiescent stem cells (10) similar to bulge cells of the hair follicle, cycling position 4 LRCs of the intestinal crypt, and endosteal hematopoietic stem cells of the bone marrow (2). Recent studies have demonstrated that Shh signaling in neurovascular bundles regulates the maintenance of MSCs in mouse incisors (16) and that the Bmp–Shh signaling network regulates the fate of ESCs via the maintenance of stem cell niche in the developing molars (17). Canonical hedgehog (Hh) signaling is activated by the binding of Hh ligands to Patched (Ptch), a transmembrane receptor. Gli1 and Gli2 are known as mediators of Hh signaling, regulating the expression of their target genes (18). In the continuously growing incisors, the Hh signaling cascade regulates both Sox2+ stem cells (17) and ameloblast differentiation (19). Hh ligands bind Ptch1 or Ptch2 to release Smoothened (Smo), which activates the transcription of downstream targets such as Gli1 and Gli2 (20–22). Dense BrdU-LRCs, Gli1+ cells, and Ptch1+ cells were actually co-localized in the outer enamel epithelium of the apical bud, the mesenchyme beneath the bud, and the perivascular niche in the central pulp in developing teeth (10). It is known that Shh signaling regulates the production of ameloblast progenitors of mouse incisors (19) and that Shh–Patch–Gli1 signaling cascade plays a crucial role in the maintenance of quiescent stem cells in the apical mesenchyme (16). However, the direct role of Shh signaling in the maintenance of quiescent stem cells in the apical bud is not fully understood.
Adult stem cells in the dental pulp are considered quiescent stem cells and contribute to pulpal healing following tooth injury by proliferating actively under pathological conditions (5, 7, 8). We succeeded in labeling the putative stem cells in the developing teeth by the prenatal BrdU labeling method in Wistar rats and Crlj:CD1 (ICR) mice (5, 6). However, the BrdU labeling method possesses several inherent problems; first, non-proliferating cells at the timing of BrdU injection are never labeled, because BrdU is incorporated into the nucleus of proliferating cells in the cell cycle. Second, the labeled cells must be fixed for visualization of the BrdU deposition. Third, the toxic effects of BrdU on host cells or animals cannot be excluded (23). Finally, transit-amplifying cells such as odontoblast-lineage cells are also densely labeled with BrdU, because the timing of BrdU injection overlaps with their final cell division (5–7, 24, 25). To overcome this issue, we used doxycycline (dox)-inducible histone 2B–green fluorescent protein (H2B–GFP) transgene (26, 27). The ingestion of dox by the pregnant mother at prenatal day 14–15 labels all cells in the pups that express GFP and decreases the intensity of GFP with the number of cell divisions. This method demonstrated the existence of periodontal H2B-GFP-LRCs in the mature teeth, whereas the BrdU labeling method failed to label the periodontal BrdU-LRCs (28). This study aimed to elucidate the role of the Shh–Ptch–Gli signaling pathway in maintaining dental ESCs and MSCs and in regulating the function of odontoblasts in the incisors and molars using dox-inducible H2B-GFP transgenic mice, immunohistochemistry, and in situ hybridization.
All animal experiments complied with the guidelines of the Ministry of Education, Culture, Sports, Science, and Technology; the Ministry of the Environment; and the Science Council of Japan and were carried out in accordance with the Act on Welfare and Management of Animals. All animal experiments were conducted in compliance with a protocol that was reviewed by the Institutional Animal Care and Use Committee and approved by the President of Niigata University (Permit Number: #27 Niigata Univ. Res.282-4). WT C57BL/6J (B6) mice were obtained from Charles River Laboratories of Japan (Yokohama, Japan). TetOP-H2B-GFP mice [B6; 129S4-Gt(ROSA)26Sor < tm1(rtTA*M2) Jae>Colla1 < tm7(tetO-HIST1H2BJ/GFP)Jae>/J] were purchased from The Jackson Laboratory (Bar Harbor, ME, USA) (26). For GFP transgene expression, dox (Sigma D9891, 2 mg/mL, supplemented with sucrose at 50 mg/mL) was ingested via a pregnant mother to label cells of the pups at embryonic day (E) 14.5 or E15.5.
Postnatal mice at day 1 (P1), week 1 (P1W), week 2 (P2W), and week 3 (P3W) were used for in vivo experiments (Table 1). The animals were transcardially perfused with physiological saline, followed by 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4) under deep anesthesia via an intraperitoneal injection of chloral hydrate (maximum dose of 350 mg/kg). The heads, including incisors and molars, were removed en bloc and immersed in the same fixative for an additional 12 h. Following decalcification at 4°C in a 10% ethylenediamine tetraacetic acid (EDTA)-2Na solution, the samples were processed for embedding in paraffin. The paraffin-embedded teeth were cut sagittally into 5 μm sections. The paraffin sections were mounted on Matsunami adhesive silane-coated glass slides (Matsunami Glass, Osaka, Japan).
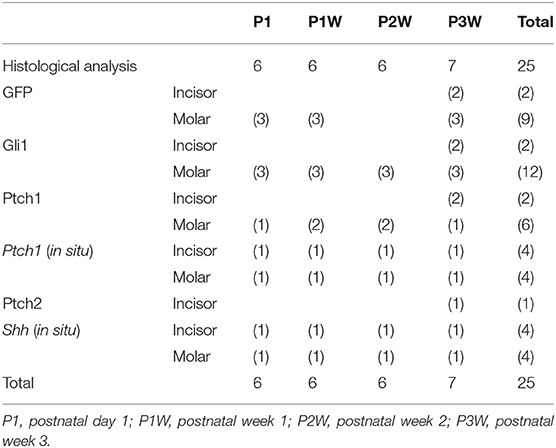
Table 1. Number of animals for histological, immunohistochemical, and statistical analyses of chronological changes in GFP, Gli1-, Ptch1-, and Shh-positive cells.
