
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cardiovasc. Med. , 10 November 2022
Sec. Cardiovascular Genetics and Systems Medicine
Volume 9 - 2022 | https://doi.org/10.3389/fcvm.2022.1014961
This article is part of the Research Topic Epigenetic Mechanisms in Cardiovascular Disease View all 4 articles
 Sara Fadaei1†
Sara Fadaei1† Fatemeh Zarepour2,3†
Fatemeh Zarepour2,3† Mehrnoosh Parvaresh4
Mehrnoosh Parvaresh4 Alireza Motamedzadeh5
Alireza Motamedzadeh5 Seyed Saeed Tamehri Zadeh6
Seyed Saeed Tamehri Zadeh6 Amirhossein Sheida2,3
Amirhossein Sheida2,3 Mohammad Shabani2,7
Mohammad Shabani2,7 Michael R. Hamblin8
Michael R. Hamblin8 Mehdi Rezaee9*
Mehdi Rezaee9* Maryam Zarei10*
Maryam Zarei10* Hamed Mirzaei11*
Hamed Mirzaei11*Myocardial infarction (MI) is one of the leading causes of deaths globally. The early diagnosis of MI lowers the rate of subsequent complications and maximizes the benefits of cardiovascular interventions. Many efforts have been made to explore new therapeutic targets for MI, and the therapeutic potential of non-coding RNAs (ncRNAs) is one good example. NcRNAs are a group of RNAs with many different subgroups, but they are not translated into proteins. MicroRNAs (miRNAs) are the most studied type of ncRNAs, and have been found to regulate several pathological processes in MI, including cardiomyocyte inflammation, apoptosis, angiogenesis, and fibrosis. These processes can also be modulated by circular RNAs and long ncRNAs via different mechanisms. However, the regulatory role of ncRNAs and their underlying mechanisms in MI are underexplored. Exosomes play a crucial role in communication between cells, and can affect both homeostasis and disease conditions. Exosomal ncRNAs have been shown to affect many biological functions. Tissue-specific changes in exosomal ncRNAs contribute to aging, tissue dysfunction, and human diseases. Here we provide a comprehensive review of recent findings on epigenetic changes in cardiovascular diseases as well as the role of ncRNAs and exosomal ncRNAs in MI, focusing on their function, diagnostic and prognostic significance.
Cardiovascular disease (CVD) is a leading cause of mortality around the world, accounting for 18.6 million deaths in 2019, with coronary heart disease (CHD) being the most common type of CVDs. About 697,000 persons in the United States died from heart disease in 2020 that is one in every five deaths (1, 2). Myocardial infarction (MI), a common manifestation of CHD, is caused by acute or chronic deprivation of nutrients, oxygen, and other important elements necessary for the myocardium survival. MI triggers a chain of severe biochemical and metabolic perturbations in the cardiomyocytes. This imbalance primarily results from acute or chronic ischemia. Cell death in the ischemia area is the initial key event of MI, however a chain of events leads to other heart diseases. Furthermore, MI is associated with several structural and functional consequences leading to permanent heart damage or death (3). This is because cardiomyocytes are terminally differentiated cells and thus have a poor regenerative capacity (4). The infarct size is considered to be the best predictor of future cardiac dysfunction and mortality following acute MI (3).
In biology, epigenetics is a field of study focused on the chemical modification of specific genes or gene-associated proteins that do not involve alterations in the DNA sequence. Growing evidence indicated that epigenetic mechanisms, including DNA methylation, histone modification, and non-coding RNA, are closely related to cardiovascular disease development and regression. Epigenetics mainly regulates cardiovascular disease-related genes function and expression level through DNA methylation, histone modification, and non-coding RNA regulation, thus affecting cardiovascular disease progression. Epigenetic markers are important molecular markers of cardiovascular disease because they occur early in the disease and involve key cardiovascular pathologically related pathways. Most importantly, it can be used as cardiovascular disease biomarkers for cardiovascular disease diagnosis, treatment response prediction and evaluation. As we all know, the pathogenesis of cardiovascular disease remains intricate and complex. Clinically, some cases are still difficult to cure, and the prevalence rate increases with age. Interestingly, because of the reversibility of epigenetic modifications, genes and proteins that control these changes have become new targets for cardiovascular disease treatment. Recently, non-coding RNAs (ncRNAs) and their regulatory functions have aroused great interest (5). There is increasing evidence that ncRNAs, such as microRNAs (miRNAs), circular RNAs (circRNAs), and long non-coding RNAs (lncRNAs) all have regulatory functions and diagnostic value for many types of CVD (6). Regulation of numerous ncRNAs modifies pathophysiological processes in cardiac development, cardiovascular remodeling, and cardiovascular diseases, such as ischemic heart disease, hypertrophy, atherosclerosis, and cardiac fibrosis. MicroRNAs are short-ncRNA molecules ~21–23 nucleotides in length, and are expressed by almost every cell (7). A mature miRNA usually binds to its mRNA 3′ untranslated region by complementary base-pairing, leading to inhibition of translation or mRNA degradation (8–10). From 2008 onwards, scientists around the world reported that circulating miRNAs have good potential as markers for the diagnosis of a variety of diseases (11–13). In this context, emerging evidence has confirmed the diagnostic value of miRNAs in MI (14). Moreover, circRNAs and lncRNAs have also been proposed as biomarkers for MI. Recently, a plethora of papers implicated the key role of exosomal ncRNAs in mediating intercellular and inter-organ communication and CADs development (15, 16).
