- 1Department of Systems Biology, Beckman Research Institute of City of Hope, Monrovia, CA, United States
- 2City of Hope Comprehensive Cancer Center, City of Hope, Duarte, CA, United States
- 3Gehr Family Center for Leukemia Research, City of Hope, Duarte, CA, United States
Cancer stem cells (CSCs) are a small population of less differentiated cells with robust self-renewal ability. CSCs have been recognized as the root cause of tumor initiation, progression, relapse, and drug resistance. Recent studies from us and others have highlighted that N6-methyladenosine (m6A), the most prevalent modification in mRNA, plays a crucial role in carcinogenesis and CSC homeostasis. Dysregulation of the m6A modification machinery has been implicated in CSC survival and self-renewal, thereby regulating cancer progression and therapeutic resistance. In this review, we provide an overview of the roles and molecular mechanisms of the RNA m6A modification machinery in CSC survival and self-renewal. Additionally, we summarize the currently known small-molecule inhibitors targeting the dysregulated m6A modification machinery and discuss proof-of-concept studies focusing on the efficacy of these compounds in eliminating CSCs and cancers.
1 Introduction
Over the past decade, RNA modifications have attracted widespread attention not only because these chemical modifications play crucial roles in determining the fate of various RNA types but also because the dysregulation of RNA modifications can lead to pathogenesis, including tumorigenesis (Barbieri and Kouzarides, 2020). Thus far, over 170 distinct chemical modifications have been identified. Current methods for mapping and sequencing RNA and its modifications - also known as the epitranscriptome–have revealed the landscape of RNA modifications, including N6-methyladenosine (m6A), pseudouridine (Ψ), 5-methylcytidine (m5C), N1-methyladenosine (m1A), and N7-methylguanosine (m7G). Among these chemical modifications, m6A is the most abundant internal mRNA modification and is also present in other RNA types, such as long non-coding RNA (lncRNA) (Fazi and Fatica, 2019). m6A was first reported in bacterial DNA as early as 1955 (Dunn and Smith, 1955) and was later identified in the mRNA of mammalian cells in 1974 (Perry and Kelley, 1974). However, the potential biological functions of m6A methylation in determining mRNA fate were not appreciated until 2011 when fat mass and obesity associated (FTO) protein was characterized as a demethylase of RNA m6A modification (Jia et al., 2011). This identification suggests that akin to epigenetic modifications at DNA and histone levels, RNA modification is also reversible and dynamic. Subsequently, transcriptome-wide m6A mapping revealed that “DRACH” (D = A, G or U; H = A, C or U) as the canonical motif of mRNA m6A modification (Dominissini et al., 2012; Meyer et al., 2012). Since then, a significant wave of studies has focused on characterizing the m6A machinery, including “writers”, “erasers”, and “readers”, and determining their biological functions in various biological contexts. Moreover, considering the crucial role of m6A modification in tumorigenesis, several small-molecule inhibitors targeting m6A machinery have been developed. Proof-of-concept studies have shown that pharmacological inhibition of the dysregulated m6A machinery exhibits robust anti-tumor efficacy.
Cancer stem cells (CSCs), similar to normal stem cells in healthy tissue, are a small population of tumor cells with a potent self-renewal ability and the potential to differentiate into multiple cellular subtypes. While the concept of CSCs was proposed in the 1970s, these cells were not identified until the 1990s, initially in leukemia (Lapidot et al., 1994). Subsequently, CSCs have been definitively identified in many other tumor types, including breast cancer (Al-Hajj et al., 2003), brain cancer (Singh et al., 2004), liver cancer (Ma et al., 2007), colon cancer (O’Brien et al., 2007), lung cancer (Eramo et al., 2008), pancreatic cancer (Li et al., 2007), and prostate cancer (Patrawala et al., 2006; Dalerba et al., 2007; Walcher et al., 2020; Yang et al., 2020b; Zhou H.-M. et al., 2021). The CSC hypothesis suggests that many cancers are not merely monoclonal expansions of cells but may actually resemble abnormal organs, sustained by a diseased stem cell population. CSCs can initiate tumorigenesis and cause drug resistance, metastasis, and relapse, often leading to the failure of conventional cancer treatments (Clarke, 2019). Consequently, targeting CSCs represents one of the most promising strategies to eliminate or potentially cure cancers.
Current studies have reported that dysregulation of m6A modification is strongly implicated in modulating tumor stemness and CSC maintenance (Ma and Ji, 2020; Kumari et al., 2023; Feng et al., 2024). Better understanding the biological functions of RNA m6A modification and machinery could substantially advance the development of innovative and effective strategies to eradicate CSCs, marking a new frontier in cancer treatment. Herein, we review the molecular mechanisms of RNA m6A modifications in post-transcriptional regulation of gene expression. Additionally, we provide an overview of the implication of m6A modification in CSC survival and self-renewal. Furthermore, we summarize current knowledge on small-molecule inhibitors targeting the dysregulated m6A modification machinery as well as the anti-tumor efficacy of these inhibitors.
2 Fundamental mechanisms and biological functions of m6A modification
m6A modification is installed onto mRNAs at the consensus DRACH motifs by m6A methyltransferases (called “writers”), removed by m6A demethylases (called “erasers”), and recognized by “readers” that directly recognize the modification and mediate its downstream impacts and biological functions. Transcriptome-wide mapping of m6A sites revealed a biased distribution of m6A modification on mRNAs–it is usually enriched in the 3′-untranslated region (3′UTR) and near the stop codon area of mRNAs (Meyer et al., 2012), whereas the distribution of the DRACH motifs is relatively even. m6A modification is involved in almost all stages of mRNA metabolism and gene expression regulation, including transcriptional regulation, alternative splicing, mRNA export, mRNA turnover, and translation (Shi et al., 2019; Deng et al., 2023; Qing et al., 2023).
2.1 The m6A “writers”
The deposition of most m6A sites on mRNA is catalyzed by the m6A methyltransferase complex (MTC) (Liu et al., 2014) (Figure 1). This complex contains methyltransferase like 3 (METTL3), METTL14, and Wilms tumor 1-associating protein (WTAP) (Ping et al., 2014). As the first discovered m6A writer, METTL3 serves as the primary catalytic subunit and is implicated in various physiological processes, including controlling the speed of the circadian clock (Fustin et al., 2013), embryogenesis (Wang Y. et al., 2014), the DNA damage response (Xiang et al., 2017), spermatogenesis (Xu et al., 2017), immune cell homeostasis (Li et al., 2017), and tumorigenesis (Deng et al., 2023). Although METTL14 is a homolog of METTL3, it lacks methyltransferase activity due to the absence of an S-adenosylmethionine (SAM)-binding domain. Importantly, METTL14 directly interacts with METTL3 to maintain its stability and catalytic activity of the MTC (Wang et al., 2016). METTL14 also plays an important role in maintaining cardiac (Wang et al., 2022) and kidney homeostasis (Lu et al., 2021), the development of osteoporosis (He et al., 2022), and antiviral innate immunity (Qin et al., 2021). The third core component, WTAP, facilitates the binding of the METTL3-METTL14 complex to mRNA targets and guides the localization of METTL3-METTL14 into nuclear speckles (Ping et al., 2014). In addition to WTAP, other accessory RNA-binding subunits include VIRMA (vir like m6A methyltransferase associated) (Yue et al., 2018), RBM15 (RNA binding motif protein 15) and its paralogues RBM15B (Patil et al., 2016), ZC3H13 (zinc finger CCCH-type containing 13) (Wen et al., 2018), and HAKAI/CBLL1 (Cbl proto-oncogene like 1) (Růžička et al., 2017; Bawankar et al., 2021). Those proteins contribute to determining the substrate RNAs and/or the precise sites of m6A methyltransferase activity.
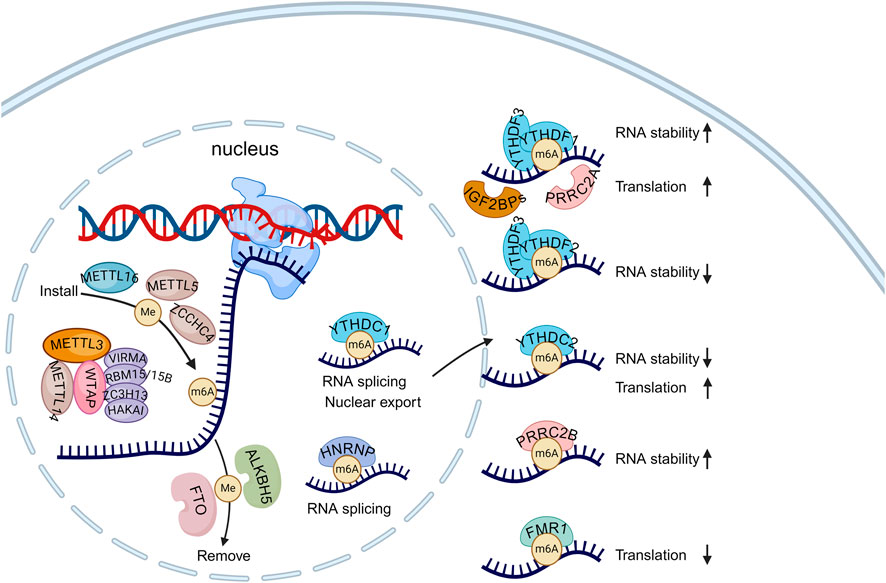
Figure 1. The regulators of m6A modification and its roles in determining RNA fates. The m6A modification is predominantly installed to mRNA by the methyltransferase complex (MTC) in the nucleus. Besides METTL3, METTL14 and WTAP, MTC also comprises a set of accessory subunits, such as VIRMA, RBM15/15B, ZC3H13, and HAKAI. Additionally, other m6A writers like METTL16, METTL5, and ZCCHC4 specifically deposit the m6A modification on certain RNAs. The m6A modification can be removed by demethylase FTO and ALKBH5. In the nucleus, YTHDC1 and HNRNP families recognize m6A-marked transcripts and affect mRNA splicing and export. Once the m6A-modified transcripts are exported to the cytoplasm, they can be recognized by cytoplasmic readers, which determine their fate. Created with BioRender.com.
Besides the MTC, other methyltransferases such as METTL16, METTL5, and ZCCHC4 (zinc-finger CCHC domain-containing protein 4) also deposit m6A on various RNA subtypes. METTL16 is responsible for methylating U6 small nuclear RNAs (snRNAs) as well as several mRNAs, such as MAT2A, BCAT1, and BCAT2 (Pendleton et al., 2017; Warda et al., 2017; Han et al., 2023). METTL5 (van Tran et al., 2019; Ignatova et al., 2020; Sepich-Poore et al., 2022) and ZCCHC4 (Ma et al., 2019; Ren et al., 2019; Pinto et al., 2020) are responsible for the m6A modification of 18S and 28S rRNA, respectively.
2.2 The m6A “erasers”
m6A methylation in mRNAs can be reversibly removed by m6A demethylases such as FTO and alkB homologue 5 (ALKBH5). Despite both belonging to the AlkB family of α-ketoglutarate (α-KG)-dependent dioxygenases, they exhibit distinct expression patterns in various cell types, suggesting their involvement in different biological pathways (Chen Y. et al., 2020; Shen et al., 2020; Tang et al., 2020; Zhou J. et al., 2021; Gao et al., 2024). FTO was first reported in 2011 to localize in nuclear speckles and remove m6A residues from RNA (Jia et al., 2011). It regulates the splicing of runt-related transcription factor 1 (RUNX1T1) by modulating the RNA binding ability of serine and arginine rich splicing factor 2 (SRSF2), thereby influencing adipogenesis (Zhao et al., 2014). Additionally, FTO can regulate dopaminergic signaling pathways through the demethylation of specific mRNAs, affecting neuronal activity (Hess et al., 2013). FTO is also capable of binding and demethylating other RNA modifications, including cap N6,2′-O-dimethyladenosine (m6Am) in mRNA and snRNAs, as well as N1-methyladenosine (m1A) in tRNA (Mauer et al., 2017; Wei et al., 2018). In contrast, ALKBH5 specifically removes m6A modifications. It can modulate mRNA export, the assembly of mRNA processing factors, and RNA metabolism in nuclear speckles by removing m6A modifications (Zheng et al., 2013). ALKBH5 also participates in antiviral and antibacterial innate responses by erasing the m6A modification of DDX46-binding antiviral transcripts in the nucleus (Zheng et al., 2017) and by regulating the protein expression of neutrophil migration-related molecules, including CXCR2 and NLRP12, to enhance the intrinsic migratory ability of neutrophils (Liu Y. et al., 2022).
2.3 The m6A “readers”
RNA m6A can be specifically recognized by multiple reader proteins, crucial for mediating various biological functions, including regulating mRNA stability, splicing, export, translation efficiency, and miRNA biogenesis. The primary readers of m6A are the YTH family proteins. This family comprises five members: YTHDF1, YTHDF2, YTHDF3, YTHDC1, and YTHDC2, all of which possess a YTH domain responsible for directly recognizing m6A sites. YTHDF2 was the first characterized reader known to promote mRNA decay via the CCR4-NOT complex in the cytosol (Du et al., 2016). YTHDF2 interacts with m6A-modified mRNAs, guiding them from the translatable pool to mRNA degradation sites, such as processing bodies (Wang X. et al., 2014). In contrast, YTHDF1 enhances the translation efficiency of its target mRNAs. Interestingly, YTHDF1 and YTHDF2 share a set of common target mRNAs, with YTHDF1 binding earlier than YTHDF2 (Wang et al., 2015). YTHDF3, partnering with both YTHDF1 and YTHDF2, assists in the translation and decay of m6A-modified RNA (Shi et al., 2017). The nuclear-localized m6A reader, YTHDC1, influences the fate of m6A-modified mRNAs through regulation of nuclear export, alternative splicing, and RNA degradation (Yan et al., 2022). While YTHDC2 primarily localizes in the cytosol and enhances the translation efficiency of its targets (Hsu et al., 2017).
