- 1Department of Neurology, Seventh People’s Hospital of Shanghai University of Traditional Chinese Medicine, Shanghai, China
- 2Department of Neurology, Shanghai Tenth People’s Hospital, Tongji University School of Medicine, Shanghai, China
- 3Department of Infectious Diseases, Shanghai Tenth People’s Hospital, Tongji University School of Medicine, Shanghai, China
Background: Polymorphisms of the apolipoprotein E (APOE) gene are related to the efficacy of statin therapy. The biological functions of the APOE subtypes determine the metabolism of blood plasma lipids and the progression of atherosclerosis. This study aimed to explore the impact of APOE gene polymorphisms on the effect of atorvastatin on lipid regulation and plaque stabilization.
Methods: The study was a prospective cohort study that consecutively included patients with acute ischemic stroke (AIS) in the Department of Neurology, Shanghai Tenth People’s Hospital, from December 2018 to December 2019. The patients were divided into E2, E3, and E4 groups according to their APOE genotype. Atorvastatin (20 mg) was administrated to all patients. Changes in blood lipid levels over 3 months and plaque size and stability over 12 months were analyzed.
Results: We enrolled 253 consecutive patients with AIS, of whom, 136 had carotid atherosclerotic plaques. Two patients with genotype E2/E4 were excluded. There were 30 patients in the E2 group (12.0%), 191 patients in the E3 group (76.0%), and 30 patients in the E4 group (12.0%). The lowest percentage reduction in low-density lipoprotein cholesterol (LDL-C) was observed in the E4 group (41.2%), while the highest percentage reduction was observed in the E2 group (17.6%). The plaques in the E2 group showed slower progression, while those in the E4 group showed more rapid progression.
Conclusion: APOE gene polymorphisms affect the biological functions of atorvastatin. Compared to the ε3 or ε4 allele, the ε2 allele exerted a greater lipid-lowering effect on LDL-C levels, enhanced the ability of atorvastatin to stabilize carotid artery plaques, and slowed carotid artery plaque progression.
Introduction
Atherosclerosis is one of the leading causes of stroke. Low-density lipoprotein cholesterol (LDL-C) is closely related to the progression of atherosclerosis (1). With every 1 mmol/L reduction in LDL-C, the relative risk of stroke decreases by 21.1%, thus showing the importance of reducing LDL-C levels for stroke prevention (2). Statins that competitively inhibit critical enzymes in cholesterol synthesis are the most widely used lipid-lowering drugs and have become one of the three cornerstones of acute ischemic stroke (AIS) treatment. However, apparent individual differences in lipid-lowering effects have been observed with the widespread use of statins. Statin metabolism is affected by a variety of genes, and gene polymorphisms are related to the lipid-lowering effects of statins. Genetic factors contribute to approximately 70% of the efficacy of statin treatment (3), and the apolipoprotein E (APOE) gene is closely related (4, 5).
APOE is mainly synthesized in the periphery of the liver. On the one hand, as a structural protein of chylomicron (CM), LDL-C, very low-density lipoprotein cholesterol (VLDL-C), and part of high-density lipoprotein cholesterol (HDL-C), APOE is beneficial in stabilizing the structure of these lipoproteins. On the other hand, as a ligand and member of the LDL receptor family, it regulates blood CM, LDL-C, VLDL-C, and HDL-C levels (6, 7). Gene polymorphisms determine the transport and regulation of blood lipids. The gene coding APOE is located on chromosome 19 and is approximately 3.7 kb in length (8). It comprises two loci, rs429358T > C3,937 and rs7412C > T4,075, and includes four alleles, ε2 (T3,937–T4,075), ε3 (T3,937–C4,075), ε4 (C3,937–C4,075), and ε3r (C3,937–T4,075), of which ε3r is extremely rare. To date, only two Caucasian families in Italy and one Yoruba family in Ibadan have been reported with this allele, which is why it is generally excluded from clinical studies (9). The ε3 is the most common in the general population (10) and has a frequency of 85% in Asia, 82% in North America, and 77% in South America (11). ε3 is considered “wild type” due to its high frequency in the general population, while ε2 and ε4 alleles are mutations of ε3. The three alleles comprise six common genotypes, including three homozygotes (E2/E2, E3/E3, and E4/E4) and three heterozygotes (E2/E3, E2/E4, and E3/E4).
Due to differences in protein conformations, the affinities for cholesterol receptors in people with distinct genotypes are different, affecting the efficacy of statin therapy. Some studies have shown that statins may confer reduced benefits in APOE ε4 carriers (12) and that ε2 gene carriers may experience superior lipid-lowering effects (13). However, other studies have suggested that the APOE genotype is not significantly associated with the lipid-lowering effect of statins (14). In another study, polymorphisms of the APOE gene determined baseline LDL-C levels, but not the lipid-lowering effect of statins (15). In the Chilean population, patients with the E3/E4 genotype had a smaller reduction in cholesterol levels after statin therapy than those with the E3/E3 genotype (16). However, the association between APOE gene polymorphisms and the progression of atherosclerotic plaques with statin therapy is not well described. Thus, we aimed to explore the differences in lipid-lowering effects and the progression of atherosclerotic plaques with atorvastatin in different APOE genotypes.
Materials and methods
Patients and study design
We prospectively and consecutively collected data from a cohort that included all hospitalized patients with AIS in the Department of Neurology, Shanghai Tenth People’s Hospital, between December 2018 and December 2019. Patients were enrolled if they met the following criteria: (1) patients diagnosed with AIS using MRI within 7 days after stroke onset; (2) no previous history of lipid-lowering drug use, such as statins, fibrins, and PCSK9 inhibitors, or lipid-lowering drug withdrawal for more than 1 month; (3) willing to receive 20 mg atorvastatin; (4) patients gave informed consent and participated voluntarily. Exclusion criteria included (1) severe liver or kidney dysfunction or major cardiovascular or respiratory diseases; (2) allergic or intolerant to atorvastatin; (3) severe trauma or major surgery recently; (4) patients with non-compliance or poor compliance. The genotype was confirmed at the study’s inception. Previous studies have indicated that the E2 and E4 mutant alleles may have opposite effects on treatment. Therefore, we excluded patients with the E2/E4 genotype. Patients were divided into three groups according to their APOE genotype: E2 (E2/E2 + E2/E3), E3 (E3/E3), and E4 (E3/E4 + E4/E4). All eligible patients received atorvastatin (atorvastatin calcium tablets, Pfizer Pharmaceuticals Limited, 20 mg*7) 20 mg daily and were followed up for 3 months. Patients with carotid plaques were followed up for 12 months. Blood lipids were tested at 3 months and carotid artery ultrasonography was performed at 12 months. The basic characteristics of every patient, including gender, age, height, weight, past medical history, and personal history, were recorded from the electronic medical records and by direct communication with the patients by two doctors at the beginning of the study, and the information was double checked by two doctors at the end.
