- School of Health Sciences, Massey University, Wellington, New Zealand
Cardiovascular diseases (CVDs) are the leading cause of death worldwide. Currently, cardiovascular disease risk algorithms play a role in primary prevention. However, this is complicated by a lack of powerfully predictive biomarkers that could be observed in individuals before the onset of overt symptoms. A key potential biomarker for heart disease is the vascular endothelial growth factor (VEGF-A), a molecule that plays a pivotal role in blood vessel formation. This molecule has a complex biological role in the cardiovascular system due to the processes it influences, and its production is impacted by various CVD risk factors. Research in different populations has shown single nucleotide polymorphisms (SNPs) may affect circulating VEGF-A plasma levels, with some variants associated with the development of CVDs, as well as CVD risk factors. This minireview aims to give an overview of the VEGF family, and of the SNPs reported to influence VEGF-A levels, cardiovascular disease, and other risk factors used in CVD risk assessments.
Introduction
Cardiovascular diseases (CVDs) are defined by the World Health Organization as a group of disorders that affect the heart and blood vessels in terms of structure or blood supply (1). Notable examples of CVDs that are a leading cause of death globally include coronary heart disease (CHD), acute coronary syndrome (ACS) and congenital heart disease (2). CHD involves inadequate coronary blood supply, which may arise from a blockage in the coronary arteries usually following progressive narrowing of the lumen of atherosclerotic blood vessels (3). Given the multifactorial nature of CVDs, there are reviews available that explore in greater detail specific diseases such as coronary artery disease (CAD) (4), CHD (5), the underlying mechanism of atherosclerosis (6, 7) and the relationship of these diseases with specific variables (8, 9).
Overall, risk factors for CVDs can be grouped as modifiable or non-modifiable. The modifiable risk factors involve lifestyle circumstances that can be behavioral (diet, physical activity, exercise, smoking, alcohol) or metabolic (circulating lipid levels and glucose levels) in nature (8). Whereas, age, genetics and ethnicity of individuals are the non-modifiable risk factors. This distinction informs the diagnosis of CVDs by determining which critical variables should be included in CVD risk assessments. Critical factors employed have included age, hypercholesterolemia, high density lipoprotein (HDL) cholesterol levels, gender, smoking, diabetes, and systolic blood pressure (10).
Genetic determinants are important non-modifiable risk factors for CVDs that have been studied intensively since the early 21st century (11–13). The influence of genetic factors on CVD development was initially explored through family history studies focused on single gene disorders during the 1980 s (4, 14). Most CVDs are now considered to be polygenic disorders impacted by susceptibility and disease-linked genes, with major impacts from lifestyle and environmental factors (14). Susceptibility genes are associated with an increase or reduction in the risk of developing a disease. Comparatively, disease-linked genes are those whose expression is linked to a pathological phenotype (4). Both susceptibility and disease-linked genes can influence the regulation of other genes and/or factors that are directly involved in the pathobiology of different CVDs. The genetic basis for CVDs such as CAD and CHD has been reviewed in greater detail elsewhere (11, 15–17).
Considering this genetic complexity, numerous studies have focused on identifying associations between genetic variants and common cardiovascular disease traits (15, 18–21). This has been supported with the establishment of genome wide association studies (GWAS), which employ technologies that detect many gene variants simultaneously (22). The predominant variants identified through GWAS are single nucleotide polymorphisms (SNPs) (15, 22–24). SNPs can be located within a protein-coding region, where they may display a functional effect, but they can also be in non-coding and regulatory areas of the genome (e.g., introns, enhancer, etc.). Moreover, SNPs can play a regulatory role by impacting gene expression and protein concentration if they are located within genetic elements such as transcription factor binding sites, splicing regions, enhancer, promoter, or silencer regions (23, 25). These are often called expression quantitative trait loci (eQTLs) and explain a proportion of the genetic variance of a particular phenotype (26). SNPs can also influence coding regions located within the same loci (cis-acting) or interact with coding regions of other chromosomes or distant loci on the same chromosome (trans-acting) (27, 28). Specifically, SNP variants can influence CVD risk through traditional risk factors, such as plasma lipid levels and blood pressure (11, 27, 28). Overall, SNP variants can have several potential effects on any given gene as summarized in Figure 1. One example covered in this review is VEGFA, which impacts the cardiovascular system through angiogenesis and increased endothelial cell activity.
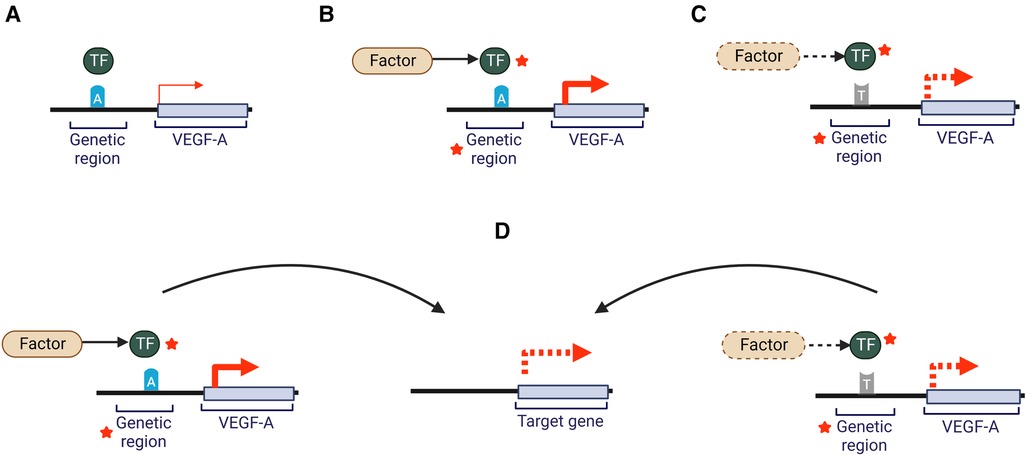
Figure 1. Potential effects of a SNP on VEGF-A gene expression. Gene regions where a SNP may be located include gene promoters, enhancers, introns, transcription factor binding motifs or non-coding RNAs. Dotted lines indicate hypothetical changes that could decrease or increase interactions or activity. Red arrows represent VEGF-A expression. Red stars indicate potential for an increase or decrease in activity through a variety of molecular mechanisms (A) an individual carrying the common allele “A” will show normally regulated levels of VEGF-A. (B) In the presence of a risk factor (e.g., elevated LDL-cholesterol plasma levels) and the common allele “A” there may be altered activity of genomic regions or transcription factors leading to altered VEGF-A expression and potentially impact circulating VEGF-A levels and/or activity. (C) Presence of a non-normal range risk factor (e.g., elevated plasma glucose levels) and the alternate allele “T”, may impact activity of genomic regions or transcription factors leading to altered VEGF-A expression and potentially influence VEGF-A levels and/or activity (D) The changes illustrated in scenario B and C may impact expression of genes located at a distance, which may impact other pathways (e.g., lipid metabolism). Adapted from (29). Created with BioRender.com.
