- 1Department of Cardiovascular Medicine, The First Hospital of Jilin University, Changchun, China
- 2Department of Cardiovascular Medicine, The Second People's Hospital of Yibin, Yibin, China
Dilated cardiomyopathy (DCM) is one of the most common primary myocardial diseases. However, to this day, it remains an enigmatic cardiovascular disease (CVD) characterized by ventricular dilatation, which leads to myocardial contractile dysfunction. It is the most common cause of chronic congestive heart failure and the most frequent indication for heart transplantation in young individuals. Genetics and various other factors play significant roles in the progression of dilated cardiomyopathy, and variants in more than 50 genes have been associated with the disease. However, the etiology of a large number of cases remains elusive. Numerous studies have been conducted on the genetic causes of dilated cardiomyopathy. These genetic studies suggest that mutations in genes for fibronectin, cytoskeletal proteins, and myosin in cardiomyocytes play a key role in the development of DCM. In this review, we provide a comprehensive description of the genetic basis, mechanisms, and research advances in genes that have been strongly associated with DCM based on evidence-based medicine. We also emphasize the important role of gene sequencing in therapy for potential early diagnosis and improved clinical management of DCM.
1. Introduction
Dilated cardiomyopathy (DCM) is characterized by the enlargement of the left ventricle (LV) and global or regional systolic dysfunction, which cannot be solely attributed to abnormal loading conditions such as hypertension, valve disease, congenital heart disease, or coronary artery disease (1, 2). It is considered one of the main causes of heart failure with reduced ejection fraction (HFrEF) worldwide. Genetic defects are the primary cause of DCM, with approximately 30%–35% of idiopathic cardiomyopathy being attributed to genetic defects. The majority of genes associated with genetic DCM exhibit autosomal dominant transmission, while a minority follow an autosomal recessive, X-linked, or mitochondrial inheritance pattern (3). There are about 60 genes associated with DCM, and evidence-based medicine has identified 12 genes that are highly associated with the condition (4, 5). Familial DCM can be inherited as a recessive or X-linked trait, although autosomal dominant inheritance is the most common (6). Non-genetic factors also play a significant role in DCM, and there is overlap between genetic and non-genetic causes (7). The objective of this review is to provide a comprehensive summary of the genes that are clearly associated with DCM and to recommend sequencing of known cardiomyopathy genes for all DCM patients. It is important to note that genetic causes of DCM cannot be ruled out in patients with no family history, as de novo mutations may be responsible for the development of the condition (4, 8–10). The standard treatment for heart failure resulting from DCM involves drug therapy, such as loop diuretics, angiotensin-converting enzyme inhibitors (ACEIs), beta blockers, and sodium-glucose cotransporter 2 inhibitors (SGLT2i), as well as cardiac resynchronization therapy (CRT) (11, 12). However, current medications do not halt myocardial degeneration, and heart transplantation remains the only option for patients in the advanced stages of heart failure (13, 14). Nevertheless, the precise pathological mechanisms responsible for variations in disease susceptibility and phenotypic expression, including the risk of heart failure (HF) or sudden cardiac death (SCD), remain elusive (15, 16). As the genetic dimension of DCM becomes better understood, gene therapy is emerging as a promising treatment strategy for DCM (17).
2. Epidemiology
Reliable epidemiological data on cardiomyopathy is primarily sourced from developed countries, where the accuracy of prevalence data is dependent on the use of established diagnostic evaluations and criteria. However, there is still a dearth of epidemiological data concerning DCM in Asia (7). The Olmsted County study concluded that the prevalence of idiopathic DCM was 1 in 2,500. However, it is possible that the incidence and prevalence of DCM may be greatly underestimated due to various biases, such as misclassification and missing or incomplete data. Recent studies suggest that the prevalence of asymptomatic idiopathic DCM may be equal to or greater than 1 in 250 (3, 18, 19). The incidence of DCM is slightly higher in males than in females, with an average sex ratio of 1.7:1 for males with hereditary DCM and 2.5:1 for females with non-hereditary DCM. The long-term prognosis for females is better than that for males (20–22). However, these data are based on estimates, and a formal, population-based epidemiological study is still needed to determine the true prevalence and incidence of DCM. Researches have shown that 26% of children with dilated cardiomyopathy experience either death or the need for a heart transplant within one year of diagnosis, with an additional 1% per year thereafter (23). DCM is also the most common cause of chronic congestive heart failure and sudden cardiac death in individuals aged 20–60, as well as the leading cause of heart transplants (24).
3. Clinical manifestation
Symptoms of DCM, such as dyspnea, fatigue, dizziness, syncope, and edema, may intermittently manifest in some patients during the early stages of DCM. However, these symptoms become more pronounced as the disease progresses to its severe stage (25). Uncommon yet significant signs and symptoms like abnormal skin pigmentation, skeletal myopathy, and neurosensory disorders (e.g., deafness, blindness) may indicate a specific form of multisystem disease or a unique DCM genotype. These symptoms are considered “red flags” for DCM diagnosis (26).
4. Etiology
The etiology of dilated cardiomyopathy can be categorized into genetic causes, which lead to primary dilated cardiomyopathy, and acquired factors resulting in secondary dilated cardiomyopathy. It is imperative for clinicians to rule out secondary causes prior to confirming a diagnosis of “idiopathic DCM”, as there might be potential reversible causes (27, 28). Single gene mutations account for 25%–50% of all DCM cases. Genes linked to DCMs can be categorized into various groups, which encompass those encoding nuclear envelope proteins, sarcomere proteins, structural proteins, ion channels, and proteins yet to be classified (29). Jan Haas and his team identified a notably higher mutation rate in familial instances compared to sporadic ones. Even when genetic investigations fail to elucidate a familial disease, alternative mechanisms like epigenetic modifications—including microRNAs, histone modifications, and DNA methylation—should be considered (30). There are also certain genetic mutations that can indirectly lead to DCM by affecting the stability of crucial cardiac structures. For instance, The Z-disc in cardiac myocytes is a crucial region where numerous proteins interact within the Z-disc to facilitate force transmission and intracellular signaling in both the heart and skeletal muscles (31, 32). Kindlin-2 collaborates with α-actinin-2 and β1 integrin to preserve the structural integrity of the Z-disc in cardiac muscle tissue (33). The elimination of Kindlin-2 in murine models results in the disruption of the Z-disc structure, subsequently causing cardiac malfunction (34, 35). Increasingly, research studies are documenting the existence of multiple potentially causative mutations in patients diagnosed with Dilated Cardiomyopathy (DCM). While a significant number of these are likely silent variants, an emerging model of oligogenic inheritance—a disease provoked by a limited number of mutations across multiple genes—is being recognized (36). These acquired factors encompass infections, excess alcohol consumption, exposure to toxins, cancer therapies, endocrine disorders, pregnancy, tachyarrhythmias, and immune-mediated diseases (37).
5. Genes strongly associated with DCM
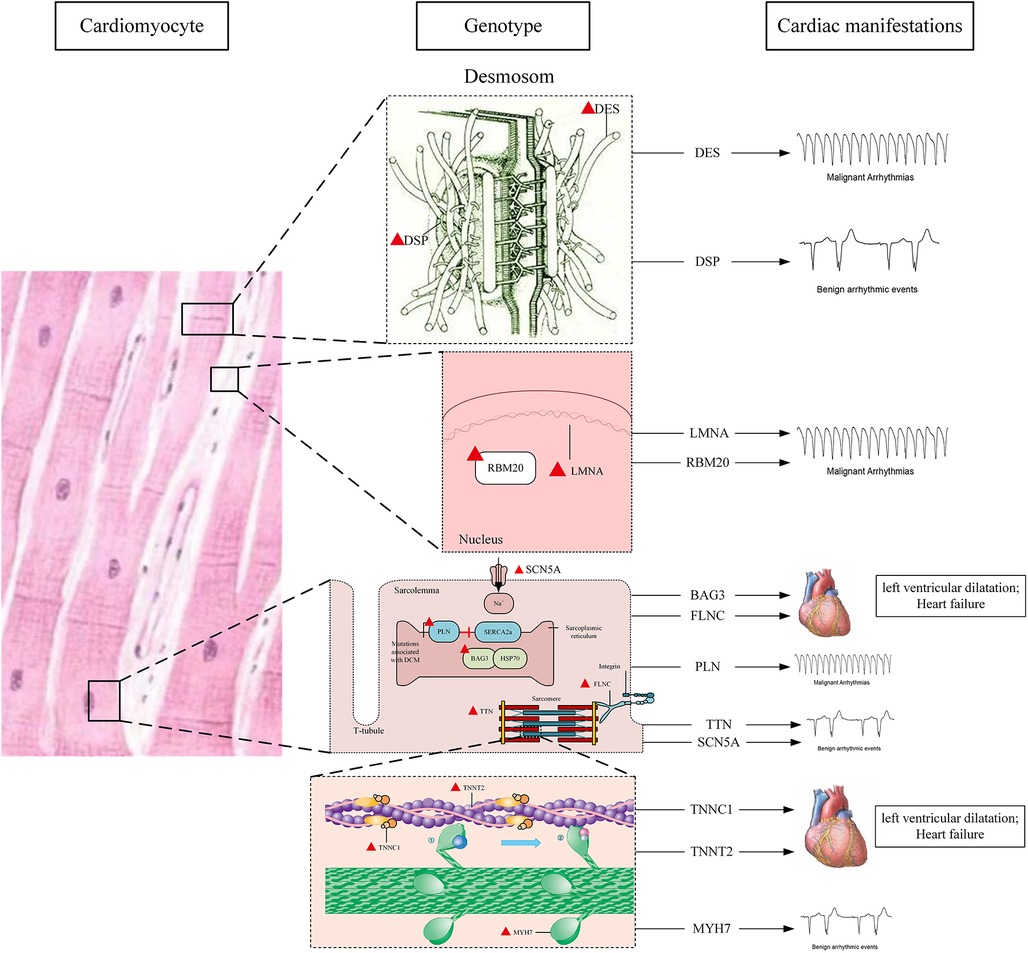
The location and major cardiac manifestations of DCM related genes are shown in Figure 1.
