
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cardiovasc. Med. , 29 September 2023
Sec. Atherosclerosis and Vascular Medicine
Volume 10 - 2023 | https://doi.org/10.3389/fcvm.2023.1247434
Vascular diseases are the leading cause of morbidity and mortality worldwide and are urgently in need of diagnostic biomarkers and therapeutic strategies. Circular RNAs (circRNAs) represent a unique class of RNAs characterized by a circular loop configuration and have recently been identified to possess a wide variety of biological functions. CircRNAs exhibit exceptional stability, tissue specificity, and are detectable in body fluids, thus holding promise as potential biomarkers. Their encoding function and stable gene expression also position circRNAs as an excellent alternative to gene therapy. Here, we briefly review the biogenesis, degradation, and functions of circRNAs. We summarize circRNAs discovered in major vascular diseases such as atherosclerosis and aneurysms, with a particular focus on molecular mechanisms of circRNAs identified in vascular endothelial cells and smooth muscle cells, in the hope to reveal new directions for mechanism, prognosis and therapeutic targets of vascular diseases.
The vasculature is the most extensive organ system in the body that delivers oxygen and nutrition to all tissues. Vascular diseases are the leading cause of morbidity and mortality worldwide. Therefore, there is an urgent need to identify novel diagnostic biomarkers and therapeutic strategies for vascular diseases.
Circular RNAs (circRNAs) are a type of single-stranded RNA molecules characterized by their covalently closed loop configuration, making them highly resistant to ribonuclease (RNase) digestion. Initially considered as junk RNAs produced by alternative splicing error (1), circRNAs have recently been shown to possess a wide variety of biological functions, including acting as miRNA/RNA binding protein (RBP) sponges, regulating parental genes, and even encoding proteins. This protein-encoding function suggests a potential role of circRNAs in gene therapy. Moreover, circRNAs are also promising biomarker candidates for diseases due to their stable expression, cell type specificity, and relatively high levels in body fluid (2).
Here, we briefly review circRNA biogenesis, degradation, and functions. We summarize circRNAs discovered in major vascular diseases such as atherosclerosis and aneurysm, with a focus on elucidating the molecular mechanisms of circRNAs identified in vascular endothelial cells (VECs) and vascular smooth muscle cells (VSMCs) in the hope to reveal new directions to identify mechanisms, prognosis method and therapeutic targets for vascular diseases.
In 1976, plant viroids were first identified as circRNAs (3). Later, circRNAs were observed in the cytoplasm of eukaryotic cell lines through electron microscope (4), which was suspected to be viral RNA genome. In the subsequent years, only a handful of mammalian genes were found to transcribe into circRNAs without clearly defined functions. Over a long period of time, circRNAs are considered as error-spliced junk RNA byproducts without critical biological functions (1).
With the rapid development of next-generation sequencing technologies and bioinformatic tools, thousands of circRNAs have been discovered in diverse species, including virus (5), archaea (6), protists (7), zebrafish (8), mice (9) and human (10). Notably, more than 300,000 circRNAs have been identified in human (11).
Compared with mRNAs and lncRNAs, circRNAs possess several crucial and unique properties. Firstly, they are highly stable compared with linear RNAs. They can resist RNase digestion due to their covalent loop structure (12), thereby escaping from canonical linear RNA degradation. Secondly, the sequences of most circRNAs are highly conserved among species (10, 13). Thirdly, circRNAs expression is tissue specific (14, 15) during development (16). They are highly enriched in the mammalian brain (17) and human platelets (18). Moreover, they are also present in body fluids, including blood, saliva, and urine (19, 20). Fourthly, the expression of circRNAs changes during the transition from physiological to pathological conditions (21, 22).
In the 1990s, circRNAs were discovered to generate through backsplicing of precursor mRNAs (pre-mRNAs) (9, 23). Both linear mRNAs and circRNAs originate from pre-mRNAs transcribed by RNA polymerase II (Pol II). Linear mRNAs are generated through the spliceosome-involved canonical splicing using splice sites (5'-GC and 3'-AG at introns) (24). In contrast, circRNAs derive from pre-mRNA backsplicing to form a covalent loop structure (10). CircRNAs can generally be divided into three categories based on the sequences they contain: circular intronic RNAs (ciRNAs, circRNAs that solely consist of intron-derived sequence) (25), exon-intron circRNAs (EIciRNAs, circRNAs that contain both exon and intron derived sequences) (26) and exonic circRNAs (ecircRNAs, circRNA that exclusively contain exon derived sequence) (27). Four models of circRNA biogenesis (Figures 1, 2) have been proposed, including lariat-driven circularization (10), intron-paring-driven circularization (10), protein-driven circularization (28), and intron cyclization (25).
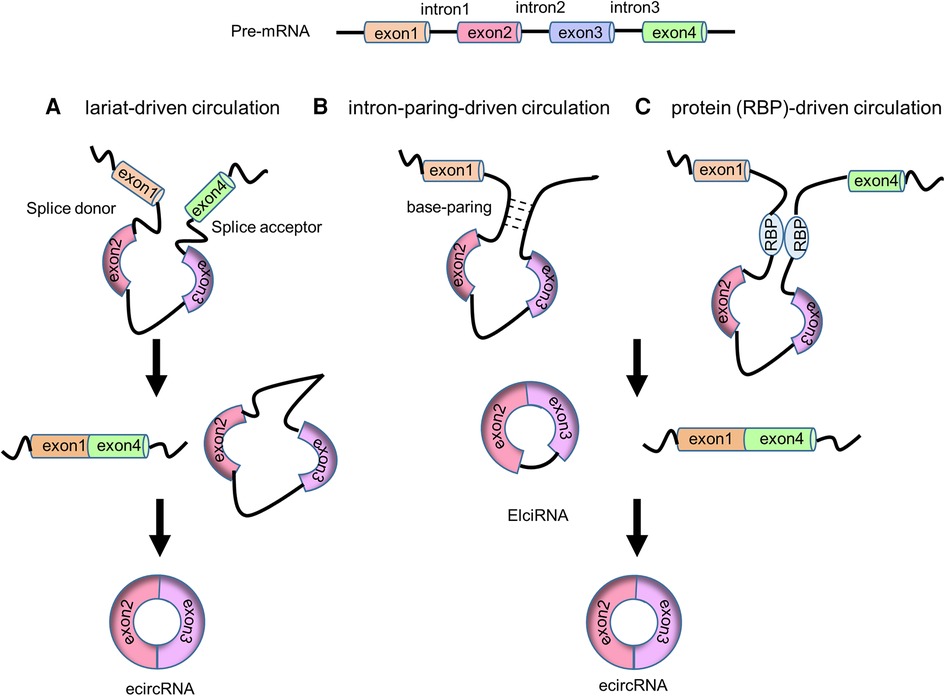
Figure 1. The biogenesis of circRNAs. (A) Lariat-driven circulation model. (B) Intron-paring-driven circulation model. (C) Protein (RBP)-driven circulation model.
In the lariat-driven circularization model, the downstream 3’ splice donor site ligates to the upstream 5’ splice acceptor site to form a 3’ → 5’ phosphodiester bond and generating a lariat consisting of the skipping exons. This lariat then undergoes canonical splicing, releasing the intron and producing an ecircRNA (10) (Figure 1A).
In intron-paring-driven circulation model, the pairing of flanking introns brings the splice sites into close proximity, facilitating backsplicing to create ElciRNAs. Once backsplicing is completed, ElciRNAs can eliminate the introns through canonical splicing, ultimately producing ecircRNAs (10) (Figure 1B). RNA paring can occur at either the repetitive complementary sequence (such as Alu elements) or nonrepetitive complementary sequences (27). These sequences are typically located at the upstream and downstream introns.
