
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Digit. Health , 03 January 2022
Sec. Health Informatics
Volume 3 - 2021 | https://doi.org/10.3389/fdgth.2021.799067
This article is part of the Research Topic Human-Centred Computer Audition: Sound, Music, and Healthcare View all 11 articles
Since the COronaVIrus Disease 2019 (COVID-19) outbreak, developing a digital diagnostic tool to detect COVID-19 from respiratory sounds with computer audition has become an essential topic due to its advantages of being swift, low-cost, and eco-friendly. However, prior studies mainly focused on small-scale COVID-19 datasets. To build a robust model, the large-scale multi-sound FluSense dataset is utilised to help detect COVID-19 from cough sounds in this study. Due to the gap between FluSense and the COVID-19-related datasets consisting of cough only, the transfer learning framework (namely CovNet) is proposed and applied rather than simply augmenting the training data with FluSense. The CovNet contains (i) a parameter transferring strategy and (ii) an embedding incorporation strategy. Specifically, to validate the CovNet's effectiveness, it is used to transfer knowledge from FluSense to COUGHVID, a large-scale cough sound database of COVID-19 negative and COVID-19 positive individuals. The trained model on FluSense and COUGHVID is further applied under the CovNet to another two small-scale cough datasets for COVID-19 detection, the COVID-19 cough sub-challenge (CCS) database in the INTERSPEECH Computational Paralinguistics challengE (ComParE) challenge and the DiCOVA Track-1 database. By training four simple convolutional neural networks (CNNs) in the transfer learning framework, our approach achieves an absolute improvement of 3.57% over the baseline of DiCOVA Track-1 validation of the area under the receiver operating characteristic curve (ROC AUC) and an absolute improvement of 1.73% over the baseline of ComParE CCS test unweighted average recall (UAR).
Since the year 2019, the coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has become a global pandemic1. As of August 2021, there have been more than 202, 000, 000 confirmed cases of COVID-19 worldwide, including more than 4, 000, 000 deaths, reported by the World Health Organization (WHO)2. The daily 1 2 increasing COVID-19 cases and deaths have resulted in global lockdown, quarantine, and many restrictions (1). Along with the above measures, a set of following problems have appeared, including the economic downturn (2) and mental health problems (e.g., depression and stress) (1).
Swift and accurate diagnosis of COVID-19 is essential to give patients appropriate treatments and effectively control its transmission (3). The reverse transcription PCR (RT-PCR) from oral-nasopharyngeal swabs identifies viral RNA and is a commonly used instrument for the diagnosis of COVID-19. Nevertheless, high false negative rate and stability issues have been reported (4). In contrast to RT-PCR, chest CT was proven to have high sensitivity and be expedited for diagnosing COVID-19(4). Serological instruments are utilised to diagnose/confirm late COVID-19 cases by measuring antibody responses to the corresponding infection (5). Compared to the above laboratory instruments, which require professionals and special medical equipment, rapid antigen and molecular tests using nasopharyngeal swabs are commercially available due to their swift and simple test procedures, reduced mortality of COVID-19 patients, internal hospital costs, and in-hospital transmission (6). However, rapid tests are still hard-to-follow for non-specialists and are not environment-friendly.
Artificial intelligence has been widely applied to respiratory sounds in the healthcare area (7–9). In a study by (8), a multilayer perceptron based classifier was developed on features extracted from respiratory sounds to screen lung health. Random forests are applied on the filter bank energy-based features to pre-screen the lung health abnormalities (9). COVID-19 patients were reported to have seven common symptoms, including fever, cough, sore throat, headache, myalgia, nausea/vomiting, and diarrhea (10). Among these symptoms, the first two symptoms of COVID-19 are fever and cough (10). As a fast and non-invasive way to detect potential infections in public areas, body temperature measurement has been commonly employed (11). Traditional body temperature measurement with a thermometer usually requires relatively close contact with potential COVID-19 positive individuals (12). Although infrared (IR) thermal cameras provide a non-contact way for mass fever detection, they may not be valid because of the absence of calibration, non-homogeneous devices/protocols, and poor correlation between skin temperature and core body temperature (11). The reading of IR thermal cameras could also be affected by the environmental temperature (11). On the other hand, cough, as a common symptom in many respiratory diseases, is a worthwhile consideration when diagnosing a disease (13). Cough sounds have been used to diagnose asthma, bronchitis, pertussis, pneumonia, etc. (13). Recent studies have also investigated the feasibility of detecting COVID-19 infections from cough sounds. For instance, cough sounds were shown to contain latent features distinguishable between COVID-19 positive individuals and COVID-19 negative individuals (i.e., normal, bronchitis, and pertussis) (14). In Brown et al.'s study (15), cough sounds from COVID-19 positive individuals were reported to have a longer duration, more onsets, higher periods, lower RMS, and MFCC features with fewer outliers. Due to the development of the internet-of-things (IoT), the algorithms for detecting potential COVID-19 positive individuals from cough sounds can be integrated into mobile phones, wearable devices, and robots. Such a rapid, easy-to-use, and environment-friendly instrument will be helpful for real-time and remote pre-screening of COVID-19 infections, thereby supplementing clinical diagnosis and reducing the medical burden.
Since the outbreak of COVID-19, several studies have collected cough samples from COVID-19 positive patients (and COVID-19 negative individuals) to detect COVID-19 infections. Coswara (16) is a crowd-sourced database consisting of various kinds of sounds, including breathing (shallow and deep), coughing (shallow and deep), sustained vowel phonation (/ey/ as in made, /i/ as in beet, /u:/ as in cool), and number counting from one to twenty (normal and fast-paced). Another crowd-sourced database, COUGHVID with cough sounds only (17), was collected via a web interface. To date, the latest version of COUGHVID is publically released with 27, 550 cough recordings.3 The crowd-sourced University of Cambridge COVID database was reported to have more than 400 cough and breathing recordings (15). The Virufy datasets consist of a Latin American crowd-sourced dataset (31 individuals) and two South Asian clinical datasets (362 and 63 individuals, respectively). Due to the difficulty of collecting cough sounds of confirmed COVID-19 patients and multi-sound (non-cough)/noise in crowd-sourced datasets, most of the above databases are small-scale, leading to a challenge for training robust machine learning models.
With this in mind, we propose a hybrid transfer learning framework for robust COVID-19 detection, where several convolutional neural networks (CNNs) are trained on large-scale databases and fine-tuned on several small-scale cough sound databases for verification. Note that the focus of this paper is not to outperform the state-of-the-art neural networks models for COVID-19 detection from cough sounds; rather, the aim of this study is to provide a framework for mitigating the effect of noise or irrelevant sounds in the crowd-sourcing datasets applied to COVID-19 by training robust CNN models with the transferred knowledge from Flusense and/or COUGHVID. The workflow of this study is indicated in Figure 1. The code of this paper is publicly available on GitHub4.
