
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cardiovasc. Med., 31 October 2022
Sec. Cardiovascular Biologics and Regenerative Medicine
Volume 9 - 2022 | https://doi.org/10.3389/fcvm.2022.1045455
This article is part of the Research TopicCellular and Molecular Mechanisms of Cardiovascular and Metabolic DiseasesView all 5 articles
Studies in the past decades have uncovered an emerging role of the nucleolus in stress response and human disease progression. The disruption of ribosome biogenesis in the nucleolus causes aberrant nucleolar architecture and function, termed nucleolar stress, to initiate stress-responsive pathways via nucleolar release sequestration of various proteins. While data obtained from both clinical and basic investigations have faithfully demonstrated an involvement of nucleolar stress in the pathogenesis of cardiomyopathy, much remains unclear regarding its precise role in the progression of cardiac diseases. On the one hand, the initiation of nucleolar stress following acute myocardial damage leads to the upregulation of various cardioprotective nucleolar proteins, including nucleostemin (NS), nucleophosmin (NPM) and nucleolin (NCL). As a result, nucleolar stress plays an important role in facilitating the survival and repair of cardiomyocytes. On the other hand, abnormalities in nucleolar architecture and function are correlated with the deterioration of cardiac diseases. Notably, the cardiomyocytes of advanced ischemic and dilated cardiomyopathy display impaired silver-stained nucleolar organiser regions (AgNORs) and enlarged nucleoli, resembling the characteristics of tissue aging. Collectively, nucleolar abnormalities are critically involved in the development of cardiac diseases.
Ribosome biogenesis is a highly conserved biological process essential to all living organisms (1). In eukaryotic cells, the nucleolus is a prominent sub-nuclear compartment central to ribosome biogenesis (2). The nucleolus is organized around ribosomal DNA (rDNA) distributed within acrocentric chromosomes. rDNA is transcribed into 47S pre-ribosomal RNA (pre-rRNA) by RNA polymerase I (Pol I) transcriptional machinery. Then, 47S rRNA is cleaved and processed into mature ribosomal RNAs, including 28 S, 18 S and 5.8 S rRNAs, via the assistance of heterogeneous nuclear ribonucleoproteins (hnRNPs) (3). Finally, the rRNAs were assembled, together with ribosomal proteins and 5S rRNA, into 40S and 60S ribosomal subunits, followed by the export of these subunits into cytoplasm. rRNA transcription is fundamental to nucleolar dynamics and integrity. Mammalian oocytes and zygotes contain morphologically distinct and transcriptionally inactive nucleoli termed nucleolus precursor bodies (NPBs), which consist of ribosomal proteins, fibrillarin and hnRNPs (4). rRNA transcription serves as a seeding mechanism to initiate nucleolar assembly (5, 6). Thus, the dynamics of nucleolar architecture is tightly controlled by the transcription of ribosomal RNA (rRNA) and the demand of ribosome synthesis.
Accumulating data have suggested a critical involvement of ribosome biogenesis and nucleolar dynamics in stress responses and human diseases. In this regard, nucleolar stress, a term indicating the disruption of ribosome biogenesis and nucleolar integrity, has been increasingly recognized as an important modulator of stress responses. Nucleolar stress is initiated under various stress conditions, including genotoxic stress, hypoxia, nutrient deprivation and thermal stress, and plays a critical role in stress-induced signaling transductions. Notably, data obtained from both clinical and basic researches have pointed to a pivotal role of nucleolar malfunction in the development of cardiac diseases. Of importance, nucleolar stress may elicit both beneficial and adverse effects during cardiac injury. In this review, we summarize current findings linking nucleolar function to cardiovascular diseases, and discuss both the protective and detrimental effects of the nucleolus and nucleolar stress.
While the canonical role of the nucleolus as a ribosome factory has been well-characterized, studies in recent years have suggested that the nucleolus possesses various non-ribosomal functions. The nucleolus is broadly implicated in the regulation of transfer RNA (tRNA) expression and processing, centromere assembly, nuclear architecture, X-chromosome inactivation and DNA damage responses (7–10). These studies indicated that, besides a central role in ribosome biogenesis, the nucleolus is critically involved in the regulation of signaling transduction and cellular homeostasis. Many nucleolar proteins and non-coding RNAs have been reportedly involved in various cellular responses and metabolic control. These studies highlight the importance of the non-canonical function of the nucleolus in the development of various human diseases, including cardiac damage and failure.
Multiple proteomic studies revealed that the nucleolus contains thousands of proteins, of which only a small proportion have been associated with ribosome biogenesis (11). The dynamics of nucleolar proteome and morphology are closely associated with cellular homeostasis and proliferative status (12). During cell cycle progression, the nucleolus is disassembled at the end of prophase as a result of nucleolar protein phosphorylation by Cdk1-cyclin B complex (13). The reassembly of the nucleolus during telophase is initiated by rRNA transcription and subsequent recruitment of various nucleolar proteins. Recent investigations revealed that the nucleolus is assembled through liquid–liquid phase separation (LLPS), displaying dynamic exchange with the surrounding nucleoplasm (14, 15). Nucleolar proteins may constitutively shuttle within and outside the nucleolus, and the dynamics of nucleolar proteins are regulated by metabolic and genotoxic stresses (16–18). Moreover, nucleolar accumulation of various proteins critical for intracellular signaling transduction and cell physiology has been increasingly characterized in the past decades (19). Mechanistic investigations revealed that proteins critical to signaling transduction may be sequestered in the nucleolus, especially under stress conditions. For instance, hypoxia and acidosis may trigger nucleolar sequestration of E3 ubiquitin ligase VHL, which is key to HIF-1α stabilization and the transcription of hypoxia-responsive genes (20, 21). Some recent studies uncovered that the nucleolus plays a central role in nuclear protein quality control in response to heat shock (22, 23). It was revealed that nuclear proteins, such as the Polycomb group protein (PcG) complex, may accumulate in the nucleolus under heat shock, and undergo recovery and refolding via an Hsp70-dependent mechanism after exiting from heat shock. Furthermore, mounting investigations suggested that the nucleolus serves as a compartment of protein ubiquitination and degradation. Nucleolar translocation of various nuclear proteins, including histones, cell cycle regulators and transcriptional factors, leads to their ubiquitination and proteasomal degradation (24–27). These findings highlight the importance of non-ribosomal functions of nucleolar proteins in cellular homeostasis.
In addition, small nucleolar RNAs (snoRNAs) have also been reportedly involved in various biological processes. snoRNAs are 60–300 nucleotide non-coding RNAs primarily localized in the nucleolus (28). snoRNAs play indispensable roles in rRNA chemical modifications, but also exert various non-ribosomal functions. Carlos Michel et al. identified three snoRNAs in ribosomal protein L13a (rpL13a) locus, including U32a, U33 and U35a, play critical roles in transmitting metabolic stress pathways (29). Further studies revealed that these snoRNAs modulate mitochondrial metabolism and the production of reactive oxygen species (ROS), leading to alterations in pancreatic insulin secretion and systemic glucose metabolism (30). Nuclear snoRNA may function as a component of spliceosome to regulate pre-mRNA splicing (31, 32). snoRNAs may also be released from the nucleolus into the cytoplasm under metabolic stress and doxorubicin exposure (33, 34). In the cytoplasm, snoRNAs may form mRNA-snoRNA to regulate mRNA translation and decay (35, 36). snoRNAs may also directly bind to various proteins to modulate the activation of signaling pathways (37, 38). Notably, altered expression of snoRNAs has also been observed in cardiomyopathic conditions, implicating an involvement of snoRNAs in cardiac disease progression (39–41).
