- 1Institute of Health Policy Management and Evaluation, University of Toronto, Toronto, ON, Canada
- 2Data Science for Health Unit, Fondazione Bruno Kessler, Trento, Italy
1. Introduction
Supervised machine learning has become pervasive in the biomedical sciences nowadays (Larrañaga et al., 2006; Tarca et al., 2007), and its validation has obtained a key role in all these scientific fields. We therefore read with great interest the article by Walsh et al. (2021), which reported a list of DOME recommendations to properly validate results achieved with supervised machine learning, according to the authors. In the past, several studies already listed common best practices and recommendations for the proper usage of machine learning (Bhaskar et al., 2006; Domingos, 2012; Chicco, 2017; Cearns et al., 2019; Stevens et al., 2020; Artrith et al., 2021; Cabitza and Campagner, 2021; Larson et al., 2021; Whalen et al., 2021; Lee et al., 2022) and computational statistics (Benjamin et al., 2018; Makin and de Xivry, 2019), but the comment by Walsh et al. (2021) has the merit to highlight the importance of computational validation, which is a key step perhaps even more important than the machine learning algorithm design itself.
Although interesting and complete, that article describes numerous of steps and aspects in a way that we find complicated, especially for beginners. We believe that the 21 questions of the Box 1 of the DOME article (Walsh et al., 2021) can be adequate for a data mining expert, but they might scare and discourage an inexperienced practitioner. For example, the recommendations about the meta-predictions and about the hyper-parameters' optimization might not be understandable by a machine learning beginner or by a wet lab biologist. And it should not be a problem: a robust machine learning analysis can be performed, in fact, without using meta-predictions or hyper-parameters, too. A beginner, in front of so many guidelines of that article, some of which being so complex, might even decide to abandon the computational intelligence analysis, to avoid making any mistake in their scientific project. Moreover, the DOME (Walsh et al., 2021) authors present the 21 questions of the article Box 1 with the same level of importance. In contrast, we think that three key aspects to keep in mind for computational validation are pivotal and can be sufficient, if verified correctly. So we believe that a practitioner would better focus all their attention and energy on accurately respecting these three recommendations.
We therefore wrote this note to propose our own recommendations for the computational validation of supervised machine learning results in the biomedical sciences: just three, explained easily and clearly, that alone can pave the way for a successful machine learning validation phase. We designed these simple quick tips from our experience gained on tens of biomedical projects involving machine learning phases. We call these recommendations ABC to highlight their essential role in any computational validation (Figure 1).
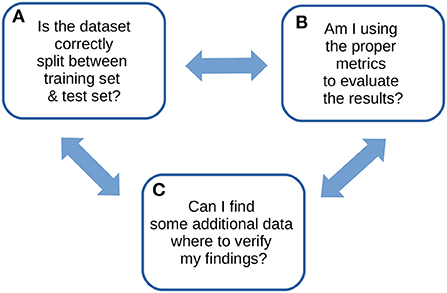
Figure 1. ABC recommendations checklist. An overview of our ABC recommendations, to keep in mind for any machine learning study.
2. The ABC recommendations
(A) Always divide the dataset carefully into separate training set and test set
This rule must become your obsession: verify and double-check that no data element is shared by both the training set and the test set. They must be completely independent.
You then can do anything you want on the training set, including the hyper-parameter optimization, but make sure you do not touch the test set. Leave the test set alone until your supervised machine learning model training has finished (and its hyper-parameters are optimized, if any). If you have enough data, consider also allocating a subset of it (such as 10% of data elements, randomly selected) as a holdout set (Skocik et al., 2016), to use as an alternative test set to confirm your findings and to avoid over-validation (Wainberg et al., 2016).
This important separation will allow you to avoid data snooping (White, 2000; Smith, 2021), that is a common mistake in multiple studies involving computational intelligence (Jensen, 2000; Sewell, 2021). Data snooping, also known as data dredging and called “the dark side of data mining” (Jensen, 2000), happens in fact when some data elements of the training set are present in the test set, too, and therefore over-optimistically improve the results obtained by the trained machine learning model on the test set. Sometimes, this problem can happen even when different data elements of the same patients (for example, radiography images in digital pathology) are shared between training set and test set, and is usually called data leakage (Bussola et al., 2021). This mistake is dangerous for every machine learning study, because it can give the illusion of success to an unaware researcher. In this situation, you need to keep in mind the famous quote by Richard Feynman: “The first principle is that you must not fool yourself, and you are the easiest person to fool” (Chicco, 2017).