All patients provided written informed consent to participate in this study. The Ethics Committee of Shanghai Tenth People’s Hospital approved this study (No. 22k205).
Apolipoprotein E genotyping
Genomic DNA was extracted from whole blood using the whole blood DNA Extraction Kit (Beijing Jingzhun Medical Technology Co., Ltd.). Polymerase chain reaction [Honglong Biotechnology (Shanghai) Co., Ltd.] was used for DNA amplification, and the product fragments were subjected to capillary electrophoresis sequencing analysis. According to the peak characteristics performing APOE genotyping.
Detection and classification of plaques
Two ultrasound specialists performed carotid ultrasonography using a color Doppler ultrasound system (Logiq E9, GE, USA). Each patient was placed in a quiet supine position, with the head tilted back and turned to the opposite side. The probe was not pressurized, following the lateral border of the sternocleidomastoid muscle from bottom to top. The patient’s common carotid artery trunk, common carotid artery bifurcation, and neck were observed in turn. The internal and external carotid arteries were observed and recorded for plaque length, thickness, and echogenicity. Information on the largest plaque was recorded if a patient had multiple plaques simultaneously. Carotid artery intima-media thickness (CIMT) was measured in a 1 cm segment at the bulb of the common carotid artery and 1 cm each from its proximal and distal segments, and the average value of three points was taken as the final value of CIMT. The contralateral side was observed in the same way.
Atherosclerotic plaque formation is defined as a condition in which the intima-media thickness (IMT) is ≥ 1.5 mm, and it protrudes from the vascular lumen, or the localized intimal thickening is > 50% of the surrounding IMT. According to the morphology and echo characteristics of ultrasound, plaques can be divided into (I) hypoechoic lipid soft plaques, (II) fibrous flat plaques with medium echoes rich in collagen tissue, (III) hyperechoic calcifications with acoustic shadows, and (IV) ulcerative mixed plaques with varying echo intensity (17). Among them, (I), (II), and (IV) are vulnerable plaques, and (III) are stable plaques.
Statistical analysis
Statistical analyses were performed using IBM SPS Statistics (version 26.0; IBM, Armonk, NY, USA). Normally distributed measurement data are presented as mean ± standard deviation, and non-normally distributed data are expressed as median and quartile. To compare the three groups, one-way analysis of variance or the Kruskal Wallis test was used, and differences between the two groups were analyzed using the independent samples T-test or the Mann-Whitney U-test. The count data are expressed as frequency (percentage), and differences between groups were compared using the chi-square test. To explore the factors affecting LDL-C reduction and changes in plaque length, univariate and multivariate linear regression analyses were performed. The Hardy-Weinberg genetic balance test of APOE was performed using the chi-square test. A two-tailed value of p < 0.05 was considered statistically significant.
Results
Polymorphisms of apolipoprotein E
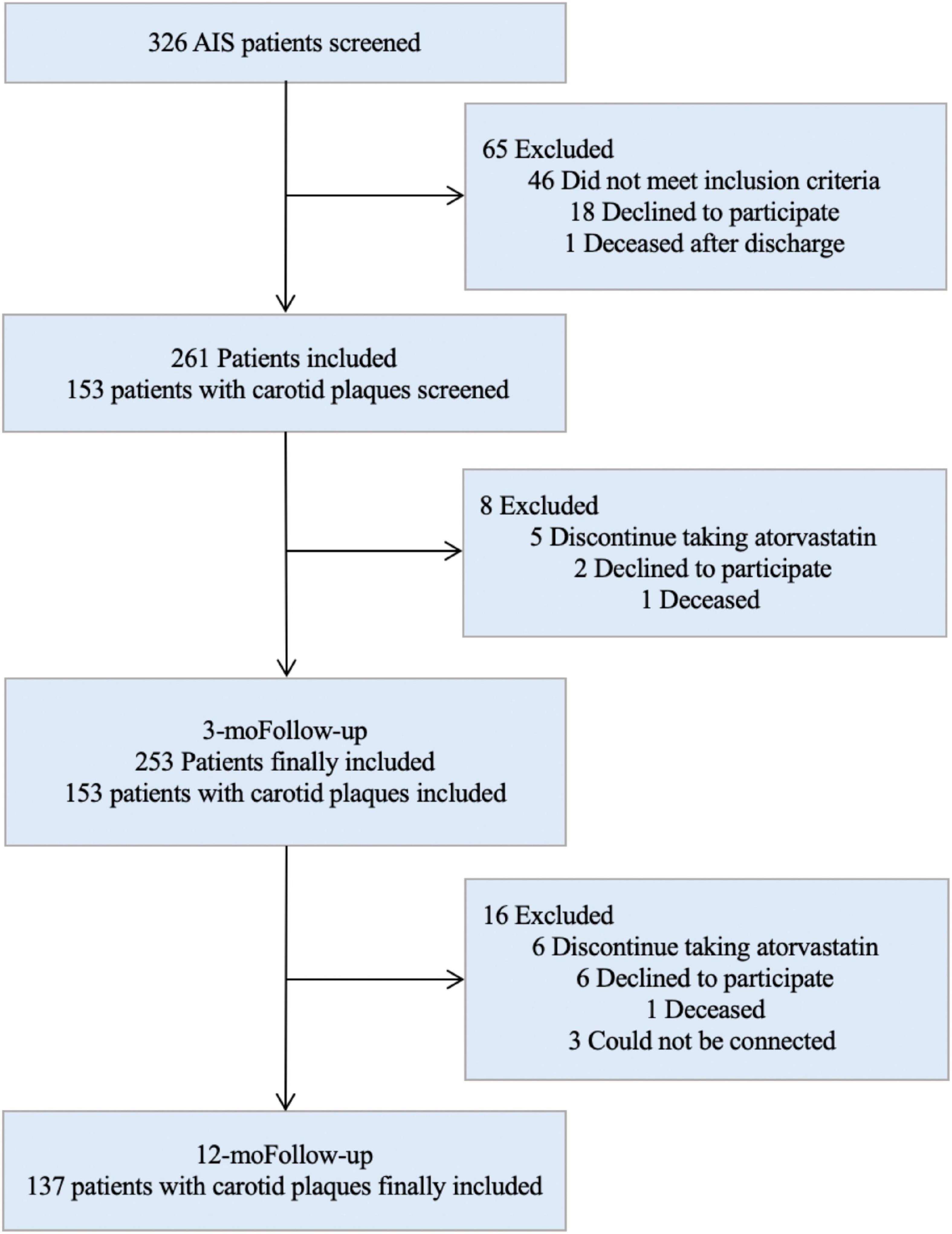
A total of 253 AIS patients with complete follow-up data were included in the study (Figure 1). Six genotypes were detected: three cases with E2/E2 (1.2%), 27 cases with E2/E3 (10.7%), two cases with E2/E4 (0.8%), 191 cases with E3/E3 (75.5%), 28 cases with E3/E4 (11.1%), and two cases with E4/E4 (0.8%).