Herein, we provide a comprehensive review of recent findings on the interplay between ncRNAs and epigenetic machinery in cardiovascular diseases. Furthermore, this paper aims to review and enhance the understanding of the mechanisms and roles of ncRNAs and exosomal ncRNAs in MI, and to provide a basis for clinical diagnosis and new therapeutic strategies for this disease.
Epigenetic regulatory processes include DNA methylation (DNAm), histone modifications, microRNAs, and lncRNAs (17). The expression of the epigenome differs between various cell types, and modulates single cell gene expression by organizing the nuclear architecture in chromosomes, suppressing or promoting the access of transcription factors to the DNA, and regulating gene expression (18). Epigenetic alterations are involved in the cause of many human diseases, including severe CVD, owing to the role of differential gene regulation in cellular differentiation and biological functions (1, 19, 20). There is much evidence in support of the assertion that epigenetic alterations play a crucial role in a variety of CVDs, such as hypertension, cardiac hypertrophy, heart failure (HF), myocardial I/R damage (21–25). Thus, epigenetics can be considered as a valuable target for the management of myocardial I/R damage.
An important mechanism for epigenetic regulation is histone modification and post-transcriptional modification, and the most frequent histone modifications include acetylation, phosphorylation, ubiquitination, and methylation (26). Researchers reported that histone methylation primarily takes place on arginine and lysine residues, and is mediated by histone demethylase and histone methyltransferase (HMTs) enzymes (27). Histone acetylation by histone acetyltransferases (HATs) is the introduction of acetyl groups into histones, leading to nucleosome relaxation and activation of transcription. The reverse progress is the elimination of acetyl groups from histones by histone deacetylase enzymes (HDACs), which causes the accumulation of nucleosomes and contributes to inhibition of gene transcription. Hence, HATs and HDACs have been likened to writers and erasers (28–30).
The main site of histone methylation in studies focusing on myocardial ischemia/reperfusion (I/R) injury is histone 3 lysine 9 (H3K9). The first confirmed H3K9 methyltransferase was Suv39h1. Since then, several other methyltransferases have been discovered, such as G9a and its related proteins Ctrl4 and Suv39h2. In contrast, PHF8 (PHD Finger Protein 8), JHDM2 (Jmjc domain-containing histone demethylase 2) and JHDM3 enzymes can demethylate H3K9 (27). Yang et al. showed that ischemic or oxidative stress induces Suv39H expression in parallel with the class III protein deacetylase sirtuin 1 (SIRT1) repression, whereas deletion/inhibition of SUV39H ameliorates MI-related damage (31).
According to report by Sung et al., histone methyltransferase G9a plays a significant role in acute myocardial infarction-induced heart failure (32). Flow cytometry analysis clarified the protective impact of G9a inhibitor (i.e., UNC0638) on H9C2 cardiomyocyte cells against H2O2-indcued apoptosis and oxidative stress. In addition, the combination of G9a inhibitor and erythropoietin (EPO) therapy effectively protected heart against damage from acute myocardial through regulating of several factors. This combined treatment promoted angiogenesis by upregulating the expressions of SDF-1α, VEFG and CXCR4, three angiogenesis indicator. Besides, combination of G9a inhibitor and EPO decreased expressions of apoptotic cleaved caspase 3 and cleaved PARP, fibrotic biomarkers (Samd3, TGF-ß and fibronectin), and DNA-damage marker, and suppressed protein expressions of inflammatory biomarkers [matrix metalloproteinase (MMP)-2 and MMP-9] and anti-oxidative-stress indicators (Sirt1 and Sirt3). The authors also discovered that combining G9a inhibitor and EPO treatment improved autophagy and cell-stress signaling (32).
