
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cardiovasc. Med. , 23 March 2022
Sec. Cardiovascular Genetics and Systems Medicine
Volume 9 - 2022 | https://doi.org/10.3389/fcvm.2022.808929
Being activated by endogenous and exogenous ligands, nuclear receptor peroxisome proliferator-activated receptor gamma (PPARγ) enhances insulin sensitivity, promotes adipocyte differentiation, stimulates adipogenesis, and has the properties of anti-atherosclerosis, anti-inflammation, and anti-oxidation. The Human PPARγ gene (PPARG) contains thousands of polymorphic loci, among them two polymorphisms (rs10865710 and rs7649970) in the promoter region and two polymorphisms (rs1801282 and rs3856806) in the exonic region were widely reported to be significantly associated with coronary artery disease (CAD). Mechanistically, PPARG polymorphisms lead to abnormal expression of PPARG gene and/or dysfunction of PPARγ protein, causing metabolic disorders such as hypercholesterolemia and hypertriglyceridemia, and thereby increasing susceptibility to CAD.
Coronary artery disease (CAD) is the most common type of cardiovascular disease globally, and is often caused by stenosis of coronary arteries due to atherosclerosis (1, 2). Under the action of various cardiovascular risk factors, atherosclerotic plaques gradually form, enlarge and ultimately block blood-vessel cavity, resulting in myocardial ischemia, hypoxia, and necrosis (3). According to the summary of the 2018 Report on Cardiovascular Diseases in China, cardiovascular disease is currently ranked as the first cause of death in China, and the morbidity and mortality are still on the rise (4). As a complex disease with multiple risk factors and being closely related to glucose and lipid metabolism, genetic variations in metabolism-related genes play an essential role in the pathogenesis of CAD (5). In recent decades, more and more genetic susceptibility genes and polymorphic loci for cardiovascular disease were explored and identified (6). Peroxisome proliferator-activated receptors (PPARs) are ligand-inducible transcription factors, belonging to the nuclear receptor superfamily (7). PPARs have multiple and complex physiological functions, involving lipid and glucose metabolism, (8, 9) inflammatory response, (10, 11) oxidative stress, (11, 12) cell differentiation and apoptosis, (13, 14) and even cognitive function (15). PPARs have been implicated in the pathogenesis of a number of major human diseases, such as cardiovascular and cerebrovascular diseases, (16, 17) malignant tumors, (18, 19) diabetes mellitus, (20, 21) metabolic syndrome, (22) and neurodegenerative disorders (23). PPARs have three subtypes: PPARα, PPARβ/δ, and PPARγ. Among them, PPARγ has the most in-depth exploration. According to NCBI’s reference sequence (RefSeq) database1, eight PPARγ isoforms have been identified in humans. Being activated by endogenous and exogenous ligands, PPARγ works in concert with retinoid X receptor (RXR) and is able to increase the insulin sensitivity, (24) promote adipocyte differentiation, (25) and stimulate adipogenesis, (26) and has the properties of anti-atherosclerosis, (27, 28) anti-inflammation, (27, 28) and anti-oxidation (29). This review focuses on recent progress in the association studies between polymorphisms in PPARγ gene (PPARG) and CAD, as well as the underlying mechanisms.
Human PPARG is located on the chromosome 3p25.3 and composed of nine exons: exon A1, exon A2, exon B, and exons 1–6 (Figure 1). There is alternative splicing in the maturation process of PPARG mRNA (30). According to the NCBI’s RefSeq database, sixteen PPARG mRNA splice variants have been identified so far in humans due to differential promoter usage and alternative splicing. PPARG is highly polymorphic (31). A large number of PPARG genetic variants have been recorded in the NCBI’s dbSNP database,2 and most of which are distributed in the intronic region. These loci can be divided into promoter region polymorphisms (e.g., rs10865710 and rs7649970), (32, 33) exonic region polymorphisms (e.g., rs1801282 and rs3856806), (32, 33) and intronic region polymorphisms (e.g., rs1152002 and rs709158) (33, 34) according to their distribution in the gene.
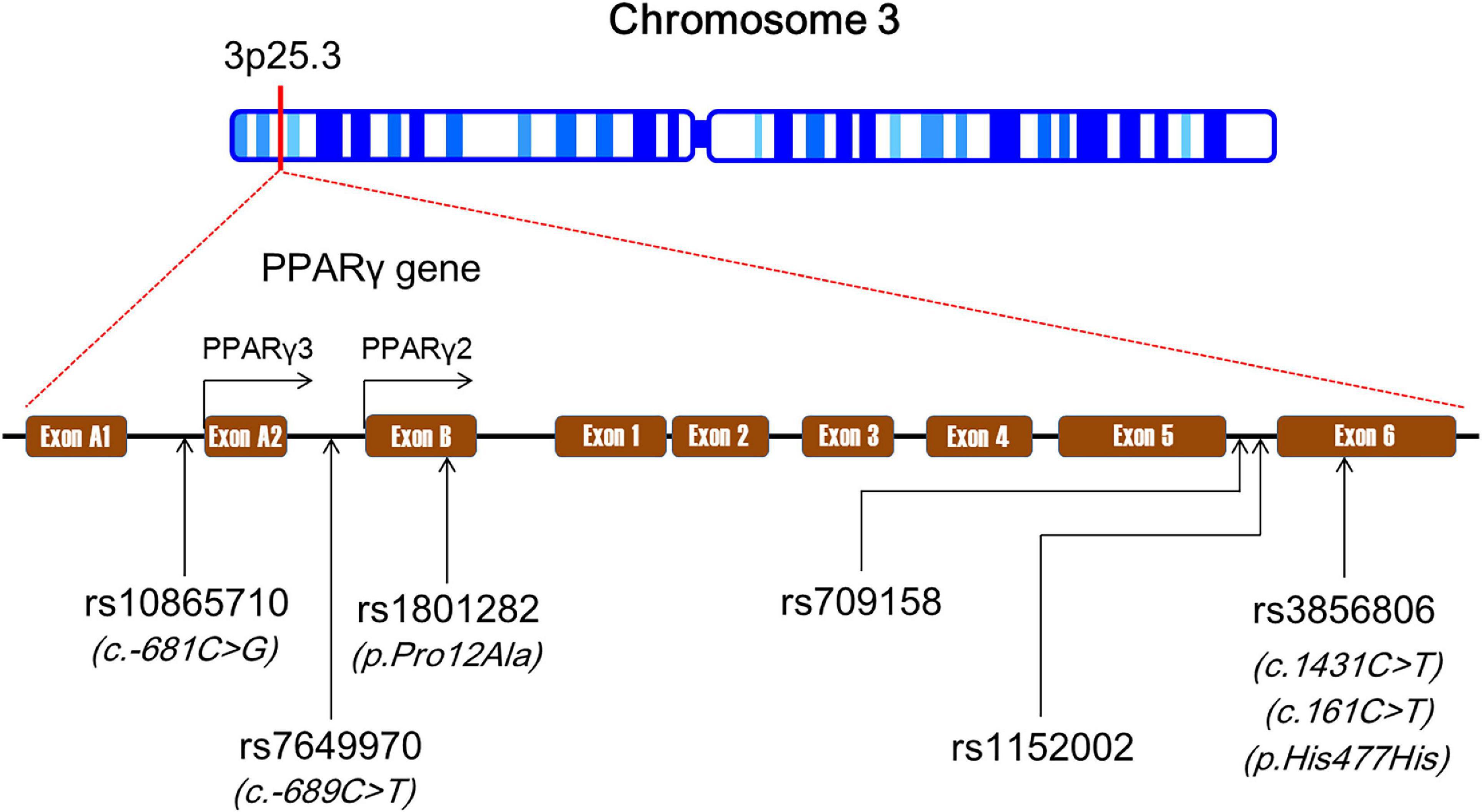
Figure 1. The genomic landscape of single-nucleotide polymorphisms (SNPs) in peroxisome proliferator-activated receptor gamma gene (PPARG). SNPs, single-nucleotide polymorphisms; PPARG, peroxisome proliferator-activated receptor gamma gene.
A large body of evidence indicated that some polymorphisms in PPARG are associated with CAD (34–63). Among them, rs1801282 (35, 41–51) and rs3856806 (52–63) in the exonic region were extensively reported to be significantly associated with CAD.
Two promoter polymorphisms rs10865710 (35–37) and rs7649970 (38, 39) have been indicated to be correlated with CAD. Some other promoter variants such as c.93640T > C, c.93673T > C, and c.93695C > T have been investigated as well, and among them c.93695C > T was detected to be significantly associated with CAD (40).
The rs10865710 polymorphism (also known as c.-681C > G) is located in the upstream promoter region of PPARγ3 gene and formed by a transversion from cytosine (C) to guanine (G) (35–37) (Figure 1). According to the NCBI’s dbSNP and VannoPortal3 databases, G is the minor allele of the rs10865710 polymorphism with frequencies ranging from 0.23 to 0.33 among Caucasian populations, 0.25 to 0.36 among Asian populations, and 0.21 to 0.24 among African populations. The results of two case–control studies (35, 36) in Chinese populations suggested that G allele of the rs10865710 polymorphism was associated with an increased risk of CAD. Zhang et al. (35) found that G allele carriers of the rs10865710 polymorphism had a higher risk of CAD than non-carriers in a Chinese population (odds ratio [OR], 1.47; 95% CI, 1.15–1.92; p < 0.001). Ding et al. (36) confirmed this finding in another Chinese population, and demonstrated that G allele of the rs10865710 polymorphism was associated with a higher risk of CAD (OR, 1.31; 95% CI, 1.16–1.95; p < 0.01). Our research team recently conducted a case–control study to assess the association between the rs10865710 polymorphism and CAD severity among Chinese patients, and observed that G allele carriers had higher Gensini scores (an indicator of CAD severity) (p < 0.05) and more diseased coronary branches than patients with CC genotype (p < 0.05) (37). However, several case–control studies carried out in Caucasians did not detect any significant association between the rs10865710 polymorphism and CAD risk (40, 60, 64). Hence, the impact of the rs10865710 polymorphism on susceptibility to CAD remains undetermined, and it may be modulated by ethnicity, living region, and/or eating habits. More studies are needed to elucidate the relationship of the rs10865710 polymorphism to CAD.
