- Cardiac Department, Aerospace Center Hospital, Beijing, China
Background: The risk of atrial fibrillation (AF) is increased in individuals with gastroesophageal reflux disease (GERD), according to observational research. The causal significance of this association is still unclear. This study sought to assess GERD's role as a potential contributing factor in AF.
Methods: With the use of a two-sample Mendelian randomization (MR) technique, we assessed the causal relationship between GERD and AF. The association of genetic variants with GERD was examined using data from a recent genome-wide association study (GWAS) that included 602,604 people. Data on the association between genetic variations and AF was obtained from a second GWAS with 1,030,836 participants. The effect sizes were examined based on the inverse-variance weighted method. Additional statistical techniques, including MR-Egger, simple mode, weighted mode, MR Pleiotropy Residual Sum, outlier, and weighted median were used in the sensitivity analysis.
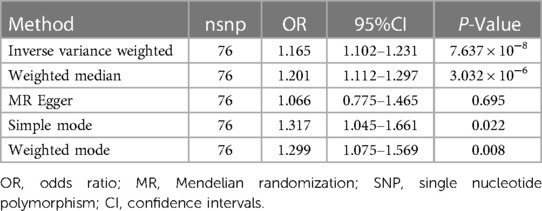
Results: MR analyses in inverse-variance weighted models, using 76 single nucleotide polymorphisms (SNPs) as markers, revealed a relationship between genetically predicted GERD and a greater AF incidence [odds ratio (OR): 1.165, 95% CI 1.102–1.231; P = 7.637 × 10−8]. According to MR-Egger, there was no evidence of gene pleiotropy that could be found (intercept = 0.003, P = 0.581). The findings of the sensitivity study, which used several MR methods, were found to be reliable.
Conclusion: The MR analysis revealed a correlation between GERD and increased AF incidence, supporting the idea that treating patients with GERD as early as possible might reduce their chance of developing AF.
Background
Presently, 2%–3% of population all around the world are affected by atrial fibrillation (AF). The electrical signals of AF in the atria are rapid and disorganized producing an irregular heartbeat (1). Smoking, cardiac surgery, sleep apnea, obesity, thyroid disease, valvular heart disease, and chronic renal disease are risk factors that increase the risk of developing AF. In addition to a higher risk of heart failure and stroke, those with AF may experience fatigue, dyspnea, dizziness, and heart palpitations (1). AF can contribute to considerable societal and medical expenses with major health and economic consequences.
Gastroesophageal reflux disease (GERD) is a condition that affects the digestive tract and is a prevalent presenting symptom. GERD occurs when acidic stomach juices, fluids, and food are backed up from the stomach into the esophagus, causing extreme discomfort. GERD and its correlation with atrial fibrillation (AF) are being studied (2). There is likely a link between GERD and AF due to their shared risk factors, including sleep apnea, obesity, and local inflammation that worsens with age (2). The prospective observational studies and retrospective database analysis suggest an association between GERD and AF (3–6). A systematic review showed the summary-adjusted relative risk for GERD-induced AF was 1.06 (95% CI, 0.86–1.31) (7). The studies also demonstrated that proton pump inhibitors may reduce the burden of symptomatic arrhythmia (8). However, an objective evaluation of GERD showed a temporal correlation between episodes of reflux and cardiac arrhythmia is low (9). Whether GERD performs a casual function in the development of AF is uncertain since the link between the two has not been extensively explored due to the possibility of biases such as confounding or reverse causality (10).
Mendelian randomization (MR) is a strategy that employs instrumental variable (IV) approaches to determine the causative relationship between complex human characteristics and genetic risk factors (11, 12). MR Studies can determine the causal relationship between exposure and disease outcomes by removing unmeasured confounders and reverse causality since IVs exposed to random allocation before conception are not expected to be impacted by disease status. Variants that influence GERD can also, to some degree, have some bearing on AF if a risk factor, such as GERD, may have a direct impact on an outcome, such as AF. Nonetheless, due to the presence of horizontal pleiotropy, any other potential pathways for this variant's influence on AF must be ruled out (13). In this investigation, we aimed to conduct an MR study to verify the causative effect of GERD on the pathogenesis of AF from a major genome-wide association study (GWAS) (14).
