Abstract
Improving the survival rate of cardiomyocytes is the key point to treat most of the heart diseases, and targeting autophagy is a potential advanced therapeutic approach. Monitoring autophagic activity in cardiomyocytes in situ will be useful for studying autophagy-related heart disease and screening autophagy-modulating drugs. Zebrafish, Danio rerio, has been proven as an animal model for studying heart diseases in situ. Taken the advantage of zebrafish, especially the imaging of intact animals, here we generated two stable transgenic zebrafish lines that specifically expressed EGFP-map1lc3b or mRFP-EGFP-map1lc3b in cardiomyocytes under the promoter of myosin light chain 7. We first used a few known autophagy-modulating drugs to confirm their usefulness. By quantifying the density of autophagosomes and autolysosomes, autophagy inducers and inhibitors showed their regulatory functions, which were consistent with previous studies. With the two lines, we then found a significant increase in the density of autophagosomes but not autolysosomes in zebrafish cardiomyocytes at the early developmental stages, indicating the involvement of autophagy in early heart development. To prove their applicability, we also tested five clinical statins by the two lines. And we found that statins did not change the density of autophagosomes but reduced the density of autolysosomes in cardiomyocytes, implying their regulation in autophagic flux. Our study provides novel animal models for monitoring autophagic activity in cardiomyocytes in situ, which could be used to study autophagy-related cardiomyopathy and drug screening.
Introduction
Cardiomyocytes, driving heart contraction, are incapable of cell division in adult mammals (1, 2). Various types of diseases, such as myocardial infarction, hypertension, and hyperlipidemia, result in irreversible loss of cardiomyocytes in acute or chronic ways (3, 4). The subsequent progression stage of cardiomyocyte deficiency is heart failure, which is the leading cause of mortality worldwide and imposes the greatest burden on the healthcare system (5, 6). Due to cardiomyocytes with high demand for energy, many studies reveal that mitochondria and mitochondria-associated ER membranes (MAMs) play important roles in maintaining the metabolism and function of cardiomyocytes, including ATP supply, and the homeostasis of ROS, lipid, and calcium (7–9). And it also has been proven that MAM proteins are involved in the occurrence of heart diseases, thus developing a strategy to eliminate the disordered mitochondria and regulate MAM formation and function might be a promising way to treat those heart diseases (9).
Autophagy, a lysosomal self-degradative process, plays a vital role in maintaining cellular homeostasis (10, 11). It is rapidly activated when cells undergo different stresses, such as starvation, oxidative stress, metabolic stress, and so on (12). In physiological conditions, autophagy maintains cardiac structure and function by degrading misfolded proteins and dysfunctional organelles (13, 14). Autophagy that targets mitochondria, named mitophagy, is critical to eliminate disordered mitochondria and regulate MAMs in cardiomyocytes both at the baseline level and in response to stress (15). In many pathological conditions, autophagy reduces the death of cardiomyocytes by supplying substrates for ATP regeneration in a non-selective or selective bulk degradation pathway (13, 16–18). However, in some other pathological conditions, autophagy in cardiomyocytes is suppressed or activated excessively, and it also contributes to the progression of these heart diseases (19–22). Thus, autophagy plays an essential role in the homeostasis and survival of cardiomyocytes under both basal and stress conditions, which promote the investigators to develop it as a promising therapeutic target (23–25).
Autophagy involves in the formation of double-membrane vesicles called the autophagosome, which sequesters cellular cargo, such as protein aggregates, organelles, ribosomes, and so on. Then, the fusion of the autophagosome with lysosome forms autolysosome, which degrades autophagosomal contents by lysosomal acid proteases (26, 27). The entire process from initiation to the eventual cargo degradation is defined as autophagic flux (28). Microtubule-associated protein 1 light chain 3 (MAP1LC3/LC3) is commonly used as an autophagic marker (29), and its phosphatidylethanolamine (PE) modified counterpart (LC3-PE/LC3-II) specifically localizes to the autophagosomal membranes. LC3-II on the outer autophagosomal membrane is removed in a process called deconjugation, and LC3-II on the inner autophagosomal membrane is degraded by lysosomal enzymes after fusion with lysosome (30). According to such characteristics, LC3-II can be used to indicate the formation and clearance of autophagosomes.
Green fluorescent protein (GFP)-tagged LC3 (GFP-LC3) has been applied to monitor the autophagosomes in living cells and model organisms (31–33). In zebrafish, enhanced GFP (EGFP)-tagged map1lc3b (EGFP-LC3), the zebrafish homolog of human MAP1LC3B, has been reported to study the autophagic function successfully in situ (34–36). These studies also point out that the increase in GFP-positive autophagosomes does not necessarily represent the increase in autophagy flux. Interestingly, the tandem monomeric red fluorescent protein (mRFP)-GFP-LC3 can be used to assess the autophagic flux partially, because GFP, not mRFP, is pH sensitive and the fluorescence intensity of GFP dramatically decreases when the fusion of the autophagosome with lysosome happens (30, 37). However, this method for assessing the autophagic flux has not been tested in zebrafish.
Due to the partial similarities with human heart in development, genetics, and physiology, the zebrafish heart is increasingly used as a model of mammalian cardiac studies (38, 39). Myosin light chain 7 (myl7) is expressed specifically in cardiomyocytes and its promoter is wildly used for driving the strong expression of exogenous genes in cardiomyocytes (40, 41). To study the autophagy in cardiomyocytes, we generated two novel stable transgenic zebrafish lines that expressed EGFP-LC3 and mRFP-EGFP-LC3 under the myl7 promoter. With the two transgenic lines, we first showed that the density of autophagosomes and autolysosomes could be faithfully quantified in zebrafish cardiomyocytes, which was proven by treatment with known autophagy-modulating drugs. Second, compared with 2-dpf (days post fertilization) zebrafish larvae, the density of autophagosomes but not autolysosomes were upregulated at 3 dpf, indicating the roles of autophagy in early heart development. Furthermore, statins, 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase inhibitors, can reduce the synthesis of cholesterol in the liver, and are broadly used in clinical treatment for kinds of heart diseases by reducing the level of LDL cholesterol in plasma (42, 43). However, statins have been reported to be pleiotropic drugs, such as involvement in autophagic regulation in cardiomyocytes (44–46). Here, we tested five statins by the two lines and found that statins did not change the density of autophagosomes, but significantly decreased the density of autolysosomes in cardiomyocytes of zebrafish larvae, suggesting their regulatory roles in autophagic flux. Thus, we described two novel animal models to monitor the autophagic activity in cardiomyocytes in situ that can also be used for screening the potential cardiac therapeutic compounds.
Results
Strategy of monitoring autophagic activity in zebrafish cardiomyocytes in situ
To examine the autophagic activity in zebrafish cardiomyocytes in situ, we first constructed the tol2 transposon-mediated transgenic plasmid, in which the expression of EGFP-LC3 was driven by the promoter of zebrafish myl7 (Figure 1A). Using the plasmid, we generated the stable line Tg(myl7:EGFP-LC3) and found EGFP was only expressed in the heart of zebrafish larvae, which recapitulated the expression of endogenous myl7 transcripts (Figure 1B and Supplementary Figure 1) (40). Using this stable transgenic larvae, we treated them with different drugs (Figure 1C) and then imaged hearts at high resolution via confocal microscopy (Figure 1D). In live animals, the imaging of autophagic puncta in cardiomyocytes was largely affected by rhythmic heart beating. Before imaging, we mounted zebrafish larvae with low-melting agarose (LMA) and then used 4% paraformaldehyde (PFA) to fix the larvae or use 2,3-butanedione monoxime (BDM) to stop the heartbeat. Immediately, we imaged the whole heart of zebrafish larvae using Z-stack imaging methods (Figure 1D). At a single optical imaging slice, we could observe distant EGFP-positive puncta that had been characterized as autophagosomes previously in zebrafish, which was compared with broadly EGFP/DsRed expression pattern in Tg(myl7:EGFP) or Tg(myl7:DsRed) transgenic lines (Figure 1E and Supplementary Figure 2) (34). And we also noticed that heart tissue was also labeled by a weak EGFP signal, which could be used for quantifying the area of heart tissue by ImageJ software (Figure 1E). To assess the autophagic activity in a relatively precise way, we chose those slices with at least one EGFP-positive puncta in multiple larvae for the analysis. And then we calculated the density of autophagosomes by dividing the number of autophagosomes by the area of heart tissue (Figure 1F).
