
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cardiovasc. Med. , 04 May 2023
Sec. Hypertension
Volume 10 - 2023 | https://doi.org/10.3389/fcvm.2023.1167346
This article is part of the Research Topic Insights in Hypertension: 2022 View all 9 articles
Background & Aims: The pathogenesis of hypertension involves a diverse range of genetic, environmental, hemodynamic, and more causative factors. Recent evidence points to an association between the gut microbiome and hypertension. Given that the microbiota is in part determined by host genetics, we used the two-sample Mendelian randomization (MR) analysis to address the bidirectional causal link between gut microbiota and hypertension.
Methods: We selected genetic variants (P < 1 × 10−5) for gut microbiota (n = 18,340) from the MiBioGen study. Genetic association estimates for hypertension were extracted from genome-wide association study (GWAS) summary statistics on 54,358 cases and 408,652 controls. Seven complementary MR methods were implemented, including the inverse-variance weighted (IVW) method, followed by sensitivity analyses to verify the robustness of the results. Reverse-direction MR analyses were further conducted to probe if there was a reverse causative relationship. Bidirectional MR analysis then examines a modulation of gut microbiota composition by hypertension.
Results: At the genus level, our MR estimates from gut microbiome to hypertension showed that there were 5 protective factors Allisonella, Parabacteroide, Phascolarctobacterium, Senegalimassilia, and unknowngenus (id.1000000073), while 6 genera Clostridiuminnocuum, Eubacteriumcoprostanoligenes, Eubacteriumfissicatena, Anaerostipes, LachnospiraceaeFCS020, and unknowngenus (id.2041) are risk factors. The Alcaligenaceae and ClostridialesvadinBB60 were detrimental and beneficial at the family level, respectively. In contrast, the MR results of hypertension-gut flora showed hypertensive states can lead to an increased abundance of Eubacteriumxylanophilum, Eisenbergiella, and Lachnospiraceae and a lower abundance of Alistipes, Bilophila, Butyricimonas, and Phascolarctobacterium.
Conclusion: Altered gut microbiota is a causal factor in the development of hypertension, and hypertension causes imbalances in the intestinal flora. Substantial research is still needed to find the key gut flora and explore the specific mechanisms of their effects so that new biomarkers can be found for blood pressure control.
Hypertension is a worldwide problem with major health implications, and its incidence continues to rise with unsatisfactory control rates (1). High blood pressure (BP) is a primary risk factor for cardiovascular, cerebrovascular, and kidney disease (2–4). BP control is considerable, with even a 2-mmHg drop reducing mortality from cardiovascular disease by 7% (5). The etiology of hypertension concerns genetics and the complex interaction of environmental and pathophysiological factors affecting multiple systems, and the regulatory mechanism of BP has yet to be entirely elucidated (6–8). Researchers incidentally noted a relatively higher pressure level in mice with gut microbiota colonization compared to germ-free mice (9). Alternatively, fecal matter from hypertensive patients was transplanted into germ-free mice and an increase in BP was observed (10). Variations in intestinal flora can perhaps directly influence hypertension.
Over past decades, gut microbes have attracted intense attention from scientists for promising roles in altering human organismal health. The gut microbiota synthesizes and facilitates the uptake of key nutrients and ions, sculpts the mucosa's immune system, and produces multiple metabolites and chemical mediators for action (11–13). Host genetic variation intervenes in differences amongst individuals for gut microbiota composition, likewise environmental exposure (14, 15). The ecological dysbiosis of the intestinal microbiota is implicated in both intestinal and extraintestinal diseases, including hypertension (16, 17). Several cross-sectional analyses proved that changes in human BP correlate with intestinal flora. The BP showed significant separation based on the abundance of intestinal microorganisms, which exhibits a negative correlation (18). Alleviation of hypertension also appears to involve specific bacteria taxa such as those that produce short-chain fatty acids (SCFAs) (19, 20). Such microbial flora as, Faecalibacterium, Ruminococcaceae Eubacterium rectale, and Roseburia have a high butyrate production capacity, which contributes to the decrease in systolic BP (21). Crucially, taking probiotics improves hypertension to some extent (22). That means it is not just about the abundance of flora, a lack of healthy flora or dysbiosis may promote higher BP (23). However, studies of clinical hypertension causation are particularly challenging in that the major pathophysiological mechanisms may be obscured by confounding effects of compensatory pathways, diet, or medication, just as Clostridiales shows opposing effects in the different hypertensive population studies (24, 25). Even more importantly, dysbiosis may also be a concomitant symptom of hypertension. Larger animal studies or clinical cohorts are still required to identify the gut flora associated with hypertension and how they are causally connected.
We used Mendelian randomization (MR) analysis to establish a causal relationship between exposure and outcome achieved through an instrumental variable using genetic variations. Such an approach exploits the random classification of genetic variation during gamete formation, consequently minimizing the possible effects of confounding factors and reverse causality (26). Here, we performed bidirectional MR analyses of causal inference between hypertension and gut microbiota. Clues for potential hazards of hypertension can be found in the hypertension-regulated flora. What's more, this causal inquiry provides suspected biomarkers for further research into hypertension pathogenesis and therapeutic targets for better BP control.
The study used the two-sample bidirectional MR design that assessed the cause-effect interaction between gut flora and hypertension with single nucleotide polymer-phisms (SNPs) as an instrumental variable. Three criteria need to be met for SNPs: associated with exposure, with no confounding of the exposure-outcome association, and affecting outcome only through exposure (27). Figure 1 illustrates the flow chart of the overall process. We used STROBE-MR as the reporting specification for reference (28).