Methods
The two-sample MR was conducted using published pooled data on characteristics of interest from major European individuals from GWAS (11, 13). Each cohort participating in the GWAS was ethically approved and agreed upon by the participants and provided summary-level information for analysis. In particular, The two-sample MR approach was used to examine the possible causal effect that GERD characteristics have on AF (Figure 1).
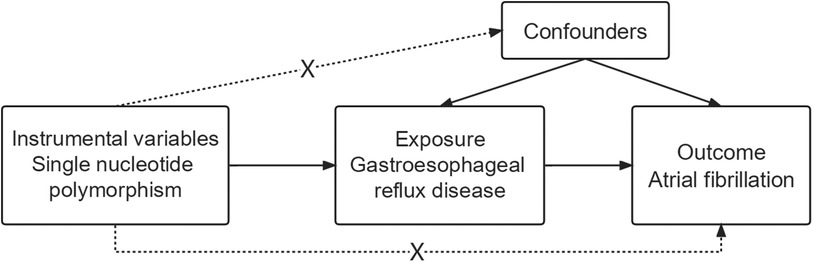
Figure 1. Flowchart depicting the analyses and core hypotheses for a two-sample Mendelian randomization (MR) study. Prediction of the association between GERD and the risk of atrial fibrillation in an MR model. The strategy assumes that GERD-related genetic variations have an effect via the condition itself and are unrelated to confounders.
GWAS summary data for GERD
An extensive European GWAS Meta-analysis was conducted to search for genes associated with GERD. The study included 602,604 individuals (129,080 patients with GERD and 473,524 controls) (15). A broader definition of GERD was used in the study since it relied on patient reports, physician assessments, and examination of medical records. A total of 80 single-nucleotide polymorphisms (SNPs) were found to have a significant association with GERD (p < 5 × 10−8), and the prerequisite for using a two-sample MR approach is that the instrument is devoid of any linkage disequilibrium (LD) (r2 < 0.001, distance threshold >10,000 kb). A detailed illustration of the 80 SNPs may be found in Supplementary Table S1. Notably, these 80 SNPs accounted for 0.51% points of the variance in GERD (16).
Summary data from the GWAS for AF
Only summary data from Europeans were utilized in this study so that we could avoid demographic heterogeneity. A second large European GWAS Meta-analysis was mined for AF genes; it included information from 1,030,836 people, of whom 60,620 with AF and 970,216 acted as controls (16). Six studies consisting of participants of European ancestry made up the sample (the Nord-Trøndelag Health Study [HUNT], deCODE, Michigan Genomics Initiative [MGI], DiscovEHR, UK Biobank, and AFGen Consortium). Cases and controls having genotype data were identified using a mix of inpatient, outpatient, and emergency department discharge diagnoses (ICD-9 and ICD-10). There were 142 genes and 111 genomic areas discovered to have independent risk mutations, and 151 functional candidate genes were ranked according to their probable involvement in AF. Genotyping data, imputation, and quality assurance are among the additional resources available (16).
Selection of genetic instrumental variables
When an individual exposure SNPS is missing from the final data, it is not replaced with a proxy SNPS using LD tagging. Summary data for each of the 80 SNPs linked to GERD were collected from the GWAS Meta-analysis. It is crucial to determine if the effect of SNPS on exposure and the influence of alleles on outcomes are consistent in two-sample MR. The two GWAS findings’ SNPs were consistent in terms of allele frequency. SNPs were omitted if there was inconsistency. Palindromicity and the presence of frequent intermediate alleles led to the elimination of three of the 80 SNPs (rs2145318, rs2358016, and rs957345).