Besides the YTH family, another important group of m6A readers is the insulin-like growth factor 2 mRNA-binding protein (IGF2BP) family, which includes IGF2BP1, IGF2BP2, and IGF2BP3 (Huang et al., 2018). These proteins contain six canonical RNA-binding domains: two RNA recognition motif (RRM) domains and 4 K homology (KH) domains. Notably, the KH3 and KH4 domains are essential for m6A recognition. IGF2BPs target thousands of mRNA transcripts that contain the consensus motif GG (m6A)C. IGF2BPs recruit known mRNA stabilizers, such as ELAV-like RNA binding protein 1 (ELAVL1), matrin 3 (MATR3), and poly(A) binding protein cytoplasmic 1 (PABPC1), to promote the stability and storage of m6A-modified mRNA targets (Huang et al., 2018). IGF2BPs can also recruit eIF proteins to their target mRNAs, increasing their translation (Weng et al., 2022).
The heterogeneous nuclear ribonucleoproteins (hnRNPs) family also functions as m6A-binding proteins, such as hnRNPA2B1, and hnRNPC. HNRNPA2B1 was reported to bind to m6A on primary microRNA (pri-miRNAs) transcripts and induce pri-miR to premature-miR processing (Alarcon et al., 2015; Alarcón et al., 2015). HNRNPC regulates the processing of m6A-modified RNA transcripts, harboring m6A as a structural switch to make those transcripts more accessible for binding (Liu et al., 2015). The proline-rich and coiled-coil-containing protein (PRRC) family is another novel m6A reader family, including PRRC2A, PRRC2B, and PRRC2C. PRRC2A promotes the translation efficiency of transcripts involved in meiotic cell division (Tan et al., 2023). During oligodendrocyte development, PRRC2A stabilizes Oligodendrocyte Transcription Factor 2 (Olig2) mRNA by binding to a consensus GGm6ACU motif (Wu et al., 2019). PRRC2B was reported to stabilize SRY-Box transcription factor 2 (SOX2) mRNA, displaying separate and cooperative functions with PRRC2A (Zhang Y. et al., 2024). Additionally, other proteins have been identified to preferentially bind m6A-modified RNA, including fragile X messenger ribonucleoprotein 1 (FMR1) and its paralogs FXR1 and FXR2 (Edupuganti et al., 2017). Furthermore, eukaryotic translation initiation factor 3 (eIF3) recognizes a subset of transcripts with m6A modifications in their 5′UTR and recruits the 43S translation initiation complex, thereby facilitating cap-independent translation (Patil et al., 2018). RNA Binding Fox-1 Homolog 2 (RBFOX2) preferentially recognizes m6A-modified chromatin-associated RNAs (caRNAs) and regulates chromatin silencing and transcription suppression by recruiting the RBM15/YTHDC1/PRC2 axis (Dou et al., 2023).
3 m6A modification in CSCs
Despite significant advancements in chemotherapy, immunotherapy, and surgery, cancer remains the leading cause of death globally. In certain cancer types, CSCs are recognized as the root cause of tumorigenesis and are also responsible for relapse and drug resistance. Thus, eradicating CSCs holds the potential to ultimately cure cancers. Evidence is emerging that aberrant m6A modification is associated with the survival and self-renewal of CSCs (Ma and Ji, 2020). Importantly, m6A plays a crucial role in pluripotency and reprogramming. Many key pluripotent genes, such as nanog homeobox (NANOG), octamer-binding transcription factor 3/4 (OCT3/4), kruppel-like factor 4 (KLF4), and SOX2, have abundant m6A modifications in their transcripts. Here, we summarize the reported functions and molecular mechanisms of m6A modification and its machinery in CSCs across various cancer types (Figure 2).
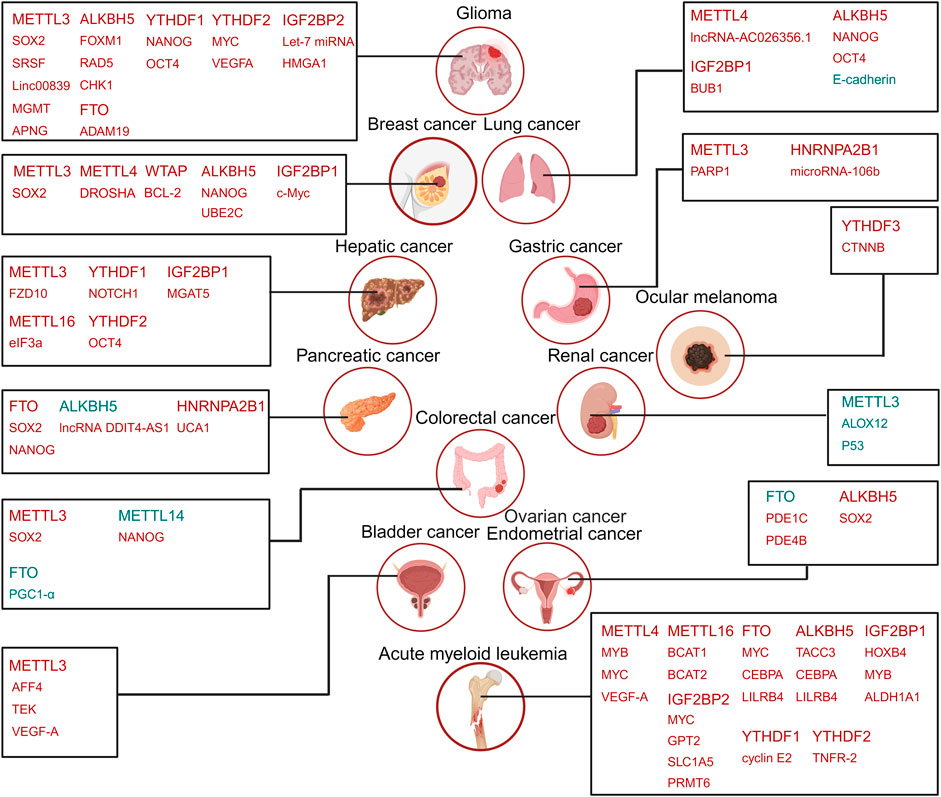
Figure 2. The roles of m6A regulators in CSCs. The m6A regulators, including METTL3, METTL14, WTAP, FTO, ALKBH5, YTHDF1, YTHDF2, IGF2BP1, IGF2BP2. YTHDF3, and HNRNPA2B1, along with their respective downstream targets, are listed for the relevant cancer types’ CSCs. Highly expressed m6A in CSCs are highlighted in red, while those decreased expression in CSCs are marked in blue. Similarly, targets positively regulated (upregulated) by m6A regulators are shown in red, and negatively regulated (downregulated) targets are indicated in blue. Created with BioRender.com.
3.1 Writers and CSCs
3.1.1 METTL3 and CSCs
METTL3 is implicated in the development of breast cancer, colorectal cancer, glioma, and bladder cancer. In these cancers, METTL3 expression is significantly upregulated and its high expression correlates with poor prognosis. In breast cancer, METTL3 directly catalyzes the formation of m6A modification in SOX2 mRNA, thereby increasing the protein levels of SOX2 and stem cell markers CD133 and CD44. This enhances the stemness of breast CSCs (Xie et al., 2021). In colorectal cancer (CRC), METTL3 knockdown significantly suppresses the self-renewal and frequency of CRC cells and inhibits tumorigenesis in vivo. Mechanistically, METTL3 promotes SOX2 expression through an m6A-IGF2BP2-dependent mechanism in CRC cells (Li T. et al., 2019). Consistently, in glioblastoma, SOX2 acts as a bona fide m6A target of METTL3. The m6A modification of SOX2 mRNA by METTL3 enhances its stability, thereby promoting the growth and self-renewal of glioma stem cells (GSCs) (Visvanathan et al., 2018). Additionally, METTL3 knockout (KO) significantly suppresses the growth and self-renewal of GSCs via targeting serine- and arginine-rich splicing factors (SRSF). METTL3 KO decreases m6A levels of SRSF transcripts as well as protein levels of SRSF through a YTHDC1-dependent nonsense-mediated mRNA decay (NMD) mechanism (Li F. et al., 2019). LncRNAs such as LINC00839 are also implicated in GSCs’ stemness maintenance. METTL3-mediated m6A modification on LINC00839 enhanced its expression via a YTHDF2-dependent manner. LINC00839, in turn, facilitates GSCs’ stemness and radiation resistance by activating the Wnt/β-catenin pathway through c-Src-mediated β-catenin phosphorylation (Yin et al., 2023). Another independent study reported that METTL3-mediated m6A modification enhances the mRNA stability of DNA repair genes, O6-methylguanine-DNA methyltransferase (MGMT) and alkylpurine-DNA-N-glycosylase (APNG), thereby reducing the sensitivity of GSCs to temozolomide (Shi et al., 2023). In gastric cancer, METTL3 promotes oxaliplatin resistance in CD133+ gastric CSCs by stabilizing PARP1 mRNA through recruiting YTHDF1 to its 3′-UTR (Li et al., 2022). In bladder cancer and hepatocellular carcinoma (HCC), METTL3 enhances the self-renewal and tumorigenicity of CSCs by regulating the expression of key factors such as AFF4, TEK, VEGF-A, and FZD10, which are extensively implicated in cancer progression and stemness maintenance (Wang J. et al., 2023). Conversely, METTL3 exhibits an opposing role in renal cell carcinoma, where its upregulation, induced by Eriani treatment, enhances the expression of ALOX12 and p53 in an m6A-dependent manner. This upregulation promotes the expression of ferroptosis-related protein, thereby inhibiting the tumorigenicity of human renal CSCs (CD44+/CD105+) (Shen et al., 2023).
3.1.2 METTL14 and CSCs
METTL14 is highly expressed in acute myeloid leukemia (AML) cells and is essential for leukemia stem cell (LSC) self-renewal. METTL14 plays an oncogenic role by suppressing myeloid differentiation and promoting LSC self-renewal through positively regulating MYB and MYC mRNA via m6A modification. Notably, METTL14 expression is negatively regulated by SPI1, a potent inducer of granulocytic/monocytic differentiation (Weng et al., 2018). In breast cancer stem-like cells (BCSCs; CD44+/CD24−), aurora kinase A (AURKA) stabilizes METTL14 by inhibiting its ubiquitylation and degradation, thereby promoting DROSHA mRNA methylation and overall expression. Specifically, METTL14-mediated m6A methylation in DROSHA mRNA can be recognized by IGF2BP2 to maintain its stability. DROSHA interacts with β-Catenin to transactivate STC1 in an RNA cleavage-independent manner, thereby maintaining BCSC phenotype (Peng et al., 2021). In lung cancer, METTL1 methylates and stabilizes lncRNA-AC026356.1 to maintain CSC stemness and mediate drug resistance. Silencing AC026356.1 blocks the oncogenicity of lung cancer stem-like cells (Zhang et al., 2023). Conversely, METTL14 plays an opposite role in CRC. It is downregulated and negatively correlated with poor prognosis in CRC patients. Mechanistically, METTL14 inhibits the self-renewal of colorectal SCSs by regulating β-catenin, thereby decreasing the expression of NANOG, a key regulator of stemness, self-renewal, metastasis, invasiveness, and chemoresistance in cancer cells (Sun et al., 2023).
3.1.3 WTAP and CSCs
In breast cancer, the transmembrane glycoprotein Neuropilin-1 (NRP1) increases the expression of WTAP to enhance CSC properties and potentiate radioresistance. Furthermore, WTAP reduces the mRNA level of Bcl-2 via an m6A-dependent mechanism (Wang Y. et al., 2023).
3.1.4 METTL16 and CSCs
Our studies showed that METTL16 is aberrantly overexpressed in human LSCs (CD34+) (Han et al., 2023) and liver CSCs (CD133+) (Xue et al., 2024) in contrast to their bulk cancer cell counterparts. Branched-chain amino acid (BCAA) transaminase 1 (BCAT1) and BCAT2 reprogram cancer metabolism to facilitate the malignant and clonal expansion of tumor cells through regulation of the TCA cycle, oxidative phosphorylation, and nucleotide biosynthesis (Wei et al., 2020). In AML, METTL16 promotes the expression of BCAT1 and BCAT2 in an m6A-dependent manner, thereby reprogramming BCAA metabolism to sustain leukemogenesis and LSC self-renewal (Han et al., 2023). METTL16 KO significantly suppresses HCC initiation and progression, and decreases the frequency of liver CSCs. These effects are largely attributed to the suppressed translation efficacy of mRNAs, especially eIF3a, following METTL16 KO (Su et al., 2022; Xue et al., 2024).
3.2 Erasers and CSCs
3.2.1 FTO and CSCs
Dysregulation of m6A demethylase FTO has been reported in various CSCs. In AML, FTO is highly expressed in LSCs and maintains the stability of MYC CEBPA mRNAs via removing m6A sites, thereby sustaining LSC frequency. Notably, genetic depletion or pharmacological inhibition of FTO significantly reduces the self-renewal ability of LSCs (Su et al., 2020). High expression of FTO is also observed in pancreatic cancer, where it is essential for maintaining pancreatic CSCs and their self-renewal (Garg et al., 2022). In glioblastoma, pharmacological inhibition of FTO suppresses GSC self-renewal and frequency. This effect is likely mediated through FTO-mediated demethylation of m6A in ADAM metallopeptidase domain 19 (ADAM19) mRNA, a member of the metalloproteinase family with pro-tumorigenic functions (Cui et al., 2017).