In protein-driven circulation model, RBPs can interact with the specific donor and acceptor splice sites, bringing them closer to promote backsplicing, resulting in the formation of EIciRNA and ecircRNA (28) (Figure 1C). For example, RBP Quaking (QKI) has been demonstrated to bind to sites flanking circRNA-forming exons to promote circRNA formation. Insertion of QKI binding sites into linear RNA can induce exon circularization (29).
In intron cyclization model, a consensus motif containing a 7 nt GU-rich element near the 5' splice site and an 11 nt C-rich element close to the branchpoint site can promote the branching enzyme to escape and a debranching failure, ultimately leading to ciRNA formation (25) (Figure 2).
CircRNAs, based on the specific sequence motif they contain, can bind to and function as molecular sponges for miRNA and RBP. Through binding and quenching of miRNA/RBP, circRNAs reduce the availability of these molecules to their natural targets, indirectly modulating the expression of the targeted genes.
In 2013, two independent studies reported that circRNA ciRS-7/CDR1as acted as a miR-7 sponge, revealing that circRNAs could function as miRNA sponges (16, 30). CiRS-7/CDR1as exhibits diverse expression patterns in various tissues, with particularly high expression levels in the brain. It originates from the antisense transcript of vertebrate cerebellar degeneration-related 1 (CDR1) and contains 63 miR-7 binding sites. Expression of human ciRS-7/CDR1as in zebrafish impaired midbrain development, resembling the phenotype observed upon miR-7 knockdown, indicating that ciRS-7/CDR1as inhibited miR-7 activity by acting as miR-7 sponge. Additionally, miR-671 could cleave ciRS-7/CDR1as in an Argonaute-dependent manner and release miR-7 (30). Another example is the circRNA known as testis-specific sex-determining region Y (sry), which contains 16 binding sites for miR-138 and serves as a miR-138 sponge (30).
Subsequently, it was revealed that certain circRNAs can act as sponges for multiple miRNAs. For instance, circCHIPK3 can sponge nine different miRNAs through 18 potential binding sites, thereby promoting cell proliferation (22). These miRNA-sponging circRNAs are primarily located in the cytoplasm and can selectively bind to specific miRNAs or groups of miRNAs with their miRNA binding sequences. These binding quenches miRNAs and reduces their binding to target gene transcript, and indirectly enhances the expression of miRNA-targeted genes (16, 30).
CircRNAs can also function as sponges of RBPs and indirectly regulate gene expression. For example, the second exon of RBP muscleblind (MBL/MBNL1) can circularize to form circMbl in flies and humans. This circMbl and its flanking introns could bind strongly and specifically to MBL proteins to decrease Mbl mRNA production (31). Another example is circPABPN1, which shares HuR binding sequences with linear PABPN1 mRNA. CircPABPN1 competes with linear PABPN1 mRNA for HuR binding, an essential RBP recognizing AU-rich elements in the 3' UTR of its target mRNAs. This competition enhances mRNA stability and translation (32).
CircRNAs can regulate their parental gene expression and these circRNAs are primarily located in the nucleus. It was previously reported that intron retention would functionally interfere with the transcriptomes (33), but the exact mechanism remained unclear. So far, both ciRNAs and EIciRNAs, which contain intron sequences, have been reported to locate in the nucleus and regulate their parental gene expression.
CiRNAs, which derive from lariat introns, are abundant in the nucleus and could regulate their parental gene expression through interaction with Pol Ⅱ (25). For instance, the depletion of ci-ankrd52 and ci-sirt7, which are ciRNAs derived from ANKRD52 and SIRT7 introns, leads to a reduction in the transcription efficiency of ANKRD52 and SIRT7 pre-mRNA, respectively (25), suggesting that these ciRNAs exert influence on parental gene transcription by interacting with Pol Ⅱ.
Similarly, EIciRNAs, arising from exon-intron sequences, also possess this regulatory function. EIciRNAs can interact with U1 small nuclear ribonucleoprotein (snRNP), further recruit Pol Ⅱ to its gene promoter region, and enhance parental gene expression (26). For example, knocking down ElciEIF3J and ElciPAIP2 decreased their binding with U1 snRNP and Pol Ⅱ, eventually decreasing EIF3J and PAIP2 transcription (26).
CircRNAs lack linear mRNA's 5' cap structure and thus cannot use the highly efficient cap-dependent translation mechanism (34) to produce protein. As a result, the majority of circRNAs do not encode peptides. However, they can employ cap-independent mechanisms for translation. Studies have shown that naturally occurring circRNAs can utilize internal ribosome entry site (IRES) elements (35) or m6A-dependent translation mechanism (36) to initiate protein synthesis.
In 1986, it was discovered that the hepatitis delta virus possessed a circular RNA genome and could encode proteins in mammalian cells (5). Later, in 1995, the construction of virus IRES into circRNA could initiate the protein coding of ORF through eukaryotic translational mechanism (37). Some naturally occurring circRNAs even contain endogenous IRES sequences that can be recognized by eIF4G2 to initiate translation (35).
Furthermore, the RNA base modification N-methyladenosine (m6A) also promotes circRNA translation. The m6A reader YTH domain family protein 3 (YTHDF3) can recognize m6A modifications and recruit eIF4G2 to initiate translation, a process further enhanced by the methyltransferases METTL3/14 (36). The m6A modification can enhance the translation efficiency of circRNAs (38, 39).
CircRNAs, unlike mRNA with a linear open end, cannot be directly degraded by exoribonuclease. Thus, circRNAs are stable and have a longer half-life in cells. However, the mechanisms underlying circRNA degradation are not fully elucidated. It was initially shown that circRNA rpsT could be degraded by RNase E (40, 41). A subunit, Rrp44, of yeast exosome has been shown to harbor the endonuclease activity and can cleave circRNAs; however, the degradation efficiency of Rrp44 for circRNAs is lower compared to linear RNAs (42).
Furthermore, in response to viral infection and activation, the widely expressed cytoplasmic endoribonuclease RNase L can globally degrade circRNAs, thereby contributing to the activation of the double-stranded RNA (dsRNA)-activated protein kinase (PKR) and autoimmunity (43). CircRNAs that carry m6A modification can be endoribonucleolytic cleaved by YTHDF2 (m6A reader protein)-HRSP12 (adaptor protein)-RNase P/MRP (endoribonucleases) axis (44).
Vasculature systems harbor the heavy duty to deliver oxygen and nutrients to all the tissues and organs in the body. Any disruptions in this intricate system can result in vascular diseases. Two critical cellular components of blood vessels are vascular ECs and SMCs. ECs form the innermost layer of blood vessels and are responsible for secreting various vasoactive substances, such as nitric oxide and angiogenic factors. ECs thus play a pivotal role in regulating vascular tone, maintaining vascular homeostasis, and promoting angiogenesis (45). Vascular SMCs are the main component of the vascular medial layer and are crucial for maintaining vascular structure. The constriction and dilation of VSMC layers regulates vascular tone, blood flow, and blood pressure (46). It has been established that endothelial dysfunction and the phenotypic switch of SMCs contribute to the development and progression of numerous vascular diseases.
With the rapid advancements in next-generation sequencing technologies and bioinformatic tools, researchers have identified dysregulated circRNAs in various vascular diseases, including conditions like atherosclerosis and aneurysms. These circRNAs are appealing due to their stability, presence in body fluids and plasma, and specific tissue expression patterns. Consequently, there is growing anticipation that circRNAs hold the potential to serve as valuable biomarkers for predicting, diagnosing, assessing treatment effectiveness, and determining prognosis in vascular diseases (2). While circRNAs have multiple functions, most of those identified in the context of vascular diseases have primarily been demonstrated to function as miRNA sponges. Functions such as parental gene regulation and peptide encoding have not been reported.