• The FluSense database (18) was collected in a platform to track influenza-related indicators, such as cough, sneeze, sniffle, and speech. Since it contains various types of sounds existing in crowd-sourced cough datasets, the FluSense dataset is applied in this study.
• Due to the gap in sound type between FluSense and databases with cough sounds only, the COUGHVID database is considered as the target data when CNNs are trained on FluSense as the source data. The trained models on COUGHVID are further adapted to the other two smaller test databases, i.e., Computational Paralinguistics challengE (ComParE) 2021 COVID-19 cough sub-challenge (CCS) (19) and DiCOVA 2021 Track-1 (20).
• We propose two transfer learning pipelines, i.e., transferring parameters from the source database to the target database for fine-tuning models and incorporating embeddings for expanding models' capability of extracting useful features.
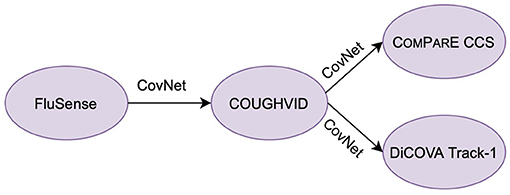
Figure 1. The workflow of this study. CovNet is the proposed transfer learning framework, which includes transferring parameters and incorporating embeddings. CovNet is first applied on the Flusense as the source data, COUGHVID as the target data. Afterwards, to further validate the effectiveness of CovNet, the CovNet based pre-trained COUGHVID models are applied on two smaller Computational Paralinguistics challengE (ComParE) 2021 COVID-19 cough sub-challenge (CCS) dataset and DiCOVA 2021 Track-1 dataset.
In the following sections, the transfer learning framework is first introduced in section 2, followed by the architecture of the models for COVID-19 detection in section 3. Next, the experimental details are described, and the results are presented and discussed in section 4. Finally, our study is summarised, and the outlook is given in section 5.
Transfer learning aims at applying the knowledge learnt from source data to different but related target data and achieving better performance in a cost-effective way (21–23). The source data and target data should be similar, otherwise negative transfer may happen (22, 24). Transfer learning has been successfully applied to COVID-19 detection based on acoustic data (14, 15). In Imran et al.'s study (14), the knowledge was transferred from the cough detection model to the COVID-19 diagnosis model. Brown et al. (15) discovered that VGGish pre-trained on a large-scale YouTube dataset was utilised to extract audio features from raw audio samples for COVID-19 diagnosis.
In this study, two ways of transfer learning are applied. One is to fine-tune the parameters of the networks with the target data. The other is extracting the embeddings from the pre-trained network and applying the embeddings when training the new network for the target dataset. Since the crowd-sourced cough recordings usually contain non-cough audio signals other than cough sounds, such as speech and breathing, the FluSense dataset and the COUGHVID dataset contain similar sound types. Therefore, the knowledge learnt from FluSense data can be employed to improve the performance of models trained on the COUGHDVID dataset. In Figure 2, DFluSense is the FluSense dataset, and DCOUGHVID means the COUGHVID dataset; convs0 and convs1 represent the convolutional layers/blocks in the neural networks on the FluSense dataset and the COUGHVID dataset, respectively; FCFluSense and FCCOUGHVID denotes the fully-connected (FC) layer of corresponding models. When separating the left part with the right part in Figures 2A,B, with the training data (x0, y0) and (x1, y1), we separately train the CNNs on the FluSense and COUGHVID datasets to produce the predicted values and , respectively.
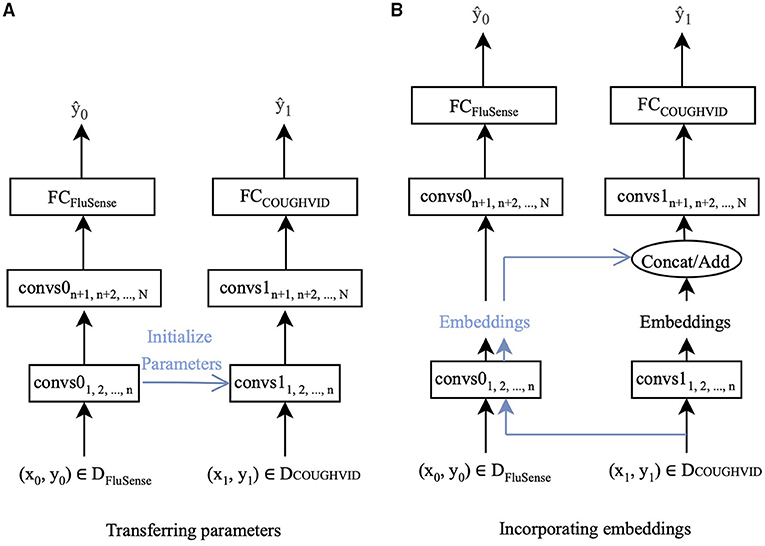
Figure 2. The proposed transfer learning framework :CovNet. (A) Parameters of the first n convolutional layers/blocks (convs1) of the current COUGHVID model are frozen and initialised by the corresponding first n convolutional layers/blocks (convs0) of the pre-trained FluSense model. (B) Embeddings are extracted after the n-th convs0 of the pre-trained FluSense model. The extracted embeddings are concatenated or added to the current embeddings generated after the n-th convs1 of the COUGHVID model.
With the parameters and embeddings from the pre-trained FluSense models, as highlighted in blue in Figure 2, the COUGHVID models are given the potential to discriminate between the various audio signals, which further helps its COVID-19 detection from crowd-sourced cough signals. Notably, the predicted value is the final output of the proposed transfer learning framework.
To further investigate the generalisation ability of CovNet, we apply it to some other small-scale crowd-sourced datasets for COVID-19 detection. In the following, we introduce the two transfer learning methods in greater detail.
Fine-tuning pre-trained models is an effective transfer learning method by sharing some parameters across tasks (21, 22). In the computer vision area, parameters of pre-trained models on ImageNet (25) are often applied for transfer learning on a wide range of image-related tasks (26–29). Similarly, parameters of pre-trained models on the Audio Set are transferred to many audio-related tasks (30–32). Parameters of pre-trained CNN models on the Audio Set are transferred to the adapting networks for acoustic event recognition (30, 31). Several pre-trained audio neural networks trained on the Audio Set dataset were proposed for other audio pattern recognition tasks (32).
In this study, as indicated in Figure 2A, the parameters of the first n convolutional layers/blocks, convs11, 2,..., n, of models trained on the COUGHVID dataset, are initialised by the corresponding layers/blocks convs01, 2,..., n of models pre-trained on FluSense dataset. The parameters of convs11, 2,..., n are frozen and not trained, and only the remaining randomly initialised parameters of convs1n+1, n+2,..., N and FCCOUGHVID are updated during the training procedure.
The embeddings generated by the convolutional layers carry either low-level edge information or high-level discrimination-related features (22, 23). Moreover, the performance of embeddings appears to be highly scalable with the amount of training data (33). In this study, the pre-trained FluSense models produce embeddings representing high-level or low-level characteristics of various audio types, which can be applied as an additional input to help develop the target model.