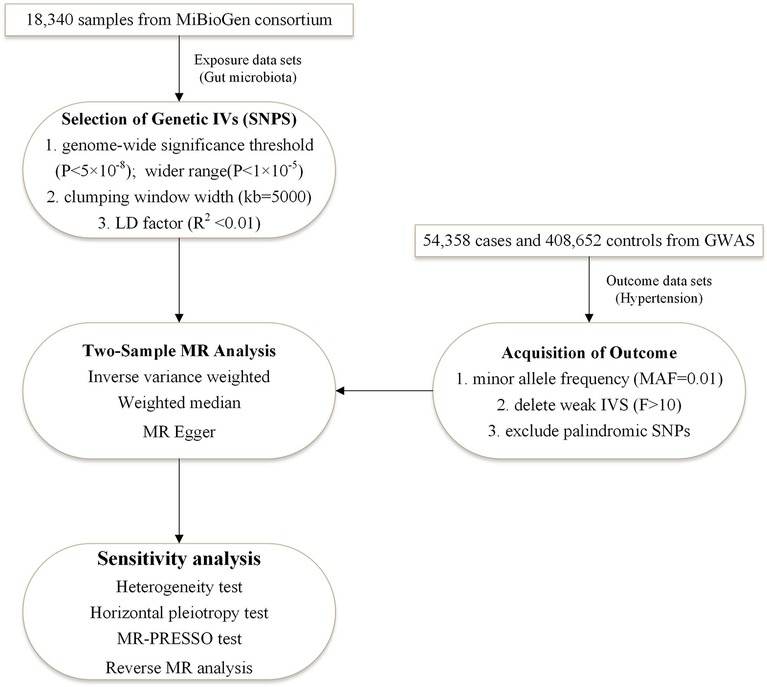
Figure 1. The two-sample MR-analysis process. Furthermore, we performed a bidirectional MR analysis in the same way, with hypertension as the exposure and intestinal flora as the outcome. It was designed to investigate whether hypertension causes imbalances in the specific intestinal flora. IVs, instrumental variables; LD factor, linkage disequilibrium factor; MR-PRESSO, MR Pleiotropy RESidual Sum and Outlier.
An existing ethical clearance from the respective institutional review board is available for all published genome-wide association studies (GWAS). Only summary-level data were used in this study, where no additional ethical approval was required.
All data used in this MR study are publicly available. GWAS data for gut microbes were derived from 16S rRNA gene sequencing profiles and genotyping data curated and analyzed by the MiBioGen International Consortium (29). The study population was mainly adults or adolescents of European origin from 11 countries, with a total sample of 18,340 individuals (24 cohorts). We accessed data on 211 gut microbial taxa at 5 taxonomic levels. Published GWAS of hypertension from the UK Biobank (id: UKB-b-12493) was then used as a data source for outcome events in 463,010 participants of European descent (30). A total of 54,358 patients with hypertension, defined as the presence of essential (primary) hypertension (International Classification of Diseases 10 diagnosis code I10) and 408,652 individuals without primary hypertension or any other hypertensive disorder, were included in the study.
We chose SNPs up to the genome-wide significance threshold (P < 5 × 10−8) as instrumental variables (IVs). Because the number of SNPs that met the criteria was too small, we selected IVs with a threshold of P < 1 × 10−5 to obtain more comprehensive results. To ensure the independence of each SNP, we set the linkage disequilibrium (LD) factor r2 to 0.01 and the clumping window width kb = 5,000. Then the information on SNPs related to the above intestinal flora was extracted from the summary GWAS data of hypertension, removing the missing SNPs, and the MAF was put at 0.01. In addition, all SNPs with palindromic structures were excluded to prevent the effect of alleles on the results. F value was used to examine the presence of bias in the causal relationship between intestinal flora and hypertension due to weak IVs. The used SNP is considered a weak IV and deleted when the F-statistic was less than 10.
All MR analysis was conducted using R software (version 4.2.1) by the “Two Sample MR” package (https://mrcieu.github.io/TwoSampleMR). P < 0.05 was accepted as a statistically significant result for MR analysis. Wald method was used to estimate the causal relationship between exposure and outcome for the bacterial taxa with only one instrumental variable. We utilized MR analysis for gut microbial taxa with multiple IVs using inverse-variance-weighted (IVW), weighted median, and MR-Egger. Fixed or random-effects IVW method was applied as the primary method for MR analysis. Cochrane's Q test was performed to assess the heterogeneity of IVs. If heterogeneity was present (P < 0.05), the random-effects IVW test was used to estimate the MR statistic. Since the IVW method requires all SNPs to be valid instrumental variables, its results would be subject to large errors in the presence of invalid IVs. While the significant results of the Weighted median method are still applicable as long as at least 50% of the IVs are valid instrumental variables. Among the three major MR assumptions, MR studies' results are subject to inaccuracy once the assumption is violated that IVs are associated with other confounders in addition to exposure factors. Horizontal pleiotropy was used to weigh the effect. Intercept terms of MR-Egger were one of the most important methods to evaluate the existence of horizontal pleiotropy among IVs. Horizontal polymorphism exists when the intercept is not zero, of which the statistical significance will be determined by the P-value of the intercept. Together with MR-Egger, the MR pleiotropy residual sum and outlier method (MR-PRESSO) were also commonly used to detect horizontal pleiotropy. MR-PRESSO consists of the following three main steps, detecting the presence of pleiotropy, calculating the corrected predicted values after removing outliers, and comparing whether the difference between the two predicted values before and after correction is statistically significant. Finally, MR results were evaluated for robustness using a leave-one-out analysis.