Coupling our understanding of CVD pathogenesis with associations of regulatory SNPs with coronary biomarkers, there is potential for the combined use of CVD-relevant genetic risk scores (cvdGRS) in risk prevention (30). This involves using multiple SNPs identified from GWAS studies in different populations and these SNP variants can be associated with clinical outcomes or risk factors (30, 31). Overall, the goal of cvdGRS is to aid in patient risk stratification and treatment (22, 28, 31–33). The functional effects of the SNP variants may provide evidence to underpin a clinical framework for prevention, treatment, and in severe cases, genetic counselling in primary care (22, 31, 34, 35).
VEGF overview
A molecule of interest in the development and progression of CVDs is the vascular endothelial growth factor (VEGF-A), a member of the platelet-derived growth factor (PDGF)/VEGF family (36, 37). This growth factor is involved in blood vessel formation, with reported impacts on the development of CVDs, as well as potential recovery (38, 39). VEGF-A was originally referred to as a vascular permeability factor, with activity observed in tumor cells from rodents (40). In 1989 several research groups identified that this factor selectively promoted the migration of vascular endothelium and induced angiogenesis in vivo (41–43). Based on these findings, factors with this activity were renamed and classified as members of the VEGF family (43).
The VEGF family are glycoproteins expressed under the regulation of soluble mediators such as growth factors or cytokines (39, 44, 45). They are involved in the regulation of blood vessel formation through endothelial cell differentiation or from existing blood vessels (44, 46). Additionally, the VEGF family is involved in lymphangiogenesis, endothelial cell survival and vascular permeability regulation, amongst other functions (44, 47). However, alterations in their functionality have also been associated with the development of atherosclerosis, CHD, tumor formation, neovascularization, and other pathologies including cancer, diabetic retinopathy, preeclampsia, and endometriosis (23, 39, 47, 48).
There are five VEGF family members that directly influence the human cardiovascular system. The archetype member is VEGF-A, a potent stimulator of vasculogenesis and angiogenesis (44, 48, 49). VEGF-A production is influenced by oxygen tension, hormones (e.g., estrogen) and proinflammatory cytokines (47, 49, 50). VEGF-B induces the development of the cardiovascular system, embryonic angiogenesis and the formation of embryonic myocardium as well as participating in blood vessel survival (51). VEGF-C and VEGF-D are primarily involved in lymphangiogenesis, while the placental growth factor (PIGF) participates in both angiogenesis and wound healing (39, 47, 49).
These VEGF proteins act through one or more of three tyrosine kinase VEGF receptors (VEGFRs) found on the surface of endothelial and non-endothelial cells (44). VEGFR1 (Flt-1) and VEGFR2 (KDR) participate in angiogenesis. VEGFR2 is the primary inducer of VEGF-mediated blood vessel growth, while VEGFR3 is involved in lymphangiogenesis (47, 52, 53). Additionally, VEGFR1 has the co-receptor neuropilin-1 (NRP1), which selectively potentiates VEGFR2-mediated vascular permeability, and endothelial cell motility in vascular development (49, 54). Once activated, the signaling pathways of these receptors have the downstream effect of influencing vascular tone, blood vessel formation, endothelial cell proliferation and migration (47). VEGFR signaling is reported to also be activated in a non-VEGF-dependent manner through receptor phosphorylation due to shear stress, or recognition of alternative ligands such as lactate and low-density lipoproteins (LDLs) (36, 53, 55).
Specifically, the VEGF-A canonical pathway occurs when it binds to either VEGFR1 or VEGFR2. This promotes receptor homodimerization or heterodimerization that leads to the phosphorylation of the receptor's intracellular domains (53, 55). VEGFR1 has a soluble splice variant (sFlt-1) that acts as a decoy receptor, decreasing VEGF-A plasma concentration and limiting its binding to KDR (36, 44, 47). Also, VEGF-A activity can be potentiated when PIGF displaces it from VEGFR1 to VEGFR2 (39). These and other mechanisms surrounding the regulation of VEGF receptors have been reviewed in greater detail elsewhere (37, 55).
VEGFA gene and related SNPs in a cardiovascular context
The VEGFA gene has a 16.3 kb coding region located at 6p21.1 on the long arm of chromosome 6, including eight exons and seven introns (56, 57). The first five exons are constitutively present among VEGF-A isoforms, since they encode the signal sequence for protein processing and residues that bind to VEGF receptors (54, 58). Meanwhile, exons 6 and 7 contain the heparin binding domains that allow some isoforms to bind to cell surfaces and impact their activity or bioavailability depending on which are present (59, 60). Lastly, exon 8 undergoes post-translational readthrough due to a non-canonical stop codon, leading to the production of sub-exons 8a and 8b, with the latter being reported to be present in a unique isoform with anti-angiogenic activity observed in bone disorders and brain diseases (54, 61–63). So far, 16 distinct VEGF-A isoforms have been identified (47, 54). The different isoforms depend on the presence or absence of exons 6 and 7, which affect the affinity for heparin or heparan sulfate proteoglycans. For example, the most prevalent VEGFA isoform is VEGFA165, which lacks exon 6, but has moderate heparin affinity allowing the isoform to remain bound to cell surfaces (64). Comparatively the isoform subtype VEGFA121 lacks exon 6 and 7 so it is found only in free form (64). Despite their size difference most of the VEGF-A isoforms act as endothelial cell mitogens, upregulate the endothelial expression of adhesion molecules and present pro-angiogenic activity (36, 51, 64). Pathologies caused by increased angiogenesis include inflammatory diseases, cancers, retinopathy and atherosclerosis, while reduced angiogenesis has been observed in bone disorders and brain diseases (61). The overall VEGFA gene structure including SNPs with reported influence on VEGF-A expression levels (discussed below and in Supplementary Tables S1, S2) is presented in Figure 2.
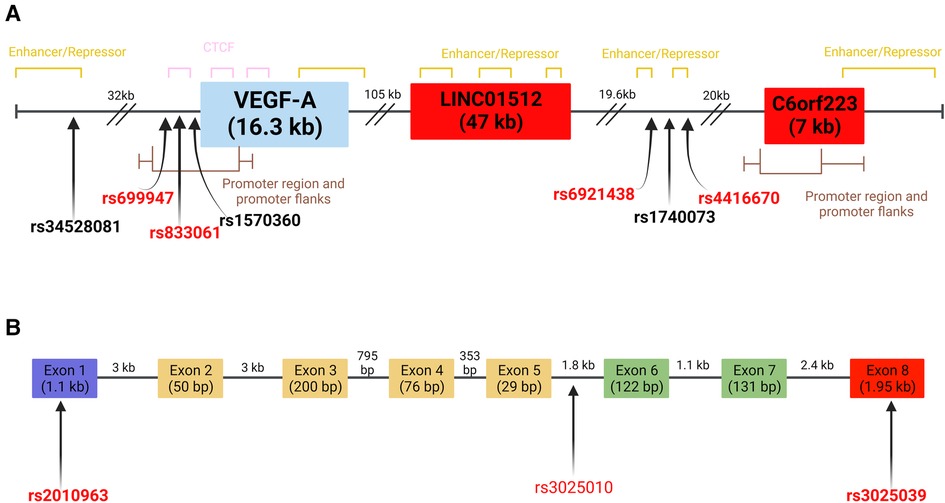
Figure 2. VEGFA gene and select SNPs on chromosome 6 (A) location of VEGF-A related SNPs at chromosome 6 loci. SNPs in bold have been reported to influence VEGF-A levels. SNPs in red have reported associations to CVD risk factors or biomarkers. Red boxes indicate regions that represent lncRNA. Full gene length is indicated individually. Distances between SNPs and genes are indicated above dashes. (B) Schematic representation of SNPs located within exons and introns of VEGFA. Individual exon and intron lengths are shown (65). The purple exon contains the signal sequence of the gene. The red exon confers anti-angiogenic capabilities when present. Green exons contain heparin-binding domains with extracellular matrix components. Yellow exons are involved in VEGF receptor binding. Location of SNPs and genomic regions determined using data from the UCSC genome browser and Ensembl databases (GRCh38-/hg38). Created with BioRender.com.
