- 1National Institute of Biological Sciences, Beijing, China
- 2Tsinghua Institute of Multidisciplinary Biomedical Research, Tsinghua University, Beijing, China
The lung is constantly exposed to the external environment, making it highly susceptible to infections and injuries caused by airborne pollutants and pathogens. Understanding the cellular players and molecular mechanisms underlying post-injury lung repair is essential for elucidating the repair processes following injury. Single-Cell Sequencing (sc-seq) offers unprecedented single-cell resolution, enabling researchers to dissect the complex biological profiles that drive diverse lung pathologies and to uncover the signaling pathways crucial for lung regeneration. This review will explore the latest findings in lung diseases and regeneration revealed by sc-seq. Additionally, we will highlight how continuous advancements in sc-seq technology are paving the way for the development of novel drugs aimed at targeting pathways involved in lung regeneration and treating lung diseases. By offering single-cell resolution, recent advancements in sc-seq have enabled researchers to dissect multiple layers of biological profiles underlying diverse lung pathogenesis and uncover signaling pathways critical for lung regeneration. In this review, we will discuss recent sc-seq findings in lung diseases and regeneration. Continuous advancements in sc-seq technology hold great promise for facilitating the development of novel drugs targeting lung regeneration pathways and lung diseases. These repair processes are mediated by resident epithelial stem cells and their niche cells.
1 Introduction
The lung is constantly exposed to a variety of air pollutants and pathogens, resulting in a high susceptibility to injuries. Despite this vulnerability, the lung exhibits a remarkable regenerative capacity. This regeneration process is coordinated by resident stem cells and their niche cells (Barkauskas et al., 2013; Desai et al., 2014; Hogan et al., 2014). After lung injury, these cells activate a regenerative program that involve cell proliferation and differentiation, coordinating tissue remodeling for restoring lung function (Parekh et al., 2020).
sc-seq technology have revolutionized our ability to understand complex cellular heterogeneity and address the challenge of gene expression variability by providing high-resolution analysis of individual cells (Tang et al., 2009; Amezquita et al., 2020; Kester and Van Oudenaarden, 2018; Stuart et al., 2019; Wen and Tang, 2022). This technology includes methodologies such as single-cell RNA sequencing, spatial transcriptomics, single-cell epigenomics, and proteomics. These approaches allow for multi-omics integration, offering a comprehensively understanding of cellular function and regulation (Chen et al., 2023). As a powerful tool for studying cellular heterogeneity, sc-seq has profoundly enhanced our understanding of the expression dynamics required for lung regeneration. It has identified rare cell populations, transitional cell states, and lineage-specific markers (Wang W. J. et al., 2023).
sc-seq studies have provided unprecedented insights into the spatial organization and regulation of resident stem cells, stromal cells, and immune cells during lung regeneration, elucidating the regulatory networks and signaling cascades that control responses to lung injury and repair. Signaling pathways such as Wnt/β-catenin, Notch, and Hippo/YAP have been identified as playing a critical role in regulating stem cell activation, proliferation, and differentiation during lung repair (Rock et al., 2011; Brechbuhl et al., 2011; Volckaert et al., 2017; Chung et al., 2018; Finn et al., 2019; LaCanna et al., 2019; Liu et al., 2016). To explore additional signaling pathways involved in lung development, regeneration, and post-injury repair, please refer to these reviews (Dupont et al., 2011; Kotton and Morrisey, 2014; Raslan and Yoon, 2020).
Therefore, sc-seq has significantly advanced our understanding of the cellular and molecular mechanisms underlying lung disease and regeneration. It has not only identified new cell types and subtypes associated with the pathogenesis of lung diseases, but also promoted the decoding of lung diseases and the development of regenerative therapy drugs. Knowledges obtained has been instrumental in studying diseases such as chronic obstructive pulmonary disease (COPD), idiopathic pulmonary fibrosis (IPF) and COVID-19 and provides key information for drug target discovery in the treatment lung diseases and regenerative medicine.
2 Resident epithelial stem cells and their niche cells coordinate lung regeneration and post-injury repair
Insults can directly invade the respiratory tract, leading to inflammation, tissue damage, and eventually impaired lung function. Respiratory infections such as influenza, pneumonia, and COVID-19 can have serious consequences, particularly for aging population and individuals with impaired lung function. To maintain an intact barrier, the lung possesses remarkable regenerative capacities, coordinated by resident lung stem cells and their stromal cells.
The lung contains a large number of resident epithelial stem cells, capable of proliferating and differentiating into various epithelial cell types necessary for lung repair and regeneration. These stem cell populations include those that participate in bronchial airway homeostasis and regeneration, as well as those that participate in alveolar regeneration. For more detailed information regarding these stem cell populations in the lung, please refer to review (Kotton and Morrisey, 2014; Barkauskas et al., 2017; Hogan et al., 2018). In addition, lung stromal cells, such as fibroblasts, endothelial cells, and immune cells, play a vital role in supporting the functions of stem cells by providing structural support, immunomodulation, and secreting growth factors. Studies have demonstrated that resident lung stem cells and stromal niche cells together respond to lung injury by activating regenerative programs, promoting cell proliferation, and coordinating tissue remodeling, thereby restoring lung function (Chung et al., 2018; Green et al., 2016; Zepp et al., 2017; Zepp and Morrisey, 2019). Therefore, the interactions between resident stem cells and these stromal niche cells are crucial for coordinating the complex process of lung injury repair (Figure 1). This review primarily focuses on alveolar regeneration and post-injury alveolar repair.
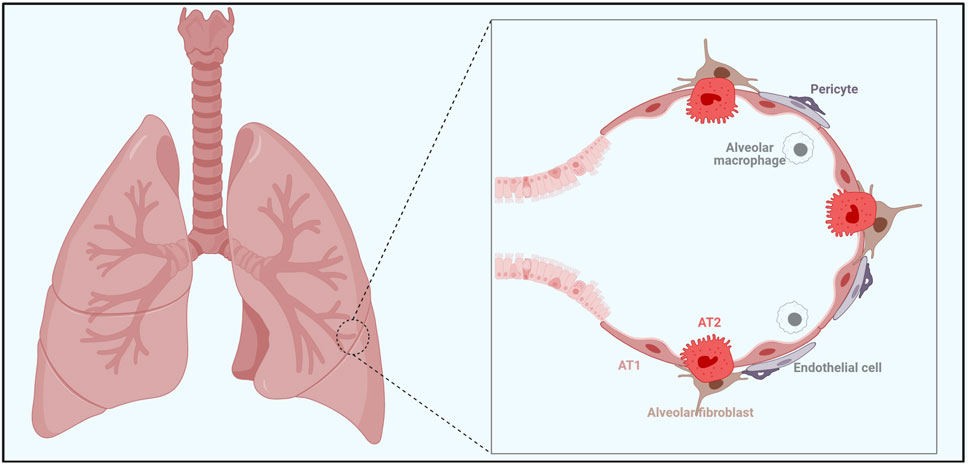
Figure 1. A complext interplay among various cellular players is crucial for maintaining alveolar homeostasis and facilitating alveolar regeneration. Alveolar epithelial cells in the alveoli of the lung consist of AT1 cells and AT2 cells. AT2 cells serve as alveolar stem cells with the ability to self-renew and differentiate into AT1 cells, especially following lung injury. Alveolar fibroblasts and pericytes provide structural support, while endothelial cells line the blood vessels, facilitating gas exchange between the alveoli and the blood vessel. The alveolar macrophages are the key player in immune defense within the alveoli. Together, all these cells form a supportive niche for AT2 cells for their stem cell activity.
2.1 Alveolar epithelial cells in alveolar function and repair
The alveolar epithelial cells contain two types: alveolar type 1 (AT1) cells and alveolar type 2 (AT2) cells. AT1 cells cover 95% of the alveolar surface area and are primarily responsible for gas exchange. AT2 cells produce pulmonary surfactant, which is essential for reducing alveolar surface tension and preventing alveolar collapse during the respiratory cycle. Studies have shown that in adults, AT2 cells are able to self-renew and differentiate into AT1 cells following lung injury (Barkauskas et al., 2013; Kathiriya et al., 2020; Desai et al., 2014). In addition, recent studies have identified that respiratory airway secretory cells (RAS) can act as unidirectional progenitors of alveolar type 2 (AT2) cells, particularly when AT2 stem cell function was impaired (Chapman, 2011; Barkauskas et al., 2013; Nabhan et al., 2018; Zacharias et al., 2018; Basil et al., 2022).
2.2 Niche cells in alveolar function and repair
Alveolar fibroblasts, immune cells, and endothelial cells function as essential niche cells in the lung, playing crucial roles in maintaining lung homeostasis and facilitating post-injury repair.