As shown by an increased ratio of intestinal Firmicutes/Bacteroidetes in spontaneously hypertensive rats, the dysbiosis can be rebalanced after oral administration of minocycline (31). Indeed, there are changes in the balance of intestinal flora species depending on environmental conditions and individual status. To investigate whether hypertension leads to an imbalance of specific intestinal flora, a bidirectional MR analysis was performed with hypertension as exposure and intestinal flora as the outcome. SNPs with a threshold of P < 1 × 10−5 were screened as instrumental variables to obtain more comprehensive MR analysis results. Meanwhile, in the bidirectional MR analysis, we gave particular attention to the role of hypertension on the causal effect of the identified statistically significant flora as reverse MR analysis.
In the genetic analysis of the bacterial taxa on hypertension, a total of 2,427 SNPs were selected as IVs associated with 211 bacterial taxa by removing SNPs with linkage disequilibrium and the presence of palindromic structures with a locus-wide significance level (P < 1 × 10−5) as the threshold, including 9 phyla (108 SNPs), 16 class (194 SNPs), 20 order (237 SNPs), 35 families (427 SNPs), and 131 genera (1,461 SNPs). The F-statistics of IVs were all greater than 10, suggesting no bias caused by weak instrumental variables (Supplementary Table S1). A series of 32 SNPs were identified as IVs (genome-wide significance, P < 5 × 10−8), and Supplementary Table S2 specifies the information on these IVs.
Figure 2 is an overview of the primary results from the Two-Sample MR study. It presents that 13 bacterial taxa were statistically significant in MR analysis with hypertension on locus-wide significance level, where 2 were family part and 11 were genera part. Among them, Alcaligenaceae, Clostridiuminnocuum, Eubacteriumcoprostanoligenes, Eubacteriumfissicatena, Anaerostipes, LachnospiraceaeFCS020, unknowngenus (id.2041) were risk factors concerning hypertension, ClostridialesvadinBB60, Allisonella, Parabacteroides, Phascolarctobacterium, Senegalimassilia, and unknowngenus (id.1000000073) were protectors for hypertension. In addition, Senegalimassilia was more statistically significant in terms of protection as its P statistic was less than 0.01.
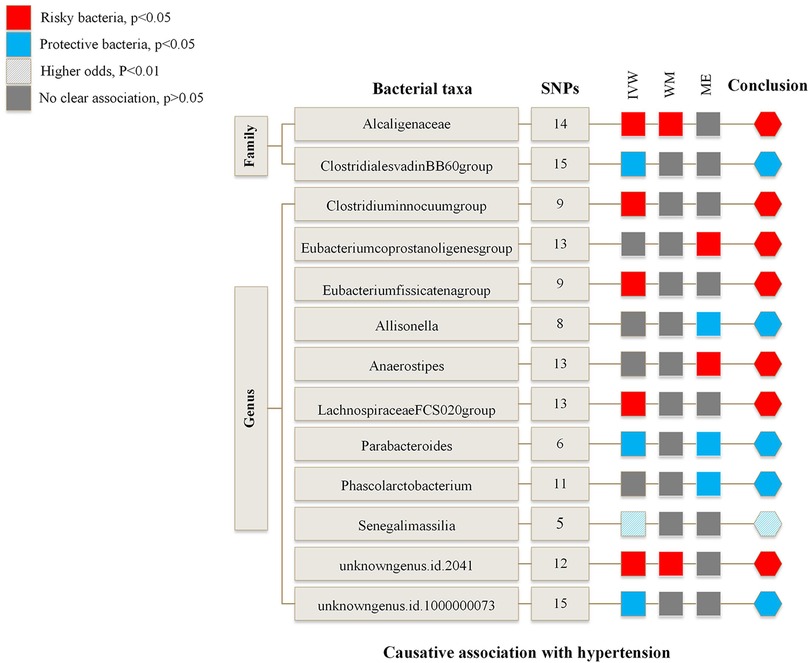
Figure 2. Risk and protective gut microbial genera underlying hypertension—an overview of primary results from the two-sample MR-study. Full data for all results depicted in the figure can be found in the Supplementary. IVW, inverse variance weighted method; WM, weighted median estimator; ME, MR Egger.
Supplementary Table S3 and Supplementary Table S4 summarize the results of the MR study and sensitivity analysis for all genera. IVW results indicate that Alcaligenaceae [odds ratio (OR) = 1.007; 95% confidence interval (CI), 1.000–1.014], Clostridiuminnocuum (OR = 1.005; 95%CI, 1.000–1.009), Eubacteriumfissicatena (OR = 1.005; 95%CI, 1.001–1.009), LachnospiraceaeFCS020 (OR = 1.006; 95%CI, 1.000–1.012), unknowngenus (id.2041) (OR = 1.006; 95%CI, 1.001–1.011) were positively correlated with hypertension; ClostridialesvadinBB60 (OR = 0.994; 95%CI, 0.989–0.999), Parabacteroides (OR = 0.987; 95%CI, 0.978–0.997), Senegalimassilia (OR = 0.990; 95%CI, 0.983–0.997), unknowngenus (id.1000000073) (OR = 0.994; 95%CI, 0.989–0.999) were negatively related to hypertension. The P statistics of heterogeneity regarding the above bacterial taxa were all >0.05, and no evidence of horizontal pleiotropy was found by MR Egger. Further assessment of pleiotropy with MRPRESSO found no outliers, indicating the results' reliability with IVW.
