- 1Department of Primary Care and Public Health, Brighton and Sussex Medical School, Brighton, United Kingdom
- 2Swansea Medical School, University of Swansea, Swansea, United Kingdom
- 3King's College London, London, United Kingdom
- 4South London and Maudsley NHS Foundation Trust, London, United Kingdom
Background: The analysis of clinical free text from patient records for research has potential to contribute to the medical evidence base but access to clinical free text is frequently denied by data custodians who perceive that the privacy risks of data-sharing are too high. Engagement activities with patients and regulators, where views on the sharing of clinical free text data for research have been discussed, have identified that stakeholders would like to understand the potential clinical benefits that could be achieved if access to free text for clinical research were improved. We aimed to systematically review all UK research studies which used clinical free text and report direct or potential benefits to patients, synthesizing possible benefits into an easy to communicate taxonomy for public engagement and policy discussions.
Methods: We conducted a systematic search for articles which reported primary research using clinical free text, drawn from UK health record databases, which reported a benefit or potential benefit for patients, actionable in a clinical environment or health service, and not solely methods development or data quality improvement. We screened eligible papers and thematically analyzed information about clinical benefits reported in the paper to create a taxonomy of benefits.
Results: We identified 43 papers and derived five themes of benefits: health-care quality or services improvement, observational risk factor-outcome research, drug prescribing safety, case-finding for clinical trials, and development of clinical decision support. Five papers compared study quality with and without free text and found an improvement of accuracy when free text was included in analytical models.
Conclusions: Findings will help stakeholders weigh the potential benefits of free text research against perceived risks to patient privacy. The taxonomy can be used to aid public and policy discussions, and identified studies could form a public-facing repository which will help the health-care text analysis research community better communicate the impact of their work.
Introduction
Electronic Health Records (EHRs) are revolutionizing health care at the point of delivery, but also offer huge potential for discovery and research worldwide. The United Kingdom (UK) with its single-payer free-to-access universal health care provision, is particularly well-placed to advance health data science, and the government has invested widely in infrastructure for availability of data, dataset linkage, and analytical capability (1). In the UK, data contained in EHRs is widely used for health research, drug safety analyses, and service planning (2). EHRs have been demonstrated to be a particularly important data source in the UK for conducting epidemiological research within general practice (2–4), mental health (5, 6), hospital episodes (7), and specialist conditions such as cancer (8).
In many cases, data are entered into EHRs in free text natural language in the form of clinic notes, letters and reports. This is particularly true of UK-based mental health records, GP clinic notes and letters, and hospital communications such as pathology or scan reports and discharge letters. These free text natural language data are considered unstructured, in comparison to clinical data stored in preset fields in records or entered in the form of clinical codes, in which a numeric or alphanumeric string represents a unique clinical concept such as a diagnosis or process of care (9). Natural Language Processing (NLP), alongside other methods, can be used to extract information from free text contained within EHRs (10), and therefore, unstructured EHR data can contribute to health research.
However, clinical data and health records are considered highly personal, and thus fall under data protection laws of most jurisdictions, such as the General Data Protection Regulation (2018) in Europe, the UK Data Protection Act (2018) and HIPAA in the USA (11). Data need to be de-identified or anonymized before they can be shared outside the clinical environment for any secondary purposes such as research or service planning or improvement. De-identification of structured records happens when fields containing identifiers are stripped out before data-sharing; it is a fairly straightforward process to comply with current data governance regulations. However, de-identifying free text clinic notes, letters and reports is more complex, and is a rapidly evolving field. These data contain names of patients, health-care professionals and family members, they may contain addresses, dates of birth, and other identifying pieces of information, occurring potentially anywhere in the text. Globally, many groups have worked on bespoke algorithms for de-identifying local corpora of free text records using approaches such as rule-based algorithms, pattern matching, conditional random fields, neural networks, recurrent neural networks, and bidirectional transformers (12–19). One drawback of machine learning models is that they require very large marked-up datasets from which to learn, which are not readily available outside of clinical environments, and the lack of available training data hampers progress in this field.
There is a lot of concern that de-identification of free text does not produce a perfect result, and some patient identifiers may slip through. Surveys and reviews have suggested that the best applications work at or above 90% accuracy for all types of personal health information, with systems working best on redacting name, date and age information and worst on profession and ID numbers (20–23). Portability of de-identification algorithms to new datasets is likely to be poor. Many teams now use a “hiding in plain sight” method where redacted identifiers are replaced with plausible substitutes, thus masking the few identifiers which slip through (24). A few studies have assessed the possibility of re-identification of patients from leaks in de-identified free text corpora. Two studies evaluated manual examination of de-identified notes by the original clinicians or by researchers with access to the data, in neither case was any patient correctly re-identified (25, 26). In a third study, using a complex reverse synthesis of de-identification methods, a simulated adversary could have identified 68% of data leaks correctly (27). Authors acknowledged that this mode of attack on de-identified data requires considerable effort and ingenuity from an adversary. At the current time, data custodians have few ways of quantifying or communicating the privacy risks to individuals associated with sharing automatically de-identified data. For this reason, text data which is used for research is often kept behind clinical firewalls or in specially designed data safe-havens.
Because of this lack of clarity of how well privacy can be protected, in the UK, free text notes, letters and reports are generally stripped out of clinical datasets before they are shared for research outside of the clinical environment, for example with university-based research teams. Some teams have created bespoke de-identified free text clinical datasets within clinical firewalls, and bring researchers with NLP skills within the clinical environment to conduct the work (28) but these multi-disciplinary analytics teams are hard to assemble and fund, and are by no means the norm. Within advanced infrastructures, sometimes text data is accessed by sending NLP algorithms into safe-haven data storage and extracting only relevant clinical information in short excerpts or as structured data (29), but usually these algorithms need marked-up training data to develop and evaluate, so do not get round the problem of needing access to full free text entirely.
There has been a concerted, UK-wide effort to clarify policy on sharing or re-using UK clinical free text, through funded research networks such as Healtex, Health Data Research-UK and others. It is clear that a society wide debate is needed on the balance between giving weight to the potential health benefits which could be realized from research analysis of clinical free text data, vs. prioritizing patient privacy. In the research literature and in policy and regulatory discourse around the use of patient data, there has been a notable failure to balance the privacy risks of data-sharing against the ethics of data non-use: not sharing data may be actively harmful, and cost lives, if progress in research is not made (30). We have worked on a portfolio of research to understand the wider context of governance that would be needed for free text data from UK NHS patient records to be shared at scale for research in a way that is acceptable to the public. We have engaged in deliberative research to elicit informed public opinion (31), scoped the data protection landscape and current governance practice for data-sharing, and engaged with other key stakeholders such as researchers, patient representatives, and regulators (32). Throughout our engagement work with UK-based stakeholders, it has been made clear that they want greater understanding of the benefits that improving access to medical free text for research could bring to patients or health-care services. Previous work has shown that adding free text to coded data can improve the accuracy of case detection in research and thus may improve research quality (9). But no study has yet brought together the available body of research using free text to produce a resource to describe to relevant stakeholders what the possible benefits to patients might be of conducting research using their clinical free text data, or what harms may be avoided. An understanding of the range of possible benefits in healthcare, achievable in the UK, available from research which uses clinical free text data, would enable stakeholders to weigh benefits against privacy risk when assessing whether they endorse researchers having access to free text data, and contribute to UK-wide policy on sharing of these data.