Mouse incisor organ culture was performed as previously described (29). Specifically, incisors were dissected from the lower jaws of B6 or TetOP-H2B-GFP mice at P2 (Table 2) and put onto membrane filters (0.1 μm pore size, OMNIPORE™, Millipore, Bedford, MA, USA) in a Trowell system and then cultured for 4 days at 37°C in Dulbecco's Modified Eagle's Medium (Gibco BRL, Grand Island, NY, USA) with 10% fetal bovine serum, 100 μg/mL ascorbic acid (Seikagaku Kogyo, Tokyo, Japan), and 100 U/mL penicillin–streptomycin (Gibco BRL). The incisor germs of the experimental group were added with Shh antibody 5E1 (26 μg/mL, the Developmental Studies Hybridoma Bank, Iowa City, IA, USA). For histological analysis of incisor explants, serial sections were made of whole explants in the mesial–distal direction.
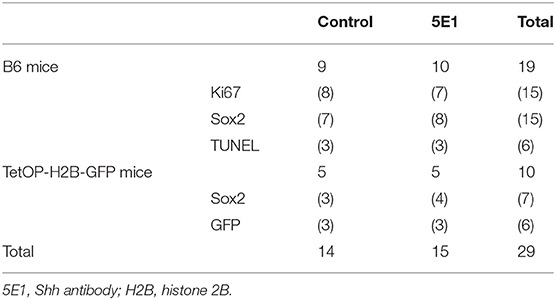
Table 2. Number of incisor explants for histological, immunohistochemical, and statistical analyses of Ki67-, Sox2-, TUNEL, and GFP-positive cells.
Apoptosis was quantified by terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) with the ApopTag Peroxidase in situ Apoptosis Detection Kit (catalog no. S7100, Merck Millipore). Immunohistochemistry for Ki67, Sox2, Gli1, Ptch1, Ptch2, and Shh was conducted as previously reported (6). A rabbit anti-Ptch2 polyclonal antibody diluted to 1:100 (Novus Biologicals; catalog number: NB200-119) was used for Ptch2 immunohistochemisty. Immunohistochemical negative controls were included lacking primary antibodies, showing no specific immunoreaction. The sections were examined with a confocal laser microscope (FV300; Olympus, Tokyo, Japan) or a light microscope.
In situ hybridization was performed as described previously (30). Digoxigenin-labeled probes for Shh (31) and Ptch1 (32) were prepared according to the manufacturer's protocol (Roche). Sense probes transcribed as negative controls did not show any significant staining.
The number of dense and granular GFP+ and Gli1+ cells in the dental pulp and apical bud [a grid of (0.24–3.5 × 104 μm2) was selected] were counted in developing teeth (Table 1). The number of Ki67+, Sox2+, TUNEL+, and GFP+ cells [a grid of (8.7–9.1 × 104 μm2) was selected] were counted in the incisor explants (Table 2). Furthermore, the number of cells/unit area at different time points in the developing teeth was compared using one-way analysis of variance after the confirmation of data normality and homogeneity of variance, followed by Bonferroni's test for multiple comparisons. The proportion of cells was compared between control and experimental groups using Student's t-test by statistical software (SPSS 20.0J for Windows; SPSS Japan, Tokyo, Japan). The samples showing non-normal distribution were compared using Kruskal–Wallis or Mann–Whitney tests.
GFP+ cells were distributed through the dental follicle and apical dental papilla in addition to the outer enamel epithelium of the apical bud (Figures 1A,B,D,E,G,H). Gli1+ cells were localized in the inner enamel epithelium and ameloblast layer in addition to the apical bud (Figures 1A–C). Ptch1+ cells were intensely localized in the apical dental papilla beneath the apical bud (Figures 1D–F). Intense Ptch2 expression was observed in the maturation ameloblasts (Figures 1G–J).
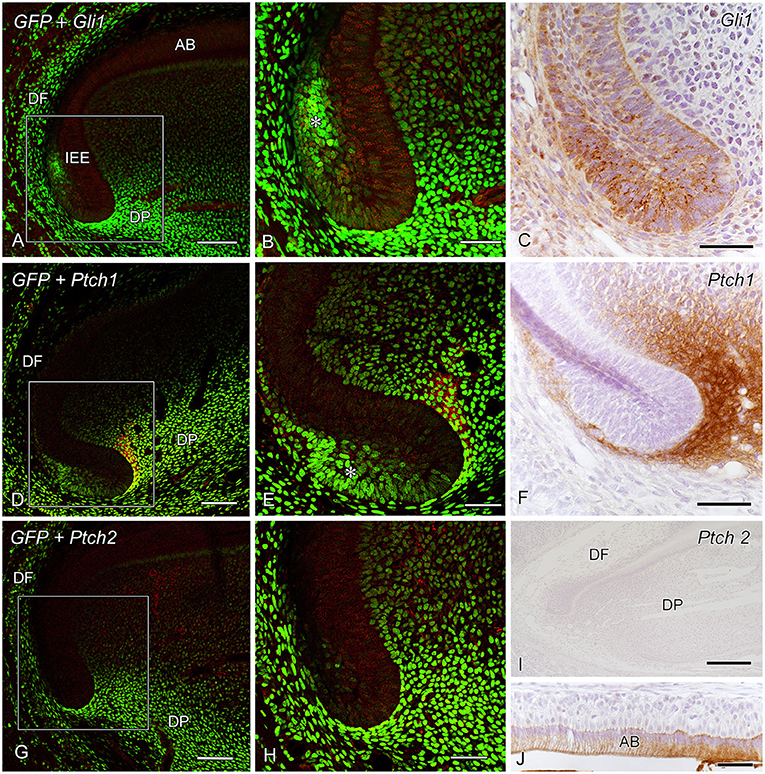
Figure 1. Green fluorescent protein (GFP) expression and Gli1 immunoreactivity (IR) (A,B), Gli1 IR (C), GFP expression and Patched1 (Ptch1) IR (D,E), Ptch1 IR (F), GFP expression and Ptch2 IR (G,H), and Ptch2 IR (I,J) in the apical end of incisors at postnatal week 3 (P3W) (DF dental follicle, DP dental papilla). (A,B,D,E,G,H) GFP+ cells are observed throughout the dental follicle (DF) and the apical dental papilla (DP) in addition to the outer enamel epithelium of the apical bud (*). (B,E,H) Higher magnification of the boxed areas in (A,D,G). (A–C) Gli1+ cells intensely localize in the inner enamel epithelium (IEE) and ameloblast layer (AB) in addition to the apical bud (*). (D–F) Ptch1+ cells are intensely localized in the apical dental papilla beneath the apical bud (*). (G–I) Intense Ptch2 expression is observed in the maturation ameloblasts (AB). Scale bars: 250 μm (I), 100 μm (A,D,G), and 50 μm (B,C,E,F,H,J).