FIGURE 1
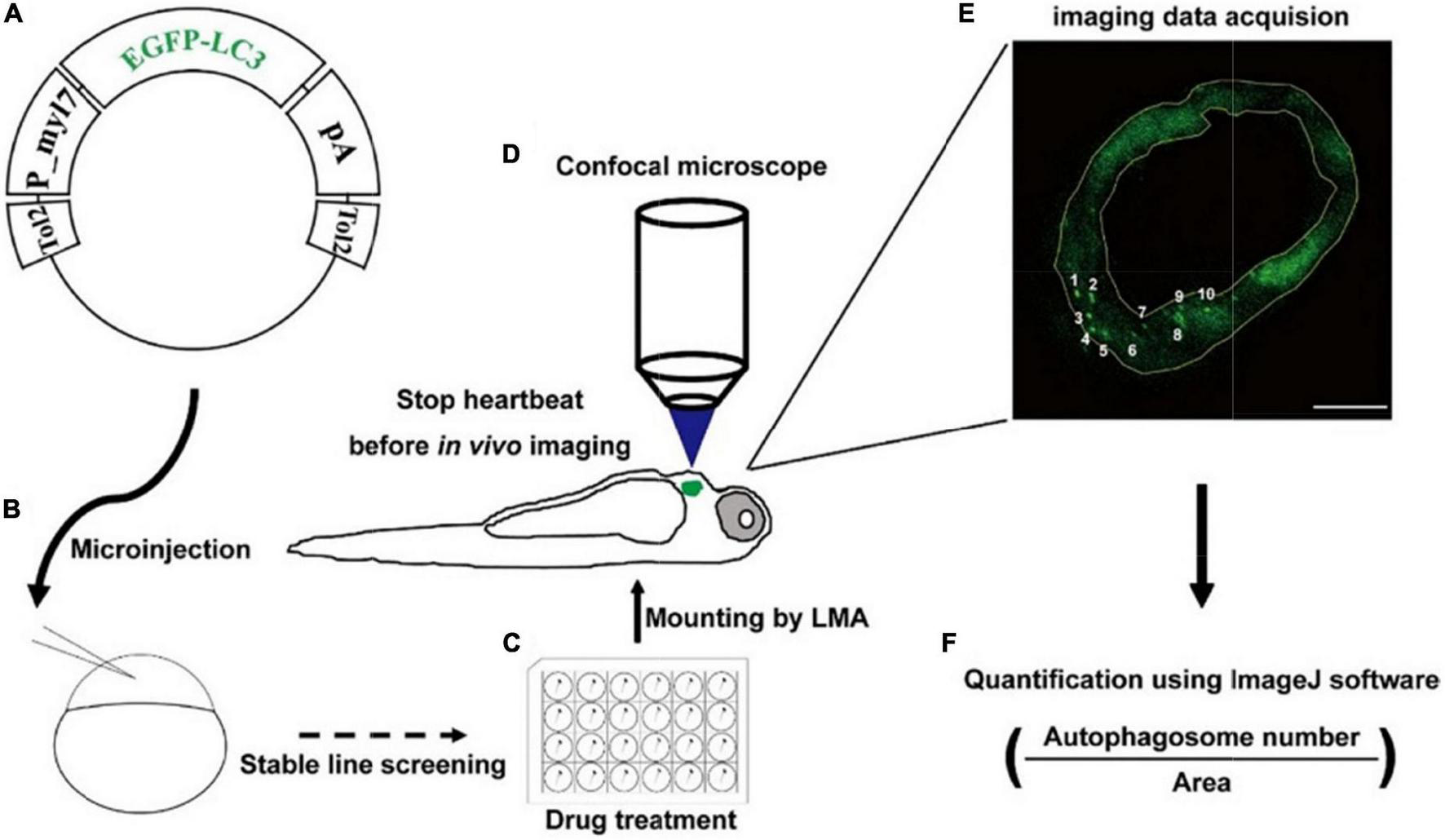
Strategy of monitoring autophagic activity in zebrafish cardiomyocytes in situ. (A) The structure of transgenic plasmid for labeling autophagosomes. (B) Microinjection of the transgenic mixture into one-cell staged embryos for generating stable lines. (C) Drug treatment of zebrafish larvae using cell culture plates. (D) Transgenic larvae are mounted, stopped heartbeat, and then imaged under a confocal microscope. (E) Confocal image shows the EGFP-positive puncta that are characterized as autophagosomes. The area of heart tissue is selected by a yellow polygon using the ImageJ software. Arabic numerals show autophagosomes. Scale bar: 20 μm. (F) The density is calculated by dividing the number of autophagosomes by the area of heart tissue.
Enhanced green fluorescent protein-labeled autophagosomes in zebrafish cardiomyocytes were regulated by autophagy-modulating drugs
To determine the usefulness of the Tg(myl7:EGFP-LC3) line, we first used several small molecules to regulate different stages of autophagic machinery. Using 2-dpf-old transgenic zebrafish larvae, we bathed these molecules, respectively, for 24 h (h). At 3 dpf, EGFP-positive puncta in control zebrafish larvae were mainly located in ventricular cardiomyocytes, and the number of autophagosomes per mm2 in cardiomyocytes was 4076.5 ± 323.1 (average ± s.e.m; Figures 2A,B). We first tested 3-methyladenine (3MA), a selective PI3K inhibitor that had been reported to block autophagosome formation (47). Compared with control group, the z-stack projection images showed that 3MA treatment obviously reduced the number of autophagosomes in heart (Figure 2A). The density of autophagosomes in 3MA-treated larvae was significantly decreased by 82% compared to the control (720.6 ± 44.3 per mm2, p-value = 1.2E-26; Figures 2A,B). Next, we tested other autophagic inhibitor Bafilomycin A1 (BafA), a vacuolar type H + –ATPase (V-ATPase) inhibitor that suppressed autophagosome-lysosome fusion (48), and we found the density of autophagosomes was increased by 73% (7051.8 ± 204.7 per mm2, p-value = 3.2E-10; Figures 2A,B). And we also noticed that BafA treatment resulted in an obviously larger size of autophagosomes than control (increased by 446%; Figure 2C). We then tested the inhibitors of lysosomal proteases, pepstatin A and E64d (P/E), which prevent the degradation of autophagic cargo inside autolysosomes (27). Similar to BafA, P/E treatment also significantly increased the density of autophagosomes by 65% (6735.5 ± 392.3 per mm2, p-value = 8.5E-4; Figures 2A,B). Although 3MA, BafA, and P/E could inhibit the autophagic activity in cardiomyocytes of zebrafish larvae, the characteristics of autophagosomes were shown differently including the density and size.
FIGURE 2
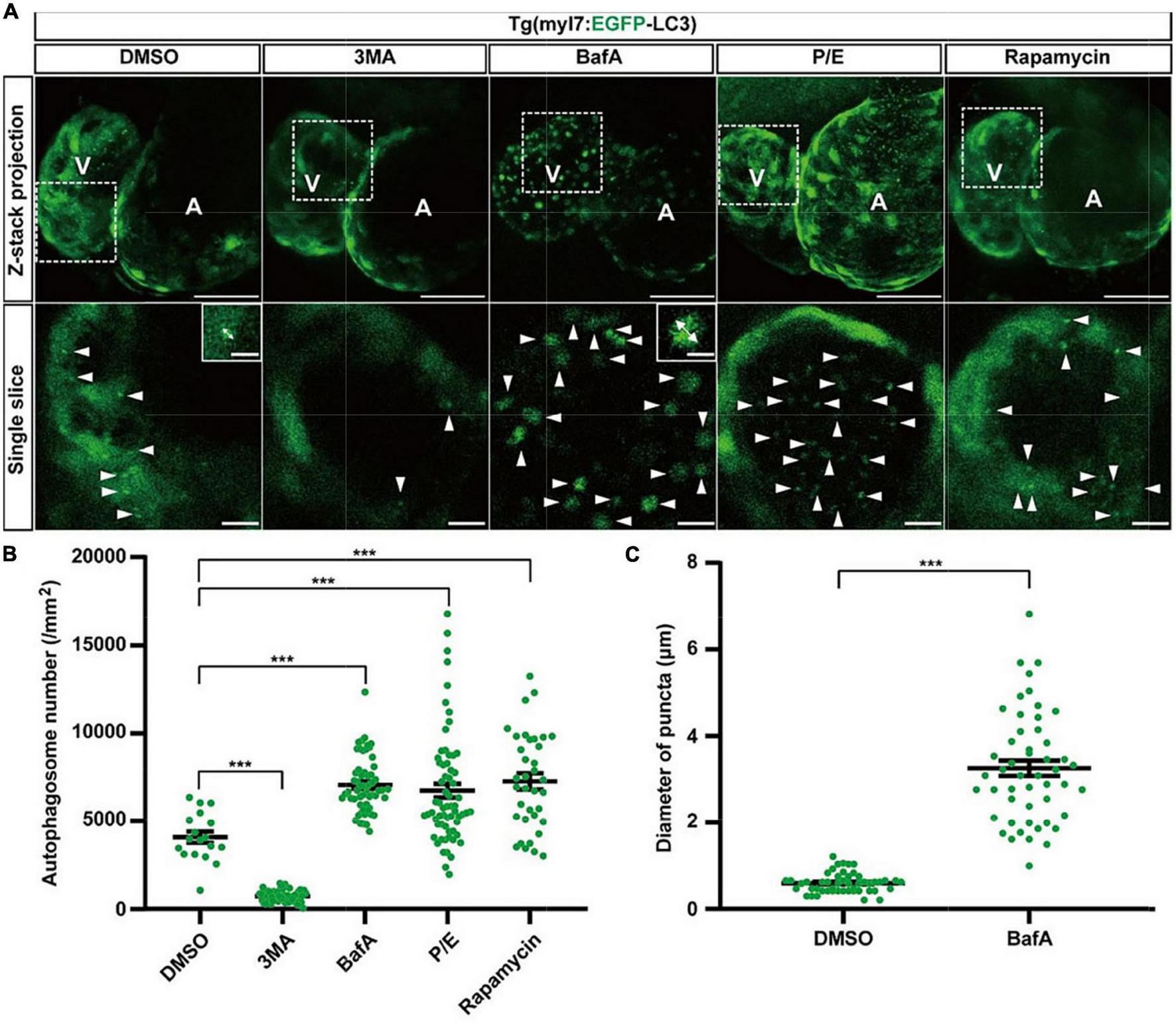
The density of autophagosomes in zebrafish heart is regulated by known autophagy-modulating drugs. (A) Representative images of 3-dpf zebrafish heart of Tg(myl7:EGFP-LC3) line when treating with indicated drugs for 24 h. The top row shows the Z-stack maximum projections (scale bar: 50 μm), the bottom row shows the single slice (scale bar: 10 μm), inset in the bottom row shows the single punctum in indicated treated groups (scale bar: 3 μm). Arrowhead represents the autophagosome, double arrow line represents the diameter of the autophagosome. (B) Quantification of the density of autophagosomes in 3-dpf zebrafish heart of Tg(myl7:EGFP-LC3) line when treating with indicated drugs for 24 h. The data are shown as mean ± s.e.m for each group. n = 18 in control group, n = 55 in 3MA group, n = 58 in BafA group, n = 65 in P/E group, and n = 36 in Rapamycin group. (C) Quantification of the diameter of puncta in 3-dpf zebrafish heart when treated with BafA compared with the control group. n = 50 in the control group, and n = 50 in the BafA group. ***p < 0.001. A, atrium; V, ventricle.