Specifically, we feed the crowd-sourced cough recordings from the COUGHVID into the pre-trained Flusnese model and extract the embeddings after certain convolutional layers/blocks. Figure 2B exhibits this strategy. Data-point (x1, y1) enters the pre-trained FluSense model, and the output embeddings of the n-th convolutional layer/block convs0n are extracted to be concatenated (on the channel dimension) or added with the embeddings generated by the corresponding convs1n. The concatenated or added embeddings enter the next convolutional layer/block convs1n+1 for the task of COVID-19 detection.
Convolutional neural networks have been successfully applied in image-related areas, such as image classification (34–37). When processing audio signals, CNNs have demonstrated their capabilities in extracting effective representations from the log Mel spectrograms (38, 39). In this study, we choose four typical CNN models: base CNN (34), VGG (40), residual network (ResNet) (41), and MobileNet (42). We focus on the proposed transfer learning framework, CovNet, instead of competing with the state-of-the-art models on COVID-19 detection. Therefore, in order to highlight the effectiveness of CovNet, we construct four simple CNN models (i.e., CNN-4, VGG-7, ResNet-6, and MobileNet-6), each of which only has three convolutional layers/blocks. A detailed description of each model is given and analysed in the following subsections.
The log Mel spectrograms are calculated by Mel filter banks and logarithmic operation worked on the spectrograms, which are produced by the Short-Time Fourier Transforms (STFTs) on the original waveforms. In this section, to better evaluate the effectiveness of the proposed transfer learning framework and compare the performance differences among different CNN architectures, four CNNs are employed to deal with the extracted log Mel spectrograms: CNN-4, VGG-7, ResNet-6, and MobileNet-6. Log Mel spectrograms (T,F) are extracted from the audio signals as the input to the CNNs, where T represents the sequence length, and F denotes the log Mel frequency. Before entering the final FC layer, the matrix has the dimension (CN, N), where CN is the output channel number of the last convolutional layer, and N is the class number. Specifically, for the FluSense database, N is set to be 9; for the other datasets used in this study, N equals 2. For comparison convenience, we regard the convolutional layers and blocks equally when ordering them in a specific model. In this notation, ResNet-6 and MobileNet-6 have “block2” following the first convolutional layer.
As shown Figure 3A, we propose a simple 4-layer CNN, CNN-4, constructed by three 5 × 5 convolutional layers. To speed up and stabilise the training procedure, each convolutional layer is followed by batch normalisation (43) and the Rectified Linear Unit (ReLU) activation function (44). Afterwards, we apply max pooling for downsampling. The first three local max pooling operations are conducted over a 2 × 2 kernel, and the last max pooling is a global one to summarise the features along the dimension of the sequence length and frequency. Before the final FC layer for the final predicted result, a dropout (45) layer is utilised to address the overfitting issue.
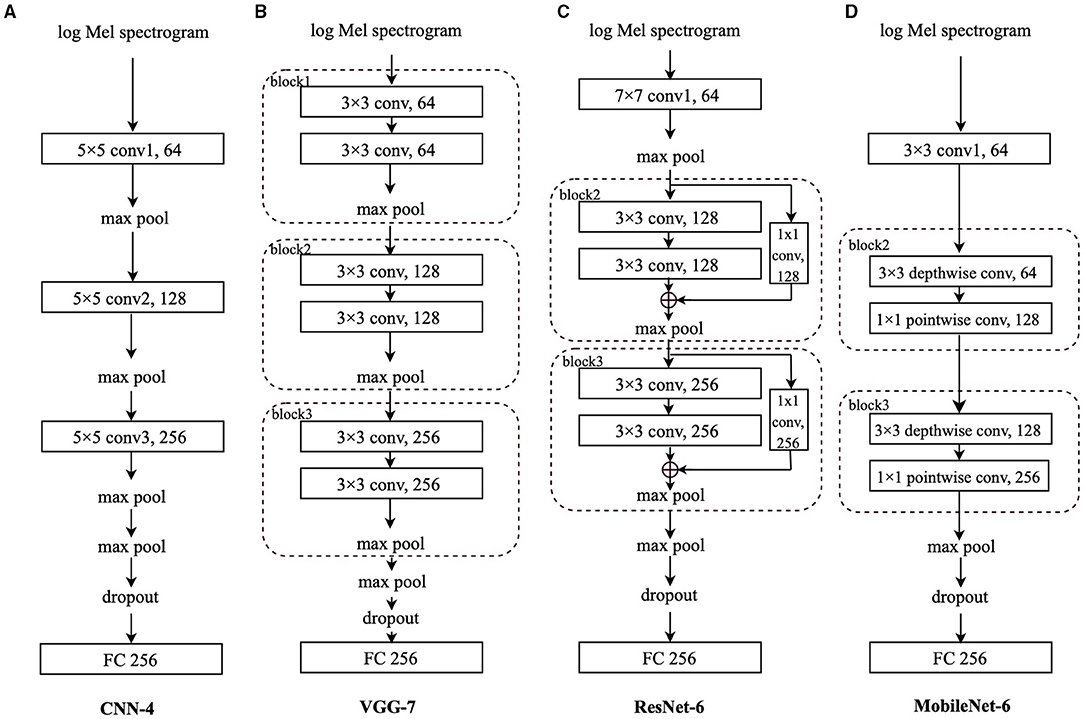
Figure 3. Models' architecture: (A) Convolutional neural network-4 (CNN-4), (B) VGG-7, (C) residual network-6 (ResNet-6), (D) MobileNet-6. “conv” stands for the convolutional layer, and “block” indicates the convolutional block. The number before the “conv” is the kernel size; the number after the “conv” is the output channel number. The number after “FC” is the input neurons' size.
Very deep CNN, known as VGG, were originally designed with up to 19 weight layers and achieved great performance on the large-scale image classification task (40, 46). VGG or VGG-like architectures were applied to extract audio features from respiratory sound data for COVID-19 detection and obtained good performances (15, 47).
As indicated in Figure 3B, we adapt the VGG (40) with 7 layers, VGG-7, which is composed of three convolutional blocks and a final FC layer. Although the VGG-7 is simple, different from its original “deep” design, it is still worthwhile to include it for fair comparison with other CNNs in this study. Each block contains two 3 × 3 convolutional layers, each of which is followed by batch normalisation (43) and the ReLU function (44) to stabilise and speed up the training process. Afterwards, a local max pooling layer with a kernel size of 2 × 2 is applied. Following the three blocks, there is also a global max pooling layer working on the sequence length and log Mel frequency dimensions. Before the FC layer, a dropout (45) layer is applied.
The Deep ResNet is proposed to address the degradation problem existing in training deeper networks (41) by incorporating shortcut connections between convolutional layers. In Hershey et al.'s (48) study,ResNet has outperformed other CNNs for audio classification on the Audio Set (49). A ResNet based model is constructed for COVID-19 detection from breath and cough audio signals (50).
