
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Drug Deliv. , 20 July 2023
Sec. Technological and Methodological Advances in Drug Delivery
Volume 3 - 2023 | https://doi.org/10.3389/fddev.2023.1168287
Osteoarthritis (OA) is a chronic inflammatory disease that causes synovial hyperplasia, cartilage destruction, and the formation of bone spurs. Macrophages play an indispensable role in the pathogenesis of OA by producing proinflammatory cytokines. To achieve the effect of arthritis, hormones can effectively inhibit the progression of inflammation by inhibiting the secretion of inflammatory cytokines by macrophages in traditional therapy. However, the drug is quickly cleared from the joint space, and the high injection site infection rate and low local drug concentration make the clinical efficacy of corticosteroids greatly reduced. We described the design and preparation of Polyethylene Glycol-grafted Poly Alpha-lipoic Acid-dexamethasone Nanoparticles (NPDXM/PPLA), elucidated the mechanism of action of NPDXM/PPLA in the treatment of OA in mice, and provided an experimental basis for investigating the treatment of OA with polymer nanoparticles loaded with dexamethasone. Flow cytometry and confocal laser scanning microscopy were used to confirm that NPDXM/PPLA was well absorbed and released by macrophages, and it was discovered that NPDXM/PPLA could efficiently reduce the proliferation of activated macrophages (RAW 264.7 cells). Enzyme-linked immunosorbent assay revealed that NPDXM/PPLA could efficiently reduce the expression of proinflammatory cytokines IL-1β, IL-6, and TNF-α. The knee bone structure of OA mice was investigated by MicroCT, and it was discovered that intraarticular injection of NPDXM/PPLA effectively alleviated the bone damage of the articular cartilage. Therefore, NPDXM/PPLA is a potential therapeutic nanomedicine for the treatment of OA.
Osteoarthritis is commonly an age-related disease with a degenerative illness of the joints. The most common symptoms of osteoarthritis are articular cartilage wear and synovitis, which causes pain and swelling in the joint, as well as limited activity (French et al., 2013; Fernanda et al., 2018). OA affects not only articular cartilage but also the entire joint, including the subchondral bone, ligament, synovium, meniscus, and even the muscles around the joint (Martel-Pelletier et al., 2012). Although the pathogenesis of OA is not fully understood, the disease is characterized by the gradual degeneration of articular cartilage. Recent studies found that this progressive degeneration is related to oxidative stress, and reactive oxygen species play a significant role in this procedure (Lepetsos and Papavassiliou, 2016). At the same time, the inflammatory microenvironment plays a significant role in the occurrence and development of OA. The severe inflammatory response of macrophages results in the recruitment of a large number of inflammatory cells and the secretion of high levels of proinflammatory cytokines such as IL-1β, IL-6, TNF-A, and matrix metalloproteinases (MMPs) at the lesion site. Reduces proteoglycan synthesis and type II collagen, and aggravates cartilage erosion and degeneration (Bondeson et al., 2006; Blom et al., 2007; Crielaard et al., 2012; Sonderegger et al., 2012; Agarwal et al., 2016; Sang et al., 2016).
In clinical practice, intraarticular injection of corticosteroids or hyaluronic acid is often employed to relieve pain and control inflammation (Jones et al., 2018; Conaghan et al., 2019). Although corticosteroids can alleviate pain and other symptoms, their related side effects and the rate of joint cavity clearance severely limit their clinical application (Evans et al., 2014; Brown et al., 2019). Corticosteroids injected into the joint cavity are cleared with a half-life of 1–4 h (Brown et al., 2019). Multiple intraarticular injections are needed to achieve therapeutic effects. However, repeated intraarticular injections can cause joint infection, and long-term corticosteroids use can destroy articular cartilage and hasten joint degeneration (McAlindon et al., 2018). With the continuous development of medical chemistry, researchers have used nanomaterials as drug delivery carriers. Nanomaterials have consistently demonstrated improved drug retention properties in the joint cavity and drug delivery to the joint when compared to free drug injection. Furthermore, active and passive targeting strategies can be used to modify nanomaterials to promote interaction and localization with specific articular tissues such as cartilage and synovium (Brown et al., 2019). In addition, α-lipoic acid (αLA) is a natural antioxidant synthesized in the human body and an important cofactor of mitochondrial metabolism, which has been employed in the treatment of Alzheimer’s illness and diabetes (Maczurek et al., 2008; Singh and Jialal, 2008; Solmonson and Deberardinis, 2017). It has been discovered that by heating above its melting point, αLA can be polymerized to poly α-lipoic acid (PαLA) without the use of a catalyst or solvent. PαLA is a drug carrier with great potential for development. PαLA and its degradation products are safe and biocompatible. Disulfide bonds in its main chain play an antioxidant role in osteoarthritis (Packer et al., 1995; Shimoda et al., 2007; Shay et al., 2009; Li et al., 2013; Yang et al., 2018). Given the use of nanomaterials as corticosteroids carriers and PLA’s excellent anti-inflammatory properties, we speculated that NPDXM/PPLA prepared by the electrostatic and hydrophobic action of the carboxyl group on mPEG-g-PαLA carrying DXM may be a promising new drug for the treatment of OA.
To test this hypothesis, we created and tested an NPDXM/PPLA for the effective treatment of OA (Figure 1.). NPDXM/PPLA was prepared from αLA by the polymerization reaction. The hydrophilic polymer mPEG was electrostatically linked to the NPDXM/PPLA by the hydrophobic DXM of the carboxyl group on mPEG-g-PαLA. The anti-inflammatory effects of NPDXM/PPLA nanoparticles on OA mice induced by monosodium iodoacetate (MIA) were investigated further. The outcome revealed that NPDXM/PPLA could effectively carry DXM and had a better effect on OA treatment than traditional DXM injection alone.
