- 1Centre for Cardiovascular Science, The Queen's Medical Research Institute, University of Edinburgh, Edinburgh, United Kingdom
- 2Department of Renal Medicine, Royal Infirmary of Edinburgh, Edinburgh, United Kingdom
RNA-based therapies are currently used for immunisation against infections and to treat metabolic diseases. They can modulate gene expression in immune cells and hepatocytes, but their use in other cell types has been limited by an inability to selectively target specific tissues. Potential solutions to this targeting problem involve packaging therapeutic RNA molecules into delivery vehicles that are preferentially delivered to cells of interest. In this review, we consider why the kidney is a desirable target for RNA-based therapies in cardiovascular disease and discuss how such therapy could be delivered. Because the kidney plays a central role in maintaining cardiovascular homeostasis, many extant drugs used for preventing cardiovascular disease act predominantly on renal tubular cells. Moreover, kidney disease is a major independent risk factor for cardiovascular disease and a global health problem. Chronic kidney disease is projected to become the fifth leading cause of death by 2040, with around half of affected individuals dying from cardiovascular disease. The most promising strategies for delivering therapeutic RNA selectively to kidney cells make use of synthetic polymers and engineered extracellular vesicles to deliver an RNA cargo. Future research should focus on establishing the safety of these novel delivery platforms in humans, on developing palatable routes of administration and on prioritising the gene targets that are likely to have the biggest impact in cardiovascular disease.
Introduction
RNA-based therapies are emerging as an effective and versatile class of medications in a range of diseases. So far, they have been largely limited to small interfering RNAs (siRNAs) that mediate gene silencing in hepatocytes and mRNA vaccines. Their full potential will be unlocked when we learn how to target them effectively to other cell types. In this review, we consider recent progress towards developing RNA-based therapies that can selectively target kidney cells and in particular renal tubular epithelial cells (rTECs). Given the central role of the renal tubules in maintaining cardiovascular homeostasis, such novel therapies could have the greatest impact in treating cardiovascular disease (CVD).
We begin by briefly introducing the concepts of RNA-based therapy and reviewing the causative relationship between kidney disease and CVD. We then discuss attempts to deliver RNA-based therapies selectively to rTECs in animal models and humans. We conclude by positing the potential clinical impact that these novel therapeutics could have in cardiovascular disease.
The advantages of RNA-based drugs
How RNA-based drugs work
There are several different classes of RNA therapeutic, including mRNAs, siRNAs, RNA aptamers and CRISPR guide RNAs (1). In principle, any class of coding or non-coding RNA could form the basis of a drug (2), as could novel engineered RNAs such as self-amplifying RNAs (3). However, three strategies are currently most advanced in terms of their clinical development.
In the first, siRNA, shRNA (short hairpin RNA) or anti-sense oligonucleotides (ASOs) silence the expression of a pathogenic mRNA through RNA interference (RNAi). These molecules directly bind their cognate mRNA and inhibit protein synthesis. This process is the mammalian correlate of a phenomenon that was first characterised in plants and C elegans (4). In the second, ASOs are used to inhibit microRNAs (miRNAs): the endogenous negative regulators of mRNA expression. Such “antagomirs” thus de-repress expression of beneficial mRNAs. In the third major class, mRNA molecules increase the expression of a gene of interest (5). This can be a successful strategy for immunisation, as the Moderna Covid-19 vaccine powerfully demonstrated (6). It could also restore expression of deficient protein or mediate gene editing in a range of diseases.
Why RNA-based drugs offer advantages over conventional small molecule drugs
Our current approach to treating kidney and cardiovascular disease relies almost entirely on conventional small molecule drugs. These drugs have been developed over decades, often more by accident than design. They tend to have rapid metabolism—and so require daily or twice-daily dosing—and to target receptors that have been chosen predominantly because they are easily “druggable”, rather than because they are necessarily the most important points in a disease pathway. Drug development is slow, in part because of the relatively unpredictable safety profile of small molecules.
RNA-based therapies could potentially solve all of these problems (7, 8). RNA-based therapies can be designed to target, in principle, any gene of interest. Defined therapeutic classes, such as siRNA, have standard pipelines for development and manufacture, allowing for rapid, relatively inexpensive drug development. They also have predictable pharmacokinetic properties. Fortuitously, liver-targeting siRNAs appear to induce long-lasting therapeutic gene silencing (9, 10), so that dosing might be required every few months, potentially improving drug adherence.
However, for the successful clinical application of RNA-based therapies in kidney disease, we must first learn how to direct these drugs selectively to kidney cells. After intravenous injection, naked RNA is widely distributed amongst different body tissues (11). There are also additional challenges: ensuring biological safety, determining the most effective route of administration and overcoming off-target effects.
The kidney is a therapeutic target in cardiovascular disease
The kidneys maintain cardiovascular homeostasis
The kidneys maintain cardiovascular homeostasis by regulating the circulating volume and blood pressure (12). Therefore, many extant drugs used for preventing or treating cardiovascular disease target the kidney and specifically the most abundant kidney cell, the renal tubular epithelial cell (rTEC). Renin-angiotensin system inhibitors, sodium-glucose co-transporter inhibitors (SGLT2i), diuretics and mineralocorticoid inhibitors, which have all been shown to reduce cardiovascular disease in various clinical contexts, act predominantly by altering kidney solute transport (13). Therefore, the concept of targeting the kidney to treat or prevent cardiovascular disease is well-established.
Kidney disease causes cardiovascular disease
The other rationale for targeting the kidney to treat cardiovascular disease, is that chronic kidney disease (CKD) is a leading risk factor for cardiovascular disease. Therefore, treating kidney disease per se will reduce cardiovascular disease.
CKD is the eighth leading cause of death worldwide (14), with just over 10% of the adult population affected to some degree (13, 15). It is associated with excess morbidity and mortality, an association that is not fully explained by confounding from co-morbid cardiovascular disease (CVD), diabetes and hypertension (16). The leading cause of death in CKD is cardiovascular disease (CVD), accounting for 40%–50% of deaths (17), compared to ∼33% of deaths in the general population (18). Both GFR and albuminuria are independent risk factors for mortality and myocardial infarction (19). The mechanisms whereby loss of glomerular filtration (GFR) or albuminuria cause cardiovascular disease are still relatively poorly understood, but likely involve volume expansion, increases in cardiac pre-load and after-load, perturbed calcium-phosphate homeostasis and increased exposure to circulating “uraemic” metabolites (20).
How could RNA therapies treat kidney and cardiovascular disease?
