- 1Key Laboratory of Birth Defects and Related Diseases of Women and Children of MOE, Department of Pediatrics, West China Second University Hospital, Sichuan University, Chengdu, China
- 2Department of Cardiovascular Surgery, West China Hospital, Sichuan University, Chengdu, China
- 3Department of Nursing, West China Second University Hospital, Sichuan University, Chengdu, China
- 4State Key Laboratory of Biocatalysis and Enzyme Engineering, School of Life Science, Hubei University, Wuhan, China
Background: Malignant hypertrophic cardiomyopathy (HCM) phenotypes have potential risks of severe heart failure, fatal arrhythmia, and sudden cardiac death. Therefore, it is critical to predict the clinical outcomes of these patients. It was reported recently that the alpha kinase 3 (ALPK3) gene was involved in the occurrence of HCM. Herein we reported a girl with HCM, while whole-exome sequencing found novel compound heterozygous variants in ALPK3 gene, which identified a potential association.
Case presentation: We reported a 14-year-girl who suffered from clinical manifestations of cardiac failure, with sudden cardiac arrest before admission. The heartbeat recovered after cardiopulmonary resuscitation, though she remained unconscious without spontaneous breath. The patient stayed comatose when she was admitted. Physical examination indicated enlargement of the heart boundary. Laboratory results revealed a significant increment of myocardial markers, while imaging demonstrated hypertrophy of the left heart and interventricular septum. Whole-exome sequencing (WES) identified a compound heterozygous variant in ALPK3 gene consisting of c.3907_3922del and c.2200A>T, which was inherited from her parents. Both variants (p.G1303Lfs*28 and p.R734*) were disease-causing evaluated by MutationTaster (probability 1.000). The crystal structure of the complete amino acid sequence is predicted and evaluated by AlphaFold and SWISS-MODEL software (July, 2022), which revealed three domains. Moreover, both variants resulted in a wide protein-truncating variant and damaged protein function. Thus, a novel compound heterozygous variant in ALPK3 associated with HCM was diagnosed.
Conclusion: We described a young patient with ALPK3-associated HCM who experienced sudden cardiac arrest. Through WES, we identified a compound heterozygous variant in the ALPK3 gene, c.3907_3922del and c.2200A>T, which were inherited from the patient's parents and resulted in a truncated protein, indirectly causing the symptoms of HCM. In addition, WES provided clues in evaluating potential risks of gene variants on fatal clinical outcomes, and the nonsense and frameshift variants of ALPK3 were related to adverse clinical outcomes in HCM patients, which required implantable cardioverter defibrillator (ICD) timely.
1. Introduction
Cardiomyopathies constitute a diverse group of disorders with clinical and genetic heterogeneity that primarily affect the ventricular myocardium, resulting in impaired cardiac function and heightened morbidity and mortality. According to existing practice and guidelines, cardiomyopathies can be classified into five subtypes based on their clinical phenotypes, including morphologic and functional features: hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM), restrictive cardiomyopathy (RCM), arrhythmogenic right ventricular cardiomyopathy (ARVC), and unclassified cardiomyopathy (1). Among children, HCM and DCM are the most frequently encountered cardiomyopathy phenotypes (2). HCM is distinguished by symmetric or asymmetric left ventricular hypertrophy, particularly in the interventricular septum, obstructing the left ventricular outflow tract (3). Additionally, HCM is an inherited disease, with a predicted prevalence of 1/500 in adulthood (4). Given the high prevalence of HCM, it is crucial to differentiate between benign and malignant phenotypes and predict the risk of cardiac failure and sudden cardiac death, as some patients may be asymptomatic. In contrast, others present with atrial fibrillation, dyspnea, chest pain, fatigue, or syncope (4–6). The challenge of HCM diagnosis lies not only in making a definitive diagnosis but also in predicting or assessing the heterogeneity of phenotypes and associated clinical outcomes that may require implantable cardioverter defibrillator (ICD) implantation or heart transplantation (7–9). Although the etiology of HCM is highly diverse, it can be summarized as genetic or environmental factors (10). With the rapid development of genetic sequencing, over 900 genes have been identified as involved in the pathogenesis of HCM, with dominant molecules in the sarcomere or sarcomere-associated proteins being implicated in an autosomal dominant manner (11). Recent research has utilized genetic or polygenetic scores to predict clinical risks of HCM on a molecular level, aiding in the clinical management of high-risk patients and guiding the administration of ICD implantation or interventional treatment. The decreasing cost of next-generation sequencing has significantly promoted the application of genetic assessments in cardiomyopathies. Furthermore, hundreds of newly identified HCM-related genes or variant sites have been recorded, providing evidence of the association between rare genetic variants and strategies for diagnosis or treatment.
The alpha kinase 3 (ALPK3) gene, located on chr15:85360587-85416710, is a member of the family of atypical protein kinases (12), recently has been implicated in some cases of HCM, highlighting its potential role in the disease (10, 13, 14). In previous studies, ALPK3 has been identified as a potential factor associated with myocardial cell differentiation, and mice with functional deficiency of ALPK3 exhibit significant ventricular hypertrophy (15, 16). The ALPK3 protein consists of two immunoglobulin (Ig)-like domains and an alpha-type protein kinase domain. Although its specific function in the heart remains unclear, it is believed to play a critical role in cardiac development and transcriptional regulation (10, 17). In this report, we present a case of a 14-year-old female with HCM who initially presented with symptoms of heart failure and experienced multiple cardiac arrests. Whole-exome sequencing (WES) revealed novel compound heterozygous variants on the ALPK3 gene, underscoring the importance of ICD placement in ALPK3-related HCM patients. Furthermore, we provide a comprehensive review of the existing literature and discuss the molecular function of the ALPK3 protein.
2. Case presentation
2.1. History of illness and physical examination
The study was approved by the ethics committee of the West China Second Hospital of Sichuan University (approval no. 2014–034). In addition, we obtained written, informed consent from the patient's parents prior to performing WES and for the inclusion of the patient's clinical and imaging details in publications.
