
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Drug Discov., 08 March 2024
Sec. Anti-Infective Agents
Volume 4 - 2024 | https://doi.org/10.3389/fddsv.2024.1326121
This article is part of the Research TopicTackling Antimicrobial ResistanceView all 4 articles
Antimicrobial resistance poses an urgent and formidable global public health threat. The escalation of bacterial multidrug resistance to antibiotics has the potential to become a leading cause of global mortality if there is no substantial improvement in antimicrobial development and therapy protocols. In light of this, it is imperative to identify the molecular determinants responsible for the reduced antibiotic activity associated with RND efflux pumps. This comprehensive study meticulously examines Minimum Inhibitory Concentration (MIC) data obtained from in vitro tests for various antibiotic families and non-active dye compounds, sourced from diverse literature references. The primary focus of this study is to assess the susceptibility of these agents to efflux-resistant Escherichia coli strains, integrating both MIC data and relevant physicochemical properties. The central objective is to unveil the specific substituents that significantly influence the uptake process mediated by the AcrAB-TolC efflux system. This exploration seeks to reveal the consequences of these substituents on pharmacodynamic responses, providing valuable insights into Structure-Activity Relationships. It is noteworthy that this analysis represents a pioneering effort, with prospective implications for RND efflux pump-producing strains. Ultimately, deciphering efflux markers is crucial to effectively mitigate the emergence of specific resistance and to better monitor the role of this primary resistance mechanism in Gram-negative bacteria, particularly as observed in clinical antibiotic therapy practice.
When antibacterials such as antibiotics and biocides began to be used to combat bacterial infectious diseases, it marked the onset of a new era in modern medicine. Their usage became widespread since 1945. Unfortunately, this new therapeutic arsenal has given rise to the reality of drug resistance.
The 2014 WHO report and the Review on Antimicrobial Resistance (AMR) highlighted disturbing epidemiological evidence regarding the acquired drug resistance of persistent bacterial pathogens in both hospitals and communities. These findings underscore the alarming projection that the death toll due to AMR could reach 10 million people annually by 2050. Recently, a study on the global burden of bacterial AMR was published, focusing on AMR-associated deaths in the year 2019 (Antimicrobial Resistance Collaborators, 2022). Numerous health-environment investigations highlight the growing concern regarding antibiotic resistance, which stands as one of the leading causes of death worldwide (Sodhi and Singh, 2022; Singh et al., 2023; Sodhi et al., 2023). This concern is particularly pronounced as the estimated global death rate for 2050 has been reached over the past 5 years, surpassing 50 per cent of the predictions. These phenomena contribute significantly to the loss of antimicrobial activity for compounds that are still expected to be effective. Bacteria, like any living organism, continually adapt to their environment to survive and evolve over time and space (Muteeb et al., 2023). The molecular underpinnings of antimicrobial resistance emergence in bacteria trace their origins to antiquity. Pre-existing mechanisms can be considered as precursors to contemporary resistance determinants. These molecular pathways, strengthened through evolutionary processes, have endowed bacteria with the capacity to deploy survival strategies when confronted with natural antimicrobial agents from their environment (Christaki et al., 2020). This adaptive process has resulted in selective pressure and the emergence of antibiotic-resistant mutants in clinical contexts (Lerminiaux and Cameron, 2019; Smith et al., 2023).
The evolution of antibiotic resistance in common bacterial strains has extended to a wide range of antibiotics, contributing to the Multi-Drug Resistance (MDR) phenotype, especially in Gram-negative bacteria (Darby et al., 2023; Salam et al., 2023). This is evident in strains exhibiting acquired and intrinsic resistance mechanisms. They can be categorized in two main types, based on their nature and impact on antibiotic activity. The first type involves enzymatic degradation, particularly affecting β-lactams, aminoglycosides and some macrolides through pharmacophore hydrolysis or biotransformation, resulting in an inactive molecular structure (Tooke et al., 2019; Sawa et al., 2020). The second type encompasses intracellular target alterations in bacterial DNA and mRNA, affecting penicillin and quinolone derivatives in both Gram-positive and Gram-negative bacteria, leading to a poorly targeted molecule (Schaenzer and Wright, 2020; Spencer and Panda, 2023). These resistance mechanisms are specifically associated with one or two groups of antibiotics based on pharmacodynamic characteristics. They may also be coupled with other membrane effects that enable bacteria to extend the lag period in response during antibiotic exposure. At the membrane level, decreased porin expression (Vergalli et al., 2020) and, significantly, the overproduction of efflux pump are major contributors to the loss of effective intracellular antibiotic concentrations. These effects manifest early in exposure and involve all antibiotic classes, leading to the MDR phenotype. Consequently, they constitute the primary resistance barrier that initiates specific alternative mechanisms following exposure to toxic molecules (Krishnamoorthy et al., 2017).
Efflux pumps are ubiquitous protein complexes that actively extrude substances damaging to bacteria such as antibiotics, detergents, antiseptics, and toxins from the cytoplasm or periplasm into the external environment. These transporters recognize a wide range of physicochemically varied substrates (Blair et al., 2015; Zgurskaya et al., 2021). Currently, there are six families, including ABC (ATP-Binding Cassette), SMR (Small Multidrug Resistance), MFS (Major Facilitator Superfamily), MATE (Multidrug And Toxic compound Extrusion), PACE (Proteobacterial Antimicrobial Compound Efflux) and RND (Resistance-Nodulation-Division) transporters (Du et al., 2018; Hassan et al., 2018).
The last two classes, but more specifically RND transporters, which exhibit an energy-dependent tripartite system provided by a proton pump initially discovered in Tetracycline resistant Escherichia coli, were isolated similarly to P-glycoproteins in mammalian cells (Waghray and Zhang, 2018).
The intricate architecture of RND pumps comprises an elaborate assembly, including an inner membrane transporter, periplasmic adaptor proteins, and an outer membrane channel protein (Alav et al., 2021). These tripartite systems, prominently observed in Gram-negative bacteria, notably within the ESKAPE group, wield a pivotal role in conferring resistance. Their influence is particularly significant in the context of the escalating prevalence of antimicrobial resistance-related infections, standing as the primary cause of nosocomial infections on a global scale (Gauba and Rahman, 2023). The chief clinical concern associated with Resistance-Nodulation-Division (RND) pumps stems from their intrinsic linkage between expression levels and broad polyselectivity, evident in both the proximal and distal binding pockets of the inner membrane protein (Jang, 2023). This interconnection poses a formidable challenge in precisely identifying pharmacophore groups within the chemical structures of drugs implicated in the recognition and transport processes governed by these pumps.
Among the extensively studied efflux systems, AcrAB-TolC stands out as a focal point, particularly within enterobacteria such as E. coli, Enterobacter spp., K. pneumoniae, and K. aerogenes. This well-explored system exhibits a diverse substrate profile encompassing various antibiotic classes, including chloramphenicol, tetracyclines, quinolones, macrolides, and β-lactams (Du et al., 2018). The elucidation of the intricate workings of these efflux systems is crucial for comprehending antibiotic resistance mechanisms and devising targeted strategies to counteract the rising tide of antimicrobial resistance in clinical settings. The AcrAB-TolC pump, a critical component in Gram-negative bacteria resistance, comprises three integral parts: the homotrimeric outer membrane channel TolC, the homotrimeric inner membrane transporter AcrB, and the periplasmic protein AcrA, present as six protomers, bridging the connection between these two integral membrane proteins (McNeil et al., 2019; Gaurav et al., 2023). State-of-the-art cryo-electron microscopy technologies, as employed by various research teams, have unveiled the intricate details of asymmetric trimeric structures characterizing AcrB. These studies span from seminal investigations (Murakami et al., 2006) to the latest advancements (El Omari et al., 2023), providing insights into a dynamic cyclic mechanism facilitated by a proton gradient. In this mechanism, each protomer assumes distinct conformations, aligning with the functional states of the transport cycle—Loose (L), Tight (T), and Open (O).
The peristaltic rotation mechanism, comprising three sequential steps, orchestrates the orderly binding of substrates within the periplasmic domain of one of the three protomers. This dynamic process has been elucidated through meticulous studies, highlighting a nuanced interplay of conformational changes (Tam et al., 2021). The structural intricacies and the complexities of substrate recognition and uptake render the AcrAB-TolC pump a complex subject of study, further compounded by the involvement of intricate regulatory cascades in the expression of all RND efflux pumps. Notably in the context of E. coli, marA, ramA, and soxS emerge as robust activators responsible for inducing the expression of the AcrAB-TolC tripartite efflux pump systems. In the regulatory landscape, AcrR assumes the role of a specific repressor, playing a central and influential role in fine-tuning the expression of the associated efflux pump. Additionally, it is noteworthy that various bactericidal or bacteriostatic agents have been identified as potential triggers for this intricate regulatory cascade (Ferrand et al., 2020; De Gaetano et al., 2023). This multifaceted regulatory landscape adds another layer of complexity to the study of AcrAB-TolC, underscoring the need for comprehensive investigations to decipher the molecular determinants for efflux in the same way as regulatory elements.
Molecular modeling enables an assessment of the activity of new compounds using models that define physicochemical features connected with pharmacological and pharmacokinetic properties of bioactive agents. Notwithstanding the paramount significance of understanding the mechanisms governing substrate efflux for the enhanced design of antibiotics, an in-depth understanding remains elusive, primarily due to the extraordinary complexity inherent in these proteins (Athar et al., 2023). However, to limit efflux resistance outbreaks, it is imperative to understand the molecular determinants of the physicochemical properties involved in substrate uptake, particularly noteworthy with RND efflux pumps, which emerge as pivotal players in the context of antimicrobial resistance mechanisms (Li et al., 2015; Gil-Gil et al., 2023). Since the 90s, many efforts have been focused on studies on Structure Activity Relationships (SAR) of antibiotics. In most research papers, antibiotic activity is determined with Minimum Inhibitory Concentration (MIC), a valuable tool for diagnostics allowing clinicians to select the most appropriate antibiotic against a specific bacteria based on current and under development antibiotic susceptibility tests (Puttaswamy et al., 2018; Gajic et al., 2022). Nevertheless, only a small number of reviews consider the target site. As a result, their interpretation is less significant concerning efflux due to the diversity of membrane transporters, their complex component structure and function, particularly for RND pumps (Mouton et al., 2018). In this case, the evaluation of antibiotic concentration inside cells is crucial.
In this review, the goal is not to be exhaustive, but pharmacochemical data have been selected based on their relevance to acquire a comprehensive array of antibiotics studied in connection with the same target. They are directly involved in the evolution of antibiotic structure compared to efflux resistance, aiming to promote an improvement in the use or design of antimicrobial agents. Additionally, the goal is to encourage further research on antibiotics and raise awareness within the scientific community about the progress that can still be made in this field. A better understanding of the molecular characteristics and the underlying mechanisms limiting uptake and intrabacterial accumulation would enable the discovery of novel and improved antimicrobial chemical structures and the prevention of early resistance due to non-specific efflux impacts (Davin-Regli et al., 2021).
Antibiotics are integral to the therapeutic arsenal that has revolutionized the clinical treatment of infected patients. While some, like penicillin produced by the fungus Penicillium, tetracyclines, macrolides and aminoglycosides isolated from actinomycetes (Mullis et al., 2019), can be found in nature, others derived from natural products are semi-synthetic (e.g., ampicillin and amikacin) or fully synthetic (e.g., norfloxacin). They have specific actions on bacteria or protozoa, acting as bactericidal or bacteriostatic agents depending on their mechanism of action (Slonczewski and Foster, 2017; Halawa et al., 2024). The AcrAB-TolC archetype efflux pump represents a substantial database source in the scientific literature. We selected publications evaluating a broad range of antibiotics against Gram-negative efflux-resistant bacterial strains and matched the effects of AcrAB-TolC with the physicochemical properties of antibiotics. This work can provide a better understanding of RND efflux contribution to antibiotic activity, especially with MIC measures, depending on efflux resistance levels and the chemical structure of antimicrobials. This pioneering study gives essential insight into various Gram-negative bacteria that produce AcrAB-TolC or other RND efflux pumps, significantly influencing drug activity at the target level without changing its multi-specific nature.
We analyzed the pharmacological descriptor MIC, as it constitutes an appropriate index of bacterial strain resistance status and antibiotic sensitivity, along with several physicochemical properties. Among pharmacochemical descriptors, we used as structural parameters (Table 1) such as rotatable bonds, ring counts, including aromatic ones, stereocenter counts and complexity. As thermodynamic parameters, we considered octanol/water partition coefficient logarithm (LogP) and as electronic parameters, we included the negative acidity constant logarithm (pKa), Hydrogen Bond Donor (HBD) and Acceptor (HBA), aromatic and non-aromatic ring counts. All the values described were calculated using software mentioned in the graph legend. Experimental physicochemical data were obtained for LogP and pKa. Initially, LogP values were obtained for certain (fluoro)-quinolones and fluoro-naphthyridines (Ross and Riley, 1992; Takács-Novák et al., 1992; Hansch et al., 1995; Zhidong et al., 2006; Avdeef and Tam, 2010), β-lactams and cyclins (Sangster, 1993; Hansch et al., 1995; Al Bakain et al., 2011), macrolides (McFarland et al., 1997; Grabowski et al., 2010), aminoglycosides, and dyes (Hansch et al., 1995; Sarmah et al., 2006; Sanderson and Thomsen, 2009; Grabowski et al., 2010). Subsequently, using a similar method, pKa values were obtained for certain (fluoro)-quinolones and fluoro-naphthyridines (Budavari et al., 1996; Torniainien et al., 1996; Park et al., 2000; Tolls, 2001; Hopfinger et al., 2009; Singh et al., 2009; Avdeef and Tam, 2010; Fujita et al., 2011), β-lactams (Sangster, 1993; Budavari et al., 1996; Khan and Ongerth, 2004; Huang and Hsieh, 2008; Fujita et al., 2011), cyclins (Serjeant and Dempsey, 1979; Avdeef and Tam, 2010; Fujita et al., 2011), macrolides (Budavari et al., 1996; McFarland et al., 1997; Qiang and Adams, 2004) and aminoglycosides (Budavari et al., 1996; Sarmah et al., 2006; Mucha et al., 2008; Fujita et al., 2011).