In this study, we mainly adopt the above mentioned shortcut connections to construct a 6-layer ResNet, ResNet-6. In Figure 3C, after the first convolutional layer with a kernel size of 7 × 7 followed by batch normalisation (43) and the ReLU function (44), we apply two convolutional blocks, each of which contains the “shortcut connections” to add the identity mapping with the outputs of two stacked 3 × 3 convolutional layers.
Inside “block2” and “block3,” after the first 3 × 3 convolutional layer, the batch normalisation (43) and ReLU function (44) are applied, whereas only the batch normalisation is utilised after the second 3 × 3 convolutional layer. For the channel number consistency, the identity is processed by a 1 × 1 convolutional layer followed by batch normalisation (43); after the addition of the identity and the output of two stacked convolutional layers, we apply the ReLU function (44). The max pooling after the 7 × 7 convolutional layer is a local one with a kernel size of 3 × 3 and the max pooling layers in “block2” and “block3” are also local with a kernel size of 2 × 2; similarly, the last max pooling is a global one, followed by a dropout (45) layer and the FC layer.
Based on depthwise separable convolutions, light-weight MobileNets have been widely applied in mobile and embedded image related applications (42, 51). MobileNets are cost-effective and are explored herein for potential solutions embedded in mobile devices for COVID-19 detection.
We adapt the MobileNet with 6 layers only. As shown in Figure 3D, after the first 3 × 3 convolutional layer followed by batch normalisation (43) and the ReLU function (44), each of “block2” and “block3” contains a 3 × 3 depthwise convolutional layer and a 1 × 1 pointwise convolutional layer, respectively. Similarly, batch normalisation (43) and ReLU function (44) are applied after each convolutional layer. Similar to the original MobileNet architecture, we only set one global max pooling layer before the dropout (45) layer and the final FC layer.
With the aforementioned transfer learning framework, the experiments will be presented in this section, including the databases, experimental setup, results, and discussions.
To verify the proposed transfer learning framework in this study, the following four datasets are employed.
The FluSense (18) project applied a part of the original Audio Set dataset (49), which includes weakly labelled 10-s audio clips from YouTube. After the re-annotation by two human raters for more precise labels in the FluSense (18) project, there are a total of 45, 550 seconds samples in Audio Set that are considered in this study, and they are labelled with the classes of breathe, burp, cough, gasp, hiccup, other, silence, sneeze, sniffle, snore, speech, throat-clearing, vomit, and wheeze. To mitigate the effect of data imbalance on the classification performance, those classes with a number of samples less than 100 are not considered in our experiments. Therefore, the audio samples labelled with the following nine classes are employed: breathe, cough, gasp, other, silence, sneeze, sniffle, speech, and throat-clearing. For all audio recordings in the above nine classes, we first re-sampled them into 16 kHz. Second, as the audio samples have various time lengths, we split the original samples with a length of greater than or equal to 0.5 s into one or more 1 s segment(s). In particular, for audio samples with a length between 0.5 and 1 s, the audio repeats itself until a full 1 s segment is reached. For those samples with a length greater than 1 s, after a certain number of 1 s segments are split, the remaining signals repeat themselves until a full segment is reached if the remaining one has a length of greater than or equal to 0.5 s; otherwise, the remaining signals are simply abandoned. Furthermore, we split the segments into train/val subsets with a ratio of 0.8/0.2 in a stratified manner. The data distribution of FluSense before and after the pre-processing is shown in Table 1.
The on-going crowd-sourced COUGHVID dataset (17) is collected via a web interface5. All participants voluntarily record and upload their cough sounds lasting for up to 10 s. In the meantime, the COVID-19 status of each cough sample is self-reported by each participant: healthy, symptomatic without COVID-19 diagnosis, and COVID-19. The information of each participant is optionally self-reported, including the geographic location (latitude, longitude), age, gender, and whether she/he has other pre-existing respiratory conditions, and muscle pain/fever symptoms. As there might be some low-quality audio samples (e.g., noise, speech, etc.), the data collectors trained an extreme gradient boosting (XBG) classifier on 215 audio samples (121 cough and 94 non-cough) to predict the probability of a recording containing cough sounds. For all audio recordings, the sampling frequency is 48 kHz.
In this study, only the classes of healthy (i.e., COVID-19 negative) and COVID-19 (i.e., COVID-19 positive) are considered, as the audio samples with symptomatic status were not explicitly reported by the participants as to whether they were diagnosed with COVID-19 or not. Furthermore, only audio samples with cough sound probabilities greater than 0.9 are included to ensure each audio sample contains cough sounds. Finally, 7, 774 audio samples (COVID-19 negative: 7, 075, COVID-19 positive: 699) are selected for our experiments. Similarly, we split the selected samples into train/test subsets with a ratio of 0.8/0.2, respectively in a stratified manner. Table 2 shows the data distribution of COUGHVID.
In the INTERPSEECH 2021 ComParE (19), the CCS provides a dataset from the crowd-sourced Cambridge COVID-19 Sound database (15). The participants are asked to provide one to three forced coughs in each recording via one of the following multiple platforms: A web interface, an Android app, and an iOS app.6 The CCS dataset consists of 929 cough recordings (1.63 h) from 397 participants. The data distribution of CCS is shown in Table 3. All recordings from the CCS dataset were resampled and converted into 16 kHz. The official training, validation, and test sets in the ComParE challenge are used in this study.
Table 3. Data distribution of the Computational Paralinguistics challengE (ComParE) COVID-19 cough sub-challenge (CCS) data.
The Track-1 of the DiCOVA challenge 2021 (20) provides cough recordings from 1, 040 participants (COVID-19 negative: 965, COVID-19 positive 75). In the challenge, the dataset was split into five train-validation folds. Each training set consists of 822 cough samples (COVID-19 negative: 772, COVID-19 positive: 50), and each validation set contains 218 cough samples (COVID-19 negative: 193, COVID-19 positive: 25). The additional test set is not used in this study, as it is blind. All cough recordings are sampled at 44.1 kHz. The data distribution of DiCOVA 2021 Track-1 is indicated in Table 4.
For faster progress (38), all audio files in the four datasets are re-sampled into 16 kHz. The log Mel spectrograms are extracted with a sliding window size of 512, an overlap of 256 units, and 64 Mel bins.
As for the evaluation metrics, we mainly use unweighted average recall (UAR), since it is more adequate for evaluating the classification performance on imbalanced datasets than accuracy,—the weighted average recall (52, 53). Apart from the UAR, we also calculate the area under the receiver operating characteristic curve (ROC AUC) score.
The proposed CNNs consist of three convolutional layers/blocks. The number of output channels for the three convolutional layers/blocks is 64, 128, and 256, respectively. During the training procedure of the neural networks, the cross-entropy loss is utilised as the loss function. To overcome the class imbalance issue, we re-scale the weight parameter for each class in the loss function. Since this study focuses on the transfer learning framework, we do not further mitigate the class imbalance issue through down-/up-sampling.