Altered plasma and tissue levels of VEGF-A have been observed in various conditions including ischemic heart disease (IHD), CAD, strokes, heart failure, and myocardial infarction (38, 66–68). Due to its impact on angiogenic processes, the effect of high VEGF-A circulating levels on CVD onset varies. High VEGF-A levels are associated with various CVD risk factors including smoking, hypercholesterolemia, diabetes, hypertension, and hyperglycemia (36). Additionally, increased VEGF-A activity has been associated with inflammation, increased blood pressure and an increase in the formation of atherosclerotic lesions, leading to CHD (20, 69–71). The impact of angiogenic molecules on atherosclerosis has been reviewed elsewhere (39).
Expression of VEGFA can be upregulated by the hypoxia inducible factor, p53 allele polymorphisms, thyroid stimulating hormone, estrogen levels and oxygen tension (45, 47). This matches studies that show VEGF-A production is influenced by elements associated with atherosclerosis including LDL concentration, hypoxia, and interleukin activity (38, 49, 72). The increased production of VEGF-A can negatively impact human health by influencing the development of atherosclerotic plaques, by affecting vascular dilation, adhesion protein expression, monocyte migration, endothelium permeability and increased trans-endothelial lipid migration (38, 39). High levels of VEGF-A in plasma have been associated with increased plaque growth and subsequent lesion vulnerability that can cause intraplaque hemorrhage (73). There is evidence that proinflammatory cytokines (e.g., IL-1, IL-6, and IL-18) present during CVD onset can enhance VEGF-A production, thus exacerbating atherosclerotic lesion development (74–76).
VEGF-A is considered a highly polymorphic gene because of the 148 untranslated region (UTR), 209 exon, 779 intron, and 124 near-gene variants that have been identified (77). At least 30 SNPs within the untranslated, exon, intron and promoter regions may have the potential to influence variation in VEGF-A expression (78, 79). This genetic influence over VEGF-A circulating levels has been explored in various studies. Debette et al. (80) investigated the heritability of VEGF-A levels in healthy individuals without a cancer diagnosis. This study identified four common variants (rs6921438, rs4416670, rs6993770 and rs10738760) distributed across three chromosomes that were independently associated with circulating VEGF-A levels and explained up to 48% of the heritability of serum VEGF-A levels (80). A meta-analysis of GWAS data evaluated the association of variants with circulating VEGF-A levels (81). Choi et al. (81) found a total of ten SNPs contributed up to 52% of the variability in circulating VEGF-A levels with some SNPs associated with increased or decreased VEGF-A levels compared to median. Additional information on the study details of SNPs identified by these groups and other studies are presented in Supplementary Tables S1, S2. The Supplementary Material also includes SNPs that have been studied in relation to VEGF-A levels in healthy individuals, CVDs, or comorbidities related to the risk of CVD (e.g., diabetes, metabolic syndrome, hypertension).
Some of the SNPs that have been studied are located within exonic regions of VEGFA (82). One noteworthy eQTL is rs2010963 from exon 1 of VEGFA (Figure 2B). The CC genotype has been associated with increased VEGF-A levels in type 2 diabetes mellitus (T2DM) (83, 84). Furthermore, the rs2010963 CC genotype has been linked to risk factors including heart rate (83), blood glucose levels (77), blood pressure, cholesterol and HDL levels (83, 85). There is also evidence for this variant influencing VEGF-A levels in non-CVDs such as glioma (86) and diabetic retinopathy (48, 87).The variant rs3025039, located within exon 8 of VEGFA, has similar effects (Figure 2B). Dong et al. (88) observed that patients diagnosed with gestational diabetes mellitus carrying the TT genotype had higher levels of VEGFA compared to healthy pregnant women (88). Meanwhile Ruggiero et al. (89) reported that the TT genotype was associated with lower median levels of VEGFA in healthy population samples from villages in Southern Italy. Some studies showed the CT genotype of rs3025039 is associated with reduced VEGFA levels as well as reducing risk of presenting with CHD and T2DM (77, 89). The associations reported for the rs3025039 variant demonstrate its link to the cardiovascular system, but the variety of findings suggest additional studies are needed. An additional variant that has shown association to the cardiovascular system is rs3025010 located in intron 5 (Figure 2B). In a Chinese cohort diagnosed with hypertension the C allele of this variant was observed to be associated with lower systolic and diastolic blood pressure measurements (20). Furthermore, in a Chinese case-control study, it was observed that the CC genotype of rs3025010 reduced risk of brain arteriovenous malformation (91). This evidence shows a clear link to CVD risk which could be further explored in additional ethnic groups to validate or identify other biomarker associations.
Other variants of interest can be found at the same loci, but outside the intron and exon regions of VEGFA (25, 80). rs69214328, is located within an enhancer region found between two long non-coding RNA genes (Figure 2A). The GWAS findings of Choi et al. (81) and Debette et al. (80) identified that the A allele of rs69214328 is associated with lower serum levels of the VEGF-A protein. Additionally, the same allele has also been reported to influence the variability of HDL and LDL in individuals of European ancestry (92). The A allele of rs6921438 appears to have eQTL activities since increased serum levels of IL-6, TNF-α and VEGF-A were observed in interaction with SNPs rs6993770 (Chr8), rs4416670 (Chr6) and rs10738760 (Chr9), respectively (93). Two additional variants (rs1740073 and rs34528081) located on chromosome 6 (Figure 2A) were identified by Choi et al. (81) to be associated with serum levels of VEGF-A (Supplementary Table S1). Furthermore, the T allele of rs34528081 was observed to be associated with increased VEGF-A serum levels in an additional GWAS study (Supplementary Table S1). Meanwhile, the T allele of rs1740073 has been reported to associate with increased VEGF-A serum in a GWAS study while analysis of IHD using 1,000 Genomes European data reported that the same allele could contribute to VEGF variance (66).