The rs7649970 polymorphism (also known as c.-689C/T) is located in the upstream promoter region of PPARγ2 gene and formed by a transition from C to thymine (T) (38, 39) (Figure 1). According to the NCBI’s dbSNP and VannoPortal databases, T is the minor allele of the rs7649970 polymorphism with frequencies ranging from 0.11 to 0.17 among Caucasian populations, 0.03 to 0.06 among Asian populations, and 0.17 to 0.19 among African populations. A couple of case–control studies demonstrated that T allele of the rs7649970 polymorphism is associated with a higher risk of CAD (38, 39). Li et al. (38) examined the relationship between the rs7649970 polymorphism and CAD in a non-diabetic Chinese Han population, and the results showed that T allele is an independent risk factor for CAD after adjustment for conventional risk factors such as smoking, hypertension, and dyslipidemia (OR, 1.67; 95% CI, 1.03–2.71; p = 0.04). In another Chinese population, T allele of the rs7649970 polymorphism was found to be an independent risk factor for myocardial infarction (MI) after adjustment for traditional risk factors (OR, 2.13; 95% CI, 1.21–3.74; p < 0.01) (39). Dallongeville et al. (60) observed that TT genotype of the rs7649970 polymorphism was correlated with a marginally insignificantly higher risk of CAD in a large group of middle-aged men recruited from Lille, Strasbourg, and Toulouse in France and Belfast in Northern Ireland (OR, 3.34; 95% CI, 0.98–11.45; p = 0.05).
Relationships of several rare variants in PPARG such as c.25924C > T and c.26233T > A in PPARγ3 promoter, and c.93640T > C, c.93673T > C, and c.93695C > T in PPARγ4 promoter to CAD were explored in an Italian population, and the explorers found that the c.93695C > T polymorphism was significantly correlated with acute coronary syndrome (ACS); T allele conferred a protective effect against ACS at both univariate (OR, 0.45; 95% CI, 0.29–0.69; p < 0.001) and multivariate (OR, 0.44; 95% CI, 0.25–0.76; p < 0.01) analyses (40).
The association studies between polymorphisms in the exonic region of PPARG and CAD were heavily focused on the rs1801282 (35, 41–51, 60, 65–78) and rs3856806 (52–64, 79–86) polymorphisms. Other PPARG exonic polymorphisms in the association studies with CAD were scarcely investigated and rarely reported in the literature.
The rs1801282 polymorphism (also known as p.Pro12Ala) is located in exon B of PPARG and is a missense variant in PPARγ2 resulting in a proline-to-alanine substitution (41–51) (Figure 1). This polymorphism is formed by a single-nucleotide change from C to G. G is the minor allele of the rs1801282 polymorphism with frequencies ranging from 0.11 to 0.17 among Caucasian populations, 0.02 to 0.06 among Asian populations, and 0.01 to 0.02 among African populations according to the NCBI’s dbSNP and VannoPortal databases. Researchers from various laboratories around the world have suggested that G allele of the rs1801282 polymorphism was associated with a higher risk of CAD (35, 41–46). Zhang et al. (35) evaluated the association between the rs1801282 polymorphism and CAD risk in a hospital-based study in Beijing, China, and observed that G allele carriers had a higher risk of CAD than non-carriers (OR, 1.69; 95% CI, 1.27–2.09; p < 0.001). In a case–control study carried out in Inner Mongolia, China, the investigators concluded that G allele of the rs1801282 polymorphism was an independent risk factor for MI after adjustment for conventional risk factors (OR, 2.68; 95% CI, 1.04–6.95; p = 0.04) (42). Hasan et al. (43) demonstrated that G allele carriers of the rs1801282 polymorphism were three times more likely to have CAD than non-carriers (OR, 3.0; 95% CI, 1.5–6.0; p = 0.001) among Egyptian patients with type 2 diabetes mellitus (T2DM). Maciejewska-Skrendo et al. (44) found that patients with unstable angina had a higher frequency of G allele of the rs1801282 polymorphism than healthy controls among European Caucasians (17.28 vs. 9.26%; p < 0.001). The association between the rs1801282 polymorphism and CAD appeared to be gender-dependent. Vogel et al. (45) demonstrated that GG genotype of the rs1801282 polymorphism was associated with a higher risk of ACS among Danish men (hazard ratio [HR], 2.12; 95% CI, 1.00–4.48; p = 0.05), but not among women. Similarly, Schneider et al. (46) observed that G allele of the rs1801282 polymorphism was significantly associated with CAD severity among German male patients (β, 0.32; p = 0.001). In a prospective cohort study involving middle-aged French men, the subjects with GG genotype of the rs1801282 polymorphism had a marginally insignificantly higher risk of CAD than those with CC genotype (OR, 3.32; 95% CI, 0.97–11.39; p = 0.06) (60).
Somehow, several case–control and prospective cohort studies have come to an opposite conclusion, demonstrating that G allele of the rs1801282 polymorphism was significantly associated with a reduced risk of CAD (47–51). Ho et al. (47) conducted a prospective case–control study among Hong Kong Chinese patients with T2DM and found that patients with CC genotype of the rs1801282 polymorphism had a higher risk of CAD than G allele carriers (HR, 4.38; 95% CI, 1.03–18.57; p = 0.05). A few studies involving American and European Caucasians confirmed that G allele of the rs1801282 polymorphism was associated with a lower risk of CAD (48–50). In an African population, Youssef and teammates (51) demonstrated that ACS patients had a lower frequency of G allele than control subjects (12.3 vs. 19.3%; p < 0.01), and the ACS patients carrying one or two G alleles had lower Gensini scores (p < 0.001) and less number of diseased coronary arteries (p < 0.001) than those with CC genotype.
Many well-designed studies have failed to detect a significant association between the rs1801282 polymorphism and CAD (65–76). Three case–control studies, respectively, conducted in Zhejiang University, (65) Shanghai Jiao Tong University, (66) and Chinese Medical University in China (67) could not find any significant association between the rs1801282 polymorphism and CAD. Furthermore, no significant association was detected between the rs1801282 polymorphism and CAD in various populations such as British, (68) Germans, (69) Canadians, (70) Koreans, (71) Turks, (72, 73) Dutch, (74) Indians, (75) and Thais (76).
The inconsistencies and contradictions among the association studies between the rs1801282 polymorphism and CAD may be due to the interaction of PPARG polymorphisms with environmental factors on cardiovascular risk factors, i.e., different alleles have different impacts on the expression patterns of PPARG under different environmental conditions. Abaj et al. (87) examined the interaction of the rs1801282 polymorphism with diet indices such as Dietary Quality Index-International, dietary phytochemical index, and healthy eating index on cardiovascular risk factors in T2DM patients, and found that these diet patterns did have a significant impact on cardiovascular risk factors in patients with different rs1801282 genotypes. In addition, small sample size, racial differences, and population heterogeneity may also be responsible for the inconsistencies and contradictions among studies. A meta-analysis combines data of the same type of studies to reduce the impact of confounding factors such as sample size and ethnicity on research results; so conclusions from meta-analyses are relatively more reliable. Wu and teammates (77) performed a meta-analysis with 22 studies and 23,375 subjects enrolled, and found that GG genotype of the rs1801282 polymorphism conferred a higher risk of CAD than CC genotype in the total population (OR, 1.30; 95% CI, 1.01–1.68; p = 0.04) and in Caucasians (OR, 1.44; 95% CI, 1.07–1.93; p = 0.02), but not in Asians. However, the results from several other meta-analyses did not support this finding and concluded that the rs1801282 polymorphism was not associated with CAD in overall and subgroup analyses (60, 61, 78). Therefore, it is difficult to reach a consistent conclusion referring to the relationship between the rs1801282 polymorphism and CAD based on the existing research data. Further studies are needed to clarify this issue.
The rs3856806 polymorphism (also known as c.1431C > T, c.161C > T, or p.His477His) is located in exon 6 of PPARG and is a synonymous variant in PPARγ2 (52–64) (Figure 1). The c.1431C > T is named according to the position of this variant in PPARγ2 cDNA, as this variant is located at 1431 bp downstream of the start codon (ATG). This c.161C > T is defined based on the position of this variant in exon 6 of PPARG gene since it is located at 161 bp downstream of the first nucleotide of exon 6 of PPARG. The rs3856806 polymorphism is formed by a single-nucleotide substitution from C to T. T is the minor allele of the rs3856806 polymorphism with frequencies ranging from 0.10 to 0.22 among Caucasian populations, 0.18 to 0.28 among Asian populations, and 0.05 to 0.07 among African populations according to the NCBI’s dbSNP and VannoPortal databases. A number of case–control and cross-sectional studies have shown that T allele of the rs3856806 polymorphism was associated with a reduced risk of CAD and was a protective allele for CAD (52–59). In a hospital-based case–control study of Chinese patients with CAD and chest pain syndrome, Liu et al. (52) found that the T allele carriers of the rs3856806 polymorphism had a reduced CAD risk compared with CC homozygotes (OR, 0.55; 95% CI, 0.33–0.83; p = 0.01). Zhou and the other two teammates (53) made a similar finding in the Chinese Han population that the T allele carriers of the rs3856806 polymorphism had a 39% decreased risk of CAD relative to CC homozygotes (OR, 0.61; 95% CI, 0.49–0.76; p < 0.001). The protective effect of T allele on CAD was also reported by other explorers in Chinese populations (54–59). It seems that the correlation between the rs3856806 polymorphism and CAD is stronger in the presence of T2DM as Wan et al. (58) observed that the T allele was significantly correlated with a lower degree of coronary stenosis (<75%) among CAD patients combined with T2DM (p = 0.02), but not among patients free of T2DM (p = 0.70).
