- 1Department of Spine Surgery, Ganga Hospital, Coimbatore, India
- 2Department of Biotechnology, Ganga Research Centre, Coimbatore, India
- 3Department of Plant Biotechnology, Tamil Nadu Agricultural University, Coimbatore, India
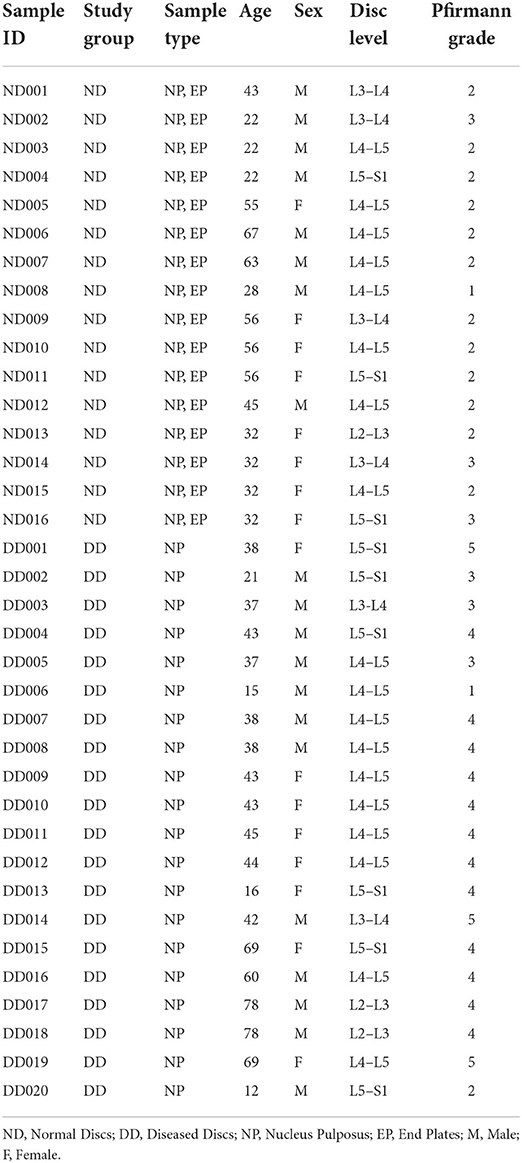
Study design: The diversity of microflora inhabiting endplate (EP) and nucleus pulposus (NP) tissues of human intervertebral disc (IVD) was profiled through NGS-supported 16S rRNA amplicon sequencing. Sixteen EP and their corresponding NP were excised from the brain-dead voluntary organ donors with no clinical history of low back pain, and 12 herniated and 8 degenerated NP tissues isolated from the patients undergoing spinal surgery were subjected to study the alteration in the microbial diversity.
Objective(s): To understand in normal IVD, whether the colonization of bacteria to the NP is through the EP in discs with intact annulus fibrosus. To identify significantly differing microbial population(s) between normal and diseased IVD (NP).
Background of the study: There is increasing evidence for subclinical infection by fastidious low, growing bacteria to be a cause of disc degeneration. Although the presence of bacteria in NP has been reported well in literature, the source of bacteria is not clearly proved as the disc is avascular in healthy condition. Documentation of similar bacterial populations in the EP and NP may add proof that bacterial inoculation of NP occurs via the EP.
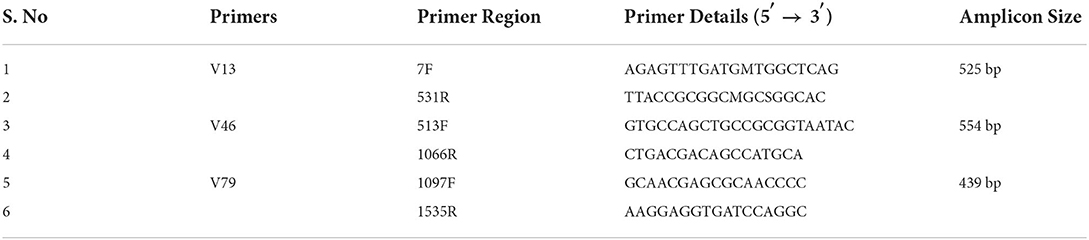
Materials and methods: Sixteen EP and their corresponding NP excised from brain-dead voluntary organ donors with no history of back pain and 20 diseased discs collected from patients undergoing microdiscectomy/fusion surgery were used for profiling microbiome through 16S rRNA amplicon sequencing using primers specific for V1-V9 hypervariable regions. Changes in bacterial diversity and abundance were analysed to identify the key microbial populations in normal IVD NP and EP tissues and those significantly altered in diseased IVD (NP).
Results: NP and EP shared a similar spectrum of microbiome but with varying abundance. The five dominant phyla identified were Proteobacteria, Firmicutes, Actinobacteria, OD1, and Bacteroidetes. Proteobacteria was found to be the most abundant phyla in both NP (62%) and EP (53%) of the normal IVD. This was followed by Firmicutes (16%), Actinobacteriota (11%), OD1 (Parcubacteria) (7.6%), and Bacteroidetes (2%) in NP and Firmicutes (23.4%), OD1 (Parcubacteria) (17.6%), Actinobacteriota (2.8%), and Bacteroidetes (2.6%) in EP, respectively. Under diseased conditions, Proteobacteria (68%) was dominant when compared with other phyla. However, there was no significant difference in the abundance of Proteobacteria between the normal and diseased discs. Interestingly, the other dominant phyla such as Firmicutes (Normal-NP: 16.2%; Diseased-NP: 4.02%) and Actinobacteria (Normal-NP: 11%; Diseased-NP: 0.99%) showed a significant reduction in degenerated discs. To understand the key microbial populations that are significantly altered during disease, correlation analysis was performed among the three phyla, which revealed a negative correlation in the ratio of Actinobacteria + Firmicutes vs. Proteobacteria (p = 0.001) in DD.
Conclusion: Results of our study clearly demonstrated a similar bacterial diversity but with varying abundance between the EP and NP, suggesting the existence of the endplate–nucleus pulposus axis in the normal IVD microbiome. Further, our results have indicated that the changes in the abundance of Actinobacteria + Firmicutes vs. Proteobacteria during DDD need further investigation.