The proband was a 14-year-old female who presented with a two-year history of reduced tolerance to physical exertion and subsequently experienced severe dyspnea, respiratory distress, and fatigue following exertion. The patient suffered a sudden cardiac arrest 2 h before arrival at the hospital, during which carotid pulsation and respiratory movement were absent. CPR and defibrillation were promptly administered by first-aid personnel, resulting in the return of heartbeats after 15 min and restoration of sinus rhythm. Nevertheless, the patient remained unconscious and exhibited no spontaneous breathing. The patient was transferred to the emergency department while receiving laryngeal mask ventilation and subsequently underwent tracheal intubation, positive pressure ventilation, fluid infusion, sedation, and analgesia. The patient was later transferred to the cardiac intensive care unit one hour after the cardiac arrest. The patient's parents denied any history of illness, especially cardiovascular disorders, and any family history of cardiac arrest or cardiovascular disease. Notably, no family member had a history of hypertension or coronary artery disease.
Upon arrival at the cardiac intensive care unit, the patient's blood pressure was approximately 92/63 mmHg, and arterial oxygen saturation was maintained at 97% through mechanical ventilation. Physical examination revealed the patient to be comatose with significant cardiac enlargement, thoracolumbar scoliosis, and muscle weakness.
2.2. Laboratory and imaging evaluation
The results of the blood gas analysis indicated extremely severe respiratory alkalosis (pH = 7.52, PCO2 = 24 mmHg, PO2 = 155 mmHg), electrolyte disturbance (K+ = 4.0 mmol/L, Na+ = 136 mmol/L, Cl− = 104 mmol/L, and Ca2+ = 0.99 mmol/L), and high lactate levels (4.8 mmol/L). Peripheral blood counts revealed an increased leukocyte count of 13.2 × 109/L. Blood biochemical tests demonstrated an elevated level of lactic dehydrogenase [494 U/L; normal range (NR) 109–245 U/L], while other renal and hepatic function parameters showed no apparent abnormalities. Thyroid function test results showed a decreased free triiodothyronine level at 3.2 pmol/L (NR > 4.3 pmol/L). Myocardial markers revealed significantly increased levels of troponin I (0.791 µg/L; NR < 0.2 µg/L), creatine kinase MB isoenzyme (11.75 µg/L; NR < 5 µg/L), and b-type natriuretic peptide (>5,000.00 pg/ml; NR < 100 pg/ml).
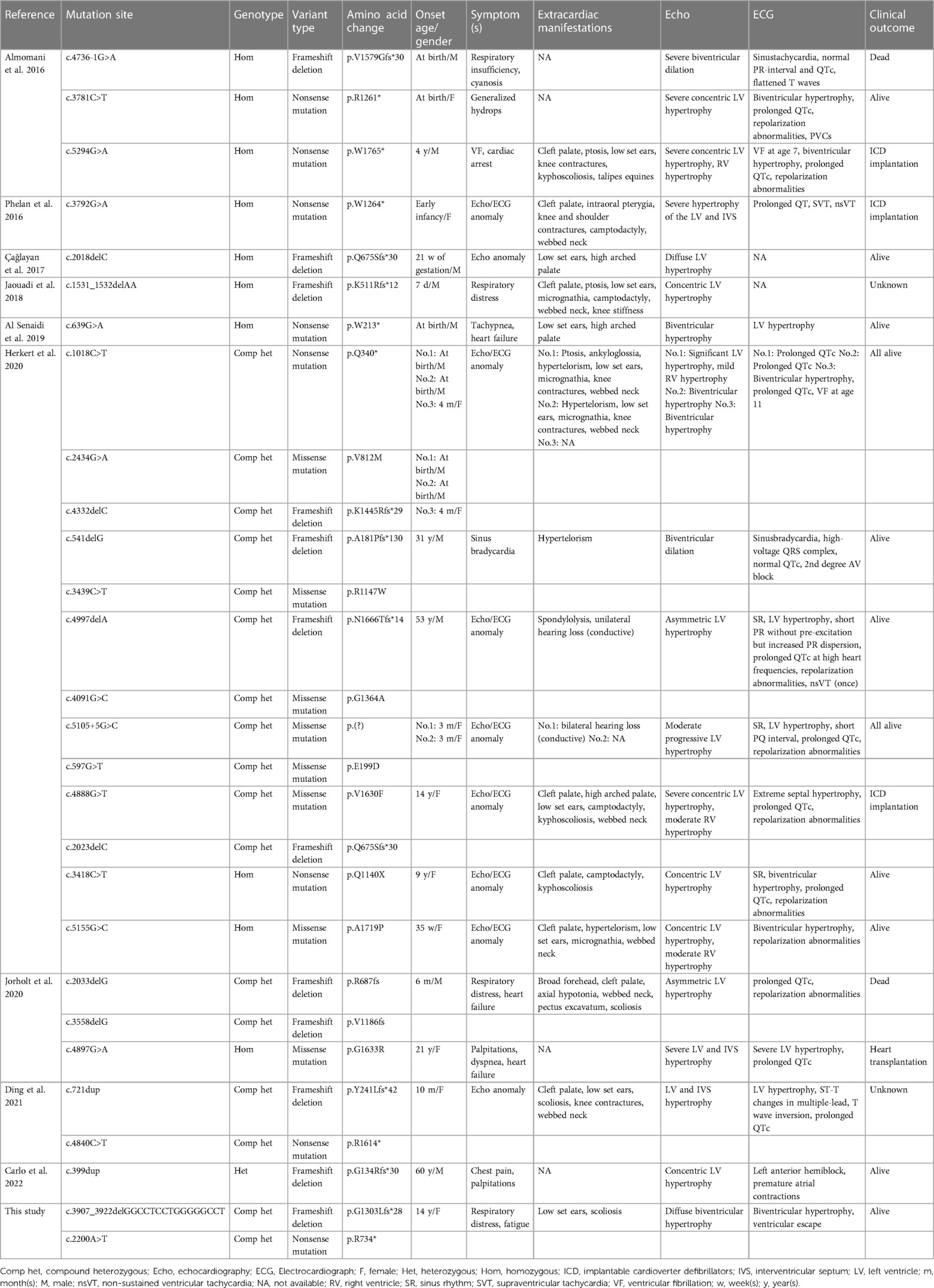
The electrocardiogram (ECG) revealed right axis deviation, biventricular hypertrophy, and ventricular escape beats (Figure 1A). Transthoracic echocardiography revealed significant hypertrophy of the left ventricular wall, particularly the interventricular septum (IVS) and left posterior ventricular wall (LVPW) (Figure 1C). In addition, cardiac magnetic resonance imaging (CMR) demonstrated diffuse hypertrophy of the ventricular myocardium. The thickness of each part during the diastolic period was as follows: LVPW, 28.3 mm; right ventricular anterior wall (RVAW), 8.3 mm; and IVS, 32.2 mm (Figure 1B, left panel). Additionally, the left ventricular ejection fraction (LVEF) measured by CMR was decreased (40.2%). Furthermore, the T2-weighted image revealed an abnormal signal intensity of the left ventricular subendocardial myocardium indicating myocardial ischemia (Figure 1B, right panel). It is noteworthy that the parents of the patient also underwent physical examinations and echocardiographic assessments conducted by cardiologists, revealing no indications associated with HCM.
