
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cardiovasc. Med. , 06 February 2024
Sec. General Cardiovascular Medicine
Volume 11 - 2024 | https://doi.org/10.3389/fcvm.2024.1349376
This article is part of the Research Topic Insights in Cardiology from Caring for a Diverse Community: Perspectives from Inova Schar Heart and Vascular View all 22 articles
Despite developments in circulating biomarker and imaging technology in the assessment of cardiovascular disease, the surveillance and diagnosis of heart transplant rejection has continued to rely on histopathologic interpretation of the endomyocardial biopsy. Increasing evidence shows the utility of molecular evaluations, such as donor-specific antibodies and donor-derived cell-free DNA, as well as advanced imaging techniques, such as cardiac magnetic resonance imaging, in the assessment of rejection, resulting in the elimination of many surveillance endomyocardial biopsies. As non-invasive technologies in heart transplant rejection continue to evolve and are incorporated into practice, they may supplant endomyocardial biopsy even when rejection is suspected, allowing for more precise and expeditious rejection therapy. This review describes the current and near-future states for the evaluation of heart transplant rejection, both in the settings of rejection surveillance and rejection diagnosis. As biomarkers of rejection continue to evolve, rejection risk prediction may allow for a more personalized approach to immunosuppression.
The field of heart transplantation has experienced many remarkable innovations including (1) expansion of the donor pool with Hepatitis C-positive donors and donation after circulatory death (DCD) via normothermic regional perfusion and ex vivo perfusion; (2) assessment and understanding of donor-specific antibodies (DSA) and their contribution to antibody-mediated rejection (AMR); and (3) molecular diagnostics including gene expression profiling (GEP), donor-derived cell-free DNA (dd-cfDNA), and DSAs (1–5). Despite these advances, the evaluation of clinical rejection has continued to rely on repetitive invasive procedures, namely the histologic evaluation of endomyocardial biopsy (EMB) specimens to identify and diagnose acute cellular rejection (ACR) and AMR (Table 1). Despite EMB's risk of complications and lack of reproducibility in the diagnosis of rejection, EMB has remained a hallmark of post-transplant care (6, 7).
Rejection evaluations occur in two settings (Figure 1): surveillance in patients without signs and symptoms of allograft dysfunction (∼98% of EMBs) and for-cause in the setting of clinical signs or symptoms of allograft dysfunction (∼2% of EMBs). While transplant centers now forego many surveillance EMBs with reassuring molecular markers, such as dd-cfDNA and/or a lack of DSAs, the high prevalence of abnormal molecular markers in patients without rejection precludes the ability to diagnose and treat patients for rejection based on molecular profiling alone. As these biomarkers are combined with newer rejection evaluation modalities, including cardiac magnetic resonance (CMR) imaging and microRNA profiling, EMB may no longer be necessary to diagnose rejection. The rapid expansion of non-invasive evaluations of heart transplant rejection has resulted in a lack of clear clinical guidance on how to best incorporate them into post-transplant care. This review highlights the molecular and imaging tools available to the clinician in both the surveillance and for-cause settings. We also present a frontier of rejection risk prediction, identifying pre- and post-transplant rejection risk to allow risk stratification and individualization of post-transplant care.
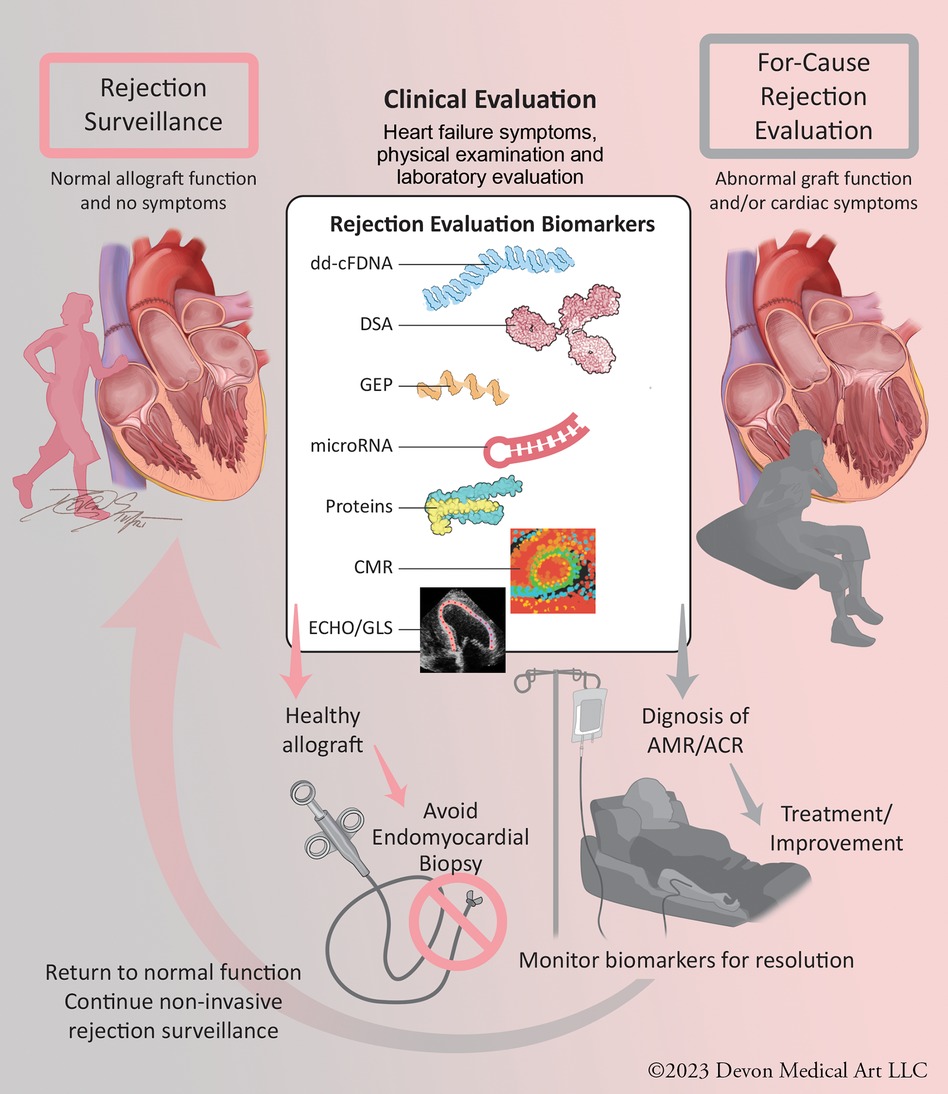
Figure 1. Central figure: evaluation of heart transplant rejection in surveillance and for-cause settings. Non-invasive molecular and imaging biomarkers can provide clinical reassurance of non-rejection, allowing avoidance of endomyocardial biopsy. With the continued validation of these technologies, they can be used in for-cause rejection evaluation settings to diagnose ACR and AMR and monitor for resolution, allowing clinical improvement and resumption of surveillance status. ACR, acute cellular rejection; AMR, antibody-mediated rejection.