Dexamethasone (DXM) (average molecular weight Mn = 206.362 g mol−1), Poly (ethylene glycol) monomethyl ether (mPEG), α-lipoic acid (average molecular weight Mn = 2000 g mol−1), 4′, 6-diamidine-2-phenylindole (DAPI), 4-dimethylamino pyridine (DMAP), 3-(4, 5-dimethyl-thiazole-2-yl)-2, 5-diphenyltetrazolium bromide (MTT), Sigma–Aldrich (Sigma, Germany) provided the 1-(3-(dimethylamino) propyl) -3-ethylcarbondiimine hydrochloride (EDCHCl), and sodium iodoacetate (MIA). Thermo Fisher Scientific (China) Co., Ltd. provided cell culture substrates such as Dulbecco’s modified Eagle’s medium (DMEM) and fetal bovine serum (FBS) (Shanghai, China). Penicillin and streptomycin were purchased from Shijiazhuang Huabei Pharmaceutical Co., Ltd. Biosharp Co., LTD. provided the lipopolysaccharide (LPS). Sinopharm Chemical Reagent Co., LTD. supplied N, N-dimethylformamide (DMF), and Sinopharm Chemical Reagent Co., LTD., supplied tetrahydrofuran (THF). All other chemicals were purchased commercially.
First, PαLA was prepared by a reaction between αLA with diethyl ether. Then, 1.68 g PαLA and 4.0 g mPEG were dissolved in 50 mL Tetrahydrofuran (THF) and 50 mL dimethyl sulfoxide (DMSO), respectively. Then mixed the two solutions slowly with a pipette gun, then 333.4 mg EDCHCl and 56.09 mg DMAP were added. After 24 h of stirring at room temperature, adding the same catalyst to the reaction solution, and continuing the reaction for another 24 h, the mixed solution was dialyzed in Milli-Q water [molecular weight cut-off (MWCO) = 3,500 Da], the dialysis time was 5 days, and the deionized water was changed 3 times per day. Also, the mPEG-g-PαLA was obtained by lyophilization (Scientz-12ND lyophilizer, Ningbo Scientz Equipment limited by share Ltd., China). Bruker AV 300 NMR spectrometer (AVANCE III, Bruker corporation, Switzerland) was used to measure the proton nuclear magnetic resonance (1H NMR) spectra of PαLA and mPEG-g-PαLA.
First, 100 mg mPEG-g-PαLA was dissolved in 8 mL DMSO, then dropped into 100 mL deionized water and stirred for 2 h to prepare mPEG-g-PαLA. The solution was then transferred to an MWCO 3500 dialysis bag for 10 h to remove DMSO before being lyophilized to obtain NPPPLA. Again, 100 mg mPEG-g-PαLA and 20 mg DXM were dissolved in 8 mL DMSO, dropped into 100 mL deionized water, stirred for 2 h, then transferred to the MWCO 3500 dialysis bag for 10 h to remove DMSO, and NPDXM/PPLA NPs were lyophilized. The Dio-labeled NPPPLA were prepared following the same method and were denoted as NPDio/PPLA.
Transmission electron microscopy (TEM) and dynamic laser scattering were used to determine the hydrodynamic radius (Rh) of NPDXM/PPLA NPs (DLS). TEM test: First, weigh 2 mg NPPPLA and NPDXM/PPLA respectively, and configure them into 0.1 mg/mL solution. The solution should then be pipetted onto a clean copper net with a 20 μL pipette and allowed to dry at room temperature for 24 h. TEM was also performed using a JEOL JEM-1011 transmission electron microscope (JEOL, Ltd., Tokyo, Japan) with a 100 kV accelerating voltage. DLS test: First, weigh 2 mg NPPPLA and NPDXM/PPLA, respectively, and dissolved them in PBS solution with pH 7.4 for later use. The Wyatt QELS instrument was then set up to measure the fluid dynamic radius (Rh) of the NPPPLA and NPDXM/PPLA.
In vitro DXM release of NPDXM/PPLA was evaluated in PBS containing 200 U mL−1 esterase at pH = 7.4. In short, NPDXM/PPLA is released in PBS buffers with or without H2O2. To begin, 3.0 mg NPDXM/PPLA was weighed and dissolved in 15 mL pH= 7.4 PBS buffer before being transferred to an MWCO 3500 dialysis bag for standby. The solution should then be added to a different medium every 200 mL. Medium (I) was 100 mL PBS buffer with pH = 7.4; Medium (II) was 100 mL PBS buffer containing 10 mol H2O2 with pH = 7.4; Medium (III) was 100 mL PBS buffer containing 1 mol H2O2 with pH = 7.4. The dialysis bags were then placed in beakers containing various release media and placed in a 37°C constant temperature oscillating box that was continuously shaking at 100 RPM to simulate the human body environment. The release solution in the 2.0 mL beaker was removed with a pipetting gun at a predetermined time point, and 2.0 mL of the media in the beaker was added. The content of DXM was determined by high-performance liquid chromatography (HPLC) (Flexar, PerkinElmer, Shelton, United States of America). The mobile phase was methanol-water (60:40, V/V) at a flow rate of 1.0 mL min−1 in an analytical C18 column (5 m, 250 4.6 mm, PerkinElmer Brownlee, United States). The detection wavelength was 240 nm, and the column temperature was 25°C. The injection volume was 20 μL. The limits of detection and quantitation were 26.2 and 87.2 ng mL−1, respectively. The calibration curve was linear in the range of 110–7,780 ng mL−1 (r2 = 0.999). The release curve was drawn using the standard curve method, which is used to calculate the DXM concentration released at each time point.
NPDXM/PPLA stability test: 0.5 mg NPDXM/PPLA was dissolved in 5 mL PBS with pH = 7.4 to prepare 0.1 mg/mL solution. The solution was then transferred to the DLS sample bottle, which was oscillated in a 37°C constant temperature oscillating chamber at a constant temperature of 100 rpm to simulate the internal environment of the human body. The change in particle size was detected and plotted by the Wyatt QELS device at specified time points.