Atherosclerosis (AS) is a chronic inflammatory disorder characterized by lipid deposition and fibrous cap formation in the arterial wall (47). The initiation of AS is closely associated with low-density lipoprotein (LDL), a particle containing the apolipoprotein B component (48, 49). Endothelial dysfunction and SMC phenotypic switch play critical roles in atherosclerosis initiation and progression (50).
Oxidized LDL (oxLDL) is widely used in in vitro cell culture systems to replicate the pathological processes involved in AS initiation. Numerous studies have confirmed that oxLDL contributes to endothelial dysfunction. OxLDL exposure can lead to reduced viability, suppressed migration, apoptosis, inflammatory responses, and oxidative stress in ECs (50). Furthermore, oxLDL can induce a transition in VSMCs from a quiescent, contractile state to a proliferative and synthetic state, both of which play pivotal roles in the formation of atherosclerotic lesions (51).
In recent years, numerous circRNAs have been reported to be dysregulated during the development of AS, with their roles in endothelial dysfunction and SMC phenotypic switching explored. These findings offer a novel direction for potential therapeutic strategies in the treatment of AS, as summarized in Table 1. While many immune cells, such as macrophages and lymphocytes, also play critical roles in AS, they are not discussed here.
In a study by Wen et al. (52), a comparison of circRNA profiles was conducted in serum exosomes obtained from patients with stable plaque atherosclerosis (SA) and those with unstable/vulnerable plaque atherosclerosis (UA). Their findings revealed a positive correlation between circKIAA1429 (circRNA-0006896) levels in serum exosomes and triglyceride, LDL cholesterol, and C-reactive protein levels in UA patients. Moreover, it was observed that serum exosomes derived from UA patients could upregulate circKIAA1429 expression in HUVECs, reduce microRNA-1264 levels, elevate DNMT1 levels and STAT3 phosphorylation to reduce SOCS3 expression, and notably enhance HUVEC proliferation and migration when compared to both mock and SA groups.
CircGNAQ interacted with miR-146a-5p to upregulate PLK2 expression, inhibiting EC senescence and AS progression (53). AAV-Tie2-circGNAQ, specifically targeting ECs, could curb vascular EC senescence and reduce aortic AS in mice (53). Additionally, circRSF1 overexpression facilitated ECs proliferation and repressed oxLDL-treated HUVE apoptosis and inflammation through the miR-135b-5p/HDAC1 axis (54).
CircSPARC (circ_0004104) is highly expressed in oxLDL-treated HUVECs. Knocking down circSPARC alleviated oxLDL-treated endothelial injury by upregulating miR-328-3p and downregulating TRIM14 (55). Liang et al. found that overexpression of circSMARCA5 (hsa_circ_0001445) activated SRSF1/β-catenin/EMT axis to recover proliferation of oxLDL-treated HUVECs (56). CircZNF532 (circ_0003423) was found to be protective against oxLDL-treated endothelial dysfunction by interacting with miR-142-3p and activating SIRT3/SOD2 pathway (57).
CircGNB4 (circ_0068087) overexpressed in oxLDL-treated HUVECs. CircGNB4 silencing facilitated proliferation of oxLDL-treated HUVECs and reduced oxLDL-treated HUVECs injury through downregulating ROBO1 expression via releasing miR-186-5p (58). CircROBO2 (circ_0124644) promoted oxLDL-treated HUVECs injury by regulating PAPP-A through sponging miR-149-5p (59). Knockdown of circROBO2 suppressed apoptosis and motivated the abilities of cell proliferation and cycle in oxLDL-treated HUVECs (59). CircCHMP5 inhibited cell cycle, proliferation and angiogenesis and facilitated apoptosis to accelerate oxLDL-treated HUVECs injury through upregulating ROCK2 via binding with miR-532-5p (60). CircNMD3 enhanced oxLDL-treated HUVECs proliferation but restrained apoptosis by upregulating BAMBI expression via regulating miR-498 (61). CircUSP9X (circ_0090231) was increased in oxLDL-treated HAECs. Knockdown of circUSP9X could reduce oxLDL-treated HAECs injury and pyroptosis and enhanced their viability through miR-635/NLRP3 axis (62). Inhibition of circNOL12 (hsa_circ_0004543) facilitated HUVECs proliferation, migration, and invasion, significantly reducing their apoptotic rate following oxLDL treatment (63). CircNOL12 knockdown activated PI3K/AKT/eNOS pathway in oxLDL-treated HUVECs to participate in angiogenesis (63).
CircHIF1ɑ (hsa_circ_0032139) promoted HAECs proliferation, motility, and neovascularization through binding with miR-199a-5p to upregulate SIRT1 expression (64). CircDLGAP4 facilitated oxLDL-treated HUVECs proliferation and autophagy, inhibited apoptosis and inflammation, and aggravated dysfunction via interacting with miR-134-5p to increase the expression of PTPN4 (65). CircFOXO1 (hsa_circ_0030042) promoted oxLDL-treated HUVECs proliferation, suppressed apoptosis, and decreased inflammation by targeting with miR-616-3p to upregulate RFX7 levels (66).
CircTEX14 (hsa_circ_0107197) overexpression attenuated oxLDL-treated HASMCs proliferation and promoted apoptosis via regulating miR-6509-3p/THAP1 axis (67). CircTM7SF3 (hsa_circ_0007478) facilitated proliferation, migration and invasion in HVSMCs through ROCK2 upregulation by binding with miR-638 under oxLDL treatment (68). CircARHGAP12 targeted miR-630 to upregulate EZH2 expression, thereby contributing to the oxLDL-treated proliferation and migration of MASMCs (69). In HUVSMCs, knockdown of circUSP36 modulated oxLDL-treated injury via interacting with miR-182-5p to reduce the expression of KLF5 (70).
CircMAPK1 (mmu_circ_0000668) promoted the proliferation and migration of VSMCs through upregulating MECP2 expression via sponging to miR-22-3p (71). CircPPAPDC1A (hsa_circ_0008896) accelerated AS by enhancing the proliferation, migration, and invasion of VSMCs via binding with hsa-miR-633 to upregulate the expression of CDC20B (72). CircTNPO1 (hsa_circ_0072951) expression levels were significantly increased in the serum of AS patients compared with control. CircTNPO1 promoted the oxLDL-treated proliferation and migration of VSMCs through the miR-181b/Notch1 axis (73). CircCHFR (hsa_circ_0029589) modulates VSMCs phenotypic change. Knockdown of circCHFR suppressed VSMCs proliferation and migration via miR-370/FOXO1/cyclin D1 pathway in AS (74). Downregulation of circCHFR inhibited the proliferation, migration, and invasion of VSMCs by modulating miR-214-3p/STIM1 axis (75).
CircPTPRA expression was upregulated in serum of AS patients and oxLDL-treated VSMCs. CircPTPRA promoted VSMC proliferation and inhibited cell apoptosis through repressing miR-636 to upregulate SP1 (76). CircUBR4 (circ_0010283) interacted with miR-370-3p to upregulate the expression of HMGB1 and regulated the viability and migration of oxLDL-induced VSMCs (77). In addition, circUBR4 acted as miR-107 sponges as well. Blocking circUBR4 could attenuate oxLDL-induced excessive proliferation, migration, and cell cycle progression in HUVSMCs through miR-107/ROCK1 axis (78). CircARHGAP32 (circ_0002984) regulated oxLDL-induced VSMCs proliferation, migration, and inflammation by modulating miR-326-3P to upregulate VAMP3 in AS (79). CircMTO1 expression was decreased in serum of AS patients. CircMTO1 suppressed oxLDL-treated proliferation and migration of VSMCs through increasing RASA1 expression via miR-182-5p sequestration (80). CircCOL1A1 exacerbated VSMCs phenotype switch through miR-30a-5p/SMAD1/TGF-β axis (81). CircTBC1D1 (hsa_circ_0001402) promoted FKBPL expression by targeting miR-183-5p to suppress VSMC proliferation and migration. Additionally, circTBC1D1 enhanced VSMC autophagy by binding with miR-183-5p to increase BECN1 levels (82).