Another variant that has been studied is rs699947, which is located in the promoter region of VEGFA (Figure 2A). Various groups report that the AA genotype of rs699947 is associated with increased risk in cardiovascular pathologies including CAD, CHD, stroke and congenital heart diseases (Supplementary Table S1). The A allele of rs699947 has been associated with total cholesterol, LDL and apolipoprotein B (77, 83, 94). These associations have been observed across different ethnic groups, which further suggests rs699947 is a potential genetic risk marker for CVDs (89, 95, 96). For its part, rs833061 is another variant that is located within the promoter region of VEGFA (Figure 2A) whose CT genotype has been observed to reduce VEGF-A levels in a T2DM cohort (77). Other reports have also shown this variant is associated with hypertension and a meta-analysis of 3 cohorts implies this variant can influence congenital heart disease risk in individuals of Asian ancestry (Supplementary Table S1). A variant located further from the promoter region that presents a similar array of findings related to lipid metabolism and inflammatory molecules is rs4416670 (Figure 2A). Both its alleles have been linked to CVD risk factors and biomarkers (Supplementary Table S1). Specifically, the T allele was reported by Debette et al. (80) to increase VEGF-A serum levels while a study by Azimi-Nezhad et al. (93) reported the same allele could decrease IL-6 levels by interacting with rs6921438 (Chr6) and rs10738760 (Chr9). However, Azimi-Nezhad et al. (93) also report that the C allele of rs4416670 can increase TNF-α and IL-6 levels by interacting with the A allele of rs6921438 thus implying a link between both VEGF-A related SNPs and inflammatory molecules. Additionally, the C allele has also been observed in other studies to be associated with apolipoprotein E levels, hypertension and metabolic syndrome (92, 97). These findings demonstrate links between VEGF-A related SNPs and lipid metabolism, inflammatory biomarkers and CVD risk factors.
Some gene variants have findings of associations with molecules used in CVD risk assessment. For example, the rs1570360 variant located in the promoter region of VEGF-A (Figure 2A), was observed to contribute to an increased risk of congenital heart disease (98). Some reports showed that the GA genotype of this variant is associated with a reduced left ventricular ejection fraction and extracranial internal carotid artery (ECICA) stenosis which are both risk factors for systemic hypertension and ischemic stroke, respectively (Supplementary Table S1). However, in a Chinese study the GG genotype was observed to increase susceptibility for coronary heart disease in patients with high smoking habits and diagnosed with hypertension. As such, this variant shows consistent links to CV risk factors which, given its location, could be attributed to a potential influence on VEGF as observed in variants located within the promoter region (rs699947 and rs833061).
Similar studies have been reported for other SNPs located across the genome, often denoted as trans-acting SNPs (Supplementary Table S2). Broadly, these eQTL SNPs have been associated with increased risk of CVDs (e.g., CAD, CHD, IHD) (66, 89, 99, 100) or metabolic syndrome (81). One example rs1870377, located on chromosome 4 in exon 11 of the VEGFR2 (KDR) gene (Supplementary Figure S1) can influence cardiovascular outcomes. Li et al. (72) reported that the AA genotype reduces risk of unfavorable CVD outcomes, particularly those related with disability, in an Asian ancestry cohort. Marks et al. (99) also reported that the AA genotype associated with reduced risk of heart failure readmission and the A allele associated with high levels of VEGF system components, specifically sFlt-1 and KDR (101), and increased the risk of ischemic stroke in a Korean cohort (100). The TA and TT genotypes were both associated with increased CHD prevalence in Han Chinese populations (89, 99). Location of additional SNPs influencing VEGF-A expression levels within the VEGFR2 (KDR) gene is presented in Supplementary Figure S1. Additional associations observed for trans-acting SNPs are presented in Supplementary Table S2.
rs6993770 is located on chromosome 8 in intron 4 of the ZPFM2 gene, which codes for a protein involved in heart morphogenesis and coronary vessel development. Broadly, studies on this variant have shown relationships with VEGF-A, CVD and CVD risk factors (Supplementary Table S2). In the GWAS findings of Choi et al. (81) and the Mendelian Randomization study done by Au Yeung (66), the A allele correlated with increased VEGF-A serum levels. The GWAS findings of Debette et al. (80) showed the T allele was associated with increased VEGF-A serum levels. Other studies involving individuals of European and Iranian ancestry observed the T allele was also associated with risk biomarkers of CVD, particularly fasting blood glucose, triglyceride levels, systolic blood pressure and HDL levels (92, 102). The TA genotype has been reported to increase the risk of metabolic syndrome (102), and impacts the expression of adhesion molecules (ICAM-1, E-selectin) as well as IL-6 levels (93). Meanwhile, the TT genotype appears to contribute to metabolic syndrome risk in individuals with low iron intake (97). This spectrum of reports demonstrates the range of associations that alleles and genotypes of trans-acting SNPs, such as rs1870377 or rs6993770, may have within the cardiovascular system. Additional trans-acting SNPs (e.g., rs2071559, rs114694170, rs6993770, rs10738760, rs10761741, rs4782371 have been reported to be capable of influencing VEGF-A circulating levels (81) or soluble VEGFR levels (rs1870377) (101). Specific study details and overall findings are presented in Supplementary Table S2. Notably, two SNPs (rs2305948 and rs7667298) have associations with potential CVD risk, but their direct impact on VEGF system components was observed in cancer related studies (103, 104). Interestingly, trans-acting SNPs most likely involve interactions with molecules or homeostatic mechanisms that have known roles in CVD onset, including inflammatory interleukins (70, 93), triglycerides, adhesion molecules, blood cell count and blood pressure (66, 102, 105, 106). There are cases of specific variants that correlate with increased risk of presenting major adverse coronary events (rs2305948, rs7667298) (106), CHD (rs2305948, rs1870377, rs2071559, rs7667298) (89, 99), ischemic stroke (rs1870377) (100) and metabolic syndrome (rs6993770) (102). As such, some SNPs appear to be potential contributors to phenotypes (IHD, CAD, CHD) while others may increase or reduce disease risk depending on the presence or absence of risk factors (72, 89, 101).
Conclusion
Overall, the impact of VEGF-A related SNPs in various forms of heart disease has been explored in many different types of studies. The collective evidence reveals a critical subset of cis-acting SNPs mapping to the region of VEGFA (Figure 2 and Supplementary Table S1), several trans-acting SNPs mapping in the region of the VEGFR2 gene (Supplementary Figure S1) and elsewhere on the human genome (Supplementary Table S2), with repeatable associations with circulating levels of VEGF-A. A small group of SNPs reproducibly associate with established biomarkers and risk factors for heart disease (rs2010963, rs3025039, rs1570360, rs699947, rs6921438) or with increased susceptibility to common heart disease pathologies (rs2010963, rs3025039, rs1570360, rs699947, rs2305948, rs1870377). This minireview highlights that these SNPs can be potential markers for CVDs and may influence significant biological pathways that impact the cardiovascular system (e.g., lipid metabolism). The wide range of pathologies that VEGF-A and its related SNPs impact emphasizes the complexity of VEGF-A interactions within the cardiovascular system. Both cis- and trans-acting SNP eQTLs can affect expression levels, but there remain many unknowns around the specific mechanisms involved. There is a clear link between SNPs and VEGF-A levels as well as established cardiovascular disease biomarkers (HDL, LDL, BNP, NTproBNP). Together these have the potential to act synergistically on the development of CVDs.
The complexity of SNP influences on CVD and CVD risk factors reinforces the importance of studying them in relation to VEGF-A. Particularly considering how altered levels of VEGF-A contribute to disease onset or exacerbate an individual's health depending on the risk factors they present with. Exploring the link between CVDs, SNPs, and VEGF-A may contribute to improved cardiovascular disease risk assessment, prevention, treatment, and. prognosis.
Author contributions
JCMA, BP, and RP: conceived the concept of the mini review. JM-A: canvassed the literature and drafted the manuscript. JM-A, BP, BM, CB, and RP: refined and extended the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2023.1190513/full#supplementary-material.
References
1. World Health Organization. Cardiovascular diseases (CVDs) (2021). Available at: https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (Accessed June 11, 2021).