Ribosome biogenesis is an energy-intensive process, and is thus subjected to delicate regulation by various upstream signals (42). Coordinate regulation of rRNA transcription is achieved mainly through modulating the activity of key components of Pol I transcriptional machinery by various upstream regulators (43). Pol I transcriptional machinery is a multi-subunit protein complex, composed of Pol I and various essential transcriptional co-factors, including SL1, TIF-IA and UBF (44). The expression and posttranscriptional modification of these proteins are tightly regulated by signaling pathways involved in cell proliferation, mitogenic growth, nutrient and energy sensing, as well as stress responses. For instance, TIF-IA is phosphorylated on Ser635 residue by AMPK following energy deprivation, and on Thr 200 residue by JNK2 following oxidative stress (45, 46). Both modifications cause the dissociation between SL1 and TIF-IA, consequently leading to the disruption of rRNA transcription. On the other hand, phosphorylation of TIF-IA by growth-related kinases, including EKR, mTOR and CK2 enhances TIF-IA activity and rRNA synthesis (47–49). Likewise, the activity of SL1, UBF and Pol I are also tightly regulated by posttranscriptional modifications, particularly phosphorylation, in response to upstream signals (43). In addition to the regulation of Pol I transcriptional machinery, epigenetic regulation of rDNA accessibility also plays a pivotal role in nucleolar dynamics (50). For instance, owing to open chromatin conformation and high rDNA accessibility, embryonic stem cells (ESCs) typically contain one large nucleolus. The exit from pluripotency in ESCs triggers epigenetic silencing of rDNA, leading to the formation of heterochromatic shell that compartments the large nucleolus into smaller nucleoli (51, 52). Notably, data obtained from multiple organisms revealed that epigenetic silencing of rDNA is tightly correlated with biological aging (53, 54). Thus, rRNA transcription and nucleolar morphology are closely associated with cellular physiological status.
Mounting studies have revealed that nucleolar integrity serves as a stress sensor and signaling hub following stress conditions (55). Nucleolar stress caused by aberrant rRNA transcription disrupts nucleolar integrity, leading to the aberrant release of nucleolar proteins, such as ribosomal proteins, NPM and NS, into the nucleoplasm, where they bind to various signaling proteins and alter their stability and activity. A most prominent stress pathway downstream of nucleolar stress is the p53 pathway. A pioneer work conducted by Carlos Rubbi and Jo Milner revealed that nucleolar disruption plays a causal role in p53 activation under DNA damage and other stress conditions, and that the disruption of nucleolar integrity is sufficient to activate p53 in the absence of stress stimuli (56). Later investigations uncovered that multiple nucleolar proteins, including NPM, p14Arf, ribosomal proteins RPL11 and RPL5, are key to p53 stabilization following nucleolar disruption (57). For instance, the blockade of rRNA transcription causes nucleoplasmic accumulation of 5S ribonucleoprotein particle (5S RNP), which comprises the 5S rRNA, ribosomal proteins RPL11 and RPL5 (58). This complex binds to and inactivates mouse double minute 2 homolog (MDM2), an E3 ligase responsible for p53 ubiquitination and degradation under quiescent conditions, leading to p53 accumulation and activation (59). Nucleolar stress-p53 pathway has been associated with a variety of cellular events, including senescence, apoptosis, autophagy and differentiation (60–63).
In addition to mdm2-p53 pathway, nucleolar stress also triggers stress events through p53-independent mechanisms (57, 64). Nucleolus and nucleolar stress have been reportedly involved in the regulation of a growing number of signaling pathways, such as NF-κB, HIF-1α and CDK4/6 (Figure 1) (20, 65, 66). It is speculated that nucleolar stress may participate in the regulation of a majority of non-ribosomal nucleolar functions discussed in section 2. A variety of nucleolar stress-inducing stimuli, such as hypoxia, heat shock and oxidative stress, cause altered distribution and function of nucleolar proteins and snoRNAs (55, 67). Accordingly, the pivotal role of the nucleolus in the development of various human diseases has increasingly attracted research and therapeutic attention in recent years. While the role of nucleolar stress in some chronic diseases, such as cancer and neurodegenerative diseases, has been intensively investigated, the connection between nucleolar stress and cardiovascular disease remains largely obscure (68). Moreover, previous studies suggest that nucleolar stress appears to elicit both protective and detrimental roles in cardiovascular disorders. These facts indicate a complicated role of the nucleolus in the development of cardiovascular diseases.
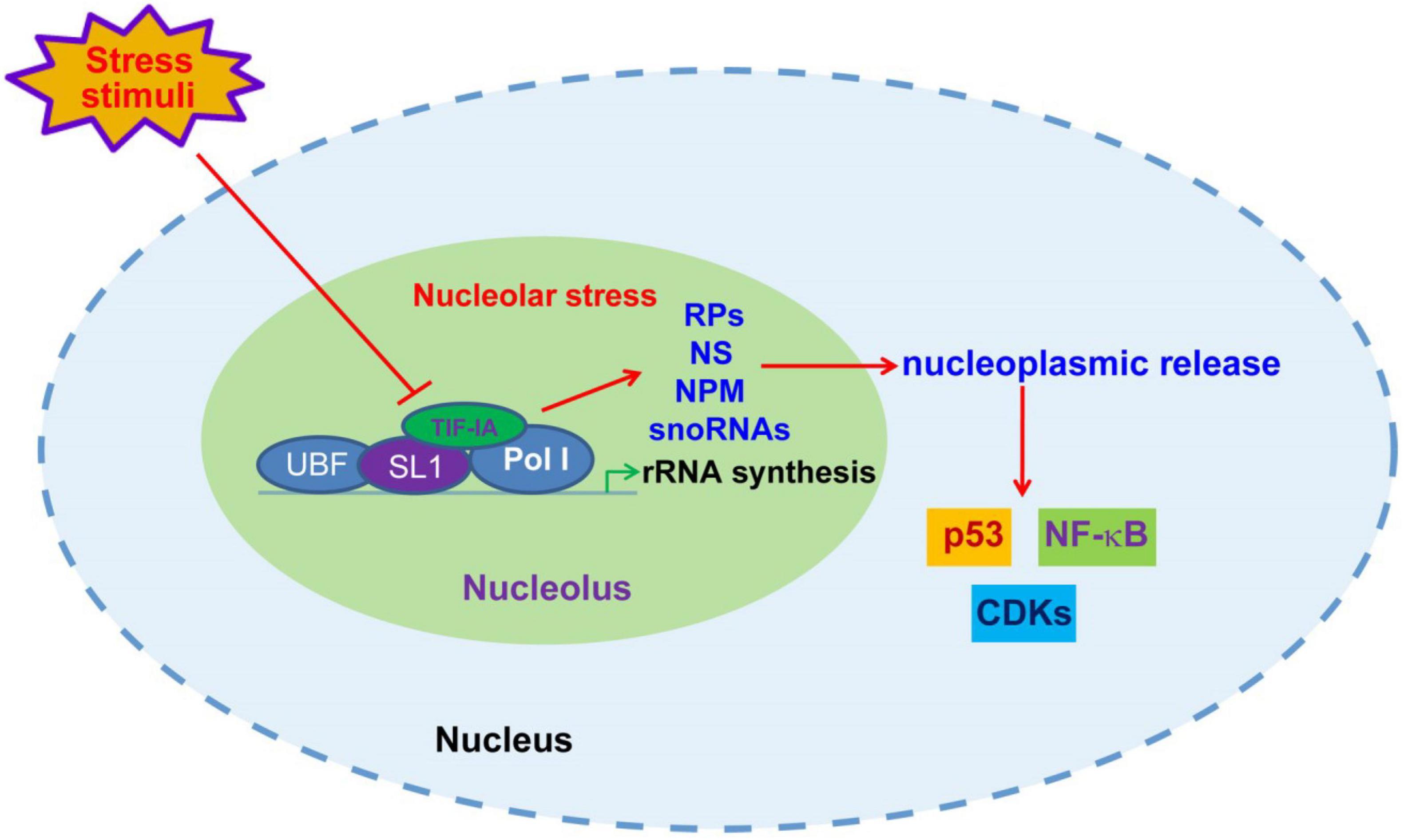
Figure 1. Nucleolar stress-initiated signaling transduction. Upon unfavorable stress stimuli, including DNA damages, nutrient deprivation, hypoxia and oxidative stress, the disruption of rRNA synthesis through posttranscriptional modification of Pol I transcriptional machinery components causes nucleolar malfunction and stress, leading to nucleoplasmic release of ribosomal proteins (RPs), NS, NPM and snoRNAs, and consequent modulation of p53, NF-κB and CDKs.