Besides miRNA sponging, circRNA has also been reported to bind with proteins in SMCs. CircANRIL impaired exonuclease-mediated pro-rRNA processing and ribosome biogenesis in VSMCs and macrophages by binding to 60S-ribosome assembly factor PES (83). CircANRIL induced nuclear stress and p53 activation, further induced apoptosis, and inhibited proliferation of VSMCs (83).
CircRNAs have been reported to regulate both EC and SMC functions. Overexpression of circGRN (circ_0044073) promoted the proliferation of HUVECs and HUVSMCs by interacting with miR-107 and activating the JAK/STAT (84) signaling pathway (85). CircGRN enhanced oxLDL-treated VSMC dysfunction by serving as miR-377-3p sponge to increase AURKA expression (86).
Aneurysm refers to the local or diffuse dilation or bulge of a blood vessel (87). The thin and weakened vessel wall is more susceptible to dissection or rupture. Most aneurysms are asymptomatic, but the rupture of aortic aneurysms (AA) or intracranial aneurysms (IA) can be life-threatening (88, 89). Although the molecular mechanism of aneurysm formation is not completely understood, dysregulation of VSMCs and aorta wall matrix degradation are critical pathological changes in AA development (90). This often involves the transition of VSMCs from a contractile phenotype to a proliferative and inflammatory one. Furthermore, the destruction of the extracellular matrix (ECM) is a significant feature of AA, mediated by an imbalance between metalloproteinases (MMPs) and tissue inhibitors of metalloproteinases (TIMPs).
CircRNAs have been reported to regulate SMC proliferation and apoptosis, mainly through their regulation of miRNA expression or miRNA sponge functions (Table 2). CircBTBD7 (hsa-circ-000595) was increased in AA tissue as well as in hypoxic aortic SMCs. Knockdown of circBTBD7 could increase miR-19a expression and reduce hypoxia-induced apoptosis of VSMCs (91). CircCCDC66 was upregulated in AA. Depletion of circCCDC66 enhanced VSMC proliferation and inhibited apoptosis via acting as a miR-342-3p sponge to promote CCDC66 transcription (92). CircChordc1 promoted the VSMCs contractive phenotype and enhanced their growth by vimentin degradation and GSK3β/β-catenin signaling activation, extenuating vascular wall remodeling, and reversing aneurysm progression (93). CircCDR1as served as an inhibitor of miR-7, leading to increased expression of the miR-7 target CKAP4. This promotes the proliferation and reduces the apoptosis of VSMCs (94). CircCBFB functioned as a sponge for miR-28-5p, releasing GRIA4 and LYPD3 from miR-28-5p suppression to promote VSMC growth (95). CircEIF2S2 (hsa_circ_0092291) reduced angiotensin II-induced damage in HAVSMCs by serving as a miR-626 sponge and upregulated COL4A1 expression (96). CircTMEM189-UBE2V1 (hsa_circ_0002168) interacted with miR-545-3p to upregulate CKAP4 levels, facilitating proliferation and restraining apoptosis in VSMCs (97).
CircRNAs have also been reported to regulate ECM degradation (Table 2), suggesting their critical involvement in AA. Wang et al. identified 65 differentially expressed circRNA in abdominal AA tissues and found that circRBM33 was upregulated in AA samples and angiotensin II stimulated VSMCs. CircRBM33 overexpression increased MMP2 expression and reduced TIMP2 expression, leading to ECM degradation. CircRBM33 acting as miR-4268 sponges to upregulate EPHB2 and inhibit TIMP-1 expression to mediate ECM degradation (98). Circ-FNDC3B (hsa_circ_0006156) enhances cytotoxicity in VSMCs triggered by angiotensin II, partly through its role as a miR-143-3p sponge and by upregulating ADAM10 (99).
Hemodynamic disturbances, gene changes, arterial wall degeneration, aging, and infection have been reported as risk factors for IA (111). CircARFIP2 (hsa_circ_0021001) promoted the proliferation, migration, and invasion of human umbilical artery SMCs (HUASMC) by increasing kinase inserts domain receptor (KDR) expression via interacting with the miR-338-3p (100). CircLIFR enhanced the proliferation, migration, invasion, and inhibited apoptosis of HUASMC via binding with miR-1299 to upregulate KDR expression (101). Depletion of circDOCK1 (circ_0020397) involved in decreasing of VSMC proliferation by reducing KDR expression in IA via binding with miR-138 (102). In addition, circDOCK1 could promote VSMC viability via miR-502-5p/GREM1 axis (103). CircDOCK1 alleviated the hydrogen peroxide-induced apoptosis and proliferation inhibition of human brain microvascular SMCs (HBVSMC) by interacting with miR-409-3p to upregulate MCL1 (104). CircATL1 was found overexpressed in IA patients. CircATL1 silencing inhibited VSMCs migration, proliferation and contractility through regulating miR-455/SIRT5 pathway (105). Chen et al. analyzed the differentially expressed circRNAs between unruptured IA (UIA) and ruptured IA (RIA) tissues and found that circIRAK3 (hsa_circ_0005505) upregulated in RIA tissues. Knocking down circIRAK3 inhibited the proliferation and migration of HBVSMCs while inducing apoptosis. Depletion of circIRAK3 reduced expression of HBVSMC phenotype switch marker, including OPN, YAP1 and reduced MMP2 and MMP expression (106).
CircRNAs implicated in endothelial dysfunction also play a role in IA. Zhang et al. analyzed circRNA microarray of ECs isolated from RIA and UIA and identified that circIGF2BP3 (circ_0079586) and circRanGAP1 expression were upregulated in RIA. CircIGF2BP3 and circRanGAP1 increased the myeloperoxidase (MPO) expression through binding with miR-183-5p and miR-877-3p, respectively (107). Notably, MPO has been associated with aneurysm rupture and may serve as a biomarker for IA (112).
In a study by Huang et al., 216 IA patients and 186 healthy volunteers were selected to assess the expression of circITGAL (hsa_circ_0000690) in their peripheral blood. They discovered that circITGAL expression was lower in individuals with multiple IAs than in healthy volunteers, indicating its role as a potential biomarker for IA diagnosis and is closely related to the volume of hemorrhage (108). In another study, Wu et al. analyzed circRNA expression profiles in peripheral blood using circRNA microarrays to compare healthy volunteers with patients harboring saccular aneurysm wall enhancement (AWE). This research revealed that circPGAP3 (hsa_circ_0007990) expression upregulated in UIA patients compared with healthy people. And circPGAP3 expression was significantly higher in UIA patients with AWE than those without. Thus, circPGAP3 could be a novel biomarker for UIA (109). Chen et al. analyzed differentially expressed circRNAs between normal superficial temporal arteries and IA samples and found that circDUS2 was upregulated in IA tissues, suggesting its potential role as biomarker (110).
Over the past decade, our understanding of the functions of circRNAs has begun to emerge. Increasing evidence supports the notion that circRNAs, far from being regarded as “junk RNA”, play pivotal roles in a wide array of biological processes. In the context of vascular diseases like atherosclerosis and aneurysms, numerous circRNAs have been identified within EC and SMC, and their involvement in the pathological progression of these vascular diseases has been indicated. Furthermore, some circRNAs have exhibited differential expression levels in the serum or exosomes of patients with atherosclerosis or aneurysms compared to the general population, suggesting the potential utility of circRNAs as biomarkers for these vascular diseases and for predicting disease prognosis.