RNA-based therapies are already being used in the clinic to treat kidney disease, but not by directly targeting kidney cells. For example, Lumasiran is an siRNA that targets glycolate oxidase in hepatocytes that is effective at treating primary hyperoxaluria (21). Similarly, RNA-based therapies for cardiovascular disease target gene expression in the liver: angiotensinogen for hypertension, or PCSK9 or apolipoprotein B-100 for hypercholesterolaemia (22–24). Underscoring the tight association between kidney and cardiovascular health, liver-targeting angiotensinogen siRNA protected against kidney damage in a rat subtotal nephrectomy model of CKD (25).
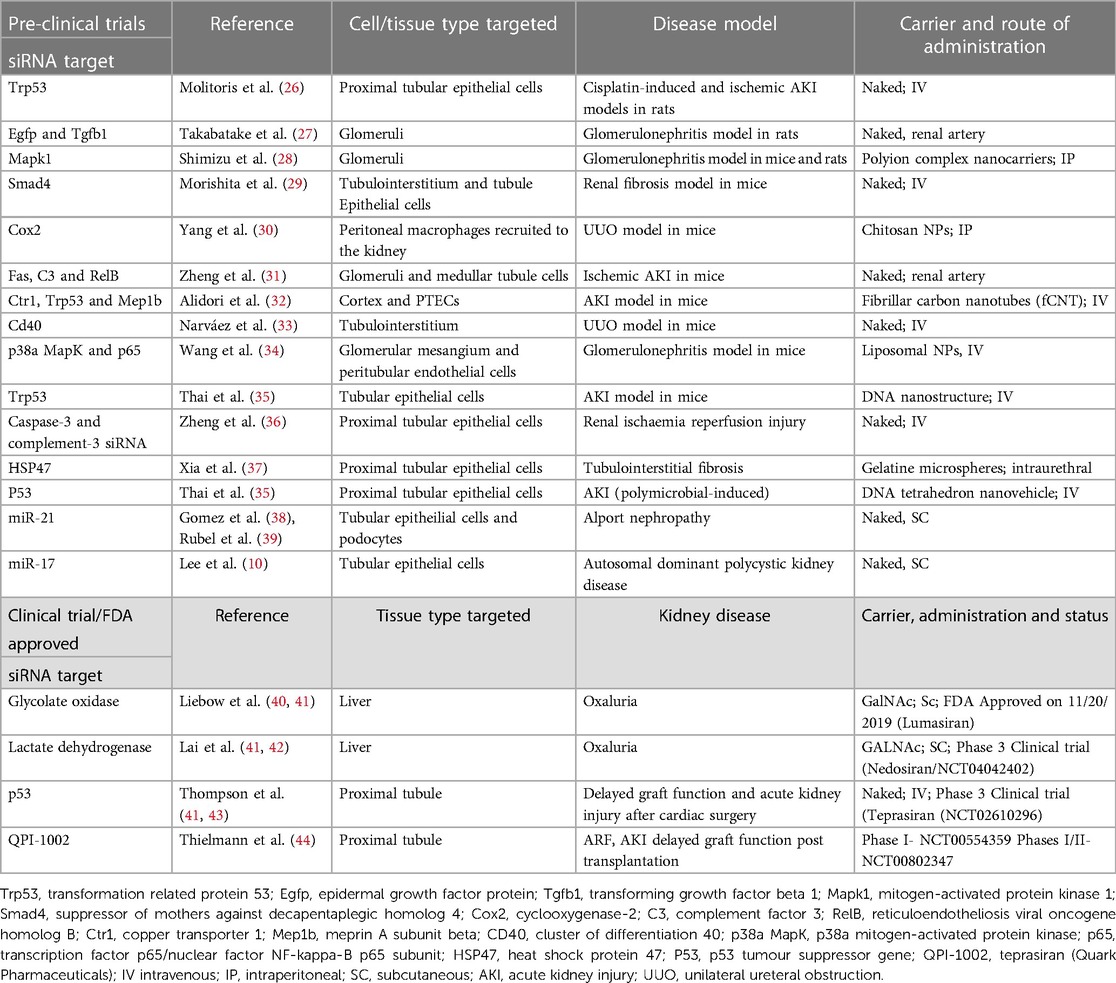
There are some other metabolic diseases in which targeting gene expression in the liver will improve kidney outcomes, but for the vast majority of kidney diseases, it will be necessary to develop therapies that can directly regulate gene expression in kidney cells. The capacity for treating cardiovascular disease will be hugely augmented if we can develop drugs that target the kidneys, given their central role in cardiovascular homeostasis. Therefore, potential strategies for kidney-specific RNA delivery are being tested extensively in pre-clinical models. These are summarised in Table 1 and we chose some interesting examples to discuss in the text below.
How can we target kidney cells with siRNA?
The challenge
Naked RNA is a negatively-charged macromolecule that does not easily cross the hydrophobic, negatively-charged cell membrane. In the absence of modifications, naked RNA is subject to nuclease degradation and clearance by the kidneys and reticuloendothelial system (45, 46). The challenge is to learn how we can modify RNA molecules, or package them within delivery vehicles, so as to enhance delivery to kidney cells. An ideal delivery vehicle would selectively deliver its RNA cargo to rTECs and preserve the functional effects of the RNA in recipient cells (e.g., gene silencing). It should be able to deliver RNA across the cell membrane and not into endosomal degradation pathways. It should also be non-immunogenic, non-toxic, biologically inert, cheap to manufacture and delivered through a patient-friendly route (e.g., subcutaneous injection) (47).
Broadly, RNA can be delivered as “naked” nucleic acid or in delivery systems that are classified as viral or non-viral (Figure 1).
