- 1College of Life Sciences, Gwangju Institute of Science and Technology, Gwangju, South Korea
- 2Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, United States
- 3Basic Research Laboratory, Chonnam National University Medical School, Gwangju, South Korea
- 4Department of Molecular and Life Science, College of Science and Convergence Technology, Hanyang University-ERICA, Ansan, South Korea
Duchenne muscular dystrophy (DMD) is a genetic disorder characterized by progressive muscle degeneration due to dystrophin gene mutations. Patients with DMD initially experience muscle weakness in their limbs during adolescence. With age, patients develop fatal respiratory and cardiac dysfunctions. During the later stages of the disease, severe cardiac fibrosis occurs, compromising cardiac function. Previously, our research showed that the matricellular protein CCN5 has antifibrotic properties. Therefore, we hypothesized that CCN5 gene transfer would ameliorate cardiac fibrosis and thus improve cardiac function in DMD-induced cardiomyopathy. We utilized mdx/utrn (±) haploinsufficient mice that recapitulated the DMD-disease phenotypes and used an adeno-associated virus serotype-9 viral vector for CCN5 gene transfer. We evaluated the onset of cardiac dysfunction using echocardiography and determined the experimental starting point in 13-month-old mice. Two months after CCN5 gene transfer, cardiac function was significantly enhanced, and cardiac fibrosis was ameliorated. Additionally, running performance was improved in CCN5 gene-transfected mice. Furthermore, in silico gene profiling analysis identified utrophin as a novel transcriptional target of CCN5. This was supplemented by a utrophin promoter assay and RNA-seq analysis, which confirmed that CCN5 was directly associated with utrophin expression. Our results showed that CCN5 may be a promising therapeutic molecule for DMD-induced cardiac and skeletal dysfunction.
Introduction
Duchenne muscular dystrophy (DMD) is an X-linked genetic disorder characterized by progressive muscle weakness. In patients with DMD, a mutation in the dystrophin gene causes non-functional dystrophin proteins. Dystrophin proteins are essential for stabilizing the sarcolemma by connecting cytoskeletal F-actin to the extracellular matrix (1). The pathophysiological consequences of the mutation begin early, with two- to three-year-old patients experiencing a waddling gait and difficulty climbing steps. In their early teenage years, they begin to need wheelchairs and develop respiratory problems (2). Previously, respiratory issues were the most common cause of death in patients with DMD, but improved respiratory support devices have prolonged life expectancy. Currently, cardiac failure is the leading cause of death in patients (1–3). Cardiac fibrosis and LV dilation are observed in approximately 90% of patients over 18 years of age, and progress to severe cardiac dysfunction over time (4, 5).
CCN5, also known as WNT1-inducible signaling pathway protein 2 (WISP-2), is a matricellular protein in the cell communication network (CCN) family. The CCN family is involved in diverse cell behaviors by regulating proliferation, differentiation, and migration (6–9). Our group previously demonstrated that CCN5 prevents cardiac fibrosis (CF) by acting as an antagonist of connective tissue growth factor (CTGF), also known as CCN2, which has been widely studied as a profibrotic molecule (10). We have also shown that CCN5 reverses established cardiac fibrosis by two distinct mechanisms: (1) induction of apoptosis in myofibroblasts (MyoFBs), but not in fibroblasts (FBs) or myocytes, and (2) reverse trans-differentiation of MyoFBs to FBs (11).
Adeno-associated viruses (AAVs) are utilized as low-pathogenicity vectors for gene transfer (12). AAV serotypes (AAV.1–9) have different organ specificities; thus, choosing an appropriate vector for high transduction and tissue specificity is necessary. AAV.9 has been shown to be specific to the muscle, heart, liver, lung, and brain (13). Our group previously showed high expression of CCN5 in mouse cardiac tissue when using an AAV.9 vector (11). Other groups have successfully delivered microdystrophin using AAV.9 to the cardiac tissue in mdx mice (14).
In this study, we attempted to reduce cardiac fibrosis, which in turn improved cardiac function in a DMD mouse model, through CCN5 gene transfer. Several mouse models have recapitulated the pathophysiology of human DMD (15). Among them, mdx/utrn (±) haploinsufficient and mdx mouse models are widely used. The mdx mouse model has a mild muscular dystrophy phenotype compared to human DMD and a near-normal life span. Conversely, mdx/utrn KO mice show extremely early onset of muscle dystrophy and premature death (16). Thus, we chose the mdx/utrn (±) mouse model for this study. This model has a total deficiency of the dystrophin gene, but retains half an allele of the autosomal homologous utrophin gene. Thus, it has a relatively normal lifespan, with a relevant degree of muscle dystrophy, fibrosis, and cardiac dysfunction (17–21). Therefore, many groups have used this model and have suggested that it is ideal for testing gene therapies for DMD (17, 19–21). In fact, our characterization of aged mdx/utrn (±) mice (>13-month-old) showed a relevant degree of muscular dystrophic phenotype and cardiac dysfunction with severe fibrosis.
In this study, we hypothesized that CCN5 gene transfer may have a therapeutic effect in the context of cardiac function and fibrosis because we previously reported that CCN5 gene therapy reversed pre-established cardiac fibrosis and prevented further deterioration of cardiac function in a TAC-induced heart failure model (11). Moreover, recent in silico analysis led us to identify an additional molecular target of CCN5, utrophin. Luciferase assay analysis using the utrophin promoter confirmed that utrophin expression was also significantly restored by CCN5 gene transfer.
In conclusion, we showed in this study that CCN5 gene transfer ameliorates cardiac fibrosis and function not only by the reduction of cardiac fibrosis, but also by the upregulation of utrophin expression. Since utrophin gene transfer has been suggested as an alternative therapy for DMD (22–24), our results provide a new modality to treat severe cardiac dysfunction and fibrosis in patients with DMD.
Materials and Methods
Animal Care
All experimental procedures were approved by the Animal Care Committee of the Gwangju Institute of Science and Technology (approval number: GIST-2020-008). C57BL/10ScSn (Dmdmdx/Utrntm1Jrs) mice were purchased from The Jackson Laboratory (Bar Harbor, ME, #016622). We maintained the heterozygous mdx/utrn (±) mouse line by mating two male and female heterozygous mdx/utrn (±) mice and subsequent genotyping of every generation. To ensure an authentic dystrophic cardiomyopathic model, only aged mdx/utrn (±) mice (13–14-month-old; weight, approximately 35 g) were included in our study.
Genotyping
Genomic DNA was obtained from each mouse tail using the DNeasy Blood and Tissue kit (Qiagen, #69504, MD, United States) following the manufacturer’s protocol. Standard PCR was performed using primers provided by Jackson Lab protocol #27099. Primer 9290: 5′-CTG AGT CAA ACA GCT TGG AAG CCT CC-3′ (utrn common reverse); primer 9291: 5′-TTG CAG TGT CTC CCA ATA AGG TAT GAA C-3′ (utrn wild type forward); primer 9292: 5′-TGC CAA GTT CTA ATT CCA TCA GAA GCT G-3′ (utrn mutant forward). The WT and mut utrn PCR product sizes were 646 and 450 bp, respectively. Agarose gel electrophoresis was performed to confirm the individual mouse genotype, and only heterozygote mice, which had two PCR products using the primer sets above, were used in our study.
AAV.9-CCN5 Production and Injection
Recombinant AAV.9 was produced by the transfection of HEK293T cells. AAV.9 particles suspended in culture media were precipitated with ammonium sulfate and purified by ultracentrifugation on an iodixanol gradient. The particles were concentrated using a Vivaspin centrifugal concentrator (Merck, #Z614645). In this step, iodixanol was diluted using lactated Ringer’s solution via multiple washes. The AAV.9 titer was determined by quantitative real-time PCR and SDS-PAGE. AAV.9-Control (Con) and -CCN5 were injected into the tail vein of WT or mdx/utrn (±) mice. Mice received 5 × 1011 viral genomes and were analyzed 4 weeks later. AAV.9-Con encodes the virus-like particle (VLP) gene, which has no effect on fibrosis or cardiac function. AAV.9-CCN5 encodes the human CCN5.