Entosis and the release of NPDio/PPLA from RAW 264.7 cells were detected using confocal laser scanning microscopy (CLSM) and flow cytometry. In brief, RAW 264.7 cells were seeded into a six-well plate at a density of 5 × 104 cells per well, add 2 mL DMEM medium containing 10% (V/V) FBS and 1% (W/V) penicillin-streptomycin, and cultured for 12 h in a 5% carbon dioxide (CO2) incubator at 37 °C in a humid environment. Then, RAW 264.7 cells were activated with 1 μg ML−1 LPS for 24 h. Following activation of the macrophages, the medium was replaced with DMEM medium containing NPDio/PPLA at a Dio concentration of 1 μg ML−1. Then, the medium was incubated at 37 °C for 0.5 h, 2 h, 4 h, 6 h, 8 h, and 10 h. Following that, the medium was removed, the cells were washed three times with PBS, fixed with 4% paraformaldehyde, and the nuclei stained with 4’,6-diamidine-2-phenylindole (DAPI) Aladdin. Finally, using an LSM 543 CLSM microscope (Carl Zeiss, Jena, Germany) with a 20-eyepiece objective lens, microscopic images of cell uptake were obtained. For FCM analysis, activated RAW 264.7 cells were co-cultured with NPDio/PPLA. Following that, the medium was removed, and the cells were washed three times with PBS, digested with trypsin, and centrifuged for 5 minutes to collect the cells. Finally, they were suspended in 500 μL PBS, and the fluorescence intensity of cell uptake was measured using a Guava EasyCyte 12 flow cytometer (Millipore, Billerica, MA, United States).
The toxicity of NPPPLA, NPDXM/PPLA, and free DXM to RAW 264.7 cells was determined using MTT. In brief, RAW 264.7 cells were sown into 96-well plates at a density of 7,000 cells per well. Following a 24-h culture with or without LPS (1 μg mL−1), the medium was replaced with 200 μL of DMEM medium containing free DXM, NPPPLA, and NPDXM/ PPLA. The concentration in the medium containing DXM was from 0.048 to 50 μg mL−1. After 24 h, add 20 μL of 5% MTT solution and incubate for another 4 h. Following that, 150.0 μL DMSO was added to the medium, and the absorbance of each well was measured at 490 nm using a multifunctional microplate reader (Spark, TECAN, Switzerland).
The expression of proinflammatory cytokines were determined by enzyme-linked immunosorbent assay (ELISA) in activated RAW 264.7 cells. In brief, the cells were seeded at a density of 4 105 cells/well in 6-well plates and cultured for 24 h. RAW 264. For 24 h, 7 cells were activated with 1 μg mL−1 LPS. Following that, 2 mL of DMEM containing saline, NPPPLA, NPDXM/PPLA, or free DXM at a final TA concentration of 1 μg mL−1 was added to the cell culture medium. After 24 h, IL-1β, TNF-α, and IL-6 levels in the supernatants were determined using ELISA kits (Spark® multimode microplate reader, TECAN, Switzerland).
50 BALB/c mice (six to eight weeks old) were purchased from the Chinese Academy of Medical Sciences Institute of Experimental Animals. All animal procedures were following the “Jilin University Laboratory Animal Care” and Use Guidelines verified by the Animal Ethics Committee of the First Hospital of Jilin University. First, 40 mice were given general anesthesia with pentobarbital sodium (2%). To induce OA, MIA (5 mg kg−1, Sigma–Aldrich, st. Louis, MO, United States of America) was injected intraarticular through the subpatellar ligament of the left knee with a 30G needle (5 mg kg−1, Sigma–Aldrich, st. Louis, MO, United States of America) in mice (Chung et al., 2015; Sun et al., 2018). The successful induction of OA was confirmed by significantly reducing the load-bearing and withdrawal point stimulus thresholds of the hind paw (Bove et al., 2003; Nwosu et al., 2016). Mice without OA induction were used as a negative control.
Three days after OA induction, the mice were randomly divided into four groups and given different treatments: normal saline, DXM, NPPPLA, and NPDXM/PPLA, respectively. The OA knee was tested by intraarticular injection in each group. The preparations concentration in the DXM, NPPPLA, and NPDXM/PPLA groups remained constant at 1.0 mg kg−1. Day zero was the first day of treatment. All groups were given treatments every 4 days until the mice were put down at the end of the third week.
The knee joint was harvested and treated for further analysis after the animals were killed. First, the knee joint specimens of mice were fixed in 10% formalin buffer at 4 °C for 24 h and then decalcified with EDTA for 1 h. Next, the decalcified specimens were buried in paraffin and cut into 5 μm sections. Immunohistochemistry was used to stain sections with hematoxylin and eosin (H&E), IL-1, TNF-, and IL-6 (Affinity Biosciences, OH, United States of America). Knee specimens were also used to assess the shape of the knee and to measure skeletal characteristics with Micro CT.
All results are presented as the means ± standard deviations, and the important statistical data were analyzed using one-way ANOVA in the GraphPad Prism Software (GraphPad Software Inc., San Diego, United States of America).* indicates that p < 0.05 was considered statistically significant.; ** represents 0.01 < p < 0.05 and *** represents p < 0.001 was considered to be more highly statistically significant.
As shown in Figure 2A, PαLA was synthesized by ring-opening polymerization of the aLA monomer at 80°C. The structure of PαLA was confirmed by 1H NMR. The mPEG-g-PαLA polymer was created by esterifying the hydroxyl group of mPEG with the carboxyl group of PαLA and coupling the mPEG with the PαLA side chain. 1H NMR spectroscopy confirmed the structure of the obtained mPEG-g-PαLA. As shown in Figure 2B, the positions of the proton peaks H, I, and J of polymer mPEG-g-PαLA in the 1H NMR spectrum indicate that mPEG has successfully connected with the PαLA side group. Furthermore, software integration of H and B peaks revealed that mPEG modified 50% of the carboxyl groups on the PαLA side chain. In addition, the structure of the product was analyzed by gel permeation chromatography (GPC). The peak time of GPC in Figure 2C is 17.9 min, indicating the weight of mPEG-g-PαLA is Mw = 7.76 × 104 Da, proving that mPEG-g-PαLA has been successfully grafted. In summary, this means that mPEG successfully connects to the side chain of PαLA.
