
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cardiovasc. Med. , 25 June 2024
Sec. Atherosclerosis and Vascular Medicine
Volume 11 - 2024 | https://doi.org/10.3389/fcvm.2024.1388025
 Anton Fliri*
Anton Fliri* Shama Kajiji
Shama Kajiji
Among the leading causes of natural death are cardiovascular diseases, cancer, and respiratory diseases. Factors causing illness include genetic predisposition, aging, stress, chronic inflammation, environmental factors, declining autophagy, and endocrine abnormalities including insufficient vitamin D levels. Inconclusive clinical outcomes of vitamin D supplements in cardiovascular diseases demonstrate the need to identify cause-effect relationships without bias. We employed a spectral clustering methodology capable of analyzing large diverse datasets for examining the role of vitamin D's genomic and non-genomic signaling in disease in this study. The results of this investigation showed the following: (1) vitamin D regulates multiple reciprocal feedback loops including p53, macrophage autophagy, nitric oxide, and redox-signaling; (2) these regulatory schemes are involved in over 2,000 diseases. Furthermore, the balance between genomic and non-genomic signaling by vitamin D affects autophagy regulation of macrophage polarization in tissue homeostasis. These findings provide a deeper understanding of how interactions between genomic and non-genomic signaling affect vitamin D pharmacology and offer opportunities for increasing the efficacy of vitamin D-centered treatment of cardiovascular disease and healthy lifespans.
Atherosclerosis is one of the prominent pathological conditions that lead to myocardial infarction and stroke. A significant risk factor for its development is chronological age (1), accelerated by epigenetic aging (2). Aging is associated with declines in cellular homeostasis causing inflammation (3–6) and organ system dysfunction (7–9), thereby resulting in the distortion of the healthy balance between inflammation and tissue homeostasis (10). Aging affects this balance in multiple ways including decreased efficiency of cellular housekeeping and tissue repair functions (11–13), which leads to the accumulation of cellular debris (14), increased endoplasmic reticulum stress (15) defective tissue homeostasis (16, 17), declining endocrine functions (18) changes in RNA compositions (19) immune senescence (20, 21), and increased inflammation (22, 23). Understanding how interactions between these systems can affect the outcomes of pharmacological studies (24), offering opportunities to slow the effects of aging (25) and prevent the development of associated diseases (26–28).
The central actors in maintaining tissue homeostasis are macrophages (29, 30), dendritic cells (31, 32), stem cells (33), and vitamin D (34–38). However, our understanding of their interaction remains incomplete, as evidenced by multiple inconclusive clinical trials seeking to halt the progression of cardiovascular disease associated with vitamin D deficiency (39–43). In previous reports, we have reported that vitamin D deficiency affects the tipping points that control network–network interactions in over 500 diseases, including atherosclerosis (44).
Cause–effect relationships in complex biological network systems are determined using an information theory-based analysis methodology that is developed to ascertain information flows induced by drugs and diseases (probes) in biological networks (45). This cause–effect relationship-driven analysis approach identifies global network architectures mediating probe-induced information transfers within and between tissues and ascertains how probes affect network linkage (46–54). The statistical value of solutions resulting from this analysis is extremely significant because the global network architecture discovered in system-wide cause–effect analysis links information in large, orthogonal, and decentralized data sets. Furthermore, no single data point can distort the global network architecture imposed by the functional and structural interdependence of subunits linked in cause–effect relationships.
Functional analysis of genomic and non-genomic signaling by vitamin D in 4,821 diseases as illustrated in Figure 1 involved use of (1) the data mining algorithm developed by SystaMedic Inc. in collaboration with the University of Connecticut, (2) the databases and data analysis tools of the STRING platform (55), (3) tissue-associated protein expression data identified in the Human Protein Atlas (56), (4) information published in Medline (57), and (5) data analysis and visualization tools of Spotfire (58). Hierarchical clustering of similarity matrices obtained through Medline data extraction was used for identifying global network architectures linking probe-induced information flows in tissue–protein networks and cause–effect relationships (59–63).
Tissue-specific protein expression data (tissue proteomes) extracted from the Human Protein Atlas are processed using gene enrichment analysis in the STRING platform. The output of this analysis allows the identification of tissue-associated protein network fragments that contain no more than five proteins capable of interacting in a swarm-like fashion with other proteins in response to probe-induced system perturbations. Henceforth, the term protein swarms in lieu of protein network fragments is used in this analysis. Identifying and collecting protein swarms representing all tissues in the Human Protein Atlas provides descriptor sets that can be used for determining co-citation frequencies of probes and protein swarms in Medline. Viewing co-citation frequencies measurements as estimates of a probe's capacity to affect information transfers within and between protein swarms, hierarchical clustering of similarity matrices obtained through Medline data extraction identifies global network architectures linking probe-induced information flows in tissue–protein networks and cause–effect relationships.,
The term vitamin D herein refers to the hormone 1,25-dihydroxyvitamin D3 (calcitriol) and not its precursor, calcidiol, which is commonly used for measuring vitamin D deficiencies (64). In turn, genomic and non-genomic signaling refers to the catenation of functions resulting from (1) the binding of calcitriol at picomolar concentrations to the nuclear vitamin D receptor (VDR) (65) and (2) the binding of calcitriol at nanomolar concentrations to the membrane receptor PDIA3, also known as ERp57 and grp58 (66, 67). PDIA3 is an endoplasmic reticulum chaperone with protein disulfide isomerase activity. It regulates vitamin D genomic signaling (68); apoptosis (69, 70); efferocytosis (71), mitochondrial functions (72); functions of macrophages (73), dendritic cells (74, 75), and stem cells (76); progranulin (77); and functions of the innate and adaptive immune system (78).
Identification of co-citation frequencies of 4,821 diseases with 7,371 protein swarms in Medline, construction of a 4,821 × 7,371 similarity matrix, and UPGMA hierarchical clustering of the similarity matrix using cosine as similarity measure were used to identify and select swarm clusters linked within a confidence in cluster similarity value (CCSV)of >0.92 with swarms containing vitamin D receptors (79). This process identified 29 discrete clusters containing in aggregate 656 protein swarms holding 363 proteins (disease-associated vitamin D interactome; Table 1).
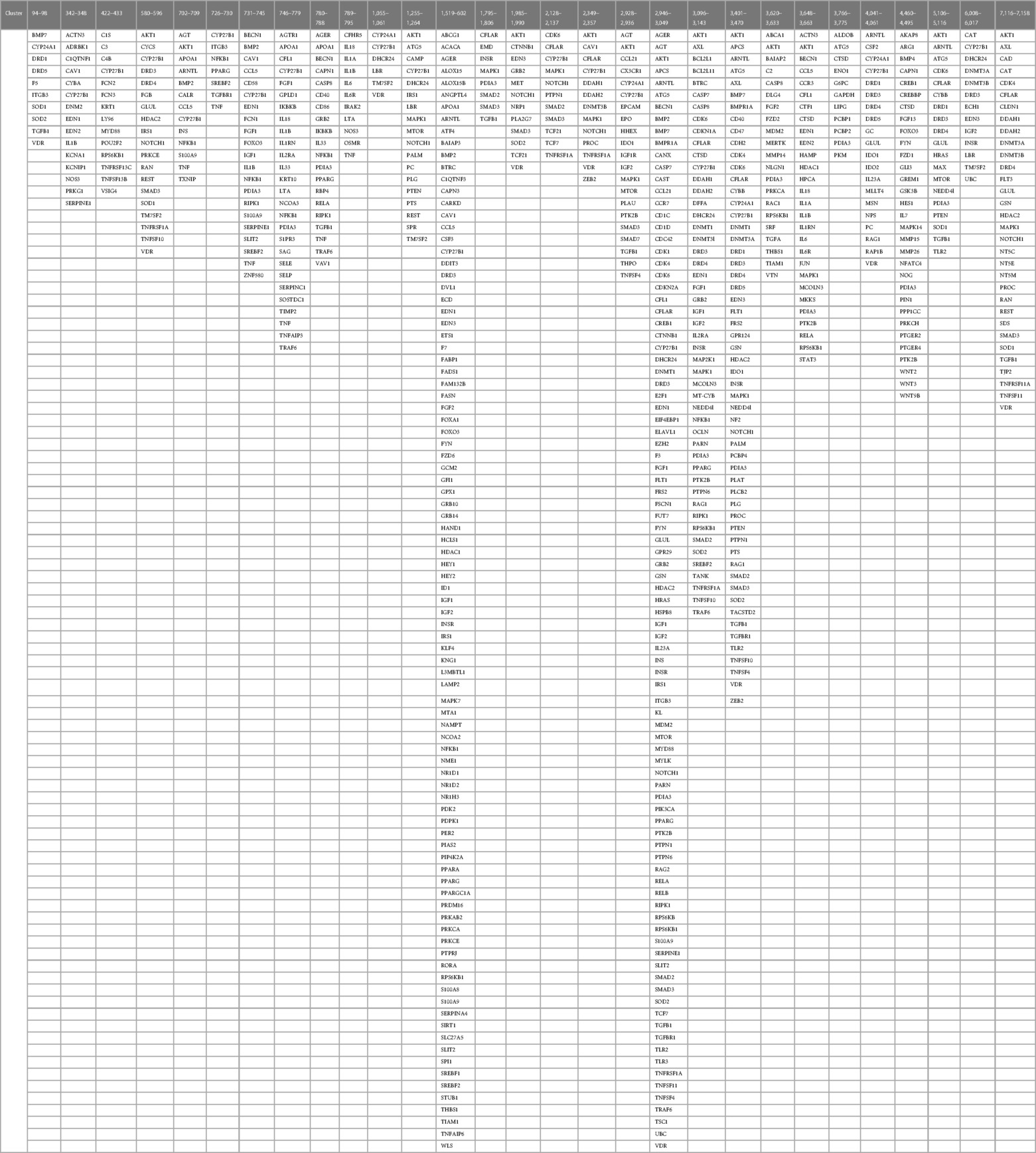
Table 1 The proteins regulating vitamin D's genomic and non-genomic pharmacologies in 4,821 diseases isolated from 29 discrete swarm clusters containing in aggregate 363 proteins (disease-associated vitamin D interactome).
Figure 2 shows a protein network constructed from 363 proteins that were identified in 656 protein swarms described in Table 1. Further analysis to determine known disease associations of these proteins showed that 234 out of the 363 proteins in the vitamin D interactome have statistically significant disease associations (p-value 2.06 × 10−33). These findings establish the linkage between vitamin D signaling and the 4,821 diseases studied
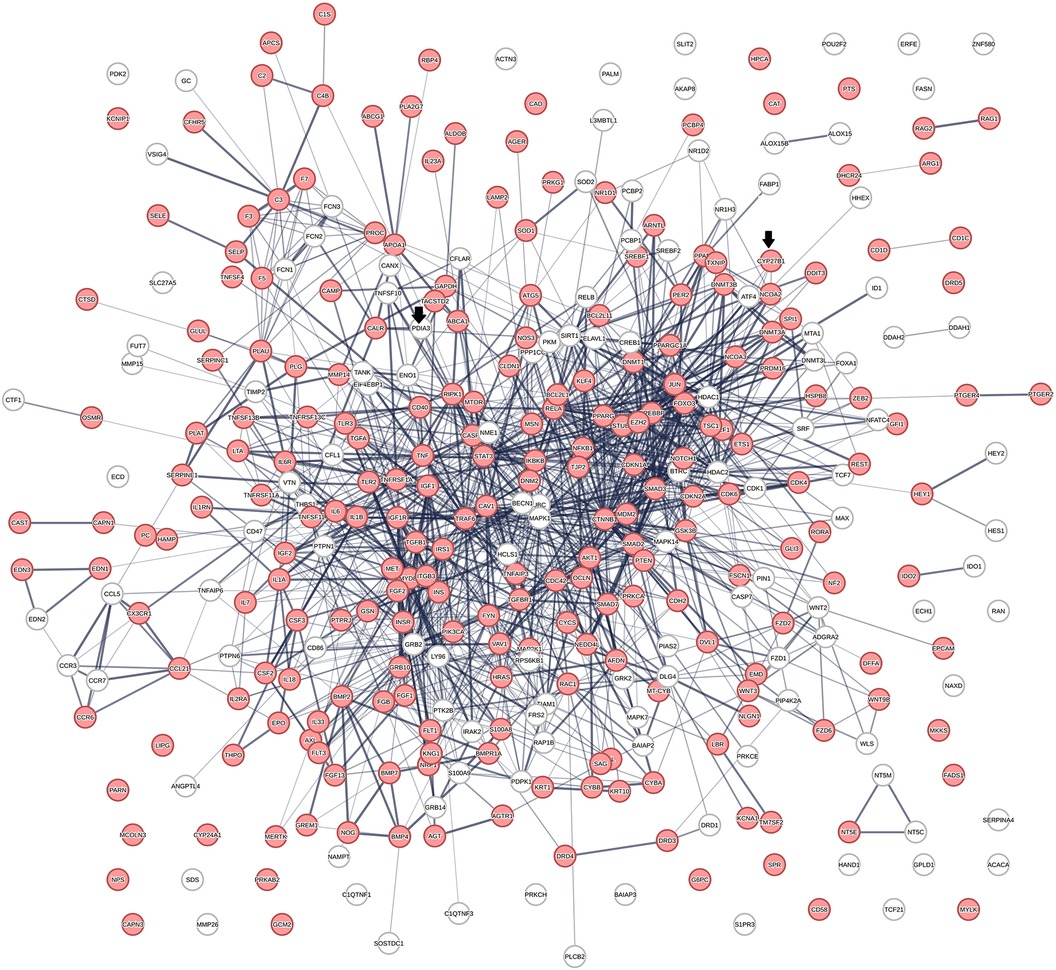
Figure 2 The physical interactions between 363 proteins captured in 29 vitamin D receptors containing swarm clusters. A total of 234 proteins (red) have known disease associations.