2. Joseph P, Leong D, McKee M, Anand SS, Schwalm J-D, Teo K, et al. Reducing the global burden of cardiovascular disease, part 1. Circ Res. (2017) 121(6):677–94. doi: 10.1161/CIRCRESAHA.117.308903
3. Sanchis-Gomar F, Perez-Quilis C, Leischik R, Lucia A. Epidemiology of coronary heart disease and acute coronary syndrome. Ann Transl Med. (2016) 4(13):256. doi: 10.21037/atm.2016.06.33
4. Malakar AK, Choudhury D, Halder B, Paul P, Uddin A, Chakraborty S. A review on coronary artery disease, its risk factors, and therapeutics. J Cell Physiol. (2019) 234(10):16812–23. doi: 10.1002/jcp.28350
5. Khamis RY, Ammari T, Mikhail GW. Gender differences in coronary heart disease. Heart. (2016) 102(14):1142–9. doi: 10.1136/heartjnl-2014-306463
6. Libby P. The changing landscape of atherosclerosis. Nature. (2021) 592(7855):524–33. doi: 10.1038/s41586-021-03392-8
7. Libby P, Buring JE, Badimon L, Hansson GK, Deanfield J, Bittencourt MS, et al. Atherosclerosis. Nat Rev Dis Primers. (2019) 5(1):56. doi: 10.1038/s41572-019-0106-z
8. Akyuz A. Exercise and coronary heart disease. Adv Exp Med Biol. (2020) 1228:169–79. doi: 10.1007/978-981-15-1792-1_11
9. Hartman J, Frishman WH. Inflammation and atherosclerosis. Cardiol Rev. (2014) 22(3):147–51. doi: 10.1097/CRD.0000000000000021
10. Damen JAAG, Hooft L, Schuit E, Debray TPA, Collins GS, Tzoulaki I, et al. Prediction models for cardiovascular disease risk in the general population: systematic review. Br Med J. (2016) 353:i2416. doi: 10.1136/bmj.i2416
11. Assimes TL, Roberts R. Genetics: implications for prevention and management of coronary artery disease. J Am Coll Cardiol. (2016) 68(25):2797–818. doi: 10.1016/j.jacc.2016.10.039
12. Roberts R. Genetics-current and future role in the prevention and management of coronary artery disease. Curr Atheroscler Rep. (2016) 18(12):78. doi: 10.1007/s11883-016-0628-8
13. Roberts R. A breakthrough in genetics and its relevance to prevention of coronary artery disease in LMIC. Glob Heart. (2017) 12(3):247–57. doi: 10.1016/j.gheart.2017.04.001
14. Roberts R, Fair J. Genetics, its role in preventing the pandemic of coronary artery disease. Clin Cardiol. (2021) 44(6):771–9. doi: 10.1002/clc.23627
15. McPherson R, Tybjaerg-Hansen A. Genetics of coronary artery disease. Circ Res. (2016) 118(4):564–78. doi: 10.1161/CIRCRESAHA.115.306566
16. Khera AV, Emdin CA, Drake I, Natarajan P, Bick AG, Cook NR, et al. Genetic risk, adherence to a healthy lifestyle, and coronary disease. N Engl J Med. (2016) 375(24):2349–58. doi: 10.1056/NEJMoa1605086
17. Roberts R. A genetic basis for coronary artery disease. Trends Cardiovasc Med. (2015) 25(3):171–8. doi: 10.1016/j.tcm.2014.10.008
18. LeBlanc M, Zuber V, Andreassen BK, Witoelar A, Zeng L, Bettella F, et al. Identifying novel gene variants in coronary artery disease and shared genes with several cardiovascular risk factors. Circ Res. (2016) 118(1):83–94. doi: 10.1161/CIRCRESAHA.115.306629
19. Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, et al. Genomic atlas of the human plasma proteome. Nature. (2018) 558(7708):73–9. doi: 10.1038/s41586-018-0175-2
20. Zhao Z, Gong C, Gao Y, Liu X, Wu S, Zhao H, et al. Association between single nucleotide polymorphisms in cardiovascular developmental critical genes and hypertension: a propensity score matching analysis. Int J Hypertens. (2020) 2020:1–8. doi: 10.1155/2020/9185697
21. Earle N, Yeo Han D, Pilbrow A, Crawford J, Smith W, Shelling AN, et al. Single nucleotide polymorphisms in arrhythmia genes modify the risk of cardiac events and sudden death in long QT syndrome. Heart Rhythm. (2014) 11(1):76–82. doi: 10.1016/j.hrthm.2013.10.005
22. Roberts R, Chang CC, Hadley T. Genetic risk stratification: a paradigm shift in prevention of coronary artery disease. JACC: Basic Transl Sci. (2021) 6(3):287–304. doi: 10.1016/j.jacbts.2020.09.004
23. Geiger M. Fundamentals of Vascular Bology. Learning Materials in Biosciences. Vienna, Austria: Springer (2019).
24. Zhang X, Johnson AD, Hendricks AE, Hwang SJ, Tanriverdi K, Ganesh SK, et al. Genetic associations with expression for genes implicated in GWAS studies for atherosclerotic cardiovascular disease and blood phenotypes. Hum Mol Genet. (2014) 23(3):782–95. doi: 10.1093/hmg/ddt461
25. Buroker NE. VEGFA rSNPs, transcriptional factor binding sites and human disease. J Physiol Sci. (2014) 64(1):73–6. doi: 10.1007/s12576-013-0293-4
26. Nica AC, Dermitzakis ET. Expression quantitative trait loci: present and future. Philos Trans R Soc Lond B Biol Sci. (2013) 368(1620):20120362. doi: 10.1098/rstb.2012.0362
27. Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. (2012) 489(7414):57–74. doi: 10.1038/nature11247
28. Roberts R, Campillo A, Schmitt M. Prediction and management of CAD risk based on genetic stratification. Trends Cardiovasc Med. (2020) 30(6):328–34. doi: 10.1016/j.tcm.2019.08.006
29. Wang K, Basu M, Malin J, Hannenhalli S. A transcription-centric model of SNP-age interaction. PLoS Genet. (2021) 17(3):e1009427. doi: 10.1371/journal.pgen.1009427
30. Yun H, Noh NI, Lee EY. Genetic risk scores used in cardiovascular disease prediction models: a systematic review. RCM. (2022) 23(1):1–9. doi: 10.31083/j.rcm2301008
31. O'Sullivan J, Raghavan S, Marquez-Luna C, Luzum JA, Damrauer SM, Ashley EA, et al. Polygenic risk scores for cardiovascular disease: a scientific statement from the American heart association. Circulation. (2022) 146(8):e93–e118. doi: 10.1161/CIR.0000000000001077
32. Gladding PA, Legget M, Fatkin D, Larsen P, Doughty R. Polygenic risk scores in coronary artery disease and atrial fibrillation. Heart Lung Circ. (2020) 29(4):634–40. doi: 10.1016/j.hlc.2019.12.004
33. Roberts R, Campillo A. Genetic stratification for primary prevention of coronary artery disease. Curr Opin Cardiol. (2018) 33(5):529–34. doi: 10.1097/HCO.0000000000000542
