- 1Epigenetic Memory Group, MRC London Institute of Medical Sciences, Imperial College London, Hammersmith Hospital Campus, London, United Kingdom
- 2Department of Biochemistry, University of Oxford, Oxford, United Kingdom
The naturally occurring phenomenon of bioluminescence has intrigued on-lookers for decades and is now being developed as a powerful tool for medical research and preclinical imaging. Luciferase enzymes emit light upon substrate encounter, enabling their activity to be visualised and dynamically tracked. By inserting luciferase genes into specific sites in the genome, it is possible to engineer reporters to monitor gene expression in its native context, and to detect epigenetic change in vivo. Endogenous bioluminescent reporters provide a highly sensitive, quantitative read-out of gene expression that is both well suited to longitudinal studies and can be adapted for high-throughput drug screens. In this article we outline some of the applications and benefits of bioluminescent reporters for epigenetic research, with a particular focus on revealing new therapeutic options for treating genetic and epigenetic disorders.
Introduction
Epigenetics is the study of heritable modifications to gene expression that are not caused by alterations to the underlying genetic code. Classically, three broad types of epigenetic regulation have been described: DNA methylation, regulatory RNAs and histone modifications (Jaenisch and Bird, 2003; Allis and Jenuwein, 2016; Statello et al., 2021). Unlike genetic mutation, where changes in DNA sequence are notoriously difficult to reverse, epigenetic information can be viewed as intrinsically reversible, and a plethora of enzymes and inhibitors have been described that can alter chromatin states and thereby gene expression. This capacity for reversibly altering gene expression has sparked new interest in using “epidrugs”, particularly where enhancing or suppresing gene expression is of therapeutic benefit (Heerboth et al., 2014; Salarinia et al., 2016; Wolff et al., 2017; Miranda Furtado et al., 2019).
Reporter genes provide a mechanism to examine gene regulation and the effect of these epidrugs in a preclinical setting. The first reporter genes to be described utilised enzymes from Escherichia coli (E. coli) to visualise colorimetric changes: ß-glucuronidase (GUS) was used in plant systems (Jefferson et al., 1987) and ß-galactosidase (lacZ) in mammals (An et al., 1982) and eukaryotic cells (Casadaban et al., 1983). ß-galactosidase has been extensively used to examine gene expression in tissue sections, for optical projection tomography (OPT) (Sharpe et al., 2002) and fluorescence activated cell sorting (FACS) (Nolan et al., 1988), offering sensitive cellular resolution (Serganova and Blasberg, 2019). However, as such assays compromise cell viability, fluorescent and bioluminescent systems have been developed to enable longitudinal studies. Free from the requirement of an enzymatic substrate, fluorescent proteins are ideal for obtaining cellular and sub-cellular resolution in living cells but are limited by low sensitivity (Serganova and Blasberg, 2019). As an example, endogenous autofluorescence in a typical mammalian cell is equivalent to 1 µM concentration of green fluorescent protein (GFP), and thereby detection, particularly of low level signals, is limited (Niswender et al., 1995). In contrast, bioluminescent reporters offer high sensitivity, combined with a capacity to dynamically monitor signal within living cells and tissues, rendering them ideal as tools for epigenetic research (Mezzanotte et al., 2017; Dimond et al., 2020). In this mini review we describe how bioluminescent imaging can be harnessed for epigenetic research and preclinical drug discovery. Bioluminescent reporter genes inserted into endogenous loci are subject to the same genomic and epigenomic control as the gene of interest and can be used to track gene expression longitudinally and across generations.
Bioluminescent systems and in vivo imaging
Bioluminescence is a naturally occurring chemiluminescent phenomenon, where a luciferase enzyme produces light as a by-product of oxygenation of its luciferin substrate (Sharifian et al., 2018; Dimond et al., 2020). Hundreds of luciferin/luciferase pairs have been identified throughout the natural world, used to deter predators, attract prey and as a form of communication, as previously discussed in more detail (Kaskova et al., 2016; Syed and Anderson, 2021; Baljinnyam et al., 2023). The majority of bioluminescent organisms are found in marine life (Sharifian et al., 2018), where coelenterazine is used as a substrate, with oxygen (O2) as a cofactor, to produce light in the blue spectrum (Mezzanotte et al., 2017; Serganova and Blasberg, 2019). The most common examples are Renilla luciferase (RLuc) found in sea pansy (Ward and Cormier, 1979; Loening et al., 2007) and Gaussia luciferase (GLuc) from the marine copepod Gaussia princepsin (Tannous et al., 2005). In addition to O2, terrestrial bioluminescent organisms require adenosine triphosphate (ATP) and magnesium (Mg2+) as cofactors (Mezzanotte et al., 2017; Serganova and Blasberg, 2019). Firefly luciferase (FLuc) from the North American firefly (Gould and Subramani, 1988), and click beetle luciferases (CBGLuc/CBRLuc) from Pyrophorus plagiophthalamus, are commonly used, with D-luciferin as their substrate (Mezzanotte et al., 2017; Serganova and Blasberg, 2019; Dimond et al., 2020). In certain bacteria and fungi, the genetic pathways encoding both the luciferase enzyme (encoded by luxAB genes in bacteria or nnLuz in tropical fungi) and the metabolic enzymes required to naturally generate the luciferin substrate have been fully characterised (Kotlobay et al., 2018; Gregor et al., 2019; Dimond et al., 2020).
Luciferase reporters have been prevalent since the 1980s, when Fluc was first used to study E.coli (Nussbaum and Cohen, 1988) and circadian rhythm in plants (Millar et al., 1992). Upon exposure to the substrate luciferin, cells or organisms expressing luciferase can be imaged with a charge-coupled device camera to produce a quantitative and localised signal (Dimond et al., 2020). As a reporter, luciferase benefits from a high signal to noise ratio, a wide dynamic range and rapidly quantifiable measurements (Roda et al., 2003; Mezzanotte et al., 2017; Dimond et al., 2020). It is estimated to be 10–1,000 times more sensitive than fluorescence reporters (Allard, 2008). This can be attributed to the fact that bioluminescence is the byproduct of a chemical reaction; oxidation of luciferin is catalysed by luciferase to form an excited-state species (oxyluciferin) that relaxes to its ground state by giving off a photon of light [outlined in more detail in Kaskova et al. (2016)]. Conversely, fluorescent proteins require excitation from an external source to produce light. External excitation generates significant background (autofluorescence) and struggles to penetrate tissue below the surface. Bioluminescence imaging (BLI), however, requires no excitation, resulting in minimal background autoluminescence and permitting a more accurate measurement of change. Indeed, imaging of fluorescent and bioluminescent reporters with similar emission wavelengths proved that bioluminescence is 500 times more sensitive than fluorescence in vivo (Yan et al., 2013). Furthermore, the substrate, luciferin, is non-toxic, has a relatively short half-life and there are no adverse issues associated with phototoxicity or bleaching (Serganova and Blasberg, 2019). This makes BLI ideal for longitudinal and transgenerational studies where repeated sampling is required (Figure 1A, left hand panels).
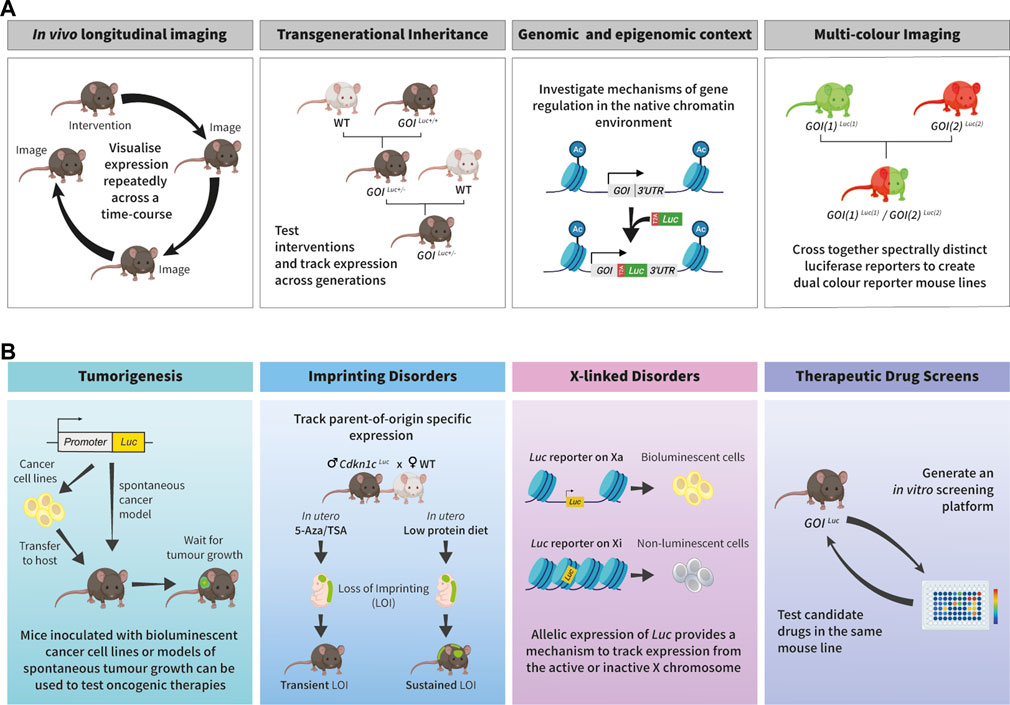
FIGURE 1. Applications of bioluminescence imaging to study epigenetic processes and unveil therapeutic options for disease. (A). Schematic outlining some of the benefits of the endogenous bioluminescent reporter systems for epigenetic research. First panel: BLI is non-toxic and reporter signal does not get diluted through cellular generations, therefore it is ideal for longitudinal studies as the same sample can be repeatedly imaged. Second panel: Bioluminescent reporter mice provide an ideal platform for tracking reporter expression across generations. Third panel: When inserted into the endogenous locus, bioluminescent reporters are subject to the same mechanisms of genetic and epigenetic control as the gene of interest (GOI). Fourth panel: Spectrally distinct luciferases can be used to visualise expression of two genes within the same animal. (B). Examples of how in vivo bioluminescence imaging can be exploited for pre-clinical research and drug discovery. First panel (tumorigenesis): Luciferase is expressed under the control of either a constitutive promoter or the regulatory sequences of a cancer-related gene. These can be used to generate bioluminescent mouse lines which exhibit spontaneous tumour growth, or a host mouse can be inoculated with bioluminescent cancer cell lines. Both can be used to track tumour progression and response to treatments. Adapted from (Dimond et al., 2020). Second panel (imprinting disorders): Endogenous bioluminescent reporters can be utilised to track parent-of-origin specific expression across generations and in response to external stimuli. An example is shown of how this system was used to compare the effects of in utero exposure to epigenetic therapy (5-Azacytidine/Trichostatin A) or low protein diet on Cdkn1c imprinting. This panel is adapted from (Van de Pette et al., 2017). Third panel (X-linked disorders): Heterozygous bioluminescent reporter models can be used to track allele specific expression of X-linked genes to investigate escape from X-inactivation. If the reporter resides on the active (Xa), the luciferase will be expressed and bioluminescence signal can be detected, if the reporter is on the inactive (Xi), then no luminescence will be produced. Fourth panel (drug screening): An advantage of endogenous bioluminescent reporters is that the same model can be used for each stage in drug discovery. In vitro screening platforms can be developed from endogenous bioluminescent reporter mouse lines. Following in vitro screens, candidate drugs can be tested in the corresponding animal model. Some aspects of this figure were created with BioRender.com.
