
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cardiovasc. Med., 18 August 2022
Sec. Cardiovascular Biologics and Regenerative Medicine
Volume 9 - 2022 | https://doi.org/10.3389/fcvm.2022.944393
This article is part of the Research TopicMyocardium Regeneration and CardioprotectionView all 6 articles
Cardiovascular diseases remain the leading cause of death worldwide, particularly ischemic heart disease (IHD). It is also classified as incurable given the irreversible damage it causes to cardiomyocytes. Thus, myocardial tissue rejuvenation following ischemia is one of the global primary research concerns for scientists. Interestingly, the mammalian heart thrives after an injury during the embryonic or neonatal period; however, this ability disappears with increasing age. Previous studies have found that specific non-coding (nc) RNAs play a pivotal role in this process. Hence, the review herein summarizes the research on cardiomyocyte regenerative medicine in recent years and sets forth the biological functions and mechanisms of the micro (mi)RNA, long non-coding (lnc)RNA, and circular (circ)RNA in the posttranscriptional regulation of cardiomyocytes. In addition, this review summarizes the roles of ncRNAs in specific species while enumerating potential therapeutic strategies for myocardial infarction.
Ischemic heart disease (IHD) is a significant health concern worldwide (1) and has been associated with increased morbidity and mortality over time (2, 3). IHD has also been incurable, given the irreversible damage to cardiomyocytes. In addition, minimal progress for myocardial infarction (MI) treatment on coronary reopening, increased blood flow to the infarcted area, and coronary artery bypass grafts (CABG) usage has been achieved (4–6).
Given that that cardiomyocytes hardly regenerate in mammals distinguishes them from other lower vertebrates (7, 8). However, recent studies have found that cardiomyocytes are not completely non-renewable (4, 9, 10). In contrast, neonatal mouse cardiomyocytes could proliferate to some extent (11, 12). During myocardium development, cardiomyocytes gradually break away from the cell cycle and become non-renewable over time (13, 14).
Cardiomyocytes undergo extensive proliferation during embryogenesis but exit the proliferating cell cycle shortly after birth (9, 13). Most cardiomyocytes undergo the last round of DNA replication and nuclear division without cytokinesis from the 5th day after birth in rodents resulting in approximately 90% of cardiomyocytes stopping proliferation in the 2nd week after birth (15, 16). At this stage (2 weeks after birth), the proliferation potential is limited to a small fraction of mono-nuclear cardiomyocytes (15).
Nonetheless, recent studies have shown that adult cardiomyocytes can re-enter the cell cycle and proliferate under certain mitogen actions (such as fibroblast growth factor, periosteal protein, and neuregulin) (17–20), which seems to be limited to a small number of mono-nuclear cardiomyocytes.
In the first 10-years of life, the human cardiomyocytes turnover rate, mainly ventricular myocytes, is estimated to be 0.3–1% yearly (21), which is lower than other human body tissues, particularly when tissues are damaged after Mi. Despite several study results indicating that mouse and neonatal fetal hearts myocardium could regenerate after tissue damage (15, 16, 22), G1/S and G2/M cyclin expression and cyclin-dependent kinases in the cell cycle are usually downregulated, while the associated cell cycle inhibitor levels are increased (10, 15).
However, when transitioning between the juvenile or adult phase, cardiomyocytes break out of the cell cycle and enter a quiescence phase (G0) (23). During the MI process or myocardial ischemia-reperfusion (I/R) injury, both regulated and non-regulated cell death processes are involved. Later, scar tissue formation will follow this initial injury response (24). Hence, understanding the heart's regenerative capacity is the primary goal of cardiac regenerative medicine.
MicroRNA (miRNA) is an endogenous non-coding single-stranded RNA composed of approximately 20–24 nucleotides (25). It was first described in nematodes in 1993 (25, 26). Since then, miRNA has been investigated in multiple contexts, confirming that it plays a regulatory role in various organisms and associated diseases. The miRNA mechanism binds to the 3' UTR at the end of the messenger RNA (mRNA) and then targets the gene through posttranscriptional regulation, thereby inhibiting the target gene expression in most cases (26). miRNA is widely present in eukaryotes, and there are traces of its existence from lower organisms to humans (8). Its biological characteristics include a high degree of conservation and temporal and tissue expression specificity (27).
With miRNA, there is a high degree of evolutionary conservation among various species with its sequence structure (28). Researchers believe that miRNAs conservation has an important biological significance, suggesting that miRNAs have the same regulatory mechanism during several organisms' development (29). It thus provides additional evidence for the conclusions derived using miRNAs in cross-species studies (28). Likewise, it also provides a basis for the homology of early biological evolution.
In addition, there are significant differences in expression levels for the same miRNA in different tissues and developmental stages (30). Some miRNAs demonstrate temporal expression during development (31). Several studies have pointed out that miRNAs can specifically regulate cell cycle-related proteins, such as CDK and P21, and the proliferation of juvenile cardiomyocytes is also inseparable from the regulation of cyclins and related pathways. miRNAs that directly act on the cell cycle and the relative signal pathway are summarized in Figures 1, 2, respectively.
This review article focuses on the most recent research concerning miRNAs and their role in cardiomyocyte regeneration. miRNAs that enhance or inhibit cardiomyocytes proliferation are summarized in Tables 1, 2.

Table 1. A summary of the in vitro and in vivo effects of miRNAs on enhanced cardiomyocyte proliferation and the potential mechanism.
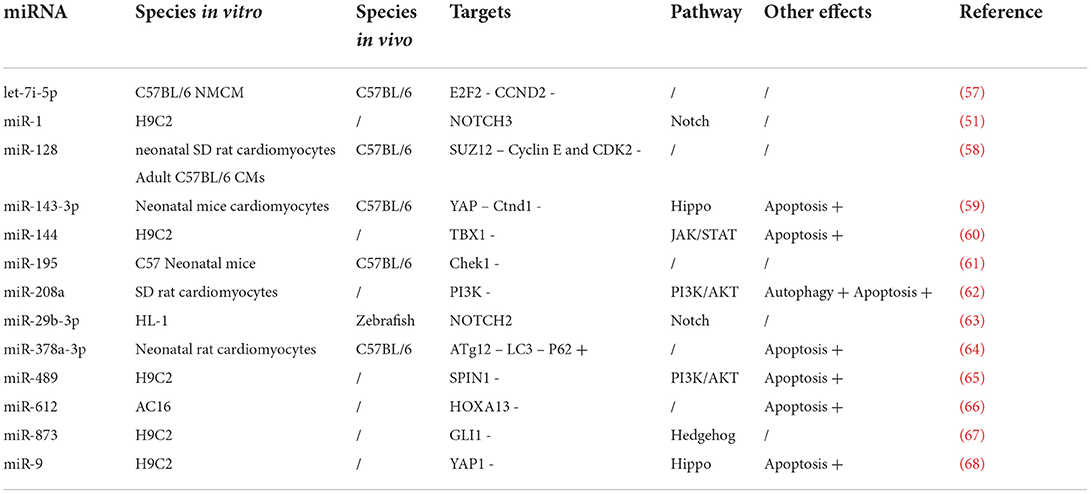
Table 2. A summary of the in vitro and in vivo effects of miRNAs on inhibited cardiomyocyte proliferation and the potential mechanism.
Researchers found that miR-294 was overexpressed in mouse embryos and was consistent with embryonic heart development. This miR changed rapidly after the heart tissue transitioned from postnatal to adult heart stages (49). After transfection of neonatal rat ventricular myocytes (NRVM) with the miR-294 mimic, CyclinB1, Cdk2, and CyclinE1 expressions were higher than the control cells, indicating that the cells changed to the G1/S phase. The transfected cells had an altered cell cycle and an advantage in energy metabolism. The extracellular acidification rate (ECAR), glycolysis ability, glycolysis, and glycolysis reserve were significantly increased 24 h after miR-294 treatment than the control group.
In addition, the researchers used a targeting scanner to predict the miR-294 target as WEE1. After, luciferase analysis was used to determine if miR-294 bound to the 3' UTR end of WEE1 to regulate NRVM proliferation (49). WEE1, a cell cycle regulatory protein, inactivates the CDK1-CyclinB1 complex and prevents CDK1 phosphorylation at Tyr1530, thus preventing it from entering the G2/M phase (13). Hence miR-294 inhibits WEE1, indicating that miR-294-mediated NRVM cell cycle reentry may be related to CDK1-CyclinB1 complex release and its downstream signal.
P27 is a cyclin-dependent kinase inhibitor that promotes the cell cycle by inhibiting various cyclins and CDK activities so that cells in the G0/G1 phase cannot enter the S phase of the cell cycle. miR-24 expression was higher in transverse aortic constricted (TAC) rats than in sham-operated rats, and researchers speculated that this might be associated with cardiac hypertrophy (47). The target scan predicted that p27 was a potential target in rats and humans. In a subsequent luciferase reporter assay experiment, 3'-UTR bounds to p27 to inhibit P27 activity, which shows that p27 is a miR-24 target.
Likewise, miR-24 can affect cardiac myocyte hypertrophy (47). miR-24, also known as a junctophilin-2 suppressing miRNA (although it could cause cardiac hypertrophy), treatment with antagomir prevents decreased L-type calcium channel/receptor signaling fidelity or efficiency and whole-cell calcium transients. Also, miR-24 decrease shows that suppression of miR-24 could effectively prevent the myocardial transition from compensated to decompensated state (36).