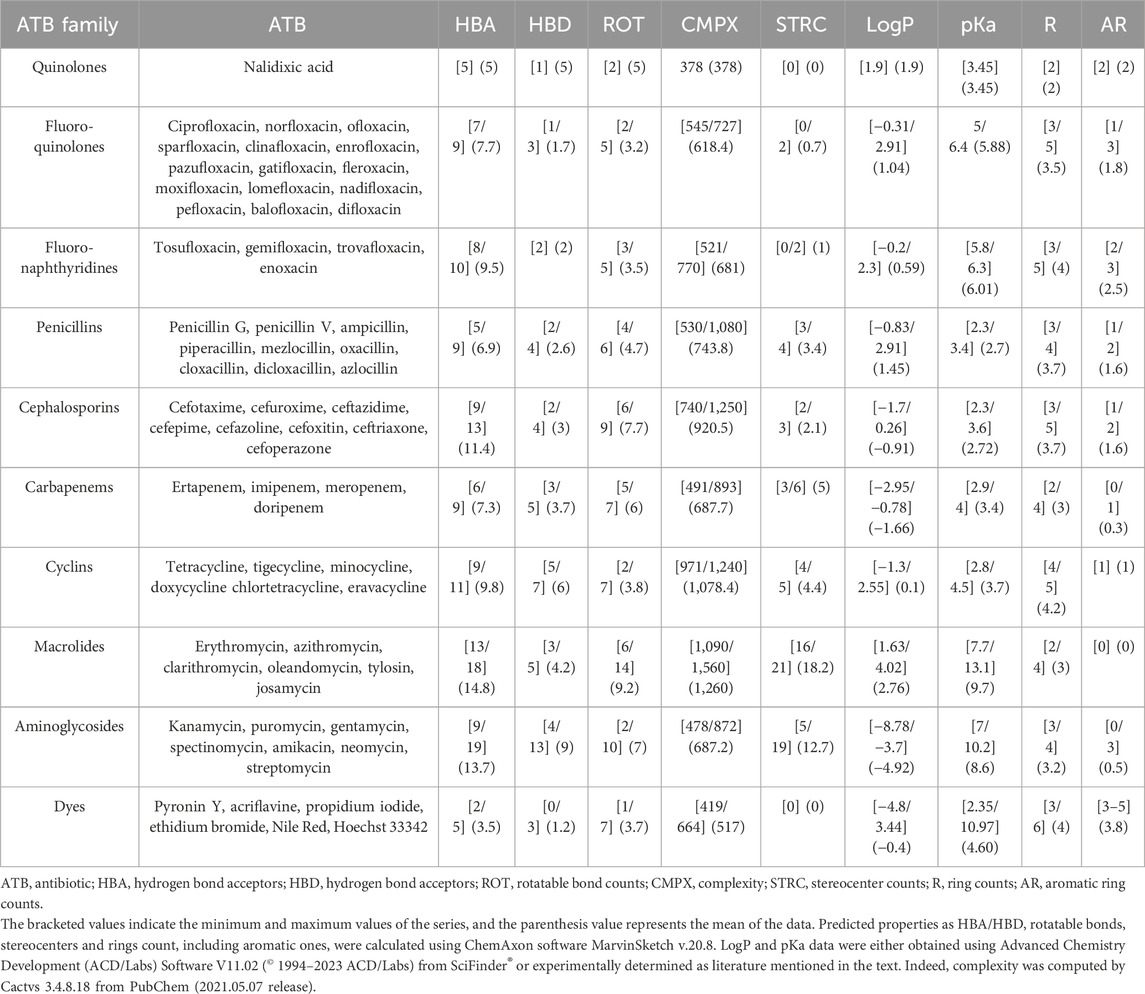
TABLE 1. Summary table of predicted or experimentally determined physicochemical properties of antibiotics and dyes included in this review.
To enhance the comprehensibility of the MIC distribution based on antibiotics within the same family, we utilized boxplots (Figure 1). Each colored rectangle within these figures depicts the median MIC values, providing a clear representation of compound efficacies against the selected E. coli strains under investigation. Each examined antibiotic family is delineated within its respective quantitative statistical series for ease of interpretation. Moreover, this integrative approach not only facilitates cross-referencing pharmacological data with the physicochemical properties of each antibiotic, contingent on the added substituents as delineated in Table 1, but also ensures comprehensive elucidation. This meticulous presentation allows for a nuanced exploration of the relationships between pharmacological effectiveness and the structural attributes of the antibiotics under consideration.
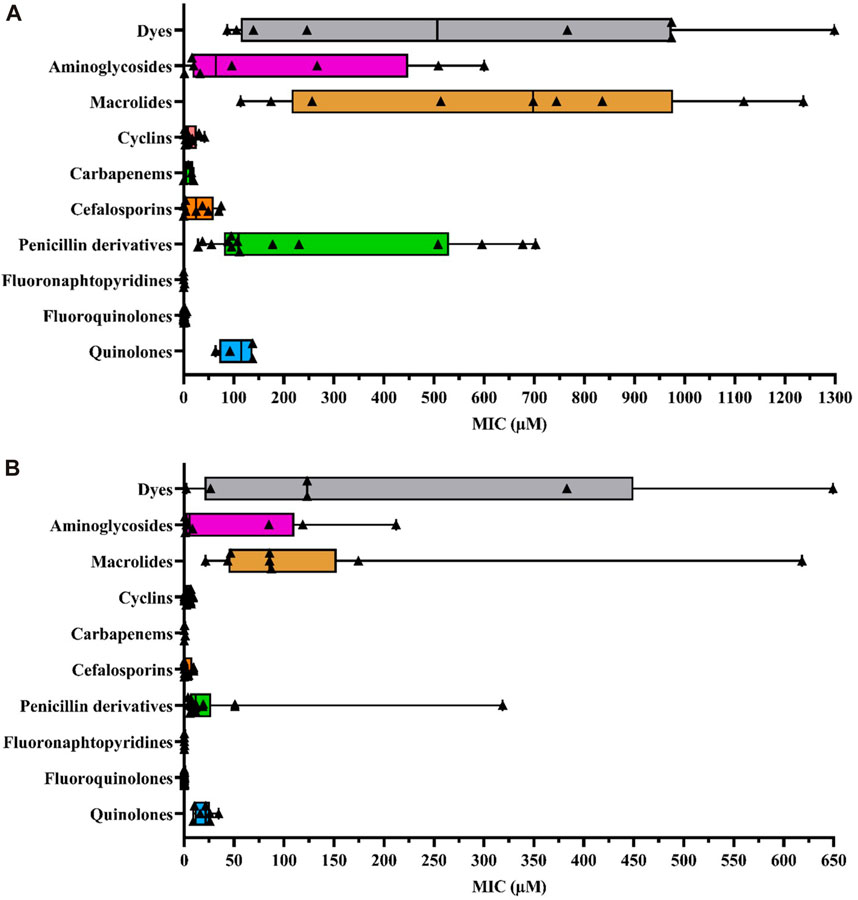
FIGURE 1. (A) Box plot representing MIC values according to different antibiotic families and other dye compounds in efflux-resistant E. coli strains (B) Box plot representing MIC values for different antibiotic families and other dye compounds in wild-type E. coli strains. The data points corresponding to each selected antibiotic, as extracted from the literature, are denoted by bold triangles (▲) for each family to construct the diagram. The statistical series include minimum, quartiles, mean, and maximal values of MICs (µM). Box plots were generated using GraphPad Prism 8.4.2 software.
Since E. coli is the most representative AMR pathogen and is frequently exposed to widely used antibiotics (Antimicrobial Resistance Collaborators, 2022), we studied the following efflux-resistant strains from literature MIC data, comparing them within a single antibiotic family (Figure 1A): 1) marR mutants: AG102 (point mutations correlated with the loss of marR repressor function by inhibiting marO promoter binding, leading to marA overexpression) (Cohen et al., 1988; Alekshun and Levy, 1997; Randall and Woodward, 2002; Whalen et al., 2015; Yilmaz et al., 2015; Vergalli et al., 2020; Ferrand et al., 2023), AG102B (Okusu et al., 1996; Aires and Nikaido, 2005) and 3-AG100 (Bohnert et al., 2008; Wehmeier et al., 2009; Bohnert et al., 2016; Schuster et al., 2016); 2) marA mutants: AG112 and CH164 (marO deletion mutation leading to an affinity loss with marR repressor and therefore marA overexpression) (Oethinger et al., 2000; Chollet et al., 2004; Nicoloff et al., 2006). Additionally, we selected some recombined strains with plasmids: HN1157/pHSGoxa7 with acrR deletion and penicillinase OXA-7 gene plasmid (operon acrR and acrAB transcription repressor) (Lim and Nikaido, 2010), GC7368/pCLL3431, an AG100A E. coli strain with cloned AcrAB gene leading to efflux pump overexpression (Visalli et al., 2003), JM101/pMAQ43, an MDR-derived strain with ramA (acrA overexpression in absence of marA or at least in reduced production) (George et al., 1995) and susceptible AG100A/pET28a-AcrB strain with acrB (plasmid containing the gene encoding AcrB leading to AcrAB-TolC overexpression as in GC7368/pCLL3431) (Li et al., 2011; Song et al., 2016). We also included clinical isolates with AcrAB-TolC overexpression, such as G5049 (Keeney et al., 2008; Sutcliffe et al., 2013). MIC data were collected from these efflux-resistant strains and compared between antibiotics and other agents to investigate variations in efflux response related to physicochemical properties.
Because the average MIC values for different antibiotic families can vary considerably in their efficacy across different experiments, it is necessary to assess whether the observed variations are attributable to the impact of AcrAB-TolC efflux pumps. For this purpose, we also plotted MIC data from wild-type E. coli strains (Figure 1B). We selected different wild-type strains, such as: AG100 (Cohen et al., 1988; Okusu et al., 1996; Alekshun and Levy, 1997; Oethinger et al., 2000; Randall and Woodward, 2002; Visalli et al., 2003; Nicoloff et al., 2006; Bohnert et al., 2008; Keeney et al., 2008; Wehmeier et al., 2009; Yilmaz et al., 2015; Schuster et al., 2016; Song et al., 2016; Vergalli et al., 2020; Ferrand et al., 2023), ATCC25922 reference strain from EUCAST 2023 (Chollet et al., 2004; Aires and Nikaido, 2005; Lim Nikaido, 2010; Li et al., 2011; Sutcliffe et al., 2013; Whalen et al., 2015; Société Française de Microbiologie, 2023), JM101 (George et al., 1995), BW1556 (Munro and Kell, 2022) and BW25113 (Plé et al., 2022). Not all antibiotics have been assessed against strains overproducing AcrAB-TolC. Nevertheless, this strain provides the most exhaustive data regarding the wide range of tested antibiotic families compared to other AcrAB-TolC producer strains.
Thus, a SAR study can be defined for this purpose thanks to various structural parameter descriptors mentioned above and used in the corpus of this review. Fluoro-quinolones and fluoro-naphthyridines are the most active antibiotic class on efflux-resistant strains, with MIC concentrations ranging from 0.39 to 2 μM and 0.25–0.3 µM, respectively, compared to cyclins, carbapenems, and cephalosporins. These three classes share the same MIC concentration range from 3.1 to 50.8 µM. The lowest values are represented by cyclins (8.9 µM), followed by carbapenems (15.6 µM), and finally cephalosporins (20.9 µM). Aminoglycosides display a narrower concentration range from 16.8 to 449.7 µM, which includes quinolones ranging from 85.4 to 137.8 µM. Penicillins have a wider concentration range from 88.9 to 537.5 µM, sharing a common feature with other antibiotic classes such as quinolones and aminoglycosides. Finally, macrolides represent the least active compounds in the overall E. coli series, with a MIC range between 247 and 982.9 µM. They have an activity almost twice as low as the least active penicillins. Thus, we find an overall disparity from 600 to 2,900 between the macrolides described as the weakest compounds and the most active ones such as fluoro-quinolones and fluoro-naphthyridines. To improve understanding of the discussed SAR, we depicted all the most representative molecules of each class of drugs in terms of chemical structure (Figure 2).