For single learning (i.e., training from scratch) on the FluSense and the COUGHVID datasets, the optimiser is set to “Adam” with an initial learning rate of 0.001, which is scheduled to be reduced by a factor of 0.4 when there is less than 0.01 improvement of the UAR after every 4 of 30 epochs in total. When transferring parameters, we set the initial learning rate as 0.0001; for incorporating embeddings, the initial learning rate is set to be 0.001.
When applying the strategy of transferring parameters introduced in section 2.1 to training the COUGHVID model, we experiment with only setting the following layer(s) trainable: the FC layer, the convolutional layer/block (conv/block) 3 & FC layer, conv/block 2−3 & FC layer, and conv/block 1−3 & FC layer, respectively. The remaining layer(s)/block(s) are initialised based on the pre-trained FluSense models' corresponding parameters and are frozen during the whole training procedure. As for the incorporating embeddings strategy described in section 2.2, we investigate the concatenation and addition of two embeddings generated from the conv/block 3, conv/block 2, and conv/block 1, respectively. One embedding is from the pre-trained FluSense model, and the other one is the COUGHVID model trained from scratch.
To further validate the effectiveness of the CovNet, we apply the pre-trained COUGHVID models on the ComParE CCS dataset and the DiCOVA Track-1 dataset. Specifically, we train the four CNNs introduced in section 3 from scratch. Afterwards, we choose up to two COUGHVID models with the best performance (best AUC or best UAR) as the pre-trained models. With the chosen pre-trained COUGHVID models and their strategies (layer(s)/block(s) number and transfer learning strategies), we transfer the parameters or embeddings of the above chosen COUGHVID models to the current train-from-scratch models on the ComParE and DiCOVA datasets during the training. Finally, we choose the best results to compete with official baselines: the average validation AUC 68.81% (20) for the DiCOVA Track-1 dataset, and test UAR without fusion 64.7% (19) for ComParE CCS. Similarly, when training models from scratch or applying the incorporating embeddings method, we set the initial learning rate as 0.001, whereas if the transferring parameters are utilised, the initial learning rate is set as 0.0001.
In Table 5, we focus on performance differences on the COUGHVID test dataset between single learning (training from scratch) models and the models produced by the proposed transfer learning strategies in section 2. For convenience, the best test AUC and test UAR of every model under three transfer learning strategies are shown in bold face. We can see that there are some improvements in test AUC/UAR, especially for the VGG-7 and MobileNet-6. In the following analysis, we compare the absolute difference between performances. On the COUGHVID test dataset, with the transfer learning, the VGG-7 obtains an improvement of 2.62% AUC (p < 0.1 in a one-tailed z-test) and an improvement of 3.75% UAR (p < 0.05 in a one-tailed z-test); the MobileNet-6 achieves 3.77% improvement in AUC (p < 0.05 in a one-tailed z-test) and 4.88% improvement in UAR (p < 0.005 in a one-tailed z-test). Moreover, for all constructed CNN models, only setting the FC layer trainable and freezing other layers with parameters transferred from pre-trained FluSense models achieves almost the lowest AUC/UAR among all transfer learning settings.
For the transferring parameters strategy, we can see that most best test AUC/UAR cases are obtained by only setting the convolutional layer/block (conv/block) 2−3 & FC layer trainable or the conv/block 1−3 & FC layer trainable. With the embeddings cat method, models' performances are mostly better than single learning models' and the most best results are achieved by concatenating the embeddings output by the conv/block 3. With the embeddings addition method, models also mostly outperform the single learning ones, and similarly, most best results are obtained by adding embeddings after the conv/block 3.
In Table 6, first, we can see that with the proposed transfer learning strategies on the pre-trained COUGHVID models generated by the CovNet, most of the models' performances improve a lot compared with the single learning models' performance. Specifically, transferring parameters improves the test UAR on ComParE by 9.05% for the VGG-7 (p < 0.05 in a one-tailed z-test); the transferring parameters improves the validation AUC on DiCOVA by 1.12, 3.86, and 5.22 % for the CNN-4, ResNet-6, and VGG-7, respectively (in a one-tailed z-test, not significant, p < 0.05, and p < 0.005, respectively). The incorporating embeddings improves the test UAR on ComParE data by 1.47, and 1.11% for the CNN-4, and VGG-7, respectively; the incorporating embeddings improves the validation AUC of DiCOVA by 3.62%, 8.85, 7.46, and 2.20% for the CNN-4, ResNet-6, VGG-7, and MobileNet-6, respectively (in a one-tailed z-test, p < 0.05, p < 0.001, p < 0.001 and not significant, respectively).
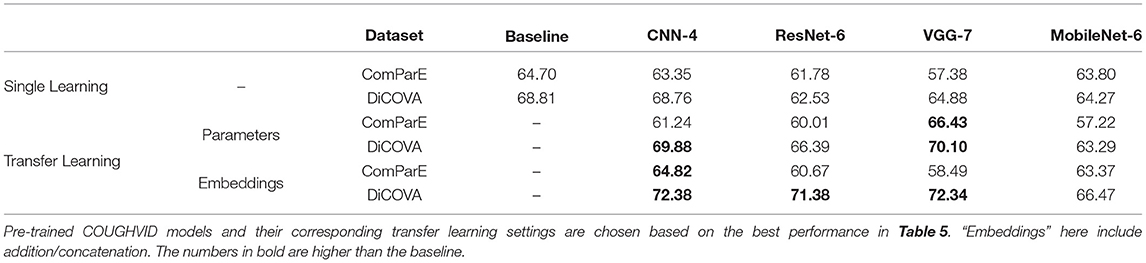
Table 6. Models' performances [ %], validation AUC on the DiCOVA Track-1 dataset, and test UAR on the ComParE dataset, with single learning (train from scratch), and the proposed transfer learning strategies.
Second, as the numbers in bold indicate better performance than the baseline, we can see that most models learnt through the transfer learning framework outperform the official baselines, even though the models here are quite simple. Notably, the best test UAR 66.43% on ComParE CCS data is achieved by the VGG-7 with transferring parameters, which is 1.73% above the official baseline; the CNN-4 with incorporating embeddings the achieves the best validation AUC 72.38% on the DiCOVA Track-1, which is 3.57% higher than the baseline (p < 0.05 in a one-tailed z-test). Figure 4 displays the confusion matrices for above-mentioned best UAR on the ComParE CCS dataset and best validation AUC on the DiCOVA Track-1 dataset. We can see that the models recognise negative samples very well, but the positive ones are frequently confused with the negative ones.