Data snooping does exactly that: it makes you fool yourself and makes you believe you obtained excellent results, while actually machine learning performance was flawed. Once you make sure the training set and the test set are independent from each other, you can use traditional cross-validation methods such as k-fold cross-validation, leave-one-out cross-validation, and nested cross-validation (Yadav and Shukla, 2016), or bootstrap validation (Efron, 1992; Efron and Tibshirani, 1994), to mitigate over-fitting (Dietterich, 1995; Chicco, 2017). Moreover, over-fitting can be tackled through calibration methods such as calibration curves (Austin et al., 2022) or calibration-in-the-large (Crowson et al., 2016), which can also help measuring the robustness of model performance.
Moreover, it is important to notice that sometimes splitting the dataset into two subsets (training set and test set) might not be enough (Picard and Berk, 1990). Even for shallow machine learning models, a correct splitting methodology should be enforced: for instance, see the Data Analysis Protocol strategy introduced by the MAQC/SEQC initiatives led by the US Food and Drug Administration (FDA) (MAQC Consortium, 2010; Zhang et al., 2015). And when there are hyper-parameters to optimize (Feurer and Hutter, 2019), such as the number of hidden layers and the number of hidden units in artificial neural networks, it is advisable to split the dataset into three subsets: training set, validation set, and test set (Chicco, 2017). In these cases, sometimes in scientific literature the names validation set and test set are used interchangeably; in this report, we call validation set the part of the dataset employed to evaluate the algorithm configuration with a particular hyper-parameter value, and we call test set the portion of the dataset to keep untouched and eventually use to verify the algorithm with the optimal hyper-parameters' configuration.
(B) Broadly use multiple rates to evaluate your results
Evaluate your results with various rates, and definitely include the Matthew's correlation coefficient (MCC) (Matthews, 1975) for binary classifications (Chicco and Jurman, 2020; Chicco et al., 2021a) and the coefficient of determination (R2) (Wright, 1921) for regression analyses (Chicco et al., 2021b). Moreover, make sure you include at least accuracy, F1 score, sensitivity, specificity, precision, negative predictive value, Cohen's Kappa, and the area under the curve (AUC) of the receiving operating characteristic curve (ROC) and of the prediction-recall curve (PR) for binary classifications. For regression analyses, make sure you incorporate at least mean absolute error (MAE), mean absolute percentage error (MAPE), mean square error (MSE), root mean square error (RMSE), and symmetric mean absolute percentage error (SMAPE), in addition to the already-mentioned R2. We recap our suggestions in Table 1.
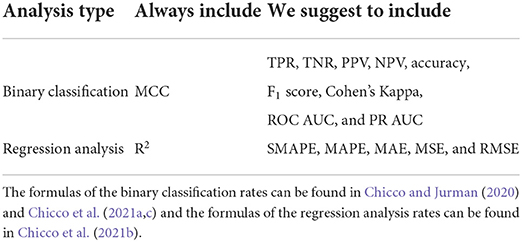
Table 1. Recap of the suggested metrics for evaluating results of binary classifications and regression analyses.
It is necessary to include all these scores because each of them provides a singular, useful piece of information about your supervised machine learning results. The more statistics you include, the more chances you have to spot any possible flaw in your predictions. All these rates work like dashboard indicator lamps in a car: if something somewhere in your machine (learning) did not work out the way it was supposed to, a lamp (rate) will inform you about it.
The Matthew's correlation coefficient, in particular, has a fundamental role in binary classification evaluation: it has a high score only if the classifier correctly predicted most of the positive elements and of the negative elements, and only if the classifier made mostly correct positive predictions and mostly correct negative predictions (Chicco and Jurman, 2020, 2022; Chicco et al., 2021; Chicco et al., 2021a). That means, a high MCC corresponds to a high score for all the four basic rates of a 2 × 2 confusion matrix: sensitivity, specificity, precision, and negative predictive value (Chicco et al., 2021a). Because of its efficacy, the MCC has been employed as the standard metric in several scientific projects. For example, the USFDA agency used the MCC as the main evaluation rate in the MicroArray II/Sequencing Quality Control (MAQC/SEQC) projects (MAQC Consortium, 2010; SEQC/MAQC-III Consortium, 2014).