Luciferase assays have been extensively used in vitro, both in cell-free systems where the target molecule is purified and activity of a drug tested directly, and in cell-based systems where the activity of a drug is evaluated in a genetically-modified cell (Conway et al., 2000; Goetz et al., 2000; Roda et al., 2003; Campana et al., 2016). In vivo, bioluminescence systems have traditionally been used for cell tracking where the growth of a luminescent tumour, or progression of infection, is measured in a living organism to provide spatial, longitudinal and quantitative information (Edinger et al., 2003; Cao et al., 2008; Rabinovich et al., 2008; Eun Kim et al., 2013; Lee et al., 2013; Yang et al., 2013; Kim et al., 2015). A unique advantage of bioluminescence for in vivo imaging is the lack of autoluminescence produced from mammalian cells, providing extremely low background (Mezzanotte et al., 2017; Dimond et al., 2020), whilst imaging at depth is aided by the lack of requirement for excitation by an external source. Nonetheless, as with any optical method, detection is limited by signal attenuation from deep and dark tissues; the scatter of light by cells, lipids and other components increases with depth, and substances such as haemoglobin absorb blue-green light (Choy et al., 2003; Zhao et al., 2005). These issues are being addressed through development of novel luciferase/luciferin variants designed for deeper and more sensitive signal detection (Zambito et al., 2021). For example, mutant luciferases have been designed which shift the emission spectrum of FLuc and CBRLuc into the far-red or near-infrared wavelengths, reducing attenuation of signal by haemoglobin and expanding the possibility of dual-coloured BLI (Mezzanotte et al., 2017; Zambito et al., 2021). Unmixing algorithms can be used to distinguish signal from spectrally distinct luciferases (Mezzanotte et al., 2011; Mezzanotte et al., 2013), as demonstrated by Mezzanotte and others using a red shifted firefly luciferase (PpyRE8) and a green click beetle luciferase (CBG99Luc) (Mezzanotte et al., 2011), allowing multiple reporters to be utilised in the same model.
Using bioluminescent reporters to monitor gene expression
Bioluminescence can be used to visualise gene activity, by inserting a luciferase reporter gene downstream of the regulatory elements of a gene of interest (GOI) (Figure 1A, right-hand panels). Advances in gene editing now enables reporter genes to be routinely inserted directly into the endogenous allele (Dimond et al., 2020). Insertion of the reporter into the endogenous allele provides the full genomic and epigenomic context of the GOI, including DNA and histone modifications, and accessibility of regulatory regions. This is particularly useful for studying epigenetic regulation, including epidrug interventions, since the reporter resides within the same chromatin environment as the GOI, and post-transcriptional regulation via the 3′ or 5’ untranslated regions (UTRs) can be retained. Changes to the chromatin regulatory environment can be transient or stable and heritable and since BLI allows individual samples to be imaged multiple times, the durability of epigenetic changes can be assessed across cell and organismal generations (Figure 1A). To interrogate mechanisms of GOI regulation, endogenous bioluminescent reporters could be combined with mice deficient in epigenetic regulators; for example, bioluminescence imaging of an endogenous reporter crossed with a conditional knockout of Dnmt1 (Golshani et al., 2005), Dicer (Cobb et al., 2005) or Ezh2 (Su et al., 2003) could be used to assess the impact of altered miRNA, H3K27me3 or DNA methylation on GOI expression.
A wide range of endogenous bioluminescent reporters have been generated so far (Table 1) and fall into two main classes. In the first, luciferase replaces the GOI at one or both alleles and is expressed under the control of the endogenous promoter. This approach was used to visualise p16INKa expression during aging and tumorigenesis (Burd et al., 2013) and p21 following TP53 mutation (Tinkum et al., 2011). The main limitation of this approach is that the resulting cells or animals lack at least one copy of the endogenous gene, whilst internal regulatory elements may also be lost. A second ‘non-disruptive’ approach aims to couple the expression of the luciferase gene with the GOI, while preserving its function. This can be achieved using either an internal ribosomal entry site (IRES), as demonstrated for Vegfr3 (Olmeda et al., 2017) and Utrn (Vuorinen et al., 2021), or using a 2A self-cleaving peptide sequence (T2A site) (Van de Pette et al., 2017; Son et al., 2021; Pham et al., 2022; Van de Pette et al., 2022; Gleneadie et al., 2023). During translation, T2A cleavage results in expression of endogenous protein and a separate luciferase reporter. This is particularly beneficial for multi-reporter models where a GOI is coupled to more than one reporter gene. Dual reporter models have been developed for Cidea (Son et al., 2021), Utrn (Gleneadie et al., 2023), Cdkn1c (Van de Pette et al., 2017), Dlk1 (Van de Pette et al., 2022), Fgf21 (Pham et al., 2022) and Dpd (Smith et al., 2022) (Table 1) where the reporters were separated from each other and from the endogenous gene by T2A sites. These models combine the high sensitivity and real-time dynamics of bioluminescence for in vitro screening and in vivo imaging with the cellular resolution of lacZ or fluorescent reporters (Day et al., 1998; Contag et al., 2000). The ever-expanding diversity of luciferin/luciferase systems provides the opportunity to visualise expression of more than one gene of interest in the same animal using luciferase proteins which either require different substrates for activation (Jones et al., 2017) or produce spectrally distinct luminescence signal (Mezzanotte et al., 2011; Gleneadie et al., 2023; McMorrow et al., 2023) (exemplified in Figure 1A, right-hand panel, red and green).

TABLE 1. Examples of how endogenous bioluminescent reporter systems can be exploited to answer different biological questions. Column two shows schematics depicting the endogenous GOI following insertion of the reporter gene(s). T2A = 2A self-cleaving peptide sequences; IRES = internal ribosome entry site; 3′UTR = 3′ untranslated region; Luc = luciferase; FLuc = firefly luciferase; RFluc = red shifted firefly luciferase; CBG99Luc = Green click beetle luciferase; CBRLuc = Red click beetle luciferase; Luc2 = optimised firefly luciferase; Hygro = hygromycin resistance gene; lacZ = ß-galactosidase gene; tdTomato = tdTomoto fluorescence gene; loxP = locus of X-over P1, excision site for Cre recombinase; Neo = neomycin resistance cassette. In column 5 (Class), the reporter mouse lines are separated into two classes based on whether the endogenous gene is inactive (class 1) or expressed (class 2) following insertion of the transgene.
Preclinical disease modelling using bioluminescence
BLI offers a powerful tool for preclinical research. Reporters can be used to visualise progression of a tumour (O’Farrell et al., 2013) or pathogen (Chang et al., 2014; Vande Velde et al., 2014; Daniel et al., 2015), or can represent expression of a disease-relevant gene, including epigenetically-silenced imprinted (Van de Pette et al., 2017; Van de Pette et al., 2022; Dimond et al., 2023) or X-linked genes (Tinsley et al., 2011; Moorwood and Khurana, 2013; Sripathy et al., 2017; Lee et al., 2020; Grimm and Lee, 2022; Gleneadie et al., 2023). A key benefit of using bioluminescent reporters for drug screening is that one genetic model can be used for each stage of preclinical research, from in vitro drug screening to in vivo testing of candidate molecules (as illustrated in Figure 1B).