To date, dd-cfDNA remains the molecular modality with the most robust evidence in the non-invasive identification of both ACR and AMR. The evaluation of cell-free DNA in transplantation stems from preclinical work where donor and recipient plasma whole genome sequencing was performed to identify single nucleotide polymorphisms (SNPs) allowing differentiation and quantification of dd-cfDNA (8). A 2019 publication by Khush et al. describes an initial dd-cfDNA clinical validation: a multicenter, prospective study of 740 heart transplant recipients whose plasma samples were evaluated for dd-cfDNA. A donor:recipient cell-free DNA percentage >0.2% had 54% sensitivity, 76% specificity, PPV of 12%, NPV of 97%, and AUC of 0.64 for identifying a composite of ACR or AMR (Table 2) (9). Another commercially available dd-cfDNA assay utilized a greater number of SNPs in a separate cohort and demonstrated 79% sensitivity, 77% specificity, PPV of 25.1%, NPV of 97.3%, and AUC of 0.86 for the detection of ACR/AMR. Finally, the multicenter Genomic Research Alliance for Transplantation (GRAfT) consortium described results of dd-cfDNA shotgun sequencing, with dd-cfDNA >0.25% having 81% sensitivity, 85% specificity, 20% PPV, 99% NPV, and AUC of 0.92 for diagnosing a composite of ACR or AMR (5).
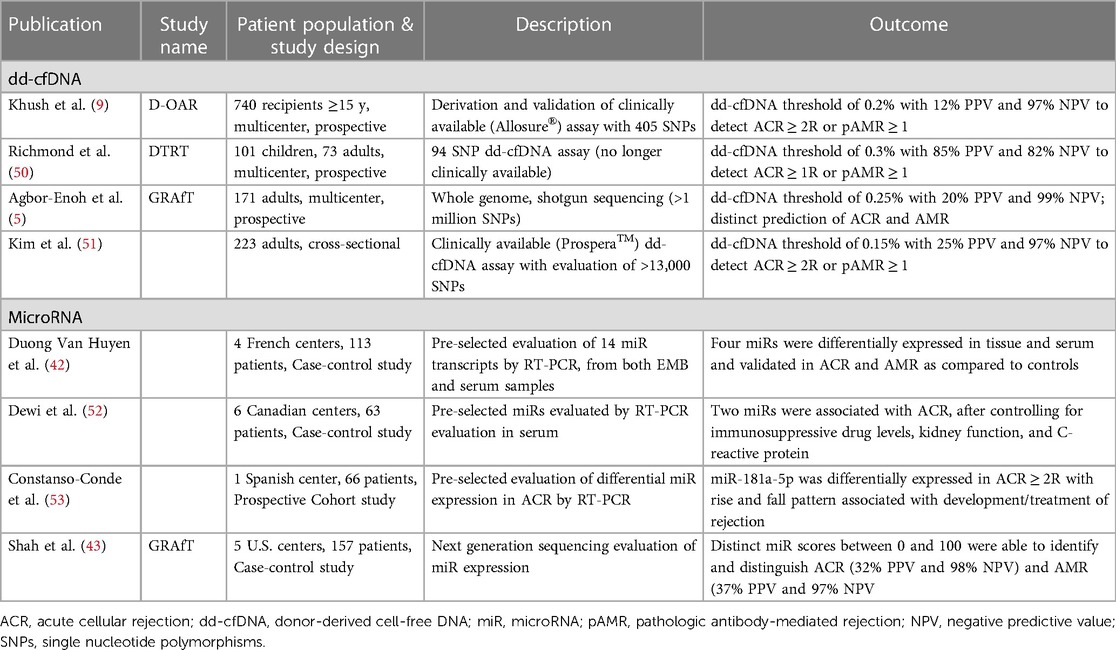
Table 2. Significant publications evaluating donor-derived cell-free DNA and microRNA in the evaluation of heart transplant rejection.