MR Egger reveals that there was a relatively greater increase in hypertension among participants with bacterial taxa including Eubacteriumcoprostanoligenes (OR = 1.042; 95%CI, 1.007–1.079), Anaerostipes (OR = 1.032; 95%CI, 1.008–1.056), while Allisonella (OR = 0.952; 95%CI, 0.918–0.987) and Phascolarctobacterium (OR = 0.968; 95%CI, 0.941–0.995) were in a negative correlation with hypertension. Besides, MR Egger shows there was horizontal pleiotropy (P < 0.05) between the IVs of these four genera and the outcome. We assumed the results of the MR Egger method were more reliable in the above four genera. The pleiotropy of Eubacteriumcoprostanoligenes and Allisonella was also confirmed by MRPRESSO. However, there was no significant change in results after removing outliers.
The outcomes of the MR study and sensitivity analysis with genome-wide significance threshold are described in Supplementary Table S5 and Supplementary Table S6. Wald ratio indicates that the Eubacteriumcoprostanoligenes (OR = 1.026; 95%CI, 1.005–1.048) was a hazard element for hypertension, which is consistent with the previous results of MR analysis at locus-wide significance level. Leave-one-out sensitivity test demonstrated no remarkable difference in the estimation of the causal roles of ClostridialesvadinBB60 and Senegalimassilia on hypertension no matter which IV was excluded, accounting for the robustness of the MR results (Figure 3). The leave-one-out analysis results of the 13 bacterial taxa with statistical significance in MR analysis were displayed in Supplementary Figure S1.
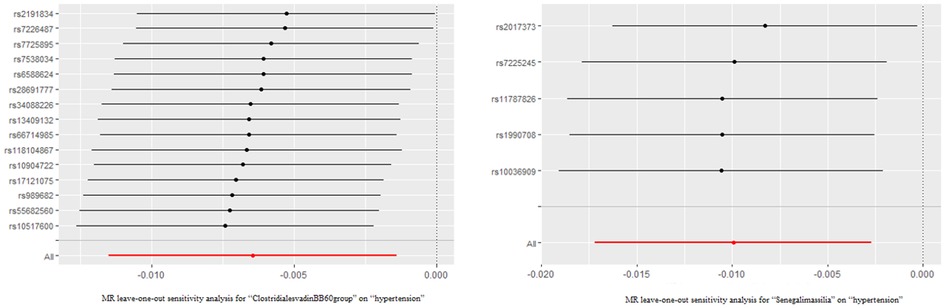
Figure 3. The leave-one-out analysis results of clostridialesvadinBB60 and senegalimassilia. Red lines represented the MR results of IVW analysis, and there was no significant difference in the causal estimation between the two bacteria and hypertension after removing either IV.
To prevent reverse causality from interfering with the above results, we performed a reverse MR analysis with significant gut flora in Two-sample MR studies as the outcome and hypertension as the exposure on locus-wide significance level (Supplementary Table S7). Supplementary Figure S2 depicts the reverse MR results for the 13 genera, where elevated BP was negatively correlated with Phascolarctobacterium (OR = 0.113, 95%CI, 0.018–0.724). There was no additional evidence for a causal effect of hypertension on the other 12 genera.
Further exploration of the causal relationship of hypertension on bacterial taxa is summarized in Figure 4. It illustrates all statistically significant results of bidirectional MR analysis for a total of 7 genera causally affected by hypertension. There was horizontal pleiotropy among Eubacteriumxylanophilum, Alistipes, Eisenbergiella, LachnospiraceaeUCG001, and Phascolarctobacterium (id.2168), which pleiotropy P statistics were less than 0.05 (Supplementary Table S8). In this light, we used MR Egger to identify the causal effect of hypertension on these bacterial taxa. MR Egger indicates that those increased attributable to hypertension are Eubacteriumxylanophilum (OR = 7.583; 95%CI, 1.310–43.906), Eisenbergiella (OR = 14.648; 95%CI, 1.169–183.606), and LachnospiraceaeUCG001 (OR = 8.696; 95%CI, 1.357–55.736), while the opposite was true for Alistipes (OR = 0.164, 95%CI, 0.040–0.679) and Phascolarctobacterium (id.2168) (OR = 0.113; 95%CI, 0.018–0.724). IVW suggests that hypertension was negatively associated with Bilophila (OR = 0.625, 95%CI, 0.391–0.997), Butyricimonas (OR = 0.482; 95%CI, 0.295–0.787) (Supplementary Table S9), and neither heterogeneity nor pleiotropy was significant for these 2 genera (Supplementary Table S10).
Hypertension is a modifiable risk factor for cardiovascular disease development and adverse prognosis (32). Although numerous studies have shown that lifestyle and environmental factors are important drivers for the induction of hypertension, a substantial proportion can nevertheless be explained by genetics (33, 34). Previous large cross-sectional studies have shown significant differences in the overall abundance of gut microbes, and some specific flora in hypertensive patients compared to normotensive individuals (35, 36). From data on hypertension levels using 463,010 UK Biobank participants, we used bidirectional MR analysis and found that 13 gut microbial taxa at 2 classification levels influence hypertension risk, whereas individuals in a hypertensive state could induce alterations in the abundance of 7 intestinal flora. Our study used genetic instrumental variables to demonstrate that onset of hypertension and gut microbiota shifts are mutually causal.
