
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cardiovasc. Med. , 20 June 2023
Sec. Cardio-Oncology
Volume 10 - 2023 | https://doi.org/10.3389/fcvm.2023.1181806
 Marwa Tantawy1
Marwa Tantawy1 Guang Yang2
Guang Yang2 Raghunandan Reddy Algubelli3
Raghunandan Reddy Algubelli3 Gabriel DeAvila3
Gabriel DeAvila3 Samuel M. Rubinstein4
Samuel M. Rubinstein4 Robert F. Cornell5
Robert F. Cornell5 Michael G. Fradley6
Michael G. Fradley6 Erin M. Siegel7
Erin M. Siegel7 Oliver A. Hampton8
Oliver A. Hampton8 Ariosto S. Silva9
Ariosto S. Silva9 Daniel Lenihan10
Daniel Lenihan10 Kenneth H. Shain3
Kenneth H. Shain3 Rachid C. Baz3
Rachid C. Baz3 Yan Gong1,11*
Yan Gong1,11*
Background: Proteasome inhibitor Carfilzomib (CFZ) is effective in treating patients with refractory or relapsed multiple myeloma (MM) but has been associated with cardiovascular adverse events (CVAE) such as hypertension, cardiomyopathy, and heart failure. This study aimed to investigate the contribution of germline genetic variants in protein-coding genes in CFZ-CVAE among MM patients using whole-exome sequencing (WES) analysis.
Methods: Exome-wide single-variant association analysis, gene-based analysis, and rare variant analyses were performed on 603,920 variants in 247 patients with MM who have been treated with CFZ and enrolled in the Oncology Research Information Exchange Network (ORIEN) at the Moffitt Cancer Center. Separate analyses were performed in European Americans and African Americans followed by a trans-ethnic meta-analysis.
Results: The most significant variant in the exome-wide single variant analysis was a missense variant rs7148 in the thymosin beta-10/TraB Domain Containing 2A (TMSB10/TRABD2A) locus. The effect allele of rs7148 was associated with a higher risk of CVAE [odds ratio (OR) = 9.3 with a 95% confidence interval of 3.9—22.3, p = 5.42*10−7]. MM patients with rs7148 AG or AA genotype had a higher risk of CVAE (50%) than those with GG genotype (10%). rs7148 is an expression quantitative trait locus (eQTL) for TRABD2A and TMSB10. The gene-based analysis also showed TRABD2A as the most significant gene associated with CFZ-CVAE (p = 1.06*10−6).
Conclusions: We identified a missense SNP rs7148 in the TMSB10/TRABD2A as associated with CFZ-CVAE in MM patients. More investigation is needed to understand the underlying mechanisms of these associations.
Multiple Myeloma (MM) is a malignancy of the plasma cells. According to yearly incidence rates and prevalence figures, it ranks third among hematologic cancers in the United States (1). Proteasome inhibitors (PIs) are one of the most effective drugs for the treatment of MM and are the backbone therapies for MM treatment (2). Three PIs have been approved by the United States Food and Drug Administration: bortezomib, carfilzomib, and ixazomib (3). Carfilzomib (Kyprolis®) is a second-generation, irreversible PI approved for treating relapsed and refractory MM due to its survival benefit and overall response rate in refractory MM patients (4, 5). The National Comprehensive Cancer Network (NCCN) Guideline recommends carfilzomib-based therapy for newly diagnosed MM patients with pre-existing neuropathy or high-risk patients and patients with relapsed and refractory disease (6). Despite its effectiveness, carfilzomib (CFZ) has been shown to have significant cardiovascular adverse events (CVAE) (20%–25%), including 7.2% incident HF, in the clinical trials that excluded patients with pre-existing cardiovascular disorders (7). In the previous two meta-analyses studies, CFZ was associated with a high incidence of CVAE (8%–18%) (2, 3) including hypertension, heart failure, cardiomyopathy, and arrhythmia (8, 9). The rate of CVAE was even higher (∼50%) in an observational study when MM patients with pre-existing cardiovascular conditions were not excluded (10). A severe clinical implication developed from this cardiotoxicity is treatment interruption, which could lead to disease progression (11).
Studies have shown that early detection of and early intervention for cardiotoxicity induced by other therapies (i.e., anthracyclines, Trastuzumab) can improve long-term outcomes (12, 13). Therefore, stratifying patients before the carfilzomib treatment might provide opportunities for early intervention to optimize patient outcomes. Pharmacogenomics, or the identification of genetic determinants of drug response and adverse effects, is a tool that has been useful in individualizing medication therapy (14, 15). This study aims to identify germline genetic variants in protein-coding genes associated with CFZ-CVAE in MM patients using whole-exome sequencing (WES) analysis.
Patients included in this study were admitted to the Moffitt Cancer Center's Total Cancer Care (TCC) Protocol with IRB approval (MCC#14690; Advarra IRB Pro00014441) (16). Patients consented to contribute blood specimens and medical information for research purposes in collaboration with the Oncology Research Information Exchange Network (ORIEN), a network of seventeen cancer centers that have agreed to implement a common TCC biospecimen collection protocol to follow patients throughout their lifetime (17). Clinical and epidemiological data were collected for select TCC-consented patients, and the molecular data were produced as described below. This study included a total of 247 patients who have been diagnosed with MM and treated with carfilzomib at the H. Lee Moffitt Cancer Center and have germline DNA WES data available through ORIEN. This study was also approved by the University of Florida Institutional Review Board (IRB202003031).
