- 1Computer Solutions SA, Research & Development Dpt., Athens, Greece
- 2School of Electrical and Computer Engineering, National Technical University of Athens, Athens, Greece
Introduction: Electronic Health Records (EHRs) are essential data structures, enabling the sharing of valuable medical care information for a diverse patient population and being reused as input to predictive models for clinical research. However, issues such as the heterogeneity of EHR data and the potential compromisation of patient privacy inhibit the secondary use of EHR data in clinical research.
Objectives: This study aims to present the main elements of the MODELHealth project implementation and the evaluation method that was followed to assess the efficiency of its mechanism.
Methods: The MODELHealth project was implemented as an Extract-Transform-Load system that collects data from the hospital databases, performs harmonization to the HL7 FHIR standard and anonymization using the k-anonymity method, before loading the transformed data to a central repository. The integrity of the anonymization process was validated by developing a database query tool. The information loss occurring due to the anonymization was estimated with the metrics of generalized information loss, discernibility and average equivalence class size for various values of k.
Results: The average values of generalized information loss, discernibility and average equivalence class size obtained across all tested datasets and k values were 0.008473 ± 0.006216252886, 115,145,464.3 ± 79,724,196.11 and 12.1346 ± 6.76096647, correspondingly. The values of those metrics appear correlated with factors such as the k value and the dataset characteristics, as expected.
Conclusion: The experimental results of the study demonstrate that it is feasible to perform effective harmonization and anonymization on EHR data while preserving essential patient information.
Introduction
Electronic Health Record (EHR) systems are being increasingly adopted to represent various data types, such as patient medical histories, laboratory test results, medication, demographics, billing records and diagnosis codes. EHR systems are the building blocks of Health Information Exchange (HIE) networks, enabling the sharing of data and information about patients' medical and health history (1–3).
EHRs surpass many existing registries and data repositories in volume, offering a window into the medical care information of a diverse population. Their effectiveness when reused for the purpose of clinical research is proven in various instances (4–7). However, their reuse has been limited due to issues such as its high dimensionality, heterogeneity, incompleteness, noise and errors, and redundant terminology (4, 5).
Interoperability is a crucial requirement for the efficiency of healthcare information systems and the utilization of health data for clinical research. The related concept of data harmonization aims to transform heterogeneous data into a standard format using computational approaches such as lexical and semantic mapping, enabling the integrative analysis of the data and, therefore, enhancing the statistical power of the clinical studies which make use of such data. Health Level Seven (HL7) is currently the most widely used set of standards for the structure and exchange of clinical data (8).
Anonymization is another essential issue regarding the secondary use of clinical data. Patient data must be disseminated without compromising their privacy against threats such as identity, membership and attribute disclosure (2). Data privacy protection can be pursued with methods such as encryption, authentication, and de-identification, which however can be inapplicable or insufficient in preserving confidential information. For example, the removal of data identifiers such as each individual's name and social security number does not prohibit their possible reidentification through the linkage of other data attributes. To prevent such attacks, the concept of k-anonymity, as well as its extensions l-diversity and t-closeness, have been proposed (9, 10).
The k-anonymity concept, introduced by Samarati and Sweeny (11), focuses on reducing data granularity. A dataset is k-anonymous if each record is indistinguishable from at least k−1 records with respect to specific identifying attributes. A quasi-identifier (QI) set is a minimal set of dataset attributes that can be joined with external information to re-identify individual records. K-anonymity requires that each equivalence class EQ (i.e., a set of records that are indistinguishable from each other with respect to the QI set) contains at least k records. K-anonymity can be provided using suppression and generalization techniques. Suppression involves replacing a portion of the original data with a special selected value to suggest its nondisclosure, while generalization focuses on replacing the values of an attribute with less specific but consistent values. K-anonymity is considered as the “bedrock” anonymization algorithm and is used as a foundation process, even in the rare case that the overall privacy it provides could be considered inadequate, allowing the potential disclosure of sensitive attributes that lack diversity through the use of background knowledge (11–16).