Like the rs1801282 polymorphism, there was also a contradiction in the relation between the rs3856806 polymorphism and CAD. Several studies suggested that the T allele of the rs3856806 polymorphism was associated with an increased risk of CAD (40, 60–63). Chao et al. (62) demonstrated that the TT genotype conferred a higher risk of MI as compared to CC genotype in a Taiwanese population (OR, 2.7; 95% CI, 1.1–6.5). A significant association between T allele of the rs3856806 polymorphism and higher risk of ACS was detected among Chinese mainland residents (OR, 1.63; 95% CI, 1.00–2.65; p = 0.05) (61) and Italians (p = 0.03) (40). In a French male population, TT homozygotes had a higher risk of CAD than CC homozygotes (OR, 5.93; 95% CI, 1.19–29.45; p = 0.03), (60) and in an Iranian population, T allele carriers had a higher risk of CAD than CC homozygotes (OR, 2.28; 95% CI, 1.20–4.35; p = 0.01) (63). Recent experimental results from our laboratory showed that the T allele of the rs3856806 polymorphism was correlated with an increased risk of T2DM complicated with CAD (p = 0.03) (37).
Some researchers failed to detect any significant association of the rs3856806 polymorphism with CAD (64, 65, 79–82). Results from three independent studies conducted in China consistently indicated that the rs3856806 polymorphism was not correlated with CAD (65, 79, 80). Neither Yilmaz-Aydogan (81) nor Yongsakulchai (82) did in a Turkish population and in a Thai population, respectively. We did not find any significant association of the rs3856806 polymorphism with CAD or CAD severity as well (37).
A couple of meta-analyses have been carried out in order to clarify the relationship between the rs3856806 polymorphism and CAD, but still no consistent results were obtained to date. Qian et al. (61) did a meta-analysis with 9 studies and a total of 3,878 subjects enrolled, and the result suggested that T allele carriers of the rs3856806 polymorphism had a lower CAD risk than CC homozygotes (OR, 0.69; 95% CI, 0.59–0.82; p < 0.001). Gonzlez-Castro et al. (83) expanded the sample size to 21 studies and 15,980 subjects, and arrived at a similar conclusion (OR, 0.33; 95% CI, 0.20–0.52). However, Ding et al. (78) demonstrated that T allele carriers of the rs3856806 polymorphism had a higher risk of CAD than non-carriers by meta-analysis (OR, 1.18; 95% CI, 1.02–1.34; p < 0.01). In addition, the results from several other meta-analyses indicated that the rs3856806 polymorphism was not correlated with CAD at all (84–86). Hence, the relationship between the rs3856806 polymorphism and CAD is not possible to be determined based on the existing research data, and it needs to be further explored.
A few studies have been carried out to explore the associations between PPARG intronic polymorphisms and CAD. The rs1152002 polymorphism is located in intron 5 of PPARG and formed by a transition from G to adenine (A). Tian et al. (34) reported that A allele of the rs1152002 polymorphism was associated with a higher risk of CAD in a Chinese population (OR, 2.92; 95% CI, 1.44–5.94; p < 0.01). The rs709158 polymorphism is located in intron 5 of PPARG and formed by a transition from A to G. Gallicchio et al. (88) prospectively examined the association of the rs709158 polymorphism with cardiovascular morbidity and mortality in a community-based cohort study, and demonstrated that there was no statistically significant association between them.
In terms of mechanisms of action by which PPARG polymorphisms influence on the susceptibility to CAD, the first thing that comes to mind is that polymorphisms in PPARG lead to abnormal expression of this gene and/or dysfunction of PPARγ protein, resulting in aberrant expressions of PPARγ-targeted genes, metabolic disorders, and arteriosclerotic cardiovascular disease (Figure 2).
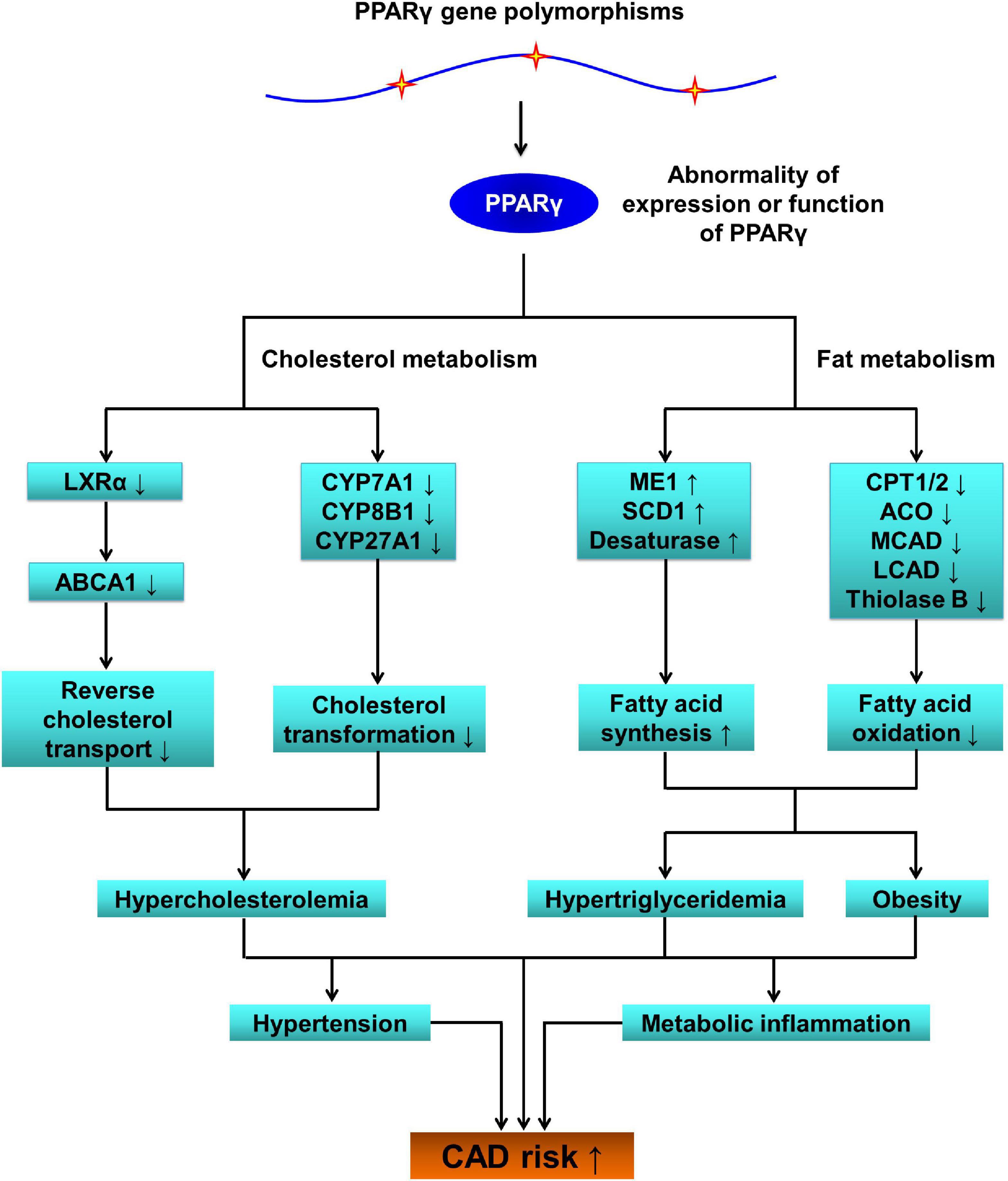
Figure 2. The pathophysiological role of PPARG SNPs in coronary artery disease (CAD). PPARG, peroxisome proliferator-activated receptor gamma gene; PPARγ, peroxisome proliferator-activated receptor gamma; LXRα, liver X receptor alpha; ABCA1, ATP-binding cassette transporter A1; CYP7A1, cytochrome P450 family 7 subfamily A member 1; CYP8B1, cytochrome P450 family 8 subfamily B member 1; CYP27A1, cytochrome P450 family 27 subfamily A member 1; ME1, malic enzyme 1; SCD1, stearoyl-coenzyme A desaturase 1; CPT1/2, carnitine palmitoyltransferase 1/2; ACO, acyl-coenzyme A oxidase; MCAD, medium-chain acyl-coenzyme A dehydrogenase; LCAD, long-chain acyl-coenzyme A dehydrogenase; SNPs, single-nucleotide polymorphisms; CAD, coronary artery disease.
Being activated by endogenous and exogenous ligands, PPARγ mainly up-regulates gene expressions of enzymes and transporters that play key roles in lipid and glucose metabolic pathways such as reverse cholesterol transport, (89, 90) cholesterol transformation, (89, 90) lipogenesis, (91, 92) fatty acid oxidation, (93, 94) and gluconeogenesis (95). By using luciferase reporter and electrophoretic mobility shift assays, Lu et al. (96) observed that G allele of the rs10865710 polymorphism significantly inhibited the DNA-binding activity of transcription factor cAMP-response element-binding protein 2 (CREB2) to PPARγ3 promoter. The rs948820149 polymorphism (c.-807A > C) is located in PPARγ2 promoter and C allele was found to significantly down-regulate PPARγ2 expression by affecting the DNA-binding activity of transcription factor glucocorticoid receptor β (GRβ) to PPARγ2 promoter (97). Another two PPARG promoter polymorphisms c.-1633C > T and c.-1572G > A were verified to modulate the expression efficiency of PPARG in Erhualian pigs as well (98). Pihlajamäki et al. (99) compared PPARγ2 mRNA expression as well as its two target genes (lipid phosphate phosphohydrolase 1 [LPIN1] and sterol-regulatory-element-binding protein 1c [SREBP-1c]) between PPARG rs1801282 genotypes in human adipose tissues, and observed that Ala12Ala genotype was associated with a significantly higher mRNA expression compared to Pro12Pro genotype. By using a computational analysis of SNPs in PPARG, researchers found that mutations in PPARG impaired functions of PPARγ, leading to serious complications such as obesity, diabetes, and cancer in humans (31).