We queried the electronic health records data of Moffitt's health research informatics platform to determine if a CVAE had occurred after initiation of carfilzomib treatment. We used the International Classification of Disease (ICD) revisions 9 and 10 to define cardiovascular events. A complete list of the ICD-9 and -10 codes used is listed in the supplementary materials Supplementary Table S1. Moffitt's Pentecost Myeloma Research Center clinical database was utilized to identify if the eligible TCC patients receiving carfilzomib as part of their treatment had one of these cardiac events between the start and end date. We performed a manual chart review on 10% of patients to verify the accuracy of the billing records in terms of the definition of the CVAE. All records reviewed for cardiovascular adverse events matched the determination by the billing codes.
Germline DNA was extracted from peripheral blood samples with buffy coat using the QIAsymphony SP instrument (QIAGEN, Hilden, Germany) following standard protocols implemented across the ORIEN. The WES of germline DNA was performed for each patient using SeqCap EZ Exome Enrichment Kit v3.0 (Roche NimbleGen, Pleasanton, CA) or xGen Exome Hybridization Panel with supplement probes (integrated Data Technologies, Inc., Coralville, IA), with 100 × coverage. Capture kits covered variants for limited regions; each captured library was loaded onto Illumina-HiSeq 4,000 (Illumina, San Diego, CA). Over 26,000 protein-coding genes were sequenced. The raw sequencing data underwent a rigorous analysis pipeline for alignment, variant calling, quality control steps, and annotation algorithms (18–20). Before the genetic association analysis, the WES data underwent additional quality control steps: variant call rate > 95%, sample call rate > 95%, sex check, and Hardy Weinberg Equilibrium analysis. Principle component analysis was performed on a subset of variants after more stringent quality control steps [variant rate and sample call rate > 99% and minor allele frequency (MAF) >10%] to evaluate the genetic ancestry of these patients.
Germline WES data was available on 605,446 variants in 247 patients. Of the 247 patients, 228 were genetically clustered with individuals of European ancestry, and 19 were clustered with African ancestry. A total of 603,920 variants passed the quality control steps and were included in the WES analysis.
All association analyses were performed in genetically clustered European Americans and African Americans separately, adjusting for age, gender, and principal components for ancestry. Trans-ethnic meta-analyses were then performed to combine the results from both groups.
Exome-wide association analysis of single variants with a MAF ≥ 1% was performed to estimate the odds ratio (OR) and 95% confidence interval (CI) for each variant on chromosomes 1–22 for the development of CFZ-CVAE using multivariable logistic regression assuming an additive model of inheritance using PLINK (21). All variants with p < 5*10−8 were considered statistical significance. Variants with p < 5*10−4 were considered suggestive (22).
Following the exome-wide association analysis, the summary statistics were functionally annotated using Functional Mapping and Annotation of Genome-Wide Association Studies (FUMA GWAS) for a gene-based and gene set analysis to recognize potential genes of interest (23). The Genotype-Tissue Expression (GTEx) database was queried to identify tissue-specific gene expression and regulation. ANNOVAR (24) was used to annotate the genetic variants appropriately.
Rare variants analysis was performed using the sequence kernel association test (SKAT) using the SKAT package (25, 26) to evaluate the association of the joint effect of multiple rare variants (MAF < 1%) with CFZ-CVAE.
Functional assignment and pathway analysis of the association results was performed on the top variants from the WES analyses (p < 0.001). IPA uses a network generation algorithm to create multiple networks and uses hypergeometric distribution to create scores for each network (27). The statistical significance level is generated using Fisher's Exact test. Any pathway enriched by genes more than by chance would be statistically significant.
Overall, 247 MM patients were included in the analysis. The mean age was ∼59 years, and 57% were men. Thirty-eight (15.5%) developed CVAE after initiation and during carfilzomib-based therapy. Table 1 summarizes the characteristics of the 38 patients who developed CVAE and the 209 who did not. The baseline demographics and medical history were similar between the two groups of patients. A total of 228 (92.3%) patients were genetically clustered with individuals of European ancestry (EA), and 19 clustered with individuals of African ancestry (AA). The baseline characteristics and medical history of the 228 EA MM patients were summarized in Supplementary Table S2.
The results of the exome-wide association analysis of the common variants in the EA patients are summarized in the Manhattan plot (Figure 1A) and the QQ plot (Figure 1B). None of the variants were genome-wide significant. However, eleven SNPs from five loci reached the suggestive significance level with p < 5*10−4 (Table 2). The top SNP rs7148 is a missense variant in the thymosin beta-10 (TMSB10) gene, with OR of 9.33% and 95% CI of 3.90 – 22.35 (p = 5.42*10−07) (Table 2, Figure 2). The minor allele frequency rs7148 was ∼7% in EA patients. Amongst patients with the rs7148 variant, 50% (99/198) of patients with the AG or AA genotype developed a CVAE compared with 10.1% (20/198) with the GG genotype (p < 0.0001). GTEx analysis revealed that the rs7148 A allele was associated with higher TraB Domain Containing 2A (TRABD2A) gene expression in the left ventricle tissue with a p-value 1.9 × 10−7 (Supplementary Figure S1). The second most significant SNP, rs12471929, is an intronic variant in the TMSB10 gene that is in perfect linkage disequilibrium (LD) (r2 = 1, D'=1) with rs7148, with OR of 8.95 (3.78–21.18), p = 6.17*10−7 (Table 2, Figure 2). A few other SNPs in LD with rs7148 in the 1,000 genomes database are shown in Supplementary Table S3.