Rejection diagnosis via dd-cfDNA and other molecular markers are evaluated against the histopathologic interpretation of an EMB; however, there remains question about the accuracy of this comparison due to the variability of EMB readings across pathologists (4, 6). Given this concern, the GRAfT investigators analyzed episodes of elevated dd-cfDNA without rejection by EMB. Among the 135 cases classified as “false positive” dd-cfDNA (“false negative” EMB) episodes, 21% were observed in patients exhibiting allograft dysfunction by echocardiography (defined by a decline in left ventricular ejection fraction ≥5%), and 44% of these cases were identified prior to a rejection diagnosis, with AMR being more prevalent than ACR (5). These results suggest that dd-cfDNA is an important biomarker of graft injury and rejection regardless of the EMB results. In addition, the PPV which is calculated in prediction of positive EMB, may actually be substantially higher. Combining dd-cfDNA with DSA, clinical, and imaging evaluations may be sufficient in surveillance of rejection.
The initial biomarker developed to non-invasively identify ACR was GEP, which measures 20 genes (11 reporter genes and 9 house-keeping genes). Individual expression of the 11 reporter genes is quantified and a GEP “score” is calculated with a score >30 (maximum score 40) having a positive predictive value (PPV) of 6.8% and negative predictive value (NPV) of >99% in the evaluation of ACR (4). In the Invasive Monitoring Attenuation through Gene Expression (IMAGE) study patients randomized as early as 6-months post-transplant to a strategy of GEP surveillance vs. EMB-based surveillance had similar outcomes (10). While the inclusion of GEP evaluation into post-transplant care marked the beginning of the “molecular era” of post-transplant evaluation, this biomarker is limited by its inability to detect AMR and lack of reliability when patients are on higher doses of corticosteroids, receive blood transfusions, or have a cytomegalovirus infection (11, 12). In addition, studies evaluating combination testing with GEP and dd-cfDNA show that patients with a positive GEP test and negative dd-cfDNA do not experience rejection, suggesting GEP as a false-positive in this setting (13, 14). As such, GEP is now of historical value in contemporary heart transplant practice, given the availability of dd-cfDNA in resource rich settings.
The diagnosis of acute cardiac rejection differs from other cardiac conditions, such as acute cardiac ischemia and myocardial infarction, in which a sensitive and specific evaluation can be conducted with high-sensitivity cardiac troponin (hs-cTn) assays and EKG. Both troponin and natriuretic peptides have been extensively studied as predictors of heart transplant rejection, with neither showing reproducible sensitivity and specificity in diagnosing rejection (15–17). Despite hs-cTn greatly improving accuracy over earlier generation troponin assays in ischemic heart disease, a recent evaluation showed no association between hs-cTn and ACR (18). Authors have also queried whether markers of inflammation, such as C-reactive protein and interleukin-6 (IL-6) can diagnose rejection, but neither have proven to be accurate markers (17, 19). There have been recent investigations into proteomic (expression assays of multiple protein biomarkers) evaluations of rejection and graft dysfunction, though rigorous evaluations of proteomic profiling in rejection have yet to be completed (20, 21). While abnormal troponins and/or natriuretic peptides can signal allograft dysfunction related to rejection, their lack of significant evidence in diagnosing rejection limits their use in rejection surveillance.
Many heart transplant centers now forego routine surveillance biopsy as early as one month after transplant in patients with favorable molecular evaluations (13). One single-center study evaluated this practice in 153 recipients where reassuring dd-cfDNA results allowed the cancellation of 84% of surveillance biopsies (14). This evaluation included a protocol recommending EMB cancellation with dd-cfDNA <0.2%. Of 172 biopsies performed during the one-year study period, only 2 patients had ACR and 2 patients had AMR: all episodes of rejection occurred in the setting of dd-cfDNA >0.2%. In another single-center evaluation, 64 recipients underwent 475 dd-cfDNA evaluations within the first year post-transplant, with molecular evaluation prompting EMB in only 22 cases (with 2 biopsies yielding ACR), while clinical evaluation led to EMB in 3 cases (with 1 biopsy yielding ACR) (13). As many centers are eliminating surveillance EMB with reassuring molecular and clinical evaluations, there are multiple ongoing trials assessing the effectiveness of these non-invasive rejection surveillance strategies.
Within five years of heart transplantation, over 40% of recipients are hospitalized for acute rejection, and repeat episodes of rejection increase the risk of graft dysfunction and cardiac allograft vasculopathy (CAV, a manifestation of chronic rejection), the leading cause of mortality in the late phase post-transplant (22, 23). The relationship between the patient and the transplant team involves frequent monitoring for signs and symptoms of rejection, which include fatigue, decreased appetite, dyspnea, edema, and weight gain. These symptoms would prompt an in-person assessment with clinical evaluation to include physical examination, echocardiography, and laboratory assessment of end-organ function and cardiac biomarkers (troponins or natriuretic peptides). Despite a clinical picture that suggests rejection, these medical and laboratory evaluations remain non-specific in the diagnosis of rejection, and patients are typically referred for a diagnostic EMB, which is evaluated for ACR and AMR based on the International Society for Heart and Lung Transplantation (ISHLT) grading nomenclature (24, 25). Increasingly, these for-cause evaluations include molecular testing such as DSAs and/or dd-cfDNA, which may provide rejection diagnosis without EMB. As these evaluations and other emerging biomarkers gain clinical utility, it may be possible to diagnose and treat rejection without EMB.
Immunologists and transplant physicians have worked together to advance the field of immuno-evaluation, where antibody data is used to assess humoral immune system activity before and after transplant. Given the limited donor pool, patient acuity, and in contrast to kidney transplantation, heart transplantation does not rely on human leukocyte antigen (HLA) matching. Rather, recipients are evaluated to ensure limited cross reactivity between pre-formed HLA antibodies and donor HLA antigens at the time of a donor offer through a process known as virtual crossmatch. As the complexity of heart transplant recipients increases (including mechanical circulatory support use, congenital heart disease, and multiparous females), heart transplant candidates may carry significant levels of pre-formed HLA antibodies, reducing the number of potential donors and increasing the risk of DSA development post-transplantation. DSA development post-transplant is associated with increased risk for AMR, CAV, allograft dysfunction, and mortality (26–29). In the past, evaluation of HLA antibodies (both pre-formed to the potential donor pool and post-transplant, specific to the donor organ—DSA) required physical complement dependent cytotoxicity (CDC) crossmatching where donor cells and recipient serum were combined to assess cross reactivity and cell lysis. This has evolved into virtual assessments of cross reactivity using solid-phase immunoassays (SPIs) where recipient serum is incubated with multiplex bead assays containing HLA antigens (30, 31).
