- 1West African Center for Cell Biology of Infectious Pathogens, University of Ghana, Accra, Ghana
- 2Department of Biochemistry, Cell and Molecular Biology, University of Ghana, Accra, Ghana
The emergence of drug-resistant malaria parasites to artemisinin and its partner drugs highlights the need to increase the arsenal of new antimalarials with novel mechanisms of action. To help achieve this aim, this study tested the potency of three Malaria Box compounds (MMV006087, MMV085203, and MMV008956) against five laboratory strains and twenty clinical isolates of Plasmodium falciparum using optimized in vitro growth inhibitory assays. The results were compared to the response from four standard antimalarials-artesunate, chloroquine, mefloquine, and halofantrine. From the results, MMV006087 was the most potent compound with an average IC50 of 22.13 nM compared to MMV085203 (average IC50 of 137.90 nM) and MMV008956 (average IC50 of 262.30 nM). On average, the laboratory strains were also less susceptible to the three Malaria Box compounds (average IC50 of 162.30 nM) compared to the clinical isolates (average IC50 of 135.40 nM). Additionally, MMV006087 was less potent than artesunate but twice more efficacious than chloroquine against the laboratory strains and clinical isolates. The data from this study validate the potency of MMV006087 and MMV085203 as promising antimalarials worthy of further exploration. This study further substantiates the need to include clinical isolates in antimalarial compound screening activities.
1 Introduction
Although the burden of malaria in sub-Saharan Africa is still high, the rate of development of drug resistance to the current antimalarials is slow compared to South-East Asia and South America (Bushman et al., 2016; WHO, 2022). Artemisinin and its partner drugs are relatively effective in treating malaria in Africa (Mathieu et al., 2021; Ndwiga et al., 2021; Stokes et al., 2021). However, there are reports of the emergence of drug-resistant malaria parasite strains in South-East Asia and the slow clearance of malaria parasites by artemisinin-based drugs in some East African countries (Ndwiga et al., 2021). This highlights the need to accelerate the arsenal of new antimalarials in the drug-development pipeline to counter antimalarial drug resistance. The World Health Organization (WHO) estimates that a reduction of about 25% of the effectiveness of antimalarials in sub-Saharan Africa can result in about 46,000 additional deaths (WHO, 2020). Additionally, the recent detection of Anopheles stephensi in several African countries coupled with an increase in the number of parasites carrying Plasmodium falciparum Kelch13 (Pfkelch13) mutations and the detection of several non-falciparum species may undermine the current treatment strategies and transmission dynamics in sub-Saharan Africa (Sinka et al., 2020; Ahmed et al., 2021; Ansah et al., 2021; Mnzava et al., 2022; WHO, 2022). Therefore, to eliminate malaria globally, it is prudent to have drugs with high efficacy at all times.
The slow pace of developing new antimalarials, in addition to the frequent emergence of drug-resistant malaria parasites to all frontline antimalarials, has created the need for new antimalarials with novel mechanisms of action. To answer this call, the Medicines for Malarial Venture (MMV) with its partners have created the Malaria Box (now called Pathogen Box) set of compounds to provide an accelerated path to the development of new, affordable and easily accessible antimalarial drugs for malaria-endemic regions (Spangenberg et al., 2013; Diagana, 2015). The Malaria Box compound library contained 400 bioactive compounds comprising 200 drug-like and 200 probe-like compounds. Several studies have been conducted to validate the potency of these compounds, and identify their targets and mechanisms of action (Plouffe et al., 2016; Tiwari et al., 2016; Van Voorhis et al., 2016). However, the majority of these screens have mostly been conducted using laboratory strains of P. falciparum. Therefore, additional studies prioritizing the most critical compounds in the Malaria Box compound library using clinical isolates will further help in prioritizing compounds in the antimalarial drug discovery and development pipeline.
Thomson Reuters Life Science Consulting indicates that more than half of the compounds in clinical trials often fail because of insufficient efficacy (Arrowsmith, 2011). Therefore, compounds with the likelihood of clinical efficacy must have novel molecular targets and exhibit high potency in both in vitro and in vivo studies. As opposed to laboratory strains, clinical isolates often have multiple parasite genotypes (clones) with different drug-resistant phenotypes that can affect the efficacy of an antimalarial compound. Consequently, it is essential to validate their potency using clinical isolates to prioritize these compounds for clinical applications (Chirawurah et al., 2017; Partey et al., 2022).
The three Malaria Box compounds -MMV006087, MMV085203, and MMV008956- were previously identified to be potent against clinical isolates of P. falciparum (Chirawurah et al., 2017). However, these were only tested using very few clinical isolates. Therefore, this study further validated the potency of MMV006087, MMV085203, and MMV008956 by screening them against five laboratory strains and twenty (20) P. falciparum clinical isolates. Additionally, the potency of the three Malaria Box compounds was also compared to four standard antimalarials-artesunate, chloroquine, mefloquine, and halofantrine. Finally, the number of parasite clones in the clinical isolates was determined using the merozoite surface protein 1 (MSP1) and the merozoite surface protein 2 (MSP2) genotyping, and the response of the three Malaria Box compounds was compared among the parasite genotypes.
2 Materials and methods
2.1 P. falciparum parasites and parasite culture
For this study, cryopreserved P. falciparum laboratory-adapted parasites, 3D7, W2mef, Dd2, NF54, and K1 and twenty (20) randomly selected clinical isolates (Figures 2A, B) were cultured with human group O+ erythrocytes using standard methods (Trager and Jensen, 1976) with slight modifications. The clinical isolates were collected as part of an ongoing study on erythrocyte invasion mechanisms (EIM) (Mensah-Brown et al., 2017). The samples were cryopreserved using the glycerolyte method (Moll et al., 2008) and stored in liquid nitrogen tanks upon arrival from the field. Samples are thawed and grown whenever an assay is to be performed. For the growth inhibition assays, P. falciparum parasites were cultured in RPMI supplement with naïve human serum and propagated to 5% parasitemia of ring-stage parasites and used for the in vitro growth inhibition assays. These parasites were diluted to the required parasitemia for the assays
2.2 Synchronisation of P. falciparum parasites
To allow for stage-specific effects to be measured, mixed-stage cultures are synchronized to rings and allowed to grow for at least one parasite cycle. Using 5% Sorbitol (Sigma-Aldrich) treatment, a synchronized culture of ring-stage parasites (>5% parasitemia) (Lambros and Vanderberg, 1979) was obtained and diluted to 1% parasitemia and 2% hematocrit for the growth inhibition assays as previously described (Chirawurah et al., 2017). Briefly, 5 mL of the parasite cultures were first transferred into 15 mL falcon tubes and centrifuged at 1,500 rpm for 5 min. The supernatant was then discarded, and 5 mL of sorbitol solution was added. The mixture was then incubated for 10 min at 37°C with gentle agitation, followed by centrifugation at 1,500 rpm. The supernatant was then discarded, followed by three washes with RPMI. To prevent a possible loss of some of the clones in the clinical isolates, sorbitol synchronization was conducted for only the laboratory strains. However, the clinical isolates were all screened using parasites with more than 90% rings.