Controversially, FTO exhibits a tumor-suppressor role in ovarian cancer and colorectal cancer. Decreased expression of FTO is identified in ovarian CSCs and FTO reduced their self-renewal and tumorigenesis through its demethylase activity. Specifically, FTO enhances 3′, 5′-cyclic adenosine monophosphate (cAMP) signaling by demethylating and destabilizing PDE1C and PDE4B mRNA, which regulate cAMP degradation (Huang et al., 2020). In colorectal cancer, Berberine transcriptionally increases FTO expression to suppress stemness and tumorigenicity of colorectal cancer stem-like cells (Zhao et al., 2021). Berberine treatment inhibits sphere formation of colorectal CSCs and reduces the expression of stem cell markers (CD44 and CD133). These effects can be reversed by pharmacological inhibition of FTO.
3.2.2 ALKBH5 and CSCs
ALKBH5 is highly expressed and exerts tumor-promoting effects in various CSCs, including GSCs, LSCs, breast CSCs, and non-small-cell lung CSCs. In glioblastoma, ALKBH5 upregulates the expression of the transcription factor forkhead box protein M1 (FOXM1) by removing the m6A site from its transcript. FOXM1 is known to regulate GSC proliferation, self-renewal, and tumorigenicity through activating β-catenin and STAT3 or inducing the expression of SOX2 (Zhang et al., 2017). Furthermore, ALKBH5 promotes radioresistance in GSCs by increasing the expression of DNA repair proteins involved in homologous recombination, such as RAD51 and CHK1 (Kowalski-Chauvel et al., 2020). ALKBH5 is overexpressed in AML, where it plays a crucial role in promoting leukemogenesis and LSC self-renewal as an m6A demethylase. ALKHB5 post-transcriptionally regulates the expression of its functionally important targets, such as Transforming Acidic Coiled-Coil Containing Protein 3 (TACC3). Genetic depletion of ALKBH5 leads to the accumulation of m6A methylation in TACC3 mRNA, resulting in decreased expression of TACC3 at both mRNA and protein levels. This, in turn, decreases MYC level and increases P21 to suppress LSC self-renewal (Shen et al., 2020). Exposure of breast cancer cells hypoxia, a hallmark of the tumor microenvironment, activates ALKBH5 gene transcription to induce m6A demethylation and stabilize NANOG mRNA, thereby promoting the specification and maintenance of breast CSCs (Zhang et al., 2016). In endometrial CSCs (CD133+ALDH1+), hypoxia promotes a stem-like state in ECSCs and hypoxia-inducing factors (HIFs) are crucial for hypoxia-induced pluripotency. Moreover, hypoxia induces ALKBH5 expression in a HIF-dependent manner and ALKBH5 promotes SOX2 expression as an m6A demethylase to sustain the stem-like state of ECSCs (Chen G. et al., 2020). It is worth mentioning that NANOG and SOX2 are well-characterized pluripotency factors that facilitate CSC maintenance and self-renewal (Novak et al., 2020; Vasefifar et al., 2022). In triple-negative breast cancer, ALKBH5 demethylates, stabilizes, and upregulates ubiquitin-conjugating enzyme E2C (UBE2C), which in turn induces p53 ubiquitination to facilitate its degradation and enhances cancer stemness, growth, and metastasis (Hu et al., 2022). ALKBH5 is highly expressed in CSCs isolated from non-small-cell lung cancer (NSCLC). Knockdown of ALKBH5 increases global m6A level, decreases stem markers, Nanog and Oct4, and inhibits stemness of CSCs. Furthermore, p53 transcriptionally enhances ALKBH5 expression and promotes CSCs’ malignancies (Liu X. et al., 2022).
However, ALKBH5 is reported to exert a tumor suppressor in pancreatic cancer. lncRNA DDIT4-AS1, encoded by DNA damage-inducible transcript 4 (DDIT4), contributes to tumorigenesis and maintains cancer stemness. ALKBH5 depletion-mediated m6A modification leads to DDIT4-AS1 overexpression in pancreatic cancer, and DDIT-AS1 increased cancer stemness and suppressed chemosensitivity to Gemcitabine (Zhang et al., 2022).
3.3 Reader and CSCs
3.3.1 YTH family and CSCs
YTHDF1 has been reported to play a pivotal oncogenic role in CSCs of colorectal cancer, AML, glioblastoma, and HCC. In colorectal cancer, YTHDF1 is elevated in colonospheres and it promotes tumorigenicity and cancer stem cell-like activity by activating the Wnt/β-catenin pathway (Bai et al., 2019). In LSCs, YTHDF1 promotes their self-renewal capacity by facilitating the translation efficacy of cyclin E2 in an m6A-dependent manner (Hong et al., 2023). In glioblastoma, high levels of YTHDF1 are associated with poor patient survival. YTHDF1 is required for maintaining cancer stem cell properties of glioblastoma cell line and its depletion reduces the expression of CSC markers CD133, NANOG, OCT4, and REX1 (Yarmishyn et al., 2020). Similarly, in HCC, elevated YTHDF1 levels correlate positively with stemness signatures. YTHDF1 promotes liver CSC renewal and resistance to the multiple tyrosine kinase inhibitors lenvatinib and sorafenib. Mechanistically, YTHDF1 recognizes the m6A modification in NOTCH1 mRNA to enhance its stability and translation, thereby increasing expression of NOTCH1 target genes (Zhang X. et al., 2024).
YTHDF2 is upregulated in advanced-stage HCC and its high expression is associated with poor overall survival of HCC patients. Moreover, YTHDF2 promotes liver CSC stemness and HCC metastasis by modulating the m6A methylation in the 5′UTR of OCT4 mRNA, thereby facilitating OCT4 mRNA translation (Zhang et al., 2020). OCT4, a pluripotency factor, plays a crucial role in promoting cancer stemness (Mohiuddin et al., 2020). Another study focusing on glioblastoma reported elevated levels of YTHDF2 GSCs compared to normal neural stem cells (NSC). YTHDF2 stabilizes, rather than destabilizes, the oncogenes required for GSCs survival, such as MYC and VEGFA transcripts, in an m6A-dependent manner (Dixit et al., 2021). Dysregulated cholesterol metabolism is a hallmark of glioblastoma. YTHDF2 promotes mRNA decay of LXRA and HIVEP2 which regulate cholesterol homeostasis, and then impacts the GSCs survival (Fang et al., 2021). YTHDF2 is also overexpressed in a broad spectrum of AML patient samples and it is necessary for AML initiation and progression. YTHDF2 shortens the half-life of diverse m6A-modified transcripts contributing to the overall integrity of LSC function, such as the tumor necrosis factor receptor TNFRSF2. Thus YTHDF2-mediated suppression of TNFRSF2 leads to resistance against TNF-induced apoptosis in LSCs (Paris et al., 2019).
Interestingly, depletion of YTHDF2 enhances the expansion of healthy hematopoietic stem cells (HSCs) by increasing the levels of key transcription factors such as Gata2, Etv6, Stat5, and Tal1, which are critical for HSCs self-renewal (Li et al., 2018). Thus, inhibiting YTHDF2 not only decreases the frequency of LSCs but also promotes the expansion of HSCs. These dual effects suggest a potentially promising therapeutic strategy to selectively eliminate LSCs while supporting healthy hematopoiesis by targeting YTHDF2.
In ocular melanoma, YTHDF3 is associated with poor prognosis and is required for tumor propagation. YTHDF3 is highly expressed in melanoma CSCs (CD133+) compared to non-CSC melanoma cells (CD133−). The mechanistic study demonstrates that YTHDF3 promotes the translation of catenin beta 1 (CTNNB1) mRNA by binding to its m6A methylation site (Xu et al., 2022a). CTNNB1 serves as a crucial regulator of the Wnt signaling pathway and enhances CSC properties through activation of Wnt/β-catenin signaling.
3.3.2 IGF2BP family and CSCs
IGF2BP1 is implicated in regulating the stemness of CSCs in HCC, NSCLC, and leukemia. In HCC, IGF2BP1 is upregulated in liver CSCs and promotes the liver CSC phenotypes by enhancing the stability of alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase (MGAT5) mRNA stability through recognition of m6A modification (Yang et al., 2021). IGF2BP1 expression is increased in NSCLC cells, especially in the enriched CSCs. Depletion of IGF2BP1 suppresses proliferation, mobility and epithelial–mesenchymal transition activity of NSCLC cells and CSCs, and reduces CSCs’ stemness, self-renewal ability, and xenograft tumorigenesis. IGF2BP1 affects stem cell properties of the CSCs by upregulating BUB1 mitotic checkpoint serine/threonine kinase B (BUB1B) expression via m6A RNA methylation (Hu et al., 2023). In leukemia, IGF2BP1 maintains LSC properties and promotes leukemogenesis by upregulating critical regulators of self-renewal such as HOXB4 and MYB, as well as the expression of aldehyde dehydrogenase 1A1 (ALDH1A1) (Elcheva et al., 2020). In breast cancer, under hypoxic conditions, the lncRNA KB1980E6.3 is upregulated and recruits IGF2BP1 to stabilize c-Myc mRNA through m6A modification. This lncRNA KB-1980E6.3/IGF2BP1/c-Myc signaling axis enhances the self-renewal and tumorigenic potential of breast CSCs in the hypoxic microenvironment (Zhu et al., 2021).
We reported the high expression of IGF2BP2 in AML, particularly in LSCs. IGF2BP2 drives AML pathogenesis and enhances the self-renewal ability of LSCs as an m6A reader protein. Mechanistically, IGF2BP2 stabilizes and upregulates the expression of critical targets such as MYC, GPT2, and SLC1A5, which activate glutamine metabolism pathways to provide energy for the proliferation and survival of AML cells (Weng et al., 2022). In addition, another independent study revealed that IGF2BP2 upregulates protein arginine methyltransferase PRMT6 in an m6A-dependent manner. PRMT6 impairs the expression of the lipid transporter MFSD2A and reduces docosahexaenoic acid levels, thereby increasing LSC self-renewal and frequency (Cheng et al., 2023). In glioma, IGF2BP2 supports the maintenance of CSCs by modulating miRNA-mediated mRNA silencing. Specifically, IGF2BP2 binds to the let-7 miRNA and prevents let-7-mediated target gene silencing to maintain GSC stemness (Degrauwe et al., 2016). Moreover, IGF2BP2 interacts with lncRNA to increase the maintenance of mesenchymal GSCs. lncRNA HIF1A antisense RNA 2 (HIF1A-AS2) is highly expressed in GSCs and enhances the expression of high mobility group A1 (HMGA1), promoting cancer occurrence and metastasis through interactions with IGF2BP2 and ATP-dependent RNA helicase A (Mineo et al., 2016). Despite the crucial role of IGF2BP2 in GSCs, it remains unclear to what extent these reported functions rely on IGF2BP2-m6A recognition.
3.3.3 HNRNPs family and CSCs
Studies focusing on HNRNPs as m6A “readers” in CSCs are limited. HNRNPA2B1 is upregulated lung adenocarcinoma CSCs and promotes cell stemness, migration, and tumor growth. Mechanistically, HNRNPA2B1 facilitates the maturation of miR-106b-5p by recognizing the m6A site on primary microRNA-106b. miR-106b-5p represses the level of secreted frizzled-related protein 2 (SFRP2), thereby activating Wnt/β-catenin signaling, which is crucial for maintaining CSC traits and promoting tumor progression (Rong et al., 2022). Another study reported that hnRNPA2B1 may enhance the stemness of pancreatic ductal adenocarcinoma cells via interacting with a long noncoding RNA UCA1, thereby increasing the activity and expression of oncogenic KRAS (Liu et al., 2019). However, it is unclear whether this function is related to hnRNPA2B1’s m6A recognition ability.
3.3.4 RBFOX2 and CSCs
In AML, the level of RBFOX2 is upregulated and is crucial for LSC self-renewal and inhibiting AML cell differentiation. Mechanistically, RBFOX2 increases m6A level in caRNA and promotes the binding of YTHDC1 and PRC2 to the TGFB1 promoter region. This interaction inhibits the expression of TGFB1, a gene that plays a tumor suppressor role in hematologic malignancies (Dou et al., 2023).
4 Development of small molecular inhibitors targeting m6A modification
As reiterated above, the majority of m6A regulators are upregulated in various CSCs, exerting an oncogenic role in tumor pathogenesis. Furthermore, these m6A regulators play crucial roles in cancer cell resistance to chemotherapy and radiotherapy. These findings suggest that targeting these oncogenic m6A regulators represents a promising therapeutic strategy to eradicate CSC. Currently, multiple small molecule inhibitors targeting m6A modifiers, including “writer”, “eraser”, and “reader”, have been developed, offering attractive insights and guidance for future clinical treatments. These inhibitors could potentially overcome therapeutic resistance by specifically targeting the m6A modification pathways critical for CSC maintenance and survival, thus improving the efficacy of cancer treatments. Here, we summarize the currently known inhibitors of m6A machinery and highlight proof-of-concept studies that support the promise of targeting m6A modification as a promising and exciting strategy to eradicate CSCs and combat cancers (Figure 3).
