- Institute of Pharmacy, Nirma University, Ahmedabad, India
Human heart development is a complex and tightly regulated process, conserving proliferation, and multipotency of embryonic cardiovascular progenitors. At terminal stage, progenitor cell type gets suppressed for terminal differentiation and maturation. In the human heart, most cardiomyocytes are terminally differentiated and so have limited proliferation capacity. MicroRNAs (miRNAs) are non-coding single-stranded RNA that regulate gene expression and mRNA silencing at the post-transcriptional level. These miRNAs play a crucial role in numerous biological events, including cardiac development, and cardiomyocyte proliferation. Several cardiac cells specific miRNAs have been discovered. Inhibition or overexpression of these miRNAs could induce cardiac regeneration, cardiac stem cell proliferation and cardiomyocyte proliferation. Clinical application of miRNAs extends to heart failure, wherein the cell cycle arrest of terminally differentiated cardiac cells inhibits the heart regeneration. The regenerative capacity of the myocardium can be enhanced by cardiomyocyte specific miRNAs controlling the cell cycle. In this review, we focus on cardiac-specific miRNAs involved in cardiac regeneration and cardiomyocyte proliferation, and their potential as a new clinical therapy for heart regeneration.
Introduction
An uninterrupted supply of oxygen and nutrients by the heart is vital for the function of every cell, tissue, and organ in our body. Therefore, acquired and congenital heart diseases are a leading cause of death in developed and developing countries (1). Irreversible damage to cardiomyocytes is caused by myocardial infarction, ischemic heart disease, hypertension, and genetic mutations. Thereby, deteriorating heart functioning to heart attack and eventually death (2). Unlike, neonatal mammalian heart, adult mammalian heart regeneration is inadequate and forms scar tissue instead. Scar formation hinders the cardiac blood supply, worsening cardiac damage to heart failure (3). Current therapies and medical devices focus on symptomatic relief, however, undermines the replacement of lost cardiac muscle. Stem cell-based therapies targeted toward cardiac repair are being tested for more than a decade, showing insignificant improvement in cardiac function (4–8). Additionally, the molecular basis of stem cells based cardiac function improvement is not evident. Also, the retention of stem cells at the injured heart tissue is questionable. Thereby, suggesting the unmet clinical application of therapeutic strategies to replace cardiomyocytes and ameliorate cardiac regeneration.
Prenatal mammals possess a unique potential to regenerate their heart post-injury. But this regeneration capacity remains for a very short period following birth (9). In neonatal mammals, transitory cellular and molecular response initiated from pre-existing cardiomyocytes drive the myocardial regeneration (10, 11). Various signaling pathways like thyroid hormone signaling (12), Hedgehog (13), Notch (14), ErbB2/4 (15, 16), and Hippo/YAP signaling pathways (17, 18) are being studied as a potential for adult heart regeneration. Similarly, non-muscle cells reprogramming to cardiac muscle cell fate in the injured heart to potentially integrate and repopulate it is being investigated as an alternative approach (19–22).
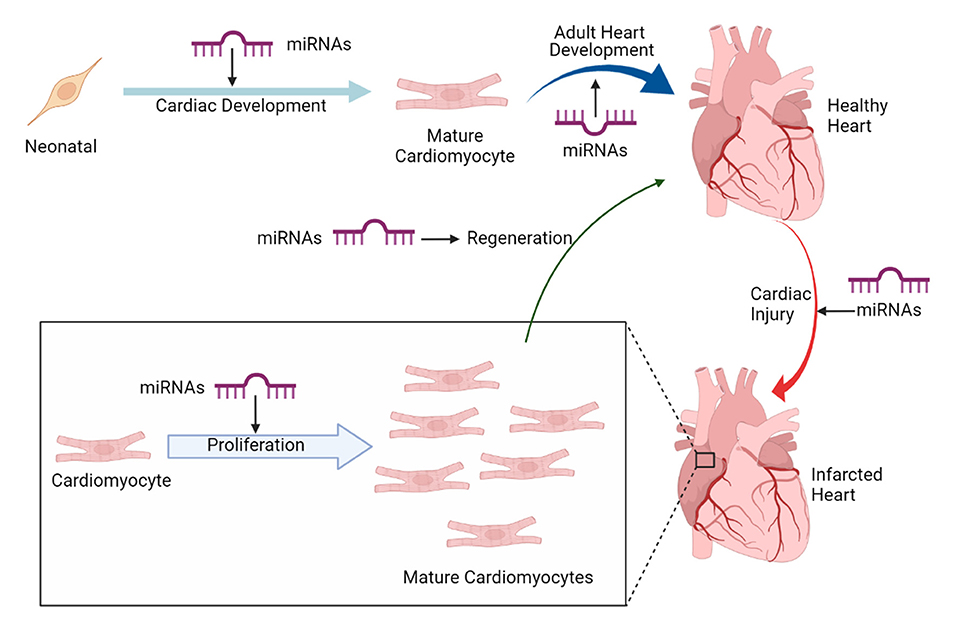
Recently, miRNAs (small non-coding RNAs, that regulate target gene expression post-translationally) have emerged as a potential therapeutic candidate for cardiomyocyte proliferation and heart regeneration (23). Several miRNAs have been shown to regulate and maintain cardiac cell fate specification of cardiac progenitor cells (24). Thus, studies pertaining to understand miRNA transcriptome and regulatory pathways in cardiac progenitors, regulated by miRNAs are gaining momentum. Furthermore, research in regenerative therapeutics of adult heart to develop embryonic cardiovascular progenitor miRNAs and adult cardiac-specific miRNAs based miRNA therapy is being tested. Figure 1 implicates on the role of miRNAs in cardiac development and regeneration. This review focuses on molecular regulatory mechanisms governed by miRNAs that induce cardiac regeneration and how these mechanisms can be targeted to potentially achieve adult mammalian heart regeneration.
miRNA in Development
miRNA mechanism of action and biogenesis has been reviewed extensively, as the field has evolved over the decade (25). Briefly, miRNA controls post-translational gene expression and has been shown to play a pivotal role in embryonic development and pluripotency maintenance (26). For instance, in mice the heterozygous mutant of Oct4 showed reduced protein expression, suggesting a decrease in the stem cell pool in the embryo. Furthermore, embryonic lethality was observed in mice with homozygous Dicer 1 knockout. Thereafter, several miRNAs involved in pluripotency maintenance of the embryonic stem cells were discovered such as the miR- 290 family (miR-295, miR-294, miR-291-3p) and miR-34a (27–29). Also, miR-372 and miR-302 showed increased efficiency in pluripotent stem cell reprogramming (30, 31). Early embryogenesis entails the differentiation of different cell types from single totipotent cells, through morphogen gradients in order to transmit positional information within the embryo (32). miRNAs also play an important role in lineage differentiation and stem cell migration into different tissues and organs. miR-430 monitors the nodal morphogen gradient by acting on both the agonist and antagonist of nodal, thereby avoiding patterning mistakes in the embryo (33). miR-430 also enables degeneration of active morphogen mRNAs that are not vital such as sdf1a transcript is degenerated in all cells, except the cells actively transcribing new sdf1a (34, 35). Conversely, several tissue-specific miRNAs have been shown to promote trans-differentiation of the cells (36, 37). Furthermore, feedback and feed-forward loops between transcriptional factors and miRNAs, reinforce or counterbalance cellular decisions toward differentiation and controls cell fate (38). These findings have established the miRNAs as a promising candidate for the development of regenerative therapies.
miRNA in Mammalian Cardiogenesis
Myocardial cells (Fibroblasts and Cardiomyocytes) derived from mesoderm at gastrulation contribute toward the formation of the heart (39, 40). Early cardiac progenitors derived from the anterior mesoderm forms the primary heart field, also called the cardiac crescent. On the other hand, pharyngeal mesoderm contributes toward the development of secondary heart field between and anterior to cardiac crescent. In mice by the embryonic day 8, linear heart tube is formed by the cells migrated from the cardiac crescent in order to provide a scaffold for heart development. Furthermore, cells migrated from the secondary heart field contribute toward the development of venous and arterial poles. This leads to a beating heart tube, composed of cardiomyocytes and underlying endothelial cells. By embryonic day 8.5 primitive atria and ventricles are formed by uneven remodeling and growth of linear heart tube. Heart maturation leads to a separation between atria and ventricles by embryonic day 15. Also, positioning the outflow tract and inflow tract on the anterior pole of the heart. The right and left atria are formed by both primary and secondary heart fields. Left ventricles are formed from the primary heart field, while the right ventricle is formed by the secondary heart field (41–45). Mammalian heart development is well-orchestrated crosstalk between primary and secondary heart field, along with extrinsic and intrinsic signals. This cardiogenesis is tightly regulated by the network of signaling pathways and transcriptional regulation, which is in turn highly regulated by the miRNAs (41, 46) (Figure 2).