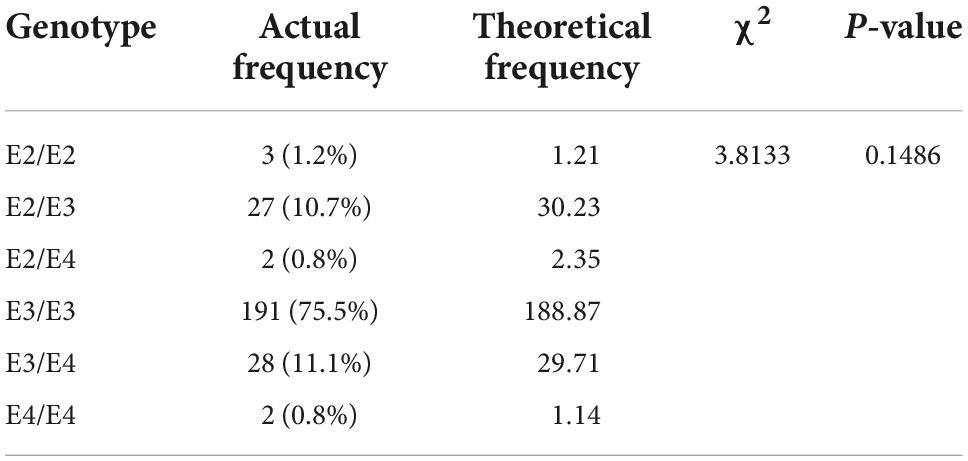
According to the Hardy-Weinberg genetic balance test, we calculated the theoretical frequency of the included population and compared it with the actual frequency in Table 1. The results indicated that the population in our study was in line with the Hardy-Weinberg genetic balance (P = 0.1486), which means that it had good group representation.
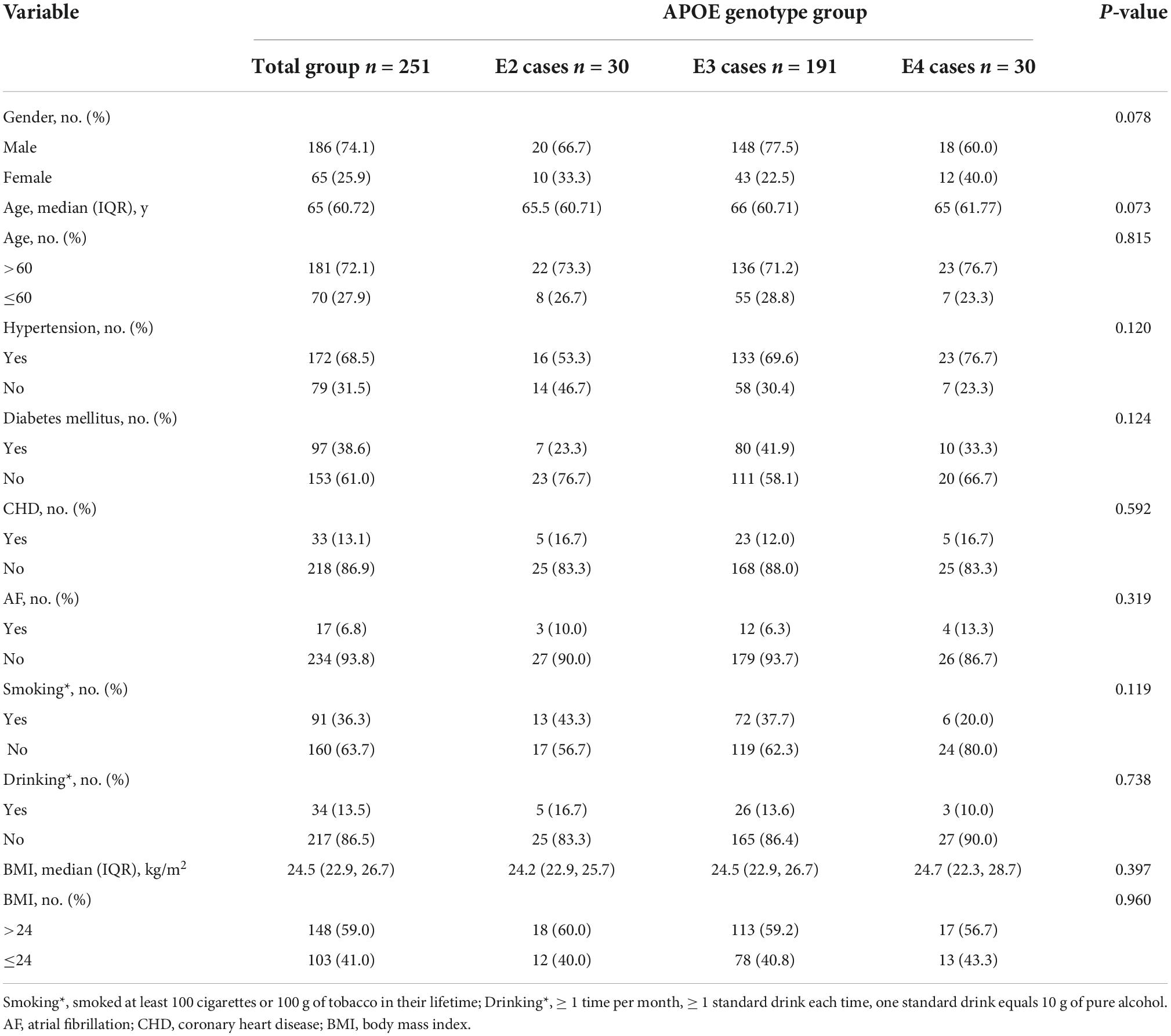
Two patients with the E2/E4 genotype were excluded and the remaining 251 patients were divided into three groups: 30 patients in the E2 group (12%), 191 in the E3 group (76%), and 30 in the E4 group (12%). The baseline characteristics of the patients were similar among the three groups (Table 2).