Determination of the role of vitamin D signaling in 4,821 diseases and identification of cause–effect relationships associated with cardiovascular disease phenotypes
Vitamin D receptor-associated protein swarms were labeled according to cluster membership and used for collecting co-citation frequencies associated with 4,821 diseases in a second round of Medline data mining. Hierarchical clustering of the resulting 4,821 × 656 similarity matrix followed by data visualization provided the heatmap shown in Figure 3.
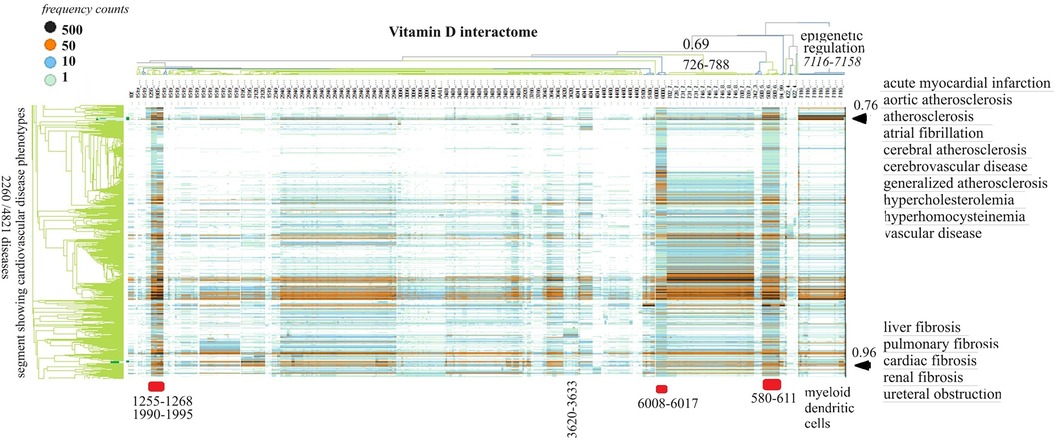
Figure 3 The horizontal dendrogram identifies the clusters of protein swarms that contain proteins engaged in similar functions. The vertical axis identifies the groups of diseases with similar cause–effect relationships. The hierarchical organization of protein swarms (CCSV > 0.69) shows that vitamin D-regulated processes are tightly coupled and subject to epigenetic regulation involving functions of myeloid dendritic cells (cluster 7,116–7,158).
Inspection of co-citation frequencies associated with disease and swarm clusters suggests that proteins in swarm clusters 580–611, 1,255–1,268, 1,990–1,995, and 6,008–6,017 (underscored in red) play a prominent role in 2,260 diseases. Furthermore, the dominant position of swarm cluster 7,117–7,158, in the hierarchical organization of protein swarms, indicates that vitamin D-mediated signaling is subject to epigenetic regulation involving myeloid dendritic cells (80, 81). Protein network construction using proteins in clusters 7,116–7,158 followed by functional analysis identified that protein interactions in these clusters regulate nitric oxide signaling (82), redox signaling (83), DNA methylation (84), and histone deacetylation (85). Similarly, the evaluation of cause–effect relationship similarities between diseases revealed that atherosclerosis, acute myocardial infarction, atrial fibrillation, cerebrovascular disease, hypercholesterolemia, hyperhomocysteinemia, and vascular disease are subject to similar regulatory mechanisms involving swarm clusters 580–611, 1,255–1,268, 1,990–1,995, 6,008–6,017, and 7,116–7,158 (CCSV > 0.76) (29).
The effects of vitamin D signaling in cardiovascular diseases were further examined in three separate categories: (1) genomic signaling, (2) a mixture of genomic and non-genomic, and (3) non-genomic signaling.
The initial focus was to determine functions regulated by proteins in swarms 7,117–7,158, 580–611, 1,255–1,268, 1,990–1,995, and 6,008–6,017 involved in 2,260 diseases containing the nuclear vitamin D receptor. Network construction using proteins in these clusters and retrieval of 10,000 publications with statistically significant protein network overlaps identified cardiovascular disease-associated functions regulated by the nuclear vitamin D receptor. Network overlaps between these functions are shown in Table 2. The results indicate that vitamin D's genomic signaling regulates protein–protein interactions involved in atherosclerosis, efferocytosis (B) (86, 87), macro-autophagy (C) (88), macrophage autophagy (D) (89, 90), macrophage polarization (E) (91, 92), cellular senescence (F) (93), ER stress response (G) (94), tissue homeostasis (H) (95), innate immunity (I) (96). caveola-mediated endocytosis (J) (97), and apoptosis (K) (98).
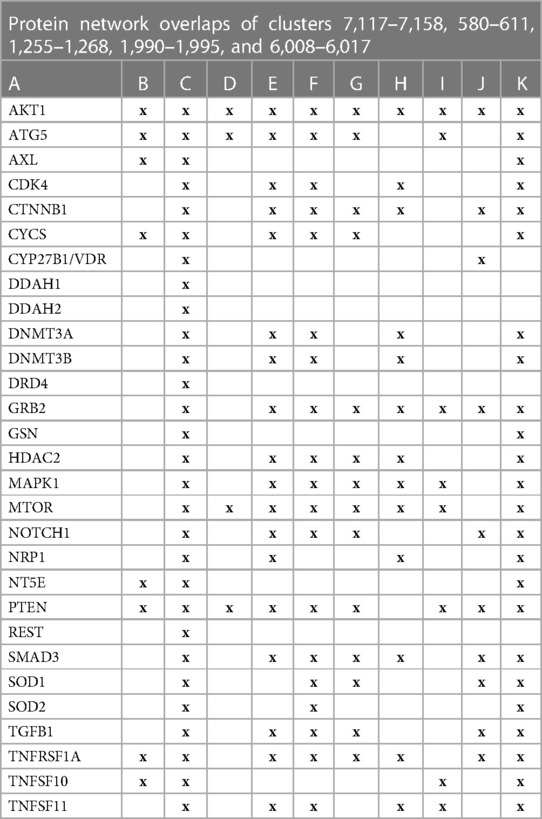
Table 2 The interactions between proteins in swarm clusters 7,117–7,158, 580–611, 1,255–1,268, 1,990–1,995, and 6,008–6,017 and overlaps of protein networks associated with atherosclerosis (A), efferocytosis (B), macro-autophagy (C), macrophage autophagy (D), macrophage polarization (E), cellular senescence (F), ER stress response (G), tissue homeostasis (H), innate immunity (I), caveola-mediated endocytosis (J), and apoptosis (K).
A protein network-centered view of results shown in Table 2 demonstrates that genomic signaling of vitamin D regulates at least 10 different functions involved in atherosclerosis (99–102) (Figure 4). Moreover, extensive protein network overlaps between these functions suggest that vitamin D's genomic signaling modulates a monitoring scheme that responds to nitric oxide signaling (DDAH1, DDAH2) (103), redox signaling (SOD1, SOD2, PDIA3) (104), environmental signals (DNMT3A, DNMT3B) (105, 106), histone acetylation (HDAC2) (107), inflammatory signals (TNFRSF1A, TNFSF10) (108), growth factor signaling (TGFB1, GRB2, NRP1) (109–111), mitochondrial status (CYS) (112), metabolic changes (MTOR) (113), and calcium signaling (GLSN) (114). Reflecting the importance of macrophages in atherosclerosis, Figure 4 shows that this regulatory scheme integrates macrophage polarization (115) and macrophage autophagy (88).
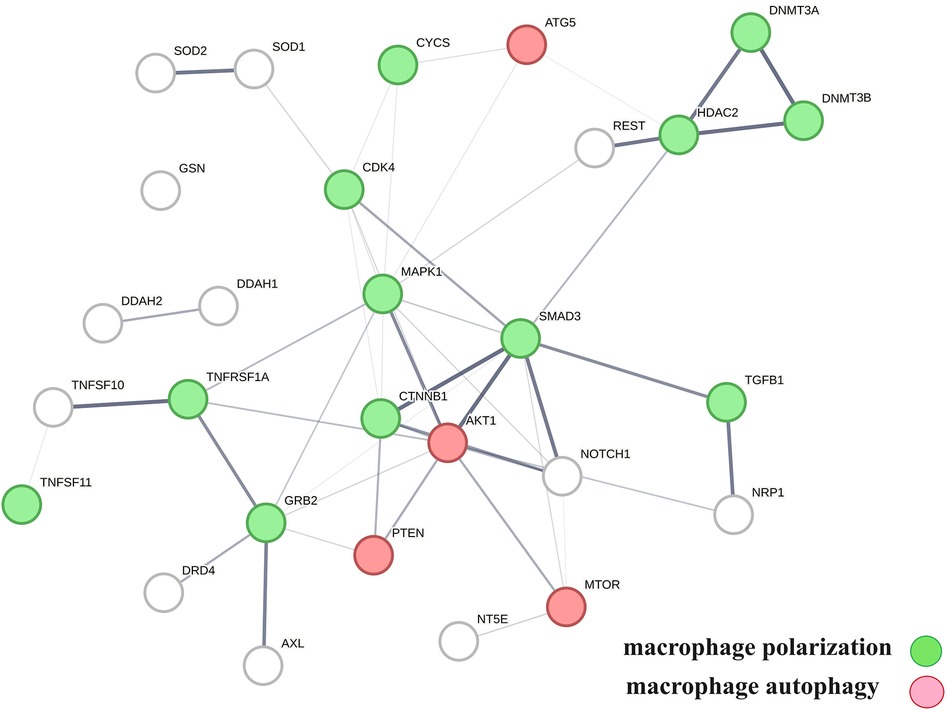
Figure 4 The physical interactions between proteins in swarm clusters 7,117–7,158, 580–611, 1,255–1,268, 1,990–1,995, and 6,008–6,017, highlighting the overlaps between protein networks associated with atherosclerosis (no color), macrophage polarization (green), and macrophage autophagy (red). Proteins ATG5, AKT1, and PTEN are involved in the regulation of all functions.
To evaluate how the interaction between vitamin D's genomic and non-genomic signaling affects atherosclerosis, we focused on swarm cluster 726–788 (CCSV 0.99) containing the nuclear and non-nuclear vitamin D receptors. Protein network construction and retrieval of publications with statistically significant protein network overlaps identified cardiovascular disease-associated functions (Table 3).
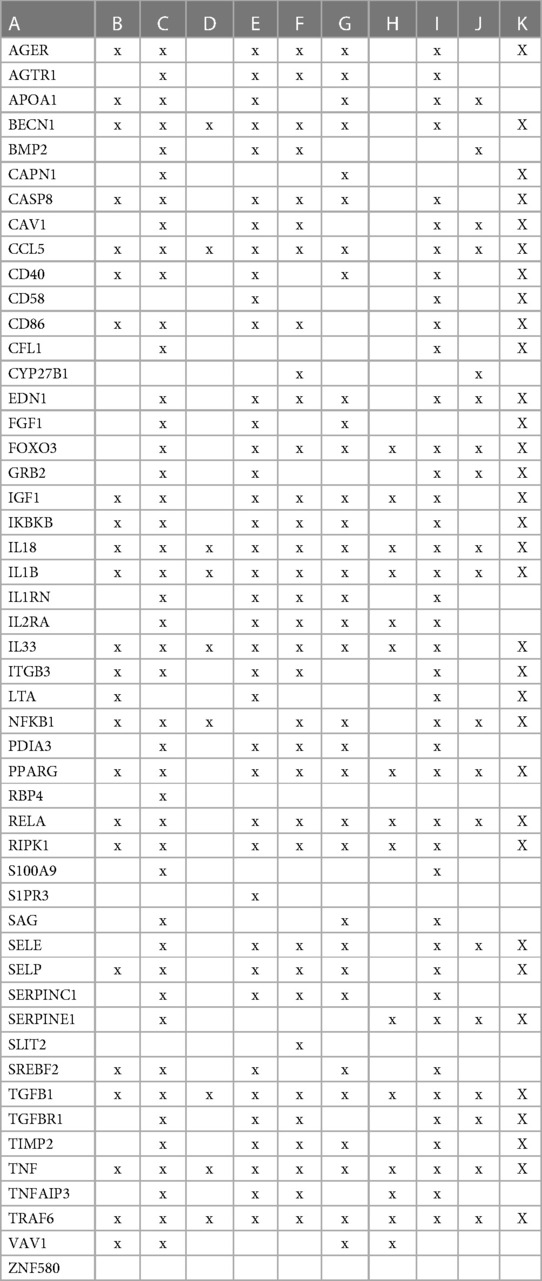
Table 3 The interactions between proteins in swarm cluster 726–788 and overlaps of protein networks associated with atherosclerosis (A), efferocytosis (B), macro-autophagy (C), macrophage autophagy (D), macrophage polarization (E), cellular senescence (F), ER stress response (G), tissue homeostasis (H), innate immunity (I), caveola-mediated endocytosis (J), and apoptosis (K).