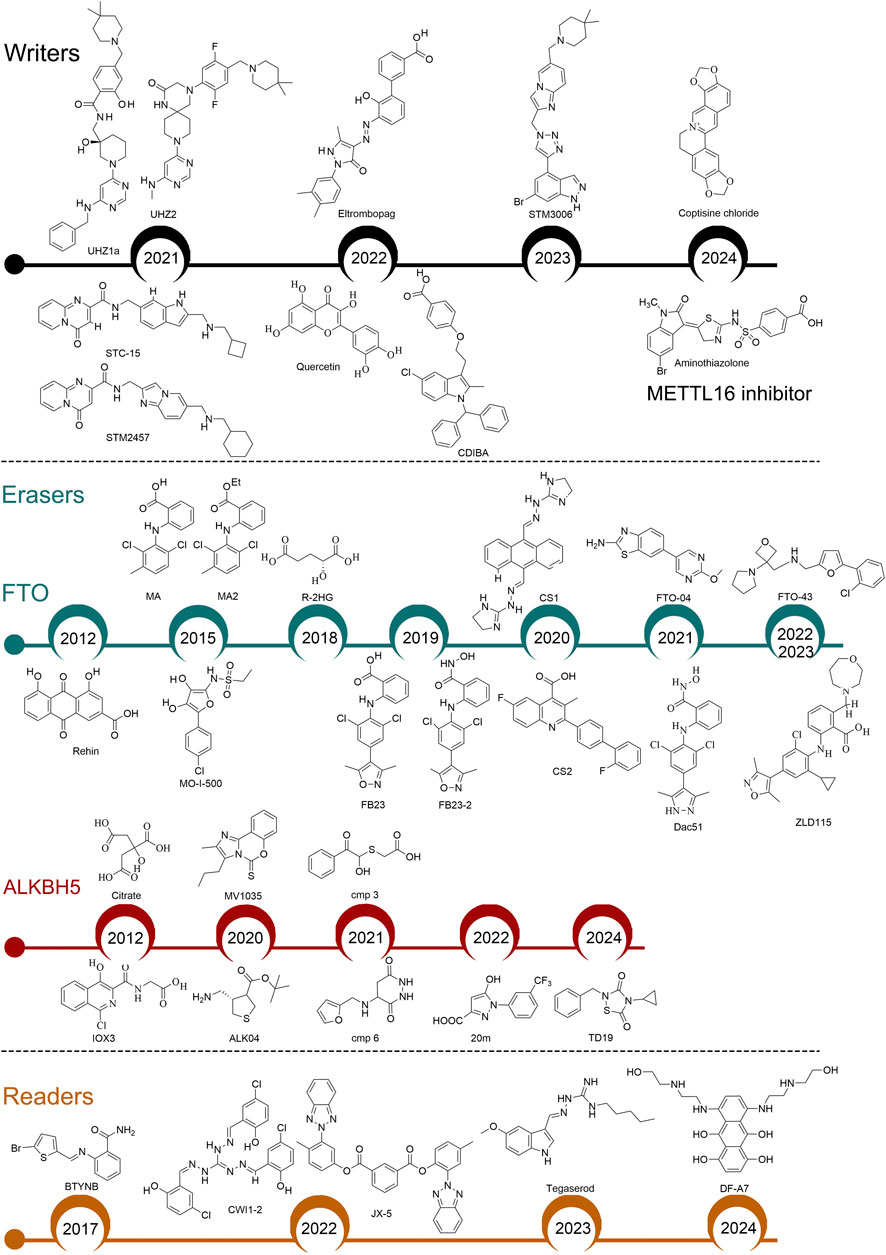
Figure 3. Identification of small-molecular inhibitors targeting m6A regulators. Writers: All these inhibitors target METTL3, except for Aminothiazolone which targets METTL16. Readers: BTYNB targets IGF2BP1; CWI1-2 and JX-5 target IGF2BP2; Tegaserod targets YTHDF1; DF-A7 targets YTHDF2.
4.1 Targeting m6A writers
4.1.1 METTL3 inhibitors
During the methyltransferase assay, METTL3 catalyzes the transfer of a methyl group from its cofactor S-adenosyl-L-methionine (SAM) to the N6 atom of adenine. Adenosine, the competitor of SAM, was the first characterized METTL3 inhibitor (Bedi et al., 2020). However, adenosine as a METTL3 inhibitor has serious drawbacks, such as poor selectivity and low permeability. In 2021, Dr. Caflisch’s team identified two non-nucleoside derivatives, UZH1a and UZH2, via a homogeneous time-resolved fluorescence (HTRF) enzyme inhibition assay. Both compounds reduced m6A levels in mRNAs of different cell lines (Moroz-Omori et al., 2021). Dr. Kouzarides’s team discovered a potent catalytic inhibitor of METTL3, STM2457, through high-throughput screening of 250,000 drug-like compounds combined with synthetic optimization. The crystal structure of STM2457 in complex with METTL3–METTL14 reveals that it binds competitively in the SAM binding pocket of the METTL3 (Yankova et al., 2021). STM2457 treatment reduces m6A levels in METTL3’s mRNA substrates, such as MYC and HOXA10, and effectively eliminates AML. The anti-tumor effects of STM2457 have also been reported in solid tumors, including oral squamous cell carcinoma, intrahepatic cholangiocarcinoma, renal cell carcinoma, NSCLC, and HCC (Gao et al., 2023; Liu et al., 2023; Xiao H. et al., 2023; Chen et al., 2024; Liu L. et al., 2024). Furthermore, a derivative of STM2457, STC-15, has advanced to phase I clinical trials (NCT05584111). By modifying STM2457’s structure, a new compound, STM3006, was created with improved binding affinity and enzymatic inhibition (Guirguis et al., 2023). STM3006-mediated inhibition of METTL3 enhances anti-tumor immunity and synergizes with anti-PD-1 immunotherapy. Natural products such as coptisine chloride (COP) and quercetin also inhibit METTL3 m6A methyltransferase activity. COP has demonstrated therapeutic effects on periodontitis by targeting METTL3, although its impact on tumor progression and CSCs remains unclear (Zhou et al., 2024). Quercetin reduces intracellular m6A methylation and suppresses tumor cell proliferation in a dose-dependent manner, but further investigation is needed to understand its selective inhibition of METTL3 (Du et al., 2022). Dr. Kim’s team reported two allosteric inhibitors, CDIBA (Lee et al., 2022b) and eltrombopag (Lee et al., 2022a), which directly interact with the METTL3/METTL14 complex and suppress the proliferation of AML cell lines by decreasing m6A levels.
4.1.2 METTL16 inhibitor
In 2024, Dr. Wu’s team reported the first METTL16 inhibitor. They identified Aminothiazolone from an in-house compound library screening as a potential METTL16 inhibitor. Although this compound can bind to METTL16 and disrupt its interaction with RNA, it does not display antiproliferative effects in human cancer cells (Liu Y. et al., 2024). Current knowledge suggests that METTL16 may play a more critical role in the survival of cancer cells compared to METTL3 and METTL14 (Campeanu et al., 2021; Su et al., 2022; Han et al., 2023). Thus, further extensive studies are necessary to develop small-molecule inhibitors targeting METTL16 with robust anti-tumor effects.
4.2 Targeting m6A erasers
4.2.1 FTO inhibitors
The crystal structure of FTO was reported in 2010 (Han et al., 2010). Since then, extensive efforts from different groups have been devoted to developing FTO inhibitors. In 2012, Dr. Yang’s team identified the first FTO inhibitor, rhein, through structure-based virtual screening. Rhein, a natural compound, competitively binds to the catalytic domain of FTO, increasing m6A levels in mRNA (Chen et al., 2012). In AML, rhein effectively inhibits the proliferation and migration of AML cells by targeting FTO and impairing the AKT/mTOR signaling pathway (Zhang S. et al., 2024). Nevertheless, rhein also targets DNA damage repair enzymes ALKBH2 and ALKBH3 in vitro, underscoring the unmet need for FTO inhibitors with increased selectivity. Dr. Yang’s team further identified meclofenamic acid (MA) and MA2 (the ethyl ester form of MA) as selective FTO inhibitors (Huang et al., 2015). MA2 treatment increases mRNA m6A levels in HeLa cells and suppresses the growth and self-renewal of GSCs (Cui et al., 2017). Since the discovery of MA as a selective FTO inhibitor, several MA-derived FTO inhibitors with increased sensitivity and/or anti-tumor efficacy have been developed, such as FB23, FB23-2, Dac51, and ZLD115 (Huang et al., 2019; Liu et al., 2021; Xu et al., 2022b; Xiao P. et al., 2023). In AML and clear cell renal cell carcinoma, FB23-2 significantly suppresses tumor growth and extends survival by targeting FTO (Huang et al., 2019; Xu et al., 2022b). Dac51, optimized from FB23-2, inhibits FTO activity, reduces glycolytic metabolites in B16-OVA cells, and increases CD8+ T cell infiltration in the tumor microenvironment (Liu et al., 2021). 44/ZLD115, a flexible alkaline side chain-substituted benzoic acid FTO inhibitor derived from FB23, demonstrates improved anti-tumor capacity in AML (Xiao P. et al., 2023). To enhance the permeability of FTO inhibitor FB23, Dr. Rana’s team develops two additional inhibitors, FTO-04 and FTO-43, via a combination of structure-based drug design and molecular docking. Consistent with FTO inhibition, FTO-04 increases m6A and m6Am levels in GSCs (Huff et al., 2021). FTO-43, derived from FTO-04, elevates m6A levels and suppresses the Wnt/PI3K-Akt signaling pathways in gastric cancer cells, thereby inhibiting the proliferation of AML and glioblastoma cells (Huff et al., 2022).
FTO belongs to Fe(II)/α-KG-dependent dioxygenases. R-2-hydroxyglutarate (R-2HG), produced at high levels by mutant isocitrate dehydrogenase 1/2 (IDH1/2), is structurally close to α-KG and competitively inhibits the enzymatic activity of Fe(II)/α-KG-dependent dioxygenases (Dang et al., 2009). In 2018, we reported R-2HG as a competitive inhibitor of FTO (Su et al., 2018). Via targeting FTO/m6A/MYC/CEBPA signaling, R-2HG exhibits a broad and intrinsic anti-tumor activity. As an FTO inhibitor, R-2HG suppresses aerobic glycolysis in AML (Qing et al., 2021). Another independent study showed that through inhibiting FTO, R-2HG suppresses glioma growth and induces m6A hypermethylation (Pianka et al., 2024). Dr. Olsen’s team synthesized an FTO inhibitor, MO-I-500, which inhibits the survival and colony formation of breast cancer cells by inhibiting FTO enzymatic activity (Zheng et al., 2014; Singh et al., 2016). Despite the identification of multiple FTO inhibitors, their clinical implication is limited due to mild biological function or low sensitivity. To address this question, we conducted a high throughput structure-based screening and characterized two potent FTO inhibitors, CS1 (NSC337766) and CS2 (NSC368390). Compared to FB23-2 and MO-I-500, CS1 and CS2 exhibit much higher efficiency in killing AMLs. Additionally, CS1 and CS2 repress LSC self-renewal and reprogram the immune response by suppressing FTO activity (Su et al., 2020). Dr. Feng’s team developed a method to encapsulate an FTO inhibitor into GSH-imprinted gold nanoclusters (GNPIPP12MA) for targeted delivery to AML cells and LSCs. Their study showed that GNPIPP12MA suppresses FTO, increases m6A levels, and induces ferroptosis by depleting GSH. This “nano-epidrug” offers new insight into anti-cancer therapies (Cao et al., 2022).
4.2.2 ALKBH5 inhibitors
The first characterized ALKBH5 inhibitors were α-KG analogs, such as IOX3 and citrate, but these inhibitors lack selectivity, sensitivity, or crucial biological functions (Aik et al., 2014). In 2020, Dr. Nicolini’s team discovered a novel ALKBH5 inhibitor, MV1035, through 3D proteome-wide scale screening. This compound reduces the proliferation, migration, and invasion of glioblastoma cells (Malacrida et al., 2020). In the same year, Dr. Rana’s group identified another inhibitor, ALK-04, through in silico screening of compounds. Although ALK-04 shows a moderate impact on suppressing tumor cell proliferation in vitro, it enhances the efficacy of anti-PD-1 immunotherapy in vivo (Li et al., 2020). In 2021, Dr. Karelson’s team reported two ALKBH5 inhibitors (Cmp 3 and Cmp 6) with promising anti-tumor activity in leukemia cell lines, discovered through high-throughput virtual screening of a library of 144,000 compounds (Selberg et al., 2021). In 2022, Dr. Xiang’s team reported a new class of ALKBH5 inhibitor through fluorescence polarization-based screening, naming it 20 m. This compound displayed high selectivity for ALKBH5 over FTO, not only increasing global m6A levels but also stabilizing ALKBH5 in HepG2 cells (Fang et al., 2022). In 2024, Dr. Yang’s team identified a covalent inhibitor, TD19, which specifically targets ALKBH5. TD19 exhibited potent anticancer efficacy in AML and glioblastoma (Lai et al., 2024).
4.3 Targeting m6A readers
4.3.1 Inhibitors of IGF2BP family
In 2017, Dr. Shapiro’s team reported BTYNB as an inhibitor of IGF2BP1. This compound displayed high selectivity and antiproliferative effects by reducing c-Myc and E2F1 target gene expression in IGF2BP1-expressing ovarian cancer and melanoma cells (Mahapatra et al., 2017). However, it is unknown whether BTYNB suppresses IGF2BP1’s ability to recognize m6A modification. In 2022, we reported an effective small-molecule inhibitor, CWI1-2, which preferentially binds to IGF2BP2 and inhibits its interaction with m6A-modified mRNA targets, such as MYC, SLC1A5, and GPT2. Pharmacological inhibition of IGF2BP2 by CWI1-2 shows promising therapeutic efficacy in treating AML and eradicating LSCs (Weng et al., 2022). Moreover, Dr. Ji’s team identified another IGF2BP2 inhibitor, JX5. Treatment with JX5 suppresses the activation of NOTCH1 signaling and the progression of T-ALL (Feng et al., 2022).