Alveolar fibroblasts are crucial for lung homeostasis and repair following injury. Under the homeostatic condition, alveolar fibroblast produce extracellular matrix (ECM) components and growth factors that maintain tissue structure and regulate the function of surrounding epithelial cells (Green et al., 2016; Barkauskas et al., 2013; Sirianni et al., 2003). However, during pulmonary fibrosis, alveolar fibroblasts differentiate into myofibroblasts, depositing excessive ECM and ultimately disrupting normal alveolar structure and function (Burgstaller et al., 2017).
Immune cells are integral in contributing to immune defense and various biological processes in both homeostatic and pathological conditions. It has been shown that immune cells participate post-injury lung repair by secreting cytokines and chemokines and through direct cell interactions (Hogan et al., 2014; Lechner et al., 2017; Danahay et al., 2015; Katsura et al., 2019; Byrne et al., 2015; Iwasaki et al., 2017). The normal functions of immune cells are vital for resolving inflammation and promoting alveolar regeneration.
Endothelial cells directly impact the tissue microenvironment by regulating vascular permeability, supplying nutrients, secreting growth factors, and participating in inflammation (Lee et al., 2014). Altered endothelial cell function can cause vascular leakage and additional tissue damage in pathological conditions such as lung injury and inflammation. In addition, factors from endothelial cells also participate the alveolar regeneration process (Ding et al., 2011; Desai et al., 2014; Rafii et al., 2015; Niethamer et al., 2020).
Taken together, multiple cellular players within alveoli, including endothelial cells, alveolar fibroblasts, and immune cells, work in concert to regulate the alveolar microenvironment, ensuring proper lung function and facilitating an efficient repair process following lung injury.
3 sc-seq technology facilitates decoding mechanisms underlying lung regeneration and diseases
Previous sequencing technologies typically analyzes the average gene expression profile of cell populations in biological samples containing millions of cells. However, they often masked the heterogeneity of gene expression between individual cells. In recent years, the newly developed sc-seq technology has revolutionized our understanding of cellular heterogeneity. sc-seq enables high-resolution analysis of single cells, addressing the issue of gene expression heterogeneity and becoming a powerful tool for studying cellular diversity. The emergence of sc-seq technology has profoundly transformed our understanding of the cellular heterogeneity and transcriptional dynamics involved in the lung regeneration process (Treutlein et al., 2014; Travaglini et al., 2020).
By analyzing gene expression at single-cell resolution, scientists have identified different cell populations and molecular features involved in lung post-injury repair, and revealed cell cross-talks that drive lung regeneration (Wang Z. et al., 2023; Wu et al., 2020; Travaglini et al., 2020; Habermann et al., 2020; Sauler et al., 2022; Xi et al., 2017). sc-seq studies provide unprecedented insights into the spatial organization of resident stem cells and their niche cells in regenerating lung tissue, highlighting the complexity of the regeneration process. Additionally, sc-seq technology can identify rare cell populations, transitional cell states, and specific lineage markers, which are crucial for understanding the cellular dynamics of lung regeneration (Figure 2) (Table 1). Combining sc-seq data with functional studies and computational analysis allows for a more comprehensive elucidation of the regulatory networks, signaling cascades, and epigenetic modifications that govern the regenerative response to lung injury.
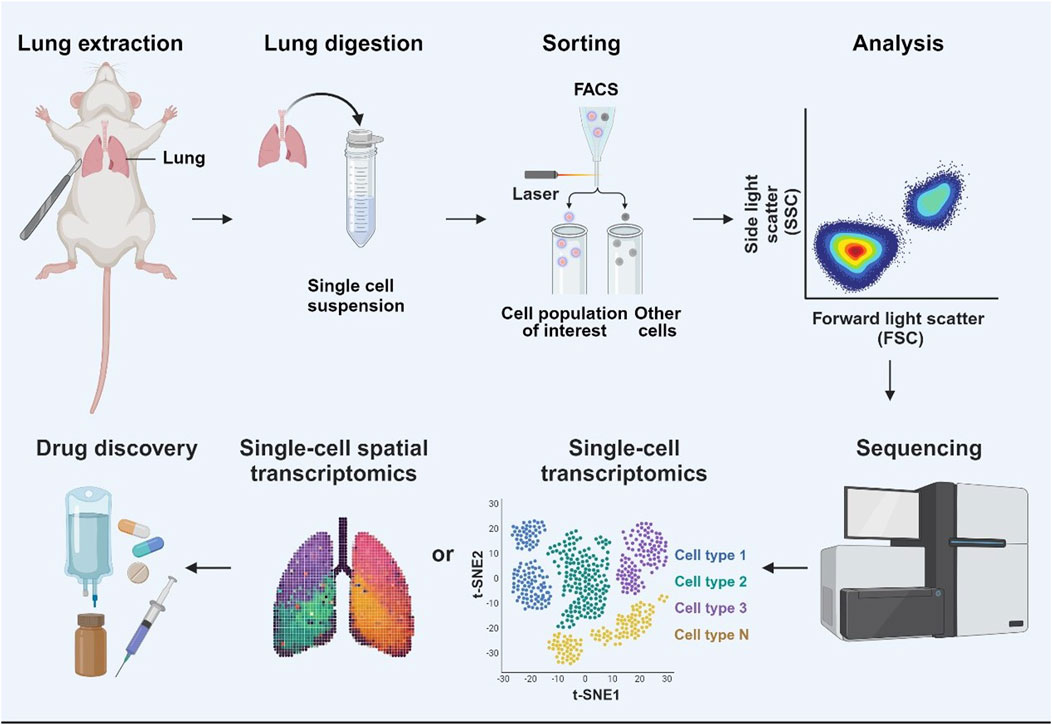
Figure 2. Single-cell sequencing (sc-seq) and its application in lung disease drug development. Sc-seq workflow for drug development in lung diseases. Lung tissue is extracted, digested, and processed into a single-cell suspension. Specific cell populations of interest in the cell suspension are isolated by FACS. These cells can be then analyzed using spatial transcriptomics to map their distribution within the tissue, and single-cell transcriptomics to classify cell types based on the gene expression profiles. The sequencing results provide a comprehensive understanding of cellular function and disease mechanisms, providing the foundation for targeted drug development in lung diseases.
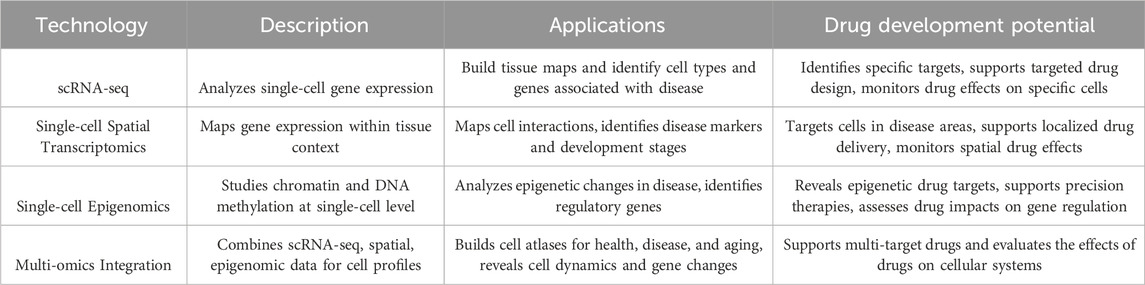
Table 1. Overview of single-cell sequencing technologies and their applications in drug development.
3.1 Single-cell RNA sequencing (scRNA-seq)
scRNA-seq enable the analysis of gene transcription at the level of individual cells, providing insights into cellular heterogeneity and the specific responses of different cell types. This technology is instrumental in identifying new cell types and subtypes in the lungs and describing their gene expression profiles.
Using scRNA-seq, researchers have constructed a molecular cell atlas of the human lung, defining the gene expression profiles of 58 cell populations in human lung tissue, which include 41 known cell types and 14 previously unknown cell types (Travaglini et al., 2020). This comprehensive effort has been instrumental in elucidating the biochemical functions and transcription factors of lung cells and their related markers. Moreover, it has enabled the identification of target cell populations and cell types affected by lung diseases. For instance, 21 genes associated with 15 diseases were identified, highlighting key cellular players in pulmonary diseases.
In addition, by comparing scRNA-seq data from humans and mice, scientists have uncovered extensive plasticity of cell types and cell type-specific gene expression during organ evolution (Jovic et al., 2022). This includes expression switching between cell types, providing a molecular basis for exploring the types, functions, and interactions of new cells in the lungs during development and disease states.