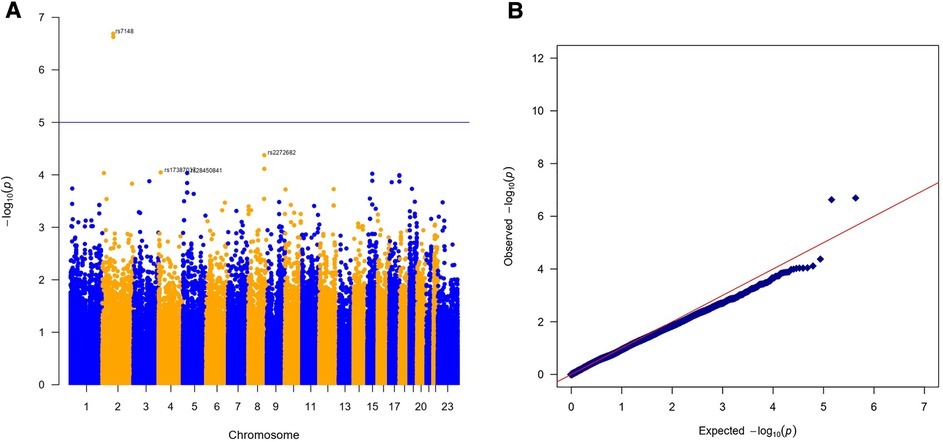
Figure 1. Results of exome-wide association analysis in European Americans. (A) Manhattan plot representing the association between single-nucleotide polymorphism (SNP) genotype and CVAE in MM patients. (B) The quantile-quantile (QQ) plot of association results.
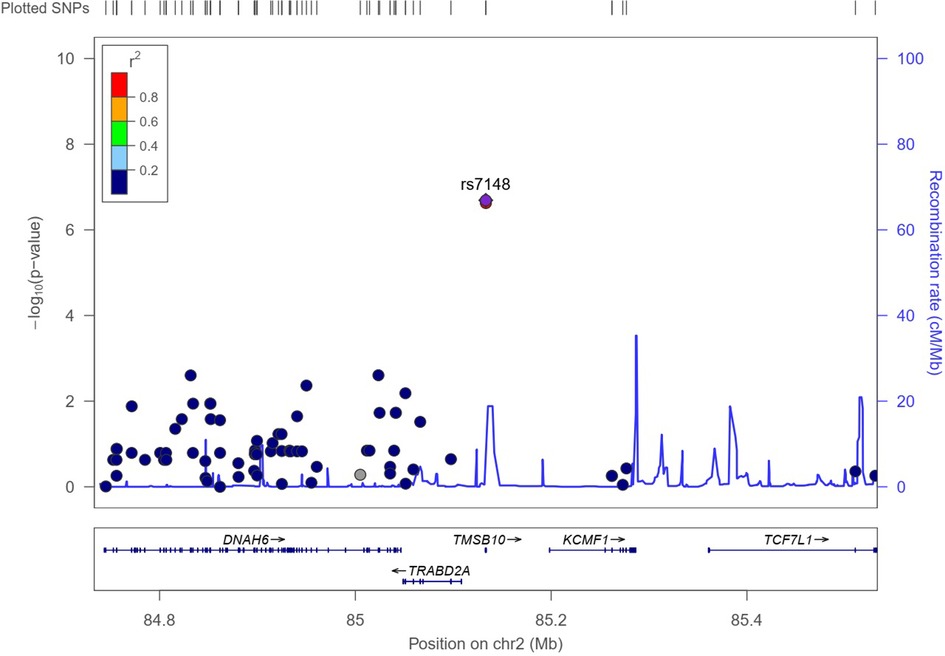
Figure 2. Regional association plot for variant rs7148. The X-axis represents a 1Mb region, 500 kb on either side of the variant, and the y-axis shows –the log10 P-value for individual SNPs. Pairwise LD (r2) with the variant is based on 1,000 Genome phase 3 v5 European reference samples and described using the color scale in the bar. The bottom panel shows the genes located within the region.
Among the other SNPs with a suggestive level of significance were: five SNPs in the PDZD2 gene, which encodes PDZ Domain Containing Protein 2; an intronic variant in the Prominin 1, CD133 (PROM1) gene; a synonymous variant on GREB1 (Growth Regulating Estrogen Receptor Binding 1) gene on Chromosome 2, and two variants in WASHC5/SQLE gene on Chromosome 8 (Table 2). The minor allele frequencies, allele counts, and Hardy Weinberg Equilibrium test results by CVAE status are summarized in Supplementary Table S4.
We also performed an exploratory analysis on AA patients. No SNPs reached statistical significance as a result of the small sample size. The four SNPs with nominal significance (p < 0.05) are shown in Supplementary Table S5. The two most frequent SNPs in EA (rs7148, rs1247192) were not observed in AA patients (MAF = 0).