In addition to being a target for miR-24, the P27 is also a target for miR-152. Wang et al. demonstrated that Toll-like receptor 3 (TLR3) promotes myocardial regeneration and repair in neonatal mice, while TLR3-deficient mice showed greater heart function loss and huge infarct size (18). TLR3 ligands can activate the YAP1 pathway in neonatal mouse cardiomyocytes, which can induce cardiomyocyte proliferation. Further experiments confirmed that YAP1 activates miR-152 downstream expression. miR-152 can inhibit p27 and DNMT1 expression and regulate the cell cycle of cardiac myocytes from G1/G0 phase to the S phase (18).
Moreover, miR-25 can promote cardiomyocyte proliferation by targeting the cyclin family of proteins (48). Previous studies showed that cardiomyocytes' ability to proliferate decreased over time in cultured hPSC-CM. In response, investigators examined gene expression at the genome-wide level and screened for significant changes on the sixth and 18th days. Finally, it was determined that miR-25 could significantly promote hESC-CMs and hiPSC-CMs proliferation.
Similarly, potential targets were predicted using a target scan, and finally, miR-25 was reported to target FBXW7 via a dual-luciferase reporter (48). Fbxw7 is a cell cycle regulator that mediates the ubiquitin-dependent proteolysis of several positive cell cycle regulators (69). In addition, miR-25 regulates the FBXW7/CyclinE pathway in cardiomyocyte proliferation, which was observed by RNA sequencing and KEGG enrichment analysis. Finally, in vivo experiments in zebrafish confirmed that miR-25 could also regulate FBXW7 to promote proliferation (48) (Table 1).
In 2017, the researchers measured the expression of miR-17-92 cluster members and miRNAs in two different mouse models and concluded that miR-17-3p increased in mouse ischemic/reperfusion injury (IRI) and TAC exercise models (37). The effects on cardiac myocyte hypertrophy, proliferation, and survival were demonstrated in vitro and in vivo. A target scan was also used to predict the target. After that, the luciferase reporter analysis indicated that metalloproteinase 3 (TIMP3) tissue inhibitor is a direct target gene for miR-17-3p, and phosphatase and angiotensin homolog (PTEN) could be indirectly suppressed by miR-17-3p (37).
These results suggest that miRNA can activate the Akt pathway to achieve cardiac myocyte proliferation. Liu et al. (70) also demonstrated that human serum-derived small exosomes could promote cell proliferation and increase miR-17-3p expression in H9C2 cells, confirming TIMP3 as a direct target gene. Moreover, two other members of the miR-17-92 cluster (miR-19a and miR-19b, which had the same seed sequence) reduced MI size in the MI mouse model, and fractional shortening (FS) was improved. Likewise, adding miR-19a/19b to NRVMs stimulated the proliferation of rat cardiomyocytes in vitro (40). Increased expression levels were also detected in patients' blood with dilated cardiomyopathy and coronary heart disease. It thus showed that miR-19a/19b significantly stimulated the proliferation of cardiomyocytes in infancy and adulthood.
In addition, BIM1 and PTEN are both direct targets of miR-19a/19b. miR-19a/19b can also inhibit apoptosis by directly inhibiting apoptosis-related genes (40). miR-301a and miR-221-3p are also direct targets for PTEN (46, 50). Increasing miR-301a expression in H9C2 cells can promote cell proliferation and increase the G1/S transition of the cell cycle. It can also increase the proliferation rate of primary newborn cardiac myocytes. Furthermore, the apoptosis rate of H9C2 cells transfected with miR-301a decreased under hypoxic conditions. Also, miR-301a can increase the proliferation of cardiomyocytes in vivo. Further experiments, such as the luciferase reporter assay, showed that the miR-301a could bind to the 3' UTR end of PTEN (50).
Similarly, miR-221-3p from small hMSC exosomes can enhance AKT kinase activity by inhibiting PTEN, thus enabling H9C2 cells to proliferate (46). Researchers have found that the small exosomes released by Young-Exo hMSCs can promote myocardial endothelial cell formation after MI, reduce myocardial fibrosis, inhibit cardiomyocyte apoptosis, and effectively improve cardiac structure and function in vivo than Age-Exo cells. Also, after sequencing, it was found that miR-221-3p was upregulated, while miRNA activated the Akt pathway by targeting PTEN (46).
A study by Cheng et al. found that MSC-derived small exosomes were significantly reduced in a rat MI model after treatment, which prevented fibrosis after infarction (71). Using microarray analysis, miR-210 was found to be abundant in MSC-derived small exosomes. With prediction and dual-luciferase analysis, AIFM3 was the downstream target of miR-210. However, the AIFM3 level was up-regulated in hypoxic cardiomyocytes, while p-AKT, PPI3K and p-p53 expressions were down-regulated using western blotting and qPCR analysis. These results suggest that miR-210 may activate the PI3K/Akt pathway (71).
Per bioinformatics analysis and target gene prediction database, Hook3 is a key target gene for Rno-miR-496 and is closely related to cell proliferation. After H9C2 cells transfection with a miR-496 mimic, it was suggested that miR-496 upregulation might be related to H9C2 hypoxia/reoxygenation (H/R-induced apoptosis) and human cardiomyocytes reduction (55). Also, the dual-luciferase reporter assay system confirmed that miR-496 targets Hook3. Subsequently, Hook3 overexpression stimulated apoptosis in H/R-treated cells, thereby inhibiting cell proliferation. miR-496-activated PI3K/Akt signaling was upregulated, while Hook3 showed the opposite trend. Thus, it shows that miR-496 directly regulates apoptosis and the proliferation-related gene expression levels by targeting Hook3 and activating the PI3K/Akt signaling pathway (55).
Programmed cell death 4 (PDCD4) can be involved in various cellular processes, including cell proliferation, autophagy, and apoptosis (43, 72). Researchers have found that it is a direct target gene of several miRNAs and is involved in cardiomyocyte proliferation. Small exosomes isolated from explant-derived cardiac stromal cells from patients with heart failure (FEXO), microRNA array, and PCR analysis showed that miR-21-5p was abnormally expressed in FEXO. miR-21-5p overexpression can restore the impaired cardiac function caused by FEXO (45). Further studies on the mechanism have shown that miR-21-5p inhibited cardiomyocyte apoptosis by targeting PDCD4, promoting angiogenesis by activating the PTEN/Akt signaling pathway, and VEGF expression in vascular endothelial cells.
In addition, Sun and Zhang (43) demonstrated the lncRNA MALAT1 mechanism in myocardial infarction. The study also confirmed that miR-200a-3p could bind to PDCD4, while MALAT1 acts on miR-200a-3p by sponge as the competitive endogenous RNA (ceRNA), upregulating PDCD4 (43). Rescue experiments showed that the MALAT1/miR-200A-3p/PDCD4 axis regulates hypoxia-induced cardiomyocyte proliferation, cell cycle progression, and apoptosis (Figure 1).
Astragaloside IV (AS/IV), an active component of Astragalus Membranaceous (AM), has been used as one of the traditional drugs to treat cardiovascular diseases. AS/IV can effectively ameliorate myocardial cell injury induced by hypoxia/reoxygenation (H/R), and it has a significant protective effect on myocardial cells after H/R (32). miR-101a expression was increased in H/R cells treated with AS/IV, while dual-luciferase reporter gene analysis showed that miR-101a could target TGFBR1 in H/R cardiomyocytes. Furthermore, AS/IV could promote cell proliferation and upregulate miR-101a expression, thus inhibiting TGFBR1 and TLR2 expression in myocardial cells injured by H/R. Moreover, western blot analysis showed that the downstream genes (p-ERK and p-p38) of the MAPK signaling pathway were inhibited, indicating that AS/IV could inhibit the MAPK signaling pathway of H/R-injured cardiomyocytes (32).
miRNAs, including miR-708, were abundant in embryonic and neonatal cardiomyocytes and less abundant in adult cardiomyocytes (56). Thus, miR-708 overexpression could promote H9C2 cell proliferation or primary cardiomyocytes in vitro. miR-708 can also promote myocardial regeneration and cardiac function recovery in vivo. In addition, miR-708 protects cardiomyocytes from stress-induced apoptosis under hypoxia or Isoprenaline treatment. Luciferase assay confirmed the direct interaction between miR-708 and MAPK14, which showed that miR-708 partially depended on MAPK14 expression (56). Moreover, miR-199a-3p was a potent activator of rodent cardiomyocyte proliferation. Ying et al. hypothesized that the miR-199a-3p direct target gene was CD151, which activates the negative proliferation regulators by expressing p38, thereby inhibiting the cell cycle (38). Gain-of-function and loss-of-function experiments confirmed that CD151 is essential in inhibiting the cardiomyocyte cell cycle. Thus, it shows that miR-199a-3p achieves cardiomyocyte proliferation by regulating the MAPK pathway.
Zhao et al. discovered the distinct miR-374 expression in a thoracic epidural anesthetic model and a myocardial I/R injured mouse model (53). TEA is a common anesthesia method that can block the sympathetic afferent and efferent nervous systems. Therefore, it is speculated that it may prevent malignant ventricular arrhythmia caused by acute myocardial infarction. In a mouse myocardial I/R injured model, low miR-374 and high DTNA expressions were found. The TEA model has a protective effect on myocardial I/R injury, achieved by increasing miR-374 and decreasing DTNA expression.
The dual-luciferase reporter assay confirmed that DTNA is the target gene for miR-374, and cardiomyocytes overexpressing miR-374 have downregulated DTNA levels that blocked the Notch1 axis. miR-374 overexpression also promoted cardiomyocyte viability, inhibited apoptosis, and increased the cell number arrested in the S phase of the cell cycle. Correspondingly, miR-374 upregulation and DTNA downregulation reduced myocardial infarct size in vivo. These results suggest that miR-374 prevents myocardial I/R injury in mice after TEA by inhibiting DTNA-mediated Notch1 axis activity (53).