Introduction
Low back pain (LBP) due to degenerative disc disease (DDD) is fast becoming the most common cause of musculoskeletal disability, resulting in a huge socioeconomic burden across all nations (1). The prevalence of LBP has increased from 377.5 million in 1990 to 577.0 million in 2017, and greater attention is needed to mitigate the global burden of LBP (2). While mechanical, traumatic, genetic susceptibility, smoking, inflammaging, physiological, and work profile have been postulated as various causes of DDD and LBP, there is mounting evidence for microbial etiology as well (3).
In our previous study, we documented for the first time that human lumbar intervertebral discs (IVD), even in normal asymptomatic subjects, were not sterile and harbored a distinct microbiome (4). While the human microbiome has a crucial role in supporting and maintaining health, alteration in relative abundance or diversity (dysbiosis) of microflora causes changes in the accumulation of microbial proteins, peptides, and metabolites, disturbing the intense crosstalk mechanisms between the host and microbiome, affecting health. Dysbiosis in both the gut and skin has clearly been linked to many disorders, such as inflammatory bowel diseases (IBD), irritable bowel syndrome (IBS), diabetes, obesity, cancer, and cardiovascular and central nervous system disorders (5). More recently, the gut microbiota has been associated with chronic musculoskeletal pain syndromes, including LBP and fibromyalgia (6, 7).
The disc being an avascular structure, the source of bacteria is unclear when the annulus fibrosis is still intact (8). In the current study, we investigate for the first time the endplate–nucleus pulposus axis in the normal human IVD microbiome. We further analyzed the microbial diversity of IVD between health and disease conditions which has thrown further insights into the discovery of metagenomic biomarkers for the diagnosis of IVD disease.
Materials and methods
Sample population
Lumbar discs from a brain-dead voluntary organ donor(s) and patients undergoing microdiscectomy/spinal fusion were obtained. The normal discs (ND) group had 16 NP (Nucleus Pulposus) tissues and 16 EP (Cartilaginous Endplates) tissues excised from MRI normal discs of brain-dead voluntary organ donors with no history of back pain. The diseased discs (DD) group had NP tissues of 20 diseased discs excised from 20 patients undergoing spinal surgery as per the ethical norms laid by the Indian Council of Medical Research (ICMR; Table 1).
Ethical clearance and IRB approval
Intervertebral discs from brain-dead organ donors were collected in full agreement with TRANSTAN-214/2016 (Transplant Authority Government of Tamil Nadu, India). Institutional review board (IRB) approval (10/09/2020) for the study was obtained from the IRB of Ganga Medical Center and Hospital, Coimbatore. Ethical clearance for the study was obtained from the ethics committee (NECRBHR Regn No: EC/NEW/INST/2020/1146), and written consent was obtained from all the patients.
Extraction of genomic DNA
Disc tissues were collected under sterile operating conditions and snap frozen in sterile cryovials using liquid nitrogen (−196°C). DNA was extracted from 200 mg of intervertebral disc tissues—NP and EP tissues of ND group and NP from DD group using QIAamp® DNA Mini Kit—and enrichment of bacterial DNA was performed using NEBNext® Microbiome DNA Enrichment Kit, #E612S/L (New England BioLabs, Ipswich, MA), as per the manufacturer's instructions.
Quality controls
To check for potential contamination of foreign DNA during PCR amplification, process control [PBS buffer, No template control (NTC)] was substituted for template DNA, and positive control (E. coli sample) was used. No spurious DNA amplification was observed for buffers used for processing tissue samples procured from patients for the study (Supplementary Figure 1).
16S rRNA amplicon sequencing
The DNA samples were purified, assessed for their quality, and further used for library construction and amplification of 16S rRNA using V1-V9 primers (Table 2). About 25 ng of DNA was used to amplify bacterial 16S rRNA hypervariable region V1–V9 using DNA KAPA HiFi HotStart Ready Mix and 100 nm of the three primer pairs 7F-531R, 513F-1066R, and 1097F-1535R. Amplified fragments were sequenced using the Illumina MiSeq platform as described earlier (4).
Bioinformatic analysis
Identification of ASVs
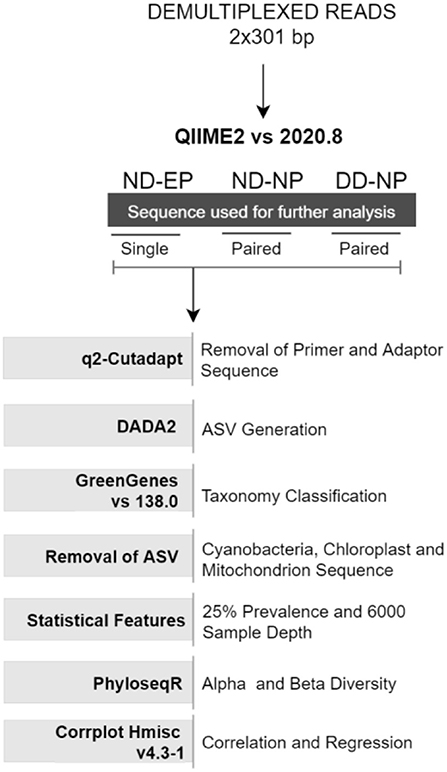
Demultiplexed paired-end (2 × 301bp) raw reads were imported into the Quantitative Insights Into Microbial Ecology-2 QIIME2 (v 2020.8) pipeline (9). Further, primers and adaptor sequences were trimmed using the q2-cutadapt plugin. Single-end reads were used for comparing the microbiome colonizing NP and EP of normal IVD, and paired-end reads were used for profiling the microbiome in the NP tissues of ND and DD groups. DADA2 (10) plugin in QIIME2 was used to produce amplicon sequence variants (ASV) feature tables and representative sequences.
Normalization of identified ASVs
Taxonomic classification was assigned using the Greengenes database (version 138) integrated within QIIME2 followed by phylogenetic diversity analysis. Unassigned reads and chloroplast (cyanobacteria) and mitochondria reads were also removed from ASVs. ASVs were normalized via simple division to their sample size and then multiplied by the size of the smaller sample and low prevalence ASVs (<0.025%) were also filtered from the feature table to strengthen the analysis (11). A rarefaction curve was built at the sampling depth of 16,000, which retained 36.91% (688,000) of sequences in 100% of samples. This sampling depth was selected based on the maximum depth that retained all samples for further analysis.