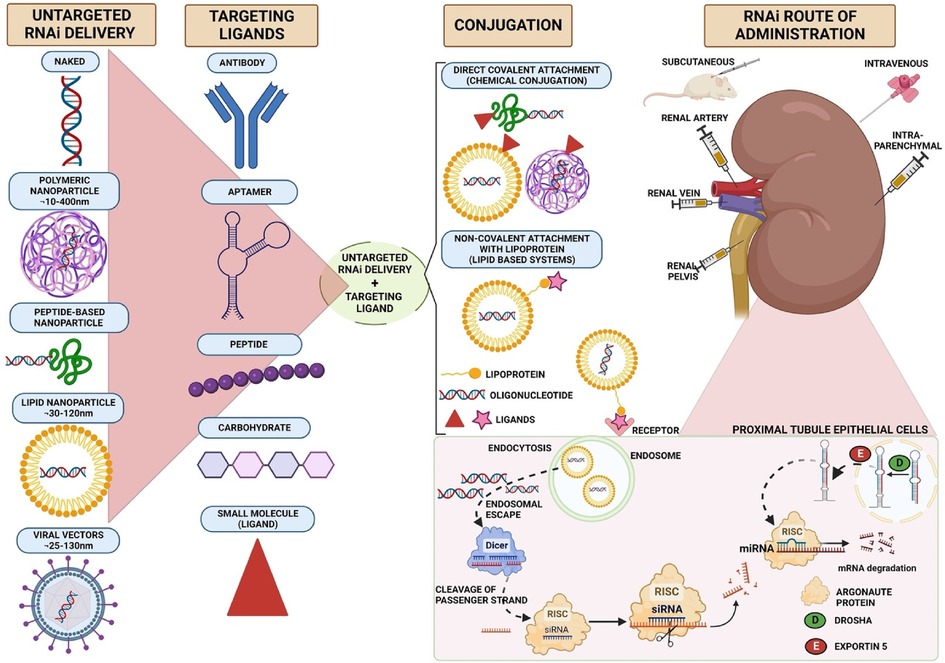
Figure 1. Strategies for selective delivery of siRNA to kidney cells. siRNA can be packaged into lipid-based, polymer-based, viral-based systems or extracellular vesicles. Viral vectors or nanoparticles conjugated to different targeting ligands, such as antibodies, aptamers, peptides or small molecules, form different carrier systems that are preferentially internalised by kidney cells. These targeting delivery vehicles bind to their cognate receptor on kidney cells and are endocytosed. Exogenous siRNA enters the cytosol by endosomal escape. siRNA is then cleaved by dicer enzyme into a passenger strand, which is recycled, and the guide strand loaded onto the RISC complex. siRNA degrades mRNA strand by full complementarity and inhibits protein synthesis. Diagram created in Biorender.
Naked RNA
Naked nucleic acids, injected systemically, are excreted in the urine and accumulate within the kidney, particularly in renal tubular cells (48–51). This kidney delivery is not selective; naked RNA is also delivered to liver, spleen, lung, heart and adipose tissue (11, 48, 49, 52, 53). Nor is naked RNA very stable: the systemic half-life of unmodified siRNA is measured in seconds to minutes (11, 53).
The stability and biodistribution of siRNA can be altered by chemical modification of the RNA backbone. Phosphorothioate siRNA accumulates preferentially in the kidneys, relative to other organs (11). Phosphorothioate modification enhances uptake from the peritubular capillaries in isolated perfused rat kidneys (54). There is also uptake from the urinary space. This might, in part, be mediated through endosomal receptors in proximal rTECs; in monkeys, 2′-O-methyl phosphorothioate oligonucleotides appear to compete with low-molecular proteins for uptake into rTECs (55).
In contexts where rapid clearance and off-target RNA delivery is not problematic, naked RNA could be used to therapeutic effect. Naked siRNA targeting the tumour suppressor gene p53 has been tested in pre-clinical and clinical trials of acute kidney injury. Intravenous administration gave effective delivery to rTECs and prevented kidney injury in rat models of ischemia-reperfusion injury and cisplatin toxicity (26). This siRNA is now in phase three clinical trials for the treatment of delayed graft function after transplantation (NCT02610296), and acute kidney injury after cardiac surgery (NCT03510897) (43).
Some oligonucleotides appear to be selectively delivered to kidney cells when delivered in their naked form. For example, an ASO directed against miRNA-17 performed well in pre-clinical trials of polycystic kidney disease (ADPKD) (10). Intriguingly, this oligonucleotide was preferentially delivered to the kidney as opposed to the liver, and specifically delivered to renal tubular epithelial cells. The mechanism directing this specific delivery is unknown, although the authors of that study speculate that it may be related to the short length of the ASO (9-nucleotides). This is now being tested in a phase I human trial (NCT04536688). Similarly, the miR-21 antagomir, Lademirsen, accumulates in kidney cells and was effective in mouse models of Alport nephropathy (38, 39). However, a phase 2 clinical trial was terminated after a futility analysis (NCT02855268), highlighting the need for careful selection of candidate oligonucleotides for clinical testing and appropriate trial design—as for any other drug class.
Viral systems
Viral vectors are highly effective at delivering genetic material to target cells (56). Viral vectors can be chosen based on an inherent tropism for a target cell type or tissue or, engineered to enhance this tropism. For example, delivery of shRNA to kidney cells has been achieved using AAV2 in rats (57). Viral vectors may be inactivated by removing the viral genome to generate a non-infectious delivery vector, circumventing the safety concerns of live virus. One such envelope vector, derived from the haemagglutinating virus of Japan (HVJ) delivered cDNA for a mitochondrial membrane protein to glomerular and renal tubular cells in diabetic mice (58).
Whilst the risks of cellular toxicity and immune reactions are minimised with third generation adenoviral vectors (59), there is a concerted effort to develop synthetic carriers for RNA such as nanoparticles or extracellular vesicles. These could eliminate the safety concerns of viral vectors and offer advantages in terms of cheap, scalable manufacturing and flexibility in design.
Lipid-based nanoparticles
Lipid-based nanoparticles (LNPs) are the most established non-viral RNA delivery system (60–63). These are typically <100 nm in diameter, can be engineered to traverse the phospholipid cell membrane and have very little toxicity and immunogenicity (63).
One issue that has faced investigators developing LNPs to target organs other than the kidney, is that renal elimination is the major route of LNP clearance. Therefore LNPs are typically modified, e.g., by including PEG-lipids, to minimise renal clearance (63). This might of course be a counterproductive strategy in the development of any kidney-targeting therapy. Indeed, there are very few examples of LNPs that have been developed to deliver RNA to kidney cells, and certainly to rTECs. As proof of principle that this could be possible, one group were able to effect gene silencing in mesangial and glomerular endothelial cells in a mouse model of IgA nephropathy, using polyethyleneimine-containing, octa-arginine-coated liposomes (34).
Polymer-based nanoparticles
Synthetic polymers have been used to stabilise and deliver RNA (64). Negatively charged nucleic acids coupled with positively charged polymers to form polyplexes (60). Polymers that have been employed include polyethylenimine (PEI), poly-(ethylene-glycol)-poly-(L-lysine) (PEG-PLL), poly (lactide-co-glycolide) (PLGA), poly-amidoamine dendrimers (PAMAMs) and the fungal polysaccharide chitosan (47, 65).