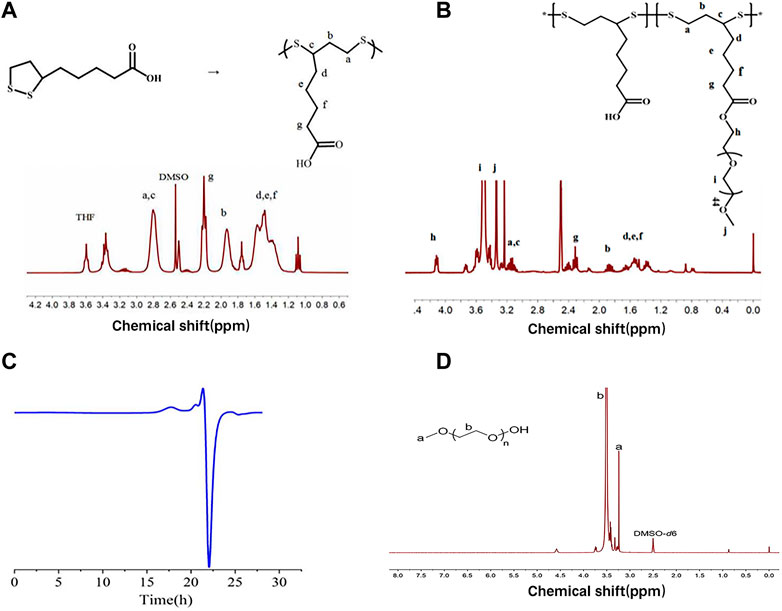
FIGURE 2. (A) Polymerization of αLA and 1H NMR spectra of PαLA (DMSO-D6) (B) 1H NMR spectra of PαLA (DMSO-D6) of mPEG-g-PαLA (DMSO). (C) GPC analysis of DMF phase of mPEG-g-PαLA.
The Rh of NPPPLA measured by DLS was (21.4 ± 1.7) nm, and TEM images showed an irregular shape of NPPPLA (Figure 3A). Meanwhile, the Rh and TEM of NPDXM/PPLA were also tested. NPDXM/PPLA had an Rh of (24.0 ± 1.2) nm, and TEM images revealed a quasi-circular structure (Figure 3B). We found a slight increase in the size of NPDXM/PPLA, which also indicates that DXM is successfully encapsulated in nanoparticles. DXM’s drug loading content and drug loading efficiency were both as expected at 10.1% and 80.7%, respectively. Due to the amphiphilic properties of polymers, drug delivery efficiency is improved. Furthermore, DLS was used to test the particle size change of NPDXM/PPLA at different times in PBS with pH = 7.4 (Figure 3C), and it was found that the particle size change of NPDXM/PPLA was not obvious within 7 days, indicating that the nanoparticles were relatively stable under normal physiological state. And can maintain the shaped structure for a long time.
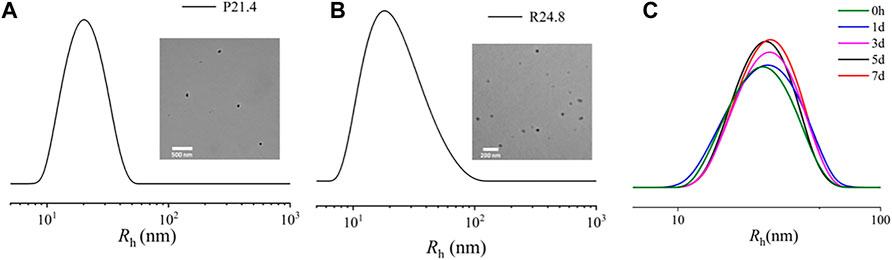
FIGURE 3. (A) Sizes of NPPPLA measured by DLS and TEM images of NPPPLA. (B) Sizes of NPDXM/PPLA measured by DLS and TEM images of NPDXM/PPLA. (C) The particle size of NPDXM/PPLA at different times tested by DLS.
During the development of OA, activated macrophages are relevant to the unregulated expression of matrix metalloproteinases (MMPs), proinflammatory cytokines, and other tissue-degrading enzymes (Bondeson et al., 2006; Bondeson et al., 2010; Sellam and Berenbaum, 2010). It has been reported that matrix metalloproteinases break ester bonds (Xia et al., 2001; Giannelli et al., 2004). As a result, detecting DXM release from NPDXM/PPLA in PBS (pH 7.4, containing 200 U ML−1 esterase) can mimic DXM release in OA (Joshi et al., 2018). In vitro drug release of NPDXM/PPLA was performed in PBS buffer with or without H2O2, and the release curve was determined by HPLC. Figure 4 depicts the results. When the pH is 7.4, we can divide the NPDXM/PPLA release into three periods: (1) rapid release stage; DXM released in PBS at pH = 7.4, PBS containing 10 mmol H2O2 at pH = 7.4 and PBS containing 1 mol H2O2 at pH = 7.4 was 33%, 53%, and 68%, respectively, in the first 10 h (2) Slow-release stage: 38%, 62%, and 73% were released within 20 h (3) Plateau stage: DXM was rarely released. The amount of DXM released increased significantly as the concentration of H2O2 in the release medium increased. The increase in DXM release should be attributed to the H2O2-mediated cleavage of NPDXM/PPLA. As a result, we concluded that NPDXM/PPLA can release DXM stably under physiological conditions, but can accelerate DXM release under the condition of higher oxidant concentration.
The endocytosis of NPDio/PPLA to activated RAW 264.7 cells was studied using FCA and CLSM to demonstrate the endocytosis and intracellular drug release ability of activated RAW 264.7 cells to NPDio/PPLA. When activated RAW 264.7 cells were treated with NPDio/PPLA, the fluorescence intensity increased with increasing incubation time as shown in Figures 5A,B. In addition, the fluorescence intensity reached a stable state after 8 h of incubation, indicating that macrophages’ internalization of NPDio/PPLA nanoparticles reached saturation after 8 h. In contrast, activated RAW 264 showed an extremely low fluorescence signal. NPPPLA or non-LPS-activated RAW 264. Was applied to 7 cells. Seven cell, implying that macrophages could release NPDio/PPLA nanoparticles could be released by macrophages. RAW 2647 cells were further examined by CLSM (Figure 5C). DAPI nuclei were blue and Dio fluorescence was green. Dio is a lipophilic membrane dye that can only be diffused laterally into cells, staining the entire cell membrane gradually. Green fluorescence surrounds blue fluorescence, indicating that the released Dio enters the cell, and green fluorescence intensity increases significantly over time. The results were consistent with the FCA results. In conclusion, NPDio/PPLA can be effectively endocytosis and released into macrophages by activated RAW 264.7 cells.