A protein network-centered view of Table 3 is shown in Figure 5. It highlights the interactions between proteins regulating macrophage polarization and macrophage autophagy in atherosclerosis (21). Specifically, the proteins colored in green regulate macrophage polarization, and the proteins highlighted in red are constituents of the NF-κB complex, which regulates inflammatory responses (116), autophagy, and macrophage autophagy (117). IL18 and TRAF6 nodes in the macrophage autophagy network regulate inflammatory processes and plaque instability in atherosclerosis (118, 119). Thus, this indicates that a mix of genomic and non-genomic signaling is involved in the regulation of atherosclerotic plaque stability.
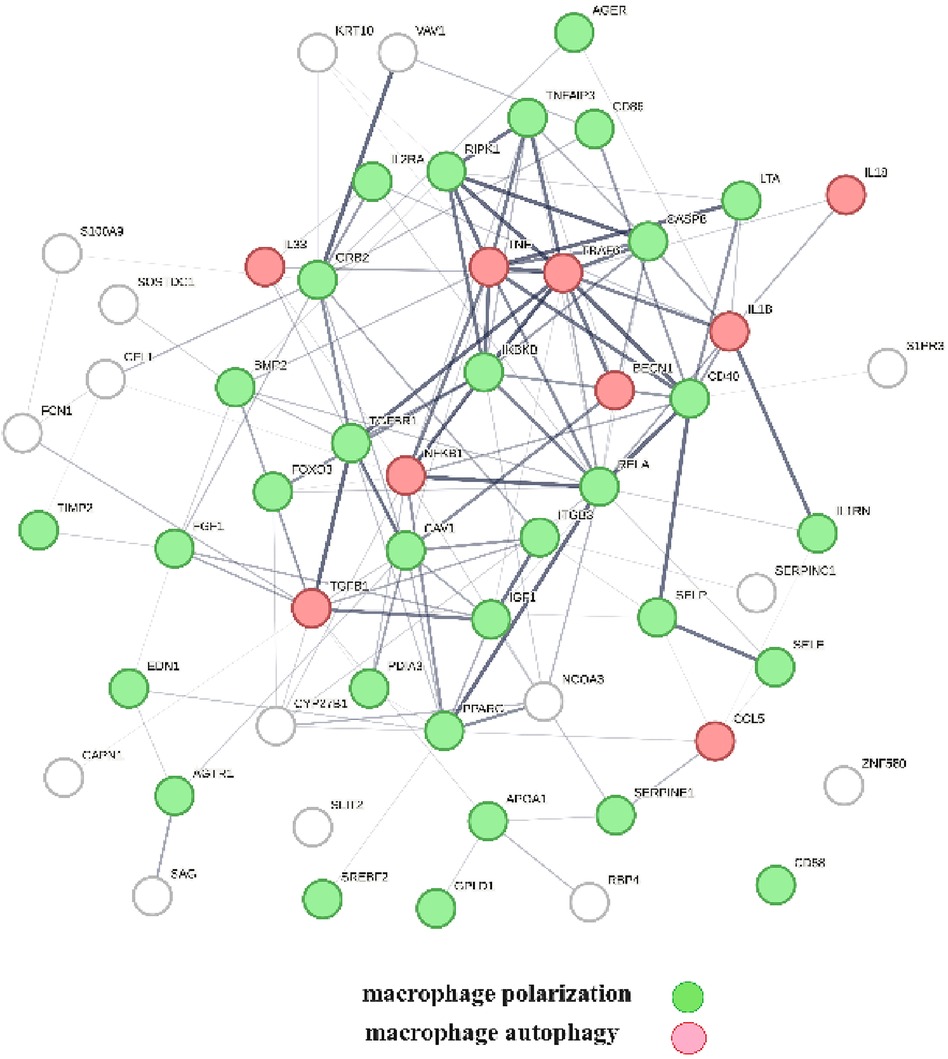
Figure 5 The protein interactions regulating vitamin D's genomic and non-genomic signaling in atherosclerosis. Proteins colored in green regulate macrophage polarization, and proteins colored in red regulate macrophage autophagy.
The first step prior to conducting cause–effect analysis was to identify clusters that only contained the non-nuclear vitamin D receptors. Specifically, there are clusters 3,780–788, 1,795–1,806, 3,620–3,633, 3,648–3,663, 3,766–3,775, 4,460–4,495, and 5,106–5,116. The next step was to evaluate if proteins in these clusters could physically interact with vitamin D's non-genomic receptor, i.e., PDIA3. This analysis identified that PDIA3 can physically interact with AKT1, BAIAP2, ENO1, GAPDH, GSK3B, MTOR, NFKB1, PCBP2, PCBP2, PDIA3, PPARG, SOD1, STAT3, and TGFB1. Protein network construction using this group of proteins and retrieval of publications with statistically significant network overlap identified that the interactions between these proteins modulate PI3K/AKT/mTOR signaling and that the natural product rutin, targeting this regulatory scheme, displays a U-shaped (non-linear) dose–response relationship in cardiomyocytes (120).
The involvement of the PDIA3 interactome in atherosclerosis was further assessed by identifying potential network–network interactions regulated by proteins in swarm cluster 3,620–3,633 (CCSV0.99) containing PDIA3 and proteins expressed in all tissues. Protein network construction using proteins in 3,620–3,633 clusters and retrieval of publications with statistically significant protein network overlaps identified cardiovascular disease-associated functions shown in Table 4. In contrast to the observations in Tables 2, 3, the absence of proteins regulating macrophage autophagy in Table 4 suggests that vitamin D's genomic and non-genomic signaling play different roles in regulating autophagy.
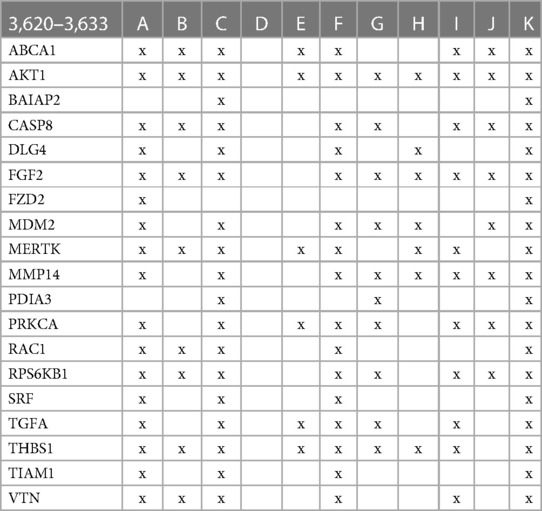
Table 4 The interactions between proteins in swarm cluster 3,620–3,633 and overlaps of protein networks associated with atherosclerosis (A), efferocytosis (B), macro-autophagy (C), macrophage autophagy (D), macrophage polarization (E), cellular senescence (F), ER stress response (G), tissue homeostasis (H), innate immunity (I), caveola-mediated endocytosis (J), and apoptosis (K).
Figure 6 provides a protein network-centered view of Table 4 and shows that proteins regulating vitamin D's non-genomic pharmacology can physically interact and affect atherosclerosis by regulating the balance between autophagy (121), apoptosis (122), endoplasmic reticulum stress response (94), cellular senescence (123, 124), and tissue homeostasis (125).
A key finding based on analysis of protein interactions in this network is that PDIA3 interactome (Figure 6) plays a key role in atherosclerosis. For example, interactions of PDIA3 with AKT1 (126) and interactions of AKT1with TIAM1 regulate the expression of the multicellular protein osteopontin (127, 128), which is involved in generating the atheromatous pathology of atherosclerosis (129). Furthermore, interactions of PDIA3 with brain-specific angiogenesis inhibitor 1-associated protein 2 (BAIAP2; a.k.a. IRSP53) regulate apoptotic cell clearance (130), plasma membrane shape homeostasis, and endocytosis. Mutations in tyrosine kinase-defective Mertk receptor MERTK decreases macrophage autophagy and increases the size of necrotic plaque in atherosclerosis (131, 132). Interactions of BAIAP2 with TIAM1, RAC1, and DLG4 regulate filopodia extension via actin polymerization (133) and regulate LDL metabolism by macrophages (134, 135). In addition, interactions of BAIAP2 with the WAVE regulatory complex (136, 137) control highly dynamic processes involved in cholesterol clearance, embryogenesis, neuron morphogenesis and plasticity, immune cell activation, chemotaxis, fibrosis, and cancer invasion and metastasis (138–141).
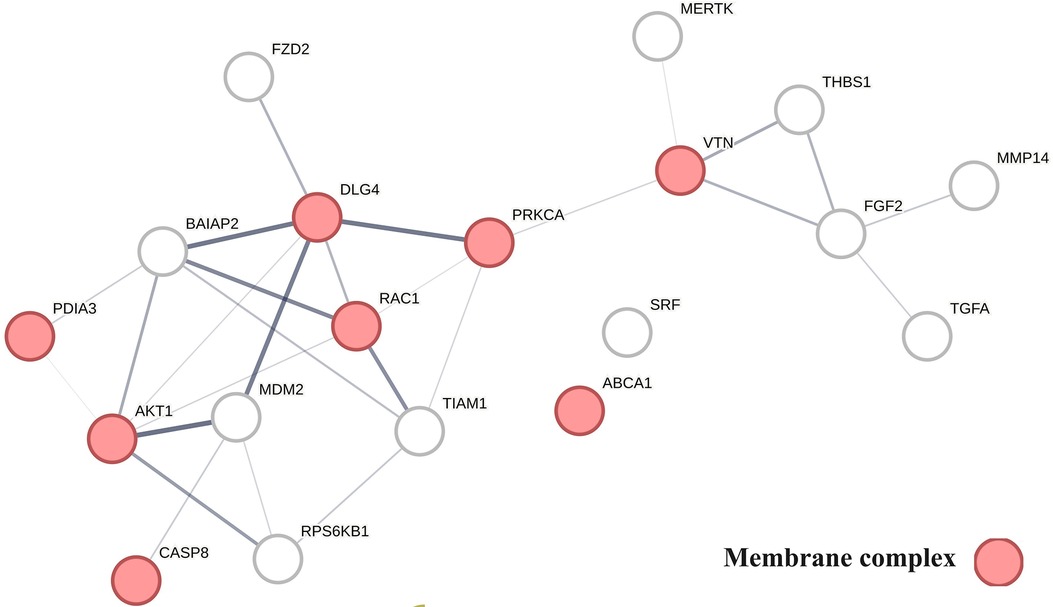
Figure 6 The physical interactions between proteins regulating vitamin D's non-genomic pharmacology in atherosclerosis.
The finding that PDIA3 interactome regulates multiple reciprocal feedback loops is supported by observations showing that calcitriol, in combination with p53 aka TP53 regulates the expression of MDM2 (142), occupying a central position as shown in Figure 6. Further, the regulation of MDM2 expression creates an autoregulatory feedback loop wherein the expression of MDM2 is coregulated by TP53 protein levels and MDM2 protein levels decrease the ability of TP53 to function as a positive transcription factor for MDM2 (143). Since the regulation of TP53 expression regulates the inactivation of SREBP2, this feedback loop regulates the activities of mevalonate and PCSK9 pathways, which are key players in atherosclerosis (144, 145). Similarly, reciprocal regulation of TP53 expression and nitric oxide levels via regulation of expression of nitric oxide synthases (NOS2, NOS3) (146, 147) and methylation and demethylation of arginine (swarm cluster 7,116–7,158) creates additional feedback loops (148, 149). Closing these feedback loops, MDM2 regulates the expression of the nuclear vitamin D receptor (150, 151), and calcitriol, in coordination with TGFB1, regulates PDIA3 expression (152, 153). Thus, these two feedback loops regulate the ratio of nuclear and non-nuclear expression of vitamin D receptors. This finding suggests that this form of regulation can fine-tune the ratio between genomic and non-genomic signaling of vitamin D and can operate independently from the binding affinities of calcitriol. Since vitamin D effects are antifibrotic whereas PDIA3 effects are profibrotic, these feedback loops may be involved in balancing processes involved in tissue homeostasis.