Regulating the Hippo signaling pathway to promote endogenous CM proliferation has emerged as a promising strategy for cardiac regeneration (33, 39, 51). LATS1 is the main component of the Hippo pathway. miR-10b was found in human embryonic stem cell-derived CMs (hESC-CMs) and was highly expressed in the early stage, with expression gradually decreasing over time (33). Cell cycle-related genes were higher than the control group, indicating that ESC-CM proliferation was promoted. In addition, ESC-CM transfected with miR-10b also had a significant inhibitory effect on H2O2-induced apoptosis. Dual-luciferase reporter assay showed that LATS1 was a target for 10b, and the pathway showed that CM proliferation was increased by inhibiting the Hippo pathway (33). Like LATS1, LATS2 is also a component of the Hippo pathway.
Xu et al. studied the fourth day of hPSC differentiation into CM and confirmed that miR-302d expression was significantly reduced (51). miR-302d overexpression significantly promoted hPSC-CMs proliferation. Target scan prediction and luciferase reporter analysis concluded that LATS2 was a direct target gene for miR-302d, promoting CM's proliferation by targeting LATS2 in the Hippo pathway. TAOK1, LATS upstream gene in the Hippo pathway, also has a miRNA targeting effect. miR-199a-3p can target CD151 to activate cardiomyocyte proliferation via the MAPK pathway (38). Thus, it targets the upstream YAP inhibitory kinase (TAOK1) and the E3 ubiquitin ligase (b-TrCP), causing YAP degradation (39), thereby inducing rat cardiomyocytes proliferation.
Ding et al. investigated the miR-1180 mechanism on cardiomyocytes and found that the miR-1180 observed in the rat's heart at embryonal day 9.5 decreased over time and completely disappeared at the seven postnatal days (34). In H/R-induced cardiomyocytes, miR-1180 expression was downregulated. On the other hand, miR-1180 transfection significantly mimics attenuated myocardial injury and apoptosis, while miR-1180 overexpression promoted cell proliferation through cell cycle processes. In addition, miR-1180 was found to target NKIRAS2, directly regulating the NF-κB pathway. Meanwhile, miR-410-3p attenuated apoptosis in hypoxia-treated cardiomyocytes, while miR-410-3p overexpression promoted hypoxia-preconditioned AC16 cells proliferation (73).
The dual-luciferase assay showed that miR-410-3p could bind to the 3' UTR TRAF5 region. The result was confirmed by inhibiting TRAF5 expression. TRAF5, a member of the tumor receptor-associated factor (TRAF) protein family, is involved in various tumor activities and usually mediates NFκB pathway activation (73). Researchers compared the differentially expressed genes (DEGs) in human myocardial infarction and normal myocardial tissue through the bioinformatics database and identified five hub genes.
One of the genes, SOCS3, was significantly elevated in the MI group (52). Next, it was confirmed by bioinformatics prediction and luciferase reporter assays that miR-324 was its upstream regulatory miRNA, which could both bind to inhibit SOCS3 expression. In in vitro experiments, it was clear that miR-324 overexpression promoted cardiomyocyte proliferation, protected cells from apoptosis in the H/R model, and reduced cardiomyocytes, such as TNF-α, p-p65, and p-IκBα. Thus, it showed that miR-324 might improve cardiomyocyte H/R injury by regulating NFκB (52).
miR-19b belongs to the miR-17-92 cluster (41). Previous studies demonstrated that miR-19b could participate in the PTEN/AKT pathway to activate cardiomyocyte proliferation (40), stimulate the WNT pathway, and interact with the gene β-Catenin-related pathway (41, 42). Arif et al. demonstrated (by western blot analysis) that cardiomyocytes in the transfected group had significantly increased proliferation but decreased cell death and downregulated APC (adenomatous polyposis coli) expression. Simultaneously, the group used murine miR-210 to transfect adult rat cardiomyocytes (44). Using a bioinformatics prediction and luciferase reporter assay, Arif et al. confirmed that the APC cell cycle repressor is a direct target for miR-210 in rodents. APC is a WNT signaling pathway antagonist involved in tumor cell migration, adhesion, and apoptosis (74). These showed that miR-210 overexpression rescued cardiac function after cardiac injury in adult mice by promoting CM proliferation, cell survival, and angiogenesis.
Physiologically, the ventricular myocytes are stretched when the heart beats normally. Researchers hypothesized that the cardiomyocyte stretch-response pathways are closely related to cardiac development and that perturbations in biomechanical stimuli may cause conditions such as hypoplastic left heart syndrome (HLHS) (54). miR-486 expression was increased in cardiomyocytes under cyclic stretch and in HLHS patients with increased right ventricular (RV) stretch in vivo. Furthermore, increasing miR-486 for 3 days in neonatal mice was sufficient to promote ventricular growth and cardiomyocyte proliferation significantly.
Subsequent studies confirmed that miR-486 inhibited TGF-β/Smad signaling to promote cardiomyocyte proliferation. Yang et al. found that with hypoxia prolongation, lncRNA TUG1 and miR-133a exhibited time-dependent variation. Both expressions were complementary, meaning TUG1 expression was increased while miR-133a expression gradually decreased (35). TUG1 silencing enhanced miR-133a and miR-133b expression. A luciferase reporter assay was subsequently performed, indicating that miR-133a is a direct target for TUG 1 in AC16 cells. In addition, TUG1 acts as a competitive endogenous RNA (ceRNA) to inhibit miR-133a expression. Hence, it shows that miR-133a can promote AC16 cell proliferation and inhibit cell apoptosis (Table 1).
Let-7i-5p affects cardiomyocyte cell proliferation and interferes with cardiomyocyte repair after injury (57). Let-7i-5p transfection mimics in mouse cardiomyocytes can significantly reduce cardiomyocyte count, indicating that let-7i-5p inhibits proliferation. Additionally, target scan prediction and luciferase reporter assay found that let-7i-5p is a target relationship with E2F2 and CCND2, which binds their 3' UTR end and inhibits E2F2 and CCND2 proteins expression (57). Huang et al. observed that miR-128 is upregulated in murine cardiomyocytes during the postnatal period, switching from proliferation to terminal differentiation (58). Furthermore, miR-128 overexpression impairs CM proliferation and cardiac function by downregulating SUZ12 expression. The outcome could suppress cyclin-dependent kinase inhibitor-p27 expression and activate the positive cell cycle regulators Cyclin E and CDK2.
miR-195 and its expression, a member of the miR-15 family, gradually decreased over time in neonatal mouse ventricular myocytes. Also, the expression level was almost 6-fold higher on the 10th day of life than on the 1st day of life (61). miR-195 was also confirmed to affect cardiomyocyte proliferation in vitro and in vivo. Subsequently, in the study of target genes, it was found that miR-195 can bind to checkpoint kinase 1 (Chek1) and inhibit the increase in cardiomyocyte count by affecting the cardiomyocyte cell cycle normal operation. Chek1, one of the key regulatory enzymes of the cell cycle, is mainly involved in regulating mitotic stages and coordinates progression via the G2/M, spindle checkpoints, chromosome segregation, and cell division (75). Therefore, miR-195 primarily regulates mouse cardiomyocyte proliferation by affecting Chek1 expression (Table 2).
Cao et al. studied the miR-144 effect on rat cardiomyocytes and confirmed that it is essential in regulating cardiomyocyte proliferation (60). miR-144 inhibits cell proliferation by blocking H9C2 cells in the G1 phase. Bioinformatics prediction and luciferase reporter gene analysis indicated that miR-144 directly targets TBX1. miR-144 and TBX1 co-overexpression promotes the transition from the G1 to the S phase, upregulates cell proliferation, and downregulates apoptosis by inhibiting the JAK2/STAT1 signaling pathway.
Researchers found that the miR-873 expression was upregulated in patients' serum with congenital heart defects (67). miR-873 overexpression can inhibit H9C2 cell proliferation and induce cell cycle arrest, which clarifies miR-873 role in cardiomyocytes. miR-873 has a predicted target site in the 3'-untranslated region (3'-UTR) of GLI1, as shown by the bioinformatics algorithm and verified by the dual-luciferase reporter experiment (67). Glioblastoma-associated oncogene 1 (Gli1) is a nuclear effector of the Hedgehog pathway and plays an essential role in embryonic development and organogenesis.
Similar to the TEA model, sevoflurane is a common gas anesthetic that blocks the sympathetic (afferent/efferent) effects and may reduce myocardial infarction (53). Shi et al. studied the protection by sevoflurane in a rat cardiomyocyte IRI model. They concluded that sevoflurane application could effectively reduce autophagy and apoptosis-related cytokines expression in IRI cells, while miR-208a could reverse sevoflurane effects on IRI cells (62). The induced effect reduces cardiomyocyte proliferation and arrests the cardiomyocytes in the G0/G1 phase. When studying the pathway, the PI3K and AKT mRNA expressions were significantly increased in the post-treatment group with sevoflurane, while sevoflurane-preconditioning combined with the miR-208a inhibitor increased PI3K and AKT mRNA levels.