5.1. TTN
The Titin Gene (TTN) is one of the largest genes found in humans, encoding the protein titin. With a molecular weight of 3,816 kDa, titin is the largest known polypeptide and a giant muscle protein that spans half of the myotome from the Z line to the M line. It serves as the fundamental structural and functional unit of striated muscle, with its presence in both the heart and skeletal muscle (25, 38, 39). The TTN gene expresses two major isoforms in the heart, namely N2B and N2BA. These isoforms contain four distinct regions referred to as z, i, a, and m lines (40). Titin proteins not only provide structural, mechanical, and regulatory support but also play a pivotal role in the passive and active contractility of skeletal muscles (41). Truncation mutations of TTN (TTNtv) are most prevalent in DCM, contributing to approximately 25% of familial DCM cases and 18% of sporadic DCM cases (40, 42). A significant number of TTN mutations, responsible for DCM, are heterozygous truncation variants of TTNtv. These include frame-shift mutations, nonsense mutations, and critical splice-site mutations, which are predominantly overexpressed in the A-band region of the titin protein. Additionally, a minor percentage of DCM cases may also be attributed to missense mutations in titin (43–45). The clinical implications of TTNtv are largely contingent on exon expression and the location of the mutation. TTNtv is predominantly located in the A-band, with the severity of the disease increasing as the mutation approaches the C-terminal, indicating a location dependence (43, 44). The exact mechanism through which titin truncation mutations induce cardiac phenotypes remains uncertain. Several mechanisms have been proposed to elucidate TTNtv-induced DCM, including haploinsufficiency, poison peptide/dominant negative mechanism, disruption of cardiac metabolism and signaling, and loss of function. However, these mechanisms warrant further validation through subsequent studies (43, 46–48). The mutation in TTNtv instigates a metabolic transition in the cardiac system from fatty acids to glycolysis (49). However, a chronical elevation in glycolytic intermediates and branched-chain amino acids may trigger the activation of the serine/threonine protein kinase mTOR complex 1 (mTORC1) signaling pathway. This activation subsequently stimulates inefficient protein synthesis cycles and suppresses autophagy (47, 50). TTNtv is characterized by frequent arrhythmias, mainly atrial fibrillation and ventricular arrhythmias (51, 52). Patients with DCM who are TTNtv-positive have a significantly heightened risk of persistent ventricular tachycardia compared to TTNtv-negative patients (44). Studies suggest that in a certain percentage of patients, TTNtv may interact synergistically with other factors (e.g., cardiotoxic chemotherapy, pregnancy), implying that treating these exacerbating factors could lead to substantial recovery (47). As our understanding of gene mutations improves, the potential for genetic engineering-based therapies becomes apparent. Techniques such as reverse-mediated exon skipping, targeted therapy, and genome editing strategies could provide promising therapeutic opportunities (53–55).
5.2. LMNA
The Lamin A and Lamin C Gene (LMNA) is comprised of 2,400 base pairs of DNA and 12 exons. Lamins A and C, intermediate filamentous nuclear envelope proteins, are encoded by the LMNA gene. This nuclear membrane protein is situated in the inner part of the nuclear membrane and is generally expressed in differentiated cells (56). The LMNA gene is the second most frequently mutated gene in DCM。The occurrence rate of Lamin A/C variations in patients diagnosed with DCM is approximately 6% (57, 58). Patients with pathogenic LMNA mutations exhibit a high rate of sudden cardiac death due to malignant arrhythmias and a poor prognosis (59, 60). In laminopathy, frameshift mutation is often associated with heart disease, splice site mutation is an independent risk factor for sudden cardiac death, and non-missense mutations (deletions/truncations or mutations affecting splicing) are major independent risk factors for malignant ventricular arrhythmias (MVAs) (61). The exact pathogenesis of DCM caused by LMNA mutation remains unclear, but three hypotheses—mechanical, gene expression, and cytotoxicity—have been proposed to explain the cardiac dysfunction associated with it. The mechanical hypothesis suggests that the destruction of the nuclear layer increases nuclear fragility and sensitivity to mechanical stress, making myocardial tissue more susceptible to pathological effects (62). One proposed mechanism views the nucleus as a mechanosensor that modulates gene expression in response to mechanical perturbations. Consequently, external forces transmitted via the cytoskeleton induce nuclear deformation. Simultaneously, contingent on lamin composition, there can be an impact on its transcriptional activity by altering chromatin's organization and positioning. Concurrently, any modifications in chromatin's structure and organization can potentially influence the nucleus's mechanical attributes (63). Given that the nuclear lamina possesses a higher degree of stiffness compared to the nuclear membrane, it serves to shield the latter from substantial mechanical forces. This buffering capability can notably influence the stretch response of the nuclear membrane, subsequently altering the distribution and organization of membrane-bound proteins. The force-induced expansion of nuclear membranes could potentially represent another mechanism activated by disrupted mechanotransduction (64). Studies indicate that decoupling the mechanical forces of the nuclear/nuclear skeleton and cytoskeletal transduction can significantly extend the lifespan of LMNA deficient mice (65). The gene expression hypothesis posits that defective lamins impede signal transduction and chromatin organization, thereby altering signal transduction, a key driver of LMNA-associated dilated cardiomyopathy. This directly affects and disrupts gene transcription and other intracellular signaling pathways, significantly increasing myocardial fibrosis and leading to left ventricular dysfunction and heart failure (66, 67). The cytotoxic hypothesis suggests that mutated prelamin A protein, also known as presenilin, contributes to the disease by disrupting nuclear morphology, heterochromatin distribution, and DNA damage repair pathways, leading to premature aging (28, 62). Due to the risk of sudden cardiac death in LMNA associated DCM being linked with heart block and bradycardia, the use of Implantable Cardioverter Defibrillators (ICDs) has been recommended for all indications (68). Various other pathways downstream of the LMNA gene have also been explored as potential therapeutic pathways, such as the use of rapamycin/rapalog to inhibit mTOR and MEK1/2 kinase pathway inhibitors, the inhibition of the activation of brominated domain protein 4 (BRD4), and the destruction of LINC complex protein SUN1 to inhibit LMNA mutations (65, 69–71). However, no specific and effective treatment is presently available.
5.3. DSP
Desmoplakin (DSP), encoded by the DSP gene, is a major component of desmosomes and is highly abundant in myocardial tissue. The DSP protein exhibits a tripartite structure, which includes a spherical N-terminal patch domain, a central α-helical rod domain, and a C-terminal tail domain. The DSP gene, located on chromosome 6p24.3, undergoes alternative splicing to produce three subtypes: DSP-I (long), DSP-IA (intermediate), and DSP-II (short). DSP-I is the primary cardiac subtype and plays a crucial role in intercellular adhesion within cardiomyocytes (72, 73). Gene-targeting studies in mice have revealed that mice with ablated DSP genes die during early embryonic development (74). In an assessment of genes associated with DCM, DSP emerged as the highest-scoring gene. However, the arrhythmia phenotype's potential to complicate the interpretation of experimental data led to questions about the trial score. Consequently, the DSP gene was identified as a strong contributor to DCM, rather than the definitive cause (4). Some research suggests that DSP mutations are unique to adult DCM (75). Palmoplantar keratoderma may serve as an early clinical symptom of DCM associated with a DSP mutation (76). Carriers of DSP variants exhibit a higher rate of arrhythmia events, similar to those with LAMA variants, even in the absence of significant left ventricular dysfunction or dilation (77). Current research has also classified desmoprotein cardiomyopathy as an arrhythmic cardiomyopathy triggered by DSP mutations. This condition is characterized by paroxysmal myocardial injury, left ventricular fibrosis preceding systolic dysfunction, and a high incidence of ventricular arrhythmia (73, 78). Typical electrocardiogram (ECG) abnormalities include limb lead QRS depression (peak <0.5 mV) and lateral or inferior lead T wave inversion (73, 79). In arrhythmogenic cardiomyopathy (ACM) caused by DSP variation, cardiomyocytes release a large number of inflammatory cytokines and chemotactic molecules (80). Histological analysis of left ventricular myocaroma in DSP patients has also revealed inflammatory infiltration and scarring (81). Consequently, inflammation is considered a key feature of the disease, and regulating inflammatory signaling pathways may present a new therapeutic target for desmosome-mediated cardiomyopathy.
5.4. DES
DES encodes the primary intermediate filament (IF) protein, desmin, in human heart and skeletal muscle (82). Desmin serves as a structural component of the extramuscular cytoskeleton, which forms a three-dimensional scaffold around the Z disk of the myofibril. This structure connects adjacent myofibrils and the myofibril apparatus to the nucleus, submuscular cytoskeleton, and cytoplasmic organelles such as mitochondria (83). Desmin proteins exhibit a tripartite structure, comprising a conserved central rod domain flanked by non-alpha helical head and tail domains. The central rod domain consists of four helical segments (2A, 1B, 12A, and 2B) separated by three short polypeptide junctions (L1, L1, and L2) (84). There are over 73 different IF proteins, with Desmin being the most abundantly expressed IF protein in muscle-specific tissues and cardiomyocytes (85). IFs perform numerous tissue-specific functions, including providing mechanical support to cells and regulating intracellular tissues, stress responses, cell growth, proliferation, migration, and death (84–86). In patients with DCM, the incidence of desmin gene mutations is less than 1.6% (87). Most DES mutations are missense mutations within the central domain; nonsense, insertion, deletion, or combination insertion and deletion mutations are rare (88). Approximately 74% of DES mutation carriers exhibit cardiac symptoms, and about 50% develop cardiomyopathy, with dilated cardiomyopathy (DCM) being the most common type (89). Researches involving mice with DES gene knockout have demonstrated that desmin deficiency not only impacts heart structure but is also associated with severe abnormalities in myocardial metabolism of glucose, fatty acids, and amino acids (90, 91). Therefore, it may be prudent to avoid drugs that could potentially exacerbate mitochondrial function in patients with desmin deficiency (92). The α-crystallin Β-chain (αB-crystallin), encoded by Desmin and CRYAB, has a potential compensatory interaction in cardiac protection. Overexpression of heart-specific αB-crystallin improves mitochondrial dysfunction in desmin-deficient mouse models, suggesting a potential new treatment approach (93).
5.5. MYH7
The MYH7 gene, encoding the cardiac beta-myosin heavy chain, is situated on chromosome 14q11.2-q13. This gene consists of 40 exons that produce a MYH7 protein containing 1,935 amino acids. This protein is primarily expressed in ventricular muscle and type 1 skeletal muscle fibers, and it is a significant component of human ventricular myosin. The MYH7 protein plays a crucial role in the energy supply for cardiomyocytes and in maintaining the Ca2+ concentration inside and outside these cells (94, 95). MYH7 mutations account for 1%–5.3% of DCM cases (96). These mutations are predominantly missense variants, inherited in a chromosomal dominant pattern, with high penetrance in families and a relatively high proportion among children (95, 97). Mutations in the MYH7 gene can damage the integrity of the sarcomere structure or function, affecting the contraction of the heart muscle (95). Atrial fibrillation and atrial fibrosis are considered early clinical manifestations of MYH7-related cardiomyopathy, which may provide valuable insights for disease diagnosis (98). It has been reported that combined mutations in MYH7 and TNNT2, MYH7 and LAMA4, or MYH7 and TPM1 can result in severe DCM (99–101). These findings highlight the importance of comprehensive screening of DCM-related genes, even after identifying a single disease-causing mutation. Studies have shown that telomere length in mice can offer protection against heart disease in humans. Mutations in proteins that are critical to cardiomyocyte function, such as MYH7, TTN, and MYBPC3, lead to shorter telomeres. Consequently, significant telomere shortening can serve as a biomarker for premature aging of cardiomyocytes in hereditary Hypertrophic Cardiomyopathy (HCM) and DCM (102, 103). The Telomere Repeat Binding Factor 2 (TRF2) has been demonstrated to prevent telomere attrition, thereby improving cell morphological defects, activation of DNA damage response, and premature cell death (104).