The underpinning outliers were filtered out before MR analysis utilizing Mendelian randomized multiplicity residual sums and outliers (MR-PRESSO) tests, which are most useful upon observing horizontal pleiotropy <50% of the instruments (17). In the MR-PRESSO analysis, one outlier (rs9940128) was identified (p = 0.662). Finally, 76 SNPs were involved in the MR analysis.
Owing to the MR analysis's use of overlapping sample individuals to ascertain the heritability of exposure and outcomes, a slight increment in the weak instrumental bias is possible. We used the F-statistic of SNP to determine how significant the exposure IVs were (18). The F-statistic of 38 for the instruments was significantly greater than the average of 10, indicating excellent prediction power for GERD (19). To determine R2, the following formula was utilized: R2 = [2 × EAF (1- EAF) × β2], whereby EAF denotes the effect of allele frequency and β denotes the presumed genetic impact on GERD. We calculated the F-statistic, based on the following formula, to rank the significance of each SNP: F = [R2 × (n−1−k)]/ [(1−R2) × k], where k is the total count of SNPs, n is the sample size, and R2 indicates the proportion of phenotypic variation that can be attributable to underlying genetic variants (20).
Testing Mendelian randomization assumptions
Three fundamental presumptions must be true for MR studies: (1) Genetic variants and exposure have a strong correlation; (2) The genetic variants and potential confounders were not linked; (3) Apart from the exposure modality, genetic variation had no effect on the outcomes (Figure 1). The inclusion of unidentified possible confounders makes it challenging to test hypotheses 2 and 3. Therefore, to ascertain if there is horizontal pleiotropy—whether hereditary variables impact AF in addition to GERD characteristics—we computed the regression coefficient of MR Egger and assessed the significant intercept.
Statistical analyses
We divided the Beta of the associated SNPS in the outcome dataset by the Beta of the same SNPS in the exposure dataset to get the Wald ratio and evaluate the causal effect of each IV. To determine the direction of any possible causation between GERD and AF risk, we conducted an MR-Steiger analysis. We used a multiplicative inverse-variance weighted (IVW) model in the case of significant Cochran's Q value (P < 0.05), and heterogeneity is acceptable (21). We applied a fixed-effects model in every other scenario. In the primary study, the characteristics linked to genetically predicted exposures and the risk of AF were compared using the IVW approach. Wald ratios for every SNP were analyzed in a meta-analysis. The foundation of IVW was the assumption that the findings could only have been affected by the exposure of interest.
The findings of single and multi-SNP studies are merged and displayed using scatter and forest plots. The 95% confidence intervals for single-SNP and multi-SNP impact estimates are displayed next to one another in a graphic that resembles a forest plot. Regression lines determined by multiple SNP analyses were added to the scatter plot to compare the effects of a single SNPS on exposure and outcomes (having matching standard deviations in either direction).
We estimate that our MR analysis, with a sample size of 602,604 and an alpha of 5%, will detect an OR of 1.10 for AF per odds of GERD (22). The analyses were completed utilizing Two-Sample MR (v0.5.6) (23) and MRPRESSO (v1.0) in R (v4.1.2). The strength of the association between a projected genetic risk for GERD and the risk of AF is described using ORs with 95% CIs. We concluded that the causal results of multiple MR methods were consistent in direction and magnitude (see below) and passed nominal significance in IVW. It was determined that p < 6.579 × 10−4 (0.05/76) provided statistically significant proof of a causal relationship. P-values < 0.05 but above the Bonferroni correction threshold were considered suggestive evidence of potential causality.