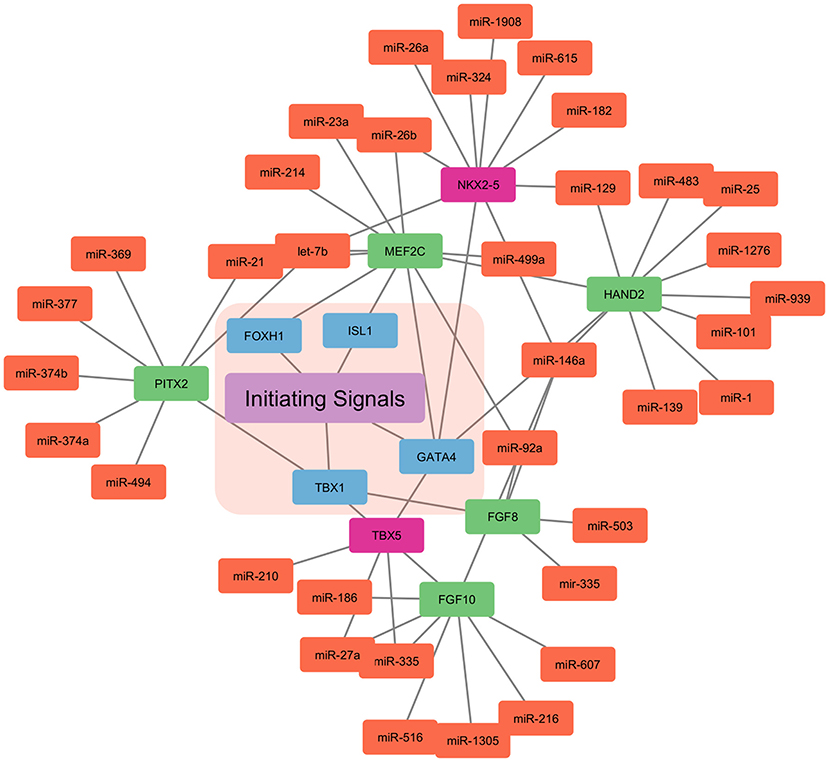
Figure 2. miRNA based regulation of cardiogenesis. The initiating signals, i.e., extrinsic and intrinsic signals in the primary and secondary heart field regulate the cardiogenesis. The nodes in blue represent the central transcription factor orchestrating the developmental signals. Nodes in red represent the transcription factor only expressed in the primary heart field. Whereas, nodes in green represent the transcription factor only expressed in the secondary heart field. Nodes in light coral color represent the miRNAs involved in the control of the overall cardiogenesis.
WNT signaling pathway from neural tube and notochord suppress differentiation into cardiomyocytes, whereas Bone morphogenetic protein (BMP) derived from endoderm enhances the cardiomyocytes specification (47). Furthermore, Pitx2 transcription factor modulates wnt signaling and miRNAs (miR-1, miR-29, and miR-200) which, in turn leads to Ca+2 remodeling (48). Other signals such as Notch, sonic hedgehog (SHH), Hand1/2, and FGF are involved in the downstream activation of genes encoding transcriptional activators (45, 49, 50). Thereby, inducing cardiac progenitor migration and setting up of primary and secondary heart fields. Transcription factor T-box 1(TBX1), centrally regulates the secondary heart field, by controlling the cardiac outflow tract development. NKX2−5 (NK2 homeobox 5) and GATA4 are central transcription factors in the primary and secondary heart fields. NKX2−5 and Isl1 contribute to heart tubes, chambers, and nodes formation (51–54). These transcription factors are also cell type-specific, such as transcription factor T-box 5 (TBX5) is solely expressed in the primary heart field. Similarly, Isl1 is only expressed in the secondary heart field.
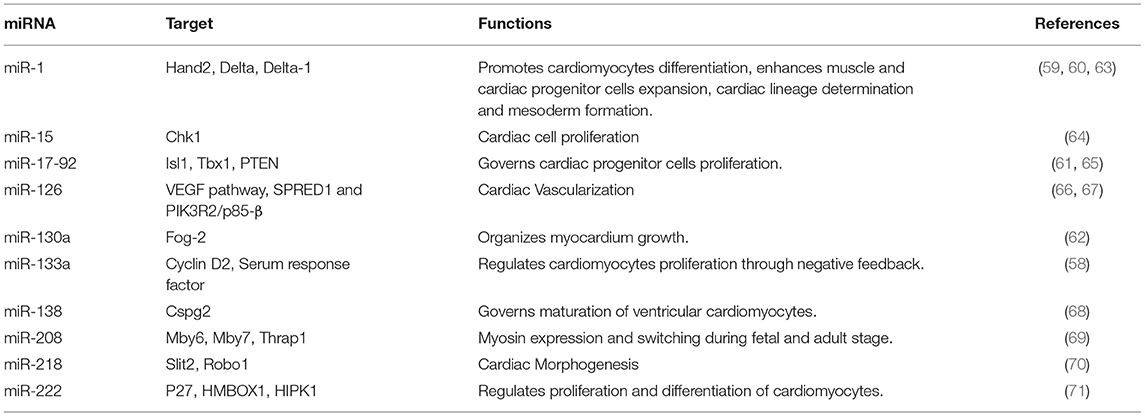
miRNA machinery is co-opted to repress inhibitors of cardiac differentiation, thus increasing the expression of cardiac specific-genes during cardiogenesis. Cardiac tissue-specific Dicer knockout ceased the cardiac development. Also, acquired pathological abnormalities like fibrosis, biventricular enlargement, hypertrophy, and heart failure (55). Several cardiac-specific miRNAs like miR-133 and miR-1, suppress ectoderm and endoderm lineage expression and mesoderm formation (56, 57). Furthermore, miR-133 acts on cyclin D2 and serum response factors to aid cardiomyocytes proliferation (58, 59). miR-1 inhibits Delta-1 from embryonic stem cells and promotes differentiation into cardiac lineage cells (59). miR-1 targets the miRNA response element (MRE) in the 3′ UTR region of the Hand2 and Irx5, which has been shown to have a key effect on cardiac development (60). Cardiac progenitors in the second heart field express the BMP signals that co-ordinate with miR-17−92 and repress the progenitor gene expression of Isl1 and Tbx1. Thereby, promoting myocardial lineage differentiation toward the cardiac outflow tract (61). Furthermore, miR-130a targets Friend-of-GATA 2 (FOG2) and regulates cardiac development in mice (62). Other miRNAs having a vital role in cardiac development are enlisted in Table 1. Currently, there is dearth of knowledge about the miRNA involved in human cardiac development. Although studies have shown the involvement of miRNAs in heart chamber, outflow tract, and septum development (72). Our current understanding is particularly reliant on in vitro human pluripotent stem cell differentiation into cardiac lineage cells like cardiomyocytes, cardiac progenitors, smooth muscle cells, and endothelial cells (73). miR-1 promotes differentiation to cardiomyocytes and repress endothelial cell fate by inhibiting WNT and FGF signaling pathways via. suppressing the expression of FZD7 and FRS2, respectively (74). miR-1 also represses gene expression of Hand2, KCNJ, HDAC4, GJA1 that are crucial for cardiac function and development (75). miR-499 through SOX6 inhibition governs the differentiation of cardiac progenitor cells to cardiomyocytes. Other cardiac-specific miRNAs like miR-669, miR-204, and miR-23 are shown to enhance cardiac progenitor differentiation, however, their mechanism needs to be studied (76, 77). Therefore, precise and orchestrated expression of miRNA is viral for proper morphological and electrophysiological development of the heart.
