- 1Department of Cardiology, Fourth Affiliated Hospital of China Medical University, Shenyang, China
- 2Key Laboratory of Liaoning Provincial Medicine and Engineering of Cardiovascular Fluid Dynamics, China Medical University, Shenyang, China
- 3Department of Cardiology, North China Medical Treatment Health Group, Fengfeng General Hospital, Handan, China
- 4Department of Intensive Care, North China Medical Treatment Health Group, Fengfeng General Hospital, Handan, China
Objective: This study aims to explore the association of methyltransferase-like protein 3 (METTL3) expression with severity of coronary artery stenosis in patients with coronary heart disease (CHD).
Methods: A total of 100 patients administrated in the Fourth Affiliated Hospital of China Medical University between October 2022 and June 2023 with primary symptoms of chest pain or tightness, or cardiac discomfort, and who underwent coronary angiography for a definitive diagnosis, were included in the study. The baseline characteristics, including TG, TC, LDL-C, HDL-C, uric acid and past history were recorded. Peripheral blood samples were collected to assess the expression levels of METTL3, YT521-B homology domains 1 (YTHDF1), YT521-B homology domains 2 (YTHDF2), and YT521-B homology domains 3 (YTHDF3) using the PCR method. Relative expression levels of METTL3 protein were determined by Western blotting. Correlation analysis were conducted to evaluate the relationship between METTL3/YTHDF1 gene expression and clinical data. Receiver operating characteristic (ROC) curve analysis was employed to assess the predictive value of METTL3 and YTHDF1 for CHD. Binary logistic regression was used to determine whether the expression of METTL3 and YTHDF1 in peripheral blood were risk factors for CHD.
Results: The study found no significant differences in baseline characteristics between CHD patients and controls, except for length of stay, Lymphocytes, Neutrophils, AST, HDL-C and modified Gensini score. The gene expression levels of METTL3 and YTHDF1 were significantly higher in CHD patients compared to controls (p < 0.05). Furthermore, METTL3 protein expression was also significantly elevated in the CHD group compared to the control group (p < 0.05). METTL3 gene expression correlated with HDL-C and Gensini score, while YTHDF1 gene expression correlated with Age, WBC, Neutrophils, RDW-CV, modified Gensini score. ROC curve analysis demonstrated an AUC of 0.692 for METTL3 in CHD, with a sensitivity of 66.7% and a specificity of 69.8% at a cut-off value of >0.052. The AUC for YTHDF1 in CHD was 0.623, with a sensitivity of 47.4% and a specificity of 74.4% at a cut-off value of >0.027. Binary logistic regression revealed that only increased METTL3 expression in peripheral blood was an independent risk factor for CHD (p < 0.05).
Conclusions: The increased expression of METTL3 in peripheral blood may serve as a potential biomarker and predictive factor for CHD.
1 Introduction
Coronary heart disease (CHD) is a prevalent cardiovascular disease associated with high morbidity and mortality, however, its pathogenesis is still not fully understood (1, 2). Therefore, exploring the molecular mechanisms underlying CHD pathogenesis is crucial for the development of effective diagnostic and therapeutic strategies (3). However, the current non-invasive methods used to diagnose CHD have low sensitivity and specificity, while coronary angiography, the gold standard for the diagnosis of CHD, is invasive (4). Consequently, the identification of specific biomarkers is essential for the early diagnosis of CHD (5).
N6-methyladenosine (m6A) modification is a common epigenetic modification in eukaryotes. The m6A modification is regulated by a group of enzymes, including the “writers” methyltransferase complex responsible for N6-adenylate methylation, the “erasers” demethyltransferases that remove N6-adenylate methylation, and the “readers” m6A RNA-binding proteins that are responsible for the biological function (6). Among them, METTL3 and methyltransferase-like 14 (METTL14) serve as key methyltransferases, fat mass and obesity-associated protein (FTO) and alkylation repair homolog protein 5 (ALKBH5) mainly function as demethyltransferases, while YTH protein family including YTHDF1, YTHDF2, and YTHDF3 are RNA-binding proteins which are capable of recognizing m6A modifications and control RNA fate (7). Studies have shown that abnormal expression of key m6A regulatory factors could be used as diagnostic and therapeutic targets in various diseases (7–9). For instance, Guo found a positive correlation between METTL14 levels and inflammatory markers, establishing it as an independent predictor of CHD risk (10). However, the correlation between the levels of m6A regulatory proteins in peripheral blood of patients with coronary artery stenosis severity remains unclear.
