- 1Department of Pharmacy, Affiliated Hospital of Shaoxing University, Shaoxing, China
- 2Department of Cardiology, Affiliated Hospital of Shaoxing University, Shaoxing, China
Background: Heart failure (HF) is a main consequence of cardiovascular diseases worldwide. Abnormal expression levels of microRNAs (miRNAs) in HF are observed in current studies. Novel biomarkers miRNAs may play an important role in the development of HF. Nevertheless, the inconsistency of miRNA expression limits the clinical application. We thus perform this systematic review of the miRNAs expression profiling to identify potential HF biomarkers.
Methods: The electronic databases of Embase, Medline, and Cochrane Library were systematically searched to identify the miRNA expression profiles between HF subjects and non-HF controls before May 26th, 2021. The pooled results were shown as log10 odds ratios (logORs) with 95% confidence intervals (CI) using random-effect models. Subgroup analyses were conducted according to species, region, and sample source. The quality assessment of included studies was independently conducted based on Diagnostic Accuracy Study 2 (QUADAS-2). The sensitivity analysis was conducted based on sample size.
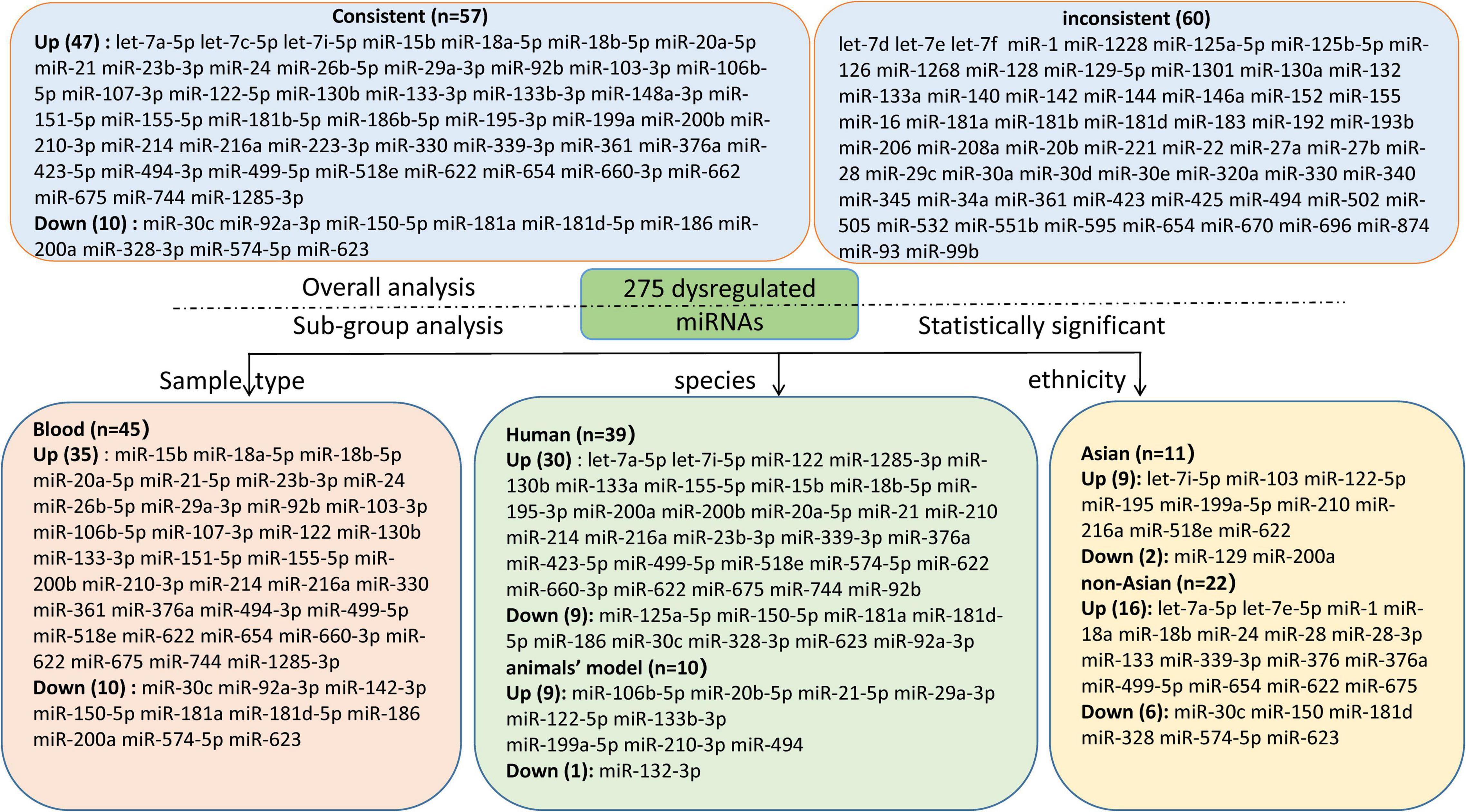
Results: A total of 55 miRNA expression articles reporting 276 miRNAs of HF were included. 47 consistently up-regulated and 10 down-regulated miRNAs were identified in the overall analysis, with the most up-regulated miR-21 (logOR 8.02; 95% CI: 6.76–9.27, P < 0.001) and the most down-regulated miR-30c (logOR 6.62; 95% CI: 3.04–10.20, P < 0.001). The subgroup analysis of sample source identified 35 up-regulated and 10 down-regulated miRNAs in blood sample, the most up-regulated and down-regulated miRNAs were miR-210-3p and miR-30c, respectively. In the region sub-groups, let-7i-5p and miR-129 were most up-regulated and down-regulated in Asian countries, while in non-Asian countries, let-7e-5p and miR-30c were the most dysregulated. It’s worth noting that miR-622 was consistently up-regulated in both Asian and non-Asian countries. Sensitivity analysis showed that 46 out of 58 (79.31%) miRNAs were dysregulated.
Conclusion: A total of 57 consistently dysregulated miRNAs related to HF were confirmed in this study. Seven dysregulated miRNAs (miR-21, miR-30c, miR-210-3p, let-7i-5p, miR-129, let-7e-5p, and miR-622) may be considered as potential non-invasive biomarkers for HF. However, further validation in larger-scale studies are needed to verify our conclusions.
Introduction
Heart failure (HF), a terminal stage of most cardiovascular diseases, is a major cause of hospitalizations and mortality worldwide (1, 2). The incidence of HF is raising rapidly because of increasing risk factors including hypertension and diabetes, and it is more common in elderly people over 80 years old (3, 4). Currently, the serum brain natriuretic peptide is the only routinely used biomarker for HF recommended in guideline (5), but its clinical value is still uncertain. Therefore, the predictive biomarkers are urgently needed for the early diagnosis of HF. In recent years, a growing number of researches have reported transcriptomic changes are associated with the pathophysiological mechanism of HF (6). Circulating microRNAs (miRNAs) gained significant interest as potential novel HF biomarkers because of high stability, sequence conservation, and non-invasive detection (7). However, prognostic value of microRNAs in HF is still unclear. Thus, the systematic review of miRNAs may contribute to identifying biomarker for early clinical diagnosis of HF.
MiRNAs are a class of conserved non-coding RNAs that regulate protein synthesis on the post-transcriptional level (8). MiRNAs are involved in the pathophysiology of various diseases (9–14), and the aberrant expression of miRNAs were potential biomarker for cardiovascular diseases (15, 16). Increasing evidence has demonstrated dysregulated miRNAs could alter the cellular response of cardiomyocytes, leading to cardiac concentric hypertrophy (17, 18). For example, miR-127 aggravates myocardial failure by regulating the expression of TGF-β1/Smad3 signaling pathway (19), miR-129-5p improves cardiac function in rats with chronic HF through targeting HMGB1 (20), and upregulated miR-132 improved the cardiac dysfunction by inhibiting PTEN expression (21). In general, understanding the pathophysiologic mechanisms in HF could provide the chance to develop new therapies (22, 23).
In previous studies, numerous studies have verified that the circulating miRNAs could be serve as potential biomarkers. For example, miRNA-208 and miRNA-150 have been identified as potential biomarkers for cardiac hypertrophy and remodeling (24, 25), miRNA-21 may serve as a biomarker for coronary heart diseases in the elderly patients (26). MiRNAs change in certain diseases, especially circulating miRNAs due to their stability makes them possible biomarkers in HF (27). Previous studies have reported that the dysregulated miRNAs can serve as the potential diagnostic biomarker in HF (16, 28, 29). Additionally, miRNA-19b, miR-21, miR-423-5p, and miR-92b-5p were observed to be promising biomarker for HF (16, 29–31). Given the pathogenic role of miRNAs in HF, the present study explored an association between miRNAs expression signatures and HF.
