
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cardiovasc. Med. , 04 June 2024
Sec. Aortic Surgery and Endovascular Repair Archive
Volume 11 - 2024 | https://doi.org/10.3389/fcvm.2024.1394889
This article is part of the Research Topic Decoding Epigenetics in Vascular Health: From Basic Science to Therapeutics View all articles
 YuChen Liu1,†
YuChen Liu1,† XiaoYun Sun1,†
XiaoYun Sun1,† Zhen Gou1,†
Zhen Gou1,† ZhenKun Deng1
ZhenKun Deng1 YunRui Zhang1
YunRui Zhang1 PingPing Zhao1
PingPing Zhao1 Wei Sun1
Wei Sun1 Yang Bai1*
Yang Bai1* YuChen Jing2*
YuChen Jing2*
Abdominal Aortic Aneurysm (AAA) is a disease characterized by localized dilation of the abdominal aorta, involving multiple factors in its occurrence and development, ultimately leading to vessel rupture and severe bleeding. AAA has a high mortality rate, and there is a lack of targeted therapeutic drugs. Epigenetic regulation plays a crucial role in AAA, and the treatment of AAA in the epigenetic field may involve a series of related genes and pathways. Abnormal expression of these genes may be a key factor in the occurrence of the disease and could potentially serve as promising therapeutic targets. Understanding the epigenetic regulation of AAA is of significant importance in revealing the mechanisms underlying the disease and identifying new therapeutic targets. This knowledge can contribute to offering AAA patients better clinical treatment options beyond surgery. This review systematically explores various aspects of epigenetic regulation in AAA, including DNA methylation, histone modification, non-coding RNA, and RNA modification. The analysis of the roles of these regulatory mechanisms, along with the identification of relevant genes and pathways associated with AAA, is discussed comprehensively. Additionally, a comprehensive discussion is provided on existing treatment strategies and prospects for epigenetics-based treatments, offering insights for future clinical interventions.
Abdominal Aortic Aneurysm (AAA) are aneurysm-like bulbous protrusions that develop on the wall of the abdominal aorta, patients with AAA typically do not exhibit significant symptoms in the early stages of the disease. There may be mild pulsation or pressure sensations, but these symptoms are not pronounced. It is only when the aneurysm reaches a certain ruptured size that the rupture of the abdominal aorta becomes the patient's sole apparent symptom, leading to life-threatening abdominal hemorrhage (1, 2). Annually, at least 150,000 people die from AAA, with the majority of deaths attributed to aneurysm rupture, carrying a high mortality rate of approximately 70%–80% after rupture (3, 4).
Under normal circumstances, the aorta of a healthy individual is elastic. However, due to the combined effects of genetic, environmental, and various complex factors, the vascular wall in the abdominal aortic region may weaken. In such cases, the gradual force of blood against the damaged vascular wall initiates the formation of AAA. As the disease progresses, the arterial wall becomes weak and swollen, unable to withstand the stress of blood flow within the artery, ultimately leading to the rupture of the AAA (2, 5). Clinical treatment for AAA generally involves surgical methods, with Open Aneurysm Repair (OAR) or Endovascular Aneurysm Repair (EVAR) being commonly used. OAR is suitable for patients with a longer life expectancy and lower morbidity rates. Although EVAR has, to some extent, reduced short-term mortality after AAA repair, operated patients experience an increased burden on the aortic vasculature, leading to an elevated risk of death (6). However, surgery has its drawbacks, as it requires complex screening and risk assessment before AAA surgery. Postoperatively, there may be instances of endoleaks, and the long-term care entails significant economic costs (7). Therefore, we may need to find a more reliable or universal treatment than surgery for AAA. An alternative treatment method is pharmaceutical intervention. Risk factors for AAA in diagnostic screening include hypertension, atherosclerosis, diabetes, etc, medications targeting these risk factors related diseases may help slow the progression of AAA, reducing the risk of rupture. In light of these information, the results of preclinical studies in animal models suggest that statins, antihypertensive drugs, doxycycline, metformin, and others have delaying effect on AAA progression (8–10). However, the three main AAA models currently available are elastase, CaCl2, and angiotensin II (AngII)/apolipoprotein E (AapoE)-deficient mouse models, each of which has defects that may not translate the findings to human AAA, and there is currently insufficient evidence from multiple clinical drug trials to conclusively prove the complete efficacy of these drugs in treatment (11, 12). At the same time, these drugs may have side effects in the treatment of AAA, and may have contraindications, for example, statins can cause related muscle symptoms (13), and β-blockers in antihypertensive drugs may reduce cardiac output and vasodilation (14), which indicate that the search for a new treatment option is essential.
Over the past few decades, with the accumulation of molecular biology knowledge and the rapid development of molecular biology techniques, epigenetics, as a significant branch of genetics, has gradually gained widespread attention and in-depth research. Its role in physiological and pathological conditions has sparked a keen interest in personalized medicine (15). Research in the cardiovascular field indicates that the three major epigenetic modifications—DNA methylation, histone modification, and non-coding RNA modification—may play a crucial role in the occurrence and development of cardiovascular diseases (16). Recent studies suggest a correlation between epigenetics and the pathogenesis of cardiovascular diseases and AAA (17–19), including the role of DNA methyltransferases, the modification of histones, the mechanisms of non-coding RNA and RNA modification, which will be described in detail below. The progress of AAA is a complex and multi-layered issue, involving inflammation, matrix metalloproteinase (MMP) activation, oxidative stress, intraluminal thrombosis, smooth muscle apoptosis, and extracellular matrix (ECM) changes, among others, the characteristics of AAA described above may be closely related to epigenetic regulatory outcomes (1, 20). By elucidating the current research on epigenetics related to AAA and discussing the potential involvement of epigenetic regulation in AAA progression, rupture, and repair, this paper aims to provide insights and perspectives for identifying new therapeutic targets.
This section focuses on the mechanisms of four epigenetic modifications in AAA, including DNA methylation, histone modification, non-coding RNA action, and RNA modification At present, DNA methylation, histone modification, non-coding RNA modification and RNA modifications, are commonly studied epigenetic mechanisms in AAA. The mechanisms have been briefly visualized in figure and detailed at the beginning of each part. However, epigenetic mechanisms also encompass other types that have not been extensively explored in AAA, and their roles in diseases are yet to be fully elucidated. Examples include chromatin remodeling and other mechanisms Figure 1.

Figure 1. Abstract figure. Current research on epigenetics in AAA focuses on DNA methylation, histone modification, non-coding RNA action, and RNA modification.
DNA methylation in cardiovascular diseases is gaining attention; however, its role in AAA has not been comprehensively studied. It is well known that smoking is a significant risk factor for AAA, and smoking has a crucial impact on the growth and rupture of AAA (21). As mentioned earlier, DNA methylation is controlled by DNMTs. Nicotine itself has been shown to not only affect the expression of DNMT1 but also influence promoter methylation levels in GABA-ergic neurons (22). Atherosclerosis is a complex pathological process involving vascular wall cells and inflammatory cells. As one of the risk factors for AAA, the mechanisms of atherosclerosis may be correlated with AAA. Endothelial dysfunction is the pathological basis of atherosclerosis and is accompanied by changes in vascular wall permeability. Its pathological process is closely related to abnormal DNA methylation (23). Other risk factors for AAA, such as hypertension and diabetes, may also involve similar mechanisms in AAA, but further correlational research is needed Figure 2.
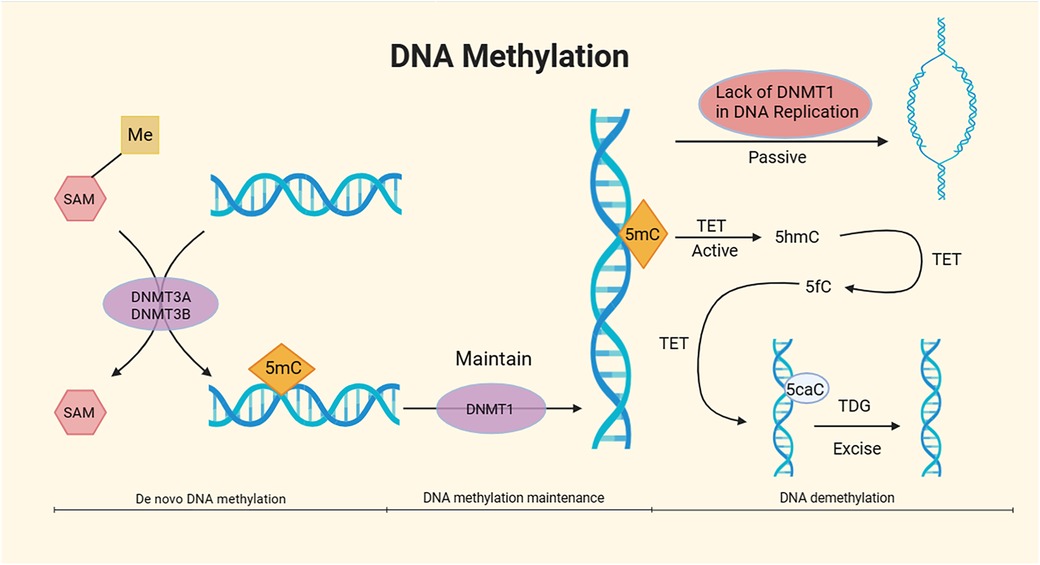
Figure 2. Mechanism of DNA methylation. DNA methylation, under the catalytic action of DNA methyltransferases (DNMTs) using S-adenosyl methionine (SAM) as the methyl donor, involves the covalent binding of a methyl group to specific DNA base positions, resulting in the formation of 5-methylcytosine (5mC) at the fifth carbon of cytosine. The overall process of DNA methylation is dynamic and can be divided into three stages: de novo DNA methylation, DNA methylation maintenance, and DNA demethylation. The DNMT3A and DNMT3B subtypes in DNMTs are responsible for de novo methylation of 5mC, while the DNMT1 subtype maintains methylation (24, 25). DNA demethylation mainly comprises passive demethylation and active demethylation. In the process of passive DNA demethylation, the absence of DNMT1 due to multiple DNA replications prevents the maintenance of DNA methylation (26). In active DNA demethylation, active DNA demethylation involves the Ten-Eleven translocation (TET) enzymes, which oxidize 5mC to 5-hydroxymethylcytosine (5hmC). TET further oxidizes 5hmC to 5-formylcytosine (5fC). Under the action of TET, 5fC is then oxidized to 5-carboxylcytosine (5caC), which is subsequently excised by thymine DNA glycosylase (TDG) in the base excision repair (BER) pathway (27, 28).
Early studies have demonstrated a significant elevation in plasma homocysteine (Hcy) levels in AAA patients, and hyperhomocysteinemia (HHcy) is associated with the expansion rate of AAA (29). AAt the same time, a study's findings indicate that Hcy does not alter the protein levels of DNMT1 and DNMT3 but selectively reduces the activity of DNMT1 by 29% (30). Elevated Hcy levels may promote the development of AAA through multiple interrelated mechanisms, including endothelial dysfunction, protein hydrolysis, endoplasmic reticulum stress, enhanced inflammation, and increased cell apoptosis (31), which may indicate that HHcy in AAA may play a crucial role in AAA progression by reducing DNA methylation. Although research has shown an association between Hcy level and the growth rate of AAA, the conclusions drawn are relatively weak due to AAA being a multifactorial disease. However, a studies have indicated that patients with HHcy exhibit a faster expansion rate compared to those with normal Hcy levels, with a considerable number of patients demonstrating rapid expansion (>10 mm/year), thereby increasing the risk of rupture (32). Another study aimed at exploring the direct causal relationship between AAA and HHcy also suggests that HHcy may exacerbate AAA formation at least partially through the activation of peripheral fibroblast NADPH oxidase 4 (33). These two studies further confirm the high association of AAA with HHcy. A cohort study by Kristina Sundquist et al. provides an alternative perspective by confirming overall high methylation in AAA cases and concluding a significant correlation between overall DNA methylation, Hcy, and the baseline diameter of AAA (34). However, this study did not establish a correlation between overall DNA methylation and plasma Hcy levels. In the future, further studies may be carried out to further explore the specific mechanism of HHcy in promoting the growth and rupture of AAA by mediating DNA methylation.
Moreover, the diminished inhibitory effect of regulatory T cells (Tregs) may contribute to the progression of AAA, which is closely related to DNA methylation. Research suggests a reduced expression of FOXP3 (a transcriptional regulatory factor) in peripheral CD4CD25 Tregs of AAA patients, leading to functional deficiencies in overall CD4CD25 Tregs, indicating impaired immune regulation by Tregs, which may contribute to the disease progression of AAA (35). Another study confirms significantly higher DNA methylation levels in Tregs of AAA patients compared to healthy subjects, indicating a transcriptionally suppressed state affecting cellular functions (36). Furthermore, changes in DNA methylation have been shown to impact the expression of various genes related to vascular smooth muscle cell (VSMC) apoptosis, inflammation, and ECM degradation. This imbalance may be associated with the dysregulation of MMPs and TIMPs, closely linking them to AAA (17). These findings warrant further research for a comprehensive understanding of the mechanistic role of DNA methylation in AAA.