The rapid development of macrogenomics and various sequencing technologies, such as 16S rRNA gene amplicon sequencing and shotgun metagenome sequencing, has allowed researchers to analyze variations in the gut microflora of hypertensive and normotensive populations with great ease (37). We selected the study, which was organized and analyzed by the MiBioGen International Consortium, as the source of data related to the human gut microbiome (29). It provided the largest host genomics and microbiome-based data across multiple races to date and examined the impact of methodological differences on microbiome data. Although the statistical power of microbiome-wide analysis was diminished due to microbiome heterogeneity and inter-individual variability, there still provided a favorable basis for exploring the causal relationship between bacterial taxa and disease traits. In numerous studies, a consistent finding is that lower microbial abundance and diversity or dysbiosis are associated with higher BP (10, 25, 31). An increased Firmicutes/Bacteroidetes ratio has widely been recognized as an indicator of intestinal ecological dysregulation, and, interestingly, this also applies to hypertension (31, 38). There was a positive association between higher systolic BP and intestinal flora composition with a higher abundance of genera Klebsiella, Anaerotruncus, Porphyromonas, Eubacteriumsiraeum, Actinomyce, Prevotellabivia, Clostridium cluster IV, Streptococcus, Eggerthella, and Sporobacter, etc., while bacteria such as Roseburia, Bifidobacterium, Coprococcus, Synergistetes, and Butyrivibrio, etc. were reduced in hypertensive patients (10, 18, 21, 25, 39, 40). However, the changed genus may not be the candidate intestinal strain affecting hypertension. Currently, relevant studies are yet to be explored in-depth on the exact correlation and specific mechanisms.
Succeeding studies were undertaken in terms of gut flora products to identify the gut flora associated with causing hypertension. Specifically, SCFAs are potentially involved in BP homeostasis (18, 25). Prospective ambulatory BP monitoring found that higher levels of SCFAs such as acetate, butyrate, and propionate in the stool followed higher BP (41). Also, decreased bacterial taxa of butyrate production were observed in rats with obstructive sleep apnea-induced hypertension (42). The major butyrate-producing bacteria in the intestine include Faecalibacterium, Ruminococcaceae, Coprococcus, and Roseburia (43, 44). The results of Clostridiaceae and Phascolarctobacterium as beneficial factors may be relevant to their butyrate secretion (45). The list of bacteria producing acetate, propionate, and hexanoate is longer, which includes Parabacteroides and Allisonella mentioned in our MR analysis (46–48). It is believed that these higher rates of SCFAs excretion may have reduced circulating SCFAs uptake and that SCFAs-producing microbiota is usually accompanied by lower BP (19). In the case of spontaneously hypertensive rats (SHR), this idea is supported by experiments in which butyrate injections stimulated the center to lower BP (49). To complicate matters, it is possible that SCFAs do not fully exert a hypotensive effect in BP regulation, depending on the role of different SCFA receptors (50, 51). Identifying these receptors may be important to elucidate the pathogenesis of SCFAs-microbiome and hypertension.
On a different note, the differential analysis findings of gut microbiota associated with hypertension were not always consistent. After matching and comparing 67 hypertensive and 62 corresponding normotensive cases by propensity score, Dan et al. found that Clostridium sensu stricto, showed a higher abundance in the hypertension group, and the same results were obtained by Huart et al. (23, 41). While the opposite was found by Verhaar et al. (20). Bacteroides, Clostridiales, Oscillibacter, and Lactobacillaceae also show varying performances in diverse studies (10, 18, 21, 23, 52). The discrepancy in results may be attributed to several reasons, starting with the fact that the study may not adjust for important confounding factors in the analysis, such as age, ethnic differences in the recruited population, diet, and medications. Alternatively, the taxonomic level at which studies focus on the flora may differ. The reverse causality might be a factor as well. A lack of evidence is available to determine their causative or improvement role in the development of hypertension for the dysregulated intestinal florae. We choose to cite hypertension as the driver and probe whether any gut flora was subsequently altered. Bidirectional MR analysis displayed a significant increase in the abundance of Eubacteriumxylanophilum, in agreement with the findings of Kim et al. (21). At the genus level, there was also an increase in Eisenbergiella with Lachnospiraceae on account of hypertension, and a decrease in Alistipes, Bilophila, Butyricimonas, and Phascolarctobacterium. Certain evidence exists that angiotensin I converting enzyme 2 (ACE2) mutant modifies the gut microbial composition (53). G-protein-coupled estrogen receptor (Gper1), a pro-hypertensive agent, has a similar effect that Gper1−/− rats exhibited significantly lower levels of Clostridiales beneath the phylum Firmicutes, implying that hypertension susceptibility genes act on gut microbes (54, 55).