We aimed to systematically review the literature of studies using clinical free text, conducted using data generated in UK healthcare systems, which reported direct or potential benefits to patients in terms of informing or improving the quality of their care. Given our aim to describe use of free text in the studies and create a taxonomy of potential clinical benefits to aid communication about the reasons for using free text data for research, we used a qualitative approach. This allowed us to synthesize results into an easily communicable set of findings for discussion with key stakeholders in this space and highlight potential case studies. We did not attempt to quantify the level of benefit achieved by the inclusion of free text in data used in the eligible studies.
Methods
A study protocol was registered on PROSPERO (No. CRD42019141504) (33). The study is reported according to PRISMA reporting guidelines (34).
Search Strategy
Two systematic database searches were carried out. The first involved a search of articles indexed in PubMed and Web of Science (WoS), conducted in July 2019, using the following search terms:
(1) “electronic health records” or “electronic medical records” or “electronic patient records” or “hospital records” or “personal health records” or “computerized patient records” or “computerized medical records”; combined with (2) “text mining” or “natural language processing” or “free text” or “narrative.”
No limits of date or location were put on this search. The second updated search was performed in March 2020. The same search string defined above was used to search PubMed and WoS. This search was limited to publications after July 2019, in order to update the previous search. Forwards and backwards searching of identified papers led to the identification of the Maudsley Biomedical Research Center (BRC) bibliography of CRIS publications which we examined (35). We further searched the published bibliographies of the Clinical Practice Research Datalink (CPRD) (36), The Health Improvement Network (THIN) (37) and the Secure Anonymized Information Linkage (SAIL) databank (38). All papers identified from searching these bibliographies were included if they met the eligibility criteria.
Eligibility Criteria
To be eligible for this review, published research had to meet the following criteria: (1) Primary research using free text records, published in English; (2) UK health record databases or hospital data; (3) Information extracted from the text of (human, not veterinary) electronic medical records, medical letters or medical reports; and (4) report a benefit or potential benefit for patients, actionable in a clinical environment (i.e., not solely methods development or data/database quality evaluation). We defined direct or potential benefit as that the study produced a result or finding which could be translated into a change in care delivery or service access or design; or any other action in a clinical environment; which would improve the care for, or outcomes of, patients.
Screening and Study Selection
Search results were screened first by title, then abstract, then full text against the eligibility criteria. All full text articles were examined and discussed between two authors (KC and one other member of the study team EF, KHJ, ES, or LG) to establish if they met inclusion criteria, where a decision could not be made, a third author arbitrated.
Critical Appraisal of Included Studies
Because of the heterogeneity of study designs in the included papers we chose the Joanna Briggs Institute (JBI) Critical Appraisal Checklist for Analytical Cross Sectional Studies (39). The articles meeting eligibility criteria above were dual-screened by the authors. Any differences in scoring between the two authors were adjudicated by EF in discussion with KC, until a consensus was reached. Publication bias and selective reporting within studies were not assessed.
Data Extraction
Characteristics of the included studies were extracted by multiple authors, including author(s), year of publication, country of publication, clinical research question, data source and type, number of patients and number of documents included in analysis, extraction method and purpose of free text in the study, summary of clinical findings, and any statement by authors of actual or potential clinical impact.
Data Synthesis
Information about clinical benefit was thematically analyzed to create a taxonomy of benefits, according to the following research questions: (1) what are the main types of benefits achieved; (2) are benefits achieved for mental and physical health; and separately (3) is there any evidence that more benefit is achieved by the use or addition of free text compared to structured or coded clinical data?
Firstly, data were identified by authors from the introduction, results or discussion sections which described a direct or potential benefit from the study for patients in terms of improvements to their health or to healthcare provision; these were then copied and pasted into a spreadsheet. The proposed benefits were iteratively examined and grouped (by EF and KC) until authors were content that themes covered all benefits emerging from the data. Papers were then re-read looking for evidence in support of these themes and examining whether they reported on physical or mental health. Themes were then reported in a narrative synthesis; examples from papers are given to support understanding and communication of themes.
Finally, results sections were examined to identify if they reported any statistics showing accuracy or information extraction improvements comparing free text models with models that used only clinical coded data. Where found, these results were reported in a narrative synthesis.
Results
Search Results
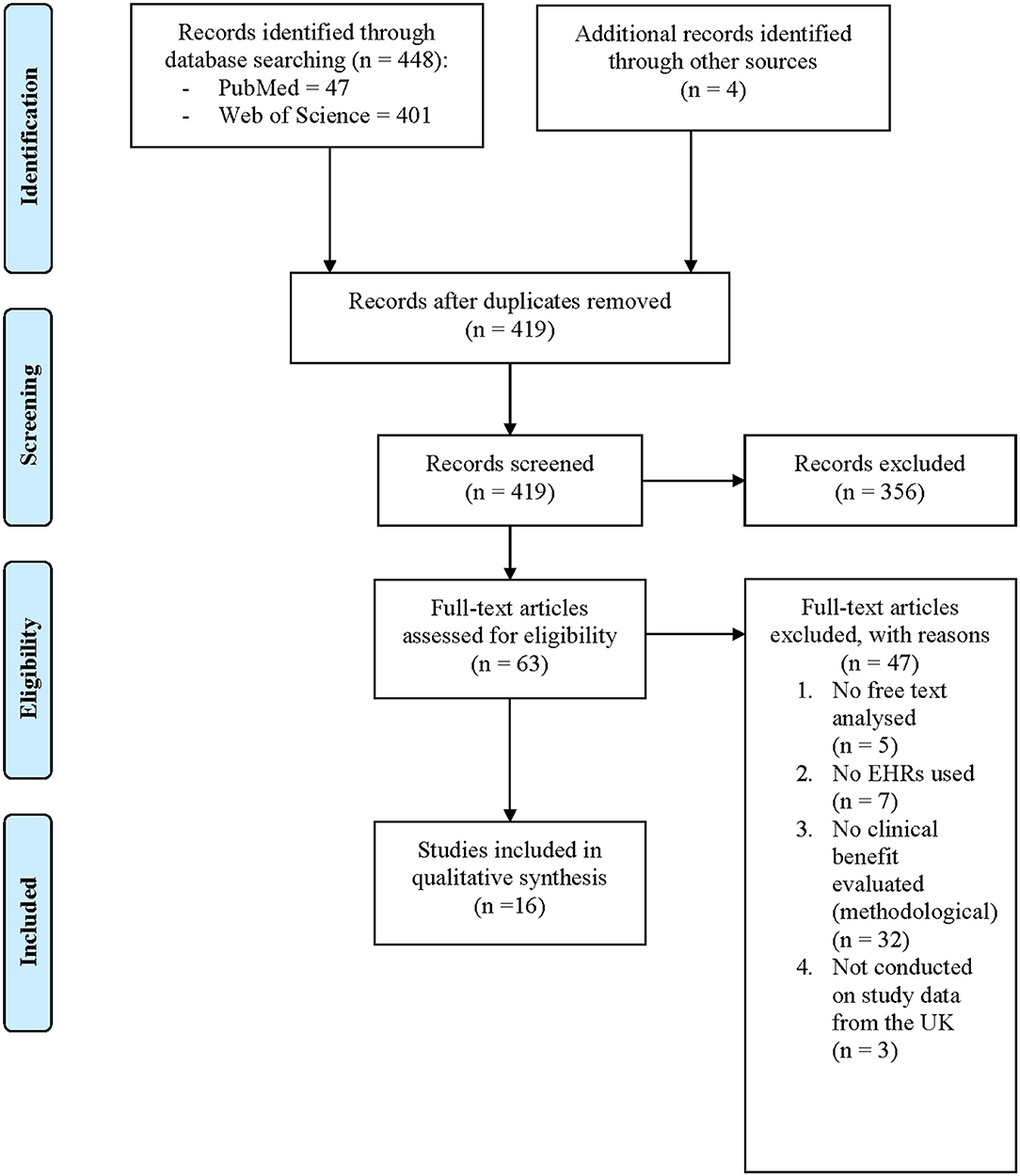
The two searches identified 448 papers. A further 179 papers were identified from lateral searching of the CRIS online database, no further papers were identified from CPRD, THIN, or SAIL bibliographies. After duplicates were removed, 598 unique papers had their titles and abstracts screened for relevance. Irrelevant papers were excluded, leaving 94 papers that underwent dual full-text review. From these papers, 51 were excluded for reasons displayed in Figure 1. Overall, 43 papers met the full eligibility criteria and were included in this systematic review.