Many polymer particles are sufficiently small (<20 nm) that they ought to cross the glomerular filtration barrier. In mice, a PEG-PLL carrier significantly enhanced delivery of fluorescently-tagged siRNA to kidneys in mice, compared to a naked siRNA control (28). A MAPK-targeting siRNA delivered through this system mediated gene silencing in glomerular cells and exerted a therapeutic effect in a glomerulonephritis model.
Chitosan particles showed promise as a kidney-targeting strategy in an early murine biodistribution study, with siRNAs accumulating in the kidney, 24 h after intravenous administration (11). The likely mechanism of this effect was subsequently demonstrated using an elegant transgenic model. AQP1-targeting siRNA delivery was restricted to megalin-expressing rTECs in a mosaic megalin knockout mouse, providing compelling evidence that these nanoparticles cross the glomerular filtration barrier and are then internalised by the multi-ligand endocytic receptor megalin (66).
Branched, symmetrical “dendrimers” are a polymer subclass with particularly attractive properties with respect to RNA delivery (67). They have multiple surface groups to which targeting ligands could be attached. For example, dendrimers decorated with folic acid or the small cyclo peptide cRGDfC have been used to deliver siRNA selective to proximal tubular cells (expressing the folic acid receptor) or podocytes (expressing αvβ3 integrin) respectively in murine kidney disease models (68, 69).
Recent developments in kidney-targeting RNA delivery vehicles have involved nanoparticles with sophisticated molecular structures. In mice, modified carbon nanotubules have been used to deliver siRNA into rTECs: a strategy that induced therapeutic effects when silencing p53 and the metalloprotease Meprin-1β in cisplatin-induced AKI (32). The authors of that study speculate that carbon nanotubes are able to traverse the glomerular filtration barrier by virtue of being long and thin, whereas more globular nanoparticles tend to accumulate in the liver. Nanosized tetrahedra, constructed from DNA (10 bp per side), were able to deliver siRNA specifically into rTECs and again conferred protection from cisplatin injury via p53 silencing (35). Although these strategies clearly show great promise, as selective delivery relies on the physiochemical properties of the delivery vehicle—rather than recognition of a specific ligand by a cell-surface receptor—they may be limited in their ability to target specific rTEC subtypes.
Extracellular vesicles
Extracellular vesicles are released by all cells into the extracellular space. They are either derived from the plasma membrane or the endosomal pathway (“exosomes”) and carry with them a subset of cellular macromolecules, including RNA (47, 70, 71). Their cellular origin endows them with inherently low toxicity and immunogenicity. They may also offer a natural solution to the targeting problem as they are preferentially internalised by cells of the same type from which they were derived (72).
Furthermore, EVs can be readily engineered to express surface receptors to enhance delivery to specific target cells. For example, EVs engineered to express the rabies virus glycoprotein (RGV) peptide gave enhanced delivery of an siRNA cargo to kidney cells (73, 74), because kidney cells express the RGV ligand, the nicotinic acetyl choline receptor (75). Similarly, EV delivery can be directed by modulating surface immunoglobulin (76) or integrin expression (77).
One elegant demonstration of the power and flexibility of this approach was provided by an experiment designed to deliver siRNA specifically to diseased rTECs. When erythrocyte-derived vesicles were engineered to express a KIM-1 ligand, they delivered cargo siRNA preferentially to injured rTECs in murine ischaemia-reperfusion and ureteral obstruction models (78). Delivery of siRNAs targeting p65 and Snail ameliorated tubulointerstitial fibrosis. The ability to target a defined subset of tubular epithelial cells (or indeed other kidney cell types) would open up a range of subtle therapeutic options with minimal off-target side-effects. For example, depletion of senescent rTECs has been shown to improve recovery from ischaemia-reperfusion injury in a mouse model (79); could a selective senolytic therapy be achieved by delivering pro-apoptotic RNAs selective to senescent rTECs?
As RNA carriers can be engineered to express surface ligands that direct them to kidney cells, the obvious question is: which ligands will deliver these vehicles most effectively to kidney target cells (e.g., rTECs)? A variety of ligand classes have been tested: peptides, antibodies, aptamers, carbohydrates, and small molecules (Figure 1). Ligands for the major proximal rTEC receptors megalin, cubulin and transferrin are obvious candidates (and we have already discussed some above). Antibodies or antibody-like molecules have been used to target kidney endothelial cells (anti-VCAM1) and proximal tubular cells (anti-CD11b and anti-CD163); however, their use may be limited by their large size which leads to deposition of immune complexes on the glomerular basement membrane and therefore a risk of glomerulonephritis (60, 80–85).
Administration routes for kidney-targeting RNA therapies
Nanoparticles delivered into the systemic circulation face significant hurdles to reaching the kidney. They may be degraded by the reticuloendothelial system or—depending on their chemical composition—be preferentially delivered to the liver. Furthermore, there is uncertainty as to the extent to which nanoparticles can cross an intact glomerular filtration barrier. Some provocative studies suggest that fluorescently-labelled extracellular vesicles can traverse this barrier in healthy mice and humans, even though their size and charge profile ought to be prohibitive (86, 87).
In pre-clinical models, direct administration into the renal artery, renal vein, ureter or kidney parenchyma have been used to deliver RNA to the kidney, but the translational potential of these routes is obviously limited (37, 60). One intriguing research question is whether physical methods could be used to enhance selective delivery of a systemically-administered RNA drug. It may be possible to modify delivery of RNA vehicles using ultrasound, electromagnetic stimuli or light (60, 88). For example, after systemic delivery of the drug delivery vehicle, focussed ultrasound beams can direct the vehicle to specific cell types and, via acoustic cavitation, induce localised cargo release. This approach was successful for enhancing kidney delivery of a DNA plasmid in mice (89). Alternatively, electroporation of the kidney also enhanced delivery of siRNA and shRNA in mice, after intra-arterial administration (27, 90).
Conclusions and perspectives
RNA-based therapies offer a powerful opportunity to target new pathways in cardiovascular disease. Existing RNA-based therapies largely target gene expression in hepatocytes and are therefore limited in their ability to treat cardiovascular disease. The kidney is an attractive target in cardiovascular disease because the kidney plays a central role in maintaining cardiovascular homeostasis and because CKD is a leading risk factor for cardiovascular disease.
Various approaches to delivering RNA selectively to kidney cells are being tested in pre-clinical models. Polymer and vesicle-based delivery systems show promise, particularly in their ability to deliver siRNA selectively to rTECs, without inciting toxicity or immune activation. Extracellular vesicles can be engineered to express surface ligands that interact with receptors on specific kidney cells, permitting highly selective delivery of RNA to target cells. Several lines of evidence suggest that megalin (and perhaps also cubulin) mediates uptake of exogenous RNA from the tubular space, whether the RNA is delivered as naked nucleic acid or complexed to polymeric delivery vehicles. Delineating the precise molecular details of this pathway and its relative contribution to total kidney RNA uptake will be important areas of study.