4.3.2 Inhibitors of YTHDF family
In 2023, Dr. Chen’s team identified tegaserod as a potential YTHDF1 inhibitor through structure-based virtual screening of FDA-approved drugs. Tegaserod reduced the proliferation of AML cells and inhibited AML progression in vivo by targeting YTHDF1 (Hong et al., 2023). Most recently, Dr. Yu’s team reported DF-A7 as an inhibitor of YTHDF2. DF-A7 induces the degradation of the YTHDF2 protein, thereby suppressing tumorigenesis and enhancing cancer immunotherapy (Xiao et al., 2024).
5 Discussion
CSCs are a rare population of immortal cells within the tumors that can self-renew through division and differentiate into many cell types, leading to high heterogeneity. CSCs not only initiate and drive tumorigenesis but also contribute to metastasis, relapse, and drug resistance. CSC populations have been functionally validated across multiple cancers, and targeting CSC has been the focus of therapeutic development efforts, although it is still in the early stages of clinical evaluation (Bayik and Lathia, 2021). The maintenance and self-renewal of CSCs are mediated by multiple factors, including SOX2, NANOG, OCT4, KLF4, and MYC. Additionally, intracellular signaling pathways, such as Wnt/β-catenin, Notch, JAK-STAT, and PI3K/AKT, as well as the tumor microenvironment, play crucial roles in sustaining CSC frequency. Investigating CSC regulatory mechanisms can substantially improve the identification of novel strategies against various types of cancers. Of note, certain drugs, antibodies, and vaccines targeting these pathways have also been developed to eradicate CSCs (Yang et al., 2020a).
As the most prevalent internal modification in eukaryotic mRNA, m6A participates in almost every stage of mRNA metabolism. Numerous studies have reported that the dysregulation of m6A modification and its machinery contribute to pathogenesis in different types of cancer. Moreover, m6A modification is extensively implicated in regulating the self-renewal and function of CSCs by post-transcriptionally controlling the expression of key pluripotent transcription factors and oncogenic genes. Based on current knowledge, the majority of these m6A regulators are demonstrated to be highly expressed in CSCs in contrast to the bulk cancer cells and/or healthy controls. This presents a promising therapeutic opportunity to target m6A regulators for eliminating CSCs and treating cancers.
Indeed, over the past few years, scientists have developed a series of potent and selective small-molecule inhibitors targeting m6A regulators such as METTL3, METTL14, FTO, ALKBH5, and IGF2BP. Multiple proof-of-concept studies strongly support the promise of “RNA-modification as druggable target”: pharmacologically targeting m6A regulators with small-molecule inhibitors demonstrates promising and robust anti-tumor efficacy. Moreover, these inhibitors targeting METTL3, FTO, and IGF2BPs show robust impacts on eradicating CSCs. More encouragingly, the METTL3 inhibitor STC-15 has advanced to phase I clinical trials. Considering that m6A modification mediates CSCs’ resistance to chemotherapy, radiotherapy, and immunotherapy, it would be beneficial to synergize inhibitors of m6A modification with other first-line therapeutic drugs to achieve better efficacy. Notably, m6A modification is highly dynamic and the biological functions of m6A regulators are context-dependent. For instance, FTO acts as an oncogene to maintain AML and glioma CSC frequency while inhibiting ovarian CSC self-renewal. This suggests that a given small-molecule inhibitor of m6A regulators may be applicable only for certain cancer types. To date, the US Food and Drug Administration (FDA) has approved multiple epigenetic drugs for cancer therapy, such as Azacitidine and Decitabine, which target DNA methyltransferases, and Vorinostat and Romidepsin, which target histone deacetylases (Davalos and Esteller, 2023). However, there are no FDA-approved drugs targeting epitranscriptomic modifications.
Despite the identification of various small-molecule inhibitors targeting the RNA m6A machinery, most of these inhibitors have not progressed to further development, except for STC-15, which is currently undergoing Phase I clinical trials. Potential reasons for the lack of advancements include, but are not limited to, low sensitivity or specificity, poor solubility, or limited bioavailability in vivo. Another issue we observed is that, while many compounds targeting m6A methylation have shown promising anti-tumor effects, their potential toxicity to normal cells or normal tissues remains underexplored. Pre-clinical studies suggested that METTL3 inhibitor STM2457 shows a nuanced and manageable effect on normal hematopoiesis in vivo (Sturgess et al., 2023). Specifically, while STM2457 showed robust anti-leukemia activity in animal models, it significantly impacts erythropoiesis during toxicity evaluation. To address this challenge, utilizing nanomaterial-based carriers for specific drug delivery could be a viable solution. These carriers can be loaded with inhibitors, allowing for targeted delivery to tumor cells and selective accumulation at the tumor site in vivo (Pérez-Herrero and Fernández-Medarde, 2015; Li et al., 2024). For many other compounds targeting the m6A machinery, such as ALKBH5 (Lai et al., 2024), YTHDF2 (Xiao et al., 2024) and IGF2BP2 (Weng et al., 2022) inhibitors, their potential toxicity and pharmacokinetics are unclear. Therefore, to advance further, more extensive toxicity evaluations are necessary. Moreover, to promote the clinical application of “epitranscriptomic drugs”, it is crucial to further optimize currently available inhibitors targeting m6A machinery to increase biological availability, specificity, and/or anti-tumor efficacy, and to develop novel inhibitors with improved biological functions.
In conclusion, the recent discoveries underpin the crucial roles of mRNA m6A modification in modulating CSC survival and frequency across a spectrum of cancer types. Convincing proof-of-concept studies highlight that pharmacologically targeting m6A regulators holds promising efficacy in eliminating CSCs and cancers.
Author contributions
HZ: Writing–review and editing, Data curation, Investigation, Writing–original draft. XW: Data curation, Writing–review and editing. JC: Supervision, Writing–review and editing. RS: Supervision, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The work is partly supported by the US National Institutes of Health (NIH) grants R37 CA292678 (RS), and U54 CA285116 (RS). RS is a Double Deckers Destroy AML St. Baldrick’s Scholar.
Acknowledgments
Figures 1 and 2 were created with BioRender.com and are licensed for publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aik, W., Scotti, J. S., Choi, H., Gong, L., Demetriades, M., Schofield, C. J., et al. (2014). Structure of human RNA N⁶-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation. Nucleic Acids Res. 42 (7), 4741–4754. doi:10.1093/nar/gku085
Alarcon, C. R., Goodarzi, H., Lee, H., Liu, X., Tavazoie, S., and Tavazoie, S. F. (2015). HNRNPA2B1 is a mediator of m(6)a-dependent nuclear RNA processing events. Cell 162 (6), 1299–1308. doi:10.1016/j.cell.2015.08.011
Alarcón, C. R., Lee, H., Goodarzi, H., Halberg, N., and Tavazoie, S. F. (2015). N6-methyladenosine marks primary microRNAs for processing. Nature 519 (7544), 482–485. doi:10.1038/nature14281
Al-Hajj, M., Wicha, M. S., Benito-Hernandez, A., Morrison, S. J., and Clarke, M. F. (2003). Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. U. S. A. 100 (7), 3983–3988. doi:10.1073/pnas.0530291100
Bai, Y., Yang, C., Wu, R., Huang, L., Song, S., Li, W., et al. (2019). YTHDF1 regulates tumorigenicity and cancer stem cell-like activity in human colorectal carcinoma. Front. Oncol. 9, 332. doi:10.3389/fonc.2019.00332
Barbieri, I., and Kouzarides, T. (2020). Role of RNA modifications in cancer. Nat. Rev. Cancer 20 (6), 303–322. doi:10.1038/s41568-020-0253-2
Bawankar, P., Lence, T., Paolantoni, C., Haussmann, I. U., Kazlauskiene, M., Jacob, D., et al. (2021). Hakai is required for stabilization of core components of the m(6)A mRNA methylation machinery. Nat. Commun. 12 (1), 3778. doi:10.1038/s41467-021-23892-5
Bayik, D., and Lathia, J. D. (2021). Cancer stem cell-immune cell crosstalk in tumour progression. Nat. Rev. Cancer 21 (8), 526–536. doi:10.1038/s41568-021-00366-w
Bedi, R. K., Huang, D., Eberle, S. A., Wiedmer, L., Sledz, P., and Caflisch, A. (2020). Small-molecule inhibitors of METTL3, the major human epitranscriptomic writer. ChemMedChem 15 (9), 744–748. doi:10.1002/cmdc.202000011
Campeanu, I. J., Jiang, Y., Liu, L., Pilecki, M., Najor, A., Cobani, E., et al. (2021). Multi-omics integration of methyltransferase-like protein family reveals clinical outcomes and functional signatures in human cancer. Sci. Rep. 11 (1), 14784. doi:10.1038/s41598-021-94019-5
Cao, K., Du, Y., Bao, X., Han, M., Su, R., Pang, J., et al. (2022). Glutathione-bioimprinted nanoparticles targeting of N6-methyladenosine FTO demethylase as a strategy against leukemic stem cells. Small 18 (13), e2106558. doi:10.1002/smll.202106558
Chen, B., Ye, F., Yu, L., Jia, G., Huang, X., Zhang, X., et al. (2012). Development of cell-active N6-methyladenosine RNA demethylase FTO inhibitor. J. Am. Chem. Soc. 134 (43), 17963–17971. doi:10.1021/ja3064149
Chen, G., Liu, B., Yin, S., Li, S., Guo, Y., Wang, M., et al. (2020a). Hypoxia induces an endometrial cancer stem-like cell phenotype via HIF-dependent demethylation of SOX2 mRNA. Oncogenesis 9 (9), 81. doi:10.1038/s41389-020-00265-z
Chen, Y., He, Y., Li, Z., Zhang, N., Zhou, C., He, X., et al. (2024). METTL3 facilitates renal cell carcinoma progression by PLOD2 m(6)A-methylation under prolonged hypoxia. Cell Death Dis. 15 (1), 62. doi:10.1038/s41419-023-06411-w
Chen, Y., Zhao, Y., Chen, J., Peng, C., Zhang, Y., Tong, R., et al. (2020b). ALKBH5 suppresses malignancy of hepatocellular carcinoma via m(6)A-guided epigenetic inhibition of LYPD1. Mol. Cancer 19 (1), 123. doi:10.1186/s12943-020-01239-w
Cheng, Y., Gao, Z., Zhang, T., Wang, Y., Xie, X., Han, G., et al. (2023). Decoding m(6)A RNA methylome identifies PRMT6-regulated lipid transport promoting AML stem cell maintenance. Cell Stem Cell 30 (1), 69–85.e7. doi:10.1016/j.stem.2022.12.003
Clarke, M. F. (2019). Clinical and therapeutic implications of cancer stem cells. N. Engl. J. Med. 380 (23), 2237–2245. doi:10.1056/NEJMra1804280
Cui, Q., Shi, H., Ye, P., Li, L., Qu, Q., Sun, G., et al. (2017). m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 18 (11), 2622–2634. doi:10.1016/j.celrep.2017.02.059
Dalerba, P., Cho, R. W., and Clarke, M. F. J. A. R. M. (2007). Cancer stem cells: models and concepts. Cancer stem cells models concepts 58 (1), 267–284. doi:10.1146/annurev.med.58.062105.204854
Dang, L., White, D. W., Gross, S., Bennett, B. D., Bittinger, M. A., Driggers, E. M., et al. (2009). Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462 (7274), 739–744. doi:10.1038/nature08617
Davalos, V., and Esteller, M. (2023). Cancer epigenetics in clinical practice. CA Cancer J. Clin. 73 (4), 376–424. doi:10.3322/caac.21765
Degrauwe, N., Schlumpf, T. B., Janiszewska, M., Martin, P., Cauderay, A., Provero, P., et al. (2016). The RNA binding protein IMP2 preserves glioblastoma stem cells by preventing let-7 target gene silencing. Cell Rep. 15 (8), 1634–1647. doi:10.1016/j.celrep.2016.04.086
Deng, X., Qing, Y., Horne, D., Huang, H., and Chen, J. (2023). The roles and implications of RNA m(6)A modification in cancer. Nat. Rev. Clin. Oncol. 20 (8), 507–526. doi:10.1038/s41571-023-00774-x