Dense GFP+ cells were distributed throughout the dental pulp during P1–P2W and decreased in number after P2W, whereas the number of dense GFP+ cells in the subodontoblastic layer increased in number at P2W (Figures 2A,C,E–G,I,K,L, 6A,B). In contrast, granular GFP+ cells increased in number at P3W in the dental pulp. Gli1+ cells were distributed throughout the enamel organ and the dental pulp, including the odontoblast and subodontoblastic layers (Figures 2, 6A,B). Intense Ptch1+ cells were located in the enamel organ, odontoblast layer, and dental pulp (Figure 3).
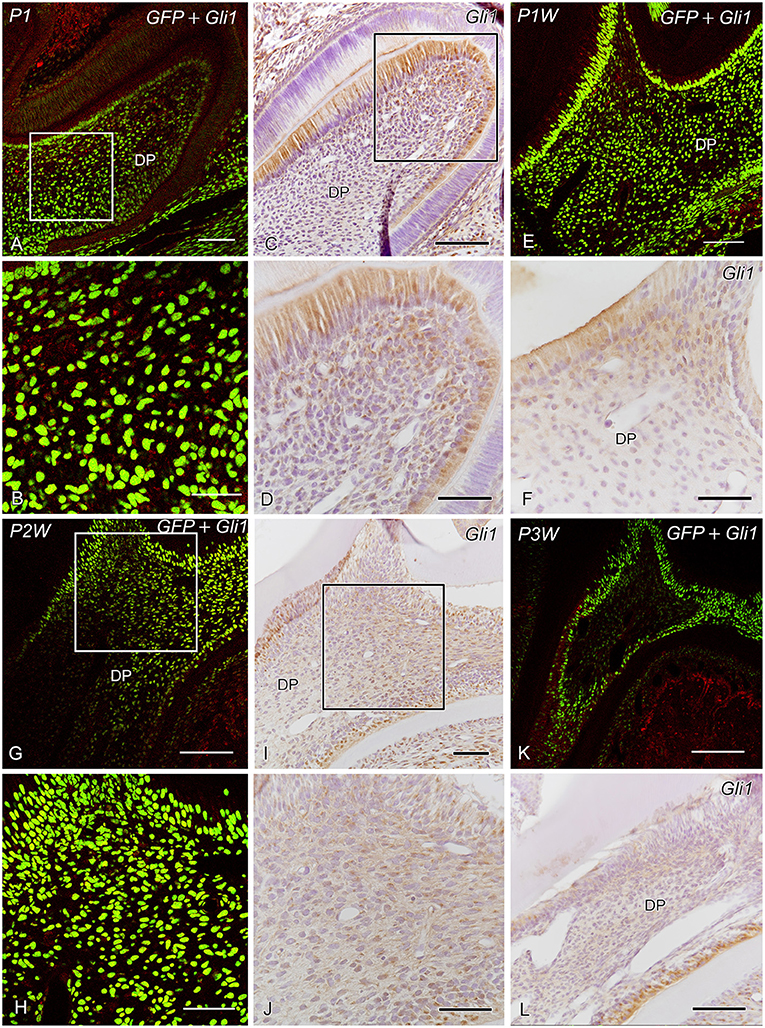
Figure 2. GFP expression and Gli1 IR (A,B,E,G,H,K) and Gli1 IR (C,D,F,I,J,L) in the developing molars at postnatal day 1 (P1) (A–D), P1W (E,F), P2W (G–J), and P3W (K,L) (DP dental pulp). (A,B,E,G,H,K) Dense GFP+ cells are observed throughout the dental pulp during P1–P2W and decrease in number at P2W, whereas the number of dense GFP+ cells in the subodontoblastic layer increases in number at P2W. In contrast, granular GFP+ cells increase in number at P3W in the dental pulp. (B,D,H,J) Higher magnification of the boxed areas in panels (A,C,G,I). (A–L) Gli1+ cells are distributed throughout the enamel organ and dental pulp, including the odontoblast and subodontoblastic layers. Scale bars: 100 μm (A,C,E,G,I,K,L) and 50 μm (B,D,F,H,J).
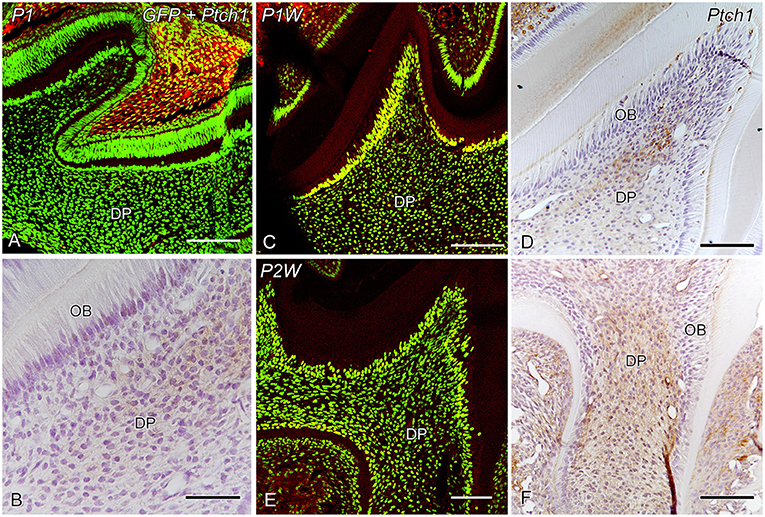
Figure 3. GFP expression and Ptch1 IR (A,C,E) and Ptch1 IR (B,D,F) in the developing molars at P1 (A,B), P1W (C,D), and P2W (E,F) (DP dental pulp, OB odontoblasts). (A–F) Intense Ptch1+ cells are located in the enamel organ and dental pulp. (B) Higher magnification of the boxed area in (A). Scale bars: 100 μm (A,C,D,E,F) and 50 μm (B).