3.2 Spatial transcriptomics
Spatial transcriptomics technology maps gene expression within the context of tissues, providing spatial information about cell interactions and microenvironmental cues. This technology is particularly valuable to identify the spatial heterogeneity and cell-cell interactions during tissue regeneration and disease progression.
By utilizing single-cell RNA and ATAC sequencing, high-throughput spatial transcriptomics, and single-cell imaging, scientists have built the first multi-omics cell atlas of the developing human lung. This comprehensive dataset offering a deep understanding of human lung development (He et al., 2022). They identified 144 cell states in early life, including those intermediate cell types and transitional cell types. Among these, they discovered a cell subtype potentially associated with human small cell lung cancer that appears later in life (Han et al., 2024).
Their research also identifies genes specifically activated at different stages of lung development, uncovering new links between developmental cells and lung cancer. By integrating spatial techniques, they precisely mapped the location of cells in the developing lung and constructed a developmental lung cell atlas (Han et al., 2024). Their findings proposed the formation process of the respiratory system, highlighting the stages and signaling pathways of lung development and how these early-stage lung cells influence later disease development.
3.3 Single-cell epigenomics
Single-cell epigenomics technology studies chromatin accessibility, DNA methylation, and histone modification at the single-cell level. By analyzing these epigenetic changes, key regulatory factors or sites that control gene expression can be identified, shedding light on the molecular mechanism of cell fate determination.
In the early stages of adenocarcinoma in situ, the somatic mutations detected are mainly concentrated in changes in key driver genes and key proliferation factors. In addition, the development of adenocarcinoma in situ involves changes in gene copy number and extensive DNA hypomethylation. Recent multi-omics analysis of transposable element methylation in the progression of lung adenocarcinoma has further revealed that when DNA methylation in lung cancer cells is disordered, some transposable elements remain unmethylated (Haga et al., 2023). This finding reflects the changes in the whole genome and multimodal molecular characteristics of lung adenocarcinoma from adenocarcinoma in situ to more advanced cases. It comprehensively shows the changes in DNA, RNA and protein levels of lung adenocarcinoma from early to late stages, particularly highlighting the distribution and change characteristics of RNA and protein at the spatial level. These analyses allow for a better understanding of the molecular mechanisms underlying lung adenocarcinoma progression and the epigenetic regulation of gene expression.
Therefore, the combined use of single-cell epigenomics and other omics can comprehensively analyze the molecular-level changes and spatial phenotypic characteristics in the progression of lung diseases. This approach helps to gain a deeper understanding of the epigenetic regulation of gene expression in lung diseases and reveal new regulatory mechanisms.
3.4 Multi-omics integration
By integrating multiple omics data sets, such as scRNA-seq, single-cell epigenomics, spatial transcriptomics, and proteomics, the state of cells and regulatory interaction networks can be comprehensively described. This integrated application and analysis of multi-omics play a significant role in understanding the occurrence and progression of diseases.
DNA damage response-related genes play a key role in repairing DNA damage and maintaining genome stability, closely related to the tumor malignancy and drug sensitivity of tumors. In lung adenocarcinoma, the expression of DNA damage response-related genes is notable. Integrating bulk RNA-seq, scRNA-seq, and spatial transcriptomics can analyze the expression and functional changes of DNA damage response genes in lung adenocarcinoma at multiple levels, allowing for the screening of these DNA damage response genes during lung adenocarcinoma progression (Sun et al., 2024).
In addition, researchers have used single-cell transcriptomics and proteomics-based mass spectrometry analysis to quantify the changes in the activity status of 30 cell types in the lungs of young and old mice, creating a comprehensive atlas of lung aging (Angelidis et al., 2019). The cellular sources of regulatory proteins in the lung aging process were predicted through an integrated analysis of single-cell transcriptomes and proteomes. This analysis revealed that aging leads to increased transcriptional noise and a relaxation of epigenetic control.
Major signs of lung aging include increased cholesterol synthesis in alveolar epithelial type 2 cells and lipofibroblasts, as well as a relative increase in ciliated cells to club cell in the airway epithelium (Angelidis et al., 2019; Burgstaller et al., 2017; Jia et al., 2023; Lee et al., 2021; Penkala et al., 2021). These findings imply that the state of senescent cells and the cellular sources of regulatory proteins can be well characterized by integrated multi-omics analyses, contributing to the creation of a cellular atlas of lung aging. Taking together, these approaches offer a deeper understanding of the complex regulatory mechanisms involved in lung diseases, cancer, and aging, providing valuable insights for developing targeted therapies and interventions.
4 sc-seq reveals the regulatory molecular networks during alveolar regeneration
Knowledges of the mechanisms that coordinate cellular functions and cell-cell interactions during lung development, homeostasis, and regeneration is essential for future developing new treatments for lung diseases. The impact of sc-seq in studying lung biology is profound. This technology allows us to dissect the multi-layered biological processes and reveal regulatory signaling pathways underlying lung regeneration (Treutlein et al., 2014; Travaglini et al., 2020). By analyzing gene expression at the single-cell level, we have been able to identify key regulators of lung regeneration, such as growth factors, cytokines, and transcription factors.
Many signaling pathways and molecules have been identified as key regulators of lung regeneration after injury. Among others, the Wnt/β-catenin signaling pathway, Notch signaling pathway, Hippo/YAP pathway, are identified as key regulatory signaling pathways involved in the activation, proliferation, and differentiation of stem cells during lung regeneration. These pathways interact with transcription factors, growth factors, and extracellular matrix components to coordinate complex cellular processes necessary for lung regeneration.
4.1 Wnt/β-catenin pathway in lung regeneration
The activation of the Wnt/β-catenin pathway, through binding with Wnt receptors such as Frizzled and LRP5/6, plays a critical role in alveolar regeneration by regulating the proliferation and differentiation of alveolar stem cells, known as AT2 cells (Nabhan et al., 2023). After lung injury, the expression of Wnt genes is induced in AT2 cells to promote the alveolar repair process (Nabhan et al., 2018; Zacharias et al., 2018). Single-cell RNA sequencing has identified an Axin2+ Wnt-responsive AT2 subpopulation in the adult murine lung, accounting for 20% of the AT2 cell population and exhibit robust stem cell self-renewal and differentiation capabilities. These Axin2+ AT2 cells are referred to as alveolar epithelial progenitor cells (AEP). AEPs have a unique transcriptional profile, characterized by enriched expression of Wnt pathway genes. Following lung injuries, the AEP pool expands rapidly to regenerate the damaged alveolar epithelium by replenishing large numbers of AT2 and AT1 cells. Activating Wnt signaling leads to AEPs into AT2 cells, while inhibiting Wnt signaling suppresses AT2 cell fate and promotes AT1 cell development (Figure 3A) (Table 2).
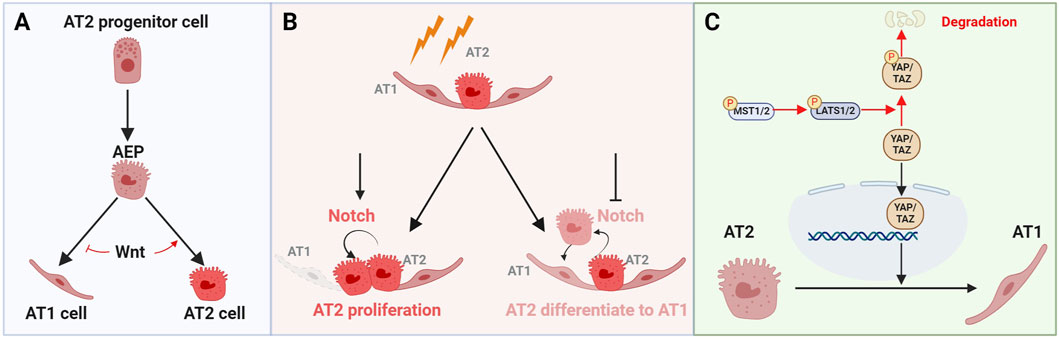
Figure 3. The roles of key signaling pathways Wnt, Notch, and Hippo in regulating the fate of alveolar epithelial cells. (A) The alveolar epithelial progenitor (AEP) promotes the proliferation of AT2 cells and their differentiation into AT1 cells. Activating Wnt signaling leads to AT2 cell proliferation, while inhibiting Wnt signaling induces AT2 cells to differentiate into AT1 cells. (B) Following lung injury, activating Notch pathway promotes AT2 cell proliferation, whereas inactivating the Notch pathway induces the differentiation of AT2 cells into AT1 cells. (C) In the Hippo signaling pathway, MST1/2 kinases activate LATS1/2 kinases, which in turn, phosphorylated YAP/TAZ. Phosphorylation YAP/TAZ leads to their degradation or retention in the cytoplasm. When YAP/TAZ are not phosphorylated, they translocate into the nucleus and promote the expression of genes that support the transformation or maintenance of AT2 cells.
scRNA-seq has also identified a novel population of RAS cells in human transitional bronchioles. These RAS cells can serve as unidirectional progenitors of AT2 cells, with Wnt signaling being a key driver of their differentiation into AT2 cells. Using an induced RAS (iRAS) cell culture system, scientists found that Wnt agonists promote the differentiation of iRAS cells into induced AT2 (iAT2) cells and activate the human AT2 gene program (Basil et al., 2020; Basil et al., 2022). This result suggests that the activation of Wnt signaling plays a critical role in driving the differentiation of RAS cells into AT2 cells. Take together, these findings underscore the vital role of Wnt signaling in the maintenance and specification of AT2 cell fate.