Dixit, D., Prager, B. C., Gimple, R. C., Poh, H. X., Wang, Y., Wu, Q., et al. (2021). The RNA m6A reader YTHDF2 maintains oncogene expression and is a targetable dependency in glioblastoma stem cells. Cancer Discov. 11 (2), 480–499. doi:10.1158/2159-8290.CD-20-0331
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485 (7397), 201–206. doi:10.1038/nature11112
Dou, X., Xiao, Y., Shen, C., Wang, K., Wu, T., Liu, C., et al. (2023). RBFOX2 recognizes N(6)-methyladenosine to suppress transcription and block myeloid leukaemia differentiation. Nat. Cell Biol. 25 (9), 1359–1368. doi:10.1038/s41556-023-01213-w
Du, H., Zhao, Y., He, J., Zhang, Y., Xi, H., Liu, M., et al. (2016). YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 7, 12626. doi:10.1038/ncomms12626
Du, Y., Yuan, Y., Xu, L., Zhao, F., Wang, W., Xu, Y., et al. (2022). Discovery of METTL3 small molecule inhibitors by virtual screening of natural products. Front. Pharmacol. 13, 878135. doi:10.3389/fphar.2022.878135
Dunn, D. B., and Smith, J. D. (1955). Occurrence of a new base in the deoxyribonucleic acid of a strain of bacterium coli. Nature 175 (4451), 336–337. doi:10.1038/175336a0
Edupuganti, R. R., Geiger, S., Lindeboom, R. G. H., Shi, H., Hsu, P. J., Lu, Z., et al. (2017). N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 24 (10), 870–878. doi:10.1038/nsmb.3462
Elcheva, I. A., Wood, T., Chiarolanzio, K., Chim, B., Wong, M., Singh, V., et al. (2020). RNA-binding protein IGF2BP1 maintains leukemia stem cell properties by regulating HOXB4, MYB, and ALDH1A1. Leukemia 34 (5), 1354–1363. doi:10.1038/s41375-019-0656-9
Eramo, A., Lotti, F., Sette, G., Pilozzi, E., Biffoni, M., Di Virgilio, A., et al. (2008). Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 15 (3), 504–514. doi:10.1038/sj.cdd.4402283
Fang, R., Chen, X., Zhang, S., Shi, H., Ye, Y., Shi, H., et al. (2021). EGFR/SRC/ERK-stabilized YTHDF2 promotes cholesterol dysregulation and invasive growth of glioblastoma. Nat. Commun. 12 (1), 177. doi:10.1038/s41467-020-20379-7
Fang, Z., Mu, B., Liu, Y., Guo, N., Xiong, L., Guo, Y., et al. (2022). Discovery of a potent, selective and cell active inhibitor of m(6)A demethylase ALKBH5. Eur. J. Med. Chem. 238, 114446. doi:10.1016/j.ejmech.2022.114446
Fazi, F., and Fatica, A. (2019). Interplay between N (6)-methyladenosine (m(6)A) and non-coding RNAs in cell development and cancer. Front. Cell Dev. Biol. 7, 116. doi:10.3389/fcell.2019.00116
Feng, G., Wu, Y., Hu, Y., Shuai, W., Yang, X., Li, Y., et al. (2024). Small molecule inhibitors targeting m(6)A regulators. J. Hematol. Oncol. 17 (1), 30. doi:10.1186/s13045-024-01546-5
Feng, P., Chen, D., Wang, X., Li, Y., Li, Z., Li, B., et al. (2022). Inhibition of the m(6)A reader IGF2BP2 as a strategy against T-cell acute lymphoblastic leukemia. Leukemia 36 (9), 2180–2188. doi:10.1038/s41375-022-01651-9
Fustin, J. M., Doi, M., Yamaguchi, Y., Hida, H., Nishimura, S., Yoshida, M., et al. (2013). RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155 (4), 793–806. doi:10.1016/j.cell.2013.10.026
Gao, J., Fang, Y., Chen, J., Tang, Z., Tian, M., Jiang, X., et al. (2023). Methyltransferase like 3 inhibition limits intrahepatic cholangiocarcinoma metabolic reprogramming and potentiates the efficacy of chemotherapy. Oncogene 42 (33), 2507–2520. doi:10.1038/s41388-023-02760-0
Gao, Z., Zha, X., Li, M., Xia, X., and Wang, S. (2024). Insights into the m6A demethylases FTO and ALKBH5: structural, biological function, and inhibitor development. Cell and Biosci. 14 (1), 108. doi:10.1186/s13578-024-01286-6
Garg, R., Melstrom, L., Chen, J., He, C., and Goel, A. (2022). Targeting FTO suppresses pancreatic carcinogenesis via regulating stem cell maintenance and EMT pathway. Cancers (Basel) 14 (23), 5919. doi:10.3390/cancers14235919
Guirguis, A. A., Ofir-Rosenfeld, Y., Knezevic, K., Blackaby, W., Hardick, D., Chan, Y. C., et al. (2023). Inhibition of METTL3 results in a cell-intrinsic interferon response that enhances antitumor immunity. Cancer Discov. 13 (10), 2228–2247. doi:10.1158/2159-8290.Cd-23-0007
Han, L., Dong, L., Leung, K., Zhao, Z., Li, Y., Gao, L., et al. (2023). METTL16 drives leukemogenesis and leukemia stem cell self-renewal by reprogramming BCAA metabolism. Cell Stem Cell 30 (1), 52–68 e13. doi:10.1016/j.stem.2022.12.006
Han, Z., Niu, T., Chang, J., Lei, X., Zhao, M., Wang, Q., et al. (2010). Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature 464 (7292), 1205–1209. doi:10.1038/nature08921
He, M., Lei, H., He, X., Liu, Y., Wang, A., Ren, Z., et al. (2022). METTL14 regulates osteogenesis of bone marrow mesenchymal stem cells via inducing autophagy through m6A/IGF2BPs/beclin-1 signal Axis. Stem Cells Transl. Med. 11 (9), 987–1001. doi:10.1093/stcltm/szac049
Hess, M. E., Hess, S., Meyer, K. D., Verhagen, L. A., Koch, L., Brönneke, H. S., et al. (2013). The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 16 (8), 1042–1048. doi:10.1038/nn.3449
Hong, Y. G., Yang, Z., Chen, Y., Liu, T., Zheng, Y., Zhou, C., et al. (2023). The RNA m6A reader YTHDF1 is required for acute myeloid leukemia progression. Cancer Res. 83 (6), 845–860. doi:10.1158/0008-5472.Can-21-4249
Hsu, P. J., Zhu, Y., Ma, H., Guo, Y., Shi, X., Liu, Y., et al. (2017). Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27 (9), 1115–1127. doi:10.1038/cr.2017.99
Hu, S., Yan, X., Bian, W., and Ni, B. (2023). The m6A reader IGF2BP1 manipulates BUB1B expression to affect malignant behaviors, stem cell properties, and immune resistance of non-small-cell lung cancer stem cells. Cytotechnology 75 (6), 517–532. doi:10.1007/s10616-023-00594-y
Hu, Y., Liu, H., Xiao, X., Yu, Q., Deng, R., Hua, L., et al. (2022). Bone marrow mesenchymal stem cell-derived exosomes inhibit triple-negative breast cancer cell stemness and metastasis via an ALKBH5-dependent mechanism. Cancers (Basel) 14 (24), 6059. doi:10.3390/cancers14246059
Huang, H., Wang, Y., Kandpal, M., Zhao, G., Cardenas, H., Ji, Y., et al. (2020). FTO-dependent N (6)-methyladenosine modifications inhibit ovarian cancer stem cell self-renewal by blocking cAMP signaling. Cancer Res. 80 (16), 3200–3214. doi:10.1158/0008-5472.Can-19-4044
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20 (3), 285–295. doi:10.1038/s41556-018-0045-z
Huang, Y., Su, R., Sheng, Y., Dong, L., Dong, Z., Xu, H., et al. (2019). Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell 35 (4), 677–691. doi:10.1016/j.ccell.2019.03.006
Huang, Y., Yan, J., Li, Q., Li, J., Gong, S., Zhou, H., et al. (2015). Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 43 (1), 373–384. doi:10.1093/nar/gku1276
Huff, S., Kummetha, I. R., Zhang, L., Wang, L., Bray, W., Yin, J., et al. (2022). Rational design and optimization of m(6)A-RNA demethylase FTO inhibitors as anticancer agents. J. Med. Chem. 65 (16), 10920–10937. doi:10.1021/acs.jmedchem.1c02075
Huff, S., Tiwari, S. K., Gonzalez, G. M., Wang, Y., and Rana, T. M. (2021). m(6)A-RNA demethylase FTO inhibitors impair self-renewal in glioblastoma stem cells. ACS Chem. Biol. 16 (2), 324–333. doi:10.1021/acschembio.0c00841
Ignatova, V. V., Stolz, P., Kaiser, S., Gustafsson, T. H., Lastres, P. R., Sanz-Moreno, A., et al. (2020). The rRNA m(6)A methyltransferase METTL5 is involved in pluripotency and developmental programs. Genes Dev. 34 (9-10), 715–729. doi:10.1101/gad.333369.119
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7 (12), 885–887. doi:10.1038/nchembio.687
Kowalski-Chauvel, A., Lacore, M. G., Arnauduc, F., Delmas, C., Toulas, C., Cohen-Jonathan-Moyal, E., et al. (2020). The m6A RNA demethylase ALKBH5 promotes radioresistance and invasion capability of glioma stem cells. Cancers (Basel) 13 (1), 40. doi:10.3390/cancers13010040
Kumari, S., Kumar, S., and Muthuswamy, S. (2023). RNA N6-methyladenosine modification in regulating cancer stem cells and tumor immune microenvironment and its implication for cancer therapy. J. Cancer Res. Clin. Oncol. 149 (4), 1621–1633. doi:10.1007/s00432-022-04158-z
Lai, G. Q., Li, Y., Zhu, H., Zhang, T., Gao, J., Zhou, H., et al. (2024). A covalent compound selectively inhibits RNA demethylase ALKBH5 rather than FTO. RSC Chem. Biol. 5 (4), 335–343. doi:10.1039/d3cb00230f
Lapidot, T., Sirard, C., Vormoor, J., Murdoch, B., Hoang, T., Caceres-Cortes, J., et al. (1994). A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 367 (6464), 645–648. doi:10.1038/367645a0
Lee, J. H., Choi, N., Kim, S., Jin, M. S., Shen, H., and Kim, Y. C. (2022a). Eltrombopag as an allosteric inhibitor of the METTL3-14 complex affecting the m(6)A methylation of RNA in acute myeloid leukemia cells. Pharm. (Basel) 15 (4), 440. doi:10.3390/ph15040440
Lee, J. H., Kim, S., Jin, M. S., and Kim, Y. C. (2022b). Discovery of substituted indole derivatives as allosteric inhibitors of m(6) A-RNA methyltransferase, METTL3-14 complex. Drug Dev. Res. 83 (3), 783–799. doi:10.1002/ddr.21910
Li, C., Heidt, D. G., Dalerba, P., Burant, C. F., Zhang, L., Adsay, V., et al. (2007). Identification of pancreatic cancer stem cells. Cancer Res. 67 (3), 1030–1037. doi:10.1158/0008-5472.Can-06-2030
Li, F., Wang, H., Ye, T., Guo, P., Lin, X., Hu, Y., et al. (2024). Recent advances in material technology for leukemia treatments. Adv. Mater. 36, 2313955. doi:10.1002/adma.202313955
Li, F., Yi, Y., Miao, Y., Long, W., Long, T., Chen, S., et al. (2019a). N6-Methyladenosine modulates nonsense-mediated mRNA decay in human glioblastoma. Cancer Res. 79 (22), 5785–5798. doi:10.1158/0008-5472.CAN-18-2868
Li, H., Wang, C., Lan, L., Yan, L., Li, W., Evans, I., et al. (2022). METTL3 promotes oxaliplatin resistance of gastric cancer CD133+ stem cells by promoting PARP1 mRNA stability. Cell Mol. Life Sci. 79 (3), 135. doi:10.1007/s00018-022-04129-0
Li, H. B., Tong, J., Zhu, S., Batista, P. J., Duffy, E. E., Zhao, J., et al. (2017). m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 548 (7667), 338–342. doi:10.1038/nature23450
Li, N., Kang, Y., Wang, L., Huff, S., Tang, R., Hui, H., et al. (2020). ALKBH5 regulates anti-PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc. Natl. Acad. Sci. U. S. A. 117 (33), 20159–20170. doi:10.1073/pnas.1918986117
Li, T., Hu, P. S., Zuo, Z., Lin, J. F., Li, X., Wu, Q. N., et al. (2019b). METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol. Cancer 18 (1), 112. doi:10.1186/s12943-019-1038-7
Li, Z., Qian, P., Shao, W., Shi, H., He, X. C., Gogol, M., et al. (2018). Suppression of m(6)A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res. 28 (9), 904–917. doi:10.1038/s41422-018-0072-0
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10 (2), 93–95. doi:10.1038/nchembio.1432
Liu, L., Zhao, T., Zheng, S., Tang, D., Han, H., Yang, C., et al. (2024a). METTL3 inhibitor STM2457 impairs tumor progression and enhances sensitivity to anlotinib in OSCC. Oral Dis. doi:10.1111/odi.14864
Liu, N., Dai, Q., Zheng, G., He, C., Parisien, M., and Pan, T. (2015). N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518 (7540), 560–564. doi:10.1038/nature14234
Liu, Q., Qi, J., Li, W., Tian, X., Zhang, J., Liu, F., et al. (2023). Therapeutic effect and transcriptome-methylome characteristics of METTL3 inhibition in liver hepatocellular carcinoma. Cancer Cell Int. 23 (1), 298. doi:10.1186/s12935-023-03096-1
Liu, X., Wang, Z., Yang, Q., Hu, X., Fu, Q., Zhang, X., et al. (2022a). RNA demethylase ALKBH5 prevents lung cancer progression by regulating EMT and stemness via regulating p53. Front. Oncol. 12, 858694. doi:10.3389/fonc.2022.858694
Liu, Y., Feng, W., Gu, S., Wang, H., Zhang, Y., Chen, W., et al. (2019). The UCA1/KRAS axis promotes human pancreatic ductal adenocarcinoma stem cell properties and tumor growth. Am. J. Cancer Res. 9 (3), 496–510.