HDACs are a large family of enzymes consisting of 18 enzymes in humans. They regulate chromatin remodeling and consequent gene transcription mostly by controlling histone acetylation status. Based on sequence homology and cellular localizations, HDACs are divided into four classes: class I (HDAC1, 2, 3, and 8); class II (HDAC4, 5, 6, 7, 9, and 10); class III (SIRT1- 7); and the class IV protein (HDAC11) (33).
Among the HDACs, class IIa (HDAC4, 5, 7 and 9) are thought to be cardiac protective because overexpression of HDAC4, HDAC5, or HDAC9 in cardiac myocytes suppresses expression of a pro-hypertrophy transcription factor, Mef2 (myocyte enhancer factor-2), and reduces stress-induced cardiac hypertrophy (34). Conversely, silencing HDAC5 or HDAC9 leads to an exacerbation of the hypertrophic response to pressure overload (35).
HDAC4 belongs to class II, plays an important role in a number of physiological and pathological processes of the heart. Under normal conditions, HDAC4 activity is very low, while it is activated under pathological conditions, including cardiac injury. In animal models, overexpression of HDAC4 increased myocardial infarct size, on the other hand, inhibition of HDAC4 stimulates regeneration and restoration of cardiac function and reduces myocardial infarction in ischemic HF (36, 37). Zhang et al. created myocyte-specific active HDAC4 transgenic mice to investigate the functional role of activated HDAC4 in regulating myocardial I/R injury. They found that active HDAC4 in the heart is critical to promote myocardial I/R injury. Moreover, delivery of HDAC inhibitor can diminish HDAC4-induced I/R injury (36). Additionally, activated HDAC4-elicited cardiac injury was related the decreased SOD-1 and increased apoptosis and autophagy (36). Recently, Li et al. concluded that HDAC4 silencing can upregulate miR-206 expression, reducing cardiomyocyte apoptosis and inhibiting oxidative stress, and protecting myocardial I/R injury through the mitogen-activated protein kinase kinase kinase 1 (MEKK1)/JNK pathway (38).
In contrast to class IIa HDACs, class I HDACs have shown pro-hypertrophy effects via a variety of mechanisms, such as reducing autophagy through activation of mTOR signaling or suppressing the expression of Inpp5f (inositol polyphosphate-5-phosphatase f) and later inhibiting GSK3β (glycogen synthase kinase 3β) signaling, or inhibiting of DUSP5 (dual-specificity phosphatase 5) that negatively regulates ERK1/2-induced cardiac hypertrophy (39).
It was shown that HDAC1 is present in cardiomyocyte mitochondria, and stimulates myocardial damage in the initial phase of reperfusion (40). According to recent researches, overexpression of HDAC2 and HDAC3 induces severe cardiac hypertrophy and hyperplasia, respectively (41, 42). Furthermore, class I HDAC inhibitors diminishes cardiac hypertrophy (43). A number of studies have shown a cardioprotective effect of class III HDACs (e.g., sirtuins) against myocardial I/R damage (44). Overall, it seems that class IIa and class III of HDACs have protective roles not only in heart injury but also in vessel injury, whereas class I HDACs protect against vessel damage but have harmful effects on the myocardium.
Enhancer of zeste homolog 2 (EZH2), an enzymatic subunit of polycomb repressive complex 2 (PRC2), is evolutionarily conserved in many species with comparable structural motifs and domains (45). It is responsible for catalyzing histone H3K27me3 to silence its target gene expression and being involved in various biological functions from cell cycle and cell proliferation to cell differentiation etc. (46, 47). EZH2 represents a major target of ncRNAs inside nucleus (48). Studies showed that in Ezh2-deficient adult hearts, fetal genes are upregulated, causing cardiac hypertrophy (49, 50). In ischemic heart disease (IHD), elevated EZH2 epigenetically represses cardiac sodium channel NaV1.5, which is one of the underlying mechanisms of arrhythmias in IHD patients (51). Recently, Jiao et al. done a study to investigate the therapeutic effects of mesenchymal stem cell-secreted exosomes (MSC-EXO) on myocardial fibrosis after MI (52). They found EZH2 alleviates the cardioprotective effects of MSC-EXO in MI rats through inhibiting HMGA2 (High mobility group AT-Hook 2) expression and disrupting the PI3K/AKT pathway (52).