Study Characteristics
Included papers were published between 2010 and 2020, and all were based on data in the UK as required by inclusion criteria. They analyzed data from South London and the Maudsley (SLaM) Biomedical Research Center (BRC) CRIS database (36 papers), Clinical Practice Research Datalink (CPRD) (four papers), The Health Improvement Network (THIN) (two papers), and hospital patient records (one paper). Details of all included studies are shown in Table A1. All papers met at least four of eight criteria on the JBI Critical Appraisal checklist for analytical cross-sectional studies (final agreed scoring on this tool is supplied in Appendix 1). No papers were rejected on the basis of study quality.
Analysis and Purpose of Free Text in the Studies
Studies described several methods for extracting information from free text. The simplest form of information extraction was manual review of free text (40–45) or a keyword search for relevant information, which was then modified by an algorithm (46) or manual review (47–49) to check for negation and uncertainty. Shah et al. described a bespoke algorithm for converting general practice text data into categorical data for analysis (50). Papers drawn from the SLAM CRIS database used a bespoke set of data extraction techniques, supplied in the CRIS system using the Generalized Architecture for Text Engineering (GATE) (51) which allows integration of a range of NLP algorithms for specific purposes, such as identification of medications or Mini-Mental State Examination (MMSE) scores for dementia. The Maudsley BRC separately publish a list of clinical concepts which can be extracted from free text by this system and the technical details, specifications and standardized performance metrics for each NLP algorithm (52). Some papers reported software used which overlaid CRIS or GATE, such as TextHunter (53), allowing development of searches for novel clinical concepts. All information extracted from text in these studies was converted to categorical data for statistical analysis.
Free text was used often to supplement ICD diagnosis codes (in the SLAM BRC CRIS case register) and Read codes (in general practice data) to improve identification of patients with the diagnosis of interest, for inclusion in the study (42–44, 54–65). Medication information was often extracted from free text, particularly in studies using the SLaM BRC CRIS case register (57, 60, 63, 64, 66–73). Also extracted from free text were disease symptoms and drug reactions (46, 47, 50, 60, 67, 72, 74–76); test scores, such as for the MMSE (58, 59, 61, 63, 64, 77, 78), and angiogram results (50); treatments such as cognitive behavioral therapy (CBT) (60, 79); substance use behaviors such as cannabis (49, 72, 80), alcohol (43, 44, 49) or smoking status (81); housing status (45); and information on symptom severity and functional status (61, 73).
Thematic Analysis and Taxonomy of Clinical Benefits
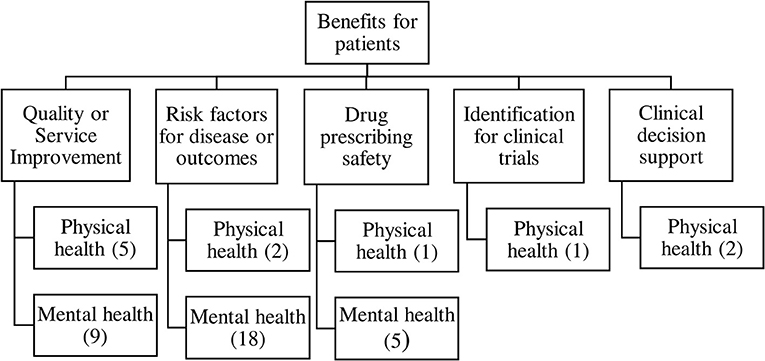
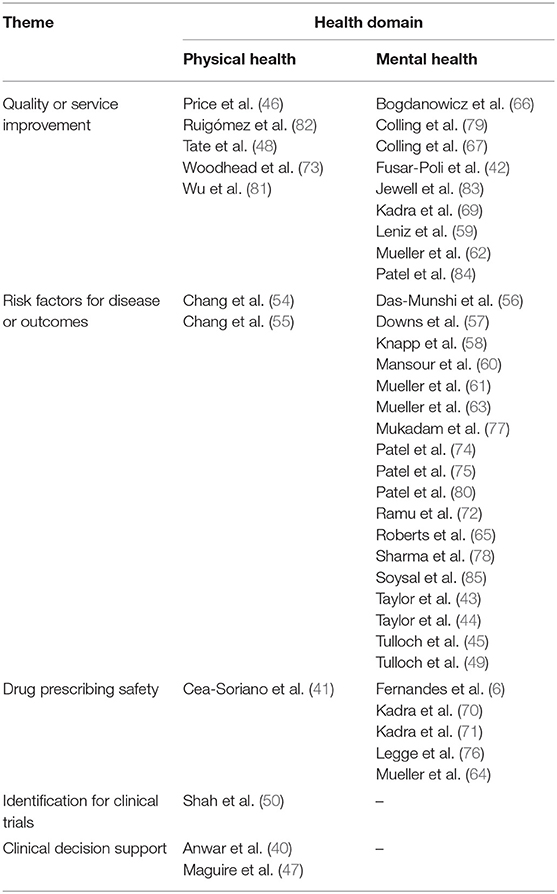
Thematic analysis of benefits showed five main types of clinical benefits reported, resulting in the taxonomy shown in Figure 2. The contribution of each study to the themes is shown in Table 1.
Quality or Safety Improvements in Healthcare
Fourteen papers reported healthcare or service quality or safety improvements that were enabled or augmented with the addition of free text information to coded data in health records, five studies focused on physical health (46, 48, 73, 81, 82) mainly drawn from general practice records; and nine on mental health (42, 59, 62, 66, 67, 69, 79, 83, 84). Authors suggested that the increase in information accuracy extracted from health records would result in service improvements such as better service planning due to more accurate prevalence or risk factor estimates, as well as better understanding of symptoms of diseases pre-diagnosis, which might speed up recognition of conditions in primary care. One example is Fusar-Poli et al. (42) who used free text data about people with first episode psychosis to show that they have a better outcome if first seen in services for people at high risk of psychosis before diagnosis, which authors argue may help health trusts to plan and prioritize services. A second example is Woodhead et al. (73) who examined whether breast and cervical cancer screening uptake is lower in women with serious mental illness. Authors stated that understanding this health disparity would help development of policies which encourage “greater screening uptake among women with other markers of severity or risk, beyond SMI status alone” (73). A final example is Patel et al. (84) who showed that individuals with bipolar disorder experienced delay in diagnosis and treatment if they had prior diagnoses of alcohol and substance misuse disorders. The authors suggested that these findings would enable “strategies to better identify underlying symptoms and offer appropriate treatment sooner in order to facilitate improved clinical outcomes” (84).