5.6. BAG3
Bcl2-associated athanogene 3 (BAG3) codes for an anti-apoptotic protein located on the Z disc of myotomes. As a member of the anti-apoptotic BAG protein family, BAG3 is abundantly expressed in the heart, skeletal muscle, various types of tumor cells, as well as in the brain and peripheral nervous system (105, 106). BAG3 plays vital roles in anti-apoptosis, protein homeostasis maintenance, mitochondrial stability regulation, myocardial contraction regulation, and arrhythmia reduction (107, 108). The multifunctionality of BAG3 in cardiomyocytes is attributed to the presence of multiple functional domains, including the WW domain, the IPV (Ile-Pro-Val) motif, the proline-rich motif, and the BAG domain (109). However, studies indicate that all known or probable pathogenic variants impact at least the WW domain, IPV domain, or BAG domain, with none of the missense pathogenic or possibly pathogenic variants affecting the proline-rich motifs. These three protein domains play significant roles in BAG3's function in the heart (108). Large multicenter cohort studies reveal that DCM resulting from BAG3 mutations is characterized by early-onset in most patients, a high risk of progressing to end-stage heart failure, and a worse prognosis in males (110). BAG3, an anti-apoptotic constituent of the BAG protein family, possesses an inhibitory function against apoptosis. A notable surge in apoptosis was observed in mice lacking BAG3 (111). As a cochaperone protein, BAG3 interacts with ATP-dependent high molecular weight heat shock proteins and ATP-independent small heat shock proteins (sHSPs) in large, functionally different multichaperone protein complexes (112). And after BAG3 is lost, the affected sHSPs levels are reduced due to protein instability (112). BAG3-mediated macrophage recruitment can maintain protein homeostasis, autophagic flux was suppressed in BAG3-deficient hearts, which might result in misfolded protein aggregates (113). Consequently, there is an increase in the content of insoluble proteins, which accelerates the process of cellular senescence. MicroRNAs (miRNAs) are small non-coding RNAs (20–25 nucleotides) that function as epigenetic regulators in cardiovascular system development and physiology. Dysregulation of their expression is directly associated with the pathophysiology of numerous cardiovascular diseases (114, 115). Studies demonstrate that the co-expression of miR-154-5p and miR-182-5p holds diagnostic value in DCM of BAG3 mutation carriers (116). The transcriptional adaptation of gene expression triggered by harmful gene mutations, known as genetic compensation, has been observed in BAG3-knockout zebrafish to protect against heart and skeletal muscle damage. This biological phenomenon may also be active in some human carriers of BAG3 mutations (117). Further investigation of the relevant molecular mechanisms may offer fresh insights for the development of therapeutic interventions.
5.7. FLNC
The filamin family comprises three isomers: filamin A (FLNA), filamin B (FLNB), and filamin C (FLNC) (118). FLNA and FLNB are commonly expressed, while FLNC is most prevalent in skeletal and cardiac muscle (119). FLNC plays a crucial role in the regulation of cellular mechanics, Z-disk arrangement and orientation, and intermyofibrillar connections in mammalian hearts (120, 121). Deficiency of FLNC in cardiomyocytes can lead to fetal death. Furthermore, adult mice deficient in FLNC develop rapid and fulminant DCM within two weeks (122). These studies underscore the significant role of FLNC in both developing and adult cardiomyocytes. A truncation mutation in FLNC (FLNCtv) is closely associated with DCM (123). Patients with FLNCtv often exhibit left ventricular dilatation with systolic dysfunction and myocardial fibrosis. Ventricular arrhythmias are prevalent, and families carrying these mutations have a high incidence of sudden cardiac death (124, 125). β-catenin (CTNNB1) has been identified as the downstream target of FLNC through co-immunoprecipitation and proteomic analysis. FLNC is unable to induce nuclear translocation of CTNNB1, which subsequently activates the platelet-derived growth factor receptor-α (PDGFRA) pathway. Inhibition of PDGFRA can partially reverse the pathological gene expression profile of FLNC patient-specific cardiocytes, cardiac insufficiency, and arrhythmia (126). Therefore, inhibition of this pathway presents a potential therapeutic approach for FLNC-associated cardiomyopathy.
5.8. PLN
Phospholamban (PLN) is a 6.1 kDa protein situated on the sarcoplasmic and endoplasmic reticulum (SR/ER) membrane. PLN is responsible for the encoding of a crucial regulatory protein associated with Ca2+ cycling. It serves as a principal mediator of beta-adrenergic effects, which subsequently leads to an augmentation of cardiac output (127). It is regulated by protein kinase-mediated phosphorylation and serves as an inhibitor of the sarcoplasmic/endoplasmic reticulum Ca2+ ATPase (SERCA2a). Homeostasis and cardiac contractility are achieved by reversibly inhibiting SERCA2a activity (128). The interaction between SERCA2a and PLN determines the rate of diastole and contraction of cardiomyocytes (129). Impaired SR function in diastolic and systolic Ca2+ circulation is a critical factor in cardiac cardiomyocyte failure (130). Among all known PLN mutants, the PLN-R14DEL mutation appears to be the most prevalent (131, 132). The mutation of the PLN gene is not a common cause of cardiomyopathy in our population, with a mutation occurrence rate of less than 1% (133, 134). A large multicentre study with long-term follow-up of PLN mutation carriers found that early ventricular arrhythmia and end-stage heart failure were common in PLN-R14DEL mutation carriers, resulting in a significant increase in cohort mortality (135). From the perspective of a cardiomyocyte, it is recognized that PLN R14del significantly affects its function. Research has shown that the R14del mutation results in the aggregation of PLN protein, an escalation in the activity of the unfolded protein response, dysregulation of calcium, as well as contractile and metabolic dysfunction (136). Despite the well-defined genetic etiology, the molecular mechanism driving the pathogenesis of PLN R14del-cardiomyopathy remains elusive (137). Low-voltage electrocardiograms are more common in women carrying PLN mutations, but their prognostic value is higher in men (138). It has been proposed that eplerenone treatment can prevent or slow disease progression in presymptomatic PLN mutation carriers. Further multicentre randomized double-blind trials are being conducted to confirm this (139). As our understanding of the PLN-R14del mutation mechanism continues to improve, precision medicine, including gene editing and targeted gene therapy, may represent a new direction for future treatment (136, 140, 141). However, most current research remains in the animal testing phase, and it is unclear whether these findings will be applicable to human patients (142).
5.9. RBM20
The RNA-binding motif protein 20 (RBM20) primarily functions as a splicing regulator, predominantly expressed in the heart and skeletal muscle, where it orchestrates both constitutive splicing and alternative splicing of pre-messenger RNA (143, 144). RBM20 gene mutations, which predominantly manifest as missense mutations that alter conserved residues, are a leading cause of DCM (145, 146). This mutations account for approximately 3% of all DCM cases (143).
The pathophysiology of these RBM20 mutations stems from a combination of functional loss and pathogenic functional gain (147). Of the genes regulated by RBM20, TTN is the most significant (146, 148). Diminished RBM20 activity also results in the altered expression of protein subtypes that sustain muscle structure and cardiac function, such as CAMKIIδ, LDB3, and CACNA1C. These alterations can induce changes in biomechanics, electrical activity, and signal transduction, potentially leading to cardiomyopathy, fibrosis, arrhythmia, and sudden death (149, 150). Patients with DCM who carry RBM20 mutations often exhibit impaired cardiac function and are susceptible to atrial fibrillation, ventricular arrhythmia, and sudden cardiac death (151). All-trans retinoic acid (ATRA) has been identified as a potential regulator of RBM20, with studies showing that ATRA can increase RBM20 expression and partially restore the in vitro DCM phenotype. Therefore, pharmacological upregulation of RBM20 expression could be a promising therapeutic strategy for DCM patients with heterozygous RBM20 mutations (152). Most RBM20 mutations are clustered in an arginine/serine (RS) -rich domain, suggesting that precision gene editing using adenine base editing (ABE) and primer editing (PE) might offer potential treatments (147).
5.10. SCN5A
The sodium channel family is comprised of nine genes (SCN1A-SCN5A, SCN7A-SCN11A). Among these, the SCN5A gene, located on human chromosome 3p22, encodes the cardiac sodium channel pore-forming α subunit Nav1.5. SCN5A/Nav1.5 is predominantly expressed in the atrial and ventricular myocardium, His bundle, bundle branch, and Purkinje fibers (153). The prevalence of SCN5A-mediated cases in patients with dilated cardiomyopathy (DCM) is approximately 2% (154). Frameshift mutations in SCN5A can result in a loss of function of the heart's sodium channels (155). The molecular pathway through which these mutations cause ventricular dilation and dysfunction is yet to be fully understood (156). Mutations in SCN5A may interfere with the interaction between Nav1.5 and other components of the complex, leading to structural deformities and contractile damage. Two Nav1.5 mutations (R222Q and R225W) in the voltage sensor domain (VSD), situated in the voltage-gated ion channel, are hypothesized to generate gated hole currents that may be linked with arrhythmia and ventricular dilation in humans (157). Occult myocardial injury may also result from the impaired function of the mutated SCN5A immune sensor (158). DCM typically exhibits age-dependent penetrance, with the phenotype becoming more pronounced with age (159). Clinically, it often manifests as severe arrhythmias (including atrial fibrillation and ventricular tachycardia) and conduction block (154, 160). The initiation of sodium channel blockers can prevent significant morbidity and mortality (161). Research has revealed that the mRNA stabilizing protein HuR protects SCN3A by binding to 5'-UTR mRNA, preventing its decay. The risk of arrhythmia can be reduced by enhancing mRNA stability to preserve decreased SCN5A expression (162).