miRNA and Cardiac Proliferation
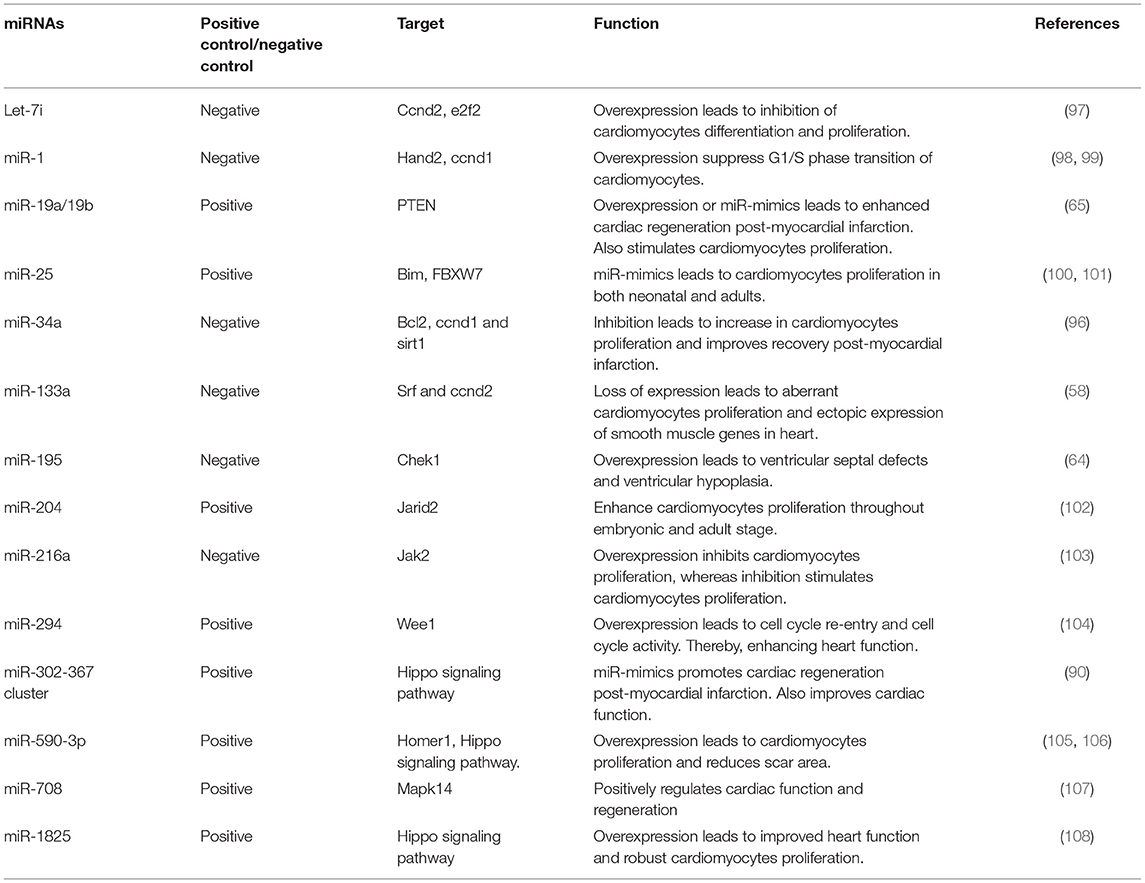
Cardiomyocytes are the most abundant cell mass in the heart but are outnumbered by the total of endothelial cells, fibroblast, smooth muscle cells, and inflammatory cells (78). This group of cells forms a microenvironment that affects cardiac cells proliferation by paracrine signaling of the secreted molecules like growth factors, cyclins, cyclin-dependent kinase (CDKs), and cytokines and thus, are vital for cardiac repair and cell cycle reentry (79–83). Overexpression of cell cycle activators like SV40 T antigen, cyclinD, cyclinA2 promotes proliferation and dedifferentiation of post-mitotic cardiomyocytes (84–87). miR-195 has been shown to be overexpressed in cardiomyocytes, leading to cell cycle arrest at the G2 phase. miR-195 targets checkpoint kinase 1 (Chek1), which encodes protein kinase promoting mitotic progression and G2-M transition (64). In mice, overexpression has also led to ventricular septal defects and hypoplasia like defects, and post-myocardial infarction lead heart regeneration (64). On the other hand, anti-miRNAs having a similar seed sequence as miR-195 showed increased cardiomyocytes in the mitotic phase. The mice treated with anti-miR-195 showed improved contractile function after ischemic reperfusion injury (88). miR-199a-3p and miR-590-3p target mRNA of HOP homeobox (HOPX), its protein product suppresses cardiomyocytes proliferation. Both miRNAs also target mRNA HOMER1, whose protein product alters Ca+2 signaling by interacting with the ryanodine receptor (RyR). Furthermore, miR-590-3p targets chloride intracellular channel 5 (CLIC5), which is a cell proliferation inhibitor. In neonatal mice, miR-199a-3p and miR-590-3p expression in cardiomyocytes improved cardiac function and reduced fibrotic scar formation after cardiomyocytes loss following injury. miR-302-367 cluster promotes cardiomyocytes mitosis by either targeting the Hippo pathway or by upregulating the expression of homeodomain transcription factor Pitx2. Pitx2 in turn activates gene expression of ROS scavengers, Nrf2, and electron transport chain to promote cardiomyocytes mitosis (89). Mechanistically miR-302-367 targets 3′ untranslated region (3′ UTR) of LATS2 and Mst1, which are key components of Hippo signaling and regulators of cardiomyocytes proliferation (90, 91). However, the prolonged expression of miR-302-367 leads to a decrease in cardiac function and cardiomyocytes dedifferentiation (90). miR-128 deletion induce cell cycle re-entry by upregulating chromatin regulator SUZ12 (Polycomb protein SUZ12), which in turn activates CDK2 and cyclin E and repress p27 (92).
miR-214, miR-17-92 cluster, miR-222 cluster have also been shown to enhance in vivo cardiac repair (65, 71, 93, 94). Furthermore, cardiac-specific overexpression of miR-17-92 cluster leads to increased cardiomyocytes proliferation, heart weight, and hyperplasia without cardiac hypertrophy (65). In vitro studies showed miR-17-92 cluster induced downregulation of PTEN expression leads to cardiomyocytes proliferation. Anti-miR-34a in adult mice has been shown to reduce fibrosis and necrosis post-myocardial infarction additionally boosts cardiac regeneration and myocardial function (95, 96). miR-34a acts on cyclin D1, Bcl2, Sirt1, and PPP1R10 that by cardiomyocytes cell cycle arrest, apoptosis, DNA damage response, and telomerase shortening to improve cardiac function. Other several miRNAs that promote/repress cardiomyocytes proliferation and heart regeneration are listed in Table 2. These findings demonstrate the therapeutic potential of miRNAs in increasing cardiomyocytes proliferation, improving cardiac function, cell-cycle reentry, and decreasing scar formation in injured heart tissue.
miRNA in Cardiovascular Disorders
miRNAs disturbance associated with cardiac development and progenitors leads to structural abnormalities of the heart. Several miRNAs associated with cardiogenesis and cardiac progenitors showed dysregulation in congenital heart defects. Whereas, adult heart-specific miRNAs are co-related with cardiovascular pathologies like myocardial infarction, ventricular hypertrophy, and arrhythmias (109–111). miRNAs are also being investigated as a diagnostic biomarker to detect cardiac diseases and understand disease pathology (112). Defects in cardiomyocytes proliferation and migration pathways are associated with congenital heart disease. miRNA transcriptome shows distinct expression of miRNAs in different pathological conditions, suggesting dysregulation of the pathways. Several studies depicts disease specific distinct miRNAs expression. For instance, low expression of miR-30b, miR-103, miR-142-3p, miR-342-3p is observed in heart failure patients as compared to chronic pulmonary obstructive disease (113). Further several studies stated distinguishing cardiac pathologies like dilated cardiomyopathies, ischemic cardiomyopathy, and aortic stenosis based on miRNAs levels. Ikeda et al. showed 43 differentially expressed miRNAs, of which at least one miRNA was distinct to a specific disease group (114). mRNA expression profile when compared with miRNA expression could provide a promising diagnostic tool to differentiate diverse cardiac pathologies. Cardiac disease-specific miRNAs identification also provides a platform for a new drug target which, can be harnessed to treat currently incurable heart disease. A list of miRNAs, their expression profile in cardiomyocytes, along with their targets, and associated cardiovascular diseases are listed in Table 3.
In mice, overexpression of miR-21 has shown to reduce apoptosis and myocardial infarction by 36.9%. miR-21 also enhances cardiomyocytes proliferation and viability post-myocardial infarction by targeting PTEN and the Akt pathway (128). Circulating miRNAs are widely studied to harness the potential of biomarkers as a risk assessment tool for heart failure. About 30 differentially expressed miRNA has been identified for heart failure, miR-199a, miR-27a, miR-26b, miR-18a, miR-652, miR-30, and miR-106a are significantly less expressed in patients with heart failure. Also, miR-1 is downregulated and miR-21 is upregulated in patients with symptomatic HF (129). miR-210 levels are directly correlated with the severity of heart disease. Inversely, patients recovering from heart failure show a decrease in miR-210 levels (130). Furthermore, miR-423 negatively correlates while, miR-208b, miR-499, miR-1306, and miR-1254 positively correlate with the mortality of heart failure patients (127, 131–133).
Plasma levels of miR-21 are significantly higher in patients with acute myocardial infarction (AMI). Additionally, miR-21 levels are correlated with creatine kinase and cardiac troponin I (cTnI) that are traditional biomarkers for AMI (111, 134). Similarly, miR-208 upregulation was observed in patients with AMI and had a high mortality rate within 6 months (135, 136). Post-cardiomyocyte injury, the high levels of miR-499 are highly associated with cTnI in patients with AMI (137). The combination of miR-1, miR-208, and miR-499 has significantly higher predictive value for AMI patients as compared to conventional biomarkers (138).
miR-150 levels were observed to be reduced by 3.2 folds in atrial fibrillation patients, affecting pathways associated with fibrosis, platelet function, and inflammation (121, 139). Dysregulation of miR-29 and miR-208b is observed in patients with atrial fibrillation (AF). Their downstream targets involve genes implicated in apoptosis and cardiac fibrosis (140–142). Also, miR-328 controls atrial remodeling by acting on Ca+2 channel protein expression, specifically L-type Ca+2 channels α1c (CACNA1C) and β1 (CACNB2) subunit (123, 143). Similarly, upregulation of miR-21 in atria affects the mitogen-activated protein kinase pathway. Thereby, governing the level of cardiac hypertrophy and interstitial fibrosis (144).