We next test rapamycin, an inhibitor of mTOR, which was reported as an inducer of autophagy (49). Similar to previous studies, rapamycin treatment also increased the density of autophagosomes in cardiomyocytes by 78% compared to the control (7247.4 ± 455.5 per mm2, p-value = 2.4E-05; Figures 2A,B). To confirm such regulation roles of the autophagy-modulating drugs, we also examined the expression levels of LC3 in WT zebrafish larvae by western blot. We found that 3MA treatment decreased the expression of LC3 significantly, while rapamycin treatment increased, which was consistent with the imaging data by the transgenic line (Supplementary Figure 3). These results show autophagosomes are labeled faithfully by EGFP in the Tg(myl7:EGFP-LC3) line, which could be used for assessing the autophagic activity in cardiomyocytes of zebrafish larvae in situ.
The density of autophagosomes changed in cardiomyocytes during zebrafish’s early developmental stages
To understand autophagy in zebrafish’s early heart development, we imaged the heart of Tg(myl7:EGFP-LC3) zebrafish larvae at different developmental stages. EGFP expression driven by myl7 promoter could be detected as early as 16 h post-fertilization (hpf) (40). However, the EGFP-LC3 signal was too weak to detect before 1 dpf in Tg(myl7:EGFP-LC3) zebrafish larvae. At 2 dpf, the EGFP-LC3 signal could be detected quite easier, and we found the density of autophagosomes was 1601.6 ± 95.7 per mm2 (Figure 3). At 3 dpf, the density of autophagosomes was significantly increased by 172% (4143.7 ± 279.6, p-value = 7.5E-14; Figure 3) compared to that at 2 dpf. After 3 dpf, the density of autophagosomes is relatively stable (3849.3 ± 132.7 per mm2 at 4 dpf; 3314.3 ± 220.6 per mm2 at 5 dpf; Figure 3). These data show that autophagic activity changes during zebrafish early heart development, implying the autophagic role in heart development.
FIGURE 3
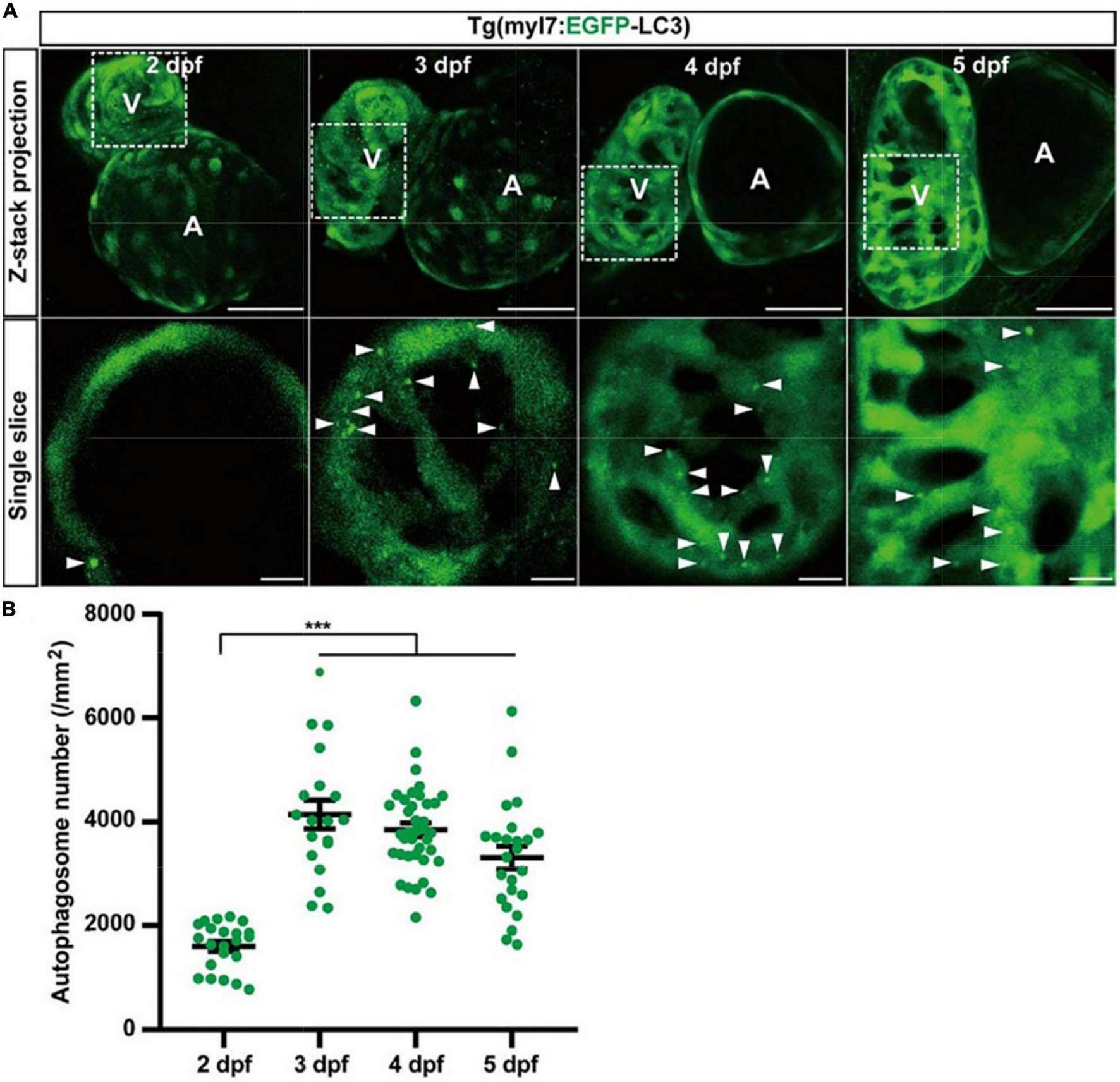
The density of autophagosomes in zebrafish cardiomyocytes is quantified during 2–5 dpf. (A) Representative images of zebrafish heart of Tg(myl7:EGFP-LC3) line at indicated developmental stages. Top row shows the Z-stack maximum projections (scale bar: 50 μm), and the bottom row shows the single slice (scale bar: 10 μm). Arrowhead represents the autophagosome. (B) Quantification of the density of autophagosomes in zebrafish cardiomyocytes of indicated developmental stages. The data are shown as mean ± s.e.m for each group. n = 22 at 2 dpf, n = 19 at 3 dpf, n = 38 at 4 dpf, and n = 24 at 5 dpf. ***p < 0.001. A, atrium; V, ventricle.