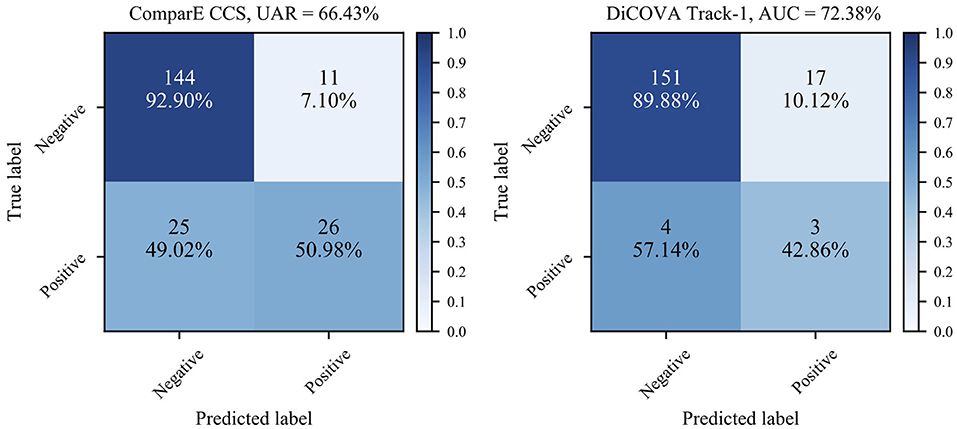
Figure 4. Confusion matrices for the best performance on the ComParE CCS test set and the DiCOVA validation set. For the DiCOVA dataset, since its test dataset is not accessible, the numbers are averaged over the five cross-validation folds.
In Table 5, if comparing the performance of single learning CNNs and transfer learning CNNs, we find that there is no improvement or even slightly worse performance of transfer learning methods on the ResNet-6 model. ResNet gains accuracy from increased neural network depth (41), which may explain the performance of the simple ResNet-6 in this study. Apart from fine-tuning the parameters of FC layers only, almost all other CNN models obtain better performance after the transfer learning, proving the usefulness of the knowledge transferred from the FluSense dataset for recognising COVID-19 on the COUGHVID dataset. Setting FC layers trainable only limits the generalisation of the pre-trained FluSense models.
For fine-tuning parameters of different layers, fine-tuning the weights of the convolutional layers/blocks 2−3 & FC layer obtains better performance. Since the target dataset COUGHVID is not large-scale enough compared with the FluSense one, fine-tuning the entire network (convolutional layers/blocks 1−3 & FC layer) might encounter an overfitting issue (23). Specifically, earlier layers/blocks generate low-level, generic features, which do not change significantly during the training procedure (23). Conversely, the convolutional layer/block 3 herein generates more high-level, domain-dependent representations. As for the embeddings incorporation, concatenation and addition of the embeddings achieve similar results, which indicates that both operations equally transfer the knowledge learnt from the FluSense dataset. Furthermore, we find that incorporating the embeddings after the convolutional layer/block 3 mostly outperforms the operations on other layers/blocks. This can be caused by more discrimination power obtained by applying the pre-trained FluSense models.
From Table 6, we further validate the generalisation ability of the proposed CovNet with the DiCOVA Track-1 and ComParE CCS datasets. By competing with the official baselines, even simple CNNs can also achieve better performance with the proposed transfer learning methods. Therefore, the considered CovNet appears robust and can provide useful knowledge when detecting COVID-19 from crowd-sourced cough recordings. However, the performance improvement over the ComParE CCS baseline by incorporating the embeddings method is not obvious, which might be caused by the inherent data difference between the FluSense and COUGHVID datasets and the ComParE CCS dataset. Moreover, the CovNet works very well on the DiCOVA track-1 dataset, especially the incorporating embeddings. Perhaps, the embeddings from the pre-trained COUGHVID models carry more beneficial knowledge compared with parameters of convolutional layers on the DiCOVA dataset.
The main purpose of this study is to introduce and prove the usefulness of the transfer learning framework CovNet, instead of competing with the state-of-the-art performance on the DiCOVA Track-1 dataset (54–56) and ComParE CCS dataset (19). The constructed four CNN models are so simple that each of them only contains three convolutional layers/blocks; we do not apply any data augmentation techniques and the only input to the networks are the original log Mel spectrograms.
In this study, we proposed a transfer learning framework, CovNet, containing transferring parameters and incorporating embeddings. Transferring parameters indicate fine-tuning the models by initialising and freezing some parameters with the pre-trained model; incorporating embeddings describe concatenating or adding the embeddings generated by a pre-trained model with the embeddings produced by the current model.
The effectiveness and generalisation ability of the proposed transfer learning framework was demonstrated when developing simple CNNs for COVID-19 detection from crowd-sourced cough sounds. In the future, one should consider deeper neural networks to further improve performance through transfer learning. Moreover, other knowledge transfer architectures, such as multi-task learning (57) and domain adaption (58) can be explored.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
YC contributed to the study design, experimenting, manuscript drafting, and editing. XJ contributed to the experimenting and manuscript editing. ZR contributed to the study design, manuscript drafting, and editing. BS supervised the whole process, from study design, overall implementation, to manuscript drafting, and editing. All authors approved the submitted version.
This study was partially supported by the Horizon H2020 Marie Skłodowska-Curie Actions Initial Training Network European Training Network (MSCA-ITN-ETN) project under grant agreement No. 766287 (TAPAS), the DFG's Reinhart Koselleck project No. 442218748 (AUDI0NOMOUS), and the Federal Ministry of Education and Research (BMBF), Germany under the project LeibnizKILabor (Grant No. 01DD20003).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors would like to express their gratitude to the holders of the FluSense, COUGHVID, ComParE 2021 CCS, and DiCOVA challenge 2021 Track-1 datasets for providing collected data for research purposes. Moreover, thanks to all the participants for their data points.
1. ^https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it; retrieved 10 August 2021.
2. ^https://covid19.who.int/; retrieved 10 August 2021.
3. ^https://zenodo.org/record/4498364#.YRKa3IgzbD4
4. ^https://github.com/ychang74/CovNet
5. ^https://COUGHVID.epfl.ch/; retrieved 09 July 2021.