Adding further complexity to the TP53-mediated regulatory scheme is that ER stress-induced posttranslational modifications of P53 affect the sensitivity of MDM2 and TP53 feedback loops (154, 155). The impact of these feedback loops on the outcomes of pharmacological studies using vitamin D is manifested in clinical and preclinical observations showing U-shaped dose–response relationships (156–160).
This study provides deeper insights into the role of vitamin D in cardiovascular disease development. The use of a spectral clustering method for identifying vitamin D-regulated processes in 4,821 diseases provides a global effect positioning system that can be used to determine cause–effect relationships in atherosclerosis. Examination of vitamin D-associated genomic and non-genomic pharmacologies using this positioning system revealed origins of non-linear dose–response relationships observed in preclinical and clinical studies with vitamin D. In addition, it also identified functional relationships between multiple reciprocal feedback loops that sometimes regulate the opposing functions activated by nuclear and membrane-associated vitamin D receptors. Among the processes of relevance for the treatment of cardiovascular diseases are p53-mediated regulation of atherosclerotic plaque stability and macrophage autophagy, which are known to safeguard the plasticity of macrophages in tissue homeostasis. We have used an unsupervised machine learning approach for deciphering these complex cause–effect relationships and used protein network analysis for identifying interactions between protein networks that regulated the functions that are of relevance in atherosclerosis. The initial step was aimed at establishing a global effect positioning system (Table 1) using information theory-based methodology to allow for an unbiased analysis of vitamin D pharmacology in diseases. Information flow-based cause–effect analysis of 4,821 diseases led to the identification of the vitamin D interactome (Figure 2) comprising 363 proteins, where 64% of proteins were found to have highly statistically significant (p-value = 2.06 × 10−33) disease associations. This observation establishes the value of our protein swarm-based framework for investigating cause–effect relationships in protein–protein interaction (PPI) networks and complements previous work reported by Silverbush and Sharan (161) who used drug response and cancer genomic data to orient the human PPI network.
Examination of the role of the vitamin D interactome in 4,821 diseases provided two key findings: (1) vitamin D-regulated processes are involved in over 2,000 diseases including cardiovascular disease, and (2) all functions regulated by vitamin D are subject to epigenetic regulation (Figure 3). Epigenetic regulation of atherosclerosis is well established (162), and our finding of the involvement of vitamin D signaling contributes to the development of diagnostics or therapeutic agents in atherosclerosis. Also depicted in Figure 3 is the observation that cardiovascular diseases including atherosclerosis are affected by PPI in swarms 580–611, 1,255–1,268, 1,990–1,995, 6,008–6,017, and 7,116–7,158.
The effects of vitamin D signaling in cardiovascular diseases were further examined as three separate categories: (1) genomic signaling mediated by PPI in swarms 580–611, 1,255–1,268, 1,990–1,995, 6,008–6,017, and 7,116–7,158 (Table 2), (2) mixture of genomic and non-genomic signaling mediated by PPI in swarms 726–788 (Table 3), and (3) non-genomic signaling mediated by PPI in swarms 3,620–3,633 (Table 4). Tables 2–4 show the results of evidence-based cause–effect analysis using network overlaps of proteins in the three swarm categories with PPI networks of atherosclerosis and the 10 biological functions affected by aging and involved in atherosclerosis. The significance of vitamin D's genomic, mixed genomic, and non-genomic signaling in atherosclerosis related to the 10 biological functions associated with atherosclerosis (Figures 4–6) was determined by measuring the extent of network overlap, ability to engage in physical interactions, and centrality of shared network nodes. Hence, the main inference from the results shown in Tables 2, 3 and Figures 4, 5 is that genomic and mixed genomic signaling by vitamin D modulates the 10 important biological functions associated with atherosclerosis. Additionally, examination of network overlaps between these 10 biological functions indicated that they are part of an integrated regulatory scheme that includes macrophage polarization and macrophage autophagy as central elements.
The effects of vitamin D's non-genomic signaling are summarized in Table 4 and Figure 6. The absence of overlap between proteins in swarm cluster 3,620–3,633 and macrophage autophagy in combination with the observation that these clusters are enriched with proteins involved in fibrosis suggests that non-genomic signaling by vitamin D plays an important role in tissue homeostasis.
The identification of protein network–network interactions described above allows the generation of a comprehensive view detailing how interactions between the 10 biological functions affect atherosclerosis.
A critical factor in atherosclerosis-associated mortality is plaque stability (163), where stable plaque forms carry a low risk of sudden mortality while rupture of unstable forms causes myocardial infarction and stroke. Our analysis (Tables 2, 3; Figures 4, 5) indicates that vitamin D signaling plays a key role in balancing plaque stability (164) by regulating macrophage autophagy (90) and macrophage polarization, a key determinant of the M1/M2 ratio, where macrophages with M1-like characteristics are associated with the inflammatory stages of atherosclerosis while macrophages with M2-like characteristics have anti-inflammatory characteristics and promote plaque regression (91, 165). Declining macrophage autophagy increases macrophage apoptosis, decreases macrophage efferocytosis, increases oxidative stress, and destabilizes plaque (132, 166).
A unique property of macrophages is that they can directly generate calcitriol, which is the most active form of vitamin D, in response to a variety of input signals (167). Calcitriol, in turn, activates macrophage autophagy and shifts the macrophage polarization toward anti-inflammatory M2-like macrophages (168, 169). Thus, by shifting macrophage polarization toward M2 (170), calcitriol can decrease inflammation (171) and increase the clearance of cellular debris by efferocytosis (172).
Additionally, of relevance to the development of atherosclerosis is aging. Aging shifts macrophage polarization toward M1 phenotypes (173), increases cellular senescence (174) and inflammation (175), and decreases macrophage functions (176). Calcitriol can counter aging-induced acceleration of atherosclerosis at the plaque initiation (177–179), progression (179, 180), rupture (181), regression (182–184), and healing stages of atherosclerosis (179, 185). Accordingly, shifting macrophage ratios from M1 to M2 controls plaque size and stability (170). For example, low shear stress or high endoplasmic reticulum negatively affects autophagy, and shifting macrophage polarization toward inflammatory M1 accelerates atherosclerosis (186–190). Vitamin D deficiency exacerbates this development by increasing M1 macrophage populations and decreasing autophagy (115, 191). Calcitriol administration can reverse this trend by shifting macrophage populations toward M2 phenotypes, which increases macrophage autophagy (192–195).
However, an important finding of our investigation is that the over-enrichment of M2 macrophage populations can result in excessive fibrosis. In other words, vitamin D's non-genomic signaling via PDIA3 at higher calcitriol concentrations promotes fibrosis, leading potentially to plaque destabilization (196). Maintenance of the delicate balance between M1 and M2 macrophage subpopulations is critical as it affects tissue homeostasis. Experimental data suggest that too much or too little on either side of the equation results in detrimental outcomes (197). This balance can be affected in multiple ways. For example, the involvement of immune responses in this delicate balancing act is implicated by observations that cathelicidins produced by vitamin D's genomic response to infections activate autophagy and shift macrophage populations toward M2 (198). In addition, the identification of regulatory schemes clearly establishes that vitamin D regulates multiple feedback loops (76, 199–203) involved in tissue homeostasis, which are subject to additional regulation by nitric oxide (204) and redox signaling (205). Evidence of these types of non-linear (U-shaped) cause–effect relationships/dose responses, expected due to opposing pharmacologies, is indeed observed in numerous preclinical and clinical studies with vitamin D [see references (156–160)].
The dynamics of protein–protein interactions impart functional diversity to proteomes and balance functions separating health and diseases. From a whole organism perspective, single-cell approaches do not provide the system-level insight that is needed to understand how interactions between proteomes at tissue levels affect the progression of a disease or the responses of organisms to treatments (206). To generate the required insights, integrated approaches are needed that can determine relationships between large heterogeneous, unstructured, and noisy data stored in different databases. By taking advantage of the modular designs of proteomes and principles governing information transfers in integrated systems (207, 208), protein swarm-based cause–effect analysis delivers unbiased insight into how interactions between tissue- and cell-associated proteomes regulate the physiological and pathological states of organisms.
“Swarm Intelligence” based analysis reveals cause-effect relationship between the many variables involved and provides understanding that is very difficult to ascertain with other means.
The reach of the current vitamin D-centered cause–effect analysis is limited by its focus on determining the system pharmacology of calcitriol. Therefore, our study neglects the potential contributions of calcidiol or the various vitamin D metabolites. Furthermore, technical constraints limit the resolution of network–network interactions by setting upper limits to the number of protein swarms used for sampling tissue proteomes. Lastly, the limiting capability of machine learning approaches in the biological cause–effect analysis is the scarcity of data on the concentration time dependency of dynamic cause–effect relationships at a system-wide scale.
Using a global framework for proteomics-based cause–effect analysis, our analysis identified a broad vitamin D-regulated scheme involved in tissue homeostasis of over 2,000 diseases. By providing a perspective on how interactions between vitamin D's genomic and non-genomic actions, nitric oxide and redox signaling, macrophage autophagy, macrophage polarization, and p53 signaling shape tissue homeostasis, our analysis addresses a long-standing enigma presented by observations that vitamin D supplementation, despite projected benefits, produces little benefits in clinical studies (209). Recognizing the importance of p53, nitric oxide, and redox signaling in vitamin D-regulated processes offers the opportunity to increase the efficacy of vitamin D supplementation in cardiovascular diseases. Toward this end, modalities integrating nitric oxide redox signaling and vitamin D-centered pharmacologies have already shown promising outcomes (210). Refinement of these approaches is expected to improve the treatment of cardiovascular diseases and increase the quality of life in aging (211).
The original contributions presented in the study are included in the article/Supplementary Materials; further inquiries can be directed to the corresponding author.
AF: Writing – original draft. SK: Writing – review & editing.
The authors declare that no financial support was received for the research, authorship, and/or publication of this article.
Ron Sostek contributed valuable feedback on use of Swarms intelligence in complex System analysis.