Over the past years, the dysregulated miRNAs were investigated in multiple studies, which come from different pathophysiological process, sample size, and inconsistent inclusion criteria, and the expression profiles of miRNAs in HF remain elusive (32, 33). Therefore, we aim to gather the currently available data about dysregulated miRNAs in HF, and to explore promising biomarker for diagnosis and prognosis of HF.
Methods
Literature Sources and Search Strategy
Two investigators (N.S. and J.W.) independently searched the PubMed, Embase, and Cochrane Library databases to identify relevant miRNA expression profiling studies with English restrictions from inception to May 26th, 2021. The search terms in the title/abstract were used as follows: (microRNA or miR- or miRNA), (HF or heart failure), (expression or profiling or profile). The detailed search strategies are shown in Supplementary Table 1. All inconsistency was discussed to reach consensus by the corresponding authors (Y-PF and J-LW).
Literature Selection
The inclusion criteria were as follows: (1) the original studies that investigated miRNA expression level between HF and non-HF subjects; (2) the obtained miRNAs were derived from blood or tissue specimens; (3) sample sizes were represented for dysregulated miRNAs; (4) the expression level of miRNAs were evaluated by qPCR, real-time PCR, and microarray, etc.; (5) only English-written papers were included. The exclusion criteria were as follows: (1) HF patients combined with other diseases; (2) studies based on cell experiments; (3) the literature were letters, comments, case reports, meta-analyses, editorials, or reviews; (4) lack of sufficient data. For duplicate studies from the same research, the one most similar to the inclusion criteria was involved. Two investigators (N.S. and J.W.) separately assessed all the studies to determine their eligibility, and any discrepancies were resolved with consensus by a third reviewer (Y.F.).
Data Collection and Quality Assessment
Two researchers (N.S. and J.W.) separately extracted the following data from all eligible articles: the name of the first author, year of publication, study population ethnicity, baseline characteristics of patients, species, miRNA detection methods, sample source, number of specimen, direction, and number of dysregulated miRNAs. The quality of included articles was independently assessed using the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2), which was conducted with eight questions by two investigators (N.S. and J.W.). Any discrepancies were resolved by a third reviewer (Y.F.), and final consensus was achieved through discussion.
Data Synthesis and Statistical Analysis