6. ^https://www.covid-19-sounds.org/; retrieved 15 July 2021
1. Atalan A. Is the lockdown important to prevent the COVID-19 pandemic? Effects on psychology, environment and economy-perspective. Ann Med Surg. (2020) 56:38–42. doi: 10.1016/j.amsu.2020.06.010
2. Inoue K, Hashioka S, Kawano N. Risk of an increase in suicide rates associated with economic downturn due to COVID-19 pandemic. Asia Pac J Public Health. (2020) 32:367. doi: 10.1177/1010539520940893
3. Schuller BW, Schuller DM, Qian K, Liu J, Zheng H, Li X. COVID-19 and computer audition: an overview on what speech & sound analysis could contribute in the SARS-CoV-2 corona crisis. Front Digit Health. (2021) 3:14. doi: 10.3389/fdgth.2021.564906
4. Li Y, Yao L, Li J, Chen L, Song Y, Cai Z, et al. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J Med Virol. (2020) 92:903–8. doi: 10.1002/jmv.25786
5. Tang YW, Schmitz JE, Persing DH, Stratton CW. Laboratory diagnosis of COVID-19: Current issues and challenges. J Clin Microbiol. (2020) 58:e00512-20. doi: 10.1128/JCM.00512-20
6. Dinnes J, Deeks JJ, Berhane S, Taylor M, Adriano A, Davenport C, et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev. (2021) 3:1–15. doi: 10.1002/14651858.CD013705.pub2
7. Santosh KC. Chapter 1: Speech processing in healthcare: can we integrate? In: Dey N, editor. Intelligent Speech Signal Processing. New York, NY: Academic Press (2019). p. 1–4. doi: 10.1016/B978-0-12-818130-0.00001-5
8. Mukherjee H, Sreerama P, Dhar A, Obaidullah SM, Roy K, Santosh KC, et al. Automatic lung health screening using respiratory sounds. J Med Syst. (2021) 45:19. doi: 10.1007/s10916-020-01681-9
9. Mukherjee H, Salam H, Santosh K. Lung health analysis: adventitious respiratory sound classification using filterbank energies. Int J Pattern Recogn Artif Intell. (2021) 2021:2157008. doi: 10.1142/S0218001421570081
10. Larsen JR, Martin MR, Martin JD, Kuhn P, Hicks JB. Modeling the onset of symptoms of COVID-19. Front Public Health. (2020) 8:473. doi: 10.3389/fpubh.2020.00473
11. Buoite Stella A, Manganotti P, Furlanis G, Accardo A, Ajčević M. Return to school in the COVID-19 era: considerations for temperature measurement. J Med Eng Technol. (2020) 44:468–71. doi: 10.1080/03091902.2020.1822941
12. Wei W, Wang J, Ma J, Cheng N, Xiao J. A real-time robot-based auxiliary system for risk evaluation of COVID-19 infection. In: Proc. Interspeech. Shanghai (2020). p. 701–5. doi: 10.21437/Interspeech.2020-2105
13. Alqudaihi KS, Aslam N, Khan IU, Almuhaideb AM, Alsunaidi SJ, Ibrahim NM, et al. Cough sound detection and diagnosis using artificial intelligence techniques: challenges and opportunities. IEEE Access. (2021). doi: 10.1109/ACCESS.2021.3097559
14. Imran A, Posokhova I, Qureshi HN, Masood U, Riaz MS, Ali K, et al. AI4COVID-19: AI enabled preliminary diagnosis for COVID-19 from cough samples via an app. Inform Med Unlocked. (2020) 20:100378. doi: 10.1016/j.imu.2020.100378
15. Brown C, Chauhan J, Grammenos A, Han J, Hasthanasombat A, Spathis D, et al. Exploring automatic diagnosis of COVID-19 from crowdsourced respiratory sound data. In: Proc. ACM SIGKDD. New York, NY: ACM (2020). p. 3474–84. doi: 10.1145/3394486.3412865
16. Sharma N, Krishnan P, Kumar R, Ramoji S, Chetupalli SR, R N, et al. Coswara–a database of breathing, cough, and voice sounds for COVID-19 diagnosis. In: Proc. Interspeech. Shanghai: Interspeech (2020). p. 4811–5. doi: 10.21437/Interspeech.2020-2768
17. Orlandic L, Teijeiro T, Atienza D. The COUGHVID crowdsourcing dataset, a corpus for the study of large-scale cough analysis algorithms. Sci Data. (2021) 8:1–10. doi: 10.1038/s41597-021-00937-4
18. Al Hossain F, Lover AA, Corey GA, Reich NG, Rahman T. FluSense: a contactless syndromic surveillance platform for influenza-like illness in hospital waiting areas. In: Proceedings of the ACM on Interactive, Mobile, Wearable and Ubiquitous Technologies. New York, NY: ACM (2020). p. 1–28. doi: 10.1145/3381014
19. Schuller B, Batliner A, Bergler C, Mascolo C, Han J, Lefter I, et al. The INTERSPEECH 2021 computational paralinguistics challenge: COVID-19 cough, COVID-19 speech, escalation & primates. In: Proc. Interspeech. Brno (2021). p. 431–5. doi: 10.21437/Interspeech.2021-19
20. Muguli A, Pinto L, R N, Sharma N, Krishnan P, Ghosh PK, et al. DiCOVA challenge: dataset, task, and baseline system for COVID-19 diagnosis using acoustics. In: Proc. Interspeech. Brno (2021). p. 901–5. doi: 10.21437/Interspeech.2021-74
21. Torrey L, Shavlik J. Transfer learning. In: Ganchev T, Sokolova M, Rada R, Garcia-Laencina PJ, Ravi V, editors. Handbook of Research on Machine Learning Applications and Trends: Algorithms, Methods, and Techniques. Hershey, PA: IGI Publishing (2010) p. 242–64. doi: 10.4018/978-1-60566-766-9.ch011
22. Pan SJ, Yang Q. A survey on transfer learning. IEEE Trans Knowledge Data Eng. (2010) 22:1345–59. doi: 10.1109/TKDE.2009.191
23. Mehdipour Ghazi M, Yanikoglu B, Aptoula E. Plant identification using deep neural networks via optimization of transfer learning parameters. Neurocomputing. (2017) 235:228–35. doi: 10.1016/j.neucom.2017.01.018
24. Cao B, Pan SJ, Zhang Y, Yeung DY, Yang Q. Adaptive transfer learning. In: Proceedings of the AAAI Conference on Artificial Intelligence. Atlanta, AL: AAAI (2010). p. 407–12.
25. Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Commun ACM. (2017) 60:84–90. doi: 10.1145/3065386
26. Kornblith S, Shlens J, Le QV. Do better imagenet models transfer better? In: Proc. CVPR. Long Beach, CA (2019). doi: 10.1109/CVPR.2019.00277
27. Morid MA, Borjali A, Del Fiol G. A scoping review of transfer learning research on medical image analysis using ImageNet. Comput Biol Med. (2021) 128:104115. doi: 10.1016/j.compbiomed.2020.104115
28. Raghu M, Zhang C, Kleinberg J, Bengio S. Transfusion: Understanding Transfer Learning for Medical Imaging. Red Hook, NY: Curran Associates Inc. (2019).