Multiple nucleolar proteins, such as nucleostemin (NS), nucleophosmin (NPM) and nucleolin (NCL), possess pro-survival and pro-renewal properties (69, 70). Their expression and activity are of great importance to the fate decision of cardiomyocytes. Pioneer works by Mark Sussman’s lab revealed that the induction of nucleolar proteins, including NS and NPM, is an early event following cardiomyopathic injury (71, 72). The upregulation of NS and NPM not only occurred following exposure to nucleolar stress inducer doxorubicin and actinomycin D (ActD), but also was observed in hypoxia/hypoxia-reperfusion conditions. Given the fact that these proteins are highly expressed in cancer and stem cells and elicit pro-survival and pro-repair roles, their upregulation are supposed to prevent the injury and death of cardiac cells. As predicted, forced expression of either NS or NPM inhibit caspase activation and cardiac cell death. The opposite is also true, as depletion of NS or NPM significantly enhance damage-induced cell death (72). Notably, NS and NPM also displayed apparent nucleoplasmic distribution following stress stimuli, suggesting an initiation of nucleolar stress in cardiac cells. In line with these findings, our recent RNA-seq data confirmed that NS is significantly upregulated in hypoxia-conditioned cardiac progenitors (Unpublished data). It is worth of notice that p53 activation was only mildly observed in these conditions, potentially as a result of NS-mediated stabilization of mdm2 in the nucleoplasm (73). More recent data uncovered that NPM is also secreted into the extracellular space via an autophagy-dependent mechanism in human cardiac mesenchymal progenitors (74). Extracellular NPM functions as a ligand of TLR4 to initiate TLR4/NF-κB inflammatory response, potentially facilitating cardiac repair (75). These studies implicated protective roles of NS and NPM in the regulation of the survival and repair of cardiomyocytes (Figure 2).
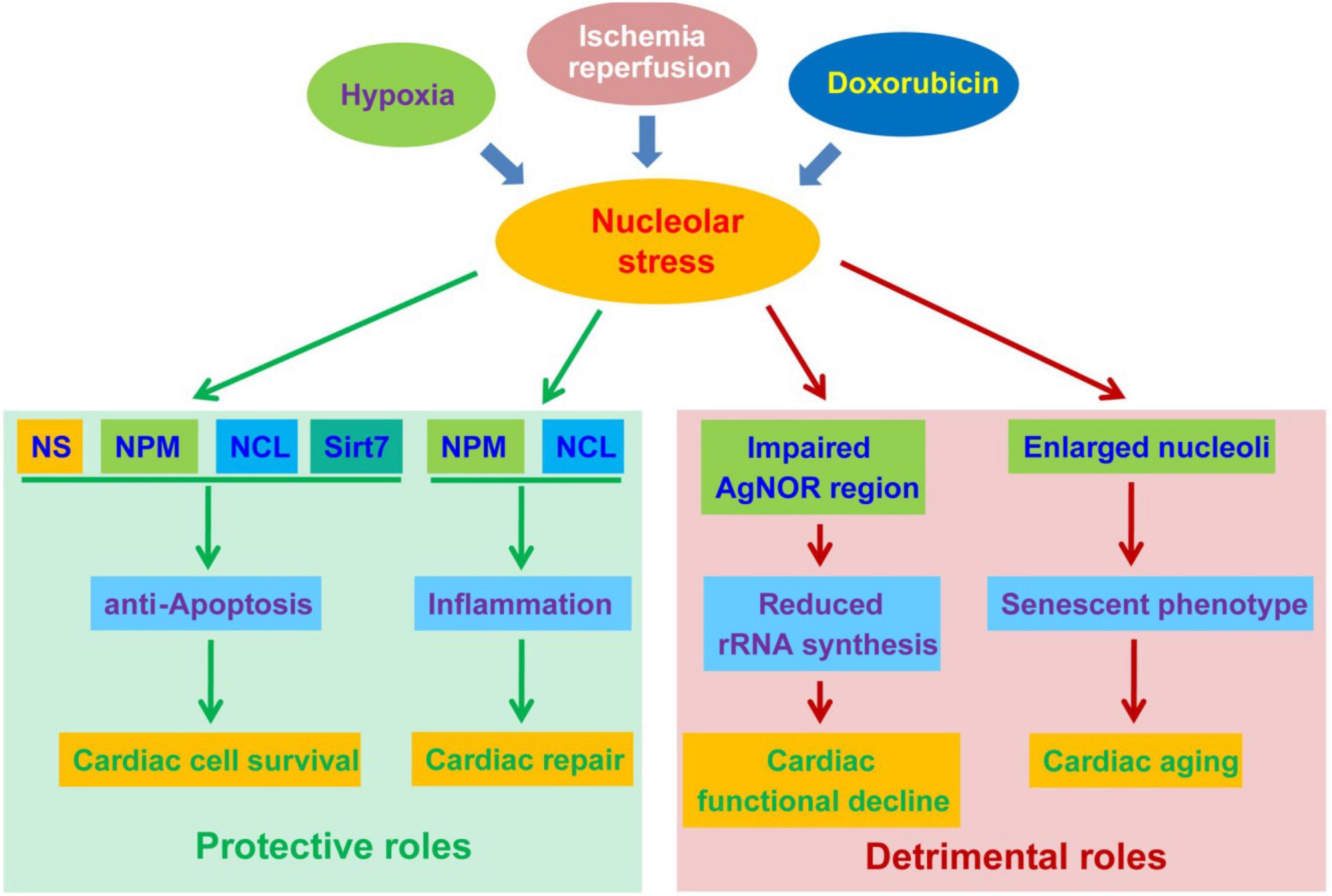
Figure 2. The protective and detrimental roles of nucleolar stress in cardiac disease progression. Nucleolar stress, as a result of hypoxia, ischemia-reperfusion (IR) and doxorubicin exposure, function as a double-edged sword in the development of cardiac diseases. On the one hand, nucleolar stress triggers the upregulation and altered distribution of cardioprotective nucleolar proteins. These proteins exert pro-survival functions in cardiomyocytes, and meanwhile activate inflammatory responses to promote cardiac repair. On the other hand, nucleolar stress may be associated with impaired rRNA transcription and enlarged nucleoli during the progression of cardiac diseases, leading to cardiac functional decline and aging.