It, therefore, suggests that miR-208a low expression can activate the PI3K/AKT signaling pathway and inhibit the expression of the autophagy-related factors in myocardial SI/RI cells (62). miR-489 can be highly expressed in H/R-treated rat cardiomyocytes, and miR-489 overexpression can accelerate its apoptosis. Chen et al. found that the SPIN1 gene is a downstream target gene for miR-489, and this miRNA can be combined with its 3' UTR to downregulate SPIN1 expression. Additionally, western blots showed that miR-489 overexpression inhibited the PI3K/AKT pathway (65). Thus, miR-489 could induce cardiomyocyte apoptosis by inhibiting the AKT pathway (Figure 2).
Yang et al. applied next-generation sequencing (NGS) to screen patients' entire miRNA expression profile with congenital heart disease (63). It was found that the of miR-29b-3p expression in the patients' right ventricular outflow tract (RVOT) was significantly increased, while NOTCH2 was downregulated in the RVOT. In vivo experiments using miR-29b-3p mimics injected into zebrafish embryos caused several malformations, including corporeal and cardiac malformations. In vitro experiments, where miR-29b-3p was transfected into HL1 cells showed that the expression of the proliferation-related proteins was downregulated, while expression was upregulated in the miR-29b-3p inhibitor group.
On the other hand, luciferase reporter analysis indicated that miR-29b-3p was an upstream regulatory miRNA for NOTCH2, indicating that miR-29b-3p could regulate the NOTCH pathway and inhibit mouse cardiomyocyte proliferation (63). Meanwhile, miR-1 was confirmed to regulate the NOTCH pathway and inhibit cardiomyocyte proliferation (76). In rat cardiomyocytes, hypoxia inhibited cell proliferation but promoted cell apoptosis and miR-1 expression, while miR-1 expression downregulation had the opposite effect. Subsequently, bioinformatics analysis, luciferase reporter, and RNA immunoprecipitation assays indicated that NOTCH3 was a direct target for miR-1. Upregulation was shown to reverse the miR-1 effects on the proliferation, apoptosis, and autophagy of the hypoxia-treated H9C2 cells (76).
Ma et al. studied the melatonin effect on mice cardiomyocytes and found that it could significantly reduce oxidative stress and apoptosis in cardiomyocytes after myocardial infarction (59). In vitro, melatonin stimulated neonatal mouse cardiomyocytes to re-enter the cell cycle and significantly inhibited miR-143-3p levels. In contrast, miR-143-3p overexpression inhibited melatonin-induced cardiomyocyte mitosis. Subsequent experiments proved that miR-143-3p target genes were YAP and Ctnd1, suggesting that miR-143-3p could regulate the YAP signaling pathway under the influence of melatonin and promotes cellular regeneration in myocardial infarction (59). With hypoxia prolongation in H9C2 cells, miR-9 expression was upregulated, while YAP1 expression was downregulated. miR-9 knockdown increased the hypoxic H9C2 cells' proliferation and inhibited apoptosis and caspase 3/7 activity, while miR-9 overexpression had the opposite effect on hypoxic H9C2 cells (68).
In addition, a luciferase reporter showed that YAP1 is a direct target for miR-9. YAP1 gene knockdown inhibited the hypoxia-exposed H9C2 cells proliferation and promoted apoptosis. Also, the YAP1 knockout attenuated the anti-miR-9 effects on the proliferation and apoptosis of the hypoxic H9C2 cells. The results showed that in hypoxia-exposed H9C2 cells, miR-9 increased cardiomyocyte apoptosis and inhibited cardiomyocyte proliferation by affecting the YAP pathway.
Li et al. investigated patients with chronic heart failure (CHF) and compared these patients with a normal control group. The result showed that the lncRNA LUCAT1 in blood was reduced by 1.7 times, suggesting that it may be related to prognostic factors (66). LUCAT1 inhibitor transfection into the AC16 cells can accelerate cardiomyocytes' apoptosis. The study also found that miR-612 expression was increased. Thus, follow-up experiments determined that LUCAT1 can compete with miR-612 as a ceRNA in AC16 cells, showing that they are directly related. Simultaneously, the HOXA13 target for miR-612 was also identified. Rescue experiments showed that the lncRNA LUCAT1 promotes cell proliferation and inhibits apoptosis in CHF patients through via miR-612/HOXA13 axis (66).
LncRNA NEAT1 is involved in various disease progression, but the mechanism involved in myocardial infarction is still unclear. LncRNA NEAT1 expression in more than 100 patients' blood with unstable angina pectoris and ischemic cardiomyopathy/MI was significantly higher than the normal controls (64). After the I/R injury model was established in vivo, the NEAT1 level significantly increased over time. NEAT1 knockdown ameliorated hypoxia-induced cardiomyocyte injury. NEAT1 was subsequently predicted to be a target for miR-378a-3p, where subsequent luciferase assay results showed that the miR-378a-3p expression significantly reduced the NEAT1 reporter gene (wild-type non-mutated) luciferase activity (64). It was also noted that the NEAT1/miR-378a-3p axis existed in rat cardiomyocytes under hypoxia (Table 2).
In the mammalian genome sequence, 4%−9% of transcripts sequentially produced are lncRNAs, with the corresponding proportion of protein-coding RNA being approximately 1%. Though lncRNAs were thought to be secondary RNA polymerase-II-transcription products with no biological functions (77), long non-coding RNAs (lncRNAs) are a cluster of functional non-translated RNA molecules with a length >200 nucleotides. Recent studies have shown that lncRNAs are widely involved in several critical regulatory processes, such as chromosome silencing, genome imprinting, chromatin modification, transcription activation, transcription interference, and nuclear transport (78, 79). Major LncRNAs are classified into four types per their position in the genome relative to the protein-coding gene, including Antisense lncRNAs, Bidirectional lncRNAs, Intergenic lncRNAs, and Sense-intronic lncRNAs. (17, 80, 81). LncRNAs play an essential role in regulating cardiomyocyte proliferation by enhancing or suppressing cell cycle progression.
On the one hand, some lncRNAs act as sponges for inhibitory miRNAs to inhibit cardiomyocyte proliferation. The lncRNA AZIN2-sv, AZIN2 gene splicing variant, directly binds to miR-214 and acts on the PTEN target gene. In addition, AZIN2-sv inhibit the AKT pathway by blocking its inhibitory effect on PTEN. Therefore, AZIN2-sv loss can promote cell survival and proliferation (80). Similarly to AZIN2-sv, the lncRNA CRRL binds to miR-199a-3p and thereby increases the target gene expression of Hopx, a critical negative regulatory factor of CM proliferation, suppressing CM proliferation (82). However, the lncRNA CAREL acted as a ceRNA for miR-296 to downregulate Trp53inp1 and Itm2a, the miR-296 target gene. The Trp53inp1 has been found to cause cell cycle arrest in G1 and to increase p53-mediated apoptosis, while Itm2a was associated with G2/M cell cycle arrest as a tumor suppressor. Hence, CAREL negatively controls cardiomyocyte proliferation.
Cardiac-specific CAREL overexpression in mice reduced cardiomyocyte division and proliferation and blunted neonatal heart regeneration after injury. Similarly, CAREL reduced human-induced pluripotent stem cell-derived cardiomyocyte proliferation (83). LncRNA-specific transcript 5 (GAS5) expression affects cardiomyocyte proliferation, cell cycle, and apoptosis. LncRNA Gas5 regulates myocardial infarction by targeting the miR-525-5p/CALM2 axis. Studies have shown that Gas5 and CALM2 expression in cardiomyocytes after infarction is significantly upregulated, while miR-525-5p expression is significantly downregulated. Gas5 and CALM2 overexpression significantly promotes cardiomyocyte apoptosis and inhibits cardiomyocyte proliferation (84).
In addition, Gas5 stimulates PDCD4, inhibiting the PI3K/AKT signal pathway. LncRNA-Gas5 regulates the PDCD4 expression by targeting miR-21, mediating myocardial infarction-induced cardiomyocyte apoptosis. Thus, it implies that Gas5 may become a therapeutic target for MI (72). However, LncRNA GAS5 inhibits NLRP3 (inflammasome) activation-mediated pyroptosis in diabetic cardiomyopathy by targeting miR-34b-3p/AHR. GAS5 acts as a competing endogenous RNA to enhance AHR expression by sponging miR-34b-3p and repressing NLRP3 (inflammasome) activation-mediated pyroptosis to improve diabetic cardiomyopathy (85). Additionally, lncRNA XIST overexpression reduces cardiac regeneration and promotes apoptosis by negatively targeting the miR-130a-3p/PDE4D axis (86).
On the other hand, some lncRNAs directly act on mRNAs and proteins or act as guides for chromatin modifiers to supress cardiomyocyte proliferation. The LncRNA CPR targets minichromosome maintenance 3 (MCM3), a DNA replication promoter, and cell cycle progression to inhibit cardiomyocyte proliferation. CPR- knocked down cardiomyocytes and CPR-knocked out mice exhibit increased CM renewal after MI (87). Moreover, the lncRNA TUC40- overexpression reduced the target gene Pbx1 expression, cardiomyocyte induction and differentiation, inhibited proliferation and promoted apoptosis. Proliferation was possibly inhibited due to G2/M cell cycle arrest and the increased induced apoptosis rate (88).