A bidirectional MR approach was used for the first time in our study to investigate the causal relationship between primary hypertension and intestinal flora, not only by performing various sensitivity analyses to evaluate the robustness of the analyses but also to exclude the interference of reverse causality. As described in the above results, 6 beneficial and 7 risky microbiota suggested a potentially causal relationship to hypertension, and higher BP also caused disruptions of 7 microbiota abundance. Although the two cohort studies have a fairly large sample size, only recipients of European ancestry were included in the GWAS data source, discounting the extrapolation of the study results. Theoretical causal interventions exist for some bacteria whose specific mechanisms of action are not yet clear. It requires single gut flora transplantation or more animal experiments to confirm the function of microbial metabolites and their contribution to BP modulation. Our team is also conducting related studies in hopes of finding potential strategic targets to control BP.
In summary, our comprehensive bidirectional MR analysis supports that intestinal flora and hypertension are mutually causal.
Publicly available datasets were analyzed in this study. This data can be found here: https://mibiogen.gcc.rug.nl/ UK Biobank (id: UKB-b-12493).
All authors contributed to the article and approved the submitted version YL: methodology, software, writing-original draft preparation. RF: conceptualization, data curation, and writing-original draft preparation. RL: visualization. JZ: data curation. TL: validation. XL and WJ: funding acquisition, project administration. All authors contributed to the article and approved the submitted version.
The study was supported by the Key Research and Development program of Hunan Province (No. 2022SK2029), and the National Natural Science Foundation of China Projects (No. 81800271).
We are grateful to the dedicated participants, the researchers who shared their findings, the GWAS data platform, and the major research consortia, all of which provided leads and possibilities for subsequent studies.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2023.1167346/full#supplementary-material.
1. Forouzanfar MH, Liu P, Roth GA, Ng M, Biryukov S, Marczak L, et al. Global burden of hypertension and systolic blood pressure of at least 110 to 115 mm Hg, 1990–2015. JAMA. (2017) 317(2):165–82. doi: 10.1001/jama.2016.19043
2. Rapsomaniki E, Timmis A, George J, Pujades-Rodriguez M, Shah AD, Denaxas S, et al. Blood pressure and incidence of twelve cardiovascular diseases: lifetime risks, healthy life-years lost, and age-specific associations in 1.25 million people. Lancet. (2014) 383(9932):1899–911. doi: 10.1016/S0140-6736(14)60685-1
3. Li L, Scott CA, Rothwell PM. Association of younger vs older ages with changes in incidence of stroke and other vascular events, 2002–2018. JAMA. (2022) 328(6):563–74. doi: 10.1001/jama.2022.12759
4. Szczech LA, Granger CB, Dasta JF, Amin A, Peacock WF, McCullough PA, et al. Acute kidney injury and cardiovascular outcomes in acute severe hypertension. Circulation. (2010) 121(20):2183–91. doi: 10.1161/CIRCULATIONAHA.109.896597
5. Lewington S, Clarke R, Qizilbash N, Peto R, Collins R, Prospective Studies C. Age-specific relevance of usual blood pressure to vascular mortality: a meta-analysis of individual data for one million adults in 61 prospective studies. Lancet. (2002) 360(9349):1903–13. doi: 10.1016/s0140-6736(02)11911-8
6. Evangelou E, Warren HR, Mosen-Ansorena D, Mifsud B, Pazoki R, Gao H, et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. (2018) 50(10):1412–25. doi: 10.1038/s41588-018-0205-x
7. Slomski A. Effect of modified Chinese diet is comparable to antihypertensives. JAMA. (2022) 328(8):700. doi: 10.1001/jama.2022.14322
8. Gruber T, Pan C, Contreras RE, Wiedemann T, Morgan DA, Skowronski AA, et al. Obesity-associated hyperleptinemia alters the gliovascular interface of the hypothalamus to promote hypertension. Cell Metab. (2021) 33(6):1155–70 e10. doi: 10.1016/j.cmet.2021.04.007
9. Moghadamrad S, McCoy KD, Geuking MB, Sagesser H, Kirundi J, Macpherson AJ, et al. Attenuated portal hypertension in germ-free mice: function of bacterial flora on the development of mesenteric lymphatic and blood vessels. Hepatology. (2015) 61(5):1685–95. doi: 10.1002/hep.27698
10. Li J, Zhao F, Wang Y, Chen J, Tao J, Tian G, et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome. (2017) 5(1):14. doi: 10.1186/s40168-016-0222-x
11. Chen RY, Mostafa I, Hibberd MC, Das S, Lynn HM, Webber DM, et al. Melding microbiome and nutritional science with early child development. Nat Med. (2021) 27(9):1503–6. doi: 10.1038/s41591-021-01451-1