Clinical Application Potential of miRNAs
miRNAs by the virtue of their small size, ability to control the expression of various mRNA, and relatively pleiotropic effects have emerged as a promising therapeutic candidate to treat cardiac diseases. Besides post-translational silencing by miRNAs, gene expression can be altered by other methods like epigenetic modification, inhibition or degradation of translated protein, decreasing transcription factor levels, and entire gene knockout (145–149). However, drugs inhibiting and degrading transcription factors or translated protein are small molecules, which require large-scale screening to check the efficacy (150). Additionally, certain protein targets are non-druggable, i.e., without small molecule interaction specific binding pockets (151). Targeted gene mutagenesis with CRISPR system is into clinical trials but the field is very naive and requires supportive data (152). Small molecules used for epigenetic modification of DNA are typically unspecific in their interaction and tend to cause global modification of the target cell's genomic DNA. miRNAs are versatile and provide a transient control over gene expression. Since the last decade, several cardiac specific miRNAs have been elucidated. Once miRNAs have been identified, complementary oligonucleotide sequence synthesis is trifling to develop drugs for treatment. Currently, miRNA based drug substances are based on locked nucleic acid (LNA) to develop antisense oligonucleotide for target mRNA sequence, thereby silencing target gene (153). A large number of miRNAs regulatory target predicting databases have emerged in the last decade (154, 155). Several independent algorithms like TargetScan that predict miRNA binding sites based on seed regions, that are critical for protein-coding mRNA binding and its associated biological networks are developed. Other databases like Kegg and Ingenuity pathway analysis are being used for disease state and putative biological pathways. Furthermore, in silico programs that estimate free energy between RNA sequences, lower free energy suggests stable and strong binding (156, 157).
miRNA based therapeutics can be beneficial by inducing differentiation, proliferation, and migration of terminally differentiated cardiomyocytes. Reversal of myocardium undergoing mitotic arrest toward proliferative progenitor cells can enable tissue repair and repopulation. MGN-1374 is an anti-miR of the miR-15 family seed region. It stimulates cardiomyocyte proliferation of post-myocardial infarction heart (158, 159). Cardiac specific miRNA like miR-208 has been shown to be critical for heart failure as it prevents myosin switching and improves cardiac function by targeting MED13 (Metabolic regulator and insulin sensor). Anti-miR-208 potential as a therapeutic option is being investigated for heart failure (159). Wills tumor gene 1 (Wt1) expression in the epicardial layer promotes proliferation, differentiation into cardiac lineage cells, and neovascularization (160–162). Post-myocardial infarction, Wt1+ epicardial cells undergoes epithelial to mesenchymal transition (EMT) to enable cardiac repair (119). Let-7 miRNA expression inhibition has been linked with increased expression of EMT-related genes in epicardial cells (163). Thus, can be a promising therapeutic candidate to stimulate cardiac regeneration by providing a proliferative niche in the infarcted heart.
Targeted delivery of miRNAs to the cardiac tissue needs to overcome certain barriers such as nuclease degradation, endothelial barrier, cell membrane transfer, and minimal toxicity. Certain delivery systems like viral lipid based delivery systems are under validation. Viral-based delivery, particularly with Adeno-Associated Virus (AAV) is studied extensively. AAV9 based delivery of hsa-miR-590 and has-miR-199 showed stable expression with increased cardiomyocytes proliferation in mice heart (106). Similarly, lipid based delivery of miR-199 was able to activate cardiomyocyte proliferation and provide robust cardiac regeneration in mice (164). Both the delivery system has some limitations that need to be addressed before their clinical application. For instance, the viral delivery system prohibits single mature miRNA specific expression and the long term persistence of viral vector in transduced cardiomyocytes leads to compounding of overall therapeutic effect. Thus, leads to several unwanted downstream consequences (23, 90). On the other hand, lipid based system particle size is comparatively larger (>1 μm) that has been linked to severe toxicity and inflammation in animal studies (165). Exosomes and biocompatible injectable hydrogels-based delivery of miRNAs in vivo have shown efficient transduction in cardiomyocytes (108, 166, 167). Taking into account that most of our current understanding is based on small animals in which miRNA delivery is quite easy as well as their cardiomyocyte biology and cardiac physiology are markedly distinct from humans. Additional preclinical studies elucidating the appropriate treatment dosage, location, and duration in models, which represent human physiology are warranted to enable the clinical application. Although no miRNA based drug candidate has entered clinical trial phase 3 in clinicaltrials.gov database, there are several clinical trials in early phase trials. Clinical trial for LNA-modified antisense oligonucleotide (MRG 110) that antagonize miR-92 is under study for heart failure (Clinicaltrials.gov). Overall, these studies and reports indicate a promising future of miRNA-based therapeutics for cardiac diseases.
Conclusion and Future Perspective
The end goal of restoring ravaged heart is complete heart regeneration through cardiomyocytes renewal, scar reduction, and neovascularization. Cardiac regeneration is governed by a network of complex and strictly controlled processes. Studies pertaining to the role of the regulatory network and signaling pathway critical for cardiac development have resulted in novel strategies to induce cardiac repair and regeneration. Central to this development is the miRNAs based gene regulation, which has heralded next-generation in situ regenerative therapies for the heart. miRNA inhibitors and mimics are easily synthesized and delivered by viral and non-viral transfection methods in small animals. Current strategies for cardiomyocytes proliferation are vastly inefficient and have been primarily tested in rodents. Therefore, preclinical trials on large animals, organoids are vital to demonstrate safety and efficacy of the therapeutic strategies. Also, with the advent of single-cell sequencing techniques, characterization of cell type specific function and expression of miRNAs will further enhance our understanding. In near future, the combination of developmental regulatory mechanism and cellular transplantation along with artificial matrices and decellularized tissue scaffolds can drive toward successful adult heart repair and regeneration.