Phylogenetic diversity analysis
Alpha diversity indices (Shannon and Observed OTU) and beta diversity analyses were performed using the unifrac weighted and unweighted phyloseq R package (12). Non-metric multidimensional scaling (NMDS) was used to investigate the (dis)similarities between bacterial communities for weighted unifrac methods, as found in Figure 1.
Correlation and regression analysis
Correlation between changes in the relative abundance of different phyla during health and disease conditions was estimated using Spearman's rank correlation coefficient test using the rcorr function in Hmisc v4.3-1 R package and visualized using the corrplot function in R with a significance level set at 0.05.
Data availability
Illumina paired metagenomic sequencing data generated for all samples in this study are deposited in the NCBI Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/sra) under BioProject ID PRJNA577202 and PRJNA831905 for NP and EP tissues, respectively.
Results
This study attempted to analyze the possible route of microbial entry into the NP tissues of IVD by profiling the bacterial population(s) in the EP and NP of the intervertebral disc and to analyze the alterations in the diversity and abundance of bacterial species during health and disease conditions. To the best of our knowledge, this is the first systematic report on the tissue-specific microbial profiling in the NP and EP tissues of the human intervertebral disc (IVD). One of our earlier studies reported a detailed analysis of the microbiome inhabiting nucleus pulposus (NP) tissues of IVD (4). This study documented changes in the diversity and abundance of bacteria between ND (16 NP and 16 EP tissues excised from the brain-dead voluntary organ donors) and DD (20 NP tissues excised from patients undergoing spinal surgery) groups using NGS-supported 16S rRNA amplicon sequencing.
PCR quality controls
Process controls (PBS and NTC) were used to evaluate contamination from extraneous DNA materials present in reagents or buffers used for metagenome sequencing. Process controls had shown no amplification while amplifying with universal primers whereas positive control showed amplification (Supplementary Figure 1).
Microbial profile of NP and EP of normal IVD
Microbiome profiling in the NP and EP tissues of normal IVD through 16S rRNA amplicon sequencing generated 141,747 and 136,569 reads, respectively. The quality of raw reads was relatively comparable between the tissues (Supplementary Table 1). Raw reads were subjected to the removal of primer and adaptor sequences using the DADA2 denoising algorithm and the obtained ASVs were mapped against Greengenes vs. 138 database for assigning OTUs. Nearly 99.5% of the reads were classified at the kingdom level, 51.7% of sequences were classified at the phylum level, 21.9% of the reads were assigned at the genus level, and <1% of reads remained uncharacterized.
At the Phylum level, five different phyla, namely, Proteobacteria, Firmicutes, Actinobacteriota, OD1 (Parcubacteria), and Bacteroidetes, were present with >1% relative abundance. Proteobacteria remained as the most abundant phylum in both NP (62%) and EP (53%) tissues of normal IVD. This was followed by Firmicutes (16%), Actinobacteriota (11%), OD1 (Parcubacteria) (7.6%), and Bacteroidetes (2%) in NP and Firmicutes (23.4%), OD1 (Parcubacteria) (17.6%), Actinobacteriota (2.8%), and Bacteroidetes (2.6%) in EP, respectively (Figure 2). It was interesting to note that the top five phyla were similar between NP and EP.
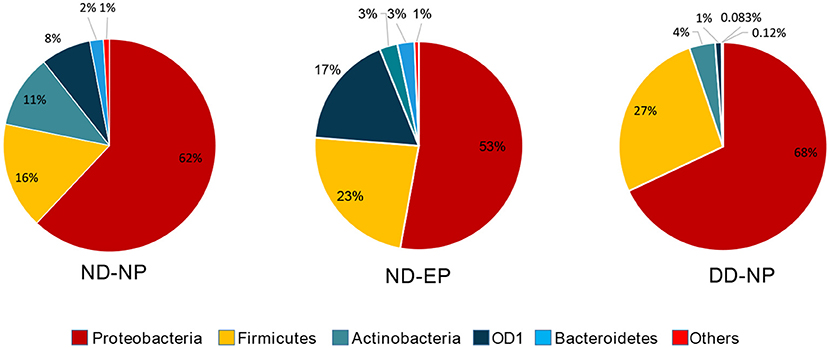
Figure 2. Abundance at Phylum level. Donut plots represent the relative abundance of phylum across ND (NP, EP) and DD (NP) groups. Proteobacteria was the most abundant phyla compared to other phyla.
Essentially, a t-test was calculated to find the significance between the NP and EP. As the study was performed based on QCA (Qualitative Comparative Analysis), FDR-control procedures were calculated to signify incorrect rejection from the comparison between NP and EP. We found an FDR <0.01 in the QCA of Proteobacteria (p < 0.001), Actinobacteria (p < 0.015), Firmicutes (p < 0.03) and OD1 (p < 0.001) in the calculated parametric test. Proteobacteria, Actinobacteria were significantly decreased whereas Firmicutes and OD1 were significantly increased in EP comparatively (Figure 3).
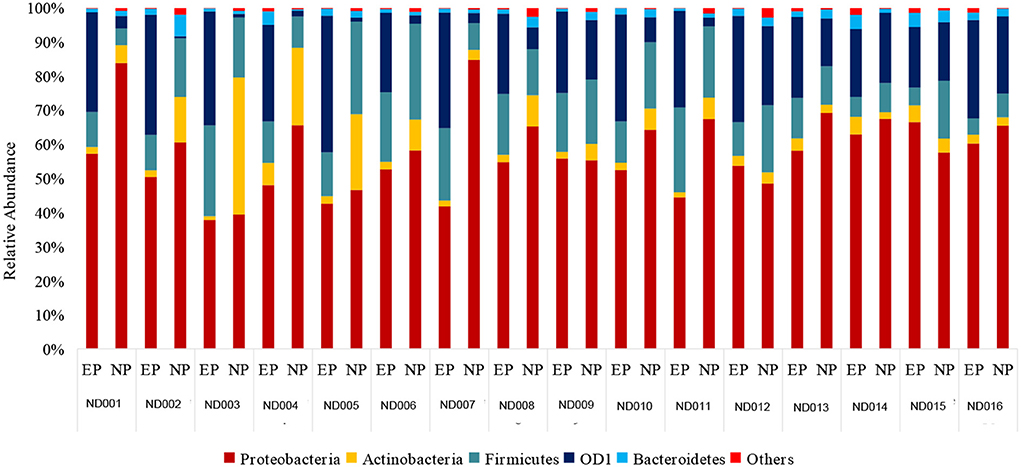
Figure 3. Comparison of bacterial phyla in NP and their respective EP of normal discs. Proteobacteria and Firmicutes were slightly abundant in NP when compared to EP.