Tumorigenesis
Cancer is a disease categorised by changes to both the genetic code and the epigenetic environment, with epigenetic drugs offering an appealing therapeutic option (Liu et al., 2022). Traditional methods to investigate tumour burden in animal models involve calliper measurements of subcutaneous tumours or post-mortem weighing. Alternatively, cancer cell lines with constitutive luciferase expression can be injected into animals to visualise tumour growth in real-time, greatly reducing the number of animals needed (Choy et al., 2003; Contag et al., 2000; O'Farrell et al., 2013; Edinger et al., 1999) (Figure 1B, left panel). Using this technique, even a single cell could be detected (Kim et al., 2010; Iwano et al., 2018), with a linear relationship between cell number and signal over 5 logarithmic scales (Contag et al., 2000; Rehemtulla et al., 2000). As the tumour progresses studies have suggested that the luciferase signal begins to plateau, likely due to ongoing necrosis and the low oxygen content in tumour cores (Soling et al., 2004; Jurczok et al., 2008; McMorrow et al., 2023). Bioluminescence has been used extensively for in vivo (O'Farrell et al., 2013; Manni et al., 2019; Alsawaftah et al., 2021) and in vitro (Wang et al., 2006; Badr et al., 2011; Blanquart et al., 2011; Improgo et al., 2011) cancer drug screening, often using the same bioluminescent cancer cell lines for each stage of the study (Qian et al., 2005; Wang et al., 2006; Wang et al., 2023). A particular benefit of this system is that the responses to treatment, cancer recurrence, and metastases can be assessed within the same animal. This is particularly appealing for epigenetic therapies, as chromatin modifications are often reversible (Ishi et al., 2022; Sanchez et al., 2022; Wang et al., 2023). For example, established ovarian tumours can be detected in mice 10 days after inoculation with bioluminescent ovarian cancer cells, and longitudinal tracking of lung metastasis over the next 3 weeks revealed that romidepsin (a histone deacetylase inhibitor (HDACi)) significantly inhibits metastatic growth (Wang et al., 2023).
To better understand cancer progression and treatment, researchers have developed bioluminescence models of spontaneous tumour formation. This approach involves crossing a tissue specific luciferase mouse with a mouse expressing a tissue specific oncogene. For example, in a model of breast cancer, Luc2 and the polyomavirus middle T antigen (PyVT) were both placed under the control of the MMTV promoter and the resulting MMTV-Luc2/MMTV-PyVT mice developed bioluminescent mammary adenocarcinomas (Zagozdzon et al., 2012). Equivalent models have been developed for pancreatic (Zhang et al., 2009), brain (Hawes and Reilly, 2010) and pituitary (Vooijs et al., 2002) cancers. These mice, and cell lines derived from them, can be used to investigate each stage of tumorigenesis and screen for drugs to target them. Similarly, commonly overexpressed genes can be used to target cancer cells for BLI imaging or ultimately, treatment. For example, an EGFR affibody-GLuc fusion protein was recently used to identify prostate cancer cells in vivo (Hersh et al., 2023) and an artificial ErbB2 ligand was used to target metastatic ovarian cancer cells for BLI and drug delivery (Han et al., 2014).
In addition to providing a valuable tool for the visualisation of cancer growth, progression and metastasis, bioluminescence has been harnessed to better understand cancer related processes such as metabolism (Indraccolo and Mueller-Klieser, 2016), apoptosis (Coppola et al., 2008), hypoxia (Miyabara et al., 2023), angiogenesis (Olmeda et al., 2017) and the immune response (McMorrow et al., 2023). This can help develop more targeted therapies and better predict response to treatment. The location of T-cells during tumour progression is important for predicting the response to immunotherapy. To model this, a mouse line was generated with two bioluminescent reporters, CBG99Luc was expressed in all T-cells (under the control of the constitutive CD2 promoter) while PpyRE9 was only expressed in activated T-cells (regulated by the LCD2 promoter) (McMorrow et al., 2023). These mice were injected with pancreatic cancer cell lines known to have either high or low T-cell infiltration and the location and function of T-cells in and surrounding the tumour were visualised longitudinally using BLI. Importantly, total bioluminescence stratified based on the two cell types and increased upon anti-PD1 and anti-CTLA-4 treatment, with more signal from activated T-cells in the tumour-draining lymph nodes (McMorrow et al., 2023). BLI has also been used in a recent study on the effect of inhibiting the histone demethylase, JMJD3, in prostate cancer. Different prostate cancer cell lines with constitutive bioluminescent reporters were inoculated into mice and treated with the JMJD3 inhibitor GSK-J4. Interestingly, upon treatment, bioluminescence signal increased in tumours developed from androgen-independent cell lines but decreased in those from androgen-dependent cells (Sanchez et al., 2022). Such models may be useful in developing methods of patient stratification and to predict response to specific therapies.
Imprinting disorders
Genomic imprinting is an epigenetic phenomenon that results in parent-of-origin specific gene expression, where a small subset of mammalian genes are preferentially expressed from either the maternal or the paternal allele (John and Surani, 2000; Ferguson-Smith, 2011; Barlow and Bartolomei, 2014). This is regulated, in part, by differential DNA methylation established in the germline (Surani, 1998; Barlow and Bartolomei, 2014). A number of rare developmental diseases such as Silver Russell syndrome, Beckwith-Wiedemann syndrome and Angelman’s syndrome, and more common conditions such as cancer, obesity and diabetes are associated with alterations at imprinted loci (Peters, 2014; Monk et al., 2019). Research into the regulation of imprinted loci is limited by the fact that investigating mono-allelic expression requires detectable differences between the parental genomes (Van de Pette et al., 2017). Endogenous single-allele bioluminescent reporters for imprinted genes allow parent-of-origin specific expression to be measured (Van de Pette et al., 2017). To visualise transmission of the paternally expressed gene Dlk1 (Van de Pette et al., 2022) and the maternally expressed gene Cdkn1c (Van de Pette et al., 2017), we previously inserted Fluc and lacZ into the endogenous loci, separated by T2A sites (Table 1). In utero exposure to high fat diet caused a sustained loss of imprinting at Dlk1 (Van de Pette et al., 2022), resulting in deregulated expression which could be tracked by BLI into the next (F2) generation, while maternal low protein diet induced loss of imprinting at Cdkn1c (Van de Pette et al., 2017). Interestingly, combined treatment with the epidrugs trichostatin A (TSA) and 5-Azacytidine (5-Aza) caused a transient loss of Cdkn1c imprinting, visible in the embryo but not maintained postpartum (Van de Pette et al., 2017) (Figure 1B, blue panel). An in vitro system developed from the same construct allowed for screening of other chromatin modifying drugs which could reverse paternal Cdkn1c silencing (Dimond et al., 2023). This screen determined that TSA increased paternal Cdkn1c expression, but only transiently, consistent with results in vivo, whilst also revealing that BET inhibitors could induce similar loss of imprinting (Dimond et al., 2023). This provides a potential therapeutic option for the 5%–10% of cases of Beckwith-Wiedemann syndrome (BWS) associated with loss of maternal Cdkn1c (Chang and Bartolomei, 2020). These examples highlight how endogenous bioluminescent reporter systems can be exploited for research into epigenetic imprint regulation and can aid in the search for new therapeutic pathways for imprinting disorders.
X-linked diseases
X chromosome inactivation is an example of epigenetic repression on a chromosome-wide scale, allowing expression of only one X chromosome per female XX cell (Panning, 2008). On the inactive X, the long non-coding RNA Xist spreads from its site of transcription to coat the chromosome, recruiting drivers of DNA methylation and heterochromatin formation to establish transcriptional silencing (Fang et al., 2019). X chromosome inactivation is a random process, tissues are a mosaic of ∼50% of cells expressing the paternal X chromosome and ∼50% expressing the maternal X chromosome (Grimm and Lee, 2022). Therefore, for X-linked disorders, in roughly half of all cells, the genetic mutation resides on the active X (Xa) whilst the wild type (WT) copy is on the inactive X chromosome (Xi) (Grimm and Lee, 2022). The majority of X-linked disorders primarily affect males; in females, WT-expressing cells can compensate or out-compete mutant-expressing cells (Grimm and Lee, 2022). For example, Duchenne Muscular Dystrophy (DMD) is fatal to males but only displays symptoms in ∼10% of female carriers (Bushby et al., 2010). A subset of X-linked disorders affect females, in these cases a potential cure can be found through reactivation of the silenced healthy allele (Grimm and Lee, 2022). Bioluminescent reporters linked to the Xi or the Xa can be used to screen for compounds which reverse this silencing (Figure 1B, pink panel). Rett syndrome is a severe neurodevelopmental disorder caused by mutations in MECP2 (Grimm and Lee, 2022). Researchers inserted Fluc into the endogenous Mecp2 locus to generate a mouse line and two clonal MEF cell lines: one in which reporter expression was confined to the Xi and one where Fluc was on the Xa (Wang et al., 2015; Sripathy et al., 2017) (Table 1). These lines have been used to screen for small molecules or siRNAs which increase Mecp2 expression from the inactive X and have led to greater understanding of Xi regulation (Sripathy et al., 2017; Carrette et al., 2018; Lee et al., 2020).
Duchenne muscular dystrophy (DMD) is an X-linked muscle wasting disease caused by loss of function mutations in the protein dystrophin (DMD/Dmd) (Guiraud et al., 2015a; Crisafulli et al., 2020). Dystrophin is expressed throughout postnatal muscle to provide support during contraction and relaxation. In the absence of dystrophin, progressive damage to muscle fibres leads to muscle degeneration, loss of ambulation and premature death. To monitor disease progression and response to therapy, a bioluminescent mouse model of muscle degeneration was generated in a Dmd-deficient background. These mice express an inducible CRE-responsive luciferase which is only active in mature myofibers. Accordingly, decreased bioluminescence directly correlates with muscle degeneration while treatment with a microdystrophin construct restored signal (Filareto et al., 2018). Some treatment options for DMD involve gene correction strategies such as exon skipping and nonsense suppression therapies. To investigate the efficacy of such strategies in vivo, luciferase was inserted prior to the 3′UTR of wild type Dmd, separated from the endogenous gene by a T2A site (Table 1) (Amoasii et al., 2019). As expected, luminescent signal mirrored the musculature of the mouse and was abolished by DMD associated mutations, suggesting these mice are useful as preclinical models to assess DMD-corrective therapy in vivo (Amoasii et al., 2019).
