- 1Department of Pulmonology and Respiratory Care, Cathay General Hospital, Taipei City, Taiwan
- 2Cardiovascular Center, Cathay General Hospital, Taipei City, Taiwan
- 3Department of Medical Imaging, Chung Shan Medical University Hospital, Taichung City, Taiwan
- 4Department of Public Health and Institute of Public Health, Chung Shan Medical University, Taichung City, Taiwan
- 5Superintendent Office, Institute of Medicine, Cathay General Hospital, Taipei City, Taiwan
- 6Institute of Medicine, Chung Shan Medical University, Taichung City, Taiwan
Background: The aetio-pathologenesis of hypertension is multifactorial, encompassing genetic, epigenetic, and environmental factors. The combined effect of genetic and epigenetic changes on hypertension is not known. We evaluated the independent and interactive association of MTHFR rs1801133 single nucleotide polymorphism (SNP) and MTHFR promoter methylation with hypertension among Taiwanese adults.
Methods: We retrieved data including, MTHFR promoter methylation, MTHFR rs1801133 genotypes (CC, CT, and TT), basic demography, personal lifestyle habits, and disease history of 1,238 individuals from the Taiwan Biobank (TWB).
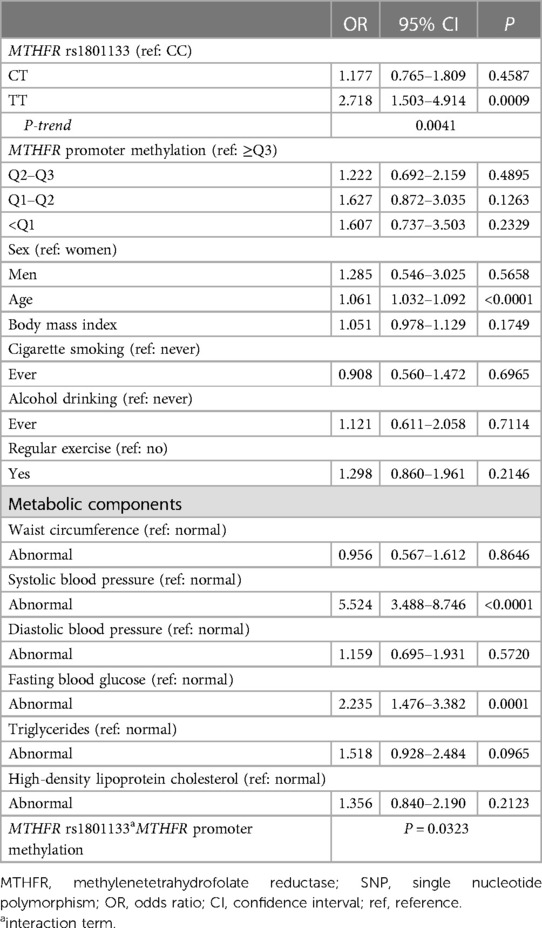
Results: The distributions of hypertension and MTHFR promoter methylation quartiles (β < 0.1338, 0.1338 ≤ β < 0.1385, 0.1385 ≤ β < 0.1423, and β ≥ 0.1423 corresponding to <Q1, Q1–Q2, Q2–Q3, and ≥Q3) among individuals with the rs1801133 genotypes (CC, CT, and TT) were significantly different (P < 0.05). The risk of hypertension was significantly higher among individuals with the TT genotype compared to the reference genotype (CC): odds ratio (OR); 95% confidence interval (CI) = 2.718; 1.503–4.914. The trend of the association of the CT and TT genotypes with hypertension was dose-dependent (P-trend = 0.0041). MTHFR promoter methylation (lower quartiles compared to ≥Q3) was not significantly associated with hypertension. However, its interaction with MTHFR rs1801133 was significant (P = 0.0323). After stratification by rs1801133 genotypes, lower MTHFR promoter methylation quartiles (<Q1, Q1–Q2, Q2–Q3) compared to ≥Q3 were significantly associated with a higher risk of hypertension among individuals carrying the CC genotype: ORs (95% CIs) = 3.225 (1.140–9.124), 4.177 (1.424–12.247), and 8.645 (2.513–29.739) for Q2–Q3, Q1–Q2, and <Q1, respectively. The trend test was significant (P-trend = 0.0009).
Conclusion: Independently, rs1801133 TT was associated with a higher risk of hypertension, but methylation was not. Based on genotypes, lower methylation was dose-dependently associated with a higher risk of hypertension in individuals with the CC genotype. Our findings suggest that MTHFR rs1801133 and MTHFR promoter methylation could jointly influence hypertension susceptibility.
Introduction
A single nucleotide polymorphism (SNP) is a genetic change at a specific location in a DNA sequence, where a single nucleotide (A, T, C, or G) is replaced by another in at least 1% of the population (1). Epigenetic changes are modifications in gene expression due to physiological or environmental stimuli (2, 3). DNA methylation is a heritable epigenetic regulatory mechanism that affects gene transcription and expression (4–8). Promoter DNA methylation is the addition of a methyl group to the C-5 position of a cytosine in the promoter region, forming 5-methylcytosine (7).
Hypertension is a life-threatening non-communicable disease characterized by abnormally higher blood pressure. Hypertension is the leading cause of premature death worldwide (9). Globally, approximately 1.278 billion adults aged between 30 and 79 were hypertensive in 2019 (10). The disease could affect about 1.56 billion adults worldwide in 2025 (11). Hypertensive patients do not present with typical symptoms, prompting the need for early identification and regular follow-up of high-risk individuals for effective implementation of preventive and therapeutic management measures (12, 13).
Hypertension has a multifaceted pathophysiology, originating from genetic, epigenetic, and environmental sources, alongside a complex interplay of factors (2, 14–19). So far, only about 10% of the hypertension etiology is known (20). Some known risk determinants of hypertension include SNP, DNA methylation, alcohol drinking, cigarette smoking, inactive lifestyle, sex, body mass index (BMI), and age (20–27). Genome-wide association studies —GWAS— (28–31) and subsequent studies including meta-analyses (26, 27, 32–34) have identified several hypertension-related single nucleotide polymorphisms (SNPs). The known loci explain only about 41% of hypertension heritability (35). Since DNA methylation also affects hypertension (2), gene-environment interactions could account for the remaining heritability (3). Moreover, since the genome and epigenome interwind (36), combining the genetic and epigenetic biomarkers could improve risk stratification and identify potential targets for pharmacological and lifestyle interventions (32, 37, 38).
MTHFR is a regulatory enzyme that plays a pivotal role in folate and homocysteine metabolism by catalyzing the synthesis of the main circulatory folate (5methylenetetrahydrofolate) from 5 to 10-methylenetetrahydrofolate (39). This MTHFR-catalyzed folate metabolism pathway plays a role in DNA methylation through methylenetetrahydrofolate, which acts as a co-substrate for the methylation of homocysteine to methionine, generating the methyl donor, S-adenosylmethionine (39, 40). Hypermethylation of MTHFR and the resulting decrease in gene expression is associated with several conditions and disorders, including but not limited to oxidative stress, diabetes, diabetic complications, ischemic stroke, and cancer (41–43).
A1298C (rs1801131) and C677T (rs1801133) are the major MTHFR variants that impair the gene’s activities (44, 45). A1298C results from adenine (A) to cytosine (C) substitution at position 1,298 in exon 7 of MTHFR while C677T (rs1801133) results when thymidine (T) replaces cytosine (C) at position 677 (45). C677T is the most common and broadly studied MTHFR variant (46). Even though several studies found MTHFR rs1801133 as an independent risk promoter for hypertension in different populations (10, 19, 47–52), a study among Mexicans found a significant protective effect of the variant on hypertension (21). Other studies involving Danish (53), Chinese (54), Caucasians (55), and Japanese (56) found no significant association between rs1801133 and hypertension. To our knowledge, only one study (with only 173 participants) assessed the association between rs1801133 and hypertension among Taiwanese (48). Moreover, no study determined the risk of hypertension in relation to both MTHFR methylation and SNP among Taiwanese.