AF and SK are founders of Emergent System Analytics LLC.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2024.1388025/full#supplementary-material
1. Sniderman AD, Furberg CD. Age as a modifiable risk factor for cardiovascular disease. Lancet. (2008) 371:1547–9. doi: 10.1016/S0140-6736(08)60313-X
2. Sánchez-Cabo F, Fuster V, Silla-Castro JC, González G, Lorenzo-Vivas E, Alvarez R, et al. Subclinical atherosclerosis and accelerated epigenetic age mediated by inflammation: a multi-omics study. Eur Heart J. (2023) 44(29):2698–709. doi: 10.1093/eurheartj/ehad361
3. Spagnoli LG, Bonanno E, Sangiorgi G, Mauriello A. Role of inflammation in atherosclerosis. J Nucl Med. (2007) 48(11):1800–15. doi: 10.2967/jnumed.107.038661
4. Ribeiro ASF, Zerolo BE, López-Espuela F, Sánchez R, Fernandes VS. Cardiac system during the aging process. Aging Dis. (2023) 14(4):1105–22. doi: 10.14336/AD.2023.0115
5. North BJ, Sinclair DA. The intersection between aging and cardiovascular disease. Circ Res. (2012) 110(8):1097–108. doi: 10.1161/CIRCRESAHA.111.246876
6. Gogulamudi VR, Durrant JR, Adeyemo AO, Ho HM, Walker AE, Lesniewski LA. Advancing age increases the size and severity of spontaneous atheromas in mouse models of atherosclerosis. Geroscience. (2023) 45(3):1913–31. doi: 10.1007/s11357-023-00776-8
7. Boss GR, Seegmiller JE. Age-related physiological changes and their clinical significance. West J Med. (1981) 135(6):434–40. PMID: 7336713; PMCID: PMC12733167336713
8. Lv J, Zhang C, Liu X, Gu C, Liu Y, Gao Y, et al. An aging-related immune landscape in the hematopoietic immune system. Immun Ageing. (2024) 21(1):3. doi: 10.1186/s12979-023-00403-2
9. Ambale-Venkatesh B, Liu CY, Liu YC, Donekal S, Ohyama Y, Sharma RK, et al. Association of myocardial fibrosis and cardiovascular events: the multi-ethnic study of atherosclerosis. Eur Heart J Cardiovasc Imaging. (2019) 20(2):168–76. doi: 10.1093/ehjci/jey140
10. Meizlish ML, Franklin RA, Zhou X, Medzhitov R. Tissue homeostasis and inflammation. Annu Rev Immunol. (2021) 39:557–81. doi: 10.1146/annurev-immunol-061020-053734
11. Hu H, Cheng X, Li F, Guan Z, Xu J, Wu D, et al. Defective efferocytosis by aged macrophages promotes STING signaling mediated inflammatory liver injury. Cell Death Discov. (2023) 9(1):236. doi: 10.1038/s41420-023-01497-9
12. Zhong W, Rao Z, Xu J, Sun Y, Hu H, Wang P, et al. Defective mitophagy in aged macrophages promotes mitochondrial DNA cytosolic leakage to activate STING signaling during liver sterile inflammation. Aging Cell. (2022) 21(6):e13622. doi: 10.1111/acel.13622
13. Tran M, Reddy PH. Defective autophagy and mitophagy in aging and Alzheimer’s disease. Front Neurosci. (2021) 14:612757. doi: 10.3389/fnins.2020.612757
14. Gladyshev VN, Kritchevsky SB, Clarke SG, Cuervo AM, Fiehn O, de Magalhães JP, et al. Molecular damage in aging. Nat Aging. (2021) 1(12):1096–106. doi: 10.1038/s43587-021-00150-3
15. Zhang Z, Guo Q, Zhao Z, Nie M, Shi Q, Li E, et al. DNMT3B activates FGFR3-mediated endoplasmic reticulum stress by regulating PTPN2 promoter methylation to promote the development of atherosclerosis. FASEB J. (2023) 37(8):e23085. doi: 10.1096/fj.202300665R
16. Jones L, Passegue E. Stem cells homeostasis and aging. Innov Aging. (2023) 7(Suppl 1):259–60. doi: 10.1093/geroni/igad104.0862
17. Li Y, Li Q, Fan GC. Macrophage efferocytosis in cardiac pathophysiology and repair. Shock. (2021) 55(2):177–88. doi: 10.1097/SHK.0000000000001625
18. van den Beld AW, Kaufman JM, Zillikens MC, Lamberts SWJ, Egan JM, van der Lely AJ. The physiology of endocrine systems with ageing. Lancet Diabetes Endocrinol. (2018) 6(8):647–58. doi: 10.1016/S2213-8587(18)30026-3
19. Cookson MR. Aging–RNA in development and disease. Wiley Interdiscip Rev RNA. (2012) 3(1):133–43. doi: 10.1002/wrna.109
20. Smit V, de Mol J, Schaftenaar FH, Depuydt MAC, Postel RJ, Smeets D, et al. Single-cell profiling reveals age-associated immunity in atherosclerosis. Cardiovasc Res. (2023) 119(15):2508–21. doi: 10.1093/cvr/cvad099
21. Popa-Fotea NM, Ferdoschi CE, Micheu MM. Molecular and cellular mechanisms of inflammation in atherosclerosis. Front Cardiovasc Med. (2023) 10:1200341. doi: 10.3389/fcvm.2023.1200341
22. Vidak S, Serebryannyy LA, Pegoraro G, Misteli T. Activation of endoplasmic reticulum stress in premature aging via the inner nuclear membrane protein SUN2. Cell Rep. (2023) 42(5):112534. doi: 10.1016/j.celrep.2023.112534
23. Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. (2008) 454(7203):455–62. doi: 10.1038/nature07203
24. Lopez DV, Al-Jaberi FAH, Woetmann A, Ødum N, Bonefeld CM, Kongsbak-Wismann M, et al. Macrophages control the bioavailability of vitamin D and vitamin D-regulated T cell responses. Front Immunol. (2021) 12:722806. doi: 10.3389/fimmu.2021.722806
25. Sendama W. The effect of ageing on the resolution of inflammation. Ageing Res Rev. (2020) 57:101000. doi: 10.1016/j.arr.2019.101000
26. Rochette L, Dogon G, Rigal E, Zeller M, Cottin Y, Vergely C. Interplay between efferocytosis and atherosclerosis. Arch Cardiovasc Dis. (2023) 116(10):474–84. doi: 10.1016/j.acvd.2023.07.007
27. Guo J, Huang X, Dou L, Yan M, Shen T, Tang W, et al. Aging and aging-related diseases: from molecular mechanisms to interventions and treatments. Signal Transduct Target Ther. (2022) 7(1):391. doi: 10.1038/s41392-022-01251-0
28. Malainou C, Abdin SM, Lachmann N, Matt U, Herold S. Alveolar macrophages in tissue homeostasis, inflammation, and infection: evolving concepts of therapeutic targeting. J Clin Invest. (2023) 133(19):e170501. doi: 10.1172/JCI170501
29. Kojima Y, Weissman IL, Leeper NJ. The role of efferocytosis in atherosclerosis. Circulation. (2017) 135(5):476–89. doi: 10.1161/CIRCULATIONAHA.116.025684
30. Xie L, Chen J, Wang Y, Jin C, Xie Y, Ma H, et al. Emerging roles of macrophages in heart failure and associated treatment approaches. Ther Adv Chronic Dis. (2023) 14:20406223231168755. doi: 10.1177/20406223231168755
31. Malik JA, Zafar MA, Lamba T, Nanda S, Khan MA, Agrewala JN. The impact of aging-induced gut microbiome dysbiosis on dendritic cells and lung diseases. Gut Microbes. (2023) 15(2):2290643. doi: 10.1080/19490976.2023.2290643
32. Suwa Y, Nagafuchi Y, Yamada S, Fujio K. The role of dendritic cells and their immunometabolism in rheumatoid arthritis. Front Immunol. (2023) 14:1161148. doi: 10.3389/fimmu.2023.1161148
33. Ahmed AS, Sheng MH, Wasnik S, Baylink DJ, Lau KW. Effect of aging on stem cells. World J Exp Med. (2017) 7(1):1–10. doi: 10.5493/wjem.v7.i1.1
34. Wang H, Chen W, Li D, Yin X, Zhang X, Olsen N, et al. Vitamin D and chronic diseases. Aging Dis. (2017) 8(3):346–53. doi: 10.14336/AD.2016.1021
35. Khammissa RAG, Fourie J, Motswaledi MH, Ballyram R, Lemmer J, Feller L. The biological activities of vitamin D and its receptor in relation to calcium and bone homeostasis, cancer, immune and cardiovascular systems, skin biology, and oral health. Biomed Res Int. (2018) 2018:9276380. doi: 10.1155/2018/9276380
36. Shi H, Duan J, Wang J, Li H, Wu Z, Wang S, et al. 1,25(OH)2D3 promotes macrophage efferocytosis partly by upregulating ASAP2 transcription via the VDR-bound enhancer region and ASAP2 may affect antiviral immunity. Nutrients. (2022) 14(22):4935. doi: 10.3390/nu14224935
37. Oh J, Riek AE, Darwech I, Funai K, Shao J, Chin K, et al. Deletion of macrophage vitamin D receptor promotes insulin resistance and monocyte cholesterol transport to accelerate atherosclerosis in mice. Cell Rep. (2015) 10(11):1872–86. doi: 10.1016/j.celrep.2015.02.043
38. Kumar S, Nanduri R, Bhagyaraj E, Kalra R, Ahuja N, Chacko AP, et al. Vitamin D3-VDR-PTPN6 axis mediated autophagy contributes to the inhibition of macrophage foam cell formation. Autophagy. (2021) 17(9):2273–89. doi: 10.1080/15548627.2020.1822088
39. Thompson B, Waterhouse M, English DR, McLeod DS, Armstrong BK, Baxter C, et al. Vitamin D supplementation and major cardiovascular events: D-Health randomised controlled trial. BMJ. (2023) 381:e075230. doi: 10.1136/bmj-2023-075230
40. Manolis AA, Manolis TA, Melita H, Manolis AS. Role of vitamins in cardiovascular health: know your facts-part 2. Curr Vasc Pharmacol. (2023) 21(6):399–423. doi: 10.2174/1570161121666230911115725
41. Patriota P, Guessous I, Rezzi S, Marques-Vidal P. Vitamin D levels are associated with cardiovascular disease events but not with cardiovascular disease or overall mortality: a prospective population-based study. Nutrients. (2023) 15(18):4046. doi: 10.3390/nu15184046
42. Cassard SD, Fitzgerald KC, Qian P, Emrich SA, Azevedo CJ, Goodman AD, et al. High-dose vitamin D3 supplementation in relapsing-remitting multiple sclerosis: a randomised clinical trial. EClinicalMedicine. (2023) 59:101957. doi: 10.1016/j.eclinm.2023.101957
43. Tobias DK, Luttmann-Gibson H, Mora S, Danik J, Bubes V, Copeland T, et al. Association of body weight with response to vitamin D supplementation and metabolism. JAMA Netw Open. (2023) 6(1):e2250681. doi: 10.1001/jamanetworkopen.2022.50681
44. Anton FF, Shama K. Vitamin D deficiency-associated comorbidities: a protein network dynamics perspective. Med. Res. Archives. (2023) 11(6):2023. doi: 10.18103/mra.v11i6.3996
45. Fliri AF, Loging WT, Volkmann RA. Analysis of information flows in interaction networks: implication for drug discovery and pharmacological research. Discov Med. (2011) 11(57):133–43. PMID: 2135616821356168
46. Fliri AF, Loging WT, Volkmann RA. Drug effects viewed from a signal transduction network perspective. J Med Chem. (2009) 52(24):8038–46. doi: 10.1021/jm901001p
47. Fliri AF, Kajiji S. Functional characterization of nutraceuticals using spectral clustering: centrality of caveolae-mediated endocytosis for management of nitric oxide and vitamin D deficiencies and atherosclerosis. Front Nutr. (2022) 9:885364. doi: 10.3389/fnut.2022.885364
48. Fierro-Monti I, Wright JC, Choudhary JS, Vizcaíno JA. Identifying individuals using proteomics: are we there yet? Front Mol Biosci. (2022 Nov 29) 9:1062031. doi: 10.3389/fmolb.2022.1062031
49. Zhu X, Shen X, Qu J, Straubinger RM, Jusko WJ. Multi-scale network model supported by proteomics for analysis of combined gemcitabine and birinapant effects in pancreatic cancer cells. CPT Pharmacometrics Syst Pharmacol. (2018 Sep) 7(9):549–61. doi: 10.1002/psp4.12320
50. Kustatscher G, Collins T, Gingras AC, Guo T, Hermjakob H, Ideker T, et al. Understudied proteins: opportunities and challenges for functional proteomics. Nat Methods. (2022) 19(7):774–9. doi: 10.1038/s41592-022-01454-x
51. Ivanov PC, Bartsch RP. Network physiology: mapping interactions between networks of physiologic networks. In: D’Angostino G, Scala A, editors. Networks of Networks: The Last Frontier of Complexity. Cham: Springer (2014). p. 203–22.
52. Broido AD, Clauset A. Scale-free networks are rare. Nat Commun. (2019) 10:1017. doi: 10.1038/s41467-019-08746-5
53. Woessmann J, Kotol D, Hober A, Uhlén M, Edfors F. Addressing the protease bias in quantitative proteomics. J Proteome Res. (2022 Oct 7) 21(10):2526–34. doi: 10.1021/acs.jproteome.2c00491
54. Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem. (2007) 389(4):1017–31. doi: 10.1007/s00216-007-1486-6
55. Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, et al. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. (2021) 49(D1):D605–12. doi: 10.1093/nar/gkaa1074
56. Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. (2015) 347(6220):1260419. doi: 10.1126/science.1260419
57. National Center for Biotechnology Information (NCBI). Bethesda, MD: National Library of Medicine (US), National Center for Biotechnology Information (1988). Available online at: https://www.ncbi.nlm.nih.gov/ (cited April 06, 2017).
58. TIBCO Statistica, v. 13.3.0, TIBCO Software Inc, Palo Alto, CA, USA (2017). Available online at: https://www.tibco.com/products/tibco-statistica (Accessed December 20, 2023).