Given the sensitive nature and complexity of clinical data, a systematic overall approach is needed for their secondary use, examples of which can be found in the literature. Ciampi et al. (17) proposed an architecture for the extraction, transformation and loading of clinical data, which incorporates de-identification and standardization to the HL7 CDA and FHIR formats (17). Somolinos et al. (18) proposed a pseudonymizing system developed according to the ISO/EN 13606 standard for facilitating the exchange and secondary use of data, allowing the total or partial anonymization of EHR extracts (18, 19). Quiroz et al. (20) developed an SQL-based ETL framework for the conversion of health databases to the OMOP CDM (20, 21). Ong et al. (22) developed a GUI-based ETL system for the conversion of data to the OMOP CDM (22).
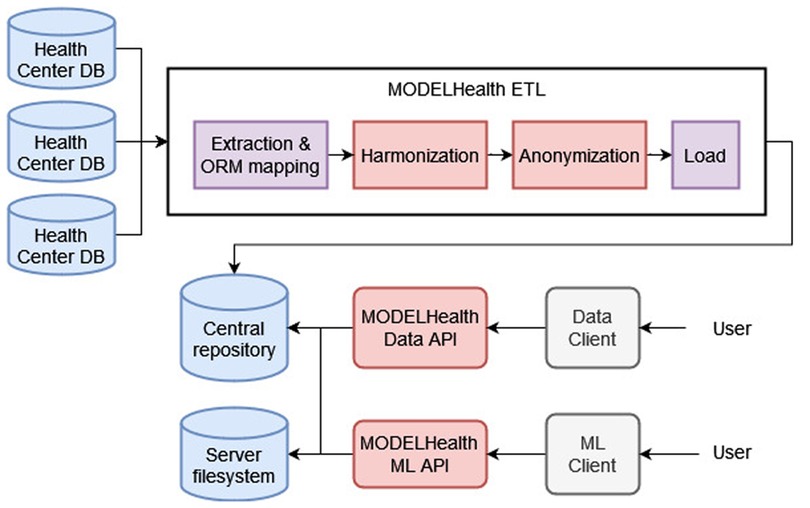
This paper proposes an integrated solution to the problem of clinical data reuse that has been implemented in the context of the MODELHealth project. The project is based on an ETL system that extracts EHR data from several hospital databases (Section 2.1), transforms the data by performing harmonization to the HL7 FHIR standard and anonymization with the k-anonymity method (Sections 2.2, 2.2.1, 2.2.2), and loads the transformed data to a central, document-based repository (Section 2.3) (23, 24). The data used is raw EHRs from selected Greek hospital databases regarding patients, hospitalization encounters, medical procedures and observations, diagnostic reports and locations. An essential objective of the MODELHealth project has been the utilization of the transformed clinical data as input to predictive models. This goal was met by developing two public-facing REST Application Program Interfaces (Data API, Machine Learning API) and client software (Data Client, ML Client). The Data API and Client serve the purpose of making the transformed data stored in the central repository available to the interested users, while the Machine Learning API exposes the functionality of trained and validated machine learning models to the interested users. The information loss that occurred due to the anonymization was evaluated using three metrics, described in Section 2.4. The components of the MODELHealth project were developed in the Python programming language, and are depicted in Figure 1.
Methods
Extraction
The data extraction process involves the automated extraction of data from three hospital databases and their mapping to relational objects that reflect the database schema with the use of the SQLAlchemy Object Relational Mapper software. Implementing the MODELHealth ETL process included versioning, allowing the additive extraction, processing and loading of the data in several points in time. Each version includes all the data extracted from a health unit database until that time point. The primary key value of the last extracted record is stored for every version and every database table so that future execution of the ETL process will take into account only the new records. The detailed ER diagrams of the relational database tables from which the EHR data originated can be seen in Supplementary Figure S1.
Transformation
Harmonization
The harmonization process refers to mapping the extracted data from the form of relational objects to FHIR (Fast Healthcare Interoperability Resources) ontology objects. FHIR is a RESTful API using the HTTP protocol and leveraging the HL7 Reference Information Model (RIM). FHIR defines a system of clinical, administrative, financial and infrastructure resources, its ontologies being organized in the clinical, financial, specialized, base and foundation categories (25–30).
The harmonization of the extracted data has been achieved with in-house software. First, the relational data are converted to the corresponding FHIR ontologies through custom specialized programming libraries and transformative functions related to the database schema from which the data originated. FHIR data were converted to the JSON (JavaScript Object Notation) format, as this is the preferred representation of the standard. The main FHIR entities incorporated were the Patient, Observation, DiagnosticReport, Encounter and Location ontologies. Supplementary Figure S2 depicts the FHIR entities according to which the relational data were harmonized.