In addition to NS and NPM, NCL also exhibits cardioprotective function during the progression of cardiomyopathy. Proteomic data revealed that NCL has higher chromatin-binding in the mouse heart during hypertrophy and failure (76). NCL was subjected to protein cleavage in the very early phases of cardiac ischaemia-reperfusion injury (77). Further investigations indicated that NCL expression was dramatically reduced in the early stage of experimental myocardial infarction, and then recovered at late stages (78). Overexpression of NCL protects cultured cardiomyocytes from hypoxia- and H2O2-induced death (77). These studies demonstrated that NCL may facilitate the recovery of cardiomyocytes following exposure to stress stimuli. Moreover, NCL has been reportedly implicated in inflammatory induction in experimental myocardial infarction, as well as in cultured cardiomyocytes following hypoxia-reoxygenation injury (78, 79). NCL plays a key role in the induction of a variety of cytokines important for inflammatory responses and cardiac repair, such as IL-6 and IL-1β (79). In vivo data revealed that NCL expression is critical for alternative (M2) polarization of macrophages, and depletion of NCL reduced M2 polarization but had no effect on general macrophage infiltration (78). Given the fact that macrophage M2 polarization is of vital importance to cardiac tissue repair following injury, it is anticipated that NCL expression may critically contribute to cardiac recovery via the initiation of alternative inflammatory responses (80).
Sirtuin 7 (Sirt7) is mainly localized in the nucleus, where it regulates rRNA transcription and nucleolar homeostasis via its deacetylase activity (81). Up on nucleolar stress, Sirt7 is released from the nucleolus to modulate stress-related signaling (82, 83). Being a member of sirtuin family proteins, Sirt7 has been implicated in the regulation of tissue repair, aging and metabolism. Similarly with NS and NPM, Sirt7 is upregulated in the early stages of acute cardiovascular injury (84). Sirt7 knockout increased the risk of cardiac rupture following myocardial infarction as a result of impaired activation of transforming growth factor-β (TGF-β) pathway. Another study demonstrated that Sirt7 ablation led to the development of heart hypertrophy and inflammatory cardiomyopathy, likely through the deacetylation and inactivation of p53 (85). These studies suggested that Sirt7 has emerged as a cardioprotective nucleolar protein.
Overall, these findings provide conceivable evidence indicating that nucleolar stress is initiated in cardiac cells following acute cardiovascular injury, which leads to the release of cardioprotective nucleolar proteins into nucleoplasm, where they may elicit cardioprotective function via diverse mechanisms, such as counteracting p53-mediated apoptosis. In addition to nucleoplasmic release, nucleolar proteins are also upregulated following cardiovascular injury, and participate in the repair and survival of cardiac cells. These data provide convincing demonstration of a cardioprotective role of nucleolus and nucleolar proteins in acute cardiomyopathy.
The initiation of nucleolar stress triggers various types of stress responses via stress-responsive pathways, especially mdm2-p53 pathway (86, 87). Depending on cell lineages, nucleolar stress has been associated with the induction of cell apoptosis, senescence, autophagy and differentiation. In cardiac cells, the cellular responses following nucleolar stress remain largely elusive. While nucleolar stress has been shown to inhibit apoptotic death of cardiomyocytes following myocardial damage, it is unclear how nucleolar stress influences cardiac cell fate decision under stress conditions (72). Particularly, nucleolar stress-inducing stimuli, such as hypoxia, ischemia/reperfusion and doxorubicin treatment, have been shown to facilitate the senescence of cardiomyocytes (88–90). It raises the question whether there is a link between nucleolar stress and senescent phenotype under these stress conditions.
Accumulating data have indicated a critical involvement of nucleolus and nucleolar stress in cell senescence and tissue aging. Genetic or pharmacological induction of nucleolar stress has been reported to trigger cell senescence and tissue degeneration (91, 92). Aberrant nucleolar morphology has been well-recognized as a hallmark of cell senescence, suggesting a mechanistic link between nucleolar function and senescence induction (93). In this regard, it is speculated that nucleolar stress may facilitate myocardial aging, in spite of a protective role during the early stages of acute myocardial damage (72). Notably, multiple nucleolar stress-inducing stimuli, including doxorubicin exposure, hypoxia-reoxygenation and ischemia-reperfusion, have been shown to facilitate the senescence of cardiomyocytes (89, 90, 94). Interestingly, cardiomyocyte senescence is partially attributed to the activation of the p53 pathway, implicating a link between nucleolar stress induction and p53-driven senescence in cardiomyocytes (89, 95). Currently, the evidence linking nucleolar stress and cardiac cell senescence is limiting. Heterozygous deletion of NS has been shown to induce the senescence of cardiac progenitors, suggesting that disturbing nucleolar proteins induces cardiac senescence (96). Disruption of rRNA transcription through TIF-IA ablation causes p53 accumulation and senescent phenotypes in smooth muscle cells (91). Whether similar phenotypes can be observed in cardiac cells is worthy of further investigations.
Studies also suggested that nucleolar disorder is correlated with cardiac functional decline. An early clinical study revealed that cardiomyocytes from patients with severe ischaemic heart disease complicated with heart failure displayed significantly reduced silver-stained nucleolar organiser regions (AgNORs) (97). AgNORs are mainly confined to the fibrillar center, where some nucleolar proteins can be specifically stained by silver nitrate (98, 99). Given the fact that the fibrillar center is responsible for rRNA transcription, the reduction of AgNOR suggests a negative correlation between nucleolar rRNA transcription and the severity of ischaemic heart disease (100). Because aberrant nucleolar function has been associated with genomic instability, the decline of nucleolar staining may potentially be indicative of increased genotoxic stress in cardiomyocytes (101, 102). In contrast to reduced nucleolar staining, NCL expression and overall size of nucleolus were increased both in ischemic and dilated cardiomyopathy (103). The mechanism underlying this difference remains unclear. Of note, enlarged nucleoli are widely regarded as a hallmark of senescence and aging (93). On the other hand, studies suggested that tissue aging is associated with hypermethylation and reduced transcriptional activity of rDNA (53, 104). Thus, the morphological changes observed in these studies may be associated with aging-like functional decline of cardiomyocytes (Figure 2). The causal relationship between nucleolar change and cardiac aging requires further mechanistic studies.