However, despite these significant strides, the biological functions and molecular mechanisms of circRNAs in vascular diseases remain incompletely understood. To date, the majority of circRNAs reported to play a role in EC and SMC function in atherosclerosis and aneurysms primarily function as miRNA sponges or regulators of miRNAs. In-depth studies using in vivo animal models are scarce, and the roles of circRNAs in functions such as binding to RBPs, regulating parental genes, or encoding peptides have yet to be identified. Further research in these areas is needed to unravel circRNA functions in vascular diseases and to exploit their potential therapeutic and diagnostic applications.
Studying circRNA functions at the DNA level, both in terms of gain and loss of function, is historically challenging, as these approaches can inadvertently affect the levels of their linear RNA counterparts. However, recent advancements in RNA circularization techniques, such as the improved PIE (permuted intron exon) methods (113), have demonstrated high efficiency and the ability to synthesize bulk circRNAs in vitro. This has opened up new avenues for circRNA research, particularly in overexpression studies, as it allows for manipulating circRNA levels without altering the linear RNA transcript from their parental genes.
Furthermore, due to the inherent stability and longer half-life of circRNAs, in vitro synthesized circRNAs containing specific gene open reading frames hold great promise for gene therapy in the treatment of vascular diseases. These synthesized circRNAs can serve as valuable tools for exploring novel therapeutic strategies.
By integrating bioinformatic methodologies, extensive circRNA profiling, and cutting-edge circRNA synthesis techniques, a more comprehensive understanding of the roles played by circRNAs in vascular diseases can be achieved. This enhanced comprehension of circRNA mechanisms will pave the way for the development of innovative therapies for vascular diseases.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
This work is supported by grants to SM (National Natural Science Foundation of China Grant 82170512 and Startup Grant from Guangzhou Laboratory) and to TZ (Guangzhou Science and Technology Plan Project 202201011255).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. (1993) 7(1):155–60. doi: 10.1096/fasebj.7.1.7678559
2. Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. (2018) 34:267–74. doi: 10.1016/j.ebiom.2018.07.036
3. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. (1976) 73(11):3852–6. doi: 10.1073/pnas.73.11.3852
4. Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. (1979) 280(5720):339–40. doi: 10.1038/280339a0
5. Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. (1986) 323(6088):558–60. doi: 10.1038/323558a0
6. Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome-wide discovery of circular RNAs in archaea. Nucleic Acids Res. (2012) 40(7):3131–42. doi: 10.1093/nar/gkr1009
7. Broadbent KM, Broadbent JC, Ribacke U, Wirth D, Rinn JL, Sabeti PC. Strand-specific RNA sequencing in plasmodium falciparum malaria identifies developmentally regulated long non-coding RNA and circular RNA. BMC Genom. (2015) 16(1):454. doi: 10.1186/s12864-015-1603-4
8. Shen Y, Guo X, Wang W. Identification and characterization of circular RNAs in zebrafish. FEBS Lett. (2017) 591(1):213–20. doi: 10.1002/1873-3468.12500
9. Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, et al. Circular transcripts of the testis-determining gene sry in adult mouse testis. Cell. (1993) 73(5):1019–30. doi: 10.1016/0092-8674(93)90279-y
10. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. (2013) 19(2):141–57. doi: 10.1261/rna.035667.112
11. Liu Y, Chen L, Liu T, Su X, Peng L, Chen J, et al. Genome-wide circular RNA (circRNA) and mRNA profiling identify a circmet-mir-410-3p regulatory motif for cell growth in colorectal cancer. Genomics. (2022) 114(1):351–60. doi: 10.1016/j.ygeno.2021.11.038
12. Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNAse R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. (2006) 34(8):e63. doi: 10.1093/nar/gkl151
13. Santos-Rodriguez G, Voineagu I, Weatheritt RJ. Evolutionary dynamics of circular RNAs in primates. eLife. (2021) 10:e69148. doi: 10.7554/eLife.69148
14. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. (2012) 7(2):e30733. doi: 10.1371/journal.pone.0030733
15. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. (2013) 9(9):e1003777. doi: 10.1371/journal.pgen.1003777
16. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. (2013) 495(7441):333–8. doi: 10.1038/nature11928
17. Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. (2015) 58(5):870–85. doi: 10.1016/j.molcel.2015.03.027
18. Preußer C, Hung LH, Schneider T, Schreiner S, Hardt M, Moebus A, et al. Selective release of circRNAs in platelet-derived extracellular vesicles. J Extracell Vesicles. (2018) 7(1):1424473. doi: 10.1080/20013078.2018.1424473
19. Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One. (2015) 10(10):e0141214. doi: 10.1371/journal.pone.0141214
20. Wang S, Zhang K, Tan S, Xin J, Yuan Q, Xu H, et al. Circular RNAs in body fluids as cancer biomarkers: the new frontier of liquid biopsies. Mol Cancer. (2021) 20(1):13. doi: 10.1186/s12943-020-01298-z
21. Werfel S, Nothjunge S, Schwarzmayr T, Strom TM, Meitinger T, Engelhardt S. Characterization of circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol. (2016) 98:103–7. doi: 10.1016/j.yjmcc.2016.07.007
22. Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant Circhipk3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. (2016) 7:11215. doi: 10.1038/ncomms11215
23. Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, et al. Scrambled exons. Cell. (1991) 64(3):607–13. doi: 10.1016/0092-8674(91)90244-s
24. Darnell JE Jr. Reflections on the history of pre-mRNA processing and highlights of current knowledge: a unified picture. RNA. (2013) 19(4):443–60. doi: 10.1261/rna.038596.113
25. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell. (2013) 51(6):792–806. doi: 10.1016/j.molcel.2013.08.017
26. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. (2015) 22(3):256–64. doi: 10.1038/nsmb.2959
27. Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. (2014) 159(1):134–47. doi: 10.1016/j.cell.2014.09.001
28. Aufiero S, Reckman YJ, Pinto YM, Creemers EE. Circular RNAs open a new chapter in cardiovascular biology. Nat Rev Cardiol. (2019) 16(8):503–14. doi: 10.1038/s41569-019-0185-2
29. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. (2015) 160(6):1125–34. doi: 10.1016/j.cell.2015.02.014
30. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. (2013) 495(7441):384–8. doi: 10.1038/nature11993
31. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. CircRNA biogenesis competes with pre-mRNA splicing. Mol Cell. (2014) 56(1):55–66. doi: 10.1016/j.molcel.2014.08.019
32. Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. (2017) 14(3):361–9. doi: 10.1080/15476286.2017.1279788
33. Braunschweig U, Barbosa-Morais NL, Pan Q, Nachman EN, Alipanahi B, Gonatopoulos-Pournatzis T, et al. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. (2014) 24(11):1774–86. doi: 10.1101/gr.177790.114
34. Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. (2009) 136(4):731–45. doi: 10.1016/j.cell.2009.01.042
35. Shi Y, Jia X, Xu J. The new function of circRNA: translation. Clin Transl Oncol. (2020) 22(12):2162–9. doi: 10.1007/s12094-020-02371-1
36. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. (2017) 27(5):626–41. doi: 10.1038/cr.2017.31
37. Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. (1995) 268(5209):415–7. doi: 10.1126/science.7536344
38. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-Znf609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. (2017) 66(1):22–37.e9. doi: 10.1016/j.molcel.2017.02.017
39. Di Timoteo G, Dattilo D, Centrón-Broco A, Colantoni A, Guarnacci M, Rossi F, et al. Modulation of circRNA metabolism by M(6)a modification. Cell Rep. (2020) 31(6):107641. doi: 10.1016/j.celrep.2020.107641
40. Mackie GA. Ribonuclease E is a 5'-end-dependent endonuclease. Nature. (1998) 395(6703):720–3. doi: 10.1038/27246
41. Mackie GA. Stabilization of circular rpst mrna demonstrates the 5'-end dependence of rnase e action in vivo. J Biol Chem. (2000) 275(33):25069–72. doi: 10.1074/jbc.C000363200
42. Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, et al. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol. (2009) 16(1):56–62. doi: 10.1038/nsmb.1528
43. Liu CX, Li X, Nan F, Jiang S, Gao X, Guo SK, et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. (2019) 177(4):865–80.e21. doi: 10.1016/j.cell.2019.03.046
44. Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, et al. Endoribonucleolytic cleavage of M(6)a-containing RNAs by RNAse P/MRP complex. Mol Cell. (2019) 74(3):494–507.e8. doi: 10.1016/j.molcel.2019.02.034
45. Naito H, Iba T, Takakura N. Mechanisms of new blood-vessel formation and proliferative heterogeneity of endothelial cells. Int Immunol. (2020) 32(5):295–305. doi: 10.1093/intimm/dxaa008
46. Shi J, Yang Y, Cheng A, Xu G, He F. Metabolism of vascular smooth muscle cells in vascular diseases. Am J Physiol Heart Circ Physiol. (2020) 319(3):H613–31. doi: 10.1152/ajpheart.00220.2020
47. Hansson GK, Libby P. The immune response in atherosclerosis: a double-edged sword. Nat Rev Immunol. (2006) 6(7):508–19. doi: 10.1038/nri1882
48. Ference BA, Ginsberg HN, Graham I, Ray KK, Packard CJ, Bruckert E, et al. Low-density lipoproteins cause atherosclerotic cardiovascular disease. 1. Evidence from genetic, epidemiologic, and clinical studies. A consensus statement from the European atherosclerosis society consensus panel. Eur Heart J. (2017) 38(32):2459–72. doi: 10.1093/eurheartj/ehx144
49. Borén J, Chapman MJ, Krauss RM, Packard CJ, Bentzon JF, Binder CJ, et al. Low-density lipoproteins cause atherosclerotic cardiovascular disease: pathophysiological, genetic, and therapeutic insights: a consensus statement from the European atherosclerosis society consensus panel. Eur Heart J. (2020) 41(24):2313–30. doi: 10.1093/eurheartj/ehz962
50. Libby P, Buring JE, Badimon L, Hansson GK, Deanfield J, Bittencourt MS, et al. Atherosclerosis. Nat Rev Dis Primers. (2019) 5(1):56. doi: 10.1038/s41572-019-0106-z
51. Bennett MR, Sinha S, Owens GK. Vascular smooth muscle cells in atherosclerosis. Circ Res. (2016) 118(4):692–702. doi: 10.1161/circresaha.115.306361
52. Wen Y, Chun Y, Lian ZQ, Yong ZW, Lan YM, Huan L, et al. CircRNA-0006896-Mir1264-Dnmt1 axis plays an important role in carotid plaque destabilization by regulating the behavior of endothelial cells in atherosclerosis. Mol Med Rep. (2021) 23(5):311. doi: 10.3892/mmr.2021.11950
53. Wu WP, Zhou MY, Liu DL, Min X, Shao T, Xu ZY, et al. Circgnaq, a circular RNA enriched in vascular endothelium, inhibits endothelial cell senescence and atherosclerosis progression. Mol Ther Nucleic Acids. (2021) 26:374–87. doi: 10.1016/j.omtn.2021.07.020
54. Zhang X, Lu J, Zhang Q, Luo Q, Liu B. CircRNA RSF1 regulated ox-LDL induced vascular endothelial cells proliferation, apoptosis and inflammation through modulating Mir-135b-5p/HDAC1 axis in atherosclerosis. Biol Res. (2021) 54(1):11. doi: 10.1186/s40659-021-00335-5
55. Zhang C, Wang L, Shen Y. Circ_0004104 knockdown alleviates oxidized low-density lipoprotein-induced dysfunction in vascular endothelial cells through targeting Mir-328-3p/TRIM14 axis in atherosclerosis. BMC Cardiovasc Disord. (2021) 21(1):207. doi: 10.1186/s12872-021-02012-7
56. Liang G, Chen S, Xin S, Dong L. Overexpression of hsa_circ_0001445 reverses oxLDL-induced inhibition of HUVEC proliferation via SRSF1. Mol Med Rep. (2021) 24(1):507. doi: 10.3892/mmr.2021.12146
57. Wang P, Zhang H, Wang Y. Circ_0003423 alleviates oxidized low-density lipoprotein-induced endothelial cell injury by sponging miR-142-3p and activating sirtuin 3/superoxide dismutase 2 pathway. J Surg Res. (2022) 277:384–97. doi: 10.1016/j.jss.2022.04.006
58. Li S, Huang T, Qin L, Yin L. Circ_0068087 silencing ameliorates oxidized low-density lipoprotein-induced dysfunction in vascular endothelial cells depending on mir-186-5p-mediated regulation of roundabout guidance receptor 1. Front Cardiovasc Med. (2021) 8:650374. doi: 10.3389/fcvm.2021.650374
59. Wang G, Li Y, Liu Z, Ma X, Li M, Lu Q, et al. Circular RNA circ_0124644 exacerbates the ox-LDL-induced endothelial injury in human vascular endothelial cells through regulating PAPP-A by acting as a sponge of mir-149-5p. Mol Cell Biochem. (2020) 471(1-2):51–61. doi: 10.1007/s11010-020-03764-0
60. Li X, Kang X, Di Y, Sun S, Yang L, Wang B, et al. Circchmp5 contributes to ox-LDL-induced endothelial cell injury through the regulation of mir-532-5p/ROCK2 axis. Cardiovasc Drugs Ther. (2022). doi: 10.1007/s10557-022-07316-0 [online ahead of print]
61. Xiu J, Yang Z, Sui Y, Zhang L, Zhou Y. CircNMD3 relieves endothelial cell injury induced by oxidatively modified low-density lipoprotein through regulating miR-498/bmp and activin membrane-bound inhibitor (BAMBI) axis. Bioengineered. (2022) 13(5):12558–71. doi: 10.1080/21655979.2022.2065813