All statistical analyses were done using STATA software (version 13; Statacorp, College Station, Texas, United States) by the random-effects model. The results were shown as logORs according to the number of dysregulation between HF and non-HF subjects. The results were shown as logORs according to the number and direction of dysregulation between HF and non-HF subjects. In comparison with those obtained for the non-HF group, logOR values obtained for the HF group higher than 1 revealed upregulation. In comparison with the HF, a marked logOR obtained for the non-HF group higher than 1 revealed downregulation. The P-value < 0.05 was considered as statistically different. The importance of ranking of dysregulated miRNAs in HF were based on: (1) number of consistent sub-studies; (2) total sample size; (3) logOR values. Subgroup analysis was performed according to the species, tissue types, and ethnicity. The serum, plasma, or whole blood was classified to blood sample. The sensitivity analysis was performed to explore the heterogeneity. As a crucial determinant for sample size, the sensitivity analysis was repeatedly performed after excluding studies with the sample size of 10 or less.
Results
Literature Retrieval, Screening, and Features of Included Articles
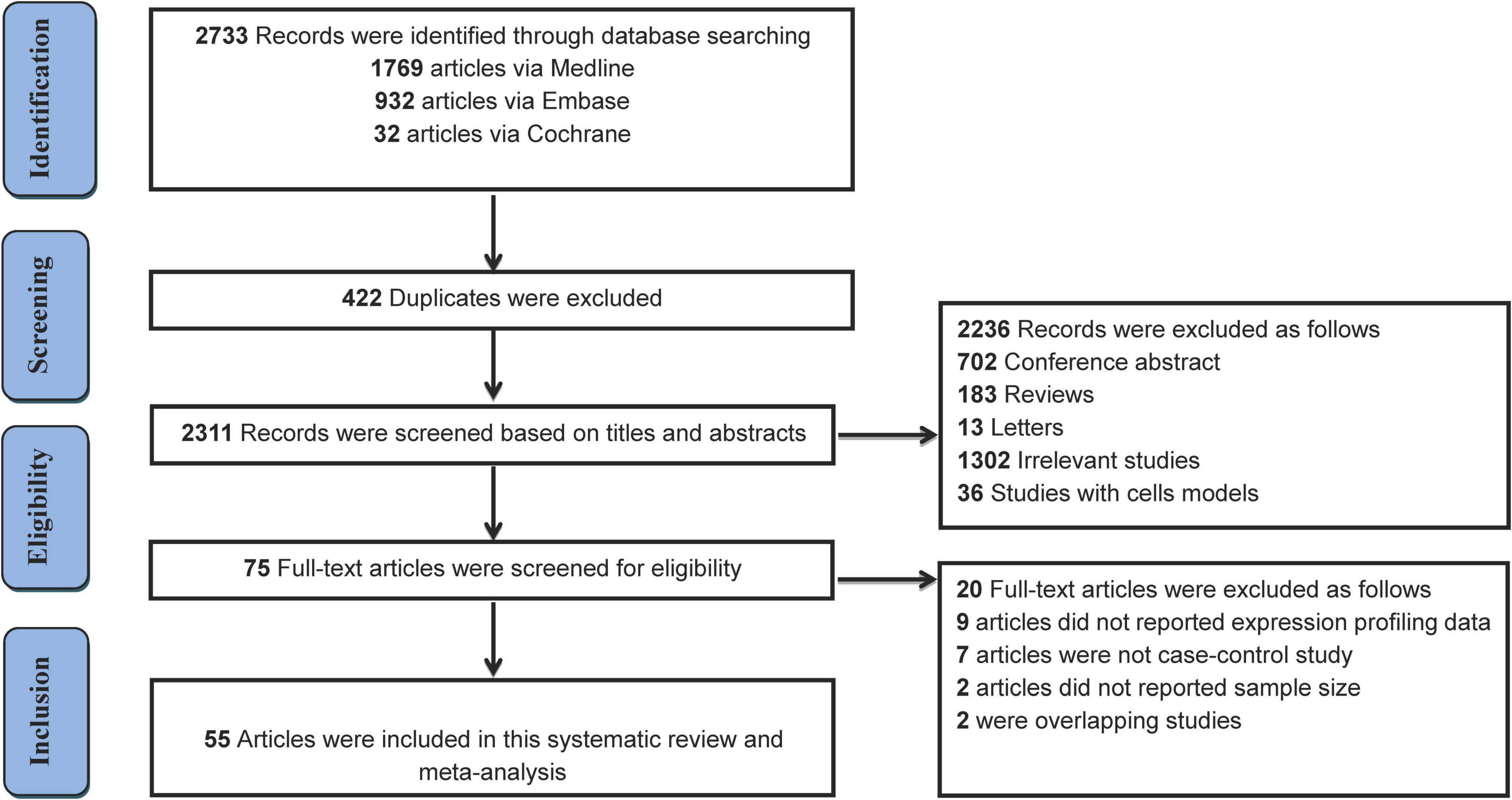
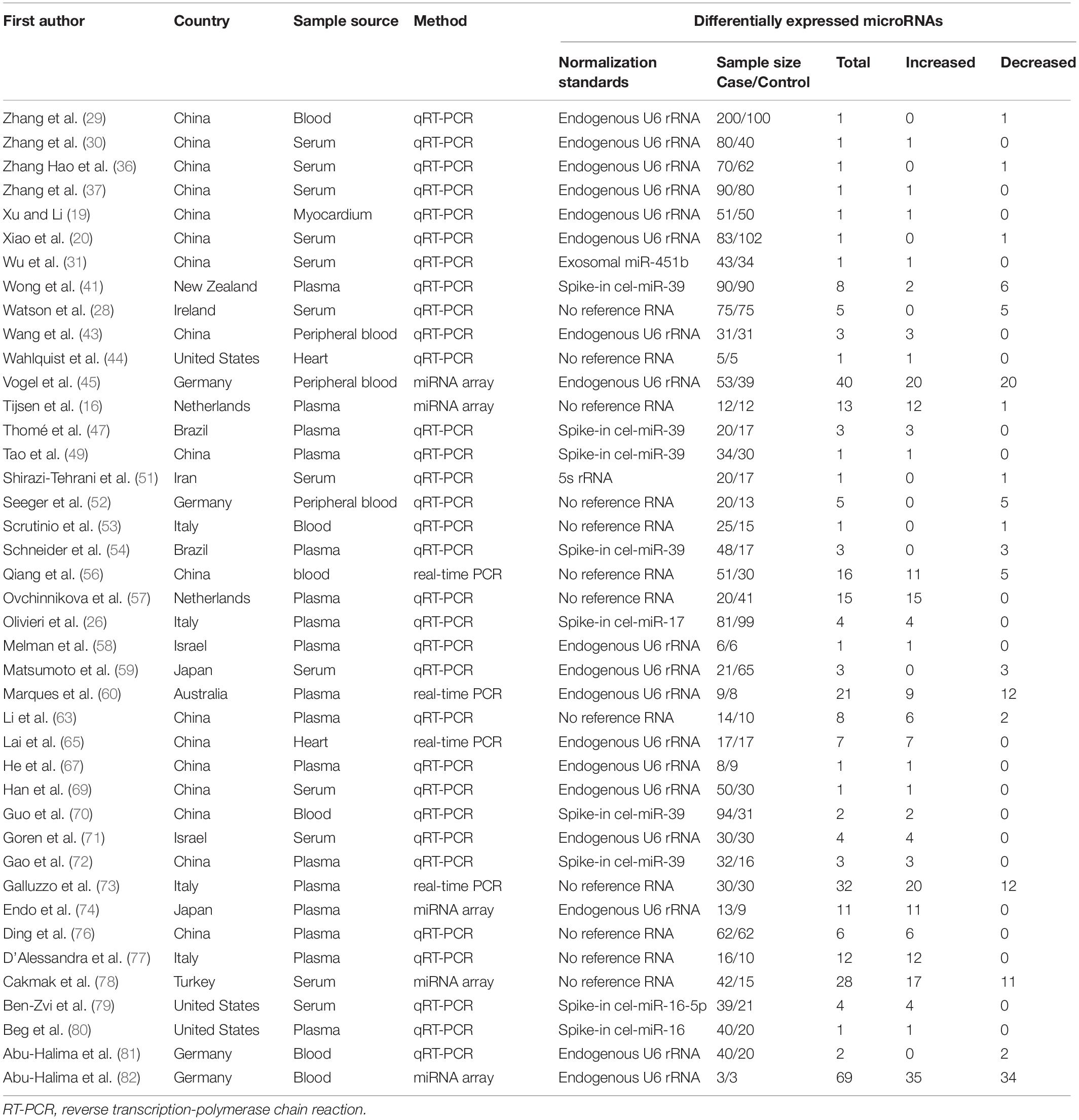
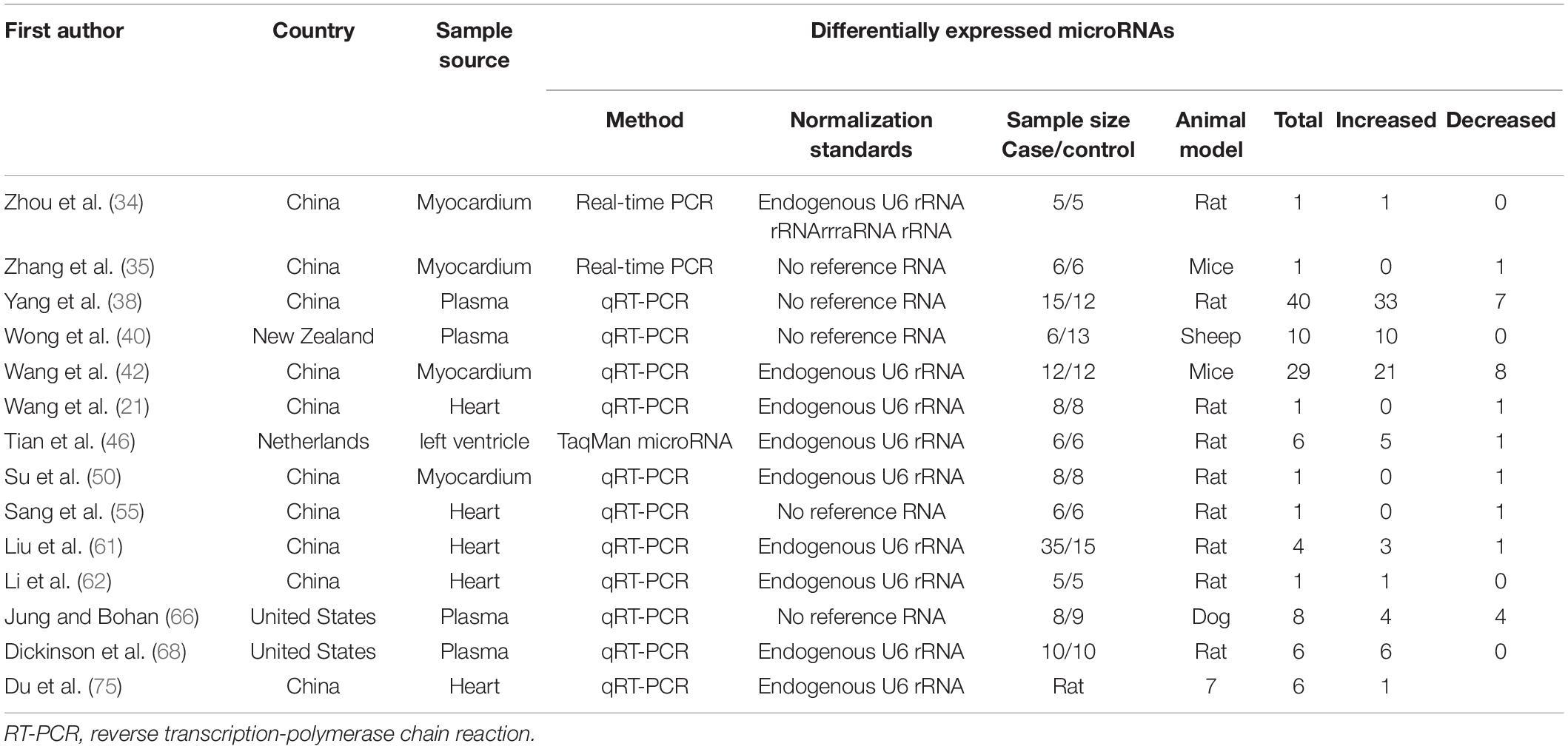
The literature selection process is shown in Figure 1. A total of 2,733 studies were initially retrieved from the databases according to the eligibility criteria (Supplementary Table 1). After excluding 422 duplicates, 2,311 studies were further screened based on the eligibility criteria. Ultimately, 55 articles were identified for this meta-analysis (16, 19–21, 26, 28–31, 34–82). The detailed reasons for the excluded studies were shown in Supplementary Table 2. The characteristics of the 55 included studies (41 human and 14 animal articles) were given in Tables 1, 2, respectively. The number of differentially expressed miRNAs in single article ranged from 1 to 69. The patient characteristics of the human studies are given in Supplementary Table 3. The sample sizes varied from 6 to 300 among the studies, and the mean year and the average age ranged 50.4–81.3.
Study Quality
The QUADAS-2 tool was used to assess the quality of all included literature. The detailed context was mentioned in “Methods” section. The results of quality assessment is given in Supplementary Table 4, the included studies satisfied a majority of the items on the QUADAS list. The evaluation results showed that the overall quality of articles was high.
Differentially Expressed MicroRNAs in Overall Analysis
We included the pooled analysis of 55 studies involving 275 differentially expressed miRNAs in that compared HF subjects with non-HF (Figure 2). Among these miRNAs, 47 were up-regulated and 10 were down-regulated (Supplementary Tables 5, 6). The details of each miRNA are summarized in Supplementary Figures 1–57. According to the results of 10 sub-studies with 709 samples and 3 sub-studies with 173 samples, miR-21 (logOR 8.02; 95% CI: 6.76–9.27, P < 0.001) was identified to be the most upregulated miRNA, and miR-30c (logOR 6.62; 95% CI: 3.04–10.20, P < 0.001) being the most downregulated one (Figures 3, 4). In addition, 60 miRNAs were identified in at least two articles with inconsistently dysregulated direction. And the inconsistently dysregulated miRNAs in the overall analysis classified by different subgroups are shown in Supplementary Table 7.