Shh expression and Ki67+ cells were observed in the apical bud and inner enamel epithelium in both the control and 5E1 groups (Figures 4A–D) in B6 mice. TUNEL+ cells of the stellate reticulum increased in number in the 5E1 group compared with those in the control group, whereas Sox2+ cells decreased in number in the 5E1 group (Figures 4E–H, 6C). TUNEL+ dental papilla cells increased in number in the 5E1 group, although there was no significant difference between the control and 5E1 groups (Figures 4E,F, 6C). A significant decrease in the number of Sox2+ cells in the 5E1 group was also observed in TetOP-H2B-GFP mice (Figures 5, 6D).
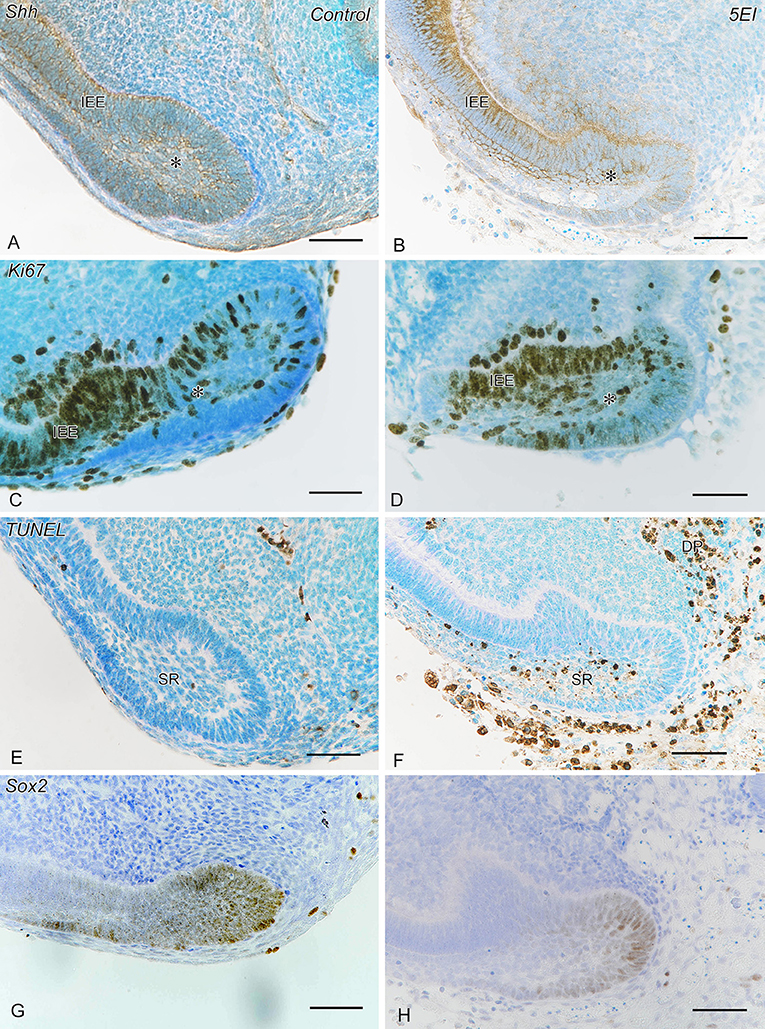
Figure 4. Shh IR (A,B), Ki67 IR (C,D), terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) (E,F), and Sox2 IR (G,H) in control (A,C,E,G) and experimental (Shh antibody: 5E1) explants (B,D,F,H). (A–D) Shh expression and Ki67+ cells are observed in the apical bud (*) and the inner enamel epithelium (IEE) in both the control and 5E1 groups. (E,F) TUNEL+ cells of stellate reticulum (SR) and dental papilla (DP) increase in number in the 5E1 group compared with those in the control. (G,H) Sox2+ cells decrease in number in the 5E1 group compared with those in the control. Scale bars: 50 μm.
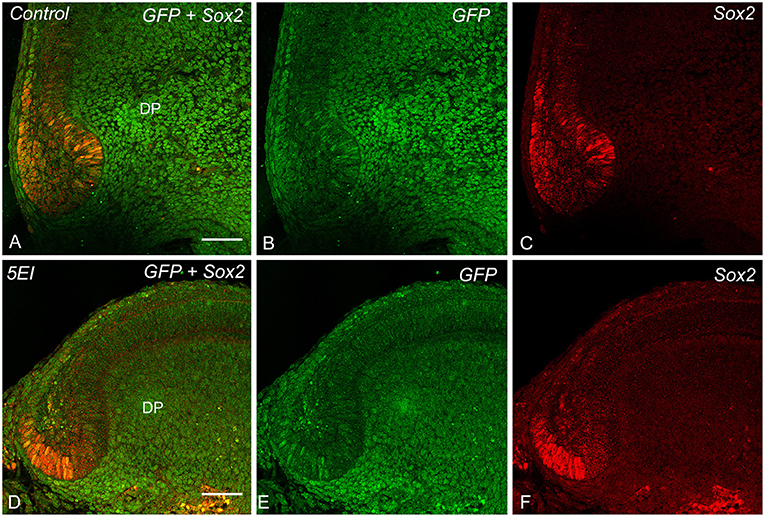
Figure 5. GFP expression and Sox2 IR (A,D), GFP expression (B,E), and Sox2 IR (C,F) in control (A–C) and 5E1 explants (D–F). Sox2+ cells decrease in number in the 5E1 group in the TetOP-H2B-GFP explants compared with those in the control. Scale bars: 50 μm.