In this research, we measured the level of METTL3, YTHDF1, YTHDF2, and YTHDF3 in the peripheral blood of patients with CHD. The study revealed that the expression of METTL3 and YTHDF1 were increased in the CHD group. Furthermore, the expression of METTL3 and YTHDF1 was significantly correlated with various clinical parameters including HDL-C, Gensini score, Age, WBC, neutrophil, and RDW-CV. The Receiver Operating Characteristic (ROC) curve demonstrated that METTL3 had a strong predictive value in distinguishing the CHD group from the control group. Binary Logistic regression analysis indicated that increased METTL3 expression is an independent risk factor for CHD. These findings may provide new insights into new diagnostic biomarker for CHD.
2 Materials and methods
2.1 Patients enrollment
A total of 100 consecutive patients who administrated in the Cardiology department of 4th Affiliated Hospital of China Medical University between October 2022 and June 2023 with primary symptoms of chest pain or tightness, or cardiac discomfort, and who underwent coronary angiography for a definitive diagnosis, were included in this study. This study was approved by the hospital Ethics Committee, and all participants provided written informed consent.
Based on the results of coronary angiography, the patients were categorized into CHD group and control group. The inclusion criteria for the CHD group included the presence of more than 50% stenosis in at least one of the coronary arteries (left main, left circumflex, left anterior descending, or right coronary artery). The control group consisted of patients with no more than 50% coronary artery stenosis or only myocardial bridge changes. Patients who had undergone coronary stent implantation and coronary artery bypass grafting were excluded, as well as those with hematological diseases, tumors, drug abuse, pregnant, precancerous lesions, severe hepatic and renal insufficiency, active bleeding of various causes, and chronic obstructive pulmonary disease.
2.2 Peripheral blood samples collection and double-stranded DNA (dsDNA) extraction
In this study, trained staff collected all subject laboratory indicators. 6 ml of peripheral venous blood was obtained from the patient prior to coronary angiography. 5 ml of the blood sample was sent to the hospital laboratory for blood routine test, blood glucose, blood lipids, uric acid, and other indicators. The remaining 1 ml of venous blood was collected in an anticoagulant tube containing EDTA for dsDNA extraction. The extraction of Peripheral Blood dsDNA was carried out using the FastPure® Blood DNA Isolation Mini Kit V2 (Vazyme, Nanjing, China) following the manufacturer's protocol. The concentration and purity of the dsDNA were assessed using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, USA).
2.3 Polymerase chain reaction (PCR)
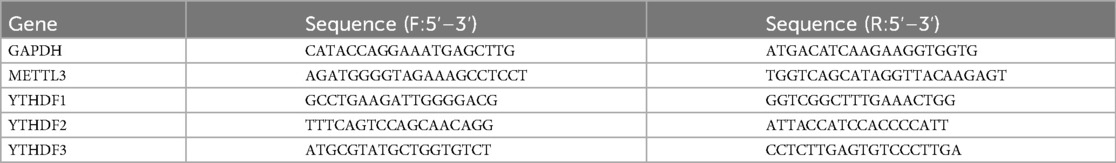
PCR was conducted using the Applied Biosystems QuantStudio 3 (Thermo Fisher Scientific, USA) using ChamQ Universal SYBR qPCR master mix (Vazyme, Q711-02). The primers for METTL3, YTHDF1, YTHDF2, YTHDF3, and GAPDH can be found in Table 1. All PCR experiments were conducted in triplicate. The relative expression of the aforementioned genes was calculated using the 2−ΔCt method (11).
2.4 Protein extraction from whole blood and western blot (WB) analysis
Whole blood proteins were extracted from the collected blood samples using the Solarbio Whole Blood Protein Extraction Kit, following the manufacturer's instructions. An appropriate volume of whole blood was mixed with the provided lysis buffer to ensure complete cell lysis. The mixture was then incubated on ice for 30 min to facilitate protein solubilization. After centrifugation at 14,000 × g for 10 min at 4°C, the supernatant containing the extracted proteins was collected and stored at −80°C for further analysis.