Associations Between Risk Factors and Disease or Disease Outcome
Closely related to the service improvement theme, 20 papers reported associations between exposures or risk factors and risk of disease or clinical outcomes. Two papers reported on physical health outcomes, these assessed the impact of serious mental illness on cancer diagnosis and survival (55), and the impact of intellectual disability on hospital admission and treatment outcomes for severe respiratory diseases (54). A further 18 papers reported on risk factors for mental health disorders and their outcomes (43–45, 49, 56–58, 60, 61, 63, 65, 72, 74, 75, 77, 78, 80, 85). These papers often described risk factors for disease which could enable services to be planned more appropriately to make early diagnosis or meet patients' needs. One example is Taylor et al.'s study of pregnant women who also had severe mental illness (SMI) (43) describing their socio-demographic characteristics and prescribing history, and concluding:
“A significant proportion of women, particularly those with non-affective psychoses, have modifiable risk factors requiring tailored care to optimize pregnancy outcomes.” (43)
Authors argue these findings would enable better planning of maternity services to meet the needs of pregnant women with SMI. Several studies looked at risk factors for outcomes of patients with dementia. Knapp et al. identified a range of risk factors for adverse outcomes such as admission to care homes for patients with Alzheimer's disease (58), and Sharma et al. (78) examined predictors of falls and fractures in people with dementia. They concluded:
“Clinicians should consider that besides established demographic and physical health related factors, the risk of hospitalization due to a fall or fractures in dementia is largely determined by environmental and socioeconomic factors.” (78)
Soysal et al., used free text data to understand polypharmacy in dementia patients and examined its impact on cognitive decline, finding that:
“Polypharmacy defined by the number of drugs does not appear to predict cognitive decline in a naturalistic cohort of patients with dementia” (85).
Taken together these studies give a range of insights of risk factors for developing disease, and for positive and adverse outcomes of diseases which could inform improvements and reduce inequity in healthcare planning and services.
Drug Prescribing Safety
Six papers reported on associations between prescriptions of drugs and their outcomes, either in terms of adverse drug reactions (ADRs), or in terms of clinical outcomes. One focused on physical health (41), examining the safety of non-insulin drugs for diabetes used during pregnancy. A further five focused on mental health (64, 68, 70, 71, 76). For example, Legge et al. (76) examined reasons for discontinuation of clozapine in patients with schizophrenia. Clozapine is effective but often the last drug to be tried in treatment-resistant schizophrenia and therefore failure of this drug is associated with adverse outcomes for the patient. The study identified that ADRs are an important cause of discontinuation of clozapine and authors suggest these should be treated more aggressively in the treatment onset phase to reduce discontinuation. Other studies examined antipsychotic poly-pharmacy (APP), and risk of readmission to hospital (71) as well as long term outcomes of APP such as mortality risk (70), including several different adjustments for confounding:
“Our results suggest that patients discharged on APP are more likely to be readmitted into hospital within 6 months in comparison to those discharged on monotherapy. This needs to be considered in treatment decisions.” (71)
These studies have the potential to change prescribing practice in the clinic, leading to safer or more effective management of patients' conditions.
Methods for Identifying Patients for Clinical Trials
This theme was sparsely populated by included papers. One paper focused on GP patient records, and identified that inclusion of free text increased the recorded proportion of patients with chest pain in the week prior to MI compared to structured data, and enabled differentiation between MI subtypes (50).
“Free text contained a large number of records of suspected conditions, for which the clinical system does not provide a facility for structured recording.” (50)
It is therefore possible to extrapolate from this study that when searching records for potential patients for clinical trials, more patients may be identifiable in the early stages of illness when algorithms have access to the free text in the records, in combination with structured data.
Development of Clinical Decision Support
Two papers reported on using free text data from patient records to contribute to clinical decision support systems, both papers focused on physical conditions (40, 47). Maguire et al. (47) suggested that the addition of free text to coded data in GP patient records could improve the sensitivity of clinical support systems to help clinicians identify rare diseases, in this case allergic bronchopulmonary aspergillosis (ABPA). The second study, by Anwar et al., used all the data in a patient's record in a hospital audiology department, including free text, to predict what type of hearing aid should be prescribed to the patient (40). Wide adoption of accurate clinical support systems may improve consistency or quality of care for patients in some healthcare clinics.
Does Inclusion of Free Text Enable Better Quality Research Than Using Structured Data Alone?
While the majority of papers used a combination of analyzing both Read or ICD codes, structured demographic data, and free text information from their respective databases, few reported on the change in quality or accuracy of case identification or data extraction by the addition of free text data.
Most commonly this was reported in papers using GP patient records, where much clinical information is recorded in codes. Five studies using GP patient records reported improved accuracy in detecting symptoms or cases, or that greater clinical information was available for the research study, with the addition of free text (46–48, 50, 82). A further three papers using mental health data reported that more data on variables of interest were available in the free text as compared to structured data, and used free text mentions of diagnoses to augment case finding, but did not quantify the additional patient numbers found by this method (67, 79, 81).
Discussion
We identified 43 UK studies which reported results of research using clinical free text which would result in direct or potential benefits to patients in terms of informing or improving the quality of their healthcare. The majority of papers used data drawn from patient records in a secondary care mental health trust in South London. This skewed our results to focus in the majority on benefits to patients in a mental health setting in terms of health condition. However, we have no reason to assume this skewed the results in terms of themes identified. We were able to classify the benefits reported in these papers into five clear themes which may aid public audiences in the UK in understanding why researchers want to access free text data from UK-based EHRs.
Our first three themes were focused on issues pertaining to patient care in the clinic and were well-supported with a range of studies; these themes were healthcare quality and service improvements; understanding risk factors for disease and disease outcomes; and improving the safety of drug prescribing. These three themes show the potential for clinical free text to contribute to research which changes clinicians' practice and contributes to the evidence-base for setting up equitable services appropriate to patients' needs, understanding who is at risk of adverse outcomes, and how drugs can be used to best treat symptoms and prevent adverse outcomes.
Two more themes related to the potential development of technology for use in the clinic, one showing case identification methods which illustrated the potential use of free text to contribute to earlier or wider recruitment of patients for clinical trials. This kind of technology may enable more patients to benefit from trial participation, as well as future patients benefiting from higher quality and faster trials, meaning new treatments are available sooner. The other theme showcased examples where free text data could be used to develop automated clinical decision support, helping clinicians make decisions on patients' diagnosis or treatment on the basis of a range of information in their records. These two technology themes were less well-populated than the more clinically focused themes, suggesting the production of well-implemented and integrated healthcare technology which draws on clinical free text may be some way off. However, if we look at wider literature on this topic, including papers which report a purely methodological focus (86, 87), or discuss more widely the future applications of analyzing data from mental health records (88), we can discover more examples of these types of technologies under development.
These themes of clinical benefit are largely in line with other reviews on similar topics. For example, Velupillai et al. (10) also identified mental health as a clinical domain which would richly benefit from the application of NLP to clinical records, and determined that prescribing outcomes, risk factors and prognoses were all features which could be extracted from clinical text and examined in research. Of interest, these authors suggested that the self-reported patient experience could also be examined using clinical free text, but we did not find this theme in our analysis of published papers.
The majority of our papers drew on one UK-based data source, curated by a secondary care trust in south London (SLaM), and focusing on mental health. This reflects that when a safe and secure research infrastructure is developed, allowing NLP researchers to work closely together with clinicians, a very productive and profitable research pipeline can be formed. Of note, SLaM uses a participatory governance model with a service user and carer advisory group which advises researchers on the development of their studies and on requesting linkages (89). This format ensures that research conducted using mental health datasets is directed by priorities identified by service users, that there is a route by which service users can find out about and become involved in research. This reduces the separation between service users and researchers, helping to increase trust. This model of transparency in the use of patient data was favored by members of the Brighton citizens' jury who were asked about their views on sharing their medical free text for research (31).