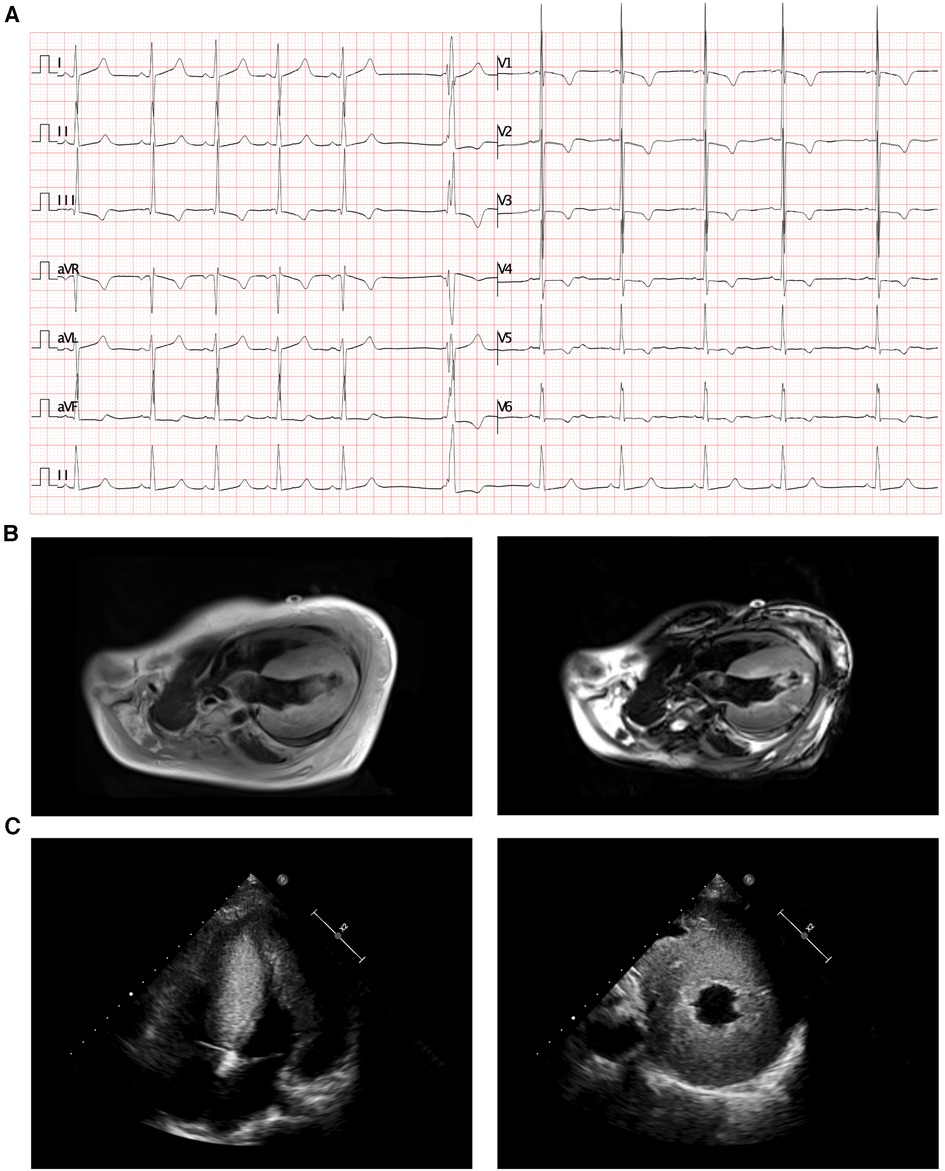
Figure 1. Radiology manifestation in the current proband. (A) Electrocardiographic examination demonstrated right axis deviation, enlargement of bi-ventricles and ventricular escape (arrow). (B) Cardiac magnetic resonance (CMR) demonstrated diffuse hypertrophy of ventricular myocardium, T2-weighting imaging revealed abnormal signal intensity of left ventricular subendocardial myocardium. (C) Transthoracic echocardiography (TTE) demonstrated significant hypertrophy of left ventricle and the interventricular septum.
2.3. Molecular results
We obtained a peripheral blood sample in an EDTA anticoagulant blood sample tube from the patient and stored it at 4°C for less than 6 h. DNA extraction was performed using the Blood Genome Column Medium Extraction Kit (Tiangen Biotech, Beijing, China) according to the instruction. Protein-coding exome enrichment was performed using the xGen Exome Research Panel v1.0. WES was performed using the Illumina NovaSeq 6000 platform (Illumina, San Diego, CA, USA), while primary quality control was performed using FastP, comprising process of the raw data and removement of filter low-quality reads. Variants were annotated in accordance with database-sourced minor allele frequencies (MAFs) and practical guidelines on pathogenicity issued by the American College of Medical Genetics. The sequencing data have been deposited in GSA database (http://ngdc.cncb.ac.cn/gsub/). MutationTaster software and combined annotation dependent depletion (CADD) scaled c-scores were used to predict the pathogenicity of variants, while GRCh37 reference genome was used for alignment. We searched database including gnomAD, ExAC and 1000G to identify prevalence of variants. Effects of genetic variants on protein structure were evaluated via PROVEAN protein batch software with Provean score. As there is no available protein crystal structure for ALPK3, AlphaFold database (https://alphafold.ebi.ac.uk/) tool is used to predict protein crystal structure. Within the structure, three important domains have been revealed with analyzed crystal structure. PyMOL software was used to annotate domains and variant sites of the protein. Then we performed modeling analysis and compared three domains with the 6c6m.2.A, 3uto.2.A, and 1ia9.1.A template via SWISS-MODEL database (https://swissmodel.expasy.org/), to visualize and analyze the altered amino acid sequence and stability of ALPK3. And other identified variants had been presented in Supplementary Table S1.