Immunohistochemistry
Cardiac tissues from mice were cryopreserved with Tissue-Tek OCT Compound (Sakura, #4583) and sectioned at 6 μm. Tissues were fixed with 4% paraformaldehyde and stained with Masson-Trichrome (Sigma, #HT15-1KT). The fibrotic areas were visualized using a Zeiss Axiophot microscope.
Fibrotic Area Measurement
Fibrotic tissue was stained blue and non-fibrotic tissue was stained red. The percentage of fibrotic area was calculated as the ratio of the area of fibrosis to the total area of the section using the ImageJ plug-in software (NIH, United States).
Cardiac Fibroblasts and Myocytes Isolation
Cardiac fibroblasts (FBs) and myocytes were isolated from 13–14-month-old WT or mdx/utrn (±) mice. Hearts were immediately resected and retrogradely perfused with a Langendorff system filled with Tyrode’s buffer (137 mM NaCl, 5.4 mM KCl, 1 mM MgCl2, 10 mM glucose, 10 mM HEPES, pH 7.4) for 3 min at 37°C. Next, the buffer was switched to an enzyme solution mixture of 250 U/mL type II collagenase (Worthington, # LS004174) and 60 U/mL hyaluronidase (Worthington, # LS002594) and perfused for 20 min. The digested hearts were transferred to a Petri dish with 5% bovine serum albumin containing Tyrode’s buffer. The cardiac tissue was separated and filtered using a 100 μm pore size strainer (Fisher Scientific, # 352098). The filtered fraction was centrifuged at 50 × g for 1 min. The supernatant, which contained cardiac FBs, was centrifuged at 250 × g for 15 min and resuspended in DMEM (HyClone, # C838R16) containing 10% fetal bovine serum and 1% Penicillin-Streptomycin antibiotics (Thermo Fisher, United States). The cells were seeded in a culture dish; after 4 h, the culture medium was replaced with fresh medium. The precipitated cardiomyocyte fraction was resuspended in phosphate buffered saline and seeded in a 5 μg/mL laminin-coated culture dish. PBS was replaced with M199 medium containing 0.1% bovine serum albumin, 1 × insulin-transferrin-selenium (ITS), 10 mM BDM, 1 × CD lipid (chemically defined lipid concentrate), and 1 × penicillin/streptomycin after 1 h. Cells were incubated at 37°C in a 5% CO2 incubator.
Cardiomyocyte Contractility Analysis and Intracellular Ca2+ Transient Measurement
The isolated cardiomyocytes were plated on laminin-coated coverslips. The culture medium was changed to fresh medium after 1 h and incubated at 37°C in a 5% CO2 incubator. The next day, cardiomyocytes were treated with 1 μM Fura-2 AM for 10 min before mounting. The cover slip mounted with cardiomyocytes attached was mounted on the stage of an inverted microscope (Nikon Eclipse TE-100F) and perfused (approximately 1 mL/min at 25°C) with Tyrode’s buffer [137 mM NaCl, 5.4 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 10 mM glucose, 0.5 mM Taurin, and 10 mM HEPES (pH 7.4)]. Cardiomyocytes were field-stimulated at a frequency of 1 Hz and 40 V using a stimulator (Grass Technologies, United States). The cardiomyocytes were displayed on a monitor screen using an IonOptix MyoCam camera, and changes in cell length during shortening and lengthening were analyzed using IonWizard software.
A dual-excitation spectrofluorometer (IonOptix Inc., United States) was used to record the fluorescence measurements. The cardiomyocytes, mounted on the stage of an inverted microscope, were exposed to light from a 360 or 380 nm wavelength-filtered 75 W halogen lamp. Fluorescence emissions were detected after initial excitation at 360 nm for 0.5 s and then at 380 nm for the duration of the recording protocol. The 340/380 ratio of the fluorescence intensity (FFI) of the Fura indicated the intracellular Ca2+ concentration.
Echocardiography
Ketamine (100 μg/g) was injected intraperitoneally to anesthetize mice. Short-axis 2D images and M-mode tracings were recorded at the papillary muscle level to measure fractional shortening (FS), left ventricular internal dimension in diastole (LVIDd), and left ventricular internal dimension in systole (LVIDs) (GE Vivid 7 Vision, United States).
Treadmill and Grip Strength Test
A treadmill test was performed as previously described, with some modifications (25). Prior to beginning measurements, WT and mdx/utrn (±) mice were placed on an unmoving treadmill for 2 min for adjustment and then run at 4 m/min for 2 min and 8 m/min for 2 min to warm up. The main exercise was performed at a speed of 12 m/min, until the mice were exhausted. Exhaustion was determined when the mice stopped exercising and did not respond to electrical stimuli for 3–5 s. The aversive electric stimuli were set at 3 Hz, 1.22 mA to maintain the mice running. There was no inclination during this test. The total exercise distance was calculated by multiplying the exercise time and velocity. At the end of the test, mice were allowed to rest for 24 h and the harvested for molecular analysis.
The forearm grip strength of the WT and mdx/utrn (±) mice was assessed using an automated grip strength meter (Bioseb, BIO-GS3). The mice grasp a grid with their forepaws instinctively and try to hold while being pulled backward by the tail, releasing when they are unable to maintain grip. Five measurements within 2 min were taken from each animal at 1 week interval after the AAV.9 virus injection.
Western Blotting
Cardiac tissues, thigh skeletal muscle, and cell lysates were solubilized in RIPA buffer (50 mM Tris, 150 mM NaCl, 0.1% SDS, 1% Triton X-100, pH 8.0) mixed with protease inhibitor cocktail set III (Merck Millipore, #535140). Lysate concentrations were quantified using a Pierce BCA Protein Assay Kit (Thermo Scientific, #23227). Lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes (Merck Millipore, #IPVH00010). Transferred blots were blocked with 5% non-fat skim milk and incubated with antibodies against mouse monoclonal CCN5 (1:1,000, Sigma, #WH0008839M9), mouse monoclonal α-SMA (1:1,000, Sigma-Aldrich, #A5228), goat polyclonal collagen I (1:1,000, Abcam, ab34710), rabbit polyclonal SERCA2a (1:1,000, a custom antibody from 21st Century Biochemicals), rabbit polyclonal RyR2 (1:1,000, Alomone Lab, ARR-002), rabbit monoclonal NCX1 (1:1,000, Abcam, EPR12739), rabbit polyclonal p-PLN (1:1,000, Badrilla, A010-12AP), mouse monoclonal t-PLN (1:1,000, Badrilla, A010-14), and mouse monoclonal α-tubulin (1:3,000, Santa Cruz, #sc-8035) for 12–16 h at 4°C. After washing the blots with Tris-buffered saline containing 0.1% Tween 20 (TBS-T), they were incubated with secondary antibodies conjugated with horseradish peroxidase (Thermo Fisher Scientific, #31430) and washed again. The band signal was developed using chemiluminescence solution (Millipore, #WBKLS0500). ImageJ plug-in software was used to quantify the western blot results.
qRT-PCR
The mRNA expression level of utrophin was determined by real-time PCR using the QuantiTect SYBR Green real-time PCR Kit (Qiagen, #204243). Total RNA was isolated from cardiac tissues using TRIzol reagent (Invitrogen, #15596), according to the manufacturer’s instructions. Reverse transcription was performed at 50°C for 20 min, and cDNA was amplified in 20 μL reaction volumes using 10 pmol of primers for 37 cycles: 94°C for 10 s, 57°C for 15 s, and 72°C for 5 s. To calculate the relative abundance of the mRNAs, 18S rRNA was used as an internal control. The primer sequences used in this study were as follows: utrophin, 5′-TCA TGC TAG CCT GGA CCA TTT T-3′ (forward) and 5′-CAC TGA TGG GTG GTT TCC CA-3′ (reverse); 18S rRNA, 5′-TAA CGA ACG AGA CTC TGG CAT-3′ (forward) and 5′-CGG ACA TCT AAG GGC ATC ACA G-3 (reverse).