The inconsistent results and inadequate studies in some populations pave the way for further investigations (50). Moreover, despite being the most common and broadly studied variant, the mechanistic insights into the causative role of MTHFR rs1801133 in hypertension require investigations (46). Hence, conducting large-scale studies to determine the risk of hypertension based on MTHFR methylation and single nucleotide polymorphism in Taiwanese and other populations is worthwhile. Therefore, in the current, we evaluated the independent and interactive association of MTHFR rs1801133 single nucleotide polymorphism and MTHFR promoter methylation with hypertension among Taiwanese adults.
Materials and methods
Study participants
The Taiwan Biobank (TWB) collected the data used in the current study. The TWB project is a community-based study aimed at identifying disease biomarkers and underlying mechanisms through the integration of lifestyle, environmental, and genetic data (57, 58). This human biological database provides data for biomedical research in Taiwan (58). The TWB project enrolls only Taiwanese adults aged 30–70 years with no previous diagnosis of cancer (58). We initially included 1,241 individuals with methylation data in our study. However, we excluded 3 people from the final analysis because of missing data on exercise, BMI, and waist circumference. The final study sample included 1,238 participants, comprising 157 hypertensive and 1,081 non-hypertensive individuals. All participants signed informed consent forms before data collection. The Institutional Review Boards of Cathay General Hospital (CGH-P109032 and CGH-P10941) and Chung Shan Medical University Hospital (CS1–20009) approved this study.
Genetic and epigenetic data
The Axiom Genome-Wide Array Plate chip system (Affymetrix Inc., Santa Clara, CA, USA) TWB (V2.0) chip was used for whole-genome genotyping. We used MTHFR rs1801133 (with genotypes CC, CT, and TT and minor allele T) in the current study because it is a well-established candidate for hypertension in several populations (10, 19, 47–52). The Hardy-Weinberg Equilibrium for MTHFR rs1801133 in the control group was 0.5999. The Illumina Infinium Methylation EPIC Bead Chip (Illumina Inc. San Diego, CA, USA) was used to determine whole blood-DNA methylation. We used MTHFR CpG sites at TSS1500 and TSS200 as promoter methylation data in our study (59, 60). Analysis of methylation data, including normalization, correction for batch effect, and cell-type heterogeneity was done using the ChAMP package.
Definition of hypertension and covariates
We obtained information on physician-diagnosed hypertension, sex, age, cigarette smoking, alcohol intake, and exercise using self-reported responses to the TWB questionnaires.
Cigarette smokers were those who have ever smoked cigarettes continuously for at least six months. Alcohol drinkers included those individuals who reported a regular habit of drinking alcohol continuously for at least six months. Having a regular exercise habit meant engaging in physical activities (lasting more than thirty minutes) at least three times per week. We calculated the BMI using the standard formula: weight/height squared (kg/m2). Metabolic components were divided into normal and abnormal values based on the cutoffs used by the Taiwan Ministry of Health and Welfare and the American Heart Association (AHA)/National Heart, Lung, and Blood Institute (NHLBI) (61, 62). Waist circumferences <90 cm in men and <80 cm in women were considered normal, while values ≥90 cm in men and ≥80 cm in women were considered abnormal. Systolic blood pressure (SBP) <130 mmHg was normal, while ≥130 mmHg was abnormal. Diastolic blood pressure (DBP) <85 mmHg was normal, while ≥85 mmHg was abnormal. Fasting blood glucose (FBG) <100 mg/dl was normal, while ≥100 mg/dl was abnormal. Triglyceride <150 mg/dl was normal and ≥150 mg/dl was abnormal. High-density lipoprotein cholesterol levels ≥40 mg/dl in men and ≥50 mg/dl in women were normal, and <40 mg/dl in men and <50 mg/dl in women were abnormal.
Statistical analyses
We evaluated the distribution of demographic data among individuals with the rs1801133 genotypes (CC, CT, and TT) using the student’s t-test (for continuous variables) and the chi-square test (for categorical variables). We used multiple logistic regression in determining the association of MTHFR rs1801133 and MTHFR promoter methylation quartiles (≥Q3 vs. Q2–Q3, Q1–Q2, and <Q1) with hypertension. We also used multiple logistic regression analysis to assess the interaction between MTHFR rs1801133 and MTHFR promoter methylation. We assessed the relationship between MTHFR rs1801133 and hypertension using the dominant (CC vs. CT + TT), recessive (CC + CT vs. TT), additive (CC/CT/TT), and log additive models (CC vs. CT and TT). The adjusted covariates were sex, age, body mass index, cigarette smoking, alcohol drinking, exercise, and metabolic components (waist circumference, systolic blood pressure, diastolic blood pressure, fasting blood glucose, triglycerides, and high-density lipoprotein cholesterol). SAS 9.4 software (SAS Institute, Cary, NC, USA) and PLINK 1.90 beta (Shaun Purcell & Christopher Chang, URL: www.cog-genomics.org/plink/1.9/) were used for data management and analyses. P < 0.05 was the threshold for statistical significance.
Results
Table 1 illustrates the basic characteristics of the study participants stratified by the MTHFR rs1801133 genotypes (CC, CT, and TT). The proportion of individuals with hypertension was significantly different according to MTHFR rs1801133 genotypes (P < 0.0001). The MTHFR promoter methylation levels (means ± standard deviations) were also significantly different in terms of rs1801133 genotypes (P = 0.0065). The CC genotype had the lowest level (0.1375 ± 0.0064) followed by CT (0.1379 ± 0.0068) and TT (0.1395 ± 0.0063). The MTHFR promoter methylation quartiles were β < 0.1338 (<Q1), 0.1338 ≤ β < 0.1385 (Q1–Q2), 0.1385 ≤ β < 0.1423 (Q2–Q3), and β ≥ 0.1423 (≥Q3). The distribution (proportion) of individuals in the various quartiles was significantly different based on MTHFR rs1801133 genotypes (P = 0.0317).