Based on the clinical manifestations and laboratory analyses, HCM induced by genetic anomaly was strongly suspected. Thus, WES was performed, which identified a novel compound heterozygous variant of c.3907_3922del (p.G1303Lfs*28) and c.2200A>T (p.R734*) in ALPK3 gene, while genomic coordinates of these two variants are chr15:85401269-85401285delGGCCTCCTGGGGGCCT and chr15:85384104A>T (depth of coverage is 236.34, percent of exome captured is 98.34%). The patient's parents presented normal cardiac morphology, thus, we employed Sanger sequencing to validate the genotypes of the parents of the patient (forward primer “agcccacacactccttgacc” and reverse primer “tacatcagagctgctgctgg” for c.2200A>T and forward primer “ctgtacctcccgccgcctca” and reverse primer “tcccctgggaacttctcctc” for c.3907_3922del), which revealed that each parent carries a heterozygous variant of the ALPK3 gene. The variant of c.3907_3922del was maternal inherit, and the variant of c.2200A>T was paternal inherit (Figures 2A,B). According to the American College of Medical Genetics, both variants have pathogenicity as PVS1+PM2_Supporting+PM3 (Trans), and both were related to familial HCM. According to updated data in gnomAD, ExAC and 1000G, these two variants have not been reported in any populations, that means it is the first report of these variants (Figure 2C). Analysis performed with MutationTaster revealed that variant of c.3907_3922del in ALPK3 was considered pathogenic (probability 1.000) due to nonsense-mediated mRNA decay (NMD), amino acid sequence changed, frameshift, protein features affected and splice site changes, while c.2200A>T was also considered pathogenic because of NMD, acid sequence changed, and protein features affected (probability 1.000). Besides, CADD scaled c-scores of variant c.2200A>T is 36, which implies that the predicted pathogenicity of the variant is extremely high. PROVEAN protein batch software indicated that the p.R734* protein was deleterious with the PROVEAN score of −4.79, due to frameshift and NMD of p.G1303Lfs*28, PROVEAN and SIFT prediction were not applicable. While all the reported variants of ALPK3 had been listed in Figure 2D.
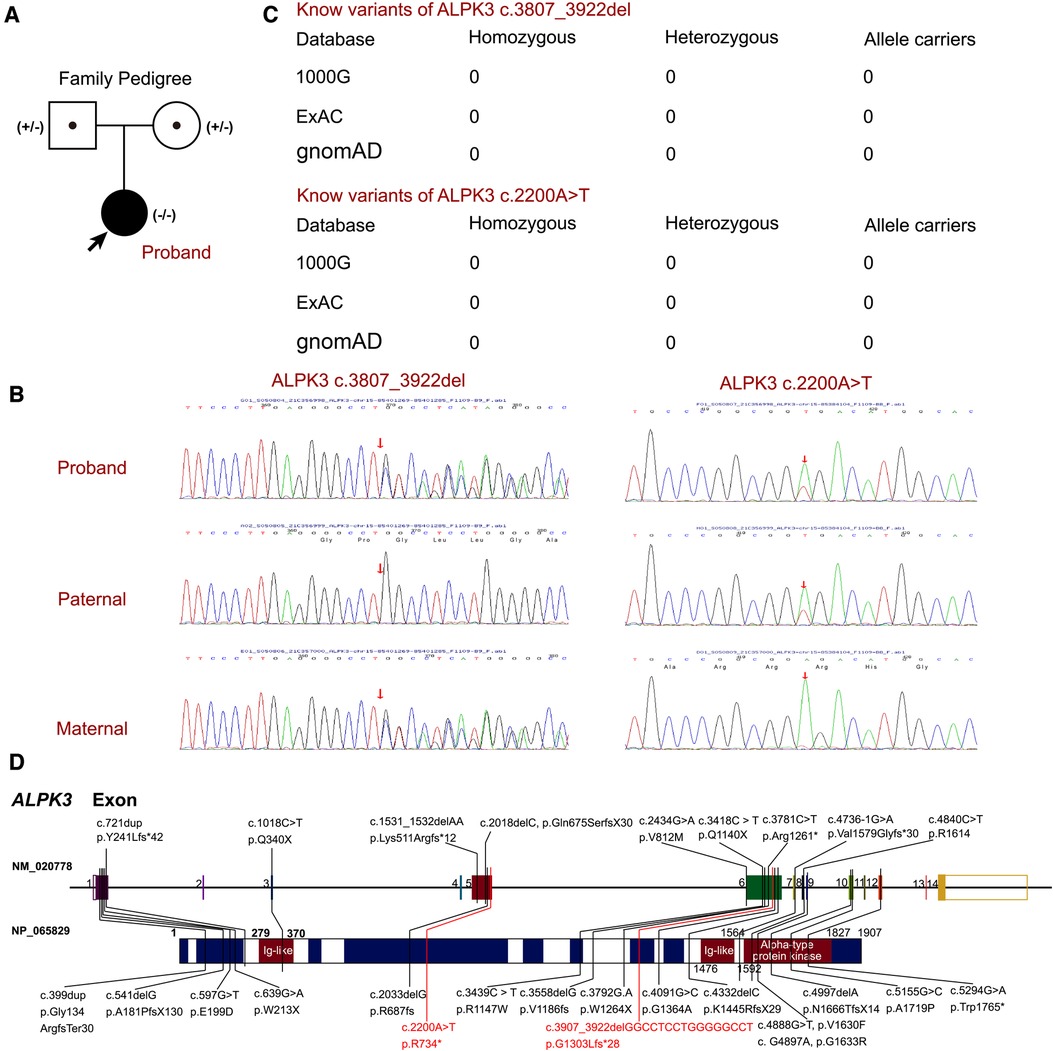
Figure 2. The ALPK3 variants in this family. (A) Family pedigree revealed the maternal carrier of c.3907_3922del (p.G1303Lfs*28) and the paternal carrier of c.2200A>T (p.R734*). The current proband exhibited significant hypertrophic cardiomyopathy with compound heterozygous variants of ALPK3. circles represent females, squares represent males, and arrow indicates the proband. Black symbols indicate the clinical presentation of hypertrophic cardiomyopathy, grey symbols indicate carriers. (B) Sanger sequencing validation of the current proband and his parents. (C) The prevalence of ALPK3 variants of c.3907_3922del and c.2200A>T. (D) Summary of current reports on individual ALPK3 variants resulting in hypertrophic cardiomyopathy.