34. Knowles JW, Ashley EA. Cardiovascular disease: the rise of the genetic risk score. PLoS Med. (2018) 15(3):e1002546. doi: 10.1371/journal.pmed.1002546
35. Rotter JI, Lin HJ. An outbreak of polygenic scores for coronary artery disease. J Am Coll Cardiol. (2020) 75(22):2781–4. doi: 10.1016/j.jacc.2020.04.054
36. Smith GA, Fearnley GW, Harrison MA, Tomlinson DC, Wheatcroft SB, Ponnambalam S. Vascular endothelial growth factors: multitasking functionality in metabolism, health and disease. J Inherit Metab Dis. (2015) 38(4):753–63. doi: 10.1007/s10545-015-9838-4
37. Shibuya M. Vascular endothelial growth factor and its receptor system: physiological functions in angiogenesis and pathological roles in various diseases. J Biochem. (2013) 153(1):13–9. doi: 10.1093/jb/mvs136
38. Braile M, Marcella S, Cristinziano L, Galdiero MR, Modestino L, Ferrara AL, et al. VEGF-A in cardiomyocytes and heart diseases. Int J Mol Sci. (2020) 21(15):1–18. doi: 10.3390/ijms21155294
39. Camaré C, Pucelle M, Nègre-Salvayre A, Salvayre R. Angiogenesis in the atherosclerotic plaque. Redox Biol. (2017) 12:18–34. doi: 10.1016/j.redox.2017.01.007
40. Senger DR, Galli SJ, Dvorak AM, Perruzzi CA, Harvey VS, Dvorak HF. Tumor cells secrete a vascular permeability factor that promotes accumulation of ascites fluid. Science. (1983) 219(4587):983–5. doi: 10.1126/science.6823562
41. Keck PJ, Hauser SD, Krivi G, Sanzo K, Warren T, Feder J, et al. Vascular permeability factor, an endothelial cell mitogen related to PDGF. Science. (1989) 246(4935):1309–12. doi: 10.1126/science.2479987
42. Connolly DT, Heuvelman DM, Nelson R, Olander JV, Eppley BL, Delfino JJ, et al. Tumor vascular permeability factor stimulates endothelial cell growth and angiogenesis. J Clin Invest. (1989) 84(5):1470–8. doi: 10.1172/JCI114322
43. Leung DW, Cachianes G, Kuang W-J, Goeddel DV, Ferrara N. Vascular endothelial growth factor is a secreted angiogenic mitogen. Science. (1989) 246(4935):1306–9. doi: 10.1126/science.2479986
44. Niklander S, Bordagaray MJ, Fernandez A, Hernandez M. Vascular endothelial growth factor: a translational view in oral non-communicable diseases. Biomolecules. (2021) 11(1):1–27. doi: 10.3390/biom11010085
45. Singh P, Singh M, Khinda R, Valecha S, Kumar N, Singh S, et al. Genetic scores of eNOS, ACE and VEGFA genes are predictive of endothelial dysfunction associated osteoporosis in postmenopausal women. Int J Environ Res Public Health. (2021) 18(3):1–18. doi: 10.3390/ijerph18030972
46. Holmes DI, Zachary I. The vascular endothelial growth factor (VEGF) family: angiogenic factors in health and disease. Genome Biol. (2005) 6(2):209. doi: 10.1186/gb-2005-6-2-209
47. Shaik F, Cuthbert GA, Homer-Vanniasinkam S, Muench SP, Ponnambalam S, Harrison MA. Structural basis for vascular endothelial growth factor receptor activation and implications for disease therapy. Biomolecules. (2020) 10(12):1–25. doi: 10.3390/biom10121673
48. Hu L, Gong C, Chen X, Zhou H, Yan J, Hong W. Associations between vascular endothelial growth factor gene polymorphisms and different types of diabetic retinopathy susceptibility: a systematic review and meta-analysis. J Diabetes Res. (2021) 2021:7059139. doi: 10.1155/2021/7059139
49. Le THV, Kwon SM. Vascular endothelial growth factor biology and its potential as a therapeutic target in rheumatic diseases. Int J Mol Sci. (2021) 22(10):1–17. doi: 10.3390/ijms22105387
50. Gu H, Chen W, Yin J, Chen S, Zhang J, Gong J. Methionine sulfoxide reductase a rs10903323 G/A polymorphism is associated with increased risk of coronary artery disease in a Chinese population. Clin Biochem. (2013) 46(16-17):1668–72. doi: 10.1016/j.clinbiochem.2013.07.011
51. Zhou Y, Zhu X, Cui H, Shi J, Yuan G, Shi S, et al. The role of the VEGF family in coronary heart disease. Front Cardiovasc Med. (2021) 8:1–16. doi: 10.3389/fcvm.2021.738325
52. Oszajca K, Szemraj J, Wyrzykowski D, Chrzanowska B, Salamon A, Przewratil P. Single-nucleotide polymorphisms of VEGF-A and VEGFR-2 genes and risk of infantile hemangioma. Int J Dermatol. (2018) 57(10):1201–7. doi: 10.1111/ijd.14127
53. Apte RS, Chen DS, Ferrara N. VEGF in signaling and disease: beyond discovery and development. Cell. (2019) 176(6):1248–64. doi: 10.1016/j.cell.2019.01.021
54. Peach CJ, Mignone VW, Arruda MA, Alcobia DC, Hill SJ, Kilpatrick LE, et al. Molecular pharmacology of VEGF-A isoforms: binding and signalling at VEGFR2. Int J Mol Sci. (2018) 19(4):1264. doi: 10.3390/ijms19041264
55. Simons M, Gordon E, Claesson-Welsh L. Mechanisms and regulation of endothelial VEGF receptor signalling. Nat Rev Mol Cell Biol. (2016) 17(10):611–25. doi: 10.1038/nrm.2016.87
56. Pagès G, Pouysségur J. Transcriptional regulation of the vascular endothelial growth factor gene–a concert of activating factors. Cardiovasc Res. (2005) 65(3):564–73. doi: 10.1016/j.cardiores.2004.09.032
57. Tischer E, Mitchell R, Hartman T, Silva M, Gospodarowicz D, Fiddes JC, et al. The human gene for vascular endothelial growth factor. Multiple protein forms are encoded through alternative exon splicing. J Biol Chem. (1991) 266(18):11947–54. doi: 10.1016/S0021-9258(18)99049-6
58. Keyt BA, Berleau LT, Nguyen HV, Chen H, Heinsohn H, Vandlen R, et al. The carboxyl-terminal domain (111–165) of vascular endothelial growth factor is critical for its mitogenic potency. J Biol Chem. (1996) 271(13):7788–95. doi: 10.1074/jbc.271.13.7788
59. Krilleke D, DeErkenez A, Schubert W, Giri I, Robinson GS, Ng Y-S, et al. Molecular mapping and functional characterization of the VEGF164 heparin-binding domain. J Biol Chem. (2007) 282(38):28045–56. doi: 10.1074/jbc.M700319200
60. Stevens M, Oltean S. Modulation of receptor tyrosine kinase activity through alternative splicing of ligands and receptors in the VEGF-A/VEGFR axis. Cells. (2019) 8(4):288. doi: 10.3390/cells8040288