Regarding regression analysis assessment, the coefficient of determination R-squared (R2) is the only rate that generates a high score only if the predictive algorithm was able to correctly predict most of the elements of each data class, considering their distribution (Chicco et al., 2021b). Additionally, R2 allows the comparison of models applied to datasets having different scales (Chicco et al., 2021b). Because of its effectiveness, the coefficient of determination has been employed as the standard evaluation metric for several international scientific projects, such as the Overhead Geopose DrivenData Challenge (DrivenData.org, 2022) and the Breast Cancer Prognosis DREAM Education Challenge (Bionetworks, 2021).
(C) Confirm your findings with external data, if possible
If you can, use data coming from a different data source and made of a different data type from the main dataset to verify your discoveries. Obtaining the same results you achieved on the main original dataset on an external dataset coming from another scientific research centre would be a strong confirmation of your scientific findings. Moreover, if this external data were in a data type different from the original data, it would even increase the level of independence between the two datasets, and even more strongly confirm your scientific outcomes.
In a bioinformatics study, for example, Kustra and Zagdanski (2008) employed a data fusion approach to cluster microarray gene expression data and associate the derived clusters to Gene Ontology annotations (Gene Ontology Consortium, 2019). For validating their results, instead of using a different microarray dataset, the authors decided to take advantage of an external database made of a different data type: a protein–protein database called General Repository for Interaction Data Sets (GRID) (Breitkreutz et al., 2003). This way, the authors were able to find in external data a strong confirmation of the results they obtained on the original data, and therefore were able to claim their study outcomes as robust and reliable in their manuscript's conclusions.
Moving from bioinformatics to health informatics, a call for external data validation has recently been raised in machine learning and computational statistics applied to heart failure prediction as well (Shin et al., 2021).
That being said, we are aware that obtaining compatible additional data and integrating them might be difficult for some biomedical studies, but we still invite all the machine learning practitioners to make an attempt and to try to collect confirmatory data for their analyses anyway. In some cases, there are plenty of public datasets available for free use that can be downloaded and integrated easily.
Bioinformaticians working on gene expression analysis, for example, can take advantage of the thousands of different datasets available on the Gene Expression Omnibus (GEO) (Edgar et al., 2002). Tens of compatible datasets of a particular cancer type can be found by specifying the microarray platform, for example, through the recently released geoCancerPrognosticDatasetsRetriever (Alameer and Chicco, 2022) bioinformatics tool. Researchers can take advantage of these compatible datasets (for example, built on the GPL570 Affymetrix platform) to verify their findings, after applying some quality-control and preprocessing steps such as batch correction (Chen et al., 2011) and data normalization, if needed.
Moreover, public data repositories for biomedical domains, such as ophthalmology images (Khan et al., 2021), cancer images (Clark et al., 2013), or neuroblastoma electronic health records (Chicco et al., in press), can provide additional datasets that can be used as validation cohorts. Additional public datasets can be found on the University of California Irvine Machine Learning Repository (University of California Irvine, 1987), on the DREAM Challenges platform (Kueffner et al., 2019; Sage Bionetworks, 2022), or on Kaggle (Kaggle, 2022), for example.
When using external data, an aspect to keep in mind is checking and correcting issues like dataset shift (Finlayson et al., 2021) and model underspecification (D'Amour et al., 2020), which might jeopardize the coherence of the learning pipeline when moving from training and testing and validation.
3. Discussion
Computational intelligence makes computers able to identify trends in data that otherwise would be difficult or impossible to notice by humans. With the spread of new technologies and electronic devices able to save and store large amounts of data, data mining has become a ubiquitous tool in numerous scientific studies, especially in biomedical informatics. In these studies, the validation of the results obtained through supervised machine learning has become a crucial phase, especially because of the high risk of achieving over-optimistic, inflated results, that can even lead to false discoveries (Ioannidis, 2005).