62. Ge Y, Liu W, Yin W, Wang X, Wang J, Zhu X, et al. Circular RNA circ_0090231 promotes atherosclerosis in vitro by enhancing NLR family pyrin domain containing 3-mediated pyroptosis of endothelial cells. Bioengineered. (2021) 12(2):10837–48. doi: 10.1080/21655979.2021.1989260
63. Han L, Li D, Hang Y, Zong X, Lv J, Bai X, et al. Downregulation of hsa_circ_0004543 activates oxLDL-induced vascular endothelial cell proliferation and angiogenesis. Front Genet. (2021) 12:632164. doi: 10.3389/fgene.2021.632164
64. Qiao S, Wang X, Li H, Zhang C, Wang A, Zhang S. Atherosclerosis-associated endothelial dysfunction is promoted by mir-199a-5p/SIRT1 axis regulated by circhif1ɑ. Nutr Metab Cardiovasc Dis. (2023) 33(8):1619–31. doi: 10.1016/j.numecd.2023.05.007
65. Xiong Y, Huang H, Chen F, Tang Y. CirCDLGAP4 induces autophagy and improves endothelial cell dysfunction in atherosclerosis by targeting PTPN4 with mir-134-5p. Environ Toxicol. (2023). doi: 10.1002/tox.23930 [online ahead of print]
66. Yu L, Ma W, Song B, Wang S, Li X, Wang Z. Hsa_circ_0030042 ameliorates oxidized low-density lipoprotein-induced endothelial cell injury via the mir-616-3p/RFX7 axis. Int Heart J. (2022) 63(4):763–72. doi: 10.1536/ihj.22-065
67. Kou L, Yang N, Dong B, Yang J, Song Y, Li Y, et al. Circular RNA testis-expressed 14 overexpression induces apoptosis and suppresses migration of ox-LDL-stimulated vascular smooth muscle cells via regulating the microRNA 6509-3p/thanatos-associated domain-containing apoptosis-associated protein 1 axis. Bioengineered. (2022) 13(5):13150–61. doi: 10.1080/21655979.2022.2070582
68. Guan Z, Lu R, Sun Y, Wang X, Yu C, Song T. Regulation of oxidized LDL-induced proliferation and migration in human vascular smooth muscle cells by a novel circ_0007478/mir-638/ROCK2 ceRNA network. Vasc Med. (2023) 28(1):6–17. doi: 10.1177/1358863-221137617
69. Miao R, Qi C, Fu Y, Wang Y, Lang Y, Liu W, et al. Silencing of circarhgap12 inhibits the progression of atherosclerosis via mir-630/EZH2/TIMP2 signal axis. J Cell Physiol. (2022) 237(1):1057–69. doi: 10.1002/jcp.30598
70. Zhao Q, Lu YH, Wang X, Zhang XJ. Circ_Usp36/mir-182-5p/KLF5 axis regulates the ox-LDL-induced injury in human umbilical vein smooth muscle cells. Am J Transl Res. (2020) 12(12):7855–69.33437365
71. Fu X, Niu T, Yang T, Li X. Circmapk1 promotes the proliferation and migration of vascular smooth muscle cells through mir-22-3p/methyl-CpG binding protein 2 axis. Nutr Metab Cardiovasc Dis. (2021) 31(7):2189–98. doi: 10.1016/j.numecd.2021.04.005
72. Hou X, Dai H, Zheng Y. Circular RNA hsa_circ_0008896 accelerates atherosclerosis by promoting the proliferation, migration and invasion of vascular smooth muscle cells via hsa-mir-633/CDC20b (cell division cycle 20b) axis. Bioengineered. (2022) 13(3):5987–98. doi: 10.1080/21655979.2022.2039467
73. Chen M, Li F, Jiang Q, Zhang W, Li Z, Tang W. Role of mir-181b/Notch1 axis in circ_TNPO1 promotion of proliferation and migration of atherosclerotic vascular smooth muscle cells. J Healthc Eng. (2022) 2022:4086935. doi: 10.1155/2022/4086935
74. Yang L, Yang F, Zhao H, Wang M, Zhang Y. Circular RNA circCHFR facilitates the proliferation and migration of vascular smooth muscle via mir-370/FOXo1/cyclin D1 pathway. Molecular Therapy Nucleic Acids. (2019) 16:434–41. doi: 10.1016/j.omtn.2019.02.028
75. Huang Z, Li P, Wu L, Zhang D, Du B, Liang C, et al. Hsa_circ_0029589 knockdown inhibits the proliferation, migration and invasion of vascular smooth muscle cells via regulating mir-214-3p and STIM1. Life Sci. (2020) 259:118251. doi: 10.1016/j.lfs.2020.118251
76. Zhang LL. CircRNA-PTPRA promoted the progression of atherosclerosis through sponging with mir-636 and upregulating the transcription factor SP1. Eur Rev Med Pharmacol Sci. (2020) 24(23):12437–49. doi: 10.26355/eurrev_202012_24039
77. Ding P, Ding Y, Tian Y, Lei X. Circular RNA circ_0010283 regulates the viability and migration of oxidized low-density lipoprotein-induced vascular smooth muscle cells via an mir-370-3p/HMGB1 axis in atherosclerosis. Int J Mol Med. (2020) 46(4):1399–408. doi: 10.3892/ijmm.2020.4703
78. Zhang Y, Zhang C, Chen Z, Wang M. Blocking circ_Ubr4 suppressed proliferation, migration, and cell cycle progression of human vascular smooth muscle cells in atherosclerosis. Open Life Sci. (2021) 16(1):419–30. doi: 10.1515/biol-2021-0044
79. Li R, Jiang Q, Zheng Y. Circ_0002984 induces proliferation, migration and inflammation response of VSMCs induced by ox-LDL through mir-326-3p/VAMP3 axis in atherosclerosis. J Cell Mol Med. (2021) 25(16):8028–38. doi: 10.1111/jcmm.16734
80. Ji N, Wang Y, Gong X, Ni S, Zhang H. Circmto1 inhibits ox-LDL-stimulated vascular smooth muscle cell proliferation and migration via regulating the mir-182-5p/RASA1 axis. Mol Med. (2021) 27(1):73. doi: 10.1186/s10020-021-00330-2
81. Ye M, Ni Q, Wang H, Wang Y, Yao Y, Li Y, et al. CircRNA circcol1a1 acts as a sponge of mir-30a-5p to promote vascular smooth cell phenotype switch through regulation of Smad1 expression. Thromb Haemost. (2023) 123(1):97–107. doi: 10.1055/s-0042-1757875
82. Lin JJ, Chen R, Yang LY, Gong M, Du MY, Mu SQ, et al. Hsa_circ_0001402 alleviates vascular neointimal hyperplasia through a mir-183-5p-dependent regulation of vascular smooth muscle cell proliferation, migration, and autophagy. J Adv Res. (2023) :S2090-1232(23)00201-1. doi: 10.1016/j.jare.2023.07.010
83. Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. (2016) 7:12429. doi: 10.1038/ncomms12429
84. Wang R, Zhang Y, Xu L, Lin Y, Yang X, Bai L, et al. Protein inhibitor of activated STAT3 suppresses oxidized LDL-induced cell responses during atherosclerosis in apolipoprotein E-deficient mice. Sci Rep. (2016) 6:36790. doi: 10.1038/srep36790
85. Shen L, Hu Y, Lou J, Yin S, Wang W, Wang Y, et al. CircRNA-0044073 is upregulated in atherosclerosis and increases the proliferation and invasion of cells by targeting mir-107. Mol Med Rep. (2019) 19(5):3923–32. doi: 10.3892/mmr.2019.10011
86. Wang Q, Wang T, Liang S, Zhou L. Ox-LDL-induced vascular smooth muscle cell dysfunction partly depends on the circ_0044073/mir-377-3p/AURKA axis in atherosclerosis. Int Heart J. (2023) 64(2):252–62. doi: 10.1536/ihj.22-148
87. Johnston KW, Rutherford RB, Tilson MD, Shah DM, Hollier L, Stanley JC. Suggested standards for reporting on arterial aneurysms. Subcommittee on reporting standards for arterial aneurysms, ad hoc committee on reporting standards, society for vascular surgery and north American chapter, international society for cardiovascular surgery. J Vasc Surg. (1991) 13(3):452–8. doi: 10.1067/mva.1991.26737