The top SNPs in the trans-ethnic meta-analysis combing EA and AA are listed in Table 3. Only two of these eleven SNPs were observed in AA. Therefore, the results of these SNPs in the meta-analysis were almost identical to those in the EA analysis. The only top EA SNPs observed in AA were the Chromosome 8 SNPs in the WASCH5/SQLE locus. While these two SNPs had minor allele frequencies of 6%–9% in EA, the frequencies were much higher in AA (42%–45%). The directions of associations with CVAE were consistent in AA patients compared to those in EA patients (Table 3).
The gene-based analysis in EA patients using FUMA revealed that the TRABD2A gene that encodes TraB Domain Containing 2A is significantly associated with CFZ-CVAE (p = 1.06*10−6) (Figure 3).
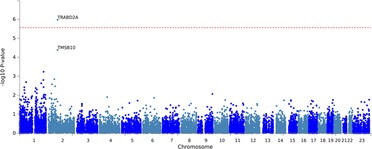
Figure 3. The gene-based association testing results from the FUMA analysis of CFZ-CVAE yielded TRABD2A as a significant gene (p = 1.06*10−6).
SKAT analysis was performed on 40,969 gene sets and 620,661 SNPs, and a significant association was determined by comparing CFZ-CVAE after correcting for multiple testing. The genes—Chromosome 1 Open Reading Frame 116 (C1orf116), LOC102724084 (DYNLRB2 antisense RNA1), Diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2 (NM_0013)), and Transmembrane Protein 183A (TMEM183A) were statistically significantly associated with CFZ-CVAE (p-value = 1.1*10−5, 4.1*10−5, 6.2*10−5, and 6.2*10−5, respectively) (Supplementary Table S6).
Using IPA, the pathway enrichment analysis showed that the lowest p-value and most significant genes overlapped with cardiotoxicity. The functional toxicity annotation of genes related to cardiotoxicity implicated cardiac arteriopathy with three variants from WES results: rs3750765 located in leucine-rich repeat-containing 20 (LRRC20) gene, rs72713436 in sterile alpha motif domain-containing 4A (SAMD4A) gene, and rs75454001 in CUB-Sushi multiple domains 1 (CSMD1) gene. All of these genes were associated with human coronary artery disease (P < 0.001) (28).
In this first genetic association analysis of CFZ-related CVAE in MM patients, we conducted a WES of germline DNA samples from patients who received CFZ in the ORIEN network. In this retrospective cohort study of patients in a real-world clinical setting, we identified a missense SNP rs7148 in the TMSB10/TRABD2A gene locus on chromosome 2 to be associated with a higher risk for CVAE in MM patients treated with CFZ.
In order to reflect real-world practices, all MM patients treated with CFZ were included in this study regardless of their cardiovascular disease history. We found that 15.4% of the MM patients treated with CFZ developed CVAE, which is in line with the event rates of 8%–18% reported in prior meta-analyses (2, 3). Due to the retrospective nature of this study and a lack of clinical guidelines on proteome inhibitor monitoring at the time of this study, these patients were not actively monitored for cardiotoxicity by cardiologists. Not surprisingly, the event rate we observed is lower than the CFZ-CVAE event rate of 50% reported in a prospective study, the Prospective Observation of Cardiac Safety with Proteasome Inhibitor (PROTECT) study (10). The recently published 2022 Cardio-Oncology guideline by the European Society of Cardiology (29) recommends testing and surveillance of MM patients at baseline and every cycle during the first 6 cycles under proteasome inhibitor treatment based on the risk. The guideline recommends first stratifying MM patients into low, moderate, high, or very high-risk groups based on clinical factors prior to the treatment. Considering genetic variants may improve the risk stratification of these patients.
To our knowledge, this is the first genetic association study of CFZ-CVAE in humans. We found that the variant carriers of missense SNP rs7148 in the TMSB10 gene were at higher risk for CFZ-CVAE. We also identified an intronic variant rs12471929 which is in perfect LD with rs7148 in EA. Both of these SNPs are also eQTLs for the nearby gene TRABD2A in that the variant alleles are associated with higher expression of TRABD2A in the left ventricle heart tissue (30).
Thymosins are a family of small peptides initially identified from the thymus and consist of three groups, alpha-, beta-, and gamma-thymosins, according to their isoelectric points (31). The beta-thymosins (TMSB4, TMSB10, TMSB15), found in the cytoplasm, interact with G-actin and produces a large pool of actin monomers (32). TMSB10 has been reported to function in cytoskeleton organization, cell morphology, proliferation, motility, and interaction with Ras and angiogenesis, cell growth, and apoptosis (33). The two isoforms, TMSB10 and TMSB4, are identified as significant actin monomer sequestering proteins that may regulate actin filament assembly (34). A recent study showed that the actin-binding protein TMSB10 was upregulated in dysfunctional endothelial dysfunction in acute myocardial infarction (AMI), and endothelial dysfunction is considered one of the primary factors in the progression of atherothrombosis in AMI (35). Another study on mice found that TMSB10 can inhibit vascular endothelial growth factor (VEGF) expression by inhibiting the Ras-ERK signaling pathway leading to the suppression of vascular formation, especially in tumor formation (36).