Utrophin (UTRN/Utrn), a homologue of dystrophin (Love et al., 1989; Perkins and Davies, 2002), functions similarly during development, however, its expression becomes restricted after birth (Ohlendieck et al., 1991; Pons et al., 1993). Therefore, it has been proposed that activation of utrophin in postnatal muscle could compensate for dystrophin loss. Bioluminescent reporter myoblast lines have been engineered to screen for compounds that elevate Utrn expression. Initial reporters contained luciferase under the control of the Utrn promoter (Moorwood et al., 2011; Tinsley et al., 2011; Guiraud et al., 2015b) or the UTRs (Moorwood et al., 2013). To better represent the full genomic context of endogenous Utrn, we recently developed a Utrn reporter mouse with luciferase and lacZ inserted directly into the 3’ UTR of endogenous Utrn, linked by T2A sites (Gleneadie et al., 2023) (Table 1). A screen of chromatin modifying drugs using a myoblast cell line derived from these mice determined that inhibition of EZH2, in combination with ERK inhibition, causes increased Utrn expression, outlining a potential route for supplementing Dmd deficiency with Utrn expression (Figure 1B, right) (Gleneadie et al., 2023). In this study a Utrn reporter, which utilises a red shifted Fluc, was used in combination with a green click beetle luciferase (CBG99Luc) reporter for Dmd and spectral differences enabled both to be visualised simultaneously (Gleneadie et al., 2023). This dual reporter system can be used to screen for drugs that increase Utrn expression in DMD, and the approach could be applied more generally to investigate whether other developmentally regulated gene paralogues can be exploited for therapeutic benefit.
Current limitations, emerging technologies and future improvements to bioluminescent reporters
Bioluminescence offers an exciting tool for in vivo imaging and drug development, with the repertoire of bioluminescent tools continuously expanding. However, a number of limitations should be considered when designing such experiments. The oxygenation of luciferin varies in response to the environment. The temperature, pH, and, importantly, the oxygen content of a cell, or extracellular space, determines the rate of luciferin turnover. This is a particular problem in cancer research, where conditions within the tumour can differ greatly from the norm (Alsawaftah et al., 2021). Due to the absorbance and scatter of light by biological materials, bioluminescent signal can also be attenuated at depth in vivo, particularly for wavelengths below 600 nm. Using directed evolution of firefly luciferase, a novel variant, termed AkaLuc was developed and paired with a D-luciferin analogue, Akalumine-HCL, to produce signal 100–1,000X higher than conventional D-luc/Fluc imaging, with an emission peak at 650 nm (Iwano et al., 2018). Excitingly, AkaBLI allows visualisation of small numbers of cells in free moving animals, in deep tissue in mice and in a female marmoset (Iwano et al., 2018). Similarly, as D-luciferin has poor permeability in the brain (Kim et al., 2022), the substrate for NanoLuc was optimised to produce cephalofurimazine (CFz), which, when paired with the NanoLuc-CyOFP fusion protein, Antares, emits 20X more luminescence than conventional D-luc/Fluc and, at high doses, outperforms AkaBLI threefold (Su et al., 2023). The development of new luciferin/luciferase pairs, such as these, improves sensitivity in vivo, and expands the options for luciferase multiplexing (Mezzanotte et al., 2017; Zambito et al., 2021). Using multiple spectrally distinct luciferases, different target genes can be visualised simultaneously, paving the way for research into gene expression networks and drug targets.
Another limitation of BLI is poor resolution. As mentioned previously, although a single bioluminescent cell can be visualised in vivo (Iwano et al., 2018), the exact position of these cells is difficult to determine with current BLI approaches. These limitations have led to the development of multimodal reporter models, where luciferase is expressed in conjunction with different reporters. In this context, photoacoustic tomography (PAT), magnetic resonance imaging (MRI) and position emission tomography (PET) offer high spatial resolution but lower sensitivity than BLI (Dimond et al., 2020) and fluorescent and colorimetric reporters, which are not well suited to in vivo imaging, provide cellular resolution ex vivo. Therefore, multimodal reporter models have been developed using combinations of these reporters to complement the benefits of luciferase (Levin et al., 2014; Van de Pette et al., 2017; Liu et al., 2019; Son et al., 2021; Pham et al., 2022; Gleneadie et al., 2023). When directly coupled with fluorescent probes, luciferase signal can be enhanced; bioluminescence resonance energy transfer (BRET) systems involve a luciferase, such as Rluc, fused to a near-infrared fluorescent protein (iRFP). Light produced from the Rluc excites iRFP resulting in a near-infrared signal, optimised for deep-tissue imaging (Schaub et al., 2015).
Currently, BLI in mammalian cells requires administration of luciferin. This can be a limiting factor in experimental design, particularly in vivo where animal welfare regulations limit the number of injections per animal and the permeability of luciferin differs between tissues (Syed and Anderson, 2021). Therefore, the development of autonomously bioluminescent animal models is an appealing concept. In plants, expression of the entire fungal bioluminescence system resulted in self-sustained luminescence which was visible to the naked eye (Khakhar et al., 2020; Mitiouchkina et al., 2020). Likewise, in mammalian cells in vitro, transfection of all components of the bacterial lux (Xu et al., 2014) or fungal nnLuz (Mitiouchkina et al., 2020) systems led to autoluminescence. However, toxicity has been associated with some lux pathway intermediates and the luciferase emits light in the blue spectrum, making it unsuitable for in vivo research. While nnLuz is reported to lose activity at temperatures above 30°C, detectable bioluminescence was emitted from mice injected with nnLuz expressing cells (Kotlobay et al., 2018). Therefore, the fungal bioluminescence system could be adapted to generate autonomously bioluminescent mice, expanding the imaging toolkit for epigenetic research and genetic modelling. Removing the need for substrate administration, coupled with recent advances improving luminescence signal (Iwano et al., 2018), one could envision continuous monitoring of gene expression in free moving animals throughout lifespan or in response to stimuli.
In previous sections we have outlined how endogenous bioluminescent reporters can be used in epigenetic research and inhibitor screening. Such systems report gene expression changes, inferring corresponding alterations to the epigenetic landscape. Bioluminescence is also being harnessed to look at mechanisms of epidrug activity by developing novel split luciferase systems, which directly measure changes in epigenetic modifications (Badran et al., 2011; Shekhawat and Ghosh, 2011; Sekar et al., 2015). For example, to monitor methylation of histone H3 tails at lysine 9 and 27 (H3K9, H3K27), fusion proteins were developed where H3K9 or H3K27 were connected to the methyl-lysine binding domains of Suv31H1 or Pc2 by a flexible linker and inserted between the N- and C- terminals of RLuc (Sekar et al., 2015). Luminescence signal can be detected when H3K9/H3K27 become methylated and bind to the Suv31H1/Pc2 domains, bringing the two-halves of the split RLuc together (Sekar et al., 2015). Global DNA methylation levels have been similarly measured using split luciferases (Badran et al., 2011) and other bioluminescent reporter systems (Wang et al., 2021; Taka et al., 2022). Interestingly, the split protein concept has been further adapted to determine site specific DNA methylation changes by fluorescence; a zinc finger attached to one-half of a GFP molecule targets a specific DNA sequence and a methyl binding domain (MBD) attached to the other half targets an adjacent CpG site (Stains et al., 2006). Fluorescence is therefore only measurable when the site is methylated. It is conceivable to envision an adaption to this technique using a split luciferase, to improve sensitivity for drug screening and in vivo imaging. Moreover, using spectrally distinct luciferases, endogenous bioluminescent reporters could be combined with the tools described above to simultaneously measure target gene expression and alterations to the epigenetic landscape.
Conclusion
Altering gene expression without mutating the underlying DNA sequence is an increasingly appealing therapeutic option for many genetic diseases, paving the way for extensive research into the design and use of epigenetic therapies. As discussed above, luciferase reporter genes provide a unique opportunity to examine the effects of these drugs. Visualising the reactivation of imprinted, X-inactivated or developmentally regulated genes with bioluminescence allows epigenetic changes to be accurately quantified at the molecular, cellular and organismal level, and across generations. Future developments, such as engineering new autonomous bioluminescent models that self-report activity throughout life, and generating multiple, spectrally-distinct reporters to image gene expression during ontogeny and in disease, promise an even brighter future for epigenetic research.
Author contributions
HG, AD and AF contributed to the development of themes outlined in this review. HG wrote the manuscript with input from AD and AF. All authors contributed to article revision, read, and approved the submitted version.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by funding from the Medical Research Council (MC_UP_1605/12).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Allard, S. T. M. (2008). Luciferase reporter assays: POWERFUL, adaptable tools for cell biology research. Cell. NOTES 21, 23–26.