5.11. TNNC1, TNNT2
The TNNC1 gene (3p21.1) encodes cardiac troponin C (cTnC) in heart tissue, while the TNNC2 gene (1q32.1) encodes cardiac troponin T (cTnT) (163). The trimer filament Tn complex, involved in muscle contraction, is formed by the combination of cTnT, cTnI, and cTnC (164). Point mutations on TNNC1 can alter the function of cTnC in two ways: by changing its binding affinity for Ca2+ or by modifying the interaction of cTnC with its binding partner (165). Troponin complex mutations are present in approximately 6% of familial DCM cases (166). The occurrence of mutations in the TNNC1 gene is roughly 1% (167). The frequency of TNNT2 mutations in DCM is around 3% (168). Patients with TNNC1 gene mutations are typically diagnosed at a younger age and have a higher risk of experiencing potentially fatal events, which often manifest as early severe systolic heart failure, necessitating heart transplant surgery (163, 169). Research indicates that a reduced sensitivity of myofilaments to Ca2+ plays a critical role in the pathophysiology of filament-associated DCM. Enhancing myofilament sensitivity to Ca2+ in the early stages of DCM might be an effective treatment strategy (170). Xin actin-binding repeat containing proteins (XIRPs) are a group of rhabdom-specific proteins. The XIN protein, encoded by the XIRP1 gene and also known as HXin-α or CMYA1, is a rhabdom-specific gene in the XIRP family. Overexpression of the repeating isomer XINB can ameliorate DCM remodeling induced by TNNT2-ΔK210 mutations in mice, partially reversing cardiac dilation, systolic dysfunction, and cardiac fibrosis. Therefore, XIN could be a potential therapeutic target (171).
6. Conclusion
This review systematically summarizes the genes and mechanisms implicated in dilated cardiomyopathy, as well as the latest research directions in understanding its causes. It should be noted that with the advancement of medical technology, the diagnosis rate of dilated cardiomyopathy has been increasing. Nonetheless, patients often present with early onset, severe clinical manifestations, and poor prognosis. The standard approach for preventing or treating heart failure is currently the first-line treatment for patients with dilated cardiomyopathy. Cardiac resynchronization therapy and implantable cardioverter-defibrillators may be necessary to prevent life-threatening arrhythmias. It is recommended that all patients with dilated cardiomyopathy undergo sequencing of known cardiomyopathy genes. Gene-level therapy may represent a new approach for future treatments, although our current understanding of disease pathogenesis and gene therapy is primarily derived from preclinical animal models. This review also has some limitations, primarily that it only encompasses genes with substantial supporting evidence within the realm of evidence-based medicine. Due to the constraints of the review's length, there is a limited number of genes currently being researched and a lack of supporting experimental data. Consequently, some genes pertinent to “moderate classification” and “limited classification” have not been included in this review. Further research in this area is warranted.
Author contributions
SW: Conceptualization, Writing – original draft. ZZ: Methodology, Writing – original draft. JH: Writing – original draft. JL: Formal analysis, Writing – original draft. XG: Writing – original draft. HC: Visualization, Writing – original draft. HX: Writing – original draft, Visualization. YW: Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2023.1296389/full#supplementary-material
References
1. Seferović PM, Polovina M, Bauersachs J, Arad M, Ben Gal T, Lund LH, et al. Heart failure in cardiomyopathies: a position paper from the heart failure association of the European society of cardiology. Eur J Heart Fail. (2019) 21(5):553–76. doi: 10.1002/ejhf.1461
2. Arbelo E, Protonotarios A, Gimeno JR, Arbustini E, Barriales-Villa R, Basso C, et al. 2023 ESC guidelines for the management of cardiomyopathies. Eur Heart J. (2023) 44(37):3503–626. doi: 10.1093/eurheartj/ehad194
3. Hershberger RE, Morales A, Siegfried JD. Clinical and genetic issues in dilated cardiomyopathy: a review for genetics professionals. Genet Med. (2010) 12(11):655–67. doi: 10.1097/GIM.0b013e3181f2481f
4. Jordan E, Peterson L, Ai T, Asatryan B, Bronicki L, Brown E, et al. Evidence-based assessment of genes in dilated cardiomyopathy. Circulation. (2021) 144(1):7–19. doi: 10.1161/CIRCULATIONAHA.120.053033
5. Mazzarotto F, Tayal U, Buchan RJ, Midwinter W, Wilk A, Whiffin N, et al. Reevaluating the genetic contribution of monogenic dilated cardiomyopathy. Circulation. (2020) 141(5):387–98. doi: 10.1161/CIRCULATIONAHA.119.037661
6. Fatkin D, MacRae C, Sasaki T, Wolff MR, Porcu M, Frenneaux M, et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N Engl J Med. (1999) 341(23):1715–24. doi: 10.1056/NEJM199912023412302
7. McKenna WJ, Maron BJ, Thiene G. Classification, epidemiology, and global burden of cardiomyopathies. Circ Res. (2017) 121(7):722–30. doi: 10.1161/CIRCRESAHA.117.309711
8. Hershberger RE, Givertz MM, Ho CY, Judge DP, Kantor PF, McBride KL, et al. Genetic evaluation of cardiomyopathy: a clinical practice resource of the American college of medical genetics and genomics (ACMG). Genet Med. (2018) 20(9):899–909. doi: 10.1038/s41436-018-0039-z
9. Merlo M, Cannatà A, Gobbo M, Stolfo D, Elliott PM, Sinagra G. Evolving concepts in dilated cardiomyopathy. Eur J Heart Fail. (2018) 20(2):228–39. doi: 10.1002/ejhf.1103
10. Wilde AAM, Semsarian C, Márquez MF, Sepehri Shamloo A, Ackerman MJ, Ashley EA, et al. European Heart rhythm association (EHRA)/heart rhythm society (HRS)/Asia pacific heart rhythm society (APHRS)/Latin American heart rhythm society (LAHRS) expert consensus statement on the state of genetic testing for cardiac diseases. J Arrhythm. (2022) 38(4):491–553. doi: 10.1002/joa3.12717
11. Altinier A, Paldino A, Gigli M, Pappalardo A, Sinagra G. Current management and treatment. In: Sinagra G, Merlo M, Pinamonti B, editors. Dilated cardiomyopathy: From genetics to clinical management. Cham, CH: Springer Copyright 2019, The Author(s). (2019). p. 199–215.
12. Yancy CW, Jessup M, Bozkurt B, Butler J, Casey DE Jr, Colvin MM, et al. 2017 ACC/AHA/HFSA focused update of the 2013 ACCF/AHA guideline for the management of heart failure: a report of the American college of cardiology/American heart association task force on clinical practice guidelines and the heart failure society of America. Circulation. (2017) 136(6):e137–e61. doi: 10.1161/CIR.0000000000000509
13. Harding D, Chong MHA, Lahoti N, Bigogno CM, Prema R, Mohiddin SA, et al. Dilated cardiomyopathy and chronic cardiac inflammation: pathogenesis, diagnosis and therapy. J Intern Med. (2023) 293(1):23–47. doi: 10.1111/joim.13556
14. Irion CI, Dunkley JC, John-Williams K, Condor Capcha JM, Shehadeh SA, Pinto A, et al. Nuclear osteopontin is a marker of advanced heart failure and cardiac allograft vasculopathy: evidence from transplant and retransplant hearts. Front Physiol. (2020) 11:928. doi: 10.3389/fphys.2020.00928
15. Kayvanpour E, Sedaghat-Hamedani F, Amr A, Lai A, Haas J, Holzer DB, et al. Genotype-phenotype associations in dilated cardiomyopathy: meta-analysis on more than 8000 individuals. Clin Res Cardiol. (2017) 106(2):127–39. doi: 10.1007/s00392-016-1033-6
16. Sedaghat-Hamedani F, Rebs S, El-Battrawy I, Chasan S, Krause T, Haas J, et al. Identification of SCN5a p.C335R variant in a large family with dilated cardiomyopathy and conduction disease. Int J Mol Sci. (2021) 22(23):12990. doi: 10.3390/ijms222312990
17. Helms AS, Thompson AD, Day SM. Translation of new and emerging therapies for genetic cardiomyopathies. JACC Basic Transl Sci. (2022) 7(1):70–83. doi: 10.1016/j.jacbts.2021.07.012
18. Hershberger RE, Hedges DJ, Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol. (2013) 10(9):531–47. doi: 10.1038/nrcardio.2013.105
19. Schultheiss HP, Fairweather D, Caforio ALP, Escher F, Hershberger RE, Lipshultz SE, et al. Dilated cardiomyopathy. Nat Rev Dis Primers. (2019) 5(1):32. doi: 10.1038/s41572-019-0084-1
20. Roth GA, Mensah GA, Johnson CO, Addolorato G, Ammirati E, Baddour LM, et al. Global burden of cardiovascular diseases and risk factors, 1990-2019: update from the GBD 2019 study. J Am Coll Cardiol. (2020) 76(25):2982–3021. doi: 10.1016/j.jacc.2020.11.010
21. Fairweather D, Beetler DJ, Musigk N, Heidecker B, Lyle MA, Cooper LT Jr, et al. Sex and gender differences in myocarditis and dilated cardiomyopathy: an update. Front Cardiovasc Med. (2023) 10:1129348. doi: 10.3389/fcvm.2023.1129348
22. Jain A, Norton N, Bruno KA, Cooper LT Jr, Atwal PS, Fairweather D. Sex differences, genetic and environmental influences on dilated cardiomyopathy. J Clin Med. (2021) 10(11):2289. doi: 10.3390/jcm10112289
23. Alexander PM, Daubeney PE, Nugent AW, Lee KJ, Turner C, Colan SD, et al. Long-term outcomes of dilated cardiomyopathy diagnosed during childhood: results from a national population-based study of childhood cardiomyopathy. Circulation. (2013) 128(18):2039–46. doi: 10.1161/CIRCULATIONAHA.113.002767
24. Qu XK, Yuan F, Li RG, Xu L, Jing WF, Liu H, et al. Prevalence and spectrum of LRRC10 mutations associated with idiopathic dilated cardiomyopathy. Mol Med Rep. (2015) 12(3):3718–24. doi: 10.3892/mmr.2015.3843
25. Sarohi V, Srivastava S, Basak T. A comprehensive outlook on dilated cardiomyopathy (DCM): state-of-the-art developments with special emphasis on OMICS-based approaches. J Cardiovasc Dev Dis. (2022) 9(6):174. doi: 10.3390/jcdd9060174