Strengths and Limitations
Our review is strengthened by our “cast the net wide” approach to finding papers, by our team approach to sifting, screening and quality-appraising the papers, and by adhering to reporting guidelines. However, we note a few limitations. Our search strategy did not pick up all eligible papers and several were found only after examination of a dataset's published bibliography. This suggests our keywords were not perfectly aligned to the papers we sought, or that papers using free text are not well-labeled as such in their title or abstract. This is something that could be addressed within the research community. Due to our aim being to inform UK-wide discussion, public engagement and policy, we limited our review to UK data only. Therefore, the themes of benefit identified indicate only the historical work that has been possible to conduct in the UK to date. An international search, encompassing differing research groups, health care structures, and data access regulations and policies would likely have yielded additional themes showing types of patient benefit. However, this review still provides a case study of international interest, and which may be used as a resource for policy discussions internationally. Due to the differing rates of progress in accessing free text data between research groups and healthcare settings in the UK, and due to the types of data recording in different part of the health system, the majority of papers so far published in the UK are on mental health. This reflects the UK's early adoption of EHRs in mental health compared to other specialist or hospital care, the national reticence around free text access for primary care EHRs and the current lack of workable governance solutions to free text access outside of mental health. These are influences of national policy with demonstrable impact on the field as reviewed. This will have introduced bias into our understanding of what possible benefits can be accrued from free text data. This bias will not be quantifiable until further work can be done to extend understanding across international research, and until access to free text data is improved in the UK for researchers aiming to study physical health conditions.
This heterogeneous group of studies was also difficult to appraise for quality and for risk of bias across studies. As we were not searching for effect sizes relating to interventions, it was not clear how to evaluate publication bias or selective reporting in our identified body of research.
The derivation of themes for our taxonomy was subjective and certain papers were felt to fit into more than one theme. The two themes of clinical decision support and recruitment to clinical trials were poorly supported by included papers, although they are the focus of methodological work known to the authors. The included papers using GP patient records data were less directly focused on clinical benefits to patients, often reporting data quality improvements as the main outcome of examining free text, although they did suggest potential clinical benefits when discussing their results. Their inclusion was debated among the authors. We aimed to be inclusive so that the maximum number of exemplar studies which described potential benefits could be showcased. Future work should revisit the taxonomy derived here, which may not have reached saturation, and which may be modified by the emergence of future work in this fast developing field.
The majority of studies identified did not present any evaluation of the impact that free text had on the study, as they primarily focused on the clinical benefits. We were unable to categorize the impact that the addition that free text had on improving study sample size or quality or influencing results. Future studies should consider clearly reporting the extent to which free text contributed in a quantifiable way so that this can be evaluated, and studies which provide a standalone assessment of the value of text data for a particular problem should be published alongside.
Finally, while we were able to synthesize potential benefits here, we did not find any reports in the studies on potential harms or costs associated with using free text data. We assume this is because we explicitly excluded papers which evaluated methodology as a primary focus of the study. Our aim is to contribute to discourse on safe and acceptable access to free text clinical data for researchers, and the hope is that when governance structures are developed and implemented, costs and harms of accessing free text data will be minimized for all stakeholders.
Future Research Directions and Conclusions
We believe these findings will be a useful resource for the public discourse on negotiating access to patient free text data in the UK and provide a methodological template for similar discussions in other international settings. Members of the research community, data-sharing regulators (32) and the public (31) have all expressed a desire to understand more about the potential benefits to health of sharing free text data from clinical records, and this review offers the first resource where a body of relevant evidence has been pulled together. As a next step, we intend to set up an online repository through an openly available website (such as www.healtex.org), which will list the papers identified in this review and give a short summary of expected patient benefit from the research for each one. This resource could be “live” and continuously updated. Open access approaches to dissemination such as project websites can be excellent ways of communicating with a range of stakeholders. We hope these results will contribute to widespread understanding of the value of using clinical free text in research to improve the health and healthcare of the UK population, and to progressive data governance regulation which works for the good of the whole population.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author.
Author Contributions
EF conceived the project, screened articles, analyzed data, and drafted the manuscript. KC conducted searches, screened articles, extracted data, analyzed data, and helped to draft the manuscript. KJ, ES, and LG screened articles, extracted data, and commented on drafts of the manuscript. RS commented on drafts of the manuscript. All authors read and approved the final manuscript.
Funding
This work was funded by the Engineering and Physical Sciences Research Council via Healtex, the UK health care text analytics research network (Grant Number EP/N027280/1).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This paper draws on studies which used data provided by patients and collected by the NHS as part of their care and support. #datasaveslives.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fdgth.2021.606599/full#supplementary-material
Appendix 1. Joanna Briggs Institute critical appraisal checklist screening results.
Appendix 2. PRISMA checklist.
References
1. Health Data Research UK. Green Shoots: Health Data Research UK's Annual Review 2019/2020. (2020). Available online at: https://www.hdruk.ac.uk/wp-content/uploads/2020/07/Annual-Review-2020_020720.pdf
2. Wolf A, Dedman D, Campbell J, Booth H, Lunn D, Chapman J, et al. Data resource profile: Clinical Practice Research Datalink (CPRD) Aurum. Int J Epidemiol. (2019) 48:1740-g. doi: 10.1093/ije/dyz034
3. Herrett E, Gallagher AM, Bhaskaran K, Forbes H, Mathur R, van Staa T, et al. Data resource profile: clinical practice research datalink (CPRD). Int J Epidemiol. (2015) 44:827–36. doi: 10.1093/ije/dyv098
4. Blak BT, Thompson M, Dattani H, and Bourke A. Generalisability of The Health Improvement Network (THIN) database: demographics, chronic disease prevalence and mortality rates. Inform Prim Care. (2011) 19:251–5. doi: 10.14236/jhi.v19i4.820
5. Jackson R, Patel R, Velupillai S, Gkotsis G, Hoyle D, and Stewart R. Knowledge discovery for deep phenotyping serious mental illness from electronic mental health records. F1000RES. (2018) 7:210. doi: 10.12688/f1000research.13830.1
6. Fernandes AC, Dutta R, Velupillai S, Sanyal J, Stewart R, and Chandran D. Identifying suicide ideation and suicidal attempts in a psychiatric clinical research database using natural language processing. Sci Rep. (2018) 8:1–10. doi: 10.1038/s41598-018-25773-2
7. Herbert A, Wijlaars L, Zylbersztejn A, Cromwell D, and Hardelid P. Data resource profile: hospital episode statistics admitted patient care (HES APC). Int J Epidemiol. (2017) 46:1093-i. doi: 10.1093/ije/dyx015
8. Henson KE, Elliss-Brookes L, Coupland VH, Payne E, Vernon S, Rous B, et al. Data resource profile: national cancer registration dataset in England. Int J Epidemiol. (2020) 49:16-h. doi: 10.1093/ije/dyz076
9. Ford E, Carroll JA, Smith H, Scott D, and Cassell J. Extracting information from the text of electronic medical records to improve case detection: a systematic review. J Am Med Inform Assoc. (2016) 23:1007–15. doi: 10.1093/jamia/ocv180
10. Velupillai S, Suominen H, Liakata M, Roberts A, Shah AD, Morley K, et al. Using clinical Natural Language Processing for health outcomes research: overview and actionable suggestions for future advances. J Biomed Informatics. (2018) 88:11–9. doi: 10.1016/j.jbi.2018.10.005
11. U.S. Department of Health & Human Services. Health Insurance Portability and Accountability Act of 1996. (1996). Available online at: https://aspe.hhs.gov/report/health-insurance-portability-and-accountability-act-1996
12. Dalianis H, and Velupillai S. De-identifying Swedish clinical text-refinement of a gold standard and experiments with conditional random fields. J Biomed Semantics. (2010) 1:6. doi: 10.1186/2041-1480-1-6
13. Pantazos K, Lauesen S, and Lippert S. De-identifying an EHR database-anonymity, correctness and readability of the medical record. Stud Health Technol Informat. (2011) 169:862–6.