Future research should also focus on establishing the safety of these novel delivery platforms in humans and on developing palatable routes of administration. In particular, strategies that induce long-lasting gene silencing could offer advances in drug adherence, which would help to reduce the prevalence of cardiovascular disease. Therefore, the potential targets of novel RNA-based therapies include established targets of conventional small-molecule drugs (e.g., renin-angiotensin and SGLT2 inhibitors) as well as a theoretically unlimited array of novel targets.
Author contributions
TP wrote the initial draft of this article and RH wrote the final version. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sullenger B, Smita N. From the RNA world to the clinic. Science. (2016) 352(6292):1417–20. doi: 10.1126/science.aad8709
2. Pastor F, Berraondo P, Etxeberria I, Frederick J, Sahin U, Gilboa E, et al. An RNA toolbox for cancer immunotherapy. Nat Rev Drug Discov. (2018) 17(10):751–67. doi: 10.1038/nrd.2018.132
3. Bloom K, van den BF, Arbuthnot P. Self-amplifying RNA vaccines for infectious diseases. Gene Ther. (2021) 28(3–4):117–29. doi: 10.1038/s41434-020-00204-y
4. Fire A, Xu S, Montgomery M, Kostas S, Driver S, Mello C. Potent and specific genetic interference by double-stranded RNA in caenorhabditis elegans. Nature. (1998) 391(6669):806–11. doi: 10.1038/35888
5. Maruggi G, Zhang C, Li J, Ulmer J, Yu D. mRNA as a transformative technology for vaccine development to control infectious diseases. Mol Ther. (2019) 27(4):757–72. doi: 10.1016/j.ymthe.2019.01.020
6. Jackson L, Anderson E, Rouphael N, Roberts P, Makhene M, Coler R, et al. An mRNA vaccine against SARS-CoV-2—preliminary report. N Engl J Med. (2020) 383(20):1920–31. doi: 10.1056/NEJMoa2022483
7. Damase T, Sukhovershin R, Boada C, Taraballi F, Pettigrew R, Cooke J. The limitless future of RNA therapeutics. Front Bioeng Biotechnol. (2021) 9:628137. doi: 10.3389/fbioe.2021.628137
8. Dowdy S, Levy M. RNA therapeutics (almost) comes of age: targeting, delivery and endosomal escape. Nucleic Acid Ther. (2018) 28(3):107–8. doi: 10.1089/nat.2018.29001.dow
9. Uijl E, Mirabito Colafella K, Sun Y, Ren L, van Veghel R, Garrelds I, et al. Strong and sustained antihypertensive effect of small interfering RNA targeting liver angiotensinogen. Hypertension. (2019) 73(6):1249–57. doi: 10.1161/HYPERTENSIONAHA.119.12703
10. Lee EC, Valencia T, Allerson C, Schairer A, Flaten A, Yheskel M, et al. Discovery and preclinical evaluation of anti-miR-17 oligonucleotide RGLS4326 for the treatment of polycystic kidney disease. Nat Commun. (2019) 10(1):4148. doi: 10.1038/s41467-019-11918-y
11. Gao S, Dagnaes-Hansen F, Nielsen E, Wengel J, Besenbacher F, Howard K, et al. The effect of chemical modification and nanoparticle formulation on stability and biodistribution of siRNA in mice. Mol Ther. (2009) 17(7):1225–33. doi: 10.1038/mt.2009.91
12. Guyton A. Blood pressure control–special role of the kidneys and body fluids. Science. (1991) 252(5014):1813–6. doi: 10.1126/science.2063193
13. Shabaka A, Cases-Corona C, Fernandez-Juarez G. Therapeutic insights in chronic kidney disease progression. Front Med (Lausanne). (2021) 8:645187. doi: 10.3389/fmed.2021.645187
14. Burden of kidney diseases: level by country USA: PAHO/WHO. (2023). Available at: https://www.paho.org/en/enlace/burden-kidney-diseases.
15. Kalantar-Zadeh K, Jafar T, Nitsch D, Neuen B, Perkovic V. Chronic kidney disease. Lancet. (2021) 398(10302):786–802. doi: 10.1016/S0140-6736(21)00519-5
16. Sarnak MJ, Levey AS, Schoolwerth AC, Coresh J, Culleton B, Hamm LL, et al. Kidney disease as a risk factor for development of cardiovascular disease: a statement from the American heart association councils on kidney in cardiovascular disease, high blood pressure research, clinical cardiology, and epidemiology and prevention. Hypertension. (2003) 42(5):1050–65. doi: 10.1161/01.HYP.0000102971.85504.7c
17. Jankowski J, Floege J, Fliser D, Böhm M, Marx N. Cardiovascular disease in chronic kidney disease. (2021).
18. WHO. Cardiovascular diseases (CVD) world health organization. (2021). Available at: https://www.who.int/en/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds).