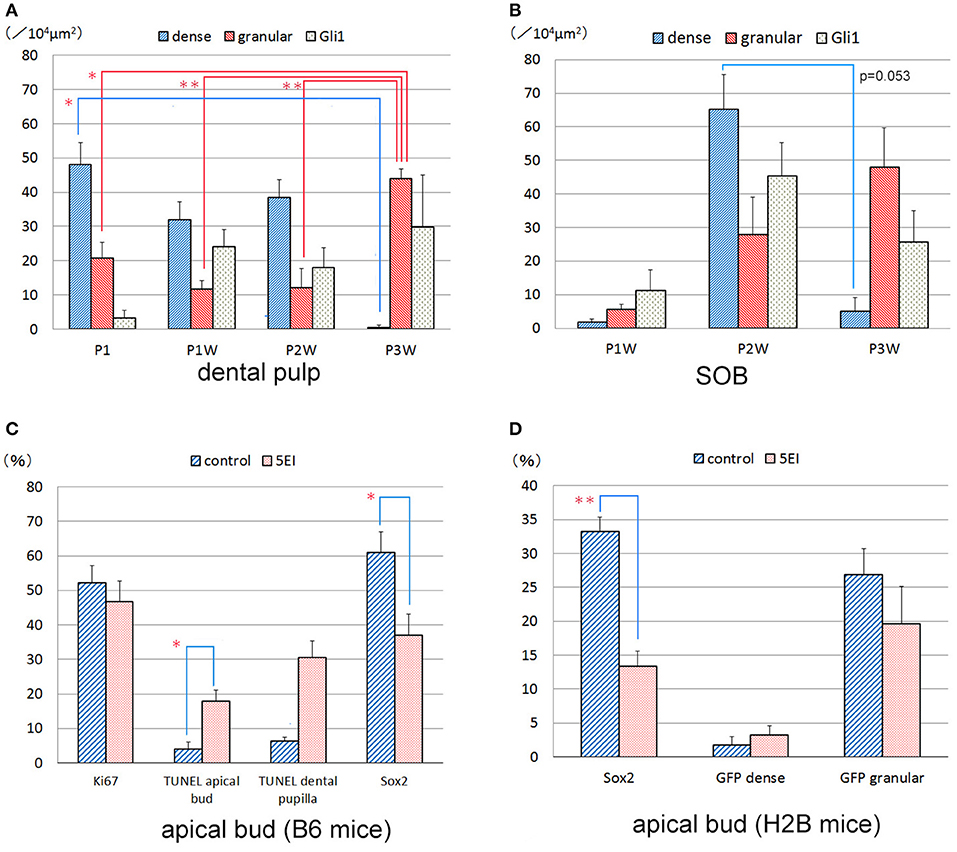
Figure 6. Quantitative analysis of dense GFP+, granular GFP+, Gli1+, Ki67+, TUNEL+, and Sox2+ cells in the dental pulp (A) and subodontoblastic layer (SOB) (B) of the developing molars (A,B) and the incisor explants (C,D). *P < 0.05, **P < 0.01.
In the apical end of the incisors, Shh was expressed in the inner enamel epithelium (Figure 7A), whereas Ptch1 expression was observed in the epithelial and mesenchymal tissues (Figure 7B). Intense Shh was exclusively expressed in the inner enamel epithelium at P1 (Figure 7C), and its expression decreased in intensity in the ameloblasts in the developing molars during P1W–P3W (Figures 7E,G). In contrast, the odontoblasts and other pulp cells began to express Shh during P1W–P3W. Intense Ptch1 expression was observed in the inner enamel epithelium and dental pulp, including odontoblasts at P1 (Figure 7D) and decreased in intensity during P1W–P3W (Figures 7F,H).
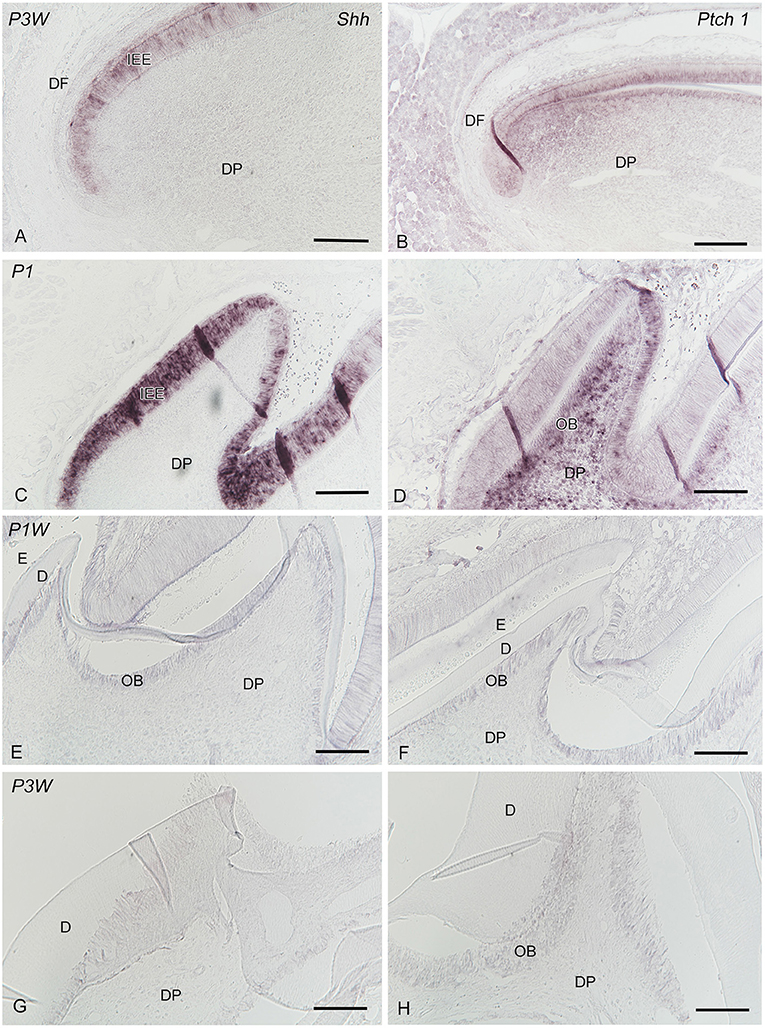
Figure 7. In situ hybridization of Shh (A,C,E,G) and Ptch1 (B,D,F,H) in the incisors (A,B) and developing molars (C–H) at P1 (C,D), P1W (E,F), and P3W (G,H) (D dentin, DP dental papilla or papilla, DF dental follicle, E enamel or enamel space, IEE inner enamel epithelium, OB odontoblasts). (A) Shh is intensely expressed in the inner enamel epithelium. (B) The epithelial and mesenchymal tissues demonstrate Ptch1 expression. (C) Shh is exclusively expressed in the inner enamel epithelium. (E,G) Shh expression in the ameloblasts decreases in intensity, whereas the odontoblasts and the other pulp cells begin to express Shh during P1W-P3W. (D) Ptch1 expression is observed in the inner enamel epithelium and the dental pulp, including odontoblasts (F,H) Ptch1 expression decreases in intensity during P1W–P3W. Scale bars: 100 μm.