In addition, there have been some exciting advances in genome-wide DNA methylation. Evan J. Ryer et al. determined the genome-wide DNA methylation in peripheral blood mononuclear cells (PBMCs) of abdominal aortic aneurysm (AAA) patients, discovering significant differences in DNA methylation at specific CpG islands (CGIs). However, the observed differential methylation was noted to be potentially age-related rather than AAA-related (37). Additionally, the correlation between DNA methylation and gene silencing increases with the density of CpG dinucleotides at the promoter region (38). The human genome has fewer CpGs, but more CGIs. CGIs, relatively small 5′ end promoter regions, may experience gene suppression due to methylation of the fifth carbon of cytosine in this region (39). Aberrant CGI hypermethylation is common in tumor progression and may lead to abnormal gene silencing (40). Furthermore, the variations in 5mC, 5hmC, TET family proteins, and DNMTs at the genome-wide level differ depending on the type of cancer (24, 41) potentially serving as relevant interrogation sites for DNA methylation. Toghill BJ et al. used next-generation sequencing (NGS) on VSMCs collected from individual aortic tissues, pioneering the determination of CpG methylation status in regulatory regions of genes located at AAA risk loci identified in genome-wide association studies (GWAS) (42). With technological advancements, epigenome-wide association studies (EWAS) have evolved after GWAS, providing a systematic approach to revealing epigenetic variations underlying common diseases. EWAS, applied for a decade to analyze DNA methylation variations in complex diseases, has unveiled new molecular mechanisms for various common diseases, such as rheumatoid arthritis, metabolic syndrome, breast cancer, Alzheimer's disease, etc. It has made the application of epigenetic variations as biomarkers possible (43). This approach may lead to innovative research perspectives on the mechanisms of DNA methylation in AAA, offering molecular-level advancements in understanding the pathogenesis of AAA.
Research on histone acetylation and methylation modifications in abdominal aortic aneurysm (AAA) has been extensive. In the related studies of histone acetylation, Han et al. compared human AAA tissue with healthy aortic tissue, analyzing the differences in histone acetylation and the expression of corresponding HATs. The results showed significant overexpression of three lysine acetyltransferase (KAT) family members in the AAA vascular wall. Some lysine acetyltransferases, such as KAT2B, were found to be correlated with AAA diameter. High expression of KAT2B, KAT3B, and KAT6B was also observed in inflammatory cells (44). Another study found higher levels of H3 and H3K14 acetylation in T lymphocytes of AAA patients (45). However, contrasting results have been reported. As mentioned earlier, HDAC/HAT constitutes a reversible enzyme pair regulating histone acetylation PTM. Galán M et al. found that HDACs corresponding to KAT were significantly upregulated in AAA. In a mouse model, HDAC inhibitors were observed to restrict aneurysm progression. The study suggested that HDAC may promote processes such as ECM degradation, increased inflammatory mediators, and VSMC apoptosis (46). While some studies in mouse models have demonstrated inhibitory effects of HDAC inhibitors on MMP expression, the specific mechanisms require further investigation (47). Qian Xia et al. also identified a decrease in acetylation rates in H3 and H3K9, attributing it to the elevated influence of HDAC1 and HDAC5. HDAC1 and HDAC5 were found to inhibit the transcriptional activity of regulatory T cells (Tregs) (36). Additionally, Jacob Greenway et al. detected downregulation of H3K4me1, H3K9me3, and H3K56ac levels in two animal AAA models. The study revealed dynamic changes in histone H3 modifications during AAA formation, with no observed upregulation in H3 modifications (48). The discrepancies in results may be attributed to the levels of HAT and HDAC expression and their alterations during AAA development. Moreover, differences in modifications at specific sites vs. global modifications, as well as species variations, risk factors, and control tissue, need further in-depth research to address these disparities and determine the processes regulating histone methylation and acetylation patterns in AAA Figure 3.
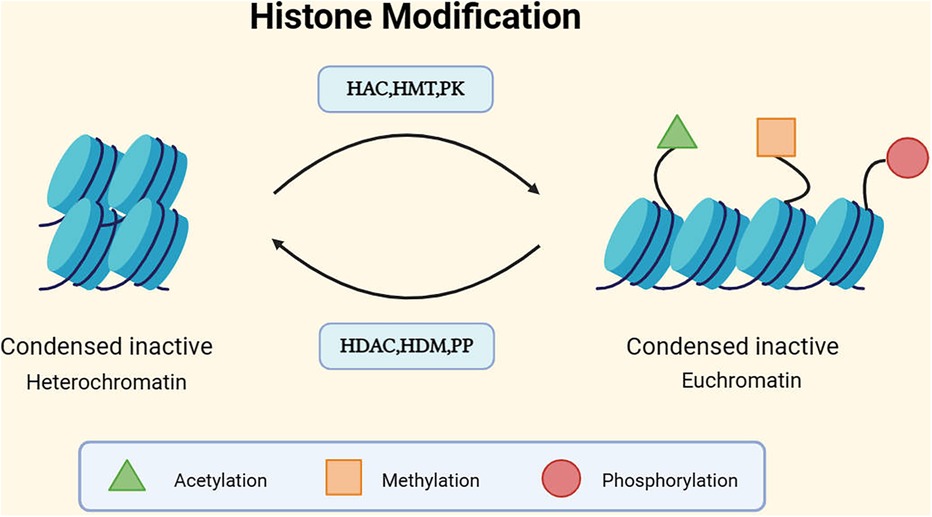
Figure 3. Mechanism of histone modification. DNA is wrapped around a histone octamer to form a nucleosome, and nucleosomes are the basic units of chromatin. The basic units of nucleosomes, after folding, constitute chromatin (49). Generally, there are two different forms of chromatin: euchromatin and heterochromatin, corresponding to the activation and silencing of gene expression, respectively. Euchromatin tends to have a more open conformation, supporting gene transcription, while the dense structure of heterochromatin tends to inhibit gene transcription (50). The structure of chromatin can be modulated through post-translational modifications (PTMs) of histone tails. Currently, the most studied histone covalent modifications include acetylation, methylation, and phosphorylation. In addition to these, there are ubiquitination, sumoylation, ADP-ribosylation, deimination, and proline isomerization, among others (51). Each modification has its corresponding enzymes that act on different histone residues. For instance, histone acetylation involves histone acetyltransferases (HATs), which are typically associated with the more open conformation of euchromatin. The action of histone deacetylases (HDACs) counteracts the effects of HATs and restores the positive charge of lysine side chains. HDAC/HAT constitutes a reversible enzyme pair that regulates histone acetylation PTM (52). Methylation is regulated by histone methyltransferases (HMTs) and demethylases (HDMs). Histone phosphorylation, similar to histone acetylation and methylation, is also a highly dynamic process, protein kinases (PKs) and phosphatases (PPs) respectively add and remove histone modifications, collectively controlling the overall level of modifications (53).
In addition, there are novel findings regarding histone methylation. Research has revealed that histone demethylase JMJD3 stimulates the pro-inflammatory monocyte/macrophage phenotype in AAA tissue by selectively removing inhibitory histone H3K27me3 methylation. This leads to vascular remodeling and aortic dilation, promoting the progression of AAA (54). Another study reported that SETDB2, a histone methyltransferase, specifically trimethylates lysine 9 on histone H3 (H3K9me3), reducing the expression of inhibitory histones. This results in the loss of TIMP expression, causing dysregulation of MMP activity, aortic wall degeneration, and AAA formation (55). Numerous studies have demonstrated that, apart from histone acetylation, methylation, and phosphorylation, histone modifications also include ubiquitination and butyrylation. These modifications not only participate in the progression of metabolic diseases (56), but also impact the tumor microenvironment (57).
Furthermore, histone modifications, particularly histone methylation and acetylation, which have been extensively studied, play a regulatory role in cardiovascular diseases, including AAA risk factors such as atherosclerosis and hypertension (16). These study of histone modifications will play some inspiring roles in the study of the mechanism of AAA disease progression. In addition, histone modifications also include phosphorylation mentioned above and some novel modifications studied in cancer, such as lactatation, ubiquitination, lysine, citrullination, etc. These modifications may be closely related to metabolites (58). Exploring other histone modifications may provide new directions for studying the pathogenesis and treatment of AAA and requires further investigation. On this basis, since nucleosomes formed after DNA binding to histones control the structure of chromatin, the levels of DNA methylation and histone modification can also be expanded, and the microscopic to macroscopic study can be carried out at the level of chromatin remodeling, so as to further explore the disease progression of AAA.
Approximately only 2% of the human genome encodes proteins, while non-coding RNAs (ncRNAs) are RNA molecules that do not encode proteins. In addition to their roles in transcription, ncRNAs may play complex and crucial epigenetic regulatory roles in higher animals. These ncRNAs, often referred to as regulatory RNAs, participate in biological processes such as gene expression regulation and chromatin structure modulation by interacting with DNA, RNA, and proteins (59). This section focuses on the above three ncRNAs that have been studied in AAA so far Figure 4.
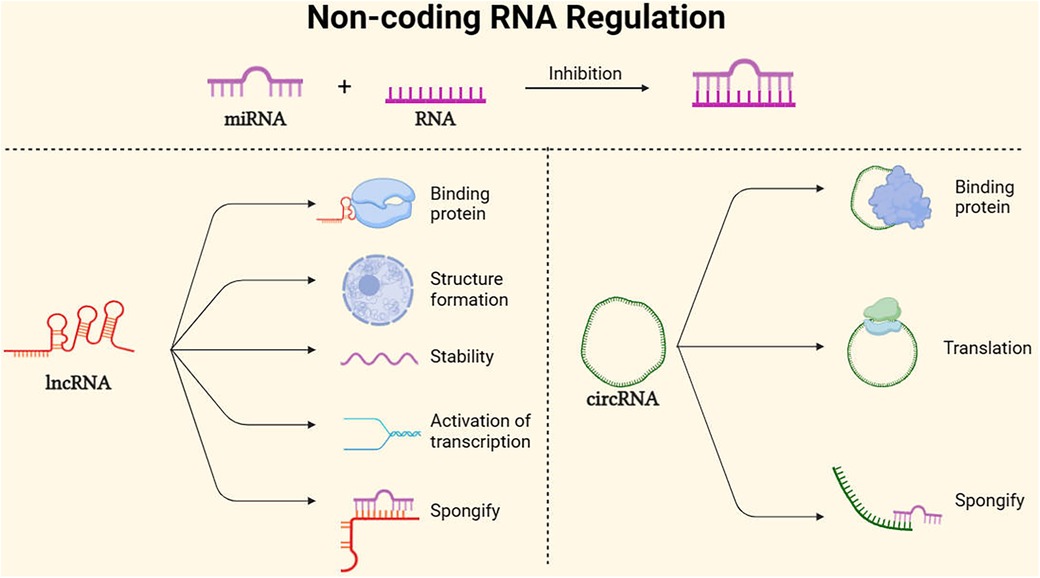
Figure 4. Main non-coding RNA regulation mode. MicroRNAs (miRNAs) are endogenous RNA molecules of approximately 23 nucleotides in length. They guide post-transcriptional expression inhibition of target genes by binding and pairing with the 3′ UTR of protein-coding RNA (mRNA) (60). Long non-coding RNAs (lncRNAs), with lengths exceeding 200 nucleotides and lacking protein-coding capacity, can act as scaffolds to regulate the expression of various genes, promote the formation of nuclear structures, and localize to specific DNA sites, In addition, lncRNAs can maintain mRNA stability, activate translation, sponging mRNA (61). Additionally, circular RNAs (circRNAs), Because of its stability, it can not only serve as a translation template for proteins, but also spongify competition to inhibit mirnas. At the same time, it can bind proteins to regulate gene expression pathways (62).
The types and functions of miRNAs in AAA are diverse, and their regulatory roles vary. The following summary outlines the current understanding of miRNA regulation in AAA based on animal models and in vitro experiments. Some common target genes and regulatory functions may provide insights into the treatment of AAA.
The main function of miRNA in gene regulation is to silence the expression of target genes by binding to mRNA. In contrast, the mechanism of action of lncRNA is more complex, as mentioned earlier, serving as a scaffold to regulate the expression of various genes, promote nuclear structure formation, and localize specific DNA sites, among other functions (61). Although there is limited research on lncRNA in AAA, studies have demonstrated its regulation in cardiovascular diseases (CVD) such as hypertension and atherosclerosis in vascular smooth muscle cells, involving various pathological and physiological changes (63).
H19 is a specific lncRNA associated with AAA. Li et al. established two mouse models of AAA and analyzed the RNA transcription expression in the models. The results indicated that the upregulation of H19 is correlated with the progression of AAA, including the content and apoptosis of SMCs. In vitro experiments demonstrated a positive correlation between H19 expression and the apoptosis rate of human aortic smooth muscle cells, suggesting that lncRNA H19 is a novel regulatory factor in the initiation and development of AAA (64). Another study demonstrated the role of H19 in enhancing vascular inflammation and inducing AAA formation. H19 was found to enhance vascular pro-inflammatory IL-6 and MCP-1 and promote AAA formation by enhancing macrophage infiltration (65). X-inactive specific transcript (XIST) is an lncRNA located on the X chromosome, and its role in certain cancers has been elucidated, including breast cancer, rectal cancer, oral cancer (66–68). XIST regulates apoptosis and proliferation through competitive miRNA mechanisms, closely related to the development and growth of tumors (69, 70). Research has also shown that XIST in thoracic aortic aneurysm (TAA) acts as a “sponge” to absorb miR-29b-3p, leading to overexpression of elastin (Eln) in VSMCs and promoting smooth muscle cell apoptosis, indicating its role in aneurysm development (71). AAA also involves abnormal proliferation and apoptosis of VSMCs. Corresponding research results are similar to those in TAA, mainly related to competitive miRNA pathways. Studies have shown that upregulation of XIST can competitively interact with miR-1264 in the WNT/β-catenin signaling pathway and competitively interact with miR-762-mediated mitogen-activated protein kinase kinase 4 (MAP2K4) pathway, thereby inhibiting apoptosis and promoting the proliferation of VSMCs, contributing to the development of AAA (72, 73).