Effects on blood lipids and plaques
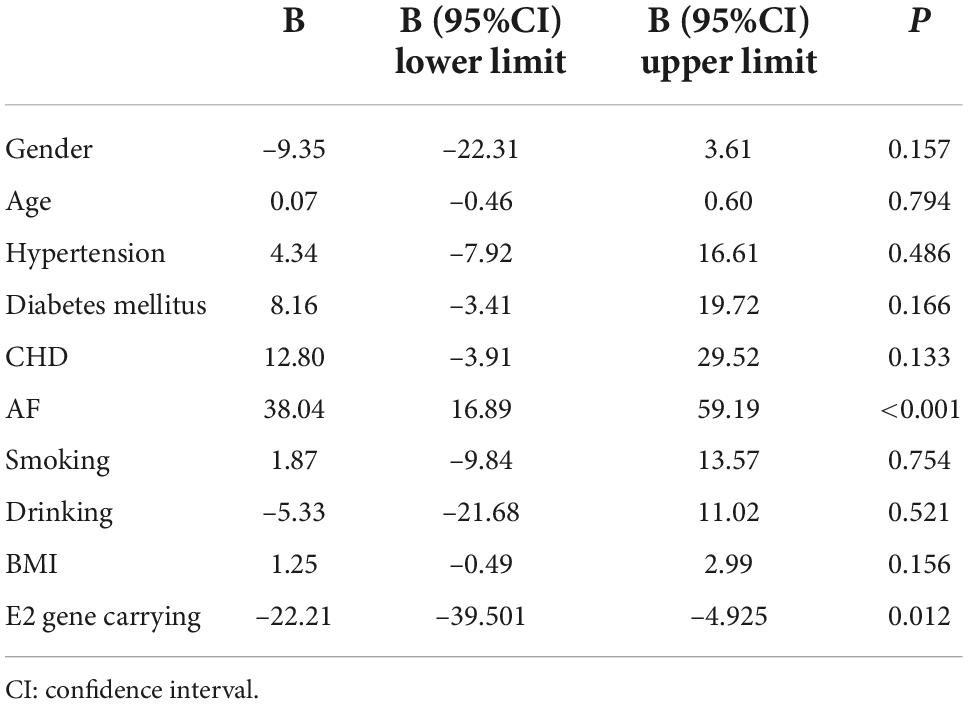
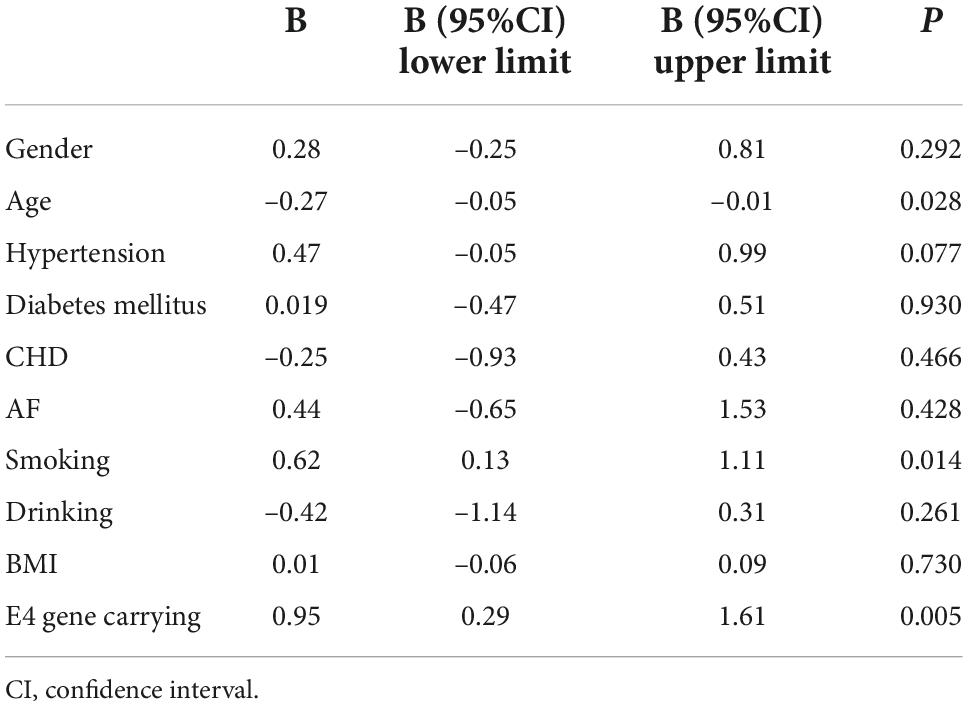
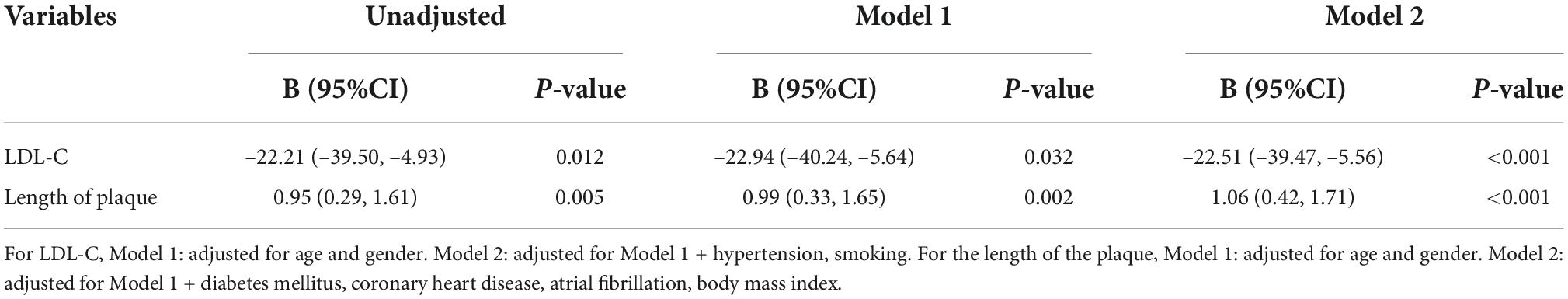
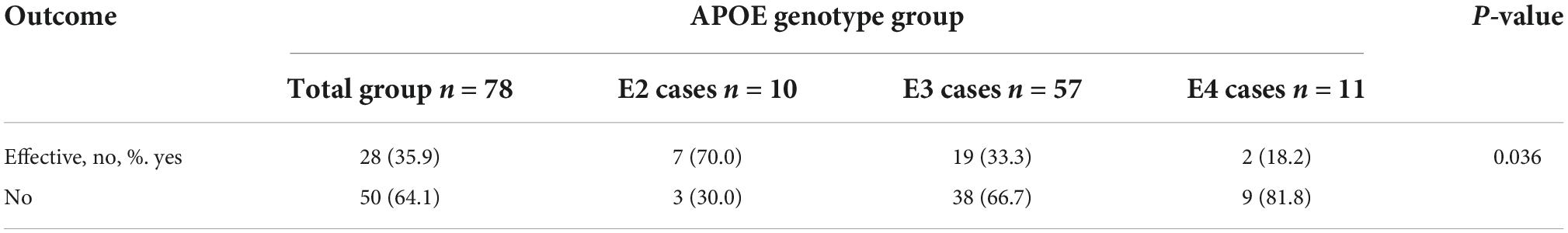
After 3 months of atorvastatin treatment, total cholesterol (TC), triglyceride (TG), and LDL-C levels were lower than baseline. There was a significant difference in the reduction rate of blood LDL-C level after treatment (Figure 2A); the E2 group had the highest reduction rate (41.2%), followed by the E3 group (19.8%), while the E4 group had the lowest (17.6%) (P = 0.020). When we stratified the cohort by gender, no differences were found in females (Table 3). We performed multiple linear regression based on univariate analysis and expertise (Tables 4, 5). The results showed that the E2 group was sensitive to atorvastatin