61. Dehghanian F, Hojati Z, Kay M. New insights into VEGF-A alternative splicing: key regulatory switching in the pathological process. Avicenna J Med Biotechnol. (2014) 6(4):192–9. PMID: 25414781
62. Eswarappa SM, Potdar AA, Koch WJ, Fan Y, Vasu K, Lindner D, et al. Programmed translational readthrough generates antiangiogenic VEGF-ax. Cell. (2014) 157(7):1605–18. doi: 10.1016/j.cell.2014.04.033
63. Bridgett S, Dellett M, Simpson DA. RNA-sequencing data supports the existence of novel VEGFA splicing events but not of VEGFAxxxb isoforms. Sci Rep. (2017) 7(1):58. doi: 10.1038/s41598-017-00100-3
64. Al Kawas H, Saaid I, Jank P, Westhoff CC, Denkert C, Pross T, et al. How VEGF-A and its splice variants affect breast cancer development–clinical implications. Cell Oncol. (2022) 45(2):227–39. doi: 10.1007/s13402-022-00665-w
65. Arcondéguy T, Lacazette E, Millevoi S, Prats H, Touriol C. VEGF-A mRNA processing, stability and translation: a paradigm for intricate regulation of gene expression at the post-transcriptional level. Nucleic Acids Res. (2013) 41(17):7997–8010. doi: 10.1093/nar/gkt539
66. Au Yeung SL, Lam H, Schooling CM. Vascular endothelial growth factor and ischemic heart disease risk: a Mendelian randomization study. J Am Heart Assoc. (2017) 6(8):1–8. doi: 10.1161/JAHA.117.005619
67. Zhang Y, Ma T, Hu H, Wang J, Zhou S. Serum vascular endothelial growth factor as a biomarker for prognosis of minor ischemic stroke. Clin Neurol Neurosurg. (2020) 196:106060. doi: 10.1016/j.clineuro.2020.106060
68. Rodríguez-Rodríguez L, García-Bermúdez M, Martín J, Fernández-Gutierrez B, González-Juanatey C, Vazquez-Rodriguez TR, et al. Vascular endothelial growth factor a and cardiovascular disease in rheumatoid arthritis patients. Tissue Antigens. (2011) 77(4):291–7. doi: 10.1111/j.1399-0039.2010.01625.x
69. Heinonen SE, Kivelä AM, Huusko J, Dijkstra MH, Gurzeler E, Mäkinen PI, et al. The effects of VEGF-A on atherosclerosis, lipoprotein profile, and lipoprotein lipase in hyperlipidaemic mouse models. Cardiovasc Res. (2013) 99(4):716–23. doi: 10.1093/cvr/cvt148
70. Ruotsalainen SE, Partanen JJ, Cichonska A, Lin J, Benner C, Surakka I, et al. An expanded analysis framework for multivariate GWAS connects inflammatory biomarkers to functional variants and disease. Eur J Hum Genet. (2021) 29(2):309–24. doi: 10.1038/s41431-020-00730-8
71. Zhao X, Meng L, Jiang J, Wu X. Vascular endothelial growth factor gene polymorphisms and coronary heart disease: a systematic review and meta-analysis. Growth Factors. (2018) 36(3-4):153–63. doi: 10.1080/08977194.2018.1477141
72. Li Z, Wang M, Gu J, Zhao L, Guo Y, Zhang Z, et al. Missense variants in hypoxia-induced VEGFA/VEGFR2 signaling predict the outcome of large artery atherosclerotic stroke. Cell Mol Neurobiol. (2021) 41(6):1217–25. doi: 10.1007/s10571-020-00890-7
73. Michel J-B, Virmani R, Arbustini E, Pasterkamp G. Intraplaque haemorrhages as the trigger of plaque vulnerability. Eur Heart J. (2011) 32(16):1977–85. doi: 10.1093/eurheartj/ehr054
74. Mitrokhin V, Shim A, Aksyonov A, Zotov A, Konev A, Ovchinnikov R, et al. Circulating interleukin-18: association with IL-8, IL-10 and VEGF serum levels in patients with and without heart rhythm disorders. Int J Cardiol. (2016) 215:105–9. doi: 10.1016/j.ijcard.2016.04.002
75. Tzeng HE, Tsai CH, Chang ZL, Su CM, Wang SW, Hwang WL, et al. Interleukin-6 induces vascular endothelial growth factor expression and promotes angiogenesis through apoptosis signal-regulating kinase 1 in human osteosarcoma. Biochem Pharmacol. (2013) 85(4):531–40. doi: 10.1016/j.bcp.2012.11.021
76. Fahey E, Doyle SL. IL-1 family cytokine regulation of vascular permeability and angiogenesis. Front Immunol. (2019) 10:1–15. doi: 10.3389/fimmu.2019.01426
77. Sellami N, Lamine LB, Turki A, Sarray S, Jailani M, Al-Ansari AK, et al. Association of VEGFA variants with altered VEGF secretion and type 2 diabetes: a case-control study. Cytokine. (2018) 106:29–34. doi: 10.1016/j.cyto.2018.03.003
78. Stevens A, Soden J, Brenchley PE, Ralph S, Ray DW. Haplotype analysis of the polymorphic human vascular endothelial growth factor gene promoter. Cancer Res. (2003) 63(4):812–6. PMID: 12591731
79. Kim Y-J, Chung WC, Jun K-H, Chin H-M. Genetic polymorphisms of vascular endothelial growth factor (VEGF) associated with gastric cancer recurrence after curative resection with adjuvant chemotherapy. BMC Cancer. (2019) 19(1):483. doi: 10.1186/s12885-019-5702-5
80. Debette S, Visvikis-Siest S, Chen MH, Ndiaye NC, Song C, Destefano A, et al. Identification of cis- and trans-acting genetic variants explaining up to half the variation in circulating vascular endothelial growth factor levels. Circ Res. (2011) 109(5):554–63. doi: 10.1161/CIRCRESAHA.111.243790
81. Choi SH, Ruggiero D, Sorice R, Song C, Nutile T, Vernon Smith A, et al. Six novel loci associated with circulating VEGF levels identified by a meta-analysis of genome-wide association studies. PLoS Genet. (2016) 12(2):e1005874. doi: 10.1371/journal.pgen.1005874
82. Cui QT, Li Y, Duan CH, Zhang W, Guo XL. Further evidence for the contribution of the vascular endothelial growth factor gene in coronary artery disease susceptibility. Gene. (2013) 521(2):217–21. doi: 10.1016/j.gene.2013.03.091
83. Skrypnik D, Mostowska A, Jagodzinski PP, Bogdanski P. Association of rs699947 (−2578 C/A) and rs2010963 (−634 G/C) single nucleotide polymorphisms of the VEGF gene, VEGF-A and leptin serum level, and cardiovascular risk in patients with excess body mass: a case-control study. J Clin Med. (2020) 9(2):1–16. doi: 10.3390/jcm9020469
84. Merlo S, Starcevic JN, Mankoc S, Letonja MS, Vujkovac AC, Zorc M, et al. Vascular endothelial growth factor gene polymorphism (rs2010963) and its receptor, kinase insert domain-containing receptor gene polymorphism (rs2071559), and markers of carotid atherosclerosis in patients with type 2 diabetes mellitus. J Diabetes Res. (2016) 2016:1–6. doi: 10.1155/2016/1482194.