Reporter Gene Assay
The utrophin (UTRN) promoter was cloned into a pEZX-PG02.1 vector from a commercial source (GeneCopeia). UTRN promoter DNA (1 μg) and pcDNA3.1-CCN5-myc-his DNA (1 μg) were mixed with 3 μg polyethylenimine (PEI) and incubated for 20 min at 25°C. HEK293T cells were treated with DNA-PEI complexes, and after 4 h the culture medium was replaced with fresh medium. After 72 h, the culture media and lysates were collected and used to detect luciferase activity using a Pierce Gaussia Luciferase assay kit (Thermo Fisher, #16160). The luciferase signal was analyzed using a GloMax microplate reader. GABPα DNA was used as a positive control for the UTRN promoter assay.
Statistical Analysis
Student’s t-test and one-way analysis of variance (ANOVA) were used for statistical analyses to determine the significance of the data. Asterisks (*p < 0.05) or double asterisks (**p < 0.01) indicate significance. Data in the figures represent the mean ± standard deviation.
Results
CCN5 Prevents Cardiac Fibrosis in mdx/utrn (±) Mice
Two AAV.9 viral vectors with a CCN5 cassette (AAV.9-CCN5) and control virus (AAV.9-Con) were generated to examine the effects of CCN5 on cardiac fibrosis. We used at least 13-month-old mdx/utrn (±) mice to ensure the establishment of cardiac fibrosis and clinical relevance prior to CCN5 gene transfer. Mice received AAV.9-CCN5 (1E11 vg/mouse) through the tail vein and were analyzed 8 weeks later (Figure 1A). First, cardiac tissues were subjected to histological analysis. Heart sections were stained with Masson’s trichrome reagent to analyze CF. Fibrotic areas were significantly developed in the mid- and sub-epicardium within the inferior and lateral walls, in line with the literature (26). AAV.9-Con did not affect fibrotic development (Figures 1B,C). However, a significant decrease in fibrosis within the walls was observed in AAV9-CCN5-administered mice (Figures 1B,C). Molecular signatures of cardiac fibrosis were also evaluated by western blot analysis, which supported this observation. For example, α-SMA, a TGF-β-induced marker of myofibroblast differentiation, was exceptionally inhibited by CCN5 gene transfer (Figures 1D,E). Furthermore, levels of major calcium regulators, such as SERCA2a, RyR2, NCX1, and phospholamban (PLN, phosphor, and total forms) in the heart were measured in the same samples. SERCA2a expression, a hallmark of calcium regulation in the heart, was markedly preserved in CCN5 gene-transfected mice (Figures 1D,E). In addition, phosphorylated PLN levels were significantly restored in the CCN5 gene transfer group. Finally, RyR2 expression was not altered in this model, although NCX1 expression was downregulated in CCN5 injected group. Moreover, the antifibrotic effect of CCN5 was confirmed in isolated cardiac fibroblasts in vitro by western blot analysis, and representative profibrotic markers, such as α-SMA, collagen I, and TGF-β, were significantly reduced by CCN5 gene transfer (Supplementary Figures 1A,B). These data prove that CCN5 ameliorates cardiac fibrosis in mdx/utrn (±) mice.
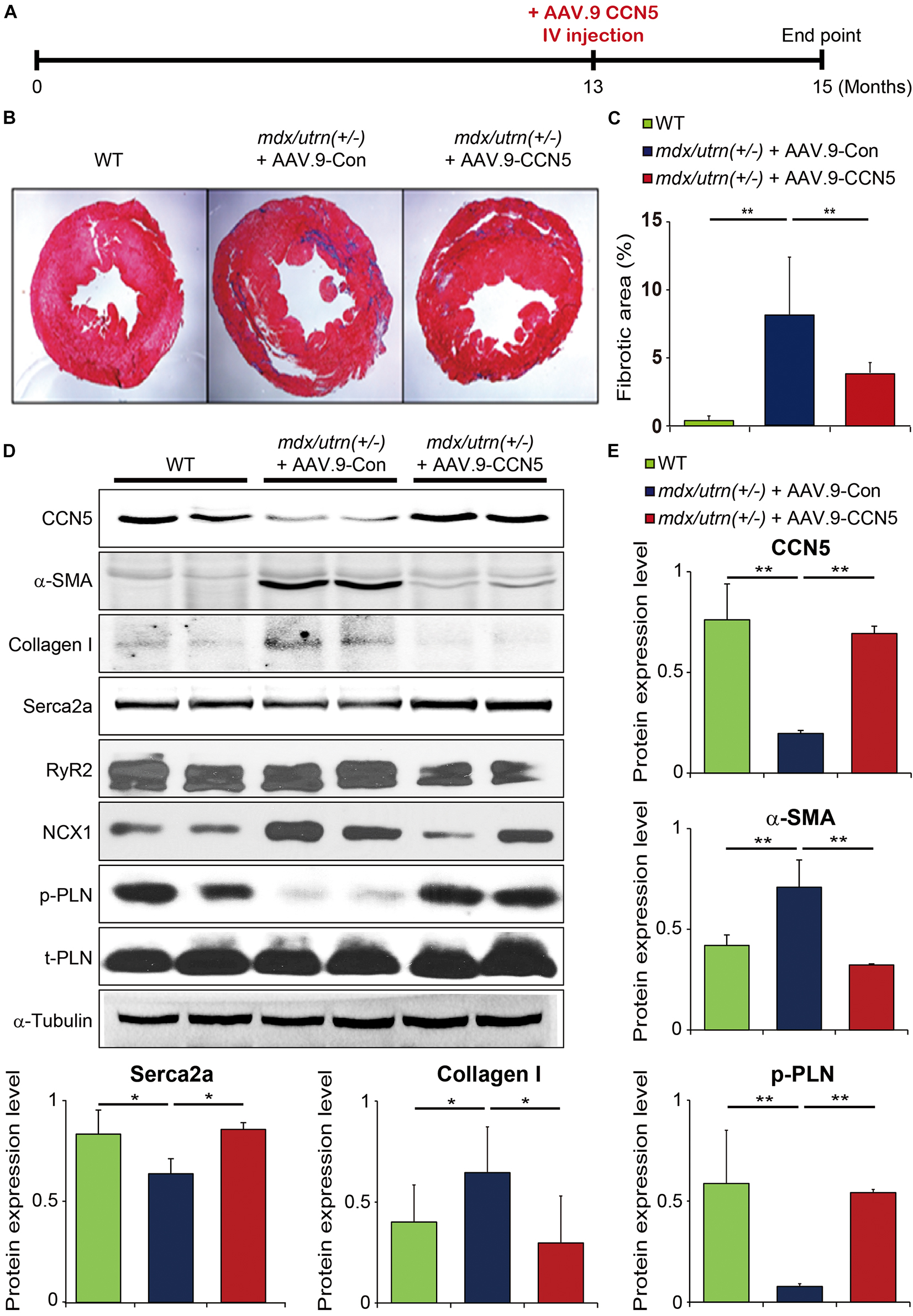
Figure 1. CCN5 prevents cardiac fibrosis in mdx/utrn (±) mice. (A) Experimental scheme for panels (B–E). mdx/utrn (±) mice were injected with AAV.9-Con or CCN5 into the tail vein, and hearts were harvested for experiments 8 weeks later. Age-matched WT mice are shown in comparison. (B) Hearts were sectioned and stained with trichrome. Blue areas indicate fibrotic tissue and red areas indicate normal tissue. (C) The ratio of fibrotic area over total tissue of the stained hearts was plotted. (D) Proteins obtained from cardiac tissue were immunoblotted with antibodies against CCN5, α-SMA, collagen I, SERCA2a, RyR2, NCX1, phosphorylated phospholamban (p-PLN), t-PLN and α-tubulin. (E) Protein bands on western blots were scanned and plotted. n = 6. *p < 0.05 and **p < 0.01.