therapy based on the LDL-C reduction rate (Table 6). The progression of carotid plaques in the E4 group was more rapid than in the E2 and E3 groups (P = 0.011) (Figure 2B). After 12 months of atorvastatin treatment, the percentage of vulnerable plaques decreased in the three groups, with significant plaque stabilization in the E2 group (Table 7).
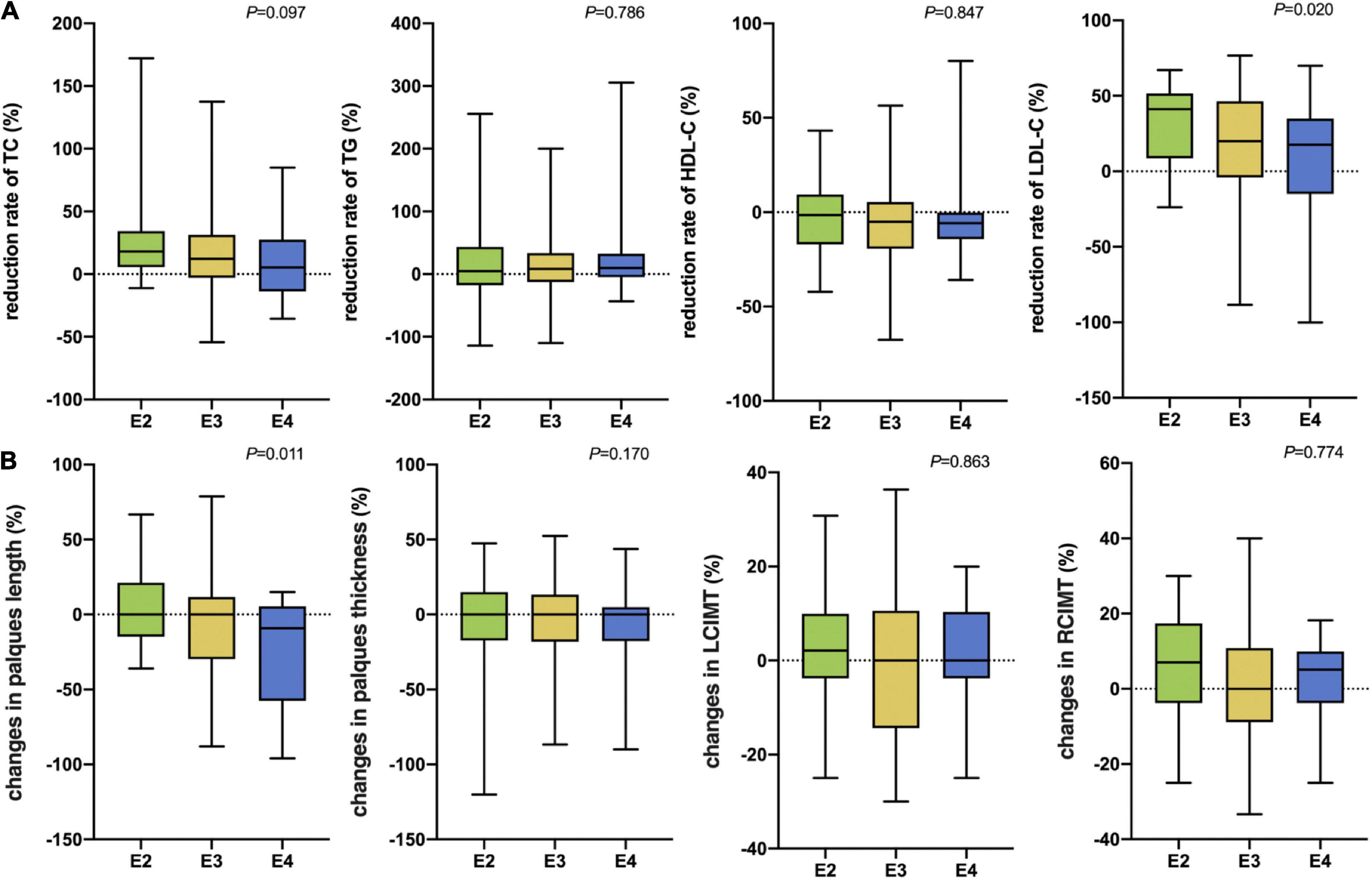
Figure 2. (A) The rate of reduction in blood lipids after 3 months of atorvastatin treatment in the three groups. (B) Changes in carotid plaque size after 12 months of atorvastatin treatment in the three groups. TC, Total cholesterol; TG, Triglyceride; HDL-C, High-density lipoprotein cholesterol; LDL-C, Low-density lipoprotein cholesterol; LCIMT, Left carotid intima-media thickness; RCIMT, Right carotid intima-media thickness.