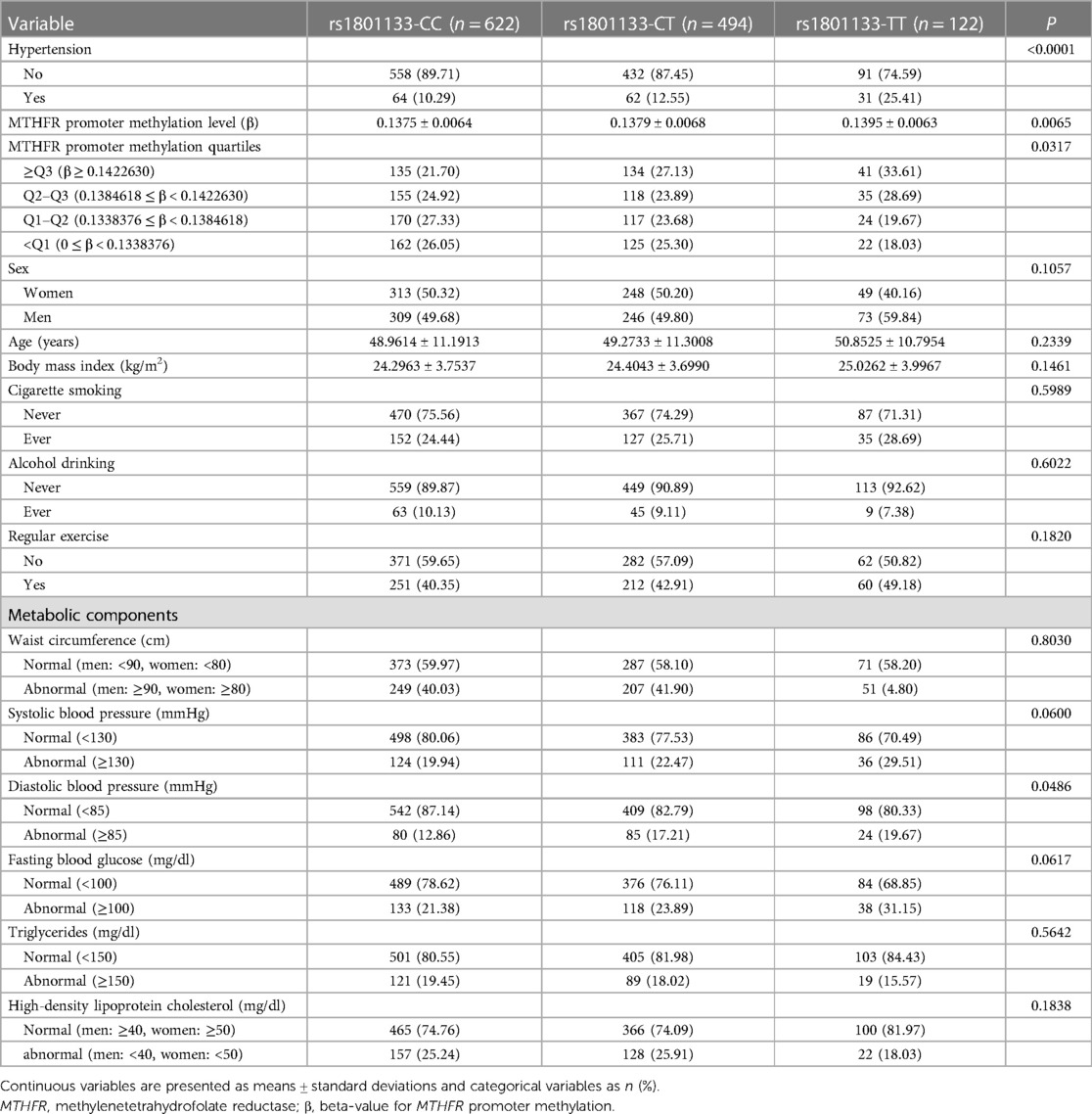
Table 1. Demographic characteristics of the study participants according to the MTHFR rs1801133 genotypes.
Table 2 shows the risk of hypertension according to the MTHFR rs1801133 genotypes (CC vs. CT and TT) and MTHFR promoter methylation (≥Q3 vs. Q2–Q3, Q1–Q2, and <Q1). The TT genotype was significantly associated with a higher risk of hypertension (OR; 95% CI = 2.718; 1.503–4.914), while the CT genotype was not. The association of CT and TT with hypertension was dose-dependent (P-trend = 0.0041). All the MTHFR methylation quartiles were not significantly associated with hypertension. However, the interaction between the MTHFR promoter methylation quartiles and rs1801133 was significant (P = 0.0323). Older age, higher SBP, and higher FBG were significantly associated with a higher risk of hypertension: OR; 95% CI = 1.061; 1.032–1.092 for age, 5.524; 3.488–8.746 for SBP, and 2.235; 1.476–3.382 for FBG.
Table 3 shows the association between MTHFR promoter methylation and hypertension stratified by the rs1801133 genotypes. Compared to the reference quartile (≥Q3), lower MTHFR promoter methylation quartiles were significantly associated with a higher risk of hypertension among individuals carrying the CC genotypes. The ORs (95% CIs) were 3.225 (1.140–9.124), 4.177 (1.424–12.247), and 8.645 (2.513–29.739) for Q2–Q3, Q1–Q2, and <Q1, respectively. The trend test was significant (P-trend = 0.0009).
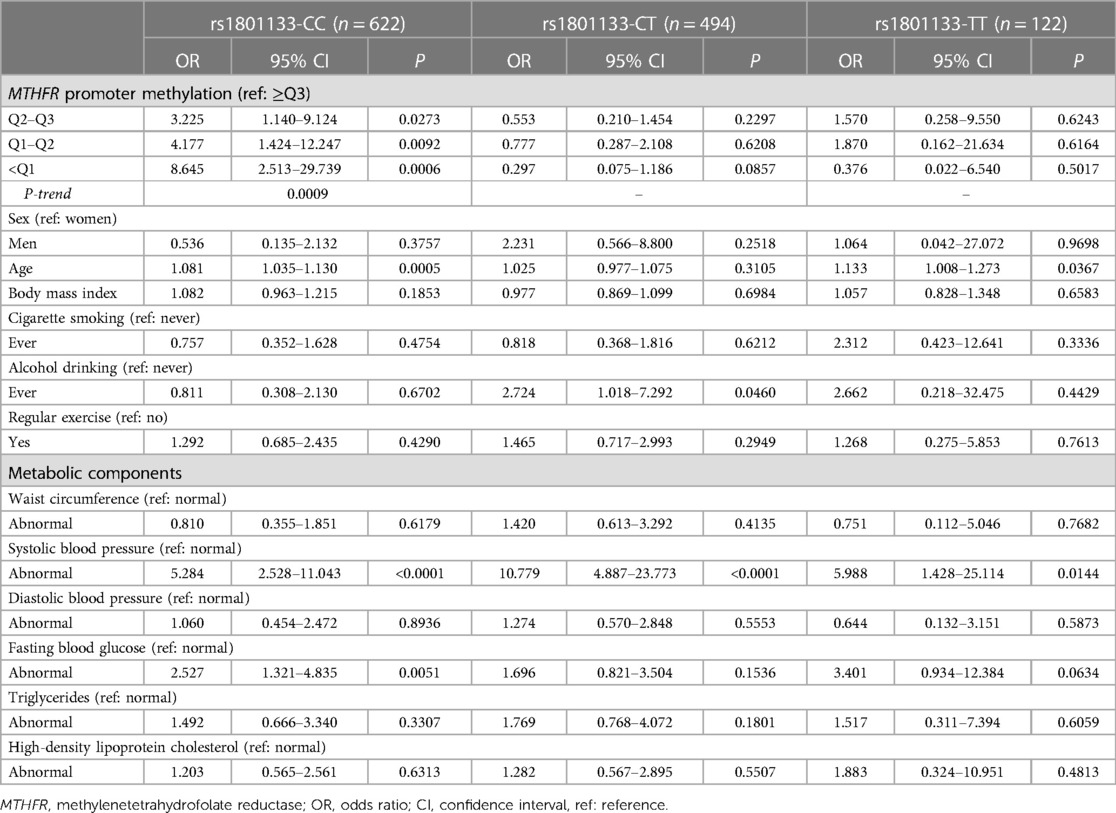
Table 3. Association between MTHFR promoter methylation and hypertension stratified by MTHFR rs1801133 genotypes.
The Supplementary Material shows the association between MTHFR rs1801133 and hypertension based on the dominant, recessive, and additive models. Using the dominant model (CC vs. CT + TT), the risk of hypertension was not significant (Supplementary Table S1-1). Nonetheless, the interaction between the MTHFR rs1801133 and promoter methylation was significant. Stratification by the genotypes revealed a significant association between hypertension and lower MTHFR promoter methylation quartiles among individuals carrying the CC genotypes (Supplementary Table S1-2). Using the recessive model (CC + CT vs. TT), the risk of hypertension was significantly higher among individuals carrying the TT genotype: OR; 95% CI = 2.513; 1.446–4.368 (Supplementary Table S2-1). The association between hypertension and MTHFR rs1801133 based on the additive model was also significant: OR; 95% CI = 1.519; 1.142–2.022 (Supplementary Table S3-1).