There are several ways to control gene expression, and one example is DNA methylation (31). DNA methylation is the well-studied epigenetic modification of the genome (53). It is essential for cellular reprogramming, normal development, tissue differentiation, and is contribute significantly in many biological processes as well as to the molecular pathology of different disease states (54). DNA methylation occurs when the methyl groups are covalently attached to the 5′ of cytosine in cytosine-guanine (CpG) dinucleotides, found mainly in CpG-rich regions known as CpG islands (CGIs) present in more than half of the promoters of mammalian genes (55). DNA methylation itself modifies transcription binding sites, preventing transcriptional activation and binding of transcription factors (56). Researchers are discovering the mammalian proteins and mediators involved in DNA methylation and demethylation, including DNA methyltransferases (DNMTs), ten-eleven translocation cytosine dioxygenases (TETs), methyl-CpG binding proteins (MeCPs), non-coding RNAs, and transcription factors (57). DNMT1 is an enzyme that tends to maintain nascent DNA methylation marks during mitosis at cytosines that are methylated on the parental strand, termed maintenance methylation. This phenomenon ensures the persistence of programmed DNA methylation patterns across cell generations. De novo DNA methyltransferase enzymes DNMT3A and DNMT3B are involved in generating new DNA methylation signatures and are essential for regulating DNA methylation patterns for embryonic or neonatal development (58, 59). DNMT3L as a cofactor is require for de novo methyltransferase activity in embryonic stem cells and a positive regulator of DNA methylation at gene bodies of housekeeping genes (60). Proteins, including TET methylcytosine deoxygenase, AID (activation-induced cytidine deaminase), and TDG (thymine DNA glycosylase), are involved in active and passive demethylation, gene activation, embryonic development, and maintenance of adult tissue homeostasis (61–63). TET enzymes catalyze the sequential oxidation of 5-methylcytosine (5 mC) to generate intermediates involved in the conversion of 5 mC to unmodified cytosines and removal of the epigenetic mark, potentially providing the first steps in a pathway for active DNA demethylation (64).
Abnormalities in DNA methylation and hypomethylation across the genome, as well as CpG island hypermethylation have been observed in CVDs (32, 33). Recent advances in epigenetic sequencing have allowed examination of associations between genomic coding, exposure to carcinogens, and disease phenotype. The DNAm epigenetic modification could provide a possible explanation for these relationships (34). It is possible to influence transcription by adding methyl groups to certain DNA nucleotide bases via the pre-transcriptional modification known as DNA methylation (35). DNA genetic sequences are essential to the natural evolution and survival of mammalian species governed by specific patterns of gene expression. Epigenetic modifications are dynamic and reversible at the same time, which is important for the regulation of genetic pathways (36). DNA methylation is a dynamic process that reflects the balance between the activity of DNMTs and TETs. The association between mutated genes DNMT3A and TET2 and development of inflammation, atherosclerosis, and heart failure has been reported (65–67). Numerous studies have focused on DNAm pattern alterations and measuring differentially methylated regions in both normal and abnormal development (37).
A number of studies have investigated DNA methylation profiles in CHD and MI, suggesting their significance as a diagnostic marker (68, 69). One study showed notable enhanced DNA methylation levels in CHD patients (70). Additionally, in a pilot study by Ma et al. for providing useful DNA methylation profiles to serve as biomarkers for detecting early-phase atherosclerosis (AS), a total of 300 persons were recruited (150 AS patients and 150 healthy subjects) for peripheral blood DNA methylation analyses at 12 gene promoter loci using nested methylation-specific polymerase chain reaction (71). Based on the test set, promoter methylation of acetyl Acetyl-CoA acetyltransferase 1 (ACAT1) was decreased whereas ATP binding cassette subfamily A member 1 (ABCA1) and tissue inhibitor of metalloproteinase-1 (TIMP1) were significantly increased in AS compared the matched controls. Thus, they suggested that methylation of the three-gene panel (TIMP1, ABCA1, and ACAT1) could serve as a valuable, high sensitivity, and reliable biomarker for the early detection of AS (71). Perinatal nicotine exposure in mice was shown to increase myocardial I/R damage, which was found to be mediated by DNA hypermethylation and increased DNMT3 expression. Conversely, the administration of the DNA methylation inhibitor 5-aza-2′-deoxycytidine (5-Aza) was able to ameliorate myocardial injury. In conclusion, the authors found out that nicotine-induced myocardial I/R damage was mediated by DNA methylation in perinatal mice (72).