19. Hemmelgarn B, Manns B, Lloyd A, James M, Klarenbach S, Quinn R, et al. Relation between kidney function, proteinuria, and adverse outcomes. JAMA. (2010) 303(5):423–9. doi: 10.1001/jama.2010.39
20. Herzog C, Asinger R, Berger A, Charytan D, Díez J, Hart R, et al. Cardiovascular disease in chronic kidney disease. A clinical update from kidney disease: improving global outcomes (KDIGO). Kidney Int. (2011) 80(6):572–86. doi: 10.1038/ki.2011.223
21. Garrelfs S, Frishberg Y, Hulton S, Koren M, O’Riordan W, Cochat P, et al. Lumasiran, an RNAi therapeutic for primary hyperoxaluria type 1. N Engl J Med. (2021) 384(13):1216–26. doi: 10.1056/NEJMoa2021712
22. Ranasinghe P, Addison M, Webb D. Small interfering RNA therapeutics in hypertension: a viewpoint on vasopressor and vasopressor-sparing strategies for counteracting blood pressure lowering by angiotensinogen-targeting small interfering RNA. J Am Heart Assoc. (2022) 11(20):e027694. doi: 10.1161/JAHA.122.027694
24. Ray K, Wright R, Kallend D, Koenig W, Leiter L, Raal F, et al. Two phase 3 trials of inclisiran in patients with elevated LDL cholesterol. N Engl J Med. (2020) 382(16):1507–19. doi: 10.1056/NEJMoa1912387
25. Bovée D, Ren L, Uijl E, Clahsen-van G MC, van Veghel R, Garrelds I, et al. Renoprotective effects of small interfering RNA targeting liver angiotensinogen in experimental chronic kidney disease. Hypertension. (2021) 77(5):1600–12. doi: 10.1161/HYPERTENSIONAHA.120.16876
26. Molitoris B, Dagher P, Sandoval R, Campos S, Ashush H, Fridman E, et al. siRNA targeted to p53 attenuates ischemic and cisplatin-induced acute kidney injury. J Am Soc Nephrol. (2009) 20(8):1754–64. doi: 10.1681/ASN.2008111204
27. Takabatake Y, Isaka Y, Imai E. In vivo transfer of small interfering RNA or small hairpin RNA targeting glomeruli. Methods Mol Biol. (2009) 466:251–63. doi: 10.1007/978-1-59745-352-3_18
28. Shimizu H, Hori Y, Kaname S, Yamada K, Nishiyama N, Matsumoto S, et al. siRNA-based therapy ameliorates glomerulonephritis. J Am Soc Nephrol. (2010) 21(4):622–33. doi: 10.1681/ASN.2009030295
29. Morishita Y, Yoshizawa H, Watanabe M, Ishibashi K, Muto S, Kusano E, et al. siRNAs targeted to Smad4 prevent renal fibrosis in vivo. Sci Rep. (2014) 4:6424. doi: 10.1038/srep06424
30. Yang C, Nilsson L, Cheema M, Wang Y, Frøkiær J, Gao S, et al. Chitosan/siRNA nanoparticles targeting cyclooxygenase type 2 attenuate unilateral ureteral obstruction-induced kidney injury in mice. Theranostics. (2015) 5(2):110–23. doi: 10.7150/thno.9717
31. Zheng X, Zang G, Jiang J, He W, Johnston N, Ling H, et al. Attenuating ischemia-reperfusion injury in kidney transplantation by perfusing donor organs with siRNA cocktail solution. Transplantation. (2016) 100(4):743–52. doi: 10.1097/TP.0000000000000960
32. Alidori S, Akhavein N, Thorek D, Behling K, Romin Y, Queen D, et al. Targeted fibrillar nanocarbon RNAi treatment of acute kidney injury. Sci Transl Med. (2016) 8(331):331ra39. doi: 10.1126/scitranslmed.aac9647
33. Narváez A, Guiteras R, Sola A, Manonelles A, Morote J, Torras J, et al. siRNA-silencing of CD40 attenuates unilateral ureteral obstruction-induced kidney injury in mice. PLoS One. (2019) 14(4):e0215232. doi: 10.1371/journal.pone.0215232
34. Wang Y, Wu Q, Wang J, Li L, Sun X, Zhang Z, et al. Co-delivery of p38α MAPK and p65 siRNA by novel liposomal glomerulus-targeting nano carriers for effective immunoglobulin a nephropathy treatment. J Control Release. (2020) 320:457–68. doi: 10.1016/j.jconrel.2020.01.024
35. Thai H, Kim K, Hong K, Voitsitskyi T, Lee J, Mao C, et al. Kidney-targeted cytosolic delivery of siRNA using a small-sized mirror DNA tetrahedron for enhanced potency. ACS Cent Sci. (2020) 6(12):2250–58. doi: 10.1021/acscentsci.0c00763
36. Zheng X, Zhang X, Sun H, Feng B, Li M, Chen G, et al. Protection of renal ischemia injury using combination gene silencing of complement 3 and caspase 3 genes. Transplantation. (2006) 82(12):1781–6. doi: 10.1097/01.tp.0000250769.86623.a3
37. Xia Z, Abe K, Furusu A, Miyazaki M, Obata Y, Tabata Y, et al. Suppression of renal tubulointerstitial fibrosis by small interfering RNA targeting heat shock protein 47. Am J Nephrol. (2008) 28(1):34–46. doi: 10.1159/000108759
38. Gomez IG, MacKenna DA, Johnson BG, Kaimal V, Roach AM, Ren S, et al. Anti-microRNA-21 oligonucleotides prevent alport nephropathy progression by stimulating metabolic pathways. J Clin Invest. (2015) 125(1):141–56. doi: 10.1172/JCI75852
39. Rubel D, Boulanger J, Craciun F, Xu EY, Zhang Y, Phillips L, et al. Anti-microRNA-21 therapy on top of ACE inhibition delays renal failure in alport syndrome mouse models. Cells. (2022) 11(4):594. doi: 10.3390/cells11040594
40. Liebow A, Li X, Racie T, Hettinger J, Bettencourt B, Najafian N, et al. An investigational RNAi therapeutic targeting glycolate oxidase reduces oxalate production in models of primary hyperoxaluria. J Am Soc Nephrol. (2017) 28(2):494–503. doi: 10.1681/ASN.2016030338
41. Zhang M, Bahal R, Rasmussen T, Manautou J, Zhong X. The growth of siRNA-based therapeutics: updated clinical studies. Biochem Pharmacol. (2021) 189:114432. doi: 10.1016/j.bcp.2021.114432
42. Lai C, Pursell N, Gierut J, Saxena U, Zhou W, Dills M, et al. Specific inhibition of hepatic lactate dehydrogenase reduces oxalate production in mouse models of primary hyperoxaluria. Mol Ther. (2018) 26(8):1983–95. doi: 10.1016/j.ymthe.2018.05.016
43. Thompson J, Kornbrust D, Foy J, Solano E, Schneider D, Feinstein E, et al. Toxicological and pharmacokinetic properties of chemically modified siRNAs targeting p53 RNA following intravenous administration. Nucleic Acid Ther. (2012) 22(4):255–64. doi: 10.1089/nat.2012.0371