Many recent studies have highlighted the involvement of various lncRNAs in AAA. For instance, lncRNA PVT1 promotes VSMC apoptosis, ECM degradation, and pro-inflammatory factor production by upregulating MMP-9. On the other hand, lncRNA PVT1 acts as a sponge for miR-3127-5p/NCKAP1l, inhibiting VSMC proliferation, inducing apoptosis, and activating inflammation, thus promoting AAA progression (74, 75). Upregulation of lncRNA NEAT1 accelerates VSMC proliferation and inhibits replicative cell apoptosis through the miR-4688/TULP3 pathway (76). The interaction between lncRNA TUG1 and transcriptional repressor KLF4 mediates impaired SMC differentiation function (77). Elevated lncRNA Sox2ot enhances oxidative stress and inflammation in VSMCs through competitive regulation of the miR-145/Egr1 pathway (78) GAS5 overexpression inhibits cell proliferation, induces SMC apoptosis, and accelerates AAA formation in a mouse model. GAS5 acts as a sponge for miR-21, inhibiting the miR-21/PTEN pathway and Akt phosphorylation. Additionally, GAS5 forms a positive feedback loop with Y-box binding protein 1 (YBX1), promoting downstream p21 expression, inhibiting SMC proliferation, and inducing apoptosis (79), GAS5 also induces SMC apoptosis through activation of the EZH2-mediated RIG-I signaling pathway (80). NUDT6, a conservative antisense transcript of FGF2, impairs SMC migration, limits proliferation, and enhances apoptosis, promoting AAA and carotid artery diseases (81). LINC00473 regulates the miR-212-5p/BASP1 pathway to exert anti-proliferative and pro-apoptotic effects in VSMCs (82). Overexpression of CRNDE upregulates Smad3 via Bcl-3, promoting VSMC proliferation and inhibiting cell apoptosis in AAA (83). SENCR overexpression may inhibit AAA formation by suppressing VSMC apoptosis and extracellular matrix degradation, but the specific pathway mechanism needs further exploration (84). These studies provide some scientific basis for understanding the pathogenesis and potential treatments of AAA involving lncRNAs. While many of these lncRNAs act on corresponding miRNAs, exploring other mechanisms of lncRNA epigenetic regulation might yield unexpected therapeutic effects in AAA treatment. Additionally, the clinical translation of these findings requires further investigation. The progression of AAA is a long-term process involving different epigenetic changes at different stages. Therefore, clinical research with sufficient sample sizes and excellent staging and indicator selection is crucial for precise and feasible AAA treatment.
CircRNAs, as a relatively novel research avenue in AAA, primarily exert their effects through miRNA sponging, possibly achieved through competitive endogenous RNA (ceRNA) mechanisms. Bioinformatics studies have explored potential molecular mechanisms in AAA by constructing ceRNA interaction networks involving circRNAs and miRNAs (85). For instance, circCBFB acts as a miR-28-5p sponge, promoting the reduction of GRIA4 and LYPD3 levels, thereby decreasing VSMC apoptosis and facilitating AAA progression (86). Novel circRNAs such as circ_0092291 might inhibit Ang II-induced smooth muscle cell damage by sponging miR-626, leading to increased levels of collagen IV alpha 1 chain (COL4A1) and suppressing AAA development (87). Circ_0002168 regulates VSMC proliferation and apoptosis by sponging miR-545-3p to enhance CKAP4 expression, potentially impacting VSMC loss in AAA (88). Another circRNA, hsa_circ_0087352, upregulated in AAA, may enhance macrophage inflammation and regulate VSMC apoptosis by sponging has-miR-149-5p and acting on LPS (89). Furthermore, circRNAs can influence protein pathways. For instance, circChordc1 induces wave protein degradation, increases GSK3β/β-catenin pathway activity, promotes VSMC phenotype transition, reduces apoptosis, and alleviates vascular remodeling to inhibit AAA progression (90). CircCdyl promotes vascular inflammation and induces M1 polarization in macrophages, contributing to AAA formation, by inhibiting interferon regulatory factor 4 (IRF4) nuclear entry and acting as a sponge for let-7c to enhance C/EBP-δ expression (91). Additionally, circRNA transcription, such as that of the circRNA of the ataxia-telangiectasia mutated gene (cATM), may represent an early characteristic change in the AAA microenvironment, triggering oxidative stress reactions in SMCs and potentially serving as a crucial molecular diagnostic indicator for AAA (92). Despite these findings, research on the role of circRNAs in AAA remains incomplete. Mechanisms involving other miRNA sponging, as well as alternative roles of circRNAs, such as protein translocation, translation, and interactions between proteins, require further exploration in subsequent studies.
Certainly, the involvement of ncRNA and epigenetic regulation extends beyond the mentioned three types. For instance, pathways related to piwi-interacting RNA (piRNA) and the target sites of small interfering RNA (siRNA) also contribute. Notably, studies have indicated the potential involvement of the piRNA pathway, specifically piRNA piRPG, in AAA (93). Further in-depth research is warranted to explore the specific roles of other ncRNAs in AAA pathogenesis.
Due to the complex and multifactorial nature of AAA, ncRNA may represent a potential factor for effective treatment. This is because ncRNAs can regulate the expression of corresponding proteins by targeting multiple mRNAs and influencing various signaling pathways related to AAA. NcRNA is currently in the exploratory stage, and when transcribing multiple genes, only a portion of them may be translated into proteins. Other genes are intricately regulated by a network of non-coding RNAs, controlling the expression of the final protein products. Numerous studies emphasize the significance of ncRNAs, as they play critical roles in the development of diseases. Moreover, these research findings are actively translating into clinical therapies. NcRNAs may serve as the foundation for clinical targeted treatments, and their role in screening and monitoring early stages of the disease is crucial for making accurate therapeutic decisions (94).
In recent years, an increasing body of research has highlighted the crucial role of epigenetic factors in the onset and progression of cardiovascular diseases (95–98). RNA methylation is a significant form of epigenetic modification, belonging to the fields of epitranscriptomics and epigenomics, which collectively regulate gene expression in eukaryotes (99, 100), this includes 6-methyladenosine (m6A), 5-methylcytosine (m5C), and N-7-methylguanosine (m7G). Currently, studies regarding the association between RNA methylation and AAA are limited, with the primary focus on m6A.
m6A modification involves methylation of the sixth nitrogen atom on the adenine base of RNA, and this modification is widely present in eukaryotes, playing a role in the regulation of certain non-coding RNA metabolism (101). The enzymes associated with m6A modification are categorized into methyltransferases (writers), demethylases (erasers), and m6A-binding proteins (readers), forming a crucial protein ensemble that can add, remove, and recognize m6A modification sites, thereby altering biological processes (102). Major methyltransferases include METTL3, METTL14, WTAP, RBM15/15B, KIAA1429, ZC3H13, and METTL16, while demethylases comprise FTO and ALKBH5. Recognized readers include YTHDF1, YTHDF2, YTHDF3, YTHDC1, YTHDC2, HNRNPC, HNRNPG, among others (103). m6A methylation profoundly impacts various aspects of RNA metabolism, including RNA expression, splicing, translation, and RNA-protein interactions (104). It participates in the regulation of multiple biological processes, such as autophagy, inflammation, oxidative stress, DNA damage, and cellular aging (105–107), all of which are closely associated with the occurrence and progression of AAA. AAA is characterized by vascular remodeling and progressive dilation (1), infiltration of lymphocytes and macrophages into the vessel wall, and smooth muscle cell apoptosis (108). Studies have revealed a significant increase in m6A modification levels in AAA, with elevated m6A posing an increased risk of AAA rupture (109, 110). Moreover, differential expression of METTL14, FTO, and YTHDF3 has been observed in various types of inflammatory cells in AAA tissue. FTO, in particular, has shown a strong correlation with the infiltration of aneurysmal smooth muscle cells and macrophages, indicating its potential pivotal role in AAA progression (111). Furthermore, METTL14, HNRNPC, and RBM15 exhibit significant expression differences in AAA and are strongly associated with infiltrative immune cells such as macrophages and mast cells (112). RBM15, by recruiting the WTAP-METTL3-METTL14 RNA methyltransferase complex, enhances m6A levels. Knocking down RBM15 can reduce the expression of m6A-dependent CASP3, inhibiting apoptosis in human abdominal aortic smooth muscle cells (113, 114). In addition, a study by Zhong et al. found that METTL3/m6A participates in AAA formation by promoting the expression of mature miR34a, thereby reducing the expression of SIRP1, providing new targets and diagnostic biomarkers for clinical treatment of AAA (115). Current research on the mechanism of m6A action in AAA mostly focuses on FTO, METTL3, and METTL14, and further investigation is needed to explore the interactions between other regulatory factors.
m5C modification refers to the methylation of the fifth carbon atom on the cytosine base of RNA molecules, a process predominantly catalyzed by enzymes of the NSUN family in eukaryotes. This modification participates in various RNA biological processes, including RNA export, translation, and ribosome assembly (116). Post-transcriptional m5C modification has been confirmed to play a crucial role in various cancer diseases such as lung cancer (117), prostate cancer (118), breast cancer (119), bladder cancer (120) among others. Currently, there is limited research on m5C modification in AAA. Several studies have indicated that NSUN2, by regulating mRNA stability and the translation process, influences a range of pathological processes, including cell proliferation, oxidative stress, and inflammatory responses (121–123). Furthermore, research has shown a significant increase in mRNA m5C modification levels in AAA compared to healthy groups (124). These findings suggest that m5C modification may play a role in the clinical mechanisms affecting AAA, providing new avenues for research in AAA treatment.
m7G modification is one of the most common post-transcriptional base modifications, widely distributed in tRNA, rRNA, and the 5′ cap region of eukaryotic mRNA (125). The identified methyltransferases responsible for m7G methylation include the yeast Trm82/Trm8 complex and its mammalian homolog METTL1/WDR4 complex (126). To date, numerous research findings suggest that m7G modification influences the occurrence and development of various cancer diseases, such as hepatocellular carcinoma (127), esophageal cancer (128), and bladder cancer (129). Additionally, reports have indicated the significant role of m7G modification in inflammation and angiogenesis (130). It can be inferred that m7G modification may also impact the development of AAA. However, the specific mechanisms through which m7G modification affects AAA remain unclear. Bioinformatic analysis has revealed a significant correlation between AAA and m7G-related genes, including CYFIP1, EIF3D, EIF4E3, NSUN2, and NUDT11 (131). Nevertheless, further in vivo and in vitro experiments are needed to validate the mechanisms through which m7G modification influences abdominal aortic aneurysm.
RNA modifications extend beyond the mentioned types. For example, 3-methylcytidine (m3C) is close to m5C sites, and 7-methylguanosine cap structure [m7Gpp(pN)] acts on dihydrouridine (D) and pseudouridine (Ψ) in tRNA and rRNA. Research on these modifications has been conducted in breast cancer, gastric cancer, liver cancer, and prostate cancer, but investigations into the relationship between RNA modifications and AAA are still ongoing (132). Studies have shown that N1-methyladenosine (m1A) may promote macrophage polarization in aortic inflammation through the reader YTHDF3, affecting target gene expression and influencing the progression of AAA (133). Further research is needed to explore the specific mechanisms and roles of RNA modifications, paving the way for understanding the pathogenesis and treatment of AAA.
In the context of epigenetic pathways related to AAA, some novel examples have caught our attention. They may be associated with the progression of AAA and could potentially regulate the expression of corresponding proteins through epigenetic mechanisms. The following outlines three examples related to AAA.
The PCSK family consists of nine proteases, with PCSK9 being demonstrated as crucial in the regulation of CVD (134). In a 2023 study, a potential therapeutic target, Proprotein Convertase Subtilisin/Kexin Type 9 (PCSK9), was identified through AAA's genome-wide association meta-analysis of 121 independent risk loci. When elastase was introduced into PCSK9 mice, AAA growth decreased, indicating a unique role for PCSK9 in AAA (135). Besides, Inhibitors of PCSK9 can reduce low-density lipoprotein (LDL) cholesterol levels, potentially decreasing the risk of severe cardiovascular disease symptoms associated with atherosclerosis (136). A large-scale study involving 100,000 individuals suggested that elevated lipoprotein levels may increase the risk of AAA 2–3 times (137). Given PCSK's role in regulating lipid metabolism, this represents the feasibility of PCSK as a therapeutic target, necessitating more preclinical research and clinical trials to assess its efficacy in AAA-related outcomes.
Meanwhile, epigenetic mechanisms likely play a role in some functional aspects of PCSK genes. For example, sirt6 deacetylase can inhibit PCSK9 gene expression by enriching the transcription factor Foxo3 in the proximal promoter region of the PCSK9 gene, leading to histone H3 deacetylation (138). Increased expression of has-miR-335 and has-miR-6825 can lower PCSK9 mRNA expression in myocardial cells (139). Additionally, the histone acetyltransferase P300 may increase PCSK9 expression (140). Non-coding RNAs (ncRNA) play a crucial role in the progression of inflammation in atherosclerosis by targeting genes related to the PCSK9 pathway at the post-transcriptional level (141). Further research is needed to uncover the pathway-related mechanisms of PCSK9 in AAA, potentially providing new insights and breakthroughs into the complex pathogenesis of AAA.
While PCSK9's Epigenetic importance is established, the specific mechanism of other PCSK family protein in AAA are still under exploration. Members of the PCSK family share structural similarities, but their functional roles vary. In cancer research, other PCSK family members have been reported to activate cytokines, influencing cell proliferation, migration, and extracellular matrix remodeling—important features in the progression of AAA (142). A study categorized PCSK analysis into plaque expression, biochemical blood parameters, morphological characteristics, and patient symptoms. The cumulative data from each section suggested the feasibility of PCSK6 as a new therapeutic target, followed by PCSK5, PCSK7, and FURIN, all showing potential for CVD treatment within this family (143). This, of course, is also closely related to the treatment of AAA. Therefore, the research on PCSK family proteins may be more in-depth and comprehensive.