Discussion
Improving hypertension prevention, diagnosis, and treatment requires pinpointing the associated genetic variants involved in the disease’s susceptibility, progression, and severity (38). As an integrated functional genomics approach, epigenetics could delineate some molecular mechanisms behind human diseases (63). However, knowledge about genetic and epigenetic events involved in disease susceptibility is scarce (36). In the current study, the risk of hypertension was significantly higher among individuals with the TT genotype compared to the CC genotype. Moreover, lower MTHFR promoter methylation levels were dose-dependently associated with a higher risk of hypertension among carriers of the CC genotype. To our knowledge, this is the first study to suggest the possible role of the MTHFR rs1801133 variant and promoter methylation in the pathogenesis of hypertension among Taiwanese. These findings add insights into some of the genetic and epigenetic mechanisms behind the disease onset.
The MTHFR rs1801133 variant interferes with the folate metabolic pathway by reducing the activity and thermostability of the MTHFR enzyme, thereby disrupting the methylation process (64). The interference in folate metabolism is associated with higher plasma homocysteine (Hcy) or hyperhomocysteinemia (65–67). Hyperhomocysteinemia increases oxidative stress and disrupts the elasticity of the vascular wall, causing endothelial damage, hypertension, and other complications (68–71).
The significantly positive association between the MTHFR rs1801133 TT genotype and hypertension observed in the current study is concurrent with previous findings. For instance, several meta-analyses involving individuals of different ethnic backgrounds found the TT genotype and the T allele of MTHFR rs1801133 as hypertension-susceptible variants (45, 72–74). A meta-analysis found a positive relationship between MTHFR C677T and hypertension but concluded that the findings were not robust enough (75). Another meta-analysis found a significantly positive relationship between the SNP and hypertension among Caucasians and East Asians (76). Several population-based studies also confirmed the positive relationship between the TT genotype or the T allele with hypertension. For instance, the T allele was associated with a higher risk of hypertension in Taiwanese (48), Chinese (47, 50, 77), Indians (78), and Australian Caucasians (52), and the TT genotype in Moroccans (51). Nonetheless, the variant was not significantly associated with hypertension in Algerians (79), South Africans (80), Chinese (54, 81), Danish (53), Caucasians (55), Japanese (56), Black Africans (76), Latinos (76), Sri Lankans (76), and Indians (76). We believe that the conflicting results between our study and those previously reported could, in part, be due to differences in ethnicities, sample sizes, and study designs.
The mechanism behind the link between MTHFR rs1801133 and hypertension is unclear. Epigenetic changes in the regulatory region, especially MTHFR promoter methylation levels are believed to affect gene expression, regardless of genetic mutations (82). Moreover, DNA methylation and epigenetic pathways are reversible and believed to be potential preventive and therapeutic targets in hypertension management (15). DNA methylation is a suggested epigenetic and pathological mechanism underpinning the role of genetic variants in disease susceptibility and progression (83). Hypomethylation related to folic acid was associated with the CC genotype of rs1801133, suggesting the possible epigenetic role of MTHFR (84). Moreover, the promoter activity of the regulatory sequence of MTHFR caused by rs1801133 was drastically reduced by in vitro methylation (21). Since MTHFR promotor methylation regulates MTHFR expression, it could be one of the mechanisms behind the effect of MTHFR variants on hypertension (21, 22). A trans-ancestry study found significant relationships between MTHFR and DNA methylation. The authors suggested that DNA methylation may be involved in the regulatory pathway connecting common genetic variants with blood pressure and multiple phenotypes (85).
In the current study, lower MTHFR promoter methylation levels were dose-dependently associated with a higher risk of hypertension among carriers of the CC genotype. So far, no studies have studied the risk of hypertension in relation to both MTHFR promoter methylation and MTHFR rs1801133. Our results support the view that DNA methylation might be involved in the association between the MTHFR gene and hypertension. MTHFR hypomethylation was associated with lower levels of MTHFR and higher levels of homocysteine (82, 86). MTHFR promoter hypermethylation was protective against ischemic stroke, implying that it could be a diagnostic marker for the disease (86). It is worth noting that hypertension is a risk factor for ischemic stroke (87). A randomized controlled trial (RCT) found riboflavin, a cofactor for MTHFR, as a modulator of global and gene-specific methylation in adults carrying the MTHFR TT genotype (88). Another RCT found that riboflavin can alter the DNA methylation profiles of some hypertension-related genes in adults carrying the MTHFR 677TT genotype (89).
In our study, alcohol drinkers with the CT genotype had a higher risk of hypertension. Alcohol drinking is positively associated with hypertension (24, 90–93). However, this relationship might differ based on the quantity consumed and frequency of intake. For instance, in a systematic meta-analysis of cohort studies among Koreans, Japanese, and Americans, alcohol intake of 50–100 g per day was associated with a higher risk of hypertension in both men and women (24). Nonetheless, an intake of <5 g per day had a linear dose-dependent relationship with hypertension in men and a J-shaped relationship in women, suggesting that low alcohol intake might protect against hypertension in women (24). In another meta-analysis of cohort studies, men who consumed at least one alcoholic drink per day had a significantly higher risk of hypertension (94). However, women who consumed 1 or 2 drinks per day did not have a significant risk (94). Similar to our results, fasting blood glucose (90, 95, 96) and age (90, 96–99) were associated with a higher risk of hypertension. Even though age is a well-documented promoter of hypertension (99), its relationship with blood pressure (a marker of hypertension) is inconsistent (90). That is, the trends in the relationship of age with SBP and DBP vary (100–104). As an example, the Framingham Heart Study showed different trends in SBP and DBP between ages 30 and 84 (103). In a study, SBP continuously increased with age while DBP increased, peaked at 50, and decreased between 60 and 80 years (100).
A limitation of the current study is that our data source did not have data on homocysteine. As such, we did not include such data in our analysis.
Conclusion
MTHFR rs1801133 TT was independently associated with a higher risk of hypertension. This suggests that the TT genotype might elevate the risk of hypertension, regardless of methylation. However, MTHFR promoter methylation was not significantly associated with hypertension. Nonetheless, its interaction with MTHFR rs1801133 was significant. Based on genotypes, lower promoter MTHFR levels were associated with a higher risk of hypertension among individuals with the CC genotype. Our findings suggest that MTHFR rs1801133 and MTHFR promoter methylation could jointly influence hypertension susceptibility. Therefore, integrating genetic and epigenetic markers of hypertension improves risk stratification, which could enhance the implementation of targeted management strategies.
Data availability statement
The data analyzed in this study is subject to the following licenses/restrictions: The data that support the findings of this study are available from Taiwan Biobank but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the corresponding author upon reasonable request and with permission of Taiwan Biobank. Requests to access these datasets should be directed to Y-PL,TGlhd3lwQGNzbXUuZWR1LnR3.