Anonymization
The anonymization process involves modifying several fields in a given dataset to prevent the individuals' reidentification. In the scope of this project, anonymization of the harmonized EHRs was carried out using Mondrian, a greedy algorithm that implements k-anonymity through multidimensional recoding and applies to both categorical and numeric data. Mondrian performs k-anonymization of a given dataset with logarithmic worst-case time complexity in two stages. The first stage focuses on partitioning the given dataset on several multidimensional regions covering its domain space by applying a recursive algorithm similar to the ones used to construct kd-trees. The second stage focuses on applying re-coding functions to the dataset, formulated using summary statistics from each region (31).
The data fields subjected to anonymization were the birthDate and address attributes of the Patient FHIR ontology and the longitude and latitude corresponding to the address. Each address was translated to longitude and latitude coordinates through the OpenStreetMap API, which were then added as numerical fields to the patient record and were included in the anonymization process (32). Supplementary Figure S3 depicts an example of the anonymization of a sample subset of male patient records, which was subjected to the ETL process and stored in the document-based database MongoDB (see Section 2.3). A sample harmonized, non-anonymized record is depicted at the top, with the FHIR id, maritalStatus fields, as well as the _id field, which serves as a primary key for MongoDB, having been suppressed for clarity. A sample anonymized record using k = 5 is displayed at the bottom, having used the FHIR fields “address”, “birthDate”, as well as the added fields “ord_latitude” and “ord_longitute” as QI attributes.
Loading
The loading process involved the transmission of the transformed data through a streaming process and their subsequent storage to the central repository. Data was streamed in predefined-sized packages through a TCP/IP connection. The central repository was implemented with the non-relational database MongoDB, in which every record is stored in the BSON format. MongoDB is a fitting choice for storing and retrieving JSON documents, as it is designed to handle effectively document-oriented, semi-structured data (33).
Information loss evaluation
The impact of the anonymization on the harmonized EHR data was estimated using the metrics of generalized information loss, discernibility and average equivalence class size.
Generalized information loss (GIL) captures the penalty incurred when generalizing a specific attribute by quantifying the fraction of the generalized domain values. GIL for an anonymized table T* was calculated according to Equation (1), where T is the original table, i = 1,…,n corresponds to an attribute, j = 1,…,|T| corresponds to a table record, Ui, Li are the upper and lower values of each arithmetic attribute i, Uij, Lij are the upper and lower values of arithmetic attribute i for the equivalence class the record j belongs in, Ni is the number of different values for each categorical attribute i and Nij is the number of different values for categorical attribute i in the equivalence class the record j belongs in (34–36).
The discernibility metric (DM) measures how indistinguishable a record is from others by assigning a penalty to each record, equal to the size of the equivalence class in which it belongs. DM for an anonymized table T* was calculated according to Equation (2), where |EQ| is the number of records of the equivalence class EQ (31, 36, 37).
The average equivalence class size (CAVG) measures how well the created equivalence classes approach the best case, where each record is generalized in an equivalence class of k records. It was calculated according to Equation (3), where |T| is the number of table records, |EQs| is the total number of equivalence classes created in the anonymized table T*, and k is the minimum equivalence class size allowed (31, 37).
class="mb15">The information loss evaluation has been applied to experimental datasets originating from three hospital databases. More specifically, the patient data populating the table CARE_PERSON of three hospital databases were subjected to the ETL process for the k values 5, 10, 15, 20. The transformed datasets S1, S2, S3 correspond to the three origin database schemas, while the dataset S123 constitutes the union of S1, S2, S3. The four datasets were evaluated in terms of the information loss that occurred during the anonymization stage using Equations (1–3). The technical characteristics of the datasets S1, S2, S3, S123 are presented in Table 1.
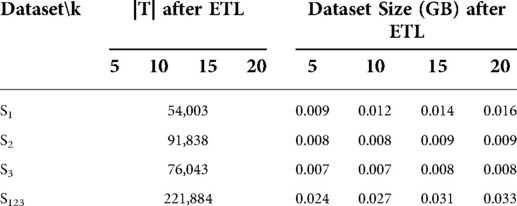
Table 1. The number of records (|T|) and the size in GBs of the tested datasets S1, S2, S3, S123 for all tested k values.