CCN5 Reduces Cardiac Dysfunction in mdx/utrn (±) Mice
Restoration of SERCA2a levels results in enhanced calcium handling, which leads to increased cardiomyocyte contractility (27, 28). Therefore, we examined the effect of CCN5 overexpression on cardiomyocyte contractility in vitro. We also assessed cardiac function using echocardiography in vivo, since cardiac fibrosis negatively affects cardiac function (29–32).
Both cardiac fibroblasts and cardiomyocytes were isolated for further measurements. For this experiment, a modified Langendorff technique was applied to isolate both cell types from the hearts of mdx/utrn (±) mice, and Ca2+ transients and cardiomyocyte contractility were assessed using the IonOptix system, as described in section “Cardiomyocyte Contractility Analysis and Intracellular Ca2+ Transient Measurement.”
CCN5 gene transfer resulted in significantly enhanced Ca2+ transients and contractility in isolated cardiomyocytes compared to those in control viral vector-delivered mice (Figures 2A–D). Furthermore, the cardiac FB fraction was analyzed by western blotting, and several profibrotic markers were significantly reduced by CCN5 overexpression, as described previously (Supplementary Figure 1).
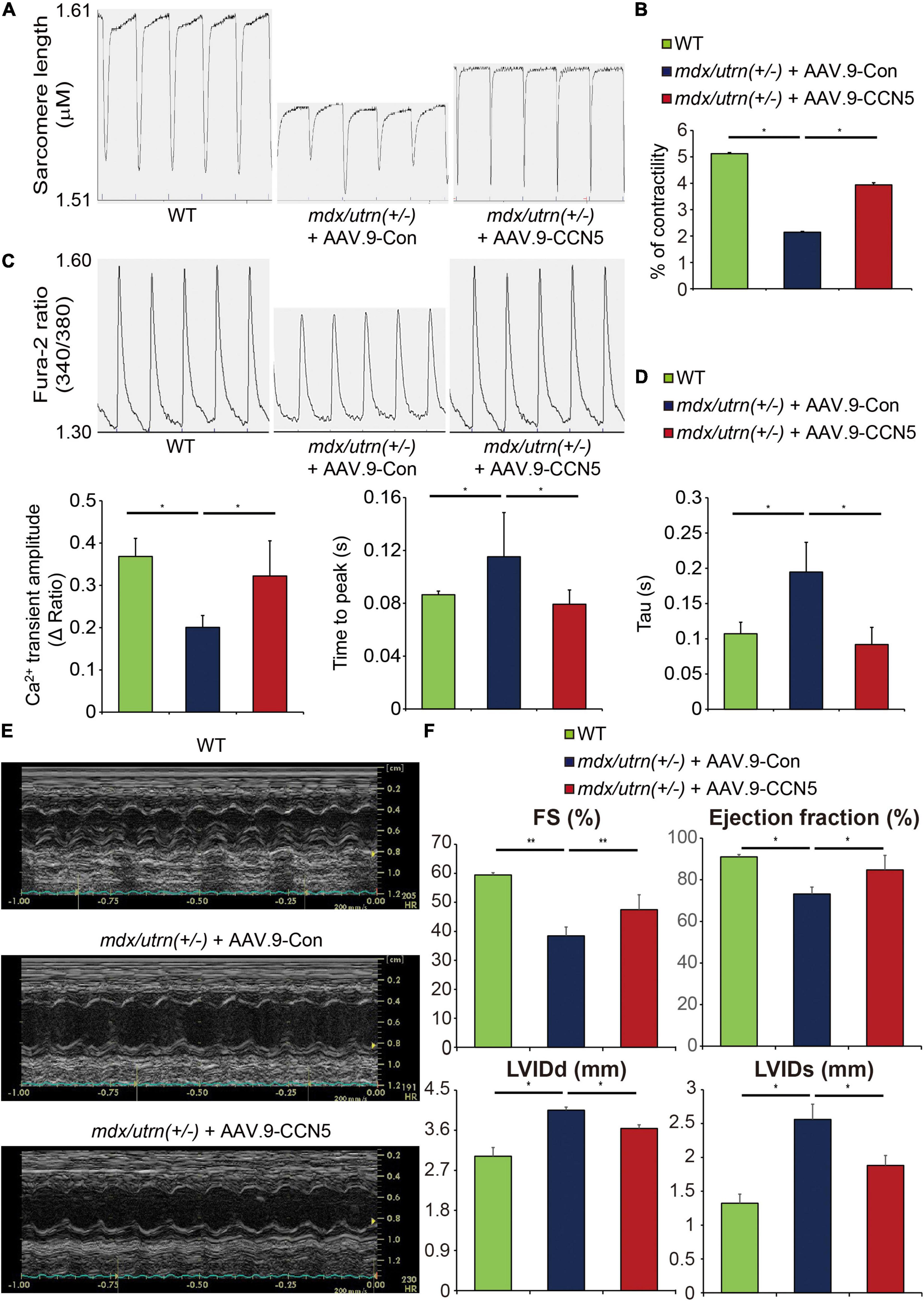
Figure 2. CCN5 reduces cardiac dysfunction in mdx/utrn (±) mice. (A) The Langendorff system was used to isolate cardiomyocytes. Traces of the sarcomere length were measured in the isolated cardiomyocytes. (B) Average contractility was calculated and analyzed. n ≥ 40 for analyzing the contractility. (C) Traces of the Fura-2 ratio of Ca2+ transients were measured in the isolated cardiomyocytes. n ≥ 40 for analyzing the Ca2+ transient parameters. (D) Average Ca2+ transient amplitude, time to peak and Tau were calculated and analyzed. (E) Representative images of echocardiogram are shown. (F) FS, LVIDd, LVIDs and EF were measured to analyze cardiac function by echocardiography. n = 6. *p < 0.05 and **p < 0.01.
To investigate in vivo cardiac function, echocardiography was performed 8 weeks after gene transfer (Figure 1A). Systolic function indices, including fractional shortening (FS) and ejection fraction (EF), were significantly enhanced, and LVIDd and LVIDs were normalized by CCN5 gene transfer (Figures 2E,F and Supplementary Table 1). These data suggest that CCN5 reduces cardiac dysfunction in mdx/utrn (±) mice by restoring calcium regulation and cardiomyocyte contractility both in vitro and in vivo.
CCN5 Enhances Exercise Performance of mdx/utrn (±) Mice
One of the primary symptoms in patients with DMD is muscle weakness throughout the body, which begins in the lower body around the thighs and pelvis (33–35). In animal models of DMD, muscle weakness has also been observed in multiple reports (36–38). In our aged mouse model, this phenomenon is heightened, and animals are unable to intake food and water properly due to their inability to cling onto the food dispenser. Therefore, food and water were placed at the bottom of the cage to make it easily accessible. However, aged mice with CCN5 gene transfer showed better mobility. Therefore, we performed exercise performance tests using a treadmill and a grip strength instrument for accurate measurements (Figure 3A) (25, 39). mdx/utrn (±) mice were exhausted at half the distance compared to the WT control, whereas mice with CCN5 gene transfer exhibited remarkably enhanced running ability (Figure 3B) as well as grip strength (Figure 3C). In addition, molecular signatures analyzed by western blotting and qRT-PCR after CCN5 gene transfer supported these observations. As shown in Figures 3D,E, the transcriptional and translational levels of α-SMA, a myofibroblast marker, were significantly reduced. Surprisingly, utrophin expression was also substantially elevated in skeletal muscle with CCN5 gene transfer, which implies that CCN5 modulates not only tissue fibrosis but also the transcription of utrophin. This result led us to further characterize the relationship between CCN5 and utrophin expression.