Specific DSA characteristics, including their persistence throughout multiple post-transplant evaluations as well as their specificity to Class II HLA (primarily at the HLA-DQ locus), have been associated with higher rates of rejection and graft failure (32, 33). While there is a significant association between DSA and AMR, DSA is also found in ACR, reflecting the interdependence between the cellular and antibody-mediated immune response in rejection (27, 29). Currently, DSA evaluation alone cannot be used as a surrogate of rejection, as many patients develop DSA but never have rejection. Rather, it is used in complement with clinical and other molecular modalities to signal potential rejection. As more is learned about DSA development and associated risk factors, its evaluation will continue to be important in the evaluation of rejection.
As CMR tissue characterization has aided in the evaluation of post-ischemic myocardial viability and inflammatory cardiomyopathies, there have now been numerous evaluations of its use in rejection diagnosis. Multiple CMR sequences including T1 and T2 mapping and extracellular volume (ECV) have been associated with acute rejection. In an evaluation of 112 simultaneous EMBs and CMRs, Imran et al. showed that native T1 mapping was able to identify a composite of ACR/AMR, with 93% sensitivity, 79% specificity, 99% NPV, and AUC of 0.89 (34). In another study by Dolan et al. of 72 subjects, global and peak T2 signals had the highest values in patients with active ACR and had sequentially lower signals in those with prior ACR followed by further decreased signal among healthy controls (35). This study also showed global and segmental ECV to be significantly higher in patients with active ACR, regardless of presence or absence of past ACR. Recent results of a randomized, non-inferiority trial of EMB vs. CMR rejection surveillance in 40 heart transplant recipients in Sydney, Australia showed 92% sensitivity, 93% specificity, 62% PPV, 92% NPV, and AUC 0.92 in the detection of composite ACR/AMR (36). In the CMR group, study protocol allowed for escalation to EMB in equivocal or clinically determined settings, which was only necessary in 6% of cases. Comparing the EMB and CMR surveillance groups, rejection rates were similar and the rates of hospitalization and infection were lower in the CMR group. Wider use of CMR for rejection diagnosis will require larger scale clinical trials as well as prompt availability of CMR in rejection suspicion (similar to rapid MRI evaluation utilized in stroke protocols). In addition, the cost of CMR varies greatly across the globe, which may limit universal availability.
Traditional 2D echocardiography can signal allograft dysfunction with the development of systolic or diastolic dysfunction. There remains question, however, about the utility of advanced echocardiography techniques, such as strain imaging, in the evaluation of rejection. Assessment of 2D global longitudinal strain (GLS) imaging has been associated with ACR, including a 2010 analysis by Kato et al. of 396 simultaneous EMB and GLS evaluations, with GLS > −27.4% having 82% sensitivity, 82% specificity, 36% PPV, and 97% NPV for ACR (37). While these results have been replicated in other studies, there have also been studies showing a lack of association between GLS and rejection (38). Additionally, there remains concern that cardiac strain imaging may not be consistently reproducible in the clinical setting, limiting its application in rejection evaluation (39). As advanced imaging modalities continue to evolve, they may complement clinical and molecular investigations of rejection, allowing non-invasive diagnosis of rejection.
With the addition of positive molecular biomarkers of rejection to a clinical picture of allograft dysfunction (such as decreased ejection fraction or signs of heart failure), standard practice has been to initiate pulse dose corticosteroids and proceed with EMB to permit histopathologic assessment for ACR and/or AMR. This includes traditional hematoxylin and eosin staining which can provide an ACR diagnosis as well as special immunohistochemical stains to facilitate the diagnosis of AMR. If ACR is identified, most patients are treated with corticosteroid therapy and, in those with graft dysfunction, anti-thymocyte globulin may be administered. The diagnosis and treatment of AMR, however, is more complex, highlighting the need for an accurate diagnosis to outline a precise therapeutic pathway. Therapies for AMR include monoclonal antibodies, intravenous immunoglobulin, and apheresis strategies that impart substantial infection risk and/or cost. The suspicion for AMR is raised in recipients with pre-transplant HLA antibody sensitization, de novo post-transplant DSA, prior AMR, or those of Black race (27, 40, 41). With the presence of these factors as well as circulating DSA and/or other molecular markers of AMR, the non-invasive diagnosis of AMR may be possible (Figure 2).

Figure 2. Utilizing molecular evaluation to diagnose and treat acute cellular rejection and antibody-mediated rejection. With continued clinical validation, molecular biomarkers may allow the diagnosis of specific rejection subtype or support addition evaluations, allowing appropriate therapy without the need for EMB. ACR, acute cellular rejection; AMR, antibody-mediated rejection; ATG, anti-thymocyte globulin; CAV, coronary artery vasculopathy; dd-cfDNA, donor-derived cell free DNA; DSA, donor-specific antibody.