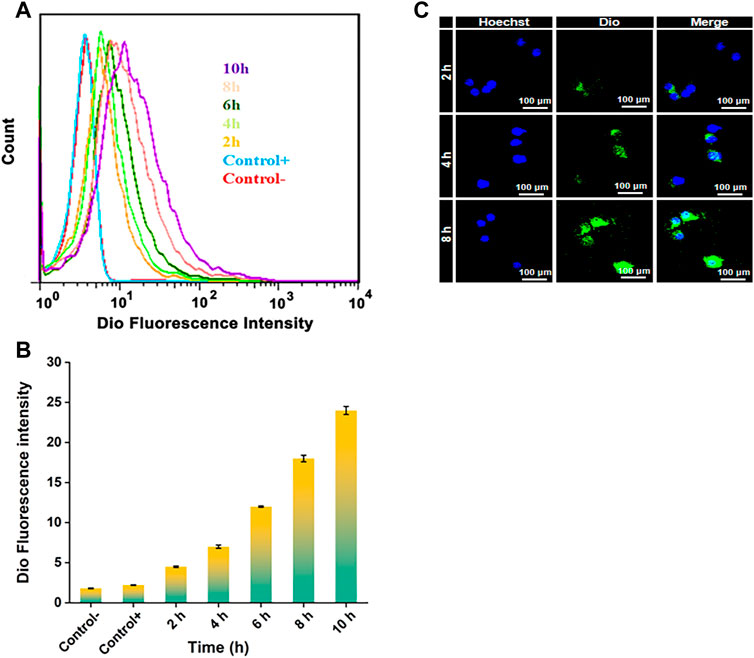
FIGURE 5. (A) FCA of RAW 264.7 cells: Cultured in DMEM medium containing NPDio/PPLA for 2 h, 4 h, 6 h, 8 h, 10 h with 1 μg mL−1 LPS. The average fluorescence intensity of Dio at each time point was detected. (B) FCA of RAW 264.7 cells: Cultured in DMEM medium containing NPDio/PPLA for 2 h, 4 h, 6 h, 8 h, 10 h without 1 μg mL−1 LPS. The average fluorescence intensity of Dio at each time point was detected (C) Confocal scanning microscope images of RAW 264.7 cells co-cultured with DMEM medium containing NPDio/PPLA with 1 μg mL−1 LPS for 2 h, 4 h, and 8 h.
PPLA was non-toxic to RAW 264.7 cells and inhibited macrophage activity, as shown in Figure 6. αLA, a natural antioxidant synthesized in the human body, does not affect RAW 264.7 cells’ activity. In addition, the poor solubility of DXM may be responsible for the decreased inhibition of RAW 264.7 cells. In the absence of LPS, with the DXM concentration increasing to 50 μg mL−1, the RAW 264.7 cell activity of the NPDXM/PPLA group was 70%, and cell inhibition was up to 30% compared to the DXM and NPPPLA groups (Figure 6A). This may be due to the endocytosis of nanoparticles by macrophages. Furthermore, the cell inhibition effect of the NPDXM/PPLA group after LPS activation was 43%, indicating that the inhibition effect of NPDXM/PPLA on the proliferation of activated RAW 264.7 cells increased (Figure 6B).
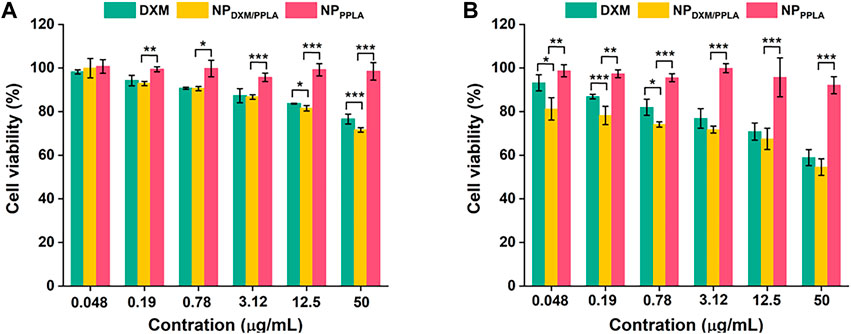
FIGURE 6. (A) Inhibitory effects of DXM, NPPPLA and NPDXM/PPLA on RAW 264.7 cells in the presence of 1 μg mL−1 LPS (B) Inhibitory effects of DXM, NPPPLA and NPDXM/PPLA on RAW 264.7 cells in the absence of 1 μg mL−1 LPS.
The MIA model is the most successful and frequent OA model, and its pathological characteristics are very similar to human OA (Pomonis et al., 2005; Liu et al., 2011; Malfait et al., 2013). Because of its good in vitro experimental results, we established an OA animal model to evaluate the therapeutic effect of NPDXM/PPLA. Three days after the successful formation of artificial OA, 40 mice were divided into 4 groups: NS group, NPPPLA group, DXM group, and NPDXM/PPLA. OA mice were injected with drugs of each group at a weight of kg every 4 days. Mice without OA induction were set as a control group. The degree of articular cartilage destruction, extracellular matrix loss, and changes in inflammatory cytokines (IL-1β, IL-6, and TNF-α) was measured after treatment. As shown in Figure 7A, H&E staining sections of chondrocytes in the normal group showed neat chondrocytes and no synovial hyperplasia around the joint. In comparison to the control group, the NPDXM/PPLA treatment group had less inflammatory cell infiltration and only minor chondrocytes degeneration, whereas the normal saline treatment group had more obvious chondrocyte destruction and severe synovial hyperplasia with a high number of inflammatory cell infiltration.