14. Srivastava A, Ekbal A, Saha S, and Bhattacharyya P (eds.). A recurrent neural network architecture for de-identifying clinical records. In: Proceedings of the 13th International Conference on Natural Language Processing. Varanasi (2016).
15. Menger V, Scheepers F, van Wijk LM, and Spruit M. DEDUCE: a pattern matching method for automatic de-identification of Dutch medical text. Telematics Informatics. (2018) 35:727–36. doi: 10.1016/j.tele.2017.08.002
16. Kajiyama K, Horiguchi H, Okumura T, Morita M, and Kano Y. De-identifying free text of japanese electronic health records. EMNLP. (2018) 2018:65. doi: 10.18653/v1/W18-5608
17. Sang ETK, de Vries B, Smink W, Veldkamp B, Westerhof G, and Sools A (eds.). De-identification of Dutch Medical Text. In: 2nd Healthcare Text Analytics Conference (HealTAC2019). Cardiff (2019).
18. Perez-Diez I, Perez-Moraga R, Lopez-Cerdan A, Salinas-Serrano J-M, and de la Iglesia-Vaya M. De-identifying Spanish medical texts-Named Entity Recognition applied to radiology reports. medRxiv. (2020). doi: 10.1101/2020.04.09.20058958
19. Johnson AE, Bulgarelli L, and Pollard TJ (eds.). Deidentification of free-text medical records using pre-trained bidirectional transformers. In: Proceedings of the ACM Conference on Health, Inference, and Learning. Toronto, ON (2020).
20. Yogarajan V, Mayo M, and Pfahringer B. A survey of automatic de-identification of longitudinal clinical narratives. arXiv [Preprint] arXiv:181006765. (2018).
21. Meystre SM, Friedlin FJ, South BR, Shen S, and Samore MH. Automatic de-identification of textual documents in the electronic health record: a review of recent research. BMC Med Res Methodol. (2010) 10:70. doi: 10.1186/1471-2288-10-70
22. Kushida CA, Nichols DA, Jadrnicek R, Miller R, Walsh JK, and Griffin K. Strategies for de-identification and anonymization of electronic health record data for use in multicenter research studies. Med Care. (2012) 50(Suppl.):S82–101. doi: 10.1097/MLR.0b013e3182585355
23. Uzuner O (ed.). Second i2b2 workshop on natural language processing challenges for clinical records. In: AMIA Annual Symposium Proceedings/AMIA Symposium AMIA Symposium. Chicago, IL (2007).
24. Carrell D, Malin B, Aberdeen J, Bayer S, Clark C, Wellner B, et al. Hiding in plain sight: use of realistic surrogates to reduce exposure of protected health information in clinical text. J Am Med Informatics Assoc. (2013) 20:342–8. doi: 10.1136/amiajnl-2012-001034
25. Meystre S, Shen S, Hofmann D, and Gundlapalli A. Can physicians recognize their own patients in de-identified notes? Stud Health Technol Informat. (2014) 205:778–82.
26. Grouin C, Griffon N, and Névéol A (eds.). Is it possible to recover personal health information from an automatically de-identified corpus of French EHRs? In: Proceedings of the Sixth International Workshop on Health Text Mining and Information Analysis. Lisbon (2015).
27. Carrell DS, Cronkite DJ, Li M, Nyemba S, Malin BA, Aberdeen JS, et al. The machine giveth and the machine taketh away: a parrot attack on clinical text deidentified with hiding in plain sight. J Am Med Informatics Assoc. (2019) 26:1536–44. doi: 10.1093/jamia/ocz114
28. Stewart R, Soremekun M, Perera G, Broadbent M, Callard F, Denis M, et al. The South London and Maudsley NHS foundation trust biomedical research centre (SLAM BRC) case register: development and descriptive data. BMC Psychiatry. (2009) 9:51. doi: 10.1186/1471-244X-9-51
29. Fonferko-Shadrach B, Lacey AS, Roberts A, Akbari A, Thompson S, Ford DV, et al. Using natural language processing to extract structured epilepsy data from unstructured clinic letters: development and validation of the ExECT (extraction of epilepsy clinical text) system. BMJ Open. (2019) 9:e023232. doi: 10.1136/bmjopen-2018-023232
30. Jones KH, Laurie G, Stevens L, Dobbs C, Ford DV, and Lea N. The other side of the coin: harm due to the non-use of health-related data. Int J Med Informatics. (2017) 97:43–51. doi: 10.1016/j.ijmedinf.2016.09.010
31. Ford E, Oswald M, Hassan L, Bozentko K, Nenadic G, and Cassell J. Should free-text data in electronic medical records be shared for research? A citizens' jury study in the UK. J Med Ethics. (2020) 46:367–77. doi: 10.1136/medethics-2019-105472
32. Jones KH, Ford EM, Lea N, Griffiths LJ, Hassan L, Heys S, et al. Toward the development of data governance standards for using clinical free-text data in health research: position paper. J Med Internet Res. (2020) 22:e16760. doi: 10.2196/16760
33. Ford E, Curlewis K, Jones K, Squires E, and Griffiths L. The Clinical Benefits of the Use of Free Text Medical Data From Patient Records: A Systematic Review. (2019). Available online at: https://www.crd.york.ac.uk/prospero/display_record.php?ID=CRD42019141504
34. Moher D, Liberati A, Tetzlaff J, and Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Internal Med. (2009) 151:264–9. doi: 10.7326/0003-4819-151-4-200908180-00135
35. NIHR Maudsley Biomedical Research Centre. CRIS Publications (2020). Available online at: https://www.maudsleybrc.nihr.ac.uk/facilities/clinical-record-interactive-search-cris/cris-publications/
36. CPRD. CPRD Bibliography. Available online at: https://www.cprd.com/Bibliography/Researchpapers.asp
37. The Health Improvement Network (THIN). Available online at: https://www.ucl.ac.uk/pcph/research-groups-themes/thin-pub/publications
38. SAIL Databank. SAIL Publications (2020). Available online at: https://saildatabank.com/saildata/sail-publications/
39. Joanna Briggs Institute. Checklist for Analytical Cross Sectional Studies: Critical Appraisal Tools for Use in JBI Systematic Reviews. (2020). Available online at: https://joannabriggs.org/sites/default/files/2020-08/Checklist_for_Analytical_Cross_Sectional_Studies.pdf
40. Anwar MN, and Oakes MP (eds.). Data mining of audiology patient records: factors influencing the choice of hearing aid type. In: Proceedings of the ACM Fifth International Workshop on Data and Text Mining in Biomedical Informatics. Glasgow (2011).