The relative abundance of the top 15 genera across NP and EP tissues of normal IVD is depicted in Figure 4. Pseudomonas belonging to the phyla Proteobacteria were dominant with higher abundance in both ND-NP (26%) and ND-EP (28%) compared to all other identified genera. In NP discs, the second most abundant genus was Brevundimonas (12%) which was relatively lower in EP discs (9%). In contrast, EP discs had Anoxybacillus (19%) to be the second most abundant genus when compared to NP (5%). To validate the NGS results, qPCR analysis was performed for the abundant bacterial genus in five representative EP and NP components of the human intervertebral disc tissue. As Pseudomonas is the abundant genus identified in our 16S NGS data, we used the Pseudomonas genus-specific primer for amplification using SYBR green-based chemistry on Bio-Rad CFX Opus 96 Real-Time PCR system. A Cq value for each sample was determined (Cq range 26.05–29.57), indicating the abundance of Pseudomonas in all sample types (Supplementary Data).
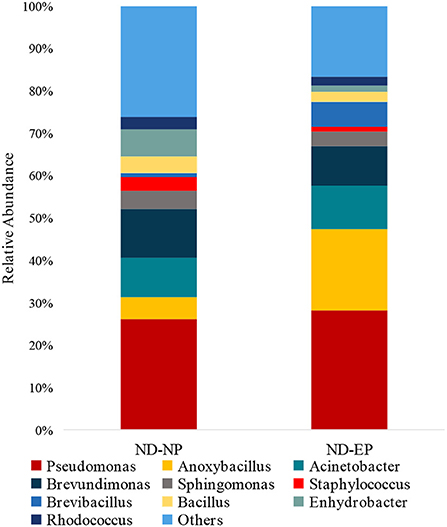
Figure 4. Abundance of microbiome at Genus level (ND-NP vs. ND-EP). Stacked bar plots representing the relative abundance of phylum across ND-NP and ND-EP groups. Pseudomonas belonging to the phyla Proteobacteria were found to be dominant with higher abundance in both ND-NP (26%) and ND-EP (28%).
Diversity of bacterial communities across EP and NP (ND)
In comparing the relative abundance of the bacterial phyla between NP and EP discs of normal donors, we found that Proteobacteria and Actinobacteria were slightly decreased in EP compared to NP, whereas an enriched microbial abundance of OD1 and Firmicutes in EP was found (Figure 3).
To assess the diversity of bacteria colonizing NP and EP tissues of ND, alpha diversity was determined through estimating observed OTUs and Shannon indices (Figure 5A), and significance was tested using a non-parametric test and the Wilcoxon rank-sum test. NP and EP tissues did not differ much in terms of their observed number of OTUs (p = 0.15) and Shannon index (p = 0.8). Beta diversity estimates, that is, weighted UniFrac distance, indicated the significant difference between the EP and NP tissues (p = 0.006) (Figure 5B).
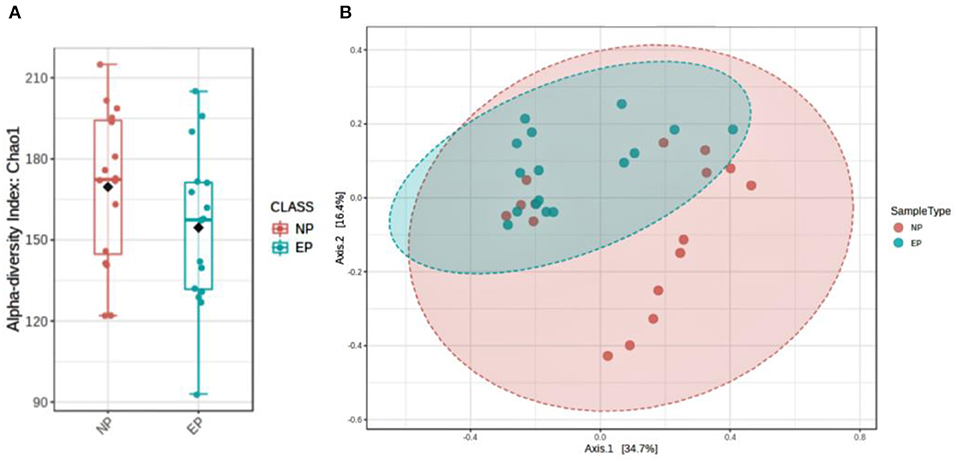
Figure 5. Assessment of microbial diversity. (A) Two indices depict alpha diversity: observed OTUs and Shannon indices showing insignificant differences tested using a non-parametric test and the Wilcoxon rank-sum test. (B) Beta diversity calculated using phylogenetic methods (UniFrac distance) shows a significant difference in the weighted (p, 0.006) index.
The alteration of NP microbiome during disease
There was a shift in the abundance of dominant bacteria colonizing NP tissues of IVD in degenerated discs. The dominant phyla Proteobacteria remained at the top in the NP tissues of both ND and DD (67.99% in DD as against 62% in the ND), as shown in Figure 2. This was followed by OD1 (Parcubacteria) (26.7%), Firmicutes (4.02%), Actinobacteriota (1.0%), and Bacteroidetes (0.08%). OD1 (Parcubacteria) showed a significant increase (7.6% in ND to 26.7% in DD) during disease progression. The other three Phyla, Firmicutes (16% to 4%), Actinobacteria (11–1%), and Bacteroidetes (2–0.08%), showed several-fold reduction during disease.
In comparing bacterial genera between ND-NP and DD-NP tissues, Pseudomonas was the most abundant genus in DD-NP (84%) and is strikingly higher than ND-NP (26%) as shown in Figure 6. In contrast, other genera, including Brevundimonas, Anoxybacillus, and Bacillus, were strikingly lesser in DD than ND.