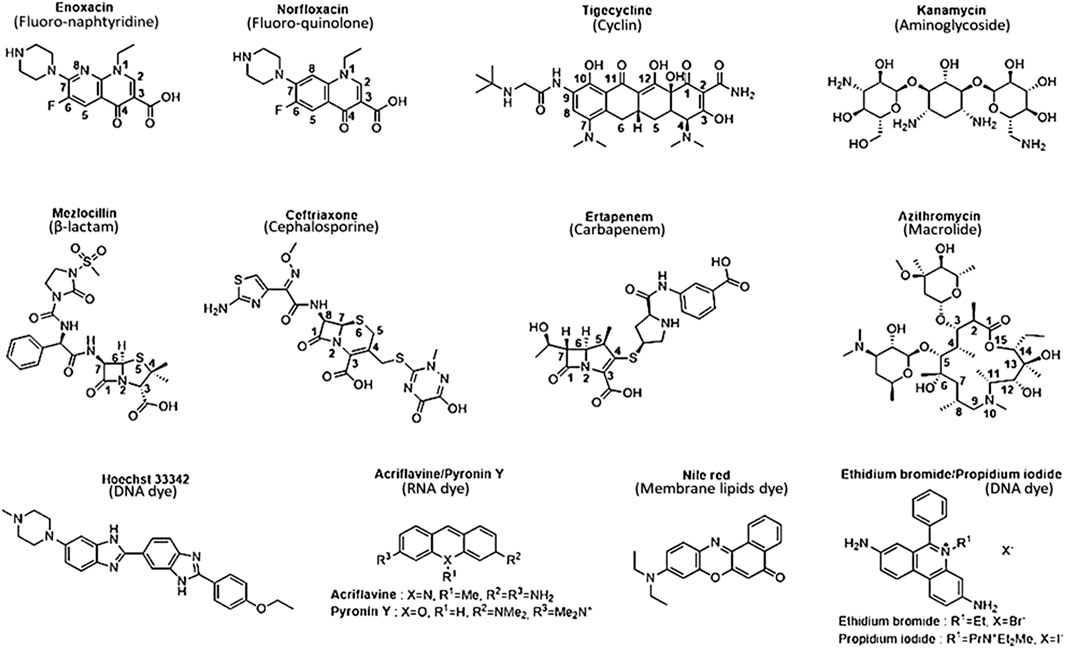
FIGURE 2. Chemical structure of the most representative antibiotics and dyes exhibit the best activity and weakest uptake by AcrAB-TolC efflux pumps. Selected antibiotics show lower MIC values in both wild-type and efflux-resistant E. coli strains, indicating heightened activity against resistant strains compared to wild-type strains. They possess physicochemical properties well-suited for strong activity against resistant-efflux strains, displaying optimal characteristics consistent with robust activity against such strains.
Due to the repetitive testing of antibiotics on laboratory strains or clinical isolates, the scope of these compounds is constrained. Frequently, the same compounds are consistently chosen to assess efflux resistance in strains derived from patients or to compare the therapeutic efficacy of synthetic molecules with one or several existing antibiotics. To address this challenge, this review focuses on a carefully restricted selection of publications that present and provide a variety of antibiotics tested against a specific bacterial strain. This analytical approach aims to explore the connections between chemical structure and substrate properties in E. coli. Integrating data from other AcrAB-TolC-producing bacteria would yield limited insights, as each strain possesses the same efflux pump with the same substrate recognizing function. Essentially, if bacteria fail to efficiently assimilate a particular substrate, they respond by producing a novel efflux pump with a broader specificity. Consequently, variations may arise due to differences in membrane composition and expression levels, significantly influencing the duration of residence within the intracellular medium without altering the antibiotic’s target affinity. The nature and composition of the targets also impose limitations in this comparison. Consequently, the molecular determinants involved in substrate recognition can vary due to alterations in amino acid composition and, more broadly, due to the distinct degrees of similarity among species that share a common efflux pump.
Quinolones represent the most powerful and widely used antibiotic family today. This class originated from nalidixic acid, the first quinolone discovered by accident as a by-product during the synthesis of chloroquine (Lesher et al., 1962). Alongside other quinolones such as piromidic acid and cinoxacin, they constitute the first-generation quinolones. These bactericidal compounds act on DNA replication and mRNA transcription machinery in nuclear sites such as gyrases and topoisomerases, primarily for Gram-positive strains (Collin et al., 2011). The interaction of the enzyme with the molecule results in the formation of a stable complex between the antibiotic-bound enzyme and the DNA strands via covalent bonds (Klostermeier, 2021). Consequently, the double strand of DNA is broken, leading to the cessation of DNA replication and subsequent cell death. Although these first-generation quinolones lacked significant pharmacological activity against Gram-negative bacteria, they served as models for the synthesis of second-generation quinolones or fluoro-quinolones, which exhibit more substantial activities. Subsequent generations, such as ciprofloxacin and fleroxacin for second-generation quinolones or fluoro-quinolones, and levofloxacin and sparfloxacin for third-generation antibiotics, were synthesized based on their chemical structure. Moxifloxacin represents the latest generation (Dube et al., 2023). Despite their efficacy, all these compounds exhibit resistance phenomena often manifested through mutations at the level of gyrases and topoisomerases, resulting in a loss of affinity of fluoro-quinolones for their enzymatic target (Ezelarab et al., 2018). In Gram-negative bacteria, the efflux phenomenon is a major cause of resistance, particularly with the expression of RND-type pumps. These mechanisms, affecting many strains, lead to a restriction of their use over time and their enduring existence within the environment exerts a sustained influence as a driving force for natural selection (Krishnamoorthy et al., 2017; Bhatt and Chatterjee, 2023). However, recent developments in influx and efflux monitoring methods may aid in designing new fluoroquinolones, as demonstrated in the case of E. coli, and could potentially be extended to other Enterobacteriaceae and Gram-negative bacteria (Vergalli et al., 2020). In this context, a comprehensive study of a large set of 6-fluoro-quinolones and 6-fluoro-1,8-naphthyridines has been conducted in isogenic K12 strains of E. coli expressing different levels of AcrAB pumps. The results, particularly on the AG102 strain that overexpresses AcrAB, draw particular attention when comparing the impact of substituents in positions N1, C7, and C8 (Figure 2) on the MIC, which is inversely proportional to the level of efflux pump expression.
Firstly, it is noteworthy that there is no difference in activity regarding the structure of the 6-fluoro-quinolone series and the 6-fluoro-1,8-naphthyridines (Figure 2). For example, norfloxacin and enoxacin (Figure 2), which have the same functional groups, exhibit similar activity. The impact of changing the substituents at the C7 and C8 positions is also not significant. However, an additional effect is observed related to the nature of the chemical function in position N1 (Figure 3). The MICs are markedly higher for compounds with aliphatic alkyl groups (ethyl in enoxacin, norfloxacin, lomefloxacin, pefloxacin, or fluoro-ethyl in fleroxacin) in this position or those forming a tricyclic structure with the C8 position (for pazufloxacin, ofloxacin, nadifloxacin) compared to cyclopropyl or difluoro-phenyl groups of the other molecules in the two series. These results indicate that the study of the SAR of the N1 chemical functions, crucial for the inhibition of the DNA gyrase, aligns with those obtained on a bacterial strain overexpressing efflux. Therefore, the N1 substituent appears to be a common pharmacophore for both gyrase and AcrB efflux pump targets. Alkyl functional groups are hydrophobic, and the decisive interaction site reported for AcrB substrates is a hydrophobic trap (Nishino et al., 2021), explaining the strong relationship between the N1 substituent and the efflux pump behavior of molecules. It is established that the AcrAB-TolC efflux pump is responsible for resistance to structurally unrelated antimicrobial agents (Lomovskaya et al., 2007; Zgurskaya et al., 2021) due to its broad substrate specificity (Alibert et al., 2017). However, increasing selectivity towards a therapeutic target requires ligand-target complementarity by limiting the molecular degrees of freedom to reach the “active” conformation. This can be achieved by rigidifying the molecular structure or inserting asymmetric centers (Figure 3). The overall number of rings and the number of aromatic rings tend to be less important for efflux-sensitive molecules. Additionally, the fewer asymmetric centers there are, the more the molecule’s sensitivity to efflux increases. Thus, the more selective the molecular therapeutic target is compared to the efflux target, the less effective the polyselectivity of the pump will be.
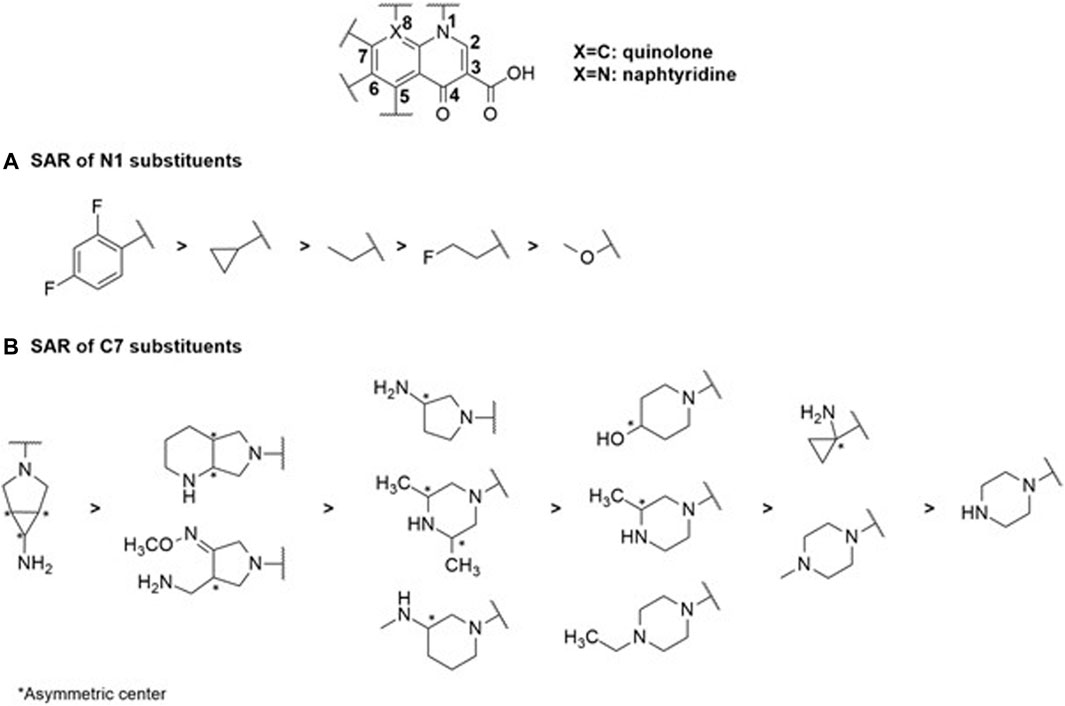
FIGURE 3. SAR study centered on the N1 (A) and C7 (B) substituents within the selected quinolone family of antibiotics reveals their substantial impact on MIC values and corresponding physicochemical properties.
Structural complexity is, therefore, an important characteristic as it contributes to selective and specific target binding. The complexity index of this family of antibiotics is the lowest. The 1,8-naphthyridine derivatives are the most complex structures of this class of drugs. Except for enoxacin, the average complexity index of naphthyridines is 734. As observed, no difference can be found in their scaffolds, so the molecular complexity lies in their substituents. Nalidixic acid (a naphthyridine with only alkyl groups in N1 and C7 positions) is particularly sensitive to efflux. Thus, increasing scaffold diversity could enhance the overall structural diversity of these types of antimicrobial agents. Diversity oriented synthesis could be the best solution to rejuvenate them.
β-lactams constitute the oldest class of antibiotic employed in combating bacterial infections, encompassing various compound families such as penicillins, cephalosporins and carbapenems (Bush and Bradford, 2016). These molecules function by inhibiting peptidoglycan synthesis, a crucial process for membrane integrity that ensures the bacterial wall’s structural integrity in response to external environmental pressures (Bugg and Walsh, 1992). In Gram-negative bacteria, peptidoglycan layers appear as a thin line within the periplasmic space and are cross-linked through the catalysis of transpeptidases, specifically penicillin-binding proteins (Park and Uehara, 2008). These enzymes become bound to a β-lactam, forming a highly stable penicilloyl-transpeptidase complex through the amide function of the β-lactam core (Figure 2). This core exhibits a structure closely resembling that of D-alanyl-D-alanine carboxypeptidase, a peptide located on the terminal chain of the peptidoglycan. As a result, the enzyme becomes obstructed and fails to fulfill its catalytic role. This obstruction leads to the destabilization of the bacterial wall, ultimately resulting in its degradation (Ghuysen et al., 1984).
Within this category of antibiotics, various classes have been developed. Penicillins were the primary β-lactam antibiotics used to target enzymatic degradations (Bradford, 2001). Initially effective against Methicillin Resistant Staphylococcus aureus (MRSA) strains and other Gram-positive bacteria, new compounds broadened the spectrum of action by enhancing the antibiotic’s impact on certain Gram-negative bacteria through the synthesis of aminopenicillins (e.g., amoxicillin). Subsequently, the development of carboxypenicillins (e.g., ticarcillin) marked increased resistance to β-lactamases, expending the spectrum of action to include several enteric anaerobic bacteria and P. aeruginosa (Kaminski et al., 2011). In this context, the design of β-lactamase inhibitors has improved the stability of penicillins against enzymatic degradation (e.g., amoxicillin and clavulanic acid), enhancing activity against resistant pathogens (Bruyère et al., 2016). These inhibitors are valuable therapeutic agents, as many types of penicillins are no longer commonly used as monotherapy (Bush, 2018; Patel et al., 2019).
For the penicillin class of antibiotics, structural variations are only found in the C7 aminoacyl side chain of the penam scaffold (Figure 4). Its length is variable, featuring a substituted phenyl group at its end. The first-generation penicillins have the shortest aminoacyl chain (Penicillins G and V, ampicillin, and amoxicillin).