While nucleolar stress has been implicated in assorted human diseases, its role in cardiovascular diseases received insufficient attention. Recent data obtained from pathological, animal model and cell culture studies all point to a critical involvement of nucleolar stress in the development of cardiovascular diseases. Moreover, different from its role in most other diseases, nucleolar stress is supposed to play both beneficial and detrimental roles in the progression of cardiovascular disease. In early stages of cardiac injury, nucleolar stress leads to the upregulation of various cardioprotective nucleolar proteins, such as NS, NPM, and NCL. The expression of these proteins plays a key role in facilitating the survival of cardiomyocytes. On the other side of the coin, cardiac functional decline and failure are correlated with apparent nucleolar abnormalities, including reduced AgNORs and enlarged nucleoli. It remains unclear whether these nucleolar alterations are merely a consequence of cardiomyopathy, or may contribute actively to the injury and death of cardiac cells. A deeper understanding of nucleolar alterations will undoubtedly provide novel insights into the pathogenesis of cardiac diseases.
Because of the complicated roles of the nucleolus in cardiac diseases, nucleolar intervention may be a promising strategy to protect against acute and chronic cardiac injury. Of importance, current data suggest that acute cardiac injury mostly initiate rapid upregulation of cardioprotective nucleolar proteins to facilitate the survival and repair of cardiomyocytes. However, chronic damage may profoundly impair rRNA transcription and nucleolar function, probably through impacting the epigenetic landscape of rDNA. These alterations within the nucleolus are tightly associated with cardiac aging and functional decline. We speculate that there are connections between of protein and epigenetic alterations in the nucleolus, given the fact that the nucleolus and nucleolar stress play a crucial role in epigenetic modulation (101). These discoveries suggest that both proteomic and epigenetic changes are involved in cardiac disease progression. In other words, intervening strategies both at protein and epigenetic levels may be beneficial to the amelioration of cardiac diseases. While upregulating the expression of nucleolar proteins has been proven to be feasible approaches, other therapeutic strategies, such as epigenetic approaches, remain to be developed. Moreover, the activities of nucleolar proteins, including NS, NPM and NCL, are also regulated at posttranscriptional levels, providing an additional option of therapeutic intervention. Taken together, we speculate that nucleolar intervention is a potential therapeutic strategy against the progression of cardiac diseases.
DY: conducting the original draft preparation. LH: reviewing and editing the manuscript. Both authors contributed to the article and approved the submitted version.
This study was supported by the National Natural Science Foundation of China (no. 81902406) and Taizhou People’s Hospital Research Project (ZL201916).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Moss T, Langlois F, Gagnon-Kugler T, Stefanovsky V. A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cell Mol Life Sci. (2007) 64:29–49. doi: 10.1007/s00018-006-6278-1
2. Gupta S, Santoro R. Regulation and roles of the nucleolus in embryonic stem cells: from ribosome biogenesis to genome organization. Stem Cell Rep. (2020) 15:1206–19. doi: 10.1016/j.stemcr.2020.08.012
3. Gallagher JE, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, et al. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. (2004) 18:2506–17. doi: 10.1101/gad.1226604
4. Lavrentyeva E, Shishova K, Kagarlitsky G, Zatsepina O. Localisation of RNAs and proteins in nucleolar precursor bodies of early mouse embryos. Reprod Fertil Dev. (2017) 29:509–20. doi: 10.1071/RD15200
5. Engel W, Zenzes MT, Schmid M. Activation of mouse ribosomal RNA genes at the 2-cell stage. Hum Genet. (1977) 38:57–63. doi: 10.1007/BF00295808
6. Savino TM, Gebrane-Younes J, De Mey J, Sibarita JB, Hernandez-Verdun D. Nucleolar assembly of the rRNA processing machinery in living cells. J Cell Biol. (2001) 153:1097–110. doi: 10.1083/jcb.153.5.1097
7. Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science. (2003) 302:1399–401. doi: 10.1126/science.1089814
8. Bertrand E, Houser-Scott F, Kendall A, Singer RH, Engelke DR. Nucleolar localization of early tRNA processing. Genes Dev. (1998) 12:2463–8. doi: 10.1101/gad.12.16.2463
9. Cerqueira AV, Lemos B. Ribosomal DNA and the nucleolus as keystones of nuclear architecture, organization, and function. Trends Genet. (2019) 35:710–23. doi: 10.1016/j.tig.2019.07.011
10. Harding SM, Boiarsky JA, Greenberg RA. ATM dependent silencing links nucleolar chromatin reorganization to DNA damage recognition. Cell Rep. (2015) 13:251–9. doi: 10.1016/j.celrep.2015.08.085
11. Ahmad Y, Boisvert FM, Gregor P, Cobley A, Lamond AI. NOPdb: nucleolar proteome database – 2008 update. Nucleic Acids Res. (2009) 37:D181–4. doi: 10.1093/nar/gkn804
12. Grummt I. The nucleolus-guardian of cellular homeostasis and genome integrity. Chromosoma. (2013) 122:487–97. doi: 10.1007/s00412-013-0430-0
13. Hernandez-Verdun D. Assembly and disassembly of the nucleolus during the cell cycle. Nucleus. (2011) 2:189–94. doi: 10.4161/nucl.2.3.16246
14. Lafontaine DLJ, Riback JA, Bascetin R, Brangwynne CP. The nucleolus as a multiphase liquid condensate. Nat Rev Mol Cell Biol. (2021) 22:165–82. doi: 10.1038/s41580-020-0272-6
15. Riback JA, Zhu L, Ferrolino MC, Tolbert M, Mitrea DM, Sanders DW, et al. Composition-dependent thermodynamics of intracellular phase separation. Nature. (2020) 581:209–14. doi: 10.1038/s41586-020-2256-2
16. Boisvert FM, Lam YW, Lamont D, Lamond AI. A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol Cell Proteomics. (2010) 9:457–70. doi: 10.1074/mcp.M900429-MCP200
17. Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, et al. Nucleolar proteome dynamics. Nature. (2005) 433:77–83. doi: 10.1038/nature03207