The present analysis of LRCs in the apical end of incisors of TetOP-H2B-GFP mice demonstrated that dense H2B-GFP-LRCs were distributed through the dental follicle and apical dental papilla in addition to the outer enamel epithelium of the apical bud. These findings are similar to the distribution pattern of BrdU-LRCs in ICR mice with prenatal BrdU labeling, except for the LRCs observed in the dental follicle (10). Prenatal BrdU labeling by three intraperitoneal injections of BrdU (once per day from E15 to 17) also failed to detect BrdU-labeled LRCs in the periodontal ligament (PDL) of the developing molars (28). These findings indicate that dense H2B-GFP-LRCs, having not proliferated during E15–17, remain in the mature PDL. These dense LRCs were primarily distributed in the tooth-related zone. Thus, the static stem/progenitor cell population in the PDL are distinct from these cells in the apical bud and dental pulp, undergoing cell division during E15–17. The coincidence of the distribution patterns of H2B-GFP-LRCs and Gli1-IR in the dental follicle of incisors is similar to the findings in the PDL in molars (28). This study demonstrates that Shh is expressed in the inner enamel epithelium, whereas Ptch1 expression is observed in the epithelial and mesenchymal tissues. Thus, this study using B6 mice confirmed that the Shh-Ptch1-Gli1 signaling cascade plays a crucial role in the maintenance of quiescent stem cells that respond to Shh signals from the inner enamel epithelium via Ptch1 localized in the outer enamel epithelium and stellate reticulum, as previously demonstrated in ICR mice (10).
Dense GFP+ cells were distributed throughout the dental pulp during P1–P2W and decreased in number at P3W, whereas the number of dense H2B-GFP-LRCs in the subodontoblastic layer increased in number at P2W in this study. Gli1+ and Ptch1+ cells were also distributed throughout the dental pulp. These findings were similar to the distribution pattern of BrdU-LRCs in ICR mice with prenatal BrdU labeling, except for the LRCs in the subodontoblastic layer (6, 10, 33). H2B-GFP-LRCs in the subodontoblastic layer also express insulin-like growth factor (IGF)-binding protein 5 (IGFBP5)/Igfbp5, which plays a crucial role in regulating the survival and apoptosis of dental pulp stem/progenitor cells during both tooth development and pulpal healing following tooth injury (33). The expression pattern of Ptch1 was similar to that of Gli1, and this also overlapped with H2B-GFP-LRCs. These findings suggest that the quiescent dental pulp stem/progenitor cells are regulated by Shh signaling.
This study demonstrated that inhibition of Shh signaling with anti-Shh antibodies (5E1) induced an increase in apoptotic cells and a decrease in Sox2+ cells in the apical bud. These findings are similar to the recent report that inhibition of Hh signaling in the incisor stem cell niche with cyclopamine decreased the number of Sox2+ stem cells as a direct consequence of increased cell death (34). Two types of Hh, Shh and desert hedgehog, react with Ptch1 and Ptch2 independently in the vicinity of the apical bud, and subsequently multiple modes of Hh signaling regulate the maintenance of stem cells and their differentiation via the complex relationship between Hh ligands and their receptors. Interestingly, Shh expression and cell proliferation in the inner enamel epithelium seemed normal, and apoptotic cells appeared mainly in the stellate reticulum in this study. Furthermore, inhibition of Shh signaling did not influence the number of H2B-GFP-LRCs and Sox2+ cells in the outer enamel epithelium. These findings suggest that Shh signaling plays crucial roles in the transition between stem and transit-amplifying cells, resulting in a decreased number of Sox2+ cells in the inner enamel epithelium. Although there was no significant difference between the control and 5E1 groups, apoptotic cells increased in number in the apical mesenchyme of incisors, suggesting that inhibition of Shh signaling also affects the stem cell niche of the apical mesenchyme.
This study demonstrated that dense H2B-GFP-LRCs were distributed throughout the dental pulp during P1–P2W and decreased in number at P3W, whereas the number of dense H2B-GFP-LRCs in the subodontoblastic layer increased in number at P2W in the developing molars. Gli1+ and Ptch1+ cells were distributed throughout the dental pulp, whereas Shh mRNA was expressed in the inner enamel epithelium and shifted into odontoblasts after dentin deposition. Ptch1 mRNA was expressed in the inner enamel epithelium and cuspal pulpal tissue on P1 and decreased in intensity from P1W to P3W. The apical bud contained H2B-GFP-LRCs, Gli1+ cells, and Ptch1+ cells in continuously growing incisors. The addition of 5E1 to the explants in vitro experiments induced a decrease in the number of Sox2+ cells due to increased cell death in the apical bud. Thus, the Shh–Ptch–Gli signaling pathway plays a role in maintaining quiescent adult stem cells and regulating the function of odontoblasts. There may be some discrepancies between proteins and mRNAs regarding the expression patterns of the Shh–Ptch–Gli signaling pathway compared with the previous studies (16, 17, 34–37). Whether this is attributed to the specificity of antibodies or the reduced translation of mRNA in the restricted cell types remains unsolved. Furthermore, the functional significance of dense H2B-GFP-LRCs in the subodontoblastic layer and the PDL remains to be clarified under pathological conditions following exogenous injury. Recent studies demonstrate that the subodontoblastic cell population may differ from odontoblasts in their origin (38) and that odontoblasts forming the primary dentin are different from odontoblast-like cells responsible for reparative dentinogenesis in their phenotype (39). Further experimental studies using TetOP-H2B-GFP and nestin-enhanced GFP transgenic mice (38) are needed to clarify the responses of subodontoblastic cells to tooth injuries in the future.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee and the President of Niigata University.