26. Rapezzi C, Arbustini E, Caforio AL, Charron P, Gimeno-Blanes J, Heliö T, et al. Diagnostic work-up in cardiomyopathies: bridging the gap between clinical phenotypes and final diagnosis. A position statement from the ESC working group on myocardial and pericardial diseases. Eur Heart J. (2013) 34(19):1448–58. doi: 10.1093/eurheartj/ehs397
27. Orphanou N, Papatheodorou E, Anastasakis A. Dilated cardiomyopathy in the era of precision medicine: latest concepts and developments. Heart Fail Rev. (2022) 27(4):1173–91. doi: 10.1007/s10741-021-10139-0
28. Fu Y, Eisen HJ. Genetics of dilated cardiomyopathy. Curr Cardiol Rep. (2018) 20(11):121. doi: 10.1007/s11886-018-1061-0
29. Garfinkel AC, Seidman JG, Seidman CE. Genetic pathogenesis of hypertrophic and dilated cardiomyopathy. Heart Fail Clin. (2018) 14(2):139–46. doi: 10.1016/j.hfc.2017.12.004
30. Haas J, Frese KS, Peil B, Kloos W, Keller A, Nietsch R, et al. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur Heart J. (2015) 36(18):1123–35a. doi: 10.1093/eurheartj/ehu301
31. Wadmore K, Azad AJ, Gehmlich K. The role of Z-disc proteins in myopathy and cardiomyopathy. Int J Mol Sci. (2021) 22(6):3058. doi: 10.3390/ijms22063058
32. Frank D, Kuhn C, Katus HA, Frey N. The sarcomeric Z-disc: a nodal point in signalling and disease. Journal of Molecular Medicine (Berlin, Germany). (2006) 84(6):446–68. doi: 10.1007/s00109-005-0033-1
33. Dowling JJ, Gibbs E, Russell M, Goldman D, Minarcik J, Golden JA, et al. Kindlin-2 is an essential component of intercalated discs and is required for vertebrate cardiac structure and function. Circ Res. (2008) 102(4):423–31. doi: 10.1161/CIRCRESAHA.107.161489
34. Qi L, Yu Y, Chi X, Xu W, Lu D, Song Y, et al. Kindlin-2 interacts with α-actinin-2 and β1 integrin to maintain the integrity of the Z-disc in cardiac muscles. FEBS Lett. (2015) 589(16):2155–62. doi: 10.1016/j.febslet.2015.06.022
35. Qi L, Yu Y, Chi X, Lu D, Song Y, Zhang Y, et al. Depletion of kindlin-2 induces cardiac dysfunction in mice. Sci China Life Sci. (2016) 59(11):1123–30. doi: 10.1007/s11427-016-0025-0
36. Pinto YM, Elliott PM, Arbustini E, Adler Y, Anastasakis A, Böhm M, et al. Proposal for a revised definition of dilated cardiomyopathy, hypokinetic non-dilated cardiomyopathy, and its implications for clinical practice: a position statement of the ESC working group on myocardial and pericardial diseases. Eur Heart J. (2016) 37(23):1850–8. doi: 10.1093/eurheartj/ehv727
37. Heymans S, Lakdawala NK, Tschöpe C, Klingel K. Dilated cardiomyopathy: causes, mechanisms, and current and future treatment approaches. Lancet (London, England). (2023) 402(10406):998–1011. doi: 10.1016/S0140-6736(23)01241-2
38. Labeit S, Kolmerer B. Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science (New York, NY). (1995) 270(5234):293–6. doi: 10.1126/science.270.5234.293
39. Tabish AM, Azzimato V, Alexiadis A, Buyandelger B, Knöll R. Genetic epidemiology of titin-truncating variants in the etiology of dilated cardiomyopathy. Biophys Rev. (2017) 9(3):207–23. doi: 10.1007/s12551-017-0265-7
40. Gigli M, Begay RL, Morea G, Graw SL, Sinagra G, Taylor MR, et al. A review of the giant protein titin in clinical molecular diagnostics of cardiomyopathies. Front Cardiovasc Med. (2016) 3:21. doi: 10.3389/fcvm.2016.00021
41. Eldemire R, Tharp CA, Taylor MRG, Sbaizero O, Mestroni L. The sarcomeric spring protein titin: biophysical properties, molecular mechanisms, and genetic mutations associated with heart failure and cardiomyopathy. Curr Cardiol Rep. (2021) 23(9):121. doi: 10.1007/s11886-021-01550-y
42. Herman DS, Lam L, Taylor MR, Wang L, Teekakirikul P, Christodoulou D, et al. Truncations of titin causing dilated cardiomyopathy. N Engl J Med. (2012) 366(7):619–28. doi: 10.1056/NEJMoa1110186
43. Kellermayer D, Smith JE 3rd, Granzier H. Titin mutations and muscle disease. Pflugers Arch. (2019) 471(5):673–82. doi: 10.1007/s00424-019-02272-5
44. Roberts AM, Ware JS, Herman DS, Schafer S, Baksi J, Bick AG, et al. Integrated allelic, transcriptional, and phenomic dissection of the cardiac effects of titin truncations in health and disease. Sci Transl Med. (2015) 7(270):270ra6. doi: 10.1126/scitranslmed.3010134
45. Begay RL, Graw S, Sinagra G, Merlo M, Slavov D, Gowan K, et al. Role of titin missense variants in dilated cardiomyopathy. J Am Heart Assoc. (2015) 4(11):e002645. doi: 10.1161/JAHA.115.002645
46. Hinson JT, Chopra A, Nafissi N, Polacheck WJ, Benson CC, Swist S, et al. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science (New York, NY). (2015) 349(6251):982–6. doi: 10.1126/science.aaa5458
47. Ware JS, Cook SA. Role of titin in cardiomyopathy: from DNA variants to patient stratification. Nat Rev Cardiol. (2018) 15(4):241–52. doi: 10.1038/nrcardio.2017.190
48. Tharp CA, Haywood ME, Sbaizero O, Taylor MRG, Mestroni L. The giant protein titin’s role in cardiomyopathy: genetic, transcriptional, and post-translational modifications of TTN and their contribution to cardiac disease. Front Physiol. (2019) 10:1436. doi: 10.3389/fphys.2019.01436
49. Schafer S, de Marvao A, Adami E, Fiedler LR, Ng B, Khin E, et al. Titin-truncating variants affect heart function in disease cohorts and the general population. Nat Genet. (2017) 49(1):46–53. doi: 10.1038/ng.3719
50. Schisler JC, Grevengoed TJ, Pascual F, Cooper DE, Ellis JM, Paul DS, et al. Cardiac energy dependence on glucose increases metabolites related to glutathione and activates metabolic genes controlled by mechanistic target of rapamycin. J Am Heart Assoc. (2015) 4(2):e001136. doi: 10.1161/JAHA.114.001136
51. Akhtar MM, Lorenzini M, Cicerchia M, Ochoa JP, Hey TM, Sabater Molina M, et al. Clinical phenotypes and prognosis of dilated cardiomyopathy caused by truncating variants in the TTN gene. Circ Heart Fail. (2020) 13(10):e006832. doi: 10.1161/CIRCHEARTFAILURE.119.006832
52. Vissing CR, Rasmussen TB, Dybro AM, Olesen MS, Pedersen LN, Jensen M, et al. Dilated cardiomyopathy caused by truncating titin variants: long-term outcomes, arrhythmias, response to treatment and sex differences. J Med Genet. (2021) 58(12):832–41. doi: 10.1136/jmedgenet-2020-107178
53. McAfee Q, Chen CY, Yang Y, Caporizzo MA, Morley M, Babu A, et al. Truncated titin proteins in dilated cardiomyopathy. Sci Transl Med. (2021) 13(618):eabd7287. doi: 10.1126/scitranslmed.abd7287
54. Gramlich M, Pane LS, Zhou Q, Chen Z, Murgia M, Schötterl S, et al. Antisense-mediated exon skipping: a therapeutic strategy for titin-based dilated cardiomyopathy. EMBO Mol Med. (2015) 7(5):562–76. doi: 10.15252/emmm.201505047
55. Romano R, Ghahremani S, Zimmerman T, Legere N, Thakar K, Ladha FA, et al. Reading frame repair of TTN truncation variants restores titin quantity and functions. Circulation. (2022) 145(3):194–205. doi: 10.1161/CIRCULATIONAHA.120.049997
56. Azibani F, Muchir A, Vignier N, Bonne G, Bertrand AT. Striated muscle laminopathies. Semin Cell Dev Biol. (2014) 29:107–15. doi: 10.1016/j.semcdb.2014.01.001
57. van Rijsingen IA, Arbustini E, Elliott PM, Mogensen J, Hermans-van Ast JF, van der Kooi AJ, et al. Risk factors for malignant ventricular arrhythmias in lamin a/c mutation carriers a European cohort study. J Am Coll Cardiol. (2012) 59(5):493–500. doi: 10.1016/j.jacc.2011.08.078
58. Parks SB, Kushner JD, Nauman D, Burgess D, Ludwigsen S, Peterson A, et al. Lamin A/C mutation analysis in a cohort of 324 unrelated patients with idiopathic or familial dilated cardiomyopathy. Am Heart J. (2008) 156(1):161–9. doi: 10.1016/j.ahj.2008.01.026
59. Wang Y, Dobreva G. Epigenetics in LMNA-related cardiomyopathy. Cells. (2023) 12(5):783. doi: 10.3390/cells12050783
60. Yamada S, Ko T, Ito M, Sassa T, Nomura S, Okuma H, et al. TEAD1 trapping by the Q353R-lamin A/C causes dilated cardiomyopathy. Sci Adv. (2023) 9(15):eade7047. doi: 10.1126/sciadv.ade7047
61. Nishiuchi S, Makiyama T, Aiba T, Nakajima K, Hirose S, Kohjitani H, et al. Gene-based risk stratification for cardiac disorders in LMNA mutation carriers. Circ Cardiovasc Genet. (2017) 10(6):e001603. doi: 10.1161/CIRCGENETICS.116.001603
62. Brayson D, Shanahan CM. Current insights into LMNA cardiomyopathies: existing models and missing LINCs. Nucleus (Austin, Tex). (2017) 8(1):17–33. doi: 10.1080/19491034.2016.1260798
63. Guilluy C, Osborne LD, Van Landeghem L, Sharek L, Superfine R, Garcia-Mata R, et al. Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nat Cell Biol. (2014) 16(4):376–81. doi: 10.1038/ncb2927
64. Stephens AD, Banigan EJ, Adam SA, Goldman RD, Marko JF. Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol Biol Cell. (2017) 28(14):1984–96. doi: 10.1091/mbc.e16-09-0653
65. Chai RJ, Werner H, Li PY, Lee YL, Nyein KT, Solovei I, et al. Disrupting the LINC complex by AAV mediated gene transduction prevents progression of lamin induced cardiomyopathy. Nat Commun. (2021) 12(1):4722. doi: 10.1038/s41467-021-24849-4
66. Quarta G, Syrris P, Ashworth M, Jenkins S, Zuborne Alapi K, Morgan J, et al. Mutations in the lamin A/C gene mimic arrhythmogenic right ventricular cardiomyopathy. Eur Heart J. (2012) 33(9):1128–36. doi: 10.1093/eurheartj/ehr451
67. Chatzifrangkeskou M, Le Dour C, Wu W, Morrow JP, Joseph LC, Beuvin M, et al. ERK1/2 Directly acts on CTGF/CCN2 expression to mediate myocardial fibrosis in cardiomyopathy caused by mutations in the lamin A/C gene. Hum Mol Genet. (2016) 25(11):2220–33. doi: 10.1093/hmg/ddw090
68. Hershberger RE, Jordan E. LMNA-related dilated cardiomyopathy. In: Adam MP, Mirzaa GM, Pagon RA, Wallace SE, Bean LJH, Gripp KW, et al., editors. Genereviews(®). Seattle, WA: University of Washington, Seattle Copyright © 1993-2023, University of Washington, Seattle. GeneReviews is a registered trademark of the University of Washington, Seattle. All rights reserved. (1993). 20301717.