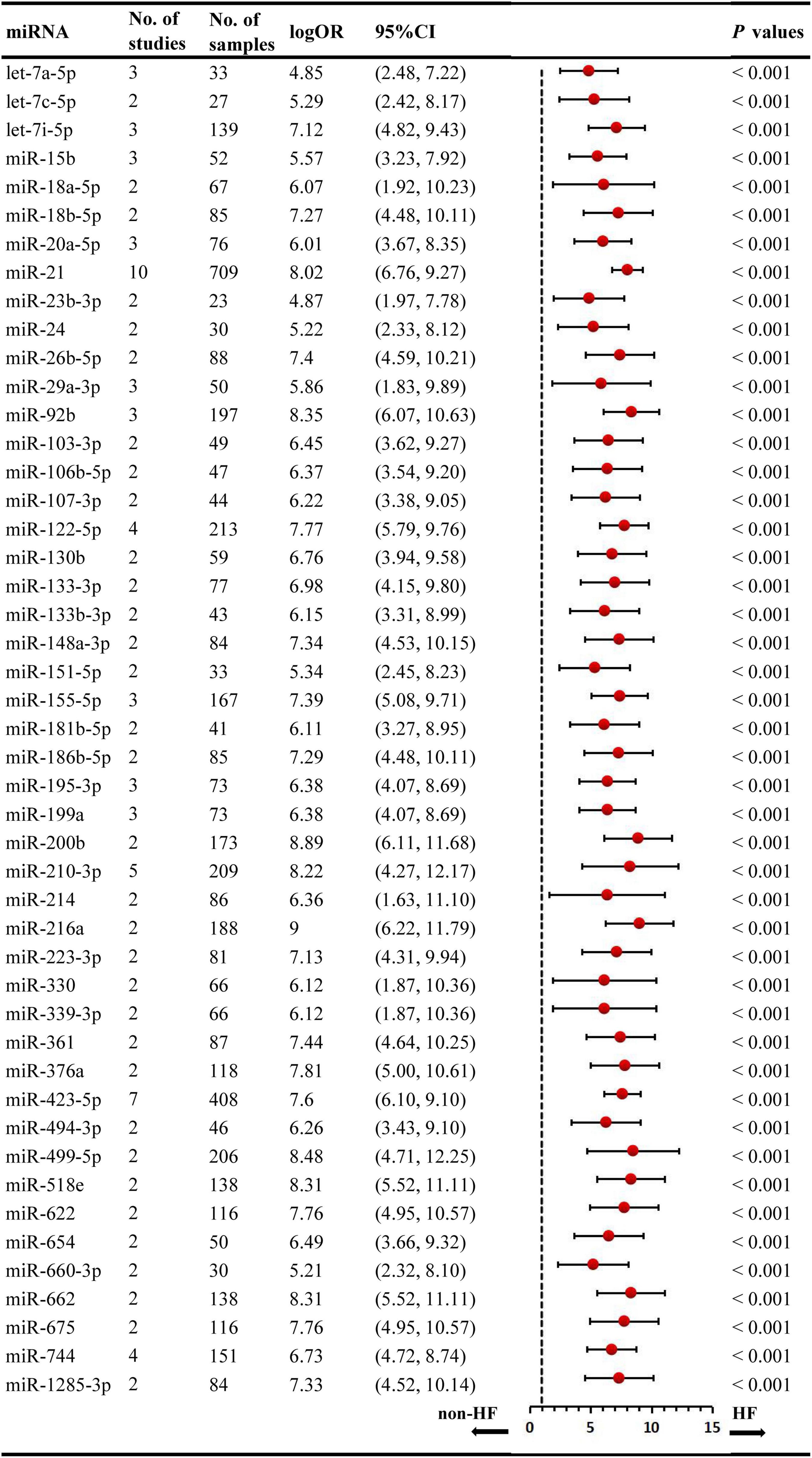
Figure 3. Statistically significant up-regulated miRNAs in overall analysis. miR, microRNA; No, number; HF, heart failure.
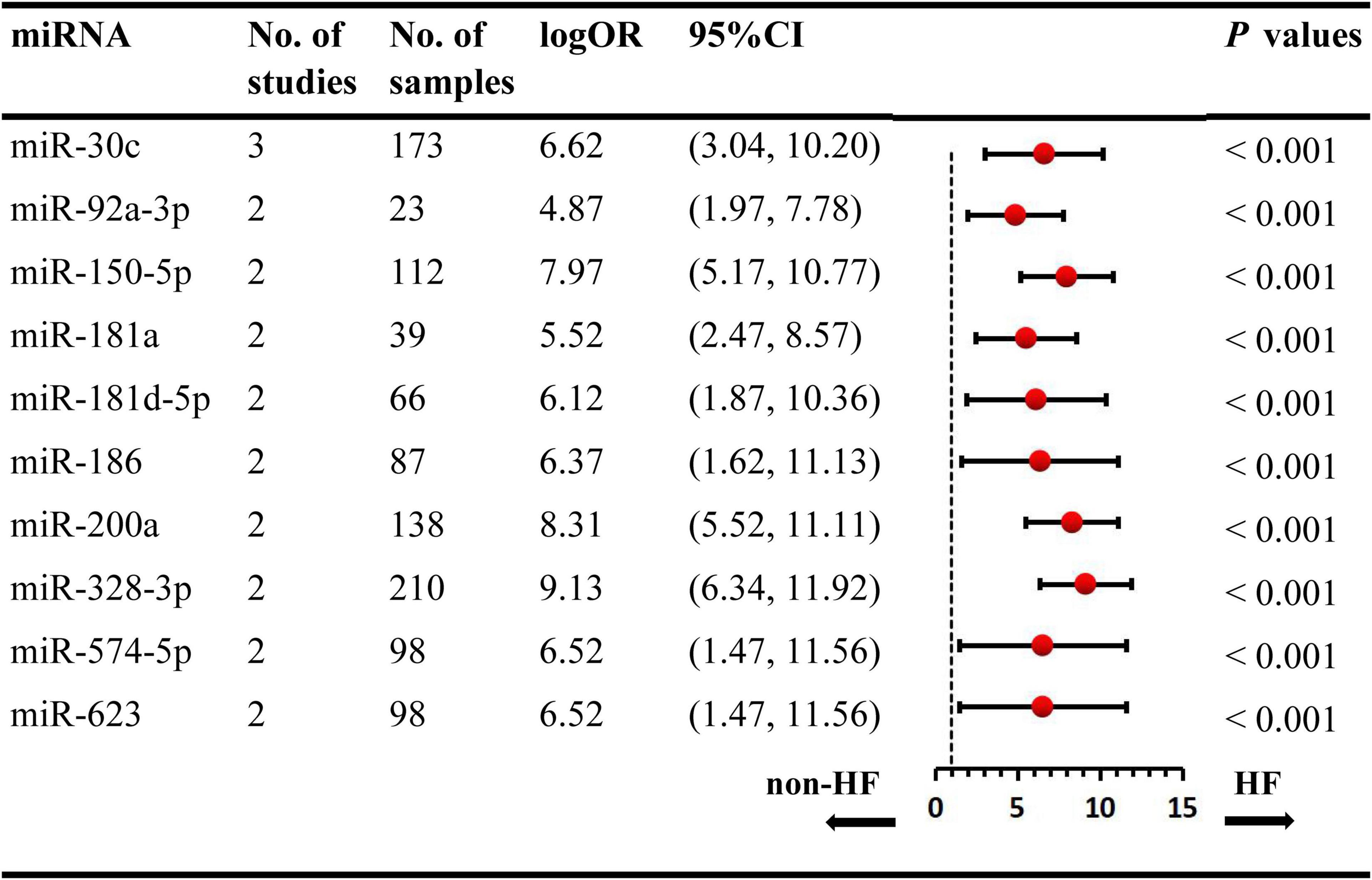
Figure 4. Statistically significant down-regulated miRNAs in overall analysis. miR, microRNA; No, number; HF, heart failure.
Subgroup Analysis
The subgroup analysis was conducted according to sample source including blood and tissue. 12 studies investigated miRNAs in tissue including myocardium, heart, and left ventricle, 43 studies detected circulating miRNAs in blood source including plasma, peripheral blood, serum, and whole blood. Overall, 45 miRNAs (35 up-regulation and 10 down-regulation) were identified to be aberrantly expressed in blood sample, with the most upregulated and downregulated miRNA being miR-210-3p (logOR 7.16; 95% CI: 5.38–8.94, P < 0.001) and miR-30c (logOR 6.62; 95% CI: 3.04–10.20, P < 0.001), respectively (Supplementary Table 8). While, no miRNAs were observed to be dysregulated statistically in tissue sample source. The results in the sub-analysis based on serum and plasma were similar to that of the overall analysis (Supplementary Table 9).
The subgroup analyses of ethnicity were conducted based on Asian and non-Asian countries. The subgroup analysis included 33 Asian studies and 22 non-Asian studies, respectively. In the subgroup of Asian countries, there were 9 upregulated and 2 downregulated miRNAs (Supplementary Table 10). For non-Asian subgroup, 16 upregulated miRNAs and 6 downregulated miRNAs were identified (Supplementary Table 11). Remarkably, the expression level of miR-622 was consistently increased in both Asian and non-Asian studies. The expression signature of miRNAs is region-specific. For example, let-7i-5p was significantly upregulated, and miR-129 was downregulated in Asian countries. While, in non-Asian countries, let-7e-5p was significantly upregulated, and miR-30c was downregulated.