The transforming growth factor-beta (TGF-β) family members include TGF-beta, pathway factors, and bone morphogenetic proteins (BMP), among other genes, with extensive research in cancer. TGFβs can activate various pathways, including the Sma- and Mad-related family proteins (Smad), mitogen-activated protein kinase (MAPK), and phosphoinositide 3-kinase (PI3K) pathways. Their association with AAA may lie in their ability to ultimately inhibit inflammatory cell infiltration, reduce ECM degradation, limit VSMC apoptosis, promote ECM formation, and be involved in tissue repair, fibrosis, extracellular matrix remodeling, cell proliferation, and migration (144, 145). It's worth mentioning that the Smad pathway, in terms of epigenetics, is regulated by various miRNAs during the AAA process. Notably, miRNA-29b-mediated Smad targeting facilitates AAA by influencing key matrix metalloproteinases (MMP-2 and MMP-9). Inhibiting miRNA-26a increases gene expression of SMAD-1 and SMAD-4, promoting vascular smooth muscle cell proliferation, inhibiting cell differentiation and apoptosis, altering TGF-β pathway signaling. Additionally, miR-424/322 analogs modulate Smad2/3/runt (146–149). Moreover, miRNA-195, miRNA-155, and miR-143/145 intersect with the Smad pathway in the TGF-β pathway (150). A genetic association study in a Dutch population analyzed single nucleotide polymorphisms (SNPs) in TGF-β receptor genes TGFBR1 and TGFBR2, revealing potential correlations similar to TAA in AAA (151). This may be evidence that some TGF-β is related to genetics or epigenetics. In the TGF-β pathway, multiple genes coordinate final outcomes, and aside from the Smad pathway, the other two pathways may form a complex and mutually influencing regulatory system with various non-coding RNAs. Furthermore, there is limited research on histone modification and DNA methylation in this context, necessitating further exploration of the pathways and discovery of new targets.
Receptor tyrosine kinases (RTKs) form the largest class of enzyme-linked receptors, serving both as receptors for growth factors and as enzymes that catalyze downstream target protein phosphorylation. RTKs play crucial regulatory roles not only in normal cells but also have pivotal functions in the progression of various cancers, including lung cancer, gastric cancer, thyroid cancer, etc (152). Mutations in receptor tyrosine kinases lead to the activation of a series of signal cascades, impacting protein expression, including epigenetic effects, although specific research in this regard is currently lacking. RTKs can modulate various downstream signaling pathways such as MAPK, PI3K/Akt, and JAK/STAT. Various types of RTKs may be overexpressed in AAA, including epidermal growth factor receptors (EGFRs) (153), vascular endothelial growth factor receptors (VEGFRs) (154), platelet-derived growth factor receptors (PDGFRs) (155), insulin-like growth factor receptors (IGFRs) (156) and fibroblast growth factor receptors (FGFRs) (157). Research indicates that caspase recruitment domain and membraneassociated guanylate kinaselike domain protein 3 (CARMA3) recruits two downstream signaling molecules, BCL10 and MALT1, through its N-terminal effector CARD domain, forming the CARD11-BCL10-MALT1 (CBM) complex (158), involved in the NF-kB signaling pathway induced by G protein-coupled receptor (GPCR) and RTK. Inflammation is a critical feature in the pathogenesis of cardiovascular diseases, including Abdominal Aortic Aneurysm (159). During the inflammatory process, blood vessels and the surrounding connective tissue are important regulatory factors (160). In endothelial cells of mice, CARMA3 assembles BCL10 and MALT1 to form the CBM signalosome, triggering NF-kB activation induced by CXCL8/IL8. CXCL8/IL8 is a crucial chemokine involved in promoting angiogenesis and inflammation. This process controls VEGF expression and promotes autocrine activation of VEGF receptors (161). The CBM complex also mediates coagulation-induced NF activation (162). However, how CARMA3 connects to GPCR and regulates the NF-kB signaling pathway remains a puzzle. Future studies can explore how RTKs, through epigenetic mechanisms, influence NF-kB to impact the occurrence and development of AAA, thereby identifying new therapeutic targets.
The three aforementioned gene pathways represent only a minute fraction of the vast gene regulatory network. In addition to the previously discussed pathways, such as the inflammatory response pathways, including the Interleukin-6 (IL-6) pathway, and the MMP pathway responsible for degrading cellular tissue matrix, there is limited research on the interplay with epigenetics. Besides refining the epigenetic aspects of these pathways, further exploration and research are needed on gene pathways related to cellular autophagy, ECM, and VSMC proliferation, and their associations with epigenetic modifications.
In preclinical studies of abdominal aortic aneurysm (AAA), three main rodent models are commonly employed: the elastase model, the CaCl2 model, and AngII/ApoE deficient mouse model. The elastase model emphasizes the role of inflammation-related mechanisms in AAA but may ultimately lead to healing unrelated to AAA rupture. The CaCl2 model can induce a smaller degree of aneurysm and, similar to the elastase model, does not result in AAA rupture. The AngII/ApoE deficient mouse model is the most commonly used, causing rupture but potentially having a higher correlation with aortic dissection, and its results may not necessarily translate to human AAA outcomes (163). As a matter of fact, a challenge in current experiments related to abdominal aortic aneurysm lies in the difficulty of translating some of the existing animal model results into clinical trial outcomes. On the one hand, animal models need continued optimization due to the physiological and biochemical differences between rodents and mammals. On the other hand, inherent differences exist between animal models and humans in various aspects. The pathogenesis of human AAA is highly complex, requiring interdisciplinary scientific approaches to prove the efficacy of drug treatments in the progression from small to large AAA after the discovery of small AAAs Figure 5.
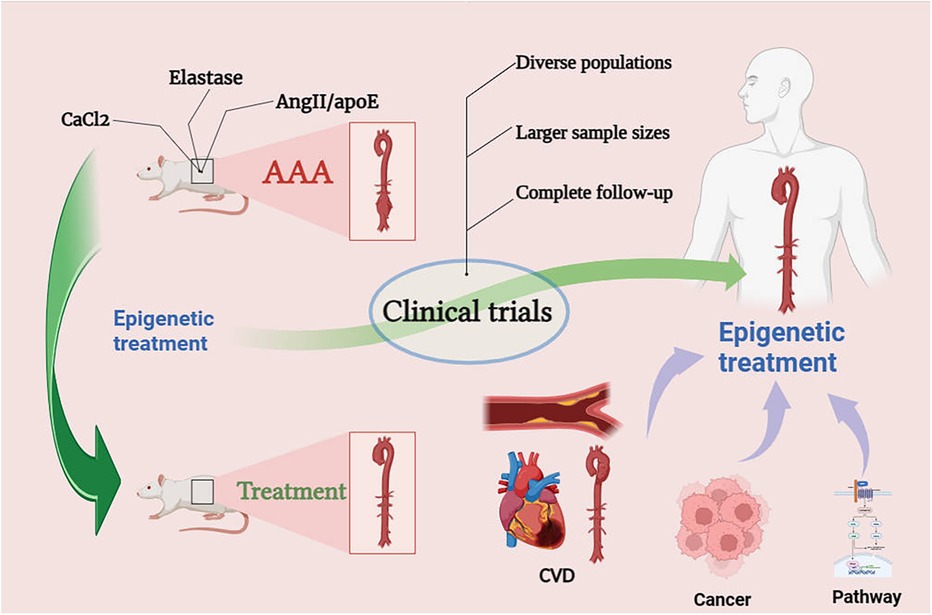
Figure 5. Prospects for epigenetic treatment of AAA. Epigenetic drugs need to go through clinical trials, we may be able to look for therapeutic ideas for AAA through epigenetics in the therapeutic mechanisms of CVD, cancer and pathways.
Due to studies demonstrating a common coexistence and positive correlation between Chlamydia pneumoniae infection and AAA progression (164, 165), antibiotics were once considered as potential candidates for clinical treatment. However, results from two large clinical studies showed that doxycycline treatment did not reduce aneurysm growth, nor did it delay the need for AAA surgery or the timing of surgical repair (166, 167). The most commonly used AAA animal model involves inducing abdominal aortic aneurysm formation with angiotensin II, and clinical trials are often conducted using ACE inhibitors (ACEIs) and angiotensin II receptor blockers (ARBs). Additionally, there are numerous clinical trials for anti-inflammatory drugs targeting immune cell inflammatory infiltration in AAA, statin drugs selected from observational experiments for their potential correlation with AAA growth and rupture, and antiplatelet drugs that weaken blood clots. It is noteworthy that metformin, targeting the risk factor of diabetes for AAA, is still under clinical trial. However, AAA growth is a long-term process influenced by multiple factors, including statistical considerations. Since age is also a risk factor for AAA, elderly AAA patients often end up with the outcome of loss to follow-up due to death. As a result, the statistical results of clinical trials are often censored, making them less convincing (9, 168, 169). Currently, most drug clinical trials involve repurposing basic drugs from other diseases. These trials test the impact of drugs on limiting AAA growth, but there is still no clinical trial data providing conclusive evidence of drug efficacy in restricting AAA progression. Further research may require larger sample sizes, diverse populations, and clinical trial designs that account for gender differences. Future exploration may also involve personalized targeting of drug development for AAA to translate the effects of AAA-related drugs into therapeutic outcomes.
Since the cellular aberrations observed in abdominal aortic aneurysms (AAA) involving proliferation, apoptosis regulation, inflammatory infiltration, and fibrosis are similar to those observed in cancer, the therapeutic significance of epigenetic modification in cancer may be instructive for AAA. It is plausible that similar treatment approaches could be applied. For example, Brd4, as an epigenetic regulator of multiple programmed cell death, has some untapped potential in cancer. This may indicate that it may serve as a potential direction for epigenetic therapy of AAA (170). In addition, m6A in RNA modification may play some crucial roles in tumor glycolysis, and the glycolytic reprogramming of cancer cells is a major characteristic of cancer, which may become a special therapeutic mechanism affecting the occurrence and development of tumors. It may represent that AAA may also have similar epigenetic mechanisms related to proliferation to be further studied (171). Immunological treatments targeting cell-specific epigenetic modifiers such as histone demethylase JMJD3 and SET domain bifurcated protein lysine methyltransferase 2 (SETDB2) have shown promise in effectively intervening in AAA progression (54, 55).
Exploring well-established pathways in cancer research for potential treatment targets is anotheravenue. For instance, the Wnt/β-catenin pathway has been found to be significantly activated in human and experimental animal AAA models, suggesting the presence of potential therapeutic targets, despite the exclusion of specific anti-tumor drugs for arterial disease in animal models (172). Due to the intricate genetic actions of pathways, in-depth research is necessary to determine whether improving arterial aneurysm development is achievable by targeting specific components of these pathways. The presence of ncRNAs may fulfill this role. Recent studies on lncRNA have shown that inhibiting H19 expression could serve as a novel molecular therapeutic target, preventing or slowing AAA progression by suppressing IL-6 expression (65). The role of miRNAs in AAA treatment has been demonstrated (Table 1), but a combination with specific pathways and target gene functions is still required. It is essential to explore the intersections of various pathways and ncRNAs in detail to identify critical therapeutic targets for AAA progression.
Another perspective for epigenetic treatment is drawing inspiration from risk factor diseases or other cardiovascular diseases similar to AAA. For example, research has shown that stem cells have potential in treating cardiovascular diseases, with mesenchymal stem cells (MSCs) effectively inhibiting cell senescence in cardiovascular disease (196). Recent results in epigenetic research indicate that upregulating miR-19b-3p can enhance the anti-aging effect of extracellular vesicles from MSCs (MSC-EXO) isolated from AAA patients. This occurs through the regulation of the MST4/ERK/Drp1 pathway, inhibiting mitochondrial fission in VSMCs (177). In addition, there are a number of epigenetic therapeutic drugs in clinical trials in cardiovascular diseases, such as DNMT inhibitors (azacytidine, decitabine, and hydroalazine), HDAC inhibitors (vorinostat, sodium valproate), histone methylation inhibitors (GSK126, EPZ-5676), and histone methylation inhibitors (azacytidine, decitabine, and hydroalazine). Bromodomain and exo-terminal motif (BET) inhibitor (RVX208) (197). They have different effects on cancer and cardiovascular diseases, and may be used as a treatment for AAA in the future.
Our exploration of epigenetic modifications extends beyond this. In addition to translating established epigenetic modification mechanisms observed in animals into clinical trial directions, other types of epigenetic modifications that have not been well-explored in AAA, such as chromatin remodeling and other types of RNA modifications, are also considered. Two studies on chromatin remodeling focused on the unique subunits BAF60a and BAF60c of the SWI/SNF chromatin remodeling complex, determining that BAF60a promotes epigenetic regulation of VSMC inflammation and BAF60c plays a crucial role in maintaining VSMC homeostasis (198, 199). However, there are still few studies on the specific mechanisms of chromatin remodeling in AAA and even cardiovascular diseases. In the future, more comprehensive studies are expected to make the effects of histone and DNA remodeling at the chromatin level on AAA more clear. With the development of epigenetic technologies, new sequencing and editing methods can reveal complex network regulations of many genes controlled by epigenetic mechanisms. The application of these epigenetic technologies in AAA research can provide a genetic-level understanding of epigenetic modifications as targets and signaling pathways for new drug development and offer critical insights into potential therapeutic strategies for AAA.
However, despite the potential therapeutic advantages of epigenetic therapy, it also comes with limitations. Epigenetic treatments may induce unnecessary changes or off-target effects outside the intended target, potentially leading to adverse reactions or even other diseases. Furthermore, uncertainties remain regarding the long-term impacts and safety of epigenetic therapy, necessitating further research and validation. It is also important to note that most epigenetic research is limited to animal models, in vitro studies on human cells, or bioinformatics-based screenings. Effective translation into clinical trials may face obstacles, such as delivery methods or cost and accessibility.
In the past two decades, there has been rapid progress in the research development of epigenetics. The role of AAA is a complex and multi-faceted issue. A comprehensive understanding of epigenetics in AAA is crucial. On one hand, it involves exploring different targets and pathways, and on the other hand, validating the feasibility of known targets and pathways. Further research is required to confirm the role of epigenetics in the treatment of AAA and the therapeutic potential in AAA animal models.
YL: Writing – review & editing, Writing – original draft. XS: Writing – original draft. ZG: Writing – original draft. ZD: Writing – review & editing. YZ: Writing – review & editing. PZ: Writing – review & editing. WS: Writing – review & editing. YB: Writing – review & editing. YJ: Writing – review & editing.