4.2 Notch signaling pathway in lung regeneration
The Notch signaling pathway is a highly evolutionarily conserved pathway that mediates interactions between adjacent cells. It plays an essential role in maintaining tissues and organs homeostasis by regulating variety of cellular processes, such as cell proliferation, differentiation, and apoptosis (Artavanis-Tsakonas et al., 1999; Kopan and Ilagan, 2009; Siebel and Lendahl, 2017). Delta-like noncanonical Notch ligand 1 (DLK1) has been identified as a key regulator of Notch signaling following lung injury. Inhibition of Notch signaling by DLK1 is essential for the differentiation of AT2 cells to AT1 cells during alveolar epithelial repair. Deletion of Dlk1 in AT2 cells impedes the differentiation of AT2 cells to AT1 cells (Finn et al., 2019), highlighting the necessity of DLK1-mediated Notch inhibition for proper AT2 to AT1 cell differentiation and subsequent epithelial repair (Figure 3B) (Table 2). Therefore, Notch signaling is required for effective epithelial regeneration by maintaining a balance between AT2 and AT1 cells.
It has been shown that club cells can serve as the origin of regeneration of AT2 cells after lung injury. scRNA-seq analysis of club cells and AT2 cells after lung injury has revealed that the Notch pathway plays an important role in the transdifferentiation of club cells to AT2 cells (Liu et al., 2024). Knocking out recombination signal binding protein for immunoglobulin kappa j region (Rbpj), the core transcription factor of Notch signaling in club cells, inhibits the fate transformation of club cells to AT2 cells.
Furthermore, dysregulated Notch signaling can lead to the aberrant expression of inflammatory cytokines, resulting in the development of pulmonary fibrosis (Wasnick et al., 2023). Pseudomonas aeruginosa (PA) infection can induce the activation of nuclear factor κB (NF-κB) signaling (Sadikot et al., 2006), which subsequently upregulates the expression of Notch ligands (Bash et al., 1999; Doi et al., 2006), resulting in pulmonary fibrosis (Aoyagi-Ikeda et al., 2011; Wang et al., 2019). Recent studies have found that Notch1 activation plays a key role in regulating the fate of alveolar epithelial AT2 cells. AT2 cell-specific overexpression of NICD1 leads to abnormal collagen deposition and a significant fibrotic phenotype (Wasnick et al., 2023). In summary, Notch signaling is pivotal in post-injury lung repair by orchestrating the complex pathophysiological processes involved in alveolar epithelial injury, inflammatory responses, and fibroblast proliferation.
4.3 Hippo/YAP pathway in lung regeneration
The Hippo/YAP pathway is a key signaling pathway in regulating tissue/organ development, homeostasis, and regeneration. This pathway consists of a cascade of key component kinases, including Mammalian Sterile 20-like Kinase 1/2 (MST1/2) and Large Tumor Suppressor Kinase 1/2 (LATS1/2) (Figure 3C) (Table 2). These kinases regulate the nuclear localization and activity of the Yes-associated protein 1/Tafazzin (YAP/TAZ) transcription co-activators (Chan et al., 2005; Zhao et al., 2007; Praskova et al., 2008). When the Hippo signaling pathway is activated, MST1/2 phosphorylates and activates LATS1/2, which then phosphorylates YAP/TAZ. Phosphorylated YAP/TAZ are retained in the cytoplasm for further degradation, preventing them from entering the nucleus and reducing the expression of their target genes (Dupont et al., 2011; Dong et al., 2007; Oh and Irvine, 2008; Liu et al., 2016).
scRNA-seq and mouse genetic studies have well established the essential function of the Hippo/YAP pathway in alveolar development and regeneration (LaCanna et al., 2019; Sun et al., 2019; Liu et al., 2016). After lung injury, the Hippo pathway is inactivated, allowing YAP/TAZ to avoid phosphorylation and translocate into the nucleus, regulating the expression of downstream genes, including cyclin D1 (Ccnd1), amphiregulin (Areg), cyclin-dependent kinase 6 (Ckd6), and connective tissue growth factor (Ctgf) (Zhou et al., 2018). The loss of YAP/TAZ in AT2 cells leads to reduced proliferation and impaired differentiation of AT2 cells into AT1 cells, resulting in pathological alveolar remodeling (Panciera et al., 2017; Nantie et al., 2018; van Soldt et al., 2019; LaCanna et al., 2019; Li et al., 2021; Tang et al., 2022; Penkala et al., 2021).
In summary, sc-seq not only reveals the complexity and heterogeneity of lung cells populations but also unveils the dynamic cell-cell interactions and signaling pathway critical for alveolar regeneration. By analyzing the expression of ligands and receptors in individual cells, researchers can further elucidate the crosstalk between various cell populations, such as epithelial cells, fibroblasts, and immune cells, in lung development, homeostasis, and regeneration. This helps to dissect the multi-layered biology underlying post-injury repair, providing insight into lung pathogenic mechanisms.
5 Research progress of sc-seq in lung disease
Lung diseases, including lung adenocarcinoma, IPF, COPD and COVID-19, are leading causes of high morbidity and mortality in humans worldwide. sc-seq technology, as an emerging tool for comprehensively understanding the cellular and molecular mechanisms of lung disease and regeneration, has played a significant role in further developing effective therapies for these diseases.
5.1 sc-seq analysis in lung adenocarcinoma
Lung adenocarcinoma, one of the most common types of lung cancer, involves in molecular changes in the genome, transcriptome, and epigenome of AT2 cells (Kim et al., 2005; Desai et al., 2014). The widespread application of sc-seq technology has significantly advance the study of lung adenocarcinoma. By sampling of patients with lung adenocarcinoma and employing sc-seq and spatial transcriptome, researcher can study changes in cell state and the tumor microenvironment, leading to a deeper understanding of the biological mechanism of lung adenocarcinoma.
Through scRNA-seq analysis, scientists have discovered a group of Keratin 8 (KRT8) positive alveolar intermediate cells (KACs) carrying Kirsten rat sarcoma viral oncogene homologue (KRAS) oncogene mutations (Han et al., 2024). These KRT8 positive cells are found in early lung adenocarcinoma tissues and adjacent areas. By lineage tracing of alveolar epithelial cells, it has been confirmed that these KACs can eventually transform into lung adenocarcinoma cells. This discovery reveals the heterogeneity of early lung adenocarcinoma tumor cells and highlights the unique transcriptional characteristics of KRAS mutant lung adenocarcinoma. This analysis also helps to identify the relationship between KRAS mutant tumor cells and normal epithelial cells. The scRNA-seq analysis discovered the important connection between the occurrence of lung adenocarcinoma and the plasticity of epithelial cells, providing potential molecular targets for the prevention and early intervention of lung adenocarcinoma.
5.2 sc-seq analysis in pulmonary fibrosis
Pulmonary fibrosis is characterized by extensive tissue remodeling and excessive deposition of extracellular matrix, leading to scarring and stiffening of lung tissue. Single-cell transcriptome sequencing technology has become invaluable in uncovering the intricate changes and diverse cell population involved in the pathogenesis of pulmonary fibrosis (Kobayashi et al., 2020). This technology has been extensively utilized to identify and analyze the roles and transformations of different lung cell populations in both human pulmonary fibrosis patients and mouse models of pulmonary fibrosis.