Some lncRNAs promote cardiomyocyte proliferation apart from lncRNAs that suppress cardiac regeneration. A novel upregulated fetal lncRNA ECRAR promoted DNA synthesis, mitosis, and cytokinesis in the seventh post-natal day and adult rat cardiomyocytes. ECRAR was transcriptionally upregulated by E2F transcription factor 1 (E2F1). After that, ECRAR bound and promoted extracellular signal-regulated kinases 1 and 2 (ERK1/2) phosphorylation, causing cyclin D1 and cyclin E1 downstream target activation, lastly activating E2F1. The E2F1-ECRAR-ERK1/2 signaling formed a positive feedback loop to drive cell cycle progression and promote CM proliferation (89). Moreover, antisense lncRNA, silent information regulator enzyme 1 (Sirt1), has been shown to enhance cardiomyocyte proliferation and synchronously attenuate cardiomyocyte apoptosis (80). Unlike other lncRNAs, Sirt1 antisense lncRNA can bind the Sirt1 3'-UTR, enhancing Sirt1 stability and increasing Sirt1 abundance at both the mRNA and protein levels.
Sirt1 was involved in Sirt1 antisense lncRNA-induced cardiomyocyte proliferation (80). The LncRNA NR_045363 was primarily expressed in cardiomyocytes and rarely in non-cardiomyocytes. NR_045363 overexpression in a 7-day-old mice heart could improve cardiac function and stimulate cardiomyocyte proliferation after myocardial infarction (90). lncRNAs that regulate cardiomyocyte proliferation are summarized in Table 3.
CircRNA is a single-stranded RNA type connected end-to-end for specific functions. It implies that the 3' and 5' ends of the RNA form a covalent bond. CircRNA is less prone to degradation by exonucleases than conventional linear RNA because it lacks of 5' and 3' tails (92). In the early 1990's, an accidental PCR amplification revealed circRNA to the world (93). 2 years later, scientists discovered the biological behavior of circular transcription while studying the key mouse gene Sry, which is dominant in sex determination during embryonic development (94).
Following extensive development and popularization of RNA sequencing technology, several achievements have been made in circRNA. Several experiments have confirmed that certain circRNAs have been proven to encode proteins, given that numerous circRNA types come from protein-coding genes (95, 96). However, the biological functions of most circular RNAs are still unclear. This article mainly introduces the relationship between circRNA, cardiomyocyte proliferation, and circRNA biological effects.
CircRNAs (circular RNAs) are emerging as powerful cardiac development regulators and diseases that regulate cardiac regeneration. CircRNA Nfix (circNfix) was overexpressed in humans, rats, and mice in the adult heart. Experiments in vitro and in vivo indicated that cardiomyocyte proliferation was increased by circNfix knockdown, whereas it was inhibited by circNfix overexpression. Mechanistically, super-enhancer-regulated circNfix inhibits cardiac regenerative repair and functional recovery after myocardial infarction by suppressing Ybx1 ubiquitin-dependent degradation and increasing miR-214 activity after MI (97).
CircHIPK3 inhibits proliferative ability and stimulates human-derived cardiomyocyte apoptosis after myocardial Ischemia-reperfusion injury by binding to miRNA-124-3p, which aggravates myocardial ischemia-reperfusion injury (98). However, another study demonstrated that circHIPK3 was overexpressed in fetal and neonatal mice hearts. CircHIPK3 promotes division and mouse cardiomyocyte proliferation by increasing the Notch1 intracellular domain (N1ICD) acetylation, significantly increasing its stability and reducing degradability. In addition, circHIPK3 acted as a sponge for miR-133a to promote connective tissue growth factor (CTGF) expression, activating endothelial cells and improving cellular function (99).
Notwithstanding, various circRNAs also show the ability to regulate the proliferation of cardiomyocytes during myocardial I/R injury positively. CircRNA-68566 participated in I/R by regulating the miR-6322/PARP2 signaling pathway, which binds to miR-6322 to enhance proliferation (100). circSNRK overexpression could also facilitate the proliferation of cardiomyocytes. In the post-infarction area induced by adeno-associated virus 9 (AAV9), circSNRK could phosphorylate GSK3β activity and miR-103-3p target, while circSNRK can act as a sponge for miR-103-3p to promote cardiomyocytes regeneration (101).
However, other researchers suggested overexpressed CircANXA2 could inhibit the H/R-treated H9C2 cell proliferation. Also, further investigation showed that CircANXA2 could be a sponge for miR-133, reversing the inhibition of proliferation and increasing apoptosis (102). Moreover, circRNA PVT1 (circPVT1) can act as a sponge for two miRNAs (miR-125b and miR-200a), increasing cell apoptosis and blocking cardiomyocyte proliferation in vitro. Luo et al., demonstrated that silencing the circPVT1 expression could prevent heart I/R injury in rats and restore cardiomyocyte viability by regulating the circPVT1/miR-125b/miR-200a axis (103). circRNAs that regulate cardiomyocyte proliferation are summarized in Table 4.
Cardiovascular diseases (CVD) are the main cause of morbidity and mortality worldwide. The adult heart cannot regenerate new cardiomyocytes during myocardial infarction and heart failure. In recent years, increasing evidence suggests that circulating ncRNAs can serve as diagnostic biomarkers and potential new therapeutic targets for several cardiovascular disease, especially myocardial infarction.
MicroRNAs are considered ideal therapeutic targets in myocardial infarction. In a reperfused AMI porcine model, a single microencapsulated anti-miR-92a intracoronary injection restrained left ventricle remodeling without adverse effects (104). Likewise, the systemic injection of a locked nucleic acid-modified anti-miR-15 effectively rendered cardiomyocytes resistant to hypoxia-induced cardiomyocyte death (105). Additionally, a miR-146a mimic selective administration reproduces some benefits for cardiosphere-derived cells (CDC) exosomes.
The findings identified exosomes as key mediators for CDC-induced regeneration and highlighted exosomes' potential utility as cell-free therapeutic candidates (104). Another study indicated that exosome cargo analysis in mice and humans identified conserved pro-regenerative miRs, which recapitulated the therapeutic effects of promoting cardiomyocyte proliferation (106). In summary, these studies show that miRNA-based therapy using modified oligonucleotides is a promising therapeutic agent for patients suffering from massive acute myocardial infarction or affecting cardiac remodeling and protecting cardiac function after ischemic injury.
Recent advances involving targeting miRNAs, while lncRNAs, a novel and challenging class of potential drug targets will be given priority. A study that collected plasma from patients with cardiac remodeling after acute myocardial infarction and performed transcriptome analysis identified LIPCAR as a potential biomarker for heart failure. In addition, LncRNA LIPCAR (long intergenic non-coding RNA predicting cardiac remodeling) is associated with cardiovascular death in patients with heart failure and a prognostic indicator in patients with heart failure (107). Another study found that LncRNA ANRIL and KCNQ1OT1 levels could predict the degree of left ventricular dysfunction (108). Recent studies have shown that LncRNAs CoroMarker and LncPPARδ are predictive markers for coronary heart disease (109). Despite these advances, circulating ncRNAs cellular origin is largely unknown, and little is currently known about the causality of the underlying disease.
Microscopically, heart failure caused by the decrease in absolute cardiomyocyte count is still the most crucial factor threatening human life. In addition, current treatment methods cannot supplement/replenish cardiomyocytes, the root cause of failed treatments. The present review collected and summarized information showing that cardiomyocytes, particularly during the neonatal or fetal period, have an inherent ability to proliferate, although gradually inhibited by different degrees of extracellular or intracellular factors.
The nature and importance of these stimuli and how they affect the myocardial cell cycle are still the subject of in-depth research. Starting with lower vertebrates and rodents is essential and effective in understanding the human heart regenerative ability in the evolutionary process and the corresponding cellular mechanism regulation. Notably, the cardiomyogenesis regulatory mechanisms identified in these animal models appear to function similarly in humans. Cell sequencing technologies will make large-scale target identification possible, and new gene therapy technologies will provide new tools for precise regulation in vivo.
From the current research, various ncRNAs, including miRNA, lncRNA, and circRNA, can control the essential genes and regulatory pathways of various cellular processes. Most non-coding RNAs in current research directly or indirectly act in a regulatory role on cell cycle-associated proteins. Therefore, enabling cardiomyocyte's' reentry into the cell cycle would be a crucial breakthrough. The studies on miRNAs in this research field of cardiomyocyte regeneration are emerging compared with other ncRNAs. Although lncRNA and circRNA are novel and unexplored, the mechanisms behind them seem to be largely inextricably linked to miRNA regulation. Hence fully understanding the miRNAs mechanism may still allow breakthroughs in solving myocardial regeneration issues. Although there is progress non-coding RNA usage for cardiomyocyte proliferation and repair, there are still significant challenges in the actual combination of clinical guidance and treatment.
KQ drafted the manuscript and were responsible for the collection of data or analysis. CF designed the study. WT, DY, JY, JP, and JG revised the manuscript. All authors read and approved the final manuscript.
This work was supported by the Science and Technology Innovation Program of Hunan Province (2021RC2106 to CF) and the Natural Science Foundation of Hunan Province (2022JJ20088 to CF).
We thanks Chukwuemeka Daniel Iroegbu for language editing of this work.
Authors JG and CF were employed by the company Hunan Fangsheng Pharmaceutical Co. Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
circRNA, circular RNA; CeRNA, competitive endogenous RNA; ECAR, Extracellular Acidification Rate; ESC, Embryonic Stem Cell; HLHS, Hypoplastic Left Heart Syndrome; IHD, Ischemic Heart Disease; iPSC, induced Pluripotent Stem Cell; I/R injury, Ischemic-Reperfusion injury; lncRNA, long non-coding RNA; miRNA, micro RNA; MI, Myocardial Infarction; mRNA, messenger RNA; MSC, Mesenchymal Stem Cell; ncRNA, non-coding RNA; NRVMs, Neonatal Rat Ventricular Myocytes; TAC, Transverse Aortic Constriction; TEA, Thoracic Epidural Anesthesia; UTR, Untranslated Region.