69. Ramos FJ, Chen SC, Garelick MG, Dai DF, Liao CY, Schreiber KH, et al. Rapamycin reverses elevated mTORC1 signaling in lamin A/C-deficient mice, rescues cardiac and skeletal muscle function, and extends survival. Sci Transl Med. (2012) 4(144):144ra03. doi: 10.1126/scitranslmed.3003802
70. Wu W, Muchir A, Shan J, Bonne G, Worman HJ. Mitogen-activated protein kinase inhibitors improve heart function and prevent fibrosis in cardiomyopathy caused by mutation in lamin A/C gene. Circulation. (2011) 123(1):53–61. doi: 10.1161/CIRCULATIONAHA.110.970673
71. Auguste G, Rouhi L, Matkovich SJ, Coarfa C, Robertson MJ, Czernuszewicz G, et al. BET Bromodomain inhibition attenuates cardiac phenotype in myocyte-specific lamin A/C-deficient mice. J Clin Invest. (2020) 130(9):4740–58. doi: 10.1172/JCI135922
72. Yuan ZY, Cheng LT, Wang ZF, Wu YQ. Desmoplakin and clinical manifestations of desmoplakin cardiomyopathy. Chin Med J. (2021) 134(15):1771–9. doi: 10.1097/CM9.0000000000001581
73. Brandão M, Bariani R, Rigato I, Bauce B. Desmoplakin cardiomyopathy: comprehensive review of an increasingly recognized entity. J Clin Med. (2023) 12(7):2660. doi: 10.3390/jcm12072660
74. Gallicano GI, Kouklis P, Bauer C, Yin M, Vasioukhin V, Degenstein L, et al. Desmoplakin is required early in development for assembly of desmosomes and cytoskeletal linkage. J Cell Biol. (1998) 143(7):2009–22. doi: 10.1083/jcb.143.7.2009
75. Pugh TJ, Kelly MA, Gowrisankar S, Hynes E, Seidman MA, Baxter SM, et al. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet Med. (2014) 16(8):601–8. doi: 10.1038/gim.2013.204
76. Karvonen V, Harjama L, Heliö K, Kettunen K, Elomaa O, Koskenvuo JW, et al. A novel desmoplakin mutation causes dilated cardiomyopathy with palmoplantar keratoderma as an early clinical sign. J Eur Acad Dermatol Venereol. (2022) 36(8):1349–58. doi: 10.1111/jdv.18164
77. Gigli M, Merlo M, Graw SL, Barbati G, Rowland TJ, Slavov DB, et al. Genetic risk of arrhythmic phenotypes in patients with dilated cardiomyopathy. J Am Coll Cardiol. (2019) 74(11):1480–90. doi: 10.1016/j.jacc.2019.06.072
78. Smith ED, Lakdawala NK, Papoutsidakis N, Aubert G, Mazzanti A, McCanta AC, et al. Desmoplakin cardiomyopathy, a fibrotic and inflammatory form of cardiomyopathy distinct from typical dilated or arrhythmogenic right ventricular cardiomyopathy. Circulation. (2020) 141(23):1872–84. doi: 10.1161/CIRCULATIONAHA.119.044934
79. Bariani R, Cason M, Rigato I, Cipriani A, Celeghin R, De Gaspari M, et al. Clinical profile and long-term follow-up of a cohort of patients with desmoplakin cardiomyopathy. Heart Rhythm. (2022) 19(8):1315–24. doi: 10.1016/j.hrthm.2022.04.015
80. Chelko SP, Asimaki A, Lowenthal J, Bueno-Beti C, Bedja D, Scalco A, et al. Therapeutic modulation of the immune response in arrhythmogenic cardiomyopathy. Circulation. (2019) 140(18):1491–505. doi: 10.1161/CIRCULATIONAHA.119.040676
81. Bauce B, Basso C, Rampazzo A, Beffagna G, Daliento L, Frigo G, et al. Clinical profile of four families with arrhythmogenic right ventricular cardiomyopathy caused by dominant desmoplakin mutations. Eur Heart J. (2005) 26(16):1666–75. doi: 10.1093/eurheartj/ehi341
82. Gomes G, Seixas MR, Azevedo S, Audi K, Jurberg AD, Mermelstein C, et al. What does desmin do: a bibliometric assessment of the functions of the muscle intermediate filament. Exp Biol Med (Maywood, NJ). (2022) 247(7):538–50. doi: 10.1177/15353702221075035
83. Schröder R, Vrabie A, Goebel HH. Primary desminopathies. J Cell Mol Med. (2007) 11(3):416–26. doi: 10.1111/j.1582-4934.2007.00057.x
84. Eldirany SA, Lomakin IB, Ho M, Bunick CG. Recent insight into intermediate filament structure. Curr Opin Cell Biol. (2021) 68:132–43. doi: 10.1016/j.ceb.2020.10.001
85. Tsikitis M, Galata Z, Mavroidis M, Psarras S, Capetanaki Y. Intermediate filaments in cardiomyopathy. Biophys Rev. (2018) 10(4):1007–31. doi: 10.1007/s12551-018-0443-2
86. Maggi L, Mavroidis M, Psarras S, Capetanaki Y, Lattanzi G. Skeletal and cardiac muscle disorders caused by mutations in genes encoding intermediate filament proteins. Int J Mol Sci. (2021) 22(8):4256. doi: 10.3390/ijms22084256
87. Tesson F, Sylvius N, Pilotto A, Dubosq-Bidot L, Peuchmaurd M, Bouchier C, et al. Epidemiology of desmin and cardiac actin gene mutations in a European population of dilated cardiomyopathy. Eur Heart J. (2000) 21(22):1872–6. doi: 10.1053/euhj.2000.2245
88. Schirmer I, Dieding M, Klauke B, Brodehl A, Gaertner-Rommel A, Walhorn V, et al. A novel desmin (DES) indel mutation causes severe atypical cardiomyopathy in combination with atrioventricular block and skeletal myopathy. Mol Genet Genomic Med. (2018) 6(2):288–93. doi: 10.1002/mgg3.358
89. van Spaendonck-Zwarts KY, van Hessem L, Jongbloed JD, de Walle HE, Capetanaki Y, van der Kooi AJ, et al. Desmin-related myopathy. Clin Genet. (2011) 80(4):354–66. doi: 10.1111/j.1399-0004.2010.01512.x
90. Elsnicova B, Hornikova D, Tibenska V, Kolar D, Tlapakova T, Schmid B, et al. Desmin knock-out cardiomyopathy: a heart on the verge of metabolic crisis. Int J Mol Sci. (2022) 23(19):12020. doi: 10.3390/ijms231912020
91. Diermeier S, Iberl J, Vetter K, Haug M, Pollmann C, Reischl B, et al. Early signs of architectural and biomechanical failure in isolated myofibers and immortalized myoblasts from desmin-mutant knock-in mice. Sci Rep. (2017) 7(1):1391. doi: 10.1038/s41598-017-01485-x
92. Koopman WJ, Willems PH, Smeitink JA. Monogenic mitochondrial disorders. N Engl J Med. (2012) 366(12):1132–41. doi: 10.1056/NEJMra1012478
93. Diokmetzidou A, Soumaka E, Kloukina I, Tsikitis M, Makridakis M, Varela A, et al. Desmin and αB-crystallin interplay in the maintenance of mitochondrial homeostasis and cardiomyocyte survival. J Cell Sci. (2016) 129(20):3705–20. doi: 10.1242/jcs.192203
94. Zheng K, Liu L, Zhang YQ. Recent research on childhood hypertrophic cardiomyopathy caused by MYH7 gene mutations. Zhongguo Dang Dai Er Ke Za Zhi. (2023) 25(4):425–30. doi: 10.7499/j.issn.1008-8830.2211044
95. Gao Y, Peng L, Zhao C. MYH7 in cardiomyopathy and skeletal muscle myopathy. Mol Cell Biochem. (2023) 2023. doi: 10.1007/s11010-023-04735-x
96. de Frutos F, Ochoa JP, Navarro-Peñalver M, Baas A, Bjerre JV, Zorio E, et al. Natural history of MYH7-related dilated cardiomyopathy. J Am Coll Cardiol. (2022) 80(15):1447–61. doi: 10.1016/j.jacc.2022.07.023
97. Khan RS, Pahl E, Dellefave-Castillo L, Rychlik K, Ing A, Yap KL, et al. Genotype and cardiac outcomes in pediatric dilated cardiomyopathy. J Am Heart Assoc. (2022) 11(1):e022854. doi: 10.1161/JAHA.121.022854
98. Zhang S, Wilson J, Madani M, Feld G, Greenberg B. Atrial arrhythmias and extensive left atrial fibrosis as the initial presentation of MYH7 gene mutation. JACC Clin Electrophysiol. (2018) 4(11):1488–90. doi: 10.1016/j.jacep.2018.07.016
99. Petropoulou E, Soltani M, Firoozabadi AD, Namayandeh SM, Crockford J, Maroofian R, et al. Digenic inheritance of mutations in the cardiac troponin (TNNT2) and cardiac beta myosin heavy chain (MYH7) as the cause of severe dilated cardiomyopathy. Eur J Med Genet. (2017) 60(9):485–8. doi: 10.1016/j.ejmg.2017.06.008
100. Abdallah AM, Carlus SJ, Al-Mazroea AH, Alluqmani M, Almohammadi Y, Bhuiyan ZA, et al. Digenic inheritance of LAMA4 and MYH7 mutations in patient with infantile dilated cardiomyopathy. Medicina (Kaunas, Lithuania). (2019) 55(1):17. doi: 10.3390/medicina55010017
101. Selvi Rani D, Nallari P, Dhandapany PS, Rani J, Meraj K, Ganesan M, et al. Coexistence of digenic mutations in both thin (TPM1) and thick (MYH7) filaments of sarcomeric genes leads to severe hypertrophic cardiomyopathy in a south Indian FHCM. DNA Cell Biol. (2015) 34(5):350–9. doi: 10.1089/dna.2014.2650
102. Chang ACY, Chang ACH, Kirillova A, Sasagawa K, Su W, Weber G, et al. Telomere shortening is a hallmark of genetic cardiomyopathies. Proc Natl Acad Sci U S A. (2018) 115(37):9276–81. doi: 10.1073/pnas.1714538115
103. Chang ACY, Blau HM. Short telomeres—a hallmark of heritable cardiomyopathies. Differentiation. (2018) 100:31–6. doi: 10.1016/j.diff.2018.02.001