Results
Data quality evaluation
The result of the ETL process regarding the data stored in the central repository was evaluated in terms of data quality. There were no duplicate entries found, which can be attributed to the origin relational database design as well as the lack of corresponding defects in the ETL process. There were null address values, which were intentionally not rejected during the transform stage since the field of patient address underwent anonymization (38, 39).
Anonymity validation
The integrity of the data anonymization process was validated through the development of a simple validation tool, the object of which is to perform queries to the central repository to retrieve the anonymized data, group them by the QI attributes in order to retrieve the equivalence classes and check if there is an equivalence class with size greater than the k value chosen during the extraction stage. The application of this method proved that the data contents of the central repository do not violate the k-anonymity condition since no equivalence class consisting of fewer than k documents was found.
Information loss evaluation
The generalized information loss (GIL), discernibility metric (DM) and average equivalence class size (CAVG) metrics (Section 2.4) were applied on the ETL output of the experimental datasets S1, S2, S3, S123 for all tested k values. The results of the evaluation can be seen in Table 2 and Figure 2.
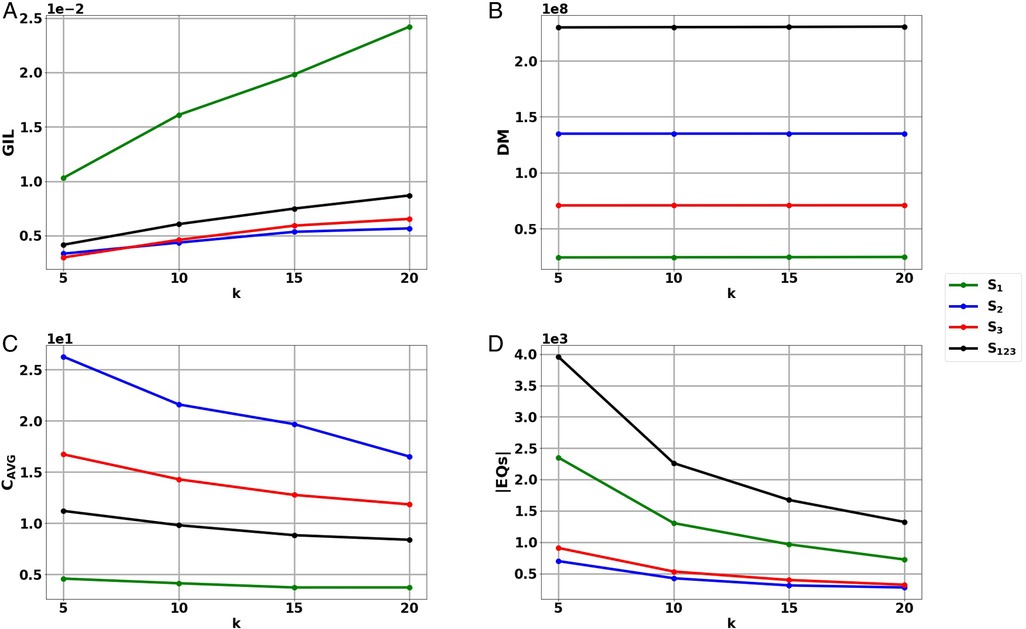
Figure 2. Results of the information loss metrics (A) Generalized Information Loss (GIL), (B) Discernibility Metric (DM) and (D) Average Equivalence Class size (CAVG), as well as (D) the number of Equivalence Classes (|EQs|) of the harmonized, anonymized data sets S1, S2, S3, S123 for the tested k values. The results are depicted in scientific format.
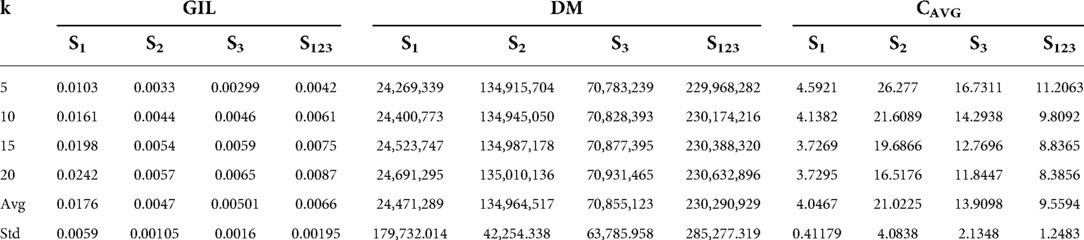
Table 2. Results of the generalized information loss (GIL), discernibility metric (DM) and average equivalence class size (CAVG) on data sets S1, S2, S3, S123 for the chosen k values. The average values (Avg) and the standard deviation (Std) of the results have been also included.