1. Maron DJ, Hochman JS, Reynolds HR, Bangalore S, O'Brien SM, Boden WE, et al. Initial invasive or conservative strategy for stable coronary disease. N Engl J Med. (2020) 382:1395–407. doi: 10.1056/NEJMoa1915922
2. Ferraro R, Latina JM, Alfaddagh A, Michos ED, Blaha MJ, Jones SR, et al. Evaluation and management of patients with stable angina: beyond the ischemia paradigm: JACC state-of-the-art review. J Am Coll Cardiol. (2020) 76:2252–66. doi: 10.1016/j.jacc.08078
3. Jenča D, Melenovský V, Stehlik J, Staněk V, Kettner J, Kautzner J, et al. Heart failure after myocardial infarction: incidence and predictors. ESC heart failure. (2021) 8:222–37. doi: 10.1002/ehf2.13144
4. Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, Zacchigna S, et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. (2012) 492:376–81. doi: 10.1038/nature11739
5. Spadaccio C, Benedetto U. (2018). Coronary artery bypass grafting (CABG) percutaneous coronary intervention (PCI) in the treatment of multivessel coronary disease: quo vadis? -a review of the evidences on coronary artery disease. Annals Cardiothor Surg. (2018) 7:506–15. doi: 10.21037/acs.0517
6. Doenst T, Haverich A, Serruys P, Bonow RO, Kappetein P, Falk V, et al. PCI and CABG for treating stable coronary artery disease: JACC review topic of the week. J Am Coll Cardiol. (2018) 73:964–76. doi: 10.1016/j.jacc.11053
7. Derks W, Bergmann O. Polyploidy in cardiomyocytes: roadblock to heart regeneration? Circ Res. (2020) 126:552–65. doi: 10.1161/CIRCRESAHA.119.315408
8. Ryan R, Moyse BR, Richardson RJ. Zebrafish cardiac regeneration-looking beyond cardiomyocytes to a complex microenvironment. Histochem Cell Biol. (2020) 154:533–48. doi: 10.1007/s00418-020-01913-6
9. Ali H, Braga L, Giacca M. Cardiac regeneration and remodelling of the cardiomyocyte cytoarchitecture. FEBS J. (2020) 287:417–38. doi: 10.1111/febs.15146
10. Vujic A, Natarajan N, Lee RT. Molecular mechanisms of heart regeneration. Sem Cell Develop Biol. (2019) 100:20–8. doi: 10.1016/j.semcdb.09003
11. Bassat E, Mutlak YE, Genzelinakh A, Shadrin IY, Baruch Umansky K, Yifa O, et al. The extracellular matrix protein agrin promotes heart regeneration in mice. Nature. (2017) 547:179–84. doi: 10.1038/nature22978
12. Nakada Y, Canseco DC, Thet S, Abdisalaam S, Asaithamby A, Santos CX, et al. Hypoxia induces heart regeneration in adult mice. Nature. (2017) 541:222–7. doi: 10.1038/nature20173
13. Mohamed TMA, Ang YS, Radzinsky E, Zhou P, Huang Y, Elfenbein A, et al. Regulation of cell cycle to stimulate adult cardiomyocyte proliferation and cardiac regeneration. Cell. (2018) 173:2014. doi: 10.1016/j.cell.02014
14. Magadum A, Singh N, Kurian AA, Munir I, Mehmood T, Brown K, et al. Pkm2 Regulates cardiomyocyte cell cycle and promotes cardiac regeneration. Circulation. (2020) 141:1249–65. doi: 10.1161/CIRCULATIONAHA.119.043067
15. Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, et al. Transient regenerative potential of the neonatal mouse heart. Science. (2011) 331:1078–80. doi: 10.1126/science.1200708
16. Lam NT, Sadek HA. Neonatal heart regeneration: comprehensive literature review. Circulation. (2018) 138:412–23. doi: 10.1161/CIRCULATIONAHA.118.033648
17. Greco S, Zaccagnini G, Fuschi P, Voellenkle C, Carrara M, Sadeghi I, et al. Increased BACE1-AS long noncoding RNA and β-amyloid levels in heart failure. Cardiovasc Res. (2017) 113:453–63. doi: 10.1093/cvr/cvx013
18. Wang X, Ha T, Liu L, Hu Y, Kao R, Kalbfleisch J, et al. TLR3 Mediates repair and regeneration of damaged neonatal heart through glycolysis dependent YAP1 Regulated miR-152 expression. Cell Death Differ. (2018) 25:966–82. doi: 10.1038/s41418-017-0036-9
19. Fan C, Oduk Y, Zhao M, Lou X, Tang Y, Pretorius D, et al. Myocardial protection by nanomaterials formulated with CHIR99021 and FGF1. JCI Insight. (2020) 5:796. doi: 10.1172./jci.insight.132796
20. Fan C, Tang Y, Zhao M, Lou X, Pretorius D, Menasche P, et al. CHIR99021 and fibroblast growth factor 1 enhance the regenerative potency of human cardiac muscle patch after myocardial infarction in mice. J Mol Cell Cardiol. (2020) 141:1–10. doi: 10.1016/j.yjmcc.03003
21. Tzahor E, Poss KD. Cardiac regeneration strategies: staying young at heart. Science. (2017) 356:1035–9. doi: 10.1126/science.aam5894
22. Foglia MJ, Poss KD. Building and re-building the heart by cardiomyocyte proliferation. Development. (2016) 143:729–40. doi: 10.1242/dev.132910
24. Wu X, Iroegbu CD, Peng J, Guo J, Yang J, Fan C, et al. Cell death and exosomes regulation after myocardial infarction and ischemia-reperfusion. Front Cell Dev Biol. (2021) 9:673677. doi: 10.3389/fcell.2021.673677
25. Lee RC, Feinbaum RL, Ambros V. The elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. (1993) 75:843–54.
26. Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans Cell. (1993) 75:855–62.
27. Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nature Rev Gen. (2007) 8:93–103. doi: 10.1038/nrg1990
28. Mor E, Shomron N. Species-specific microRNA regulation influences phenotypic variability: perspectives on species-specific microRNA regulation. BioEssays: News Rev Mol Cell Develop Biol. (2013) 35:881–8. doi: 10.1002/bies.201200157
29. Niwa R, Slack FJ. The evolution of animal microRNA function. Curr Opin Genet Dev. (2007) 17:145–50. doi: 10.1016/j.gde.2007.02.004
30. Gharanei S, Shabir K, Brown JE, Weickert MO, Barber TM, Kyrou I, et al. Regulatory microRNAs in brown, brite and white adipose tissue. Cells. (2020) 9:2489. doi: 10.3390./cells9112489
31. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. (2004) 5:522–31. doi: 10.1038/nrg1379
32. Wu Y, Fan Z, Chen Z, Hu J, Cui J, Liu Y, et al. Astragaloside IV protects human cardiomyocytes from hypoxia/reoxygenation injury by regulating miR-101a. Mol Cell Biochem. (2020) 470:41–51. doi: 10.1007/s11010-020-03743-5
33. Xie Y, Wang Q, Gao N, Wu F, Lan F, Zhang F, et al. MircroRNA-10b promotes human embryonic stem cell-derived cardiomyocyte proliferation via novel target gene LATS1. Mol Ther Nucleic Acids. (2019) 19:437–45. doi: 10.1016/j.omtn.11026
34. Ding Y, Bi L, Wang J. MiR-1180 promotes cardiomyocyte cell cycle re-entry after injury through the NKIRAS2-NFκB pathway. Biochem Cell Biol. (2020) 98:449–57. doi: 10.1139/bcb-2019-0364
35. Yang H, He X, Wang C, Zhang L, Yu J, Wang K, et al. Knockdown of TUG 1 suppresses hypoxia-induced apoptosis of cardiomyocytes by up-regulating miR-133a. Arch Biochem Biophys. (2020) 681:108262. doi: 10.1016/j.abb.2020.108262
36. Li RC, Tao J, Guo YB, Wu HD, Liu RF, Bai Y, et al. In vivo suppression of microRNA-24 prevents the transition toward decompensated hypertrophy in aortic-constricted mice. Circ Res. (2013) 112:601–5. doi: 10.1161/CIRCRESAHA.112.300806
37. Shi J, Bei Y, Kong X, Liu X, Lei Z, Xu T, et al. miR-17-3p Contributes to exercise-induced cardiac growth and protects against myocardial ischemia-reperfusion injury. Theranostics. (2017) 7:664–76. doi: 10.7150/thno.15162
38. Tao Y, Zhang H, Huang S, Pei L, Feng M, Zhao X, et al. miR-199a-3p promotes cardiomyocyte proliferation by inhibiting Cd151 expression. Biochem Biophys Res Commun. (2019) 516:28–36. doi: 10.1016/j.bbrc.05174