The entire amino acid sequence crystal structure was predicted by AlphaFold and assigned the name AF-Q96L96-F1 (Figure 3A). Although the predicted protein covered the entire length of the amino acid sequence, only three domains demonstrated high confidence (pLDDT >70). These domains were labeled red, green, and orange (Figure 3B). Other regions displayed low confidence in the crystal structure. While all potential templates were searched, only partial parts of the protein had been analyzed previously. We picked structures with the highest predictive value, which may cause several parts do not have a specific folding. In a word, the AlphaFold-predicted structure was the only model that could be utilized. The c.2200A>T and c.3907_3922del variants would result in a protein-truncating variant, typically leading to protein denaturation. The sites of truncated sequences caused by variants were labeled in yellow (Figures 3C,D). SWISS-MODEL was then employed to present the crystal structures of the variant's three domains, including two immunoglobulin-like (Ig-like) domains and an alpha-type protein kinase domain (Figure 3B). An ig-like domain superfamily is a heterogeneous group of proteins that play the role of cell recognition. The alpha-kinase domain is an atypical protein kinase catalytic domain that exhibits no detectable similarity to conventional protein serine/threonine kinases. This protein kinase recognizes protein sequences that adopt an alpha-helical conformation by its initial members, which act as the final link and effector of intracellular information transmission. The identified ALPK3 variants, c.3907_3922del and c.2200A>T would cause truncated protein, leading to the loss of an Ig-like domain and an alpha-type protein kinase domain, resulting in the dysfunction of the ALPK3 molecule. The aforementioned analyses suggested that both newly identified variants could alter the transcription of the ALPK3 gene and damage the protein structures. Therefore, the compound heterozygous variant of ALPK3 was considered to be genetically associated with HCM in this patient. In addition, several heterozygous variants were identified by WES, such as c.4639A>G in the FBN1 gene, c.3791G>A in ANKRD26, and c.1123G>T in DPYS. However, all three genes were considered to exhibit recessive inheritance, and the pathogenicity predictions for these variants were uncertain. Furthermore, all of these variants were inherited from one of her parents, unaffected by associated diseases. Consequently, they were not considered to be associated with HCM.
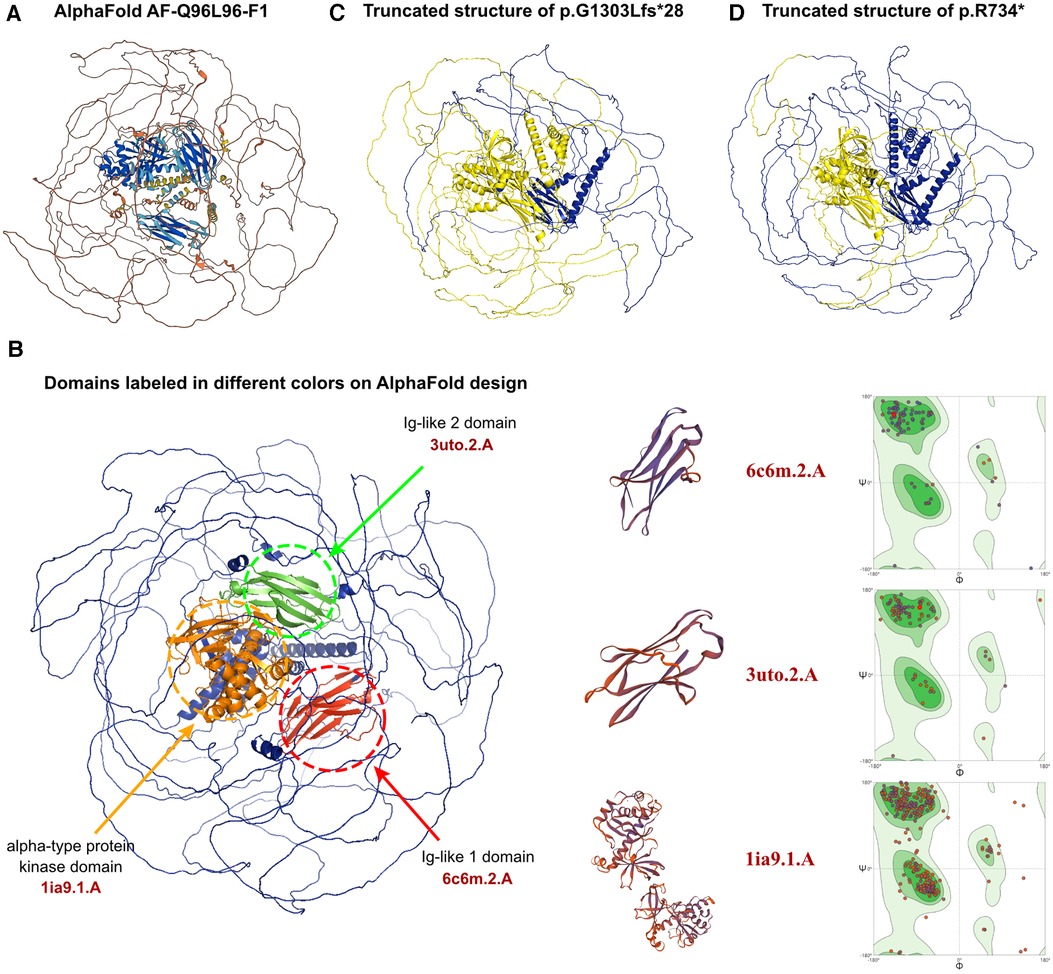
Figure 3. The effects of ALPK3 c.3907_3922delGGCCTCCTGGGGGCCT and c.2200A>T variants on the molecular structure of the protein. (A) AlphaFold protein structure database was used to predict the ALPK3 wild-type protein crystal structure. (B) Three domains of ALPK3 protein were labeled in red, green and orange individually. SWISS-MODEL presented the crystal structures of the variant's three domains, including two immunoglobulin-like (Ig-like) domains and an alpha-type protein kinase domain according to 6c6m.2.A, 3uto.2.A, and 1ia9.1.A model template. Ramachandran plots of three functional domains in wild-type sequences were displayed. (C,D) Truncating variant sites of p.G1303Lfs*28 and p.R734* caused by variants of c.3907_3922delGGCCTCCTGGGGGCCT and c.2200A>T was labelled in yellow.