12. Wastyk HC, Fragiadakis GK, Perelman D, Dahan D, Merrill BD, Yu FB, et al. Gut-microbiota-targeted diets modulate human immune status. Cell. (2021) 184(16):4137–53 e14. doi: 10.1016/j.cell.2021.06.019
13. Tanes C, Bittinger K, Gao Y, Friedman ES, Nessel L, Paladhi UR, et al. Role of dietary fiber in the recovery of the human gut microbiome and its metabolome. Cell Host Microbe. (2021) 29(3):394–407 e5. doi: 10.1016/j.chom.2020.12.012
14. Hall AB, Tolonen AC, Xavier RJ. Human genetic variation and the gut microbiome in disease. Nat Rev Genet. (2017) 18(11):690–9. doi: 10.1038/nrg.2017.63
15. Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. (2014) 159(3):514–29. doi: 10.1016/j.cell.2014.09.048
16. Wu H, Tremaroli V, Schmidt C, Lundqvist A, Olsson LM, Kramer M, et al. The gut microbiota in prediabetes and diabetes: a population-based cross-sectional study. Cell Metab. (2020) 32(3):379–90 e3. doi: 10.1016/j.cmet.2020.06.011
17. Svolos V, Hansen R, Nichols B, Quince C, Ijaz UZ, Papadopoulou RT, et al. Treatment of active Crohn's disease with an ordinary food-based diet that replicates exclusive enteral nutrition. Gastroenterology. (2019) 156(5):1354–67 e6. doi: 10.1053/j.gastro.2018.12.002
18. Sun S, Lulla A, Sioda M, Winglee K, Wu MC, Jacobs DR Jr., et al. Gut microbiota composition and blood pressure. Hypertension. (2019) 73(5):998–1006. doi: 10.1161/HYPERTENSIONAHA.118.12109
19. de la Cuesta-Zuluaga J, Mueller NT, Alvarez-Quintero R, Velasquez-Mejia EP, Sierra JA, Corrales-Agudelo V, et al. Higher fecal short-chain fatty acid levels are associated with gut microbiome dysbiosis, obesity, hypertension and cardiometabolic disease risk factors. Nutrients. (2018) 11(1):51. doi: 10.3390/nu11010051
20. Verhaar BJH, Collard D, Prodan A, Levels JHM, Zwinderman AH, Backhed F, et al. Associations between gut microbiota, faecal short-chain fatty acids, and blood pressure across ethnic groups: the HELIUS study. Eur Heart J. (2020) 41(44):4259–67. doi: 10.1093/eurheartj/ehaa704
21. Kim S, Goel R, Kumar A, Qi Y, Lobaton G, Hosaka K, et al. Imbalance of gut microbiome and intestinal epithelial barrier dysfunction in patients with high blood pressure. Clin Sci. (2018) 132(6):701–18. doi: 10.1042/CS20180087
22. Khalesi S, Sun J, Buys N, Jayasinghe R. Effect of probiotics on blood pressure: a systematic review and meta-analysis of randomized, controlled trials. Hypertension. (2014) 64(4):897–903. doi: 10.1161/HYPERTENSIONAHA.114.03469
23. Dan X, Mushi Z, Baili W, Han L, Enqi W, Huanhu Z, et al. Erratum: differential analysis of hypertension-associated intestinal Microbiota. Int J Med Sci. (2021) 18(16):3748. doi: 10.7150/ijms.65972
24. Ganesh BP, Nelson JW, Eskew JR, Ganesan A, Ajami NJ, Petrosino JF, et al. Prebiotics, probiotics, and acetate supplementation prevent hypertension in a model of obstructive sleep apnea. Hypertension. (2018) 72(5):1141–50. doi: 10.1161/HYPERTENSIONAHA.118.11695
25. Yan Q, Gu Y, Li X, Yang W, Jia L, Chen C, et al. Alterations of the gut microbiome in hypertension. Front Cell Infect Microbiol. (2017) 7:381. doi: 10.3389/fcimb.2017.00381
26. Neeland IJ, Kozlitina J. Mendelian randomization: using natural genetic variation to assess the causal role of modifiable risk factors in observational studies. Circulation. (2017) 135(8):755–8. doi: 10.1161/CIRCULATIONAHA.117.026857
27. Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. (2008) 27(8):1133–63. doi: 10.1002/sim.3034
28. Skrivankova VW, Richmond RC, Woolf BAR, Yarmolinsky J, Davies NM, Swanson SA, et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomization: the STROBE-MR statement. JAMA. (2021) 326(16):1614–21. doi: 10.1001/jama.2021.18236
29. Kurilshikov A, Medina-Gomez C, Bacigalupe R, Radjabzadeh D, Wang J, Demirkan A, et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet. (2021) 53(2):156–65. doi: 10.1038/s41588-020-00763-1
30. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-base platform supports systematic causal inference across the human phenome. Elife. (2018) 7:e34408. doi: 10.7554/eLife.34408
31. Yang T, Santisteban MM, Rodriguez V, Li E, Ahmari N, Carvajal JM, et al. Gut dysbiosis is linked to hypertension. Hypertension. (2015) 65(6):1331–40. doi: 10.1161/HYPERTENSIONAHA.115.05315
32. Bayes-Genis A, Diez J. Transition to heart failure in hypertension: going to the heart of the matter. Eur Heart J. (2022) 43(35):3332–4. doi: 10.1093/eurheartj/ehab651
33. Giri A, Hellwege JN, Keaton JM, Park J, Qiu C, Warren HR, et al. Trans-ethnic association study of blood pressure determinants in over 750,000 individuals. Nat Genet. (2019) 51(1):51–62. doi: 10.1038/s41588-018-0303-9
34. Liu X, Hu C, Bao M, Li J, Liu X, Tan X, et al. Genome wide association study identifies L3MBTL4 as a novel susceptibility gene for hypertension. Sci Rep. (2016) 6:30811. doi: 10.1038/srep30811
35. Tang WH, Kitai T, Hazen SL. Gut microbiota in cardiovascular health and disease. Circ Res. (2017) 120(7):1183–96. doi: 10.1161/CIRCRESAHA.117.309715