It can be observed that GIL, DM and CAVG follow the same trends as k increases regardless of the experimental dataset. More specifically, increasing k results in the increase of GIL, the increase of DM and the decrease of CAVG for all tested datasets S1, S2, S3, S123.
GIL depends on the dataset QI values and the record number |T| of a given dataset (Equation 1), meaning that a smaller |T| can lead to a larger GIL value. Indeed, in Figure 2A, it can be observed that GIL takes the highest values in the smallest dataset S1 and lower values in the larger datasets S2, S3, S123. The average and standard deviation GIL values obtained for datasets S1, S2, S3, S123 were 0.0176 ± 0.0059, 0.0047 ± 0.00105, 0.00501 ± 0.0016, 0.0066 ± 0.00195, respectively.
DM depends on the number of records in each EQ, as well as the number of EQs (|EQs|) created (Equation 2). As record number |T| increases, anonymization can result in more and larger EQs increasing DM, as can be seen in Figure 2B. The average and standard deviation DM values obtained for datasets S1, S2, S3, S123 were 24,471,289 ± 179,732.014, 134,964,517 ± 42,254.338, 70,855,123 ± 63,785.958, 230,290,929 ± 285,277.319, respectively.
CAVG is proportional to the record number |T| but inversely proportional to |EQs| and k (Equation 3). In Figure 2C, it can be observed that CAVG takes the smallest values in dataset S1 with the lowest record number. The highest values occur in dataset S2, which is second in terms of record number and at the same time has a rather low number of equivalence classes |EQs| (Figure 2D). The fact that CAVG does not take the highest values in the largest dataset, S123 coincides with the high |EQs| value of S123 (Figure 2D). The average and standard deviation DM values obtained for the datasets S1, S2, S3, S123 were 4.0467 ± 0.41179, 21.0255 ± 4.0838, 13.9098 ± 2.1348, 9.5594 ± 1.2483, respectively.
Discussion
In this paper, an integrated architecture for the facilitation of the secondary usage of clinical data has been proposed. The MODELHealth project has aimed to enable an organization to access real health record data in a universally accepted format and carry out research at a low cost. Data was harmonized to the HL7 FHIR standard, and anonymized according to the k-anonymity principle through the Mondrian algorithm. The effect of anonymization was quantified using the generalized information loss, discernibility metric and average class size metrics. In future work and subsequent versions of the platform, extensions of k-anonymity will be considered in order to add more privacy features to the central data repository, as well as other state-of-the-art approaches, such as differential privacy.
A noteworthy challenge that was met at the stage of transformation concerned the quality of EHR data, which were characterized by high dimensionality, heterogeneity, noise and sparseness. Different codes, measure units and terminologies were often used to represent the same clinical phenotype. Therefore, the harmonization of these EHR data, initially stored in relational health center databases, to the FHIR scheme required extensive transformations through custom software.
The development of predictive models utilizing EHRs has been proposed as a promising means towards the improvement of personalized medicine and health care quality. Numerous machine learning methods have been successfully applied to patient hospitalization metadata to accomplish meaningful prediction of medical-related outcomes. Deep neural networks, in particular, have proven their ability to handle large volumes of relatively messy clinical data and have emerged as a preferred method (5, 40–44). The applicability of the MODELHealth data as input to predictive models was reassured through the development of proof-of-concept machine learning models that utilized the transformed clinical data.
Conclusions
The secondary research use of EHR data without compromising the patients' rights to privacy is one of the most discussed topics in Health IT nowadays as well as a source of great controversy on whichever level (academic, technical, administrative, political) this discussion takes place. The results of this study add experimental data in favor of the side of the argument that adequate anonymization while preserving actionable and meaningful information can be performed on health datasets via proper utilization of network and data flow architectures and algorithmic tools already available in the respective literature.
Data availability statement
The project datasets and code cannot be made publicly available, because the submitted paper is part of the MODELHealth project, which has been co-funded by the European Regional Development Fund of the European Union and Greek national funds.