Author Contributions
VS conceptualized, formal analysis, writing—original draft, and writing—review and editing. JS conceptualized, supervision, and writing—review and editing. Both authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Roth GA, Abate D, Abate KH, Abay SM, Abbafati C, Abbasi N, et al. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980−2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. (2018) 392:1736–88. doi: 10.1016/S0140-6736(18)32203-7
2. Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. (2009) 358:1370–80. doi: 10.1056/NEJMra072139
3. Richardson WJ, Clarke SA, Alexander Quinn T, Holmes JW. Physiological implications of myocardial scar structure. Compr Physiol. (2015) 5:1877–909. doi: 10.1002/cphy.c140067
4. Ptaszek LM, Mansour M, Ruskin JN, Chien KR. Towards regenerative therapy for cardiac disease. Lancet. (2012) 379:933–42. doi: 10.1016/S0140-6736(12)60075-0
5. Passier R, Van Laake LW, Mummery CL. Stem-cell-based therapy and lessons from the heart. Nature. (2008) 453:322–9. doi: 10.1038/nature07040
6. Wollert KC, Drexler H. Cell therapy for the treatment of coronary heart disease: a critical appraisal. Nat Rev Cardiol. (2010) 7:204–15. doi: 10.1038/nrcardio.2010.1
8. Segers VFM, Lee RT. Stem-cell therapy for cardiac disease. Nature. (2008) 451:937–42. doi: 10.1038/nature06800
9. Vujic A, Natarajan N, Lee RT. Molecular mechanisms of heart regeneration. Semin Cell Dev Biol. (2020) 100:20–8. doi: 10.1016/j.semcdb.2019.09.003
10. Mahmoud AI, Porrello ER, Kimura W, Olson EN, Sadek HA. Surgical models for cardiac regeneration in neonatal mice. Nat Protoc. (2014) 9:305–11. doi: 10.1038/nprot.2014.021
11. Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, et al. Transient regenerative potential of the neonatal mouse heart. Science. (2011) 331:1078–80. doi: 10.1126/science.1200708
12. Hirose K, Payumo AY, Cutie S, Hoang A, Zhang H, Guyot R, et al. Evidence for hormonal control of heart regenerative capacity during endothermy acquisition. Science. (2019) 364:184–8. doi: 10.1126/science.aar2038
13. Singh BN, Koyano-Nakagawa N, Gong W, Moskowitz IP, Weaver C V, Braunlin E, et al. A conserved HH-Gli1-Mycn network regulates heart regeneration from newt to human. Nat Commun. (2018) 9:1–18. doi: 10.1038/s41467-018-06617-z
14. Campa VM, Gutiérrez-Lanza R, Cerignoli F, Díaz-Trelles R, Nelson B, Tsuji T, et al. Notch activates cell cycle reentry and progression in quiescent cardiomyocytes. J Cell Biol. (2008) 183:129–41. doi: 10.1083/jcb.200806104
15. Gemberling M, Karra R, Dickson AL, Poss KD. Nrg1 is an injury-induced cardiomyocyte mitogen for the endogenous heart regeneration program in zebrafish. Elife. (2015) 4:e05871. doi: 10.7554/eLife.05871
16. Bersell K, Arab S, Haring B, Kühn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. (2009) 138:257–70. doi: 10.1016/j.cell.2009.04.060
17. Xiao Y, Leach J, Wang J, Martin JF. Hippo/Yap signaling in cardiac development and regeneration. Curr Treat Options Cardiovasc Med. (2016) 18:38. doi: 10.1007/s11936-016-0461-y
18. Ikeda S, Sadoshima J. Regulation of myocardial cell growth and death by the hippo pathway. Circ J. (2016) 80:CJ-16-0476. doi: 10.1253/circj.CJ-16-0476
19. Aguirre A, Sancho-Martinez I, Izpisua Belmonte JC. Reprogramming toward heart regeneration: stem cells and beyond. Cell Stem Cell. (2013) 12:275–84. doi: 10.1016/j.stem.2013.02.008
20. Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. (2012) 485:599–604. doi: 10.1038/nature11139
21. Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. (2012) 485:593–8. doi: 10.1038/nature11044
22. Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. (2010) 142:375–86. doi: 10.1016/j.cell.2010.07.002
23. Braga L, Ali H, Secco I, Giacca M. Non-coding RNA therapeutics for cardiac regeneration. Cardiovasc Res. (2021) 117:674–93. doi: 10.1093/cvr/cvaa071
24. O'Brien JE, Kibiryeva N, Zhou XG, Marshall JA, Lofland GK, Artman M, et al. Noncoding RNA expression in myocardium from infants with tetralogy of fallot. Circ Cardiovasc Genet. (2012) 5:279–86. doi: 10.1161/CIRCGENETICS.111.961474
25. Shah V, Shah J. Recent trends in targeting miRNAs for cancer therapy. J Pharm Pharmacol. (2020) 72:1732–49. doi: 10.1111/jphp.13351
26. Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. (2002) 16:2733–42. doi: 10.1101/gad.1026102
27. Choi YJ, Lin CP, Risso D, Chen S, Kim TA, Tan MH, et al. Deficiency of microRNA miR-34a expands cell fate potential in pluripotent stem cells. Science. (2017) 355:eaag1927. doi: 10.1126/science.aag1927
28. Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell–specific microRNAs promote induced pluripotency. Nat Biotechnol. (2009) 27:459–61. doi: 10.1038/nbt.1535
29. Li MA, He L. microRNAs as novel regulators of stem cell pluripotency and somatic cell reprogramming. BioEssays. (2012) 34:670–80. doi: 10.1002/bies.201200019
30. Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, et al. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. (2011) 29:443–8. doi: 10.1038/nbt.1862
31. Heinrich EM, Dimmeler S. MicroRNAs and stem cells: control of pluripotency, reprogramming, and lineage commitment. Circ Res. (2012) 110:1014–22. doi: 10.1161/CIRCRESAHA.111.243394
32. Gregor T, Tank DW, Wieschaus EF, Bialek W. Probing the limits to positional information. Cell. (2007) 130:153–64. doi: 10.1016/j.cell.2007.05.025
33. Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of nodal agonist and antagonist by miR-430. Science. (2007) 318:271–4. doi: 10.1126/science.1147535
34. Boldajipour B, Mahabaleshwar H, Kardash E, Reichman-Fried M, Blaser H, Minina S, et al. Control of chemokine-guided cell migration by ligand sequestration. Cell. (2008) 132:463–73. doi: 10.1016/j.cell.2007.12.034
35. Staton AA, Knaut H, Giraldez AJ. miRNA regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration. Nat Genet. (2011) 43:204–11. doi: 10.1038/ng.758
36. Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. (2011) 476:228–31. doi: 10.1038/nature10323
37. Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Alan Payne J, Pandya K, et al. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. (2012) 110:1465–73. doi: 10.1161/CIRCRESAHA.112.269035
38. Ivey KN, Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. (2010) 7:36–41. doi: 10.1016/j.stem.2010.06.012
39. Garcia-Martinez V, Schoenwolf GC. Primitive-streak origin of the cardiovascular system in avian embryos. Dev Biol. (1993) 159:706–19. doi: 10.1006/dbio.1993.1276
40. Tam PPL, Parameswaran M, Kinder SJ, Weinberger RP. The allocation of epiblast cells to the embryonic heart and other mesodermal lineages: the role of ingression and tissue movement during gastrulation. Development. (1997) 124:1631–42. doi: 10.1242/dev.124.9.1631
41. Bruneau BG. Signaling and transcriptional networks in heart development and regeneration. Cold Spring Harb Perspect Biol. (2013) 5:a008292. doi: 10.1101/cshperspect.a008292
42. Garry DJ, Olson EN. A common progenitor at the heart of development. Cell. (2006) 127:1101–4. doi: 10.1016/j.cell.2006.11.031
43. Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. (2006) 126:1037–48. doi: 10.1016/j.cell.2006.09.003
44. Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. (2006) 313:1922–7. doi: 10.1126/science.1132292
45. Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. (2005) 6:826–35. doi: 10.1038/nrg1710
46. Pang JKS, Phua QH, Soh BS. Applications of miRNAs in cardiac development, disease progression and regeneration. Stem Cell Res Ther. (2019) 10:1–11. doi: 10.1186/s13287-019-1451-2
47. Xin M, Olson EN, Bassel-Duby R. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat Rev Mol Cell Biol. (2013) 14:529–41. doi: 10.1038/nrm3619
48. Lozano-Velasco E, Wangensteen R, Quesada A, Garcia-Padilla C, Osorio JA, Ruiz-Torres MD, et al. Hyperthyroidism, but not hypertension, impairs PITX2 expression leading to Wnt-microRNA-ion channel remodeling. PLoS ONE. (2017) 12:e0188473. doi: 10.1371/journal.pone.0188473
49. Bondue A, Lapouge G, Paulissen C, Semeraro C, Iacovino M, Kyba M, et al. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. (2008) 3:69–84. doi: 10.1016/j.stem.2008.06.009
50. Saga Y, Miyagawa-Tomita S, Takagi A, Kitajima S, Miyazaki JI, Inoue T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. (1999) 126:3437–47. doi: 10.1242/dev.126.15.3437
51. Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. (1999) 126:1269–80. doi: 10.1242/dev.126.6.1269
52. Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. (2003) 5:877–89. doi: 10.1016/S1534-5807(03)00363-0
53. Plageman TF, Yutzey KE. T-box genes and heart development: putting the “T” in heart. Dev Dyn. (2005) 232:11–20. doi: 10.1002/dvdy.20201
54. Habets PEMH, Moorman AFM, Clout DEW, Van Roon MA, Lingbeek M, Van Lohuizen M, et al. Cooperative action of Tbx2 and Nkx2.5 inhibits ANF expression in the atrioventricular canal: implications for cardiac chamber formation. Genes Dev. (2002) 16:1234–46. doi: 10.1101/gad.222902
55. Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci USA. (2008) 105:2111–6. doi: 10.1073/pnas.0710228105
56. Sayed D, Abdellatif M. Micrornas in development and disease. Physiol Rev. (2011) 91:827–87. doi: 10.1152/physrev.00006.2010
57. Boettger T, Braun T. A new level of complexity: the role of microRNAs in cardiovascular development. Circ Res. (2012) 110:1000–13. doi: 10.1161/CIRCRESAHA.111.247742
58. Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. (2008) 22:3242–54. doi: 10.1101/gad.1738708
59. Ivey KN, Muth A, Arnold J, King FW, Yeh RF, Fish JE, et al. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. (2008) 2:219–29. doi: 10.1016/j.stem.2008.01.016
60. Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. (2005) 436:214–20. doi: 10.1038/nature03817