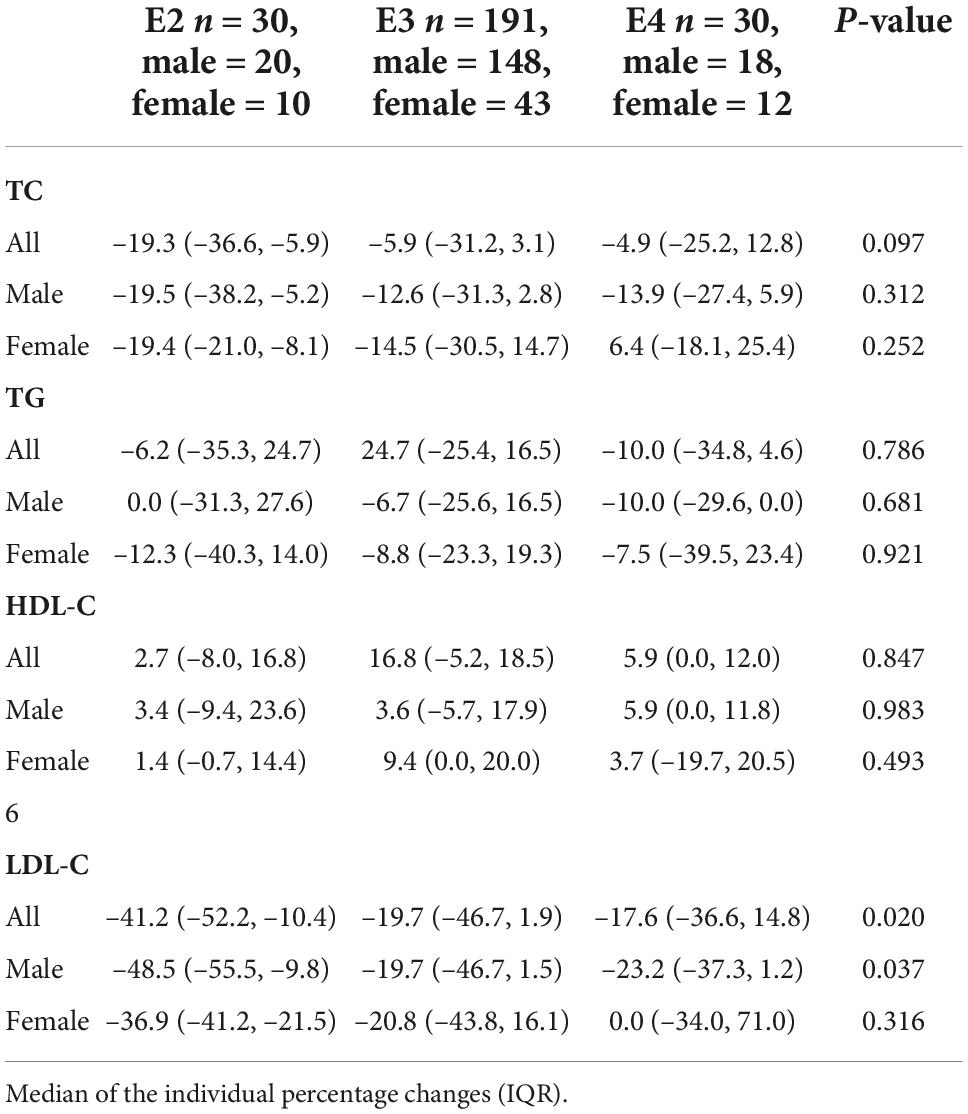
Table 3. Percentage variation in lipid concentrations after treatment with atorvastatin and stratified analyses according to gender.
Discussion
This study examined the effects of APOE genetic polymorphisms on atorvastatin therapy in regulating blood lipids and promoting plaque stabilization. Our results showed that compared to ε3 and ε4 allele carriers, ε2 allele carriers had greater lipid-lowering effect on LDL-C, enhanced carotid artery plaque stabilization by atorvastatin, and slower plaque progression.
Polymorphisms of the APOE gene change the structure and function of APOE lipoproteins, resulting in a difference in the affinity of different isomers for LDL receptors and leading to differences in blood lipid levels between carriers of different genotypes. Type E4 preferentially binds to the larger LDL and VLDL receptors, whereas types E2 and E3 preferentially bind to the smaller HDL receptor (18, 19). Additionally, the affinity of the E2 type for LDL receptors is more than 50 times weaker than that of the E3 type, and the binding force of the E4 type to VLDL is strong, which weakens the process of VLDL lipolysis in the peripheral blood (6, 20). Therefore, the APOE genotype may be associated with basal blood lipid levels, and the E4 allele may be associated with higher TC, LDL-C, TG, and VLDL-C and lower HDL-C levels (21). A study found that the LDL-C and TC levels of patients in the E4 carrier group tended to be higher than those of patients in the E2 and E3 groups (22), while a study by Xie et al. showed that the blood TC level of the E4 gene carrier group was significantly higher than that of the E2 and E3 groups (23). Our study also found that the median initial TC level in the E4 group was slightly higher than that in the E2 and E3 groups, suggesting that there may be a correlation between the APOE genotype and basal blood lipid level.