TRABD2A gene enables Wnt-protein binding activity and metalloendopeptidase activity (37). A recent genome-wide association study on cardiac troponin T (cTnT) in large cohorts identified a genetic variant rs548487604 near the TRABD2A gene to be associated with the elevation of cTnT level (38), which is related to the incidence of cardiovascular diseases, cardiovascular death, and heart failure in a general population (39, 40).
A previous study on mice proposed a pathway associated with CFZ-CVAE. This study found that treatment with CFZ induced the apoptosis pathway by activation of PP2A (protein phosphatase 2A), which inactivates AMPKα and the downstream signaling related to autophagy phosphoinositide 3′-kinase (PI3K)-Akt-endothelial nitric oxide synthase (eNOS) (PI3K/Akt/eNOS) axis. This axis is responsible for myocardial cell growth and survival and plays a vital role the cardiac dysfunction (41). Previous studies have identified a specific subtype of hematopoietic stem cells in peripheral blood called endothelial progenitor cells (EPCs) that express numerous combinations of antigens associated with hematopoietic stem and endothelial cells and play an essential role in neovascularization of ischemic tissue and reversal of endothelial dysfunction (42, 43), and in vivo studies showed that TMSB4 increases EPC migration and decreases EPC apoptosis under serum deprivation via the (PI3K/Akt/eNOS) signal transduction pathway (44, 45), and several studies showed that the telomerase length and telomerase activity of circulating EPCs and decreased in patients with coronary artery disease (46, 47). In a recent study that included 48 patients with relapsed/refractory MM and received CFZ, the brachial artery flow-mediated dilation (FMD) and 26s proteasome activity were detected to evaluate the endothelial function. This study concluded that patients who received CFZ and with low potential for proteasome activity recovery may suffer from both acute and long-term endothelial dysfunction (48).
Endothelial cell homeostasis depends on the ubiquitin-proteasome system, which induces oxidative stress in the cells and regulates the expression of endothelial nitric oxide synthase (49). The proteasome inhibitor CFZ causes the plasma of cancer cells, cardiomyocytes, and endothelial cells to accumulate with unfolded, dysfunctional proteins, which may lead to impaired vasodilation, excessive oxidative stress, inflammation, cell apoptosis, and autophagy (50, 51). Other studies have shown that the endothelial dysfunction caused by CFZ's inhibition of proteasome activity may result in CVAE or other endothelial dysfunction-related events like hypertension, heart failure, and coronary artery disease (51–53). In light of the literature, our finding of the genetic variants in the TMSB10/TRABD2A locus appears to support the role of endothelial dysfunction in CFZ-CVAE.
Among the other SNPs with a suggestive significance level, five are located on the PDZ domain-containing protein 2 (PDZD2) gene, which contains six PDZ domains and shares sequence similarity with pro-interleukin-16 (pro-IL-16). SNPs in the PDZD2 gene have been associated with heart rate in heart failure patients with reduced ejection fraction (54). PROM1 has a role in cell differentiation, proliferation, and apoptosis. PR1P is a small peptide derived from the extracellular domain of PROM1-derived peptide and improves cardiac function following ischemia. SQLE encodes squalene epoxidase, which catalyzes the first oxygenation step in sterol biosynthesis and is one of the rate-limiting enzymes in this pathway.
It is important to recognize some limitations of our study. Firstly, the patient population is predominantly European Americans. The sample size of patients of African descent was too small to have enough statistical power for any meaningful discovery. Further investigation is required to explore this phenotype and outcomes in MM patients of African ancestry. Secondly, using WES means we may have missed critical genetic variants outside the coding regions of the genome. Thirdly, using ICD codes to identify CVAE has its limitations. Even though our manual chart review of 10% of patients indicated 100% of CVAE were confirmed, it would have been ideal to review all the charts to confirm CVAE status. Fourthly, due to the small sample size, we had to combine all the CVAEs. Lastly, our study findings need to be replicated in an independent study before these genetic variants can be incorporated into the risk stratification of MM patients.
In summary, in this WES study of MM patients in a real-world clinical setting, we identified a missense variant in the TMSB10/TRABD2A locus to be associated with CFZ-CVAE among MM patients. Once validated, this association could provide the basis for a Precision Medicine approach to identify MM patients at high risk for CFZ-CVAE.
The data presented in the study are deposited in the database of Genotypes and Phenotypes (dbGaP) repository, accession number phs003308.v1.p1.
The studies involving human participants were reviewed and approved by the University of Florida Institutional Review Board (IRB202003031). The patients/participants provided their written informed consent to participate in this study.
MT and YG wrote the manuscript, and RA extracted the data from ORIEN, RCB manually reviewed the charts to validate the CVAE. GD, EMS, and KHS directed sample identification and contributed to the TCC and ORIEN project. MT, GY, and YG performed statistical analyses. YG secured the fund for this study. All authors contributed to the article and approved the submitted version.
Part of the funding for this study came from the NIH/NHLBI under grant number R01HL151659 and the UF CTSA under award number UL1TR001427. In addition to the Total Cancer Care Protocol, the H Lee Moffitt Cancer Center & Research Institute's Collaborative Data Services Core Facility, Tissue Core Facility, Molecular Genomics Core Facility, and Biostatistics and Bioinformatics Shared Resource were also involved in this research; an NCI designated Comprehensive Cancer Center (P30-CA076292). The WES data delivered by the ORIEN Avatar Project is managed and funded partially by M2Gen®, which received partial funding from third-party partners for data generated in the Avatar Project. This work was also supported by the Pentecost Family Myeloma Research Center (PMRC) at the H. Lee Moffitt Cancer Center & Research Institute.