61. Wang J, Greene SB, Bonilla-Claudio M, Tao Y, Zhang J, Bai Y, et al. Bmp signaling regulates myocardial differentiation from cardiac progenitors through a MicroRNA-mediated mechanism. Dev Cell. (2010) 19:903–12. doi: 10.1016/j.devcel.2010.10.022
62. Kim GH, Samant SA, Earley JU, Svensson EC. Translational control of FOG-2 expression in cardiomyocytes by microRNA-130a. PLoS ONE. (2009) 4:e6161. doi: 10.1371/journal.pone.0006161
63. Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci. (2005) 102:18986–91. doi: 10.1073/pnas.0509535102
64. Porrello ER, Johnson BA, Aurora AB, Simpson E, Nam YJ, Matkovich SJ, et al. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ Res. (2011) 109:670–9. doi: 10.1161/CIRCRESAHA.111.248880
65. Chen J, Huang ZP, Seok HY, Ding J, Kataoka M, Zhang Z, et al. Mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ Res. (2013) 112:1557–66. doi: 10.1161/CIRCRESAHA.112.300658
66. Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. (2008) 15:261–71. doi: 10.1016/j.devcel.2008.07.002
67. Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. (2008) 15:272–84. doi: 10.1016/j.devcel.2008.07.008
68. Morton SU, Scherz PJ, Cordes KR, Ivey KN, Stainier DYR, Srivastava D. microRNA-138 modulates cardiac patterning during embryonic development. Proc Natl Acad Sci USA. (2008) 105:17830–5. doi: 10.1073/pnas.0804673105
69. Callis TE, Pandya K, Hee YS, Tang RH, Tatsuguchi M, Huang ZP, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest. (2009) 119:2772–86. doi: 10.1172/JCI36154
70. Fish JE, Wythe JD, Xiao T, Bruneau BG, Stainier DYR, Srivastava D, et al. A Slit/miR-218/Robo regulatory loop is required during heart tube formation in zebrafish. Development. (2011) 138:1409–19. doi: 10.1242/dev.060046
71. Liu X, Xiao J, Zhu H, Wei X, Platt C, Damilano F, et al. miR-222 is necessary for exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell Metab. (2015) 21:584–95. doi: 10.1016/j.cmet.2015.02.014
72. Zhou J, Dong X, Zhou Q, Wang H, Qian Y, Tian W, et al. MicroRNA expression profiling of heart tissue during fetal development. Int J Mol Med. (2014) 33:1250–60. doi: 10.3892/ijmm.2014.1691
73. Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc. (2012) 8:162–75. doi: 10.1038/nprot.2012.150
74. Li J, Cao Y, Ma XJ, Wang HJ, Zhang J, Luo X, et al. Roles of miR-1-1 and miR-181c in ventricular septal defects. Int J Cardiol. (2013) 168:1441–6. doi: 10.1016/j.ijcard.2012.12.048
75. Sluijter JPG, Van Mil A, Van Vliet P, Metz CHG, Liu J, Doevendans PA, et al. MicroRNA-1 and-499 regulate differentiation and proliferation in human-derived cardiomyocyte progenitor cells. Arterioscler Thromb Vasc Biol. (2010) 30:859–68. doi: 10.1161/ATVBAHA.109.197434
76. Xiao J, Liang D, Zhang H, Liu Y, Zhang D, Liu Y, et al. MicroRNA-204 is required for differentiation of human-derived cardiomyocyte progenitor cells. J Mol Cell Cardiol. (2012) 53:751–9. doi: 10.1016/j.yjmcc.2012.08.024
77. Crippa S, Cassano M, Messina G, Galli D, Galvez BG, Curk T, et al. miR669a and miR669q prevent skeletal muscle differentiation in postnatal cardiac progenitors. J Cell Biol. (2011) 193:1197–212. doi: 10.1083/jcb.201011099
78. Skelly DA, Squiers GT, McLellan MA, Bolisetty MT, Robson P, Rosenthal NA, et al. Single-cell transcriptional profiling reveals cellular diversity and intercommunication in the mouse heart. Cell Rep. (2018) 22:600–10. doi: 10.1016/j.celrep.2017.12.072
79. Andreadou I, CA16225 on behalf of the E-CCA, Cabrera-Fuentes HA, CA16225 on behalf of the E-CCA, Devaux Y, CA16225 on behalf of the E-CCA, et al. Immune cells as targets for cardioprotection: new players and novel therapeutic opportunities. Cardiovasc Res. (2019) 115:1117–30. doi: 10.1093/cvr/cvz050
80. Wodsedalek DJ, Paddock SJ, Wan TC, Auchampach JA, Kenarsary A, Tsaih SW, et al. Il-13 promotes in vivo neonatal cardiomyocyte cell cycle activity and heart regeneration. Am J Physiol Hear Circ Physiol. (2019) 316:H24–34. doi: 10.1152/ajpheart.00521.2018
81. Han C, Nie Y, Lian H, Liu R, He F, Huang H, et al. Acute inflammation stimulates a regenerative response in the neonatal mouse heart. Cell Res. (2015) 25:1137–51. doi: 10.1038/cr.2015.110
82. Pasumarthi KBS, Field LJ. Cardiomyocyte cell cycle regulation. Circ Res. (2002) 90:1044–54. doi: 10.1161/01.RES.0000020201.44772.67
83. Ahuja P, Sdek P, MacLellan WR. Cardiac myocyte cell cycle control in development, disease, and regeneration. Physiol Rev. (2007) 87:521–44. doi: 10.1152/physrev.00032.2006
84. Katz EB, Steinhelper ME, Delcarpio JB, Daud AI, Claycomb WC, Field LJ. Cardiomyocyte proliferation in mice expressing alpha-cardiac myosin heavy chain-SV40 T-antigen transgenes. Am J Physiol. (1992) 262:H1867–76. doi: 10.1152/ajpheart.1992.262.6.H1867
85. Chaudhry HW, Dashoush NH, Tang H, Zhang L, Wan X, Wu EX, et al. Cyclin A2 mediates cardiomyocyte mitosis in the postmitotic myocardium. J Biol Chem. (2004) 279:35858–66. doi: 10.1074/jbc.M404975200
86. Engel FB, Schebesta M, Duong MT, Lu G, Ren S, Madwed JB, et al. p38 MAP kinase inhibition enables proliferation of adult mammalian cardiomyocytes. Genes Dev. (2005) 19:1175–87. doi: 10.1101/gad.1306705
87. Kühn B, Del Monte F, Hajjar RJ, Chang YS, Lebeche D, Arab S, et al. Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Nat Med. (2007) 13:962–9. doi: 10.1038/nm1619
88. Hullinger TG, Montgomery RL, Seto AG, Dickinson BA, Semus HM, Lynch JM, et al. Inhibition of miR-15 protects against cardiac ischemic injury. Circ Res. (2012) 110:71–81. doi: 10.1161/CIRCRESAHA.111.244442
89. Tao G, Kahr PC, Morikawa Y, Zhang M, Rahmani M, Heallen TR, et al. Pitx2 promotes heart repair by activating the antioxidant response after cardiac injury. Nature. (2016) 534:119–23. doi: 10.1038/nature17959
90. Tian Y, Liu Y, Wang T, Zhou N, Kong J, Chen L, et al. A microRNA-Hippo pathway that promotes cardiomyocyte proliferation and cardiac regeneration in mice. Sci Transl Med. (2015) 7:279ra38. doi: 10.1126/scitranslmed.3010841
91. Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, et al. Hippo pathway inhibits wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. (2011) 332:458–61. doi: 10.1126/science.1199010
92. Huang W, Feng Y, Liang J, Yu H, Wang C, Wang B, et al. Loss of microRNA-128 promotes cardiomyocyte proliferation and heart regeneration. Nat Commun. (2018) 9:1–16. doi: 10.1038/s41467-018-03019-z
93. Aurora AB, Mahmoud AI, Luo X, Johnson BA, Van Rooij E, Matsuzaki S, et al. MicroRNA-214 protects the mouse heart from ischemic injury by controlling Ca2+ overload and cell death. J Clin Invest. (2012) 122:1222–32. doi: 10.1172/JCI59327
94. Vujic A, Lerchenmüller C, Wu T Di, Guillermier C, Rabolli CP, Gonzalez E, et al. Exercise induces new cardiomyocyte generation in the adult mammalian heart. Nat Commun. (2018) 9:1–9. doi: 10.1038/s41467-018-04083-1
95. Boon RA, Iekushi K, Lechner S, Seeger T, Fischer A, Heydt S, et al. MicroRNA-34a regulates cardiac ageing and function. Nature. (2013) 495:107–10. doi: 10.1038/nature11919
96. Yang Y, Cheng HW, Qiu Y, Dupee D, Noonan M, Lin YD, et al. MicroRNA-34a plays a key role in cardiac repair and regeneration following myocardial infarction. Circ Res. (2015) 117:450–9. doi: 10.1161/CIRCRESAHA.117.305962
97. Hu Y, Jin G, Li B, Chen Y, Zhong L, Chen G, et al. Suppression of miRNA let-7i-5p promotes cardiomyocyte proliferation and repairs heart function post injury by targetting CCND2 and E2F2. Clin Sci. (2019) 133:425–41. doi: 10.1042/CS20181002
98. Wei Y, Peng S, Wu M, Sachidanandam R, Tu Z, Zhang S, et al. Multifaceted roles of miR-1s in repressing the fetal gene program in the heart. Cell Res. (2014) 24:278–92. doi: 10.1038/cr.2014.12
99. Gan J, Tang FMK, Su X, Lu G, Xu J, Lee HSS, et al. MicroRNA-1 inhibits cardiomyocyte proliferation in mouse neonatal hearts by repressing CCND1 expression. Ann Transl Med. (2019) 7:455. doi: 10.21037/atm.2019.08.68
100. Wang B, Xu M, Li M, Wu F, Hu S, Chen X, et al. miR-25 promotes cardiomyocyte proliferation by targeting FBXW7. Mol Ther Nucleic Acids. (2020) 19:1299–308. doi: 10.1016/j.omtn.2020.01.013
101. Qin X, Gao S, Yang Y, Wu L, Wang L. microRNA-25 promotes cardiomyocytes proliferation and migration via targeting Bim. J Cell Physiol. (2019) 234:22103–15. doi: 10.1002/jcp.28773
102. Liang D, Li J, Wu Y, Zhen L, Li C, Qi M, et al. miRNA-204 drives cardiomyocyte proliferation via targeting Jarid2. Int J Cardiol. (2015) 201:38–48. doi: 10.1016/j.ijcard.2015.06.163
103. Wang J, Chen X, Shen D, Ge D, Chen J, Pei J, et al. A long noncoding RNA NR_045363 controls cardiomyocyte proliferation and cardiac repair. J Mol Cell Cardiol. (2019) 127:105–14. doi: 10.1016/j.yjmcc.2018.12.005
104. Borden A, Kurian J, Nickoloff E, Yang Y, Troupes CD, Ibetti J, et al. Transient introduction of miR-294 in the heart promotes cardiomyocyte cell cycle reentry after injury. Circ Res. (2019) 125:14–25. doi: 10.1161/CIRCRESAHA.118.314223
105. Torrini C, Cubero RJ, Dirkx E, Braga L, Ali H, Prosdocimo G, et al. Common regulatory pathways mediate activity of microRNAs inducing cardiomyocyte proliferation. Cell Rep. (2019) 27:2759–71.e5. doi: 10.1016/j.celrep.2019.05.005
106. Eulalio A, Mano M, Ferro MD, Zentilin L, Sinagra G, Zacchigna S, et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. (2012) 492:376–81. doi: 10.1038/nature11739
107. Deng S, Zhao Q, Zhang C, Zhen L, Liu C, Wang G, et al. Neonatal heart-enriched miR-708 promotes proliferation and stress resistance of cardiomyocytes in rodents. Theranostics. (2017) 7:1953. doi: 10.7150/thno.16478
108. Pandey R, Velasquez S, Durrani S, Jiang M, Neiman M, Crocker JS, et al. MicroRNA-1825 induces proliferation of adult cardiomyocytes and promotes cardiac regeneration post ischemic injury. Am J Transl Res. (2017) 9:3120.
109. Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, Van Laake LW, et al. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. (2007) 116:258–67. doi: 10.1161/CIRCULATIONAHA.107.687947
110. Wang K, Lin ZQ, Long B, Li JH, Zhou J, Li PF. Cardiac hypertrophy is positively regulated by microRNA miR-23a *. J Biol Chem. (2012) 287:589–99. doi: 10.1074/jbc.M111.266940
111. Grabmaier U, Clauss S, Gross L, Klier I, Franz WM, Steinbeck G, et al. Diagnostic and prognostic value of miR-1 and miR-29b on adverse ventricular remodeling after acute myocardial infarction – the SITAGRAMI-miR analysis. Int J Cardiol. (2017) 244:30–6. doi: 10.1016/j.ijcard.2017.06.054
112. Dhingra R, Vasan RS. Biomarkers in cardiovascular disease: Statistical assessment and section on key novel heart failure biomarkers. Trends Cardiovasc Med. (2017) 27:123–33. doi: 10.1016/j.tcm.2016.07.005
113. Ellis KL, Cameron VA, Troughton RW, Frampton CM, Ellmers LJ, Richards AM. Circulating microRNAs as candidate markers to distinguish heart failure in breathless patients. Eur J Heart Fail. (2013) 15:1138–47. doi: 10.1093/eurjhf/hft078
114. Ikeda S, Kong SW, Lu J, Bisping E, Zhang H, Allen PD, et al. Altered microRNA expression in human heart disease. Physiol Genom. (2007) 31:367–73. doi: 10.1152/physiolgenomics.00144.2007
115. Caspi O, Lesman A, Basevitch Y, Gepstein A, Arbel G, Huber I, et al. Tissue engineering of vascularized cardiac muscle from human embryonic stem cells. Circ Res. (2007) 100:263–72. doi: 10.1161/01.RES.0000257776.05673.ff
116. Marfella R, Di Filippo C, Potenza N, Sardu C, Rizzo MR, Siniscalchi M, et al. Circulating microRNA changes in heart failure patients treated with cardiac resynchronization therapy: responders vs. non-responders. Eur J Heart Fail. (2013) 15:1277–88. doi: 10.1093/eurjhf/hft088
117. Jung SE, Kim SW, Jeong S, Moon H, Choi WS, Lim S, et al. MicroRNA-26a/b-5p promotes myocardial infarction-induced cell death by downregulating cytochrome c oxidase 5a. Exp Mol Med. (2021) 53:1332–43. doi: 10.1038/s12276-021-00665-0
118. Watson CJ, Gupta SK, O'Connell E, Thum S, Glezeva N, Fendrich J, et al. MicroRNA signatures differentiate preserved from reduced ejection fraction heart failure. Eur J Heart Fail. (2015) 17:405–15. doi: 10.1002/ejhf.244
119. Zhou B, Honor LB, He H, Qing M, Oh JH, Butterfield C, et al. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J Clin Invest. (2011) 121:1894–904. doi: 10.1172/JCI45529
120. Eryilmaz U, Akgüllü Ç, Beşer N, Yildiz Ö, Ömürlü IK, Bozdogan B. Circulating microRNAs in patients with ST-elevation myocardial infarction. Anatol J Cardiol. (2016) 16:392. doi: 10.5152/AnatolJCardiol.2015.6603
121. Goren Y, Meiri E, Hogan C, Mitchell H, Lebanony D, Salman N, et al. Relation of reduced expression of MiR-150 in platelets to atrial fibrillation in patients with chronic systolic heart failure. Am J Cardiol. (2014) 113:976–81. doi: 10.1016/j.amjcard.2013.11.060
122. Shen J, Xing W, Gong F, Wang W, Yan Y, Zhang Y, et al. MiR-150-5p retards the progression of myocardial fibrosis by targeting EGR1. Cell Cycle. (2019) 18:1335–48. doi: 10.1080/15384101.2019.1617614
123. Lu Y, Zhang Y, Wang N, Pan Z, Gao X, Zhang F, et al. MicroRNA-328 contributes to adverse electrical remodeling in atrial fibrillation. Circulation. (2010) 122:2378–87. doi: 10.1161/CIRCULATIONAHA.110.958967
124. Bittel DC, Kibiryeva N, Marshall JA, O'Brien JE. MicroRNA-421 dysregulation is associated with tetralogy of fallot. Cells. (2014) 3:713–23. doi: 10.3390/cells3030713
125. Harling L, Lambert J, Ashrafian H, Darzi A, Gooderham NJ, Athanasiou T. Elevated serum microRNA 483-5p levels may predict patients at risk of post-operative atrial fibrillation. Eur J Cardio Thoracic Surg. (2017) 51:73–8. doi: 10.1093/ejcts/ezw245
126. Gidlöf O, Smith JG, Miyazu K, Gilje P, Spencer A, Blomquist S, et al. Circulating cardio-enriched microRNAs are associated with long-term prognosis following myocardial infarction. BMC Cardiovasc Disord. (2013) 13:1–9. doi: 10.1186/1471-2261-13-12
127. Bayés-Genis A, Lanfear DE, de Ronde MWJ, Lupón J, Leenders JJ, Liu Z, et al. Prognostic value of circulating microRNAs on heart failure-related morbidity and mortality in two large diverse cohorts of general heart failure patients. Eur J Heart Fail. (2018) 20:67–75. doi: 10.1002/ejhf.984
128. Gu GL, Xu XL, Sun XT, Zhang J, Guo CF, Wang CS, et al. Cardioprotective effect of microRNA-21 in murine myocardial infarction. Cardiovasc Ther. (2015) 33:109–17. doi: 10.1111/1755-5922.12118
129. Ovchinnikova ES, Schmitter D, Vegter EL, Ter Maaten JM, Valente MAE, Liu LCY, et al. Signature of circulating microRNAs in patients with acute heart failure. Eur J Heart Fail. (2016) 18:414–23. doi: 10.1002/ejhf.332
130. Endo K, Naito Y, Ji X, Nakanishi M, Noguchi T, Goto Y, et al. MicroRNA 210 as a biomarker for congestive heart failure. Biol Pharm Bull. (2013) 36:48–54. doi: 10.1248/bpb.b12-00578
131. Seronde MF, Vausort M, Gayat E, Goretti E, Ng LL, Squire IB, et al. Circulating microRNAs and outcome in patients with acute heart failure. PLoS ONE. (2015) 10:e0142237. doi: 10.1371/journal.pone.0142237
132. Tijsen AJ, Creemers EE, Moerland PD, De Windt LJ, Van Der Wal AC, Kok WE, et al. MiR423-5p as a circulating biomarker for heart failure. Circ Res. (2010) 106:1035–9. doi: 10.1161/CIRCRESAHA.110.218297
133. Goren Y, Kushnir M, Zafrir B, Tabak S, Lewis BS, Amir O. Serum levels of microRNAs in patients with heart failure. Eur J Heart Fail. (2012) 14:147–54. doi: 10.1093/eurjhf/hfr155
134. Zhang Y, Liu YJ, Liu T, Zhang H, Yang SJ. Plasma microRNA-21 is a potential diagnostic biomarker of acute myocardial infarction. Eur Rev Med Pharmacol Sci. (2016) 20:323–9.