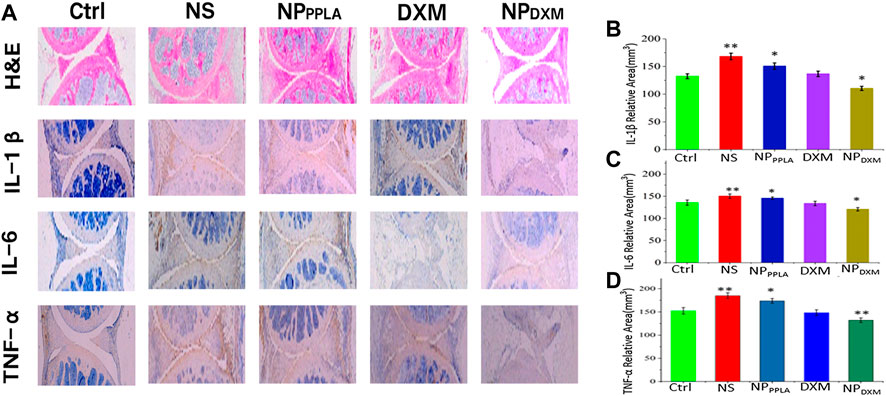
FIGURE 7. (A) The efficacies of NS, NPPPLA, DXM and NPDXM/PPLA on OA were observed by H&E staining joint sections and corresponding immunohistochemical staining sections (B–D) Expression of pro-inflammatory cytokines in all treatment groups.
Proinflammatory cytokines expressed by macrophages have been shown to play an important role in the early stages of OA and in promoting disease progression (Blanco et al., 2011; Huang and Kraus, 2016). Inhibiting the release of proinflammatory cytokines in OA tissue therefore can effectively reduce the degree of cartilage damage (Agarwal et al., 2016; Utomo et al., 2016, 2019; Chevalier and Eymard, 2019). To further investigate the anti-inflammatory effect of the NPDXM/PPLA group, the expression of proinflammatory cytokines including IL-1β, IL-6, and TNF-α in mouse knee osteoarthritis specimens was determined by an ELISA. The expressions of proinflammatory cytokines IL-1β, IL-6, and TNF-α were lower in the NPDXM/PPLA group compared to the saline group, as shown in Figures 7B–D. The expression of proinflammatory cytokines in the NPDXM/PPLA treatment group was also decreased.
Next, we evaluated the knee bone structure of OA mice by Micro CT. The comparison of bone volume fraction (BV/TV) in the subchondral region of the knee in mice, as shown in Figure 8, revealed that the bone mass in the saline group was approximately 55%, while that in the NPDXM/PPLA group was 90%. The bone mass of the knee subchondral bone in mice treated with normal saline was importantly lesser than that in mice treated with NPDXM/PPLA, whereas the bone mass of the knee in mice treated with NPDXM/PPLA was similar to that in healthy mice. Taken together, all these outcomes show that NPDXM/PPLA is promising for the treatment of OA.
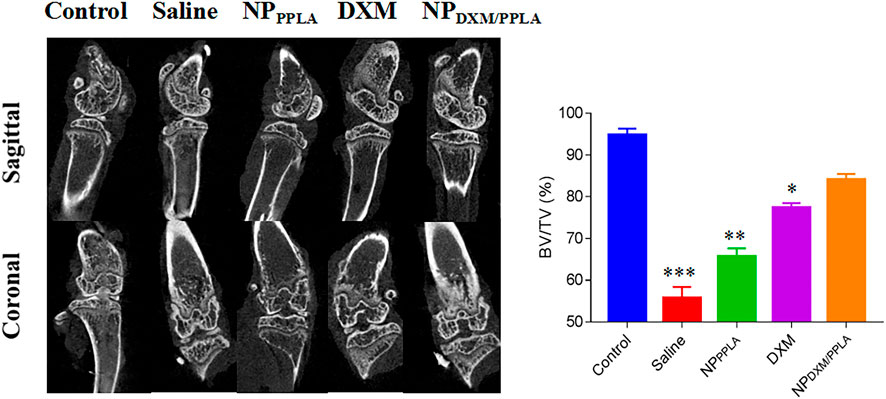
FIGURE 8. The efficacies of NS, NPPPLA, DXM and NPDXM/PPLA on OA were observed by Micro CT and analyzed the bone volume fraction (BV/TV) in subchondral region.
In this study, we synthesized NPDXM/PPLA through polymerization and hydrophilic interactions. In vitro characterization affirmed that NPDXM/PPLA can carry and release DXM effectively. It also revealed good stability under physiological conditions. FCA and CLSM confirmed that NPDXM/PPLA can be activated by macrophage endocytosis and release. Finally, synovial inflammation was reduced in the OA mice model, and cartilage destruction and bone loss were inhibited, which was closer to the knee joint of the control mice. As a result, NPDXM/PPLA can be used to treat osteoarthritis and is a potential vector material for clinical use. At present, the research on drug delivery, targeting, controlled release and other aspects is the forefront of drug delivery systems. In this study, drug delivery nanoparticles were prepared by simple and easy methods, achieving a more stable drug controlled release, which has a certain anti osteoarthritis effect, providing a certain reference for the development of more efficient drug delivery systems in the future.
The original contributions presented in the study are included in the article/supplementary materials, further inquiries can be directed to the corresponding author.
The animal study was reviewed and approved by the Animal Ethics Committee of Jilin University.
DsL, ZJ, YC, YX and JL contributed to conception and design of the study. ZJ, YC, YX, LiS performed the experiments. LS, YC, DbL and DsL analyzed data and interpreted results of experiments. JL and YX provided resources. JL and DsL supervised the study. YC and LS wrote the original draft, ZJ, DbL, JL, YX reviewed and edited it. All authors contributed to manuscript revision and approved the submitted version.