An earlier version of this study was presented in part as a poster presentation at the American Society of Clinical Pharmacology and Therapeutics (ASCPT) 2022 Virtual meeting (55).
MGF: research funding from Medtronic; Advisory board/consulting fees: Abbott, AstraZeneca, Myovant, Takeda, and Zoll. RCB: research funding from Janssen, Karyopharm, BMS, and Abbvie; Advisory board member for Janssen, Karyopharm, BMS, Shattuck labs, GSK, Pfizer, and Abbvie. RFC: Employed at AbbVie and stock shareholder. SMR: Advisory board member for Janssen, Sanofi, Roche Diagnostics, and EUSA pharma.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2023.1181806/full#supplementary-material
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. (2022) 72:7–33. doi: 10.3322/caac.21708
2. Gandolfi S, Laubach JP, Hideshima T, Chauhan D, Anderson KC, Richardson PG. The proteasome and proteasome inhibitors in multiple myeloma. Cancer Metastasis Rev. (2017) 36:561–84. doi: 10.1007/s10555-017-9707-8
3. Teicher BA, Tomaszewski JE. Competitive landscape report. Biochem Pharmacol. (2015) 96:1–9. doi: 10.1016/j.bcp.2015.04.008
4. Hasinoff BB, Patel D, Wu X. Molecular mechanisms of the cardiotoxicity of the proteasomal-targeted drugs bortezomib and carfilzomib. Cardiovasc Toxicol. (2017) 17:237–50. doi: 10.1007/s12012-016-9378-7
5. Dimopoulos MA, Goldschmidt H, Niesvizky R, Joshua D, Chng WJ, Oriol A, et al. Carfilzomib or bortezomib in relapsed or refractory multiple myeloma (ENDEAVOR): an interim overall survival analysis of an open-label, randomised, phase 3 trial. Lancet Oncol. (2017) 18:1327–37. doi: 10.1016/S1470-2045(17)30578-8
6. Callander NS, Baljevic M, Adekola K, Anderson LD, Campagnaro E, Castillo JJ, et al. NCCN Guidelines® Insights: Multiple Myeloma, Version 3.2022. J Natl Compr Canc Netw. (2022) 20(1):8–19. doi: 10.6004/jnccn.2022.0002
7. Siegel D, Martin T, Nooka A, Harvey RD, Vij R, Niesvizky R, et al. Integrated safety profile of single-agent carfilzomib: experience from 526 patients enrolled in 4 phase II clinical studies. Haematologica. (2013) 98(11):1753–61. doi: 10.3324/haematol.2013.089334
8. Kistler KD, Kalman J, Sahni G, Murphy B, Werther W, Rajangam K, et al. Incidence and risk of cardiac events in patients with previously treated multiple myeloma versus matched patients without multiple myeloma: an observational, retrospective, cohort study. Clin Lymphoma Myeloma Leuk. (2017) 17:89–96.e3. doi: 10.1016/j.clml.2016.11.009
9. Atrash S, Tullos A, Panozzo S, Bhutani M, Van Rhee F, Barlogie B, et al. Cardiac complications in relapsed and refractory multiple myeloma patients treated with carfilzomib. Blood Cancer J. (2015) 5:e272. doi: 10.1038/bcj.2014.93
10. Cornell RF, Ky B, Weiss BM, Dahm CN, Gupta DK, Du L, et al. Prospective study of cardiac events during proteasome inhibitor therapy for relapsed multiple myeloma. J Clin Oncol. (2019) 37:1946–55. doi: 10.1200/JCO.19.00231
11. Yu AF, Yadav NU, Lung BY, Eaton AA, Thaler HT, Hudis CA, et al. Trastuzumab interruption and treatment-induced cardiotoxicity in early HER2-positive breast cancer. Breast Cancer Res Treat. (2015) 149:489–95. doi: 10.1007/s10549-014-3253-7
12. Koutsoukis A, Ntalianis A, Repasos E, Kastritis E, Dimopoulos MA, Paraskevaidis I. Cardio-oncology: a focus on cardiotoxicity. European Cardiology Review. (2018) 13:64–9. doi: 10.15420/ecr.2017:17:2
13. Henry ML, Niu J, Zhang N, Giordano SH, Chavez-MacGregor M. Cardiotoxicity and cardiac monitoring among chemotherapy-treated breast cancer patients. JACC Cardiovasc Imaging. (2018) 11:1084–93. doi: 10.1016/j.jcmg.2018.06.005
14. Shuldiner AR, O’Connell JR, Bliden KP, Gandhi A, Ryan K, Horenstein RB, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. (2009) 302:849–57. doi: 10.1001/jama.2009.1232
15. Krynetski EY, Tai HL, Yates CR, Fessing MY, Loennechen T, Schuetz JD, et al. Genetic polymorphism of thiopurine S-methyltransferase: clinical importance and molecular mechanisms. Pharmacogenetics. (1996) 6:279–90. doi: 10.1097/00008571-199608000-00001
16. Fenstermacher DA, Wenham RM, Rollison DE, Dalton WS. Implementing personalized medicine in a cancer center. Cancer J. (2011) 17:528–36. doi: 10.1097/PPO.0b013e318238216e
17. Dalton WS, Sullivan D, Ecsedy J, Caligiuri MA. Patient enrichment for precision-based cancer clinical trials: using prospective cohort surveillance as an approach to improve clinical trials. Clin Pharmacol Ther. (2018) 104:23–6. doi: 10.1002/cpt.1051
18. Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics (2011) 27:2987–93. doi: 10.1093/bioinformatics/btr509
19. Depristo MA, Banks E, Poplin R, Garimella K V, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. (2011) 43:491–501. doi: 10.1038/ng.806
20. Van der Auwera GA, Carneiro MO, Hartl C. Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. (2013) 43(1110):11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43
21. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. (2007) 81:559–75. doi: 10.1086/519795
22. Fadista J, Manning AK, Florez JC, Groop L. The (in)famous GWAS P-value threshold revisited and updated for low-frequency variants. Eur J Hum Genet. (2016) 24:1202–5. doi: 10.1038/ejhg.2015.269
23. Watanabe K, Taskesen E, Van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. (2017) 8:1826. doi: 10.1038/s41467-017-01261-5
24. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. (2010) 38:e164. doi: 10.1093/NAR/GKQ603
25. Lee S, Abecasis GR, Boehnke M, Lin X. Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet. (2014) 95:5–23. doi: 10.1016/j.ajhg.2014.06.009
26. Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. (2011) 89:82–93. doi: 10.1016/j.ajhg.2011.05.029
27. Krämer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. (2014) 30(4):523–30. doi: 10.1093/bioinformatics/btt703
28. Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A, et al. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. (2007) 447:661–78. doi: 10.1038/nature05911
29. Lyon AR, López-Fernández T, Couch LS, Asteggiano R, Aznar MC, Bergler-Klein J, et al. 2022 ESC guidelines on cardio-oncology developed in collaboration with the European hematology association (EHA), the European society for therapeutic radiology and oncology (ESTRO) and the international cardio-oncology society (IC-OS). Eur Heart J. (2022) 43:4229–361. doi: 10.1093/eurheartj/ehac244
30. Carithers LJ, Ardlie K, Barcus M, Branton PA, Britton A, Buia SA, et al. A novel approach to high-quality postmortem tissue procurement: the GTEx project. Biopreserv Biobank. (2015) 13:311–7. doi: 10.1089/bio.2015.0032
31. Santelli G, Califano D, Chiappetta G, Vento MT, Bartoli PC, Zullo F, et al. Thymosin beta-10 gene overexpression is a general event in human carcinogenesis. Am J Pathol. (1999) 155:799–804. doi: 10.1016/s0002-9440(10)65178-4
32. Chen C, Li M, Yang H, Chai H, Fisher W, Yao Q. Roles of thymosins in cancers and other organ systems. World J Surg. (2005) 29:264–70. doi: 10.1007/S00268-004-7817-2
33. Sribenja S, Li M, Wongkham S, Wongkham C, Yao Q, Chen C. Cancer investigation advances in thymosin β10 research: differential expression, molecular mechanisms, and clinical implications in cancer and other conditions advances in thymosin β10 research: differential expression, molecular mechanisms, and clinical implications in cancer and other conditions. Cancer Invest. (2009) 27:1016–22. doi: 10.3109/07357900902849640
34. Yu FX, Lin SC, Morrison-Bogorad M, Yin HL. Effects of thymosin beta 4 and thymosin beta 10 on actin structures in living cells. Cell Motil Cytoskeleton. (1994) 27:13–25. doi: 10.1002/cm.970270103
35. Nukala SB, Regazzoni L, Aldini G, Zodda E, Tura-Ceide O, Mills NL, et al. Differentially expressed proteins in primary endothelial cells derived from patients with acute myocardial infarction. Hypertension. (2019) 74:947–56. doi: 10.1161/HYPERTENSIONAHA.119.13472
36. Lee SH, Son MJ, Oh SH, Rho SB, Park K, Kim YJ, Park MS, Lee JH. Thymosin β10 inhibits angiogenesis and tumor growth by interfering with ras function. Cancer Res (2005) 65:137–47. doi: 10.1158/0008-5472.137.65.1
37. Liu M, Dong J, Ouyang J, Zhao L, Liang G, Shang H. Metalloprotease TRABD2A restriction of HIV-1 production in monocyte-derived dendritic cells. AIDS Res Hum Retroviruses. (2019) 35:887–9. doi: 10.1089/AID.2019.0140
38. Nasu T, Satoh M, Hachiya T, Sutoh Y, Ohmomo H, Hitomi S, et al. A genome-wide association study for highly sensitive cardiac troponin T levels identified a novel genetic variation near a RBAK-ZNF890P locus in the Japanese general population. Int J Cardiol. (2021) 329:186–91. doi: 10.1016/j.ijcard.2020.12.019
39. Welsh P, Preiss D, Hayward C, Shah AS, McAllister D, Briggs A, et al. Cardiac troponin T and troponin I in the general population. Circulation. (2019) 139:2754–64. doi: 10.1161/CIRCULATIONAHA.118.038529