18. Moore HM, Bai B, Boisvert FM, Latonen L, Rantanen V, Simpson JC, et al. Quantitative proteomics and dynamic imaging of the nucleolus reveal distinct responses to UV and ionizing radiation. Mol Cell Proteomics. (2011) 10:M111009241. doi: 10.1074/mcp.M111.009241
19. Iarovaia OV, Minina EP, Sheval EV, Onichtchouk D, Dokudovskaya S, Razin SV, et al. Nucleolus: a central hub for nuclear functions. Trends Cell Biol. (2019) 29:647–59. doi: 10.1016/j.tcb.2019.04.003
20. Mekhail K, Gunaratnam L, Bonicalzi ME, Lee S. HIF activation by pH-dependent nucleolar sequestration of VHL. Nat Cell Biol. (2004) 6:642–7. doi: 10.1038/ncb1144
21. Mekhail K, Khacho M, Carrigan A, Hache RR, Gunaratnam L, Lee S. Regulation of ubiquitin ligase dynamics by the nucleolus. J Cell Biol. (2005) 170:733–44. doi: 10.1083/jcb.200506030
22. Frottin F, Schueder F, Tiwary S, Gupta R, Korner R, Schlichthaerle T, et al. The nucleolus functions as a phase-separated protein quality control compartment. Science. (2019) 365:342–7. doi: 10.1126/science.aaw9157
23. Azkanaz M, Rodriguez Lopez A, De Boer B, Huiting W, Angrand PO, Vellenga E, et al. Protein quality control in the nucleolus safeguards recovery of epigenetic regulators after heat shock. Elife. (2019) 8:e45205. doi: 10.7554/eLife.45205
24. Liu Y, Wang Y, Yang L, Sun F, Li S, Zhang GA, et al. The nucleolus functions as the compartment for histone H2B protein degradation. IScience. (2021) 24:102256. doi: 10.1016/j.isci.2021.102256
25. Hu B, Hua L, Ni W, Wu M, Yan D, Chen Y, et al. Nucleostemin/GNL3 promotes nucleolar polyubiquitylation of p27(kip1) to drive hepatocellular carcinoma progression. Cancer Lett. (2017) 388:220–9. doi: 10.1016/j.canlet.2016.12.008
26. Welcker M, Orian A, Grim JE, Eisenman RN, Clurman BE. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr Biol. (2004) 14:1852–7. doi: 10.1016/j.cub.2004.09.083
27. Bhaskaran N, Van Drogen F, Ng HF, Kumar R, Ekholm-Reed S, Peter M, et al. Fbw7alpha and Fbw7gamma collaborate to shuttle cyclin E1 into the nucleolus for multiubiquitylation. Mol Cell Biol. (2013) 33:85–97. doi: 10.1128/MCB.00288-12
28. Liang J, Wen J, Huang Z, Chen XP, Zhang BX, Chu L. Small nucleolar RNAs: insight into their function in cancer. Front Oncol. (2019) 9:587. doi: 10.3389/fonc.2019.00587
29. Michel CI, Holley CL, Scruggs BS, Sidhu R, Brookheart RT, Listenberger LL, et al. Small nucleolar RNAs U32a, U33, and U35a are critical mediators of metabolic stress. Cell Metab. (2011) 14:33–44. doi: 10.1016/j.cmet.2011.04.009
30. Lee J, Harris AN, Holley CL, Mahadevan J, Pyles KD, Lavagnino Z, et al. Rpl13a small nucleolar RNAs regulate systemic glucose metabolism. J Clin Invest. (2016) 126:4616–25. doi: 10.1172/JCI88069
31. Hirose T, Ideue T, Nagai M, Hagiwara M, Shu MD, Steitz JA. A spliceosomal intron binding protein, IBP160, links position-dependent assembly of intron-encoded box C/D snoRNP to pre-mRNA splicing. Mol Cell. (2006) 23:673–84. doi: 10.1016/j.molcel.2006.07.011
32. Falaleeva M, Pages A, Matuszek Z, Hidmi S, Agranat-Tamir L, Korotkov K, et al. Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre-mRNA splicing. Proc Natl Acad Sci USA. (2016) 113:E1625–34. doi: 10.1073/pnas.1519292113
33. Li MW, Sletten AC, Lee J, Pyles KD, Matkovich SJ, Ory DS, et al. Nuclear export factor 3 regulates localization of small nucleolar RNAs. J Biol Chem. (2017) 292:20228–39. doi: 10.1074/jbc.M117.818146
34. Holley CL, Li MW, Scruggs BS, Matkovich SJ, Ory DS, Schaffer JE. Cytosolic accumulation of small nucleolar RNAs (snoRNAs) is dynamically regulated by NADPH oxidase. J Biol Chem. (2015) 290:11741–8. doi: 10.1074/jbc.M115.637413
35. Shi J, Huang C, Huang S, Yao C. snoRNAs associate with mRNA 3’ processing complex: new wine in old bottles. RNA Biol. (2018) 15:194–7. doi: 10.1080/15476286.2017.1416278
36. Liu Y, Demario S, He K, Gibbs MR, Barr KW, Chanfreau GF. Splicing inactivation generates hybrid mRNA-snoRNA transcripts targeted by cytoplasmic RNA decay. Proc Natl Acad Sci USA. (2022) 119:e2202473119. doi: 10.1073/pnas.2202473119
37. Youssef OA, Safran SA, Nakamura T, Nix DA, Hotamisligil GS, Bass BL. Potential role for snoRNAs in PKR activation during metabolic stress. Proc Natl Acad Sci USA. (2015) 112:5023–8. doi: 10.1073/pnas.1424044112
38. Tian B, Liu J, Zhang N, Song Y, Xu Y, Xie M, et al. Oncogenic SNORD12B activates the AKT-mTOR-4EBP1 signaling in esophageal squamous cell carcinoma via nucleus partitioning of PP-1alpha. Oncogene. (2021) 40:3734–47. doi: 10.1038/s41388-021-01809-2
39. Rajan KS, Ramasamy S, George-William JN, Rajendhran J. Emerging cardiac non-coding landscape: the importance of meta-analysis. Biochimie. (2017) 133:87–94. doi: 10.1016/j.biochi.2016.12.018
40. Tallo CA, Duncan LH, Yamamoto AH, Slaydon JD, Arya GH, Turlapati L, et al. Heat shock proteins and small nucleolar RNAs are dysregulated in a Drosophila model for feline hypertrophic cardiomyopathy. G3 (Bethesda). (2021) 11:jkaa014. doi: 10.1093/g3journal/jkaa014
41. Schutt C, Hallmann A, Hachim S, Klockner I, Valussi M, Atzberger A, et al. Linc-MYH conSupplementary Figure 80 to regulate muscle stem cell numbers and skeletal muscle hypertrophy. EMBO J. (2020) 39:e105098. doi: 10.15252/embj.2020105098
42. Pecoraro A, Pagano M, Russo G, Russo A. Ribosome biogenesis and cancer: overview on ribosomal proteins. Int J Mol Sci. (2021) 22:5496. doi: 10.3390/ijms22115496
43. Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu Rev Pharmacol Toxicol. (2010) 50:131–56. doi: 10.1146/annurev.pharmtox.010909.105844
44. Russell J, Zomerdijk JC. The RNA polymerase I transcription machinery. Biochem Soc Symp. (2006) 73:203–16. doi: 10.1042/bss0730203
45. Hoppe S, Bierhoff H, Cado I, Weber A, Tiebe M, Grummt I, et al. Amp-activated protein kinase adapts rRNA synthesis to cellular energy supply. Proc Natl Acad Sci USA. (2009) 106:17781–6. doi: 10.1073/pnas.0909873106