Autophagic flux in zebrafish cardiomyocytes was regulated by autophagy-modulating drugs
To assess the autophagic flux of cardiomyocytes in intact animals in situ instead of lysosome staining by LysoTraker Red in fixed samples (36), we generated another transgenic line Tg(myl7:mRFP-EGFP-LC3), in which EGFP and mRFP were specifically expressed in the heart of zebrafish larvae (Supplementary Figure 1). Similar to Tg(myl7:EGFP-LC3), we used autophagy-modulating drugs to demonstrate their applicability. In the control group, we quantified the puncta with both EGFP and mRFP positive signals representing autophagosomes and found the density of colocalized puncta was 2440.0 ± 376.1 per mm2 (Figures 4A,B). The density of puncta with only mRFP positive signal representing autolysosomes was 12344.8 ± 1281.1 per mm2 (Figure 4C). Compared with the control, 3MA treatment decreased the density of autophagosomes by 76% (577.8 ± 48.2 per mm2, p-value = 2.2E-8; Figure 4B), and it also decreased the density of autolysosomes by 39% (7571.5 ± 402.1 per mm2, p-value = 3.3E-5; Figure 4C). BafA treatment increased the density of autophagosomes by 229% (8022.4 ± 504.4 per mm2, p-value = 4.8E-9; Figure 4B), but decreased the density of autolysosomes by 81% (2330.2 ± 278.9 per mm2, p-value = 2.4E-12; Figure 4C). Different from the BafA treatment, the P/E treatment increased the density of autophagosomes by 87% (4555.6 ± 587.2 per mm2, p-value = 4.3E-3; Figure 4B), and the density of autolysosomes was also increased by 37% (16904.4 ± 2114.5 per mm2, p-value = 4.9E-2; Figure 4C). Similar to Tg(myl7:EGFP-LC3) line, the BafA1 treatment also resulted in a large size of the puncta than the control (Supplementary Figure 4). Using rapamycin to induce the autophagy increased the density of autophagosomes by 186% (6986.7 ± 837.8 per mm2, p-value = 9.9E-6; Figure 4B) and autolysosomes by 32% (16347.9 ± 1130.2 per mm2, p-value = 2.3E-2; Figure 4C). According to the function of BafA and P/E, which were reported previously (27, 48), our data indicate that BafA inhibits the autophagic flux by blocking the fusion of autophagosomes and lysosomes, and P/E inhibits the autophagic flux by preventing the degradation of autophagic cargo inside autolysosomes. These data showed this transgenic line could be applied for assessing the autophagic flux by quantification the density of autophagosomes and autolysosomes simultaneously.
FIGURE 4
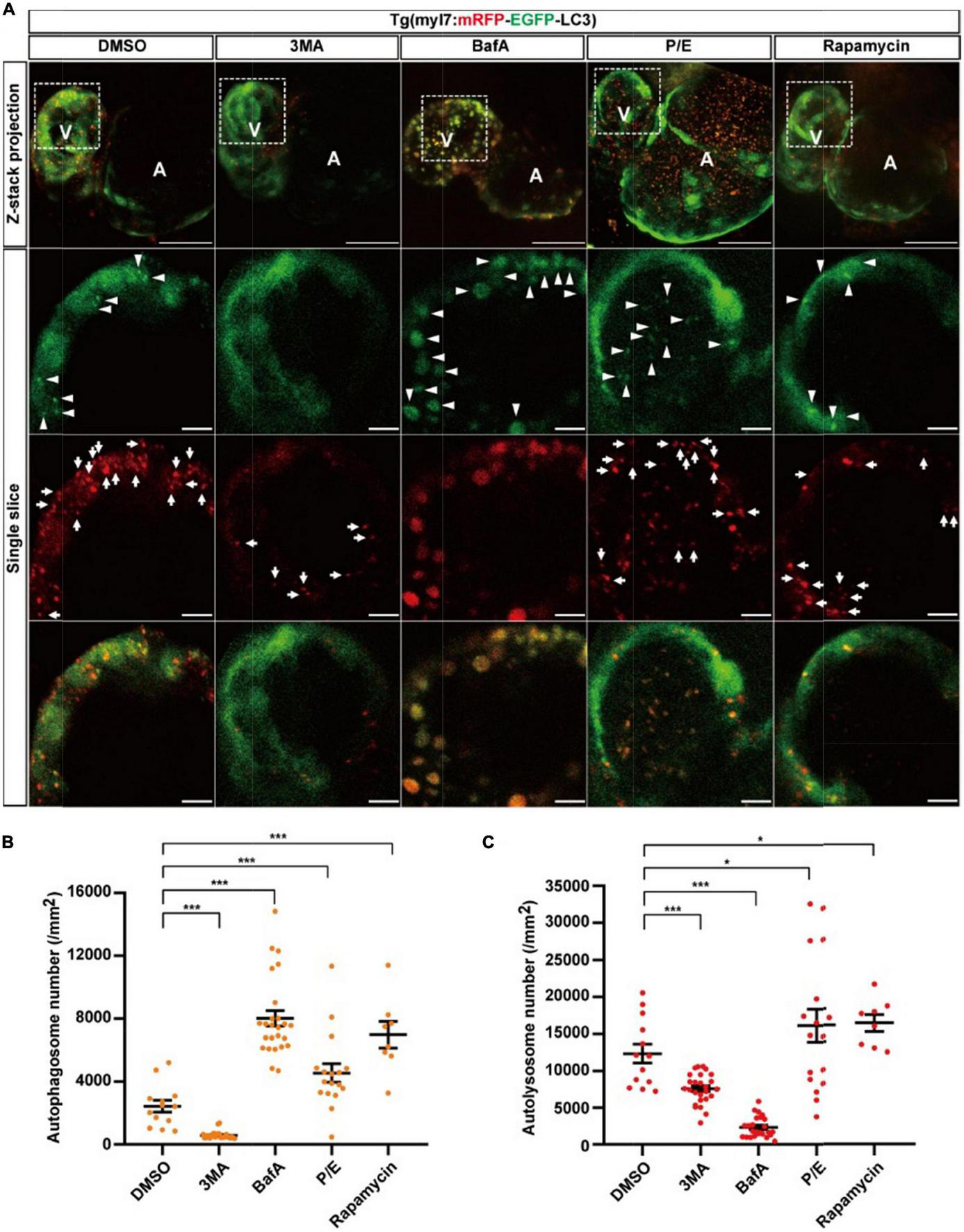
The densities of autophagosomes and autolysosomes in zebrafish cardiomyocytes are regulated by known autophagy-modulating drugs. (A) Representative images of 3-dpf zebrafish heart of Tg(myl7:mRFP-EGFP-LC3) line when treated with indicated drugs for 24 h. The top row shows the Z-stack maximum projections with merged EGFP and mRFP signals (scale bar: 50 μm), the second row shows the single slice with EGFP signal, the third row shows the single slice with mRFP signal, and the bottom row shows the single slice with merged EGFP and mRFP signals (scale bar: 10 μm). Arrowhead represents autophagosome, the arrow represents autolysosome. (B) Quantification of the density of autophagosomes in 3-dpf zebrafish heart of Tg(myl7:mRFP-EGFP-LC3) line when treated with indicated drugs for 24 h. (C) Quantification of the density of autolysosomes in 3-dpf zebrafish heart of Tg(myl7:mRFP-EGFP-LC3) line when treated with indicated drugs for 24 h. The data are shown as mean ± s.e.m for each group. n = 13 in control group, n = 26 in 3MA group, n = 25 in BafA group, n = 17 in P/E group, and n = 8 in Rapamycin group. *p < 0.05, ***p < 0.001. A, atrium; V, ventricle.
The density of autolysosomes did not change in cardiomyocytes during zebrafish’s early developmental stages
We next used Tg(myl7:mRFP-EGFP-LC3) to quantify autophagic flux at zebrafish developmental stages. At 2 dpf, the density of autophagosomes is 1351.9 ± 101.0 per mm2), and the density of autolysosomes was 14586.9 ± 999.0 per mm2 (Figure 5). Similar to the Tg(myl7:EGFP-LC3) line, the density of autophagosomes was also increased at 3 dpf (2541.2 ± 495.9 per mm2, p = 1.9E-3; Figure 5B) compared with that at 2 dpf. However, the density of autolysosomes did not change significantly at 3 dpf (14328.1 ± 1297.2 per mm2; Figure 5C). After 3 dpf, the density of autophagosomes and autolysosomes were both relatively stable (autophagosome: 1646.8 ± 103.6 per mm2 at 4 dpf; 1779.7 ± 184.0 per mm2 at 5 dpf; autolysosome: 12504.7 ± 777.2 per mm2 at 4 dpf; 13928.1 ± 831.8 per mm2 at 5 dpf; Figures 5B,C). Combing the data obtained using Tg(myl7:EGFP-LC3) line, these data reveal that autophagosome but not autophagic flux is increased in zebrafish cardiomyocytes at the early developmental stages.