104. Eguchi A, Gonzalez A, Torres-Bigio SI, Koleckar K, Birnbaum F, Zhang JZ, et al. TRF2 rescues telomere attrition and prolongs cell survival in duchenne muscular dystrophy cardiomyocytes derived from human iPSCs. Proc Natl Acad Sci U S A. (2023) 120(6):e2209967120. doi: 10.1073/pnas.2209967120
105. Brenner CM, Choudhary M, McCormick MG, Cheung D, Landesberg GP, Wang JF, et al. BAG3: nature’s quintessential multi-functional protein functions as a ubiquitous intra-cellular glue. Cells. (2023) 12(6):937. doi: 10.3390/cells12060937
106. Kirk JA, Cheung JY, Feldman AM. Therapeutic targeting of BAG3: considering its complexity in cancer and heart disease. J Clin Invest. (2021) 131(16):e149415. doi: 10.1172/JCI149415
107. Liu L, Sun K, Zhang X, Tang Y, Xu D. Advances in the role and mechanism of BAG3 in dilated cardiomyopathy. Heart Fail Rev. (2021) 26(1):183–94. doi: 10.1007/s10741-019-09899-7
108. Qu HQ, Feldman AM, Hakonarson H. Genetics of BAG3: a paradigm for developing precision therapies for dilated cardiomyopathies. J Am Heart Assoc. (2022) 11(23):e027373. doi: 10.1161/JAHA.122.027373
109. Mizushima W, Sadoshima J. BAG3 plays a central role in proteostasis in the heart. J Clin Invest. (2017) 127(8):2900–3. doi: 10.1172/JCI95839
110. Domínguez F, Cuenca S, Bilińska Z, Toro R, Villard E, Barriales-Villa R, et al. Dilated cardiomyopathy due to BLC2-associated athanogene 3 (BAG3) mutations. J Am Coll Cardiol. (2018) 72(20):2471–81. doi: 10.1016/j.jacc.2018.08.2181
111. Myers VD, Tomar D, Madesh M, Wang J, Song J, Zhang XQ, et al. Haplo-insufficiency of Bcl2-associated athanogene 3 in mice results in progressive left ventricular dysfunction, β-adrenergic insensitivity, and increased apoptosis. J Cell Physiol. (2018) 233(9):6319–26. doi: 10.1002/jcp.26482
112. Fang X, Bogomolovas J, Wu T, Zhang W, Liu C, Veevers J, et al. Loss-of-function mutations in co-chaperone BAG3 destabilize small HSPs and cause cardiomyopathy. J Clin Invest. (2017) 127(8):3189–200. doi: 10.1172/JCI94310
113. Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. (2009) 28(7):889–901. doi: 10.1038/emboj.2009.29
114. Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. (2011) 469(7330):336–42. doi: 10.1038/nature09783
115. Kalayinia S, Arjmand F, Maleki M, Malakootian M, Singh CP. MicroRNAs: roles in cardiovascular development and disease. Cardiovasc Pathol. (2021) 50:107296. doi: 10.1016/j.carpath.2020.107296
116. Zaragoza C, Saura M, Hernández I, Ramirez-Carracedo R, García-García F, Zamorano JL, et al. Differential expression of circulating miRNAs as a novel tool to assess BAG3-associated familial dilated cardiomyopathy. Biosci Rep. (2019) 39(3):BSR20180934. doi: 10.1042/BSR20180934
117. Diofano F, Weinmann K, Schneider I, Thiessen KD, Rottbauer W, Just S. Genetic compensation prevents myopathy and heart failure in an in vivo model of Bag3 deficiency. PLoS Genet. (2020) 16(11):e1009088. doi: 10.1371/journal.pgen.1009088
118. Song S, Shi A, Lian H, Hu S, Nie Y. Filamin C in cardiomyopathy: from physiological roles to DNA variants. Heart Fail Rev. (2022) 27(4):1373–85. doi: 10.1007/s10741-021-10172-z
119. Mao Z, Nakamura F. Structure and function of filamin C in the muscle Z-disc. Int J Mol Sci. (2020) 21(8):2696. doi: 10.3390/ijms21082696
120. Noureddine M, Gehmlich K. Structural and signaling proteins in the Z-disk and their role in cardiomyopathies. Front Physiol. (2023) 14:1143858. doi: 10.3389/fphys.2023.1143858
121. Powers JD, Kirkland NJ, Liu C, Razu SS, Fang X, Engler AJ, et al. Subcellular remodeling in filamin C deficient mouse hearts impairs myocyte tension development during progression of dilated cardiomyopathy. Int J Mol Sci. (2022) 23(2):871. doi: 10.3390/ijms23020871
122. Zhou Y, Chen Z, Zhang L, Zhu M, Tan C, Zhou X, et al. Loss of filamin C is catastrophic for heart function. Circulation. (2020) 141(10):869–71. doi: 10.1161/CIRCULATIONAHA.119.044061
123. Begay RL, Graw SL, Sinagra G, Asimaki A, Rowland TJ, Slavov DB, et al. Filamin C truncation mutations are associated with arrhythmogenic dilated cardiomyopathy and changes in the cell-cell adhesion structures. JACC Clin Electrophysiol. (2018) 4(4):504–14. doi: 10.1016/j.jacep.2017.12.003
124. Ortiz-Genga MF, Cuenca S, Dal Ferro M, Zorio E, Salgado-Aranda R, Climent V, et al. Truncating FLNC mutations are associated with high-risk dilated and arrhythmogenic cardiomyopathies. J Am Coll Cardiol. (2016) 68(22):2440–51. doi: 10.1016/j.jacc.2016.09.927
125. Celeghin R, Cipriani A, Bariani R, Bueno Marinas M, Cason M, Bevilacqua M, et al. Filamin-C variant-associated cardiomyopathy: a pooled analysis of individual patient data to evaluate the clinical profile and risk of sudden cardiac death. Heart Rhythm. (2022) 19(2):235–43. doi: 10.1016/j.hrthm.2021.09.029
126. Chen SN, Lam CK, Wan YW, Gao S, Malak OA, Zhao SR, et al. Activation of PDGFRA signaling contributes to filamin C-related arrhythmogenic cardiomyopathy. Sci Adv. (2022) 8(8):eabk0052. doi: 10.1126/sciadv.abk0052
127. Feyen DAM, Perea-Gil I, Maas RGC, Harakalova M, Gavidia AA, Arthur Ataam J, et al. Unfolded protein response as a compensatory mechanism and potential therapeutic target in PLN R14del cardiomyopathy. Circulation. (2021) 144(5):382–92. doi: 10.1161/CIRCULATIONAHA.120.049844
128. Cuello F, Knaust AE, Saleem U, Loos M, Raabe J, Mosqueira D, et al. Impairment of the ER/mitochondria compartment in human cardiomyocytes with PLN p.Arg14del mutation. EMBO Mol Med. (2021) 13(6):e13074. doi: 10.15252/emmm.202013074
129. MacLennan DH, Kranias EG. Phospholamban: a crucial regulator of cardiac contractility. Nat Rev Mol Cell Biol. (2003) 4(7):566–77. doi: 10.1038/nrm1151
130. Haghighi K, Gregory KN, Kranias EG. Sarcoplasmic reticulum ca-ATPase-phospholamban interactions and dilated cardiomyopathy. Biochem Biophys Res Commun. (2004) 322(4):1214–22. doi: 10.1016/j.bbrc.2004.07.164
131. Hof IE, van der Heijden JF, Kranias EG, Sanoudou D, de Boer RA, van Tintelen JP, et al. Prevalence and cardiac phenotype of patients with a phospholamban mutation. Netherlands Heart J. (2019) 27(2):64–9. doi: 10.1007/s12471-018-1211-4
132. van der Zwaag PA, van Rijsingen IA, Asimaki A, Jongbloed JD, van Veldhuisen DJ, Wiesfeld AC, et al. Phospholamban R14del mutation in patients diagnosed with dilated cardiomyopathy or arrhythmogenic right ventricular cardiomyopathy: evidence supporting the concept of arrhythmogenic cardiomyopathy. Eur J Heart Fail. (2012) 14(11):1199–207. doi: 10.1093/eurjhf/hfs119
133. Medin M, Hermida-Prieto M, Monserrat L, Laredo R, Rodriguez-Rey JC, Fernandez X, et al. Mutational screening of phospholamban gene in hypertrophic and idiopathic dilated cardiomyopathy and functional study of the PLN -42 C>G mutation. Eur J Heart Fail. (2007) 9(1):37–43. doi: 10.1016/j.ejheart.2006.04.007
134. Villard E, Duboscq-Bidot L, Charron P, Benaiche A, Conraads V, Sylvius N, et al. Mutation screening in dilated cardiomyopathy: prominent role of the beta myosin heavy chain gene. Eur Heart J. (2005) 26(8):794–803. doi: 10.1093/eurheartj/ehi193
135. van Rijsingen IA, van der Zwaag PA, Groeneweg JA, Nannenberg EA, Jongbloed JD, Zwinderman AH, et al. Outcome in phospholamban R14del carriers: results of a large multicentre cohort study. Circ Cardiovasc Genet. (2014) 7(4):455–65. doi: 10.1161/CIRCGENETICS.113.000374
136. Deiman FE, Bomer N, van der Meer P, Grote Beverborg N. Review: precision medicine approaches for genetic cardiomyopathy: targeting phospholamban R14del. Curr Heart Fail Rep. (2022) 19(4):170–9. doi: 10.1007/s11897-022-00558-x
137. Doevendans PA, Glijnis PC, Kranias EG. Leducq transatlantic network of excellence to cure phospholamban-induced cardiomyopathy (CURE-PLaN). Circ Res. (2019) 125(7):720–4. doi: 10.1161/CIRCRESAHA.119.315077
138. de Brouwer R, Meems LMG, Verstraelen TE, Mahmoud B, Proost V, Wilde AAM, et al. Sex-specific aspects of phospholamban cardiomyopathy: the importance and prognostic value of low-voltage electrocardiograms. Heart Rhythm. (2022) 19(3):427–34. doi: 10.1016/j.hrthm.2021.11.009
139. Te Rijdt WP, Hoorntje ET, de Brouwer R, Oomen A, Amin A, van der Heijden JF, et al. Rationale and design of the PHOspholamban RElated CArdiomyopathy intervention STudy (i-PHORECAST). Netherlands Heart J. (2022) 30(2):84–95. doi: 10.1007/s12471-021-01584-5