40. Saunders JT, Nambi V, de Lemos JA, Chambless LE, Virani SS, Boerwinkle E, et al. Cardiac troponin T measured by a highly sensitive assay predicts coronary heart disease, heart failure, and mortality in the atherosclerosis risk in communities study. Circulation. (2011) 123:1367–76. doi: 10.1161/CIRCULATIONAHA.110.005264
41. Efentakis P, Kremastiotis G, Varela A, Nikolaou PE, Papanagnou ED, Davos CH, et al. Molecular mechanisms of carfilzomib-induced cardiotoxicity in mice and the emerging cardioprotective role of metformin. Blood. (2019) 133:710–23. doi: 10.1182/blood-2018-06-858415
42. Shi Q, Rafii S, Wu Hong-De M, Wijelath ES, Yu C, Ishida A, et al. Evidence for circulating bone marrow-derived endothelial cells. Blood. (1998) 92:362–7. doi: 10.1182/blood.v92.2.362
43. Asahara T, Murohara T, Sullivan A, Silver M, Van Der Zee R, Li T, et al. Isolation of putative progenitor endothelial cells for angiogenesis. Science. (1997) 275:964–7. doi: 10.1126/science.275.5302.964
44. Zhao Y, Qiu F, Xu S, Yu L, Fu G. Thymosin β4 activates integrin-linked kinase and decreases endothelial progenitor cells apoptosis under serum deprivation. J Cell Physiol. (2011) 226:2798–806. doi: 10.1002/jcp.22624
45. Qiu FY, Song XX, Zheng H, Zhao YB, Fu GS. Thymosin β4 induces endothelial progenitor cell migration via PI3K/akt/eNOS signal transduction pathway. J Cardiovasc Pharmacol. (2009) 53:209–14. doi: 10.1097/FJC.0b013e318199f326
46. Murasawa S, Llevadot J, Silver M, Isner JM, Losordo DW, Asahara T. Constitutive human telomerase reverse transcriptase expression enhances regenerative properties of endothelial progenitor cells. Circulation. (2002) 106:1133–9. doi: 10.1161/01.CIR.0000027584.85865.B4
47. Vasa M, Fichtlscherer S, Aicher A, Adler K, Urbich C, Martin H, et al. Number and migratory activity of circulating endothelial progenitor cells inversely correlate with risk factors for coronary artery disease. Circ Res. (2001) 89:E1–7. doi: 10.1161/hh1301.093953
48. Kastritis E, Laina A, Georgiopoulos G, Gavriatopoulou M, Papanagnou E-D, Eleutherakis-Papaiakovou E, et al. Carfilzomib-induced endothelial dysfunction, recovery of proteasome activity, and prediction of cardiovascular complications: a prospective study. Leukemia. (2021) 35:1418–27. doi: 10.1038/s41375-021-01141-4
49. de Carvalho JE R, Verwoert MT, Vogels IMC, Reits EA, van Noorden CJF, Klaassen I, et al. Involvement of the ubiquitin-proteasome system in the expression of extracellular matrix genes in retinal pigment epithelial cells. Biochem Biophys Rep. (2018) 13:83–92. doi: 10.1016/j.bbrep.2018.01.005
50. Meiners S, Ludwig A, Stangl V, Stangl K. Proteasome inhibitors: poisons and remedies. Med Res Rev. (2008) 28:309–27. doi: 10.1002/med.20111
51. Brandes RP. Endothelial dysfunction and hypertension. Hypertension. (2014) 64:924–8. doi: 10.1161/HYPERTENSIONAHA.114.03575
52. Lerman A, Zeiher AM. Endothelial function: cardiac events. Circulation. (2005) 111:363–8. doi: 10.1161/01.CIR.0000153339.27064.14
53. Chen-Scarabelli C, Corsetti G, Pasini E, Dioguardi FS, Sahni G, Narula J, et al. Spasmogenic effects of the proteasome inhibitor carfilzomib on coronary resistance, vascular tone and reactivity. EBioMedicine. (2017) 21:206–12. doi: 10.1016/j.ebiom.2017.05.024
54. Evans KL, Wirtz HS, Li J, She R, Maya J, Gui H, et al. Genetics of heart rate in heart failure patients (GenHRate). Hum Genomics. (2019) 13:22. doi: 10.1186/s40246-019-0206-6
Keywords: cardio-oncology, proteosome inhibitors, Multiple Myeloma, carfilzomib, cardiotoxcity, whole exome sequencing
Citation: Tantawy M, Yang G, Algubelli RR, DeAvila G, Rubinstein SM, Cornell RF, Fradley MG, Siegel EM, Hampton OA, Silva AS, Lenihan D, Shain KH, Baz RC and Gong Y (2023) Whole-Exome sequencing analysis identified TMSB10/TRABD2A locus to be associated with carfilzomib-related cardiotoxicity among patients with multiple myeloma. Front. Cardiovasc. Med. 10:1181806. doi: 10.3389/fcvm.2023.1181806
Received: 7 March 2023; Accepted: 5 June 2023;
Published: 20 June 2023.
Edited by:
Fabrizio Carta, University of Florence, ItalyReviewed by:
Nadine Norton, Mayo Clinic Florida, United States© 2023 Tantawy, Yang, Algubelli, DeAvila, Rubinstein, Cornell, Fradley, Siegel, Hampton, Silva, Lenihan, Shain, Baz and Gong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Gong Z29uZ0Bjb3AudWZsLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.