Ethics statement
This article does not contain any studies involving human participants or animals performed by any of the authors.
Author contributions
SP, AF, and AA contributed to the writing of the paper. AF wrote the main part of the computer code and conducted the experiments. DK and GKM provided scientific supervision. SP and DK were the coordinator and scientific director of the MODELHealth project, respectively. All authors contributed to the article and approved the submitted version.
Funding
This research has been co-funded by the European Regional Development Fund of the European Union and Greek national funds through the Operational Program Competitiveness, Entrepreneurship and Innovation, under the call Research-Create-Innovate (Project Code: T1EDK-04066).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fdgth.2022.841853/full#supplementary-material.
References
1. Heart T, Ben-Assuli O, Shabtai I. A review of PHR, EMR and EHR integration: a more personalized healthcare and public health policy. Health Policy Technol. (2017) 6(1):20–5. doi: 10.1016/j.hlpt.2016.08.002
2. Gkoulalas-Divanis A, Loukides G, Sun J. Publishing data from electronic health records while preserving privacy: a survey of algorithms. J Biomed Inform. (2014) 50:4–19. doi: 10.1016/j.jbi.2014.06.002
3. Khokhar RH, Chen R, Fung BCM, Lui SM. Quantifying the costs and benefits of privacy-preserving health data publishing. J Biomed Inform. (2014) 50(August):107–21. doi: 10.1016/J.JBI.2014.04.012
4. Weiskopf NG, Weng C. Methods and dimensions of electronic health record data quality assessment: enabling reuse for clinical research. J Am Med Inform Assoc. (2013) 20(1):144–51. doi: 10.1136/amiajnl-2011-000681
5. Miotto R, Li L, Kidd BA, Dudley JT. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci Rep. (2016) 6(May). doi: 10.1038/srep26094
6. Bean DM, Wu H, Dzahini O, Broadbent M, Stewart R, Dobson RJB. Knowledge graph prediction of unknown adverse drug reactions and validation in electronic health records. Sci Rep. (2017) 7(1):1–11. doi: 10.1038/s41598-017-16674-x
7. Zhao J, Henriksson A, Asker L, Boström H. Predictive modeling of structured electronic health records for adverse drug event detection. BMC Med Inform Decis Mak. (2015) 15(4):S1. doi: 10.1186/1472-6947-15-S4-S1
8. “Health Level Seven International | HL7 International.” n.d. https://www.hl7.org/ (Accessed July 29, 2022.).
9. Abouelmehdi K, Beni-Hssane A, Khaloufi H, Saadi M. Big data security and privacy in healthcare: a review. Procedia Comput Sci. (2017) 113:73–80. doi: 10.1016/j.procs.2017.08.292
10. Park H, Shim K. Approximate algorithms with generalizing attribute values for K-anonymity. Inf Syst. (2010) 35(8):933–55. doi: 10.1016/j.is.2010.06.002
11. Samarati P, Sweeney L. “Protecting privacy when disclosing information: k-anonymity and its enforcement through generalization and suppresion.” In: Proceedings of the IEEE symposium on research in security and privacy (1998). p. 384–93. doi: 10.1145/1150402.1150499
12. Aggarwal CC, Yu PS. “A General Survey of Privacy-Preserving Data Mining Models and Algorithms.” In, 11–52. doi: 10.1007/978-0-387-70992-5_2. (2008).
13. Li N, Li T, Venkatasubramanian S. “T-Closeness: privacy beyond k-anonymity and l-diversity.” In: 2007 IEEE 23rd international conference on data engineering. IEEE. (2007). p. 106–15. doi: 10.1109/ICDE.2007.367856
14. Machanavajjhala A, Kifer D, Gehrke J, Venkitasubramaniam M. ℓ-Diversity: privacy beyond k-anonymity. ACM Trans Knowl Discov Data. (2007) 1(1):24. doi: 10.1145/1217299.1217302
15. Emam KE, Dankar FK. Protecting privacy using K-anonymity. J Am Med Inform Assoc. (2008) 15(5):627–37. doi: 10.1197/jamia.M2716
16. Truta TM, Vinay B. “Privacy protection: p-sensitive k-anonymity property.” In: ICDEW 2006 - Proceedings of the 22nd international conference on data engineering workshops. Institute of Electrical and Electronics Engineers Inc. (2006). doi: 10.1109/ICDEW.2006.116
17. Ciampi M, Sicuranza M, Silvestri S. A privacy-preserving and standard-based architecture for secondary use of clinical data. Information. (2022) 13(2):87. doi: 10.3390/info13020087
18. Somolinos R, Muñoz A, Elena Hernando M, Pascual M, Cáceres J, Sánchez-De-madariaga R, et al. Service for the pseudonymization of electronic healthcare records based on ISO/EN 13606 for the secondary use of information. IEEE J Biomed Health Inform. (2015) 19(6):1937–44. doi: 10.1109/JBHI.2014.2360546
19. “ISO - ISO 13606-1. Health Informatics — Electronic Health Record Communication — Part 1: Reference Model.” n.d. Accessed July 29, 2022. https://www.iso.org/standard/67868.html (2019).