140. Grote Beverborg N, Später D, Knöll R, Hidalgo A, Yeh ST, Elbeck Z, et al. Phospholamban antisense oligonucleotides improve cardiac function in murine cardiomyopathy. Nat Commun. (2021) 12(1):5180. doi: 10.1038/s41467-021-25439-0
141. Dave J, Raad N, Mittal N, Zhang L, Fargnoli A, Oh JG, et al. Gene editing reverses arrhythmia susceptibility in humanized PLN-R14del mice: modelling a European cardiomyopathy with global impact. Cardiovasc Res. (2022) 118(15):3140–50. doi: 10.1093/cvr/cvac021
142. Vafiadaki E, Glijnis PC, Doevendans PA, Kranias EG, Sanoudou D. Phospholamban R14del disease: the past, the present and the future. Front Cardiovasc Med. (2023) 10:1162205. doi: 10.3389/fcvm.2023.1162205
143. Brauch KM, Karst ML, Herron KJ, de Andrade M, Pellikka PA, Rodeheffer RJ, et al. Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J Am Coll Cardiol. (2009) 54(10):930–41. doi: 10.1016/j.jacc.2009.05.038
144. Fochi S, Lorenzi P, Galasso M, Stefani C, Trabetti E, Zipeto D, et al. The emerging role of the RBM20 and PTBP1 ribonucleoproteins in heart development and cardiovascular diseases. Genes (Basel). (2020) 11(4):402. doi: 10.3390/genes11040402
145. Refaat MM, Lubitz SA, Makino S, Islam Z, Frangiskakis JM, Mehdi H, et al. Genetic variation in the alternative splicing regulator RBM20 is associated with dilated cardiomyopathy. Heart Rhythm. (2012) 9(3):390–6. doi: 10.1016/j.hrthm.2011.10.016
146. Watanabe T, Kimura A, Kuroyanagi H. Alternative splicing regulator RBM20 and cardiomyopathy. Front Mol Biosci. (2018) 5:105. doi: 10.3389/fmolb.2018.00105
147. Nishiyama T, Zhang Y, Cui M, Li H, Sanchez-Ortiz E, McAnally JR, et al. Precise genomic editing of pathogenic mutations in RBM20 rescues dilated cardiomyopathy. Sci Transl Med. (2022) 14(672):eade1633. doi: 10.1126/scitranslmed.ade1633
148. LeWinter MM, Granzier HL. Titin is a major human disease gene. Circulation. (2013) 127(8):938–44. doi: 10.1161/CIRCULATIONAHA.112.139717
149. Guo W, Schafer S, Greaser ML, Radke MH, Liss M, Govindarajan T, et al. RBM20, A gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. (2012) 18(5):766–73. doi: 10.1038/nm.2693
150. Zhang Y, Wang C, Sun M, Jin Y, Braz CU, Khatib H, et al. RBM20 Phosphorylation and its role in nucleocytoplasmic transport and cardiac pathogenesis. FASEB J. (2022) 36(5):e22302. doi: 10.1096/fj.202101811RR
151. Ihara K, Sasano T, Hiraoka Y, Togo-Ohno M, Soejima Y, Sawabe M, et al. A missense mutation in the RSRSP stretch of Rbm20 causes dilated cardiomyopathy and atrial fibrillation in mice. Sci Rep. (2020) 10(1):17894. doi: 10.1038/s41598-020-74800-8
152. Briganti F, Sun H, Wei W, Wu J, Zhu C, Liss M, et al. iPSC modeling of RBM20-deficient DCM identifies upregulation of RBM20 as a therapeutic strategy. Cell Rep. (2020) 32(10):108117. doi: 10.1016/j.celrep.2020.108117
153. Remme CA. SCN5A Channelopathy: arrhythmia, cardiomyopathy, epilepsy and beyond. Philos Trans R Soc Lond Ser B Biol Sci. (2023) 378(1879):20220164. doi: 10.1098/rstb.2022.0164
154. Zaklyazminskaya E, Dzemeshkevich S. The role of mutations in the SCN5A gene in cardiomyopathies. Biochim Biophys Acta. (2016) 1863(7 Pt B):1799–805. doi: 10.1016/j.bbamcr.2016.02.014
155. Zegkos T, Panagiotidis T, Parcharidou D, Efthimiadis G. Emerging concepts in arrhythmogenic dilated cardiomyopathy. Heart Fail Rev. (2021) 26(5):1219–29. doi: 10.1007/s10741-020-09933-z
156. Li W, Yin L, Shen C, Hu K, Ge J, Sun A. SCN5A variants: association with cardiac disorders. Front Physiol. (2018) 9:1372. doi: 10.3389/fphys.2018.01372
157. Moreau A, Gosselin-Badaroudine P, Delemotte L, Klein ML, Chahine M. Gating pore currents are defects in common with two Nav1.5 mutations in patients with mixed arrhythmias and dilated cardiomyopathy. J Gen Physiol. (2015) 145(2):93–106. doi: 10.1085/jgp.201411304
158. Poller W, Escher F, Haas J, Heidecker B, Schultheiss HP, Attanasio P, et al. Missense variant E1295K of sodium channel SCN5A associated with recurrent ventricular fibrillation and myocardial inflammation. JACC Case Rep. (2022) 4(5):280–6. doi: 10.1016/j.jaccas.2022.01.016
159. Wilde AAM, Amin AS. Clinical Spectrum of SCN5A mutations: long QT syndrome, brugada syndrome, and cardiomyopathy. JACC Clin Electrophysiol. (2018) 4(5):569–79. doi: 10.1016/j.jacep.2018.03.006
160. McNair WP, Sinagra G, Taylor MR, Di Lenarda A, Ferguson DA, Salcedo EE, et al. SCN5A mutations associate with arrhythmic dilated cardiomyopathy and commonly localize to the voltage-sensing mechanism. J Am Coll Cardiol. (2011) 57(21):2160–8. doi: 10.1016/j.jacc.2010.09.084
161. Peters S, Thompson BA, Perrin M, James P, Zentner D, Kalman JM, et al. Arrhythmic phenotypes are a defining feature of dilated cardiomyopathy-associated SCN5A variants: a systematic review. Circ Genomic Precis Med. (2022) 15(1):e003432. doi: 10.1161/CIRCGEN.121.003432
162. Zhou A, Xie A, Kim TY, Liu H, Shi G, Kang GJ, et al. HuR-mediated SCN5A messenger RNA stability reduces arrhythmic risk in heart failure. Heart Rhythm. (2018) 15(7):1072–80. doi: 10.1016/j.hrthm.2018.02.018
163. Li MX, Hwang PM. Structure and function of cardiac troponin C (TNNC1): implications for heart failure, cardiomyopathies, and troponin modulating drugs. Gene. (2015) 571(2):153–66. doi: 10.1016/j.gene.2015.07.074
164. Keyt LK, Duran JM, Bui QM, Chen C, Miyamoto MI, Silva Enciso J, et al. Thin filament cardiomyopathies: a review of genetics, disease mechanisms, and emerging therapeutics. Front Cardiovasc Med. (2022) 9:972301. doi: 10.3389/fcvm.2022.972301
165. Kalyva A, Parthenakis FI, Marketou ME, Kontaraki JE, Vardas PE. Biochemical characterisation of troponin C mutations causing hypertrophic and dilated cardiomyopathies. J Muscle Res Cell Motil. (2014) 35(2):161–78. doi: 10.1007/s10974-014-9382-0
166. Mogensen J, Murphy RT, Shaw T, Bahl A, Redwood C, Watkins H, et al. Severe disease expression of cardiac troponin C and T mutations in patients with idiopathic dilated cardiomyopathy. J Am Coll Cardiol. (2004) 44(10):2033–40. doi: 10.1016/j.jacc.2004.08.027
167. Hershberger RE, Norton N, Morales A, Li D, Siegfried JD, Gonzalez-Quintana J. Coding sequence rare variants identified in MYBPC3, MYH6, TPM1, TNNC1, and TNNI3 from 312 patients with familial or idiopathic dilated cardiomyopathy. Circ Cardiovasc Genet. (2010) 3(2):155–61. doi: 10.1161/CIRCGENETICS.109.912345
168. Hershberger RE, Parks SB, Kushner JD, Li D, Ludwigsen S, Jakobs P, et al. Coding sequence mutations identified in MYH7, TNNT2, SCN5A, CSRP3, LBD3, and TCAP from 313 patients with familial or idiopathic dilated cardiomyopathy. Clin Transl Sci. (2008) 1(1):21–6. doi: 10.1111/j.1752-8062.2008.00017.x
169. Tadros HJ, Life CS, Garcia G, Pirozzi E, Jones EG, Datta S, et al. Meta-analysis of cardiomyopathy-associated variants in troponin genes identifies loci and intragenic hot spots that are associated with worse clinical outcomes. J Mol Cell Cardiol. (2020) 142:118–25. doi: 10.1016/j.yjmcc.2020.04.005
170. Alves ML, Warren CM, Simon JN, Gaffin RD, Montminy EM, Wieczorek DF, et al. Early sensitization of myofilaments to Ca2+ prevents genetically linked dilated cardiomyopathy in mice. Cardiovasc Res. (2017) 113(8):915–25. doi: 10.1093/cvr/cvx068
Keywords: dilated cardiomyopathy, DCM, gene mutations, molecular mechanism, therapy, targeted therapy, genetic testing
Citation: Wang S, Zhang Z, He J, Liu J, Guo X, Chu H, Xu H and Wang Y (2023) Comprehensive review on gene mutations contributing to dilated cardiomyopathy. Front. Cardiovasc. Med. 10:1296389. doi: 10.3389/fcvm.2023.1296389
Received: 18 September 2023; Accepted: 17 November 2023;
Published: 1 December 2023.
Edited by:
Lukas J. Motloch, Paracelsus Medical University, AustriaReviewed by:
Ralph Knöll, AstraZeneca, SwedenNaufal Zagidullin, Bashkir State Medical University, Russia
© 2023 Wang, Zhang, He, Liu, Guo, Chu, Xu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yushi Wang eXVzaGlAamx1LmVkdS5jbg==
 Shipeng Wang
Shipeng Wang Zhiyu Zhang
Zhiyu Zhang Jiahuan He
Jiahuan He Junqian Liu
Junqian Liu Xia Guo
Xia Guo Haoxuan Chu
Haoxuan Chu Hanchi Xu
Hanchi Xu Yushi Wang
Yushi Wang