85. Totomoch-Serra A, de Lourdes Muñoz M, Burgueño J, Revilla-Monsalve MC, Diaz-Badillo A, Muñoz MdL Association of common polymorphisms in the and genes with type 2 diabetes-related traits in Mexicans. Arch Med Sci. (2018) 14(6):1361–73. doi: 10.5114/aoms.2018.74757
86. Linhares P, Viana-Pereira M, Ferreira M, Amorim J, Nabico R, Pinto F, et al. Genetic variants of vascular endothelial growth factor predict risk and survival of gliomas. Tumour Biol. (2018) 40(3):1010428318766273. doi: 10.1177/1010428318766273
87. Petrovič MG, Korošec P, Košnik M, Osredkar J, Hawlina M, Peterlin B, et al. Local and genetic determinants of vascular endothelial growth factor expression in advanced proliferative diabetic retinopathy. Mol Vision. (2008) 14:1382–7. PMID: 18682813
88. Dong P-P. Association of vascular endothelial growth factor expression and polymorphisms with the risk of gestational diabetes mellitus. J Clin Lab Anal. (2019) 33(2):e22686. doi: 10.1002/jcla.22686
89. Ruggiero D, Dalmasso C, Nutile T, Sorice R, Dionisi L, Aversano M, et al. Genetics of VEGF serum variation in human isolated populations of cilento: importance of VEGF polymorphisms. PLoS One. (2011) 6(2):e16982. doi: 10.1371/journal.pone.0016982
90. Liu D, Song J, Ji X, Liu Z, Cong M, Hu B. Association of genetic polymorphisms on VEGFA and VEGFR2 with risk of coronary heart disease. Medicine. (2016) 95(19):e3413. doi: 10.1097/MD.0000000000003413
91. Chen H, Gu Y, Wu W, Chen D, Li P, Fan W, et al. Polymorphisms of the vascular endothelial growth factor a gene and susceptibility to sporadic brain arteriovenous malformation in a Chinese population. J Clin Neurosci. (2011) 18(4):549–53. doi: 10.1016/j.jocn.2010.08.025
92. Stathopoulou MG, Bonnefond A, Ndiaye NC, Azimi-Nezhad M, El Shamieh S, Saleh A, et al. A common variant highly associated with plasma VEGFA levels also contributes to the variation of both LDL-C and HDL-C. J Lipid Res. (2013) 54(2):535–41. doi: 10.1194/jlr.P030551
93. Azimi-Nezhad M, Stathopoulou MG, Bonnefond A, Rancier M, Saleh A, Lamont J, et al. Associations of vascular endothelial growth factor (VEGF) with adhesion and inflammation molecules in a healthy population. Cytokine. (2013) 61(2):602–7. doi: 10.1016/j.cyto.2012.10.024
94. Yadav BK, Yadav R, Chang H, Choi K, Kim JT, Park MS, et al. Genetic polymorphisms rs699947, rs1570360, and rs3025039 on the VEGF gene are correlated with extracranial internal carotid artery stenosis and ischemic stroke. Ann Clin Lab Sci. (2017) 47(2):144–55. PMID: 28442515
95. Ma W-Q, Wang Y, Han X-Q, Zhu Y, Liu N-F. Association of genetic polymorphisms in vascular endothelial growth factor with susceptibility to coronary artery disease: a meta–analysis. BMC Med Genet. (2018) 19(1):1–12. doi: 10.1186/s12881-018-0628-3
96. Palmer BR, Paterson MA, Frampton CM, Pilbrow AP, Skelton L, Pemberton CJ, et al. Vascular endothelial growth factor-A promoter polymorphisms, circulating VEGF-A and survival in acute coronary syndromes. PLoS One. (2021) 16(7):1–5. doi: 10.1371/journal.pone.0254206
97. Hoseini Z, Ghayour-Mobarhan M, Avan A, Nematy M, Safarian M, Ferns GA, et al. VEGF gene polymorphism interactions with dietary trace elements intake in determining the risk of metabolic syndrome. J Cell Biochem. (2019) 120(2):1398–406. doi: 10.1002/jcb.27171
98. Wang W, Xu A, Xu H. The roles of vascular endothelial growth factor gene polymorphisms in congenital heart diseases: a meta-analysis. Growth Factors. (2018) 36(5-6):232–8. doi: 10.1080/08977194.2018.1513505
99. Li L, Pan Y, Dai L, Liu B, Zhang D. Association of genetic polymorphisms on vascular endothelial growth factor and its receptor genes with susceptibility to coronary heart disease. Med Sci Monit. (2016) 22:31–40. doi: 10.12659/MSM.895163
100. Oh S-H, Min K-T, Jeon Y-J, Kim M-H, Moon J-S, Kim H-S, et al. Association between kinase insert domain-containing receptor gene polymorphism and haplotypes and ischemic stroke. J Neurol Sci. (2011) 308(1):62–6. doi: 10.1016/j.jns.2011.06.012
101. Marks ECA, Wilkinson TM, Frampton CM, Skelton L, Pilbrow AP, Yandle TG, et al. Plasma levels of soluble VEGF receptor isoforms, circulating pterins and VEGF system SNPs as prognostic biomarkers in patients with acute coronary syndromes. BMC Cardiovasc Disord. (2018) 18(1):169. doi: 10.1186/s12872-018-0894-1
102. Azimi-Nezhad M, Mirhafez SR, Stathopoulou MG, Murray H, Ndiaye NC, Bahrami A, et al. The relationship between vascular endothelial growth factor cis- and trans-acting genetic variants and metabolic syndrome. Am J Med Sci. (2018) 355(6):559–65. doi: 10.1016/j.amjms.2018.03.009
103. Góra-Tybor J, Szemraj J, Robak T, Jamroziak K Clinical relevance of vascular endothelial growth factor type A (VEGFA) and VEGF receptor type 2 (VEGFR2) gene polymorphism in chronic lymphocytic leukemia. Blood Cells Mol Dis. (2015) 54(2):139–43. doi: 10.1016/j.bcmd.2014.11.022
104. Mafu T, September A, Shamley D. KDR inferred haplotype is associated with upper limb dysfunction in breast cancer survivors of mixed ancestry. Cancer Manag Res. (2019) 11:3829–45. doi: 10.2147/CMAR.S191969
105. Salami A, El Shamieh S. Association between SNPs of circulating vascular endothelial growth factor levels, hypercholesterolemia and metabolic syndrome. Medicina. (2019) 55(8):1–9. doi: 10.3390/medicina55080464
Keywords: vascular endothelial growth factor, single nucleotide polymorphism, cardiovascular disease, VEGF-A eQTLs, genetic association
Citation: Meza-Alvarado JC, Page RA, Mallard B, Bromhead C and Palmer BR (2023) VEGF-A related SNPs: a cardiovascular context. Front. Cardiovasc. Med. 10:1190513. doi: 10.3389/fcvm.2023.1190513
Received: 29 March 2023; Accepted: 27 April 2023;
Published: 23 May 2023.
Edited by:
Paul Cahill, Dublin City University, IrelandReviewed by:
Sadie Slater, University of Bristol, United Kingdom© 2023 Meza-Alvarado, Page, Mallard, Bromhead and Palmer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: B. R. Palmer Yi5wYWxtZXJAbWFzc2V5LmFjLm56
 J. C. Meza-Alvarado
J. C. Meza-Alvarado R. A. Page
R. A. Page B. Mallard
B. Mallard C. Bromhead
C. Bromhead B. R. Palmer
B. R. Palmer