It is easy to understand that PPARG polymorphisms in the promoter region, as well as missense polymorphisms in the exonic region, may cause metabolic disorders such as hypercholesterolemia, hypertriglyceridemia, and hyperglycemia, which subsequently increase the risk of CAD. However, it is difficult to explain how intronic and synonymous polymorphisms are responsible for susceptibility to CAD. So far, there is no direct evidence that PPARG intronic and synonymous polymorphisms modulate PPARG gene expression efficiency. Little is known about the molecular mechanisms underlying the regulatory function of intronic and synonymous polymorphisms in PPARG on its gene expression, but several possible explanations can be put forward for intronic polymorphisms. Firstly, there are functional elements in intronic regions to regulate gene expression, such as intronic enhancer/repressor (100, 101). Secondly, intronic polymorphisms may affect the pre-mRNA splicing process (102). Thirdly, some non-coding RNAs with a wide range of regulatory effects are encoded by introns (103, 104). Regarding synonymous polymorphisms, they may alter the secondary structure of pre-mRNA, and thereby influencing mRNA splicing efficiency and protein translation (105–107).
Dyslipidemia is a major risk factor for CAD, accounting for 50% of the population attributable risk (108). Increases in the levels of triglycerides, total cholesterol and low-density lipoprotein cholesterol (LDL-C), and/or decreases in HDL-C levels confer a high risk of CAD. There is accumulating evidence indicating that PPARG exonic polymorphisms rs1801282 (41–43, 64–66, 109–112) and rs3856806 (37, 53, 62, 64, 79, 112) are significantly associated with abnormal levels of plasma lipids. The PPARG promoter polymorphisms rs10865710 and rs7649970 have been reported to be significantly correlated with plasma lipid levels as well, (37–39) although there were few studies conducted in the scientific community.
A number of observational studies suggested that G allele of the rs1801282 polymorphism was associated with increased levels of triglycerides, (110, 112) total cholesterol, (41–43, 64–66, 109) and LDL-C, (43, 64, 65, 109) and decreased levels of HDL-C, (42, 43) which is in line with the finding of several case–control studies that G allele carriers had a significantly higher risk of CAD than CC homozygotes (41–46). In a Chinese population, Wang et al. (42) found that G allele carriers of the rs1801282 polymorphism had significantly higher levels of total cholesterol and LDL-C than the subjects with CC genotype, and also that G allele carriers were at a higher risk of MI. Similarly, Hasan et al. (43) demonstrated that G allele carriers of the rs1801282 polymorphism had significantly higher levels of total cholesterol and LDL-C, and lower levels of HDL-C than CC homozygotes in an Egyptian diabetic population, and the researchers also observed that the risk of CAD was three times higher among G allele carriers than among non-carriers.
Just as there were contradictions in the associations between the rs1801282 polymorphism and CAD, some notable inconsistencies were present in the relations between the rs1801282 polymorphism and plasma lipid levels. In a Chinese longevity population (age >90 years), the levels of total cholesterol, LDL-C, and HDL-C were comparable between the rs1801282 genotypes (CG + GG vs. CC), but G allele carriers had significantly lower levels of triglycerides than the subjects with CC genotype (p < 0.001) (110). Koohdani et al. (111) also reported lower levels of triglycerides in G allele carriers of the rs1801282 polymorphism than in CC homozygotes among Iranian T2DM patients.
Several studies demonstrated that the T allele of the rs3856806 polymorphism was correlated with decreased levels of triglycerides, (64) total cholesterol, (112) and LDL-C, (112) as well as elevated levels of HDL-C (53). This may explain the phenomenon that the T allele was associated with a reduced risk of CAD in several case–control studies (52–59). Zhou et al. (53) observed that the T allele carriers of the rs3856806 polymorphism had lower levels of triglycerides, total cholesterol, and LDL-C, higher levels of HDL-C, and a 40% lower risk of CAD than non-carriers in the Chinese Han population. In the Russian population, the investigators reported that serum levels of triglycerides in T allele carriers of the rs3856806 polymorphism were significantly lower than in the subjects with CC genotype, and simultaneously the frequency of T allele tended to decrease in CAD patients compared to control subjects (64).
A few studies have produced conflicting results that the T allele of the rs3856806 polymorphism was correlated with higher levels of atherogenic lipids (37, 62, 79). Chao et al. (62) reported that the T allele carriers had significantly higher levels of oxidized low-density lipoprotein (an atherogenic lipoprotein) than the subjects with CC genotype in a Taiwanese population, and accordantly TT homozygotes were found to have a significantly higher risk of MI than C carriers. In a group of Chinese patients with CAD, our research team found that the T allele carriers of the rs3856806 polymorphism had significantly higher levels of total cholesterol, LDL-C and apolipoprotein B than non-carriers (37). In addition, triglycerides, very-low-density lipoprotein cholesterol (VLDL-C) and lipoprotein (a) were found to be higher in T allele carriers of the rs3856806 polymorphism than in non-carriers in a hospital-based study (79).
A couple of studies (38, 39) showed that the T allele of the rs7649970 polymorphism was correlated with increased levels of triglycerides and total cholesterol, and T allele frequency was also higher in CAD patients than in control subjects. We found that G allele carriers of the rs10865710 polymorphism had significantly higher levels of atherogenic lipids such as total cholesterol, lipoprotein (a) and apolipoprotein B, higher Gensini scores, and more diseased coronary branches (37).
Hypertension is a recognized risk factor for CAD. Ettehad et al. (113) reported that the risk of CAD decreased by 17% for every 10 mmHg reduction in systolic blood pressure. One population-based study (110) and three meta-analyses (114–116) collectively pointed out that G allele of the rs1801282 polymorphism was associated with significantly reduced blood pressure. It is consistent with the finding that G allele of the rs1801282 polymorphism was correlated with a reduced risk of CAD in case–control studies (47–51). Lu et al. (110) reported that G allele of the rs1801282 polymorphism appeared to have a protective effect against hypertension. This finding was validated by Regieli et al. in the Dutch population (50) and by three meta-analyses (114–116). However, some other studies had a completely different finding (41, 43). Li et al. (41) suggested that G allele carriers of the rs1801282 polymorphism had significantly higher systolic blood pressure than non-carriers in patients with MI, and accordingly, the frequency of G allele was significantly higher in MI patients than in healthy subjects. Hasan et al. (43) demonstrated that G allele of the rs1801282 polymorphism was significantly correlated with increased systolic and diastolic blood pressure compared to C allele among Egyptian T2DM patients, and as well G allele carriers had a significantly higher risk of CAD. Regarding the rs3856806 polymorphism, our research team found that T allele carriers had significantly higher systolic and diastolic blood pressure than CC homozygotes among CAD patients (37).
Body mass index (BMI), waist-to-hip ratio (WHR), and waist circumference (WC) are common indicators of obesity and are closely related to CAD (117–119). Several studies have shown that G allele of the rs1801282 polymorphism was associated with higher BMI, (43, 66, 111, 120) WC, (43, 111) and WHR, (66) higher prevalence of central obesity and higher percentage of body fat (121). This can explain from one aspect why G allele of the rs1801282 polymorphism was correlated with increased risk of CAD in several case–control studies (41–46). The association between the rs1801282 polymorphism and BMI was validated by two meta-analyses which concluded that G allele carriers had significantly higher BMI and higher prevalence of obesity than the subjects with CC genotype (122, 123). However, some studies (124–126) have come to conflicting conclusions. da Silva et al. (124) demonstrated that CC homozygotes had significantly higher BMI and WHR compared to CG heterozygotes among Brazilian adult men. A similar finding was observed in teenagers from Northern Mexico, and the researchers noted that G allele carriers of the rs1801282 polymorphism exhibited significantly lower overweight/obesity phenotype (BMI Z-score) frequency than CC homozygotes (125). Zafar et al. (126) found that BMI and WC were significantly lower in GG homozygotes compared to CC homozygotes among patients with metabolic syndrome. The rs3856806 polymorphism was also reported to be significantly correlated with BMI in Chinese populations, and T allele carriers had significantly higher BMI than the subjects with CC genotype (37, 53).
Atherosclerosis is actually an ongoing chronic inflammatory disorder, not just a simple lipid deposition on the intima and media walls of blood vessels (127, 128). A fundamental role of low-grade inflammation has been established in mediating all stages of atherosclerosis from the initiation to the formation of atherosclerotic plaques and ultimately to thrombosis in the blood vessels (129). Liu et al. (130) observed that T allele carriers of the rs3856806 polymorphism had significantly lower levels of C-reactive protein (CRP) than the subjects with CC genotype in hemodialysis patients. An interaction between the rs1801282 polymorphism and diet indices on cardiovascular risk factors was evaluated among patients with T2DM, and the highest IL-18 level was observed in G allele carriers with the highest adherence to Diet Quality Index (DQI) (76). Adiponectin has the properties of enhancing insulin sensitivity, (131) inhibiting inflammation, (132) and attenuating atherosclerosis (133). Campos-Perez et al. (134) reported that G allele carriers of the rs1801282 polymorphism had significantly higher levels of serum adiponectin than CC homozygotes in a general population, whereas Baldani et al. (135) could not find any significant correlation between the rs1801282 polymorphism and serum adiponectin levels among women with polycystic ovary syndrome.
With the increase of morbidity and mortality in patients with CAD in recent decades, researchers from all over the world have carried out a large number of observational and experimental studies on the associations between PPARG polymorphisms and CAD. Two polymorphic loci (rs1801282 and rs3856806) in the exonic region of PPARG were extensively explored in various populations and were reported to be significantly associated with CAD, but the risk alleles of these polymorphic loci are still elusive. In the future, multi-center, multi-ethnic, and large-sample case–control and cohort studies are needed to identify PPARG risk alleles for CAD. In addition, great efforts are required to assess the interactions between PPARG polymorphisms and environmental factors on the expression patterns of PPARG, the function of PPARγ, and the susceptibility to CAD.
All authors retrieved the literature and wrote and organized the manuscript.
This project was supported by Medical Science and Technology Project of Sichuan Provincial Health Commission (21PJ124).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Ginsberg HN, Packard CJ, Chapman MJ, Borén J, Aguilar-Salinas CA, Averna M, et al. Triglyceride-rich lipoproteins and their remnants: metabolic insights, role in atherosclerotic cardiovascular disease, and emerging therapeutic strategies-a consensus statement from the European Atherosclerosis Society. Eur Heart J. (2021) 42:4791–806. doi: 10.1093/eurheartj/ehab551
2. Al Rifai M, Blaha MJ, Nambi V, Shea SJC, Michos ED, Blumenthal RS, et al. Determinants of incident atherosclerotic cardiovascular disease events among those with absent coronary artery calcium: multi-ethnic study of atherosclerosis. Circulation. (2021) 145:259–67. doi: 10.1161/CIRCULATIONAHA.121.056705
3. Bergström G, Persson M, Adiels M, Björnson E, Bonander C, Ahlström H, et al. Prevalence of subclinical coronary artery atherosclerosis in the general population. Circulation. (2021) 144:916–29. doi: 10.1161/CIRCULATIONAHA.121.055340
4. Hu S, Gao R, Liu L, Zhu M, Wang W, Wang Y, et al. Summary of the 2018 report on cardiovascular diseases in China. Chin Circul J. (2019) 34:209–20.