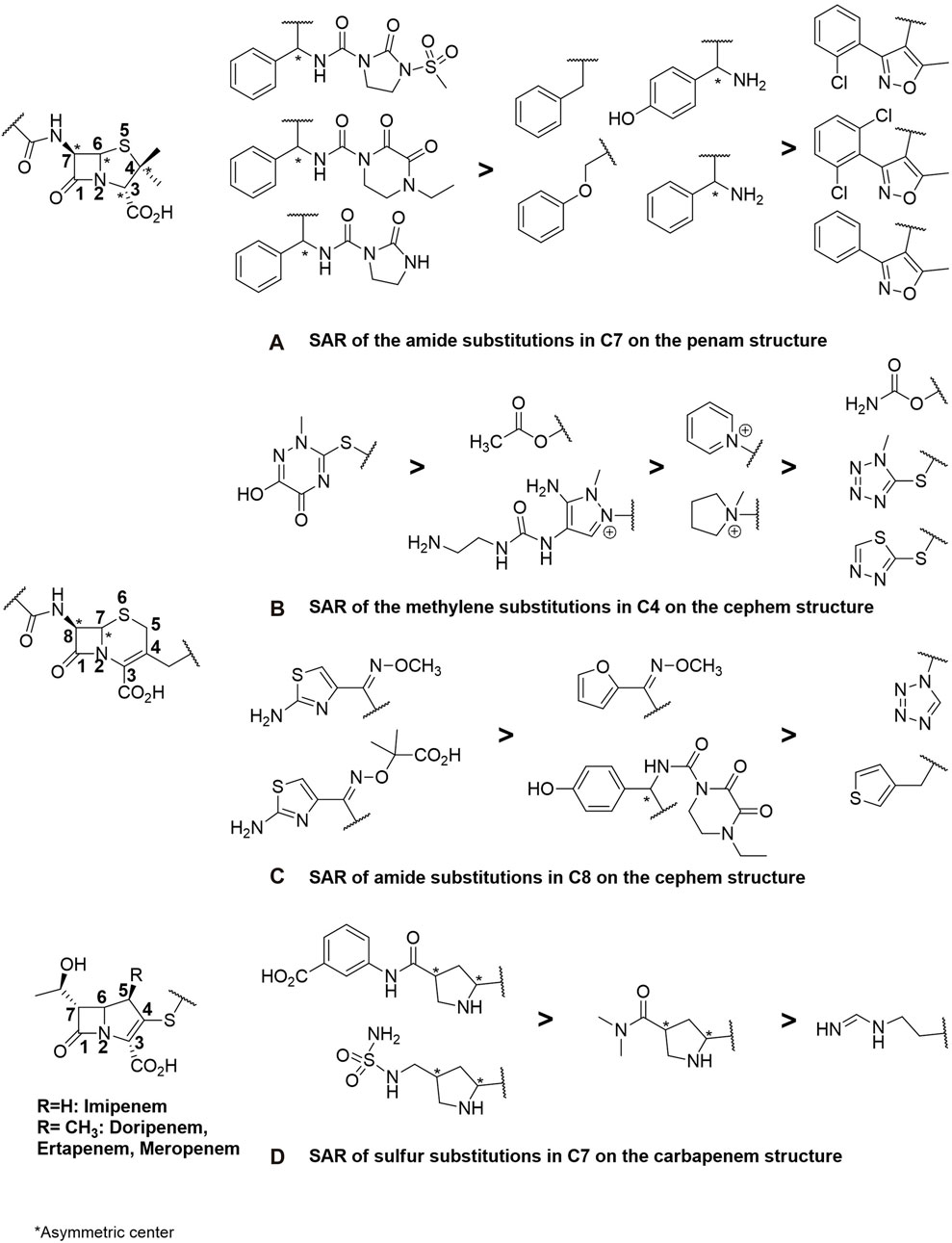
FIGURE 4. SAR study encompassing substitutions at strategic positions, specifically focusing on (A) the C7 amide in penam (penicillin derivatives), (B,C) C4 methylene and C8 amide in cephem (cephalosporins), and finally, (D) the C7 sulfur carbapenem structures included in theβ-lactam antibiotics.
In the side chain of oxacillin derivatives, the oxazole aromatic structure has been inserted between the phenyl and amide functions. These derivatives exhibit the highest MIC. The higher number of aromatic rings in them gives a greater capacity to be substrates of efflux due to the AcrB binding site rich in aromatic amino acids. Moreover, all the molecular fragments constituting it are conjugated, conferring coplanarity and thus a weak flexibility characterized by a low number of rotatable bonds (on average 5). The SAR study indicates that oxazole pharmacomodulation strategy, which improved the absorption of these antibiotics and their resistance to β-lactamase, has increased efflux resistance compared to penicillins G and V, ampicillin, and amoxicillin. Finally, penams with the lowest level of efflux transport and uptake are those with polyamide chains (Figure 4). However, the greater flexibility of penams (up to 6 rotational bonds) is relative, due to the succession of π-π-n conjugate systems that block free rotations around the amide bonds, reinforced by their cyclisation. A higher amount of HBA and HBD, greater polarizability, and a higher number of asymmetric carbons enhance the penam molecular complexity, contributing, along with lower LogP and pKa, to reduce sensitivity to efflux while providing better specificity towards their therapeutic target compared to the hydrophobic AcrB. As mentioned above, increased scaffold diversity allows increased drug potency.
Indeed, the discovery of cephalosporins has expanded the spectrum of activity of antibiotics. The incorporation of the β-lactam ring with a dihydrothiazine instead of a thiazolidine ring has allowed the introduction of a new substitution position in the structure, increasing molecular diversity. Structural enhancements have given rise to five generations of antibiotics. The first generations (e.g., cefuroxime) exhibited resistance to MRSA and some Enterobacteriaceae (E. coli, K. pneumoniae) and subsequently to β-lactamases (Ghuysen et al., 1984). However, amendments were made from the third generation onward (e.g., cefotaxime, cefepime) because, despite decreased activity against staphylococci and enterococci, they demonstrated lower β-lactamase affinity and a broader spectrum of action against Gram-negative bacteria and MRSA (Bush, 2015). The latest generation of cephalosporins, known as ceftolozane, is currently used in combination with β-lactamase inhibitors for highly potent activity against enzymes overproducing pseudomonal pathogens (Zhanel et al., 2014). Cefalexin, in fact, was prepared from the sulfoxide ester of the corresponding penicillin by ring expansion (Garbrecht, 1972). In general, cephems exhibit better antibacterial activity than penams, a fact confirmed in efflux-resistant E. coli strains. Cefazolin and cefoxitin share structural analogies with penicillins such as mezlocillin (Figure 2), piperacillin, and azlocillin, including the length and flexibility of their side chains and the number of HBA and LogP. The insertion of an O-substituted oxime function (Figure 4) has allowed for the lengthening and cross-linking of the side chain of third and fourth-generation cephalosporins, significantly reducing the MIC by enhancing the flexibility (8 rotatable bonds) of these antibiotics and the number of HBA (10–13). This results in higher molecular complexity (740–1,250), which strongly contributes to decreasing hydrophobicity and, consequently, sensitivity to efflux, leading to better inhibition of cell wall biosynthesis and enhanced bactericidal effects.
Carbapenems share similar structural characteristics, but they also exhibit differences that contribute to their broad-spectrum activity, high potency, and classification as last-resort antibiotics. Derived from penicillin through 3-position substitutions in thiol groups (Figure 4), the most representative among them is imipenem, employed in cases of resistance of Gram-negative bacilli against the third generation of cephalosporins due to enzyme overproduction (El-Gamal et al., 2017). Resistance to carbapenems can also be observed in MRSA and the emergence of Carbapenem-Resistant Enterobacteriaceae, generated by the synthesis of a specific group of β-lactamases called carbapenemases. This takes into consideration the escalating impact of stress-induced upregulation in AcrB expression and efflux pumps, notably triggered by imipenem in E. coli (Iovleva and Doi, 2017; Chetri et al., 2019). The latest widely available antibiotic is doripenem, known for its higher chemical enzyme stability and potency against Gram-negative bacteria (Nordmann et al., 2011). Firstly, their side chains differ from those found in penams and cephems, featuring a hydroxyl group at the C7 position of the β-lactam ring and an amino functional group at the C4 position of the dihydropyrrole ring, which begins with a sulfur atom (Figure 4). Hydrogen atoms at the C6 and C7 positions have a trans configuration compared to the cis configuration of penicillins and cephalosporins (Figure 2). Although the complexity index of carbapenems remains in the same order as that of other β-lactams, the higher average number of asymmetric carbons (up to 6 carbons, Figure 4), the absence of aromatic rings (except one for ertapenem, Figure 4), and very low hydrophobicity (on average −1.66 against −0.90 and 1.45, respectively, for cephems and penams) endow them with a significantly low efflux substrate character. This, in turn, enhances the duration during which the intrabacterial antibiotic concentration surpasses the MIC. These physicochemical and structural properties can be considered as the parameters that best correlate with the efficacy of antimicrobial agents.
Cyclins, macrolides, and aminoglycosides act by inhibiting protein synthesis through interactions with ribosome subunits. This step is crucial for protein biosynthesis following mRNA translation (Macé et al., 2015).
Cyclins belong to a class of antibiotics derived from tetracycline that can penetrate eukaryotic cells. This feature makes them suitable for targeting intracellular parasitic bacteria. These compounds are bacteriostatic and operate by inhibiting the 30S ribosome subunit through the inhibition of tRNA-A-site interaction. Their spectrum of action extends to combating both Gram-negative and Gram-positive bacteria, including naturally resistant Enterobacteriaceae, anaerobic pathogens, and Pseudomonas aeruginosa. Doxycycline remains the most widely used and clinically recommended therapeutic agent (Grossman, 2016). Unfortunately, mutations in the ribosomal complex, as well as efflux resistance phenomena due to their bacteriostatic activity, have contributed to the emergence of mutants with overexpression of RND (mar locus mutation) and MFS (tet protein) family pumps. Ribosomal mutations, efflux resistance, and ribosomal protection through a greater tetracyclines-A-site dissociation constant (KD) have significantly reduced the pharmacological activity of this class of antibiotics, both for past and novel derivatives (Chopra and Roberts, 2001; Warburton et al., 2016).
Compounds within the tetracycline family share a common octahydro-naphthacene-carboxamide basic structure (Figure 5), crucial for their activity. Enolized tricarbonyl-methane and phenolic diketone systems are present at positions C1-C3 and C10-C12, respectively (Figure 2). Their amphoteric behavior is attributed to the dimethylammonium group at C4 (Figure 5). Variations in activity are particularly attributed to substitutions at positions C6, C7, and C9 (Figure 5). Favorable substitutions include the replacement of the aromatic ring with a dimethylamino or fluoro group at C7 and an aminoalkyl chain branched on an acylamino function at C9. Like other antibiotic families, the presence of a long flexible chain of nitrogenous functions significantly reduces the substrate character of AcrB-type efflux pumps in E. coli (by a factor of 5 between the MIC of minocycline and tigecycline (Figure 2), for example,). Decisive physicochemical parameters include the number of HBA and HBD (optimum at 11 and 7, respectively), the number of rotational bonds (optimum at 7), and the complexity index (>1,000). In the case of tetracyclines, the number of stereocenters, LogP, and the number of rings, particularly aromatic rings, are less significant due to the common nature of the octahydro-naphthacene scaffold known to interact with various pharmacological targets. Co-crystallization of doxorubicin and minocycline in a putative AcrB binding pocket suggests that the broad substrate spectrum of AcrB results from interactions with hydrophobic amino acids [e.g., phenylalanine residues F136, F178, F610, F615, F617, and F628 (Murakami et al., 2006; Nakashima et al., 2011)] and, to a lesser extent, polar residues in the binding site. Therefore, even with improvements to its basic pharmacophore, this antibiotic class would remain limited by AcrAB-TolC uptake, necessitating the exploration of other synthesis strategies for chemical optimization.
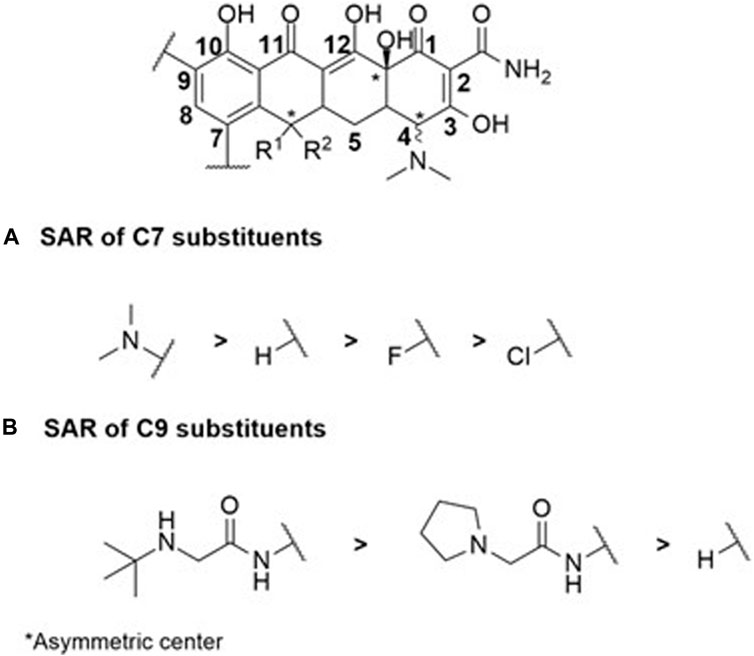
FIGURE 5. SAR study centered on C7 (A) and C9 (B) substituents within the octahydro-naphthacene-carboxamide moiety illustrates the tight correlation between the physicochemical properties of the substituents and antibiotic biological activity among the studied cyclin antibiotics.