For Western blotting, the extracted whole blood proteins were quantified using the BCA protein assay. Equal amounts of protein (typically 30 µg) were loaded onto a 10% SDS-PAGE gel and separated by electrophoresis. The proteins were subsequently transferred to a PVDF membrane. The membrane was incubated overnight at 4°C with a primary antibody against METTL3 (Cell Signaling Technology, USA), diluted at a ratio of 1:1,000. After washing, the membrane was incubated with an appropriate secondary antibody for 1 h, followed by additional washing steps. Finally, protein bands were detected using enhanced chemiluminescence (ECL), quantified with ImageJ software, and assembled into figures using Adobe Photoshop 2022. Statistical analysis and graphing were performed using GraphPad Prism 8.0.2.
2.5 Modified Gensini score calculation
According to the results of quantitative coronary angiography, we utilized the modified Gensini Score to assess the severity of coronary artery. The modified Gensini Score assigns different scores based on the degree of stenosis of the coronary artery (1 for 0%–25% stenosis, 2 for 26%–50%, 4 for 51%–75%, 8 for 76%–90%, 16 for 91%–99%, and 32 for 100%). If the lesion was ≥99%, the score was adjusted based on the presence of collateral supplying arteries and the degree of narrowing of the supplying arteries. Additionally, different multiplier factors were assigned based on the specific positions of the coronary artery tree where the lesions were located (5 for the left main coronary artery, 2.5 for the proximal left anterior descending branch and the proximal left circumflex branch, 1.5 for the midsegment of left anterior descending artery, 0.5 for the second diagonal branch and the posterolateral branch, and 1 for other branches) (12, 13). The modified Gensini score for each enrolled patient was the total of all lesion severity scores. The higher the score is, the more severe the coronary is.
2.6 Statistical analysis
All statistical analyses were performed using GraphPad Prism v8.0 software (GraphPad Software) and SPSS version 26.0 software (SPSS Inc.). For measurement data, we initially utilized the Kolmogorov-Smirnov test to assess the data distribution. Data that followed a normal distribution were presented as mean ± standard deviation, and an independent sample T-test was used to compare the two groups of data. Data that did not follow the normal distribution were presented as the median (interquartile range), and the Mann–Whitney U rank sum test was conducted for both groups. For counting data, expressed as [n (%)], the chi-square test was used for comparison between the two groups. Pearson or Spearman correlation analysis was utilized to assess the correlation between variables. ROC curves were performed to evaluate the diagnostic significance of METTL3 and YTHDF1. Binary Logistic regression analysis was employed to assess the risk factor. A P value < 0.05 was considered statistically significant.
3 Results
3.1 The baseline characteristics comparison between the two groups
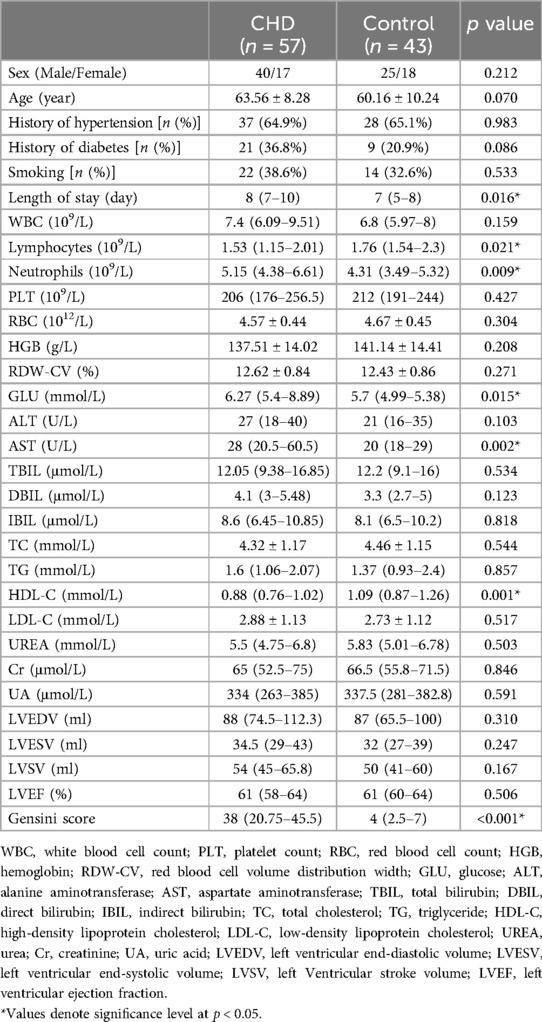
This study included a total of 100 patients, 57 patients with CHD and 43 controls. The baseline characteristics of the two groups are outlined in Table 2. No significant differences were found between the two groups in terms of age, sex, history of hypertension, history of diabetes, smoking, WBC, PLT, RBC, HGB, RDW-CV, ALT, TBIL, DBIL, IBIL, TC, TG, LDL-C, UREA, Cr, UA, LVEDV, LVESV, LVSV, LVEF. However, there were statistically significant differences between the two groups in length of hospital stay, Lymphocytes, Neutrophils, AST, HDL-C and Gensini score.