5. Hao K, Ermel R, Sukhavasi K, Cheng H, Ma L, Li L, et al. Integrative prioritization of causal genes for coronary artery disease. Circ Genom Precis Med. (2021) 15:e003365. doi: 10.1161/CIRCGEN.121.003365
6. Katz DH, Tahir UA, Bick AG, Pampana A, Ngo D, Benson MD, et al. Whole genome sequence analysis of the plasma proteome in black adults provides novel insights into cardiovascular disease. Circulation. (2021) 145:357–70. doi: 10.1161/CIRCULATIONAHA.121.055117
7. Francque S, Szabo G, Abdelmalek MF, Byrne CD, Cusi K, Dufour JF, et al. Nonalcoholic steatohepatitis: the role of peroxisome proliferator-activated receptors. Nat Rev Gastroenterol Hepatol. (2021) 18:24–39. doi: 10.1038/s41575-020-00366-5
8. Danielewski M, Kucharska AZ, Matuszewska A, Rapak A, Gomułkiewicz A, Dzimira S, et al. Cornelian cherry (Cornus mas L.) iridoid and anthocyanin extract enhances PPAR-α, PPAR-γ expression and reduces I/M ratio in aorta, increases LXR-α expression and alters adipokines and triglycerides levels in cholesterol-rich diet rabbit model. Nutrients. (2021) 13:3621. doi: 10.3390/nu13103621
9. Sun C, Mao S, Chen S, Zhang W, Liu C. PPARs-orchestrated metabolic homeostasis in the adipose tissue. Int J Mol Sci. (2021) 22:8974. doi: 10.3390/ijms22168974
10. Xu T, Liu R, Lu X, Wu X, Heneberg P, Mao Y, et al. Lycium barbarum polysaccharides alleviate LPS-induced inflammatory responses through PPARγ/MAPK/NF-κB pathway in bovine mammary epithelial cells. J Anim Sci. (2022) 100:skab345. doi: 10.1093/jas/skab345
11. Das NR, Vaidya B, Khare P, Bishnoi M, Sharma SS. Combination of Peroxisome proliferator-activated receptor gamma (PPARγ) agonist and PPAR gamma co-activator 1α (PGC-1α) activator ameliorates cognitive deficits, oxidative stress, and inflammation in rodent model of parkinson’s disease. Curr Neurovasc Res. (2021) 18:497–507. doi: 10.2174/1567202619666211217140954
12. Muzio G, Barrera G, Pizzimenti S. Peroxisome proliferator-activated receptors (PPARs) and oxidative stress in physiological conditions and in Cancer. Antioxidants (Basel). (2021) 10:1734. doi: 10.3390/antiox10111734
13. Wang H, Wang A, Wang X, Zeng X, Xing H. AMPK/PPAR-γ/NF-κB axis participates in ROS-mediated apoptosis and autophagy caused by cadmium in pig liver. Environ Pollut. (2022) 294:118659. doi: 10.1016/j.envpol.2021.118659
14. Bernardo A, Malara M, Bertuccini L, De Nuccio C, Visentin S, Minghetti L. The antihypertensive drug telmisartan protects oligodendrocytes from cholesterol accumulation and promotes differentiation by a PPAR-γ-mediated mechanism. Int J Mol Sci. (2021) 22:9434. doi: 10.3390/ijms22179434
15. Lim J, Kim HI, Bang Y, Choi HJ. Peroxisome proliferator-activated receptor gamma: a novel therapeutic target for cognitive impairment and mood disorders that functions via the regulation of adult neurogenesis. Arch Pharm Res. (2021) 44:553–63. doi: 10.1007/s12272-021-01333-7
16. Montaigne D, Butruille L, Staels B. PPAR control of metabolism and cardiovascular functions. Nat Rev Cardiol. (2021) 18:809–23. doi: 10.1038/s41569-021-00569-6
17. Bazina Martinović A, Merkler A, Ćelić I, Starčević K, Šimić M, Karmelić I, et al. Moderating effect of ppar-γ on the association of c-reactive protein and ischemic stroke in patients younger than 60. Gene. (2022) 809:146029. doi: 10.1016/j.gene.2021.146029
18. Migdalska-Sêk M, Modrzewska B, Kordiak J, Pastuszak-Lewandoska D, Kiszałkiewicz JM, Bielec F, et al. Diagnostic value of PPARδ and miRNA-17 expression levels in patients with non-small cell lung cancer. Sci Rep. (2021) 11:24136.
19. Yoon JS, Lee HJ, Sim DY, Im E, Park JE, Park WY, et al. Moracin D induces apoptosis in prostate cancer cells via activation of PPAR gamma/PKC delta and inhibition of PKC alpha. Phytother Res. (2021) 35:6944–53. doi: 10.1002/ptr.7313
20. Chandra A, Kaur P, Kumar Sahu S, Mittal A. A new insight into the treatment of Diabetes by means of pan PPAR agonists. Chem Biol Drug Des. (2022). doi: 10.1111/cbdd.14020 [Epub ahead of print].
21. Gong S, Han X, Li M, Cai X, Liu W, Luo Y, et al. Genetics and clinical characteristics of PPARγ variant-induced diabetes in a chinese han population. Front Endocrinol (Lausanne). (2021) 12:677130.
22. Li Z, Ren Q, Zhou Z, Cai Z, Wang B, Han J, et al. Discovery of the first-in-class dual PPARδ/γ partial agonist for the treatment of metabolic syndrome. Eur J Med Chem. (2021) 225:113807.
23. Sanjay SA, Lee HJ. Role of phytoconstituents as PPAR agonists: implications for neurodegenerative disorders. Biomedicines. (2021) 9:1914. doi: 10.3390/biomedicines9121914
24. Liu J, Zhao H, Yang L, Wang X, Yang L, Xing Y, et al. The role of CD36-Fabp4-PPARγ in skeletal muscle involves insulin resistance in intrauterine growth retardation mice with catch-up growth. BMC Endocr Disord. (2022) 22:10. doi: 10.1186/s12902-021-00921-4
25. Lai F, Wang J, Tang H, Bian X, Lu K, He G, et al. Adipogenic differentiation was inhibited by downregulation of PPARγ signaling pathway in aging tendon stem/progenitor cells. J Orthop Surg Res. (2021) 16:614. doi: 10.1186/s13018-021-02720-y
26. Fujimori K, Uno S, Kuroda K, Matsumoto C, Maehara T. Leukotriene C4 synthase is a novel PPARγ target gene, and leukotriene C4 and D4 activate adipogenesis through cysteinyl LT1 receptors in adipocytes. Biochim Biophys Acta Mol Cell Res. (2021) 1869:119203. doi: 10.1016/j.bbamcr.2021.119203
27. Liu F, Liu Y, Du Y, Li Y. MiRNA-130a promotes inflammation to accelerate atherosclerosis via the regulation of proliferator-activated receptor γ (PPARγ) expression. Anatol J Cardiol. (2021) 25:630–7. doi: 10.5152/AnatolJCardiol.2021.56721
28. Wang J, Xu X, Li P, Zhang B, Zhang J. HDAC3 protects against atherosclerosis through inhibition of inflammation via the microRNA-19b/PPARγ/NF-κB axis. Atherosclerosis. (2021) 323:1–12.
29. Zhuang J, Peng Y, Gu C, Chen H, Lin Z, Zhou H, et al. Wogonin accelerates hematoma clearance and improves neurological outcome via the PPAR-γ pathway after intracerebral hemorrhage. Transl Stroke Res. (2021) 12:660–75. doi: 10.1007/s12975-020-00842-9
30. Cataldi S, Costa V, Ciccodicola A, Aprile M. PPARγ and diabetes: beyond the genome and towards personalized medicine. Curr Diab Rep. (2021) 21:18.
31. Stalin A, Lin D, Josephine Princy J, Feng Y, Xiang H, Ignacimuthu S., et al. Computational analysis of single nucleotide polymorphisms (SNPs) in PPAR gamma associated with obesity, diabetes and cancer. J Biomol Struct Dyn. (2020):1–15. doi: 10.1080/07391102.2020.1835724
32. Meirhaeghe A, Cottel D, Amouyel P, Dallongeville J. Association between peroxisome proliferator-activated receptor gamma haplotypes and the metabolic syndrome in French men and women. Diabetes. (2005) 54:3043–8. doi: 10.2337/diabetes.54.10.3043
33. Li T, Xu X, Xu Y, Jin P, Chen J, Shi Y, et al. PPARG polymorphisms are associated with unexplained mild vision loss in patients with type 2 diabetes mellitus. J Ophthalmol. (2019) 2019:5284867. doi: 10.1155/2019/5284867
34. Tian J, Hu S, Wang F, Yang X, Li Y, Huang C. PPARG, AGTR1, CXCL16 and LGALS2 polymorphisms are correlated with the risk for coronary heart disease. Int J Clin Exp Pathol. (2015) 8:3138–43.