Among 55 miRNA expression profiling articles in HF, 41 were human studies, and 14 were from animal articles. There were differences in the expression level of miRNAs between animals and humans, and the species subgroups of the miRNAs were analyzed in Supplementary Tables 12, 13. In the human miRNA studies, 30 significantly upregulated and 9 downregulated miRNAs were identified, with the most upregulated miR-21 (logOR 8.02; 95% CI: 6.76–9.27, P < 0.001) and with miR-328-3p (logOR 9.13; 95% CI: 6.34–11.92, P < 0.001) being the most downregulated. Among the animal’s studies, several miRNAs were consistently upregulated in two studies, including miR-106a, miR-20b-5p, miR-21-5p, miR-29a-3p, miR-122-5p, etc. But only one miRNA, miR-132-3p, was significantly downregulated in animal research.
Sensitivity Analysis
The small sample size might influence the robustness of this systematic review, which was determined by sensitivity analysis. We systematically removed 1 study with sample size less than 10, the remaining 54 studies were reanalyzed. As a result, 46 miRNAs were identified significant, with 42 of them upregulated and 4 downregulated (Supplementary Table 14). Finally, the results of sensitivity analysis confirmed that the other 11 miRNAs were not significant, but significant in the overall analysis. The above results showed that a small sample size may lead to differences in miRNA expression profiling.
Discussion
Over the past years, miRNAs have be observed to play key roles in the physiopathology of HF (56, 72), and there is increasing number of articles reporting on the differentially expressed miRNAs in HF (21, 29, 35). The identification of the dysregulated miRNAs may lead to the novel discovery to monitor the progression of HF. Nevertheless, a major problem with miRNA expression profiling was the inconsistency among different studies. Therefore, we reassembled the data and summarized the abnormal expression signatures of multiple miRNAs in HF through comprehensive data analysis. Finally, this systematic review identified 57 aberrantly expressed miRNAs (47 upregulated, and 10 downregulated) in all included studies. Based on further analysis, several miRNAs (miR-21, miR-30c, miR-210-3p, let-7i-5p, miR-129, let-7e-5p, and miR-622) were identified as potential biomarkers of HF.
The dysregulation of miRNAs, their target genes, are related to signaling pathways of HF, including cardiac hypertrophy, oxidative stress, cardiac fibrosis, etc. (30, 34, 35). The upregulated let-7a regulates β1-adrenoceptors (AR) signaling pathway by suppressing β1-AR expression in ischemia HF (75). The target genes of the top dysregulation miRNAs identified in this systematic review are all associated with HF. MiR-21 induces fibrotic process by regulating TGFβ1-Smad3 signaling (83), and the inhibition of miR-21 by specific antagomiR leads to reduced hypertrophy and fibrosis (84). Hypoxia-inducible factor-1α (HIF-1α), the target of miR-221-3p inhibits angiogenesis in HF (62). Additionally, miR-122-5p and miR-184 were proved to promote apoptosis in post-infarction HF (61). MiR-150 regulates apoptosis signaling by targeting pro-apoptotic genes and suppressing p53 activity as a major inducer of apoptosis (85).
To date, the early diagnosis of HF is still challenging, a more novel and advanced biomarker are urgently needed. According to the previous studies, miRNAs can be used as a new method for monitoring of HF. The dysregulated miRNAs (miR-19b, miR-129-5p, miR-221-3p, miR-208a, etc.) may provide non-invasive biomarkers for the diagnosis and prognosis of HF (29, 36, 62, 64). Therefore, the practicable detection methods are essential to ensure the accuracy and precision of miRNAs (86). Currently, there are the increasing concern about normalization strategies, that may contribute to the inconsistent expression data of microRNAs in PCR detection (87). The appropriate internal controls in PCR technology were required for the quantification of miRNAs, and relevant studies have verified that the spiked exogenous C. elegans miRNAs (e.g., miR-17, miR-454, cel-miR-39, and RNU6) as assay-normalization controls, did not significantly alter assay imprecision (16, 87–90). In PCR-based microRNAs detecting, the findings were robust irrespective of the way of standardization (91). Therefore, the findings in this meta-analysis were relatively reliable. However, in the future study, the rigorous experimental verification for confirming their validity with HF is necessary.
It was investigated whether miRNAs could serve as a promising biomarker of HF. Circulating miRNAs are promising diagnostic biomarkers for HF because of their high sensitivity and specificity. The increasing numbers of studies have focused on the clinical role of miRNAs. However, due to different specimens, sample sizes and regions, the current miRNAs expression characteristics were contradictive. The inconsistent results about dysregulated miRNAs may be caused by a variety of factors, including ethnic differences, the source of samples, and sample sizes. For example, plasma let-7e was upregulated (60, 66), while was downregulated in myocardial tissue (75). MiR-125a-5p was observed to be significantly downregulated (73), but upregulated in animal sample (40). Meanwhile, effects of concomitant treatment on miRNA expression in cardiovascular diseases should not be ignored. Actually, concomitant pharmacological treatment may have an important influence on circulating miRNA signatures. As reported in previous studies, significant changes in circulating miRNAs were observed in response to antiplatelet therapy (92, 93), and antidiabetic treatment increased the levels of let-7a and let-7f (94). The pharmacological treatment should be addressed and provided in the miRNA studies. Regrettably, among our included studies, few studies reported concomitant drugs, and little is known about the influence of drug treatment on the miRNA modulation. Therefore, in future studies, concomitant treatment should be taken into account when using miRNAs as diagnostic biomarker for cardiovascular diseases.
In the present systematic review, subgroup analysis was performed to explore probable sources of heterogeneity. The findings suggest that blood sample may be more informative about HF indicator. 45 dysregulated miRNAs were identified, the most down-regulated miRNAs was miR-30c, and the most upregulated was miR-210-3p. Because the circulating miRNAs are relatively stable and easily detectable by non-invasive methodology (95), circulating miR-30c and miR-210-3p, may be ideal candidate biomarkers for HF. Besides, in Asian countries, let-7i-5p and miR-195 were the most upregulated miRNAs, and miR-129 was the most downregulated one. On the contrary, in non-Asian countries, the significantly upregulated and downregulated miRNAs were let-7e-5p and miR-30c, respectively. Further analysis reveals that more miRNAs are dysregulated in humans than in animals, including miR-21, miR-423-5p, and miR-328-3p. These miRNAs may be the most suitable non-invasive biomarkers in the human’s blood for diagnosing and monitoring of HF.
To the best of our knowledge, the present meta-analysis was the comprehensive and credible to evaluate the miRNA expression signatures in HF. We combined the data of 55 articles to enhance the statistical power and the reliability of the results. However, there are still some limitations. Firstly, individual studies included relatively few patients, which limiting the strength of the conclusions. We thus conducted the sensitivity analysis to guarantee the robustness of the results. Secondly, although a comprehensive literature search was applied, some valuable research may be missing. Additionally, the biological characteristics and mechanisms of the different miRNAs in HF may differ, which may limit the applicability of the pooled results. We thus carried out subgroup analysis to increase the validity and reliability of miRNA analysis for HF.