Ethics statement
The studies involving humans were approved by The Institutional Review Boards of Cathay General Hospital (CGH-P109032 and CGH-P10941) and Chung Shan Medical University Hospital (CS1-20009). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
Conceptualization, M-HC, C-HC, DMT, T-WH, C-HH, J-HZ, and Y-PL; Formal analysis, C-HH, J-HZ, and Y-PL; Methodology, M-HC, C-HC, DMT, T-WH, C-HH, J-HZ, and Y-PL; Supervision, Y-PL; Validation, M-HC, C-HC, DMT, T-WH, C-HH, J-HZ, and Y-PL; Writing—original draft, M-HC, C-HC, and DMT; Writing—review and editing, M-HC, C-HC, DMT, T-WH, C-HH, J-HZ, and Y-PL. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the Cathay General Hospital (CGH-MR-A10907 and CGH-MR-A10908) and the Ministry of Science and Technology, MOST (MOST 111-2121-M-040-002).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2023.1159764/full#supplementary-material
References
1. Tang B. Preview of single nucleotide polymorphism (Snp) and potential correlation between human genomes and evolutionary history as well as religious behaviors. J Adv Res Biotech. (2019) 4(1):1–11.
2. Xu G, Wang Z, Li L, Li W, Hu J, Wang S, et al. Hypermethylation of dihydrofolate reductase promoter increases the risk of hypertension in Chinese. J Res Med Sci. (2020) 25:117. doi: 10.4103/jrms.JRMS_895_19
3. Thomas D. Gene–environment-wide association studies: emerging approaches. Nat Rev Genet. (2010) 11(4):259–72. doi: 10.1038/nrg2764
4. Boks MP, Derks EM, Weisenberger DJ, Strengman E, Janson E, Sommer IE, et al. The relationship of DNA methylation with age, gender and genotype in twins and healthy controls. PLoS One. (2009) 4(8):e6767. doi: 10.1371/journal.pone.0006767
5. Kaminsky ZA, Tang T, Wang S-C, Ptak C, Oh GH, Wong AH, et al. DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet. (2009) 41(2):240–5. doi: 10.1038/ng.286
6. Handy DE, Castro R, Loscalzo J. Epigenetic modifications: basic mechanisms and role in cardiovascular disease. Circulation. (2011) 123(19):2145–56. doi: 10.1161/CIRCULATIONAHA.110.956839
7. Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. (2013) 38(1):23–38. doi: 10.1038/npp.2012.112
8. Jin Z, Liu Y. DNA methylation in human diseases. Genes Dis. (2018) 5(1):1–8. doi: 10.1016/j.gendis.2018.01.002
9. Organization WH. Hypertension. (2021). [Cited 2022 September 30]. Available at: https://www.who.int/news-room/fact-sheets/detail/hypertension
10. Zhou B, Carrillo-Larco RM, Danaei G, Riley LM, Paciorek CJ, Stevens GA, et al. Worldwide trends in hypertension prevalence and progress in treatment and control from 1990 to 2019: a pooled analysis of 1,201 population-representative studies with 104 million participants. Lancet. (2021) 398(10304):957–80. doi: 10.1016/S0140-6736(21)01330-1
11. Kearney PM, Whelton M, Reynolds K, Muntner P, Whelton PK, He J. Global burden of hypertension: analysis of worldwide data. Lancet. (2005) 365(9455):217–23. doi: 10.1016/S0140-6736(05)17741-1
12. Flack JM, Sica DA, Bakris G, Brown AL, Ferdinand KC, Grimm Jr RH, et al. Management of high blood pressure in blacks: an update of the international society on hypertension in blacks consensus statement. Hypertension. (2010) 56(5):780–800. doi: 10.1161/HYPERTENSIONAHA.110.152892
13. Ferdinand KC, Nasser SA. Management of essential hypertension. Cardiol Clin. (2017) 35(2):231–46. doi: 10.1016/j.ccl.2016.12.005
14. Zhang X, Zhao H, Zhang J, Han D, Zheng Y, Guo X, et al. Gene environment interaction of Galnt2 and apoe gene with hypertension in the Chinese Han population. Biomed Mater Eng. (2015) 26(s1):S1977–S83. doi: 10.3233/BME-151501
15. Wang J, Gong L, Tan Y, Hui R, Wang Y. Hypertensive epigenetics: from DNA methylation to micrornas. J Hum Hypertens. (2015) 29(10):575–82. doi: 10.1038/jhh.2014.132
16. Snieder H, Harshfield GA, Treiber FA. Heritability of blood pressure and hemodynamics in African-and European-American youth. Hypertension. (2003) 41(6):1196–201. doi: 10.1161/01.HYP.0000072269.19820.0D
17. Dominiczak A, Delles C, Padmanabhan S. Genomics and precision medicine for clinicians and scientists in hypertension. Hypertension. (2017) 69(4):e10–3. doi: 10.1161/HYPERTENSIONAHA.116.08252
18. Ahn S-Y, Gupta C. Genetic programming of hypertension. Front Pediatr. (2018) 5:285. doi: 10.3389/fped.2017.00285
19. Bayramoglu A, Urhan Kucuk M, Guler HI, Abaci O, Kucukkaya Y, Colak E. Is there any genetic predisposition of Mmp-9 gene C1562t and Mthfr gene C677t polymorphisms with essential hypertension? Cytotechnology. (2015) 67(1):115–22. doi: 10.1007/s10616-013-9665-0
20. Rossier BC, Bochud M, Devuyst O. The hypertension pandemic: an evolutionary perspective. Physiology. (2017) 32(2):112–25. doi: 10.1152/physiol.00026.2016
21. Pérez-Razo JC, Cano-Martínez LJ, Vargas Alarcón G, Canizales-Quinteros S, Martínez-Rodríguez N, Canto P, et al. Functional polymorphism Rs13306560 of the MTHFR gene is associated with essential hypertension in a Mexican-mestizo population. Circ: Cardiovasc Genet. (2015) 8(4):603–9. doi: 10.1161/CIRCGENETICS.114.000942
22. Tomaszewski M, Debiec R, Braund PS, Nelson CP, Hardwick R, Christofidou P, et al. Genetic architecture of ambulatory blood pressure in the general population: insights from cardiovascular gene-centric array. Hypertension. (2010) 56(6):1069–76. doi: 10.1161/HYPERTENSIONAHA.110.155721
23. Liu Y, Xu C, Wang Y, Yang C, Pu G, Zhang L, et al.. Association analysis of Mthfr (Rs1801133 and Rs1801131) and Mtrr (Rs1801394) gene polymorphisms towards the development of hypertension in the Bai population from Yunnan, China. Clin Exp Hypertens. (2023) 45(1):2206066. doi: 10.1080/10641963.2023.2206066
24. Taylor B, Irving HM, Baliunas D, Roerecke M, Patra J, Mohapatra S, et al. Alcohol and hypertension: gender differences in dose–response relationships determined through systematic review and meta-analysis. Addiction. (2009) 104(12):1981–90. doi: 10.1111/j.1360-0443.2009.02694.x
25. Ornosa-Martín G, Fernandez-Ballart JD, Ceruelo S, Ríos L, Ueland PM, Meyer K, et al. Homocysteine, the methylenetetrahydrofolate reductase 677c > T polymorphism and hypertension: effect modifiers by lifestyle factors and population subgroups. Br J Nutr. (2020) 124(1):69–79. doi: 10.1017/S0007114520000793
26. Ghafar MTA. An overview of the classical and tissue-derived renin-angiotensin-aldosterone system and its genetic polymorphisms in essential hypertension. Steroids. (2020) 163:108701. doi: 10.1016/j.steroids.2020.108701