In the past, several studies proposed rules and guidelines to develop more effective and efficient predictive models in medical informatics and computational epidemiology (Steyerberg and Vergouwe, 2014, Riley et al., 2016, 2021; Bonnett et al., 2019; Wolff et al., 2019; Navarro et al., 2021; Van Calster et al., 2021). Most of them however, provided complicated lists of steps and tips which might be hard to follow by machine learning practitioners, especially by beginners.
In this context, the article of Walsh et al. (2021) plays its part by describing thoroughly several DOME recommendations and steps for validating supervised machine learning results, but in our opinion it suffers from excessive complexity and might be difficult to follow by beginners. In this note, we propose our own simple, easy, essential ABC tips to keep in mind when validating results obtained with data mining methods.
We believe our ABC recommendations can be an effective tool to follow for all the machine learning practitioners, both by beginners and experienced ones, and can pave the way to stronger, more robust, more reliable scientific results in all the biomedical sciences.
Author contributions
DC conceived the study and wrote most of the article. GJ reviewed and contributed to the article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alameer, A., and Chicco, D. (2022). geoCancerPrognosticDatasetsRetriever, a bioinformatics tool to easily identify cancer prognostic datasets on Gene Expression Omnibus (GEO). Bioinformatics 2021:btab852. doi: 10.1093/bioinformatics/btab852
Artrith, N., Butler, K. T., Coudert, F. -X., Han, S., Isayev, O., Jain, A., et al. (2021). Best practices in machine learning for chemistry. Nat. Chem. 13, 505–508. doi: 10.1038/s41557-021-00716-z
Austin, P. C., Putter, H., Giardiello, D., and van Klaveren, D. (2022). Graphical calibration curves and the integrated calibration index (ICI) for competing risk models. Diagn. Progn. Res. 6, 1–22. doi: 10.1186/s41512-021-00114-6
Benjamin, D. J., Berger, J. O., Johannesson, M., Nosek, B. A., Wagenmakers, E. -J., Berk, R., et al. (2018). Redefine statistical significance. Nat. Hum. Behav. 2, 6–10. doi: 10.1038/s41562-017-0189-z
Bhaskar, H., Hoyle, D. C., and Singh, S. (2006). Machine learning in bioinformatics: a brief survey and recommendations for practitioners. Comput. Biol. Med. 36, 1104–1125. doi: 10.1016/j.compbiomed.2005.09.002
Bionetworks, S. (2021). Breast Cancer Prognosis DREAM Education Challenge. Available online at: https://www.synapse.org/#!Synapse:syn8650663/wiki/436447 (accessed August 12, 2021).
Bonnett, L. J., Snell, K. I. E., Collins, G. S., and Riley, R. D. (2019). Guide to presenting clinical prediction models for use in clinical settings. BMJ 365:l737. doi: 10.1136/bmj.l737
Breitkreutz, B. -J., Stark, C., and Tyers, M. (2003). The GRID: the general repository for interaction datasets. Genome Biol. 4:R23. doi: 10.1186/gb-2003-4-2-p1
Bussola, N., Marcolini, A., Maggio, V., Jurman, G., and Furlanello, C. (2021). “AI slipping on tiles: data leakage in digital pathology,” in Proceedings of ICPR 2021 – The 25th International Conference on Pattern Recognition. ICPR International Workshops and Challenges (Berlin: Springer International Publishing), 167–182.
Cabitza, F., and Campagner, A. (2021). The need to separate the wheat from the chaff in medical informatics: introducing a comprehensive checklist for the (self)-assessment of medical AI studies. Int. J. Med. Inform. 153:104510. doi: 10.1016/j.ijmedinf.2021.104510
Cearns, M., Hahn, T., and Baune, B. T. (2019). Recommendations and future directions for supervised machine learning in psychiatry. Transl. Psychiatry 9:271. doi: 10.1038/s41398-019-0607-2
Chen, C., Grennan, K., Badner, J., Zhang, D., Gershon, E., Jin, L., et al. (2011). Removing batch effects in analysis of expression microarray data: an evaluation of six batch adjustment methods. PLoS ONE 6:e17238. doi: 10.1371/journal.pone.0017238
Chicco, D. (2017). Ten quick tips for machine learning in computational biology. BioData Min. 10:35. doi: 10.1186/s13040-017-0155-3
Chicco, D., Cerono, G, and Cangelosi, D. (in press). A survey on publicly available open datasets of electronic health records (EHRs) of patients with neuroblastoma. Data Sci. J. 1–15.