36. Richards EM, Li J, Stevens BR, Pepine CJ, Raizada MK. Gut microbiome and neuroinflammation in hypertension. Circ Res. (2022) 130(3):401–17. doi: 10.1161/CIRCRESAHA.121.319816
37. Lagier JC, Khelaifia S, Alou MT, Ndongo S, Dione N, Hugon P, et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. (2016) 1:16203. doi: 10.1038/nmicrobiol.2016.203
38. Mulder M, Radjabzadeh D, Kiefte-de Jong JC, Uitterlinden AG, Kraaij R, Stricker BH, et al. Long-term effects of antimicrobial drugs on the composition of the human gut microbiota. Gut Microbes. (2020) 12(1):1795492. doi: 10.1080/19490976.2020.1791677
39. Ko CY, Liu QQ, Su HZ, Zhang HP, Fan JM, Yang JH, et al. Gut microbiota in obstructive sleep apnea-hypopnea syndrome: disease-related dysbiosis and metabolic comorbidities. Clin Sci. (2019) 133(7):905–17. doi: 10.1042/CS20180891
40. Yu Y, Mao G, Wang J, Zhu L, Lv X, Tong Q, et al. Gut dysbiosis is associated with the reduced exercise capacity of elderly patients with hypertension. Hypertens Res. (2018) 41(12):1036–44. doi: 10.1038/s41440-018-0110-9
41. Huart J, Leenders J, Taminiau B, Descy J, Saint-Remy A, Daube G, et al. Gut microbiota and fecal levels of short-chain fatty acids differ upon 24-hour blood pressure levels in men. Hypertension. (2019) 74(4):1005–13. doi: 10.1161/HYPERTENSIONAHA.118.12588
42. Durgan DJ, Ganesh BP, Cope JL, Ajami NJ, Phillips SC, Petrosino JF, et al. Role of the gut microbiome in obstructive sleep apnea-induced hypertension. Hypertension. (2016) 67(2):469–74. doi: 10.1161/HYPERTENSIONAHA.115.06672
43. Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol. (2017) 19(1):29–41. doi: 10.1111/1462-2920.13589
44. Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. (2009) 294(1):1–8. doi: 10.1111/j.1574-6968.2009.01514.x
45. Sherman SB, Sarsour N, Salehi M, Schroering A, Mell B, Joe B, et al. Prenatal androgen exposure causes hypertension and gut microbiota dysbiosis. Gut Microbes. (2018) 9(5):400–21. doi: 10.1080/19490976.2018.1441664
46. Venegas D P, De la Fuente MK, Landskron G, Gonzalez MJ, Quera R, Dijkstra G, et al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol. (2019) 10:277. doi: 10.3389/fimmu.2019.00277
47. Lei Y, Tang L, Liu S, Hu S, Wu L, Liu Y, et al. Parabacteroides produces acetate to alleviate heparanase-exacerbated acute pancreatitis through reducing neutrophil infiltration. Microbiome. (2021) 9(1):115. doi: 10.1186/s40168-021-01065-2
48. Che L, Hu Q, Wang R, Zhang D, Liu C, Zhang Y, et al. Inter-correlated gut microbiota and SCFAs changes upon antibiotics exposure links with rapid body-mass gain in weaned piglet model. J Nutr Biochem. (2019) 74:108246. doi: 10.1016/j.jnutbio.2019.108246
49. Yang T, Magee KL, Colon-Perez LM, Larkin R, Liao YS, Balazic E, et al. Impaired butyrate absorption in the proximal colon, low serum butyrate and diminished central effects of butyrate on blood pressure in spontaneously hypertensive rats. Acta Physiol. (2019) 226(2):e13256. doi: 10.1111/apha.13256
50. Pluznick JL. Gut microbiota in renal physiology: focus on short-chain fatty acids and their receptors. Kidney Int. (2016) 90(6):1191–8. doi: 10.1016/j.kint.2016.06.033
51. Le Poul E, Loison C, Struyf S, Springael JY, Lannoy V, Decobecq ME, et al. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J Biol Chem. (2003) 278(28):25481–9. doi: 10.1074/jbc.M301403200
52. Jackson MA, Verdi S, Maxan ME, Shin CM, Zierer J, Bowyer RCE, et al. Gut microbiota associations with common diseases and prescription medications in a population-based cohort. Nat Commun. (2018) 9(1):2655. doi: 10.1038/s41467-018-05184-7
53. Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, et al. ACE2 Links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. (2012) 487(7408):477–81. doi: 10.1038/nature11228
54. Gros R, Ding Q, Liu B, Chorazyczewski J, Feldman RD. Aldosterone mediates its rapid effects in vascular endothelial cells through GPER activation. Am J Physiol Cell Physiol. (2013) 304(6):C532–40. doi: 10.1152/ajpcell.00203.2012
Keywords: gut microbiome, hypertension, genome-Wide association study, causality, mendelian randomization
Citation: Li Y, Fu R, Li R, Zeng J, Liu T, Li X and Jiang W (2023) Causality of gut microbiome and hypertension: A bidirectional mendelian randomization study. Front. Cardiovasc. Med. 10:1167346. doi: 10.3389/fcvm.2023.1167346
Received: 16 February 2023; Accepted: 18 April 2023;
Published: 4 May 2023.
Edited by:
Guido Iaccarino, University of Naples Federico II, ItalyReviewed by:
Marija Vavlukis, Ss. Cyril and Methodius University in Skopje, North Macedonia© 2023 Li, Fu, Li, Zeng, Liu, Li and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaogang Li aG54eWx4Z0Bjc3UuZWR1LmNu Weihong Jiang amlhbmd3ZWlob25nZG9jQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.