FIGURE 5
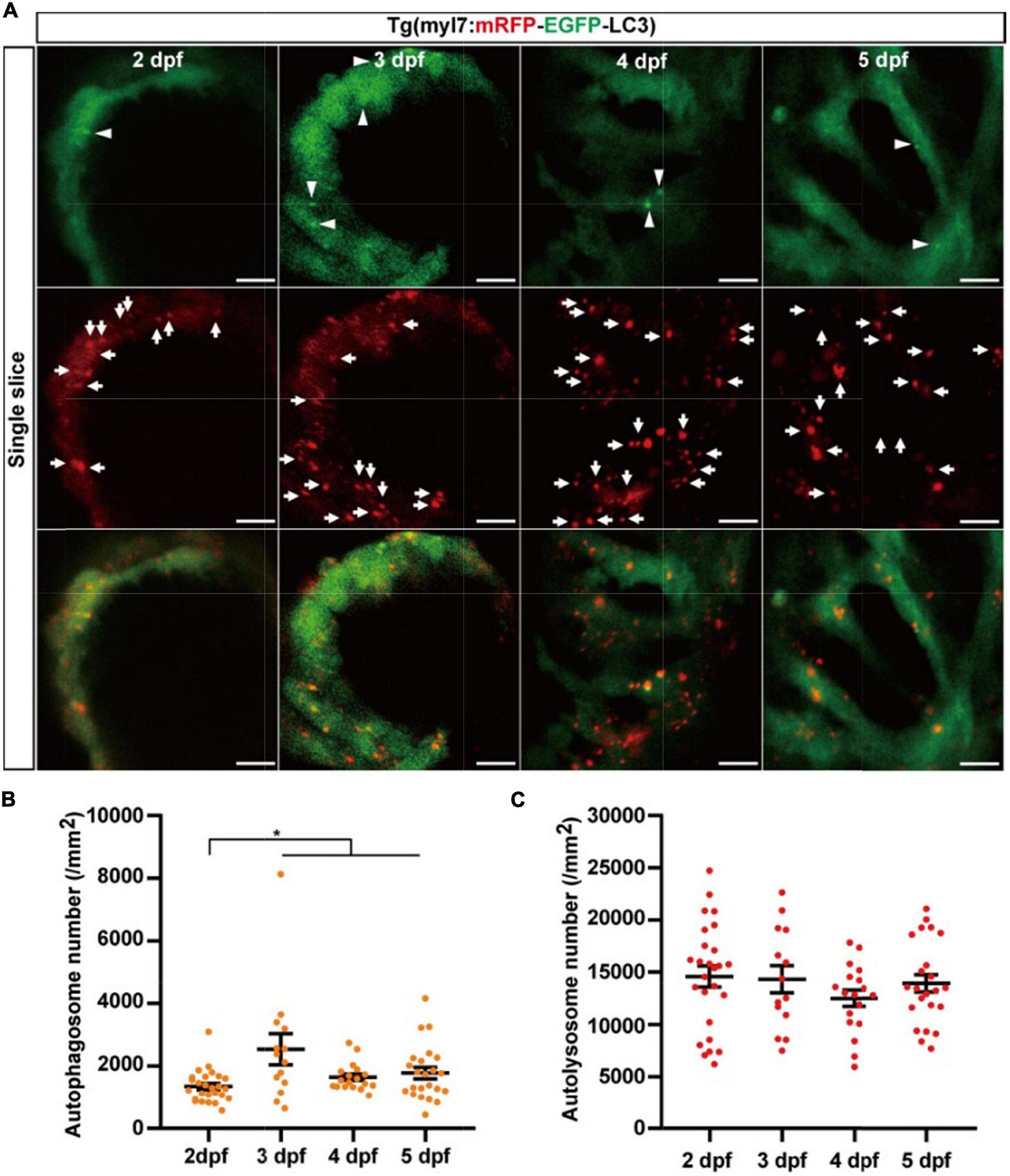
The densities of autophagosomes and autolysosomes in zebrafish hearts are quantified simultaneously during 2–5 dpf. (A) Representative images of 3-dpf zebrafish heart of Tg(myl7:mRFP-EGFP-LC3) line at indicated developmental stages. The top row shows the single slice with an EGFP signal, the second row shows the single slice with an mRFP signal, and the bottom row shows the single slice with merged EGFP and mRFP signals (scale bar: 10 μm). Arrowhead represents autophagosomes, the arrow represents autolysosomes. (B) Quantification of the density of autophagosomes in zebrafish heart of Tg(myl7:mRFP-EGFP-LC3) line at indicated developmental stages. (C) Quantification of the density of autolysosomes in zebrafish heart of Tg(myl7:mRFP-EGFP-LC3) line at indicated developmental stages. The data are shown as mean ± s.e.m for each group. n = 26 at 2 dpf, n = 14 at 3 dpf, n = 18 at 4 dpf, and n = 23 at 5 dpf. *p < 0.05. A, atrium; V, ventricle.
Statins regulate autophagic flux in zebrafish cardiomyocytes
Statins are broadly applied in clinical treatment for different heart diseases (43, 45). To test whether the two lines can be used for studying the autophagy-related drugs in cardiomyocytes, we tested five statins, including atorvastatin, fluvastatin, pitavastatin, pravastatin, and rosuvastatin. We first tested five statins by Tg(myl7:EGFP-LC3) zebrafish line and found that these five statins did not change the density of autophagosomes (4191.1 ± 299.0 per mm2 in the control group, 4256.8 ± 358.4 per mm2 in the atorvastatin group, 3964.3 ± 238.7 per mm2 in the fluvastatin group, 5050.2 ± 436.6 per mm2 in pitavastatin, 4823.1 ± 180.0 per mm2 in the pravastatin group, and 4822.9 ± 218.9 per mm2 in the rosuvastatin group; Figure 6A). Next, we test them by Tg(myl7:mRFP-EGFP-LC3) zebrafish line. Similar to Tg(myl7:EGFP-LC3) line, we also found statins did not change the density of autophagosomes (2470.9 ± 269.1 per mm2 in the control group, 2102.3 ± 124.7 per mm2 in the atorvastatin group, 1667.8 ± 373.7 per mm2 in the fluvastatin group, 1820.2 ± 148.2 per mm2 in pitavastatin, 1941.2 ± 258.5 per mm2 in the pravastatin group, and 2090.3 ± 345.0 per mm2 in the rosuvastatin group; Figure 6B). Interestingly, the density of autolysosomes in cardiomyocytes was decreased when treated with statins (12812.1 ± 1057.7 per mm2 in the control group; 7886.0 ± 761.3 per mm2 in the atorvastatin group, p = 5.3E-4; 6308.1 ± 308.2 per mm2 in the fluvastatin group, p = 4.6E-3; 8355.8 ± 855.6 per mm2 in pitavastatin, p = 4.2E-3; 7588.1 ± 582.1 per mm2 in the pravastatin group, p = 3.7E-5; and 10285.2 ± 542.2 per mm2 in the rosuvastatin group, p = 4.4E-2; Figure 6C). These results suggest that statins might play roles in the regulation of autophagic flux in cardiomyocytes.
FIGURE 6
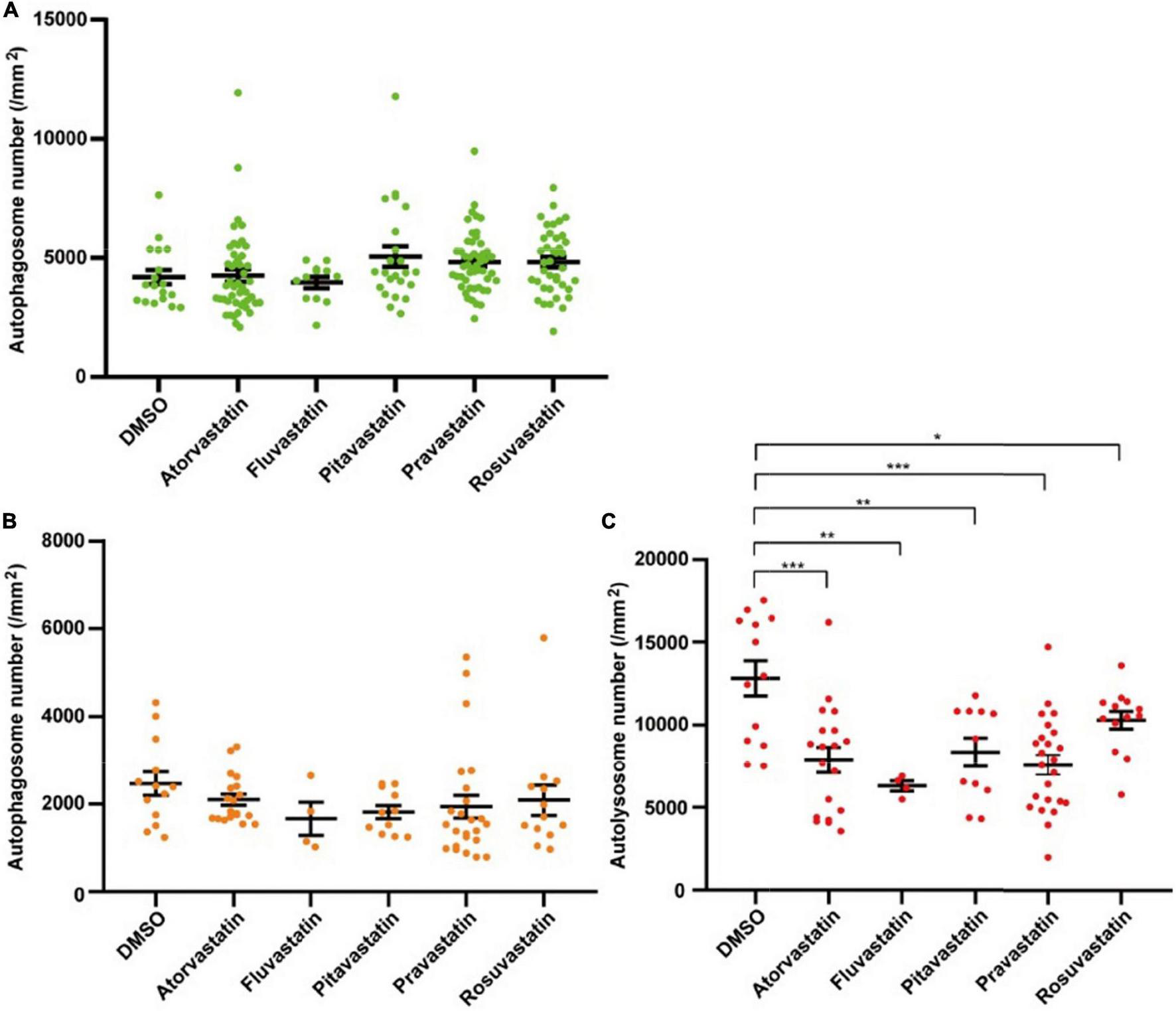
The densities of autophagosomes and autolysosomes in zebrafish cardiomyocytes are quantified when treated with statins. (A) Quantification of the density of autophagosomes in 3-dpf zebrafish heart of Tg(myl7:EGFP-LC3) line when treated with indicated statins. (B) Quantification of the densities of autophagosomes in 3-dpf zebrafish heart of Tg(myl7:mRFP-EGFP-LC3) line when treated with indicated statins. (C) Quantification of the densities of autolysosomes in 3-dpf zebrafish heart of Tg(myl7:mRFP-EGFP-LC3) line when treated with indicated statins. The data are shown as mean ± s.e.m for each group. n = 13 in the control group, n = 19 in the atorvastatin group, n = 4 in the fluvastatin group, n = 11 in pitavastatin, n = 24 in the pravastatin group, and n = 13 in the rosuvastatin group. *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
Many studies demonstrate that autophagy is involved in different heart diseases, and targeting autophagic processes has emerged as an effective therapeutic strategy (23–25). Due to the partial similarity especially in the conserved molecular and cellular pathways, the zebrafish heart is increasingly used as a model for studying the function and disease of human hearts. Besides, zebrafish have many advantages among vertebrate models, such as transparent larvae for in situ observation, ease of genetic manipulation, and large amount of embryos with low cost for drug screening. Furthermore, it has also been demonstrated that autophagy in cardiomyocytes plays essential role in heart development and regeneration in zebrafish models (50, 51). For these reasons, the developing zebrafish transgenic lines in this study will offer researchers novel models for easily assessing autophagic activities in cardiomyocytes in situ.