3.2 Differential expression levels of METTL3, YTHDF1, YTHDF2, and YTHDF3 genes and METTL3 protein in peripheral blood of CHD patients compared to controls
To compare the levels of METTL3, YTHDF1, YTHDF2, and YTHDF3 in patients with CHD and the control group, peripheral blood samples were collected for expression analysis using PCR. As depicted in Figures 1A–D, the gene expression levels of METTL3 and YTHDF1 in the peripheral blood of CHD patients were significantly higher than those in the control group (p < 0.05), whereas the levels of YTHDF2 and YTHDF3 did not differ significantly (p > 0.05). Subsequently, we conducted a WB experiment on the blood samples. As shown in Figures 1E,F, the expression of METTL3 protein was significantly elevated in the CHD group compared to the control group (p < 0.05).
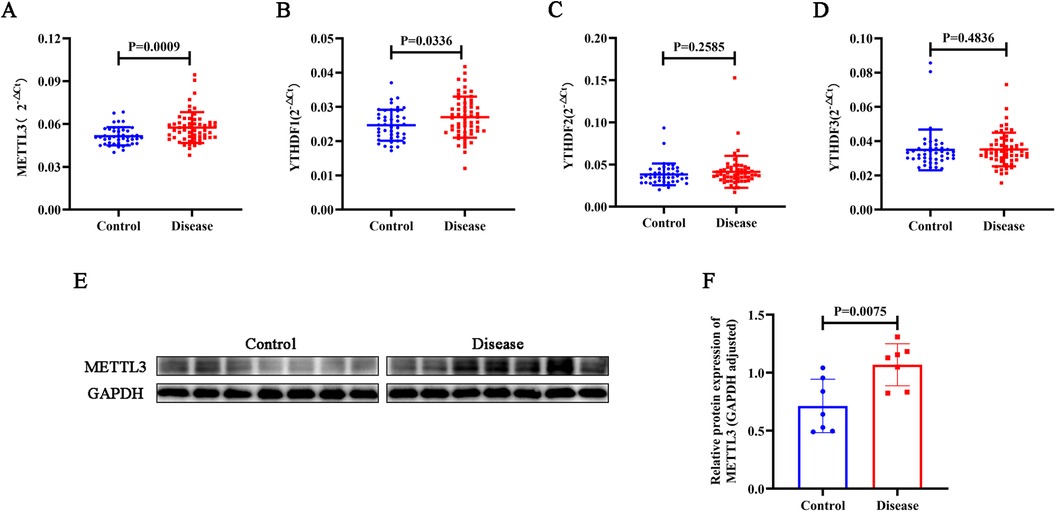
Figure 1. Using PCR, the gene expression levels of METTL3 (A), YTHDF1 (B), YTHDF2 (C), and YTHDF3 (D) were assessed in peripheral blood samples from CHD patients and controls. (E–F) The protein expression levels of METTL3 were assessed via WB analysis in CHD patients and controls, with GAPDH protein serving as an internal control.
3.3 Correlation between METTL3 and YTHDF1 in peripheral blood and clinical data in all patients
The correlation analysis was conducted to assess the relationship between the clinical data (Age, Length of stay, WBC, Lymphocytes, Neutrophils, PLT, RBC, HGB, RDW-CV, ALT, TBIL, DBIL, IBIL, TC, TG, HDL-C, LDL-C, UREA, Cr, UA, LVEDV, LVESV, LVSV, LVEF, Gensini score) and the expression of peripheral blood METTL3 and YTHDF1. As shown in Table 3 and Figure 2, it demonstrated that the expression of METTL3 in peripheral blood displayed negative correlation with HDL-C (r = −0.217, p = 0.031) and positive correlation with modified Gensini score (r = 0.320, p = 0.001). Additionally, the expression of YTHDF1 in peripheral blood was significant associated with Age (r = 0.210, p = 0.036), WBC (r = −0.234, p = 0.019), Neutrophils (r = −0.209, p = 0.037), RDW-CV (r = 0.201, p = 0.045), Gensini score (r = 0.235, p = 0.018). However, there was no correlation between the expression of METTL3 and YTHDF1 with other clinical data.