41. Cea-Soriano L, García-Rodríguez LA, Brodovicz KG, Masso Gonzalez E, Bartels DB, and Hernández-Díaz S. Safety of non-insulin glucose-lowering drugs in pregnant women with pre-gestational diabetes: a cohort study. Diabetes Obesity Metab. (2018) 20:1642–51. doi: 10.1111/dom.13275
42. Fusar-Poli P, Diaz-Caneja C, Patel R, Valmaggia L, Byrne M, Garety P, et al. Services for people at high risk improve outcomes in patients with first episode psychosis. Acta Psychiatrica Scand. (2016) 133:76–85. doi: 10.1111/acps.12480
43. Taylor CL, Stewart R, Ogden J, Broadbent M, Pasupathy D, and Howard LM. The characteristics and health needs of pregnant women with schizophrenia compared with bipolar disorder and affective psychoses. BMC Psychiatry. (2015) 15:88. doi: 10.1186/s12888-015-0451-8
44. Taylor CL, Stewart RJ, and Howard LM. Relapse in the first three months postpartum in women with history of serious mental illness. Schizophrenia Res. (2019) 204:46–54. doi: 10.1016/j.schres.2018.07.037
45. Tulloch AD, Khondoker MR, Fearon P, and David AS. Associations of homelessness and residential mobility with length of stay after acute psychiatric admission. BMC Psychiatry. (2012) 12:121. doi: 10.1186/1471-244X-12-121
46. Price SJ, Stapley SA, Shephard E, Barraclough K, and Hamilton WT. Is omission of free text records a possible source of data loss and bias in Clinical Practice Research Datalink studies? A case–control study. BMJ Open. (2016) 6:e011664. doi: 10.1136/bmjopen-2016-011664
47. Maguire A, Johnson ME, Denning DW, Ferreira GL, and Cassidy A. Identifying rare diseases using electronic medical records: the example of allergic bronchopulmonary aspergillosis. Pharmacoepidemiol Drug Safety. (2017) 26:785–91. doi: 10.1002/pds.4204
48. Tate AR, Martin AG, Ali A, and Cassell JA. Using free text information to explore how and when GPs code a diagnosis of ovarian cancer: an observational study using primary care records of patients with ovarian cancer. BMJ Open. (2011) 1:e000025. doi: 10.1136/bmjopen-2010-000025
49. Tulloch AD, Frayn E, Craig TK, and Nicholson TR. Khat use among Somali mental health service users in South London. Soc Psychiatry Psychiatr Epidemiol. (2012) 47:1649–56. doi: 10.1007/s00127-011-0471-8
50. Shah AD, Bailey E, Williams T, Denaxas S, Dobson R, and Hemingway H. Natural language processing for disease phenotyping in UK primary care records for research: a pilot study in myocardial infarction and death. J Biomed Semantics. (2019) 10:20. doi: 10.1186/s13326-019-0214-4
51. Cunningham H, Tablan V, Roberts A, and Bontcheva K. Getting more out of biomedical documents with GATE's full lifecycle open source text analytics. PLOS Comput Biol. (2013) 9:e1002854. doi: 10.1371/journal.pcbi.1002854
52. NIHR Maudsley Biomedical Research Centre. Natural Language Processing (NLP) Service (2020). Available online at: https://www.maudsleybrc.nihr.ac.uk/facilities/clinical-record-interactive-search-cris/cris-natural-language-processing/
53. Jackson RG, Ball M, Patel R, Hayes RD, Dobson RJB, and Stewart R. TextHunter–a user friendly tool for extracting generic concepts from free text in clinical research. AMIA Annu Symposium Proc AMIA Symposium. (2014) 2014:729–38. doi: 10.13140/2.1.3722.9121
54. Chang C-K, Chen C-Y, Broadbent M, Stewart R, and O'Hara J. Hospital admissions for respiratory system diseases in adults with intellectual disabilities in Southeast London: a register-based cohort study. BMJ Open. (2017) 7:e014846. doi: 10.1136/bmjopen-2016-014846
55. Chang C-K, Hayes RD, Broadbent MT, Hotopf M, Davies E, Møller H, et al. A cohort study on mental disorders, stage of cancer at diagnosis and subsequent survival. BMJ Open. (2014) 4:e004295. doi: 10.1136/bmjopen-2013-004295
56. Das-Munshi J, Chang C-K, Dutta R, Morgan C, Nazroo J, Stewart R, et al. Ethnicity and excess mortality in severe mental illness: a cohort study. Lancet Psychiatry. (2017) 4:389–99. doi: 10.1016/S2215-0366(17)30097-4
57. Downs J, Dean H, Lechler S, Sears N, Patel R, Shetty H, et al. Negative symptoms in early-onset psychosis and their association with antipsychotic treatment failure. Schizophrenia Bull. (2019) 45:69–79. doi: 10.1093/schbul/sbx197
58. Knapp M, Chua K-C, Broadbent M, Chang C-K, Fernandez J-L, Milea D, et al. Predictors of care home and hospital admissions and their costs for older people with Alzheimer's disease: findings from a large London case register. BMJ Open. (2016) 6:e013591. doi: 10.1136/bmjopen-2016-013591
59. Leniz J, Higginson IJ, Stewart R, and Sleeman KE. Understanding which people with dementia are at risk of inappropriate care and avoidable transitions to hospital near the end-of-life: a retrospective cohort study. Age Ageing. (2019) 48:672–9. doi: 10.1093/ageing/afz052
60. Mansour R, Tsamakis K, Rizos E, Perera G, Das-Munshi J, Stewart R, et al. Late-life depression in people from ethnic minority backgrounds: differences in presentation and management. J Affect Disord. (2020) 264:340–7. doi: 10.1016/j.jad.2019.12.031
61. Mueller C, Perera G, Hayes RD, Shetty H, Stewart R. Associations of acetylcholinesterase inhibitor treatment with reduced mortality in Alzheimer's disease: a retrospective survival analysis. Age Ageing. (2018) 47:88–94. doi: 10.1093/ageing/afx098
62. Mueller C, Perera G, Rajkumar AP, Bhattarai M, Price A, O'Brien JT, et al. Hospitalization in people with dementia with Lewy bodies: frequency, duration, and cost implications. Alzheimer's Dement Diagnosis Assessment Dis Monitor. (2018) 10:143–52. doi: 10.1016/j.dadm.2017.12.001
63. Mueller C, Molokhia M, Perera G, Veronese N, Stubbs B, Shetty H, et al. Polypharmacy in people with dementia: associations with adverse health outcomes. Exp Gerontol. (2018) 106:240–5. doi: 10.1016/j.exger.2018.02.011
64. Mueller C, Huntley J, Stubbs B, Sommerlad A, Carvalho AF, Perera G, et al. Associations of neuropsychiatric symptoms and antidepressant prescription with survival in Alzheimer's disease. J Am Med Direct Assoc. (2017) 18:1076–81. doi: 10.1016/j.jamda.2017.07.001
65. Roberts E, Wessely S, Chalder T, Chang C-K, and Hotopf M. Mortality of people with chronic fatigue syndrome: a retrospective cohort study in England and Wales from the South London and Maudsley NHS Foundation Trust Biomedical Research Centre (SLaM BRC) Clinical Record Interactive Search (CRIS) register. Lancet. (2016) 387:1638–43. doi: 10.1016/S0140-6736(15)01223-4
66. Bogdanowicz KM, Stewart R, Chang CK, Shetty H, Khondoker M, Day E, et al. Excess overdose mortality immediately following transfer of patients and their care as well as after cessation of opioid substitution therapy. Addiction. (2018) 113:946–51. doi: 10.1111/add.14114
67. Colling C, Khondoker M, Patel R, Fok M, Harland R, Broadbent M, et al. Predicting high-cost care in a mental health setting. BJPsych Open. (2020) 6:e10. doi: 10.1192/bjo.2019.96