44. Thielmann M, Corteville D, Szabo G, Swaminathan M, Lamy A, Lehner L, et al. Teprasiran, a small interfering RNA, for the prevention of acute kidney injury in high-risk patients undergoing cardiac surgery: a randomized clinical study. Circulation. (2021) 144(14):1133–44. doi: 10.1161/CIRCULATIONAHA.120.053029
45. Boada C, Sukhovershin R, Pettigrew R, Cooke J. RNA therapeutics for cardiovascular disease. Curr Opin Cardiol. (2021) 36(3):256–63. doi: 10.1097/HCO.0000000000000850
46. Dowdy S. Overcoming cellular barriers for RNA therapeutics. Nat Biotechnol. (2017) 35(3):222–9. doi: 10.1038/nbt.3802
47. Chen X, Mangala L, Rodriguez-Aguayo C, Kong X, Lopez-Berestein G, Sood A. RNA interference-based therapy and its delivery systems. Cancer Metastasis Rev. (2018) 37(1):107–24. doi: 10.1007/s10555-017-9717-6
48. Bartlett DW, Su H, Hildebrandt IJ, Weber WA, Davis ME. Impact of tumor-specific targeting on the biodistribution and efficacy of siRNA nanoparticles measured by multimodality in vivo imaging. Proc Natl Acad Sci U S A. (2007) 104(39):15549–54. doi: 10.1073/pnas.0707461104
49. Huang Y, Hong J, Zheng S, Ding Y, Guo S, Zhang H, et al. Elimination pathways of systemically delivered siRNA. Mol Ther. (2011) 19(2):381–5. doi: 10.1038/mt.2010.266
50. Liu N, Ding H, Vanderheyden JL, Zhu Z, Zhang Y. Radiolabeling small RNA with technetium-99m for visualizing cellular delivery and mouse biodistribution. Nucl Med Biol. (2007) 34(4):399–404. doi: 10.1016/j.nucmedbio.2007.02.006
51. van de Water FM, Boerman OC, Wouterse AC, Peters JG, Russel FG, Masereeuw R. Intravenously administered short interfering RNA accumulates in the kidney and selectively suppresses gene function in renal proximal tubules. Drug Metab Dispos. (2006) 34(8):1393–7. doi: 10.1124/dmd.106.009555
52. Braasch DA, Paroo Z, Constantinescu A, Ren G, Oz OK, Mason RP, et al. Biodistribution of phosphodiester and phosphorothioate siRNA. Bioorg Med Chem Lett. (2004) 14(5):1139–43. doi: 10.1016/j.bmcl.2003.12.074
53. Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. (2004) 432(7014):173–8. doi: 10.1038/nature03121
54. Sawai K, Mahato RI, Oka Y, Takakura Y, Hashida M. Disposition of oligonucleotides in isolated perfused rat kidney: involvement of scavenger receptors in their renal uptake. J Pharmacol Exp Ther. (1996) 279(1):284–90.8859005
55. Janssen M, Nieskens T, Steevels T, Caetano-Pinto P, den Braanker D, Mulder M, et al. Therapy with 2′-O-me phosphorothioate antisense oligonucleotides causes reversible proteinuria by inhibiting renal protein reabsorption. Mol Ther Nucleic Acids. (2019) 18:298–307. doi: 10.1016/j.omtn.2019.08.025
56. Warnock J, Daigre C, Al-Rubeai M. Introduction to viral vectors. Methods Mol Biol. (2011) 737:1–25. doi: 10.1007/978-1-61779-095-9_1
57. Wang X, Skelley L, Cade R, Sun Z. AAV delivery of mineralocorticoid receptor shRNA prevents progression of cold-induced hypertension and attenuates renal damage. Gene Ther. (2006) 13(14):1097–103. doi: 10.1038/sj.gt.3302768
58. Zhang Y, Wada J, Hashimoto I, Eguchi J, Yasuhara A, Kanwar Y, et al. Therapeutic approach for diabetic nephropathy using gene delivery of translocase of inner mitochondrial membrane 44 by reducing mitochondrial superoxide production. J Am Soc Nephrol. (2006) 17(4):1090–101. doi: 10.1681/ASN.2005111148
59. Singh S, Kumar R, Agrawal B. Adenoviral vector-based vaccines and gene therapies: current status and future prospects. (2018).
60. Bondue T, van den Heuvel L, Levtchenko E, Brock R. The potential of RNA-based therapy for kidney diseases. Pediatr Nephrol. (2022).
61. Cullis P, Hope M. Lipid nanoparticle systems for enabling gene therapies. Mol Ther. (2017) 25(7):1467–75. doi: 10.1016/j.ymthe.2017.03.013
62. Leung A, Hafez I, Baoukina S, Belliveau N, Zhigaltsev I, Afshinmanesh E, et al. Lipid nanoparticles containing siRNA synthesized by microfluidic mixing exhibit an electron-dense nanostructured core. J Phys Chem C Nanomater Interfaces. (2012) 116(34):18440–450. doi: 10.1021/jp303267y
63. Hou X, Zaks T, Langer R, Dong Y. Lipid nanoparticles for mRNA delivery. Nat Rev Mater. (2021) 6(12):1078–94. doi: 10.1038/s41578-021-00358-0
64. Zielińska A, Carreiró F, Oliveira A, Neves A, Pires B, Venkatesh D, et al. Polymeric nanoparticles: production, characterization, toxicology and ecotoxicology. Molecules. (2020) 25(16):3731. doi: 10.3390/molecules25163731
65. Mellis D, Caporali A. MicroRNA-based therapeutics in cardiovascular disease: screening and delivery to the target. Biochem Soc Trans. (2018) 46(1):11–21. doi: 10.1042/BST20170037
66. Gao S, Hein S, Dagnæs-Hansen F, Weyer K, Yang C, Nielsen R, et al. Megalin-mediated specific uptake of chitosan/siRNA nanoparticles in mouse kidney proximal tubule epithelial cells enables AQP1 gene silencing. Theranostics. (2014) 4(10):1039–51. doi: 10.7150/thno.7866
67. Kesharwani P, Banerjee S, Gupta U, Amin MCIM, Padhye S, Sarkar FH, et al. PAMAM dendrimers as promising nanocarriers for RNAi therapeutics. Mater Today. (2015) 18(10):565–72. doi: 10.1016/j.mattod.2015.06.003
68. Xie D, Wang J, Hu G, Chen C, Yang H, Ritter JK, et al. Kidney-targeted delivery of prolyl hydroxylase domain protein 2 small interfering RNA with nanoparticles alleviated renal ischemia/reperfusion injury. J Pharmacol Exp Ther. (2021) 378(3):235–43. doi: 10.1124/jpet.121.000667