Allis, C. D., and Jenuwein, T. (2016). The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17 (8), 487–500. doi:10.1038/nrg.2016.59
Alsawaftah, N., Farooq, A., Dhou, S., and Majdalawieh, A. F. (2021). Bioluminescence imaging applications in cancer: A comprehensive review. IEEE Rev. Biomed. Eng. 14, 307–326. doi:10.1109/RBME.2020.2995124
Amoasii, L., Li, H., Zhang, Y., Min, Y. L., Sanchez-Ortiz, E., Shelton, J. M., et al. (2019). In vivo non-invasive monitoring of dystrophin correction in a new Duchenne muscular dystrophy reporter mouse. Nat. Commun. 10 (1), 4537. doi:10.1038/s41467-019-12335-x
An, G., Hidaka, K., and Siminovitch, L. (1982). Expression of bacterial beta-galactosidase in animal cells. Mol. Cell. Biol. 2 (12), 1628–1632. doi:10.1128/mcb.2.12.1628
Badr, C. E., Wurdinger, T., and Tannous, B. A. (2011). Functional drug screening assay reveals potential glioma therapeutics. Assay. Drug Dev. Technol. 9 (3), 281–289. doi:10.1089/adt.2010.0324
Badran, A. H., Furman, J. L., Ma, A. S., Comi, T. J., Porter, J. R., and Ghosh, I. (2011). Evaluating the global CpG methylation status of native DNA utilizing a bipartite split-luciferase sensor. Anal. Chem. 83 (18), 7151–7157. doi:10.1021/ac2015239
Baljinnyam, B., Ronzetti, M., and Simeonov, A. (2023). Advances in luminescence-based technologies for drug discovery. Expert Opin. Drug Discov. 18 (1), 25–35. doi:10.1080/17460441.2023.2160441
Barlow, D. P., and Bartolomei, M. S. (2014). Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol. 6 (2), a018382. doi:10.1101/cshperspect.a018382
Blanquart, C., Francois, M., Charrier, C., Bertrand, P., and Gregoire, M. (2011). Pharmacological characterization of histone deacetylase inhibitor and tumor cell-growth inhibition properties of new benzofuranone compounds. Curr. Cancer Drug Targets 11 (8), 919–928. doi:10.2174/156800911797264761
Burd, C. E., Sorrentino, J. A., Clark, K. S., Darr, D. B., Krishnamurthy, J., Deal, A. M., et al. (2013). Monitoring tumorigenesis and senescence in vivo with a p16(INK4a)-luciferase model. Cell. 152 (1-2), 340–351. doi:10.1016/j.cell.2012.12.010
Bushby, K., Finkel, R., Birnkrant, D. J., Case, L. E., Clemens, P. R., Cripe, L., et al. (2010). Diagnosis and management of Duchenne muscular dystrophy, part 1: diagnosis, and pharmacological and psychosocial management. Lancet Neurol. 9 (1), 77–93. doi:10.1016/S1474-4422(09)70271-6
Campana, C., Rege, J., Turcu, A. F., Pezzi, V., Gomez-Sanchez, C. E., Robins, D. M., et al. (2016). Development of a novel cell based androgen screening model. J. Steroid Biochem. Mol. Biol. 156, 17–22. doi:10.1016/j.jsbmb.2015.11.005
Cao, F., Wagner, R. A., Wilson, K. D., Xie, X., Fu, J. D., Drukker, M., et al. (2008). Transcriptional and functional profiling of human embryonic stem cell-derived cardiomyocytes. PLoS One 3 (10), e3474. doi:10.1371/journal.pone.0003474
Carrette, L. L. G., Wang, C. Y., Wei, C., Press, W., Ma, W., Kelleher, R. J., et al. (2018). A mixed modality approach towards Xi reactivation for Rett syndrome and other X-linked disorders. Proc. Natl. Acad. Sci. U. S. A. 115 (4), E668–E75. doi:10.1073/pnas.1715124115
Casadaban, M. J., Martinez-Arias, A., Shapira, S. K., and Chou, J. (1983). Beta-galactosidase gene fusions for analyzing gene expression in escherichia coli and yeast. Methods Enzymol. 100, 293–308. doi:10.1016/0076-6879(83)00063-4
Chang, M., Anttonen, K. P., Cirillo, S. L., Francis, K. P., and Cirillo, J. D. (2014). Real-time bioluminescence imaging of mixed mycobacterial infections. PLoS One 9 (9), e108341. doi:10.1371/journal.pone.0108341
Chang, S., and Bartolomei, M. S. (2020). Modeling human epigenetic disorders in mice: beckwith-wiedemann syndrome and silver-russell syndrome. Dis. Model. Mech. 13 (5), dmm044123. doi:10.1242/dmm.044123
Choy, G., Choyke, P., and Libutti, S. K. (2003). Current advances in molecular imaging: noninvasive in vivo bioluminescent and fluorescent optical imaging in cancer research. Mol. Imaging 2 (4), 303–312. doi:10.1162/153535003322750646
Cobb, B. S., Nesterova, T. B., Thompson, E., Hertweck, A., O'Connor, E., Godwin, J., et al. (2005). T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J. Exp. Med. 201 (9), 1367–1373. doi:10.1084/jem.20050572
Contag, C. H., Jenkins, D., Contag, P. R., and Negrin, R. S. (2000). Use of reporter genes for optical measurements of neoplastic disease in vivo. Neoplasia 2 (1-2), 41–52. doi:10.1038/sj.neo.7900079
Conway, S., Canning, S. J., Howell, H. E., Mowat, E. S., Barrett, P., Drew, J. E., et al. (2000). Characterisation of human melatonin mt(1) and MT(2) receptors by CRE-luciferase reporter assay. Eur. J. Pharmacol. 390 (1-2), 15–24. doi:10.1016/s0014-2999(99)00914-0
Coppola, J. M., Ross, B. D., and Rehemtulla, A. (2008). Noninvasive imaging of apoptosis and its application in cancer therapeutics. Clin. Cancer Res. 14 (8), 2492–2501. doi:10.1158/1078-0432.CCR-07-0782
Crisafulli, S., Sultana, J., Fontana, A., Salvo, F., Messina, S., and Trifiro, G. (2020). Global epidemiology of Duchenne muscular dystrophy: an updated systematic review and meta-analysis. Orphanet J. Rare Dis. 15 (1), 141. doi:10.1186/s13023-020-01430-8
Daniel, C., Poiret, S., Dennin, V., Boutillier, D., Lacorre, D. A., Foligne, B., et al. (2015). Dual-Color bioluminescence imaging for simultaneous monitoring of the intestinal persistence of lactobacillus plantarum and lactococcus lactis in living mice. Appl. Environ. Microbiol. 81 (16), 5344–5349. doi:10.1128/AEM.01042-15
Day, R. N., Kawecki, M., and Berry, D. (1998). Dual-function reporter protein for analysis of gene expression in living cells. Biotechniques 25 (5), 848–850.
Dimond, A., Van de Pette, M., and Fisher, A. G. (2020). Illuminating epigenetics and inheritance in the immune system with bioluminescence. Trends Immunol. 41 (11), 994–1005. doi:10.1016/j.it.2020.09.001
Dimond, A., Van de Pette, M., Taylor-Bateman, V., Brown, K., Sardini, A., Whilding, C., et al. (2023). Drug-induced loss of imprinting revealed using bioluminescent reporters of Cdkn1c. Sci. Rep. 13 (1), 5626. doi:10.1038/s41598-023-32747-6
Edinger, M., Cao, Y. A., Verneris, M. R., Bachmann, M. H., Contag, C. H., and Negrin, R. S. (2003). Revealing lymphoma growth and the efficacy of immune cell therapies using in vivo bioluminescence imaging. Blood 101 (2), 640–648. doi:10.1182/blood-2002-06-1751
Edinger, M., Sweeney, T. J., Tucker, A. A., Olomu, A. B., Negrin, R. S., and Contag, C. H. (1999). Noninvasive assessment of tumor cell proliferation in animal models. Neoplasia 1 (4), 303–310. doi:10.1038/sj.neo.7900048
Eun Kim, J., Ahn, B. C., Won Lee, H., Hwang, M. H., Hyun Shin, S., Woo Lee, S., et al. (2013). In vivo monitoring of survival and proliferation of hair stem cells in a hair follicle generation animal model. Mol. Imaging 12 (5), 310–317.