39. Torrini C, Cubero RJ, Dirkx E, Braga L, Ali H, Prosdocimo G, et al. Common regulatory pathways mediate activity of micrornas inducing cardiomyocyte proliferation. Cell reports. (2019) 27:005. doi: 10.1016/jcelrep05
40. Gao F, Kataoka M, Liu N, Liang T, Huang ZP, Gu F, et al. Therapeutic role of miR-19a/19b in cardiac regeneration and protection from myocardial infarction. Nat Commun. (2019) 10:1802. doi: 10.1038/s41467-019-09530-1
41. Qin DN, Qian L, Hu DL, Yu ZB, Han SP, Zhu C, et al. Effects of miR-19b overexpression on proliferation, differentiation, apoptosis and Wnt/β-catenin signaling pathway in P19 cell model of cardiac differentiation in vitro. Cell Biochem Biophys. (2013) 66:709–722. doi: 10.1007/s12013-013-9516-9
42. Liu X, Yang L, Wang H, Xu G, Zhu S, Li M, et al. Effects of miR-19b knockdown on the cardiac differentiation of P19 mouse embryonic carcinoma cells. Mol Med Rep. (2015) 11:2504–12. doi: 10.3892/mmr.2014.3037
43. Sun R, Zhang L. Long non-coding RNA MALAT1 regulates cardiomyocytes apoptosis after hypoxia/reperfusion injury via modulating miR-200a-3p/PDCD4 axis Biomedicine and pharmacotherapy. Biomed Pharmacotherapie. (2018) 111:1036–45. doi: 10.1016/j.biopha.12122
44. Arif M, Pandey R, Alam P, Jiang S, Sadayappan S, Paul A, et al. MicroRNA-210-mediated proliferation, survival, and angiogenesis promote cardiac repair post myocardial infarction in rodents. J Mol Med. (2017) 95:1369–85. doi: 10.1007/s00109-017-1591-8
45. Qiao L, Hu S, Liu S, Zhang H, Ma H, Huang K, et al. microRNA-21-5p dysregulation in exosomes derived from heart failure patients impairs regenerative potential. J Clin Invest. (2019) 129:2237–50. doi: 10.1172/JCI123135
46. Sun L, Zhu W, Zhao P, Zhang J, Lu Y, Zhu Y, et al. Down-regulated exosomal microRNA-221 - 3p derived from senescent mesenchymal stem cells impairs heart repair. Front Cell Dev Biol. (2020) 8:263. doi: 10.3389/fcell.2020.00263
47. Gao J., Zhu M., Liu R-. F., Zhang J-. S., Xu M. Cardiac hypertrophy is positively regulated by microRNA-24 in rats. Chin Med J. (2018) 131:1333–41. doi: 10.4103/0366-6999.232793
48. Wang B, Xu M, Li M, Wu F, Hu S, Chen X, et al. miR-25 Promotes Cardiomyocyte Proliferation by Targeting FBXW7 Molecular therapy. Nucl Acids. (2020) 20:1299–308. doi: 10.1016/j.omtn.01013
49. Borden A, Kurian J, Nickoloff E, Yang Y, Troupes CD, Ibetti J, et al. Transient introduction of miR-294 in the heart promotes cardiomyocyte cell cycle reentry after injury. Circ Res. (2019) 125:14–25. doi: 10.1161/CIRCRESAHA.118.314223
50. Zhen L, Zhao Q, Lu J, Deng S, Xu Z, Zhang L, et al. (2020). miR-301a-PTEN-AKT signaling induces cardiomyocyte proliferation and promotes cardiac repair post-MI. Mol Ther Nucleic Acids. (2020) 22:251–62. doi: 10.1016/j.omtn.08033
51. Xu F, Yang J, Shang J, Lan F, Li M, Shi L, et al. MicroRNA-302d promotes the proliferation of human pluripotent stem cell-derived cardiomyocytes by inhibiting in the Hippo pathway. Clin Sci. (2019) 133:1387–99. doi: 10.1042/CS20190099
52. Han X, Chen X, Han J, Zhong Y, Li Q, An Y, et al. MiR-324/SOCS3 axis protects against hypoxia/reoxygenation-induced cardiomyocyte injury and regulates myocardial ischemia via TNF/NF-κB signaling pathway. Int Heart J. (2020) 61:1258–69. doi: 10.1536/ihj.19-687
53. Zhao Z, Zhao Y, Ying-Chun L, Zhao L, Zhang W, Yang JG, et al. Protective role of microRNA-374 against myocardial ischemia-reperfusion injury in mice following thoracic epidural anesthesia by downregulating dystrobrevin alpha-mediated Notch1 axis. J Cell Physiol. (2019) 234:10726–40. doi: 10.1002/jcp.27745
54. Lange S, Banerjee I, Carrion K, Serrano R, Habich L, Kameny R, et al. miR-486 is modulated by stretch and increases ventricular growth. JCI Insight. (2019) 4:507. doi: 10.1172./jci.insight.125507
55. Jin Y, Ni S. miR-496 remedies hypoxia reoxygenation-induced H9c2 cardiomyocyte apoptosis via Hook3-targeted PI3k/Akt/mTOR signaling pathway activation. J Cell Biochem. (2020) 121:698–712. doi: 10.1002/jcb.29316
56. Deng S, Zhao Q, Zhen L, Zhang C, Liu C, Wang G, et al. Neonatal heart-enriched miR-708 promotes proliferation and stress resistance of cardiomyocytes in rodents. Theranostics. (2017) 7:1953–65. doi: 10.7150/thno.16478
57. Hu Y, Jin G, Li B, Chen Y, Zhong L, Chen G, et al. Suppression of miRNA let-7i-5p promotes cardiomyocyte proliferation and repairs heart function post injury by targetting CCND2 and E2F2. Clinical science. (2019). 133:425–41. doi: 10.1042/CS20181002
58. Huang W, Feng Y, Liang J, Yu H, Wang C, Wang B, et al. Loss of microRNA-128 promotes cardiomyocyte proliferation and heart regeneration. Nat Commun. (2018) 9:700. doi: 10.1038/s41467-018-03019-z
59. Ma WY, Song RJ, Xu BB, Xu Y, Wang XX, Sun HY, et al. Melatonin promotes cardiomyocyte proliferation and heart repair in mice with myocardial infarction via miR-143-3p/Yap/Ctnnd1 signaling pathway. Acta Pharmacol Sin. (2021) 42:921–31. doi: 10.1038/s41401-020-0495-2
60. Cao ML, Zhu BL, Sun YY, Qiu GR, Fu WN, Jiang HK, et al. MicroRNA-144 regulates cardiomyocyte proliferation and apoptosis by targeting TBX1 through the JAK2/STAT1 pathway. Cytogen Genome Res. (2019) 159:190–200. doi: 10.1159/000505143
61. Porrello ER, Johnson BA, Aurora AB, Simpson E, Nam YJ, Matkovich SJ, et al. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ Res. (2011) 109:670–9. doi: 10.1161/CIRCRESAHA.111.248880
62. Shi HH, Wang ZQ, Zhang S. MiR-208a participates with sevoflurane post-conditioning in protecting neonatal rat cardiomyocytes with simulated ischemia-reperfusion injury via PI3K/AKT signaling pathway. Eur Rev Med Pharmacol Sci. (2020) 24:943–55. doi: 10.26355/eurrev_202001_20080
63. Yang Q, Wu F, Mi Y, Wang F, Cai K, Yang X, et al. Aberrant expression of miR-29b-3p influences heart development and cardiomyocyte proliferation by targeting NOTCH2. Cell Prolif. (2020) 53:e12764. doi: 10.1111/cpr.12764
64. Zhao J, Chen F, Ma W, Zhang P. Suppression of long noncoding RNA NEAT1 attenuates hypoxia-induced cardiomyocytes injury by targeting miR-378a-3p. Gene. (2020) 731:144324. doi: 10.1016/j.gene.2019.144324
65. Chen Y, Wu S, Zhang XS, Wang DM, Qian CY. MicroRNA-489 promotes cardiomyocyte apoptosis induced by myocardial ischemia-reperfusion injury through inhibiting SPIN1. Eur Rev Med Pharmacol Sci. (2019) 23:6683–90. doi: 10.26355/eurrev_201908_18559
66. Li T, Qian D, Guoyan J, Lei Z. Downregulated long noncoding RNA LUCAT1 inhibited proliferation and promoted apoptosis of cardiomyocyte via miR-612/HOXA13 pathway in chronic heart failure. Eur Rev Med Pharmacol Sci. (2020) 24:385–95. doi: 10.26355/eurrev_202001_19937
67. Zhang JS, Zhao Y, Lv Y, Liu PY, Ruan JX, Sun YL, et al. miR-873 suppresses H9C2 cardiomyocyte proliferation by targeting GLI1. Gene. (2017) 626:426–432. doi: 10.1016/j.gene.05062
68. Zheng J, Peng B, Zhang Y, Ai F, Hu X. miR-9 knockdown inhibits hypoxia-induced cardiomyocyte apoptosis by targeting. Yap1 Life Sci 219. (2019) 129–35. doi: 10.1016/j.lfs.01, 014.