It has been proved that EGFP-LC3 can be used for monitoring the autolysosomes when it is overexpressed in zebrafish ubiquitously, or specifically in hepatocytes or neurons (35, 36). For studying cardiomyocyte-specific autophagy, we generated the first transgenic zebrafish model that EGFP-LC3 specifically overexpressed in cardiomyocytes. To maximize the benefits of in situ autophagy-related research, we also generated another zebrafish model that mRFP-EGFP-LC3 specifically overexpressed in cardiomyocytes. After testing the known autophagy-modulating drugs, we validated the usefulness of the two lines for labeling the autophagosomes and autolysosomes. In the two lines, we found the change in autophagic activity in cardiomyocytes happened at the developmental stages, which was consistent with the notion that autophagy played a critical role in heart development (50). Especially, autophagic activity in zebrafish cardiomyocytes showed a significant change within 2 dpf and 3 dpf (Figures 3, 5). During this stage, it is reported that internal cardiac structure changes, such as the zebrafish heart valve and conduction system (38, 52, 53). During 55–72 hpf, the retrograde blood flow has ceased gradually, and pacemaker activity becomes restricted to a small area on the dorsal right quadrant of the sinoatrial ring (52, 53), which implies autophagic activity in cardiomyocytes might be regulated by hemodynamics and/or cardiac conduction system. And we also found the number of EGFP labeled autophagosomes in Tg(myl7:EGFP-LC3) line was much higher than that in Tg(myl7:mRFP-EGFP-LC3) line, but the changes in autophagic activities showed more sensitive in Tg(myl7:mRFP-EGFP-LC3) than that in Tg(myl7:EGFP-LC3) line when treating with autophagy-modulating drugs. Recently, Mareninova et al. reported that GFP-LC3 overexpression could affect autophagy itself (54). Although we did not observe any abnormal phenotype of the heart in at least five generations of the two transgenic zebrafish, it still reminds us to develop better in situ autophagic models in the future.
The rate of autophagic degradation is defined as autophagic flux, which includes the entire process of autophagy. Usually, assessing autophagic flux requires the combination of multiple techniques, but they cannot be achieved in intact animals in situ (27, 55). By examining the changes of both autophagosomes and autolysosomes simultaneously, we could partially assess the autophagic flux. After testing different autophagy-modulating drugs by Tg(myl7:mRFP-EGFP-LC3) line, we found that 3MA treatment reduced the density of both autophagosomes and autolysosomes, which was consistent with its inhibitory role of autophagosome formation. BafA treatment resulted in the increase of autophagosomes but a decrease in autolysosomes, which was also consistent with its inhibitory role of autophagosome-lysosome fusion. However, the increase of both autophagosomes and autolysosomes has resulted from increased autophagic activity or decreased degradation in autolysosomes. In the present study, treating rapamycin showed a similar increase to that of P/E. To distinguish such situations, different known autophagy inhibitors can be used in combination (27, 36). Besides, the rate of autophagosome/autolysosome accumulation also can be used for the evaluation (36). Due to the autophagic interference when blocking heartbeat (13), the methods for quantifying the rate of autophagosome/autolysosome accumulation in cardiomyocytes still need to be developed. In addition, we noticed that the size of autophagosomes is enlarged when treated with BafA, which could be a novel parameter for the assessment of autophagic flux.
One of the advantages of the zebrafish models is the application of in situ drug development. Statins can effectively lower the cholesterol level in the blood by inhibiting cholesterol synthesis, and then achieve therapeutic effects for heart diseases (45). They also have been reported to be involved in autophagic regulation, but their mechanism in autophagic regulation is still unclear (46). In the present study, we tested statins initially by the generated transgenic lines at the highest concentration with no side effects in zebrafish larvae. Although it might not be the optimal concentration to regulate the autophagy in the cardiomyocytes of zebrafish larvae, we still found their regulatory roles in our transgenic lines. Different from the known autophagy-modulating drugs, statins only significantly decreased the density of autolysosomes but not autophagosomes. Based on the data, we speculate that statins might promote autophagic flux in cardiomyocytes by regulating the degradation process in autolysosomes, suggesting the feasibility of autophagic drug development in cardiomyocytes by zebrafish transgenic models.
In this study, we developed two transgenic lines that could be used to determine the density of autophagosomes and autolysosomes in zebrafish cardiomyocytes. With these lines, we found the changes in autophagic activity that happened in cardiomyocytes at early zebrafish developmental stages. In the applicability test, we found that statins could play roles in the regulation of autophagic flux. Taken together, we believe that these zebrafish lines would be useful for autophagy-related studies for cardiomyopathy. With the help of our in situ vertebrate models, we hope it would alleviate the stress of the healthcare system by accelerating the development of autophagy-related drugs for cardiomyopathy.
Materials and methods
Zebrafish care and maintenance
Zebrafish were maintained at 28.5°C on a 14 h/10 h light/dark cycle with automatic fish housing system (Haishen, China). Embryos and larvae were raised in 10% Hank’s solution that consisted of (in mM) 140 NaCl, 5.4 KCl, 0.25 Na2HPO4, 0.44 KH2PO4, 1.3 CaCl2, 1.0 MgSO4, and 4.2 NaHCO3 (pH 7.2). To prevent pigment formation for imaging experiments, the embryos were treated with 0.003% 1-phenyl-2-thiourea (PTU, Sigma) from 24 h post-fertilization. AB wild-type was used for generating novel transgenic lines. Tg(myl7:EGFP) and Tg(myl7:DsRed) transgenic lines were used as the negative line for identifying the autophagic puncta. Zebrafish handling procedures were approved by the Animal Research Committee of Zhongshan Hospital, Fudan University.
Transgenic plasmid construction
The transgenic plasmids were constructed by Hieff Clone® Plus Multi One Step Cloning Kit (YEASEN, China). The 269-bp promoter of myl7 was amplified from zebrafish genomic DNA. The coding sequence of zebrafish map1lc3b was amplified from the cDNA of 3-dpf zebrafish larvae. EGFP or mRFP sequences were purchased from the Miaolingbio Company (China). Different fragments were ligated in the tol2mini transgenic vector (56).
Generation of Tg(myl7:EGFP-LC3) and Tg(myl7:mRFP-EGFP-LC3) transgenic zebrafish
One-cell staged AB zebrafish embryos were collected by natural mating and were injected with 25 pg of myl7-EGFP-LC3 along with 25 pg tol2 transposase mRNA. The founders of Tg(myl7:EGFP-LC3) were screened by EGFP expression of their F1 progenies, and F1 larvae were then confirmed by genotyping. Similarly, stable lines of Tg(myl7:mRFP-EGFP-LC3) were screened out. In total, we identified six different alleles of the two transgenic lines. The alleles with strong expression were selected by MVX10 stereoscopic fluorescence microscope (Olympus, Japan) for further studies. All stable lines have been outcrossed for at least five generations with no abnormalities at developmental stages or adulthood (data not shown).