Conclusion
This systematic review on miRNA expression profiling studies confirmed several valuable miRNAs, including miR-21, miR-30c, miR-210-3p, let-7i-5p, miR-129, let-7e-5p, and miR-622. These miRNAs may be used as potential non-invasive biomarkers and drug targets for HF. In the future, further investigations are required to verify the value of miRNAs in diagnostic and therapeutic approaches in HF.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
Y-PF was the guarantors of the entire manuscript. Y-PF and N-NS contributed to the study conception and design, critical revision of the manuscript for important intellectual content, and final approval of the version to be published. J-LW contributed to the data acquisition, analysis, and interpretation. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2022.856358/full#supplementary-material
References
1. Kratlian RG, Hajjar RJ. Cardiac gene therapy: from concept to reality. Curr Heart Fail Rep. (2012) 9:33–9. doi: 10.1007/s11897-011-0077-1
2. Desai AS. Intensive management to reduce hospitalizations in patients with heart failure. Circulation. (2016) 133:1704–7. doi: 10.1161/CIRCULATIONAHA.115.017594
3. Udelson JE, Stevenson LW. The future of heart failure diagnosis, therapy, and management. Circulation. (2016) 133:2671–86. doi: 10.1161/CIRCULATIONAHA.116.023518
4. Chopra VK, Mittal S, Bansal M, Singh B, Trehan N. Clinical profile and one-year survival of patients with heart failure with reduced ejection fraction: the largest report from India. Indian Heart J. (2019) 71:242–8. doi: 10.1016/j.ihj.2019.07.008
5. Ponikowski P, Voors AA, Anker SD, Bueno H, Cleland JG, Coats AJ, et al. [2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure]. Kardiol Pol. (2016) 74:1037–147. doi: 10.5603/KP.2016.0141
6. Lin D, Hollander Z, Meredith A, Stadnick E, Sasaki M, Cohen Freue G, et al. Biomarkers in transplantation T, excellence NCPCo. Molecular signatures of end-stage heart failure. J Cardiac Fail. (2011) 17:867–74. doi: 10.1016/j.cardfail.2011.07.001
7. van Rooij E, Olson EN. microRNAs put their signatures on the heart. Physiol Genomics. (2007) 31:365–6. doi: 10.1152/physiolgenomics.00206.2007
8. Topkara VK, Mann DL. Role of microRNAs in cardiac remodeling and heart failure. Cardiovasc Drugs Ther. (2011) 25:171–82. doi: 10.1007/s10557-011-6289-5
9. Xu J, Zhu X, Wu L, Yang R, Yang Z, Wang Q, et al. MicroRNA-122 suppresses cell proliferation and induces cell apoptosis in hepatocellular carcinoma by directly targeting Wnt/beta-catenin pathway. Liver Int. (2012) 32:752–60. doi: 10.1111/j.1478-3231.2011.02750.x
10. Dumortier O, Van Obberghen E. MicroRNAs in pancreas development. Diabetes Obes Metab. (2012) 14:22–8. doi: 10.1111/j.1463-1326.2012.01656.x
11. Manetti AC, Maiese A, Baronti A, Mezzetti E, Frati P, Fineschi V, et al. MiRNAs as new tools in lesion vitality evaluation: a systematic review and their forensic applications. Biomedicines. (2021) 9:1731. doi: 10.3390/biomedicines9111731
12. Manetti AC, Maiese A, Paolo MD, De Matteis A, La Russa R, Turillazzi E, et al. MicroRNAs and sepsis-induced cardiac dysfunction: a systematic review. Int J Mol Sci. (2020) 22:321. doi: 10.3390/ijms22010321
13. Liu J, Yang Y, Lu R, Liu Q, Hong S, Zhang Z, et al. MicroRNA-381-3p signatures as a diagnostic marker in patients with sepsis and modulates sepsis-steered cardiac damage and inflammation by binding HMGB1. Bioengineered. (2021) 12:11936–46. doi: 10.1080/21655979.2021.2006967
14. Cheng XJ, Li L, Xin BQ. MiR-124 Regulates the Inflammation and Apoptosis in Myocardial Infarction Rats by Targeting STAT3. Cardiovasc Toxicol. (2021) 21:710–20. doi: 10.1007/s12012-021-09661-2
15. Li S, Zhu J, Zhang W, Chen Y, Zhang K, Popescu LM, et al. Signature microRNA expression profile of essential hypertension and its novel link to human cytomegalovirus infection. Circulation. (2011) 124:175–84. doi: 10.1161/CIRCULATIONAHA.110.012237
16. Tijsen AJ, Creemers EE, Moerland PD, de Windt LJ, van der Wal AC, Kok WE, et al. MiR423-5p as a circulating biomarker for heart failure. Circ Res. (2010) 106:1035–9. doi: 10.1161/CIRCRESAHA.110.218297
17. Jeong MH, Lee JS, Kim DH, Park WJ, Yang DK. Identification of novel microRNAs negatively regulating cardiac hypertrophy. Biochem Biophys Res Commun. (2012) 428:191–6. doi: 10.1016/j.bbrc.2012.10.040
18. Han M, Yang Z, Sayed D, He M, Gao S, Lin L, et al. GATA4 expression is primarily regulated via a miR-26b-dependent post-transcriptional mechanism during cardiac hypertrophy. Cardiovasc Res. (2012) 93:645–54. doi: 10.1093/cvr/cvs001
19. Xu H, Li F. miR-127 aggravates myocardial failure by promoting the TGF-β1/Smad3 signaling. Mol Med Rep. (2018) 18:4839–46. doi: 10.3892/mmr.2018.9514
20. Xiao N, Zhang J, Chen C, Wan Y, Wang N, Yang J. miR-129-5p improves cardiac function in rats with chronic heart failure through targeting HMGB1. Mamm Genome. (2019) 30:276–88. doi: 10.1007/s00335-019-09817-0
21. Wang G, Wang R, Ruan Z, Liu L, Li Y, Zhu L. MicroRNA-132 attenuated cardiac fibrosis in myocardial infarction-induced heart failure rats. Biosci Rep. (2020) 40:BSR20201696. doi: 10.1042/BSR20201696
22. Wang J, Liew OW, Richards AM, Chen YT. Overview of MicroRNAs in cardiac hypertrophy, fibrosis, and apoptosis. Int J Mol Sci. (2016) 17:749. doi: 10.3390/ijms17050749
23. Sun XQ, Abbate A, Bogaard HJ. Role of cardiac inflammation in right ventricular failure. Cardiovasc Res. (2017) 113:1441–52. doi: 10.1093/cvr/cvx159
24. Liu L, Aguirre SA, Evering WE, Hirakawa BP, May JR, Palacio K, et al. miR-208a as a biomarker of isoproterenol-induced cardiac injury in Sod2+/- and C57BL/6J wild-type mice. Toxicol Pathol. (2014) 42:1117–29. doi: 10.1177/0192623314525684
25. Devaux Y, Vausort M, McCann GP, Zangrando J, Kelly D, Razvi N, et al. MicroRNA-150: a novel marker of left ventricular remodeling after acute myocardial infarction. Circ Cardiovasc Genet. (2013) 6:290–8. doi: 10.1161/CIRCGENETICS.113.000077
26. Olivieri F, Antonicelli R, Lorenzi M, D’Alessandra Y, Lazzarini R, Santini G, et al. Diagnostic potential of circulating miR-499-5p in elderly patients with acute non ST-elevation myocardial infarction. Int J Cardiol. (2013) 167:531–6. doi: 10.1016/j.ijcard.2012.01.075
27. Akat KM, Moore-McGriff D, Morozov P, Brown M, Gogakos T, Correa Da Rosa J, et al. Comparative RNA-sequencing analysis of myocardial and circulating small RNAs in human heart failure and their utility as biomarkers. Proc Natl Acad Sci USA. (2014) 111:11151–6. doi: 10.1073/pnas.1401724111
28. Watson CJ, Gupta SK, O’Connell E, Thum S, Glezeva N, Fendrich J, et al. MicroRNA signatures differentiate preserved from reduced ejection fraction heart failure. Eur J Heart Fail. (2015) 17:405–15. doi: 10.1002/ejhf.244
29. Zhang L, Xu RL, Liu SX, Dong SH, Zhao XX, Zhang BL. Diagnostic value of circulating microRNA-19b in heart failure. Eur J Clin Invest. (2020) 50:e13308. doi: 10.1111/eci.13308
30. Zhang J, Xing Q, Zhou X, Li J, Li Y, Zhang L, et al. Circulating miRNA-21 is a promising biomarker for heart failure. Mol Med Rep. (2017) 16:7766–74. doi: 10.3892/mmr.2017.7575
31. Wu T, Chen Y, Du Y, Tao J, Zhou Z, Yang Z. Serum exosomal MiR-92b-5p as a potential biomarker for acute heart failure caused by dilated cardiomyopathy. Cell Physiol Biochem. (2018) 46:1939–50. doi: 10.1159/000489383
32. Vegter EL, van der Meer P, de Windt LJ, Pinto YM, Voors AA. MicroRNAs in heart failure: from biomarker to target for therapy. Eur J Heart Fail. (2016) 18:457–68. doi: 10.1002/ejhf.495