Sensitivity analysis
To gauge the connections’ robustness, we conducted multiple sensitivity tests. Initially, we employed a weighted median approach to examine the linkages, assuming that at least 50% of the weights came from reliable instruments (24). Additionally, simple model, MR-Egger, and weighted model approaches are employed even though they are less effective since they may offer more precise estimates in a broader variety of scenarios (with a wider CI). Additionally, we performed a leave-one analysis based on the IVW MR technique, where each SNP was omitted from the analysis separately, to assess the probable effects of outliers and/or pleiotropy SNPS. Moreover, a funnel plot was applied to investigate heterogeneity.
Results
To determine whether the causal effect estimates were robust, we conducted a Steiger-MR analysis. According to Steiger-MR, SNPs account for greater variation in exposure than in outcome (P = 0.725). Because Cochran's Q value was insignificant (P = 0.092), the relationship between genetically determined GERD-related characteristics and the risk of AF was evaluated utilizing the fixed-effects IVW method. As indicated in Table 1, we found evidence of a probable causal relationship between GERD and AF in traditional MR analysis using the IVW approach [odds ratio (OR): 1.165, 95% CI 1.102–1.231; P = 7.637 × 10−8]. The causal estimates made from each IV were shown on the forest plot (Supplementary Figure S1) and scatter plot (Supplementary Figure S2).
We discovered that GERD and AF exhibited a causal relationship utilizing the weighted median model (OR = 1.201, 95% CI = 1.112–1.297; P = 3.032 × 10−6). Both weighted mode and simple mode techniques produced consistent results (P < 0.05).
No evidence of pleiotropy was observed (MR-Egger regression test, intercept = 0.003, P = 0.581). The leave-one-out analysis illustrated that none of the instruments could independently account for the overall effect of GERD on AF (Supplementary Figure S3). According to the IVW and MR Egger model, no statistically significant heterogeneity was observed in the causal estimates among IVs (Supplementary Figure S4).
Discussion
Our two-sample MR analysis was based on data from publically accessible GWASs to probe the link between GERD and AF risk and draw inferences regarding their causality. Genetic susceptibility to GERD was associated with an increased likelihood of developing AF, as shown by the statistical causal relationship.
Previous observational studies did correlate GERD with AF, but their findings were contradictory. Patients with GERD, and specifically those with more severe GERD-associated symptoms, were shown to have a higher chance of developing AF as opposed to those without the condition as per the findings of the epidemiological data obtained from these observational studies. However, these studies did not provide enough evidence to prove that GERD exhibits a causal relationship with AF (25). The presence of AF was associated with an increased incidence of GERD in an observational study, suggesting that this condition might be a risk factor for its development (26). Nonetheless, the causality between GERD and AF is uncertain because of the possibility of bias in observational studies related to several confounders. It is difficult to use conventional epidemiological methodologies to determine the causal relationship between GERD and AF. Moreover, we adopted a two-sample MR, a technique that accounts for the effects of potential confounders by employing genetic variations with a biological pathway separate from the exposure of relevance, in contrast to these conventional epidemiological investigations. Our findings held up across multiple sensitivity studies that probed the effect of pleiotropy on causation estimations. There is presently insufficient data to decide on anti-GERD treatment schemes for AF patients (27). These findings need to be confirmed by further research methods, such as large-scale intervention trials and prospective cohort studies.
Based on the results of our two-sample MR study, we infer that GERD may increase the risk of AF. Multiple potential biological mechanisms have been hypothesized to account for the beneficial impact of GERD on AF. Previous research has shown that proinflammatory cytokines play a role in the development of GERD (25, 28). Injuries to the esophagus may cause the release of cytokines locally, creating conditions favorable to the development of AF. Inflammatory cells have been observed infiltrating the atriums of patients with AF, indicating an inflammation-AF link (29–31). Furthermore, GERD can cause autonomic nervous system dysfunction (32–34), consequently increasing AF risk (35). To help researchers and physicians develop new preventive and therapeutic methods, further analysis into the molecular mechanisms behind these relationships is required.