Drug treatment of zebrafish larvae
Autophagy-modulating drugs and statins were all purchased from Selleck Company (China). Dechorionated 2-dpf transgenic embryos were incubated with different drugs for 24 h. We tested different concentrations of each drug, and the groups with all larvae developed normally were considered as having no side effects. And we chose the highest concentration group with no side effects of each drug for further autophagy imaging experiments (Supplementary Table 1). The larvae treated with 10 mM 3-methyladenine (3MA, S2767), 200 nM bafilomycin A1 (BafA, S1413), P/E: 10 μg/mL pepstatin A (S7381) and 5 μg/mL E64d (Aloxistatin, S7393), 1 μM rapamycin (S1039), 10 μg/mL Atorvastatin (S5715), 8 μg/mL Fluvastatin (S1909), 4 μg/mL Pitavastatin (S1759), 9 μg/mL Pravastatin (S5713), or 10 μg/mL Rosuvastatin (S2169), respectively, were used for the autophagy imaging experiments. All drugs were diluted in 10% Hank’s solution. The final concentration of DMSO in all groups including the control group was 0.1% (v/v). Six-well plates were used for the drug treatment. Not more than 10 embryos were treated in each well containing 4 mL of treatment solution.
Confocal imaging and quantification of puncta
For in situ confocal imaging, larvae were immobilized in 1.2% low melting-point agarose into 35-mm glass-bottom culture dishes without anesthetics. The heartbeat of mounted larvae was stopped by adding 4% paraformaldehyde (PFA) or 20 mM 2,3-butanedione 2-monoxime (BDM) in culture dishes. An imaging experiment of each larva would be completed within 10 min after the heartbeat stopped. Z-stack imaging of the entire heart of transgenic larvae with the ventral view was acquired using a 488/594 nm excitation lasers of Fluoview 3000 confocal microscope (Olympus, Japan). Imaging experiments were performed under a 20X dry objective with zoom in x3.0. The spatial resolution of all images was 1,024 × 1,024 pixels with a z-step size of 4 μm.
Fluorescent points (EGFP + or mRFP +) which were distinguishable from the background were considered puncta. The area of heart tissue was quantified based on the weak EGFP signals by the manual polygon selection using ImageJ software. Numbers of puncta and areas of heart tissue were quantified from each z-stack slice. The slices without EGFP + puncta were excluded from the quantification. The data point from each zebrafish larva varied from 0 to 5. EGFP + or EGFP + /mRFP + points were considered autophagosomes, and EGFP-/mRFP + points were considered autolysosomes.
Western blot
Wild-type zebrafish larvae treated with autophagic regulation drugs (3MA and rapamycin) were used for total protein extraction. About 40 3-dpf wild-type zebrafish larvae were lysed by Radio Immunoprecipitation Assay Lysis buffer (RIPA, Beyotime, China) for total protein extraction. The protein samples were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and electro-transferred on polyvinylidene fluoride membranes (Millipore, Schwalbach, Germany), then were blocked with 5% skim milk. Subsequently, the membranes were incubated with primary antibody (BOSTER, China) for LC3B/MAP1LC3B (BM4827, 1:500) or GAPDH (BM1623, 1:10,000) at 4°C overnight. Then the membranes were put into the secondary antibody (BOSTER, China) HRP Conjugated AffiniPure Goat Anti-Mouse IgG (H + L) (BA1050, 1:5000) or Anti-Rabbit (BA1054, 1:5,000) for 1 h at room temperature. After washing, the proteins were visualized with a BeyoECL plus kit (Beyotime, China).
Statistical analysis
Statistical analysis was performed using Microsoft Excel or GraphPad Prism version 8.0.1. Student’s t-tests were used to compare two groups. One-way ANOVA analyses were used for comparisons among multiple groups followed by Tukey’s multiple comparisons tests. All results are represented as mean ± SEM. GraphPad Prism software was used to generate the statistical graphs.
Statements
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
Ethics statement
This animal study was reviewed and approved by the Animal Ethics Committee, Zhongshan Hospital, Fudan University.
Author contributions
YD and HL: conceptualization, methodology, supervision, project administration, and funding acquisition. JZ and ZZ: formal analysis and investigation. JZ, ZZ, and JH: data curation. JZ, YW, and JH: writing—original draft preparation. LX and KJ: writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Animal Model Project of Shanghai Scientific Committee (grant no. 19140900900), the Shanghai Natural Science Foundation (grant no. 20ZR1439700), the Exploratory Clinical Research Projects of National Clinical Research Center for Interventional Medicine (grant no. 2021-002), the Shanghai Clinical Research Center for Interventional Medicine (grant no. 19MC1910300), National Key R&D Program of China (grant no. 2021YFC2500500), and Program of Shanghai Academic Research Leader (grant no. 22XD1423300).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2022.921829/full#supplementary-material
References
1.
Ali H Braga L Giacca M. Cardiac regeneration and remodelling of the cardiomyocyte cytoarchitecture. Febs J. (2020) 287:417–38. 10.1111/febs.15146
2.
Ponnusamy M Li PF Wang K. Understanding cardiomyocyte proliferation: an insight into cell cycle activity. Cell Mol Life Sci. (2017) 74:1019–34. 10.1007/s00018-016-2375-y
3.
Whelan RS Kaplinskiy V Kitsis RN. Cell death in the pathogenesis of heart disease: mechanisms and significance. Annu Rev Physiol. (2010) 72:19–44. 10.1146/annurev.physiol.010908.163111
4.
Del Re DP Amgalan D Linkermann A Liu QH Kitsis RN. Fundamental mechanisms of regulated cell death and implications for heart disease. Physiol Rev. (2019) 99:1763–817. 10.1152/physrev.00022.2018
5.
Virani SS Alonso A Benjamin EJ Bittencourt MS Callaway CW Carson AP et al Heart disease and stroke statistics-2020 update: a report from the American heart association. Circulation. (2020) 141:E139–596. 10.1161/CIR.0000000000000746
6.
Timmis A Townsend N Gale CP Torbica A Lettino M Petersen SE et al European society of cardiology: cardiovascular disease statistics 2019. Eur Heart J. (2020) 41:12–85.
7.
Zhang K Zhou Q Guo Y Chen L Li L. Mitochondria-associated endoplasmic reticulum membranes (MAMs) involve in the regulation of mitochondrial dysfunction and heart failure. Acta Biochim Biophys Sin. (2018) 50:618–9. 10.1093/abbs/gmy044
8.
Silva-Palacios A Zazueta C Pedraza-Chaverri J. ER membranes associated with mitochondria: possible therapeutic targets in heart-associated diseases. Pharmacol Res. (2020) 156:104758. 10.1016/j.phrs.2020.104758
9.
Gao P Yan Z Zhu Z. Mitochondria-associated endoplasmic reticulum membranes in cardiovascular diseases. Front Cell Dev Biol. (2020) 8:604240. 10.3389/fcell.2020.604240
10.
Wirawan E Berghe TV Lippens S Agostinis P Vandenabeele P. Autophagy: for better or for worse. Cell Res. (2012) 22:43–61. 10.1038/cr.2011.152
11.
Levine B Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. (2019) 176:11–42. 10.1016/j.cell.2018.09.048
12.
Parzych KR Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antiox Redox Signal. (2014) 20:460–73. 10.1089/ars.2013.5371
13.
Nakai A Yamaguchi O Takeda T Higuchi Y Hikoso S Taniike M et al The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. (2007) 13:619–24. 10.1038/nm1574
14.
Sciarretta S Maejima Y Zablocki D Sadoshima J. The role of autophagy in the heart. Ann Rev Physiol. (2018) 80:1–26. 10.1146/annurev-physiol-021317-121427
15.
Shirakabe A Ikeda Y Sciarretta S Zablocki DK Sadoshima J. Aging and autophagy in the heart. Circ Res. (2016) 118:1563–76. 10.1161/CIRCRESAHA.116.307474
16.
Sciarretta S Zhai PY Shao D Maejima Y Robbins J Volpe M et al Rheb is a critical regulator of autophagy during myocardial ischemia pathophysiological implications in obesity and metabolic syndrome. Circulation. (2012) 125:1134–46. 10.1161/CIRCULATIONAHA.111.078212
17.
Hariharan N Maejima Y Nakae J Paik J DePinho RA Sadoshima J. Deacetylation of foxo by sirt1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Circ Res. (2010) 107:1470–82. 10.1161/CIRCRESAHA.110.227371
18.
Ikeda Y Shirakabe A Maejima Y Zhai P Sciarretta S Toli J et al Endogenous Drp1 mediates mitochondrial autophagy and protects the heart against energy stress. Circ Res. (2015) 116:264–78. 10.1161/CIRCRESAHA.116.303356
19.
Matsui Y Takagi H Qu X Abdellatif M Sakoda H Asano T et al Distinct roles of autophagy in the heart during ischemia and reperfusion – Roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. (2007) 100:914–22. 10.1161/01.RES.0000261924.76669.36
20.