Fang, H., Disteche, C. M., and Berletch, J. B. (2019). X inactivation and escape: epigenetic and structural features. Front. Cell. Dev. Biol. 7, 219. doi:10.3389/fcell.2019.00219
Ferguson-Smith, A. C. (2011). Genomic imprinting: the emergence of an epigenetic paradigm. Nat. Rev. Genet. 12 (8), 565–575. doi:10.1038/nrg3032
Filareto, A., Maguire-Nguyen, K., Gan, Q., Aldanondo, G., Machado, L., Chamberlain, J. S., et al. (2018). Monitoring disease activity noninvasively in the mdx model of Duchenne muscular dystrophy. Proc. Natl. Acad. Sci. U. S. A. 115 (30), 7741–7746. doi:10.1073/pnas.1802425115
Gleneadie, H. J., Fernandez-Ruiz, B., Sardini, A., Van de Pette, M., Dimond, A., Prinjha, R. K., et al. (2023). Endogenous bioluminescent reporters reveal a sustained increase in utrophin gene expression upon EZH2 and ERK1/2 inhibition. Commun. Biol. 6 (1), 318. doi:10.1038/s42003-023-04666-9
Goetz, A. S., Andrews, J. L., Littleton, T. R., and Ignar, D. M. (2000). Development of a facile method for high throughput screening with reporter gene assays. J. Biomol. Screen 5 (5), 377–384. doi:10.1177/108705710000500510
Golshani, P., Hutnick, L., Schweizer, F., and Fan, G. (2005). Conditional Dnmt1 deletion in dorsal forebrain disrupts development of somatosensory barrel cortex and thalamocortical long-term potentiation. Thalamus Relat. Syst. 3 (3), 227–233. doi:10.1017/S1472928807000222
Gould, S. J., and Subramani, S. (1988). Firefly luciferase as a tool in molecular and cell biology. Anal. Biochem. 175 (1), 5–13. doi:10.1016/0003-2697(88)90353-3
Gregor, C., Pape, J. K., Gwosch, K. C., Gilat, T., Sahl, S. J., and Hell, S. W. (2019). Autonomous bioluminescence imaging of single mammalian cells with the bacterial bioluminescence system. Proc. Natl. Acad. Sci. U. S. A. 116 (52), 26491–26496. doi:10.1073/pnas.1913616116
Grimm, N. B., and Lee, J. T. (2022). Selective Xi reactivation and alternative methods to restore MECP2 function in Rett syndrome. Trends Genet. 38 (9), 920–943. doi:10.1016/j.tig.2022.01.007
Guiraud, S., Aartsma-Rus, A., Vieira, N. M., Davies, K. E., van Ommen, G. J., and Kunkel, L. M. (2015a). The pathogenesis and therapy of muscular dystrophies. Annu. Rev. Genomics Hum. Genet. 16, 281–308. doi:10.1146/annurev-genom-090314-025003
Guiraud, S., Squire, S. E., Edwards, B., Chen, H., Burns, D. T., Shah, N., et al. (2015b). Second-generation compound for the modulation of utrophin in the therapy of DMD. Hum. Mol. Genet. 24 (15), 4212–4224. doi:10.1093/hmg/ddv154
Han, X. J., Wei, Y. F., Wan, Y. Y., Jiang, L. P., Zhang, J. F., and Xin, H. B. (2014). Development of a novel liposomal nanodelivery system for bioluminescence imaging and targeted drug delivery in ErbB2-overexpressing metastatic ovarian carcinoma. Int. J. Mol. Med. 34 (5), 1225–1232. doi:10.3892/ijmm.2014.1922
Hawes, J. J., and Reilly, K. M. (2010). Bioluminescent approaches for measuring tumor growth in a mouse model of neurofibromatosis. Toxicol. Pathol. 38 (1), 123–130. doi:10.1177/0192623309357075
Heerboth, S., Lapinska, K., Snyder, N., Leary, M., Rollinson, S., and Sarkar, S. (2014). Use of epigenetic drugs in disease: an overview. Genet. Epigenet 6, 9–19. doi:10.4137/GEG.S12270
Hersh, J., Yang, Y. P., Roberts, E., Bilbao, D., Tao, W., Pollack, A., et al. (2023). Targeted bioluminescent imaging of pancreatic ductal adenocarcinoma using nanocarrier-complexed EGFR-binding affibody-gaussia luciferase fusion protein. Pharmaceutics 15 (7), 1976. doi:10.3390/pharmaceutics15071976
Improgo, M. R., Johnson, C. W., Tapper, A. R., and Gardner, P. D. (2011). Bioluminescence-based high-throughput screen identifies pharmacological agents that target neurotransmitter signaling in small cell lung carcinoma. PLoS One 6 (9), e24132. doi:10.1371/journal.pone.0024132
Indraccolo, S., and Mueller-Klieser, W. (2016). Potential of induced metabolic bioluminescence imaging to uncover metabolic effects of antiangiogenic therapy in tumors. Front. Oncol. 6, 15. doi:10.3389/fonc.2016.00015
Ishi, Y., Zhang, Y., Zhang, A., Sasaki, T., Piunti, A., Suri, A., et al. (2022). Therapeutic targeting of EZH2 and BET BRD4 in pediatric rhabdoid tumors. Mol. Cancer Ther. 21 (5), 715–726. doi:10.1158/1535-7163.MCT-21-0646
Iwano, S., Sugiyama, M., Hama, H., Watakabe, A., Hasegawa, N., Kuchimaru, T., et al. (2018). Single-cell bioluminescence imaging of deep tissue in freely moving animals. Science 359 (6378), 935–939. doi:10.1126/science.aaq1067
Jaenisch, R., and Bird, A. (2003). Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33, 245–254. doi:10.1038/ng1089
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 (13), 3901–3907. doi:10.1002/j.1460-2075.1987.tb02730.x
John, R. M., and Surani, M. A. (2000). Genomic imprinting, mammalian evolution, and the mystery of egg-laying mammals. Cell. 101 (6), 585–588. doi:10.1016/s0092-8674(00)80870-3
Jones, K. A., Porterfield, W. B., Rathbun, C. M., McCutcheon, D. C., Paley, M. A., and Prescher, J. A. (2017). Orthogonal luciferase-luciferin pairs for bioluminescence imaging. J. Am. Chem. Soc. 139 (6), 2351–2358. doi:10.1021/jacs.6b11737
Jurczok, A., Fornara, P., and Soling, A. (2008). Bioluminescence imaging to monitor bladder cancer cell adhesion in vivo: A new approach to optimize a syngeneic, orthotopic, murine bladder cancer model. BJU Int. 101 (1), 120–124. doi:10.1111/j.1464-410X.2007.07193.x
Kaskova, Z. M., Tsarkova, A. S., and Yampolsky, I. V. (2016). 1001 lights: lLuciferins, luciferases, their mechanisms of action and applications in chemical analysis, biology and medicine. Chem. Soc. Rev. 45 (21), 6048–6077. doi:10.1039/c6cs00296j
Khakhar, A., Starker, C. G., Chamness, J. C., Lee, N., Stokke, S., Wang, C., et al. (2020). Building customizable auto-luminescent luciferase-based reporters in plants. Elife 9, e52786. doi:10.7554/eLife.52786
Kim, J. B., Urban, K., Cochran, E., Lee, S., Ang, A., Rice, B., et al. (2010). Non-invasive detection of a small number of bioluminescent cancer cells in vivo. PLoS One 5 (2), e9364. doi:10.1371/journal.pone.0009364
Kim, J. E., Kalimuthu, S., and Ahn, B. C. (2015). In vivo cell tracking with bioluminescence imaging. Nucl. Med. Mol. Imaging 49 (1), 3–10. doi:10.1007/s13139-014-0309-x
Kim, M., Gupta, S. K., Zhang, W., Talele, S., Mohammad, A. S., Laramy, J., et al. (2022). Factors influencing luciferase-based bioluminescent imaging in preclinical models of brain tumor. Drug Metab. Dispos. 50 (3), 277–286. doi:10.1124/dmd.121.000597
Kotlobay, A. A., Sarkisyan, K. S., Mokrushina, Y. A., Marcet-Houben, M., Serebrovskaya, E. O., Markina, N. M., et al. (2018). Genetically encodable bioluminescent system from fungi. Proc. Natl. Acad. Sci. U. S. A. 115 (50), 12728–12732. doi:10.1073/pnas.1803615115
Lee, H. M., Kuijer, M. B., Ruiz Blanes, N., Clark, E. P., Aita, M., Galiano Arjona, L., et al. (2020). A small-molecule screen reveals novel modulators of MeCP2 and X-chromosome inactivation maintenance. J. Neurodev. Disord. 12 (1), 29. doi:10.1186/s11689-020-09332-3
Lee, H. W., Jeon, Y. H., Hwang, M. H., Kim, J. E., Park, T. I., Ha, J. H., et al. (2013). Dual reporter gene imaging for tracking macrophage migration using the human sodium iodide symporter and an enhanced firefly luciferase in a murine inflammation model. Mol. Imaging Biol. 15 (6), 703–712. doi:10.1007/s11307-013-0645-8
Levin, R. A., Felsen, C. N., Yang, J., Lin, J. Y., Whitney, M. A., Nguyen, Q. T., et al. (2014). An optimized triple modality reporter for quantitative in vivo tumor imaging and therapy evaluation. PLoS One 9 (5), e97415. doi:10.1371/journal.pone.0097415
Liu, S. W., Hsu, C. H., Chen, M. R., Chiu, I. M., and Lin, K. M. (2019). A tri-fusion reporter mouse reveals tissue-specific FGF1B promoter activity in vivo. Sci. Rep. 9 (1), 11143. doi:10.1038/s41598-019-47641-3
Liu, Z., Ren, Y., Weng, S., Xu, H., Li, L., and Han, X. (2022). A new trend in cancer treatment: the combination of epigenetics and immunotherapy. Front. Immunol. 13, 809761. doi:10.3389/fimmu.2022.809761
Loening, A. M., Wu, A. M., and Gambhir, S. S. (2007). Red-shifted Renilla reniformis luciferase variants for imaging in living subjects. Nat. Methods 4 (8), 641–643. doi:10.1038/nmeth1070
Love, D. R., Hill, D. F., Dickson, G., Spurr, N. K., Byth, B. C., Marsden, R. F., et al. (1989). An autosomal transcript in skeletal muscle with homology to dystrophin. Nature 339 (6219), 55–58. doi:10.1038/339055a0
Manni, I., de Latouliere, L., Gurtner, A., and Piaggio, G. (2019). Transgenic animal models to visualize cancer-related cellular processes by bioluminescence imaging. Front. Pharmacol. 10, 235. doi:10.3389/fphar.2019.00235