59. Fliri AF, Loging WT, Volkmann RA. Cause-effect relationships in medicine: a protein network perspective. Trends Pharmacol Sci. (2010) 31(11):547–55. doi: 10.1016/j.tips.2010.07.005
60. Dunn K, Marshall JG, Wells AL, Backus JEB. Examining the role of MEDLINE as a patient care information resource: an analysis of data from the Value of Libraries study. J Med Libr Assoc. (2017 Oct) 105(4):336–46. doi: 10.5195/jmla.2017.87
61. Finlayson SG, LePendu P, Shah NH. Building the graph of medicine from millions of clinical narratives. Sci Data. (2014 Sep 16) 1:140032. doi: 10.1038/sdata.2014.32
62. Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. (2011) 39(Database issue):D561–8. doi: 10.1093/nar/gkq973
63. Kaushal D, Naeve CW. An overview of spotfire for gene-expression studies. Curr Protoc Hum Genet. (2005) 45(1):11.9.1–21. doi: 10.1002/0471142905.hg1109s45
64. Della Nera G, Sabatino L, Gaggini M, Gorini F, Vassalle C. Vitamin D determinants, status, and antioxidant/anti-inflammatory-related effects in cardiovascular risk and disease: not the last word in the controversy. Antioxidants (Basel). (2023) 12(4):948. doi: 10.3390/antiox12040948
65. Daryabor G, Gholijani N, Kahmini FR. A review of the critical role of vitamin D axis on the immune system. Exp Mol Pathol. (2023) 132–133:104866. doi: 10.1016/j.yexmp.2023.104866
66. Gaucci E, Raimondo D, Grillo C, Cervoni L, Altieri F, Nittari G, et al. Analysis of the interaction of calcitriol with the disulfide isomerase ERp57. Sci Rep. (2016) 6:37957. doi: 10.1038/srep37957
67. Żmijewski MA. Nongenomic activities of vitamin D. Nutrients. (2022) 14(23):5104. doi: 10.3390/nu14235104
68. Nowak JI, Olszewska AM, Piotrowska A, Myszczyński K, Domżalski P, Żmijewski MA. PDIA3 modulates genomic response to 1,25-dihydroxyvitamin D3 in squamous cell carcinoma of the skin. Steroids. (2023) 199:109288. doi: 10.1016/j.steroids.2023.109288
69. Tu Z, Ouyang Q, Long X, Wu L, Li J, Zhu X, et al. Protein disulfide-isomerase A3 is a robust prognostic biomarker for cancers and predicts the immunotherapy response effectively. Front. Immunol. (2022) 2022(13):837512. doi: 10.3389/fimmu.2022.837512
70. Zhao G, Lu H, Li C. Proapoptotic activities of protein disulfide isomerase (PDI) and PDIA3 protein, a role of the bcl-2 protein bak. J Biol Chem. (2015) 290(14):8949–63. doi: 10.1074/jbc.M114.619353
71. Cockram TOJ, Dundee JM, Popescu AS, Brown GC. The phagocytic code regulating phagocytosis of mammalian cells. Front Immunol. (2021) 12:629979. doi: 10.3389/fimmu.2021.629979
72. Keasey MP, Razskazovskiy V, Jia C, Peterknecht ED, Bradshaw PC, Hagg T. PDIA3 Inhibits mitochondrial respiratory function in brain endothelial cells and C. elegans through STAT3 signaling and decreases survival after OGD. Cell Commun Signal. (2021) 19(1):119. doi: 10.1186/s12964-021-00794-z
73. Xiao Y, Li C, Gu M, Wang H, Chen W, Luo G, et al. Protein disulfide isomerase silence inhibits inflammatory functions of macrophages by suppressing reactive oxygen species and NF-κB pathway. Inflammation. (2018) 41(2):614–25. doi: 10.1007/s10753-017-0717-z
74. Pol JG, Plantureux C, Pérez-Lanzón M, Kroemer G. PDIA3 as a potential bridge between immunogenic cell death and autoreactivity. Oncoimmunology. (2022) 11(1):2130558. doi: 10.1080/2162402X.2022.2130558
75. Barragan M, Good M, Kolls JK. Regulation of dendritic cell function by vitamin D. Nutrients. (2015) 7(9):8127–51. doi: 10.3390/nu7095383
76. Huang T, You Q, Huang D, Zhang Y, He Z, Shen X, et al. A positive feedback between PDIA3P1 and OCT4 promotes the cancer stem cell properties of esophageal squamous cell carcinoma. Cell Commun Signal. (2024) 22(1):60. doi: 10.1186/s12964-024-01475-3
77. Liu Y, Zhao X, Jian J, Hasan S, Liu C. Interaction with ERp57 is required for progranulin protection against type 2 Gaucher disease. Biosci Trends. (2023) 17(2):126–35. doi: 10.5582/bst.2023.01022
78. Zmijewski MA, Carlberg C. Vitamin D receptor(s): in the nucleus but also at membranes? Exp Dermatol. (2020) 29(9):876–84. doi: 10.1111/exd.14147
79. Fliri AF, Loging WT, Thadeio PF, Volkmann RA. Biospectra analysis: model proteome characterizations for linking molecular structure and biological response. J Med Chem. (2005) 48(22):6918–25. doi: 10.1021/jm050494g
80. Ivashkiv LB, Park SH. Epigenetic regulation of myeloid cells. Microbiol Spectr. (2016) 4(3):1–29. doi: 10.1128/microbiolspec.MCHD-0010-2015
81. Català-Moll F, Ferreté-Bonastre AG, Godoy-Tena G, Morante-Palacios O, Ciudad L, Barberà L, et al. Vitamin D receptor, STAT3, and TET2 cooperate to establish tolerogenesis. Cell Rep. (2022) 38(3):110244. doi: 10.1016/j.celrep.2021.110244
82. Vasudevan D, Bovee RC, Thomas DD. Nitric oxide, the new architect of epigenetic landscapes. Nitric Oxide. (2016) 59:54–62. doi: 10.1016/j.niox.2016.08.002
83. Berridge MJ. Vitamin D, reactive oxygen species and calcium signalling in ageing and disease. Philos Trans R Soc Lond B Biol Sci. (2016) 371(1700):20150434. doi: 10.1098/rstb.2015.0434
84. Fetahu IS, Höbaus J, Kállay E. Vitamin D and the epigenome. Front Physiol. (2014) 5:164. doi: 10.3389/fphys.2014.00164
85. Hossain S, Liu Z, Wood RJ. Histone deacetylase activity and vitamin D-dependent gene expressions in relation to sulforaphane in human breast cancer cells. J Food Biochem. (2020) 44(2):e13114. doi: 10.1111/jfbc.13114
86. Fliri AF, Loging WT, Thadeio PF, Volkmann RA. Biological spectra analysis: linking biological activity profiles to molecular structure. Proc Natl Acad Sci U S A. (2005) 102(2):261–6. doi: 10.1073/pnas.0407790101
87. Baraniecki Ł, Tokarz-Deptuła B, Syrenicz A, Deptuła W. Macrophage efferocytosis in atherosclerosis. Scand J Immunol. (2023) 97(5):e13251. doi: 10.1111/sji.13251
88. Sergin I, Razani B. Self-eating in the plaque: what macrophage autophagy reveals about atherosclerosis. Trends Endocrinol Metab. (2014) 25(5):225–34. doi: 10.1016/j.tem.2014.03.010
89. Ni D, Mo Z, Yi G. Recent insights into atherosclerotic plaque cell autophagy. Exp Biol Med (Maywood). (2021) 246(24):2553–8. doi: 10.1177/15353702211038894
90. Shao BZ, Han BZ, Zeng YX, Su DF, Liu C. The roles of macrophage autophagy in atherosclerosis. Acta Pharmacol Sin. (2016) 37(2):150–6. doi: 10.1038/aps.2015.87
91. Hou P, Fang J, Liu Z, Shi Y, Agostini M, Bernassola F, et al. Macrophage polarization and metabolism in atherosclerosis. Cell Death Dis. (2023) 14(10):691. doi: 10.1038/s41419-023-06206-z
92. Yang S, Yuan HQ, Hao YM, Ren Z, Qu SL, Liu LS, et al. Macrophage polarization in atherosclerosis. Clin Chim Acta. (2020) 501:142–6. doi: 10.1016/j.cca.2019.10.034
93. Molnár AÁ, Pásztor DT, Tarcza Z, Merkely B. Cells in atherosclerosis: focus on cellular senescence from basic science to clinical practice. Int J Mol Sci. (2023) 24(24):17129. doi: 10.3390/ijms242417129
94. Yang S, Wu M, Li X, Zhao R, Zhao Y, Liu L, et al. Role of endoplasmic reticulum stress in atherosclerosis and its potential as a therapeutic target. Oxid Med Cell Longev. (2020) 2020:9270107. doi: 10.1155/2020/9270107
95. Immanuel J, Yun S. Vascular inflammatory diseases and endothelial phenotypes. Cells. (2023) 12(12):1640. doi: 10.3390/cells12121640
96. Aronova A, Tosato F, Naser N, Asare Y. Innate immune pathways in atherosclerosis-from signaling to long-term epigenetic reprogramming. Cells. (2023) 12(19):2359. doi: 10.3390/cells12192359
97. Shu Y, Jin S. Caveolin-1 in endothelial cells: a potential therapeutic target for atherosclerosis. Heliyon. (2023) 9(8):e18653. doi: 10.1016/j.heliyon.2023.e18653
98. Kockx MM, Herman AG. Apoptosis in atherosclerosis: beneficial or detrimental? Cardiovasc Res. (2000) 45(3):736–46. doi: 10.1016/s0008-6363(99)00235-7
99. Shafi O. Switching of vascular cells towards atherogenesis, and other factors contributing to atherosclerosis: a systematic review. Thromb J. (2020) 18:28. doi: 10.1186/s12959-020-00240-z
100. Ferreira-Martins AJ, Castaldoni R, Alencar BM, Ferreira MV, Nogueira T, Rios RA, et al. Full-scale network analysis reveals properties of the FV protein structure organization. Sci Rep. (2023) 13(1):9546. doi: 10.1038/s41598-023-36528-z
101. Wojtasińska A, Frąk W, Lisińska W, Sapeda N, Młynarska E, Rysz J, et al. Novel insights into the molecular mechanisms of atherosclerosis. Int J Mol Sci. (2023) 24(17):13434. doi: 10.3390/ijms241713434
102. Weber C, Habenicht AJR, von Hundelshausen P. Novel mechanisms and therapeutic targets in atherosclerosis: inflammation and beyond. Eur Heart J. (2023) 44(29):2672–81. doi: 10.1093/eurheartj/ehad304
103. Böger RH, Bode-Böger SM, Frölich JC. The L-arginine-nitric oxide pathway: role in atherosclerosis and therapeutic implications. Atherosclerosis. (1996) 127(1):1–11. doi: 10.1016/s0021-9150(96)05953-9
104. Madamanchi NR, Runge MS. Redox signaling in cardiovascular health and disease. Free Radic Biol Med. (2013) 61:473–501. doi: 10.1016/j.freeradbiomed.2013.04.001
105. Saccone D, Asani F, Bornman L. Regulation of the vitamin D receptor gene by environment, genetics and epigenetics. Gene. (2015) 561(2):171–80. doi: 10.1016/j.gene.2015.02.024
106. Ho SM, Johnson A, Tarapore P, Janakiram V, Zhang X, Leung YK. Environmental epigenetics and its implication on disease risk and health outcomes [published correction appears in ILAR J. 2017 Dec 15;58(3):413]. ILAR J. (2012) 53(3–4):289–305. doi: 10.1093/ilar.53.3-4.289
107. Lee HT, Oh S, Ro DH, Yoo H, Kwon YW. The key role of DNA methylation and histone acetylation in epigenetics of atherosclerosis. J Lipid Atheroscler. (2020) 9(3):419–34. doi: 10.12997/jla.2020.9.3.419
108. Bruunsgaard H, Skinhøj P, Pedersen AN, Schroll M, Pedersen BK. Ageing, tumour necrosis factor-alpha (TNF-alpha) and atherosclerosis. Clin Exp Immunol. (2000) 121(2):255–60. doi: 10.1046/j.1365-2249.2000.01281.x
109. Gómez-Bernal F, Quevedo-Abeledo JC, García-González M, Fernández-Cladera Y, González-Rivero AF, Martín-González C, et al. Transforming growth factor beta 1 is associated with subclinical carotid atherosclerosis in patients with systemic lupus erythematosus. Arthritis Res Ther. (2023) 25(1):64. doi: 10.1186/s13075-023-03046-2
110. Proctor BM, Ren J, Chen Z, Schneider JG, Coleman T, Lupu TS, et al. Grb2 is required for atherosclerotic lesion formation. Arterioscler Thromb Vasc Biol. (2007) 27(6):1361–7. doi: 10.1161/ATVBAHA.106.134007
111. Gaddis DE, Padgett LE, Wu R, Hedrick CC. Neuropilin-1 expression on CD4T cells is atherogenic and facilitates T cell migration to the aorta in atherosclerosis. J Immunol. (2019) 203(12):3237–46. doi: 10.4049/jimmunol.1900245
112. Suárez-Rivero JM, Pastor-Maldonado CJ, Povea-Cabello S, Álvarez-Córdoba M, Villalón-García I, Talaverón-Rey M, et al. From mitochondria to atherosclerosis: the inflammation path. Biomedicines. (2021) 9(3):258. doi: 10.3390/biomedicines9030258