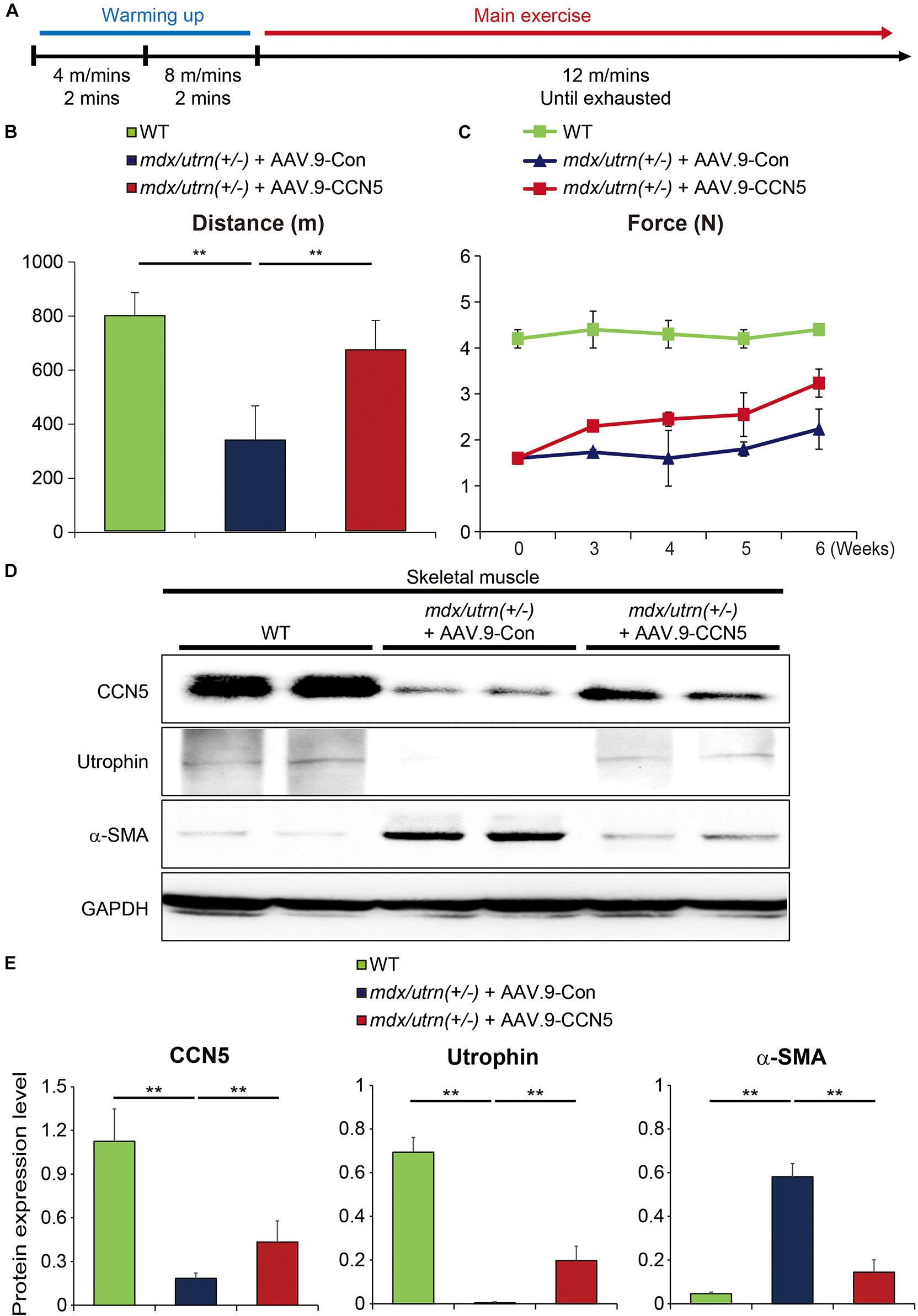
Figure 3. CCN5 enhances exercise performance of mdx/utrn (±) mice. (A) Experimental scheme for panel (B). Mice were injected with AAV.9-Con or CCN5 into the tail vein, and treadmill tests were performed 8 weeks later. Warming up exercises were serially performed at 4 m/min for 2 min and 8 m/min for 2 min. Main exercise was performed at 12 m/min until the mice were exhausted. (B) Total running distance was measured by calculating running time and velocity. (C) Comparison of grip strength tests was performed after AAV.9-Con or CCN5 injection. (D) Proteins from skeletal muscle tissue were immunoblotted with antibodies against CCN5, utrophin, α-SMA and GAPDH. (E) Protein bands on western blots were scanned and plotted. n = 6. **p < 0.01.
CCN5 Directly Regulates Utrophin Expression
Our results in section “CCN5 Enhances Exercise Performance of mdx/utrn (±) Mice” and previous literature imply that CCN5 can act as a transcriptional regulator in the nucleus (Supplementary Figure 3) (40). Our results suggest that CCN5 may modulate gene expression of certain structural molecules in cells. We identified utrophin as a novel target of CCN5 using gene expression profiling (data not shown).
To test this hypothesis, we observed utrophin levels in mdx/utrn (±) mice 8 weeks after CCN5 gene transfer. We confirmed mdx/utrn (±) genotypes following the Jackson Lab protocol, as described in section “Materials and Methods” (Supplementary Figure 2). We analyzed utrophin expression by western blotting and qRT-PCR.
Utrophin expression was significantly induced at both protein and mRNA levels (Figures 4A–C). In contrast, dystrophin expression was unaltered in all animals (Figures 4A,B). Furthermore, to confirm whether this effect was directly mediated between CCN5 and the target gene promoter, we performed a luciferase assay using the utrophin promoter with the positive control, GABPα, which is a previously reported transcriptional regulator of utrophin (Figure 4D). Our data suggest that CCN5 directly upregulates utrophin expression at the transcriptional level.