35. Zhang X, Lv S, Guo C, Shi C, Chi Y, Zhao L, et al. Gene-gene interaction between PPARG and CYP1A1 gene on coronary artery disease in the Chinese Han Population. Oncotarget. (2017) 8:34398–404. doi: 10.18632/oncotarget.16186
36. Ding X, Wang R, Liu L, Yu Q, Wang Z, Ma Z, et al. Interaction between peroxisome proliferator-activated receptor gamma and smoking on cardiovascular disease. Physiol Behav. (2016) 153:28–32. doi: 10.1016/j.physbeh.2015.10.014
37. Song Y, Raheel TM, Jia A, Dai G, Liu L, Long X, et al. rs10865710 polymorphism in PPARG promoter is associated with the severity of type 2 diabetes mellitus and coronary artery disease in a Chinese population. Postgrad Med J. (2021) 2021:140354. doi: 10.1136/postgradmedj-2021-140354
38. Li JP, Fu YP, Chang WX, Yi CR, Liu LH, Xing HY. Functional variant of C-689T in the peroxisome proliferator-activated receptor-γ2 promoter is associated with coronary heart disease in Chinese nondiabetic han people. Chin Med Sci J. (2017):177–84. doi: 10.24920/J1001-9294.2017.042
39. Li JP, Cheng LX, He MA, Wu TC. Study on the association of -689C/T polymorphism in the PPARgamma2 promoter with myocardial infarction. Chin J Med Genet. (2008) 25:19–22.
40. Evangelisti L, Attanasio M, Lucarini L, Sofi F, Marcucci R, Giglioli C, et al. PPARgamma promoter polymorphisms and acute coronary syndrome. Atherosclerosis. (2009) 205:186–91. doi: 10.1016/j.atherosclerosis.2008.11.009
41. Li L, Cheng LX, Nsenga R, He MA, Wu TC. Association between Pro12Ala polymorphism of peroxisome proliferator-activated receptor-gamma 2 and myocardial infarction in the Chinese Han population. Clin Cardiol. (2006) 29:300–4. doi: 10.1002/clc.4960290706
42. Wang LP, Zhao LR, Cui HW, Yan MR, Yang L, Su XL. Association between PPARγ2 Pro12Ala polymorphism and myocardial infarction and obesity in Han Chinese in Hohhot, China. Genet Mol Res. (2012) 11:2929–38. doi: 10.4238/2012.May.18.13
43. Hasan NS, Kamel SA, Hamed M, Awadallah E, Rahman AHA, Musa NI, et al. Peroxisome proliferator-activated receptor-γ polymorphism (rs1801282) is associated with obesity in Egyptian patients with coronary artery disease and type 2 diabetes mellitus. J Genet Eng Biotechnol. (2017) 15:409–14. doi: 10.1016/j.jgeb.2017.08.002
44. Maciejewska-Skrendo A, Pawlik A, Sawczuk M, Rać M, Kusak A, Safranow K, et al. PPARD and PPARG polymorphisms in patients with unstable angina. Gene. (2019) 711:143947. doi: 10.1016/j.gene.2019.143947
45. Vogel U, Segel S, Dethlefsen C, Tjønneland A, Saber AT, Wallin H, et al. PPARgamma Pro12Ala polymorphism and risk of acute coronary syndrome in a prospective study of Danes. BMC Med Genet. (2009) 10:52. doi: 10.1186/1471-2350-10-52
46. Schneider JG, Schiekofer S, von Eynatten M, Schlimmer P, Dugi KA. The proline 12 alanine substitution in the PPARgamma2 gene is associated with increased extent of coronary artery disease in men. Exp Clin Endocrinol Diabetes. (2009) 117:519–21. doi: 10.1055/s-0029-1216351
47. Ho JS, Germer S, Tam CH, So WY, Martin M, Ma RC, et al. Association of the PPARG Pro12Ala polymorphism with type 2 diabetes and incident coronary heart disease in a Hong Kong Chinese population. Diabetes Res Clin Pract. (2012) 97:483–91. doi: 10.1016/j.diabres.2012.03.012
48. Ridker PM, Cook NR, Cheng S, Erlich HA, Lindpaintner K, Plutzky J, et al. Alanine for proline substitution in the peroxisome proliferator-activated receptor gamma-2 (PPARG2) gene and the risk of incident myocardial infarction. Arterioscler Thromb Vasc Biol. (2003) 23:859–63. doi: 10.1161/01.ATV.0000068680.19521.34
49. Galgani A, Valdes A, Erlich HA, Mano C, Cheng S, Petrone A, et al. Homozygosity for the Ala allele of the PPARγ2 Pro12Ala polymorphism is associated with reduced risk of coronary artery disease. Dis Markers. (2010) 29:259–64. doi: 10.3233/DMA-2010-0756
50. Regieli JJ, Jukema JW, Doevendans PA, Zwinderman AH, van der Graaf Y, Kastelein JJ, et al. PPAR gamma variant influences angiographic outcome and 10-year cardiovascular risk in male symptomatic coronary artery disease patients. Diabetes Care. (2009) 32:839–44.
51. Youssef SM, Mohamed N, Afef S, Khaldoun BH, Fadoua N, Fadhel NM, et al. A Pro 12 Ala substitution in the PPARγ2 polymorphism may decrease the number of diseased vessels and the severity of angiographic coronary artery. Coron Artery Dis. (2013) 24:347–51. doi: 10.1097/MCA.0b013e328361a95e
52. Liu Y, Yuan Z, Liu Y, Zhang J, Yin P, Wang D, et al. PPARgamma gene C161T substitution is associated with reduced risk of coronary artery disease and decreased proinflammatory cytokine expression. Am Heart J. (2007) 154:718–24. doi: 10.1016/j.ahj.2007.06.009
53. Zhou X, Chen J, Xu W. Association between C1431T polymorphism in peroxisome proliferator-activated receptor-γ gene and coronary artery disease in Chinese Han population. Mol Biol Rep. (2012) 39:1863–8. doi: 10.1007/s11033-011-0931-y
54. Peng DQ, Zhao SP, Nie S, Li J. Gene-gene interaction of PPARgamma and ApoE affects coronary heart disease risk. Int J Cardiol. (2003) 92:257–63. doi: 10.1016/s0167-5273(03)00101-3
55. Wan J, Xiong S, Ma Y, Ren J, Cao M. Relationship of peroxisome proliferation-activated receptor-gamma C161→T gene polymorphism with coronary artery disease. Chin J Cardio. (2004) 32:39–42.
56. Li L, Dong Y, Ding M, Fu F, Gao W, Dong C. The association between PPARγ C161T point mutation and type 2 diabetes complicated with coronary heart disease. Chin J Diabet Mellit. (2005) 13:213–5.
57. He M, Jiang C, Wang F, Yang M, Tan H, Hu B, et al. Relationship between polymorphism of P22phox and PPAR-γ genes and coronary heart disease. J Environ Occupat Med. (2007) 24:1–4.
58. Wan J, Xiong S, Chao S, Xiao J, Ma Y, Wang J, et al. PPARgamma gene C161T substitution alters lipid profile in Chinese patients with coronary artery disease and type 2 diabetes mellitus. Cardiovas Diabet. (2010) 9:13. doi: 10.1186/1475-2840-9-13
59. Wang XL, Oosterhof J, Duarte N. Peroxisome proliferator-activated receptor gamma C161–>T polymorphism and coronary artery disease. Cardiovasc Res. (1999) 44:588–94. doi: 10.1016/s0008-6363(99)00256-4
60. Dallongeville J, Iribarren C, Ferričres J, Lyon L, Evans A, Go AS, et al. Peroxisome proliferator-activated receptor gamma polymorphisms and coronary heart disease. PPAR Res. (2009) 2009:543746.
61. Qian Y, Li P, Zhang J, Shi Y, Chen K, Yang J, et al. Association between peroxisome proliferator-activated receptor-alpha, delta, and gamma polymorphisms and risk of coronary heart disease: a case-control study and meta-analysis. Medicine (Baltimore). (2016) 95:e4299. doi: 10.1097/md.0000000000004299
62. Chao TH, Li YH, Chen JH, Wu HL, Shi GY, Liu PY, et al. The 161TT genotype in the exon 6 of the peroxisome-proliferator-activated receptor gamma gene is associated with premature acute myocardial infarction and increased lipid peroxidation in habitual heavy smokers. Clin Sci (Lond). (2004) 107:461–6. doi: 10.1042/CS20040014
63. Oladi M, Nohtani M, Avan A, Mirhafez SR, Tajbakhsh A, Ghasemi F, et al. Impact of the C1431T polymorphism of the peroxisome proliferator activated receptor-gamma (PPAR-γ) Gene on fasted serum lipid levels in patients with coronary artery disease. Ann Nutr Metab. (2015) 66:149–54. doi: 10.1159/000381358
64. Nikitin AG, Chistiakov DA, Minushkina LO, Zateyshchikov DA, Nosikov VV. Association of the CYBA, PPARGC1A, PPARG3, and PPARD gene variants with coronary artery disease and metabolic risk factors of coronary atherosclerosis in a Russian population. Heart Vessels. (2010) 25:229–36. doi: 10.1007/s00380-009-1159-9
65. Jiang J, Lan J, Du X. Association of peroxisome proliferator-activated receptor-gamma 2 variants with coronary heart disease. Zheji Med J. (2016) 38:485–8.
66. Fan L, Wu S, Yan Y. Association of PPAR gamma Pro12Ala polymorphisms with coronary heart disease. Clin Med China. (2010) 26:1043–6.
67. Wang Y, Song Y, Zhou J, Wu Z. Association of PPARγ2 gene Pro12Ala polymorphism in type 2 diabetes with coronary heart disease. J Fourth Milit Med Univ. (2009) 30:3107–9.