Idiopathic pulmonary fibrosis (IPF) is a severe, progressive pulmonary fibrosis, marked by continuous fibrotic changes from the edge of the lung lobes to the center of the lung lobes (Lederer and Martinez, 2018). scRNA-seq has revealed the presence of intermediate alveolar stem cells in post-injury lungs of both mice and humans. These intermediate alveolar stem cells represent a transient intermediate state during the differentiation of alveolar stem cells (AT2 cells) (Wu et al., 2020). Studies have demonstrated that intermediated alveolar stem cells accumulated in mouse lungs when stem cell differentiation is inhibited post injury. scRNA-seq has also identified the enrichment of these intermediate alveolar stem cells in IPF lungs, where they show increased expression level of various fibrotic factors. Understanding the mechanisms by which these intermediate alveolar stem cells contribute to progressive pulmonary fibrosis will provide deeper insights into IPF pathological mechanisms and suggest new therapeutic targets.
Recently, by analyzing published single-cell RNA sequencing datasets of healthy donor lungs and IPF lung fibroblasts (Habermann et al., 2020), scientists found that epidermal growth factor receptor (EGFR) signaling was significantly activated in IPF lung fibroblasts. Subsequent studies found that AREG expression levels were significantly increased in IPF lungs. Scientists then developed an anti-AREG-neutralizing antibody drug, PMG1015 to treat IPF (Zhao et al., 2024). This study demonstrated the application of scRNA-seq in discovering therapeutic targets in lung diseases.
In additional to alveolar stem cells, the single-cell transcriptomic analysis atlas of human pulmonary fibrosis has shed light on changes in various cell populations within fibrotic lungs (Habermann et al., 2020; Green et al., 2016; Zepp et al., 2017; Reyfman et al., 2019). This comprehensive analysis has highlighted the heterogeneity among epithelial cells and immune cells in fibrotic lungs. These findings also underscore the complexity of pulmonary fibrosis and greatly enhance our understanding of pathogenesis of pulmonary fibrosis. Therefore, single-cell transcriptome analysis has greatly improved our understanding of the mechanisms underlying pulmonary fibrosis and paved the way for developing therapeutic strategies aimed at alleviating fibrosis.
5.3 sc-seq analysis in COPD
Chronic obstructive pulmonary disease (COPD) is a progressive lung disease characterized by chronic airflow obstruction. It includes disorders such as emphysema and chronic bronchitis, both of which progressively diminish lung function and exacerbate breathing difficulties (Saetta et al., 2001). The pathogenesis of COPD involves complex biological processes that differ across various cell types.
A high-resolution single-cell atlas of alveolar microenvironment in COPD lungs has been constructed using scRNA-seq. The detailed atlas revealed previously unknown gene expression changes and cellular interactions among epithelial, endothelial, and macrophage populations. scRNA-seq studies on COPD lungs have also identified novel cell types and subtypes involved in the disease’s pathogenesis (Adams and Walls, 2020). For instance, analysis of lung tissues from late-stage COPD patients has revealed a subpopulation of alveolar epithelial type II cells, characterized by abnormal cellular metabolic functions and reduced stress tolerance (Sauler et al., 2022). Additionally, increased chemokine (C-X-C motif) ligand 1 (CXCL1) was observed in capillary endothelial cells of COPD patients, suggesting a role in mediating the inflammatory responses in COPD lungs. Furthermore, the researchers identified a subpopulation of macrophages that highly expressed metallothionein in advanced COPD patients.
In summary, sc-seq studies have unveiled previously unknown gene expression changes, cellular interactions and novel cell types in COPD lungs, highlighting the intricate and diverse nature of cellular injury and inflammation in the disease.
5.4 sc-seq analysis of COVID-19 lungs
The lungs of COVID-19 patients show significant infiltration of immune cells, including abnormally activated monocyte-derived macrophages and alveolar macrophages. scRNA-seq analysis of immune cells in bronchoalveolar lavage fluid (BALF) from COVID-19 patients with different severities and from healthy subjects has revealed abnormal responses of macrophages and T cells when compared to healthy subjects (Melms et al., 2021). In severe cases, BALF is rich in pro-inflammatory monocyte-derived macrophages, whereas BALF in mild cases shows an abundance of cloned CD8 positive T cells. Additionally, interleukin-1β from monocyte/macrophage- and epithelial cell-derived interleukin-6 from epithelial cells are key features distinguishing SARS-CoV-2 infection from pneumonia caused by other viruses.
Studies have found that macrophages in the BALF of COVID-19 patients highly expressed profibrotic genes, including myeloid cell receptor 2 (TREM2), transforming growth factor β1 (TGFB1), and secretory phosphoprotein 1 (SPP1) (Gustine and Jones, 2021). This suggests that macrophage accumulation in the BALF may contribute to the progression of lung tissue fibrosis, providing insight for potential targets for future therapies.
The metabolic characteristics of immune cells in COVID-19 patients are also notable. scRNA-seq has shown enhanced glycolysis as a significant metabolic characteristic of immune cells in BALF of COVID-19 patients (Shao et al., 2022). Specifically, CCL2 positive T cells, bone marrow dendritic cells, and macrophages with high expression of SPP1, which are involved in the cytokine storm, exhibit significantly upregulated metabolic levels, including glycolysis and bile acid synthesis. This indicates that lung tissue immune cells produce a large number of cytokines through metabolic regulation, thereby leading to inflammatory responses.
In summary, sc-seq provides crucial insights into the complexity and heterogeneity of lung cell populations in diseases. By analyzing the expression of ligands and receptors at the single-cell level, researchers can better understand the interactions between various cell populations in lung homeostasis, response to injury, and diseases. This approach elucidates the multifaceted biology underlying post-injury repair and enhances our understanding of lung pathogenic mechanisms.
6 Conclusion
Lung disease remains a major global health burden, underscore the urgent need for effective treatments. sc-seq has emerged as a powerful tool that provides unparalleled resolution in studying lung disease and lung regeneration. Advances in sc-seq technologies have enable researchers to uncover cellular heterogeneity, offering insights into the cellular and molecular mechanisms of lung diseases and the discovery of therapeutic targets (Regev et al., 2017). Integrated scRNA-seq with other omics datasets, including single-cell epigenomics and proteomics, allows for a comprehensive understanding of function and regulation of various cell populations (Luecken and Theis, 2019). In addition, sc-seq technology holds great promise to drug development by revealing the complex biological characteristics of the lungs at the single-cell level (Stuart et al., 2019). As new targets and pathways are identified, sc-seq can provide valuable information on signaling regulation in specific cell types, which is essential for developing drugs aimed at promoting lung regeneration and treating lung diseases (La Manno et al., 2018) (Table 2).
In the context of lung regeneration, sc-seq technology has identified key stem cell populations and signaling pathways, revealing crucial cell types and pathways involved in lung repair (Zappia et al., 2018). This technology has also been widely utilized to explore various aspects of lung diseases, leading to the identification of previously uncharacterized cell types and subtypes in the lungs, each with a unique gene expression profile and function, shedding light on their role in lung diseases (Kester and Van Oudenaarden, 2018). sc-seq has also been instrumental in studying the initiation and progression of lung diseases, such as chronic obstructive pulmonary disease and IPF. These studies have identified cell type-specific changes in gene expression and cell interactions that contribute to the pathogenesis of the disease (Zilionis et al., 2019). sc-seq studies have also discovered potential molecular pathways dysregulated in abnormal cells in lung diseases, facilitating the discovery of new therapeutic targets for lung diseases, paving the way for targeted therapy of lung diseases and lung regeneration strategies.
Despite its great potential in drug development for lung diseases, sc-seq technology still faces challenges such as high cost, data analysis complexity, and technical variability. To address these challenges, future study should also focus on integrating sc-seq data with other holographic datasets and functional assays, therefore to validate and translate these findings into clinical applications (Amezquita et al., 2020). By overcoming these obstacles, sc-seq can achieve its full potential in lung regenerative medicine (Zheng et al., 2017), offering new avenues for effective treatments and improved patient outcomes.