Limitation
Some drawbacks are present in this MR study. First, because this information was generated from GWAS data of European ancestry, it must be confirmed in other ethnic groups to guarantee that it is relevant to people of other racial backgrounds. Second, the results from the research cannot differentiate between AF episodes (including atrial flutter and paroxysmal, persistent, and chronic AF), disorders that may have different etiologies (36, 37). Third, it was difficult to determine the extent of overlapping between the outcome and the exposure samples since sample and diagnostic data were unavailable.
Conclusion
A higher incidence of AF was shown to be associated with GERD, as determined by the MR analysis, lending support to the idea that treating GERD patients early might reduce their risk of developing AF.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: The datasets generated and/or analysed during the current study are available in the IEU GWAS repository (https://gwas.mrcieu.ac.uk).
Ethics statement
Ethical review and approval was not required for the study, in accordance with the local legislation and institutional requirements. This study used summary-level data from the published studies and publicly available GWASs, with ethical approvals obtained from their respective institutional review boards. All these studies obtained relevant participant consent.
Author contributions
LW: Writing – original draft, Writing – review & editing. YL: Writing – original draft.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
We would like to thank the MRC IEU UK Biobank GWAS pipeline for making the GWAS datasets publicly available.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2024.1393383/full#supplementary-material
References
1. Rahman F, Kwan GF, Benjamin EJ. Global epidemiology of atrial fibrillation. Nat Rev Cardiol. (2014) 11(11):639–54. doi: 10.1038/nrcardio.2014.118
2. Mohamed A, Ochoa Crespo D, Kaur G, Ashraf I, Peck MM, Maram R, et al. Gastroesophageal reflux and its association with atrial fibrillation: a traditional review. Cureus. (2020) 12(9):e10387. doi: 10.7759/cureus.10387
3. Maret-Ouda J, Santoni G, Xie S, Rosengren A, Lagergren J. Objectively confirmed gastroesophageal reflux disease and risk of atrial fibrillation: a population-based cohort study in Sweden. Eur J Gastroenterol Hepatol. (2022) 34(11):1116–20. doi: 10.1097/MEG.0000000000002419
4. Bunch TJ, Packer DL, Jahangir A, Locke GR, Talley NJ, Gersh BJ, et al. Long-term risk of atrial fibrillation with symptomatic gastroesophageal reflux disease and esophagitis. Am J Cardiol. (2008) 102(9):1207–11. doi: 10.1016/j.amjcard.2008.06.048
5. Huang CC, Chan WL, Luo JC, Chen YC, Chen TJ, Chung CM, et al. Gastroesophageal reflux disease and atrial fibrillation: a nationwide population-based study. PLoS One. (2012) 7(10):e47575. doi: 10.1371/journal.pone.0047575
6. Kubota S, Nakaji G, Shimazu H, Odashiro K, Maruyama T, Akashi K. Further assessment of atrial fibrillation as a risk factor for gastroesophageal reflux disease: a multicenter questionnaire survey. Intern Med. (2013) 52(21):2401–7. doi: 10.2169/internalmedicine.52.0923
7. Xu L, Zhang Y, Xie J, Liu Y, Xu L. Association between gastroesophageal reflux disease and atrial fibrillation: a systematic review and meta-analysis. Rev Esp Enferm Dig. (2019) 111(11):874–9. doi: 10.17235/reed.2019.5389/2017