The lipid-lowering effect of statins is closely related to APOE gene polymorphisms. The E4 allele may attenuate the lipid-lowering effect of statins (24), while the E2 allele exerts a relatively better lipid-lowering effect (4). After 3 months of atorvastatin treatment, the effect on LDL-C reduction was highest in the E2 group, followed by the E3 and E4 groups. Although there was no significant difference between the three groups in the TC reduction rate after treatment, the reduction effect of TC was found to be in the following order: E2 group > E3 group > E4 group, indicating that E2 carriers had a higher tendency to lower lipids with statins than other carriers. Some studies have demonstrated that there are sex differences in the lipid-lowering effect of APOE on statins; that is, the E2 allele enhances the lipid-lowering effect of statins, which is more significant in male patients, but not in female patients (25); this may be related to differences in immune activation and hormone levels (26). Consistent with previous findings, our study also found that E2 allele carriers had a greater LDL-C-lowering effect on statins in males, while this difference was not apparent in females.
APOE polymorphisms can affect the occurrence and development of carotid plaques through various mechanisms. First, many previous studies have shown that APOE gene polymorphisms affect blood lipid metabolism, and dyslipidemia is a significant risk factor for the occurrence and development of atherosclerotic plaques. Therefore, APOE may affect the blood lipid metabolic pathway and carotid plaque progression. Second, APOE gene polymorphisms are associated with the development of diabetes (27), but the underlying mechanism is currently unknown. Studies have speculated that E4 carrier status may affect peripheral and central insulin metabolism (28, 29); therefore, differences in blood glucose metabolism can impact arterial plaque development. Third, studies have found that E4 carriers have higher expression of lipoprotein-related phospholipase, which can promote the body’s inflammatory response and plaque instability (30, 31).
Our results provide robust evidence for personalized lipid-lowering and plaque stabilization treatments based on APOE genotypes. This precise treatment can maximize the efficacy of statins. People can achieve lower LDL-C levels, reducing the incidence and recurrence rate of stroke. However, several limitations should also be noted. First, this is a single-center study, which limited the ability to draw major conclusions for all populations. Further studies with larger sample sizes are needed to corroborate our findings. Second, we determined the properties of plaques using ultrasound based on the echo morphology of plaques, which may be less precise than using high-resolution magnetic resonance imaging.
Conclusion
Polymorphisms of the APOE gene are related to the effects of atorvastatin on lipid lowering and the progression of carotid artery plaques. The population with the ε2 allele experienced a better lipid-lowering effect on LDL-C and slower progression of carotid artery plaques than the population with the ε3 or ε4 allele. Patients with E4 genotype need a higher statin dose or a change to another statin to achieve a better effect. A larger population is required to provide more reliable evidence to explore this relationship.
Data availability statement
The data analyzed in this study is subject to the following licenses/restrictions: The data are available from the corresponding author on reasonable request. Requests to access these datasets should be directed to YmlxaWFucWlhbjEyQDE2My5jb20=.
Ethics statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Shanghai Tenth People’s Hospital. The patients/participants provided their written informed consent to participate in this study.
Author contributions
XZ and JW revised the study. QB drafted the manuscript. QB, YL, and YW analyzed and interpreted the data. QB and WF collected the data. XZ and FW contributed to the critical revision of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the 2020 Health Science and Technology Project of the Pudong New Area Health Commission (No. PW2020D-5), the 2021 Scientific Research Project of Shanghai Municipal Commission of Health and Family Planning (No. 202140282), 2019 Scientific Research Project of Shanghai Science and Technology Commission (No. 19401972803), the 2019 Shanghai Municipal Key Clinical Specialty (No. shslczdzk06102), and the 2020 Science and Technology Development Fund of Pudong New Area Special Fund for People’s Livelihood Scientific Research (PKJ2020-Y-15). The content was solely the responsibility of the authors.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Libby P, Buring JE, Badimon L, Hansson GK, Deanfield J, Bittencourt MS, et al. Atherosclerosis. Nat Rev Dis Primers. (2019) 5:56. doi: 10.1038/s41572-019-0106-z
2. Amarenco P, Labreuche J. Lipid management in the prevention of stroke: review and updated meta-analysis of statins for stroke prevention. Lancet Neurol. (2009) 8:453–63. doi: 10.1016/S1474-4422(09)70058-4
3. Hubacek JA, Vrablik M. Effect of apolipoprotein E polymorphism on statin-induced decreases in plasma lipids and cardiovascular events. Drug Metabol Drug Interact. (2011) 26:13–20. doi: 10.1515/DMDI.2011.107
4. Zhang L, He S, Li Z, Gan X, Li S, Cheng X, et al. Apolipoprotein E polymorphisms contribute to statin response in Chinese ASCVD patients with dyslipidemia. Lipids Health Dis. (2019) 18:129. doi: 10.1186/s12944-019-1069-5
5. Kirac D, Bayam E, Dagdelen M, Gezmis H, Sarikaya S, Pala S, et al. HMGCR and ApoE mutations may cause different responses to lipid lowering statin therapy. Cell Mol Biol. (2017) 63:43–8. doi: 10.14715/cmb/2017.63.10.6
6. Phillips MC. Apolipoprotein E isoforms and lipoprotein metabolism. IUBMB Life. (2014) 66:616–23. doi: 10.1002/iub.1314
7. Marais AD. Apolipoprotein E in lipoprotein metabolism, health and cardiovascular disease. Pathology. (2019) 51:165–76. doi: 10.1016/j.pathol.2018.11.002
8. Borgaonkar DS, Schmidt LC, Martin SE, Kanzer MD, Edelsohn L, Growdon J, et al. Linkage of late-onset Alzheimer’s disease with apolipoprotein E type 4 on chromosome 19. Lancet. (1993) 342:625. doi: 10.1016/0140-6736(93)91458-x
9. Seripa D, D’Onofrio G, Panza F, Cascavilla L, Masullo C, Pilotto A. The genetics of the human APOE polymorphism. Rejuvenation Res. (2011) 14:491–500. doi: 10.1089/rej.2011.1169
10. Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. a meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. (1997) 278:1349–56. doi: 10.1001/jama.278.16.1349
11. Huebbe P, Rimbach G. Evolution of human apolipoprotein E (APOE) isoforms: gene structure, protein function and interaction with dietary factors. Ageing Res Rev. (2017) 37:146–61. doi: 10.1016/j.arr.2017.06.002
12. Cai C, Wen Z, Li L. The relationship between ApoE gene polymorphism and the efficacy of statins controlling hyperlipidemia. Am J Transl Res. (2021) 13:6772–7.