Macrolides are bacteriostatic compounds that operate by inhibiting the 50S ribosome subunit, dissociating it from tRNA. They exhibit a medium spectrum of action and demonstrate broad resistance against Enterobacteriaceae, Pseudomonas, and some Gram-positive bacilli due to methylation of the 23S ribosomal RNA subunit. The classes of compounds vary from 14 atoms (e.g., erythromycin) to 16 atoms (e.g., josamycin). Their activity is compromised by slow accumulation in Gram-negative bacteria, attributed to low permeability, not overlooking the intervention of RND and ABC efflux pumps, which significantly reduces the effective intracellular concentration of these antibiotics (Zhong and Shortridge, 2000; Kobayashi et al., 2001; Vázquez-Laslop and Mankin, 2018). Despite the distinctive physicochemical and molecular characteristics that set macrolides apart from other antibiotic families—with an average of 15 HBA and 4 HBD, 9 rotatable bonds, 18 asymmetric carbons, and a complexity index of 1,260—macrocyclic lactone (Figure 2) molecular flexibility remains crucial for their ribosomal target interactions compared to their efflux substrate qualities. Notably, 15-membered macrolides (such as azithromycin, Figure 2) have more conformational degrees of freedom than 14-membered ones (such as clarithromycin). Depending on the folded-out or the folded-in conformations, the polar groups of the lactone scaffold are directed towards one side of the molecule, while the other side is mostly hydrophobic (Jednačak et al., 2020). Considering the evident increase in MICs of 16-membered macrolides compared to 14- and 15-membered ones, it can be assumed that the predominant bound conformation is the folded-out one, allowing polar regions to bind to ribosomes. Conversely, the folded-in conformation could bring the hydrophobic macrolide region (average LogP 9.09) into contact with the hydrophobic AcrB binding pocket. The number and position of sugar moieties (Figure 6) also impact the nature of the functional groups in the exposed conformation.
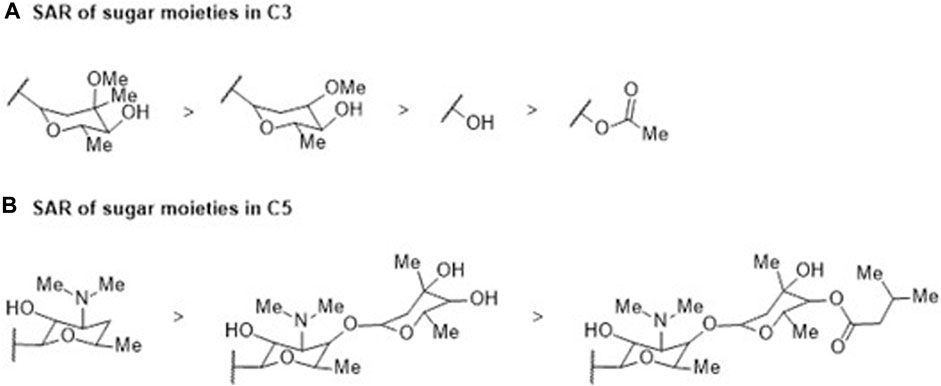
FIGURE 6. SAR study involving (A) C3 and (B) C5 sugar moieties within the macrocyclic lactone structure of the reviewed macrolide antibiotics. Variations in activity are indicated in relation to the incorporation of a sugar moiety, which subsequently influences the physicochemical properties.
Aminoglycosides are polycationic hydrophilic bactericidal antibiotics that impact protein synthesis by binding to the A-site 30S ribosome subunit. Their spectrum of action is broad, including Enterobacteriaceae, MRSA and mycobacteria. Resistance to Gram-positive bacilli has been observed, notably due to enzymatic transformation phenomena and 30S ribosome subunit mutation (McDermott et al., 2003; Ojdana et al., 2017). Gentamicin, tobramycin and amikacin are the most widely used aminoglycosides with minimal resistance in clinical practice due to their accurate activity against Gram-negative aerobic strains (Lang et al., 2023). Efflux phenomena also occur in resistance. Most aminoglycoside efflux exporters belong to the RND pump family, such as AcrAB-TolC in E. coli or MexXY-OprM and MexAB-OprM pumps in P. aeruginosa. All these mechanisms may compromise the use of the entire class of agents (Poole, 2005; Thacharodi and Lamont, 2022). The basic chemical structure required for the potency and spectrum of aminoglycosides’ activity is one or more amino sugars linked by glycosidic bonds to a 2-deoxystreptamine moiety (or streptidine in streptomycin derivatives). These molecules cross the outer membrane of the bacteria by disrupting magnesium Mg2+ bridges between lipopolysaccharide components. Their polycationic character, due to the presence of 4–7 amino groups, and their pKa around 9.6, favor this permeabilization mechanism of the external envelope, thus allowing the passage of materials (Vaara, 1992). Despite the highly polar property of amino groups, their large size does not allow them to cross through porins. Passage across the inner membrane depends on a low Energy-Dependent Phase I electron transport, followed by a large Phase II influx requiring aminoglycoside-induced translation inhibition (Davis, 1987). The particularly low LogP, up to −8.78 for amikacin, favors the antibiotic’s location in the periplasm. Thus, their uptake already demonstrates an unusual physicochemical profile compared to other classes of antibiotics that can spontaneously cross the cytoplasmic membrane barrier. Apart from the bacterial expression of enzymes modifying aminoglycoside structure, the main resistance mechanism affecting antibiotics is precisely their decreased uptake and/or accumulation. Aminoglycoside active efflux has long remained uncertain because not all RND pumps transport only lipophilic and amphiphilic substrates, as seen previously. Indeed, AcrD is responsible for resistance to very hydrophilic aminoglycoside drugs. Compared to other antibiotics, aminoglycosides have a particularly high number of HBA and HBD, respectively 14 and 9 on average. In addition, the substrate specificity between AcrB and AcrD is different despite their strong sequence similarity (Elkins and Nikaido, 2002) because it depends on the transporter domain location, and thus, the antibiotic uptake mainly takes place in the periplasm. Interactions with AcrD must be studied to identify molecular determinants to decrease efflux uptake without inducing AcrB efflux.
In general, the structure of dyes (Figure 2) is characterized by a tri- or tetracyclic skeleton with 3 aromatic or heteroaromatic rings substituted by 1 or 2 aniline groups, themselves functionalized by alkyl side chains. The succession of π-conjugated systems generates a planar skeleton with relatively flexible amino side chains, analogous to the structure of quinolones and cyclins. However, their physicochemical properties, such as the quantity of HBA and HBD, the number of rotatable bonds, the number of stereocenters, and the complexity, are generally lower than antibiotics, which does not endow them with strong antibacterial potency. Nevertheless, the higher number of aromatic rings (3–5) and their amphiphilic character (LogP = −0.4, pKa = 4,6) make dyes substrates for RND pumps (Murakami et al., 2006; Nakashima et al., 2011). They also exhibit strong fluorescent properties when in contact with DNA (e.g., ethidium bromide) or membrane lipids (e.g., Nile red). Thus, variations in fluorescence can be monitored in real time (Iyer and Erwin, 2015; Blair et al., 2016). This approach yields quantitative data, enabling the analysis of efflux kinetics in a manner impossible with bacterial growth-based efflux measurement methods such as MIC. It provides the opportunity to directly measure the consequences of environmental stressors, such as antibiotic exposure, on changes in bacterial efflux potential (Salcedo-Sora et al., 2021). These studies could elucidate the mechanisms of interaction between bacterial cell responses and their relationship to changes in efflux pump function. As the overexpression of efflux pumps is one of the major early triggers for the development of MDR phenotypes, this trend emphasizes the need to develop technologies and deploy diagnosis devices that provide meaningful (including efflux), rapid, and minimally invasive antibiotic susceptibility tests based on results relevant for therapeutic decision-making while minimizing resistance phenomena and, thus, therapeutic failure. In this methodology, it is imperative to devise and/or identify one or more inert molecules capable of being effluxed by any AcrAB-TolC-producing bacterium or other RND pumps. This approach maintains efflux pump uptake as a universal parameter, allowing manipulation solely of the interaction and specificity between an antibiotic molecule and its target. This ensures the preservation of the excretory function, adaptable to diverse structural patterns. Consequently, dyes can emerge as potent instruments for researchers and clinicians, facilitating comprehension and adaptation of antimicrobial treatments to address efflux-related challenges and enhance patient care.
Most of the clinically studied strains exhibit specific resistance combined with efflux mechanisms (Davin-Regli et al., 2021). However, efflux is typically not investigated in epidemiological studies involving clinical strains. Studies conducted in the laboratory show that inhibiting efflux mechanisms never fully restores their susceptibility to antibiotics, despite a significant reduction in MIC.
AcrAB is the major multidrug-resistant (MDR) pump in E. coli. This pump exhibits constitutive expression in wild-type strains and undergoes substantial overexpression during exposure to drugs (Teelucksingh et al., 2020). Additionally, close homologs such as AcrD, AcrEF, MdtABC, and YhiUV (MdtEF) play a secondary role, triggered only after AcrB inactivation (Anes et al., 2015). Notably, MdfA transporters, linked to quinolone resistance, belong to the MFS family (Yasufuku et al., 2011).
Despite the clinical importance of E. coli, studies on the role of efflux in hospital infections are limited. A study in Japan in 2008 revealed a correlation between overexpression of the marA gene, upregulating acrAB, and resistance to fluoro-quinolones (Yasufuku et al., 2011). Another study in China in 2018 found high resistance rates in E. coli strains isolated from urine samples, with increased expression of various efflux pump genes, including ABC transport efflux pump genes and acrB (Zeng et al., 2020).
In an international collection of MDR E. coli, Camp et al. (2021) explored efflux mechanisms using MIC tests with an AcrAB-TolC efflux inhibitor and qRT-PCR. Their findings indicated that 50% of strains had efflux mechanisms associated with antimicrobial resistance. Significant overexpression of the AcrAB-TolC system was observed in these strains, supported by whole-genome sequencing showing amino acid substitutions in genetic regulators AcrR, MarR, and SoxR.
In essence, the evidence presented underscores the crucial role of efflux pumps, particularly AcrAB, in mediating resistance in E. coli, and emphasizes the need for further research in clinical settings to understand and address the implications of efflux mechanisms in antibiotic resistance.
The crisis in anti-infective treatments exacerbated by the rapid development of resistance compared to the creation of new antimicrobials, presents a significant public health concern. While current antibiotic treatments remain effective, the primary challenge is to prevent the emergence of MDR pathogens.
Historically, medicinal chemists have sustained the antibiotic arsenal by synthetically adapting existing antibacterial structures to counteract evolving resistance. The evolution of β-lactam antibiotics into multiple generations exemplifies this method. Modification of existing structures remains the primary strategy for managing antibiotic resistance today. However, a long-term strategy for managing antibiotic resistance is essential to regain our medicinal advantage. With the increase in antibiotic resistance and the impact of broad-spectrum antibiotics, the development of specific antibiotics has become more attractive, especially when combined with rapid diagnostics.
Physicochemical properties play a crucial role in medicinal chemistry, serving as key descriptors that significantly impact drug attrition. In drug discovery, physicochemical properties define molecular features related to interactions with different media and environments.
Antibiotics, organic substances produced by living organisms, inhibit the growth of other microorganisms. Bacteria have developed resistance to antibiotics long before their human use, exemplified by the successive discoveries in the β-lactam family. Penicillin G, cephalosporin C, thienamycin and clavulanic acid broadened the spectrum of antimicrobial activity to include P. aeruginosa and enhanced resistance to β-lactamases. The environment-health link in epidemiological analyses of the AMR prevalence is crucial. Natural products play a major role in drug discovery, with half of therapeutic small molecules having a chemical structure based on a natural product. This impact is even more significant for certain therapeutic classes, such as antibiotics.
Natural products exhibit molecular scaffolds evolved to bind to biological targets, resulting in complex structures with physicochemical characteristics correlated with increased binding specificity, reduced preclinical toxicity, and improved pharmacokinetic profiles. SAR studies characterize a physicochemical profile important for improving pharmacokinetic-pharmacodynamic properties or countering resistance phenomena such as efflux.
While the causes of drug candidate attrition are multiple, an analysis of drug design practices over the past decade demonstrates that recent molecules favor high molecular weight and unnecessary hydrophobicity. “Molecular obesity” (Hann, 2011) describes common hydrophobicity-oriented practices, such as the excessive use of aromatic rings in structures seen in the design of quinolones or tetracyclines, containing fused aromatic rings or four adjacent cyclic hydrocarbon rings, leading to rapid efflux resistance. Conversely, some findings trace their origin to macrocyclic natural products such as macrolides with unique physicochemical properties, favoring intramolecular and intermolecular bond networks allowing conformational variations that optimize interactions for high potency and selectivity. This chemical adaptation to their environment is due to the high number of HBA and HBD, conferring the so-called “beyond the rule of 5” (bRo5) drugs (Danelius et al., 2020). Historically, bRo5 drugs have played an important role in the treatment of various pathologies, including infectious diseases and this trend has continued with the evolution of the chemical structure of antibiotics, regardless of the family to which they belong (Degoey et al., 2018). There is growing interest in exploring bRo5 drugs, including natural product-based drugs exhibiting lower hydrophobicity and higher stereochemical content compared to their purely synthetic counterparts. This pharmaceutically relevant chemical space offers increased structural diversity for drug discovery. Increased flexibility, higher HBA and HBD, coupled with molecular complexity make antibiotics less prone to efflux and enhancing intrabacterial concentration above the MIC and efficacy. However, bacteria express RND pumps responsible for resistance to hydrophilic drugs like aminoglycosides. SAR studies have elucidated how antibiotics can escape MDR efflux, providing insights into molecular profile of efflux substrates. Dyes, chosen as efflux substrates, could enable the monitoring of molecule accumulation based on efflux pump expression levels. This paves the way for designing new diagnostic tools for assessing bacterial efflux resistance prevalence in clinical practice, following a one-health approach.