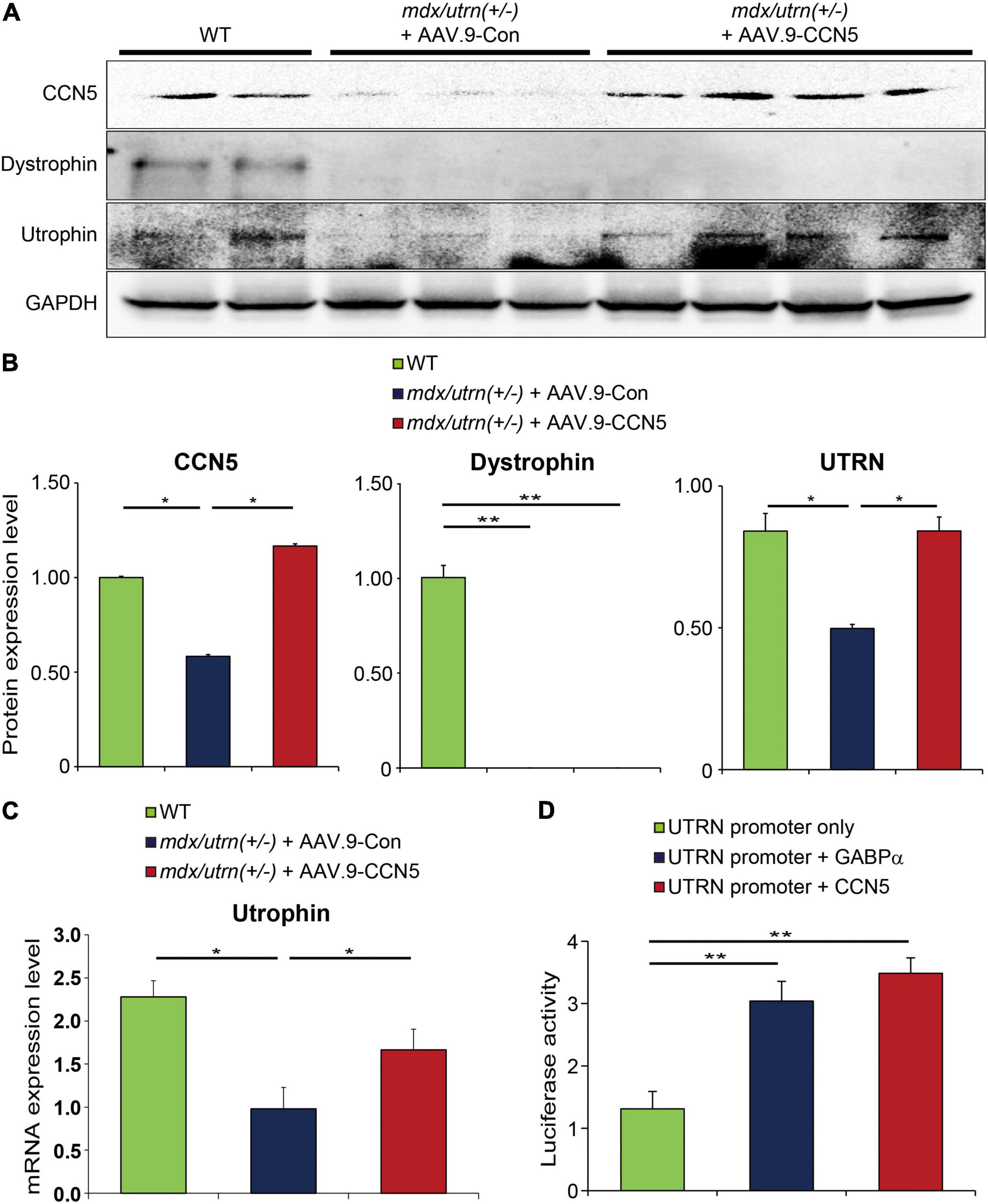
Figure 4. CCN5 directly regulates utrophin expression. (A) Proteins obtained from cardiac tissues were immunoblotted with antibodies against CCN5, dystrophin, utrophin and GAPDH. (B) Protein bands on western blots were scanned and plotted. (C) Relative mRNA expression level of utrophin in hearts. (D) Utrophin (UTRN) promoter only, UTRN promoter + GABPα, and UTRN promoter + CCN5 were transfected to 293T cells. Luciferase activity was measured 72 h later. Luciferase activity was analyzed by GloMax Microplate Reader. n = 6 for western blots. n = 4 for luciferase assay. *p < 0.05 and **p < 0.01.
Discussion
Patients with DMD suffer from deficiencies in the functional dystrophin protein, which causes a host of debilitating phenotypes. Dystrophin links actin and glycoproteins in the sarcolemma through the amino acid terminus of each protein. It stabilizes the plasma membrane, which controls the force generated by sarcomere contractions. Deficiency of dystrophin expression causes sarcolemma weakening, functional ischemia, free-radical damage, and cytosolic calcium overload, which promotes muscle damage (1). Therefore, dystrophin is a major target in DMD therapy. In fact, multiple groups have focused on dystrophin as a therapeutic target. Approaches include exon skipping, dystrophin gene transfer using an AAV vector, and Cas9-CRISPR-mediated dystrophin gene editing (41–44). However, the large size of the dystrophin protein, 427 kDa, makes gene therapy difficult because AAV vectors have cargo size limitations.
In addition to dystrophin, utrophin is a potential target for DMD treatment. Utrophin is similar in structure and function to dystrophin, and multiple reports have shown that utrophin compensates for the loss of dystrophin (45–48). In previous studies, utrophin upregulation by small molecules in DMD mouse models successfully ameliorated the symptoms of muscle weakness (49, 50). Since then, many studies have focused on developing novel small molecules that enhance utrophin expression (51). For example, “Ezutromid,” an orally administered small molecule that modulates utrophin production, was clinically tested as a novel DMD treatment despite not meeting the primary endpoint in phase 2 clinical trials (NCT02858362, PhaseOut DMD) (52, 53).
Although DMD patients show excessive fibrosis throughout all muscle fibers in the body, fibrotic pathology has not been the primary focus of treatment. However, significant fibrosis causes stiffness of the muscles and organs, further accelerating muscle deterioration. Cardiac dysfunction is highly correlated with deposition of fibrotic tissues in the hearts of DMD patients (4, 5). Previous studies have suggested that antifibrotic treatments, such as the TGF-β blocker suramin, which prevents muscle fibrosis in mdx mice, show promise in preventing DMD-related disease progression (54–56).
Previously, we showed that CCN5 is a promising treatment for cardiac fibrosis in a TAC-induced HF model (11). Therefore, we hypothesized that CCN5 might have a beneficial effect on cardiac fibrosis and function in DMD-associated cardiomyopathy. First, we evaluated whether cardiac fibrosis was reduced by CCN5 gene transfer in mdx/utrn (±) mice. As shown by the Masson trichrome tissue staining in Figure 1, cardiac fibrosis was markedly reduced by CCN5 overexpression (Figures 1B,C). The molecular signatures of profibrotic markers and calcium-handling proteins supported our observations (Figures 1D,E). Next, we characterized cardiac function with CCN5 gene transfer in vitro and in vivo in mdx/utrn (±) mice. Cardiomyocyte contractility and calcium transients were measured in cardiomyocytes isolated from the control and CCN5-treated groups. CCN5-treated groups showed significantly enhanced Ca2+ transients and contractility compared with the control group. In vivo experimental results also showed improved cardiac function, in line with the in vitro results.
Our functional data showed that CCN5 improved cardiac contractility and reduced fibrosis. Thus, we attempted to elucidate the mechanism underlying the ionotropic effects of CCN5. We hypothesized that CCN5 downregulated miRNA-25 (miR-25), which then enhanced cardiac contractility. In a previous study, we reported that miR-25 was a major regulator of SERCA2a in a TAC-induced heart failure model (57). Since then, we further characterized the regulatory mechanism of miR-25 biosynthesis and identified the major cell types that produce miR-25. As shown in Supplementary Figure 4, mature miR-25 was dominantly expressed in cardiomyocytes, whereas primary miR-25 was almost exclusively expressed in fibroblasts. This suggests that fibroblasts are the major miR-25 manufacturing cell type. Because CCN5 reverses cardiac fibrosis through the inhibition of transdifferentiation and selectively induces apoptosis in myofibroblasts, miR-25 expression may be downregulated by CCN5 overexpression. In turn, SERCA2a levels were not downregulated by miR-25, thus enhancing contractility and improving calcium handling. Further characterization is necessary to confirm our observations.
In addition, we observed that CCN5 gene transfer enhanced body mobility in mdx/utrn (±) mice (Figures 3B,C). We found that utrophin expression was substantially increased in the skeletal muscle of the CCN5-treated group (Figures 3D,E). This result led us to explore the role of CCN5 in the nucleus (Supplementary Figure 3), which has only been hinted at in previous report (40). To characterize the role of CCN5 as a transcriptional regulator, we performed a luciferase assay with a utrophin promoter in HEK-293 cells and compared the results with the in silico screening results for potential targets of CCN5 (data not shown). Although any indirect modulation is not completely ruled out, it clearly shows that there is a possible direct interaction between CCN5 and utrophin promoter (Figure 4D). Thus, further experiment such as Chromatin Immunoprecipitation (ChIP) assay will be required to support our observation. Furthermore, cardiac (Figures 4A,B) and skeletal muscle (Figures 3D,E) from CCN5 gene-transferred mice showed significantly enhanced expression of utrophin, strongly supporting our conclusions. Collectively, CCN5 gene transfer not only affects tissue fibrosis and calcium regulation but also modulates utrophin expression at the transcriptional level.