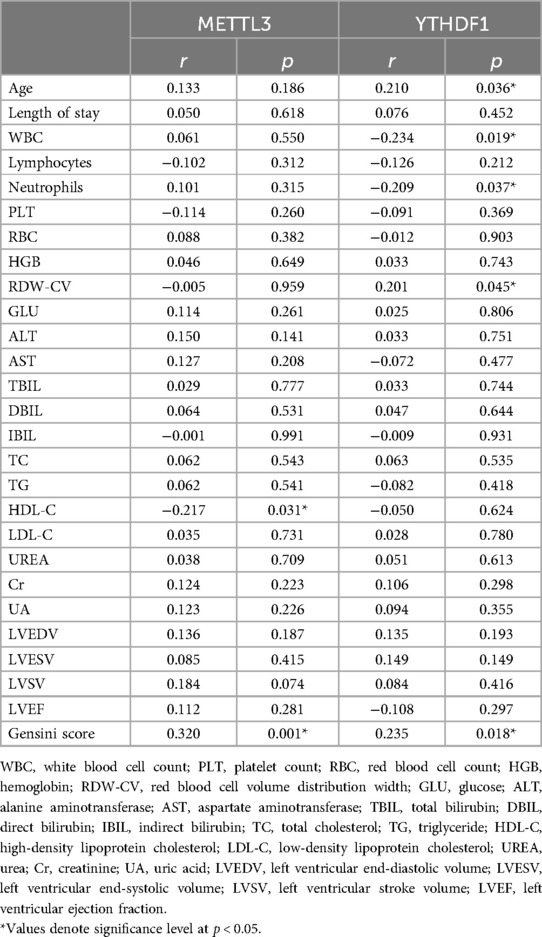
Table 3. Correlation between METTL3 and YTHDF1 in peripheral blood and clinical baseline characteristics.
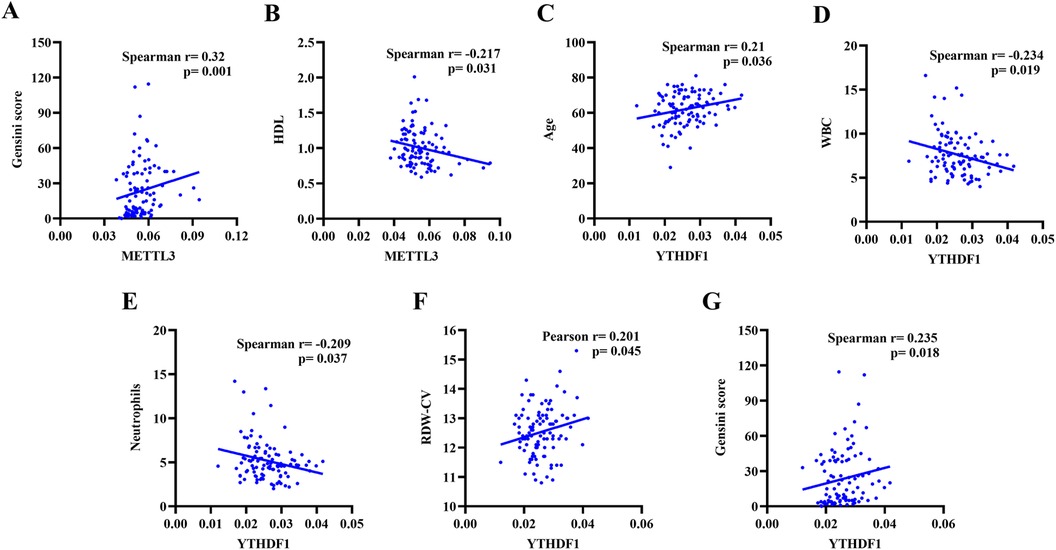
Figure 2. Correlation analysis of METTL3 gene expression level with Gensini score (A), HDL (B), and Age (C). Correlation analysis of YTHDF1 gene expression level with WBC (D), Neutrophils (E), RDW-CV (F), and Gensini score (G).