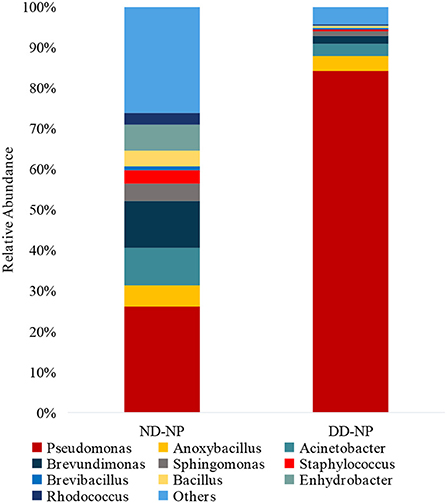
Figure 6. Abundance of microbiome at Genus level (ND vs. DD). Stacked bar plots representing the relative abundance of phylum across ND-NP and DD-NP groups. Pseudomonas was the most abundant genera (84%) compared to all other identified genera.
Altered ratio of A+F and proteobacteria during DD
To analyze the shift in the abundance of bacteria during disease (DD), Spearman correlation analysis was carried out using the rcorr function in the Hmisc R package. The correlation analysis revealed that the relative abundance of Actinobacteria and Proteobacteria exhibited a weak positive correlation (p = 0.001) in DD, whereas in ND, it demonstrated a moderate positive correlation (R = 0.345; p = 0.001) (Figure 7A). Firmicutes and Proteobacteria showed a significant negative correlation (R = −0.501) in DD, whereas, in ND, a weak positive correlation was observed (Figure 7B). Actinobacteria and Firmicutes exhibited a positive correlation and were found significant in both ND (Figure 7C). The ratio of Actinobacteria + Firmicutes: Proteobacteria showed a significant negative correlation during DD (R = −0.422). In contrast, ND showed a moderate positive correlation (R = 0.232) which was significant (Figure 7D).
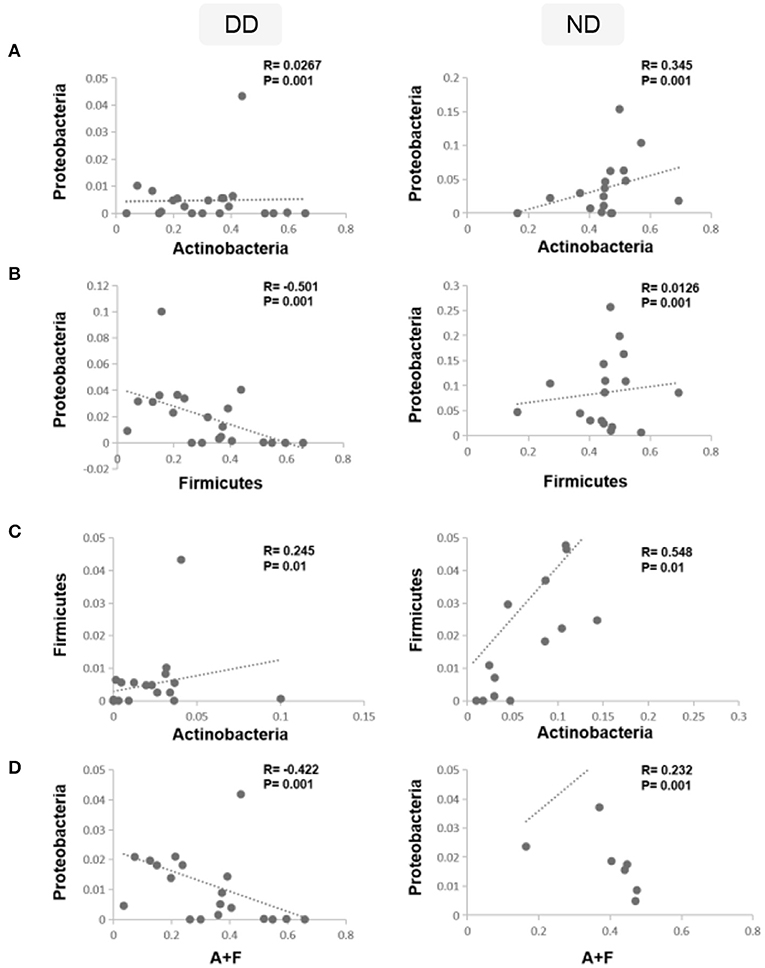
Figure 7. Correlations between dominant phyla and disease condition. Spearman's rank correlation coefficient was used to analyze the correlation between dominant phyla, ND, and DD. (A) The relative abundance of Actinobacteria and Proteobacteria was correlated positively in ND and found significant, whereas, in DD, it demonstrated a weak positive correlation. (B) Firmicutes and Proteobacteria showed a weak positive correlation in ND, whereas DD demonstrated a significant negative correlation. (C) Actinobacteria and Firmicutes exhibited a positive correlation and were found significant in both ND and DD. (D) Ratio of Actinobacteria + Firmicutes: Proteobacteria showed a significant negative correlation during DD and a significant positive correlation in ND.
Discussion
Traditionally intervertebral discs with intact annulus fibrosus were considered avascular (8). Hence, the reports of bacterial presence in intervertebral disc specimens obtained during surgery were met with criticism and deemed as contamination (13). However, growing evidence documents the bacterial presence in the IVD using various modalities (3, 4, 14). Our earlier study had proved the existence of a “Microbiome” within the intervertebral disc in both normal discs and degenerated discs using NGS techniques and led to the questions of how and when bacteria colonized the disc, what is their route of entry into IVD, and their role in health and disease (4).
The nutrition supply to NP tissue occurs via chemical diffusion through the capillaries that arise in vertebrate bodies and traverse to the cartilaginous endplates (EP) (15). The rate of nutrient transport is determined by the rate of cellular demand and the concentrations of nutrients, oxygen (< 5%), and pH (< 6.0) at the center of the discs comprising NP (16). We hypothesized that the endplate would not only be a gateway for nutrition but also the microbiome.
Bacterial presence vs. contamination
The bacterial presence in the intervertebral disc has been criticized as contamination in view of its avascular nature. However, there is growing evidence to the contrary. Recent studies have documented the existence of a microbiome within IVD (4). In our study, we procured the disc tissues in a sterile operating room and have Snap-froze them immediately to avoid contamination. In the lab, the samples were handled under sterile precautions for DNA extraction. We utilized positive and negative controls for PCR amplification and our results verified that there was no contamination during the experiment. Using metabolomics, a recent study has documented the presence of bacterial metabolites in both control and degenerated discs indicating that the bacteria in discs are not only alive but are multiplying and active metabolically ruling out the possibility of contamination (17).