33. Peterlin A, Pocivavsek K, Petrovic D, Peterlin B. The role of microRNAs in heart failure: a systematic review. Front Cardiovasc Med. (2020) 7:161. doi: 10.3389/fcvm.2020.00161
34. Zhou J, Gao J, Zhang X, Liu Y, Gu S, Zhang X, et al. microRNA-340-5p functions downstream of cardiotrophin-1 to regulate cardiac eccentric hypertrophy and heart failure via target gene dystrophin. Int Heart J. (2015) 56:454–8. doi: 10.1536/ihj.14-386
35. Zhang WB, Lai X, Guo XF. Activation of Nrf2 by miR-152 inhibits doxorubicin-induced cardiotoxicity via attenuation of oxidative stress, inflammation, and apoptosis. Oxid Med Cell Longev. (2021) 2021:8860883. doi: 10.1155/2021/8860883
36. Zhang H, Zhang N, Jiang W, Lun X. Clinical significance of the long non-coding RNA NEAT1/miR-129-5p axis in the diagnosis and prognosis for patients with chronic heart failure. Exp Ther Med. (2021) 21:512. doi: 10.3892/etm.2021.9943
37. Zhang B, Li B, Qin F, Bai F, Sun C, Liu Q. Expression of serum microRNA-155 and its clinical importance in patients with heart failure after myocardial infarction. J Int Med Res. (2019) 47:6294–302. doi: 10.1177/0300060519882583
38. Yang Y, Li R, Cao Y, Dai S, Luo S, Guo Q, et al. Plasma MIR-212-3p as a biomarker for acute right heart failure with pulmonary artery hypertension. Ann Transl Med. (2020) 8:1571. doi: 10.21037/atm-20-1653a
39. Wu T, Chen Y, Du Y, Tao J, Li W, Zhou Z, et al. Circulating exosomal miR-92b-5p is a promising diagnostic biomarker of heart failure with reduced ejection fraction patients hospitalized for acute heart failure. J Thorac Dis. (2018) 10:6211–20. doi: 10.21037/jtd.2018.10.52
40. Wong LL, Rademaker MT, Saw EL, Lew KS, Ellmers LJ, Charles CJ, et al. Identification of novel microRNAs in the sheep heart and their regulation in heart failure. Sci Rep. (2017) 7:8250. doi: 10.1038/s41598-017-08574-x
41. Wong LL, Armugam A, Sepramaniam S, Karolina DS, Lim KY, Lim JY, et al. Circulating microRNAs in heart failure with reduced and preserved left ventricular ejection fraction. Eur J Heart Fail. (2015) 17:393–404. doi: 10.1002/ejhf.223
42. Wang X, Song C, Zhou X, Han X, Li J, Wang Z, et al. Mitochondria associated microRNA expression profiling of heart failure. Biomed Res Int. (2017) 2017:4042509. doi: 10.1155/2017/4042509
43. Wang L, Liu J, Xu B, Liu YL, Liu Z. Reduced exosome miR-425 and miR-744 in the plasma represents the progression of fibrosis and heart failure. Kaohsiung J Med Sci. (2018) 34:626–33. doi: 10.1016/j.kjms.2018.05.008
44. Wahlquist C, Jeong D, Rojas-Muñoz A, Kho C, Lee A, Mitsuyama S, et al. Inhibition of miR-25 improves cardiac contractility in the failing heart. Nature. (2014) 508:531–5. doi: 10.1038/nature13073
45. Vogel B, Keller A, Frese KS, Leidinger P, Sedaghat-Hamedani F, Kayvanpour E, et al. Multivariate miRNA signatures as biomarkers for non-ischaemic systolic heart failure. Eur Heart J. (2013) 34:2812–22. doi: 10.1093/eurheartj/eht256
46. Tian C, Gao L, Zimmerman MC, Zucker IH. Myocardial infarction-induced microRNA-enriched exosomes contribute to cardiac Nrf2 dysregulation in chronic heart failure. Am J Physiol Heart Circ Physiol. (2018) 314:H928–39. doi: 10.1152/ajpheart.00602.2017
47. Thomé JG, Mendoza MR, Cheuiche AV, La Porta VL, Silvello D, Dos Santos KG, et al. Circulating microRNAs in obese and lean heart failure patients: a case-control study with computational target prediction analysis. Gene. (2015) 574:1–10. doi: 10.1016/j.gene.2015.07.068
48. Täubel J, Hauke W, Rump S, Viereck J, Batkai S, Poetzsch J, et al. Novel antisense therapy targeting microRNA-132 in patients with heart failure: results of a first-in-human Phase 1b randomized, double-blind, placebo-controlled study. Eur Heart J. (2021) 42:178–88. doi: 10.1093/eurheartj/ehaa898
49. Tao J, Wang J, Li C, Wang W, Yu H, Liu J, et al. MiR-216a accelerates proliferation and fibrogenesis via targeting PTEN and SMAD7 in human cardiac fibroblasts. Cardiovasc Diagn Ther. (2019) 9:535–44. doi: 10.21037/cdt.2019.11.06
50. Su Q, Zhang P, Yu D, Wu Z, Li D, Shen F, et al. Upregulation of miR-93 and inhibition of LIMK1 improve ventricular remodeling and alleviate cardiac dysfunction in rats with chronic heart failure by inhibiting RhoA/ROCK signaling pathway activation. Aging. (2019) 11:7570–86. doi: 10.18632/aging.102272
51. Shirazi-Tehrani E, Firouzabadi N, Tamaddon G, Bahramali E, Vafadar A. Carvedilol alters circulating MiR-1 and MiR-214 in heart failure. Pharmgenomics Pers Med. (2020) 13:375–83. doi: 10.2147/pgpm.s263740
52. Seeger T, Haffez F, Fischer A, Koehl U, Leistner DM, Seeger FH, et al. Immunosenescence-associated microRNAs in age and heart failure. Eur J Heart Fail. (2013) 15:385–93. doi: 10.1093/eurjhf/hfs184
53. Scrutinio D, Conserva F, Passantino A, Iacoviello M, Lagioia R, Gesualdo L. Circulating microRNA-150-5p as a novel biomarker for advanced heart failure: a genome-wide prospective study. J Heart Lung Transplant. (2017) 36:616–24. doi: 10.1016/j.healun.2017.02.008
54. Schneider S, Silvello D, Martinelli NC, Garbin A, Biolo A, Clausell N, et al. Plasma levels of microRNA-21, -126 and -423-5p alter during clinical improvement and are associated with the prognosis of acute heart failure. Mol Med Rep. (2018) 17:4736–46. doi: 10.3892/mmr.2018.8428
55. Sang HQ, Jiang ZM, Zhao QP, Xin F. MicroRNA-133a improves the cardiac function and fibrosis through inhibiting Akt in heart failure rats. Biomed Pharmacother. (2015) 71:185–9. doi: 10.1016/j.biopha.2015.02.030
56. Qiang L, Hong L, Ningfu W, Huaihong C, Jing W. Expression of miR-126 and miR-508-5p in endothelial progenitor cells is associated with the prognosis of chronic heart failure patients. Int J Cardiol. (2013) 168:2082–8. doi: 10.1016/j.ijcard.2013.01.160
57. Ovchinnikova ES, Schmitter D, Vegter EL, Ter Maaten JM, Valente MA, Liu LC, et al. Signature of circulating microRNAs in patients with acute heart failure. Eur J Heart Fail. (2016) 18:414–23. doi: 10.1002/ejhf.332
58. Melman YF, Shah R, Danielson K, Xiao J, Simonson B, Barth A, et al. Circulating MicroRNA-30d is associated with response to cardiac resynchronization therapy in heart failure and regulates cardiomyocyte apoptosis: a translational pilot study. J Int Med Res. (2015) 131:2202–16. doi: 10.1161/circulationaha.114.013220
59. Matsumoto S, Sakata Y, Suna S, Nakatani D, Usami M, Hara M, et al. Circulating p53-responsive microRNAs are predictive indicators of heart failure after acute myocardial infarction. Circ Res. (2013) 113:322–6. doi: 10.1161/circresaha.113.301209
60. Marques FZ, Vizi D, Khammy O, Mariani JA, Kaye DM. The transcardiac gradient of cardio-microRNAs in the failing heart. Eur J Heart Fail. (2016) 18:1000–8. doi: 10.1002/ejhf.517
61. Liu X, Meng H, Jiang C, Yang S, Cui F, Yang P. Differential microRNA expression and regulation in the rat model of post-infarction heart failure. PLoS One. (2016) 11:e0160920. doi: 10.1371/journal.pone.0160920
62. Li Y, Yan C, Fan J, Hou Z, Han Y. MiR-221-3p targets Hif-1α to inhibit angiogenesis in heart failure. Lab Invest. (2021) 101:104–15. doi: 10.1038/s41374-020-0450-3
63. Li H, Fan J, Yin Z, Wang F, Chen C, Wang DW. Identification of cardiac-related circulating microRNA profile in human chronic heart failure. Oncotarget. (2016) 7:33–45. doi: 10.18632/ONCOTARGET.6631