The authors declare financial support was received for the research, authorship, and/or publication of this article.
This work is supported by grants from the National Nature Science Foundation of China No. 81803530 (To YB), and the scientific research projects of the Education Department of Liaoning Province No. LJKQZ 20222350 (To YCJ).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Sakalihasan N, Limet R, Defawe OD. Abdominal aortic aneurysm. Lancet. (2005) 365(9470):1577–89. doi: 10.1016/S0140-6736(05)66459-8
2. Baman JR, Eskandari MK. What is an abdominal aortic aneurysm? JAMA. (2022) 328(22):2280. doi: 10.1001/jama.2022.18638
3. GBD 2017 Causes of Death Collaborators. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: a systematic analysis for the global burden of disease study 2017. Lancet. (2018) 392(10159):1736–88. doi: 10.1016/S0140-6736(18)32203-7
4. Schmitz-Rixen T, Böckler D, Vogl TJ, Grundmann RT. Endovascular and open repair of abdominal aortic aneurysm. Dtsch Arztebl Int. (2020) 117(48):813–9. doi: 10.3238/arztebl.2020.0813
5. Lasheras JC. The biomechanics of arterial aneurysms. Annu Rev Fluid Mech. (2007) 39(1):293–319. doi: 10.1146/annurev.fluid.39.050905.110128
6. Wanhainen A, Verzini F, Van Herzeele I, Allaire E, Bown M, Cohnert T, et al. Editor’s choice—European society for vascular surgery (ESVS) 2019 clinical practice guidelines on the management of abdominal aorto-iliac artery aneurysms. Eur J Vasc Endovasc Surg. (2019) 57(1):8–93. doi: 10.1016/j.ejvs.2018.09.020
7. Chaikof EL, Dalman RL, Eskandari MK, Jackson BM, Lee WA, Mansour MA, et al. The society for vascular surgery practice guidelines on the care of patients with an abdominal aortic aneurysm. J Vasc Surg. (2018) 67(1):2–77.e2. doi: 10.1016/j.jvs.2017.10.044
8. Altobelli E, Rapacchietta L, Profeta VF, Fagnano R. Risk factors for abdominal aortic aneurysm in population-based studies: a systematic review and meta-analysis. Int J Environ Res Public Health. (2018) 15(12):2805. doi: 10.3390/ijerph15122805
9. Golledge J, Moxon JV, Singh TP, Bown MJ, Mani K, Wanhainen A. Lack of an effective drug therapy for abdominal aortic aneurysm. J Intern Med. (2020) 288(1):6–22. doi: 10.1111/joim.12958
10. Puertas-Umbert L, Almendra-Pegueros R, Jiménez-Altayó F, Sirvent M, Galán M, Martínez-González J, et al. Novel pharmacological approaches in abdominal aortic aneurysm. Clin Sci. (2023) 137(15):1167–94. doi: 10.1042/CS20220795
11. Carsten CG, Calton WC, Johanning JM, Armstrong PJ, Franklin DP, Carey DJ, et al. Elastase is not sufficient to induce experimental abdominal aortic aneurysms. J Vasc Surg. (2001) 33(6):1255–62. doi: 10.1067/mva.2001.112706
12. Poulsen JL, Stubbe J, Lindholt JS. Animal models used to explore abdominal aortic aneurysms: a systematic review. Eur J Vasc Endovasc Surg. (2016) 52(4):487–99. doi: 10.1016/j.ejvs.2016.07.004
13. Ward NC, Watts GF, Eckel RH. Statin toxicity. Circ Res. (2019) 124(2):328–50. doi: 10.1161/CIRCRESAHA.118.312782
14. Laurent S. Antihypertensive drugs. Pharmacol Res. (2017) 124:116–25. doi: 10.1016/j.phrs.2017.07.026
15. Peixoto P, Cartron PF, Serandour AA, Hervouet E. From 1957 to nowadays: a brief history of epigenetics. Int J Mol Sci. (2020) 21(20):7571. doi: 10.3390/ijms21207571
16. Shi Y, Zhang H, Huang S, Yin L, Wang F, Luo P, et al. Epigenetic regulation in cardiovascular disease: mechanisms and advances in clinical trials. Signal Transduct Target Ther. (2022) 7:200. doi: 10.1038/s41392-022-01055-2
17. Toghill BJ, Saratzis A, Harrison SC, Verissimo AR, Mallon EB, Bown MJ. The potential role of DNA methylation in the pathogenesis of abdominal aortic aneurysm. Atherosclerosis. (2015) 241(1):121–9. doi: 10.1016/j.atherosclerosis.2015.05.001
18. Udali S, Guarini P, Moruzzi S, Choi SW, Friso S. Cardiovascular epigenetics: from DNA methylation to microRNAs. Mol Asp Med. (2013) 34(4):883–901. doi: 10.1016/j.mam.2012.08.001
19. Golledge J, Biros E, Bingley J, Iyer V, Krishna SM. Epigenetics and peripheral artery disease. Curr Atheroscler Rep. (2016) 18(4):15. doi: 10.1007/s11883-016-0567-4
20. Sakalihasan N, Michel JB, Katsargyris A, Kuivaniemi H, Defraigne JO, Nchimi A, et al. Abdominal aortic aneurysms. Nat Rev Dis Primers. (2018) 4(1):1–22. doi: 10.1038/s41572-018-0030-7
21. Stackelberg O, Björck M, Larsson SC, Orsini N, Wolk A. Sex differences in the association between smoking and abdominal aortic aneurysm. Br J Surg. (2014) 101(10):1230–7. doi: 10.1002/bjs.9526
22. Satta R, Maloku E, Zhubi A, Pibiri F, Hajos M, Costa E, et al. Nicotine decreases DNA methyltransferase 1 expression and glutamic acid decarboxylase 67 promoter methylation in GABAergic interneurons. Proc Natl Acad Sci U S A. (2008) 105(42):16356–61. doi: 10.1073/pnas.0808699105
23. Dai Y, Chen D, Xu T. DNA methylation aberrant in atherosclerosis. Front Pharmacol. (2022) 13:815977. doi: 10.3389/fphar.2022.815977
24. Greenberg MVC, Bourc’his D. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol. (2019) 20(10):590–607. doi: 10.1038/s41580-019-0159-6
25. Liu X, Gao Q, Li P, Zhao Q, Zhang J, Li J, et al. UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat Commun. (2013) 4:1563. doi: 10.1038/ncomms2562
26. Rougier N, Bourc’his D, Gomes DM, Niveleau A, Plachot M, Pàldi A, et al. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev. (1998) 12(14):2108–13. doi: 10.1101/gad.12.14.2108
27. Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. (2011) 333(6047):1300–3. doi: 10.1126/science.1210597
28. Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. (2013) 502(7472):472. doi: 10.1038/nature12750
29. Warsi AA, Davies B, Morris-Stiff G, Hullin D, Lewis MH. Abdominal aortic aneurysm and its correlation to plasma homocysteine, and vitamins. Eur J Vasc Endovasc Surg. (2004) 27(1):75–9. doi: 10.1016/j.ejvs.2003.09.001
30. Jamaluddin MS, Chen I, Yang F, Jiang X, Jan M, Liu X, et al. Homocysteine inhibits endothelial cell growth via DNA hypomethylation of the cyclin agene. Blood. (2007) 110(10):3648–55. doi: 10.1182/blood-2007-06-096701
31. Krishna SM, Dear A, Craig JM, Norman PE, Golledge J. The potential role of homocysteine mediated DNA methylation and associated epigenetic changes in abdominal aortic aneurysm formation. Atherosclerosis. (2013) 228(2):295–305. doi: 10.1016/j.atherosclerosis.2013.02.019
32. Halazun KJ, Bofkin KA, Asthana S, Evans C, Henderson M, Spark JI. Hyperhomocysteinaemia is associated with the rate of abdominal aortic aneurysm expansion. Eur J Vasc Endovasc Surg. (2007) 33(4):391–4. doi: 10.1016/j.ejvs.2006.10.022
33. Liu Z, Luo H, Zhang L, Huang Y, Liu B, Ma K, et al. Hyperhomocysteinemia exaggerates adventitial inflammation and angiotensin II-induced abdominal aortic aneurysm in mice. Circ Res. (2012) 111(10):1261–73. doi: 10.1161/CIRCRESAHA.112.270520
34. Vats S, Sundquist K, Wang X, Zarrouk M, Ågren-Witteschus S, Sundquist J, et al. Associations of global DNA methylation and homocysteine levels with abdominal aortic aneurysm: a cohort study from a population-based screening program in Sweden. Int J Cardiol. (2020) 321:137–42. doi: 10.1016/j.ijcard.2020.06.022
35. Yin M, Zhang J, Wang Y, Wang S, Böckler D, Duan Z, et al. Deficient CD4+ CD25+ T regulatory cell function in patients with abdominal aortic aneurysms. Arterioscler Thromb Vasc Biol. (2010) 30(9):1825–31. doi: 10.1161/ATVBAHA.109.200303
36. Xia Q, Zhang J, Han Y, Zhang X, Jiang H, Lun Y, et al. Epigenetic regulation of regulatory T cells in patients with abdominal aortic aneurysm. FEBS Open Bio. (2019) 9(6):1137–43. doi: 10.1002/2211-5463.12643
37. Ryer EJ, Ronning KE, Erdman R, Schworer CM, Elmore JR, Peeler TC, et al. The potential role of DNA methylation in abdominal aortic aneurysms. Int J Mol Sci. (2015) 16(5):11259–75. doi: 10.3390/ijms160511259
38. Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. (2007) 39(4):457–66. doi: 10.1038/ng1990
39. Larsen F, Gundersen G, Lopez R, Prydz H. CpG islands as gene markers in the human genome. Genomics. (1992) 13(4):1095–107. doi: 10.1016/0888-7543(92)90024-M
40. Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. (2012) 13(7):484–92. doi: 10.1038/nrg3230
41. Skvortsova K, Stirzaker C, Taberlay P. The DNA methylation landscape in cancer. Essays Biochem. (2019) 63(6):797–811. doi: 10.1042/EBC20190037
42. Toghill BJ, Saratzis A, Freeman PJ, Sylvius N, Bown MJ. SMYD2 promoter DNA methylation is associated with abdominal aortic aneurysm (AAA) and SMYD2 expression in vascular smooth muscle cells. Clin Epigenetics. (2018) 10:29. doi: 10.1186/s13148-018-0460-9
43. Wei S, Tao J, Xu J, Chen X, Wang Z, Zhang N, et al. Ten years of EWAS. Adv Sci. (2021) 8(20):e2100727. doi: 10.1002/advs.202100727
44. Han Y, Tanios F, Reeps C, Zhang J, Schwamborn K, Eckstein HH, et al. Histone acetylation and histone acetyltransferases show significant alterations in human abdominal aortic aneurysm. Clin Epigenetics. (2016) 8:3. doi: 10.1186/s13148-016-0169-6
45. Jiang H, Xia Q, Xin S, Lun Y, Song J, Tang D, et al. Abnormal epigenetic modifications in peripheral T cells from patients with abdominal aortic aneurysm are correlated with disease development. J Vasc Res. (2016) 52(6):404–13. doi: 10.1159/000445771
46. Galán M, Varona S, Orriols M, Rodríguez JA, Aguiló S, Dilmé J, et al. Induction of histone deacetylases (HDACs) in human abdominal aortic aneurysm: therapeutic potential of HDAC inhibitors. Dis Model Mech. (2016) 9(5):541–52. doi: 10.1242/dmm.024513
47. Vinh A, Gaspari TA, Liu HB, Dousha LF, Widdop RE, Dear AE. A novel histone deacetylase inhibitor reduces abdominal aortic aneurysm formation in angiotensin II-infused apolipoprotein E-deficient mice. J Vasc Res. (2007) 45(2):143–52. doi: 10.1159/000110041
48. Greenway J, Gilreath N, Patel S, Horimatsu T, Moses M, Kim D, et al. Profiling of histone modifications reveals epigenomic dynamics during abdominal aortic aneurysm formation in mouse models. Front Cardiovasc Med. (2020) 7:595011. doi: 10.3389/fcvm.2020.595011
49. Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. (1999) 98(3):285–94. doi: 10.1016/S0092-8674(00)81958-3
50. Jenuwein T, Allis CD. Translating the histone code. Science. (2001) 293(5532):1074–80. doi: 10.1126/science.1063127
51. Kouzarides T. Chromatin modifications and their function. Cell. (2007) 128(4):693–705. doi: 10.1016/j.cell.2007.02.005
52. Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol. (2014) 6(4):a018713. doi: 10.1101/cshperspect.a018713
53. Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. (2007) 14(11):1008–16. doi: 10.1038/nsmb1337
54. Davis FM, Tsoi LC, Melvin WJ, denDekker A, Wasikowski R, Joshi AD, et al. Inhibition of macrophage histone demethylase JMJD3 protects against abdominal aortic aneurysms. J Exp Med. (2021) 218(6):e20201839. doi: 10.1084/jem.20201839
55. Davis FM, Melvin WJ, Mangum K, Tsoi LC, Joshi AD, Cai Q, et al. The histone methyltransferase SETDB2 modulates tissue inhibitors of metalloproteinase–matrix metalloproteinase activity during abdominal aortic aneurysm development. Ann Surg. (2023) 278(3):426. doi: 10.1097/SLA.0000000000005963