Recent publications of molecular biomarkers suggest the ability to potentially discriminate between ACR and AMR without an EMB. Agbor-Enoh and Shah et al. demonstrated that dd-cfDNA has comparable diagnostic performance in assessing ACR (AUC 0.89) and AMR (AUC 0.95) individually (5). These results from the GRAfT consortium also showed distinct dd-cfDNA characteristics between rejection subtypes, with AMR having significantly higher dd-cfDNA values, shorter dd-cfDNA fragment lengths, and a higher percentage of guanosine:cytosine bases. Emerging modalities, including that of microRNAs may further allow for the diagnosis of rejection subtype without EMB. Duong van Huyen et al. showed that microRNAs were differentially expressed in ACR as compared to AMR in both the myocardium and circulation of heart transplant patients with rejection (42). More recently, a GRAfT study developed and validated microRNA clinical rejection scores for both ACR and AMR, having AUCs of 0.86 and 0.84, respectively (43).
Catheterization and EMB carry risk, especially in the setting of rejection, where these procedures can precipitate hemodynamic compromise. They also are resource-dependent, which may delay the initiation of rejection therapy or lead to non-specific therapy. As molecular and imaging biomarkers of rejection continue to evolve in a diagnosis-specific manner, EMB may not be needed to initiate rejection therapy. There remains the entity of non-specific graft failure or biopsy-negative rejection, a diagnosis which may be further elucidated and adequately treated with molecular biomarker differentiation of ACR or AMR phenotype.
It has become clear that a universal approach to post-transplant care may be less favorable than a personalized medicine approach frequently weighing each patient's rejection risk, which is dynamic as a function of time post-transplant. The 2022 ISHLT Guidelines for the Care of Heart Transplant Recipients support dd-cfDNA and DSA monitoring, without a recommendation for the elimination of EMB with reassuring evaluations (44). They include a Class IIa, Level of Evidence C recommendation to perform DSA monitoring at 1, 3, 6, and 12 months, followed by annually thereafter as well as more frequent assessments for sensitized patients. However, there are no specific schedules recommended for dd-cfDNA monitoring, citing a lack of significant evidence. The incorporation of clinical characteristics and non-invasive biomarkers may lead to multi-item risk prediction modeling that can be readily and repeatedly calculated for each individual patient, in a fashion similar to the AHA/ACC Atherosclerotic Cardiovascular Disease (ASCVD) Risk Calculator. In the long-term evaluation of CAV in heart transplant recipients, Loupy et al. described trajectory-based modeling to identify multivariable risk for CAV (45). Risk modeling of rejection, however, would require real-time rejection prediction, combining post-transplant biomarkers including DSA and dd-cfDNA with known pre-transplant predictors of rejection including HLA antibody sensitization and demographic factors. A composite risk score could help guide rejection monitoring, prompting more or less frequent evaluations given specific patient risk.
Prediction of rejection risk should also include immune system assessment, to help guide immunosuppressive therapy. Current immunosuppression strategies include induction (rapid immune state suppression to induce transplant tolerance) at time of transplant followed by lifelong maintenance immunosuppression with monitoring of circulating immunosuppression medication levels. Medications are typically tapered in a non-precise manner, decreasing doses and desired medication levels at set time intervals after transplant. In addition, levels are often reduced when a patient develops drug toxicity such as gastrointestinal intolerance, leukopenia, renal insufficiency, or neurologic dysfunction. Drug level and toxicity monitoring is important, as differences in pharmacogenomic and physiologic states require altered immunosuppression doses. However, ideal immune monitoring would include an evaluation of net state of immunosuppression, determining risk for rejection with under-immunosuppression and risk for infection or malignancy with over-immunosuppression. The Immuknow® Assay [Viracor-IBT (formerly Cylex), Lenexa, Kansas] has been used toward this goal; however, clinical evaluations of this assay have not shown a significant association with rejection (46, 47). Rather, real-time molecular evaluations such as dd-cfDNA (measuring donor allograft quiescence), torque teno virus quantification (measuring net immunosuppression load), and microRNA (measuring epigenetic factors regulating immune system transcription) may provide insight into immune state (5, 48, 49). These biomarkers have the potential to predict risk of not only rejection (with under-immunosuppression) but also complications of over-immunosuppression (such as infection and malignancy). As these modalities continue to be studied, a rejection risk prediction model may provide a comprehensive evaluation of each patient’s risk profile and the impact of specific rejection therapy and monitoring strategies.
At many heart transplant centers, patients undergo a gentler approach to rejection surveillance, with the elimination of many routine surveillance EMBs in the setting of favorable clinical, imaging, and/or molecular evaluations. As more data emerge regarding the positive predictive value of molecular and advanced imaging evaluations of ACR and AMR, many for-cause biopsies may also be eliminated. This shift may enable non-invasive rejection diagnosis and prompt initiation of rejection therapy. Molecular biomarkers continue to transform the way physicians treat patients, potentially creating rejection risk prediction tools for heart transplant recipients, allowing for less invasive evaluation and improved post-transplant longevity.
JG: Writing – original draft. AM: Writing – review & editing. RB: Writing – review & editing. SA-E: Writing – review & editing. PS: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article.
SAE: National Heart, Lung, and Blood Institute Division of Intramural Research (HHSN268201300001C); PS: NIH K23 Career Development Award 1K23HL143179.