64. Li DM, Li BX, Yang LJ, Gao P, Ma ZY, Li ZJD. Diagnostic value of circulating microRNA-208a in differentiation of preserved from reduced ejection fraction heart failure. Heart Lung. (2021) 50:71–4. doi: 10.1016/j.hrtlng.2020.07.010
65. Lai KB, Sanderson JE, Izzat MB, Yu CM. Micro-RNA and mRNA myocardial tissue expression in biopsy specimen from patients with heart failure. Int J Cardiol. (2015) 199:79–83. doi: 10.1016/j.ijcard.2015.07.043
66. Jung S, Bohan A. Genome-wide sequencing and quantification of circulating microRNAs for dogs with congestive heart failure secondary to myxomatous mitral valve degeneration. Am J Vet Res. (2018) 79:163–9. doi: 10.2460/ajvr.79.2.163
67. He X, Ji J, Wang T, Wang MB, Chen XL. Upregulation of circulating miR-195-3p in heart failure. Cardiology. (2017) 138:107–14. doi: 10.1159/000476029
68. Dickinson BA, Semus HM, Montgomery RL, Stack C, Latimer PA, Lewton SM, et al. Plasma microRNAs serve as biomarkers of therapeutic efficacy and disease progression in hypertension-induced heart failure. Expert Opin Ther Targets. (2013) 15:650–9. doi: 10.1093/eurjhf/hft018
69. Han R, Li K, Li L, Zhang L, Zheng H. Expression of microRNA-214 and galectin-3 in peripheral blood of patients with chronic heart failure and its clinical significance. Exp Ther Med. (2020) 19:1322–8. doi: 10.3892/etm.2019.8318
70. Guo M, Luo J, Zhao J, Shang D, Lv Q, Zang P. Combined use of circulating miR-133a and NT-proBNP improves heart failure diagnostic accuracy in elderly patients. Med Sci Monit. (2018) 24:8840–8. doi: 10.12659/msm.911632
71. Goren Y, Kushnir M, Zafrir B, Tabak S, Lewis BS, Amir O. Serum levels of microRNAs in patients with heart failure. Eur J Heart Fail. (2012) 14:147–54. doi: 10.1093/eurjhf/hfr155
72. Gao G, Chen W, Liu M, Yan X, Yang P. Circulating microRNAs as novel potential biomarkers for left ventricular remodeling in postinfarction heart failure. Dis Markers. (2019) 2019:5093803. doi: 10.1155/2019/5093803
73. Galluzzo A, Gallo S, Pardini B, Birolo G, Fariselli P, Boretto P. Identification of novel circulating microRNAs in advanced heart failure by next-generation sequencing. ESC Heart Fail. (2021) 8:2907–19. doi: 10.1002/ehf2.13371
74. Endo K, Naito Y, Ji X, Nakanishi M, Noguchi T, Goto Y, et al. MicroRNA 210 as a biomarker for congestive heart failure. Biol Pharm Bull. (2013) 36:48–54. doi: 10.1248/bpb.b12-00578
75. Du Y, Zhang M, Zhao W, Shu Y, Gao M, Zhuang Y, et al. Let-7a regulates expression of β1-adrenoceptors and forms a negative feedback circuit with the β1-adrenoceptor signaling pathway in chronic ischemic heart failure. Oncotarget. (2017) 8:8752–64. doi: 10.18632/oncotarget.14436
76. Ding H, Wang Y, Hu L, Xue S, Wang Y, Zhang L, et al. Combined detection of miR-21-5p, miR-30a-3p, miR-30a-5p, miR-155-5p, miR-216a and miR-217 for screening of early heart failure diseases. J Cell Mol Med. (2020) 40:BSR20191653. doi: 10.1042/bsr20191653
77. D’Alessandra Y, Chiesa M. Differential role of circulating microRNAs to track progression and pre-symptomatic stage of chronic heart failure: a pilot study. Biomedicines. (2020) 8:597. doi: 10.3390/biomedicines8120597
78. Cakmak HA, Coskunpinar E, Ikitimur B, Barman HA, Karadag B, Tiryakioglu NO, et al. The prognostic value of circulating microRNAs in heart failure: preliminary results from a genome-wide expression study. J Cardiovasc Med. (2015) 16:431–7. doi: 10.2459/jcm.0000000000000233
79. Ben-Zvi I, Volinsky N, Grosman-Rimon L, Haviv I, Rozen G, Andria N, et al. Cardiac-peripheral transvenous gradients of microRNA expression in systolic heart failure patients. ESC Heart Fail. (2020) 7:835–43. doi: 10.1002/ehf2.12597
80. Beg F, Wang R, Saeed Z, Devaraj S, Masoor K, Nakshatri H. Inflammation-associated microRNA changes in circulating exosomes of heart failure patients. BMC Res Notes. (2017) 10:751. doi: 10.1186/s13104-017-3090-y
81. Abu-Halima M, Meese E, Saleh MA, Keller A, Abdul-Khaliq H, Raedle-Hurst T. Micro-RNA 150-5p predicts overt heart failure in patients with univentricular hearts. PLoS One. (2019) 14:e0223606. doi: 10.1371/journal.pone.0223606
82. Abu-Halima M, Meese E, Keller A, Abdul-Khaliq H, Rädle-Hurst T. Analysis of circulating microRNAs in patients with repaired Tetralogy of Fallot with and without heart failure. J Transl Med. (2017) 15:156. doi: 10.1186/s12967-017-1255-z
83. Liang H, Zhang C, Ban T, Liu Y, Mei L, Piao X, et al. A novel reciprocal loop between microRNA-21 and TGFbetaRIII is involved in cardiac fibrosis. Int J Biochem Cell Biol. (2012) 44:2152–60. doi: 10.1016/j.biocel.2012.08.019
84. Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. (2008) 456:980–4. doi: 10.1038/nature07511
85. Tang Y, Wang Y, Park KM, Hu Q, Teoh JP, Broskova Z, et al. MicroRNA-150 protects the mouse heart from ischaemic injury by regulating cell death. Cardiovasc Res. (2015) 106:387–97. doi: 10.1093/cvr/cvv121
86. Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, et al. Serum microRNAs are promising novel biomarkers. PLoS One. (2008) 3:e3148. doi: 10.1371/journal.pone.0003148
87. McDonald JS, Milosevic D, Reddi HV, Grebe SK, Algeciras-Schimnich A. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem. (2011) 57:833–40. doi: 10.1373/clinchem.2010.157198
88. Duttagupta R, Jiang R, Gollub J, Getts RC, Jones KW. Impact of cellular miRNAs on circulating miRNA biomarker signatures. PLoS One. (2011) 6:e20769. doi: 10.1371/journal.pone.0020769
89. Ji X, Takahashi R, Hiura Y, Hirokawa G, Fukushima Y, Iwai N. Plasma miR-208 as a biomarker of myocardial injury. Clin Chem. (2009) 55:1944–9. doi: 10.1373/clinchem.2009.125310
90. Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. (2010) 107:810–7. doi: 10.1161/CIRCRESAHA.110.226357
91. Zampetaki A, Willeit P, Drozdov I, Kiechl S, Mayr M. Profiling of circulating microRNAs: from single biomarkers to re-wired networks. Cardiovasc Res. (2012) 93:555–62. doi: 10.1093/cvr/cvr266
92. Willeit P, Zampetaki A, Dudek K, Kaudewitz D, King A, Kirkby NS, et al. Circulating microRNAs as novel biomarkers for platelet activation. Circ Res. (2013) 112:595–600. doi: 10.1161/CIRCRESAHA.111.300539
93. de Boer HC, van Solingen C, Prins J, Duijs JM, Huisman MV, Rabelink TJ, et al. Aspirin treatment hampers the use of plasma microRNA-126 as a biomarker for the progression of vascular disease. Eur Heart J. (2013) 34:3451–7. doi: 10.1093/eurheartj/eht007
94. Santovito D, De Nardis V, Marcantonio P, Mandolini C, Paganelli C, Vitale E, et al. Plasma exosome microRNA profiling unravels a new potential modulator of adiponectin pathway in diabetes: effect of glycemic control. J Clin Endocrinol Metab. (2014) 99:E1681–5. doi: 10.1210/jc.2013-3843
Keywords: miRNAs, heart failure, systematic review, meta-analysis, biomarker
Citation: Shen N-N, Wang J-L and Fu Y-p (2022) The microRNA Expression Profiling in Heart Failure: A Systematic Review and Meta-Analysis. Front. Cardiovasc. Med. 9:856358. doi: 10.3389/fcvm.2022.856358
Received: 17 January 2022; Accepted: 27 May 2022;
Published: 15 June 2022.
Edited by:
Alexander Pott, Ulm University Medical Center, GermanyReviewed by:
Donato Santovito, Unit of Milan, Institute of Genetic and Biomedical Research (CNR), ItalyAniello Maiese, University of Pisa, Italy
Copyright © 2022 Shen, Wang and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jia-Liang Wang, MTU4NTc1ODEyMTNAcXEuY29t; Yong-ping Fu, MzgwNzk0NEBxcS5jb20=
 Nan-Nan Shen
Nan-Nan Shen Jia-Liang Wang1*
Jia-Liang Wang1*