2.3 In vitro drug susceptibility assay
To evaluate the potency of the Malaria Box compounds and standard antimalarials against the laboratory strains and clinical isolates, cryopreserved samples from liquid nitrogen were thawed using serial NaCl solutions (Moll et al., 2008). Following this, a stock of 10 mM of each of the compounds was prepared and screened against the parasites at final-well concentrations from 0.006 nM to 25 µM as previously described (Chirawurah et al., 2017). The growth inhibition assays were set up in a 96-well plate with final-well volumes of 100 μL-consisting of 10 μL of each compound and 90 μL parasite culture at 1% parasitemia and 2% hematocrit. Unmodified media (RPMI) was used as a negative control, and uninfected erythrocytes at 2% hematocrit were also used to correct the background fluorescence. The parasitemia corresponding to each culture well after 72 h of incubation was determined by replacing 80 μL of the supernatant with 1X SYBR Green I (Invitrogen, United States) stain in lysis buffer (20 mM Tris (pH 7.5), 5 mm EDTA, 0.008% (W/V) saponin and 0.08% (V/V) Triton X-100), based on established methods (Smilkstein et al., 2004). The plates were then incubated at 37°C in the dark for at least 30 min, and the total fluorescence from each of the wells was determined with the Varioskan Lux multimode microplate fluorescent plate reader (ThermoFisher Scientific, United States) at an excitation of 485 nm, emission of 520 nM and with a gain of 100.
The data from the fluorescent plate reader was formatted using Microsoft Excel software by subtracting the background fluorescence from all the data sets. Sigmoidal dose-response curves were then generated with the GraphPad Prism (Version 9.01) software by first transforming the concentrations into a logarithmic scale and normalizing the fluorescent data, which corresponds with the parasitemia reading. Because the dose-response curves typically follow a sigmoidal (S-shaped) pattern, a non-linear regression curve was used to fit the sigmoidal curve to the data. This study used the four-parameter logistic model (4 PL), which is the most frequently used model for dose-response analysis and provides information about the minimum and maximum response levels, the IC50 value and the slope of the curve. The R-squared value, which is a statistical test for the goodness-of-fit for the data was also used to examine the quality of the fit for all the data sets. R-squared values of greater than 0.9 were considered a good fit for all the curves generated. The dose-response curves were used to determine the 50% inhibitory concentration (IC50) values. Each data point on the dose-response curve represents the mean ± (SEM) of two experiments, set up in triplicate. The IC50 values and their 95% confidence intervals obtained from the drug assays were used to establish and compare the potency of the three compounds in all the laboratory strains and clinical isolates.
2.4 Evaluating the clonal diversity of the clinical isolates
The different parasite genotypes (clones) in the clinical isolates were identified by analyzing the polymorphic genetic loci of the merozoite surface protein genes, msp1 and msp2 (Snounou et al., 1999). Briefly, genomic DNA was extracted from fourteen (14) clinical isolates of P. falciparum parasites using the QIAamp DNA Extraction Mini Kit (Qiagen, United Kingdom). The purified genomic DNA were used as templates in a polymerase chain reaction (PCR) assay with nested primers targeting the msp1 block 2 and msp2 block 3 as previously reported (Supplementary Table S5). The resulting amplicons from the nest 1 assays were used as DNA in the nest 2 reactions to determine the allelic families of msp1 (MAD20, K1, and RO33) and msp2 (3D7/IC and FC27). Genomic DNA from laboratory strain was used as a positive control, and a template-free control was also included as a negative control for all the reactions. The size of the PCR product was determined using a 100 bp DNA ladder. The polymorphisms in each allelic family of the msp1 and msp2 genes were determined as previously described (Mayengue et al., 2011).
3 Results
3.1 The potency of the Malaria Box compounds against laboratory strains and clinical isolates of P. falciparum
The potency of the three compounds-MMV006087, MMV085203, and MMV008956- were first validated by screening them against five laboratory strains (Supplementary Table S1). From the IC50 values, we found MMV006087 to be the most potent with IC50 values less than 100 nM (Figure 1A; Supplementary Figure S5). Based on the average IC50 values, MMV006087 was found to be about seven and seventeen-fold more potent against the laboratory strains than MMV008956 and MMV085203, respectively (Figure 2A; Table 1).

FIGURE 1. A heatmap showing the potency of the Malaria Box compounds against P. falciparum parasites. (A) Laboratory strains (B) Clinical isolates.
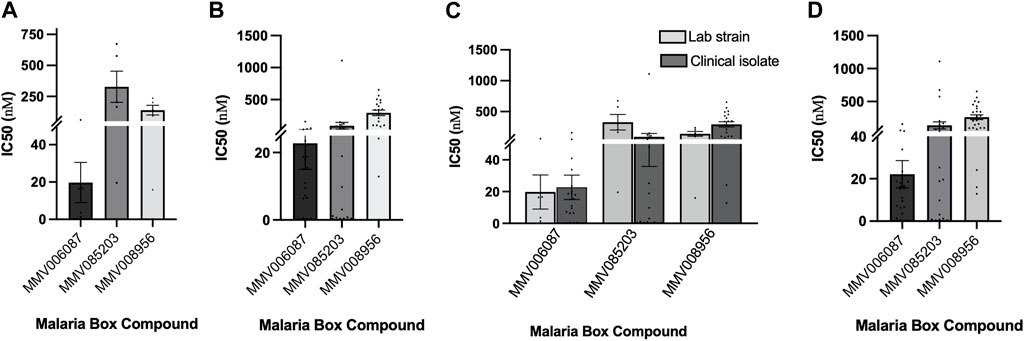
FIGURE 2. Comparing the potency of the three Malaria Box compounds in the laboratory strains (Lab) and clinical isolates (Clinical). (A) Comparing the mean IC50 values of each of the Malaria Box compounds against five laboratory strains of P. falciparum; (B) Comparing IC50 values of the Malaria Box compounds against twenty clinical isolates of P. falciparum; (C) Comparing the mean IC50 values of each of the Malaria Box compounds between the laboratory and clinical isolates of P. falciparum; (D) The mean IC50 values of each of the Malaria Box compounds in both laboratory and clinical isolates of P. falciparum. Error bars denote SEM.