88. Sakalihasan N, Limet R, Defawe OD. Abdominal aortic aneurysm. Lancet. (2005) 365(9470):1577–89. doi: 10.1016/s0140-6736(05)66459-8
89. van Donkelaar CE, Bakker NA, Veeger NJ, Uyttenboogaart M, Metzemaekers JD, Luijckx GJ, et al. Predictive factors for rebleeding after aneurysmal subarachnoid hemorrhage: rebleeding aneurysmal subarachnoid hemorrhage study. Stroke. (2015) 46(8):2100–6. doi: 10.1161/strokeaha.115.010037
90. Allaire E, Muscatelli-Groux B, Mandet C, Guinault AM, Bruneval P, Desgranges P, et al. Paracrine effect of vascular smooth muscle cells in the prevention of aortic aneurysm formation. J Vasc Surg. (2002) 36(5):1018–26. doi: 10.1067/mva.2002.127347
91. Zheng C, Niu H, Li M, Zhang H, Yang Z, Tian L, et al. Cyclic RNA hsa-circ-000595 regulates apoptosis of aortic smooth muscle cells. Mol Med Rep. (2015) 12(5):6656–62. doi: 10.3892/mmr.2015.4264
92. Yang R, Wang Z, Meng G, Hua L. Circular RNA CCDC66 facilitates abdominal aortic aneurysm through the overexpression of CCDC66. Cell Biochem Funct. (2020) 38(7):830–8. doi: 10.1002/cbf.3494
93. He X, Li X, Han Y, Chen G, Xu T, Cai D, et al. CircRNA Chordc1 protects mice from abdominal aortic aneurysm by contributing to the phenotype and growth of vascular smooth muscle cells. Molecular Therapy Nucleic Acids. (2022) 27:81–98. doi: 10.1016/j.omtn.2021.11.005
94. Zhao F, Chen T, Jiang N. CDR1as/mir-7/CKAP4 axis contributes to the pathogenesis of abdominal aortic aneurysm by regulating the proliferation and apoptosis of primary vascular smooth muscle cells. Exp Ther Med. (2020) 19(6):3760–6. doi: 10.3892/etm.2020.8622
95. Yue J, Zhu T, Yang J, Si Y, Xu X, Fang Y, et al. CircCBFB-mediated mir-28-5p facilitates abdominal aortic aneurysm via LYPD3 and GRIA4. Life Sci. (2020) 253:117533. doi: 10.1016/j.lfs.2020.117533
96. Ma M, Yang X, Han F, Wang H. Circ_0092291 attenuates angiotensin ii-induced cell damages in human aortic vascular smooth muscle cells via mediating the mir-626/COL4a1 signal axis. J Physiol Biochem. (2022) 78(1):245–56. doi: 10.1007/s13105-021-00859-0
97. Wei J, Wang H, Zhao Q. Circular RNA suppression of vascular smooth muscle apoptosis through the mir-545-3p/CKAP4 axis during abdominal aortic aneurysm formation. Vasc Med. (2023) 28(2):104–12. doi: 10.1177/1358863-221132591
98. Wang S, Yuan Q, Zhao W, Zhou W. Circular RNA RBM33 contributes to extracellular matrix degradation via mir-4268/EPHB2 axis in abdominal aortic aneurysm. PeerJ. (2021) 9:e12232. doi: 10.7717/peerj.12232
99. Liu Y, Zhong Z, Xiao L, Li W, Wang Z, Duan Z, et al. Identification of circ-FNDC3B, an overexpressed circRNA in abdominal aortic aneurysm, as a regulator of vascular smooth muscle cells. Int Heart J. (2021) 62(6):1387–98. doi: 10.1536/ihj.21-186
100. Qin K, Tian G, Zhou D, Chen G. Circular RNA circ-ARFIP2 regulates proliferation, migration and invasion in human vascular smooth muscle cells via mir-338-3p-dependent modulation of KDR. Metab Brain Dis. (2021) 36(6):1277–88. doi: 10.1007/s11011-021-00726-3
101. Zhang H, Zhang B, Chen C, Chen J. Circular RNA circLIFR regulates the proliferation, migration, invasion and apoptosis of human vascular smooth muscle cells via the mir-1299/KDR axis. Metab Brain Dis. (2022) 37(1):253–63. doi: 10.1007/s11011-021-00853-x
102. Wang Y, Wang Y, Li Y, Wang B, Miao Z, Liu X, et al. Decreased expression of circ_0020397 in intracranial aneurysms may be contributing to decreased vascular smooth muscle cell proliferation via increased expression of mir-138 and subsequent decreased KDR expression. Cell Adh Migr. (2019) 13(1):220–8. doi: 10.1080/19336918.2019.1619432
103. Yin K, Liu X. Circ_0020397 regulates the viability of vascular smooth muscle cells by up-regulating GREM1 expression via mir-502-5p in intracranial aneurysm. Life Sci. (2021) 265:118800. doi: 10.1016/j.lfs.2020.118800
104. Ding X, Wang X, Han L, Zhao Z, Jia S, Tuo Y. CircRNA DOCK1 regulates mir-409-3p/MCL1 axis to modulate proliferation and apoptosis of human brain vascular smooth muscle cells. Front Cell Dev Biol. (2021) 9:655628. doi: 10.3389/fcell.2021.655628
105. Xu J, Fang C. Circ-ATL1 silencing reverses the activation effects of SIRT5 on smooth muscle cellular proliferation, migration and contractility in intracranial aneurysm by adsorbing mir-455. BMC Mol Cell Biol. (2023) 24(1):3. doi: 10.1186/s12860-022-00461-2
106. Chen X, Yang S, Yang J, Liu Q, Li M, Wu J, et al. The potential role of hsa_circ_0005505 in the rupture of human intracranial aneurysm. Front Mol Biosci. (2021) 8:670691. doi: 10.3389/fmolb.2021.670691
107. Zhang Z, Sui R, Ge L, Xia D. CircRNA_0079586 and circRNA_RanGAP1 are involved in the pathogenesis of intracranial aneurysms rupture by regulating the expression of MPO. Sci Rep. (2021) 11(1):19800. doi: 10.1038/s41598-021-99062-w
108. Huang Y, Cao H, Qi X, Guan C, Que S. Circular RNA hsa_circ_0000690 as a potential biomarker for diagnosis and prognosis of intracranial aneurysm: closely relating to the volume of hemorrhage. Brain Behav. (2023) 13(4):e2929. doi: 10.1002/brb3.2929
109. Wu XB, Wu YT, Guo XX, Xiang C, Chen PS, Qin W, et al. Circular RNA hsa_circ_0007990 as a blood biomarker for unruptured intracranial aneurysm with aneurysm wall enhancement. Front Immunol. (2022) 13:1061592. doi: 10.3389/fimmu.2022.1061592
110. Chen X, Yang S, Yang J, Liu Q, Li M, Wu J, et al. Circular RNA CirCDUS2 is a potential biomarker for intracranial aneurysm. Front Aging Neurosci. (2021) 13:632448. doi: 10.3389/fnagi.2021.632448
111. Francis SE, Tu J, Qian Y, Avolio AP. A combination of genetic, molecular and haemodynamic risk factors contributes to the formation, enlargement and rupture of brain aneurysms. J Clin Neurosci. (2013) 20(7):912–8. doi: 10.1016/j.jocn.2012.12.003
112. Gounis MJ, Vedantham S, Weaver JP, Puri AS, Brooks CS, Wakhloo AK, et al. Myeloperoxidase in human intracranial aneurysms: preliminary evidence. Stroke. (2014) 45(5):1474–7. doi: 10.1161/strokeaha.114.004956
Keywords: circRNAs, vascular diseases, endothelial cells, smooth muscle cells, atherosclerosis, aneurysms
Citation: Liu Q, Wang Y, Zhang T, Fang J and Meng S (2023) Circular RNAs in vascular diseases. Front. Cardiovasc. Med. 10:1247434. doi: 10.3389/fcvm.2023.1247434
Received: 27 June 2023; Accepted: 18 September 2023;
Published: 29 September 2023.
Edited by:
Paul H. A. Quax, Leiden University, NetherlandsReviewed by:
Xuzhu Gao, Second People’s Hospital of Lianyungang, China© 2023 Liu, Wang, Zhang, Fang and Meng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shu Meng bWVuZ19zaHVAZ3psYWIuYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.