YI and HO: conceptualization, formal analysis, and writing – original draft. YI, HI-Y, KS, MN, and HO: data curation, investigation, methodology, validation, and writing – review and editing. HO: supervision. All authors contributed to the article and approved the submitted version.
This work was supported by Grants-in-Aid for Scientific Research (C) (no. 17K11730 to YI), Scientific Research (B) (no. 17H04366 to HO), and Challenging Research (Pioneering) (no. 20K21672 to HO) from the Japan Society for the Promotion of Science (JSPS).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors cordially thank A. P. McMahon and M. P. Scott for providing riboprobes, S. Kenmotsu for his technical assistance, and Enago (www.enago.jp) for the English language review.
2. Li L, and Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. (2010) 327:542–5. doi: 10.1126/science.1180794
3. Sasaki M, Abe R, Fujita Y, Ando S, Inokuma D, and Shimizu H. Mesenchymal stem cells are recruited into wounded skin and contribute to wound repair by transdifferentiation into multiple skin cell type. J Immunol. (2008) 180:2581–7. doi: 10.4049/jimmunol.180.4.2581
4. Nanci A. Ten Cate's Oral Histology: Development, Strucutre, and Function. St. Louis, MO: Elsevier (2018).
5. Ishikawa Y, Ida-Yonemochi H, Suzuki H, Nakakura-Ohshima K, Jung HS, Honda MJ, et al. Mapping of BrdU label-retaining dental pulp cells in growing teeth and their regenerative capacity after injuries. Histochem Cell Biol. (2010) 134:227–41. doi: 10.1007/s00418-010-0727-5
6. Ishikawa Y, Ida-Yonemochi H, Nakakura-Ohshima K, and Ohshima H. The relationship between cell proliferation and differentiation and mapping of putative dental pulp stem/progenitor cells during mouse molar development by chasing BrdU-labeling. Cell Tissue Res. (2012) 348:95–107. doi: 10.1007/s00441-012-1347-2
7. Saito K, Nakatomi M, and Ohshima H. Dynamics of bromodeoxyuridine label-retaining dental pulp cells during pulpal healing after cavity preparation in mice. J Endod. (2013) 39:1250–5. doi: 10.1016/j.joen.2013.06.017
8. Saito K, and Ohshima H. Differentiation capacity and maintenance of dental pulp stem/progenitor cells in the process of pulpal healing following tooth injuries. J Oral Biosci. (2017) 59:63–70. doi: 10.1016/j.job.2017.03.001
9. Sharpe PT. Dental mesenchymal stem cells. Development. (2016) 143, 2273–80. doi: 10.1242/dev.134189
10. Ishikawa Y, Nakatomi M, Ida-Yonemochi H, and Ohshima H. Quiescent adult stem cells in murine teeth are regulated by Shh signaling. Cell Tissue Res. (2017) 369:497–512. doi: 10.1007/s00441-017-2632-x
11. Hosoya A, Shalehin N, Takebe H, Fujii S, Seki Y, Mizoguchi T, et al. Stem cell properties of Gli1-positive cells in the periodontal ligament. J Oral Biosci. (2020) 62:299–305. doi: 10.1016/j.job.2020.08.002
12. Harada H, and Ohshima H. New perspectives on tooth development and the dental stem cell niche. Arch Histol Cytol. (2004) 67:1–11. doi: 10.1679/aohc.67.1
13. Ohshima H, Nakasone N, Hashimoto E, Sakai H, Nakakura-Ohshima K, and Harada H. The eternal tooth germ is formed at the apical end of continuously growing teeth. Arch Oral Biol. (2005) 50:153–7. doi: 10.1016/j.archoralbio.2004.09.008
14. Harada H, Kettunen P, Jung HS, Mustonen T, Wang YA, and Thesleff I. Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J Cell Biol. (1999) 147:105–20. doi: 10.1083/jcb.147.1.105
15. Juuri E, Saito K, Ahtiainen L, Seidel K, Tummers M, Hochedlinger K, et al. Sox2+ stem cells contribute to all epithelial lineages of the tooth via Sfrp5+ progenitors. Dev Cell. (2012) 23:317–28. doi: 10.1016/j.devcel.2012.05.012
16. Zhao H, Feng J, Seidel K, Shi S, Klein O, Sharpe P, et al. Secretion of shh by a neurovascular bundle niche supports mesenchymal stem cell homeostasis in the adult mouse incisor. Cell Stem Cell. (2014) 14:160–73. doi: 10.1016/j.stem.2013.12.013
17. Li J, Feng J, Liu Y, Ho TV, Grimes W, Ho HA, et al. BMP-SHH signaling network controls epithelial stem cell fate via regulation of its niche in the developing tooth. Dev Cell. (2015) 33:125–35. doi: 10.1016/j.devcel.2015.02.021
18. Pandolfi S, and Stecca B. Cooperative integration between HEDGEHOG-GLI signalling and other oncogenic pathways: implications for cancer therapy. Expert Rev Mol Med. (2015) 17:e5. doi: 10.1017/erm.2015.3
19. Seidel K, Ahn CP, Lyons D, Nee A, Ting K, Brownell I, et al. Hedgehog signaling regulates the generation of ameloblast progenitors in the continuously growing mouse incisor. Development. (2010) 137:3753–61. doi: 10.1242/dev.056358
20. Stone DM, Hynes M, Armanini M, Swanson TA, Gu Q, Johnson RL, et al. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. (1996) 384:129–34. doi: 10.1038/384129a0