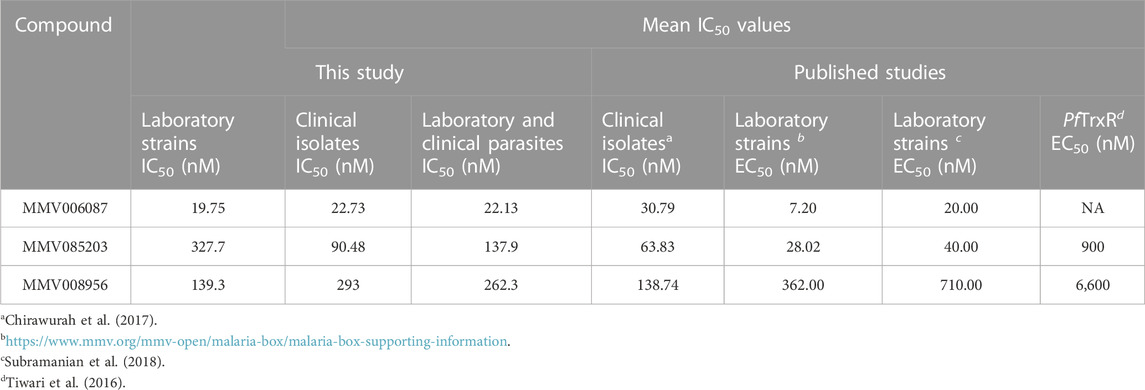
TABLE 1. A summary of the mean IC50 values of the Malaria Box compounds against the laboratory strains and clinical isolates of P. falciparum.
Following this, the three compounds were also screened against twenty P. falciparum clinical isolates. The average IC50 values also indicated that MMV006087 was about four-fold more potent against the clinical isolates than MMV085203 and fifteen-fold more potent than MMV008956 (Figure 2B; Table 1). Therefore, MMV006087 was also the most potent compound in the clinical isolates with an average IC50 value below 50 nM, followed by MMV085203 and MMV008956 (Figure 1B; Supplementary Figure S2; Table 1). The data corroborated the observations in our previous report (Chirawurah et al., 2017).
Furthermore, the response of the parasites to the three compounds was examined by comparing the potencies of the laboratory strains to those of the clinical isolates (Figure 2C; Table 1). We found that MMV006087 and MMV008956 were more potent against the laboratory strains than the clinical isolates, but the contrary was true for MMV085203 (Figure 2C; Table 1). This suggests that MMV085203 may have a novel molecular target(s) and could be a promising partner antimalarial.
The overall efficacy of the three Malaria Box compounds in both the laboratory strains and clinical isolates of P. falciparum was determined by averaging the IC50 values for each compound (Figure 3D; Table 1). The mean IC50 value for MMV006087 was about six-fold that of MMV085203 and about 12-fold that of MMV008956 (Table 1). Therefore, MMV006087 was the most potent compound of the three Malaria Box compounds in both the laboratory strains and clinical isolates of P. falciparum with IC50 values less than 100 nM as we previously reported (Chirawurah et al., 2017). The differences in the response of the parasites to the compounds highlight the need of including clinical isolates in antimalarial screening studies, which could provide an additional layer of validation for the compounds.
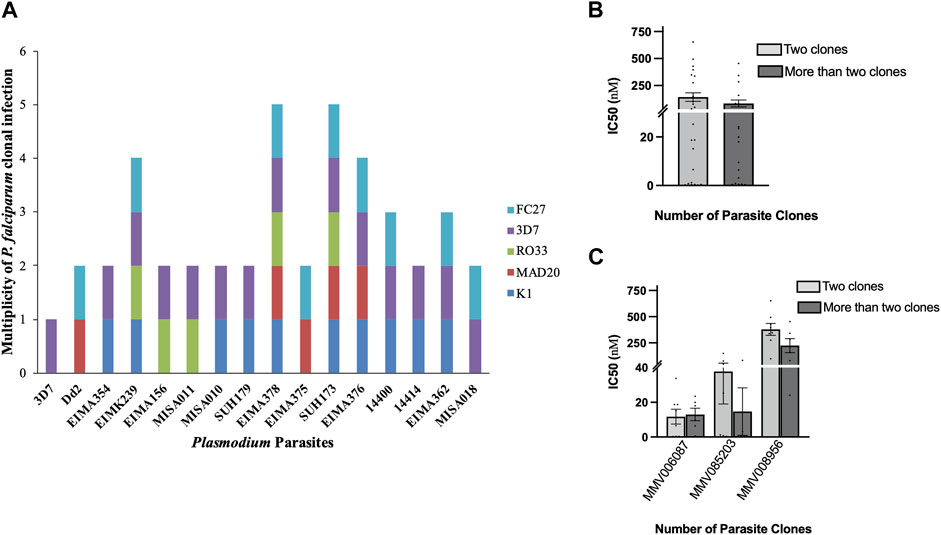
FIGURE 3. The effect of genetic diversity on the susceptibility of the clinical isolates to the Malaria Box compounds. (A) Genetic diversity of the clinical isolates of P. falciparum parasites based on msp1 and msp2 genotyping. The number of genotypes in the clinical isolates varied between two and five parasite clones; (B) The IC50 values of the three Malaria Box compounds against the clinical isolates with two and more than two P. falciparum genotypes; (C) The IC50 values of the clinical isolates with two P. falciparum genotypes against each of the three Malaria box compounds compared to those with more than two genotypes. Error bars denote SEM.