113. Poznyak AV, Sukhorukov VN, Zhuravlev A, Orekhov NA, Kalmykov V, Orekhov AN. Modulating mTOR signaling as a promising therapeutic strategy for atherosclerosis. Int J Mol Sci. (2022) 23(3):1153. doi: 10.3390/ijms23031153
114. Zhang Q, Wen XH, Tang SL, Zhao ZW, Tang CK. Role and therapeutic potential of gelsolin in atherosclerosis. J Mol Cell Cardiol. (2023) 178:59–67. doi: 10.1016/j.yjmcc.2023.03.012
115. Gunasekar P, Swier VJ, Fleegel JP, Boosani CS, Radwan MM, Agrawal DK. Vitamin D and macrophage polarization in epicardial adipose tissue of atherosclerotic swine. PLoS One. (2018) 13(10):e0199411. doi: 10.1371/journal.pone.019941
116. Oeckinghaus A, Ghosh S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol. (2009) 1(4):a000034. doi: 10.1101/cshperspect.a000034
117. Wang EJ, Wu MY, Ren ZY, Zheng Y, Ye RD, Tan CSH, et al. Targeting macrophage autophagy for inflammation resolution and tissue repair in inflammatory bowel disease. Burns Trauma. (2023) 11:tkad004. doi: 10.1093/burnst/tkad004
118. Mallat Z, Corbaz A, Scoazec A, Besnard S, Lesèche G, Chvatchko Y, et al. Expression of interleukin-18 in human atherosclerotic plaques and relation to plaque instability. Circulation. (2001) 104(14):1598–603. doi: 10.1161/hc3901.096721
119. Chu T, Xu X, Ruan Z, Wu L, Zhou M, Zhu G. miR-146a contributes to atherosclerotic plaque stability by regulating the expression of TRAF6 and IRAK-1. Mol Biol Rep. (2022) 49(6):4205–16. doi: 10.1007/s11033-022-07253-z
120. Fei J, Sun Y, Duan Y, Xia J, Yu S, Ouyang P, et al. Low concentration of rutin treatment might alleviate the cardiotoxicity effect of pirarubicin on cardiomyocytes via activation of PI3 K/AKT/mTOR signaling pathway. Biosci Rep. (2019) 39(6):BSR20190546. doi: 10.1042/BSR20190546
121. Ma C, Lu T, He Y, Guo D, Duan L, Jia R, et al. Comprehensive analysis of autophagy-related gene expression profiles identified five gene biomarkers associated with immune infiltration and advanced plaques in carotid atherosclerosis. Orphanet J Rare Dis. (2023) 18(1):66. doi: 10.1186/s13023-023-02660-2
122. Perrotta I, Aquila S. The role of oxidative stress and autophagy in atherosclerosis. Oxid Med Cell Longev. (2015) 2015:130315. doi: 10.1155/2015/130315
123. Wu CM, Zheng L, Wang Q, Hu YW. The emerging role of cell senescence in atherosclerosis. Clin Chem Lab Med. (2020) 59(1):27–38. doi: 10.1515/cclm-2020-0601
124. Grootaert MOJ, Moulis M, Roth L, Martinet W, Vindis C, Bennett MR, et al. Vascular smooth muscle cell death, autophagy and senescence in atherosclerosis. Cardiovasc Res. (2018) 114(4):622–34. doi: 10.1093/cvr/cvy007
125. Xu S, Lyu QR, Ilyas I, Tian XY, Weng J. Vascular homeostasis in atherosclerosis: a holistic overview. Front Immunol. (2022) 13:976722. doi: 10.3389/fimmu.2022.976722
126. Yamagishi N, Ueda T, Mori A, Saito Y, Hatayama T. Decreased expression of endoplasmic reticulum chaperone GRP78 in liver of diabetic mice. Biochem Biophys Res Commun. (2012) 417(1):364–70. doi: 10.1016/j.bbrc.2011.11.118
127. Liu J, Xu K, Chase M, Ji Y, Logan JK, Buchsbaum RJ. Tiam1-regulated osteopontin in senescent fibroblasts contributes to the migration and invasion of associated epithelial cells. J Cell Sci. (2012) 125(Pt 2):376–86. doi: 10.1242/jcs.089466
128. Wolak T. Osteopontin—a multi-modal marker and mediator in atherosclerotic vascular disease. Atherosclerosis. (2014) 236(2):327–37. doi: 10.1016/j.atherosclerosis.2014.07.004
129. Li M, Jiao Q, Xin W, Niu S, Liu M, Song Y, et al. Corrigendum: the emerging role of rho guanine nucleotide exchange factors in cardiovascular disorders: insights into atherosclerosis: a mini review. Front Cardiovasc Med. (2022) 9:850258. doi: 10.3389/fcvm.2022.850258. Erratum for: Front Cardiovasc Med. 2022 Jan 03;8:782098.35174236
130. Moon SY, Shin SA, Oh YS, Park HH, Lee CS. Understanding the role of the BAI subfamily of adhesion G protein-coupled receptors (GPCRs) in pathological and physiological conditions. Genes (Basel). (2018) 9(12):597. doi: 10.3390/genes9120597
131. Thorp E, Cui D, Schrijvers DM, Kuriakose G, Tabas I. Mertk receptor mutation reduces efferocytosis efficiency and promotes apoptotic cell accumulation and plaque necrosis in atherosclerotic lesions of apoe-/- mice. Arterioscler Thromb Vasc Biol. (2008) 28(8):1421–8. doi: 10.1161/ATVBAHA.108.167197
132. Liao X, Sluimer JC, Wang Y, Subramanian M, Brown K, Pattison JS, et al. Macrophage autophagy plays a protective role in advanced atherosclerosis. Cell Metab. (2012) 15(4):545–53. doi: 10.1016/j.cmet.2012.01.022
133. Galic M, Tsai FC, Collins SR, Matis M, Bandara S, Meyer T. Dynamic recruitment of the curvature-sensitive protein ArhGAP44 to nanoscale membrane deformations limits exploratory filopodia initiation in neurons. Elife. (2014) 3:e03116. doi: 10.7554/eLife.03116
134. Abou-Kheir W, Isaac B, Yamaguchi H, Cox D. Membrane targeting of WAVE2 is not sufficient for WAVE2-dependent actin polymerization: a role for IRSp53 in mediating the interaction between Rac and WAVE2. J Cell Sci. (2008) 121(Pt 3):379–90. doi: 10.1242/jcs.010272
135. Singh RK, Haka AS, Bhardwaj P, Zha X, Maxfield FR. Dynamic actin reorganization and vav/Cdc42-dependent actin polymerization promote macrophage aggregated LDL (low-density lipoprotein) uptake and catabolism. Arterioscler Thromb Vasc Biol. (2019) 39(2):137–49. doi: 10.1161/ATVBAHA.118.312087
136. Nakagawa H, Miki H, Nozumi M, Takenawa T, Miyamoto S, Wehland J, et al. IRSp53 is colocalised with WAVE2 at the tips of protruding lamellipodia and filopodia independently of Mena. J Cell Sci. (2003) 116(Pt 12):2577–83. doi: 10.1242/jcs.00462
137. Ding B, Yang S, Schaks M, Liu Y, Brown AJ, Rottner K, et al. Structures reveal a key mechanism of WAVE regulatory complex activation by Rac1 GTPase. Nat Commun. (2022) 13(1):5444. doi: 10.1038/s41467-022-33174-3
138. Kramer DA, Piper HK, Chen B. WASP family proteins: molecular mechanisms and implications in human disease [published correction appears in Eur J Cell Biol. 2023 Mar;102(1):151287]. Eur J Cell Biol. (2022) 101(3):151244. doi: 10.1016/j.ejcb.2022.151244
139. Chen B, Brinkmann K, Chen Z, Pak CW, Liao Y, Shi S, et al. The WAVE regulatory complex links diverse receptors to the actin cytoskeleton. Cell. (2014) 156(1-2):195–207. doi: 10.1016/j.cell.2013.11.048
140. Lim KB, Bu W, Goh WI, Koh E, Ong SH, Pawson T, et al. The Cdc42 effector IRSp53 generates filopodia by coupling membrane protrusion with actin dynamics. J Biol Chem. (2008) 283(29):20454–72. doi: 10.1074/jbc.M710185200
141. Cai GQ, Chou CF, Hu M, Zheng A, Reichardt LF, Guan JL, et al. Neuronal Wiskott-Aldrich syndrome protein (N-WASP) is critical for formation of α-smooth muscle actin filaments during myofibroblast differentiation. Am J Physiol Lung Cell Mol Physiol. (2012) 303(8):L692–702. doi: 10.1152/ajplung.00390.2011
142. Chen H, Reed G, Guardia J, Lakhan S, Couture O, Hays E, et al. Vitamin D directly regulates Mdm2 gene expression in osteoblasts. Biochem Biophys Res Commun. (2013) 430(1):370–4. doi: 10.1016/j.bbrc.2012.11.003
143. Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. (1993) 7(7A):1126–32. doi: 10.1101/gad.7.7a.1126
144. Moon SH, Huang CH, Houlihan SL, Regunath K, Freed-Pastor WA, Morris JP 4th, et al. p53 represses the mevalonate pathway to mediate tumor suppression. Cell. (2019) 176(3):564–580.e19. doi: 10.1016/j.cell.2018.11.011
145. Dong B, Wu M, Li H, Kraemer FB, Adeli K, Seidah NG, et al. Strong induction of PCSK9 gene expression through HNF1alpha and SREBP2: mechanism for the resistance to LDL-cholesterol lowering effect of statins in dyslipidemic hamsters. J Lipid Res. (2010) 51(6):1486–95. doi: 10.1194/jlr.M003566
146. Forrester K, Ambs S, Lupold SE, Kapust RB, Spillare EA, Weinberg WC, et al. Nitric oxide-induced p53 accumulation and regulation of inducible nitric oxide synthase expression by wild-type p53. Proc Natl Acad Sci U S A. (1996) 93(6):2442–7. doi: 10.1073/pnas.93.6.2442
147. Ambs S, Ogunfusika MO, Merriam WG, Bennett WP, Billiar TR, Harris CC. Up-regulation of inducible nitric oxide synthase expression in cancer-prone p53 knockout mice. Proc Natl Acad Sci U S A. (1998) 95(15):8823–8. doi: 10.1073/pnas.95.15.882
148. Chan GH, Chan E, Kwok CT, Leung GP, Lee SM, Seto SW. The role of p53 in the alternation of vascular functions. Front Pharmacol. (2022) 13:981152. doi: 10.3389/fphar.2022.981152
149. Jansson M, Durant ST, Cho EC, Sheahan S, Edelmann M, Kessler B, et al. Arginine methylation regulates the p53 response. Nat Cell Biol. (2008) 10(12):1431–9. doi: 10.1038/ncb1802
150. Heyne K, Heil TC, Bette B, Reichrath J, Roemer K. MDM2 binds and inhibits vitamin D receptor. Cell Cycle. (2015) 14(13):2003–10. doi: 10.1080/15384101.2015.104417
151. Zhao Y, Yu H, Hu W. The regulation of MDM2 oncogene and its impact on human cancers. Acta Biochim Biophys Sin (Shanghai). (2014) 46(3):180–9. doi: 10.1093/abbs/gmt147
152. Rohe B, Safford SE, Nemere I, Farach-Carson MC. Regulation of expression of 1,25D3-MARRS/ERp57/PDIA3 in rat IEC-6 cells by TGF beta and 1,25(OH)2D3. Steroids. (2007) 72(2):144–50. doi: 10.1016/j.steroids.2006.11.013
153. Srikuea R, Hirunsai M. TGF-β1 stimulation and VDR-dependent activation modulate calcitriol action on skeletal muscle fibroblasts and Smad signalling-associated fibrogenesis. Sci Rep. (2023) 13(1):13811. doi: 10.1038/s41598-023-40978-w
154. Meek DW, Anderson CW. Posttranslational modification of p53: cooperative integrators of function. Cold Spring Harb Perspect Biol. (2009) 1(6):a000950. doi: 10.1101/cshperspect.a000950
155. Qu L, Huang S, Baltzis D, Rivas-Estilla AM, Pluquet O, Hatzoglou M, et al. Endoplasmic reticulum stress induces p53 cytoplasmic localization and prevents p53-dependent apoptosis by a pathway involving glycogen synthase kinase-3beta. Genes Dev. (2004) 18(3):261–77. doi: 10.1101/gad.1165804
156. Zittermann A. Vitamin D and cardiovascular disease. Anticancer Res. (2014) 34(9):4641–8. PMID: 2520203925202039
157. Levita J, Wilar G, Wahyuni I, Bawono LC, Ramadaini T, Rohani R, et al. Clinical toxicology of vitamin D in pediatrics: a review and case reports. Toxics. (2023) 11(7):642. doi: 10.3390/toxics11070642
158. Zittermann A, Trummer C, Theiler-Schwetz V, Pilz S. Long-term supplementation with 3200 to 4000 IU of vitamin D daily and adverse events: a systematic review and meta-analysis of randomized controlled trials. Eur J Nutr. (2023) 62(4):1833–44. doi: 10.1007/s00394-023-03124-w
159. Leblond F, Poirier S, Yu C, Duquette N, Mayer G, Thorin E. The anti-hypercholesterolemic effect of low p53 expression protects vascular endothelial function in mice. PLoS One. (2014) 9(3):e92394. doi: 10.1371/journal.pone.0092394
160. Tuohimaa P. Vitamin D and aging. J Steroid Biochem Mol Biol. (2009) 114(1-2):78–84. doi: 10.1016/j.jsbmb.2008.12.020
161. Silverbush D, Sharan R. A systematic approach to orient the human protein-protein interaction network. Nat Commun. (2019) 10(1):3015. doi: 10.1038/s41467-019-10887-6