46. Mayer C, Bierhoff H, Grummt I. The nucleolus as a stress sensor: JNK2 inactivates the transcription factor TIF-IA and down-regulates rRNA synthesis. Genes Dev. (2005) 19:933–41. doi: 10.1101/gad.333205
47. Zhao J, Yuan X, Frodin M, Grummt I. ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol Cell. (2003) 11:405–13. doi: 10.1016/S1097-2765(03)00036-4
48. Mayer C, Zhao J, Yuan X, Grummt I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. (2004) 18:423–34. doi: 10.1101/gad.285504
49. Bierhoff H, Dundr M, Michels AA, Grummt I. Phosphorylation by casein kinase 2 facilitates rRNA gene transcription by promoting dissociation of TIF-IA from elongating RNA polymerase I. Mol Cell Biol. (2008) 28:4988–98. doi: 10.1128/MCB.00492-08
50. Srivastava R, Ahn SH. The epigenetic pathways to ribosomal DNA silencing. Microbiol Mol Biol Rev. (2016) 80:545–63. doi: 10.1128/MMBR.00005-16
51. Savic N, Bar D, Leone S, Frommel SC, Weber FA, Vollenweider E, et al. lncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in ESCs. Cell Stem Cell. (2014) 15:720–34. doi: 10.1016/j.stem.2014.10.005
52. Caudron-Herger M, Pankert T, Seiler J, Nemeth A, Voit R, Grummt I, et al. Alu element-containing Rnas maintain nucleolar structure and function. EMBO J. (2015) 34:2758–74. doi: 10.15252/embj.201591458
53. Wang M, Lemos B. Ribosomal DNA harbors an evolutionarily conserved clock of biological aging. Genome Res. (2019) 29:325–33. doi: 10.1101/gr.241745.118
54. Gensous N, Ravaioli F, Pirazzini C, Gramignoli R, Ellis E, Storci G, et al. Aging and caloric restriction modulate the DNA methylation profile of the ribosomal RNA locus in human and rat liver. Nutrients. (2020) 12:277. doi: 10.3390/nu12020277
55. Yang K, Yang J, Yi J. Nucleolar stress: hallmarks, sensing mechanism and diseases. Cell Stress. (2018) 2:125–40. doi: 10.15698/cst2018.06.139
56. Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. (2003) 22:6068–77. doi: 10.1093/emboj/cdg579
57. James A, Wang Y, Raje H, Rosby R, Dimario P. Nucleolar stress with and without p53. Nucleus. (2014) 5:402–26. doi: 10.4161/nucl.32235
58. Sloan KE, Bohnsack MT, Watkins NJ. The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep. (2013) 5:237–47. doi: 10.1016/j.celrep.2013.08.049
59. Donati G, Peddigari S, Mercer CA, Thomas G. 5S ribosomal RNA is an essential component of a nascent ribosomal precursor complex that regulates the Hdm2-p53 checkpoint. Cell Rep. (2013) 4:87–98. doi: 10.1016/j.celrep.2013.05.045
60. Nishimura K, Kumazawa T, Kuroda T, Katagiri N, Tsuchiya M, Goto N, et al. Perturbation of ribosome biogenesis drives cells into senescence through 5S RNP-mediated p53 activation. Cell Rep. (2015) 10:1310–23. doi: 10.1016/j.celrep.2015.01.055
61. Yuan X, Zhou Y, Casanova E, Chai M, Kiss E, Grone HJ, et al. Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest, and p53-mediated apoptosis. Mol Cell. (2005) 19:77–87. doi: 10.1016/j.molcel.2005.05.023
62. Liao H, Gaur A, Mauvais C, Denicourt C. p53 induces a survival transcriptional response after nucleolar stress. Mol Biol Cell. (2021) 32:ar3.
63. Le Goff S, Boussaid I, Floquet C, Raimbault A, Hatin I, Andrieu-Soler C, et al. p53 activation during ribosome biogenesis regulates normal erythroid differentiation. Blood. (2021) 137:89–102. doi: 10.1182/blood.2019003439
64. Russo A, Russo G. Ribosomal proteins control or bypass p53 during nucleolar stress. Int J Mol Sci. (2017) 18:140. doi: 10.3390/ijms18010140
65. Chen J, Stark LA. Insights into the relationship between nucleolar stress and the NF-kappaB pathway. Trends Genet. (2019) 35:768–80. doi: 10.1016/j.tig.2019.07.009
66. Lessard F, Brakier-Gingras L, Ferbeyre G. Ribosomal proteins control tumor suppressor pathways in response to nucleolar stress. Bioessays. (2019) 41:e1800183. doi: 10.1002/bies.201800183
67. Latonen L. Phase-to-phase with nucleoli – stress responses, protein aggregation and novel roles of RNA. Front Cell Neurosci. (2019) 13:151. doi: 10.3389/fncel.2019.00151
68. Nunez Villacis L, Wong MS, Ferguson LL, Hein N, George AJ, Hannan KM. New roles for the nucleolus in health and disease. Bioessays. (2018) 40:e1700233. doi: 10.1002/bies.201700233
69. Tsai RY. Turning a new page on nucleostemin and self-renewal. J Cell Sci. (2014) 127:3885–91. doi: 10.1242/jcs.154054
70. Scott DD, Oeffinger M. Nucleolin and nucleophosmin: nucleolar proteins with multiple functions in DNA repair. Biochem Cell Biol. (2016) 94:419–32. doi: 10.1139/bcb-2016-0068
71. Siddiqi S, Gude N, Hosoda T, Muraski J, Rubio M, Emmanuel G, et al. Myocardial induction of nucleostemin in response to postnatal growth and pathological challenge. Circ Res. (2008) 103:89–97. doi: 10.1161/CIRCRESAHA.107.169334
72. Avitabile D, Bailey B, Cottage CT, Sundararaman B, Joyo A, Mcgregor M, et al. Nucleolar stress is an early response to myocardial damage involving nucleolar proteins nucleostemin and nucleophosmin. Proc Natl Acad Sci USA. (2011) 108:6145–50. doi: 10.1073/pnas.1017935108
73. Meng L, Lin T, Tsai RY. Nucleoplasmic mobilization of nucleostemin stabilizes MDM2 and promotes G2-M progression and cell survival. J Cell Sci. (2008) 121:4037–46. doi: 10.1242/jcs.037952
74. Beji S, D’agostino M, Gambini E, Sileno S, Scopece A, Vinci MC, et al. Doxorubicin induces an alarmin-like Tlr4-dependent autocrine/paracrine action of nucleophosmin in human cardiac mesenchymal progenitor cells. BMC Biol. (2021) 19:124. doi: 10.1186/s12915-021-01058-5
75. Frangogiannis NG. Regulation of the inflammatory response in cardiac repair. Circ Res. (2012) 110:159–73. doi: 10.1161/CIRCRESAHA.111.243162
76. Monte E, Mouillesseaux K, Chen H, Kimball T, Ren S, Wang Y, et al. Systems proteomics of cardiac chromatin identifies nucleolin as a regulator of growth and cellular plasticity in cardiomyocytes. Am J Physiol Heart Circ Physiol. (2013) 305:H1624–38. doi: 10.1152/ajpheart.00529.2013
77. Jiang B, Zhang B, Liang P, Chen G, Zhou B, Lv C, et al. Nucleolin protects the heart from ischaemia-reperfusion injury by up-regulating heat shock protein 32. Cardiovasc Res. (2013) 99:92–101. doi: 10.1093/cvr/cvt085