Liu, Y., Goebel, G. L., Kanis, L., Hasturk, O., Kemker, C., and Wu, P. (2024b). Aminothiazolone inhibitors disrupt the protein-RNA interaction of METTL16 and modulate the m(6)A RNA modification. JACS Au 4 (4), 1436–1449. doi:10.1021/jacsau.3c00832
Liu, Y., Liang, G., Xu, H., Dong, W., Dong, Z., Qiu, Z., et al. (2021). Tumors exploit FTO-mediated regulation of glycolytic metabolism to evade immune surveillance. Cell Metab. 33 (6), 1221–1233.e11. doi:10.1016/j.cmet.2021.04.001
Liu, Y., Song, R., Zhao, L., Lu, Z., Li, Y., Zhan, X., et al. (2022b). m(6)A demethylase ALKBH5 is required for antibacterial innate defense by intrinsic motivation of neutrophil migration. Signal Transduct. Target Ther. 7 (1), 194. doi:10.1038/s41392-022-01020-z
Lu, Z., Liu, H., Song, N., Liang, Y., Zhu, J., Chen, J., et al. (2021). METTL14 aggravates podocyte injury and glomerulopathy progression through N(6)-methyladenosine-dependent downregulating of Sirt1. Cell Death Dis. 12 (10), 881. doi:10.1038/s41419-021-04156-y
Ma, H., Wang, X., Cai, J., Dai, Q., Natchiar, S. K., Lv, R., et al. (2019). N(6-)Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 15 (1), 88–94. doi:10.1038/s41589-018-0184-3
Ma, S., Chan, K. W., Hu, L., Lee, T. K., Wo, J. Y., Ng, I. O., et al. (2007). Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology 132 (7), 2542–2556. doi:10.1053/j.gastro.2007.04.025
Ma, Z., and Ji, J. (2020). N6-methyladenosine (m6A) RNA modification in cancer stem cells. Stem Cells 38, 1511–1519. doi:10.1002/stem.3279
Mahapatra, L., Andruska, N., Mao, C., Le, J., and Shapiro, D. J. (2017). A novel IMP1 inhibitor, BTYNB, targets c-myc and inhibits melanoma and ovarian cancer cell proliferation. Transl. Oncol. 10 (5), 818–827. doi:10.1016/j.tranon.2017.07.008
Malacrida, A., Rivara, M., Di Domizio, A., Cislaghi, G., Miloso, M., Zuliani, V., et al. (2020). 3D proteome-wide scale screening and activity evaluation of a new ALKBH5 inhibitor in U87 glioblastoma cell line. Bioorg Med. Chem. 28 (4), 115300. doi:10.1016/j.bmc.2019.115300
Mauer, J., Luo, X., Blanjoie, A., Jiao, X., Grozhik, A. V., Patil, D. P., et al. (2017). Reversible methylation of m(6)A(m) in the 5' cap controls mRNA stability. Nature 541 (7637), 371–375. doi:10.1038/nature21022
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 149 (7), 1635–1646. doi:10.1016/j.cell.2012.05.003
Mineo, M., Ricklefs, F., Rooj, A. K., Lyons, S. M., Ivanov, P., Ansari, K. I., et al. (2016). The long non-coding RNA HIF1A-AS2 facilitates the maintenance of mesenchymal glioblastoma stem-like cells in hypoxic niches. Cell Rep. 15 (11), 2500–2509. doi:10.1016/j.celrep.2016.05.018
Mohiuddin, I. S., Wei, S. J., and Kang, M. H. (2020). Role of OCT4 in cancer stem-like cells and chemotherapy resistance. Biochim. Biophys. Acta Mol. Basis Dis. 1866 (4), 165432. doi:10.1016/j.bbadis.2019.03.005
Moroz-Omori, E. V., Huang, D., Kumar Bedi, R., Cheriyamkunnel, S. J., Bochenkova, E., Dolbois, A., et al. (2021). METTL3 inhibitors for epitranscriptomic modulation of cellular processes. ChemMedChem 16 (19), 3035–3043. doi:10.1002/cmdc.202100291
Novak, D., Huser, L., Elton, J. J., Umansky, V., Altevogt, P., and Utikal, J. (2020). SOX2 in development and cancer biology. Semin. Cancer Biol. 67 (Pt 1), 74–82. doi:10.1016/j.semcancer.2019.08.007
O'Brien, C. A., Pollett, A., Gallinger, S., and Dick, J. E. (2007). A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 445 (7123), 106–110. doi:10.1038/nature05372
Paris, J., Morgan, M., Campos, J., Spencer, G. J., Shmakova, A., Ivanova, I., et al. (2019). Targeting the RNA m(6)A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell 25 (1), 137–148. doi:10.1016/j.stem.2019.03.021
Patil, D. P., Chen, C. K., Pickering, B. F., Chow, A., Jackson, C., Guttman, M., et al. (2016). m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537 (7620), 369–373. doi:10.1038/nature19342
Patil, D. P., Pickering, B. F., and Jaffrey, S. R. (2018). Reading m(6)A in the transcriptome: m(6)a-binding proteins. Trends Cell Biol. 28 (2), 113–127. doi:10.1016/j.tcb.2017.10.001
Patrawala, L., Calhoun, T., Schneider-Broussard, R., Li, H., Bhatia, B., Tang, S., et al. (2006). Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene 25 (12), 1696–1708. doi:10.1038/sj.onc.1209327
Pendleton, K. E., Chen, B., Liu, K., Hunter, O. V., Xie, Y., Tu, B. P., et al. (2017). The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 169 (5), 824–835. doi:10.1016/j.cell.2017.05.003
Peng, F., Xu, J., Cui, B., Liang, Q., Zeng, S., He, B., et al. (2021). Oncogenic AURKA-enhanced N(6)-methyladenosine modification increases DROSHA mRNA stability to transactivate STC1 in breast cancer stem-like cells. Cell Res. 31 (3), 345–361. doi:10.1038/s41422-020-00397-2
Pérez-Herrero, E., and Fernández-Medarde, A. (2015). Advanced targeted therapies in cancer: drug nanocarriers, the future of chemotherapy. Eur. J. Pharm. Biopharm. 93, 52–79. doi:10.1016/j.ejpb.2015.03.018
Perry, R. P., and Kelley, D. E. (1974). Existence of methylated messenger RNA in mouse L cells. Cell 1 (1), 37–42. doi:10.1016/0092-8674(74)90153-6
Pianka, S. T., Li, T., Prins, T. J., Eldred, B. S. C., Kevan, B. M., Liang, H., et al. (2024). D-2-HG inhibits IDH1mut glioma growth via FTO inhibition and resultant m6A hypermethylation. Cancer Res. Commun. 4 (3), 876–894. doi:10.1158/2767-9764.CRC-23-0271
Ping, X. L., Sun, B. F., Wang, L., Xiao, W., Yang, X., Wang, W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24 (2), 177–189. doi:10.1038/cr.2014.3
Pinto, R., Vagbo, C. B., Jakobsson, M. E., Kim, Y., Baltissen, M. P., O'Donohue, M. F., et al. (2020). The human methyltransferase ZCCHC4 catalyses N6-methyladenosine modification of 28S ribosomal RNA. Nucleic Acids Res. 48 (2), 830–846. doi:10.1093/nar/gkz1147
Qin, F., Cai, B., Zhao, J., Zhang, L., Zheng, Y., Liu, B., et al. (2021). Methyltransferase-like protein 14 attenuates mitochondrial antiviral signaling protein expression to negatively regulate antiviral immunity via N(6) -methyladenosine modification. Adv. Sci. (Weinh) 8 (15), e2100606. doi:10.1002/advs.202100606
Qing, Y., Dong, L., Gao, L., Li, C., Li, Y., Han, L., et al. (2021). R-2-hydroxyglutarate attenuates aerobic glycolysis in leukemia by targeting the FTO/m(6)A/PFKP/LDHB axis. Mol. Cell 81 (5), 922–939.e9. doi:10.1016/j.molcel.2020.12.026
Qing, Y., Wu, D., Deng, X., Chen, J., and Su, R. (2023). RNA modifications in cancer metabolism and tumor microenvironment. Cancer Treat. Res. 190, 3–24. doi:10.1007/978-3-031-45654-1_1
Ren, W., Lu, J., Huang, M., Gao, L., Li, D., Wang, G. G., et al. (2019). Structure and regulation of ZCCHC4 in m(6)A-methylation of 28S rRNA. Nat. Commun. 10 (1), 5042. doi:10.1038/s41467-019-12923-x
Rong, L., Xu, Y., Zhang, K., Jin, L., and Liu, X. (2022). HNRNPA2B1 inhibited SFRP2 and activated Wnt-β/catenin via m6A-mediated miR-106b-5p processing to aggravate stemness in lung adenocarcinoma. Pathol. Res. Pract. 233, 153794. doi:10.1016/j.prp.2022.153794
Růžička, K., Zhang, M., Campilho, A., Bodi, Z., Kashif, M., Saleh, M., et al. (2017). Identification of factors required for m(6) A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 215 (1), 157–172. doi:10.1111/nph.14586
Selberg, S., Seli, N., Kankuri, E., and Karelson, M. (2021). Rational design of novel anticancer small-molecule RNA m6A demethylase ALKBH5 inhibitors. ACS Omega 6 (20), 13310–13320. doi:10.1021/acsomega.1c01289
Sepich-Poore, C., Zheng, Z., Schmitt, E., Wen, K., Zhang, Z. S., Cui, X. L., et al. (2022). The METTL5-TRMT112 N(6)-methyladenosine methyltransferase complex regulates mRNA translation via 18S rRNA methylation. J. Biol. Chem. 298 (3), 101590. doi:10.1016/j.jbc.2022.101590
Shen, C., Sheng, Y., Zhu, A. C., Robinson, S., Jiang, X., Dong, L., et al. (2020). RNA demethylase ALKBH5 selectively promotes tumorigenesis and cancer stem cell self-renewal in acute myeloid leukemia. Cell Stem Cell 27 (1), 64–80. doi:10.1016/j.stem.2020.04.009
Shen, H., Geng, Z., Nie, X., and Liu, T. (2023). Erianin induces ferroptosis of renal cancer stem cells via promoting ALOX12/P53 mRNA N6-methyladenosine modification. J. Cancer 14 (3), 367–378. doi:10.7150/jca.81027
Shi, H., Wang, X., Lu, Z., Zhao, B. S., Ma, H., Hsu, P. J., et al. (2017). YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 27 (3), 315–328. doi:10.1038/cr.2017.15
Shi, H., Wei, J., and He, C. (2019). Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 74 (4), 640–650. doi:10.1016/j.molcel.2019.04.025
Shi, J., Zhang, P., Dong, X., Yuan, J., Li, Y., Li, S., et al. (2023). METTL3 knockdown promotes temozolomide sensitivity of glioma stem cells via decreasing MGMT and APNG mRNA stability. Cell Death Discov. 9 (1), 22. doi:10.1038/s41420-023-01327-y
Singh, B., Kinne, H. E., Milligan, R. D., Washburn, L. J., Olsen, M., and Lucci, A. (2016). Important role of FTO in the survival of rare panresistant triple-negative inflammatory breast cancer cells facing a severe metabolic challenge. PLoS One 11 (7), e0159072. doi:10.1371/journal.pone.0159072
Singh, S. K., Hawkins, C., Clarke, I. D., Squire, J. A., Bayani, J., Hide, T., et al. (2004). Identification of human brain tumour initiating cells. Nature 432 (7015), 396–401. doi:10.1038/nature03128
Sturgess, K., Yankova, E., Vijayabaskar, M. S., Isobe, T., Rak, J., Kucinski, I., et al. (2023). Pharmacological inhibition of METTL3 impacts specific haematopoietic lineages. Leukemia 37 (10), 2133–2137. doi:10.1038/s41375-023-01965-2
Su, R., Dong, L., Li, C., Nachtergaele, S., Wunderlich, M., Qing, Y., et al. (2018). R-2HG exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell 172 (1-2), 90–105. doi:10.1016/j.cell.2017.11.031
Su, R., Dong, L., Li, Y., Gao, M., Han, L., Wunderlich, M., et al. (2020). Targeting FTO suppresses cancer stem cell maintenance and immune evasion. Cancer Cell 38 (1), 79–96. doi:10.1016/j.ccell.2020.04.017
Su, R., Dong, L., Li, Y., Gao, M., He, P. C., Liu, W., et al. (2022). METTL16 exerts an m(6)A-independent function to facilitate translation and tumorigenesis. Nat. Cell Biol. 24 (2), 205–216. doi:10.1038/s41556-021-00835-2
Sun, C. L., Chen, J., Xing, Z. W., and Tao, G. S. (2023). METTL14 suppresses cancer stem cell phenotype of colorectal cancer via regulating of β-catenin/NANOG. J. Cancer 14 (8), 1407–1416. doi:10.7150/jca.82158
Tan, X., Zheng, C., Zhuang, Y., Jin, P., and Wang, F. (2023). The m6A reader PRRC2A is essential for meiosis I completion during spermatogenesis. Nat. Commun. 14 (1), 1636. doi:10.1038/s41467-023-37252-y
Tang, B., Yang, Y., Kang, M., Wang, Y., Wang, Y., Bi, Y., et al. (2020). m(6)A demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation and mediating Wnt signaling. Mol. Cancer 19 (1), 3. doi:10.1186/s12943-019-1128-6
van Tran, N., Ernst, F. G. M., Hawley, B. R., Zorbas, C., Ulryck, N., Hackert, P., et al. (2019). The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 47 (15), 7719–7733. doi:10.1093/nar/gkz619
Vasefifar, P., Motafakkerazad, R., Maleki, L. A., Najafi, S., Ghrobaninezhad, F., Najafzadeh, B., et al. (2022). Nanog, as a key cancer stem cell marker in tumor progression. Gene 827, 146448. doi:10.1016/j.gene.2022.146448
Visvanathan, A., Patil, V., Arora, A., Hegde, A. S., Arivazhagan, A., Santosh, V., et al. (2018). Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 37 (4), 522–533. doi:10.1038/onc.2017.351