Pattison JS Osinska H Robbins J. Atg7 induces basal autophagy and rescues autophagic deficiency in CryAB(R120G) cardiomyocytes. Circ Res. (2011) 109:151–60. 10.1161/CIRCRESAHA.110.237339
21.
Li DL Wang ZV Ding G Tan W Luo X Criollo A et al Doxorubicin blocks cardiomyocyte autophagic flux by inhibiting lysosome acidification. Circulation. (2016) 133:1668–87. 10.1161/CIRCULATIONAHA.115.017443
22.
Zhu HX Tannous P Johnstone JL Kong YL Shelton JM Richardson JA et al Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest. (2007) 117:1782–93. 10.1172/JCI27523
23.
Nishida K Taneike M Otsu K. The role of autophagic degradation in the heart. J Mol Cell Cardiol. (2015) 78:73–9. 10.1016/j.yjmcc.2014.09.029
24.
Wu X Liu Z Yu X-Y Xu S Luo J. Autophagy and cardiac diseases: therapeutic potential of natural products. Med Res Rev. (2021) 41:314–41. 10.1002/med.21733
25.
Kaludercic N Maiuri MC Kaushik S Fernandez AF de Bruijn J Castoldi F et al Comprehensive autophagy evaluation in cardiac disease models. Cardiovascul Res. (2020) 116:483–504. 10.1093/cvr/cvz233
26.
Mizushima N Komatsu M. Autophagy: renovation of cells and tissues. Cell. (2011) 147:728–41. 10.1016/j.cell.2011.10.026
27.
Klionsky DJ Abdel-Aziz AK Abdelfatah S Abdellatif M Abdoli A Abel S et al Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy. (2021) 17:1–382.
28.
Zhao YG Codogno P Zhang H. Machinery, regulation and pathophysiological implications of autophagosome maturation. Nat Rev Mol Cell Biol. (2021) 22:733–50. 10.1038/s41580-021-00392-4
29.
Mizushima N Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. (2007) 3:542–5. 10.4161/auto.4600
30.
Galluzzi L Green DR. Autophagy-independent functions of the autophagy machinery. Cell. (2019) 177:1682–99. 10.1016/j.cell.2019.05.026
31.
Kimura S Noda T Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. (2007) 3:452–60. 10.4161/auto.4451
32.
Mizushima N Yamamoto A Matsui M Yoshimori T Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. (2004) 15:1101–11. 10.1091/mbc.e03-09-0704
33.
Scott RC Schuldiner O Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. (2004) 7:167–78. 10.1016/j.devcel.2004.07.009
34.
He C Bartholomew CR Zhou W Klionsky DJ. Assaying autophagic activity in transgenic GFP-Lc3 and GFP-gabarap zebrafish embryos. Autophagy. (2009) 5:520–6. 10.4161/auto.5.4.7768
35.
Cui J Sim TH-F Gong Z Shen H-M. Generation of transgenic zebrafish with liver-specific expression of EGFP-Lc3: a new in vivo model for investigation of liver autophagy. Biochem Biophys Res Commun. (2012) 422:268–73. 10.1016/j.bbrc.2012.04.145
36.
Khuansuwan S Barnhill LM Cheng S Bronstein JM. A novel transgenic zebrafish line allows for in vivo quantification of autophagic activity in neurons. Autophagy. (2019) 15:1322–32. 10.1080/15548627.2019.1580511
37.
Hariharan N Zhai P Sadoshima J. Oxidative stress stimulates autophagic flux during ischemia/reperfusion. Antiox Redox Signal. (2011) 14:2179–90. 10.1089/ars.2010.3488
38.
Staudt D Stainier D. Uncovering the molecular and cellular mechanisms of heart development using the zebrafish. Annu Rev Genet. (2012) 46:397–418. 10.1146/annurev-genet-110711-155646
39.
Rayani K Lin E Craig C Lamothe M Shafaattalab S Gunawan M et al Zebrafish as a model of mammalian cardiac function: optically mapping the interplay of temperature and rate on voltage and calcium dynamics. Prog Biophys Mol Biol. (2018) 138:69–90. 10.1016/j.pbiomolbio.2018.07.006
40.
Huang CJ Tu CT Hsiao CD Hsieh FJ Tsai HJ. Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev Dynam. (2003) 228:30–40. 10.1002/dvdy.10356
41.
Zhang Y Lin Y Zhang Y Wang Y Li Z Zhu Y et al Familial atrial myopathy in a large multigenerational heart-hand syndrome pedigree carrying an LMNA missense variant in rod 2B domain (p.R335W). Heart Rhythm. (2022) 19:466–75. 10.1016/j.hrthm.2021.11.022
42.
Sirtori CR. The pharmacology of statins. Pharmacol Res. (2014) 88:3–11. 10.1016/j.phrs.2014.03.002
43.
Tang WHW Francis GS. Statin treatment for patients with heart failure. Nat Rev Cardio. (2010) 7:249–55. 10.1038/nrcardio.2010.29
44.
Correale M Abruzzese S Greco CA Concilio M Biase MD Brunetti ND. Pleiotropic effects of statin in therapy in heart failure: a review. Curr Vasc Pharmacol. (2014) 12:873–84. 10.2174/1570161112999141127161508
45.
Lee MMY Sattar N McMurray JJV Packard CJ. Statins in the prevention and treatment of heart failure: a review of the evidence. Curr Atheroscler Rep. (2019) 21:41. 10.1007/s11883-019-0800-z
46.
Gorabi AM Kiaie N Aslani S Sathyapalan T Jamialahmadi T Sahebkar A. Implications on the therapeutic potential of statins via modulation of autophagy. Oxid Med Cell Longev. (2021) 2021:9599608. 10.1155/2021/9599608
47.
Wu Y-T Tan H-L Shui G Bauvy C Huang Q Wenk MR et al Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem. (2010) 285:10850–61. 10.1074/jbc.M109.080796
48.
Mauvezin C Neufeld TP. Bafilomycin A1 disrupts autophagic flux by inhibiting both V-ATPase-dependent acidification and Ca-P60A/SERCA-dependent autophagosome-lysosome fusion. Autophagy. (2015) 11:1437–8. 10.1080/15548627.2015.1066957
49.
Kamada Y Funakoshi T Shintani T Nagano K Ohsumi M Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. (2000) 150:1507–13. 10.1083/jcb.150.6.1507
50.
Lee E Koo Y Ng A Wei Y Luby-Phelps K Juraszek A et al Autophagy is essential for cardiac morphogenesis during vertebrate development. Autophagy. (2014) 10:572–87. 10.4161/auto.27649
51.
Xie F Xu S Lu Y Wong KF Sun L Hasan KMM et al Metformin accelerates zebrafish heart regeneration by inducing autophagy. NPJ Regen Med. (2021) 6:62. 10.1038/s41536-021-00172-w
52.
Scherz PJ Huisken J Sahai-Hernandez P Stainier DYR. High-speed imaging of developing heart valves reveals interplay of morphogenesis and function. Development. (2008) 135:1179–87. 10.1242/dev.010694
53.
Arrenberg AB Stainier DYR Baier H Huisken J. Optogenetic control of cardiac function. Science. (2010) 330:971–4. 10.1126/science.1195929
54.
Mareninova OA Jia W Gretler SR Holthaus CL Thomas DDH Pimienta M et al Transgenic expression of GFP-LC3 perturbs autophagy in exocrine pancreas and acute pancreatitis responses in mice. Autophagy. (2020) 16:2084–97. 10.1080/15548627.2020.1715047
55.
Loos B du Toit A Hofmeyr J-HS. Defining and measuring autophagosome flux-concept and reality. Autophagy. (2014) 10:2087–96. 10.4161/15548627.2014.973338
56.
Suster ML Kikuta H Urasaki A Asakawa K Kawakami K. Transgenesis in zebrafish with the tol2 transposon system. Methods Mol Biol. (2009) 561:41–63. 10.1007/978-1-60327-019-9_3
Summary
Keywords
autophagy, myocardial cell, zebrafish, statins, animal model
Citation
Zhang J, Zuo Z, Li J, Wang Y, Huang J, Xu L, Jin K, Lu H and Dai Y (2022) In situ assessment of statins’ effect on autophagic activity in zebrafish larvae cardiomyocytes. Front. Cardiovasc. Med. 9:921829. doi: 10.3389/fcvm.2022.921829
Received
16 April 2022
Accepted
28 October 2022
Published
17 November 2022
Volume
9 - 2022
Edited by
Rajesh Katare, University of Otago, New Zealand
Reviewed by
Xupin Jiang, Third Military Medical University, China; Maxinne Watchon, Macquarie University, Australia
Updates
Copyright
© 2022 Zhang, Zuo, Li, Wang, Huang, Xu, Jin, Lu and Dai.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hao Lu, lu.hao@zs-hospital.sh.cnYuxiang Dai, dai.yuxiang@zs-hospital.sh.cn
†These authors have contributed equally to this work
This article was submitted to General Cardiovascular Medicine, a section of the journal Frontiers in Cardiovascular Medicine
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.