69. Raval N, Jogi H, Gondaliya P, Kalia K, Tekade RK. Cyclo-RGD truncated polymeric nanoconstruct with dendrimeric templates for targeted HDAC4 gene silencing in a diabetic nephropathy mouse model. Mol Pharm. (2021) 18(2):641–66. doi: 10.1021/acs.molpharmaceut.0c00094
70. Lu K, Zhang Y, Song E. Extracellular RNA: mechanisms of it’s transporting into target cells. ExRNA. (2019) 1(1):1–5. doi: 10.1186/s41544-018-0004-7
71. McAndrews K, Kalluri R. Mechanisms associated with biogenesis of exosomes in cancer. Mol Cancer. (2019) 18(1):1–11. doi: 10.1186/s12943-019-0963-9
72. Qiao L, Hu S, Huang K, Su T, Li Z, Vandergriff A, et al. Tumor cell-derived exosomes home to their cells of origin and can be used as Trojan horses to deliver cancer drugs. Theranostics. (2020) 10(8):3474–87. doi: 10.7150/thno.39434
73. Wang H, Wang B, Zhang A, Hassounah F, Seow Y, Wood M, et al. Exosome-mediated miR-29 transfer reduces muscle atrophy and kidney fibrosis in mice. Mol Ther. (2019) 27(3):571–83. doi: 10.1016/j.ymthe.2019.01.008
74. Wang B, Wang J, He W, Zhao Y, Zhang A, Liu Y, et al. Exogenous miR-29a attenuates muscle atrophy and kidney fibrosis in unilateral ureteral obstruction mice. Hum Gene Ther. (2020) 31(5–6):367–75. doi: 10.1089/hum.2019.287
75. Yeboah M, Xue X, Javdan M, Susin M, Metz C. Nicotinic acetylcholine receptor expression and regulation in the rat kidney after ischemia-reperfusion injury. Am J Physiol Renal. (2008) 295(3):F654–61. doi: 10.1152/ajprenal.90255.2008
76. Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood M. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. (2011) 29(4):341–5. doi: 10.1038/nbt.1807
77. Hoshino A, Costa-Silva B, Shen T, Rodrigues G, Hashimoto A, Tesic Mark M, et al. Tumour exosome integrins determine organotropic metastasis. Nature. (2015) 527(7578):329–35. doi: 10.1038/nature15756
78. Tao-Tao T, Bin W, Zuo-Lin L, Wen Y, Song-Tao F, Min W, et al. Kim-1 targeted extracellular vesicles: a new therapeutic platform for RNAi to treat AKI. (2021).
79. Mylonas K, O’Sullivan E, Humphries D, Baird D, Docherty M, Neely S, et al. Cellular senescence inhibits renal regeneration after injury in mice, with senolytic treatment promoting repair. Sci Transl Med. (2021) 13(594):eabb0203. doi: 10.1126/scitranslmed.abb0203
80. Kowalski P, Zwiers P, Morselt H, Kuldo J, Leus N, Ruiters M, et al. Anti-VCAM-1 SAINT-O-somes enable endothelial-specific delivery of siRNA and downregulation of inflammatory genes in activated endothelium in vivo. J Control Release. (2014) 176:64–75. doi: 10.1016/j.jconrel.2013.12.029
81. Hultman K, Raffo A, Grzenda A, Harris P, Brown T, O’Brien S. Magnetic resonance imaging of major histocompatibility class II expression in the renal medulla using immunotargeted superparamagnetic iron oxide nanoparticles. ACS Nano. (2008) 2(3):477–84. doi: 10.1021/nn700400h
82. Scindia Y, Deshmukh U, Thimmalapura P, Bagavant H. Anti-alpha8 integrin immunoliposomes in glomeruli of lupus-susceptible mice: a novel system for delivery of therapeutic agents to the renal glomerulus in systemic lupus erythematosus. Arthritis Rheum. (2008) 58(12):3884–91. doi: 10.1002/art.24026
83. Shirai T, Kohara H, Tabata Y. Inflammation imaging by silica nanoparticles with antibodies orientedly immobilized. J Drug Targeting. (2012) 20(6):535–43. doi: 10.3109/1061186X.2012.693500
84. Rubio-Navarro A, Carril M, Padro D, Guerrero-Hue M, Tarín C, Samaniego R, et al. CD163-macrophages are involved in rhabdomyolysis-induced kidney injury and may be detected by MRI with targeted gold-coated iron oxide nanoparticles. Theranostics. (2016) 6(6):896–914. doi: 10.7150/thno.14915
85. Hébert M, Takano T, Papayianni A, Rennke H, Minto A, Salant D, et al. Acute nephrotoxic serum nephritis in complement knockout mice: relative roles of the classical and alternate pathways in neutrophil recruitment and proteinuria. Nephrol Dial Transplant. (1998) 13(11):2799–803. doi: 10.1093/ndt/13.11.2799
86. Oosthuyzen W, Scullion K, Ivy J, Morrison E, Hunter R, Starkey L PJ, et al. Vasopressin regulates extracellular vesicle uptake by kidney collecting duct cells. J Am Soc Nephrol. (2016) 27(11):3345–55. doi: 10.1681/ASN.2015050568
87. Oosthuyzen W, Sime N, Ivy J, Turtle E, Street J, Pound J, et al. Quantification of human urinary exosomes by nanoparticle tracking analysis. J Physiol. (2013) 591(23):5833–42. doi: 10.1113/jphysiol.2013.264069
88. Rodriguez-Devora J, Ambure S, Shi Z, Yuan Y, Sun W, Xui T. Physically facilitating drug-delivery systems. Ther Deliv. (2012) 3(1):125–39. doi: 10.4155/tde.11.137
89. Omata D, Munakata L, Kageyama S, Suzuki Y, Maruyama T, Shima T, et al. Ultrasound image-guided gene delivery using three-dimensional diagnostic ultrasound and lipid-based microbubbles. J Drug Target. (2022) 30(2):200–7. doi: 10.1080/1061186X.2021.1953510
Keywords: chronic kidney disease (CKD), cardiovascular disease (CVD), ribonucleic acid (RNA)-based therapy, RNA interference (RNAi), small interfering RNA (siRNA), nanoparticles, nanocarrier
Citation: Palmer TC and Hunter RW (2023) Using RNA-based therapies to target the kidney in cardiovascular disease. Front. Cardiovasc. Med. 10:1250073. doi: 10.3389/fcvm.2023.1250073
Received: 29 June 2023; Accepted: 20 September 2023;
Published: 6 October 2023.
Edited by:
Fabio Martelli, IRCCS San Donato Polyclinic, ItalyReviewed by:
Gaia Spinetti, MultiMedica Holding SpA (IRCCS), Italy© 2023 Palmer and Hunter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert W. Hunter cm9iZXJ0Lmh1bnRlckBlZC5hYy51aw==
†ORCID Trecia C. Palmer orcid.org/0000-0001-5127-8860 Robert W. Hunter orcid.org/0000-0003-2344-8978
 Trecia C. Palmer
Trecia C. Palmer Robert W. Hunter
Robert W. Hunter