Author contributions
PW: Funding acquisition, Writing–original draft, Writing–review and editing. NT: Data curation, Funding acquisition, Supervision, Writing–original draft, Writing–review and editing, Methodology, Project administration, Resources.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the National Natural Science Foundation of China (32400698) and China Postdoctoral Science Foundation (2022M720458).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adams, J. G., and Walls, R. M. (2020). Supporting the health care workforce during the COVID-19 global epidemic. Jama 323 (15), 1439–1440. doi:10.1001/jama.2020.3972
Amezquita, R. A., Lun, A. T. L., Becht, E., Carey, V. J., Carpp, L. N., Geistlinger, L., et al. (2020). Orchestrating single-cell analysis with Bioconductor. Nat. Methods 17 (2), 137–145. doi:10.1038/s41592-019-0654-x
Angelidis, I., Simon, L. M., Fernandez, I. E., Strunz, M., Mayr, C. H., Greiffo, F. R., et al. (2019). An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat. Commun. 10 (1), 963. doi:10.1038/s41467-019-08831-9
Aoyagi-Ikeda, K., Maeno, T., Matsui, H., Ueno, M., Hara, K., Aoki, Y., et al. (2011). Notch induces myofibroblast differentiation of alveolar epithelial cells via transforming growth factor-{β}-Smad3 pathway. Am. J. Respir. Cell Mol. Biol. 45 (1), 136–144. doi:10.1165/rcmb.2010-0140oc
Artavanis-Tsakonas, S., Rand, M. D., and Lake, R. J. (1999). Notch signaling: cell fate control and signal integration in development. Science 284 (5415), 770–776. doi:10.1126/science.284.5415.770
Barkauskas, C. E., Chung, M.-I., Fioret, B., Gao, X., Katsura, H., and Hogan, B. L. M. (2017). Lung organoids: current uses and future promise. Development 144 (6), 986–997. doi:10.1242/dev.140103
Barkauskas, C. E., Cronce, M. J., Rackley, C. R., Bowie, E. J., Keene, D. R., Stripp, B. R., et al. (2013). Type 2 alveolar cells are stem cells in adult lung. J. Clin. Investigation 123 (7), 3025–3036. doi:10.1172/JCI68782
Bash, J., Zong, W., Banga, S., Rivera, A., Ballard, D. W., Ron, Y., et al. (1999). Rel/NF-kappaB can trigger the Notch signaling pathway by inducing the expression of Jagged1, a ligand for Notch receptors. EMBO J. 18, 2803–2811. doi:10.1093/emboj/18.10.2803
Basil, M. C., Cardenas-Diaz, F. L., Kathiriya, J. J., Morley, M. P., Carl, J., Brumwell, A. N., et al. (2022). Human distal airways contain a multipotent secretory cell that can regenerate alveoli. Nature 604 (7904), 120–126. doi:10.1038/s41586-022-04552-0
Basil, M. C., Katzen, J., Engler, A. E., Guo, M., Herriges, M. J., Kathiriya, J. J., et al. (2020). The cellular and physiological basis for lung repair and regeneration: past, present, and future. Cell Stem Cell 26 (4), 482–502. doi:10.1016/j.stem.2020.03.009
Brechbuhl, H. M., Ghosh, M., Smith, M. K., Smith, R. W., Li, B., Hicks, D. A., et al. (2011). β-catenin dosage is a critical determinant of tracheal basal cell fate determination. Am. J. Pathology 179 (1), 367–379. doi:10.1016/j.ajpath.2011.03.016
Burgstaller, G., Oehrle, B., Gerckens, M., White, E. S., Schiller, H. B., and Eickelberg, O. (2017). The instructive extracellular matrix of the lung: basic composition and alterations in chronic lung disease. Eur. Respir. J. 50 (1), 1601805. doi:10.1183/13993003.01805-2016
Byrne, A. J., Mathie, S. A., Gregory, L. G., and Lloyd, C. M. (2015). Pulmonary macrophages: key players in the innate defence of the airways. Thorax 70 (12), 1189–1196. doi:10.1136/thoraxjnl-2015-207020
Chan, E. H. Y., Nousiainen, M., Chalamalasetty, R. B., Schäfer, A., Nigg, E. A., and Sillje, H. H. W. (2005). The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene 24 (12), 2076–2086. doi:10.1038/sj.onc.1208445
Chapman, H. A. (2011). Epithelial-mesenchymal interactions in pulmonary fibrosis. Annu. Rev. Physiology 73 (1), 413–435. doi:10.1146/annurev-physiol-012110-142225
Chen, C., Wang, J., Pan, D., Wang, X., Xu, Y., Yan, J., et al. (2023). Applications of multi-omics analysis in human diseases. MedComm 4 (4), e315. doi:10.1002/mco2.315
Chung, M.-I., Bujnis, M., Barkauskas, C. E., Kobayashi, Y., and Hogan, B. L. M. (2018). Niche-mediated BMP/SMAD signaling regulates lung alveolar stem cell proliferation and differentiation. Development 145 (9), dev163014. doi:10.1242/dev.163014
Danahay, H., Pessotti, A. D., Coote, J., Montgomery, B. E., Xia, D., Wilson, A., et al. (2015). Notch2 is required for inflammatory cytokine-driven goblet cell metaplasia in the lung. Cell Rep. 10 (2), 239–252. doi:10.1016/j.celrep.2014.12.017
Desai, T. J., Brownfield, D. G., and Krasnow, M. A. (2014). Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature 507 (7491), 190–194. doi:10.1038/nature12930
Ding, B.-S., Nolan, D. J., Guo, P., Babazadeh, A. O., Cao, Z., Rosenwaks, Z., et al. (2011). Endothelial-derived angiocrine signals induce and sustain regenerative lung alveolarization. Cell 147 (3), 539–553. doi:10.1016/j.cell.2011.10.003
Dong, J., Feldmann, G., Huang, J., Wu, S., Zhang, N., Comerford, S. A., et al. (2007). Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130 (6), 1120–1133. doi:10.1016/j.cell.2007.07.019
Doi, H., Iso, T., Sato, H., Yamazaki, M., Matsui, H., Tanaka, T., et al. (2006). Jagged1-selective notch signaling induces smooth muscle differentiation via a RBP-Jkappa-dependent pathway. J. Biol. Chem. 281 (39), 28555–64.
Dupont, S., Morsut, L., Aragona, M., Enzo, E., Giulitti, S., Cordenonsi, M., et al. (2011). Role of YAP/TAZ in mechanotransduction. Nature 474 (7350), 179–183. doi:10.1038/nature10137
Finn, J., Sottoriva, K., Pajcini, K. V., Kitajewski, J. K., Chen, C., Zhang, W., et al. (2019). Dlk1-mediated temporal regulation of notch signaling is required for differentiation of alveolar type II to type I cells during repair. Cell Rep. 26 (11), 2942–2954. doi:10.1016/j.celrep.2019.02.046
Green, J., Endale, M., Auer, H., and Perl, A.-K. T. (2016). Diversity of interstitial lung fibroblasts is regulated by platelet-derived growth factor receptor α kinase activity. Am. J. Respir. Cell Mol. Biol. 54 (4), 532–545. doi:10.1165/rcmb.2015-0095OC
Gustine, J. N., and Jones, D. (2021). Immunopathology of hyperinflammation in COVID-19. Am. J. Pathology 191 (1), 4–17. doi:10.1016/j.ajpath.2020.08.009
Habermann, A. C., Gutierrez, A. J., Bui, L. T., Yahn, S. L., Winters, N. I., Calvi, C. L., et al. (2020). Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci. Adv. 6 (28), eaba1972. doi:10.1126/sciadv.aba1972
Haga, Y., Sakamoto, Y., Kajiya, K., Kawai, H., Oka, M., Motoi, N., et al. (2023). Whole-genome sequencing reveals the molecular implications of the stepwise progression of lung adenocarcinoma. Nat. Commun. 14 (1), 8375. doi:10.1038/s41467-023-43732-y
Han, G., Sinjab, A., Rahal, Z., Lynch, A. M., Treekitkarnmongkol, W., Liu, Y., et al. (2024). Author Correction: an atlas of epithelial cell states and plasticity in lung adenocarcinoma. Nature 628, 1–8. doi:10.1038/s41586-024-07277-4
He, P., Lim, K., Sun, D., Pett, J. P., Jeng, Q., Polanski, K., et al. (2022). A human fetal lung cell atlas uncovers proximal-distal gradients of differentiation and key regulators of epithelial fates. Cell 185 (25), 4841–4860.e25. doi:10.1016/j.cell.2022.11.005
Hogan, B., Tata, P. R., Choo-Wing, R., Savova, V., Knehr, J., Roma, G., et al. (2018). A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 560, 377–381. doi:10.1038/s41586-018-0394-6