Chicco, D., and Jurman, G. (2020). The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genomics 21:6. doi: 10.1186/s12864-019-6413-7
Chicco, D., and Jurman, G. (2022). An invitation to greater use of Matthews correlation coefficient in robotics and artificial intelligence. Front. Robot. AI 9:876814. doi: 10.3389/frobt.2022.876814
Chicco, D., Starovoitov, V., and Jurman, G. (2021). The benefits of the Matthews correlation coefficient (MCC) over the diagnostic odds ratio (DOR) in binary classification assessment. IEEE Access. 9, 47112–47124. doi: 10.1109/ACCESS.2021.3068614
Chicco, D., Tötsch, N., and Jurman, G. (2021a). The Matthews correlation coefficient (MCC) is more reliable than balanced accuracy, bookmaker informedness, and markedness in two-class confusion matrix evaluation. BioData Min. 14:13. doi: 10.1186/s13040-021-00244-z
Chicco, D., Warrens, M. J., and Jurman, G. (2021b). The coefficient of determination R-squared is more informative than SMAPE, MAE, MAPE, MSE and RMSE in regression analysis evaluation. PeerJ Comput. Sci. 7:e623. doi: 10.7717/peerj-cs.623
Chicco, D., Warrens, M. J., and Jurman, G. (2021c). The Matthews correlation coefficient (MCC) is more informative than Cohens Kappa and Brier score in binary classification assessment. IEEE Access. 9, 78368–78381. doi: 10.1109/ACCESS.2021.3084050
Clark, K., Vendt, B., Smith, K., Freymann, J., Kirby, J., Koppel, P., et al. (2013). The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J. Digit. Imaging 26, 1045–1057. doi: 10.1007/s10278-013-9622-7
Crowson, C. S., Atkinson, E. J., and Therneau, T. M. (2016). Assessing calibration of prognostic risk scores. Stat. Methods Med. Res. 25, 1692–1706. doi: 10.1177/0962280213497434
D'Amour, A., Heller, K., Moldovan, D., Adlam, B., Alipanahi, B., Beutel, A., et al. (2020). Underspecification presents challenges for credibility in modern machine learning. arXiv Preprint arXiv:2011.03395. doi: 10.48550/arXiv.2011.03395
Dietterich, T. (1995). Overfitting and undercomputing in machine learning. ACM Comput. Surveys 27, 326–327. doi: 10.1145/212094.212114
Domingos, P. (2012). A few useful things to know about machine learning. Commun. ACM 55, 78–87. doi: 10.1145/2347736.2347755
DrivenData.org (2022). Overhead Geopose Challenge. Available online at: https://www.drivendata.org/competitions/78/competition-overhead-geopose/page/372/ (accessed August 12, 2021).
Edgar, R., Domrachev, M., and Lash, A. E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucl. Acids Res. 30, 207–210. doi: 10.1093/nar/30.1.207
Efron, B. (1992). “Bootstrap methods: another look at the jackknife,” in Breakthroughs in Statistics, eds S. Kotz and N. L. Johnson (New York, NY: Springer), 569–593. doi: 10.1007/978-1-4612-4380-9_41
Efron, B., and Tibshirani, R. J. (1994). An Introduction to the Bootstrap. New York, NY: CRC Press. doi: 10.1201/9780429246593
Feurer, M., and Hutter, F. (2019). “Hyperparameter optimization,” in Automated Machine Learning, eds F. Hutter, L. Kotthoff, and J. Vanschoren (Berlin: Springer), 3–33. doi: 10.1007/978-3-030-05318-5_1
Finlayson, S. G., Subbaswamy, A., Singh, K., Bowers, J., Kupke, A., Zittrain, J., et al. (2021). The clinician and dataset shift in artificial intelligence. N. Engl. J. Med. 385, 283–286. doi: 10.1056/NEJMc2104626
Gene Ontology Consortium (2019). The Gene Ontology resource: 20 years and still GOing strong. Nucl. Acids Res. 47, D330–D338. doi: 10.1093/nar/gky1055
Ioannidis, J. P. (2005). Why most published research findings are false. PLOS Med. 2:e124. doi: 10.1371/journal.pmed.0020124
Jensen, D. (2000). Data snooping, dredging and fishing: the dark side of data mining a SIGKDD99 panel report. ACM SIGKDD Explor. Newsl. 1, 52–54. doi: 10.1145/846183.846195
Kaggle (2022). Kaggle.com – Find Open Datasets. Available online at: https://www.kaggle.com/datasets (accessed March 27, 2022).