68. Tobin MD, Braund PS, Burton PR, Thompson JR, Steeds R, Channer K, et al. Genotypes and haplotypes predisposing to myocardial infarction: a multilocus case-control study. Eur Heart J. (2004) 25:459–67. doi: 10.1016/j.ehj.2003.11.014
69. Blüher M, Klemm T, Gerike T, Krankenberg H, Schuler G, Paschke R. Lack of association between peroxisome proliferator-activated receptor-gamma-2 gene variants and the occurrence of coronary heart disease in patients with diabetes mellitus. Eur J Endocrinol. (2002) 146:545–51. doi: 10.1530/eje.0.1460545
70. Nassar BA, Rockwood K, Kirkland SA, Ransom TP, Darvesh S, MacPherson K, et al. Improved prediction of early-onset coronary artery disease using APOE epsilon4, BChE-K, PPARgamma2 Pro12 and ENOS T-786C in a polygenic model. Clin Biochem. (2006) 39:109–14. doi: 10.1016/j.clinbiochem.2005.10.002
71. Rhee EJ, Kwon CH, Lee WY, Kim SY, Jung CH, Kim BJ, et al. No association of Pro12Ala polymorphism of PPAR-gamma gene with coronary artery disease in Korean subjects. Circ J. (2007) 71:338–42. doi: 10.1253/circj.71.338
72. Yilmaz-Aydogan H, Kurnaz O, Kurt O, Akadam-Teker B, Kucukhuseyin O, Tekeli A, et al. Effects of the PPARG P12A and C161T gene variants on serum lipids in coronary heart disease patients with and without Type 2 diabetes. Mol Cell Biochem. (2011) 358:355–63. doi: 10.1007/s11010-011-0987-y
73. Aydoğan HY, Küçükhüseyin O, Tekeli A, Isbir T. Associations of receptor for advanced glycation end products -374 T/A and Gly82 Ser and peroxisome proliferator-activated receptor gamma Pro12Ala polymorphisms in Turkish coronary artery disease patients. Genet Test Mol Biomarkers. (2012) 16:134–7. doi: 10.1089/gtmb.2011.0077
74. Zafarmand MH, van der Schouw YT, Grobbee DE, de Leeuw PW, Bots ML. Peroxisome proliferator-activated receptor gamma-2 P12Apolymorphism and risk of acute myocardial infarction, coronaryheart disease and ischemic stroke: a case-cohort study and meta-analyses. Vasc Health Risk Manag. (2008) 4:427–36. doi: 10.2147/vhrm.s2397
75. AshokKumar M, Veera Subhashini NG, Kanthimathi S, SaiBabu R, Ramesh A, Cherian KM, et al. Associations for lipoprotein lipase and peroxisome proliferator-activated receptor-gamma geneand coronary artery disease in an Indian population. Arch Med Res. (2010) 41:19–25. doi: 10.1016/j.arcmed.2010.01.005
76. Satirapoj B, Tasanavipas P, Supasyndh O. Role of TCF7L2 and PPARG2 Gene Polymorphisms in Renal and Cardiovascular Complications among Patients with Type 2 Diabetes: A Cohort Study. Kidney Dis (Basel). (2019) 5:220–7. doi: 10.1159/000497100
77. Wu Z, Lou Y, Jin W, Liu Y, Lu L, Lu G. The Pro12Ala polymorphism in the peroxisome proliferator-activated receptor gamma-2 gene (PPARγ2) is associated with increased risk of coronary artery disease: a meta-analysis. PLoS One. (2012) 7:e53105. doi: 10.1371/journal.pone.0053105
78. Ding S, Liu L, Zhuge QC, Yu Z, Zhang X, Xie J, et al. The meta-analysis of the association of PPARG P12A, C161T polymorphism and coronary heart disease. Wien Klin Wochenschr. (2012) 124:671–7. doi: 10.1007/s00508-012-0223-0
79. Zheng P, Xing X, Qing Q. Effect of PPARγC161-T gene polymorphism on coronary heart disease. Ji Xu Yi Xue Jiao Yu. (2018) 32:83–5.
80. Peng D, Zhao S, Li J, Nie S. Relationship between PPARγ C161→T Substitution and coronary heart disease. Chin Circul J. (2002) 17:370–3.
81. Yilmaz-Aydogan H, Kurnaz O, Kucukhuseyin O, Akadam-Teker B, Kurt O, Eronat AP, et al. Different effects of PPARA, PPARG and ApoE SNPs on serum lipids in patients with coronary heart disease based on the presence of diabetes. Gene. (2013) 523:20–6. doi: 10.1016/j.gene.2013.03.136
82. Yongsakulchai P, Settasatian C, Settasatian N, Komanasin N, Kukongwiriyapan U, Cote ML, et al. Association of combined genetic variations in PPARγ, PGC-1α, and LXRα with coronary artery disease and severity in Thai population. Atherosclerosis. (2016) 248:140–8. doi: 10.1016/j.atherosclerosis.2016.03.005
83. González-Castro TB, Tovilla-Zárate CA, Juárez-Rojop IE, Hernández-Díaz Y, López-Narváez ML, Rodríguez-Pérez C, et al. PON2 and PPARG polymorphisms as biomarkers of risk for coronary heart disease. Biomark Med. (2018) 12:287–97. doi: 10.2217/bmm-2017-0227
84. Wang P, Wang Q, Yin Y, Yang Z, Li W, Liang D, et al. Association between Peroxisome proliferator-activated receptor gamma gene polymorphisms and atherosclerotic diseases: a meta-analysis of case-control studies. J Atheroscler Thromb. (2015) 22:912–25. doi: 10.5551/jat.26138
85. Xu W, Xu J, Sun B, Chen H, Wang Y, Huang F, et al. The effect of PPARG polymorphisms on the risk of coronary heart disease: a meta-analysis. Mol Biol Rep. (2013) 40:875–84.
86. Wu Z, Lou Y, Jin W, Liu Y, Lu L, Lu G. The C161T polymorphism in the peroxisome proliferator-activated receptor gamma gene (PPARγ) is associated with risk of coronary artery disease: a meta-analysis. Mol Biol Rep. (2013) 40:3101–12. doi: 10.1007/s11033-012-2384-3
87. Abaj F, Sotoudeh G, Karimi E, Rafiee M, Koohdani F. Interaction between the dietary indices and PPAR-γ Pro12Ala gene variants on cardiovascular risk factors in patients with type 2 diabetes mellitus. Int J Clin Pract (2021) 21:e14307. doi: 10.1111/ijcp.14307
88. Gallicchio L, Kalesan B, Huang HY, Strickland P, Hoffman SC, Helzlsouer KJ. Genetic polymorphisms of peroxisome proliferator-activated receptors and the risk of cardiovascular morbidity and mortality in a community-based cohort in washington county. Maryland. PPAR Res. (2008) 2008:276581. doi: 10.1155/2008/276581
89. Lv O, Wang L, Li J, Ma Q, Zhao W. Effects of pomegranate peel polyphenols on lipid accumulation and cholesterol metabolic transformation in L-02 human hepatic cells via the PPARγ-ABCA1/CYP7A1 pathway. Food Funct. (2016) 7:4976–83. doi: 10.1039/c6fo01261b
90. Zhang F, Liu P, He Z, Zhang L, He X, Liu F, et al. Crocin ameliorates atherosclerosis by promoting the reverse cholesterol transport and inhibiting the foam cell formation via regulating PPARγ/LXR-α. Cell Cycle. (2022) 22:1–17. doi: 10.1080/15384101.2021.2015669
91. Jie J, Ling L, Yi Y, Tao L, Liao X, Gao P, et al. Tributyltin triggers lipogenesis in macrophages via modifying PPARγ pathway. Environ Pollut. (2021) 271:116331. doi: 10.1016/j.envpol.2020.116331
92. Schubert M, Becher S, Wallert M, Maeß MB, Abhari M, Rennert K, et al. The Peroxisome proliferator-activated receptor (PPAR)-γ antagonist 2-chloro-5-nitro-N-phenylbenzamide (GW9662) triggers perilipin 2 expression via PPARδ and induces lipogenesis and triglyceride accumulation in human THP-1 macrophages. Mol Pharmacol. (2020) 97:212–25. doi: 10.1124/mol.119.117887
93. Legchenko E, Chouvarine P, Borchert P, Fernandez-Gonzalez A, Snay E, Meier M, et al. PPARγ agonist pioglitazone reverses pulmonary hypertension and prevents right heart failure via fatty acid oxidation. Sci Transl Med. (2018) 10:eaao0303. doi: 10.1126/scitranslmed.aao0303
94. Sikder K, Shukla SK, Patel N, Singh H, Rafiq K. High Fat Diet Upregulates Fatty Acid Oxidation and Ketogenesis via Intervention of PPAR-γ. Cell Physiol Biochem. (2018) 48:1317–31. doi: 10.1159/000492091
95. Ren T, Yang WS, Lin Y, Liu JF, Li Y, Yang LC, et al. A novel PPARα/γ agonist, propane-2-sulfonic acid octadec-9-enyl-amide, ameliorates insulin resistance and gluconeogenesis in vivo and vitro. Eur J Pharmacol. (2018) 826:1–8. doi: 10.1016/j.ejphar.2018.02.029
96. Lu H, Wen D, Sun J, Zeng L, Du J, Du D, et al. Enhancer polymorphism rs10865710 associated with traumatic sepsis is a regulator of PPARG gene expression. Crit Care. (2019) 23:430. doi: 10.1186/s13054-019-2707-z
97. Wu L, Song Y, Zhang Y, Liang B, Deng Y, Tang T, et al. Novel genetic variants of PPARγ2 promoter in gestational diabetes mellitus and its molecular regulation in adipogenesis. Front Endocrinol (Lausanne). (2021) 11:499788. doi: 10.3389/fendo.2020.499788
98. Wang H, Xiong K, Sun W, Fu Y, Jiang Z, Yu D, et al. Two completely linked polymorphisms in the PPARG transcriptional regulatory region significantly affect gene expression and intramuscular fat deposition in the longissimus dorsi muscle of Erhualian pigs. Anim Genet. (2013) 44:458–62. doi: 10.1111/age.12025
99. Pihlajamäki J, Schwab U, Kaminska D, Ågren J, Kuusisto J, Kolehmainen M, et al. Dietary polyunsaturated fatty acids and the Pro12Ala polymorphisms of PPARG regulate serum lipids through divergent pathways: a randomized crossover clinical trial. Genes Nutr. (2015) 10:43. doi: 10.1007/s12263-015-0493-z
100. Rigau M, Juan D, Valencia A, Rico D. Intronic CNVs and gene expression variation in human populations. PLoS Genet. (2019) 15:e1007902. doi: 10.1371/journal.pgen.1007902
101. Suen TC, Goss PE. Identification of a novel transcriptional repressor element located in the first intron of the human BRCA1 gene. Oncogene. (2001) 20:440–50. doi: 10.1038/sj.onc.1204078
102. van Kuilenburg AB, Meijer J, Mul AN, Meinsma R, Schmid V, Dobritzsch D, et al. Intragenic deletions and a deep intronic mutation affecting pre-mRNA splicing in the dihydropyrimidine dehydrogenase gene as novel mechanisms causing 5-fluorouracil toxicity. Hum Genet. (2010) 128:529–38. doi: 10.1007/s00439-010-0879-3
103. Kumari P, Singh SK, Raman R. A novel non-coding RNA within an intron of CDH2 and association of its SNP with non-syndromic cleft lip and palate. Gene. (2018) 658:123–8. doi: 10.1016/j.gene.2018.03.017
104. Wang H, Song Y, Wu Y, Kumar V, Mahato RI, Su Q. Activation of dsRNA-dependent protein kinase R by miR-378 sustains metabolic inflammation in hepatic insulin resistance. Diabetes. (2021) 70:710–9. doi: 10.2337/db20-0181
105. Bartoszewski RA, Jablonsky M, Bartoszewska S, Stevenson L, Dai Q, Kappes J, et al. A synonymous single nucleotide polymorphism in DeltaF508 CFTR alters the secondary structure of the mRNA and the expression of the mutant protein. J Biol Chem. (2010) 285:28741–8. doi: 10.1074/jbc.M110.154575
106. Bruun GH, Doktor TK, Andresen BS. A synonymous polymorphic variation in ACADM exon 11 affects splicing efficiency and may affect fatty acid oxidation. Mol Genet Metab. (2013) 110:122–8. doi: 10.1016/j.ymgme.2013.06.005
107. Bertalovitz AC, Badhey MLO, McDonald TV. Synonymous nucleotide modification of the KCNH2 gene affects both mRNA characteristics and translation of the encoded hERG ion channel. J Biol Chem. (2018) 293:12120–36. doi: 10.1074/jbc.RA118.001805
108. Yusuf S, Hawken S, Ounpuu S, Dans T, Avezum A, Lanas F, et al. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet. (2004) 364:937–52. doi: 10.1016/s0140-6736(04)17018-9
109. Shen D, Ha D. Relationship between Pro12Ala variant of peroxisome proliferator-activated Receptorγ2 gene and coronary heart disease, or lipids metabolic disorders. Med J Wuhan Univ. (2005) 26:253–6.