69. Yeh CH, Bellon M, Nicot C. FBXW7: a critical tumor suppressor of human cancers. Mol Cancer. (2018) 17:115. doi: 10.1186/s12943-018-0857-2
70. Liu Z, Zhang Z, Yao J, Xie Y, Dai Q, Zhang Y, et al. Serum extracellular vesicles promote proliferation of H9C2 cardiomyocytes by increasing miR-17-3p. Biochem Biophys Res Commun. (2018) 499:441–6. doi: 10.1016/j.bbrc.03157
71. Cheng H, Chang S, Xu R, Chen L, Song X, Wu J, et al. Hypoxia-challenged MSC-derived exosomes deliver miR-210 to attenuate post-infarction cardiac apoptosis. Stem Cell Res Ther. (2020) 11:224. doi: 10.1186/s13287-020-01737-0
72. Zhou X- H., Chai H-. X., Bai M., Zhang Z. LncRNA-GAS5 regulates PDCD4 expression and mediates myocardial infarction-induced cardiomyocytes apoptosis via targeting MiR-21. Cell cycle. (2018) 19:1363–77. doi: 10.1080/15384101.2020.1750257
73. Teng YL, Ren F, Xu H, Song HJ. Overexpression of miRNA-410-3p protects hypoxia-induced cardiomyocyte injury via targeting TRAF5. Eur Rev Med Pharmacol Sci. (2019) 23:9050–7. doi: 10.26355/eurrev_201910_19307
74. Krishnamurthy N, Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treat Rev. (2017) 62:50–60. doi: 10.1016/j.ctrv.11002
75. Gralewska P, Gajek A, Marczak A, Rogalska A. Participation of the ATR/CHK1 pathway in replicative stress targeted therapy of high-grade ovarian cancer. J Hematol= Oncol. (2020) 13:39. doi: 10.1186/s13045-020-00874-6
76. Xu J, Cao D, Zhang D, Zhang Y, Yue Y. MicroRNA-1 facilitates hypoxia-induced injury by targeting NOTCH3. J Cell Biochem. (2020) 121:4458–69. doi: 10.1002/jcb.29663
77. Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. (2005) 308:1149–54. doi: 10.1126/science.1108625
78. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. (2016) 17:47–62. doi: 10.1038/nrg.2015.10
79. Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol. (2018) 19:143–57. doi: 10.1038/nrm.2017.104
80. Li X, Sun Y, Huang S, Chen Y, Chen X, Li M, et al. Inhibition of AZIN2-sv induces neovascularization and improves prognosis after myocardial infarction by blocking ubiquitin-dependent talin1 degradation and activating the Akt pathway. EBio Med. (2018) 39:69–82. doi: 10.1016/j.ebiom.12001
81. Song KY, Zhang XZ, Li F, Ji QR. Silencing of ATP2B1-AS1 contributes to protection against myocardial infarction in mouse via blocking NFKBIA-mediated NF-κB signalling pathway. J Cell Mol Med. (2020) 24:4466–4479. doi: 10.1111/jcmm.15105
82. Chen G, Li H, Li X, Li B, Zhong L, Huang S, et al. Loss of long non-coding RNA CRRL promotes cardiomyocyte regeneration and improves cardiac repair by functioning as a competing endogenous RNA. J Mol Cell Cardiol. (2018) 122:152–64. doi: 10.1016/j.yjmcc.08013
83. Cai B, Ma W, Ding F, Zhang L, Huang Q, Wang X, et al. The long non-coding RNA CAREL controls cardiac regeneration. J Am Coll Cardiol. (2018) 72:534–50. doi: 10.1016/j.jacc.04085
84. Zhang Y, Hou YM, Gao F, Xiao JW, Li CC, Tang Y, et al. lncRNA GAS5 regulates myocardial infarction by targeting the miR-525-5p/CALM2 axis. J Cell Biochem. (2019) 120:18678–88. doi: 10.1002/jcb.29156
85. Xu Y, Fang H, Xu Q, Xu C, Yang L, Huang C, et al. LncRNA GAS5 inhibits NLRP3 inflammasome activation-mediated pyroptosis in diabetic cardiomyopathy by targeting miR-34b-3p/AHR. Cell Cycle. (2020) 19:3054–65. doi: 10.1080/15384101.2020.1831245
86. Zhou T, Qin G, Yang L, Xiang D, Li S. LncRNA XIST regulates myocardial infarction by targeting miR-130a-3p. J Cell Physiol. (2019) 234:8659–67. doi: 10.1002/jcp.26327
87. Ponnusamy M., Liu F., Zhang Y-. H., Li R-. B., Zhai M., Liu F., et al. Long Noncoding RNA CPR (Cardiomyocyte Proliferation Regulator) regulates cardiomyocyte proliferation and cardiac repair. Circulation. (2019) 139:2668–84. doi: 10.1161/CIRCULATIONAHA.118.035832
88. Li H, Jiang L, Yu Z, Han S, Liu X, Li M, et al. The role of a novel long non-coding RNA TUC40- in cardiomyocyte induction and maturation in P19 cells. Am J Med Sci. (2017) 354:608–16. doi: 10.1016/j.amjms.08019
89. Chen Y, Li X, Li B, Wang H, Li M, Huang S, et al. Long non-coding RNA ECRAR triggers post-natal myocardial regeneration by activating ERK1/2 signaling molecular therapy. J Am Soc Gene Therapy. (2019) 27:29–45. doi: 10.1016/j.ymthe.10021
90. Wang J, Chen X, Shen D, Ge D, Chen J, Pei J, et al. (2019). A long noncoding RNA NR_045363 controls cardiomyocyte proliferation and cardiac repair J Mol Cell Cardiol 127. (2019) 105–14. doi: 10.1016/j.yjmcc.12, 005.
91. Li B, Hu Y, Li X, Jin G, Chen X, Chen G, et al. Sirt1 antisense long non-coding RNA promotes cardiomyocyte proliferation by enhancing the stability of Sirt1. J Am Heart Assoc. (2018) 7:e009700. doi: 10.1161/JAHA.118.009700
92. Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. (2016) 143:1838–47. doi: 10.1242/dev.128074
93. Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, et al. Scrambled exons. Cell. (1991) 64:607–13.
94. Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. (1993) 73:1019–30.
95. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell. (2017) 66:021 doi: 10.1016/jmolcel02
96. Diallo LH, Tatin F, David F, Godet AC, Zamora A, Prats AC, et al. How are circRNAs translated by non-canonical initiation mechanisms? Biochimie. (2019) 164:45–52. doi: 10.1016/j.biochi.06015
97. Huang S, Li X, Zheng H, Si X, Li B, Wei G, et al. Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation. (2019) 139:2857–76. doi: 10.1161/CIRCULATIONAHA.118.038361
98. Bai M, Pan CL, Jiang GX, Zhang YM, Zhang Z. CircHIPK3 aggravates myocardial ischemia-reperfusion injury by binding to miRNA-124-3p. Eur Rev Med Pharmacol Sci. (2019) 23:10107–14. doi: 10.26355/eurrev_201911_19580
99. Si X, Zheng H, Wei G, Li M, Li W, Wang H, et al. circRNA Hipk3 Induces Cardiac Regeneration after Myocardial Infarction in Mice by Binding to Notch1 and miR-133a molecular therapy. Nucleic Acids. (2020) 21:636–55. doi: 10.1016/j.omtn.06024
100. Zhou HF, Xu LL, Xie B, Ding HG, Fang F, Fang Q, et al. Hsa-circ-0068566 inhibited the development of myocardial ischemia reperfusion injury by regulating hsa-miR-6322/PARP2 signal pathway. Eur Rev Med Pharmacol Sci. (2020) 24:6980–93. doi: 10.26355/eurrev_202006_21690
101. Zhu Y, Zhao P, Sun L, Lu Y, Zhu W, Zhang J, et al. Overexpression of circRNA SNRK targets miR-103-3p to reduce apoptosis and promote cardiac repair through GSK3beta/beta-catenin pathway in rats with myocardial infarction. Cell Death Discov. (2021) 7:84. doi: 10.1038/s41420-021-00467-3
102. Zong L, Wang W. CircANXA2 promotes myocardial apoptosis in myocardial ischemia-reperfusion injury via inhibiting miRNA-133 expression. Biomed Res Int. (2020) 2020:8590861. doi: 10.1155/2020/8590861
103. Luo C, Ling GX, Lei BF, Feng X, Xie XY, Fang C, et al. Circular RNA PVT1 silencing prevents ischemia-reperfusion injury in rat by targeting microRNA-125b and microRNA-200a. J Mol Cell Cardiol. (2021) 3:019. doi: 10.1016/jyjmcc05
104. Ibrahim AG, Cheng K, Marban E. Exosomes as critical agents of cardiac regeneration triggered by cell therapy. Stem Cell Reports. (2014) 2:606–19. doi: 10.1016/j.stemcr.04006
105. Hullinger TG, Montgomery RL, Seto AG, Dickinson BA, Semus HM, Lynch JM, et al. Inhibition of miR-15 protects against cardiac ischemic injury. Circ Res. (2012) 110:71–81. doi: 10.1161/CIRCRESAHA.111.244442
106. Del Campo CV, Liaw NY, Gunadasa-Rohling M, Matthaei M, Braga L, Kennedy T, et al. Regenerative potential of epicardium-derived extracellular vesicles mediated by conserved miRNA transfer. Cardiovasc Res. (2022) 118:597–611. doi: 10.1093/cvr/cvab054
107. Kumarswamy R, Bauters C, Volkmann I, Maury F, Fetisch J, Holzmann A, et al. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res. (2014) 114:1569–75. doi: 10.1161/CIRCRESAHA.114.303915
108. Skroblin P, Mayr M. “Going long”: long non-coding RNAs as biomarkers. Circ Res. (2014) 115:607–9. doi: 10.1161/CIRCRESAHA.114.304839
Keywords: cardiomyocyte, regeneration, ncRNA, miRNA, lncRNA, circRNA
Citation: Qin K, Xie X, Tang W, Yang D, Peng J, Guo J, Yang J and Fan C (2022) Non-coding RNAs to regulate cardiomyocyte proliferation: A new trend in therapeutic cardiac regeneration. Front. Cardiovasc. Med. 9:944393. doi: 10.3389/fcvm.2022.944393
Received: 15 May 2022; Accepted: 02 August 2022;
Published: 18 August 2022.
Edited by:
Serena Zacchigna, International Centre for Genetic Engineering and Biotechnology, ItalyReviewed by:
Francesco Loffredo, University of Campania “L. Vanvitelli”, ItalyCopyright © 2022 Qin, Xie, Tang, Yang, Peng, Guo, Yang and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengming Fan, ZmFuY2hlbmdtaW5nQGNzdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.