68. Fernandes AC, Chandran D, Khondoker M, Dewey M, Shetty H, Dutta R, et al. Demographic and clinical factors associated with different antidepressant treatments: a retrospective cohort study design in a UK psychiatric healthcare setting. BMJ Open. (2018) 8:e022170. doi: 10.1136/bmjopen-2018-022170
69. Kadra G, Stewart R, Shetty H, Downs J, MacCabe JH, Taylor D, et al. Predictors of long-term (≥ 6 months) antipsychotic polypharmacy prescribing in secondary mental healthcare. Schizophrenia Res. (2016) 174:106–12. doi: 10.1016/j.schres.2016.04.010
70. Kadra G, Stewart R, Shetty H, MacCabe J, Chang CK, Taylor D, et al. Long-term antipsychotic polypharmacy prescribing in secondary mental health care and the risk of mortality. Acta Psychiatrica Scand. (2018) 138:123–32. doi: 10.1111/acps.12906
71. Kadra G, Stewart R, Shetty H, MacCabe JH, Chang C-K, Kesserwani J, et al. Antipsychotic polypharmacy prescribing and risk of hospital readmission. Psychopharmacology. (2018) 235:281–9. doi: 10.1007/s00213-017-4767-6
72. Ramu N, Kolliakou A, Sanyal J, Patel R, and Stewart R. Recorded poor insight as a predictor of service use outcomes: cohort study of patients with first-episode psychosis in a large mental healthcare database. BMJ Open. (2019) 9:e028929. doi: 10.1136/bmjopen-2019-028929
73. Woodhead C, Cunningham R, Ashworth M, Barley E, Stewart RJ, and Henderson MJ. Cervical and breast cancer screening uptake among women with serious mental illness: a data linkage study. BMC Cancer. (2016) 16:819. doi: 10.1186/s12885-016-2842-8
74. Patel R, Jayatilleke N, Broadbent M, Chang C-K, Foskett N, Gorrell G, et al. Negative symptoms in schizophrenia: a study in a large clinical sample of patients using a novel automated method. BMJ Open. (2015) 5:e007619. doi: 10.1136/bmjopen-2015-007619
75. Patel R, Lloyd T, Jackson R, Ball M, Shetty H, Broadbent M, et al. Mood instability is a common feature of mental health disorders and is associated with poor clinical outcomes. BMJ Open. (2015) 5:e007504. doi: 10.1136/bmjopen-2014-007504
76. Legge SE, Hamshere M, Hayes RD, Downs J, O'Donovan MC, Owen MJ, et al. Reasons for discontinuing clozapine: a cohort study of patients commencing treatment. Schizophrenia Res. (2016) 174:113–9. doi: 10.1016/j.schres.2016.05.002
77. Mukadam N, Lewis G, Mueller C, Werbeloff N, Stewart R, and Livingston G. Ethnic differences in cognition and age in people diagnosed with dementia: a study of electronic health records in two large mental healthcare providers. Int J Geriatric Psychiatry. (2019) 34:504–10. doi: 10.1002/gps.5046
78. Sharma S, Mueller C, Stewart R, Veronese N, Vancampfort D, Koyanagi A, et al. Predictors of falls and fractures leading to hospitalization in people with dementia: a representative cohort study. J Am Med Direct Assoc. (2018) 19:607–12. doi: 10.1016/j.jamda.2018.03.009
79. Colling C, Evans L, Broadbent M, Chandran D, Craig TJ, Kolliakou A, et al. Identification of the delivery of cognitive behavioural therapy for psychosis (CBTp) using a cross-sectional sample from electronic health records and open-text information in a large UK-based mental health case register. BMJ Open. (2017) 7:e015297. doi: 10.1136/bmjopen-2016-015297
80. Patel R, Wilson R, Jackson R, Ball M, Shetty H, Broadbent M, et al. Association of cannabis use with hospital admission and antipsychotic treatment failure in first episode psychosis: an observational study. BMJ Open. (2016) 6:e009888. doi: 10.1136/bmjopen-2015-009888
81. Wu C-Y, Chang C-K, Robson D, Jackson R, Chen S-J, Hayes RD, et al. Evaluation of smoking status identification using electronic health records and open-text information in a large mental health case register. PLoS ONE. (2013) 8:e74262. doi: 10.1371/journal.pone.0074262
82. Ruigómez A, Martín-Merino E, and Rodríguez LA. Validation of ischemic cerebrovascular diagnoses in the health improvement network (THIN). Pharmacoepidemiol Drug Saf . (2010) 19:579–85. doi: 10.1002/pds.1919
83. Jewell A, Dean K, Fahy T, and Cullen AE. Predictors of Mental Health Review Tribunal (MHRT) outcome in a forensic inpatient population: a prospective cohort study. BMC Psychiatry. (2017) 17:25. doi: 10.1186/s12888-016-1188-8
84. Patel R, Shetty H, Jackson R, Broadbent M, Stewart R, Boydell J, et al. Delays before diagnosis and initiation of treatment in patients presenting to mental health services with bipolar disorder. PLoS ONE. (2015) 10:e0126530. doi: 10.1371/journal.pone.0126530
85. Soysal P, Perera G, Isik AT, Onder G, Petrovic M, Cherubini A, et al. The relationship between polypharmacy and trajectories of cognitive decline in people with dementia: a large representative cohort study. Exp Gerontol. (2019) 120:62–7. doi: 10.1016/j.exger.2019.02.019
86. Wu H, Toti G, Morley KI, Ibrahim ZM, Folarin A, Jackson R, et al. SemEHR: a general-purpose semantic search system to surface semantic data from clinical notes for tailored care, trial recruitment, and clinical research. J Am Med Informatics Assoc. (2018) 25:530–7. doi: 10.1093/jamia/ocx160
87. Clark MM, Hildreth A, Batalov S, Ding Y, Chowdhury S, Watkins K, et al. Diagnosis of genetic diseases in seriously ill children by rapid whole-genome sequencing and automated phenotyping and interpretation. Sci Transl Med. (2019) 11:eaat6177. doi: 10.1126/scitranslmed.aat6177
88. Chaturvedi J. From learning about machines to machine learning: applications for mental health rehabilitation. J Psychosoc Rehab Mental Health. (2020) 7:3–4. doi: 10.1007/s40737-020-00163-y
89. Jewell A, Pritchard M, Barrett K, Green P, Markham S, McKenzie S, et al. The Maudsley Biomedical Research Centre (BRC) data linkage service user and carer advisory group: creating and sustaining a successful patient and public involvement group to guide research in a complex area. Res Involve Engage. (2019) 5:20. doi: 10.1186/s40900-019-0152-4
Keywords: clinical free text, natural language processing, text analysis, data governance, privacy, patient benefit
Citation: Ford E, Curlewis K, Squires E, Griffiths LJ, Stewart R and Jones KH (2021) The Potential of Research Drawing on Clinical Free Text to Bring Benefits to Patients in the United Kingdom: A Systematic Review of the Literature. Front. Digit. Health 3:606599. doi: 10.3389/fdgth.2021.606599
Received: 15 September 2020; Accepted: 15 January 2021;
Published: 10 February 2021.
Edited by:
Aurélie Névéol, Université Paris-Saclay, FranceReviewed by:
Kirk Roberts, University of Texas Health Science Center at Houston, United StatesJames Cimino, University of Alabama at Birmingham, United States
Copyright © 2021 Ford, Curlewis, Squires, Griffiths, Stewart and Jones. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kerina H. Jones, ay5oLmpvbmVzQHN3YW5zZWEuYWMudWs=
 Elizabeth Ford
Elizabeth Ford Keegan Curlewis1
Keegan Curlewis1 Robert Stewart
Robert Stewart Kerina H. Jones
Kerina H. Jones