8. Maruyama T, Fukata M, Akashi K. Association of atrial fibrillation and gastroesophageal reflux disease: natural and therapeutic linkage of the two common diseases. J Arrhythm. (2019) 35(1):43–51. doi: 10.1002/joa3.12125
9. Coutinho EL, Herbella FAM, Lovato CAV, Patti MG, Schlottmann F, de Paola AAV. Objective evaluation of gastroesophageal reflux disease in patients with paroxysmal atrial fibrillation. World J Surg. (2018) 42(5):1458–62. doi: 10.1007/s00268-017-4337-4
10. Shimazu H, Nakaji G, Fukata M, Odashiro K, Maruyama T, Akashi K. Relationship between atrial fibrillation and gastroesophageal reflux disease: a multicenter questionnaire survey. Cardiology. (2011) 119(4):217–23. doi: 10.1159/000331497
11. Skrivankova VW, Richmond RC, Woolf BAR, Yarmolinsky J, Davies NM, Swanson SA, et al. Strengthening the reporting of observational studies in epidemiology using Mendelian randomization: the STROBE-MR statement. JAMA. (2021) 326(16):1614–21. doi: 10.1001/jama.2021.18236
12. Bowden J, Holmes MV. Meta-analysis and Mendelian randomization: a review. Res Synth Methods. (2019) 10(4):486–96. doi: 10.1002/jrsm.1346
13. Burgess S, Davey Smith G, Davies NM, Dudbridge F, Gill D, Glymour MM, et al. Guidelines for performing Mendelian randomization investigations. Wellcome Open Res. (2019) 4:186. doi: 10.12688/wellcomeopenres.15555.1
14. Rasooly D, Peloso GM. Two-Sample multivariable Mendelian randomization analysis using R. Curr Protoc. (2021) 1(12):e335. doi: 10.1002/cpz1.335
15. Ong J-S, An J, Han X, Law MH, Nandakumar P, Schumacher J, et al. Multitrait genetic association analysis identifies 50 new risk loci for gastro-oesophageal reflux, seven new loci for barrett’s oesophagus and provides insights into clinical heterogeneity in reflux diagnosis. Gut. (2022) 71(6):1053–61. doi: 10.1136/gutjnl-2020-323906
16. Nielsen JB, Thorolfsdottir RB, Fritsche LG, Zhou W, Skov MW, Graham SE, et al. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat Genet. (2018) 50(9):1234–9. doi: 10.1038/s41588-018-0171-3
17. Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. (2018) 50(5):693–8. doi: 10.1038/s41588-018-0099-7
18. Burgess S, Thompson SG, Collaboration CCG. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. (2011) 40(3):755–64. doi: 10.1093/ije/dyr036
19. Burgess S, Davies NM, Thompson SG. Bias due to participant overlap in two-sample Mendelian randomization. Genet Epidemiol. (2016) 40(7):597–608. doi: 10.1002/gepi.21998
20. Palmer TM, Lawlor DA, Harbord RM, Sheehan NA, Tobias JH, Timpson NJ, et al. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat Methods Med Res. (2012) 21(3):223–42. doi: 10.1177/0962280210394459
21. Yoshikawa M, Asaba K, Nakayama T. Causal effect of atrial fibrillation/flutter on chronic kidney disease: a bidirectional two-sample Mendelian randomization study. PLoS One. (2021) 16(12):e0261020. doi: 10.1371/journal.pone.0261020
22. Brion MJ, Shakhbazov K, Visscher PM. Calculating statistical power in Mendelian randomization studies. Int J Epidemiol. (2013) 42(5):1497–501. doi: 10.1093/ije/dyt179
23. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-base platform supports systematic causal inference across the human phenome. eLife. (2018) 7:e34408. doi: 10.7554/eLife.34408
24. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. (2016) 40(4):304–14. doi: 10.1002/gepi.21965
25. Roman C, des Varannes S B, Muresan L, Picos A, Dumitrascu DL. Atrial fibrillation in patients with gastroesophageal reflux disease: a comprehensive review. World J Gastroenterol. (2014) 20(28):9592–9. doi: 10.3748/wjg.v20.i28.9592