27. Ghafar MTA. Association of aldosterone synthase Cyp11b2 (-344c/T) gene polymorphism with essential hypertension and left ventricular hypertrophy in the Egyptian population. Clin Exp Hypertens. (2019) 41(8):779–86. doi: 10.1080/10641963.2018.1557679
28. Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. (2009) 41(6):666–76. doi: 10.1038/ng.361
29. Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. (2009) 41(6):677–87. doi: 10.1038/ng.384
30. Padmanabhan S, Melander O, Johnson T, Di Blasio AM, Lee WK, Gentilini D, et al. Genome-wide association study of blood pressure extremes identifies variant near umod associated with hypertension. PLoS Genet. (2010) 6(10):e1001177. doi: 10.1371/journal.pgen.1001177
31. Wang Y, Wang J-G. Genome-wide association studies of hypertension and several other cardiovascular diseases. Pulse. (2019) 6(3-4):169–86. doi: 10.1159/000496150
32. Sun D, Richard MA, Musani SK, Sung YJ, Winkler TW, Schwander K, et al. Multi-ancestry genome-wide association study accounting for gene-psychosocial factor interactions identifies novel loci for blood pressure traits. Human Genetics and Genomics Advances. (2021) 2(1):100013. doi: 10.1016/j.xhgg.2020.100013
33. Surendran P, Drenos F, Young R, Warren H, Cook JP, Manning AK, et al. Trans-ancestry meta-analyses identify rare and common variants associated with blood pressure and hypertension. Nat Genet. (2016) 48(10):1151–61. doi: 10.1038/ng.3654
34. Liu C, Kraja AT, Smith JA, Brody JA, Franceschini N, Bis JC, et al. Meta-analysis identifies common and rare variants influencing blood pressure and overlapping with metabolic trait loci. Nat Genet. (2016) 48(10):1162–70. doi: 10.1038/ng.3660
35. Warren H, Edwards T, Vaez A, Keaton J, Kamali Z, Xie T, et al. Genome-wide analysis in over 1 million individuals reveals over 2,000 independent genetic signals for blood pressure. (2022). Res Sq. doi: 10.21203/rs.3.rs-1409164/v1
36. Romanowska J, Haaland ØA, Jugessur A, Gjerdevik M, Xu Z, Taylor J, et al. Gene–methylation interactions: discovering region-wise DNA methylation levels that modify Snp-associated disease risk. Clin Epigenetics. (2020) 12(1):1–18. doi: 10.1186/s13148-020-00881-x
37. Pashayan N, Reisel D, Widschwendter M. Integration of genetic and epigenetic markers for risk stratification: opportunities and challenges. Per Med. (2016) 13(2):93–5. doi: 10.2217/pme.15.53
38. Padmanabhan S, Melander O, Hastie C, Menni C, Delles C, Connell JM, et al. Hypertension and genome-wide association studies: combining high fidelity phenotyping and hypercontrols. J Hypertens. (2008) 26(7):1275–81. doi: 10.1097/HJH.0b013e3282ff634f
39. Wiemels JL, Smith RN, Taylor GM, Eden OB, Alexander FE, Greaves MF, et al. Methylenetetrahydrofolate reductase (mthfr) polymorphisms and risk of molecularly defined subtypes of childhood acute leukemia. Proc Natl Acad Sci U S A. (2001) 98(7):4004–9. doi: 10.1073/pnas.061408298
40. Yan L, Zhao L, Long Y, Zou P, Ji G, Gu A, et al. Association of the maternal mthfr C677t polymorphism with susceptibility to neural tube defects in offsprings: evidence from 25 case-control studies. PloS One. (2012) 7(10):e41689. doi: 10.1371/journal.pone.0041689
41. dos Santos Nunes MK, Silva AS, de Queiroga Evangelista IW, Gomes CNAP, do Nascimento RAF, Luna RCP, et al. Hypermethylation in the promoter of the mthfr gene is associated with diabetic complications and biochemical indicators. Diabetol Metab Syndr. (2017) 9(1):1–9. doi: 10.1186/s13098-017-0284-3
42. Bezerra H S, de Assis C S, dos Santos Nunes MK, Wanderley de Queiroga Evangelista I, Modesto Filho J, Alves Pegado Gomes CN, et al. The mthfr promoter hypermethylation pattern associated with the A1298c polymorphism influences lipid parameters and glycemic control in diabetic patients. Diabetol Metab Syndr. (2019) 11(1):1–15. doi: 10.1186/s13098-018-0396-4
43. Wei L K, Sutherland H, Au A, Camilleri E, Haupt LM, Gan SH, et al. A potential epigenetic marker mediating serum folate and vitamin B12 levels contributes to the risk of ischemic stroke. BioMed Res Int. (2015) 2015:167976. doi: 10.1155/2015/167976
44. Clément A, Amar E, Brami C, Clément P, Alvarez S, Jacquesson-Fournols L, et al. Mthfr snps (methyl tetrahydrofolate reductase, single nucleotide polymorphisms) C677t and A1298c prevalence and Serum homocysteine levels in >2,100 hypofertile Caucasian male patients. Biomolecules. (2022) 12(8):1086. doi: 10.3390/biom12081086
45. Wu Y-L, Hu C-Y, Lu S-S, Gong F-F, Feng F, Qian Z-Z, et al. Association between methylenetetrahydrofolate reductase (mthfr) C677t/A1298c polymorphisms and essential hypertension: a systematic review and meta-analysis. Metab Clin Exp. (2014) 63(12):1503–11. doi: 10.1016/j.metabol.2014.10.001
46. Fan S, Yang B, Zhi X, Wang Y, Wei J, Zheng Q, et al. Interactions of methylenetetrahydrofolate reductase C677t polymorphism with environmental factors on hypertension susceptibility. Int J Environ Res Public Health. (2016) 13(6):601. doi: 10.3390/ijerph13060601
47. Wen C, Lv J-F, Wang L, Zhu W-F, Wan F-S, Wang X-Z. Association of a methylene tetrahydrofolate reductase C677t polymorphism with several blood chemical levels in a Chinese population. Genet Test Mol Biomarkers. (2015) 19(1):24–9. doi: 10.1089/gtmb.2014.0213
48. Lin C, Wei H. Low plasma pyridoxal 5’-phosphate concentration and mthfr 677c→ T genotypes are associated with increased risk of hypertension. Int J Vitam Nutr Res. (2008) 78(1):33–40. doi: 10.1024/0300-9831.78.1.33
49. Ilhan N, Kucuksu M, Kaman D, Ilhan N, Ozbay Y. The 677 C/T mthfr polymorphism is associated with essential hypertension, coronary artery disease, and higher homocysteine levels. Arch Med Res. (2008) 39(1):125–30. doi: 10.1016/j.arcmed.2007.07.009
50. Wu H, Huang Q, Yu Z, Zhong Z. Association of aldh2 Rs671 and mthfr Rs1801133 polymorphisms with hypertension among hakka people in southern China. BMC Cardiovasc Disord. (2022) 22(1):1–9. doi: 10.1186/s12872-021-02434-3
51. Nassereddine S, Kassogue Y, Korchi F, Habbal R, Nadifi S. Association of methylenetetrahydrofolate reductase gene (C677t) with the risk of hypertension in Morocco. BMC Res Notes. (2015) 8(1):1–5. doi: 10.1186/s13104-015-1772-x
52. Heux S, Morin F, Lea RA, Ovcaric M, Tajouri L, Griffiths LR. The methylentetrahydrofolate reductase gene variant (C677t) as a risk factor for essential hypertension in caucasians. Hypertens Res. (2004) 27(9):663–7. doi: 10.1291/hypres.27.663