2.4. Treatment and clinical outcome
Following comprehensive laboratory and echocardiographic assessments, the patient was diagnosed with HCM. A 6-week hospitalization period was instituted, during which the patient received a range of medical interventions, including invasive and noninvasive mechanical ventilation, myocardial protection, anti-arrhythmia, cerebral protection, anti-infection, anti-inflammatory, diuresis, and blood transfusion. Although there was residual muscle weakness, the patient was discharged from the hospital after partial recovery from her major concerns with respect to heart rhythm control and cardiac function. However, two weeks after discharge, the patient experienced recurrent cardiac arrest during rehabilitation training and subsequently regained consciousness. The patient was subsequently readmitted to our department, where mechanical ventilation and gastrointestinal decompression were provided to alleviate symptoms. Anti-infective therapy was initiated with cefoperazone and sulbactam, while captopril and metoprolol were prescribed to address HCM and inhibit potentially lethal arrhythmias. Nutritional and rehabilitation therapies were also administered. After a month of comprehensive medical management, including fluid infusion, diuretic therapy, and vitamin supplementation, the patient was discharged with improved symptoms. However, due to the prolonged duration of the condition and recurrent cardiac arrest, the patient had not regained consciousness and experienced severe cognitive and neurological impairment. Oral medication administration and continuous follow-up and evaluation were regularly conducted, with clinic visits scheduled every two weeks in the first month and every three months thereafter.
3. Discussion and conclusion
HCM is a primary cardiomyopathy that is commonly associated with a genetic variant. Its prevalence is high worldwide, but some subtypes of HCM with specific genetic variants can result in lethal arrhythmia and severe heart dysfunction, leading to sudden cardiac death (18). It is now commonly accepted that HCM is usually inherited with a complex genetic etiology. Studies and books have revealed that pathogenic variants in the core genes encoding sarcomeric proteins, including thick filament encoding genes MYBPC3, MYH7, MYL2, MYL3, and thin filament encoding genes TNNC1, TNNT2, TNNI3, account for over 90% of the pathogenic variants in patients with HCM (19, 20). Additionally, variants in several genes encoding non-sarcomeric proteins with diverse functions, including ACTN2, ALPK3, CSRP3, FHOD3, FLNC, JPH2, KLHL24, PLN, and TRIM63, have also been considered as genetic etiology of HCM (21). Furthermore, variants in FHL1, FXN, GAA, LAMP2, and TTR genes have an extremely low prevalence of 1/100,000–1/20,000 but have also been reported to be associated with HCM (22).
ALPK3 gene locates on chromosome 15q25.2 and contains 14 exons. It had been recently identified as a possible disease-causing gene of pediatric HCM, myopathic and dysmorphic skeletal features (10, 17). Initially, the Midori gene was discovered and named by Hosoda et al. through differential display analysis of the P19CL6 cell line (16), and later identified as the ALPK3 gene. The study revealed that expression of Midori was restricted in the fetal and adult heart and adult skeletal muscle in mice. At the same time, the overexpression of Midori could promote the differentiation of P19CL6 cells into cardiomyocytes (16). A mouse model of ALPK3 knockout by Van Sligtenhorst et al. in 2012 revealed biventricular hypertrophy in ALPK3−/− mice (15). Additionally, the electron microscopy showed impaired cardiomyocyte architecture characterized by reduced numbers of abnormal intercalated discs (15). The experimental data suggested ALPK3 could regulate the transcript of cardiomyocyte differentiation and heart development, and the loss of function of ALPK3 would lead to cardiomyopathy. Thus, the OMIM number of HCM in our manuscript is #618052, which is named familial hypertrophic cardiomyopathy-27 caused by homozygous mutation in the ALPK3 gene (OMIM 617608) on chromosome 15q25. Indeed, several studies have reported on cardiomyopathies caused by other types of kinases. For instance, Brodehl et al. identified protein mutations p.H77Y and p.P70l in integrin-linked kinase, which were found to be associated with arrhythmogenic cardiomyopathy in both humans and transgenic zebrafish (23).
After conducting a comprehensive review of the literature, we identified 22 patients with ALPK3-associated HCM, involving 28 distinct variants of the ALPK3 gene, as described in nine studies and a case report. A summary of all reported variants can be found in Table 1 and Figure 2D (10, 13, 14, 24–29). Consistent with previous reports, the majority of described patients were from consanguineous families, and nearly all patients exhibited biallelic damage, as homozygous or compound heterozygous variants of the ALPK3 gene were commonly observed (15). Notably, only patients with HCM were identified with heterozygous variants of the ALPK3 gene, and one such patient was found to have an accompanying DSP gene and was free of lethal cardiac events (29). Thus, ALPK3 variants appeared to demonstrate a recessive feature in inducing HCM.