29. Shin HC, Roth HR, Gao M, Lu L, Xu Z, Nogues I, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. (2016) 35:1285–98. doi: 10.1109/TMI.2016.2528162
30. Pons J, Serra J, Serra X. Training neural audio classifiers with few data. In: Proc. ICASSP. Brighton (2019). p. 16–20. doi: 10.1109/ICASSP.2019.8682591
31. Kumar A, Khadkevich M, Fagen C. Knowledge transfer from weakly labeled audio using convolutional neural network for sound events and scenes. In: Proc. ICASSP. Calgary, AB (2018). p. 326–30. doi: 10.1109/ICASSP.2018.8462200
32. Kong Q, Cao Y, Iqbal T, Wang Y, Wang W, Plumbley MD. PANNs: Large-scale pretrained audio neural networks for audio pattern recognition. IEEE/ACM Trans Audio Speech Lang Process. (2020) 28:2880–94. doi: 10.1109/TASLP.2020.3030497
33. Snyder D, Garcia-Romero D, Sell G, Povey D, Khudanpur S. X-vectors: robust DNN embeddings for speaker recognition. In: Proc. ICASSP. Calgary, AB (2018). p. 5329–33. doi: 10.1109/ICASSP.2018.8461375
34. Zeiler MD, Fergus R. Visualizing and understanding convolutional networks. In: Fleet D, Pajdla T, Schiele B, Tuytelaars T, editors. Computer Vision-ECCV 2014. Cham: Springer International Publishing (2014). p. 818–33. doi: 10.1007/978-3-319-10590-1_53
35. Wang J, Yang Y, Mao J, Huang Z, Huang C, Xu W. CNN-RNN: a unified framework for multi-label image classification. In: Proc. CVPR. Las Vegas, NV (2016). doi: 10.1109/CVPR.2016.251
36. Wei Y, Xia W, Lin M, Huang J, Ni B, Dong J, et al. HCP: a flexible CNN framework for multi-label image classification. IEEE Trans Pattern Anal Mach Intell. (2016) 38:1901–7. doi: 10.1109/TPAMI.2015.2491929
37. Li Q, Cai W, Wang X, Zhou Y, Feng DD, Chen M. Medical image classification with convolutional neural network. In: Proc. ICARCV. Marina Bay Sands (2014). p. 844–8. doi: 10.1109/ICARCV.2014.7064414
38. Ren Z, Baird A, Han J, Zhang Z, Schuller B. Generating and protecting against adversarial attacks for deep speech-based emotion recognition models. In: Proc. ICASSP. Barcelona (2020). p. 7184–88. doi: 10.1109/ICASSP40776.2020.9054087
39. Kong Q, Yu C, Xu Y, Iqbal T, Wang W, Plumbley MD. Weakly labelled AudioSet tagging with attention neural networks. IEEE/ACM Trans Audio Speech Lang Process. (2019) 27:1791–802. doi: 10.1109/TASLP.2019.2930913
40. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. In: Proc. ICLR. San Diego, CA (2015).
41. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proc. CVPR. Las Vegas, NV (2016). p. 770–8. doi: 10.1109/CVPR.2016.90
42. Howard AG, Zhu M, Chen B, Kalenichenko D, Wang W, Weyand T, et al. MobileNets: efficient convolutional neural networks for mobile vision applications (2017). arXiv [Preprint]. arXiv: 1704.04861. Available Online at: https://dblp.uni-trier.de/rec/journals/corr/HowardZCKWWAA17.html?view=bibtex
43. Ioffe S, Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift. In: Proc. ICML. Lille: ICML (2015). p. 448–56.
44. Nair V, Hinton GE. Rectified linear units improve restricted Boltzmann machines. In: Proc. ICML. Madison, WI (2010). p. 807–14.
45. Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res. (2014) 15:1929–58. Available online at: https://jmlr.org/papers/v15/srivastava14a.html
46. Sitaula C, Belayet Hossain M. Attention-based VGG-16 model for COVID-19 chest X-ray image classification. Appl Intell. (2021) 51:2850–63. doi: 10.1007/s10489-020-02055-x
47. Lella KK, Pja A. Automatic diagnosis of COVID-19 disease using deep convolutional neural network with multi-feature channel from respiratory sound data: cough, voice, and breath. Alexandria Eng J. (2021) 61:1319–34. doi: 10.1016/j.aej.2021.06.024
48. Hershey S, Chaudhuri S, Ellis DPW, Gemmeke JF, Jansen A, Moore RC, et al. CNN architectures for large-scale audio classification. In: Proc. ICASSP. New Orleans, LA (2017). p. 131–5. doi: 10.1109/ICASSP.2017.7952132
49. Gemmeke JF, Ellis DPW, Freedman D, Jansen A, Lawrence W, Moore RC, et al. Audio Set: An ontology and human-labeled dataset for audio events. In: Proc. ICASSP. New Orleans, LA (2017). p. 776–80. doi: 10.1109/ICASSP.2017.7952261
50. Coppock H, Gaskell A, Tzirakis P, Baird A, Jones L, Schuller B. End-to-end convolutional neural network enables COVID-19 detection from breath and cough audio: a pilot study. BMJ Innovations. (2021) 7:356–62. doi: 10.1136/bmjinnov-2021-000668
51. Nayak SR, Nayak DR, Sinha U, Arora V, Pachori RB. Application of deep learning techniques for detection of COVID-19 cases using chest X-ray images: a comprehensive study. Biomed Signal Process Control. (2021) 64:102365. doi: 10.1016/j.bspc.2020.102365
52. Schuller B, Batliner A. Computational Paralinguistics: Emotion, Affect and Personality in Speech and Language Processing. 1st ed. New York, NY: Wiley Publishing (2013). doi: 10.1002/9781118706664
53. Rosenberg A. Classifying skewed data: importance weighting to optimize average recall. In: Proc. Interspeech. Portland, OR (2012). p. 2242–5. doi: 10.21437/Interspeech.2012-131
54. Sodergren I, Nodeh MP, Chhipa PC, Nikolaidou K, Kovacs G. Detecting COVID-19 from audio recording of coughs using random forests and support vector machines. In: Proc. Interspeech. Brno: Interspeech (2021) p. 916–20. doi: 10.21437/Interspeech.2021-2191
55. Das RK, Madhavi M, Li H. Diagnosis of COVID-19 using auditory acoustic cues. In: Proc. Interspeech. Brno: Interspeech (2021) p. 921–5. doi: 10.21437/Interspeech.2021-497
56. Harvill J, Wani YR, Hasegawa-Johnson M, Ahuja N, Beiser D, Chestek D. Classification of COVID-19 from cough using autoregressive predictive coding pretraining and spectral data augmentation. In: Proc. Interspeech. (2021) p. 926–30. doi: 10.21437/Interspeech.2021-799
Keywords: transfer learning, COVID-19, cough, FluSense, COUGHVID
Citation: Chang Y, Jing X, Ren Z and Schuller BW (2022) CovNet: A Transfer Learning Framework for Automatic COVID-19 Detection From Crowd-Sourced Cough Sounds. Front. Digit. Health 3:799067. doi: 10.3389/fdgth.2021.799067
Received: 21 October 2021; Accepted: 29 November 2021;
Published: 03 January 2022.
Edited by:
Harry Hochheiser, University of Pittsburgh, United StatesReviewed by:
Samrat Kumar Dey, Bangladesh Open University, BangladeshCopyright © 2022 Chang, Jing, Ren and Schuller. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi Chang, eS5jaGFuZzIwQGltcGVyaWFsLmFjLnVr; Zhao Ren, enJlbkBsM3MuZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.