3.2 The effects of clonal diversity of P. falciparum clinical isolates on their susceptibility to antimalarials
The effects of multiple P. falciparum genotypes (clones) in the clinical isolates on their response to the three Malaria Box compounds were examined using msp1 and msp2 genotyping. For this, fourteen (14) clinical isolates were genotyped. From the results, the clinical isolates had between two and five parasite genotypes (clones) (Figure 3A). Therefore, the mean IC50 values of the isolates with two genotypes were compared to those with three or more genotypes (Figure 3B). It was observed that the clinical isolates with two parasite genotypes were less susceptible to the three Malaria Box compounds compared to those with more than two genotypes (Figure 3B). Furthermore, our data also suggested that MMV085203 and MMV008956 were more potent against the clinical isolates with more than two parasite genotypes compared to MMV006087 (Figure 3C; Table 2). This may suggest that the two compounds could be further explored as promising antimalarials or partner drugs against malaria parasites.
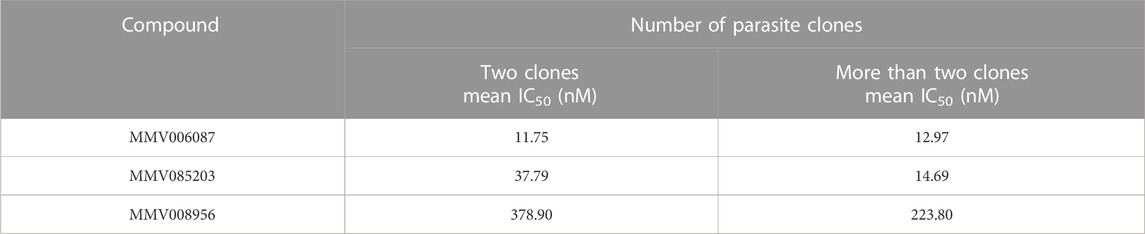
TABLE 2. Summary of the mean IC50 values of the clinical isolates with two P. falciparum genotypes against each of the three Malaria Box compounds compared to those with more than two genotypes.
3.3 Evaluating and comparing the potency of the three Malaria Box compounds to standard antimalarials
This study also evaluated and compared the potency of the three Malaria Box compounds to four standard antimalarials; artesunate, chloroquine, mefloquine, and halofantrine (Supplementary Tables S3, S4). From the IC50 values obtained for the four standard antimalarials and the three Malaria Box compounds against the laboratory strains, artesunate was the most potent compound, followed by mefloquine, MMV006087, halofantrine, chloroquine, MMV008956, and MMV085203 (Figure 4A). We also observed that while MMV006087 (average IC50 of 26.95 nM) was less potent compared to artesunate (average IC50 of 8.22 nM), its potency was twice that of chloroquine (average IC50 of 59.26 nM) (Table 3). As expected, both 3D7 and NF54 strains, which are chloroquine-susceptible strains (IC50 values < 100 nM) had IC50 values of 7.43 nM and 13.46 nM, respectively. However, Dd2, which is a chloroquine-resistant strain (IC50 > 100 nM) had an IC50 of 156.90 nM (Figure 4A; Supplementary Table S3). Similarly, MMV006087 was more potent against the two chloroquine-susceptible strains (3D7; IC50 = 16.56 nM and NF54; IC50 = 3.61 nM) than the chloroquine-resistant strain (Dd2; IC50 = 60.83 nM) (Supplementary Table S3).
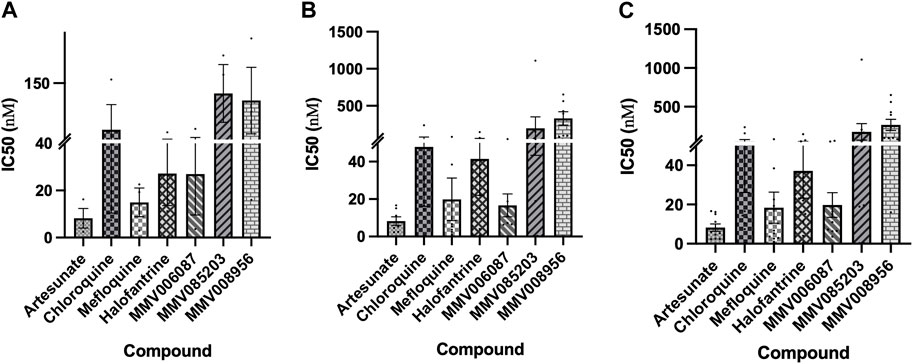
FIGURE 4. Comparing the potency of the Malaria Box compounds to standard antimalarials. (A) The potency of the standard antimalarial compounds against laboratory strains of P. falciparum; (B) The potency of the standard antimalarial compounds against the clinical isolates of P. falciparum; (C) The potency of the standard antimalarials and Malaria Box compounds against three laboratory strains and seven clinical isolates of P. falciparum.
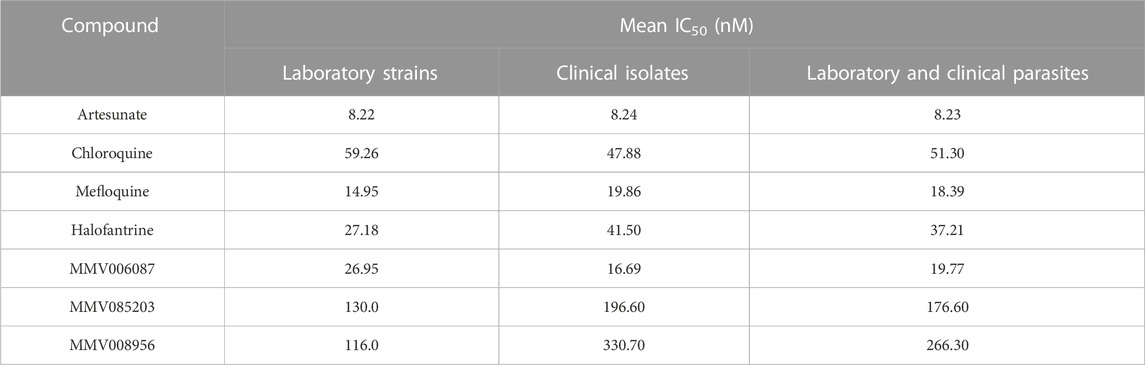
TABLE 3. The mean IC50 values of the potency of the Malaria Box compounds against the laboratory and clinical isolates compared to standard antimalarials.
For the clinical isolates, artesunate was more potent than MMV006087, mefloquine, halofantrine, chloroquine, MMV085203, and MMV008956 (Figure 5B). Thus, comparing the IC50 values of the seven antimalarials against the clinical isolates, MMV006087 (average IC50 of 16.69 nM) was also less potent than artesunate (average IC50 of 8.24 nM), but twice more efficacious than chloroquine (average IC50 of 47.88 nM) (Table 3).