Hogan, B. L. M., Barkauskas, C. E., Chapman, H. A., Epstein, J. A., Jain, R., Hsia, C. C. W., et al. (2014). Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell 15 (2), 123–138. doi:10.1016/j.stem.2014.07.012
Iwasaki, A., Foxman, E. F., and Molony, R. D. (2017). Early local immune defences in the respiratory tract. Nat. Rev. Immunol. 17 (1), 7–20. doi:10.1038/nri.2016.117
Jia, M., Agudelo Garcia, P. A., Ovando-Ricardez, J. A., Tabib, T., Bittar, H. T., Lafyatis, R. A., et al. (2023). Transcriptional changes of the aging lung. Aging Cell 22 (10), e13969. doi:10.1111/acel.13969
Jovic, D., Liang, X., Zeng, H., Lin, L., Xu, F., and Luo, Y. (2022). Single cell RNA sequencing technologies and applications: a brief overview. Clin. Transl. Med. 12 (3), e694. doi:10.1002/ctm2.694
Kathiriya, J. J., Brumwell, A. N., Jackson, J. R., Tang, X., and Chapman, H. A. (2020). Distinct airway epithelial stem cells hide among club cells but mobilize to promote alveolar regeneration. Cell Stem Cell 26 (3), 346–358. doi:10.1016/j.stem.2019.12.014
Katsura, H., Kobayashi, Y., Tata, P. R., and Hogan, B. L. M. (2019). IL-1 and TNFα contribute to the inflammatory niche to enhance alveolar regeneration. Stem Cell Rep. 12 (4), 657–666. doi:10.1016/j.stemcr.2019.02.013
Kester, L., and Van Oudenaarden, A. (2018). Single-cell transcriptomics meets lineage tracing. Cell Stem Cell 23 (2), 166–179. doi:10.1016/j.stem.2018.04.014
Kim, C. F. B., Jackson, E. L., Woolfenden, A. E., Lawrence, S., Babar, I., Vogel, S., et al. (2005). Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 121 (6), 823–835. doi:10.1016/j.cell.2005.03.032
Kobayashi, Y., Tata, A., Konkimalla, A., Katsura, H., Lee, R. F., Ou, J., et al. (2020). Persistence of a regeneration-associated, transitional alveolar epithelial cell state in pulmonary fibrosis. Nat. Cell Biol. 22 (8), 934–946. doi:10.1038/s41556-020-0542-8
Kopan, R., and Ilagan, M. X. G. (2009). The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137 (2), 216–233. doi:10.1016/j.cell.2009.03.045
Kotton, D. N., and Morrisey, E. E. (2014). Lung regeneration: mechanisms, applications and emerging stem cell populations. Nat. Med. 20 (8), 822–832. doi:10.1038/nm.3642
LaCanna, R., Liccardo, D., Zhang, P., Tragesser, L., Wang, Y., Cao, T., et al. (2019). Yap/Taz regulate alveolar regeneration and resolution of lung inflammation. J. Clin. Investigation 129 (5), 2107–2122. doi:10.1172/JCI125014
La Manno, G., Soldatov, R., Zeisel, A., Braun, E., Hochgerner, H., Petukhov, V., et al. (2018). RNA velocity of single cells. Nature 560 (7719), 494–498. doi:10.1038/s41586-018-0414-6
Lechner, A. J., Driver, I. H., Lee, J., Conroy, C. M., Nagle, A., Locksley, R. M., et al. (2017). Recruited monocytes and type 2 immunity promote lung regeneration following pneumonectomy. Cell Stem Cell 21 (1), 120–134. doi:10.1016/j.stem.2017.03.024
Lederer, D. J., and Martinez, F. J. (2018). Idiopathic pulmonary fibrosis. N. Engl. J. Med. 378 (19), 1811–1823. doi:10.1056/NEJMra1705751
Lee, J.-H., Bhang, D. H., Beede, A., Huang, T. L., Stripp, B. R., Bloch, K. D., et al. (2014). Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis. Cell 156 (3), 440–455. doi:10.1016/j.cell.2013.12.039
Lee, S., Islam, M. N., Boostanpour, K., Aran , D., Jin, G., Christenson, S., et al. (2021). Molecular programs of fibrotic change in aging human lung. Nat Commun. 12 (1), 6309.
Li, R., Li, X., Hagood, J., Zhu, M.-S., and Sun, X. (2021). Myofibroblast contraction is essential for generating and regenerating the gas-exchange surface. J. Clin. Investigation 130 (6), 2859–2871. doi:10.1172/JCI132189
Liu, K., Meng, X., Liu, Z., Tang, M., Lv, Z., Huang, X., et al. (2024). Tracing the origin of alveolar stem cells in lung repair and regeneration. Cell 187 (10), 2428–2445.e20. doi:10.1016/j.cell.2024.03.010
Liu, Z., Wu, H., Jiang, K., Wang, Y., Zhang, W., Chu, Q., et al. (2016). MAPK-mediated YAP activation controls mechanical-tension-induced pulmonary alveolar regeneration. Cell Rep. 16 (7), 1810–1819. doi:10.1016/j.celrep.2016.07.020
Luecken, M. D., and Theis, F. J. (2019). Current best practices in single cell RNA seq analysis: a tutorial. Mol. Syst. Biol. 15 (6), e8746. doi:10.15252/msb.20188746
Melms, J. C., Biermann, J., Huang, H., Wang, Y., Nair, A., Tagore, S., et al. (2021). A molecular single-cell lung atlas of lethal COVID-19. Nature 595 (7865), 114–119. doi:10.1038/s41586-021-03569-1
Nabhan, A. N., Brownfield, D. G., Harbury, P. B., Krasnow, M. A., and Desai, T. J. (2018). Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science 359 (6380), 1118–1123. doi:10.1126/science.aam6603
Nabhan, A. N., Webster, J. D., Adams, J. J., Blazer, L., Everrett, C., Eidenschenk, C., et al. (2023). Targeted alveolar regeneration with Frizzled-specific agonists. Cell 186 (14), 2995–3012.e15. doi:10.1016/j.cell.2023.05.022
Nantie, L. B., Young, R. E., Paltzer, W. G., Zhang, Y., Johnson, R. L., Verheyden, J. M., et al. (2018). Lats1/2 inactivation reveals Hippo function in alveolar type I cell differentiation during lung transition to air breathing. Development 145 (21), dev163105. doi:10.1242/dev.163105
Niethamer, T. K., Stabler, C. T., Leach, J. P., Zepp, J. A., Morley, M. P., Babu, A., et al. (2020). Defining the role of pulmonary endothelial cell heterogeneity in the response to acute lung injury. Elife 9, e53072. doi:10.7554/eLife.53072
Oh, H., and Irvine, K. D. (2008). In vivo regulation of Yorkie phosphorylation and localization. Develop. 135(6):1081–8. doi:10.1242/dev.015255
Panciera, T., Azzolin, L., Cordenonsi, M., and Piccolo, S. (2017). Mechanobiology of YAP and TAZ in physiology and disease. Nat. Rev. Mol. Cell Biol. 18 (12), 758–770. doi:10.1038/nrm.2017.87
Parekh, K. R., Nawroth, J., Pai, A., Busch, S. M., Senger, C. N., and Ryan, A. L. (2020). Stem cells and lung regeneration. Am. J. Physiology-Cell Physiology 319 (4), C675-C693–C693. doi:10.1152/ajpcell.00036.2020
Penkala, I. J., Liberti, D. C., Pankin, J., Sivakumar, A., Kremp, M. M., Jayachandran, S., et al. (2021). Age-dependent alveolar epithelial plasticity orchestrates lung homeostasis and regeneration. Cell Stem Cell 28 (10), 1775–1789.e5. doi:10.1016/j.stem.2021.04.026
Praskova, M., Xia, F., and Avruch, J. (2008). MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr. Biol. 18 (5), 311–321. doi:10.1016/j.cub.2008.02.006
Rafii, S., Cao, Z., Lis, R., Siempos, I. I., Chavez, D., Shido, K., et al. (2015). Platelet-derived SDF-1 primes the pulmonary capillary vascular niche to drive lung alveolar regeneration. Nat. Cell Biol. 17 (2), 123–136. doi:10.1038/ncb3096
Raslan, A. A., and Yoon, J. K. (2020). WNT signaling in lung repair and regeneration. Mol. Cells 43 (9), 774–783. doi:10.14348/molcells.2020.0059
Regev, A., Teichmann, S. A., Lander, E. S., Amit, I., Benoist, C., Birney, E., et al. (2017). The human cell atlas. Elife 6, e27041. doi:10.7554/eLife.27041
Reyfman, P. A., Walter, J. M., Joshi, N., Anekalla, K. R., McQuattie-Pimentel, A. C., Chiu, S., et al. (2019). Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 199 (12), 1517–1536. doi:10.1164/rccm.201712-2410OC
Rock, J. R., Gao, X., Xue, Y., Randell, S. H., Kong, Y.-Y., and Hogan, B. L. M. (2011). Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell 8 (6), 639–648. doi:10.1016/j.stem.2011.04.003