20. Quiroz JC, Chard T, Sa Z, Ritchie A, Jorm L, Gallego B. “Extract, Transform, Load Framework for the Conversion of Health Databases to OMOP.” Edited by Thomas Martin Deserno. PLOS ONE 17 (4): e0266911. doi: 10.1371/journal.pone.0266911 (2022).
21. “OMOP Common Data Model – OHDSI.” . Accessed July 29, 2022. https://www.ohdsi.org/data-standardization/the-common-data-model/ (n.d).
22. Ong TC, Kahn MG, Kwan BM, Yamashita T, Brandt E, Hosokawa P, et al. Dynamic-ETL: a hybrid approach for health data extraction, transformation and loading. BMC Med Inform Decis Mak. (2017) 17(1):134. doi: 10.1186/s12911-017-0532-3
23. Anastasiou A, Pitoglou S, Androutsou T, Kostalas E, Matsopoulos G, Koutsouris D. “Modelhealth: an innovative software platform for machine learning in healthcare leveraging indoor localization services.” In: Proceedings - IEEE international conference on mobile data management; 2019-June. Institute of Electrical and Electronics Engineers Inc. (2019). p. 443–46. doi: 10.1109/MDM.2019.000-5
24. Pitoglou S, Anastasiou A, Androutsou T, Giannouli D, Kostalas E, Matsopoulos G, et al. “MODELHealth: facilitating machine learning on big health data networks.” In: Proceedings of the annual international conference of the IEEE Engineering in medicine and biology society, EMBS. Institute of Electrical and Electronics Engineers (IEEE). (2019). p. 2174–77. doi: 10.1109/EMBC.2019.8857394
25. Bender D, Sartipi K. “HL7 FHIR: an Agile and RESTful approach to healthcare information exchange.” In: Proceedings of CBMS 2013 - 26th IEEE international symposium on computer-based medical systems. (2013). p. 326–31. doi: 10.1109/CBMS.2013.6627810
26. Pezoulas VC, Exarchos TP, Fotiadis DI. “Medical data harmonization.” In: Medical data sharing, harmonization and analytics. Elsevier. (2020). p. 137–83. doi: 10.1016/b978-0-12-816507-2.00005-0. https://www.sciencedirect.com/book/9780128165072/medical-data-sharing-harmonization-and-analytics?via=ihub=
27. Saripalle R, Runyan C, Russell M. Using HL7 FHIR to achieve interoperability in patient health record. J Biomed Inform. (2019) 94. doi: 10.1016/j.jbi.2019.103188
28. Silva RJ, Sloane EB, Cooper T. Application of HL7® FHIR for device and health information system interoperability.” In: Iadanza E, editor. Clinical engineering handbook. Elsevier. (2020). p. 611–15. doi: 10.1016/b978-0-12-813467-2.00086-9. https://www.sciencedirect.com/book/9780128134672/clinical-engineering-handbook
29. Kiourtis A, Mavrogiorgou A, Kyriazis D. “FHIR Ontology mapper (FOM): aggregating structural and semantic similarities of ontologies towards their alignment to HL7 FHIR.” In: 2018 IEEE 20th international conference on E-health networking, applications and services, Healthcom 2018. Institute of Electrical and Electronics Engineers Inc. (2018). doi: 10.1109/HealthCom.2018.8531149
30. Neumann A, Laranjeiro N, Bernardino J. “An Analysis of Public REST Web Service APIs.” IEEE Transactions on Services Computing, June 13, 2018. doi: 10.1109/TSC.2018.2847344 (2018).