This study supported by the Jilin Province Science and Technology Development Plan Project (Grant No. 20230204077YY).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Agarwal, R., Volkmer, T. M., Wang, P., Lee, L. A., Wang, Q., and García, A. J. (2016). Synthesis of self-assembled IL-1Ra-presenting nanoparticles for the treatment of osteoarthritis. J. Biomed. Mater. Res. Part A 104 (3), 595–599. doi:10.1002/jbm.a.35601
Blanco, F. J., Rego, I., and Ruiz-Romero, C. (2011). The role of mitochondria in osteoarthritis. Nat. Rev. Rheumatol. 7 (3), 161–169. doi:10.1038/nrrheum.2010.213
Blom, A. B., van Lent, P. L., Libregts, S., Holthuysen, A. E., van der Kraan, P. M., van Rooijen, N., et al. (2007). Crucial role of macrophages in matrix metalloproteinase-mediated cartilage destruction during experimental osteoarthritis: Involvement of matrix metalloproteinase 3. Arthritis rheumatism 56 (1), 147–157. doi:10.1002/art.22337
Bondeson, J., Blom, A. B., Wainwright, S., Hughes, C., Caterson, B., and van den Berg, W. B. (2010). The role of synovial macrophages and macrophage-produced mediators in driving inflammatory and destructive responses in osteoarthritis. Arthritis rheumatism 62 (3), 647–657. doi:10.1002/art.27290
Bondeson, J., Wainwright, S. D., Lauder, S., Amos, N., and Hughes, C. E. (2006). The role of synovial macrophages and macrophage-produced cytokines in driving aggrecanases, matrix metalloproteinases, and other destructive and inflammatory responses in osteoarthritis. Arthritis Res. Ther. 8 (6), R187. doi:10.1186/ar2099
Bove, S. E., Calcaterra, S. L., Brooker, R. M., Huber, C. M., Guzman, R. E., Juneau, P. L., et al. (2003). Weight bearing as a measure of disease progression and efficacy of anti-inflammatory compounds in a model of monosodium iodoacetate-induced osteoarthritis. Osteoarthr. Cartil. 11 (11), 821–830. doi:10.1016/s1063-4584(03)00163-8
Brown, S., Kumar, S., and Sharma, B. (2019). Intra-articular targeting of nanomaterials for the treatment of osteoarthritis. Acta biomater. 93, 239–257. doi:10.1016/j.actbio.2019.03.010
Chevalier, X., and Eymard, F. (2019). Anti-IL-1 for the treatment of OA: Dead or alive? Nat. Rev. Rheumatol. 15 (4), 191–192. doi:10.1038/s41584-019-0185-y
Chung, M. F., Chia, W. T., Wan, W. L., Lin, Y. J., and Sung, H. W. (2015). Controlled release of an anti-inflammatory drug using an ultrasensitive ROS-responsive gas-generating carrier for localized inflammation inhibition. J. Am. Chem. Soc. 137 (39), 12462–12465. doi:10.1021/jacs.5b08057
Conaghan, P. G., Cook, A. D., Hamilton, J. A., and Tak, P. P. (2019). Therapeutic options for targeting inflammatory osteoarthritis pain. Nat. Rev. Rheumatol. 15 (6), 355–363. doi:10.1038/s41584-019-0221-y
Crielaard, B. J., Lammers, T., Schiffelers, R. M., and Storm, G. (2012). Drug targeting systems for inflammatory disease: One for all, all for one. J. Control. release official J. Control. Release Soc. 161 (2), 225–234. doi:10.1016/j.jconrel.2011.12.014
Culemann, S., Grüneboom, A., Nicolás-Ávila, J. Á., Weidner, D., Lämmle, K. F., Rothe, T., et al. (2019). Locally renewing resident synovial macrophages provide a protective barrier for the joint. Nature 572 (7771), 670–675. doi:10.1038/s41586-019-1471-1
Evans, C. H., Kraus, V. B., and Setton, L. A. (2014). Progress in intra-articular therapy. Nat. Rev. Rheumatol. 10 (1), 11–22. doi:10.1038/nrrheum.2013.159
French, H. P., Cusack, T., Brennan, A., Caffrey, A., Conroy, R., Cuddy, V., et al. (2013). Exercise and manual physiotherapy arthritis research trial (EMPART) for osteoarthritis of the hip: A multicenter randomized controlled trial. Archives Phys. Med. rehabilitation 94 (2), 302–314. doi:10.1016/j.apmr.2012.09.030
Giannelli, G., Erriquez, R., Iannone, F., Marinosci, F., Lapadula, G., and Antonaci, S. (2004). MMP-2, MMP-9, TIMP-1 and TIMP-2 levels in patients with rheumatoid arthritis and psoriatic arthritis. Clin. Exp. rheumatology 22 (3), 335–338. doi:10.1002/art.20330
Huang, Z., and Kraus, V. B. (2016). Does lipopolysaccharide-mediated inflammation have a role in OA? Nat. Rev. Rheumatol. 12 (2), 123–129. doi:10.1038/nrrheum.2015.158
Jones, I. A., Togashi, R., Wilson, M. L., Heckmann, N., and Vangsness, C. T. (2019). Intra-articular treatment options for knee osteoarthritis. Nat. Rev. Rheumatol. 15 (2), 77–90. doi:10.1038/s41584-018-0123-4
Joshi, N., Yan, J., Levy, S., Bhagchandani, S., Slaughter, K. V., Sherman, N. E., et al. (2018). Towards an arthritis flare-responsive drug delivery system. Nat. Commun. 9 (1), 1275. doi:10.1038/s41467-018-03691-1
Lepetsos, P., and Papavassiliou, A. G. (2016). ROS/oxidative stress signaling in osteoarthritis. Biochimica biophysica acta 1862 (4), 576–591. doi:10.1016/j.bbadis.2016.01.003
Li, M., Tang, Z., Sun, H., Ding, J., Song, W., and Chen, X. (2013). pH and reduction dual-responsive nanogel cross-linked by quaternization reaction for enhanced cellular internalization and intracellular drug delivery. Polym. Chem. 4, 1199–1207. doi:10.1039/C2PY20871G