In conclusion, CCN5 is a promising and potent therapeutic molecule for DMD-induced cardiac and skeletal dysfunctions.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee (IACUC) at the Gwangju Institute of Science and Technology (GIST).
Author Contributions
WP and DJ conceptualized, designed, and supervised the study. MS and JY performed the experiments and analyzed the data. HK, WP, and DJ analyzed the data. MS, WP, and DJ wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
WP was supported by grants from the National Research Foundation of Korea (2019R1A2C2085457 and 2019R1A4A1028534) funded by the Korean government (MSIP). DJ was supported by grants from the National Research Foundation of Korea (2021R1A2C1008058 and 2021R1A4A5032463), funded by the Korean Government (MSIP) and Hanyang University (HY-202000000002749).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2022.763544/full#supplementary-material
References
1. Duan D, Goemans N, Takeda S, Mercuri E, Aartsma-Rus A. Duchenne muscular dystrophy. Nat Rev Dis Primers. (2021) 7:13.
3. Farini A, Gowran A, Bella P, Sitzia C, Scopece A, Castiglioni E, et al. Fibrosis rescue improves cardiac function in Dystrophin-deficient mice and duchenne patient-specific cardiomyocytes by immunoproteasome modulation. Am J Pathol. (2019) 189:339–53. doi: 10.1016/j.ajpath.2018.10.010
4. Kamdar F, Garry DJ. Dystrophin-deficient cardiomyopathy. J Am Coll Cardiol. (2016) 67:2533–46. doi: 10.1016/j.jacc.2016.02.081
5. Finsterer J, Stollberger C. The heart in human dystrophinopathies. Cardiology. (2003) 99:1–19. doi: 10.1159/000068446
6. Lau LF. Cell surface receptors for CCN proteins. J Cell Commun Signal. (2016) 10:121–7. doi: 10.1007/s12079-016-0324-z
7. Lau LF, Lam SC. The CCN family of angiogenic regulators: the integrin connection. Exp Cell Res. (1999) 248:44–57. doi: 10.1006/excr.1999.4456
8. Chen CC, Lau LF. Functions and mechanisms of action of CCN matricellular proteins. Int J Biochem Cell B. (2009) 41:771–83. doi: 10.1016/j.biocel.2008.07.025
9. Leask A, Abraham DJ. All in the CCN family: essential matricellular signaling modulators emerge from the bunker. J Cell Sci. (2006) 119:4803–10. doi: 10.1242/jcs.03270
10. Yoon PO, Lee MA, Cha H, Jeong MH, Kim J, Jang SP, et al. The opposing effects of CCN2 and CCN5 on the development of cardiac hypertrophy and fibrosis. J Mol Cell Cardiol. (2010) 49:294–303. doi: 10.1016/j.yjmcc.2010.04.010
11. Jeong D, Lee MA, Li Y, Yang DK, Kho C, Oh JG, et al. Matricellular protein CCN5 reverses established cardiac fibrosis. J Am Coll Cardiol. (2016) 67:1556–68. doi: 10.1016/j.jacc.2016.01.030
12. Gray SJ, Samulski RJ. Optimizing gene delivery vectors for the treatment of heart disease. Expert Opin Biol Ther. (2008) 8:911–22. doi: 10.1517/14712598.8.7.911
13. Inagaki K, Fuess S, Storm TA, Gibson GA, McTiernan CF, Kay MA, et al. Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8. Mol Ther. (2006) 14:45–53. doi: 10.1016/j.ymthe.2006.03.014
14. Shin JH, Nitahara-Kasahara Y, Hayashita-Kinoh H, Ohshima-Hosoyama S, Kinoshita K, Chiyo T, et al. Improvement of cardiac fibrosis in dystrophic mice by rAAV9-mediated microdystrophin transduction. Gene Ther. (2011) 18:910–9. doi: 10.1038/gt.2011.36
15. Yucel N, Chang AC, Day JW, Rosenthal N, Blau HM. Humanizing the mdx mouse model of DMD: the long and the short of it. NPJ Regen Med. (2018) 3:4.
16. Grady RM, Teng H, Nichol MC, Cunningham JC, Wilkinson RS, Sanes JR. Skeletal and cardiac myopathies in mice lacking utrophin and dystrophin: a model for Duchenne muscular dystrophy. Cell. (1997) 90:729–38. doi: 10.1016/s0092-8674(00)80533-4
17. Gutpell KM, Hrinivich WT, Hoffman LM. Skeletal muscle fibrosis in the mdx/utrn plus /- mouse validates its suitability as a murine model of Duchenne muscular dystrophy. PLoS One. (2015) 10:e0117306. doi: 10.1371/journal.pone.0117306
18. Deconinck AE, Rafael JA, Skinner JA, Brown SC, Potter AC, Metzinger L, et al. Utrophin-dystrophin-deficient mice as a model for Duchenne muscular dystrophy. Cell. (1997) 90:717–27. doi: 10.1016/s0092-8674(00)80532-2
19. Huang P, Cheng G, Lu HY, Aronica M, Ransohoff RM, Zhou L. IMPAIRED RESPIRATORY FUNCTION IN MDX AND MDX/UTRN(+/-) MICE. Muscle Nerve. (2011) 43:263–7. doi: 10.1002/mus.21848
20. van Putten M, Kumar D, Hulsker M, Hoogaars WM, Plomp JJ, van Opstal A, et al. Comparison of skeletal muscle pathology and motor function of dystrophin and utrophin deficient mouse strains. Neuromuscul Disord. (2012) 22:406–17. doi: 10.1016/j.nmd.2011.10.011
21. Zhou L, Rafael-Fortney JA, Huang P, Zhao XS, Cheng G, Zhou X, et al. Haploinsufficiency of utrophin gene worsens skeletal muscle inflammation and fibrosis in mdx mice. J Neurol Sci. (2008) 264:106–11. doi: 10.1016/j.jns.2007.08.029
22. Tinsley JM, Potter AC, Phelps SR, Fisher R, Trickett JI, Davies KE. Amelioration of the dystrophic phenotype of mdx mice using a truncated utrophin transgene. Nature. (1996) 384:349–53. doi: 10.1038/384349a0
23. Tinsley J, Deconinck N, Fisher R, Kahn D, Phelps S, Gillis JM, et al. Expression of full-length utrophin prevents muscular dystrophy in mdx mice. Nat Med. (1998) 4:1441–4. doi: 10.1038/4033
24. Deconinck N, Tinsley J, De Backer F, Fisher R, Kahn D, Phelps S, et al. Expression of truncated utrophin leads to major functional improvements in dystrophin-deficient muscles of mice. Nat Med. (1997) 3:1216–21. doi: 10.1038/nm1197-1216
25. Castro B, Kuang S. Evaluation of muscle performance in mice by treadmill exhaustion test and whole-limb grip strength assay. Bio Protoc. (2017) 7:e2237. doi: 10.21769/BioProtoc.2237
26. Silva MC, Meira ZMA, Giannetti JG, da Silva MM, Campos AFO, Barbosa MD, et al. Myocardial delayed enhancement by magnetic resonance imaging in patients with muscular dystrophy. J Am Coll Cardiol. (2007) 49:1874–9. doi: 10.1016/j.jacc.2006.10.078
27. Kranias EG, Hajjar RJ. Modulation of cardiac contractility by the phopholamban/SERCA2a regulatome. Circ Res. (2012) 110:1646–60. doi: 10.1161/CIRCRESAHA.111.259754
28. Park WJ, Oh JG. SERCA2a: a prime target for modulation of cardiac contractility during heart failure. BMB Rep. (2013) 46:237–43. doi: 10.5483/bmbrep.2013.46.5.077