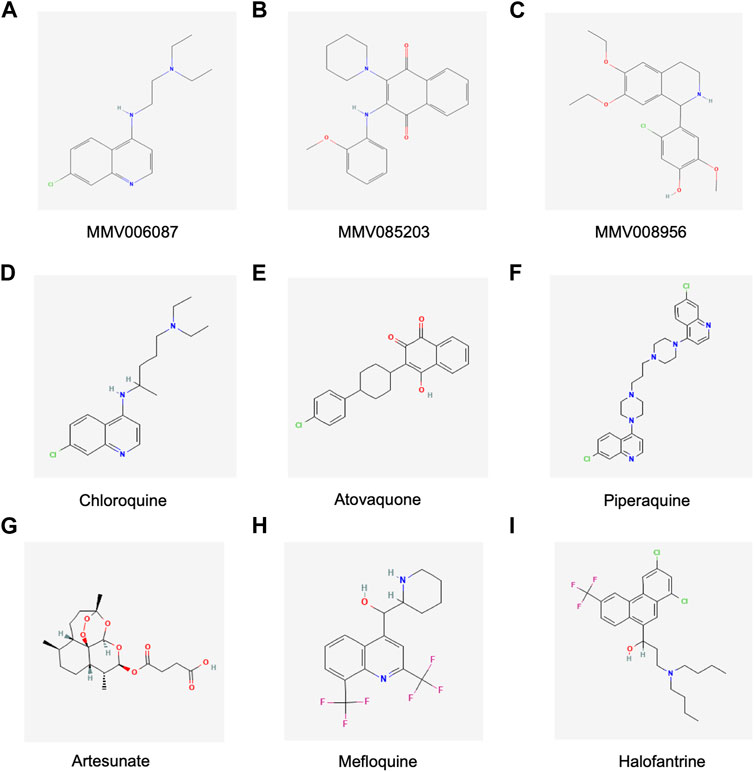
FIGURE 5. Chemical structures of the Malaria Box compounds and standard antimalarial compounds. (A) MMV006087, (B) MMV085203, (C) MMV008956, (D) Chloroquine, (E) Atovaquone (F) Piperaquine, (G)Artesunate, (H) Mefloquine and (I) Halofantrine. Figures courtesy of https://pubchem.ncbi.nlm.nih.gov MMV006087: https://pubchem.ncbi.nlm.nih.gov/compound/408190#section=2D-Structure, MMV085203: https://pubchem.ncbi.nlm.nih.gov/compound/1388836#section=2D-Structure, MMV008956: https://pubchem.ncbi.nlm.nih.gov/compound/3338861#section=2D-Structure, Chloroquine: https://pubchem.ncbi.nlm.nih.gov/compound/2719#section=2D-Structure, Atovaquone: https://pubchem.ncbi.nlm.nih.gov/compound/74989#section=2D-Structure, Artesunate: https://pubchem.ncbi.nlm.nih.gov/compound/Artesunate#section=2D-Structure, Mefloquine: https://pubchem.ncbi.nlm.nih.gov/compound/4046#section=2D-Structure, Halofantrine: https://pubchem.ncbi.nlm.nih.gov/compound/37393#section=2D-Structure.
Furthermore, the potency of the seven antimalarials in the laboratory strains and clinical isolates was determined by comparing the mean IC50 values of these compounds between the laboratory strains and clinical isolates (Figures 4A, B; Table 3). From the mean IC50 values, five of the antimalarials were more potent against the laboratory strains compared to the clinical isolates -artesunate (average IC50 of 8.22 nM and 8.24 nM, respectively), mefloquine (average IC50 of 14.95 nM and 19.86 nM, respectively), halofantrine (average IC50 of 27.18 nM and 41.50 nM, respectively), MMV008956 (average IC50 of 116.00 nM and 330.70 nM, respectively), and MMV08503 (average IC50 of 130.00 nM and 196.60 nM, respectively) (Table 3). On the contrary, two compounds were more potent against the clinical isolates than the laboratory strains -chloroquine (average IC50 of 59.26 nM and 47.88 nM, respectively) and MMV006087 (average IC50 of 26.95 nM and 16.69 nM, respectively) (Table 3). This may suggest that chloroquine and MMV006087 could have a similar mechanism of action or target.
The potency of each of the antimalarial compounds against the clinical isolates and laboratory strains was also determined by comparing the mean IC50 values in both the clinical isolates and laboratory strains (Figure 4C; Table 3). The results indicated that artesunate (average IC50 of 8.23 nM) was the most potent compound, followed by mefloquine (average IC50 of 18.39 nM), MMV006087 (average IC50 of 19.77 nM), halofantrine (average IC50 of 37.21 nM), chloroquine (average IC50 of 51.30 nM), MMV085203 (average IC50 of 176.60 nM), and MMV008956 (average IC50 of 266.30 nM) (Figure 4C; Table 3). We also found that MMV006087 was less potent against the clinical isolates and laboratory strains than artesunate. However, MMV006087 was more potent against the laboratory strains and clinical isolates than chloroquine, mefloquine, halofantrine, MMV085203, and MMV008956. Therefore, the results from this study not only validate MMV006087 as a promising antimalarial but also suggest that this compound might possess an interesting mechanism of action that is different from chloroquine and artesunate. Thus, MMV006087 and MMV085203 are two of the most promising compounds in this screen that are worthy of further exploration.
4 Discussion
Anopheles stephensi is a mosquito species that thrives well in urban settings and is capable of transmitting both P. falciparum and Plasmodium vivax malaria parasites (WHO, 2022). The recent identification of An. stephensi in many African countries coupled with the detection of parasites carrying P. falciparum Kelch13 (Pfkelch13) mutations calls for intense surveillance (Sinka et al., 2020; Ahmed et al., 2021; Ansah et al., 2021; WHO, 2022). Thus, efforts towards the global malaria eradication agenda must include consideration for both P. falciparum and non-falciparum species (Ansah et al., 2021; Dembele et al., 2021). The World Health Organization (WHO) indicates that the existence of unplanned urbanization may increase the focal transmission of malaria to the urban poor (WHO, 2022). Thus, it is important to accelerate the development of new antimalarial drugs. Currently, several new formulations such as triple ACT, ACT plus single low-dose primaquine for transmission-blocking and artemether-lumefantrine for neonates are being developed for malaria treatment (Chen and Hsiang, 2022). Additionally, several next-generation drugs such as ganaplacide-lumefantrine, M5717-pyronaridine, ZY19489-ferroquine and cipargamin are at different phases of clinical development (Abd-Rahman et al., 2022; WHO, 2022).