The clinical presentation of ALPK3-associated HCM varied, with most pediatric patients presenting symptoms or positive imaging results before age 18. In addition to typical clinical manifestations and findings on ECG and echocardiography, extracardiac manifestations, such as facial and musculoskeletal abnormalities, were observed in some patients (10, 24, 25, 27). Fatal arrhythmia, such as ventricular fibrillation, was the leading cause of death, necessitating ICDs and, in some cases, heart transplantation. Notably, only nonsense and frameshift variants among the 22 reported patients resulted in death and fatal arrhythmia, necessitating ICD implantation. Conversely, missense variants were associated with mild clinical outcomes. Thus, homozygous and compound heterozygous variants of ALPK3 with nonsense or frameshift variants were linked to adverse clinical outcomes, warranting careful follow-up and timely ICD implantation to prevent sudden cardiac death. Therefore, given the patient's clinical symptoms and history of cardiac arrest, we recommended ICD implantation. Regrettably, the patient's family declined this recommendation.
In the present study, we report the case of a 14-year-old female who suffered from ALPK3-associated HCM and experienced sudden cardiac arrest. Through molecular analysis, we identified a novel compound heterozygous variant (c.3907_3922del and c.2200A>T) in the ALPK3 gene, which was inherited from her parents and resulted in truncated protein formation. WES was employed for molecular diagnosis, which has emerged as an efficient and favorable technique for HCM diagnosis and provides valuable insights for assessing sudden cardiac death risks. The entire process, from sampling to library preparation for sequencing, usually takes about five days, and subsequent analysis requires approximately one week, enabling us to produce a report within two weeks. Therefore, we recommend the utilization of WES for the timely identification of deleterious variants in HCM patients. Furthermore, our findings suggest that the nonsense and frameshift variants of ALPK3 are associated with unfavorable clinical outcomes, and prompt implantation of an ICD is crucial for preventing sudden cardiac death.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The studies involving human participants were reviewed and approved by Ethics Committee of West China Second Hospital of Sichuan University (2014-034). Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin. Written informed consent was obtained from the individual(s) and minor(s)’ legal guardian/next of kin for the publication of any potentially identifiable images or data included in this article.
Author contributions
TL, YJ and RL contributed equally to this work. TL, YJ, RL, SL and YL were the patient's physicians. TL and RL reviewed the literature and contributed to manuscript drafting. DZ, TL and YJ performed the variant analysis. DZ, SL and YL conceptualized and designed the study, coordinated and supervised data collection, and critically reviewed the manuscript for important intellectual content. DZ, YL and SL were responsible for the revision of the manuscript for important intellectual content. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from Technology Project of Sichuan Province of China (2021YFQ0061) and the National Natural Science Foundation of China (82270249). The funding did not participate in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2023.1212417/full#supplementary-material
References
1. Elliott P, Andersson B, Arbustini E, Bilinska Z, Cecchi F, Charron P, et al. Classification of the cardiomyopathies: a position statement from the European society of cardiology working group on myocardial and pericardial diseases. Eur Heart J. (2008) 29(2):270–6. doi: 10.1093/eurheartj/ehm342
2. Kimura A. Molecular genetics and pathogenesis of cardiomyopathy. J Hum Genet. (2016) 61(1):41–50. doi: 10.1038/jhg.2015.83
3. Brouwer WP, van Dijk SJ, Stienen GJ, van Rossum AC, van der Velden J, Germans T. The development of familial hypertrophic cardiomyopathy: from mutation to bedside. Eur J Clin Invest. (2011) 41(5):568–78. doi: 10.1111/j.1365-2362.2010.02439.x
4. Geske JB, Ommen SR, Gersh BJ. Hypertrophic cardiomyopathy: clinical update. JACC Heart Fail. (2018) 6(5):364–75. doi: 10.1016/j.jchf.2018.02.010
5. Raphael CE, Cooper R, Parker KH, Collinson J, Vassiliou V, Pennell DJ, et al. Mechanisms of myocardial ischemia in hypertrophic cardiomyopathy: insights from wave intensity analysis and magnetic resonance. J Am Coll Cardiol. (2016) 68(15):1651–60. doi: 10.1016/j.jacc.2016.07.751
6. Siontis KC, Geske JB, Ong K, Nishimura RA, Ommen SR, Gersh BJ. Atrial fibrillation in hypertrophic cardiomyopathy: prevalence, clinical correlations, and mortality in a large high-risk population. J Am Heart Assoc. (2014) 3(3):e001002. doi: 10.1161/JAHA.114.001002
7. Gersh BJ, Maron BJ, Bonow RO, Dearani JA, Fifer MA, Link MS, et al. ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: executive summary: a report of the American college of cardiology foundation/American heart association task force on practice guidelines. Circulation. (2011) 124(24):2761–96. doi: 10.1161/CIR.0b013e318223e230
8. Elliott PM, Anastasakis A, Borger MA, Borggrefe M, Cecchi F, Charron P, et al. ESC guidelines on diagnosis and management of hypertrophic cardiomyopathy: the task force for the diagnosis and management of hypertrophic cardiomyopathy of the European society of cardiology (ESC). Eur Heart J. (2014) 35(39):2733–79. doi: 10.1093/eurheartj/ehu284