McMorrow, R., Zambito, G., Nigg, A., Lila, K., van den Bosch, T. P. P., Lowik, C., et al. (2023). Whole-body bioluminescence imaging of T-cell response in PDAC models. Front. Immunol. 14, 1207533. doi:10.3389/fimmu.2023.1207533
Mezzanotte, L., Aswendt, M., Tennstaedt, A., Hoeben, R., Hoehn, M., and Lowik, C. (2013). Evaluating reporter genes of different luciferases for optimized in vivo bioluminescence imaging of transplanted neural stem cells in the brain. Contrast Media Mol. Imaging 8 (6), 505–513. doi:10.1002/cmmi.1549
Mezzanotte, L., Que, I., Kaijzel, E., Branchini, B., Roda, A., and Lowik, C. (2011). Sensitive dual color in vivo bioluminescence imaging using a new red codon optimized firefly luciferase and a green click beetle luciferase. PLoS One 6 (4), e19277. doi:10.1371/journal.pone.0019277
Mezzanotte, L., van 't Root, M., Karatas, H., Goun, E. A., and Lowik, C. (2017). In vivo molecular bioluminescence imaging: new tools and applications. Trends Biotechnol. 35 (7), 640–652. doi:10.1016/j.tibtech.2017.03.012
Millar, A. J., Short, S. R., Chua, N. H., and Kay, S. A. (1992). A novel circadian phenotype based on firefly luciferase expression in transgenic plants. Plant Cell. 4 (9), 1075–1087. doi:10.1105/tpc.4.9.1075
Miranda Furtado, C. L., Dos Santos Luciano, M. C., Silva Santos, R. D., Furtado, G. P., Moraes, M. O., and Pessoa, C. (2019). Epidrugs: targeting epigenetic marks in cancer treatment. Epigenetics 14 (12), 1164–1176. doi:10.1080/15592294.2019.1640546
Mitiouchkina, T., Mishin, A. S., Somermeyer, L. G., Markina, N. M., Chepurnyh, T. V., Guglya, E. B., et al. (2020). Plants with genetically encoded autoluminescence. Nat. Biotechnol. 38 (8), 944–946. doi:10.1038/s41587-020-0500-9
Miyabara, H., Hirano, R., Watanabe, S., Soriano, J. C. C., Watanabe, H., Kuchimaru, T., et al. (2023). In vivo optical imaging of tumor stromal cells with hypoxia-inducible factor activity. Cancer Sci. doi:10.1111/cas.15907
Monk, D., Mackay, D. J. G., Eggermann, T., Maher, E. R., and Riccio, A. (2019). Genomic imprinting disorders: lessons on how genome, epigenome and environment interact. Nat. Rev. Genet. 20 (4), 235–248. doi:10.1038/s41576-018-0092-0
Moorwood, C., and Khurana, T. S. (2013). Duchenne muscular dystrophy drug discovery - the application of utrophin promoter activation screening. Expert Opin. Drug Discov. 8 (5), 569–581. doi:10.1517/17460441.2013.777040
Moorwood, C., Lozynska, O., Suri, N., Napper, A. D., Diamond, S. L., and Khurana, T. S. (2011). Drug discovery for Duchenne muscular dystrophy via utrophin promoter activation screening. PLoS One 6 (10), e26169. doi:10.1371/journal.pone.0026169
Moorwood, C., Soni, N., Patel, G., Wilton, S. D., and Khurana, T. S. (2013). A cell-based high-throughput screening assay for posttranscriptional utrophin upregulation. J. Biomol. Screen 18 (4), 400–406. doi:10.1177/1087057112465648
Niswender, K. D., Blackman, S. M., Rohde, L., Magnuson, M. A., and Piston, D. W. (1995). Quantitative imaging of green fluorescent protein in cultured cells: comparison of microscopic techniques, use in fusion proteins and detection limits. J. Microsc. 180 (2), 109–116. doi:10.1111/j.1365-2818.1995.tb03665.x
Nolan, G. P., Fiering, S., Nicolas, J. F., and Herzenberg, L. A. (1988). Fluorescence-activated cell analysis and sorting of viable mammalian cells based on beta-D-galactosidase activity after transduction of Escherichia coli lacZ. Proc. Natl. Acad. Sci. U. S. A. 85 (8), 2603–2607. doi:10.1073/pnas.85.8.2603
Nussbaum, A., and Cohen, A. (1988). Use of a bioluminescence gene reporter for the investigation of red-dependent and gam-dependent plasmid recombination in Escherichia coli K12. J. Mol. Biol. 203 (2), 391–402. doi:10.1016/0022-2836(88)90007-1
O'Farrell, A. C., Shnyder, S. D., Marston, G., Coletta, P. L., and Gill, J. H. (2013). Non-invasive molecular imaging for preclinical cancer therapeutic development. Br. J. Pharmacol. 169 (4), 719–735. doi:10.1111/bph.12155
Ohlendieck, K., Ervasti, J. M., Matsumura, K., Kahl, S. D., Leveille, C. J., and Campbell, K. P. (1991). Dystrophin-related protein is localized to neuromuscular junctions of adult skeletal muscle. Neuron 7 (3), 499–508. doi:10.1016/0896-6273(91)90301-f
Olmeda, D., Cerezo-Wallis, D., Riveiro-Falkenbach, E., Pennacchi, P. C., Contreras-Alcalde, M., Ibarz, N., et al. (2017). Whole-body imaging of lymphovascular niches identifies pre-metastatic roles of midkine. Nature 546 (7660), 676–680. doi:10.1038/nature22977
Panning, B. (2008). X-Chromosome inactivation: the molecular basis of silencing. J. Biol. 7 (8), 30. doi:10.1186/jbiol95
Perkins, K. J., and Davies, K. E. (2002). The role of utrophin in the potential therapy of Duchenne muscular dystrophy. Neuromuscul. Disord. 12 (1), S78–S89. doi:10.1016/s0960-8966(02)00087-1
Peters, J. (2014). The role of genomic imprinting in biology and disease: an expanding view. Nat. Rev. Genet. 15 (8), 517–530. doi:10.1038/nrg3766
Pham, H. T. A., Lee, S., and Lee, Y. J. (2022). Bicistronic reporter mice for monitoring of Fgf21 expression. Biochem. Biophys. Res. Commun. 619, 104–109. doi:10.1016/j.bbrc.2022.06.045
Pons, F., Nicholson, L. V., Robert, A., Voit, T., and Leger, J. J. (1993). Dystrophin and dystrophin-related protein (utrophin) distribution in normal and dystrophin-deficient skeletal muscles. Neuromuscul. Disord. 3 (5-6), 507–514. doi:10.1016/0960-8966(93)90106-t
Qian, D. Z., Ren, M., Wei, Y., Wang, X., van de Geijn, F., Rasmussen, C., et al. (2005). In vivo imaging of retinoic acid receptor beta2 transcriptional activation by the histone deacetylase inhibitor MS-275 in retinoid-resistant prostate cancer cells. Prostate 64 (1), 20–28. doi:10.1002/pros.20209
Rabinovich, B. A., Ye, Y., Etto, T., Chen, J. Q., Levitsky, H. I., Overwijk, W. W., et al. (2008). Visualizing fewer than 10 mouse T cells with an enhanced firefly luciferase in immunocompetent mouse models of cancer. Proc. Natl. Acad. Sci. U. S. A. 105 (38), 14342–14346. doi:10.1073/pnas.0804105105
Rehemtulla, A., Stegman, L. D., Cardozo, S. J., Gupta, S., Hall, D. E., Contag, C. H., et al. (2000). Rapid and quantitative assessment of cancer treatment response using in vivo bioluminescence imaging. Neoplasia 2 (6), 491–495. doi:10.1038/sj.neo.7900121
Roda, A., Guardigli, M., Pasini, P., and Mirasoli, M. (2003). Bioluminescence and chemiluminescence in drug screening. Anal. Bioanal. Chem. 377 (5), 826–833. doi:10.1007/s00216-003-2096-6
Salarinia, R., Sahebkar, A., Peyvandi, M., Mirzaei, H. R., Jaafari, M. R., Riahi, M. M., et al. (2016). Epi-drugs and epi-miRs: moving beyond current cancer therapies. Curr. Cancer Drug Targets 16 (9), 773–788. doi:10.2174/1568009616666151207110143
Sanchez, A., Penault-Llorca, F., Bignon, Y. J., Guy, L., and Bernard-Gallon, D. (2022). Effects of GSK-J4 on JMJD3 histone demethylase in mouse prostate cancer xenografts. Cancer Genomics Proteomics 19 (3), 339–349. doi:10.21873/cgp.20324
Schaub, F. X., Reza, M. S., Flaveny, C. A., Li, W., Musicant, A. M., Hoxha, S., et al. (2015). Fluorophore-NanoLuc BRET reporters enable sensitive in vivo optical imaging and flow cytometry for monitoring tumorigenesis. Cancer Res. 75 (23), 5023–5033. doi:10.1158/0008-5472.CAN-14-3538
Sekar, T. V., Foygel, K., Gelovani, J. G., and Paulmurugan, R. (2015). Genetically encoded molecular biosensors to image histone methylation in living animals. Anal. Chem. 87 (2), 892–899. doi:10.1021/ac502629r
Serganova, I., and Blasberg, R. G. (2019). Molecular imaging with reporter genes: has its promise been delivered? J. Nucl. Med. 60 (12), 1665–1681. doi:10.2967/jnumed.118.220004
Shan, Y., Abel, J. H., Li, Y., Izumo, M., Cox, K. H., Jeong, B., et al. (2020). Dual-Color single-cell imaging of the suprachiasmatic nucleus reveals a circadian role in network synchrony. Neuron 108 (1), 164–179. doi:10.1016/j.neuron.2020.07.012
Sharifian, S., Homaei, A., Hemmati, R., and Khajeh, K. (2018). The emerging use of bioluminescence in medical research. Biomed. Pharmacother. 101, 74–86. doi:10.1016/j.biopha.2018.02.065
Sharpe, J., Ahlgren, U., Perry, P., Hill, B., Ross, A., Hecksher-Sorensen, J., et al. (2002). Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science 296 (5567), 541–545. doi:10.1126/science.1068206
Shekhawat, S. S., and Ghosh, I. (2011). Split-protein systems: beyond binary protein-protein interactions. Curr. Opin. Chem. Biol. 15 (6), 789–797. doi:10.1016/j.cbpa.2011.10.014
Smith, C. B., van der Vinne, V., McCartney, E., Stowie, A. C., Leise, T. L., Martin-Burgos, B., et al. (2022). Cell-type-specific circadian bioluminescence rhythms in dbp reporter mice. J. Biol. Rhythms 37 (1), 53–77. doi:10.1177/07487304211069452