The existence of the Malaria Box compound library provides an avenue for researchers to help prioritize compounds to increase the arsenal of antimalarials in the drug development pipeline (Spangenberg et al., 2013).
This study validated the potency of three compounds-MMV006087, MMV085203, MMV008956- with low nanomolar potency, using five laboratory strains and twenty clinical isolates of P. falciparum. The data presented here is one of the few reports to have confirmed the potency of these compounds using this number of P. falciparum parasites in a single study. MMV006087 was the most potent compound among the three Malaria Box compounds with IC50 values less than 100 nM in the laboratory strains and clinical isolates. Hence, the data in this report corroborates with data from previous reports (Van Voorhis et al., 2016; Chirawurah et al., 2017). MMV006087 (Nʹ-(7-chloroquine in-4-yl)-N, N-diethyl ethane-1,2-diamine) has been shown to have early ring-stage activity in P. falciparum parasites with EC50 values below 200 nM, less toxic with a selectivity index greater than 10, and gametocidal activity (Van Voorhis et al., 2016). Therefore, the low toxicity of this compound, coupled with its blood stage and gametocidal activity, makes MMV006087 a promising candidate for further exploration as an antimalarial.
Our study observed that MMV006087 (a quinoline compound) was twice more potent than chloroquine (a 4-aminoquinoline compound), even though MMV006087 has been reported to have a similar mechanism of action as chloroquine (Creek et al., 2016). Unlike MMV006087 which has been shown to have gametocidal activity (Van Voorhis et al., 2016), there is no evidence that chloroquine has gametocidal activity against P. falciparum parasites (Van Voorhis et al., 2016). MMV006087 was also found to inhibit the egress of P. falciparum at 1 μM and affect the integrity of the P. falciparum digestive vacuole (Subramanian et al., 2018). Another study also showed that MMV006087 selectively inhibited specific aspects of protein degradation, resulting in increased dimethylarginine (DMA) levels and protein turnover (Creek et al., 2016). Considering that this process usually occurs on intact proteins, the authors further concluded that haemoglobin metabolism may not be the primary target of MMV006087 as was the case for chloroquine (Creek et al., 2016). These observations together underscore the need for further studies to determine or validate the molecular targets and the mechanism of action of MMV006087.
Furthermore, MMV006087 has been shown to have a drug-drug chemogenomic profile similar to artesunate (Van Voorhis et al., 2016). Therefore, we expected MMV006087 and artesunate to have similar potencies against the parasites. However, our data demonstrate that artesunate was more potent (more than 2-fold) compared to MMV006087. Thus, despite the similarity in drug-drug chemogenomic profile and blood-stage activity, MMV006087 and artesunate may have different molecular targets and mechanisms of action. Therefore, considering that MMV006087 behaved differently from chloroquine and artesunate, suggests it may be a promising candidate molecule with a novel mechanism of action that can be further explored for the treatment of malaria.
Another interesting observation in this study was the variability in the response of the clinical isolates and laboratory strains to the antimalarials. We found that the clinical isolates were more susceptible to MMV085203 compared to MMV006087 and MMV008956. But the contrary was true for the laboratory strains. Furthermore, we also observed that MMV085203 was more potent against parasites with more than two genotypes compared to MMV006087 and MMV008956. The chemical structures of MMV006087 (a quinoline), and MMV008956(5-chloro-4-(6,7-diethoxy-1,2,3,4-tetrahydroisoquinolin-1-yl)-2-methoxyphenol), which is an isoquinoline, are similar to chloroquine (4-N-(7-chloroquinolin-4-yl)-1-N,1-N-diethylpentane-1,4-diamine) (Figure 5) (Creek et al., 2016). Albeit the reduction in chloroquine-resistant strains in clinical settings, there is still an appreciable number of chloroquine-resistant parasites (Ofori et al., 2021). Therefore, the structural similarities between the three compounds could have accounted for the reduced susceptibility of the clinical isolates to MMV006087 and MMV008956. MMV085203 (2-(2-Methoxyanilino)-3-piperidin-1-ylnaphthalene-1,4-dione) is a naphthoquinone derivative compound (Figure 5) similar to atovaquone and is potent against both asexual and sexual stage P. falciparum parasites (Van Voorhis et al., 2016). Compounds with 1,4-naphthoquinone moiety act by interfering with NADPH-dependent disulfide reductases (such as thioredoxin reductase and glutathione reductase) resulting in the generation of reactive oxygen species that disrupt haemoglobin digestion (Müller et al., 2011; Morin et al., 2008). Other studies have also shown that MMV085203 affects the activity of the P. falciparum thioredoxin reductase (PfTrxR), which is an enzyme that is essential for maintaining redox homeostasis (Müller et al., 2001; Tiwari et al., 2016). Furthermore, MMV085203 was also found to selectively upregulates aconitrates and fumarate-intermediates of the tricarboxylic acid (TCA) cycle in the mitochondrion, suggesting the target of this compound is in this organelle (Rocamora et al., 2021). The study identified the P. falciparum mitochondrial transporter (PfMFR3) to be essential for the activity of MMV085203 (Rocamora et al., 2021). However, these proteins were not found to be the primary targets for MMV085203(Rocamora et al., 2021). Hence, the potency of this compound against clonally diverse clinical isolates, coupled with its sexual and asexual-stage activity, qualifies MMV085203 as another promising candidate that is worthy of further exploration.