Nucleus pulposus and endplate share highly similar microbiome
Our results showed that the microbiome inhabiting EP and NP tissues of normal discs were almost identical. Proteobacteria was the predominant phyla in both tissues, followed by Firmicutes. There were only minor differences between the two tissues in the abundance of Actinobacteria and OD1, which resulted in slight changes in the order of abundance of the top five phyla of microflora inhabiting IVD. Nevertheless, the overall bacterial composition was identical in EP and NP.
At the genus level, Pseudomonas was the dominant genus in both EP and NP. Pseudomonas is a gram-negative aerobe and has the ability to survive under adverse conditions. Although it is well known for its pathogenicity, it can degrade a large number of toxic compounds. It is also known for its industrial uses and beneficial effect on plants as it represses many peripheral catabolic enzymes and helps in biocontrol (18, 19). Their role in human IVD needs to be investigated. The next abundant genus was Anoxybacillus, a heat-tolerant bacteria that have been shown to have a role in human cellular metabolism and protein folding.
In addition, Anoxybacillus has been used commercially to produce maltose in the baking industry and in bioremediation to remove toxic metals and as a probiotic and stimulant of the immune system in fish against pathogens (20). Brevundimonas are found in abundance in normal NP, and they are carotenoid-producing bacteria. These carotenoids are potent human antioxidants that play an essential role in preventing tissue damage from occurring secondary to immune and inflammatory activity. The source of these antioxidants could be either dietary or bacterial (21). Hence, these antioxidants could play a role in maintaining extracellular matrix (ECM) integrity and tissue homeostasis (22).
Functional diversity of microbiome inhabiting NP and EP
To analyze the overall differences in the bacterial composition, we performed alpha diversity analysis using observed OTUs and Shannon indices. Alpha diversity represents the mean species diversity in sites or habitats. It was not surprising that there was no significant difference between NP and EP in terms of their microbial composition, which adds strength to our hypothesis that microbial colonies enter NP through EP. The minor differences in their metagenomic profile between NP and EP could have arisen as a result of the difference in their microenvironment, that is, vascularity, oxygenation, and pH between the two (23, 24).
Origin of microbiome
Although human IVD is avascular, endplate breaks resulting from microtrauma occur as early as the second decade and lead to disc-marrow contact (25). Although our study proves that endplate could be the route of entry for microbes into NP, the origin of the microbiome remains speculation. The majority of bacteria found in our study are common inhabitants of the human gut, leading us to believe that many gut bacteria via the hematogenous route could have been harbored in NP. However, this needs to be proven by a study comparing the disc and gut microbiomes of the same individuals.
Changes in the abundance of microflora between health and disease
To understand the alterations in microbial diversity in DDD, we compared the microbiome of normal discs, which showed a significant decrease in Firmicutes and Actinobacteria. They form two of the most abundant phyla in the human gut, and their decreased abundance in the human gut has been associated with dysbiosis and many inflammatory diseases (26). In addition, Vaiserman et al. reported the altered ratio of relative abundance of the top abundant bacteria, Firmicutes: Bacteroidetes, in the gut as a biomarker determining health and disease (27).
Similarly, the correlation between dominant phyla of health and disease conditions was studied. While Firmicutes and Proteobacteria showed a positive correlation in ND, they demonstrated a significant negative correlation in DD. This led us to investigate whether the ratios of the abundant phyla observed in ND are altered in DD. While evaluating the ratios of various phyla across ND and DD, the ratio of Firmicutes/Proteobacteria showed a significant negative correlation in DD compared to Actinobacteria/Proteobacteria (positive correlation) and Actinobacteria/Firmicutes (positive correlation). Interestingly, the combined ratio of Actinobacteria + Firmicutes/Proteobacteria showed a negative correlation that may be of potential significance in the progression of DD.
Bacteria are present in all areas of the human body as commensals in every individual and play a vital role in gut metabolism, immunity, and so on (28). Approximately, 98% of the microbiome of blood is primarily comprised of Proteobacteria followed by Bacteriodetes, Actinobacteria, and Firmicutes, with a higher abundance of Proteobacteria in patients with chronic kidney disease (CKD) (29). Proteobacteria encompasses a wide spectrum of bacteria which may be either commensal, pathogenic, or opportunistic pathogens (depending upon host factors, they may become pathogenic) (30). However, the hematogenous spread of bacteria from one location to other is well known and does not always result in sepsis (31). The bacterial seeding that occurs is very minimal and unless there is a suitable environment and favorable host factors, they do not result in overt infection.
Clinical significance
The last decade has witnessed a change in our understanding of human bacterial interaction and the role of bacteria in health and disease (32). Many diseases that were thought to be idiopathic or inflammatory have been proven to have a microbial origin or have a close association (33). Recent studies have also focussed on altering the gut microbiota by the use of probiotics to restore health and induce remission in many chronic illnesses (34–36). The findings of our study prove the existence of a “microbiome” and the possible route of entry into NP is via EP, and “dysbiosis” plays a possible role in disc degeneration. Our earlier study had proven that the diffusion across the endplates could be altered by drugs, such as Nimodipine (2, 37). In this context, altering the gut microbiome in restoring IVD health and its facilitation by pharmacological enhancement remains a possible avenue to be explored.
Limitations and future applications
In NGS metagenome data of EP, we used single-end reads for our analysis due to the quality of reads. The ratio of Actinobacteria + Firmicutes: Proteobacteria is of significance in DD; this finding needs to be validated in a larger number of samples. It would be interesting to employ a multi-omic approach comprising metabolomics, metaproteome, and host proteome to understand the intricate crosstalk between the host and microbial colony and its effect on disc health. Further analyzing the fetal IVD microbiome will throw further evidence to the above findings.
Conclusion
Our metagenome study has demonstrated a close similarity in the metagenomic profile of human lumbar intervertebral disc nucleus pulposus and the endplate, suggesting a possible existence of the endplate–nucleus pulposus axis in normal IVD. Minor variations between the microbiome in normal NP and EP tissues reiterate the sterility of disc, where the shift in their proportional existence could alter the health status. The increased abundance of Proteobacteria with a significant reduction in Firmicutes, Actinobacteria, and Bacteroidetes could be a potential microbial signature in the progression of DD. In addition, the altered ratio of dominant phyla Actinobacteria + Firmicutes: Proteobacteria during diseased states needs further investigation.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: BioSample, PRJNA577202 and PRJNA831905.