162. Tang H, Zeng Z, Shang C, Li Q, Liu J. Epigenetic regulation in pathology of atherosclerosis: a novel perspective. Front Genet. (2021) 12:810689. doi: 10.3389/fgene.2021.810689
163. Chen YC, Smith M, Ying YL, Makridakis M, Noonan J, Kanellakis P, et al. Quantitative proteomic landscape of unstable atherosclerosis identifies molecular signatures and therapeutic targets for plaque stabilization. Commun Biol. (2023) 6(1):265. doi: 10.1038/s42003-023-04641-4
164. Das LM, Binko AM, Traylor ZP, Peng H, Lu KQ. Vitamin D improves sunburns by increasing autophagy in M2 macrophages. Autophagy. (2019) 15(5):813–26. doi: 10.1080/15548627.2019.1569298
165. Lee SG, Oh J, Bong SK, Kim JS, Park S, Kim S, et al. Macrophage polarization and acceleration of atherosclerotic plaques in a swine model. PLoS One. (2018) 13(3):e0193005. doi: 10.1371/journal.pone.0193005
166. Chen W, Xiao W, Liu X, Yuan P, Zhang S, Wang Y, et al. Pharmacological manipulation of macrophage autophagy effectively rejuvenates the regenerative potential of biodegrading vascular graft in aging body. Bioact Mater. (2021) 11:283–299. doi: 10.1016/j.bioactmat.2021.09.027
167. Stoffels K, Overbergh L, Giulietti A, Verlinden L, Bouillon R, Mathieu C. Immune regulation of 25-hydroxyvitamin-D3-1alpha-hydroxylase in human monocytes. J Bone Miner Res. (2006) 21(1):37–47. doi: 10.1359/JBMR.050908
168. Fang M, Zhong C. Vitamin D receptor mediates liver ischemia and reperfusion injury by autophagy-regulated M2 macrophage polarization. Turk J Biol. (2023) 47(2):120–9. doi: 10.55730/1300-0152.2647
169. Jantsch J, Binger KJ, Müller DN, Titze J. Macrophages in homeostatic immune function. Front Physiol. (2014) 5:146. doi: 10.3389/fphys.2014.00146
170. Wu J, He S, Song Z, Chen S, Lin X, Sun H, et al. Macrophage polarization states in atherosclerosis. Front Immunol. (2023) 14:1185587. doi: 10.3389/fimmu.2023.1185587
171. Khanolkar S, Hirani S, Mishra A, Vardhan S, Hirani S, Prasad R, et al. Exploring the role of vitamin D in atherosclerosis and its impact on cardiovascular events: a comprehensive review. Cureus. (2023) 15(7):e42470. doi: 10.7759/cureus.42470
172. Bi Y, Chen J, Hu F, Liu J, Li M, Zhao L. M2 macrophages as a potential target for antiatherosclerosis treatment. Neural Plast. (2019) 2019:6724903. doi: 10.1155/2019/6724903
173. Becker L, Nguyen L, Gill J, Kulkarni S, Pasricha PJ, Habtezion A. Age-dependent shift in macrophage polarisation causes inflammation-mediated degeneration of enteric nervous system. Gut. (2018) 67(5):827–36. doi: 10.1136/gutjnl-2016-312940
174. Behmoaras J, Gil J. Similarities and interplay between senescent cells and macrophages. J Cell Biol. (2021) 220(2):e202010162. doi: 10.1083/jcb.202010162
175. Chung HY, Kim DH, Lee EK, Chung KW, Chung S, Lee B, et al. Redefining chronic inflammation in aging and age-related diseases: proposal of the senoinflammation concept. Aging Dis. (2019) 10(2):367–82. doi: 10.14336/AD.2018.0324
176. Linehan E, Fitzgerald DC. Ageing and the immune system: focus on macrophages. Eur J Microbiol Immunol (Bp). (2015) 5(1):14–24. doi: 10.1556/EUJMI-D-14-00035
177. Bobryshev YV, Ivanova EA, Chistiakov DA, Nikiforov NG, Orekhov AN. Macrophages and their role in atherosclerosis: pathophysiology and transcriptome analysis. Biomed Res Int. (2016) 2016:9582430. doi: 10.1155/2016/958243
178. Subramanian M, Tabas I. Dendritic cells in atherosclerosis. Semin Immunopathol. (2014) 36(1):93–102. doi: 10.1007/s00281-013-0400-x
179. Moore KJ, Sheedy FJ, Fisher EA. Macrophages in atherosclerosis: a dynamic balance. Nat Rev Immunol. (2013) 13(10):709–21. doi: 10.1038/nri3520
180. Mosser DM, Hamidzadeh K, Goncalves R. Macrophages and the maintenance of homeostasis. Cell Mol Immunol. (2021) 18(3):579–87. doi: 10.1038/s41423-020-00541-3
181. Boyle JJ. Macrophage activation in atherosclerosis: pathogenesis and pharmacology of plaque rupture. Curr Vasc Pharmacol. (2005) 3(1):63–8. doi: 10.2174/1570161052773861
182. Barrett TJ. Macrophages in atherosclerosis regression. Arterioscler Thromb Vasc Biol. (2020) 40(1):20–33. doi: 10.1161/ATVBAHA.119.312802
183. Zhao Y, Zhang J, Zhang W, Xu Y. A myriad of roles of dendritic cells in atherosclerosis. Clin Exp Immunol. (2021) 206(1):12–27. doi: 10.1111/cei.13634
184. Feig JE, Feig JL. Macrophages, dendritic cells, and regression of atherosclerosis. Front Physiol. (2012) 3:286. doi: 10.3389/fphys.2012.00286
185. Krzyszczyk P, Schloss R, Palmer A, Berthiaume F. The role of macrophages in acute and chronic wound healing and interventions to promote pro-wound healing phenotypes. Front Physiol. (2018) 9:419. doi: 10.3389/fphys.2018.00419
186. Guo FX, Hu YW, Zheng L, Wang Q. Shear stress in autophagy and its possible mechanisms in the process of atherosclerosis. DNA Cell Biol. (2017) 36(5):335–46. doi: 10.1089/dna.2017.3649
187. Seneviratne AN, Cole JE, Goddard ME, Park I, Mohri Z, Sansom S, et al. Low shear stress induces M1 macrophage polarization in murine thin-cap atherosclerotic plaques. J Mol Cell Cardiol. (2015) 89(Pt B):168–72. doi: 10.1016/j.yjmcc.2015.10.034. Erratum in: J Mol Cell Cardiol. 2016;91:10.26523517
188. Yang F, Liu Y, Ren H, Zhou G, Yuan X, Shi X. ER-stress regulates macrophage polarization through pancreatic EIF-2alpha kinase. Cell Immunol. (2019) 336:40–7. doi: 10.1016/j.cellimm.2018.12.008
189. Chipurupalli S, Samavedam U, Robinson N. Crosstalk between ER stress, autophagy and inflammation. Front Med (Lausanne). (2021) 8:758311. doi: 10.3389/fmed.2021.758311
190. Wen JH, Li DY, Liang S, Yang C, Tang JX, Liu HF. Macrophage autophagy in macrophage polarization, chronic inflammation and organ fibrosis. Front Immunol. (2022) 13:946832. doi: 10.3389/fimmu.2022.946832
191. McGillis L, Bronte-Tinkew DM, Dang F, Capurro M, Prashar A, Ricciuto A, et al. Vitamin D deficiency enhances expression of autophagy-regulating miR-142-3p in mouse and “involved” IBD patient intestinal tissues. Am J Physiol Gastrointest Liver Physiol. (2021) 321(2):G171–G184. doi: 10.1152/ajpgi.00398.2020
192. Krishna SM. Vitamin D as a protector of arterial health: potential role in peripheral arterial disease formation. Int J Mol Sci. (2019) 20(19):4907. doi: 10.3390/ijms20194907
193. Liang S, Cai J, Li Y, Yang R. 1,25-dihydroxy-vitamin D3 induces macrophage polarization to M2 by upregulating T-cell Ig-mucin-3 expression. Mol Med Rep. (2019) 19(5):3707–13. doi: 10.3892/mmr.2019.10047
194. Yin K, You Y, Swier V, Tang L, Radwan MM, Pandya AN, et al. Vitamin D protects against atherosclerosis via regulation of cholesterol efflux and macrophage polarization in hypercholesterolemic swine. Arterioscler Thromb Vasc Biol. (2015) 35(11):2432–42. doi: 10.1161/ATVBAHA.115.306132
195. Szeto FL, Reardon CA, Yoon D, Wang Y, Wong KE, Chen Y, et al. Vitamin D receptor signaling inhibits atherosclerosis in mice. Mol Endocrinol. (2012) 26(7):1091–101. doi: 10.1210/me.2011-1329
196. Singh S, Torzewski M. Fibroblasts and their pathological functions in the fibrosis of aortic valve sclerosis and atherosclerosis. Biomolecules. (2019) 9(9):472. doi: 10.3390/biom9090472
197. Ahn J, Peters U, Albanes D, Purdue MP, Abnet CC, Chatterjee N, et al. Serum vitamin D concentration and prostate cancer risk: a nested case-control study. J Natl Cancer Inst. (2008) 100(11):796–804.18505967
198. Yuk JM, Shin DM, Lee HM, Yang CS, Jin HS, Kim KK, et al. Vitamin D3 induces autophagy in human monocytes/macrophages via cathelicidin. Cell Host Microbe. (2009) 6(3):231–43. doi: 10.1016/j.chom.2009.08.004
199. Nguyen LK, Kholodenko BN, von Kriegsheim A. Rac1 and RhoA: networks, loops and bistability. Small GTPases. (2018) 9(4):316–21. doi: 10.1080/21541248.2016.1224399
200. Quiroga X, Walani N, Disanza A, Chavero A, Mittens A, Tebar F, et al. A mechanosensing mechanism controls plasma membrane shape homeostasis at the nanoscale. Elife. (2023) 12:e72316. doi: 10.7554/eLife.72316
201. Khundmiri SJ, Murray RD, Lederer E. PTH and vitamin D. Compr Physiol. (2016) 6(2):561–601. doi: 10.1002/cphy.c140071
202. Robens JM, Yeow-Fong L, Ng E, Hall C, Manser E. Regulation of IRSp53-dependent filopodial dynamics by antagonism between 14 and 3-3 binding and SH3-mediated localization. Mol Cell Biol. (2010) 30(3):829–44. doi: 10.1128/MCB.01574-08
203. Bisi S, Marchesi S, Rizvi A, Carra D, Beznoussenko GV, Ferrara I, et al. IRSp53 controls plasma membrane shape and polarized transport at the nascent lumen in epithelial tubules. Nat Commun. (2020) 11(1):3516. doi: 10.1038/s41467-020-17091-x
204. Zhang HH, Wang W, Feng L, Yang Y, Zheng J, Huang L, et al. S-nitrosylation of Cofilin-1 serves as a novel pathway for VEGF-stimulated endothelial cell migration. J Cell Physiol. (2015) 230(2):406–17. doi: 10.1002/jcp.24724
205. Acevedo A, González-Billault C. Crosstalk between Rac1-mediated actin regulation and ROS production. Free Radic Biol Med. (2018) 116:101–13. doi: 10.1016/j.freeradbiomed.2018.01.008
206. Lähnemann D, Köster J, Szczurek E, McCarthy DJ, Hicks SC, Robinson MD, et al. Eleven grand challenges in single-cell data science. Genome Biol. (2020) 21(1):31. doi: 10.1186/s13059-020-1926-6
207. Kustatscher G, Hödl M, Rullmann E, Grabowski P, Fiagbedzi E, Groth A, et al. Higher-order modular regulation of the human proteome. Mol Syst Biol. (2023) 19(5):e9503. doi: 10.15252/msb.20209503
208. Singh MS, Pasumarthy R, Vaidya U, Leonhardt S. On quantification and maximization of information transfer in network dynamical systems. Sci Rep. (2023) 13(1):5588. doi: 10.1038/s41598-023-32762-7
209. e la Guía-Galipienso F, Martínez-Ferran M, Vallecillo N, Lavie CJ, Sanchis-Gomar F, Pareja-Galeano H. Vitamin D and cardiovascular health. Clin Nutr. (2021) 40(5):2946–57. doi: 10.1016/j.clnu.2020.12.025
210. Molinari C, Morsanuto V, Polli S, Uberti F. Cooperative effects of Q10, vitamin D3, and L-arginine on cardiac and endothelial cells. J Vasc Res. (2018) 55(1):47–60. doi: 10.1159/000484928
211. Tewani GR, Silwal K, Sharma G, Yadav D, Siddiqui A, Kriplani S, et al. Effect of medically supervised prolonged fasting therapy on vitamin D, B12, body weight, body mass index, vitality and quality of life: a randomized control trial. Nutr Metab Insights. (2022) 15:11786388221130560. doi: 10.1177/11786388221130560
Keywords: aging, cardiovascular disease, inflammation, immunity, macrophage autophagy, nitric oxide, machine learning, vitamin D
Citation: Fliri A and Kajiji S (2024) Effects of vitamin D signaling in cardiovascular disease: centrality of macrophage polarization. Front. Cardiovasc. Med. 11:1388025. doi: 10.3389/fcvm.2024.1388025
Received: 19 February 2024; Accepted: 24 May 2024;
Published: 25 June 2024.
Edited by:
Xi-Ming Yuan, Linköping University, SwedenReviewed by:
Hiroya Ohta, Hokkaido University of Science, Japan© 2024 Fliri and Kajiji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anton Fliri, YW50b24uZmxpcmlAZW1lcmdlbnRzYS5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.