56. Wu YL, Lin ZJ, Li CC, Lin X, Shan SK, Guo B, et al. Epigenetic regulation in metabolic diseases: mechanisms and advances in clinical study. Signal Transduct Target Ther. (2023) 8:98. doi: 10.1038/s41392-023-01333-7
57. Yang J, Xu J, Wang W, Zhang B, Yu X, Shi S. Epigenetic regulation in the tumor microenvironment: molecular mechanisms and therapeutic targets. Signal Transduct Target Ther. (2023) 8:210. doi: 10.1038/s41392-023-01480-x
58. Sun L, Zhang H, Gao P. Metabolic reprogramming and epigenetic modifications on the path to cancer. Protein Cell. (2022) 13(12):877–919. doi: 10.1007/s13238-021-00846-7
59. Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell. (2019) 179(5):1033–55. doi: 10.1016/j.cell.2019.10.017
60. Bartel DP. MicroRNA target recognition and regulatory functions. Cell. (2009) 136(2):215–33. doi: 10.1016/j.cell.2009.01.002
61. Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol. (2016) 17(12):756–70. doi: 10.1038/nrm.2016.126
62. Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, Xu RH. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. (2020) 19:172. doi: 10.1186/s12943-020-01286-3
63. Leung A, Trac C, Jin W, Lanting L, Akbany A, Sætrom P, et al. Novel long non-coding RNAs are regulated by angiotensin II in vascular smooth muscle cells. Circ Res. (2013) 113(3):266–78. doi: 10.1161/CIRCRESAHA.112.300849
64. Li DY, Busch A, Jin H, Chernogubova E, Pelisek J, Karlsson J, et al. H19 induces abdominal aortic aneurysm development and progression. Circulation. (2018) 138(15):1551–68. doi: 10.1161/CIRCULATIONAHA.117.032184
65. Sun Y, Zhong L, He X, Wang S, Lai Y, Wu W, et al. LncRNA H19 promotes vascular inflammation and abdominal aortic aneurysm formation by functioning as a competing endogenous RNA. J Mol Cell Cardiol. (2019) 131:66–81. doi: 10.1016/j.yjmcc.2019.04.004
66. Richart L, Picod-Chedotel ML, Wassef M, Macario M, Aflaki S, Salvador MA, et al. XIST loss impairs mammary stem cell differentiation and increases tumorigenicity through mediator hyperactivation. Cell. (2022) 185(12):2164–83.e25. doi: 10.1016/j.cell.2022.04.034
67. Yang X, Zhang S, He C, Xue P, Zhang L, He Z, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. (2020) 19(1):46. doi: 10.1186/s12943-020-1146-4
68. Liu H, Wang D, Kan S, Hao M, Chang L, Lu P, et al. The role of lncRNAs and XIST in oral cancer. Front Cell Dev Biol. (2022) 10:826650. doi: 10.3389/fcell.2022.826650
69. Zhu H, Zheng T, Yu J, Zhou L, Wang L. LncRNA XIST accelerates cervical cancer progression via upregulating fus through competitively binding with miR-200a. Biomed Pharmacother. (2018) 105:789–97. doi: 10.1016/j.biopha.2018.05.053
70. Yao Y, Ma J, Xue Y, Wang P, Li Z, Liu J, et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. (2015) 359(1):75–86. doi: 10.1016/j.canlet.2014.12.051
71. Liang K, Cui M, Fu X, Ma J, Zhang K, Zhang D, et al. LncRNA XIST induces arterial smooth muscle cell apoptosis in thoracic aortic aneurysm through miR-29b-3p/Eln pathway. Biomed Pharmacother. (2021) 137:111163. doi: 10.1016/j.biopha.2020.111163
72. Zou L, Xia PF, Chen L, Hou YY. XIST knockdown suppresses vascular smooth muscle cell proliferation and induces apoptosis by regulating miR-1264/WNT5A/β-catenin signaling in aneurysm. Biosci Rep. (2021) 41(3):BSR20201810. doi: 10.1042/BSR20201810
73. Zhang D, Lu D, Xu R, Zhai S, Zhang K. Inhibition of XIST attenuates abdominal aortic aneurysm in mice by regulating apoptosis of vascular smooth muscle cells through miR-762/MAP2K4 axis. Microvasc Res. (2022) 140:104299. doi: 10.1016/j.mvr.2021.104299
74. Zhang Z, Zou G, Chen X, Lu W, Liu J, Zhai S, et al. Knockdown of lncRNA PVT1 inhibits vascular smooth muscle cell apoptosis and extracellular matrix disruption in a murine abdominal aortic aneurysm model. Mol Cells. (2019) 42(3):218–27. doi: 10.14348/molcells.2018.0162
75. Huang Y, Ren L, Li J, Zou H. Long non-coding RNA PVT1/microRNA miR-3127-5p/NCK-associated protein 1-like axis participates in the pathogenesis of abdominal aortic aneurysm by regulating vascular smooth muscle cells. Bioengineered. (2021) 12(2):12583–96. doi: 10.1080/21655979.2021.2010384
76. Cai B, Yang B, Huang D, Wang D, Tian J, Chen F, et al. STAT3-induced up-regulation of lncRNA NEAT1 as a ceRNA facilitates abdominal aortic aneurysm formation by elevating TULP3. Biosci Rep. (2020) 40(1):BSR20193299. doi: 10.1042/BSR20193299
77. Bharadhwaj RA, Kumarswamy R. Long noncoding RNA TUG1 regulates smooth muscle cell differentiation via KLF4-myocardin axis. Am J Physiol Cell Physiol. (2023) 325(4):C940–50. doi: 10.1152/ajpcell.00275.2023
78. Lin H, You B, Lin X, Wang X, Zhou D, Chen Z, et al. Silencing of long non-coding RNA Sox2ot inhibits oxidative stress and inflammation of vascular smooth muscle cells in abdominal aortic aneurysm via microRNA-145-mediated Egr1 inhibition. Aging (Albany NY). (2020) 12(13):12684–702. doi: 10.18632/aging.103077
79. He X, Wang S, Li M, Zhong L, Zheng H, Sun Y, et al. Long noncoding RNA GAS5 induces abdominal aortic aneurysm formation by promoting smooth muscle apoptosis. Theranostics. (2019) 9(19):5558–76. doi: 10.7150/thno.34463
80. Le T, He X, Huang J, Liu S, Bai Y, Wu K. Knockdown of long noncoding RNA GAS5 reduces vascular smooth muscle cell apoptosis by inactivating EZH2-mediated RIG-I signaling pathway in abdominal aortic aneurysm. J Transl Med. (2021) 19:466. doi: 10.1186/s12967-021-03023-w
81. Winter H, Winski G, Busch A, Chernogubova E, Fasolo F, Wu Z, et al. Targeting long non-coding RNA NUDT6 enhances smooth muscle cell survival and limits vascular disease progression. Mol Ther. (2023) 31(6):1775–90. doi: 10.1016/j.ymthe.2023.04.020
82. Tian Z, Sun Y, Sun X, Wang J, Jiang T. LINC00473 inhibits vascular smooth muscle cell viability to promote aneurysm formation via miR-212-5p/BASP1 axis. Eur J Pharmacol. (2020) 873:172935. doi: 10.1016/j.ejphar.2020.172935
83. Li K, Cui M, Zhang K, Wang G, Zhai S. LncRNA CRNDE affects the proliferation and apoptosis of vascular smooth muscle cells in abdominal aortic aneurysms by regulating the expression of Smad3 by bcl-3. Cell Cycle. (2020) 19(9):1036–47. doi: 10.1080/15384101.2020.1743915
84. Cai Z, Huang J, Yang J, Pan B, Wang W, Ou Y, et al. LncRNA SENCR suppresses abdominal aortic aneurysm formation by inhibiting smooth muscle cells apoptosis and extracellular matrix degradation. Bosn J Basic Med Sci. (2021) 21(3):323–30. doi: 10.17305/bjbms.2020.4994
85. Chen L, Wang S, Wang Z, Liu Y, Xu Y, Yang S, et al. Construction and analysis of competing endogenous RNA network and patterns of immune infiltration in abdominal aortic aneurysm. Front Cardiovasc Med. (2022) 9:955838. doi: 10.3389/fcvm.2022.955838
86. Yue J, Zhu T, Yang J, Si Y, Xu X, Fang Y, et al. CircCBFB-mediated miR-28-5p facilitates abdominal aortic aneurysm via LYPD3 and GRIA4. Life Sci. (2020) 253:117533. doi: 10.1016/j.lfs.2020.117533
87. Ma M, Yang X, Han F, Wang H. Circ_0092291 attenuates angiotensin II–induced cell damages in human aortic vascular smooth muscle cells via mediating the miR-626/COL4A1 signal axis. J Physiol Biochem. (2022) 78(1):245–56. doi: 10.1007/s13105-021-00859-0
88. Wei J, Wang H, Zhao Q. Circular RNA suppression of vascular smooth muscle apoptosis through the miR-545-3p/CKAP4 axis during abdominal aortic aneurysm formation. Vasc Med. (2023) 28(2):104–12. doi: 10.1177/1358863X221132591
89. Ma X, Xu J, Lu Q, Feng X, Liu J, Cui C, et al. Hsa_circ_0087352 promotes the inflammatory response of macrophages in abdominal aortic aneurysm by adsorbing hsa-miR-149-5p. Int Immunopharmacol. (2022) 107:108691. doi: 10.1016/j.intimp.2022.108691
90. He X, Li X, Han Y, Chen G, Xu T, Cai D, et al. CircRNA Chordc1 protects mice from abdominal aortic aneurysm by contributing to the phenotype and growth of vascular smooth muscle cells. Mol Ther Nucleic Acids. (2021) 27:81–98. doi: 10.1016/j.omtn.2021.11.005
91. Song H, Yang Y, Sun Y, Wei G, Zheng H, Chen Y, et al. Circular RNA Cdyl promotes abdominal aortic aneurysm formation by inducing M1 macrophage polarization and M1-type inflammation. Mol Ther. (2022) 30(2):915–31. doi: 10.1016/j.ymthe.2021.09.017
92. Fasolo F, Winski G, Li Z, Wu Z, Winter H, Ritzer J, et al. The circular RNA ataxia telangiectasia mutated regulates oxidative stress in smooth muscle cells in expanding abdominal aortic aneurysms. Mol Ther Nucleic Acids. (2023) 33:848–65. doi: 10.1016/j.omtn.2023.08.017
93. Jia D, Wang K, Huang L, Zhou Z, Zhang Y, Chen N, et al. Revealing PPP1R12B and COL1A1 as piRNA pathway genes contributing to abdominal aortic aneurysm through integrated analysis and experimental validation. Gene. (2024) 897:148068. doi: 10.1016/j.gene.2023.148068
94. Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. J Clin Invest. (2017) 127(3):761–71. doi: 10.1172/JCI84424
95. Berulava T, Buchholz E, Elerdashvili V, Pena T, Islam MR, Lbik D, et al. Changes in m6A RNA methylation contribute to heart failure progression by modulating translation. Eur J Heart Fail. (2020) 22(1):54–66. doi: 10.1002/ejhf.1672
96. Chien CS, Li JYS, Chien Y, Wang ML, Yarmishyn AA, Tsai PH, et al. METTL3-dependent N6-methyladenosine RNA modification mediates the atherogenic inflammatory cascades in vascular endothelium. Proc Natl Acad Sci U S A. (2021) 118(7):e2025070118. doi: 10.1073/pnas.2025070118
97. Qin Y, Qiao Y, Li L, Luo E, Wang D, Yao Y, et al. The m6A methyltransferase METTL3 promotes hypoxic pulmonary arterial hypertension. Life Sci. (2021) 274:119366. doi: 10.1016/j.lfs.2021.119366
98. Wu S, Zhang S, Wu X, Zhou X. m6A RNA methylation in cardiovascular diseases. Mol Ther. (2020) 28(10):2111–9. doi: 10.1016/j.ymthe.2020.08.010
99. He C. Grand challenge commentary: RNA epigenetics? Nat Chem Biol. (2010) 6(12):863–5. doi: 10.1038/nchembio.482
100. Saletore Y, Meyer K, Korlach J, Vilfan ID, Jaffrey S, Mason CE. The birth of the epitranscriptome: deciphering the function of RNA modifications. Genome Biol. (2012) 13(10):175. doi: 10.1186/gb-2012-13-10-175
101. Sun HL, Zhu AC, Gao Y, Terajima H, Fei Q, Liu S, et al. Stabilization of ERK-phosphorylated METTL3 by USP5 increases m6A methylation. Mol Cell. (2020) 80(4):633–47.e7. doi: 10.1016/j.molcel.2020.10.026
102. Meyer KD, Jaffrey SR. Rethinking m6A readers, writers, and erasers. Annu Rev Cell Dev Biol. (2017) 33:319–42. doi: 10.1146/annurev-cellbio-100616-060758
103. Wang Y, Wang Y, Patel H, Chen J, Wang J, Chen ZS, et al. Epigenetic modification of m6A regulator proteins in cancer. Mol Cancer. (2023) 22(1):102. doi: 10.1186/s12943-023-01810-1
104. Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. (2018) 28(6):616–24. doi: 10.1038/s41422-018-0040-8
105. Chen Y, Wu Y, Fang L, Zhao H, Xu S, Shuai Z, et al. METTL14-m6A-FOXO3a Axis regulates autophagy and inflammation in ankylosing spondylitis. Clin Immunol. (2023) 257:109838. doi: 10.1016/j.clim.2023.109838
106. Li N, Hui H, Bray B, Gonzalez GM, Zeller M, Anderson KG, et al. METTL3 regulates viral m6A RNA modification and host cell innate immune responses during SARS-CoV-2 infection. Cell Rep. (2021) 35(6):109091. doi: 10.1016/j.celrep.2021.109091