21. Motoyama J, Takabatake T, Takeshima K, and Hui C. Ptch2, a second mouse Patched gene is co-expressed with Sonic hedgehog. Nat Genet. (1998) 18:104–6. doi: 10.1038/ng0298-104
22. Von Ohlen T, Lessing D, Nusse R, and Hooper JE. Hedgehog signaling regulates transcription through cubitus interruptus, a sequence-specific DNA binding protein. Proc Natl Acad Sci U S A. (1997) 94:2404–9. doi: 10.1073/pnas.94.6.2404
23. Schaniel C, and Moore KA. Genetic models to study quiescent stem cells and their niches. Ann N Y Acad Sci. (2009) 1176:26–35. doi: 10.1111/j.1749-6632.2009.04608.x
24. Mutoh N, Nakatomi M, Ida-Yonemochi H, Nakagawa E, Tani-Ishii N, and Ohshima H. Responses of BrdU label-retaining dental pulp cells to allogenic tooth transplantation into mouse maxilla. Histochem Cell Biol. (2011) 136:649–61. doi: 10.1007/s00418-011-0868-1
25. Saito K, Ishikawa Y, Nakakura-Ohshima K, Ida-Yonemochi H, Nakatomi M, Kenmotsu S, et al. Differentiation capacity of BrdU label-retaining dental pulp cells during pulpal healing following allogenic transplantation in mice. Biomed Res. (2011) 32:247–57. doi: 10.2220/biomedres.32.247
26. Foudi A, Hochedlinger K, Van Buren D, Schindler JW, Jaenisch R, Carey V, et al. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat Biotechnol. (2009) 27:84–90. doi: 10.1038/nbt.1517
27. Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, et al. Defining the epithelial stem cell niche in skin. Science. (2004) 303:359–63. doi: 10.1126/science.1092436
28. Nakaki T, Nakakura-Ohshima K, Nakagawa E, Ishikawa Y, Saito K, Ida-Yonemochi H, et al. Donor-host tissue interaction in allogenic transplanted tooth germ with special reference to periodontal tissue. J Oral Biosci. (2018) 60:21–30. doi: 10.1016/j.job.2018.02.002
29. Ida-Yonemochi H, Otsu K, Ohshima H, and Harada H. The glycogen metabolism via Akt signaling is important for the secretion of enamel matrix in tooth development. Mech Dev. (2016) 139:18–30. doi: 10.1016/j.mod.2016.01.002
30. Nakatomi M, Morita I, Eto K, and Ota MS. Sonic hedgehog signaling is important in tooth root development. J Dent Res. (2006) 85:427–31. doi: 10.1177/154405910608500506
31. Bitgood MJ, and McMahon AP. Hedgehog and Bmp genes are coexpressed at many diverse sites of cell-cell interaction in the mouse embryo. Dev Biol. (1995) 172:126–38. doi: 10.1006/dbio.1995.0010
32. Milenkovic L, Goodrich LV, Higgins KM, and Scott MP. Mouse patched1 controls body size determination and limb patterning. Development. (1999) 126:4431–40.
33. Saito K, and Ohshima H. The putative role of insulin-like growth factor (IGF)-binding protein 5 independent of IGF in the maintenance of pulpal homeostasis in mice. Regen Ther. (2019) 11:217–24. doi: 10.1016/j.reth.2019.08.001
34. Binder M, Chmielarz P, McKinnon PJ, Biggs LC, Thesleff I, and Balic A. Functionally distinctive ptch receptors establish multimodal hedgehog signaling in the tooth epithelial stem cell niche. Stem Cells. (2019) 37:1238–48. doi: 10.1002/stem.3042
35. Liu Y, Feng J, Li J, Zhao H, Ho TV, and Chai Y. An Nfic-hedgehog signaling cascade regulates tooth root development. Development. (2015) 142:3374–82. doi: 10.1242/dev.127068
36. Khan M, Seppala M, Zoupa M, and Cobourne MT. Hedgehog pathway gene expression during early development of the molar tooth root in the mouse. Gene Expr Patterns. (2007) 7:239–43. doi: 10.1016/j.modgep.2006.10.001
37. Gritli-Linde A, Bei M, Maas R, Zhang XM, Linde A, and McMahon AP. Shh signaling within the dental epithelium is necessary for cell proliferation, growth and polarization. Development. (2002) 129:5323–37. doi: 10.1242/dev.00100
38. Nakatomi M, Quispe-Salcedo A, Sakaguchi M, Ida-Yonemochi H, Okano H, and Ohshima H. Nestin expression is differently regulated between odontoblasts and the subodontoblastic layer in mice. Histochem Cell Biol. (2018) 149:383–91. doi: 10.1007/s00418-018-1651-3
Keywords: adult stem cells, dental pulp, gli protein, hedgehog proteins, mice, patched-1 receptor, odontogenesis, tooth germ
Citation: Ishikawa Y, Ida-Yonemochi H, Saito K, Nakatomi M and Ohshima H (2021) The Sonic Hedgehog–Patched–Gli Signaling Pathway Maintains Dental Epithelial and Pulp Stem/Progenitor Cells and Regulates the Function of Odontoblasts. Front. Dent. Med. 2:651334. doi: 10.3389/fdmed.2021.651334
Received: 09 January 2021; Accepted: 22 March 2021;
Published: 14 April 2021.
Edited by:
Motoki Okamoto, Osaka University, JapanReviewed by:
Thimios Mitsiadis, University of Zurich, SwitzerlandCopyright © 2021 Ishikawa, Ida-Yonemochi, Saito, Nakatomi and Ohshima. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hayato Ohshima, aGlzdG9tYW5AZGVudC5uaWlnYXRhLXUuYWMuanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.