Walcher, L., Kistenmacher, A.-K., Suo, H., Kitte, R., Dluczek, S., Strauß, A., et al. (2020). Cancer stem cells-origins and biomarkers: perspectives for targeted personalized therapies, Front. Immunol Cancer stem cells—origins biomarkers Perspect. Target. personalized Ther. 11, 1280, doi:10.3389/fimmu.2020.01280
Wang, J., Yu, H., Dong, W., Zhang, C., Hu, M., Ma, W., et al. (2023a). N6-Methyladenosine-Mediated up-regulation of FZD10 regulates liver cancer stem cells' properties and lenvatinib resistance through WNT/β-Catenin and hippo signaling pathways. Gastroenterology 164 (6), 990–1005. doi:10.1053/j.gastro.2023.01.041
Wang, L., Wang, J., Yu, P., Feng, J., Xu, G. E., Zhao, X., et al. (2022). METTL14 is required for exercise-induced cardiac hypertrophy and protects against myocardial ischemia-reperfusion injury. Nat. Commun. 13 (1), 6762. doi:10.1038/s41467-022-34434-y
Wang, P., Doxtader, K. A., and Nam, Y. (2016). Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63 (2), 306–317. doi:10.1016/j.molcel.2016.05.041
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014a). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505 (7481), 117–120. doi:10.1038/nature12730
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161 (6), 1388–1399. doi:10.1016/j.cell.2015.05.014
Wang, Y., Li, Y., Toth, J. I., Petroski, M. D., Zhang, Z., and Zhao, J. C. (2014b). N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16 (2), 191–198. doi:10.1038/ncb2902
Wang, Y., Zhang, L., Sun, X. L., Lu, Y. C., Chen, S., Pei, D. S., et al. (2023b). NRP1 contributes to stemness and potentiates radioresistance via WTAP-mediated m6A methylation of Bcl-2 mRNA in breast cancer. Apoptosis 28 (1-2), 233–246. doi:10.1007/s10495-022-01784-3
Warda, A. S., Kretschmer, J., Hackert, P., Lenz, C., Urlaub, H., Hobartner, C., et al. (2017). Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 18 (11), 2004–2014. doi:10.15252/embr.201744940
Wei, J., Liu, F., Lu, Z., Fei, Q., Ai, Y., He, P. C., et al. (2018). Differential m(6)A, m(6)A(m), and m(1)A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell 71 (6), 973–985. doi:10.1016/j.molcel.2018.08.011
Wei, Z., Liu, X., Cheng, C., Yu, W., and Yi, P. (2020). Metabolism of amino acids in cancer. Front. Cell Dev. Biol. 8, 603837. doi:10.3389/fcell.2020.603837
Wen, J., Lv, R., Ma, H., Shen, H., He, C., Wang, J., et al. (2018). Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol. Cell 69 (6), 1028–1038. doi:10.1016/j.molcel.2018.02.015
Weng, H., Huang, F., Yu, Z., Chen, Z., Prince, E., Kang, Y., et al. (2022). The m(6)A reader IGF2BP2 regulates glutamine metabolism and represents a therapeutic target in acute myeloid leukemia. Cancer Cell 40 (12), 1566–1582.e10. doi:10.1016/j.ccell.2022.10.004
Weng, H., Huang, H., Wu, H., Qin, X., Zhao, B. S., Dong, L., et al. (2018). METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell 22 (2), 191–205. doi:10.1016/j.stem.2017.11.016
Wu, R., Li, A., Sun, B., Sun, J. G., Zhang, J., Zhang, T., et al. (2019). A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 29 (1), 23–41. doi:10.1038/s41422-018-0113-8
Xiang, Y., Laurent, B., Hsu, C. H., Nachtergaele, S., Lu, Z., Sheng, W., et al. (2017). RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature 543 (7646), 573–576. doi:10.1038/nature21671
Xiao, H., Zhao, R., Meng, W., and Liao, Y. (2023a). Effects and translatomics characteristics of a small-molecule inhibitor of METTL3 against non-small cell lung cancer. J. Pharm. Anal. 13 (6), 625–639. doi:10.1016/j.jpha.2023.04.009
Xiao, P., Duan, Z., Liu, Z., Chen, L., Zhang, D., Liu, L., et al. (2023b). Rational design of RNA demethylase FTO inhibitors with enhanced antileukemia drug-like properties. J. Med. Chem. 66 (14), 9731–9752. doi:10.1021/acs.jmedchem.3c00543
Xiao, S., Ma, S., Sun, B., Pu, W., Duan, S., Han, J., et al. (2024). The tumor-intrinsic role of the m(6)A reader YTHDF2 in regulating immune evasion. Sci. Immunol. 9 (95), eadl2171. doi:10.1126/sciimmunol.adl2171
Xie, J., Ba, J., Zhang, M., Wan, Y., Jin, Z., and Yao, Y. (2021). The m6A methyltransferase METTL3 promotes the stemness and malignant progression of breast cancer by mediating m6A modification on SOX2. J. buon 26 (2), 444–449.
Xu, K., Yang, Y., Feng, G. H., Sun, B. F., Chen, J. Q., Li, Y. F., et al. (2017). Mettl3-mediated m(6)A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 27 (9), 1100–1114. doi:10.1038/cr.2017.100
Xu, Y., He, X., Wang, S., Sun, B., Jia, R., Chai, P., et al. (2022a). The m(6)A reading protein YTHDF3 potentiates tumorigenicity of cancer stem-like cells in ocular melanoma through facilitating CTNNB1 translation. Oncogene 41 (9), 1281–1297. doi:10.1038/s41388-021-02146-0
Xu, Y., Zhou, J., Li, L., Yang, W., Zhang, Z., Zhang, K., et al. (2022b). FTO-mediated autophagy promotes progression of clear cell renal cell carcinoma via regulating SIK2 mRNA stability. Int. J. Biol. Sci. 18 (15), 5943–5962. doi:10.7150/ijbs.77774
Xue, M., Dong, L., Zhang, H., Li, Y., Qiu, K., Zhao, Z., et al. (2024). METTL16 promotes liver cancer stem cell self-renewal via controlling ribosome biogenesis and mRNA translation. J. Hematol. Oncol. 17 (1), 7. doi:10.1186/s13045-024-01526-9
Yan, H., Zhang, L., Cui, X., Zheng, S., and Li, R. (2022). Roles and mechanisms of the m6A reader YTHDC1 in biological processes and diseases. Cell Death Discov. 8 (1), 237. doi:10.1038/s41420-022-01040-2
Yang, L., Shi, P., Zhao, G., Xu, J., Peng, W., Zhang, J., et al. (2020a). Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target Ther. 5 (1), 8. doi:10.1038/s41392-020-0110-5
Yang, L., Shi, P., Zhao, G., Xu, J., Peng, W., Zhang, J., et al. (2020b). Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target. Ther. 5 (1), 8. doi:10.1038/s41392-020-0110-5
Yang, Y., Wu, J., Liu, F., He, J., Wu, F., Chen, J., et al. (2021). IGF2BP1 promotes the liver cancer stem cell phenotype by regulating MGAT5 mRNA stability by m6A RNA methylation. Stem Cells Dev. 30 (22), 1115–1125. doi:10.1089/scd.2021.0153
Yankova, E., Blackaby, W., Albertella, M., Rak, J., De Braekeleer, E., Tsagkogeorga, G., et al. (2021). Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 593 (7860), 597–601. doi:10.1038/s41586-021-03536-w
Yarmishyn, A. A., Yang, Y. P., Lu, K. H., Chen, Y. C., Chien, Y., Chou, S. J., et al. (2020). Musashi-1 promotes cancer stem cell properties of glioblastoma cells via upregulation of YTHDF1. Cancer Cell Int. 20 (1), 597. doi:10.1186/s12935-020-01696-9
Yin, J., Ding, F., Cheng, Z., Ge, X., Li, Y., Zeng, A., et al. (2023). METTL3-mediated m6A modification of LINC00839 maintains glioma stem cells and radiation resistance by activating Wnt/β-catenin signaling. Cell Death Dis. 14 (7), 417. doi:10.1038/s41419-023-05933-7
Yue, Y., Liu, J., Cui, X., Cao, J., Luo, G., Zhang, Z., et al. (2018). VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 4, 10. doi:10.1038/s41421-018-0019-0
Zhang, C., Huang, S., Zhuang, H., Ruan, S., Zhou, Z., Huang, K., et al. (2020). YTHDF2 promotes the liver cancer stem cell phenotype and cancer metastasis by regulating OCT4 expression via m6A RNA methylation. Oncogene 39 (23), 4507–4518. doi:10.1038/s41388-020-1303-7
Zhang, C., Samanta, D., Lu, H., Bullen, J. W., Zhang, H., Chen, I., et al. (2016). Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m⁶A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. U. S. A. 113 (14), E2047–E2056. doi:10.1073/pnas.1602883113
Zhang, S., Zhao, B. S., Zhou, A., Lin, K., Zheng, S., Lu, Z., et al. (2017). m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 31 (4), 591–606. doi:10.1016/j.ccell.2017.02.013
Zhang, S., Zhou, L., Yang, J., Lu, J., Tao, L., Feng, Y., et al. (2024a). Rhein exerts anti-multidrug resistance in acute myeloid leukemia via targeting FTO to inhibit AKT/mTOR. Anticancer Drugs 35, 597–605. doi:10.1097/cad.0000000000001608
Zhang, X., Su, T., Wu, Y., Cai, Y., Wang, L., Liang, C., et al. (2024b). N6-Methyladenosine reader YTHDF1 promotes stemness and therapeutic resistance in hepatocellular carcinoma by enhancing NOTCH1 expression. Cancer Res. 84 (6), 827–840. doi:10.1158/0008-5472.Can-23-1916
Zhang, Y., Liu, X., Wang, Y., Lai, S., Wang, Z., Yang, Y., et al. (2022). The m(6)A demethylase ALKBH5-mediated upregulation of DDIT4-AS1 maintains pancreatic cancer stemness and suppresses chemosensitivity by activating the mTOR pathway. Mol. Cancer 21 (1), 174. doi:10.1186/s12943-022-01647-0
Zhang, Y., Song, Z., Wu, R., Kong, X., Zhang, H., Li, S., et al. (2024c). PRRC2B modulates oligodendrocyte progenitor cell development and myelination by stabilizing Sox2 mRNA. Cell Rep. 43 (3), 113930. doi:10.1016/j.celrep.2024.113930
Zhang, Z., Tan, X., Wu, R., Deng, T., Wang, H., Jiang, X., et al. (2023). m6A-mediated upregulation of lncRNA-AC026356.1 promotes cancer stem cell maintenance in lung adenocarcinoma via activating Wnt signaling pathway. Aging (Albany NY) 15 (9), 3538–3548. doi:10.18632/aging.204689
Zhao, X., Yang, Y., Sun, B. F., Shi, Y., Yang, X., Xiao, W., et al. (2014). FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 24 (12), 1403–1419. doi:10.1038/cr.2014.151
Zhao, Z., Zeng, J., Guo, Q., Pu, K., Yang, Y., Chen, N., et al. (2021). Berberine suppresses stemness and tumorigenicity of colorectal cancer stem-like cells by inhibiting m(6)A methylation. Front. Oncol. 11, 775418. doi:10.3389/fonc.2021.775418
Zheng, G., Cox, T., Tribbey, L., Wang, G. Z., Iacoban, P., Booher, M. E., et al. (2014). Synthesis of a FTO inhibitor with anticonvulsant activity. ACS Chem. Neurosci. 5 (8), 658–665. doi:10.1021/cn500042t
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C. M., Li, C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49 (1), 18–29. doi:10.1016/j.molcel.2012.10.015
Zheng, Q., Hou, J., Zhou, Y., Li, Z., and Cao, X. (2017). The RNA helicase DDX46 inhibits innate immunity by entrapping m(6)A-demethylated antiviral transcripts in the nucleus. Nat. Immunol. 18 (10), 1094–1103. doi:10.1038/ni.3830
Zhou, H.-M., Zhang, J.-G., Zhang, X., Li, Q. J. S. t., and therapy, t. (2021a). Targeting cancer stem cells for reversing therapy resistance: mechanism, signaling, and prospective agents. Signal Transduct. Target. Ther. 6 (1), 62. doi:10.1038/s41392-020-00430-1
Zhou, J., Zhang, X., Hu, J., Qu, R., Yu, Z., Xu, H., et al. (2021b). m(6)A demethylase ALKBH5 controls CD4(+) T cell pathogenicity and promotes autoimmunity. Sci. Adv. 7 (25), eabg0470. doi:10.1126/sciadv.abg0470
Zhou, X., Yang, X., Huang, S., Lin, G., Lei, K., Wang, Q., et al. (2024). Inhibition of METTL3 alleviates NLRP3 inflammasome activation via increasing ubiquitination of NEK7. Adv. Sci. (Weinh) 11, e2308786. doi:10.1002/advs.202308786
Keywords: N6-methyladenosine (m6 A), RNA methylation, methyltransferase-like 3 (METTL3), METTL14, fat mass and obesity associated (FTO), alkB homolog 5 (ALKBH5), cancer stem cells (CSCs), self-renewal
Citation: Zhang H, Wang X, Chen J and Su R (2024) Combating cancer stem cells: RNA m6A methylation and small-molecule drug discovery. Front. Drug Discov. 4:1465222. doi: 10.3389/fddsv.2024.1465222
Received: 15 July 2024; Accepted: 17 September 2024;
Published: 30 September 2024.
Edited by:
Hoang V. Le, National Institute on Drug Abuse (NIH), United StatesReviewed by:
Soma Samanta, University of Michigan, United StatesSicong Zhang, The Rockefeller University, United States
Copyright © 2024 Zhang, Wang, Chen and Su. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rui Su, cnN1QGNvaC5vcmc=
 Honghai Zhang
Honghai Zhang Xueer Wang
Xueer Wang Jianjun Chen
Jianjun Chen Rui Su
Rui Su