This review aims to establish a relevant model correlating SAR studies on AcrAB-TolC- mediated efflux with inherent structure-property relationships, considering additional molecular determinants and their implications for antimicrobial and substrate activity. The study focuses on AcrAB-TolC and E. coli, emphasizing the significance of enhancing the antibiotic affinity for its target, as the pivotal factor then becomes the speed at which it engages with the target. Efflux, prevalent in all Gram-negative bacteria through various RND pumps, remains nonspecific. This approach offers a pathway to enhance the effectiveness of upcoming antibiotics by scrutinizing existing ones where efflux poses a substantial hindrance to pharmacological activity.
In conclusion, as we confront the formidable challenge of antibiotic resistance, it becomes increasingly apparent that a multifaceted approach is essential in shaping the future of antibiotic development. Beyond the considerations discussed, such as physicochemical properties, natural product-based drugs, and efflux mechanisms, several other critical aspects demand attention.
Targeted drug delivery holds promise in optimizing the efficacy of antibiotics while minimizing systemic exposure and side effects. Combination therapy, by exploring synergistic combinations of antibiotics or combining them with other therapeutic agents, offers avenues to enhance effectiveness and combat resistance. Host-directed therapy presents an opportunity to modulate host immune responses to augment infection clearance and antibiotic efficacy.
The integration of nanotechnology enables innovative strategies for drug delivery, pharmacokinetics optimization, and stability enhancement, further advancing the effectiveness of antibiotics. Surveillance and monitoring systems are indispensable for tracking antibiotic resistance patterns and identifying emerging threats in real-time, while regulatory frameworks play a pivotal role in incentivizing the development of new antibiotics and ensuring their timely approval and access.
By addressing these challenges and embracing innovative strategies, we can forge a path toward more effective and sustainable treatments to combat the growing threat of antibiotic-resistant infections.
JR-T: Writing–original draft. GB: Writing–original draft. SA: Writing–original draft.
The authors declare financial support was received for the research, authorship, and/or publication of this article. This work received support from the French government under the France 2030 investment plan, as part of the Initiative d’Excellence d’Aix-Marseille Université—A*MIDEX—AMX-21-FAE-006.
We greatly appreciate the fruitful discussion and advice provided by Jean-Marie Pagès. Additionally, we would like to express our gratitude to the Amidex Foundation at Aix Marseille Université, France, for the support through grant AMX-21-FAE-006 from the Academy of Excellence Program for the period 2021–2024.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aires, J. R., and Nikaido, H. (2005). Aminoglycosides are captured from both periplasm and cytoplasm by the AcrD multidrug efflux transporter of Escherichia coli. J. Bacteriol. 187, 1923–1929. doi:10.1128/JB.187.6.1923-1929.2005
Alav, I., Kobylka, J., Kuth, M. S., Pos, K. M., Picard, M., Blair, J. M. A., et al. (2021). Structure, assembly, and function of tripartite efflux and type 1 secretion systems in gram-negative bacteria. Chem. Rev. 121, 5479–5596. doi:10.1021/acs.chemrev.1c00055
Al Bakain, R., Rivals, I., Sassiat, P., Thiébaut, D., Hennion, M. C., Euvrard, G., et al. (2011). Comparison of different statistical approaches to evaluate the orthogonality of chromatographic separations: application to reverse phase systems. J. Chromatogr. A 1218, 2963–2975. doi:10.1016/j.chroma.2011.03.031
Alekshun, M. N., and Levy, S. B. (1997). Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob. Agents Chemother. 41, 2067–2075. doi:10.1128/AAC.41.10.2067
Alibert, S., N'gompaza Diarra, J., Hernandez, J., Stutzmann, A., Fouad, M., Boyer, G., et al. (2017). Multidrug efflux pumps and their role in antibiotic and antiseptic resistance: a pharmacodynamic perspective. Expert Opin. Drug Metab. Toxicol. 13, 301–309. doi:10.1080/17425255.2017.1251581
Anes, J., McCusker, M. P., Fanning, S., and Martins, M. (2015). The ins and outs of RND efflux pumps in Escherichia coli. Front. Microbiol. 6, 587. doi:10.3389/fmicb.2015.00587
Antimicrobial Resistance Collaborators (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. doi:10.1016/S0140-6736(21).02724-0
Athar, M., Gervasoni, S., Catte, A., Basciu, A., Malloci, G., Ruggerone, P., et al. (2023). Tripartite efflux pumps of the RND superfamily: what did we learn from computational studies? Microbiology 169, 001307. doi:10.1099/mic.0.001307
Avdeef, A., and Tam, K. Y. (2010). How well can the Caco-2/Madin-Darby canine kidney models predict effective human jejunal permeability? J. Med. Chem. 53, 3566–3584. doi:10.1021/jm901846t
Bhatt, S., and Chatterjee, S. (2023). Fluoroquinolone antibiotics: occurrence, mode of action, resistance, environmental detection, and remediation - a comprehensive review. Environ. Pollut. 315, 120440. doi:10.1016/j.envpol.2022.120440
Blair, J. M., and Piddock, L. J. (2016). How to measure export via bacterial multidrug resistance efflux pumps. mBio 7, 008400–e916. doi:10.1128/mBio.00840-16
Blair, J. M., Webber, M. A., Baylay, A. J., Ogbolu, D. O., and Piddock, L. J. (2015). Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51. doi:10.1038/nrmicro3380
Bohnert, J. A., Schuster, S., Kern, W. V., Karcz, T., Olejarz, A., Kaczor, A., et al. (2016). Novel piperazine arylideneimidazolones inhibit the AcrAB-TolC pump in Escherichia coli and simultaneously act as fluorescent membrane probes in a combined real-time influx and efflux assay. Antimicrob. Agents Chemother. 60, 1974–1983. doi:10.1128/AAC.01995-15
Bohnert, J. A., Schuster, S., Seeger, M. A., Fähnrich, E., Pos, K. M., and Kern, W. V. (2008). Site-directed mutagenesis reveals putative substrate binding residues in the Escherichia coli RND efflux pump AcrB. J. Bacteriol. 190, 8225–8229. doi:10.1128/JB.00912-08
Bradford, P. A. (2001). Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology and detection of this important resistance threat. Clin. Microb. Rev. 14, 933–951. doi:10.1128/CMR.14.4.933-951.2001
Bruyère, F., Dihn, A., and Sotto, A. (2016). Interest of amoxicillin-clavulanic acid combination in urology: an update. Prog. Urol. 26, 437–441. doi:10.1016/j.purol.2016.05.002
Budavari, S., Smith, A., and O'Neill, M. (1996). The merck index: an encyclopedia of chemicals, drugs, and biologicals. Twelth Edition. U.S.A: Chapman & Hall.
Bugg, T. D., and Walsh, C. T. (1992). Intracellular steps of bacterial cell wall peptidoglycan biosynthesis: enzymology, antibiotics, and antibiotic resistance. Nat. Prod. Rep. 9, 199–215. doi:10.1039/np9920900199
Bush, K. (2015). A resurgence of β-lactamase inhibitor combinations effective against multidrug-resistant Gram-negative pathogens. Int. J. Antimicrob. Agents 46, 483–493. doi:10.1016/j.ijantimicag.2015.08.011
Bush, K. (2018). Past and present perspectives on β-lactamases. Antimicrob. Agents Chemother. 62, 010766–e1118. doi:10.1128/AAC.01076-18
Bush, K., and Bradford, P. A. (2016). β-Lactams and β-lactamase inhibitors: an overview. Cold Spring Harb. Perspect. Med. 6, a025247. doi:10.1101/cshperspect.a025247
Camp, J., Schuster, S., Vavra, M., Schweigger, T., Rossen, J. W. A., Reuter, S., et al. (2021). Limited multidrug resistance efflux pump overexpression among multidrug-resistant Escherichia coli strains of ST131. Antimicrob. Agents Chemother. 65, 017355–e1820. doi:10.1128/AAC.01735-20
Chetri, S., Bhowmik, D., Paul, D., Pandey, P., Chanda, D. D., Chakravarty, A., et al. (2019). AcrAB-TolC efflux pump system plays a role in carbapenem non-susceptibility in Escherichia coli. BMC Microbiol. 19, 210. doi:10.1186/s12866-019-1589-1
Chollet, R., Chevalier, J., Bollet, C., Pages, J.-M., and Davin-Regli, A. (2004). RamA is an alternate activator of the multidrug resistance cascade in Enterobacter aerogenes. Antimicrob. Agents Chemother. 48, 2518–2523. doi:10.1128/AAC.48.7.2518–2523.2004
Chopra, I., and Roberts, M. (2001). Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65, 232–260. doi:10.1128/MMBR.65.2.232-260.2001
Christaki, E., Marcou, M., and Tofarides, A. (2020). Antimicrobial resistance in bacteria: mechanisms, evolution, and persistence. J. Mol. Evol. 88, 26–40. doi:10.1007/s00239-019-09914-3
Cohen, S. P., McMurry, L. M., and Levy, S. B. (1988). marA locus causes decreased expression of OmpF porin in multiple-antibiotic-resistant (Mar). mutants of Escherichia coli. J. Bacteriol. 170, 5416–5422. doi:10.1128/jb.170.12.5416-5422.1988
Collin, F., Karkare, S., and Maxwell, A. (2011). Exploiting bacterial DNA gyrase as a drug target: current state and perspectives. Appl. Microbiol. Biotechnol. 92, 479–497. doi:10.1007/s00253-011-3557-z
Danelius, E., Poongavanam, V., Peintner, S., Wieske, L. H. E., Erdélyi, M., and Kihlberg, J. (2020). Solution conformations explain the chameleonic behaviour of macrocyclic drugs. Chemistry 26, 5231–5244. doi:10.1002/chem.201905599
Darby, E. M., Trampari, E., Siasat, P., Gaya, M. S., Alav, I., Webber, M. A., et al. (2023). Molecular mechanisms of antibiotic resistance revisited. Nat. Rev. Microbiol. 21, 280–295. doi:10.1038/s41579-022-00820-y
Davin-Regli, A., Pages, J.-M., and Ferrand, A. (2021). Clinical status of efflux resistance mechanisms in gram-negative bacteria. Antibiot. (Basel) 10, 1117. doi:10.3390/antibiotics10091117
Davis, B. D. (1987). Mechanism of bactericidal action of aminoglycosides. Microb. Rev. 51, 341–350. doi:10.1128/mr.51.3.341-350.1987
De Gaetano, G. V., Lentini, G., Famà, A., Coppolino, F., and Beninati, C. (2023). Antimicrobial resistance: two-component regulatory systems and multidrug efflux pumps. Antibiot. (Basel). 12, 965. doi:10.3390/antibiotics12060965
DeGoey, D. A., Chen, H. J., Cox, P. B., and Wendt, M. D. (2018). Beyond the rule of 5: lessons learned from AbbVie's drugs and compound collection. J. Med. Chem. 61, 2636–2651. doi:10.1021/acs.jmedchem.7b00717
Du, D., Wang-Kan, X., Neuberger, A., van Veen, H. W., Pos, K. M., Piddock, L. J. V., et al. (2018). Multidrug efflux pumps: structure, function and regulation. Nat. Rev. Microbiol. 16, 523–539. doi:10.1038/s41579-018-0048-6
Dube, P. S., Legoabe, L. J., and Beteck, R. M. (2023). Quinolone: a versatile therapeutic compound class. Mol. Divers 27, 1501–1526. doi:10.1007/s11030-022-10581-8
El-Gamal, M. I., Brahim, I., Hisham, N., Aladdin, R., Mohammed, H., and Bahaaeldin, A. (2017). Recent updates of carbapenem antibiotics. Eur. J. Med. Chem. 131, 185–195. doi:10.1016/j.ejmech.2017.03.022
Elkins, C. A., and Nikaido, H. (2002). Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominantly by two large periplasmic loops. J. Bacteriol. 184, 6490–6498. doi:10.1128/JB.184.23.6490-6499.2002
El Omari, K., Duman, R., Mykhaylyk, V., Orr, C. M., Latimer-Smith, M., Winter, G., et al. (2023). Experimental phasing opportunities for macromolecular crystallography at very long wavelengths. Commun. Chem. 6, 219. doi:10.1038/s42004-023-01014-0
Ezelarab, H. A. A., Abbas, S. H., Hassan, H. A., and Abuo-Rahma, G. E. A. (2018). Recent updates of fluoroquinolones as antibacterial agents. Arch. Pharm. Weinh. 351, e1800141. doi:10.1002/ardp.201800141
Ferrand, A., Vergalli, J., Bosi, C., Pantel, A., Pagès, J.-M., and Davin-Regli, A. (2023). Contribution of efflux and mutations in fluoroquinolone susceptibility in MDR enterobacterial isolates: a quantitative and molecular study. J. Antimicrob. Chemother. 78, 1532–1542. doi:10.1093/jac/dkad122
Ferrand, A., Vergalli, J., Pagès, J. M., and Davin-Regli, A. (2020). An intertwined network of regulation controls membrane permeability including drug influx and efflux in Enterobacteriaceae. Microorganisms 8, 833. doi:10.3390/microorganisms8060833
Fujita, Y., Tokunaga, T., and Kataoka, H. (2011). Saline and buffers minimize the action of interfering factors in the bacterial endotoxins test. Anal. Biochem. 409, 46–53. doi:10.1016/j.ab.2010.10.014
Gajic, I., Kabic, J., Kekic, D., Jovicevic, M., Milenkovic, M., Mitic Culafic, D., et al. (2022). Antimicrobial susceptibility testing: a comprehensive review of currently used methods. Antibiot. (Basel) 11, 427. doi:10.3390/antibiotics11040427
Garbrecht, W. L. (1972). Cephalexin synthesis. Eli Lilly patent U.S. Patent No 3,632,850. Indianapolis: US Patent Office.