Khan, S. M., Liu, X., Nath, S., Korot, E., Faes, L., Wagner, S. K., et al. (2021). A global review of publicly available datasets for ophthalmological imaging: barriers to access, usability, and generalisability. Lancet Digit. Health 3, e51–e66. doi: 10.1016/S2589-7500(20)30240-5
Kueffner, R., Zach, N., Bronfeld, M., Norel, R., Atassi, N., Balagurusamy, V., et al. (2019). Stratification of amyotrophic lateral sclerosis patients: a crowdsourcing approach. Sci. Reports 9:690. doi: 10.1038/s41598-018-36873-4
Kustra, R., and Zagdanski, A. (2008). Data-fusion in clustering microarray data: balancing discovery and interpretability. IEEE/ACM Trans. Comput. Biol. Bioinform. 7, 50–63. doi: 10.1109/TCBB.2007.70267
Larrañaga, P., Calvo, B., Santana, R., Bielza, C., Galdiano, J., Inza, I., et al. (2006). Machine learning in bioinformatics. Brief. Bioinform. 7, 86–112. doi: 10.1093/bib/bbk007
Larson, D. B., Harvey, H., Rubin, D. L., Irani, N., Justin, R. T., and Langlotz, C. P. (2021). Regulatory frameworks for development and evaluation of artificial intelligence–based diagnostic imaging algorithms: summary and recommendations. J. Amer. Coll. Radiol. 18, 413–424. doi: 10.1016/j.jacr.2020.09.060
Lee, B. D., Gitter, A., Greene, C. S., Raschka, S., Maguire, F., Titus, A. J., et al. (2022). Ten quick tips for deep learning in biology. PLoS Comput. Biol. 18:e1009803. doi: 10.1371/journal.pcbi.1009803
Makin, T. R., and de Xivry, J.-J. O. (2019). Science forum: ten common statistical mistakes to watch out for when writing or reviewing a manuscript. eLife 8:e48175. doi: 10.7554/eLife.48175.005
MAQC Consortium (2010). The MicroArray quality control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat. Biotechnol. 28, 827–838. doi: 10.1038/nbt.1665
Matthews, B. W. (1975). Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta Prot. Struct. 405, 442–451. doi: 10.1016/0005-2795(75)90109-9
Navarro, C. L. A., Damen, J. A., Takada, T., Nijman, S. W., Dhiman, P., Ma, J., et al. (2021). Risk of bias in studies on prediction models developed using supervised machine learning techniques: systematic review. BMJ 375:n2281. doi: 10.1136/bmj.n2281
Picard, R. R., and Berk, K. N. (1990). Data splitting. Amer. Stat. 44, 140–147. doi: 10.1080/00031305.1990.10475704
Riley, R. D., Debray, T. P. A., Collins, G. S., Archer, L., Ensor, J., Smeden, M., et al. (2021). Minimum sample size for external validation of a clinical prediction model with a binary outcome. Stat. Med. 40, 4230–4251. doi: 10.1002/sim.9025
Riley, R. D., Ensor, J., Snell, K. I. E., Debray, T. P. A., Altman, D. G., Moons, K. G. M., et al. (2016). External validation of clinical prediction models using big datasets from e-health records or IPD meta-analysis: opportunities and challenges. BMJ 353:i3140. doi: 10.1136/bmj.i3140
Sage Bionetworks (2022). DREAM Challenges Publications. Available online at: https://dreamchallenges.org/publications/ (accessed January 17, 2022).
SEQC/MAQC-III Consortium (2014). A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the sequencing quality control consortium. Nat. Biotechnol. 32, 903–914. doi: 10.1038/nbt.2957
Sewell, M. (2021). Data Snooping. Available online at: http://data-snooping.martinsewell.com (accessed August 6, 2021).