3.4 Diagnostic predictive value of METTL3 and YTHDF1 for CHD
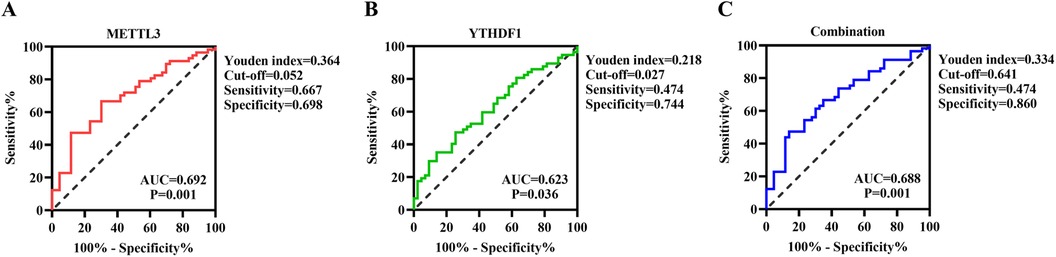
Subsequently, we conducted a ROC curve analysis to investigate the potential predictive value of METTL3 and YTHDF1 in peripheral blood for CHD. The results showed that the AUC of METTL3 for distinguishing CHD from Control was 0.692, with a cutoff value of >0.052, a sensitivity of 66.7%, and a specificity of 69.8% (Figure 3A). The AUC of YTHDF1 was 0.623, with a cutoff value of >0.027, a sensitivity of 47.4%, and a specificity of 74.4% (Figure 3B). Furthermore, the AUC of the combination of METTL3 and YTHDF1 for distinguishing CHD from Control was 0.688, with a sensitivity of 47.4% and a specificity of 86% (Figure 3C). It is noteworthy that the combination of METTL3 and YTHDF1 (0.688) did not show improvement in predicting patients with CHD compared to the sole use of METTL3 (0.692).
3.5 The increased expression of METTL3 was a risk factor for CHD
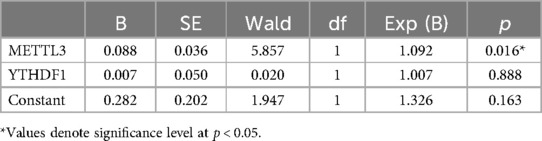
The previous results revealed that the increased expression of METTL3 and YTHDF1 in peripheral blood were associated with severity of coronary artery. To further investigate the association between the expression of METTL3 and YTHDF1 in peripheral blood and the risk of CHD, binary logistic regression was employed. The results, as shown in Table 4, yielded the equation Y = 0.282 + 0.088 X1(METTL3*1000) + 0.007 X2 (YTHDF1*1000), indicating that only the increased expression of METTL3 in peripheral blood was an independent risk factor for CHD (p < 0.05).
4 Discussion
M6A modification is a common and important type of RNA modification regulated by m6A methylation regulators (14). Dysregulation of RNA m6A modification is increasingly recognized as a contributing factor in many diseases, including cardiovascular diseases (15–17). Our study found that the expression levels of the METTL3 gene and protein in peripheral blood were significantly increased in patients with CHD, and this expression was positively associated with the severity of coronary artery disease. Furthermore, this paper revealed that METTL3 is an independent risk factor for CHD, providing valuable insights into its potential as a biomarker and risk factor.
Patients in the CHD group showed increased expression of METTL3 and YTHDF1 in peripheral blood, indicating a potential association between m6A modification and CHD. Li et al. found that the expression of METTL3 in macrophages increased with the progression of atherosclerosis and METTL3 targets BRAF mRNA and promoted its translation through YTHDF1, exacerbating macrophage-mediated inflammatory responses and atherosclerotic progression (18). Additionally, Chien et al. found that METTL3 in endothelial cells mediated inflammatory responses under pro-atherosclerotic flow treatment, including secretion of inflammatory cytokines, activation of the NF-κB signaling pathway, and adhesion of monocytes to endothelial cells, exacerbating the development of atherosclerosis (19). On the other hand, Li et al. found that METTL3 mitigates the progression of atherosclerosis by regulating m6A-dependent EGFR mRNA stability (20). While the roles of METTL3 in the above results are not entirely consistent, all findings indicate a connection between dysregulated m6A modification and the pathogenesis of CHD.