135. Lv P, Zhou M, He J, Meng W, Ma X, Dong S, et al. Circulating miR-208b and miR-34a are associated with left ventricular remodeling after acute myocardial infarction. Int J Mol Sci. (2014) 15:5774–88. doi: 10.3390/ijms15045774
136. Widera C, Gupta SK, Lorenzen JM, Bang C, Bauersachs J, Bethmann K, et al. Diagnostic and prognostic impact of six circulating microRNAs in acute coronary syndrome. J Mol Cell Cardiol. (2011) 51:872–5. doi: 10.1016/j.yjmcc.2011.07.011
137. Corsten MF, Dennert R, Jochems S, Kuznetsova T, Devaux Y, Hofstra L, et al. Circulating microRNA-208b and microRNA-499 reflect myocardial damage in cardiovascular disease. Circ Cardiovasc Genet. (2010) 3:499–506. doi: 10.1161/CIRCGENETICS.110.957415
138. Liu X, Fan Z, Zhao T, Cao W, Zhang L, Li H, et al. Plasma miR-1, miR-208, miR-499 as potential predictive biomarkers for acute myocardial infarction: An independent study of Han population. Exp Gerontol. (2015) 72:230–8. doi: 10.1016/j.exger.2015.10.011
139. Schotten U, Verheule S, Kirchhof P, Goette A. Pathophysiological mechanisms of atrial fibrillation: a translational appraisal. Physiol Rev. (2011) 91:265–325. doi: 10.1152/physrev.00031.2009
140. Hale CS, Levis WR. MicroRNA-29 and an integrated understanding of atrial fibrillation. J Drugs Dermatol. (2013) 12:1083.
141. Cushing L, Kuang PP, Qian J, Shao F, Wu J, Little F, et al. miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol. (2012) 45:287–94. doi: 10.1165/rcmb.2010-0323OC
142. Oliveira-Carvalho V, Carvalho VO, Bocchi EA. The emerging role of miR-208a in the heart. DNA Cell Biol. (2012) 32:8–12. doi: 10.1089/dna.2012.1787
143. Lu Y, Hou S, Huang D, Luo X, Zhang J, Chen J, et al. Expression profile analysis of circulating microRNAs and their effects on ion channels in Chinese atrial fibrillation patients. Int J Clin Exp Med. (2015) 8:845.
144. Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. (2008) 456:980–4. doi: 10.1038/nature07511
145. Wu P, Nielsen TE, Clausen MH. Small-molecule kinase inhibitors: an analysis of FDA-approved drugs. Drug Discov Today. (2016) 21:5–10. doi: 10.1016/j.drudis.2015.07.008
146. Fellmann C, Gowen BG, Lin PC, Doudna JA, Corn JE. Cornerstones of CRISPR–Cas in drug discovery and therapy. Nat Rev Drug Discov. (2016) 16:89–100. doi: 10.1038/nrd.2016.238
147. Toure M, Crews CM. Small-molecule PROTACS: new approaches to protein degradation. Angew Chemie Int Ed. (2016) 55:1966–73. doi: 10.1002/anie.201507978
148. Yang X, Han H, DeCarvalho DD, Lay FD, Jones PA, Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. (2014) 26:577–90. doi: 10.1016/j.ccr.2014.07.028
149. Sato T, Issa JPJ, Kropf P. DNA hypomethylating drugs in cancer therapy. Cold Spring Harb Perspect Med. (2017) 7:a026948. doi: 10.1101/cshperspect.a026948
150. Janzen WP. Screening technologies for small molecule discovery: the state of the art. Chem Biol. (2014) 21:1162–70. doi: 10.1016/j.chembiol.2014.07.015
151. Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. (2002) 1:727–30. doi: 10.1038/nrd892
152. Mullard A. Industry advances landmark CRISPR candidate into the clinic. Nat Rev Drug Discov. (2018) 17:697–7. doi: 10.1038/nrd.2018.175
153. Veedu RN, Wengel J. Locked nucleic acids: promising nucleic acid analogs for therapeutic applications. Chem Biodivers. (2010) 7:536–42. doi: 10.1002/cbdv.200900343
154. Washietl S, Will S, Hendrix DA, Goff LA, Rinn JL, Berger B, et al. Computational analysis of noncoding RNAs. Wiley Interdiscip Rev RNA. (2012) 3:759–78. doi: 10.1002/wrna.1134
155. Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. (2009) 10:126–39. doi: 10.1038/nrm2632
156. Riffo-Campos ÁL, Riquelme I, Brebi-Mieville P. Tools for sequence-based mirna target prediction: what to choose? Int J Mol Sci. (2016) 17:1987. doi: 10.3390/ijms17121987
157. Lekprasert P, Mayhew M, Ohler U. Assessing the utility of thermodynamic features for microrna target prediction under relaxed seed and no conservation requirements. PLoS ONE. (2011) 6:e20622. doi: 10.1371/journal.pone.0020622
158. Hydbring P, Badalian-Very G. Clinical applications of microRNAs. F1000Research. (2013) 2:1–16. doi: 10.12688/f1000research.2-136.v1
159. Chakraborty C, Sharma AR, Sharma G, Doss CGP, Lee SS. Therapeutic miRNA and siRNA: moving from bench to clinic as next generation medicine. Mol Ther Nucleic Acids. (2017) 8:132–43. doi: 10.1016/j.omtn.2017.06.005
160. Zhou B, Pu WT. More than a cover: epicardium as a novel source of cardiac progenitor cells. Regen Med. (2008) 3:633–5. doi: 10.2217/17460751.3.5.633
161. Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. (2008) 454:109–13. doi: 10.1038/nature07060
162. Weeke-Klimp A, Bax NAM, Bellu AR, Winter EM, Vrolijk J, Plantinga J, et al. Epicardium-derived cells enhance proliferation, cellular maturation and alignment of cardiomyocytes. J Mol Cell Cardiol. (2010) 49:606–16. doi: 10.1016/j.yjmcc.2010.07.007
163. Seeger T, Xu QF, Muhly-Reinholz M, Fischer A, Kremp EM, Zeiher AM, et al. Inhibition of let-7 augments the recruitment of epicardial cells and improves cardiac function after myocardial infarction. J Mol Cell Cardiol. (2016) 94:145–52. doi: 10.1016/j.yjmcc.2016.04.002
164. Lesizza P, Prosdocimo G, Martinelli V, Sinagra G, Zacchigna S, Giacca M. Single-dose intracardiac injection of pro-regenerative MicroRNAs improves cardiac function after myocardial infarction. Circ Res. (2017) 120:1298–304. doi: 10.1161/CIRCRESAHA.116.309589
165. Kulkarni JA, Cullis PR, Van Der Meel R. Lipid nanoparticles enabling gene therapies: from concepts to clinical Utility. Nucleic Acid Ther. (2018) 28:146–57. doi: 10.1089/nat.2018.0721
166. Yang H, Qin X, Wang H, Zhao X, Liu Y, Wo HT, et al. An in vivo miRNA delivery system for restoring infarcted myocardium. ACS Nano. (2019) 13:9880–94. doi: 10.1021/acsnano.9b03343
Keywords: miRNA, heart regeneration, cardiac development, cardiomyocyte, cardiovascular diseases
Citation: Shah V and Shah J (2022) Restoring Ravaged Heart: Molecular Mechanisms and Clinical Application of miRNA in Heart Regeneration. Front. Cardiovasc. Med. 9:835138. doi: 10.3389/fcvm.2022.835138
Received: 14 December 2021; Accepted: 17 January 2022;
Published: 10 February 2022.
Edited by:
Katherine Athayde Teixeira De Carvalho, Pelé Pequeno Príncipe Research Institute, BrazilReviewed by:
Estefania Lozano Velasco, University of East Anglia, United KingdomShun Yan, University of Alabama at Birmingham, United States
Copyright © 2022 Shah and Shah. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jigna Shah, amlnbmEuc2hhaEBuaXJtYXVuaS5hYy5pbg==
 Vandit Shah
Vandit Shah Jigna Shah
Jigna Shah