Gauba, A., and Rahman, K. M. (2023). Evaluation of antibiotic resistance mechanisms in gram-negative bacteria. Antibiot. (Basel) 12, 1590. doi:10.3390/antibiotics12111590
Gaurav, A., Bakht, P., Saini, M., Pandey, S., and Pathania, R. (2023). Role of bacterial efflux pumps in antibiotic resistance, virulence, and strategies to discover novel efflux pump inhibitors. Microbiol. Read. 169, 001333–1333. doi:10.1099/mic.0.001333
George, A. M., Hall, R. M., and Stokes, H. W. (1995). Multidrug resistance in Klebsiella pneumoniae: a novel gene, ramA, confers a multidrug resistance phenotype in Escherichia coli. Microb 141, 1909–1920. doi:10.1099/13500872-141-8-1909
Ghuysen, J. M., Frère, J. M., Leyh-Bouille, M., Nguyen-Distèche, M., Coyette, J., Dusart, J. A., et al. (1984). Bacterial wall peptidoglycan, DD-peptidases and beta-lactam antibiotics. Scand. J. Inf. Dis. 42, 17–37.
Gil-Gil, T., Laborda, P., Ochoa-Sánchez, L. E., Martínez, J. L., and Hernando-Amado, S. (2023). Efflux in Gram-negative bacteria: what are the latest opportunities for drug discovery? Expert Opin. Drug Discov. 18, 671–686. doi:10.1080/17460441.2023.2213886
Grabowski, T., Jaroszewski, J. J., and Piotrowski, W. (2010). Correlations between no observed effect level and selected parameters of the chemical structure for veterinary drugs. Toxicol. Vitro 24, 953–959. doi:10.1016/j.tiv.2010.01.003
Grossman, T. H. (2016). Tetracycline antibiotics and resistance. Cold Spring Harb. Perspect. Med. 6, a025387. doi:10.1101/cshperspect.a025387
Halawa, E. M., Fadel, M., Al-Rabia, M. W., Behairy, A., Nouh, N. A., Abdo, M., et al. (2024). Antibiotic action and resistance: updated review of mechanisms, spread, influencing factors, and alternative approaches for combating resistance. Front. Pharmacol. 14, 1305294. doi:10.3389/fphar.2023.1305294
Hann, M. M. (2011). Molecular obesity, potency and other addictions in drug discovery. Med. Chem. Commun. 2, 349–355. doi:10.1039/C1MD00017A
Hansch, C., Leo, A., and Hoekman, D. (1995). Exploring QSAR. Hydrophobic, electronic, and steric constants. Washington DC, USA: American Chemical Society.
Hassan, K. A., Liu, Q., Elbourne, L. D. H., Ahmad, I., Sharples, D., Naidu, V., et al. (2018). Pacing across the membrane: the novel PACE family of efflux pumps is widespread in Gram-negative pathogens. Res. Microbiol. 169, 450–454. doi:10.1016/j.resmic.2018.01.001
Hopfinger, A. J., Esposito, E. X., Llinàs, A., Glen, R. C., and Goodman, J. M. (2009). Findings of the challenge to predict aqueous solubility. J. Chem. Inf. Model 49, 1–5. doi:10.1021/ci800436c
Huang, H. Y., and Hsieh, S. H. (2008). Sample stacking for the analysis of penicillins by microemulsion electrokinetic chromatography. Electrophoresis 29, 3905–3915. doi:10.1002/elps.200800008
Iovleva, A., and Doi, Y. (2017). Carbapenem-resistant Enterobacteriaceae. Clin. Lab Med 37, 303–315. doi:10.1016/j.cll.2017.01.005
Iyer, R., and Erwin, A. L. (2015). Direct measurement of efflux in Pseudomonas aeruginosa using an environment-sensitive fluorescent dye. Res. Microbiol. 166, 516–524. doi:10.1016/j.resmic.2015.06.006
Jang, S. (2023). AcrAB-TolC, a major efflux pump in Gram negative bacteria: toward understanding its operation mechanism. BMB Rep. 56, 326–334. doi:10.5483/BMBRep.2023-0070
Jednačak, T., Mikulandra, I., and Novak, P. (2020). Advanced methods for studying structure and interactions of macrolide antibiotics. Int. J. Mol. Sci. 21, 7799. doi:10.3390/ijms21207799
Kaminski, C., Timsit, J. F., Dubois, Y., Zahar, J. R., Garrouste-Orgeas, M., Vesin, A., et al. (2011). Impact of ureido/carboxypenicillin resistance on the prognosis of ventilator-associated pneumonia due to Pseudomonas aeruginosa. Crit. Care 15, R112. doi:10.1186/cc10136
Keeney, D., Ruzin, A., McAleese, F., Murphy, E., and Bradford, P. A. (2008). MarA mediated overexpression of the AcrAB efflux pump results in decreased susceptibility to tigecycline in Escherichia coli. J. Antimicrob. Chemother. 61, 46–53. doi:10.1093/jac/dkm397
Khan, S. J., and Ongerth, J. E. (2004). Modelling of pharmaceutical residues in Australian sewage by quantities of use and fugacity calculations. Chemosphere 54, 355–367. doi:10.1016/j.chemosphere.2003.07.001
Klostermeier, D. (2021). Towards conformation-sensitive inhibition of gyrase: implications of mechanistic insight for the identification and improvement of inhibitors. Molecules 26 (5), 1234. doi:10.3390/molecules26051234
Kobayashi, N., Nishino, K., and Yamaguchi, A. (2001). Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J. Bacteriol. 183, 5639–5644. doi:10.1128/JB.183.19.5639-5644.2001
Krishnamoorthy, G., Leus, I. V., Weeks, J. W., Wolloscheck, D., Rybenkov, V. V., and Zgurskaya, H. I. (2017). Synergy between active efflux and outer membrane diffusion defines rules of antibiotic permeation into gram-negative bacteria. mBio 8, 011722–e1217. doi:10.1128/mBio.01172-17
Lang, M., Carvalho, A., Baharoglu, Z., and Mazel, D. (2023). Aminoglycoside uptake, stress, and potentiation in Gram-negative bacteria: new therapies with old molecules. Microbiol. Mol. Biol. Rev. 87, e0003622. doi:10.1128/mmbr.00036-22
Lerminiaux, N. A., and Cameron, A. D. S. (2019). Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 65, 34–44. doi:10.1139/cjm-2018-0275
Lesher, G. Y., Forelich, E. J., Gruett, M. D., Bailey, J. H., and Brundage, R. P. (1962). 1,8-Naphthyridine derivatives, a new class of chemotherapeutic agents. J. Med. Chem. 5, 1063–1065. doi:10.1021/jm01240a021
Li, B., Yao, Q., Pan, X. C., Wang, N., Zhang, R., Li, J., et al. (2011). Artesunate enhances the antibacterial effect of {beta}-lactam antibiotics against Escherichia coli by increasing antibiotic accumulation via inhibition of the multidrug efflux pump system AcrAB-TolC. J. Antimicrob. Chemother. 66, 769–777. doi:10.1093/jac/dkr017
Li, X. Z., Plésiat, P., and Nikaido, H. (2015). The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 28, 337–418. doi:10.1128/CMR.00117-14
Lim, S. P., and Nikaido, H. (2010). Kinetic parameters of efflux of penicillins by the multidrug efflux transporter AcrAB-TolC of Escherichia coli. Antimicrob. Agents Chemother. 54, 1800–1806. doi:10.1128/AAC.01714-09
Lomovskaya, O., Zgurskaya, H., Totrov, M., and Watkins, W. (2007). Waltzing transporters and ‘the dance macabre’ between humans and bacteria. Nat. Rev. Drug Discov. 6, 56–65. doi:10.1038/nrd2200
Macé, K., Giudice, E., and Gillet, R. (2015). Protein synthesis by the ribosome: a pathway full of pitfalls. Med. Sci. 31, 282–290. doi:10.1051/medsci/20153103014
McDermott, P. F., Walker, R. D., and White, D. G. (2003). Antimicrobials: modes of action and mechanisms of resistance. Int. J. Toxicol. 22, 135–143. doi:10.1080/10915810305089
McFarland, J. W., Berger, C. M., Froshauer, S. A., Hayashi, S. F., Hecker, S. J., Jaynes, B. H., et al. (1997). Quantitative structure-activity relationships among macrolide antibacterial agents: in vitro and in vivo potency against Pasteurella multocida. J. Med. Chem. 40, 1340–1346. doi:10.1021/jm960436i
McNeil, H. E., Alav, I., Torres, R. C., Rossiter, A. E., Laycock, E., Legood, S., et al. (2019). Identification of binding residues between periplasmic adapter protein (PAP) and RND efflux pumps explains PAP-pump promiscuity and roles in antimicrobial resistance. PLoS Pathog. 15, e1008101. doi:10.1371/journal.ppat.1008101
Mouton, J. W., Muller, A. E., Canton, R., Giske, C. G., Kahlmeter, G., and urnidge, J. (2018). MIC-based dose adjustment: facts and fables. J. Antimicrob. Chemother. 73, 564–568. doi:10.1093/jac/dkx427
Mucha, A., Bal, W., and Jezowska-Bojczuk, W. (2008). Comparative studies of coordination properties of puromycin and puromycin aminonucleoside towards copper (II) ions. J. Inorg. Biochem. 102, 46–52. doi:10.1016/j.jinorgbio.2007.06.031
Mullis, M. M., Rambo, I. M., Baker, B. J., and Reese, B. K. (2019). Diversity, ecology, and prevalence of antimicrobials in nature. Front. Microbiol. 10, 2518. doi:10.3389/fmicb.2019.02518
Munro, L. J., and Kell, D. B. (2022). Analysis of a library of Escherichia coli transporter knockout strains to identify transport pathways of antibiotics. Antibiot. (Basel) 11, 1129. doi:10.3390/antibiotics11081129
Murakami, S., Nakashima, R., Yamashita, E., Matsumoto, T., and Yamaguchi, A. (2006). Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443, 173–179. doi:10.1038/nature05076
Muteeb, G., Rehman, M. T., Shahwan, M., and Aatif, M. (2023). Origin of antibiotics and antibiotic resistance, and their impacts on drug development: a narrative review. Pharm. (Basel). 16, 1615. doi:10.3390/ph16111615
Nakashima, R., Sakurai, K., Yamasaki, S., Nishino, K., and Yamaguchi, A. (2011). Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature 480, 565–569. doi:10.1038/nature10641
Nicoloff, H., Perreten, V., McMurry, L. M., and Levy, S. B. (2006). Role for tandem duplication and lon protease in AcrAB-TolC- dependent multiple antibiotic resistance (Mar) in an Escherichia coli mutant without mutations in marRAB or acrRAB. J. Bacteriol. 188, 4413–4423. doi:10.1128/JB.01502-05
Nishino, K., Yamasaki, S., Nakashima, R., Zwama, M., and Hayashi-Nishino, M. (2021). Function and inhibitory mechanisms of multidrug efflux pumps. Front. Microbiol. 12, 737288. doi:10.3389/fmicb.2021.737288
Nordmann, P., Picazo, J., Mutters, R., Korten, V., Quintana, A., Laeuffer, J., et al. (2011). Comparative activity of carbapenem testing: the COMPACT study. J. Antimicrob. Chemother. 66, 1070–1078. doi:10.1093/jac/dkr056
Oethinger, M., Kern, W. V., Jellen-Ritter, A. S., McMurry, L. M., and Levy, S. B. (2000). Ineffectiveness of topoisomerase mutations in mediating clinically significant fluoroquinolone resistance in Escherichia coli in the absence of the AcrAB efflux pump. Antimicrob. Agents Chemother. 44, 10–13. doi:10.1128/aac.44.1.10-13.2000
Ojdana, D., Sieńko, A., Sacha, P., Majewski, P., Wieczorek, P., Wieczorek, A., et al. (2017). Genetic basis of enzymatic resistance of E. coli to aminoglycosides. Adv. Med. Sci. 63, 9–13. doi:10.1016/j.advms.2017.05.004
Okusu, H. D., Ma, D., and Nikaido, H. (1996). AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar). mutants. J. Bacteriol. 178, 306–308. doi:10.1128/jb.178.1.306-308.1996
Park, H. R., Chung, K. Y., Lee, H. C., Lee, J. K., and Bark, K. M. (2000). Ionization and divalent cation complexation of quinolone antibiotics in aqueous solution. Bull. Korean Chem. Soc. 21, 849–854. doi:10.5012/bkcs.2000.21.9.849
Park, J. T., and Uehara, T. (2008). How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol. Mol. Biol. Rev. 72, 211–227. doi:10.1128/MMBR.00027-07
Patel, H. B., Lusk, K. A., and Cota, J. M. (2019). The role of cefepime in the treatment of extended-spectrum beta-lactamase infections. J. Pharm. Pract. 32, 458–463. doi:10.1177/0897190017743134
Plé, C., Tam, H. K., Vieira Da Cruz, A., Compagne, N., Jiménez-Castellanos, J.-L., Müller, R. T., et al. (2022). Pyridylpiperazine-based allosteric inhibitors of RND-type multidrug efflux pumps. Nat. Commun. 13, 115. doi:10.1038/s41467-021-27726-2
Poole, K. (2005). Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49, 479–487. doi:10.1128/AAC.49.2.479-487.2005
Puttaswamy, S., Gupta, S., Regunath, H., Smith, L., and Sengupta, S. (2018). A comprehensive review of the present and future antibiotic susceptibility testing (AST) systems. Archives Clin. Microbiol. 9, 83. doi:10.4172/1989-8436.100083
Qiang, Z., and Adams, C. (2004). Potentiometric determination of acid dissociation constants (pKa). for human and veterinary antibiotics. Water Res. 38, 2874–2890. doi:10.1016/j.watres.2004.03.017
Randall, L. P., and Woodward, M. J. (2002). The multiple antibiotic resistance (mar). locus and its significance. Res Veter Sci. 72, 87–93. doi:10.1053/rvsc.2001.0537
Ross, D. L., and Riley, C. M. (1992). Physicochemical properties of the fluoroquinolone antimicrobials V. Effect of fluoroquinolone structure and pH on the complexation of various fluoroquinolones with magnesium and calcium ions. Int. J. Pharm. 93, 121–129. doi:10.1016/0378-5173(93).90170-K
Salam, M. A., Al-Amin, M. Y., Salam, M. T., Pawar, J. S., Akhter, N., Rabaan, A. A., et al. (2023). Antimicrobial resistance: a growing serious threat for global public health. Healthc. (Basel) 11, 1946. doi:10.3390/healthcare11131946
Salcedo-Sora, J. E., Jindal, S., O'Hagan, S., and Kell, D. B. (2021). A palette of fluorophores that are differentially accumulated by wild-type and mutant strains of Escherichia coli: surrogate ligands for profiling bacterial membrane transporters. Microbiology 167, 001016. doi:10.1099/mic.0.001016
Sanderson, H., and Thomsen, M. (2009). Comparative analysis of pharmaceuticals versus industrial chemicals acute aquatic toxicity classification according to the United Nations classification system for chemicals. Assessment of the (Q).SAR predictability of pharmaceuticals acute aquatic toxicity and their predominant acute toxic mode-of-action. Toxicol. Lett. 187, 84–93. doi:10.1016/j.toxlet.2009.02.003
Sangster, J. (1993). Logkow - a databank of evaluated octanol-water partition coefficients: fundamentals and physical chemistry. New York, USA: Wiley.