78. Tang Y, Lin X, Chen C, Tong Z, Sun H, Li Y, et al. Nucleolin improves heart function during recovery from myocardial infarction by modulating macrophage polarization. J Cardiovasc Pharmacol Ther. (2021) 26:386–95. doi: 10.1177/1074248421989570
79. Mariero LH, Torp MK, Heiestad CM, Baysa A, Li Y, Valen G, et al. Inhibiting nucleolin reduces inflammation induced by mitochondrial DNA in cardiomyocytes exposed to hypoxia and reoxygenation. Br J Pharmacol. (2019) 176:4360–72. doi: 10.1111/bph.14830
80. Kim Y, Nurakhayev S, Nurkesh A, Zharkinbekov Z, Saparov A. Macrophage polarization in cardiac tissue repair following myocardial infarction. Int J Mol Sci. (2021) 22:2715. doi: 10.3390/ijms22052715
81. Kumari P, Tarighi S, Braun T, Ianni A. SIRT7 acts as a guardian of cellular integrity by controlling nucleolar and extra-nucleolar functions. Genes (Basel). (2021) 12:1361. doi: 10.3390/genes12091361
82. Zhang PY, Li G, Deng ZJ, Liu LY, Chen L, Tang JZ, et al. Dicer interacts with SIRT7 and regulates H3K18 deacetylation in response to DNA damaging agents. Nucleic Acids Res. (2016) 44:3629–42. doi: 10.1093/nar/gkv1504
83. Lu YF, Xu XP, Lu XP, Zhu Q, Liu G, Bao YT, et al. SIRT7 activates p53 by enhancing PCAF-mediated MDM2 degradation to arrest the cell cycle. Oncogene. (2020) 39:4650–65. doi: 10.1038/s41388-020-1305-5
84. Araki S, Izumiya Y, Rokutanda T, Ianni A, Hanatani S, Kimura Y, et al. Sirt7 contributes to myocardial tissue repair by maintaining transforming growth factor-beta signaling pathway. Circulation. (2015) 132:1081–93. doi: 10.1161/CIRCULATIONAHA.114.014821
85. Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, et al. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res. (2008) 102:703–10. doi: 10.1161/CIRCRESAHA.107.164558
86. Chen J, Lobb IT, Morin P, Novo SM, Simpson J, Kennerknecht K, et al. Identification of a novel TIF-IA-NF-kappaB nucleolar stress response pathway. Nucleic Acids Res. (2018) 46:6188–205. doi: 10.1093/nar/gky455
87. Lee C, Smith BA, Bandyopadhyay K, Gjerset RA. DNA damage disrupts the p14ARF-B23(nucleophosmin) interaction and triggers a transient subnuclear redistribution of p14ARF. Cancer Res. (2005) 65:9834–42. doi: 10.1158/0008-5472.CAN-05-1759
88. Nishimura A, Shimauchi T, Tanaka T, Shimoda K, Toyama T, Kitajima N, et al. Hypoxia-induced interaction of filamin with Drp1 causes mitochondrial hyperfission-associated myocardial senescence. Sci Signal. (2018) 11:eaat5185. doi: 10.1126/scisignal.aat5185
89. Maejima Y, Adachi S, Ito H, Hirao K, Isobe M. Induction of premature senescence in cardiomyocytes by doxorubicin as a novel mechanism of myocardial damage. Aging Cell. (2008) 7:125–36. doi: 10.1111/j.1474-9726.2007.00358.x
90. Dookun E, Walaszczyk A, Redgrave R, Palmowski P, Tual-Chalot S, Suwana A, et al. Clearance of senescent cells during cardiac ischemia-reperfusion injury improves recovery. Aging Cell. (2020) 19:e13249. doi: 10.1111/acel.13249
91. Zhang W, Cheng W, Parlato R, Guo X, Cui X, Dai C, et al. Nucleolar stress induces a senescence-like phenotype in smooth muscle cells and promotes development of vascular degeneration. Aging (Albany NY). (2020) 12:22174–98. doi: 10.18632/aging.104094
92. Drygin D, Lin A, Bliesath J, Ho CB, O’brien SE, Proffitt C, et al. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. (2011) 71:1418–30. doi: 10.1158/0008-5472.CAN-10-1728
93. Pathak RU, Soujanya M, Mishra RK. Deterioration of nuclear morphology and architecture: a hallmark of senescence and aging. Ageing Res Rev. (2021) 67:101264. doi: 10.1016/j.arr.2021.101264
94. Zhang FX, Chen ML, Shan QJ, Zou JG, Chen C, Yang B, et al. Hypoxia reoxygenation induces premature senescence in neonatal SD rat cardiomyocytes. Acta Pharmacol Sin. (2007) 28:44–51. doi: 10.1111/j.1745-7254.2007.00488.x
95. Shan H, Li T, Zhang L, Yang R, Li Y, Zhang M, et al. Heme oxygenase-1 prevents heart against myocardial infarction by attenuating ischemic injury-induced cardiomyocytes senescence. EBioMedicine. (2019) 39:59–68. doi: 10.1016/j.ebiom.2018.11.056
96. Hariharan N, Quijada P, Mohsin S, Joyo A, Samse K, Monsanto M, et al. Nucleostemin rejuvenates cardiac progenitor cells and antagonizes myocardial aging. J Am Coll Cardiol. (2015) 65:133–47. doi: 10.1016/j.jacc.2014.09.086
97. Mamaev NN, Gudkova AY, Amineva KK. AgNORs in the myocardium in ischaemic heart disease complicated by heart failure: a postmortem study. Mol Pathol. (1998) 51:102–4. doi: 10.1136/mp.51.2.102
98. Bersaglieri C, Santoro R. Genome organization in and around the nucleolus. Cells. (2019) 8:579. doi: 10.3390/cells8060579
99. Trere D. AgNOR staining and quantification. Micron. (2000) 31:127–31. doi: 10.1016/S0968-4328(99)00069-4
100. Hernandez-Verdun D, Roussel P, Thiry M, Sirri V, Lafontaine DL. The nucleolus: structure/function relationship in RNA metabolism. Wiley Interdiscip Rev RNA. (2010) 1:415–31. doi: 10.1002/wrna.39
101. Lindstrom MS, Jurada D, Bursac S, Orsolic I, Bartek J, Volarevic S. Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis. Oncogene. (2018) 37:2351–66. doi: 10.1038/s41388-017-0121-z
102. Tsekrekou M, Stratigi K, Chatzinikolaou G. The nucleolus: in genome maintenance and repair. Int J Mol Sci. (2017) 18:1411. doi: 10.3390/ijms18071411
103. Rosello-Lleti E, Rivera M, Cortes R, Azorin I, Sirera R, Martinez-Dolz L, et al. Influence of heart failure on nucleolar organization and protein expression in human hearts. Biochem Biophys Res Commun. (2012) 418:222–8.
Keywords: nucleolus, nucleolar stress, cardiac disease, nucleostemin, nucleophosmin, nucleolin, senescence
Citation: Yan D and Hua L (2022) Nucleolar stress: Friend or foe in cardiac function? Front. Cardiovasc. Med. 9:1045455. doi: 10.3389/fcvm.2022.1045455
Received: 15 September 2022; Accepted: 17 October 2022;
Published: 31 October 2022.
Edited by:
Chia-Feng Liu, Cleveland Clinic, United StatesReviewed by:
Wesley H. Brooks, University of South Florida, United StatesCopyright © 2022 Yan and Hua. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lu Hua, aHVhbHUxMjE5QDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.