Liu, P., Okun, A., Ren, J., Guo, R. C., Ossipov, M. H., Xie, J., et al. (2011). Ongoing pain in the MIA model of osteoarthritis. Neurosci. Lett. 493 (3), 72–75. doi:10.1016/j.neulet.2011.01.027
Maczurek, A., Hager, K., Kenklies, M., Sharman, M., Martins, R., Engel, J., et al. (2008). Lipoic acid as an anti-inflammatory and neuroprotective treatment for Alzheimer's disease. Adv. drug Deliv. Rev. 60 (13-14), 1463–1470. doi:10.1016/j.addr.2008.04.015
Madaleno, F. O., Santos, B. A., Araújo, V. L., Oliveira, V. C., and Resende, R. A. (2018). Prevalence of knee osteoarthritis in former athletes: A systematic review with meta-analysis. Braz. J. Phys. Ther. 22 (6), 437–451. doi:10.1016/j.bjpt.2018.03.012
Malfait, A. M., Little, C. B., and McDougall, J. J. (2013). A commentary on modelling osteoarthritis pain in small animals. Osteoarthr. Cartil. 21 (9), 1316–1326. doi:10.1016/j.joca.2013.06.003
Martel-Pelletier, J., Wildi, L. M., and Pelletier, J. P. (2012). Future therapeutics for osteoarthritis. Bone 51 (2), 297–311. doi:10.1016/j.bone.2011.10.008
McAlindon, T. E., and Bannuru, R. R. (2018). Osteoarthritis in 2017: Latest advances in the management of knee OA. Rheumatology 14 (2), 73–74. doi:10.1038/nrrheum.2017.219
Nwosu, L. N., Mapp, P. I., Chapman, V., and Walsh, D. A. (2016). Relationship between structural pathology and pain behaviour in a model of osteoarthritis (OA). Osteoarthr. Cartil. 24 (11), 1910–1917. doi:10.1016/j.joca.2016.06.012
Packer, L., Witt, E. H., and Tritschler, H. J. (1995). alpha-Lipoic acid as a biological antioxidant. Free Radic. Biol. Med. 19 (2), 227–250. doi:10.1016/0891-5849(95)00017-r
Pomonis, J. D., Boulet, J. M., Gottshall, S. L., Phillips, S., Sellers, R., Bunton, T., et al. (2005). Development and pharmacological characterization of a rat model of osteoarthritis pain. Pain 114 (3), 339–346. doi:10.1016/j.pain.2004.11.008
Sang, J., Ji, E., Su, H., Sun, J., Su, J., Shkbc, D., et al. (2016). Therapeutic effects of neuropeptide substance P coupled with self-assembled peptide nanofibers on the progression of osteoarthritis in a rat model. Biomaterials 74, 119–130. doi:10.1016/j.biomaterials.2015.09.040
Sellam, J., and Berenbaum, F. (2010). The role of synovitis in pathophysiology and clinical symptoms of osteoarthritis. Nat. Rev. Rheumatol. 6 (11), 625–635. doi:10.1038/nrrheum.2010.159
Shay, K. P., Moreau, R. F., Smith, E. J., Smith, A. R., and Hagen, T. M. (2009). Alpha-lipoic acid as a dietary supplement: Molecular mechanisms and therapeutic potential. Biochimica biophysica acta 1790 (10), 1149–1160. doi:10.1016/j.bbagen.2009.07.026
Shimoda, H., Tanaka, J., Seki, A., Honda, H., Akaogi, S., Komatsubara, H., et al. (2007). [Safety and structural analysis of polymers produced in manufacturing process of alpha-lipoic acid]. J. Food Hyg. Soc. Jpn. 48 (5), 125–131. doi:10.3358/shokueishi.48.125
Singh, U., and Jialal, I. (2008). Alpha-lipoic acid supplementation and diabetes. Nutr. Rev., 66(11), 646–657. doi:10.1111/j.1753-4887.2008.00118.x
Solmonson, A., and DeBerardinis, R. J. (2018). Lipoic acid metabolism and mitochondrial redox regulation. J. Biol. Chem. 293 (20), 7522–7530. doi:10.1074/jbc.TM117.000259
Sonderegger, F. L., Ma, Y., Maylor-Hagan, H., Brewster, J., Huang, X., Spangrude, G. J., et al. (2012). Localized production of IL-10 suppresses early inflammatory cell infiltration and subsequent development of IFN-γ-mediated Lyme arthritis. J. Immunol., 188(3), 1381–1393. doi:10.4049/jimmunol.1102359
Sun, Y., Zhang, G., Liu, Q., Liu, X., Wang, L., Wang, J., et al. (2018). Chondroitin sulfate from sturgeon bone ameliorates pain of osteoarthritis induced by monosodium iodoacetate in rats. Int. J. Biol. Macromol. 117, 95–101. doi:10.1016/j.ijbiomac.2018.05.124
Utomo, L., Bastiaansen-Jenniskens, Y. M., Verhaar, J. A., and van Osch, G. J. (2016). Cartilage inflammation and degeneration is enhanced by pro-inflammatory (M1) macrophages in vitro, but not inhibited directly by anti-inflammatory (M2) macrophages. Osteoarthr. Cartil. 24 (12), 2162–2170. doi:10.1016/j.joca.2016.07.018
Xia, J., Xu, Y., Li, S., Sun, W., Yu, K., and Tang, W. (2001). Carboxy ester hydrolysis promoted by a zinc(II) 2-[bis(2-aminomethyl)amino]ethanol complex: A new model for indirect activation on the serine nucleophile by zinc(II) in zinc enzymes. Inorg. Chem. 40 (10), 2394–2401. doi:10.1021/ic000642n
Keywords: polylipoic acid, osteoarthritis, dexamethasone, nanoparticles, intraarticular injection, intraarticular injection of knee joint
Citation: Cheng Y, Jing Z, Xu Y, Sun L, Li D, Liu J and Li D (2023) Polyethylene Glycol-grafted poly alpha-lipoic acid-dexamethasone nanoparticles for osteoarthritis. Front. Drug Deliv. 3:1168287. doi: 10.3389/fddev.2023.1168287
Received: 17 February 2023; Accepted: 11 May 2023;
Published: 20 July 2023.
Edited by:
Faisal Raza, Shanghai Jiao Tong University, ChinaReviewed by:
Juan M. Colazo, Vanderbilt University, United StatesCopyright © 2023 Cheng, Jing, Xu, Sun, Li, Liu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongsong Li, lidongsong@jlu.edu.cn
†These authors contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.