Ethics statement
The studies involving human participants were reviewed and approved by the Institutional Review Board of Ganga Medical Center and Hospital, Coimbatore (No: 2020/09/10). Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
Author contributions
SR, CT, RM, and SKS were involved in conceptualization and supervision of the project. CT and SN designed and performed the experiments. MM, SN, and RS did the extensive data analysis. SN and MM wrote the first draft of the article which was further improvised by SR, CT, RM, and SKS. SR, CT, RM, SKS, RK, and AS involved in review and editing. All authors contributed to the article and approved the submitted version.
Funding
The study was funded by Ganga Orthopaedic Research and Education Foundation (GOREF) (GOREF-2019/12) (Recognized by Department of Scientific and Industrial Research (DSIR) and co-funded by Department of Biotechnology (DBT), Government of India, Grant reference no: BT/PR35631/MED/30/2186/2019.
Acknowledgments
The authors thank Dr. Vasudevan Gowdaman MSc, Ph.D., Dr. Murugesh Easwaran MSc, Ph.D., and Ms. Sri Alamelu MSc for their valuable assistance in data analysis and statistical interpretations.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2022.927652/full#supplementary-material
References
1. Hartvigsen J, Hancock MJ, Kongsted A, Louw Q, Ferreira ML, Genevay S, et al. What low back pain is and why we need to pay attention. Lancet. (2018) 391:2356–67. doi: 10.1016/S0140-6736(18)30480-X
2. Wu A, March L, Zheng X, Huang J, Wang X, Zhao J, et al. Global low back pain prevalence and years lived with disability from 1990 to 2017: estimates from the global burden of disease study 2017. Ann Transl Med. (2020) 8:299. doi: 10.21037/atm.2020.02.175
3. Rajasekaran S, Tangavel C, Soundararajan DCR, Nayagam SM, Matchado MS. Inflammaging determines health and disease in lumbar discs-evidence from differing proteomic signatures of healthy, aging, and degenerating discs. Spine J. (2020) 20:48–59. doi: 10.1016/j.spinee.2019.04.023
4. Rajasekaran S, Soundararajan DCR, Tangavel C, Muthurajan R, Sri Vijay Anand KS, Matchado MS, et al. Human intervertebral discs harbour a unique microbiome and dysbiosis determines health and disease. Eur Spine J. (2020) 29:1621–40. doi: 10.1007/s00586-020-06446-z
5. Ellis SR, Nguyen M, Vaughn AR, Notay M, Burney WA, Sandhu S, et al. The skin and gut microbiome and its role in common dermatologic conditions. Microorganisms. (2019) 7:E550. doi: 10.3390/microorganisms7110550
6. Minerbi A, Gonzalez E, Brereton NJB, Anjarkouchian A, Dewar K, Fitzcharles M-A, et al. Altered microbiome composition in individuals with fibromyalgia. Pain. (2019) 160:2589–602. doi: 10.1097/j.pain.0000000000001640
7. Shmagel A, Langsetmo L, Demmer R, Knights D, Lane NE, Ensrud K, et al. Gut microbiome associations with chronic musculoskeletal pain in older men. Osteoarthritis Cartilage. (2019) 27:S73. doi: 10.1016/j.joca.2019.02.103
8. Fournier DE, Kiser PK, Shoemaker JK, Battié MC, Séguin CA. Vascularization of the human intervertebral disc: a scoping review. JOR Spine. (2020) 3:e1123. doi: 10.1002/jsp2.1123
9. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP, et al. DADA2: high-resolution sample inference from illumina amplicon data. Nat Methods. (2016) 13:581–3. doi: 10.1038/nmeth.3869
10. DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. (2006) 72:5069–72. doi: 10.1128/AEM.03006-05
11. Lagkouvardos I, Fischer S, Kumar N, Clavel T. Rhea: a transparent and modular R pipeline for microbial profiling based on 16S rRNA gene amplicons. PeerJ. (2017) 5:e2836. doi: 10.7717/peerj.2836
12. McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. (2013) 8:e61217. doi: 10.1371/journal.pone.0061217
13. Carricajo A, Nuti C, Aubert E, Hatem O, Fonsale N, Mallaval FO, et al. Propionibacterium acnes contamination in lumbar disc surgery. J Hosp Infect. (2007) 66:275–7. doi: 10.1016/j.jhin.2007.04.007
14. Jiao Y, Lin Y, Zheng Y, Yuan Y, Chen Z, Cao P, et al. The bacteria-positive proportion in the disc tissue samples from surgery: a systematic review and meta-analysis. Eur Spine J. (2019) 28:2941–50. doi: 10.1007/s00586-019-06062-6
15. Urban JPG, Smith S, Fairbank JCT. Nutrition of the intervertebral disc. Spine. (2004) 29:2700–9. doi: 10.1097/01.brs.0000146499.97948.52
16. Grunhagen T, Wilde G, Soukane DM, Shirazi-Adl SA, Urban JP. Nutrient supply and intervertebral disc metabolism. JBJS. (2006) 88:30–5. doi: 10.2106/00004623-200604002-00007
17. Rajasekaran S, Tangavel C, Vasudevan G, Easwaran M, Muthurajan R, Murugan KSSV, et al. Bacteria in human lumbar discs–subclinical infection or contamination? Metabolomic evidence for colonization, multiplication, and cell-cell cross-talk of bacteria. Spine J. (2022) 13. doi: 10.1016/j.spinee.2022.05.001
18. Novik G, Savich V, Kiseleva E. An insight into beneficial Pseudomonas bacteria. Microbiol Agri Hum Health. (2015) 1:73–105. doi: 10.5772/60502
19. Passera A, Compant S, Casati P, Maturo MG, Battelli G, Quaglino F, et al. Not just a pathogen? Description of a plant-beneficial pseudomonas syringae strain. Front Microbiol. (2019) 10:1409. doi: 10.3389/fmicb.2019.01409
20. Goh K, Kahar U, Chai Y, Chong CS, Chai KP, Ranjani V, et al. Recent discoveries and applications of Anoxybacillus. App Microbiol Biotechnol. (2013) 97:1475–88. doi: 10.1007/s00253-012-4663-2