In conclusion, this study is one of the first to validate the potency of MMV006087, MMV085203, and MMV008956 using this many P. falciparum parasites in a single study. This study also found MMV006087 to be more potent than MMV085203 and MMV008956, which corroborated with data from our previous report. Our data suggest that MMV006087 may have molecular targets and mechanisms of action that are different from artesunate and chloroquine. Lastly, MMV085203 was more potent against clonally diverse clinical parasites, which is an essential feature for novel antimalarials against multidrug-resistant malaria parasites. Taken together, MMV006087 and MMV085203 are two promising antimalarials that need to be prioritized for further exploration in the drug development pipeline. Towards addressing antimalarial drug resistance in Africa, the WHO highlighted the need for new treatments that delay the emergence and spread of resistance. Based on our findings, further exploration of MMV006087 and MMV085203 as new antimalarials may help address emerging threats and the spread of antimalarial drug resistance in malaria-endemic regions.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
Conceptualization, JC,GA, and YA; methodology, JC, YA, CMʼa, and FA; investigation, JC, BA, EL-A, FA, and DD; resources, YA and GA; writing—original draft preparation, JC and FA; writing—review and editing, JC, HD, CMʼa, YA, GA, LA-E, BA, DD, and EL-A; supervision, YA, LA-E, and GA; project administration, YA, LA-E, and GA; funding acquisition, GA. All authors contributed to the article and approved the submitted version.
Funding
JC was supported by a DELTAS Africa grant (DEL-15-007: GA). The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)’s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust (107755/Z/15/Z: GA) and the United Kingdom government.
Acknowledgments
We thank the Medicines for Malaria Venture for giving us the Malaria Box for this work and the study participants who provided the samples for the Erythrocyte Invasion Mechanism (EIM) project.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fddsv.2023.1190471/full#supplementary-material
References
Abd-Rahman, A. N., Zaloumis, S., McCarthy, J. S., Simpson, J. A., and Commons, R. J. (2022). Scoping review of antimalarial drug candidates in phase I and II drug development. Antimicrob. Agents Chemother. 66, e0165921. doi:10.1128/AAC.01659-21
Ahmed, A., Khogali, R., Elnour, M.-A. B., Nakao, R., and Salim, B. (2021). Emergence of the invasive malaria vector Anopheles stephensi in Khartoum State, Central Sudan. Parasit. Vectors 14, 511–515. doi:10.1186/s13071-021-05026-4
Ansah, F., Suurbaar, J., Darko, D., Anabire, N. G., Blankson, S. O., Domson, B. K. S., et al. (2021). Development of cooperative primer-based Real-time PCR assays for the detection of plasmodium malariae and plasmodium ovale. J. Mol. Diagn 23, 1393–1403. doi:10.1016/j.jmoldx.2021.07.022
Arrowsmith, J. (2011). Trial watch: Phase II failures: 2008-2010. Nat. Rev. Drug Discov. 10, 328–329. doi:10.1038/nrd3439
Bushman, M., Morton, L., Duah, N., Quashie, N., Abuaku, B., Koram, K. A., et al. (2016). Within-host competition and drug resistance in the human malaria parasite Plasmodium falciparum. Proc. Biol. Sci. 283, 20153038. doi:10.1098/rspb.2015.3038
Chen, I., and Hsiang, M. S. (2022). Triple artemisinin-based combination therapies for malaria: A timely solution to counter antimalarial drug resistance. Lancet Infect. Dis. 22, 751–753. doi:10.1016/S1473-3099(21)00748-9
Chirawurah, J. D., Ansah, F., Nyarko, P. B., Duodu, S., Aniweh, Y., and Awandare, G. A. (2017). Antimalarial activity of malaria box compounds against plasmodium falciparum clinical isolates. Int. J. Parasitol. Drugs Drug Resist 7, 399–406. doi:10.1016/j.ijpddr.2017.10.005
Creek, D. J., Chua, H. H., Cobbold, S. A., Nijagal, B., MacRae, J. I., Dickerman, B. K., et al. (2016). Metabolomics-based screening of the malaria box reveals both novel and established mechanisms of action. Antimicrob. Agents Chemother. 60, 6650–6663. doi:10.1128/AAC.01226-16
Dembele, L., Diallo, N., Sogore, F., Diarra, B., Ballo, F. I., Daou, A., et al. (2021). Ex vivo plasmodium malariae culture method for antimalarial drugs screen in the field. ACS Infect. Dis. 7, 3025–3033. doi:10.1021/acsinfecdis.1c00262
Diagana, T. T. (2015). Supporting malaria elimination with 21st century antimalarial agent drug discovery. Drug Discov. Today 20, 1265–1270. doi:10.1016/j.drudis.2015.06.009
Lambros, C., and Vanderberg, J. P. (1979). Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 65, 418–420. doi:10.2307/3280287
Mathieu, L. C., Singh, P., Monteiro, W. M., Magris, M., Cox, H., Lazrek, Y., et al. (2021). Kelch13 mutations in Plasmodium falciparum and risk of spreading in Amazon basin countries. J. Antimicrob. Chemother. 76, 2854–2862. doi:10.1093/jac/dkab264
Mayengue, P. I., Ndounga, M., Malonga, F. V., Bitemo, M., and Ntoumi, F. (2011). Genetic polymorphism of merozoite surface protein-1 and merozoite surface protein-2 in Plasmodium falciparum isolates from Brazzaville, Republic of Congo. Malar. J. 10, 276. doi:10.1186/1475-2875-10-276
Mensah-Brown, H. E., Abugri, J., Asante, K. P., Dwomoh, D., Dosoo, D., Atuguba, F., et al. (2017). Assessing the impact of differences in malaria transmission intensity on clinical and haematological indices in children with malaria. Malar. J. 16, 96. doi:10.1186/s12936-017-1745-8
Mnzava, A., Monroe, A. C., and Okumu, F. (2022). Anopheles stephensi in Africa requires a more integrated response. Malar. J. 21, 156–6. doi:10.1186/s12936-022-04197-4
Moll, K., Ljungström, I., Perlmann, H., Scherf, A., and Wahlgren, M. (2008). Methods in malaria research. VA: Manassas.