110. Lu Z, Dong B, Mo X, Chen T, Wu H, Zhang Y, et al. Pro12Ala polymorphism in PPAR gamma 2 associated with essential hypertension in Chinese nonagenarians/centenarians. Exp Gerontol. (2008) 43:1108–13. doi: 10.1016/j.exger.2008.08.046
111. Koohdani F, Sotoudeh G, Kalantar Z, Mansoori A. PPARγ Pro12Ala polymorphism influences the relationship between dietary fat intake, adiposity and lipid profile in patients with type 2 diabetes. Int J Vitam Nutr Res. (2018) 88:263–9. doi: 10.1024/0300-9831/a000595
112. Rahimi Z, Chamaie-Nejad F, Saeidi S, Rahimi Z, Ebrahimi A, Shakiba E, et al. The association of PPARγ Pro12Ala and C161T polymorphisms with polycystic ovary syndrome and their influence on lipid and lipoprotein profiles. Int J Fertil Steril. (2018) 12:147–51. doi: 10.22074/ijfs.2018.5270
113. Ettehad D, Emdin CA, Kiran A, Anderson SG, Callender T, Emberson J, et al. Blood pressure lowering for prevention of cardiovascular disease and death: a systematic review and meta-analysis. Lancet. (2016) 387:957–67. doi: 10.1016/S0140-6736(15)01225-8
114. Yang W, Wang J, Ye W, Li X. Quantitative evaluation of PPAR-γ2 Pro12Ala polymorphism with hypertension. Herz. (2018) 43:719–27.
115. Cai G, Zhang X, Weng W, Shi G, Xue S, Zhang B. Associations between PPARG polymorphisms and the risk of essential hypertension. PLoS One. (2017) 12:e0181644. doi: 10.1371/journal.pone.0181644
116. Zhang M, Zhang J, Li L, Wang Q, Feng L. Association between peroxisome proliferator-activated receptor γ-2 gene Pro12Ala polymorphisms and risk of hypertension: an updated meta-analysis. Biosci Rep. (2019) 39:BSR20190022. doi: 10.1042/BSR20190022
117. Baghbani-Oskouei A, Gholampourdehaki M. Anthropometric measures and the risk of coronary artery disease. Caspian J Intern Med. (2020) 11:183–90. doi: 10.22088/cjim.11.2.183
118. Solymanzadeh F, Rokhafroz D, Asadizaker M, Dastoorpoor M. Prediction of risk of coronary artery disease based on framingham risk score in association with shift work among nurses. Int J Occup Saf Ergon. (2022) 22:1–17. doi: 10.1080/10803548.2021.2024403
119. Agrawal S, Klarqvist MDR, Emdin C, Patel AP, Paranjpe MD, Ellinor PT, et al. Selection of 51 predictors from 13,782 candidate multimodal features using machine learning improves coronary artery disease prediction. Patterns (N Y). (2021) 2:100364. doi: 10.1016/j.patter.2021.100364
120. Nishida Y, Iyadomi M, Tominaga H, Taniguchi H, Higaki Y, Tanaka H, et al. Influence of single-nucleotide polymorphisms in PPAR-δ, PPAR-γ, and PRKAA2 on the changes in anthropometric indices and blood measurements through exercise-centered lifestyle intervention in japanese middle-aged men. Int J Mol Sci. (2018) 19:703. doi: 10.3390/ijms19030703
121. Aguayo-Armendáriz J, Montalvo-Corral M, González-Martínez KA, Grijalva-Haro MI, Ballesteros-Vásquez MN, Caire-Juvera G, et al. Central obesity and body fat, but not body mass index, are associated with the Pro12Ala polymorphism in the peroxisome proliferator-activated receptor γ gene in a population with a high consumption of saturated and trans-fatty acids. Nutr Res. (2018) 57:28–35.
122. Masud S, Ye S. Effect of the peroxisome proliferator activated receptor-gamma gene Pro12Ala variant on body mass index: a meta-analysis. J Med Genet. (2003) 40:773–80. doi: 10.1136/jmg.40.10.773
123. Li M, Wang Y, Ayiguli A, Haiqiemuhan A, Liu Z. Meta-analysis of the association between PPARγ2 gene Pro12Ala polymorphism and obesity in Chinese population. Wei Sheng Yan Jiu. (2021) 50:315–9.
124. da Silva RSB, Persuhn DC, Barbosa FKL, de Souza MF, Sena KF, Costa MDS, et al. Relationship of the Pro12Ala polymorphism on the PPARy2 gene with the body composition of practitioners of cyclic exercises. Front Physiol. (2021) 11:633721. doi: 10.3389/fphys.2020.633721
125. Carrillo-Venzor MA, Erives-Anchondo NR, Moreno-González JG, Moreno-Brito V, Licón-Trillo A, González-Rodríguez E, et al. Pro12Ala PPAR-γ2 and +294T/C PPAR-δ polymorphisms and association with metabolic traits in teenagers from northern Mexico. Genes (Basel). (2020) 11:776. doi: 10.3390/genes11070776
126. Zafar U, Khaliq S, Ahmad HU, Lone KP. Peroxisome proliferator activated receptor gamma 34C>G variant and anthropometric parameters in metabolic syndrome. J Pak Med Assoc. (2019) 69:1259–65.
127. Sagris M, Theofilis P, Antonopoulos AS, Oikonomou E, Paschaliori C, Galiatsatos N, et al. Inflammation in coronary microvascular dysfunction. Int J Mol Sci. (2021) 22:13471.
128. Farahi L, Sinha SK, Lusis AJ. Roles of macrophages in atherogenesis. Front Pharmacol. (2021) 12:785220.
129. Bugger H, Zirlik A. Anti-inflammatory strategies in atherosclerosis. Hamostaseologie. (2021) 41:433–42. doi: 10.1055/a-1661-0020
130. Liu F, Mei X, Zhang Y, Qi H, Wang J, Wang Y, et al. Association of peroxisome proliferator-activated receptorγ gene Pro12Ala and C161T polymorphisms with cardiovascular risk factors in maintenance hemodialysis patients. Mol Biol Rep. (2014) 41:7555–65. doi: 10.1007/s11033-014-3645-0
131. Tsai YZ, Tsai ML, Hsu LY, Ho CT, Lai CS. Tetrahydrocurcumin upregulates the adiponectin-AdipoR pathway and improves insulin signaling and pancreatic β-cell function in high-fat Diet/streptozotocin-induced diabetic obese mice. Nutrients. (2021) 13:4552. doi: 10.3390/nu13124552
132. Ryu J, Hadley JT, Li Z, Dong F, Xu H, Xin X, et al. Adiponectin alleviates diet-induced inflammation in the liver by suppressing MCP-1 expression and macrophage infiltration. Diabetes. (2021) 70:1303–16. doi: 10.2337/db20-1073
133. Lei Y, Cui Q, Yang G, Piao L, Inoue A, Wu H, et al. Statins mitigate stress-related vascular aging and atherosclerosis in apoE-deficient mice fed high fat-diet: the role of glucagon-like peptide-1/adiponectin axis. Front Cell Dev Biol. (2021) 9:687868. doi: 10.3389/fcell.2021.687868
134. Campos-Perez W, Torres-Castillo N, Perez-Robles M, Muñoz-Valle JF, Vizmanos-Lamotte B, Martinez-Lopez E. Interaction of vitamin E intake and Pro12Ala polymorphism of PPARG with adiponectin levels. J Nutrigenet Nutrigenomics. (2017) 10:172–80. doi: 10.1159/000486160
Keywords: peroxisome proliferator-activated receptor gamma, PPARγ, PPARG, polymorphism, coronary artery disease
Citation: Song Y, Li S and He C (2022) PPARγ Gene Polymorphisms, Metabolic Disorders, and Coronary Artery Disease. Front. Cardiovasc. Med. 9:808929. doi: 10.3389/fcvm.2022.808929
Received: 18 November 2021; Accepted: 22 February 2022;
Published: 23 March 2022.
Edited by:
Liya Yin, Northeast Ohio Medical University, United StatesReviewed by:
Dora Janeth Fonseca, Rosario University, ColombiaCopyright © 2022 Song, Li and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuan He, aGVjaHVhbmNkdUBxcS5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.