26. Hwang JJ, Lee DH, Yoon H, Shin CM, Park YS, Kim N. Is atrial fibrillation a risk factor for gastroesophageal reflux disease occurrence? Medicine (Baltimore. (2015) 94(43):e1921. doi: 10.1097/MD.0000000000001921
27. Velagapudi P, Turagam MK, Leal MA, Kocheril AG. Atrial fibrillation and acid reflux disease. Clin Cardiol. (2012) 35(3):180–6. doi: 10.1002/clc.21969
28. Linz D, Hohl M, Vollmar J, Ukena C, Mahfoud F, Bohm M. Atrial fibrillation and gastroesophageal reflux disease: the cardiogastric interaction. Europace. (2017) 19(1):16–20. doi: 10.1093/europace/euw092
29. Aviles RJ, Martin DO, Apperson-Hansen C, Houghtaling PL, Rautaharju P, Kronmal RA, et al. Inflammation as a risk factor for atrial fibrillation. Circulation. (2003) 108(24):3006–10. doi: 10.1161/01.CIR.0000103131.70301.4F
30. Engelmann MD, Svendsen JH. Inflammation in the genesis and perpetuation of atrial fibrillation. Eur Heart J. (2005) 26(20):2083–92. doi: 10.1093/eurheartj/ehi350
31. La Fazia VM, Pierucci N, Mohanty S, Gianni C, Della Rocca DG, Compagnucci P, et al. Catheter ablation approach and outcome in HIV+ patients with recurrent atrial fibrillation. J Cardiovasc Electrophysiol. (2023) 34(12):2527–34. doi: 10.1111/jce.16076
32. Armaganijan L, Patel D, Lopes RD, Morillo CA, Araujo RR, Munhoz FP, et al. Gastroesophageal reflux and atrial fibrillation: is there any correlation? Expert Rev Cardiovasc Ther. (2012) 10(3):317–22. doi: 10.1586/erc.11.198
33. Malik V, Lau DH, Linz D, Middeldorp ME, Sanders P. Is the stomach a way to one’s heart? Gastric autonomic activity and AF susceptibility in gastroesophageal reflux disease. J Cardiovasc Electrophysiol. (2019) 30(11):2271–3. doi: 10.1111/jce.14179
34. Huang TC, Lo LW, Yamada S, Chou YH, Lin WL, Chang SL, et al. Gastroesophageal reflux disease and atrial fibrillation: insight from autonomic cardiogastric neural interaction. J Cardiovasc Electrophysiol. (2019) 30(11):2262–70. doi: 10.1111/jce.14181
35. Sheng X, Scherlag BJ, Yu L, Li S, Ali R, Zhang Y, et al. Prevention and reversal of atrial fibrillation inducibility and autonomic remodeling by low-level vagosympathetic nerve stimulation. J Am Coll Cardiol. (2011) 57(5):563–71. doi: 10.1016/j.jacc.2010.09.034
36. Mariani MV, Pierucci N, Piro A, Trivigno S, Chimenti C, Galardo G, et al. Incidence and determinants of spontaneous cardioversion of early onset symptomatic atrial fibrillation. Medicina (B Aires). (2022) 58(11):1513. doi: 10.3390/medicina58111513
Keywords: gastroesophageal reflux disease, atrial fibrillation, Mendelian randomization, genome-wide association study, single nucleotide polymorphisms
Citation: Wang L and Lu YW (2024) Gastroesophageal reflux disease may causally associate with the increased atrial fibrillation risk: evidence from two-sample Mendelian randomization analyses. Front. Cardiovasc. Med. 11:1393383. doi: 10.3389/fcvm.2024.1393383
Received: 29 February 2024; Accepted: 21 May 2024;
Published: 3 June 2024.
Edited by:
Danilo Menichelli, Sapienza University of Rome, ItalyReviewed by:
Carlo Lavalle, Sapienza University of Rome, ItalyVincenzo Mirco La Fazia, Texas Cardiac Arrhythmia Institute, United States
Ilaria Maria Palumbo, Sapienza University of Rome, Italy
© 2024 Wang and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi Wei Lu, bHV5dzkxOUAxNjMuY29t
 Lei Wang
Lei Wang Yi Wei Lu*
Yi Wei Lu*