53. Husemoen LLN, Skaaby T, Jørgensen T, Thuesen BH, Fenger M, Grarup N, et al. Mthfr C677t genotype and cardiovascular risk in a general population without mandatory folic acid fortification. Eur J Nutr. (2014) 53(7):1549–59. doi: 10.1007/s00394-014-0659-2
54. Zhan S, Gao Y, Yin X, Huang Y, Hu Y, Li L. A case-control study on the relationship between abnormal homocysteine metabolism and essential hypertension. Zhonghua Liu Xing Bing Xue Za Zhi. (2000) 21(3):194–7.11860783
55. Fowdar JY, Lason MV, Szvetko AL, Lea RA, Griffiths LR. Investigation of homocysteine-pathway-related variants in essential hypertension. Int J Hypertens. (2012) 2012:190923. doi: 10.1155/2012/190923
56. Lwin H, Yokoyama T, Yoshiike N, Saito K, Yamamoto A, Date C, et al. Polymorphism of methylenetetrahydrofolate reductase gene (C677t mthfr) is not a confounding factor of the relationship between serum uric acid level and the prevalence of hypertension in Japanese men. Circ J. (2006) 70(1):83–7. doi: 10.1253/circj.70.83
57. Fan C-T, Lin J-C, Lee C-H. Taiwan biobank: a project aiming to aid Taiwan’s transition into a biomedical island. Pharmacogenomics. (2008) 9(2):235–46. doi: 10.2217/14622416.9.2.235
58. Biobank T. Purpose of Taiwan Biobank. (2014). [Cited 2023]. Available at: https://taiwanview.twbiobank.org.tw/index
59. Ringh MV, Hagemann-Jensen M, Needhamsen M, Kular L, Breeze CE, Sjöholm LK, et al. Tobacco smoking induces changes in true DNA methylation, hydroxymethylation and gene expression in bronchoalveolar lavage cells. EBioMedicine. (2019) 46:290–304. doi: 10.1016/j.ebiom.2019.07.006
60. Titus AJ, Way GP, Johnson KC, Christensen BC. Deconvolution of DNA methylation identifies differentially methylated gene regions on 1p36 across breast cancer subtypes. Sci Rep. (2017) 7(1):11594. doi: 10.1038/s41598-017-10199-z
61. Grundy SM, Cleeman JI, Daniels SR, Donato KA, Eckel RH, Franklin BA, et al. Diagnosis and management of the metabolic syndrome: an American heart association/national heart, lung, and blood institute scientific statement. Circulation. (2005) 112(17):2735–52. doi: 10.1161/CIRCULATIONAHA.105.169404
62. Ministry of Health and Welfare T. 2022 Taiwan health and welfare report. (2022). [Cited 2023 August 2023]. Available at: https://www.mohw.gov.tw/dl-82936-f986905e-b09d-45e8-b87d-57ae9a7c5806.html
63. Mill J, Heijmans BT. From promises to practical strategies in epigenetic epidemiology. Nat Rev Genet. (2013) 14(8):585–94. doi: 10.1038/nrg3405
64. Frosst P, Blom H, Milos R, Goyette P, Sheppard CA, Matthews R, et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. (1995) 10(1):111–3. doi: 10.1038/ng0595-111
65. Kluijtmans L, Van den Heuvel L, Boers G, Frosst P, Stevens E, van Oost BA, et al. Molecular genetic analysis in mild hyperhomocysteinemia: a common mutation in the methylenetetrahydrofolate reductase gene is a genetic risk factor for cardiovascular disease. Am J Hum Genet. (1996) 58(1):35.8554066
66. Brattström L, Wilcken DE, Öhrvik J, Brudin L. Common methylenetetrahydrofolate reductase gene mutation leads to hyperhomocysteinemia but not to vascular disease: the result of a meta-analysis. Circulation. (1998) 98(23):2520–6. doi: 10.1161/01.CIR.98.23.2520
67. Pereira AC, Schettert IT, Morandini Filho AAF, Guerra-Shinohara EM, Krieger JE. Methylenetetrahydrofolate reductase (mthfr) C677t gene variant modulates the homocysteine folate correlation in a mild folate-deficient population. Clin Chim Acta. (2004) 340(1-2):99–105. doi: 10.1016/j.cccn.2003.09.016
68. Rodrigo R, González J, Paoletto F. The role of oxidative stress in the pathophysiology of hypertension. Hypertens Res. (2011) 34(4):431–40. doi: 10.1038/hr.2010.264
69. Lip GY, Edmunds E, Martin SC, Jones AF, Blann AD, Beevers DG. A pilot study of homocyst (E) ine levels in essential hypertension: relationship to von Willebrand factor, an index of endothelial damage. Am J Hypertens. (2001) 14(7):627–31. doi: 10.1016/S0895-7061(00)01321-2
70. Mallamaci F, Zoccali C, Tripepi G, Fermo I, Benedetto FA, Cataliotti A, et al. Hyperhomocysteinemia predicts cardiovascular outcomes in hemodialysis patients. Kidney Int. (2002) 61(2):609–14. doi: 10.1046/j.1523-1755.2002.00144.x
71. Fu L, Yn L, Luo D, Deng S, Wu B, Hu YQ. Evidence on the causal link between homocysteine and hypertension from a meta-analysis of 40 173 individuals implementing Mendelian randomization. J Clin Hypertens. (2019) 21(12):1879–94. doi: 10.1111/jch.13737
72. Qian X, Lu Z, Tan M, Liu H, Lu D. A meta-analysis of association between C677t polymorphism in the methylenetetrahydrofolate reductase gene and hypertension. Eur J Hum Genet. (2007) 15(12):1239–45. doi: 10.1038/sj.ejhg.5201914
73. Fan Y, Wu L, Zhuang W. Methylenetetrahydrofolate reductase gene Rs1801133 and Rs1801131 polymorphisms and essential hypertension risk: a comprehensive analysis. Cardiovasc Ther. (2022) 2022:2144443. doi: 10.1155/2022/2144443
74. Yang KM, Jia J, Mao LN, Men C, Tang KT, Li YY, et al. Methylenetetrahydrofolate reductase C677t gene polymorphism and essential hypertension: a meta-analysis of 10,415 subjects. Biomed Rep. (2014) 2(5):699–708. doi: 10.3892/br.2014.302
75. Meng H, Huang S, Yang Y, He X, Fei L, Xing Y. Association between mthfr polymorphisms and the risk of essential hypertension: an updated meta-analysis. Front Genet. (2021) 12:698590. doi: 10.3389/fgene.2021.698590
76. Yang B, Fan S, Zhi X, Li Y, Liu Y, Wang D, et al. Associations of mthfr gene polymorphisms with hypertension and hypertension in pregnancy: a meta-analysis from 114 studies with 15411 cases and 21970 controls. PloS one. (2014) 9(2):e87497. doi: 10.1371/journal.pone.0087497
77. Cai W, Yin L, Yang F, Zhang L, Cheng J. Association between hcy levels and the Cbs844ins68 and mthfr C677t polymorphisms with essential hypertension. Biomed Rep. (2014) 2(6):861–8. doi: 10.3892/br.2014.357
78. Markan S, Sachdeva M, Sehrawat BS, Kumari S, Jain S, Khullar M. Mthfr 677 ct/mthfr 1,298 cc genotypes are associated with increased risk of hypertension in Indians. Mol Cell Biochem. (2007) 302(1):125–31. doi: 10.1007/s11010-007-9434-5
79. Amrani-Midoun A, Kiando SR, Treard C, Jeunemaitre X, Bouatia-Naji N. The relationship between mthfr C677t gene polymorphism and essential hypertension in a sample of an Algerian population of Oran City. Int J Cardiol. (2016) 225:408–11. doi: 10.1016/j.ijcard.2016.10.027