In this study, we explored the associations between the expression of METTL3 and YTHDF1 and clinical parameters. Specifically, we observed a positive correlation between the level of METTL3 and modified Gensini score, and a negative correlation with HDL-C levels. These findings imply that METTL3 could potentially serve as a risk factor for CHD. Additionally, YTHDF1 expression was positively correlated with age, RDW-CV, and modified Gensini score, and negatively correlated with WBC and Neutrophil levels. These correlations highlight the complex interactions between m6A modification regulatory molecules and various clinical parameters, providing evidence for the potential impact of RNA m6A modification on the pathological and physiological processes of CHD.
The ROC curve analysis demonstrated the potential predictive value of METTL3 and YTHDF1 for CHD, with the AUC for METTL3 being 0.692 and for YTHDF1 being 0.688. However, the combination of METTL3 and YTHDF1 did not significantly improve predictive capability compared to the use of METTL3 alone. These findings underscore the potential utility of METTL3 as a biomarker for CHD. Building on these results, we further evaluated the expression levels of METTL3 protein in blood samples through WB analysis. The results showed that METTL3 protein expression levels were significantly elevated in the CHD group compared to the control group. Moreover, the binary logistic regression analysis revealed that only the increased expression of METTL3 in peripheral blood was identified as an independent risk factor for CHD. This observation highlights the potential significance of METTL3 as a risk factor for CHD and suggests its potential ability in risk stratification and prognostication for patients with CHD.
Although our study has produced some findings, it also has limitations that need to be acknowledged. Firstly, the research is based on a small sample from a single center, and some important information in the medical records—such as family history of premature CHD, inflammatory factors (CRP, IL-6, etc.), homocysteine levels, and drug therapy information—is incomplete. Secondly, our research may be subject to certain information biases, such as selection bias. Additionally, external factors, such as variations in treatment regimens and differences in patient populations, may have influenced our study. Therefore, in the future, it is necessary to validate the diagnostic and prognostic utility of METTL3 and YTHDF1 in larger samples and to explore their roles in the pathogenesis of CHD.
In conclusion, this study found that the expression level of METTL3 in peripheral blood is significantly increased in CHD patients and its expression level is positive associated with the severity of coronary artery. Furthermore, METTL3 is an independent risk factor of CHD, which providing valuable insights into its potential as a biomarker and risk factor for CHD.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by Ethics Committee of the Fourth Affiliated Hospital of China Medical University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
JC: Data curation, Formal Analysis, Funding acquisition, Investigation, Resources, Software, Validation, Visualization, Writing – original draft. RS: Data curation, Formal Analysis, Funding acquisition, Software, Visualization, Writing – original draft. XX: Conceptualization, Funding acquisition, Methodology, Resources, Supervision, Validation, Writing – review & editing. YJ: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded by the Natural Science Foundation of Liaoning Province (Approval No: 2022-MS-225).
Acknowledgment
We are grateful to all the members of the laboratory for their assistance and support in this research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Roth GA, Mensah GA, Johnson CO, Addolorato G, Ammirati E, Baddour LM, et al. Global burden of cardiovascular diseases and risk factors, 1990–2019: update from the GBD 2019 study. J Am Coll Cardiol. (2020) 76(25):2982–3021. doi: 10.1016/j.jacc.2020.11.010
2. Stone PH, Libby P, Boden WE. Fundamental pathobiology of coronary atherosclerosis and clinical implications for chronic ischemic heart disease management-the plaque hypothesis: a narrative review. JAMA Cardiol. (2023) 8(2):192–201. doi: 10.1001/jamacardio.2022.3926