Sadikot, R. T., Zeng, H., Joo, M., Everhart, M. B., Sherrill, T. P., Li, B., et al. (2006). Targeted immunomodulation of the NF-kappaB pathway in airway epithelium impacts host defense against Pseudomonas aeruginosa. J. Immunol. 176 (8), 4923–4930. doi:10.4049/jimmunol.176.8.4923
Saetta, M., Turato, G., Maestrelli, P., Mapp, C. E., and Fabbri, L. M. (2001). Cellular and structural bases of chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 163 (6), 1304–1309. doi:10.1164/ajrccm.163.6.2009116
Sauler, M., McDonough, J. E., Adams, T. S., Kothapalli, N., Barnthaler, T., Werder, R. B., et al. (2022). Characterization of the COPD alveolar niche using single-cell RNA sequencing. Nat. Commun. 13 (1), 494. doi:10.1038/s41467-022-28062-9
Shao, M.-M., Shi, M., Du, J., Pei, X.-B., Gu, B.-B., and Yi, F.-S. (2022). Metabolic landscape of bronchoalveolar lavage fluid in coronavirus disease 2019 at single cell resolution. Front. Immunol. 13, 829760. doi:10.3389/fimmu.2022.829760
Siebel, C., and Lendahl, U. (2017). Notch signaling in development, tissue homeostasis, and disease. Physiol. Rev. 97 (4), 1235–1294. doi:10.1152/physrev.00005.2017
Sirianni, F. E., Chu, F. S. F., and Walker, D. C. (2003). Human alveolar wall fibroblasts directly link epithelial type 2 cells to capillary endothelium. Am. J. Respir. Crit. Care Med. 168 (12), 1532–1537. doi:10.1164/rccm.200303-371OC
Stuart, T., Butler, A., Hoffman, P., Hafemeister, C., Papalexi, E., Mauck, W. M., et al. (2019). Comprehensive integration of single-cell data. Cell 177 (7), 1888–1902. doi:10.1016/j.cell.2019.05.031
Sun, S., Wang, K., Guo, D., Zheng, H., Liu, Y., Shen, H., et al. (2024). Identification of the key DNA damage response genes for predicting immunotherapy and chemotherapy efficacy in lung adenocarcinoma based on bulk, single-cell RNA sequencing, and spatial transcriptomics. Comput. Biol. Med. 171, 108078. doi:10.1016/j.compbiomed.2024.108078
Sun, T., Huang, Z., Zhang, H., Posner, C., Jia, G., Ramalingam, T. R., et al. (2019). TAZ is required for lung alveolar epithelial cell differentiation after injury. JCI Insight 4 (14), e128674. doi:10.1172/jci.insight.128674
Tang, F., Barbacioru, C., Wang, Y., Nordman, E., Lee, C., Xu, N., et al. (2009). mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6 (5), 377–382. doi:10.1038/nmeth.1315
Tang, W., Li, M., Yangzhong, X., Zhang, X., Zu, A., Hou, Y., et al. (2022). Hippo signaling pathway and respiratory diseases. Cell Death Discov. 8 (1), 213. doi:10.1038/s41420-022-01020-6
Travaglini, K. J., Nabhan, A. N., Penland, L., Sinha, R., Gillich, A., Sit, R. V., et al. (2020). A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 587 (7835), 619–625. doi:10.1038/s41586-020-2922-4
Treutlein, B., Brownfield, D. G., Wu, A. R., Neff, N. F., Mantalas, G. L., Espinoza, F. H., et al. (2014). Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509 (7500), 371–375. doi:10.1038/nature13173
van Soldt, B. J., Qian, J., Li, J., Tang, N., Lu, J., and Cardoso, W. V. (2019). Yap and its subcellular localization have distinct compartment-specific roles in the developing lung. Development 146 (9), dev175810. doi:10.1242/dev.175810
Volckaert, T., Yuan, T., Chao, C.-M., Bell, H., Sitaula, A., Szimmtenings, L., et al. (2017). Fgf10-Hippo epithelial-mesenchymal crosstalk maintains and recruits lung basal stem cells. Dev. Cell 43 (1), 48–59. doi:10.1016/j.devcel.2017.09.003
Wang, W.-J., Chu, L.-X., He, L.-Y., Zhang, M.-J., Dang, K.-T., Gao, C., et al. (2023). Spatial transcriptomics: recent developments and insights in respiratory research. Mil. Med. Res. 10 (1), 38. doi:10.1186/s40779-023-00471-x
Wang, Y.-C., Chen, Q., Luo, J.-M., Nie, J., Meng, Q.-H., Shuai, W., et al. (2019). Notch1 promotes the pericyte-myofibroblast transition in idiopathic pulmonary fibrosis through the PDGFR/ROCK1 signal pathway. Exp. and Mol. Med. 51 (3), 1–11. doi:10.1038/s12276-019-0228-0
Wang, Z., Wei, D., Bin, E., Li, J., Jiang, K., Lv, T., et al. (2023). Enhanced glycolysis-mediated energy production in alveolar stem cells is required for alveolar regeneration. Cell Stem Cell 30 (8), 1028–1042.e7. doi:10.1016/j.stem.2023.07.007
Wasnick, R., Korfei, M., Piskulak, K., Henneke, I., Wilhelm, J., Mahavadi, P., et al. (2023). Notch1 induces defective epithelial surfactant processing and pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 207 (3), 283–299. doi:10.1164/rccm.202105-1284OC
Wen, L., and Tang, F. (2022). Recent advances in single-cell sequencing technologies. Precis. Clin. Med. 5 (1), pbac002. doi:10.1093/pcmedi/pbac002
Wu, H., Yu, Y., Huang, H., Hu, Y., Fu, S., Wang, Z., et al. (2020). Progressive pulmonary fibrosis is caused by elevated mechanical tension on alveolar stem cells. Cell 180 (1), 107–121. doi:10.1016/j.cell.2019.11.027
Xi, Y., Kim, T., Brumwell, A. N., Driver, I. H., Wei, Y., Tan, V., et al. (2017). Local lung hypoxia determines epithelial fate decisions during alveolar regeneration. Nat. Cell Biol. 19 (8), 904–914. doi:10.1038/ncb3580
Zacharias, W. J., Frank, D. B., Zepp, J. A., Morley, M. P., Alkhaleel, F. A., Kong, J., et al. (2018). Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 555 (7695), 251–255. doi:10.1038/nature25786
Zappia, L., Phipson, B., and Oshlack, A. (2018). Exploring the single-cell RNA-seq analysis landscape with the scRNA-tools database. PLoS Comput. Biol. 14 (6), e1006245. doi:10.1371/journal.pcbi.1006245
Zepp, J. A., and Morrisey, E. E. (2019). Cellular crosstalk in the development and regeneration of the respiratory system. Nat. Rev. Mol. Cell Biol. 20 (9), 551–566. doi:10.1038/s41580-019-0141-3
Zepp, J. A., Zacharias, W. J., Frank, D. B., Cavanaugh, C. A., Zhou, S., Morley, M. P., et al. (2017). Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell 170 (6), 1134–1148. doi:10.1016/j.cell.2017.07.034
Zhao, B., Wei, X., Li, W., Udan, R. S., Yang, Q., Kim, J., et al. (2007). Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes and Dev. 21 (21), 2747–2761. doi:10.1101/gad.1602907
Zhao, R., Wang, Z., Wang, G., Geng, J., Wu, H., Liu, X., et al. (2024). Sustained amphiregulin expression in intermediate alveolar stem cells drives progressive fibrosis. Cell Stem Cell 31 (9), 1344–1358.e6. doi:10.1016/j.stem.2024.07.004
Zhou, B., Flodby, P., Luo, J., Castillo, D. R., Liu, Y., Yu, F. X., et al. (2018). Claudin-18-mediated YAP activity regulates lung stem and progenitor cell homeostasis and tumorigenesis. J. Clin. Invest. 128 (3), 970–984.
Zheng, G. X. Y., Terry, J. M., Belgrader, P., Ryvkin, P., Bent, Z. W., Wilson, R., et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8 (1), 14049. doi:10.1038/ncomms14049
Keywords: single-cell sequencing, lung diseases, lung regeneration, drug discovery, lung complexity
Citation: Wang P and Tang N (2024) Decoding lung complexity: single-cell sequencing in lung diseases, regeneration, and drug discovery. Front. Drug Discov. 4:1495208. doi: 10.3389/fddsv.2024.1495208
Received: 12 September 2024; Accepted: 04 November 2024;
Published: 19 November 2024.
Edited by:
Linheng Li, Stowers Institute for Medical Research, United StatesReviewed by:
Carlos A Méndez-Cuesta, Universidad Autónoma Metropolitana, MexicoXu Ma, University of California, Santa Barbara, United States
Copyright © 2024 Wang and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nan Tang, dGFuZ25hbkBuaWJzLmFjLmNu
 Peng Wang
Peng Wang Nan Tang
Nan Tang