80. Mabhida SEE, Mabhida SE, Sharma D, Rajan J, Apalata T, Masilela C, et al. The association of mthfr (Rs1801133) with hypertension in an indigenous South African population: a short communication. Front Genet. (2022) 13:937639. doi: 10.3389/fgene.2022.937639
81. Liu S, Liu M, Li Q, Liu X, Wang Y, Mambiya M, et al. Association of single nucleotide polymorphisms of mthfr, Tcn2, Rnf213 with susceptibility to hypertension and blood pressure. Biosci Rep. (2019) 39(12):BSR20191454. doi: 10.1042/BSR20191454
82. Osunkalu V, Taiwo I, Makwe C, Abiola A, Quao R, Anorlu R. Epigenetic modification in methylene tetrahydrofolate reductase (mthfr) gene of women with pre-eclampsia. J Obstet Gynecol India. (2021) 71(1):52–7. doi: 10.1007/s13224-020-01374-w
83. Shilpi A, Bi Y, Jung S, Patra SK, Davuluri RV. Identification of genetic and epigenetic variants associated with breast cancer prognosis by integrative bioinformatics analysis. Cancer Inform. (2017) 16:S39783. doi: 10.4137/CIN.S39783
84. Crider KS, Quinlivan EP, Berry RJ, Hao L, Li Z, Maneval D, et al. Genomic DNA methylation changes in response to folic acid supplementation in a population-based intervention study among women of reproductive age. PLoS One. (2011) 6(12):e28144. doi: 10.1371/journal.pone.0028144
85. Kato N, Loh M, Takeuchi F, Verweij N, Wang X, Zhang W, et al. Trans-ancestry genome-wide association study identifies 12 genetic loci influencing blood pressure and implicates a role for DNA methylation. Nat Genet. (2015) 47(11):1282–93. doi: 10.1038/ng.3405
86. Xu S, Shi Q, Li B, Han L, Xu G, Peng X, et al. High mthfr promoter methylation levels in men confer protection against ischemic stroke. Bosn J Basic Med Sci. (2020) 20(4):477–86. doi: 10.17305/bjbms.2020.4636
87. Hägg-Holmberg S, Dahlström EH, Forsblom CM, Harjutsalo V, Liebkind R, Putaala J, et al. The role of blood pressure in risk of ischemic and hemorrhagic stroke in type 1 diabetes. Cardiovasc Diabetol. (2019) 18(1):1–9. doi: 10.1186/s12933-019-0891-4
88. Amenyah SD, McMahon A, Ward M, Deane J, McNulty H, Hughes CF, et al. Riboflavin supplementation alters global and gene-specific DNA methylation in adults with the mthfr 677 Tt genotype. Biochimie. (2020) 173:17–26. doi: 10.1016/j.biochi.2020.04.007
89. Amenyah SD, Ward M, McMahon A, Deane J, McNulty H, Hughes C, et al. DNA methylation of hypertension-related genes and effect of riboflavin supplementation in adults stratified by genotype for the mthfr C677t polymorphism. Int J Cardiol. (2021) 322:233–9. doi: 10.1016/j.ijcard.2020.09.011
90. Tatsumi Y, Morimoto A, Asayama K, Sonoda N, Miyamatsu N, Ohno Y, et al. Fasting blood glucose predicts incidence of hypertension independent of Hba1c levels and insulin resistance in middle-aged Japanese: the saku study. Am J Hypertens. (2019) 32(12):1178–85. doi: 10.1093/ajh/hpz123
91. Zhao F, Liu Q, Li Y, Feng X, Chang H, Lyu J. Association between alcohol consumption and hypertension in Chinese adults: findings from the Chns. Alcohol. (2020) 83:83–8. doi: 10.1016/j.alcohol.2019.09.004
92. Wang Y, Yao Y, Chen Y, Zhou J, Wu Y, Fu C, et al. Association between drinking patterns and incident hypertension in Southwest China. Int J Environ Res Public Health. (2022) 19(7):3801. doi: 10.3390/ijerph19073801
93. Santana NMT, Mill JG, Velasquez-Melendez G, Moreira AD, Barreto SM, Viana MC, et al. Consumption of alcohol and blood pressure: results of the Elsa-Brasil study. PLoS One. (2018) 13(1):e0190239. doi: 10.1371/journal.pone.0190239
94. Roerecke M, Tobe SW, Kaczorowski J, Bacon SL, Vafaei A, Hasan OS, et al. Sex-specific associations between alcohol consumption and incidence of hypertension: a systematic review and meta-analysis of cohort studies. J Am Heart Assoc. (2018) 7(13):e008202. doi: 10.1161/JAHA.117.008202
95. Boyko E, Barr E, Zimmet P, Shaw J. Two-Hour glucose predicts the development of hypertension over 5 years: the ausdiab study. J Hum Hypertens. (2008) 22(3):168–76. doi: 10.1038/sj.jhh.1002316
96. Kuwabara M, Chintaluru Y, Kanbay M, Niwa K, Hisatome I, Andres-Hernando A, et al. Fasting blood glucose is predictive of hypertension in a general Japanese population. J Hypertens. (2019) 37(1):167–74. doi: 10.1097/HJH.0000000000001895
97. Group CPC. Association of age and blood pressure among 3.3 million adults: insights from China peace million persons project. J Hypertens. (2021) 39(6):1143–54. doi: 10.1097/HJH.0000000000002793
98. Ostchega Y, Fryar CD, Nwankwo T, Nguyen DT. Hypertension prevalence among adults aged 18 and over: United States, 2017–2018. Natl Health Stat. (2020).
99. Hameed F, Haque MU, Iqbal J, Hussain T, Memon ZH, Naz S. Effect of age on relationship between hypertension and its clinical manifestations. Pak J Med Health Sci. (2022) 16(1). doi: 10.53350/pjmhs2216146
100. Franklin S. Ageing and hypertension: the assessment of blood pressure indices in predicting coronary heart disease. J Hypertens Suppl. (1999) 17(5):S29–36.10706323
101. Chaudhry SI, Krumholz HM, Foody JM. Systolic hypertension in older persons. JAMA. (2004) 292(9):1074–80. doi: 10.1001/jama.292.9.1074
102. Franklin SS, Jacobs MJ, Wong ND, L’Italien GJ, Lapuerta P. Predominance of isolated systolic hypertension among middle-aged and elderly US hypertensives: analysis based on national health and nutrition examination survey (Nhanes) Iii. Hypertension. (2001) 37(3):869–74. doi: 10.1161/01.HYP.37.3.869
103. Franklin SS, Gustin IV W, Wong ND, Larson MG, Weber MA, Kannel WB, et al. Hemodynamic patterns of age-related changes in blood pressure: the Framingham heart study. Circulation. (1997) 96(1):308–15. doi: 10.1161/01.CIR.96.1.308
Keywords: MTHFR, promoter methylation, rs1801133, hypertension, susceptibility, interaction
Citation: Chiu M-H, Chang C-H, Tantoh DM, Hsu T-W, Hsiao C-H, Zhong J-H and Liaw Y-P (2023) Susceptibility to hypertension based on MTHFR rs1801133 single nucleotide polymorphism and MTHFR promoter methylation. Front. Cardiovasc. Med. 10:1159764. doi: 10.3389/fcvm.2023.1159764
Received: 7 February 2023; Accepted: 11 September 2023;
Published: 2 October 2023.
Edited by:
Roberto Pedrinelli, University of Pisa, ItalyReviewed by:
Ramcés Falfán-Valencia, National Institute of Respiratory Diseases-Mexico (INER), MexicoMuhammad Tarek Abdel Ghafar, Tanta University, Egypt
© 2023 Chiu, Chang, Tantoh, Hsu, Hsiao, Zhong and Liaw. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yung-Po Liaw TGlhd3lwQGNzbXUuZWR1LnR3
†These authors contributed equally and share the first authorship
 Ming-Huang Chiu1,†
Ming-Huang Chiu1,† Yung-Po Liaw
Yung-Po Liaw