31. LeFevre K, DeWitt DJ, Ramakrishnan R. “Mondrian multidimensional K-anonymity.” In: Proceedings - international conference on data engineering (2006). 25 p. doi: 10.1109/ICDE.2006.101
32. “OpenStreetMap — Geocoder 1.38.1.” (n.d). Accessed April 21, 2020. https://geocoder.readthedocs.io/providers/OpenStreetMap.html
33. “MongoDB.”. (n.d). Accessed April 21, 2020. https://www.mongodb.com/
34. Ayala-Rivera V, McDonagh P, Cerqueus T, Murphy L. A systematic comparison and evaluation of K-anonymization algorithms for practitioners. Trans Data Privacy. (2014) 7(3):337–70. doi: 10.5555/2870614.2870620
35. Iyengar VS. “Transforming data to satisfy privacy constraints.” In: Proceedings of the eighth ACM SIGKDD international conference on knowledge discovery and data mining - KDD ‘02, 279. New York, NY, USA: ACM Press. (2002). doi: 10.1145/775047.775089
36. Nergiz ME, Clifton C. “Thoughts on K-anonymization.” In: 22nd international conference on data engineering workshops (ICDEW’06). IEEE. (2006). 96–96. doi: 10.1109/ICDEW.2006.147
37. Bayardo RJ, Agrawal R. “Data privacy through optimal K-anonymization.” In: Proceedings - international conference on data engineering. IEEE. (2005). p. 217–28. doi: 10.1109/ICDE.2005.42
38. Souibgui M, Atigui F, Zammali S, Cherfi S, Yahia SB. “Data quality in ETL process: a preliminary study.” Procedia Comput Sci, 159:676–87. Elsevier B.V. (2019). doi: 10.1016/j.procs.2019.09.223
39. Theodorou V, Abelló A, Lehner W, Thiele M. Quality measures for ETL processes: from goals to implementation. Concurrency Comput Pract Exp. (2016) 28(15):3969–93. doi: 10.1002/cpe.3729
40. Gangwar PS, Hasija Y. “Deep learning for analysis of electronic health records (EHR).” In: Deep learning techniques for biomedical and health informatics. (2020). p. 149–66. doi: 10.1007/978-3-030-33966-1_8
41. Pitoglou S, Koumpouros Y, Anastasiou A. “Using electronic health records and machine learning to make medical-related predictions from non-medical data.” In: Institute of electrical and electronics engineers (IEEE). (2019). p. 56–60. doi: 10.1109/icmlde.2018.00021
42. Rajkomar A, Oren E, Chen K, Dai AM, Hajaj N, Hardt M, et al. Scalable and accurate deep learning with electronic health records. Npj Digit Med. (2018) 1(1):18. doi: 10.1038/s41746-018-0029-1
43. Ravi D, Wong C, Deligianni F, Berthelot M, Andreu-Perez J, Lo B, et al. Deep learning for health informatics. IEEE J Biomed Health Inform. (2017) 21(1):4–21. doi: 10.1109/JBHI.2016.2636665
Keywords: electronic health records, harmonization, anonymization, information loss, real data
Citation: Pitoglou S, Filntisi A, Anastasiou A, Matsopoulos GK and Koutsouris D (2022) Measuring the impact of anonymization on real-world consolidated health datasets engineered for secondary research use: Experiments in the context of MODELHealth project. Front. Digit. Health 4:841853. doi: 10.3389/fdgth.2022.841853
Received: 22 December 2021; Accepted: 10 August 2022;
Published: 1 September 2022.
Edited by:
Constantinos S. Pattichis, University of Cyprus, CyprusReviewed by:
Brijesh Mehta, Automaton AI Infosystems Pvt Ltd, IndiaManisha Mantri, Center for Development of Advanced Computing (C-DAC), India
© 2022 Pitoglou, Filntisi, Anastasiou, Matsopoulos and Koutsouris. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stavros Pitoglou c3BpdG9nbG91QGJpb21lZC5udHVhLmdy
Specialty Section: This article was submitted to Connected Health, a section of the journal Frontiers in Digital Health
 Stavros Pitoglou
Stavros Pitoglou Arianna Filntisi
Arianna Filntisi Athanasios Anastasiou
Athanasios Anastasiou George K. Matsopoulos
George K. Matsopoulos Dimitrios Koutsouris2
Dimitrios Koutsouris2