9. Kitaoka H, Tsutsui H, Kubo T, Ide T, Chikamori T, Fukuda K, et al. JCS/JHFS 2018 guideline on the diagnosis and treatment of cardiomyopathies. Circ J. (2021) 85(9):1590–689. doi: 10.1253/circj.CJ-20-0910
10. Almomani R, Verhagen JM, Herkert JC, Brosens E, van Spaendonck-Zwarts KY, Asimaki A, et al. Biallelic truncating mutations in ALPK3 cause severe pediatric cardiomyopathy. J Am Coll Cardiol. (2016) 67(5):515–25. doi: 10.1016/j.jacc.2015.10.093
11. Bos JM, Towbin JA, Ackerman MJ. Diagnostic, prognostic, and therapeutic implications of genetic testing for hypertrophic cardiomyopathy. J Am Coll Cardiol. (2009) 54(3):201–11. doi: 10.1016/j.jacc.2009.02.075
12. Middelbeek J, Clark K, Venselaar H, Huynen MA, van Leeuwen FN. The alpha-kinase family: an exceptional branch on the protein kinase tree. Cell Mol Life Sci. (2010) 67(6):875–90. doi: 10.1007/s00018-009-0215-z
13. Çağlayan AO, Sezer RG, Kaymakçalan H, Ulgen E, Yavuz T, Baranoski JF, et al. ALPK3 gene mutation in a patient with congenital cardiomyopathy and dysmorphic features. Mol Case Stud. (2017) 3(5):a001859. doi: 10.1101/mcs.a001859
14. Jorholt J, Formicheva Y, Vershinina T, Kiselev A, Muravyev A, Demchenko E, et al. Two new cases of hypertrophic cardiomyopathy and skeletal muscle features associated with ALPK3 homozygous and compound heterozygous variants. Genes (Basel). (2020) 11(10):1201. doi: 10.3390/genes11101201
15. Van Sligtenhorst I, Ding ZM, Shi ZZ, Read RW, Hansen G, Vogel P. Cardiomyopathy in α-kinase 3 (ALPK3)-deficient mice. Vet Pathol. (2012) 49(1):131–41. doi: 10.1177/0300985811402841
16. Hosoda T, Monzen K, Hiroi Y, Oka T, Takimoto E, Yazaki Y, et al. A novel myocyte-specific gene midori promotes the differentiation of P19CL6 cells into cardiomyocytes. J Biol Chem. (2001) 276(38):35978–89. doi: 10.1074/jbc.M100485200
17. Lopes LR, Garcia-Hernández S, Lorenzini M, Futema M, Chumakova O, Zateyshchikov D, et al. Alpha-protein kinase 3 (ALPK3) truncating variants are a cause of autosomal dominant hypertrophic cardiomyopathy. Eur Heart J. (2021) 42(32):3063–73. doi: 10.1093/eurheartj/ehab424
18. Huang C, Zheng Y, Zhang W, Chen Z, Huang Z, Fang Y. Case report: a Chinese family of hypertrophic cardiomyopathy caused by a novel splicing mutation in the FLNC gene. Front Genet. (2022) 13:894791. doi: 10.3389/fgene.2022.894791
19. Yamada T, Nomura S. Recent findings related to cardiomyopathy and genetics. Int J Mol Sci. (2021) 22(22):12522. doi: 10.3390/ijms222212522
20. Gerull B, Klaassen S, Brodehl A. The genetic landscape of cardiomyopathies. In: Erdmann J, Moretti A, editors. Genetic causes of cardiac disease. Lubeck: Springer Cham (2019). p. 45–91.
21. Walsh R, Offerhaus JA, Tadros R, Bezzina CR. Minor hypertrophic cardiomyopathy genes, major insights into the genetics of cardiomyopathies. Nat Rev Cardiol. (2022) 19(3):151–67. doi: 10.1038/s41569-021-00608-2
22. Bonaventura J, Polakova E, Vejtasova V, Veselka J. Genetic testing in patients with hypertrophic cardiomyopathy. Int J Mol Sci. (2021) 22(19):10401. doi: 10.3390/ijms221910401
23. Brodehl A, Rezazadeh S, Williams T, Munsie NM, Liedtke D, Oh T, et al. Mutations in ILK, encoding integrin-linked kinase, are associated with arrhythmogenic cardiomyopathy. Transl Res. (2019) 208:15–29. doi: 10.1016/j.trsl.2019.02.004
24. Phelan DG, Anderson DJ, Howden SE, Wong RC, Hickey PF, Pope K, et al. ALPK3-deficient cardiomyocytes generated from patient-derived induced pluripotent stem cells and mutant human embryonic stem cells display abnormal calcium handling and establish that ALPK3 deficiency underlies familial cardiomyopathy. Eur Heart J. (2016) 37(33):2586–90. doi: 10.1093/eurheartj/ehw160
25. Jaouadi H, Kraoua L, Chaker L, Atkinson A, Delague V, Levy N, et al. Novel ALPK3 mutation in a Tunisian patient with pediatric cardiomyopathy and facio-thoraco-skeletal features. J Hum Genet. (2018) 63(10):1077–82. doi: 10.1038/s10038-018-0492-1
26. Al Senaidi K, Joshi N, Al-Nabhani M, Al-Kasbi G, Al Farqani A, Al-Thihli K, et al. Phenotypic spectrum of ALPK3-related cardiomyopathy. Am J Med Genet A. (2019) 179(7):1235–40. doi: 10.1002/ajmg.a.61176
27. Herkert JC, Verhagen JMA, Yotti R, Haghighi A, Phelan DG, James PA, et al. Expanding the clinical and genetic spectrum of ALPK3 variants: phenotypes identified in pediatric cardiomyopathy patients and adults with heterozygous variants. Am Heart J. (2020) 225:108–19. doi: 10.1016/j.ahj.2020.03.023
28. Ding WW, Wang BZ, Han L, Li ZP, Zhang W, Wang H, et al. ALPK3 gene-related pediatric cardiomyopathy with craniofacial-skeletal features: a report and literature review. Zhonghua Er Ke Za Zhi. (2021) 59(9):787–92. doi: 10.3760/cma.j.cn112140-20210222-00150
Keywords: ALPK3, hypertrophic cardiomyopathy, novel variant, case report, whole-exome sequencing
Citation: Li T, Jin Y, Liu R, Hua Y, Zhou K, Luo S, Li Y and Zhang D (2023) A novel compound heterozygous variant in ALPK3 induced hypertrophic cardiomyopathy: a case report. Front. Cardiovasc. Med. 10:1212417. doi: 10.3389/fcvm.2023.1212417
Received: 26 April 2023; Accepted: 31 May 2023;
Published: 15 June 2023.
Edited by:
Junjie Xiao, Shanghai University, ChinaReviewed by:
Andreas Brodehl, Heart and Diabetes Center North Rhine-Westphalia, GermanyAhmet O. Caglayan, Dokuz Eylül University, Türkiye
© 2023 Li, Jin, Liu, Hua, Zhou, Luo, Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuhua Luo ZHJzaHVodWFsdW9AZ21haWwuY29t Yifei Li bGl5Zndjc2hAc2N1LmVkdS5jbg== Donghui Zhang ZG9uZ2guemhhbmdAaHVidS5lZHUuY24=
†These authors have contributed equally to this work
 Tiange Li
Tiange Li Yuxi Jin1,†
Yuxi Jin1,† Kaiyu Zhou
Kaiyu Zhou Yifei Li
Yifei Li Donghui Zhang
Donghui Zhang