3. Perrotta I. Atherosclerosis: from molecular biology to therapeutic perspective. Int J Mol Sci. (2022) 23(7):3444. doi: 10.3390/ijms23073444
4. Timmis A, Vardas P, Townsend N, Torbica A, Katus H, De Smedt D, et al. European society of cardiology: cardiovascular disease statistics 2021. Eur Heart J. (2022) 43(8):716–99. doi: 10.1093/eurheartj/ehab892
5. Nayor M, Brown KJ, Vasan RS. The molecular basis of predicting atherosclerotic cardiovascular disease risk. Circ Res. (2021) 128(2):287–303. doi: 10.1161/CIRCRESAHA.120.315890
6. Sendinc E, Shi Y. RNA M6a methylation across the transcriptome. Mol Cell. (2023) 83(3):428–41. doi: 10.1016/j.molcel.2023.01.006
7. Liu Y, Yang D, Liu T, Chen J, Yu J, Yi P. N6-methyladenosine-mediated gene regulation and therapeutic implications. Trends Mol Med. (2023) 29(6):454–67. doi: 10.1016/j.molmed.2023.03.005
8. He PC, He C. M(6) A RNA methylation: from mechanisms to therapeutic potential. EMBO J. (2021) 40(3):e105977. doi: 10.15252/embj.2020105977
9. Liu S, Chen L, Zhang Y, Zhou Y, He Y, Chen Z, et al. M6AREG: m6A-centered regulation of disease development and drug response. Nucleic Acids Res. (2023) 51(D1):D1333–D44. doi: 10.1093/nar/gkac801
10. Guo F, He M, Hu B, Li G. Levels and clinical significance of the m6A methyltransferase METTL14 in patients with coronary heart disease. Front Cardiovasc Med. (2023) 10:1167132. doi: 10.3389/fcvm.2023.1167132
11. Zhang F, Wu L, Qian J, Qu B, Xia S, La T, et al. Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. J Autoimmun. (2016) 75:96–104. doi: 10.1016/j.jaut.2016.07.012
12. Mazzotta C, Basu S, Gower AC, Karki S, Farb MG, Sroczynski E, et al. Perivascular adipose tissue inflammation in ischemic heart disease. Arterioscler Thromb Vasc Biol. (2021) 41(3):1239–50. doi: 10.1161/ATVBAHA.120.315865
13. Rampidis GP, Benetos G, Benz DC, Giannopoulos AA, Buechel RR. A guide for Gensini Score calculation. Atherosclerosis. (2019) 287:181–3. doi: 10.1016/j.atherosclerosis.2019.05.012
14. Chen YS, Ouyang XP, Yu XH, Novak P, Zhou L, He PP, et al. N6-adenosine methylation (m(6)A) RNA modification: an emerging role in cardiovascular diseases. J Cardiovasc Transl Res. (2021) 14(5):857–72. doi: 10.1007/s12265-021-10108-w
15. Liu C, Gu L, Deng W, Meng Q, Li N, Dai G, et al. N6-methyladenosine RNA methylation in cardiovascular diseases. Front Cardiovasc Med. (2022) 9:887838. doi: 10.3389/fcvm.2022.887838
16. Deng X, Qing Y, Horne D, Huang H, Chen J. The roles and implications of RNA m(6)A modification in cancer. Nat Rev Clin Oncol. (2023) 20(8):507–26. doi: 10.1038/s41571-023-00774-x
17. Fu J, Cui X, Zhang X, Cheng M, Li X, Guo Z, et al. The role of m6A ribonucleic acid modification in the occurrence of atherosclerosis. Front Genet. (2021) 12:733871. doi: 10.3389/fgene.2021.733871
18. Li Q, Yu L, Gao A, Ren R, Zhang J, Cao L, et al. METTL3 (methyltransferase like 3)-dependent N6-methyladenosine modification on braf mRNA promotes macrophage inflammatory response and atherosclerosis in mice. Arterioscler Thromb Vasc Biol. (2023) 43(5):755–73. doi: 10.1161/ATVBAHA.122.318451
19. Chien CS, Li JY, Chien Y, Wang ML, Yarmishyn AA, Tsai PH, et al. METTL3-dependent N(6)-methyladenosine RNA modification mediates the atherogenic inflammatory cascades in vascular endothelium. Proc Natl Acad Sci U S A. (2021) 118(7):e2025070118. doi: 10.1073/pnas.2025070118
Keywords: coronary heart disease, m6A, METTL3, Gensini score, peripheral blood
Citation: Chang J, Shao R, Xu X and Jin Y (2024) Clinical significance of the m6A methyltransferase METTL3 in peripheral blood of patients with coronary heart disease. Front. Cardiovasc. Med. 11:1442098. doi: 10.3389/fcvm.2024.1442098
Received: 1 June 2024; Accepted: 10 September 2024;
Published: 20 September 2024.
Edited by:
Guido Krenning, University Medical Center Groningen, NetherlandsCopyright: © 2024 Chang, Shao, Xu and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangshan Xu, eHN4dUBjbXUuZWR1LmNu; Yuanzhe Jin, eXpqaW5AY211LmVkdS5jbg==
†These authors share first authorship
 Jianshe Chang
Jianshe Chang Rui Shao4,†
Rui Shao4,† Xiangshan Xu
Xiangshan Xu Yuanzhe Jin
Yuanzhe Jin