13. Xi N, Wang Y, Zhou J, Li Y, Ruan S. [Influence of ApoE gene polymorphisms on therapeutic effects of lipid-lowering statins among patients with ischemic cerebral infarction]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. (2022) 39:94–8. doi: 10.3760/cma.j.cn511374-20201104-00775
14. Zintzaras E, Kitsios GD, Triposkiadis F, Lau J, Raman G. APOE gene polymorphisms and response to statin therapy. Pharmacogenomics J. (2009) 9:248–57. doi: 10.1038/tpj.2009.25
15. Hu M, Mak VW, Tomlinson B. Polymorphisms in apolipoprotein E and apolipoprotein A-V do not influence the lipid response to rosuvastatin but are associated with baseline lipid levels in Chinese patients with hyperlipidemia. J Clin Lipidol. (2012) 6:585–92. doi: 10.1016/j.jacl.2012.02.005
16. Lagos J, Zambrano T, Rosales A, Salazar LA. APOE polymorphisms contribute to reduced atorvastatin response in Chilean Amerindian subjects. Int J Mol Sci. (2015) 16:7890–9. doi: 10.3390/ijms16047890
17. Wang YY, Tang XB, Kou L, Wu ZM, Liu H, Wang S, et al. A comparative study of noninvasive imaging in assessing carotid atherosclerotic plaque stability. Chin J Vasc Surg. (2017) 2:209–17. doi: 10.1161/STROKEAHA.119.025496
18. Weisgraber KH. Apolipoprotein E distribution among human plasma lipoproteins: role of the cysteine-arginine interchange at residue 112. J Lipid Res. (1990) 31:1503–11.
19. Huang Y, Mahley RW. Apolipoprotein E: structure and function in lipid metabolism, neurobiology, and Alzheimer’s diseases. Neurobiol Dis. (2014) 72:3–12. doi: 10.1016/j.nbd.2014.08.025
20. Hatters DM, Peters-Libeu CA, Weisgraber KH. Apolipoprotein E structure: insights into function. Trends Biochem Sci. (2006) 31:445–54. doi: 10.1016/j.tibs.2006.06.008
21. Shafagoj YA, Naffa RG, El-Khateeb MS, Abdulla YL, Al-Qaddoumi AA, Khatib FA, et al. APOE Gene polymorphism among Jordanian Alzheimer‘s patients with relation to lipid profile. Neurosciences. (2018) 23:29–34. doi: 10.17712/nsj.2018.1.20170169
22. Sun Q, Zhou H, Guo LN. ApoE and SLCO1B1 gene polymorphisms and their correlation in dyslipidemia population. J Mod Lab Med. (2017) 2017:28–31.
23. Xie L, Hong M, Liu T, Li Q, Duan YP, Feng Y. Analysis of SLCO1B1 and ApoE gene polymorphisms in patients with hyperlipidemia and their correlation with blood lipid levels in patients. Label Immun Clin Med. (2019) 2019:1723–8.
24. Zhang Y, Wei DD, Yuan RR, Ge QX, Chen F, Yang SX, et al. [Effects of ApoE gene polymorphism on the efficacy of atorvastatin in the treatment of hyperlipidemia]. Zhonghua Yi Xue Za Zhi. (2017) 97:291–4.
25. Pedro-Botet J, Schaefer EJ, Bakker-Arkema RG, Black DM, Stein EM, Corella D, et al. Apolipoprotein E genotype affects plasma lipid response to atorvastatin in a gender specific manner. Atherosclerosis. (2001) 158:183–93. doi: 10.1016/s0021-9150(01)00410-5
26. Faubion SS, Kapoor E, Moyer AM, Hodis HN, Miller VM. Statin therapy: does sex matter? Menopause. (2019) 26:1425–35. doi: 10.1097/GME.0000000000001412
27. Gonzalez-Aldaco K, Roman S, Torres-Reyes LA, Panduro A. Association of apolipoprotein e2 allele with insulin resistance and risk of type 2 diabetes mellitus among an admixed population of Mexico. Diabetes Metab Syndr Obes. (2020) 13:3527–34. doi: 10.2147/DMSO.S268329
28. Rhea EM, Raber J, Banks WA. ApoE and cerebral insulin: trafficking, receptors, and resistance. Neurobiol Dis. (2020) 137:104755. doi: 10.1016/j.nbd.2020.104755
29. Pekkala T, Hall A, Mangialasche F, Kemppainen N, Mecocci P, Ngandu T, et al. Association of peripheral insulin resistance and other markers of type 2 diabetes mellitus with brain amyloid deposition in healthy individuals at risk of Dementia. J Alzheimers Dis. (2020) 76:1243–8. doi: 10.3233/JAD-200145
30. Corson MA. Phospholipase A2 inhibitors in atherosclerosis: the race is on. Lancet. (2009) 373:608–10. doi: 10.1016/S0140-6736(09)60378-0
Keywords: apolipoprotein E, polymorphism, lipid-lowering, atherosclerosis, carotid artery plaques
Citation: Bi Q, Zhou X, Lu Y, Fu W, Wang Y, Wang F and Wang J (2022) Polymorphisms of the apolipoprotein E gene affect response to atorvastatin therapy in acute ischemic stroke. Front. Cardiovasc. Med. 9:1024014. doi: 10.3389/fcvm.2022.1024014
Received: 20 August 2022; Accepted: 20 October 2022;
Published: 08 November 2022.
Edited by:
Jia Qi, Shanghai Jiao Tong University, ChinaReviewed by:
Genmao Cao, Second Hospital of Shanxi Medical University, ChinaZhi Geng, Shanghai Jiao Tong University, China
Xiaoqin Cheng, Fudan University, China
Copyright © 2022 Bi, Zhou, Lu, Fu, Wang, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Wang, MTM4MTY1NjY1NTZAMTYzLmNvbQ==; Jue Wang, d2FuZ2p1ZXNoaXl1YW5AMTYzLmNvbQ==
†These authors have contributed equally to this work
 QianQian Bi
QianQian Bi XiaoYu Zhou
XiaoYu Zhou YanQin Lu
YanQin Lu Wang Fu
Wang Fu YongPeng Wang
YongPeng Wang Feng Wang
Feng Wang Jue Wang
Jue Wang