29. Burlew BS, Weber KT. Cardiac fibrosis as a cause of diastolic dysfunction. Herz. (2002) 27:92–8. doi: 10.1007/s00059-002-2354-y
30. Dzeshka MS, Lip GY, Snezhitskiy V, Shantsila E. Cardiac fibrosis in patients with atrial fibrillation: mechanisms and clinical implications. J Am Coll Cardiol. (2015) 66:943–59. doi: 10.1016/j.jacc.2015.06.1313
31. Kong P, Christia P, Frangogiannis NG. The pathogenesis of cardiac fibrosis. Cell Mol Life Sci. (2014) 71:549–74. doi: 10.1007/s00018-013-1349-6
32. Segura AM, Frazier OH, Buja LM. Fibrosis and heart failure. Heart Fail Rev. (2014) 19:173–85. doi: 10.1007/s10741-012-9365-4
33. Kim HK, Merrow AC, Shiraj S, Wong BL, Horn PS, Laor T. Analysis of fatty infiltration and inflammation of the pelvic and thigh muscles in boys with Duchenne muscular dystrophy (DMD): grading of disease involvement on MR imaging and correlation with clinical assessments. Pediatr Radiol. (2013) 43:1327–35. doi: 10.1007/s00247-013-2696-z
34. Torriani M, Townsend E, Thomas BJ, Bredella MA, Ghomi RH, Tseng BS. Lower leg muscle involvement in Duchenne muscular dystrophy: an MR imaging and spectroscopy study. Skeletal Radiol. (2012) 41:437–45. doi: 10.1007/s00256-011-1240-1
36. Lefaucheur JP, Pastoret C, Sebille A. Phenotype of dystrophinopathy in old mdx mice. Anat Rec. (1995) 242:70–6. doi: 10.1002/ar.1092420109
37. Muntoni F, Mateddu A, Marchei F, Clerk A, Serra G. Muscular weakness in the MDX mouse. J Neurol Sci. (1993) 120:71–7. doi: 10.1016/0022-510x(93)90027-v
38. Tanabe Y, Esaki K, Nomura T. Skeletal muscle pathology in X chromosome-linked muscular dystrophy (MDX) mouse. Acta Neuropathol. (1986) 69:91–5. doi: 10.1007/BF00687043
39. Dougherty JP, Springer DA, Gershengorn MC. The treadmill fatigue test: a simple, high-throughput assay of fatigue-like behavior for the mouse. J Vis Exp. (2016) 31:54052. doi: 10.3791/54052
40. Sabbah M, Prunier C, Ferrand N, Megalophonos V, Lambein K, De Wever O, et al. CCN5, a novel transcriptional repressor of the transforming growth factor beta signaling pathway. Mol Cell Biol. (2011) 31:1459–69. doi: 10.1128/MCB.01316-10
41. Ousterout DG, Kabadi AM, Thakore PI, Majoros WH, Reddy TE, Gersbach CA. Multiplex CRISPR/CAS9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat Commun. (2015) 6:6244. doi: 10.1038/ncomms7244
42. Bengtsson NE, Hall JK, Odom GL, Phelps MP, Andrus CR, Hawkins RD, et al. Muscle-specific CRISPR/CAS9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat Commun. (2017) 8:14454.
43. Wang Z, Kuhr CS, Allen JM, Blankinship M, Gregorevic P, Chamberlain JS, et al. Sustained AAV-mediated dystrophin expression in a canine model of Duchenne muscular dystrophy with a brief course of immunosuppression. Mol Ther. (2007) 15:1160–6. doi: 10.1038/sj.mt.6300161
44. Duan D. Systemic AAV Micro-dystrophin gene therapy for Duchenne muscular dystrophy. Mol Ther. (2018) 26:2337–56. doi: 10.1016/j.ymthe.2018.07.011
45. Blake DJ, Tinsley JM, Davies KE. Utrophin: a structural and functional comparison to dystrophin. Brain Pathol. (1996) 6:37–47. doi: 10.1111/j.1750-3639.1996.tb00781.x
46. Hirst RC, McCullagh KJ, Davies KE. Utrophin upregulation in Duchenne muscular dystrophy. Acta Myol. (2005) 24:209–16.
47. Perkins KJ, Davies KE. The role of utrophin in the potential therapy of Duchenne muscular dystrophy. Neuromuscul Disord. (2002) 12(Suppl. 1):S78–89. doi: 10.1016/s0960-8966(02)00087-1
48. Tinsley JM, Davies KE. Utrophin: a potential replacement for dystrophin? Neuromuscul Disord. (1993) 3:537–9. doi: 10.1016/0960-8966(93)90111-v
49. Sengupta K, Loro E, Khurana TS. PMO-based let-7c site blocking oligonucleotide (SBO) mediated utrophin upregulation in mdx mice, a therapeutic approach for Duchenne muscular dystrophy (DMD). Sci Rep. (2020) 10:21492. doi: 10.1038/s41598-020-76338-1
50. Tinsley JM, Fairclough RJ, Storer R, Wilkes FJ, Potter AC, Squire SE, et al. Daily treatment with SMTC1100, a novel small molecule utrophin upregulator, dramatically reduces the dystrophic symptoms in the MDX mouse. PLoS One. (2011) 6:e19189. doi: 10.1371/journal.pone.0019189
51. Cossu G, Sampaolesi M. New therapies for Duchenne muscular dystrophy: challenges, prospects and clinical trials. Trends Mol Med. (2007) 13:520–6. doi: 10.1016/j.molmed.2007.10.003
52. Muntoni F, Maresh K, Davies K, Harriman S, Layton G, Rosskamp R, et al. PhaseOut DMD: a Phase 2, proof of concept, clinical study of utrophin modulation with ezutromid. Neuromuscular Disord. (2017) 27:S217.
53. Mah JK. An overview of recent therapeutics advances for Duchenne Muscular dystrophy. Methods Mol Biol. (2018) 1687:3–17. doi: 10.1007/978-1-4939-7374-3_1
54. Pessina P, Cabrera D, Morales MG, Riquelme CA, Gutierrez J, Serrano AL, et al. Novel and optimized strategies for inducing fibrosis in vivo: focus on Duchenne muscular dystrophy. Skelet Muscle. (2014) 4:7. doi: 10.1186/2044-5040-4-7
55. Taniguti AP, Pertille A, Matsumura CY, Santo Neto H, Marques MJ. Prevention of muscle fibrosis and myonecrosis in mdx mice by suramin, a TGF-beta1 blocker. Muscle Nerve. (2011) 43:82–7. doi: 10.1002/mus.21869
56. Zhou L, Lu H. Targeting fibrosis in Duchenne muscular dystrophy. J Neuropathol Exp Neurol. (2010) 69:771–6. doi: 10.1097/NEN.0b013e3181e9a34b
Keywords: CCN5, DMD, cardiomyopathy, utrophin, AAV9
Citation: Song MH, Yoo J, Oh JG, Kook H, Park WJ and Jeong D (2022) Matricellular Protein CCN5 Gene Transfer Ameliorates Cardiac and Skeletal Dysfunction in mdx/utrn (±) Haploinsufficient Mice by Reducing Fibrosis and Upregulating Utrophin Expression. Front. Cardiovasc. Med. 9:763544. doi: 10.3389/fcvm.2022.763544
Received: 24 August 2021; Accepted: 31 March 2022;
Published: 26 April 2022.
Edited by:
Jingyan Han, Boston University, United StatesReviewed by:
Satvik Mareedu, Rutgers University, Newark, United StatesChang-Hoon Woo, Yeungnam University, South Korea
Copyright © 2022 Song, Yoo, Oh, Kook, Park and Jeong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Woo Jin Park, d29vamlucGFya0BpY2xvdWQuY29t; Dongtak Jeong, Y29vbGpkdEBoYW55YW5nLmFjLmty
 Min Ho Song
Min Ho Song Jimeen Yoo2
Jimeen Yoo2 Jae Gyun Oh
Jae Gyun Oh Dongtak Jeong
Dongtak Jeong