Shin, S., Austin, P. C., Ross, H. J., Abdel-Qadir, H., Freitas, C., Tomlinson, G., et al. (2021). Machine learning vs. conventional statistical models for predicting heart failure readmission and mortality. ESC Heart Fail. 8, 106–115. doi: 10.1002/ehf2.13073
Skocik, M., Collins, J., Callahan-Flintoft, C., Bowman, H., and Wyble, B. (2016). I tried a bunch of things: the dangers of unexpected overfitting in classification. bioRxiv 2016:078816. doi: 10.1101/078816
Smith, M. K. (2021). Data snooping. Available online at: https://web.ma.utexas.edu/users/mks/statmistakes/datasnooping.html (accessed August 5, 2021).
Stevens, L. M., Mortazavi, B. J., Deo, R. C., Curtis, L., and Kao, D. P. (2020). Recommendations for reporting machine learning analyses in clinical research. Circ. Cardiovasc. Qual. Outcomes 13:e006556. doi: 10.1161/CIRCOUTCOMES.120.006556
Steyerberg, E. W., and Vergouwe, Y. (2014). Towards better clinical prediction models: seven steps for development and an ABCD for validation. Eur. Heart J. 35, 1925–1931. doi: 10.1093/eurheartj/ehu207
Tarca, A. L., Carey, V. J., Chen, X.-W., Romero, R., and Drăghici, S. (2007). Machine learning and its applications to biology. PLoS Comput. Biol. 3:e116. doi: 10.1371/journal.pcbi.0030116
University of California Irvine (1987). Machine Learning Repository. Available online at: https://archive.ics.uci.edu/ml (accessed January 12, 2021).
Van Calster, B., Wynants, L., Riley, R. D., van Smeden, M., and Collins, G. S. (2021). Methodology over metrics: current scientific standards are a disservice to patients and society. J. Clin. Epidemiol. 138, 219–226. doi: 10.1016/j.jclinepi.2021.05.018
Wainberg, M., Alipanahi, B., and Frey, B. J. (2016). Are random forests truly the best classifiers? J. Mach. Learn. Res. 17, 3837–3841. doi: 10.5555/2946645.3007063
Walsh, I., Fishman, D., Garcia-Gasulla, D., Titma, T., Pollastri, G., Capriotti, E., et al. (2021). DOME: recommendations for supervised machine learning validation in biology. Nat. Methods 5, 1122–1127. doi: 10.1038/s41592-021-01205-4
Whalen, S., Schreiber, J., Noble, W. S., and Pollard, K. S. (2021). Navigating the pitfalls of applying machine learning in genomics. Nat. Rev. Genet. 23, 169–181. doi: 10.1038/s41576-021-00434-9
White, H. (2000). A reality check for data snooping. Econometrica 68, 1097–1126. doi: 10.1111/1468-0262.00152
Wolff, R. F., Moons, K. G., Riley, R. D., Whiting, P. F., Westwood, M., Collins, G. S., et al. (2019). PROBAST: a tool to assess the risk of bias and applicability of prediction model studies. Ann. Intern. Med. 170, 51–58. doi: 10.7326/M18-1376
Yadav, S., and Shukla, S. (2016). “Analysis of k-fold cross-validation over hold-out validation on colossal datasets for quality classification,” in Proceedings of IACC 2016—the 6th International Conference on Advanced Computing (Bhimavaram), 78–83.
Keywords: supervised machine learning, computational validation, recommendations, data mining, best practices in machine learning
Citation: Chicco D and Jurman G (2022) The ABC recommendations for validation of supervised machine learning results in biomedical sciences. Front. Big Data 5:979465. doi: 10.3389/fdata.2022.979465
Received: 27 June 2022; Accepted: 12 September 2022;
Published: 27 September 2022.
Edited by:
Sujata Dash, North Orissa University, IndiaReviewed by:
Ashish Luhach, Papua New Guinea University of Technology, Papua New GuineaCopyright © 2022 Chicco and Jurman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Davide Chicco, ZGF2aWRlY2hpY2NvQGRhdmlkZWNoaWNjby5pdA==
†ORCID: Davide Chicco orcid.org/0000-0001-9655-7142
Giuseppe Jurman orcid.org/0000-0002-2705-5728
 Davide Chicco
Davide Chicco Giuseppe Jurman
Giuseppe Jurman