Soling, A., Theiss, C., Jungmichel, S., and Rainov, N. G. (2004). A dual function fusion protein of Herpes simplex virus type 1 thymidine kinase and firefly luciferase for noninvasive in vivo imaging of gene therapy in malignant glioma. Genet. Vaccines Ther. 2 (1), 7. doi:10.1186/1479-0556-2-7
Son, Y., Choi, C., Song, C., Im, H., Cho, Y. K., Son, J. S., et al. (2021). Development of CIDEA reporter mouse model and its application for screening thermogenic drugs. Sci. Rep. 11 (1), 18429. doi:10.1038/s41598-021-97959-0
Sripathy, S., Leko, V., Adrianse, R. L., Loe, T., Foss, E. J., Dalrymple, E., et al. (2017). Screen for reactivation of MeCP2 on the inactive X chromosome identifies the BMP/TGF-beta superfamily as a regulator of XIST expression. Proc. Natl. Acad. Sci. U. S. A. 114 (7), 1619–1624. doi:10.1073/pnas.1621356114
Stains, C. I., Furman, J. L., Segal, D. J., and Ghosh, I. (2006). Site-specific detection of DNA methylation utilizing mCpG-SEER. J. Am. Chem. Soc. 128 (30), 9761–9765. doi:10.1021/ja060681j
Statello, L., Guo, C. J., Chen, L. L., and Huarte, M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell. Biol. 22 (2), 96–118. doi:10.1038/s41580-020-00315-9
Su, I. H., Basavaraj, A., Krutchinsky, A. N., Hobert, O., Ullrich, A., Chait, B. T., et al. (2003). Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat. Immunol. 4 (2), 124–131. doi:10.1038/ni876
Su, Y., Walker, J. R., Hall, M. P., Klein, M. A., Wu, X., Encell, L. P., et al. (2023). An optimized bioluminescent substrate for non-invasive imaging in the brain. Nat. Chem. Biol. 19 (6), 731–739. doi:10.1038/s41589-023-01265-x
Surani, M. A. (1998). Imprinting and the initiation of gene silencing in the germ line. Cell. 93 (3), 309–312. doi:10.1016/s0092-8674(00)81156-3
Syed, A. J., and Anderson, J. C. (2021). Applications of bioluminescence in biotechnology and beyond. Chem. Soc. Rev. 50 (9), 5668–5705. doi:10.1039/d0cs01492c
Taka, N., Baba, Y., Iwasaki, Y., and Yoshida, W. (2022). Bioluminescence resonance energy transfer for global DNA methylation quantification. Methods Mol. Biol. 2525, 267–279. doi:10.1007/978-1-0716-2473-9_20
Tannous, B. A., Kim, D. E., Fernandez, J. L., Weissleder, R., and Breakefield, X. O. (2005). Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol. Ther. 11 (3), 435–443. doi:10.1016/j.ymthe.2004.10.016
Tinkum, K. L., Marpegan, L., White, L. S., Sun, J., Herzog, E. D., Piwnica-Worms, D., et al. (2011). Bioluminescence imaging captures the expression and dynamics of endogenous p21 promoter activity in living mice and intact cells. Mol. Cell. Biol. 31 (18), 3759–3772. doi:10.1128/MCB.05243-11
Tinsley, J. M., Fairclough, R. J., Storer, R., Wilkes, F. J., Potter, A. C., Squire, S. E., et al. (2011). Daily treatment with SMTC1100, a novel small molecule utrophin upregulator, dramatically reduces the dystrophic symptoms in the mdx mouse. PLoS One 6 (5), e19189. doi:10.1371/journal.pone.0019189
Van de Pette, M., Abbas, A., Feytout, A., McNamara, G., Bruno, L., To, W. K., et al. (2017). Visualizing changes in Cdkn1c expression links early-life adversity to imprint mis-regulation in adults. Cell. Rep. 18 (5), 1090–1099. doi:10.1016/j.celrep.2017.01.010
Van de Pette, M., Dimond, A., Galvao, A. M., Millership, S. J., To, W., Prodani, C., et al. (2022). Epigenetic changes induced by in utero dietary challenge result in phenotypic variability in successive generations of mice. Nat. Commun. 13 (1), 2464. doi:10.1038/s41467-022-30022-2
Vande Velde, G., Kucharikova, S., Schrevens, S., Himmelreich, U., and Van Dijck, P. (2014). Towards non-invasive monitoring of pathogen-host interactions during Candida albicans biofilm formation using in vivo bioluminescence. Cell. Microbiol. 16 (1), 115–130. doi:10.1111/cmi.12184
Veland, N., Gleneadie, H. J., Brown, K. E., Sardini, A., Pombo, J., Dimond, A., et al. (2023). Bioluminescence imaging of <em>Cyp1a1-</em>luciferase reporter mice demonstrates prolonged activation of the aryl hydrocarbon receptor in the lung. bioRxiv.
Vooijs, M., Jonkers, J., Lyons, S., and Berns, A. (2002). Noninvasive imaging of spontaneous retinoblastoma pathway-dependent tumors in mice. Cancer Res. 62 (6), 1862–1867.
Vuorinen, A., Wilkinson, I. V. L., Chatzopoulou, M., Edwards, B., Squire, S. E., Fairclough, R. J., et al. (2021). Discovery and mechanism of action studies of 4,6-diphenylpyrimidine-2-carbohydrazides as utrophin modulators for the treatment of Duchenne muscular dystrophy. Eur. J. Med. Chem. 220, 113431. doi:10.1016/j.ejmech.2021.113431
Wang, J., Wegener, J. E., Huang, T. W., Sripathy, S., De Jesus-Cortes, H., Xu, P., et al. (2015). Global spine congress 2015. Nature 521 (7552), E1–E4. doi:10.1055/s-0035-1552926
Wang, J., Zhong, F., Li, J., Yue, H., Li, W., and Lu, X. (2023). The epigenetic factor CHD4 contributes to metastasis by regulating the EZH2/β-catenin axis and acts as a therapeutic target in ovarian cancer. J. Transl. Med. 21 (1), 38. doi:10.1186/s12967-022-03854-1
Wang, W., Kim, S. H., and El-Deiry, W. S. (2006). Small-molecule modulators of p53 family signaling and antitumor effects in p53-deficient human colon tumor xenografts. Proc. Natl. Acad. Sci. U. S. A. 103 (29), 11003–11008. doi:10.1073/pnas.0604507103
Wang, X. X., Jia, H. J., Lv, Y. R., Sun, H. H., Wei, X. L., Tan, J. Y., et al. (2021). A luciferase-EGFP reporter system for the evaluation of DNA methylation in mammalian cells. Mol. Biol. 55 (5), 742–751. doi:10.1134/S0026893321040099
Ward, W. W., and Cormier, M. J. (1979). An energy transfer protein in coelenterate bioluminescence. Characterization of the Renilla green-fluorescent protein. J. Biol. Chem. 254 (3), 781–788. doi:10.1016/s0021-9258(17)37873-0
Wolff, F., Leisch, M., Greil, R., Risch, A., and Pleyer, L. (2017). The double-edged sword of (re)expression of genes by hypomethylating agents: from viral mimicry to exploitation as priming agents for targeted immune checkpoint modulation. Cell. Commun. Signal 15 (1), 13. doi:10.1186/s12964-017-0168-z
Xu, T., Ripp, S., Sayler, G. S., and Close, D. M. (2014). Expression of a humanized viral 2A-mediated lux operon efficiently generates autonomous bioluminescence in human cells. PLoS One 9 (5), e96347. doi:10.1371/journal.pone.0096347
Yan, X., Ray, P., Paulmurugan, R., Tong, R., Gong, Y., Sathirachinda, A., et al. (2013). A transgenic tri-modality reporter mouse. PLoS One 8 (8), e73580. doi:10.1371/journal.pone.0073580
Yang, J. J., Liu, Z. Q., Zhang, J. M., Wang, H. B., Hu, S. Y., Liu, J. F., et al. (2013). Real-time tracking of adipose tissue-derived stem cells with injectable scaffolds in the infarcted heart. Heart Vessels 28 (3), 385–396. doi:10.1007/s00380-012-0275-0
Zagozdzon, A. M., O'Leary, P., Callanan, J. J., Crown, J., Gallagher, W. M., and Zagozdzon, R. (2012). Generation of a new bioluminescent model for visualisation of mammary tumour development in transgenic mice. BMC Cancer 12, 209. doi:10.1186/1471-2407-12-209
Zambito, G., Chawda, C., and Mezzanotte, L. (2021). Emerging tools for bioluminescence imaging. Curr. Opin. Chem. Biol. 63, 86–94. doi:10.1016/j.cbpa.2021.02.005
Zhang, N., Lyons, S., Lim, E., and Lassota, P. (2009). A spontaneous acinar cell carcinoma model for monitoring progression of pancreatic lesions and response to treatment through noninvasive bioluminescence imaging. Clin. Cancer Res. 15 (15), 4915–4924. doi:10.1158/1078-0432.CCR-08-2256
Zhao, H., Doyle, T. C., Coquoz, O., Kalish, F., Rice, B. W., and Contag, C. H. (2005). Emission spectra of bioluminescent reporters and interaction with mammalian tissue determine the sensitivity of detection in vivo. J. Biomed. Opt. 10 (4), 41210. doi:10.1117/1.2032388
Nomenclature
Keywords: epigenetics, in vivo imaging, bioluminescence, endogenous reporter genes, luciferase, epidrugs, drug screening, genetic and epigenetic disorders
Citation: Gleneadie HJ, Dimond A and Fisher AG (2023) Harnessing bioluminescence for drug discovery and epigenetic research. Front. Drug Discov. 3:1249507. doi: 10.3389/fddsv.2023.1249507
Received: 24 July 2023; Accepted: 17 August 2023;
Published: 05 September 2023.
Edited by:
Shicheng Guo, University of Wisconsin-Madison, United StatesReviewed by:
Huiya Yang, Prime Medicine, United StatesYue Liu, The University of Texas at Austin, United States
Ke Wu, Wuhan University, China
Tianyi Zhang, Emory University, United States
Minghan Yang, New York University, United States
Copyright © 2023 Gleneadie, Dimond and Fisher. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hannah J. Gleneadie, aGFubmFoLmVhZGllQGxtcy5tcmMuYWMudWs=; Amanda G. Fisher, YW1hbmRhLmZpc2hlckBiaW9jaC5veC5hYy51aw==
 Hannah J. Gleneadie
Hannah J. Gleneadie Andrew Dimond
Andrew Dimond Amanda G. Fisher1,2*
Amanda G. Fisher1,2*