107. Ye G, Li J, Yu W, Xie Z, Zheng G, Liu W, et al. ALKBH5 facilitates CYP1B1 mRNA degradation via m6A demethylation to alleviate MSC senescence and osteoarthritis progression. Exp Mol Med. (2023) 55(8):1743–56. doi: 10.1038/s12276-023-01059-0
108. Raffort J, Lareyre F, Clément M, Hassen-Khodja R, Chinetti G, Mallat Z. Monocytes and macrophages in abdominal aortic aneurysm. Nat Rev Cardiol. (2017) 14(8):457–71. doi: 10.1038/nrcardio.2017.52
109. He Y, Xing J, Wang S, Xin S, Han Y, Zhang J. Increased m6A methylation level is associated with the progression of human abdominal aortic aneurysm. Ann Transl Med. (2019) 7(24):797. doi: 10.21037/atm.2019.12.65
110. Wang K, Kan Q, Ye Y, Qiu J, Huang L, Wu R, et al. Novel insight of N6-methyladenosine modified subtypes in abdominal aortic aneurysm. Front Genet. (2022) 13:1055396. doi: 10.3389/fgene.2022.1055396
111. Ke WL, Huang ZW, Peng CL, Ke YP. M6a demethylase FTO regulates the apoptosis and inflammation of cardiomyocytes via YAP1 in ischemia-reperfusion injury. Bioengineered. (2022) 13(3):5443–52. doi: 10.1080/21655979.2022.2030572
112. Li T, Wang T, Jing J, Sun L. Expression pattern and clinical value of key m6A RNA modification regulators in abdominal aortic aneurysm. J Inflamm Res. (2021) 14:4245–58. doi: 10.2147/JIR.S327152
113. Fu C, Feng L, Zhang J, Sun D. Bioinformatic analyses of the role of m6A RNA methylation regulators in abdominal aortic aneurysm. Ann Transl Med. (2022) 10(10):547. doi: 10.21037/atm-22-1891
114. Li K, Zhang D, Zhai S, Wu H, Liu H. METTL3-METTL14 complex induces necroptosis and inflammation of vascular smooth muscle cells via promoting N6 methyladenosine mRNA methylation of receptor-interacting protein 3 in abdominal aortic aneurysms. J Cell Commun Signal. (2023) 17(3):897–914. doi: 10.1007/s12079-023-00737-y
115. Zhong L, He X, Song H, Sun Y, Chen G, Si X, et al. METTL3 induces AAA development and progression by modulating N6-methyladenosine-dependent primary miR34a processing. Mol Ther Nucleic Acids. (2020) 21:394–411. doi: 10.1016/j.omtn.2020.06.005
116. Wang YY, Tian Y, Li YZ, Liu YF, Zhao YY, Chen LH, et al. The role of m5C methyltransferases in cardiovascular diseases. Front Cardiovasc Med. (2023) 10:1225014. doi: 10.3389/fcvm.2023.1225014
117. Wang Y, Wei J, Feng L, Li O, Huang L, Zhou S, et al. Aberrant m5C hypermethylation mediates intrinsic resistance to gefitinib through NSUN2/YBX1/QSOX1 axis in EGFR-mutant non-small-cell lung cancer. Mol Cancer. (2023) 22(1):81. doi: 10.1186/s12943-023-01780-4
118. Yu G, Bao J, Zhan M, Wang J, Li X, Gu X, et al. Comprehensive analysis of m5C methylation regulatory genes and tumor microenvironment in prostate cancer. Front Immunol. (2022) 13:914577. doi: 10.3389/fimmu.2022.914577
119. Huang Z, Pan J, Wang H, Du X, Xu Y, Wang Z, et al. Prognostic significance and tumor immune microenvironment heterogenicity of m5C RNA methylation regulators in triple-negative breast cancer. Front Cell Dev Biol. (2021) 9:657547. doi: 10.3389/fcell.2021.657547
120. Wang JZ, Zhu W, Han J, Yang X, Zhou R, Lu HC, et al. The role of the HIF-1α/ALYREF/PKM2 axis in glycolysis and tumorigenesis of bladder cancer. Cancer Commun. (2021) 41(7):560–75. doi: 10.1002/cac2.12158
121. Gkatza NA, Castro C, Harvey RF, Heiß M, Popis MC, Blanco S, et al. Cytosine-5 RNA methylation links protein synthesis to cell metabolism. PLoS Biol. (2019) 17(6):e3000297. doi: 10.1371/journal.pbio.3000297
122. Luo Y, Feng J, Xu Q, Wang W, Wang X. NSun2 deficiency protects endothelium from inflammation via mRNA methylation of ICAM-1. Circ Res. (2016) 118(6):944–56. doi: 10.1161/CIRCRESAHA.115.307674
123. Xu X, Zhang Y, Zhang J, Zhang X. NSun2 promotes cell migration through methylating autotaxin mRNA. J Biol Chem. (2020) 295(52):18134–47. doi: 10.1074/jbc.RA119.012009
124. He Y, Zhang H, Yin F, Guo P, Wang S, Wu Y, et al. Novel insights into the role of 5-methylcytosine RNA methylation in human abdominal aortic aneurysm. Front Biosci (Landmark Ed). (2021) 26(11):1147–65. doi: 10.52586/5016
125. Luo Y, Yao Y, Wu P, Zi X, Sun N, He J. The potential role of N7-methylguanosine (m7G) in cancer. J Hematol Oncol. (2022) 15(1):63. doi: 10.1186/s13045-022-01285-5
126. Xia X, Wang Y, Zheng JC. Internal m7G methylation: a novel epitranscriptomic contributor in brain development and diseases. Mol Ther Nucleic Acids. (2023) 31:295–308. doi: 10.1016/j.omtn.2023.01.003
127. Huang M, Long J, Yao Z, Zhao Y, Zhao Y, Liao J, et al. METTL1-mediated m7G tRNA modification promotes lenvatinib resistance in hepatocellular carcinoma. Cancer Res. (2023) 83(1):89–102. doi: 10.1158/0008-5472.CAN-22-0963
128. Han H, Yang C, Ma J, Zhang S, Zheng S, Ling R, et al. N7-methylguanosine tRNA modification promotes esophageal squamous cell carcinoma tumorigenesis via the RPTOR/ULK1/autophagy axis. Nat Commun. (2022) 13(1):1478. doi: 10.1038/s41467-022-29125-7
129. Ying X, Liu B, Yuan Z, Huang Y, Chen C, Jiang X, et al. METTL1-m7G-EGFR/EFEMP1 axis promotes the bladder cancer development. Clin Transl Med. (2021) 11(12):e675. doi: 10.1002/ctm2.675
130. Zhao Y, Kong L, Pei Z, Li F, Li C, Sun X, et al. m7G methyltransferase METTL1 promotes post-ischemic angiogenesis via promoting VEGFA mRNA translation. Front Cell Dev Biol. (2021) 9:642080. doi: 10.3389/fcell.2021.642080
131. Tian Y, Fu S, Zhang N, Zhang H, Li L. The abdominal aortic aneurysm-related disease model based on machine learning predicts immunity and m1A/m5C/m6A/m7G epigenetic regulation. Front Genet. (2023) 14:1131957. doi: 10.3389/fgene.2023.1131957
132. Esteve-Puig R, Bueno-Costa A, Esteller M. Writers, readers and erasers of RNA modifications in cancer. Cancer Lett. (2020) 474:127–37. doi: 10.1016/j.canlet.2020.01.021
133. Wu Y, Jiang D, Zhang H, Yin F, Guo P, Zhang X, et al. N1-methyladenosine (m1A) regulation associated with the pathogenesis of abdominal aortic aneurysm through YTHDF3 modulating macrophage polarization. Front Cardiovasc Med. (2022) 9:883155. doi: 10.3389/fcvm.2022.883155
134. Schmidt AF, Carter JPL, Pearce LS, Wilkins JT, Overington JP, Hingorani AD, et al. PCSK9 monoclonal antibodies for the primary and secondary prevention of cardiovascular disease. Cochrane Database Syst Rev. (2020) 2020(10):CD011748. doi: 10.1002/14651858.CD011748.pub3
135. Roychowdhury T, Klarin D, Levin MG, Spin JM, Rhee YH, Deng A, et al. Genome-wide association meta-analysis identifies risk loci for abdominal aortic aneurysm and highlights PCSK9 as a therapeutic target. Nat Genet. (2023) 55(11):1831–42. doi: 10.1038/s41588-023-01510-y
136. Sabatine MS, Giugliano RP, Keech AC, Honarpour N, Wiviott SD, Murphy SA, et al. Evolocumab and clinical outcomes in patients with cardiovascular disease. N Engl J Med. (2017) 376(18):1713–22. doi: 10.1056/NEJMoa1615664
137. Thomas PE, Vedel-Krogh S, Nielsen SF, Nordestgaard BG, Kamstrup PR. Lipoprotein(a) and risks of peripheral artery disease, abdominal aortic aneurysm, and major adverse limb events. J Am Coll Cardiol. (2023) 82(24):2265–76. doi: 10.1016/j.jacc.2023.10.009
138. Tao R, Xiong X, DePinho RA, Deng CX, Dong XC. Foxo3 transcription factor and Sirt6 deacetylase regulate low density lipoprotein (LDL)-cholesterol homeostasis via control of the proprotein convertase subtilisin/kexin type 9 (Pcsk9) gene expression. J Biol Chem. (2013) 288(41):29252–9. doi: 10.1074/jbc.M113.481473
139. Lin YK, Yeh CT, Kuo KT, Yadav VK, Fong IH, Kounis NG, et al. Pterostilbene increases LDL metabolism in HL-1 cardiomyocytes by modulating the PCSK9/HNF1α/SREBP2/LDLR signaling cascade, upregulating epigenetic hsa-miR-335 and hsa-miR-6825, and LDL receptor expression. Antioxidants. (2021) 10(8):1280. doi: 10.3390/antiox10081280
140. Kim HJ, Lee J, Chung MY, Hong S, Park JH, Lee SH, et al. Piceatannol reduces resistance to statins in hypercholesterolemia by reducing PCSK9 expression through p300 acetyltransferase inhibition. Pharmacol Res. (2020) 161:105205. doi: 10.1016/j.phrs.2020.105205
141. Al-Awsi GRL, Hadi Lafta M, Hashim Kzar H, Samieva G, Alsaikhan F, Ahmad I, et al. PCSK9 pathway-noncoding RNAs crosstalk: emerging opportunities for novel therapeutic approaches in inflammatory atherosclerosis. Int Immunopharmacol. (2022) 113:109318. doi: 10.1016/j.intimp.2022.109318
142. Seidah NG, Prat A. The biology and therapeutic targeting of the proprotein convertases. Nat Rev Drug Discov. (2012) 11(5):367–83. doi: 10.1038/nrd3699
143. Suur BE, Chemaly M, Lindquist Liljeqvist M, Djordjevic D, Stenemo M, Bergman O, et al. Therapeutic potential of the proprotein convertase subtilisin/kexin family in vascular disease. Front Pharmacol. (2022) 13:988561. doi: 10.3389/fphar.2022.988561
144. Wang Y, Krishna S, Walker PJ, Norman P, Golledge J. Transforming growth factor-β and abdominal aortic aneurysms. Cardiovasc Pathol. (2013) 22(2):126–32. doi: 10.1016/j.carpath.2012.07.005
145. Forte A, Galderisi U, Cipollaro M, De Feo M, Corte AD. Epigenetic regulation of TGF-β1 signalling in dilative aortopathy of the thoracic ascending aorta. Clin Sci. (2016) 130(16):1389–405. doi: 10.1042/CS20160222
146. Leeper NJ, Raiesdana A, Kojima Y, Chun HJ, Azuma J, Maegdefessel L, et al. MicroRNA-26a is a novel regulator of vascular smooth muscle cell function. J Cell Physiol. (2011) 226(4):1035–43. doi: 10.1002/jcp.22422
147. Maegdefessel L, Azuma J, Toh R, Merk DR, Deng A, Chin JT, et al. Inhibition of microRNA-29b reduces murine abdominal aortic aneurysm development. J Clin Invest. (2012) 122(2):497–506. doi: 10.1172/JCI61598
148. Tsai HY, Wang JC, Hsu YJ, Chiu YL, Lin CY, Lu CY, et al. miR-424/322 protects against abdominal aortic aneurysm formation by modulating the Smad2/3/runt-related transcription factor 2 axis. Mol Ther Nucleic Acids. (2021) 27:656–69. doi: 10.1016/j.omtn.2021.12.028
149. Chen KC, Wang YS, Hu CY, Chang WC, Liao YC, Dai CY, et al. OxLDL up-regulates microRNA-29b, leading to epigenetic modifications of MMP-2/MMP-9 genes: a novel mechanism for cardiovascular diseases. FASEB J. (2011) 25(5):1718–28. doi: 10.1096/fj.10-174904
150. Tang Y, Fan W, Zou B, Yan W, Hou Y, Kwabena Agyare O, et al. TGF-β signaling and microRNA cross-talk regulates abdominal aortic aneurysm progression. Clin Chim Acta. (2021) 515:90–5. doi: 10.1016/j.cca.2020.12.031
151. Baas AF, Medic J, van‘t Slot R, de Kovel CG, Zhernakova A, Geelkerken RH, et al. Association of the TGF-β receptor genes with abdominal aortic aneurysm. Eur J Hum Genet. (2010) 18(2):240–4. doi: 10.1038/ejhg.2009.141
152. Du Z, Lovly CM. Mechanisms of receptor tyrosine kinase activation in cancer. Mol Cancer. (2018) 17:58. doi: 10.1186/s12943-018-0782-4
153. Dayal S, Broekelmann T, Mecham RP, Ramamurthi A. Targeting epidermal growth factor receptor to stimulate elastic matrix regenerative repair. Tissue Eng Part A. (2023) 29(7–8):187–99. doi: 10.1089/ten.tea.2022.0170
154. Xu B, Iida Y, Glover KJ, Ge Y, Wang Y, Xuan H, et al. Inhibition of VEGF (vascular endothelial growth factor)-A or its receptor activity suppresses experimental aneurysm progression in the aortic elastase infusion model. Arterioscler Thromb Vasc Biol. (2019) 39(8):1652–66. doi: 10.1161/ATVBAHA.119.312497