PS=Unrelated grant support paid to the institution from Merck, Bayer, Roche, and Abbott. Consulting for Natera, Merck and Procyrion.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Schlendorf KH, Zalawadiya S, Shah AS, Perri R, Wigger M, Brinkley DM, et al. Expanding heart transplant in the era of direct-acting antiviral therapy for hepatitis C. JAMA Cardiol. (2020) 5:167–74. doi: 10.1001/jamacardio.2019.4748
2. Louca J, Öchsner M, Shah A, Hoffman J, Vilchez FG, Garrido I, et al. The international experience of in-situ recovery of the DCD heart: a multicentre retrospective observational study. eClinicalMedicine. (2023) 58:101887. doi: 10.1016/j.eclinm.2023.101887
3. Coutance G, Kransdorf E, Aubert O, Bonnet G, Yoo D, Rouvier P, et al. Clinical prediction model for antibody-mediated rejection: a strategy to minimize surveillance endomyocardial biopsies after heart transplantation. Circ Hear Fail. (2022) 15:e009923. doi: 10.1161/circheartfailure.122.009923
4. Deng MC, Eisen HJ, Mehra MR, Billingham M, Marboe CC, Berry G, et al. Noninvasive discrimination of rejection in cardiac allograft recipients using gene expression profiling. Am J Transplant. (2006) 6:150–60. doi: 10.1111/j.1600-6143.2005.01175.x
5. Agbor-Enoh S, Shah P, Tunc I, Hsu S, Russell S, Feller E, et al. Cell-free DNA to detect heart allograft acute rejection. Circulation. (2021) 143:1184–97. doi: 10.1161/circulationaha.120.049098
6. Crespo-Leiro MG, Zuckermann A, Bara C, Mohacsi P, Schulz U, Boyle A, et al. Concordance among pathologists in the second cardiac allograft rejection gene expression observational study (CARGO II). Transplant J. (2012) 94:1172–7. doi: 10.1097/tp.0b013e31826e19e2
7. Bermpeis K, Esposito G, Gallinoro E, Paolisso P, Bertolone DT, Fabbricatore D, et al. Safety of right and left ventricular endomyocardial biopsy in heart transplantation and cardiomyopathy patients. Jacc Hear Fail. (2022) 10:963–73. doi: 10.1016/j.jchf.2022.08.005
8. DeVlaminck I, Valantine HA, Snyder TM, Strehl C, Cohen G, Luikart H, et al. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci Transl Med. (2014) 6:241ra77. doi: 10.1126/scitranslmed.3007803
9. Khush KK, Patel J, Pinney S, Kao A, Alharethi R, DePasquale E, et al. Noninvasive detection of graft injury after heart transplant using donor-derived cell-free DNA: a prospective multicenter study. Am J Transplant. (2019) 19:2889–99. doi: 10.1111/ajt.15339
10. Pham MX, Teuteberg JJ, Kfoury AG, Starling RC, Deng MC, Cappola TP, et al. Gene-expression profiling for rejection surveillance after cardiac transplantation. New Engl J Medicine. (2010) 362:1890–900. doi: 10.1056/nejmoa0912965
11. Kobashigawa J, Patel J, Azarbal B, Kittleson M, Chang D, Czer L, et al. Randomized pilot trial of gene expression profiling versus heart biopsy in the first year after heart transplant. Circulation Hear Fail. (2015) 8:557–64. doi: 10.1161/circheartfailure.114.001658
12. FDA. FDA Device Classification, Cardiac Allograft Gene Expression Profiling Test System. Available online at: https://www.accessdata.fda.gov/cdrh_docs/reviews/k073482.pdf (accessed January 31, 2024).
13. Henricksen EJ, Moayedi Y, Purewal S, Twiggs JV, Waddell K, Luikart H, et al. Combining donor derived cell free DNA and gene expression profiling for non-invasive surveillance after heart transplantation. Clin Transplant. (2023) 37(3):e14699. doi: 10.1111/ctr.14699
14. Gondi KT, Kao A, Linard J, Austin BA, Everley MP, Fendler TJ, et al. Single-center utilization of donor-derived cell-free DNA testing in the management of heart transplant patients. Clin Transplant. (2021) 35:e14258. doi: 10.1111/ctr.14258
15. Arnau-Vives MA, Almenar L, Hervas I, Osa A, Martinez-Dolz L, Rueda J, et al. Predictive value of brain natriuretic peptide in the diagnosis of heart transplant rejection. J Hear Lung Transplant. (2004) 23:850–6. doi: 10.1016/j.healun.2003.08.005
16. Hammerer-Lercher A, Mair J, Antretter H, Ruttmann E, Poelzl G, Laufer G, et al. B-type natriuretic peptide as a marker of allograft rejection after heart transplantation. J Hear Lung Transplant. (2005) 24:1444.e5–.e8. doi: 10.1016/j.healun.2004.08.018
17. Battes LC, Caliskan K, Rizopoulos D, Constantinescu AA, Robertus JL, Akkerhuis M, et al. Repeated measurements of NT-pro-B-type natriuretic peptide, troponin T or C-reactive protein do not predict future allograft rejection in heart transplant recipients. Transplantation. (2015) 99:580–5. doi: 10.1097/tp.0000000000000378
18. Fitzsimons SJ, Evans JDW, Rassl DM, Lee KK, Strachan FE, Parameshwar J, et al. High-sensitivity cardiac troponin is not associated with acute cellular rejection after heart transplantation. Transplantation. (2022) 106:1024–30. doi: 10.1097/tp.0000000000003876
19. Eisenberg MS, Chen HJ, Warshofsky MK, Sciacca RR, Wasserman HS, Schwartz A, et al. Elevated levels of plasma C-reactive protein are associated with decreased graft survival in cardiac transplant recipients. Circulation. (2000) 102:2100–4. doi: 10.1161/01.cir.102.17.2100