Sarmah, A. K., Meyer, M. T., and Boxall, A. B. (2006). A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 65, 725–759. doi:10.1016/j.chemosphere.2006.03.026
Sawa, T., Kooguchi, K., and Moriyama, K. (2020). Molecular diversity of extended-spectrum β-lactamases and carbapenemases, and antimicrobial resistance. J. Intensive Care 8, 13. doi:10.1186/s40560-020-0429-6
Schaenzer, A. J., and Wright, G. D. (2020). Antibiotic resistance by enzymatic modification of antibiotic targets. Trends Mol. Med. 26, 768–782. doi:10.1016/j.molmed.2020.05.001
Schuster, S., Vavra, M., and Kern, W. (2016). Evidence of a substrate-discriminating entrance channel in the lower porter domain of the multidrug resistance efflux pump AcrB. Antimicrob. Agents Chemother. 7, 4315–4323. doi:10.1128/AAC.00314-16
Serjeant, E. P., and Dempsey, B. (1979). “Ionisation constants of organic acids in aqueous solution,” in IUPAC chem data (Oxford, New York, USA: Pergamon Press).
Singh, B. K., Parwate, D. V., and Shukla, S. K. (2009). Rapid color test identification system for screening of counterfeit fluoroquinolone. J. Chem. 6, 377–384. doi:10.1155/2009/870286
Singh, C. K., Sodhi, K. K., and Mubarak, M. S. (2023). Editorial: new drugs, approaches, and strategies to combat antimicrobial resistance. Front. Pharmacol. 14, 1295623. doi:10.3389/fphar.2023.1295623
Slonczewski, J. L., and Foster, J. W. (2017). Microbiology an evolving science. New York, USA: Norton & Company.
Smith, W. P. J., Wucher, B. R., Nadell, C. D., and Foster, K. R. (2023). Bacterial defences: mechanisms, evolution and antimicrobial resistance. Nat. Rev. Microbiol. 21, 519–534. doi:10.1038/s41579-023-00877-3
Société Française de Microbiologie (2023). Détermination de la sensibilité aux antibiotiques. CA-SFM/EUCAST. Paris, France: Société Française de Microbiologie. https://www.sfm-microbiologie.org/wp-content/uploads/2023/06/CASFM2023_V1.0.pdf (Accessed July 01, 2023).
Sodhi, K. K., and Singh, C. K. (2022). Recent development in the sustainable remediation of antibiotics: a review. Total Environ. Res. Themes 3-4, 100008. doi:10.1016/j.totert.2022.100008
Sodhi, K. K., Singh, C. K., Kumar, M., and Singh, D. K. (2023). Whole-genome sequencing of Alcaligenes sp. Strain MMA: insight into the antibiotic and heavy metal resistant genes. Front. Pharmacol. 14, 1144561. doi:10.3389/fphar.2023.1144561
Song, Y., Qin, R., Pan, X., Ouyang, Q., Liu, T., Zhai, Z. H., et al. (2016). Design of new antibacterial enhancers based on AcrB's structure and the evaluation of their antibacterial enhancement activity. Int. J. Mol. Sci. 17, 1934. doi:10.3390/ijms17111934
Spencer, A. C., and Panda, S. S. (2023). DNA gyrase as a target for quinolones. Biomedicines 11, 371. doi:10.3390/biomedicines11020371
Sutcliffe, J. A., O’Brien, W., Fyfe, C., and Grossman, T. H. (2013). Antibacterial activity of eravacycline (TP-434)., a novel fluorocycline, against hospital and community pathogens. Antimicrob. Agents Chemother. 57, 5548–5558. doi:10.1128/AAC.01288-13
Takács-Novák, K., Nagy, P., Józan, M., Orfi, L., Dunn, W. J., and Szász, G. (1992). Relationship between partitioning properties and (calculated) molecular surface. SPR investigation of imidazoquinazolone derivatives. Acta Pharm. Hung 62, 55–64. doi:10.1016/0378-5173(92)90099-N
Tam, H. K., Foong, W. E., Oswald, C., Herrmann, A., Zeng, H., and Pos, K. M. (2021). Allosteric drug transport mechanism of multidrug transporter AcrB. Nat. Commun. 12, 3889. doi:10.1038/s41467-021-24151-3
Teelucksingh, T., Thompson, L. K., and Cox, G. (2020). The evolutionary conservation of Escherichia coli drug efflux pumps supports physiological functions. J. Bacteriol. 202, 003677–e420. doi:10.1128/JB.00367-20
Thacharodi, A., and Lamont, I. L. (2022). Aminoglycoside resistance in Pseudomonas aeruginosa: the contribution of the MexXY-OprM efflux pump varies between isolates. J. Med. Microbiol. 71. doi:10.1099/jmm.0.001551
Tolls, J. (2001). Sorption of veterinary pharmaceuticals in soils: a review. Environ. Sci. Technol. 35, 3397–3406. doi:10.1021/es0003021
Tooke, C. L., Hinchliffe, P., Bragginton, E. C., Colenso, C. K., Hirvonen, V. H. A., Takebayashi, Y., et al. (2019). β-Lactamases and β-lactamase inhibitors in the 21st century. J. Mol. Biol. 431, 3472–3500. doi:10.1016/j.jmb.2019.04.002
Torniainien, K., Tammilehto, S., and Veikko, U. (1996). The effect of pH, buffer type and drug concentration on the photodegradation of ciprofloxacin. Int. J. Pharm. 132, 53–61. doi:10.1016/0378-5173(95).04332-2
Vaara, M. (1992). Agents that increase the permeability of the outer membrane. Microb. Rev. 56, 395–411. doi:10.1128/mr.56.3.395-411.1992
Vázquez-Laslop, N., and Mankin, A. S. (2018). How macrolide antibiotics work. Trends Biochem. Sci. 43, 668–684. doi:10.1016/j.tibs.2018.06.011
Vergalli, J., Atzori, A., Pajovic, J., Masi, M., Vargiu, A. V., Winterhalter, M., et al. (2020). The challenge of intracellular antibiotic accumulation, a function of fluoroquinolone influx versus bacterial efflux. Commun. Biol. 3, 198. doi:10.1038/s42003-020-0929-x
Visalli, M. A., Murphy, E., Projan, S. J., and Bradford, P. A. (2003). AcrAB multidrug efflux pump is associated with reduced levels of susceptibility to tigecycline (GAR-936). In Proteus mirabilis. Antimicrob. Agents Chemother. 47, 665–669. doi:10.1128/AAC.47.2.665-669.2003
Waghray, D., and Zhang, Q. (2018). Inhibit or evade multidrug resistance P-glycoprotein in cancer treatment. J. Med. Chem. 61, 5108–5121. doi:10.1021/acs.jmedchem.7b01457
Warburton, P. J., Amodeo, N., and Roberts, A. P. (2016). Mosaic tetracycline resistance genes encoding ribosomal protection proteins. J. Antimicrob. Chemother. 71, 3333–3339. doi:10.1093/jac/dkw304
Wehmeier, C., Schuster, S., Fahnrich, E., Kern, W. V., and Bohnert, J. A. (2009). Site-directed mutagenesis reveals amino acid residues in the Escherichia coli RND efflux pump AcrB that confer macrolide resistance. Antimicrob. Agents Chemother. 53, 329–330. doi:10.1128/AAC.00921-08
Whalen, K. E., Poulson-Ellestad, K. L., Deering, R. W., Rowley, D. C., and Mincer, T. J. (2015). Enhancement of antibiotic activity against multidrug-resistant bacteria by the efflux pump inhibitor 3, 4- dibromopyrrole-2, 5-dione isolated from a Pseudoalteromonas sp. J. Nat. Prod. 78, 402–412. doi:10.1021/np500775e
Yasufuku, T., Shigemura, K., Shirakawa, T., Matsumoto, M., Nakano, Y., Tanaka, K., et al. (2011). Correlation of overexpression of efflux pump genes with antibiotic resistance in Escherichia coli strains clinically isolated from urinary tract infection patients. J. Clin. Microbiol. 49, 189–194. doi:10.1128/jcm.00827-10
Yilmaz, S., Altinkanat-Gelmez, G., Bolelli, K., Guneser-Merdan, D., UfukOver-Hasdemir, M., Aki-Yalcin, E., et al. (2015). Binding site feature description of 2-substituted benzothiazoles as potential AcrAB-TolC efflux pump inhibitors in E. coli. SAR QSAR Environ. Res. 26, 853–871. doi:10.1080/1062936X.2015.1106581
Zeng, W., Xu, W., Xu, Y., Liao, W., Zhao, Y., Zheng, X., et al. (2020). The prevalence and mechanism of triclosan resistance in Escherichia coli isolated from urine samples in Wenzhou, China. Antimicrob. Infect. Control 9, 161. doi:10.1186/s13756-020-00823-5
Zgurskaya, H. I., Malloci, G., Chandar, B., Vargiu, A. V., and Ruggerone, P. (2021). Bacterial efflux transporters' polyspecificity - a gift and a curse? Curr. Opin. Microbiol. 61, 115–123. doi:10.1016/j.mib.2021.03.009
Zhanel, G. G., Chung, P., Adam, H., Zelenitsky, S., Denisuik, A., Schweizer, F., et al. (2014). Ceftolozane/tazobactam: a novel cephalosporin/β-lactamase inhibitor combination with activity against multidrug-resistant gram-negative bacilli. Drugs 74, 31–51. doi:10.1007/s40265-013-0168-2
Zhidong, L., Shufang, N., Hong, G., Weisan, P., and Jiawei, L. (2006). Effects of Transcutol P on the corneal permeability of drugs and evaluation of its ocular irritation of rabbit eyes. J. Pharm. Pharmacol. 58, 45–50. doi:10.1211/jpp.58.1.0006
Keywords: bacterial resistance, Gram-negative, Escherichia coli, RND efflux pumps, AcrAB-TolC, multidrug resistance, antibiotics, molecular determinants
Citation: Revol-Tissot J, Boyer G and Alibert S (2024) Molecular determinant deciphering of MIC-guided RND efflux substrates in E. coli. Front. Drug Discov. 4:1326121. doi: 10.3389/fddsv.2024.1326121
Received: 22 October 2023; Accepted: 19 February 2024;
Published: 08 March 2024.
Edited by:
Vinayak Singh, University of Cape Town, South AfricaReviewed by:
Kushneet Kaur Sodhi, University of Delhi, IndiaCopyright © 2024 Revol-Tissot, Boyer and Alibert. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sandrine Alibert, c2FuZHJpbmUuYWxpYmVydEB1bml2LWFtdS5mcg==
†ORCID: Johan Revol-Tissot, orcid.org/0009-0002-6203-5498; Sandrine Alibert, orcid.org/0000-0001-5811-0139
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.