Morin, C., Besset, T., Moutet, J. C., Fayolle, M., Bruckner, M., Limosin, D., et al. (2008). The aza-analogues of 1,4-naphthoquinones are potent substrates and inhibitors of plasmodial thioredoxin and glutathione reductases and of human erythrocyte glutathione reductase. Org. Biomol. Chem. 6, 2731–2742. doi:10.1039/b802649c
Müller, S., Gilberger, T., Krnajski, Z., Lüersen, K., Meierjohann, S., and Walter, R. (2001). Thioredoxin and glutathione system of malaria parasitePlasmodium falciparum. Protoplasma 217, 43–49. doi:10.1007/BF01289412
Müller, T., Johann, L., Jannack, B., Brückner, M., Lanfranchi, D. A., Bauer, H., et al. (2011). Glutathione reductase-catalyzed cascade of redox reactions to bioactivate potent antimalarial 1, 4-naphthoquinones–a new strategy to combat malarial parasites. J. Am. Chem. Soc. 133, 11557–11571. doi:10.1021/ja201729z
Ndwiga, L., Kimenyi, K. M., Wamae, K., Osoti, V., Akinyi, M., Omedo, I., et al. (2021). A review of the frequencies of Plasmodium falciparum Kelch 13 artemisinin resistance mutations in Africa. Int. J. Parasitol. Drugs Drug Resist 16, 155–161. doi:10.1016/j.ijpddr.2021.06.001
Ofori, M. F., Kploanyi, E. E., Mensah, B. A., Dickson, E. K., Kyei-Baafour, E., Gyabaa, S., et al. (2021). Ex vivo sensitivity profile of plasmodium falciparum clinical isolates to a panel of antimalarial drugs in Ghana 13 Years after national policy change. Infect. Drug Resist 14, 267–276. doi:10.2147/IDR.S295277
Partey, F. D., Frimpong, A., and Ofori, M. F. (2022). Collection and cryopreservation of plasmodium falciparum clinical isolates in the field. Methods Mol. Biol. 2470, 11. doi:10.1007/978-1-0716-2189-9_2
Plouffe, D. M., Wree, M., Du, A. Y., Meister, S., Li, F., Patra, K., et al. (2016). High-throughput assay and discovery of small molecules that interrupt malaria transmission. Cell host microbe 19, 114–126. doi:10.1016/j.chom.2015.12.001
Rocamora, F., Gupta, P., Istvan, E. S., Luth, M. R., Carpenter, E. F., Kumpornsin, K., et al. (2021). PfMFR3: A multidrug-resistant modulator in plasmodium falciparum. ACS Infect. Dis. 7, 811–825. doi:10.1021/acsinfecdis.0c00676
Sinka, M., Pironon, S., Massey, N., Longbottom, J., Hemingway, J., Moyes, C., et al. (2020). A new malaria vector in Africa: Predicting the expansion range of Anopheles stephensi and identifying the urban populations at risk. Proc. Natl. Acad. Sci. 117, 24900–24908. doi:10.1073/pnas.2003976117
Smilkstein, M., Sriwilaijaroen, N., Kelly, J. X., Wilairat, P., and Riscoe, M. (2004). Simple and inexpensive fluorescence-based technique for high-throughput antimalarial drug screening. Antimicrob. Agents Chemother. 48, 1803–1806. doi:10.1128/aac.48.5.1803-1806.2004
Snounou, G., Zhu, X., Siripoon, N., Jarra, W., Thaithong, S., Brown, K. N., et al. (1999). Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Trans. R. Soc. Trop. Med. Hyg. 93, 369–374. doi:10.1016/s0035-9203(99)90120-7
Spangenberg, T., Burrows, J. N., Kowalczyk, P., McDonald, S., Wells, T. N., and Willis, P. (2013). The open access malaria box: A drug discovery catalyst for neglected diseases. PLoS One 8, e62906. doi:10.1371/journal.pone.0062906
Stokes, B. H., Dhingra, S. K., Rubiano, K., Mok, S., Straimer, J., Gnadig, N. F., et al. (2021). Plasmodium falciparum K13 mutations in Africa and Asia impact artemisinin resistance and parasite fitness. Elife 10, e66277. doi:10.7554/eLife.66277
Subramanian, G., Belekar, M. A., Shukla, A., Tong, J. X., Sinha, A., Chu, T. T. T., et al. (2018). Targeted phenotypic screening in plasmodium falciparum and toxoplasma gondii reveals novel modes of action of Medicines for malaria venture malaria box molecules. mSphere 3, e00534. doi:10.1128/mSphere.00534-17
Tiwari, N. K., Reynolds, P. J., and Calderón, A. I. (2016). Preliminary LC-MS based screening for inhibitors of Plasmodium falciparum thioredoxin reductase (PfTrxR) among a set of antimalarials from the Malaria Box. Molecules 21, 424. doi:10.3390/molecules21040424
Trager, W., and Jensen, J. B. (1976). Human malaria parasites in continuous culture. Science 193, 673–675. doi:10.1126/science.781840
Van Voorhis, W. C., Adams, J. H., Adelfio, R., Ahyong, V., Akabas, M. H., Alano, P., et al. (2016). Open source drug discovery with the malaria box compound collection for neglected diseases and beyond. PLoS Pathog. 12, e1005763. doi:10.1371/journal.ppat.1005763
WHO (2020). World malaria report 2020: 20 years of global progress and challenges. Switzerland: World Health Organization.
Keywords: antimalarials, malaria, clinical isolates, malaria box compounds, artemisinin, medicines for malaria venture, drug-resistant, Plasmodium falciparum
Citation: Chirawurah JD, Adikah B, Ansah F, Laryea-Akrong E, Danwonno H, Morang’a CM, Dosoo D, Amenga-Etego L, Awandare GA and Aniweh Y (2023) MMV006087 is a potent Malaria Box compound against Plasmodium falciparum clinical parasites. Front. Drug Discov. 3:1190471. doi: 10.3389/fddsv.2023.1190471
Received: 20 March 2023; Accepted: 01 June 2023;
Published: 15 June 2023.
Edited by:
Mark T. Hamann, Medical University of South Carolina, United StatesReviewed by:
Rodolpho C. Braga, InsilicAll, BrazilSmith B. Babiaka, University of Buea, Cameroon
Stefanie Costa Pinto Lopes, Instituto Leônidas & Maria Deane (ILMD/Fiocruz Amazônia), Brazil
Copyright © 2023 Chirawurah, Adikah, Ansah, Laryea-Akrong, Danwonno, Morang’a, Dosoo, Amenga-Etego, Awandare and Aniweh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gordon A. Awandare, Z2F3YW5kYXJlQHVnLmVkdS5naA==; Yaw Aniweh, eWFuaXdlaEB1Zy5lZHUuZ2g=
 Jersley D. Chirawurah
Jersley D. Chirawurah Bridget Adikah1,2
Bridget Adikah1,2 Felix Ansah
Felix Ansah Collins M. Morang’a
Collins M. Morang’a Lucas Amenga-Etego
Lucas Amenga-Etego Gordon A. Awandare
Gordon A. Awandare Yaw Aniweh
Yaw Aniweh