155. Munshaw S, Bruche S, Redpath AN, Jones A, Patel J, Dube KN, et al. Thymosin beta4 protects against aortic aneurysm via endocytic regulation of growth factor signaling. J Clin Invest. (2021) 131(10):e127884. doi: 10.1172/JCI127884
156. Algul S, Schuldt M, Manders E, Jansen V, Schlossarek S, de Goeij-de Haas R, et al. EGFR/IGF1R signaling modulates relaxation in hypertrophic cardiomyopathy. Circ Res. (2023) 133(5):387–99. doi: 10.1161/CIRCRESAHA.122.322133
157. Sun X, Lu Y, Wu J, Wen Q, Li Z, Tang Y, et al. Meta-analysis of single-cell RNA-seq data reveals the mechanism of formation and heterogeneity of tertiary lymphoid organ in vascular disease. Arterioscler Thromb Vasc Biol. (2023) 43(10):1867–86. doi: 10.1161/ATVBAHA.123.318762
158. Balakrishnan MEN, Muralkar P, Ranjana Ponraj M, Nadiger S, Dhandayutham S, Justus S, et al. Recycling of saw dust as a filler reinforced cotton seed oil resin amalgamated polystyrene composite material for sustainable waste management applications. Mater Today Proc. (2022) 58:783–8. doi: 10.1016/j.matpr.2022.03.331
159. Piqueras L, Sanz MJ. Angiotensin II and leukocyte trafficking: new insights for an old vascular mediator. Role of redox-signaling pathways. Free Radic Biol Med. (2020) 157:38–54. doi: 10.1016/j.freeradbiomed.2020.02.002
160. Schwager S, Detmar M. Inflammation and lymphatic function. Front Immunol. (2019) 10:308. doi: 10.3389/fimmu.2019.00308
161. Martin D, Galisteo R, Gutkind JS. CXCL8/IL8 stimulates vascular endothelial growth factor (VEGF) expression and the autocrine activation of VEGFR2 in endothelial cells by activating NFkappaB through the CBM (Carma3/Bcl10/Malt1) complex. J Biol Chem. (2009) 284(10):6038–42. doi: 10.1074/jbc.C800207200
162. Delekta PC, Apel IJ, Gu S, Siu K, Hattori Y, McAllister-Lucas LM, et al. Thrombin-dependent NF-kappaB activation and monocyte/endothelial adhesion are mediated by the CARMA3.Bcl10.MALT1 signalosome. J Biol Chem. (2010) 285(53):41432–42. doi: 10.1074/jbc.M110.158949
163. Lindeman JH, Matsumura JS. Compendium: pharmacologic management of aneurysms. Circ Res. (2019) 124(4):631–46. doi: 10.1161/CIRCRESAHA.118.312439
164. Lindholt JS, Ashton HA, Scott RAP. Indicators of infection with Chlamydia pneumoniae are associated with expansion of abdominal aortic aneurysms. J Vasc Surg. (2001) 34(2):212–5. doi: 10.1067/mva.2001.115816
165. Karlsson L, Gnarpe J, Nääs J, Olsson G, Lindholm J, Steen B, et al. Detection of viable chlamydia pneumoniae inAbdominal aortic aneurysms. Eur J Vasc Endovasc Surg. (2000) 19(6):630–5. doi: 10.1053/ejvs.1999.1057
166. Baxter BT, Matsumura J, Curci JA, McBride R, Larson L, Blackwelder W, et al. Effect of doxycycline on aneurysm growth among patients with small infrarenal abdominal aortic aneurysms. JAMA. (2020) 323(20):2029–38. doi: 10.1001/jama.2020.5230
167. Meijer CA, Stijnen T, Wasser MNJM, Hamming JF, van Bockel JH, Lindeman JHN. Doxycycline for stabilization of abdominal aortic aneurysms. Ann Intern Med. (2013) 159(12):815–23. doi: 10.7326/0003-4819-159-12-201312170-00007
168. Golledge J, Thanigaimani S, Powell JT, Tsao PS. Pathogenesis and management of abdominal aortic aneurysm. Eur Heart J. (2023) 44(29):2682–97. doi: 10.1093/eurheartj/ehad386
169. Golledge J. Abdominal aortic aneurysm: update on pathogenesis and medical treatments. Nat Rev Cardiol. (2019) 16(4):225–42. doi: 10.1038/s41569-018-0114-9
170. Hu J, Pan D, Li G, Chen K, Hu X. Regulation of programmed cell death by Brd4. Cell Death Dis. (2022) 13(12):1059. doi: 10.1038/s41419-022-05505-1
171. Yue SW, Liu HL, Su HF, Luo C, Liang HF, Zhang BX, et al. m6A-regulated tumor glycolysis: new advances in epigenetics and metabolism. Mol Cancer. (2023) 22:137. doi: 10.1186/s12943-023-01841-8
172. Puertas-Umbert L, Varona S, Ballester-Servera C, Alonso J, Aguiló S, Orriols M, et al. Activation of Wnt/β-catenin signaling in abdominal aortic aneurysm: a potential therapeutic opportunity? Genes Dis. (2023) 10(3):639. doi: 10.1016/j.gendis.2022.05.017
173. Li T, Jing J, Sun L, Gong Y, Yang J, Ma C, et al. The SNP rs4591246 in pri-miR-1-3p is associated with abdominal aortic aneurysm risk by regulating cell phenotypic transformation via the miR-1-3p/TLR4 axis. Int Immunopharmacol. (2023) 118:110016. doi: 10.1016/j.intimp.2023.110016
174. Wågsäter D, Ramilo AB, Näsström M, Kunath A, Agic MB, Mani K, et al. miR-10b promotes aortic aneurysm formation and aortic rupture in angiotensin II-induced ApoE-deficient mice. Vasc Pharmacol. (2021) 141:106927. doi: 10.1016/j.vph.2021.106927
175. Gao P, Si J, Yang B, Yu J. Upregulation of MicroRNA-15a contributes to pathogenesis of abdominal aortic aneurysm (AAA) by modulating the expression of cyclin-dependent kinase inhibitor 2B (CDKN2B). Med Sci Monit. (2017) 23:881–8. doi: 10.12659/MSM.898233
176. Hu J, Jiang Y, Wu X, Wu Z, Qin J, Zhao Z, et al. Exosomal miR-17-5p from adipose-derived mesenchymal stem cells inhibits abdominal aortic aneurysm by suppressing TXNIP-NLRP3 inflammasome. Stem Cell Res Ther. (2022) 13:349. doi: 10.1186/s13287-022-03037-1
177. Zhang Y, Huang X, Sun T, Shi L, Liu B, Hong Y, et al. MicroRNA-19b-3p dysfunction of mesenchymal stem cell-derived exosomes from patients with abdominal aortic aneurysm impairs therapeutic efficacy. J Nanobiotechnology. (2023) 21:135. doi: 10.1186/s12951-023-01894-3
178. Maegdefessel L, Azuma J, Toh R, Deng A, Merk DR, Raiesdana A, et al. MicroRNA-21 blocks abdominal aortic aneurysm development and nicotine-augmented expansion. Sci Transl Med. (2012) 4(122):122ra22. doi: 10.1126/scitranslmed.3003441
179. Si X, Chen Q, Zhang J, Zhou W, Chen L, Chen J, et al. MicroRNA-23b prevents aortic aneurysm formation by inhibiting smooth muscle cell phenotypic switching via FoxO4 suppression. Life Sci. (2022) 288:119092. doi: 10.1016/j.lfs.2021.119092
180. Maegdefessel L, Spin JM, Raaz U, Eken SM, Toh R, Azuma J, et al. miR-24 limits aortic vascular inflammation and murine abdominal aneurysm development. Nat Commun. (2014) 5:5214. doi: 10.1038/ncomms6214
181. Zhou Y, Wang M, Zhang J, Xu P, Wang H. MicroRNA-29a-3p regulates abdominal aortic aneurysm development and progression via direct interaction with PTEN. J Cell Physiol. (2020) 235(12):9414–23. doi: 10.1002/jcp.29746
182. Shen G, Sun Q, Yao Y, Li S, Liu G, Yuan C, et al. Role of ADAM9 and miR-126 in the development of abdominal aortic aneurysm. Atherosclerosis. (2020) 297:47–54. doi: 10.1016/j.atherosclerosis.2020.01.014
183. Shi X, Ma W, Pan Y, Li Y, Wang H, Pan S, et al. MiR-126-5p promotes contractile switching of aortic smooth muscle cells by targeting VEPH1 and alleviates Ang II-induced abdominal aortic aneurysm in mice. Lab Invest. (2020) 100(12):1564–74. doi: 10.1038/s41374-020-0454-z
184. Li L, Ma W, Pan S, Li Y, Wang H, Wang B, et al. Mir-126a-5p limits the formation of abdominal aortic aneurysm in mice and decreases ADAMTS-4 expression. J Cell Mol Med. (2020) 24(14):7896–906. doi: 10.1111/jcmm.15422
185. Yang Z, Zhang L, Liu Y, Zeng W, Wang K. Potency of miR-144-3p in promoting abdominal aortic aneurysm progression in mice correlates with apoptosis of smooth muscle cells. Vasc Pharmacol. (2022) 142:106901. doi: 10.1016/j.vph.2021.106901
186. Shi X, Ma W, Li Y, Wang H, Pan S, Tian Y, et al. MiR-144-5p limits experimental abdominal aortic aneurysm formation by mitigating M1 macrophage-associated inflammation: suppression of TLR2 and OLR1. J Mol Cell Cardiol. (2020) 143:1–14. doi: 10.1016/j.yjmcc.2020.04.008
187. Spinosa M, Lu G, Su G, Bontha SV, Gehrau R, Salmon MD, et al. Human mesenchymal stromal cell–derived extracellular vesicles attenuate aortic aneurysm formation and macrophage activation via microRNA-147. FASEB J. (2018) 32(11):6038–50. doi: 10.1096/fj.201701138RR
188. Zhao L, Ouyang Y, Bai Y, Gong J, Liao H. miR-155-5p inhibits the viability of vascular smooth muscle cell via targeting FOS and ZIC3 to promote aneurysm formation. Eur J Pharmacol. (2019) 853:145–52. doi: 10.1016/j.ejphar.2019.03.030
189. Zhang Z, Liang K, Zou G, Chen X, Shi S, Wang G, et al. Inhibition of miR-155 attenuates abdominal aortic aneurysm in mice by regulating macrophage-mediated inflammation. Biosci Rep. (2018) 38(3):BSR20171432. doi: 10.1042/BSR20171432
190. Di Gregoli K, Mohamad Anuar NN, Bianco R, White SJ, Newby AC, George SJ, et al. MicroRNA-181b controls atherosclerosis and aneurysms through regulation of TIMP-3 and elastin. Circ Res. (2017) 120(1):49–65. doi: 10.1161/CIRCRESAHA.116.309321
191. Zhang H, Wang Y, Bian X, Yin H. MicroRNA-194 acts as a suppressor during abdominal aortic aneurysm via inhibition of KDM3A-mediated BNIP3. Life Sci. (2021) 277:119309. doi: 10.1016/j.lfs.2021.119309
192. Ma X, Yao H, Yang Y, Jin L, Wang Y, Wu L, et al. miR-195 suppresses abdominal aortic aneurysm through the TNF-α/NF-κB and VEGF/PI3K/Akt pathway. Int J Mol Med. (2018) 41(4):2350–8. doi: 10.3892/ijmm.2018.3426
193. Tao W, Hong Y, He H, Han Q, Mao M, Hu B, et al. MicroRNA-199a-5p aggravates angiotensin II–induced vascular smooth muscle cell senescence by targeting Sirtuin-1 in abdominal aortic aneurysm. J Cell Mol Med. (2021) 25(13):6056–69. doi: 10.1111/jcmm.16485
194. Chan CYT, Chan YC, Cheuk BLY, Cheng SWK. Clearance of matrix metalloproteinase-9 is dependent on low-density lipoprotein receptor-related protein-1 expression downregulated by microRNA-205 in human abdominal aortic aneurysm. J Vasc Surg. (2017) 65(2):509–20. doi: 10.1016/j.jvs.2015.10.065
195. Kim CW, Kumar S, Son DJ, Jang IH, Griendling KK, Jo H. Prevention of abdominal aortic aneurysm by anti-miRNA-712 or anti-miR-205 in angiotensin II infused mice. Arterioscler Thromb Vasc Biol. (2014) 34(7):1412–21. doi: 10.1161/ATVBAHA.113.303134
196. Ranganath SH, Levy O, Inamdar MS, Karp JM. Harnessing the mesenchymal stem cell secretome for the treatment of cardiovascular disease. Cell Stem Cell. (2012) 10(3):244–58. doi: 10.1016/j.stem.2012.02.005
197. Nicorescu I, Dallinga GM, de Winther MPJ, Stroes ESG, Bahjat M. Potential epigenetic therapeutics for atherosclerosis treatment. Atherosclerosis. (2019) 281:189–97. doi: 10.1016/j.atherosclerosis.2018.10.006
198. Chang Z, Zhao G, Zhao Y, Lu H, Xiong W, Liang W, et al. BAF60a deficiency in vascular smooth muscle cells prevents abdominal aortic aneurysm by reducing inflammation and extracellular matrix degradation. Arterioscler Thromb Vasc Biol. (2020) 40(10):2494–507. doi: 10.1161/ATVBAHA.120.314955
Keywords: abdominal aortic aneurysm, epigenetics, DNA methylation, histone modification, non-coding RNA
Citation: Liu Y, Sun X, Gou Z, Deng Z, Zhang Y, Zhao P, Sun W, Bai Y and Jing Y (2024) Epigenetic modifications in abdominal aortic aneurysms: from basic to clinical. Front. Cardiovasc. Med. 11:1394889. doi: 10.3389/fcvm.2024.1394889
Received: 11 March 2024; Accepted: 21 May 2024;
Published: 4 June 2024.
Edited by:
Mario D’Oria, Azienda Sanitaria Università Integrata di Trieste, ItalyReviewed by:
Jifeng Zhang, University of Michigan, United States© 2024 Liu, Sun, Gou, Deng, Zhang, Zhao, Sun, Bai and Jing. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Bai, eWJhaUBjbXUuZWR1LmNu; YuChen Jing, eWNqaW5nQGNtdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.