20. Kennel PJ, Saha A, Maldonado DA, Givens R, Brunjes DL, Castillero E, et al. Serum exosomal protein profiling for the non-invasive detection of cardiac allograft rejection. J Hear Lung Transplant. (2018) 37:409–17. doi: 10.1016/j.healun.2017.07.012
21. Truby LK, Kwee LC, Agarwal R, Grass E, DeVore AD, Patel CB, et al. Proteomic profiling identifies CLEC4C expression as a novel biomarker of primary graft dysfunction after heart transplantation. J Hear Lung Transplant. (2021) 40:1589–98. doi: 10.1016/j.healun.2021.07.024
22. Khush KK, Cherikh WS, Chambers DC, Harhay MO, Hayes D, Hsich E, et al. The international thoracic organ transplant registry of the international society for heart and lung transplantation: thirty-sixth adult heart transplantation report — 2019; focus theme: donor and recipient size match. J Hear Lung Transplant. (2019) 38:1056–66. doi: 10.1016/j.healun.2019.08.004
23. Hammond MEH, Revelo MP, Miller DV, Snow GL, Budge D, Stehlik J, et al. ISHLT pathology antibody mediated rejection score correlates with increased risk of cardiovascular mortality: a retrospective validation analysis. J Hear Lung Transplant. (2016) 35:320–5. doi: 10.1016/j.healun.2015.10.035
24. Stewart S, Winters GL, Fishbein MC, Tazelaar HD, Kobashigawa J, Abrams J, et al. Revision of the 1990 working formulation for the standardization of Nomenclature in the diagnosis of heart rejection. J Hear Lung Transplant. (2005) 24:1710–20. doi: 10.1016/j.healun.2005.03.019
25. Berry GJ, Burke MM, Andersen C, Bruneval P, Fedrigo M, Fishbein MC, et al. The 2013 international society for heart and lung transplantation working formulation for the standardization of nomenclature in the pathologic diagnosis of antibody-mediated rejection in heart transplantation. J Hear Lung Transplant. (2013) 32:1147–62. doi: 10.1016/j.healun.2013.08.011
26. Moayedi Y, Fan CS, Tinckam KJ, Ross HJ, McCaughan JA. De novo donor-specific HLA antibodies in heart transplantation: do transient de novo DSA confer the same risk as persistent de novo DSA? Clin Transplant. (2018) 32:e13416. doi: 10.1111/ctr.13416
27. Torres MF, Pando MJ, Luo C, Luikart H, Valantine H, Khush K. The role of complement-fixing donor-specific antibodies identified by a C1q assay after heart transplantation. Clin Transplant. (2017) 31:e13121. doi: 10.1111/ctr.13121
28. Wang M, Patel NJ, Zhang X, Kransdorf EP, Azarbal B, Kittleson MM, et al. The effects of donor-specific antibody characteristics on cardiac allograft vasculopathy. Clin Transplant. (2021) 35:e14483. doi: 10.1111/ctr.14483
29. Dipchand AI, Webber S, Mason K, Feingold B, Bentlejewski C, Mahle WT, et al. Incidence, characterization, and impact of newly detected donor-specific anti-HLA antibody in the first year after pediatric heart transplantation: a report from the CTOTC-04 study. Am J Transplant. (2018) 18:2163–74. doi: 10.1111/ajt.14691
30. Colvin MM, Cook JL, Chang P, Francis G, Hsu DT, Kiernan MS, et al. Antibody-mediated rejection in cardiac transplantation: emerging knowledge in diagnosis and management. Circulation. (2015) 131:1608–39. doi: 10.1161/cir.0000000000000093
31. Zachary AA, Kopchaliiska D, Jackson AM, Leffell MS. Immunogenetics and immunology in transplantation. Immunol Res. (2010) 47:232–9. doi: 10.1007/s12026-009-8154-1
32. Cole RT, Gandhi J, Bray RA, Gebel HM, Morris A, McCue A, et al. De novo DQ donor-specific antibodies are associated with worse outcomes compared to non-DQ de novo donor-specific antibodies following heart transplantation. Clin Transplant. (2017) 31:e12924. doi: 10.1111/ctr.12924
33. Irving CA, Carter V, Gennery AR, Parry G, Griselli M, Hasan A, et al. Effect of persistent versus transient donor-specific HLA antibodies on graft outcomes in pediatric cardiac transplantation. J Hear Lung Transplant. (2015) 34:1310–7. doi: 10.1016/j.healun.2015.05.001
34. Imran M, Wang L, McCrohon J, Yu C, Holloway C, Otton J, et al. Native T1 mapping in the diagnosis of cardiac allograft rejection A prospective histologically validated study. JACC Cardiovasc Imaging. (2019) 12:1618–28. doi: 10.1016/j.jcmg.2018.10.027
35. Dolan RS, Rahsepar AA, Blaisdell J, Suwa K, Ghafourian K, Wilcox JE, et al. Multiparametric cardiac magnetic resonance imaging can detect acute cardiac allograft rejection after heart transplantation. JACC Cardiovasc Imaging. (2019) 12:1632–41. doi: 10.1016/j.jcmg.2019.01.026
36. Anthony C, Imran M, Pouliopoulos J, Emmanuel S, Iliff J, Liu Z, et al. Cardiovascular magnetic resonance for rejection surveillance after cardiac transplantation. Circulation. (2022) 145:1811–24. doi: 10.1161/circulationaha.121.057006
37. Kato T-S, Oda N, Hashimura K, Hashimoto S, Nakatani T, Ueda H-I, et al. Strain rate imaging would predict sub-clinical acute rejection in heart transplant recipients. Eur J Cardiothorac Surg. (2010) 37:1104–10. doi: 10.1016/j.ejcts.2009.11.037
38. Ambardekar AV, Alluri N, Patel AC, Lindenfeld J, Dorosz JL. Myocardial strain and strain rate from speckle-tracking echocardiography are unable to differentiate asymptomatic biopsy-proven cellular rejection in the first year after cardiac transplantation. J Am Soc Echocardiogr. (2015) 28:478–85. doi: 10.1016/j.echo.2014.12.013