21. Johnson EJ. The role of carotenoids in human health. Nutr Clin Care. (2002) 5:56–65. doi: 10.1046/j.1523-5408.2002.00004.x
22. Liu H, Tan KS, Zhang X, Zhang H, Cheng D, Ting Y, et al. Comparison of gut microbiota between golden and brown noble scallop chlamys nobilis and its association with carotenoids. Front Microbiol. (2020) 11:36. doi: 10.3389/fmicb.2020.00036
23. Horner HA, Urban JP. Volvo award winner in basic science studies: effect of nutrient supply on the viability of cells from the nucleus pulposus of the intervertebral disc. Spine. (2001) 26:2543–9. doi: 10.1097/00007632-200112010-00006
24. Holm S, Maroudas A, Urban JP, Selstam G, Nachemson A. Nutrition of the intervertebral disc: solute transport and metabolism. Connect Tissue Res. (1981) 8:101–19. doi: 10.3109/03008208109152130
25. Tomaszewski KA, Saganiak K, Gładysz T, Walocha JA. The biology behind the human intervertebral disc and its endplates. Folia Morphologica. (2015) 74:157–68. doi: 10.5603/FM.2015.0026
26. Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano GAD, Gasbarrini A, et al. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. (2019) 7:14. doi: 10.3390/microorganisms7010014
27. Vaiserman A, Romanenko M, Piven L, Moseiko V, Lushchak O, Kryzhanovska N, et al. Differences in the gut Firmicutes to Bacteroidetes ratio across age groups in healthy Ukrainian population. BMC Microbiol. (2020) 20:221. doi: 10.1186/s12866-020-01903-7
28. Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, et al. The NIH human microbiome project. Genome Res. (2009) 19:2317–23. doi: 10.1101/gr.096651.109
29. Shah NB, Allegretti AS, Nigwekar SU, Kalim S, Zhao S, Lelouvier B, et al. Blood microbiome profile in CKD: a pilot study. Clin J Am Soc Nephrol. (2019) 14:692–701. doi: 10.2215/CJN.12161018
30. Rizzatti G, Lopetuso LR, Gibiino G, Binda C, Gasbarrini A. Proteobacteria: a common factor in human diseases. BioMed Res. Int. (2017) 2017:7. doi: 10.1155/2017/9351507
31. Minasyan H. Sepsis: mechanisms of bacterial injury to the patient. Scand J Trauma Res Emerg Med. (2019) 27:1–22. doi: 10.1186/s13049-019-0596-4
32. Shreiner AB, Kao JY, Young VB. The gut microbiome in health and in disease. Curr Opin Gastroenterol. (2015) 31:69–75. doi: 10.1097/MOG.0000000000000139
33. Dekker Nitert M, Mousa A, Barrett H, Naderpoor N, Courten DEB. Altered gut microbiota composition is associated with back pain in overweight and obese individuals. Front Endocrinol. (2020) 11:605. doi: 10.3389/fendo.2020.00605
34. Gagliardi A, Totino V, Cacciotti F, Iebba V, Neroni B, Bonfiglio G, et al. Rebuilding the gut microbiota ecosystem. Int J Environ Res Public Health. (2018) 15:1679. doi: 10.3390/ijerph15081679
35. Jensen OK, Andersen MH, Østgård RD, Andersen NT, Rolving N. Probiotics for chronic low back pain with type 1 Modic changes: a randomized double-blind, placebo-controlled trial with 1-year follow-up using Lactobacillus Rhamnosis GG. Eur Spine J. (2019) 28:2478–86. doi: 10.1007/s00586-019-06046-6
36. Fong W, Li Q, Yu J. Gut microbiota modulation: a novel strategy for prevention and treatment of colorectal cancer. Oncogene. (2020) 39:4925–43. doi: 10.1038/s41388-020-1341-1
37. Rajasekaran S, Venkatadass K, Naresh Babu J, Ganesh K, Shetty AP. Pharmacological enhancement of disc diffusion and differentiation of healthy, ageing and degenerated discs : results from in-vivo serial post-contrast MRI studies in 365 human lumbar discs. Eur Spine J. (2008) 17:626–43. doi: 10.1007/s00586-008-0645-6
Keywords: intervertebral disc, disc degeneration, bacteria, microbiome, endplate, dysbiosis, next-generation sequencing
Citation: Shanmuganathan R, Tangavel C, K S SVA, Muthurajan R, Nayagam SM, Matchado MS, Rajendran S, Kanna RM and Shetty AP (2022) Comparative metagenomic analysis of human intervertebral disc nucleus pulposus and cartilaginous end plates. Front. Cardiovasc. Med. 9:927652. doi: 10.3389/fcvm.2022.927652
Received: 24 April 2022; Accepted: 01 September 2022;
Published: 28 September 2022.
Edited by:
Velmurugan Ganesan, KMCH Research Foundation, IndiaReviewed by:
Dinakaran Vasudevan, SKAN Medical Research Trust, IndiaChaitanya Dende, University of Texas Southwestern Medical Center, United States
Copyright © 2022 Shanmuganathan, Tangavel, K S, Muthurajan, Nayagam, Matchado, Rajendran, Kanna and Shetty. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rajasekaran Shanmuganathan, cmFqYXNla2FyYW4ub3J0aEBnbWFpbC5jb20=
†ORCID: Rajasekaran Shanmuganathan orcid.org/0000-0001-6043-006X
Chitraa Tangavel orcid.org/0000-0002-5905-9619
Sri Vijay Anand K S orcid.org/0000-0002-8885-5411
Raveendran Muthurajan orcid.org/0000-0002-8803-7662
Sharon Miracle Nayagam orcid.org/0000-0002-0812-4468
Monica Steffi Matchado orcid.org/0000-0002-2058-7924
Sunmathi Rajendran orcid.org/0000-0002-4239-8104
Rishi Mugesh Kanna orcid.org/0000-0001-5817-4909
Ajoy Prasad Shetty orcid.org/0000-0001-5885-7152
 Rajasekaran Shanmuganathan
Rajasekaran Shanmuganathan Chitraa Tangavel
Chitraa Tangavel Sri Vijay Anand K S
Sri Vijay Anand K S Raveendran Muthurajan
Raveendran Muthurajan Sharon Miracle Nayagam
Sharon Miracle Nayagam Monica Steffi Matchado
Monica Steffi Matchado Sunmathi Rajendran
Sunmathi Rajendran Rishi Mugesh Kanna
Rishi Mugesh Kanna Ajoy Prasad Shetty
Ajoy Prasad Shetty