39. Ruiz-Ortiz M, Rodriguez-Diego S, Delgado M, Kim J, Weinsaft JW, Ortega R, et al. Myocardial deformation and acute cellular rejection after heart transplantation: impact of inter-vendor variability in diagnostic effectiveness. Echocardiography. (2019) 36:2185–94. doi: 10.1111/echo.14544
40. Cole RT, Gandhi J, Bray RA, Gebel HM, Yin M, Shekiladze N, et al. Racial differences in the development of de-novo donor-specific antibodies and treated antibody-mediated rejection after heart transplantation. J Hear Lung Transplant. (2018) 37:503–12. doi: 10.1016/j.healun.2017.11.003
41. Clarke B, Ducharme A, Giannetti N, Kim D, McDonald M, Pflugfelder P, et al. Multicenter evaluation of a national organ sharing policy for highly sensitized patients listed for heart transplantation in Canada. J Hear Lung Transplant. (2017) 36:491–8. doi: 10.1016/j.healun.2017.01.003
42. DuongVanHuyen J-P, Tible M, Gay A, Guillemain R, Aubert O, Varnous S, et al. MicroRNAs as non-invasive biomarkers of heart transplant rejection. Eur Heart J. (2014) 35:3194–202. doi: 10.1093/eurheartj/ehu346
43. Shah P, Agbor-Enoh S, Bagchi P, deFilippi CR, Mercado A, Diao G, et al. Circulating microRNAs in cellular and antibody-mediated heart transplant rejection. J Hear Lung Transplant. (2022) 41:1401–13. doi: 10.1016/j.healun.2022.06.019
44. Velleca A, Shullo MA, Dhital K, Azeka E, Colvin M, DePasquale E, et al. The international society for heart and lung transplantation (ISHLT) guidelines for the care of heart transplant recipients. J Hear Lung Transplant. (2023) 42:e1–e141. doi: 10.1016/j.healun.2022.09.023
45. Loupy A, Coutance G, Bonnet G, Keer JV, Raynaud M, Aubert O, et al. Identification and characterization of trajectories of cardiac allograft vasculopathy after heart transplantation. Circulation. (2020) 141:1954–67. doi: 10.1161/circulationaha.119.044924
46. Kobashigawa JA, Kiyosaki KK, Patel JK, Kittleson MM, Kubak BM, Davis SN, et al. Benefit of immune monitoring in heart transplant patients using ATP production in activated lymphocytes. J Hear Lung Transplant. (2010) 29:504–8. doi: 10.1016/j.healun.2009.12.015
47. Rossano JW, Denfield SW, Kim JJ, Price JF, Jefferies JL, Decker JA, et al. Assessment of the cylex ImmuKnow cell function assay in pediatric heart transplant patients. J Hear Lung Transplant. (2009) 28:26–31. doi: 10.1016/j.healun.2008.10.001
48. Rezahosseini O, Drabe CH, Sørensen SS, Rasmussen A, Perch M, Ostrowski SR, et al. Torque-teno virus viral load as a potential endogenous marker of immune function in solid organ transplantation. Transplant Rev. (2019) 33:137–44. doi: 10.1016/j.trre.2019.03.004
49. Aelst LNLV, Summer G, Li S, Gupta SK, Heggermont W, Vusser KD, et al. RNA profiling in human and murine transplanted hearts: identification and validation of therapeutic targets for acute cardiac and renal allograft rejection. Am J Transplant. (2016) 16:99–110. doi: 10.1111/ajt.13421
50. Richmond ME, Zangwill SD, Kindel SJ, Deshpande SR, Schroder JN, Bichell DP, et al. Donor fraction cell-free DNA and rejection in adult and pediatric heart transplantation. J Hear Lung Transplant. (2020) 39:454–63. doi: 10.1016/j.healun.2019.11.015
51. Kim PJ, Olymbios M, Siu A, Pinzon OW, Adler E, Liang N, et al. A novel donor-derived cell-free DNA assay for the detection of acute rejection in heart transplantation. J Hear Lung Transplant. (2022) 41:919–27. doi: 10.1016/j.healun.2022.04.002
52. Dewi IS, Celik S, Karlsson A, Hollander Z, Lam K, McManus J-W, et al. Exosomal miR-142-3p is increased during cardiac allograft rejection and augments vascular permeability through down-regulation of endothelial RAB11FIP2 expression. Cardiovasc Res. (2017) 113:440–52. doi: 10.1093/cvr/cvw244
Keywords: acute cellular rejection (ACR), antibody-mediated rejection (AMR), cardiac allograft vasculopathy (CAV), cardiac magnetic resonance (CMR), donor-derived cell-free DNA (dd-cfDNA), donor-specific antibody (DSA), endomyocardial biopsy (EMB), gene expression profiling (GEP)
Citation: Goldberg JF, Mehta A, Bahniwal RK, Agbor-Enoh S and Shah P (2024) A gentler approach to monitor for heart transplant rejection. Front. Cardiovasc. Med. 11:1349376. doi: 10.3389/fcvm.2024.1349376
Received: 4 December 2023; Accepted: 24 January 2024;
Published: 6 February 2024.
Edited by:
Lauren Cooper, Northwell Health, United StatesReviewed by:
Marc Samsky, Yale University, United States© 2024 Goldberg, Mehta, Bahniwal, Agbor-Enoh and Shah. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Palak Shah cGFsYWsuc2hhaEBpbm92YS5vcmc=
Abbreviations ACR, acute cellular rejection; AMR, antibody-mediated rejection; CAV, cardiac allograft vasculopathy; CMR, cardiac magnetic resonance; dd-cfDNA, donor-derived cell-free DNA; DSA, donor-specific antibody; EMB, endomyocardial biopsy; GEP, gene expression profiling; GLS, global longitudinal strain; HLA, human leukocyte antigen.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.