More than 90% of the human genome is composed of genes that do not encode protein. Nevertheless, these genes show considerable transcriptional activity, and code for a wide variety of ncRNAs with biological regulatory functions (73).
As discussed above, ncRNAs include three main classes: miRNAs, lncRNAs, and circRNAs. lncRNAs contain more nucleotides compared to miRNAs (>200 nucleotides vs. 21–23 nucleotides), which is the primary difference between them. MiRNAs were shown to modulate gene expression by inhibiting messenger RNA (mRNA) translation and causing mRNA degradation (6, 74, 75). It has been reported that miRNAs can affect myocardial angiogenesis and cardiomyocyte survival and proliferation by regulation of target gene expression (Figures 1A–C).
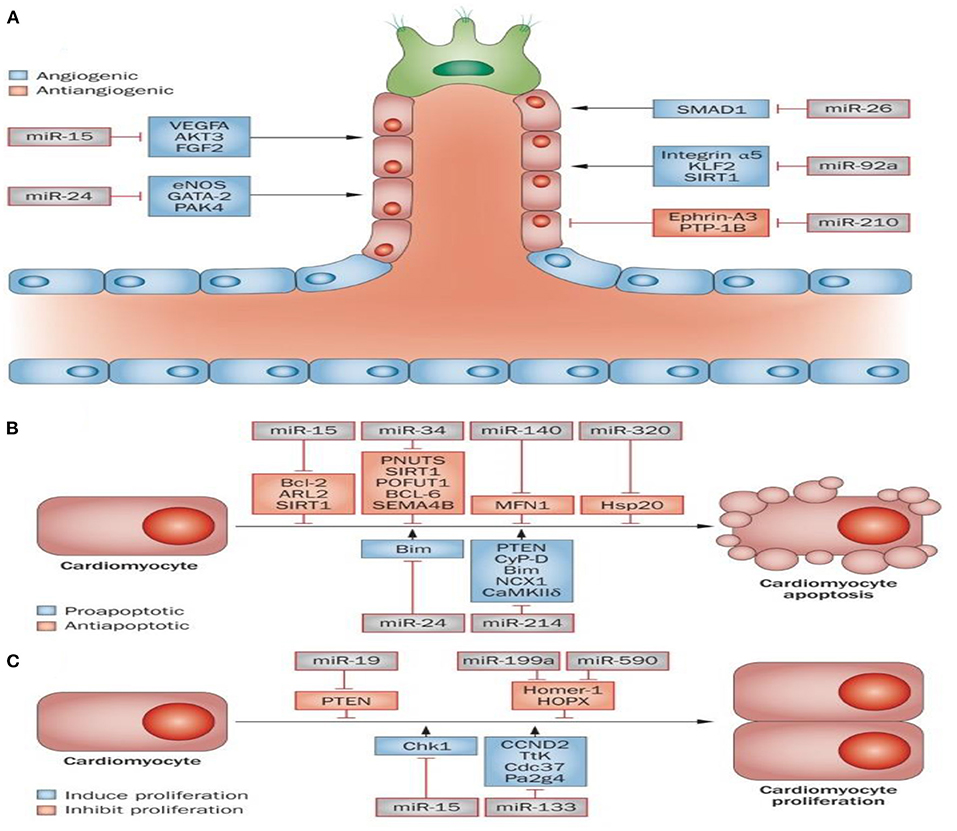
Figure 1. Role of microRNAs in MI. Some miRNAs are known to be implicated in cardiac muscle angiogenesis (A), cardiac cell survival, and proliferation (B,C). Angiogenesis is pivotal for the delivery of oxygen and essential nutrients to heart muscle. Angiogenesis can be activated or repressed by a number of microRNAs. For instance, miR-15, miR-24, miR-26, and miR-92a all repress angiogenesis through inhibition of endothelial-cell functions. In contrast, miR-210 prompts angiogenesis via inhibiting several antiangiogenic factors (A). A number of miRNAs have been identified to be involved in the apoptosis and survival of cardiac cells. Apoptosis inhibition or an increase in survival signals promotes cardiac regeneration. The miR-15 family (miR-320, miR-34, and miR-140) can serve as pro-apoptotic factors, while miR-24 and miR-214 can act as anti-apoptotic factors (B). The proliferation of cardiac cells is limited; however, cardiac regeneration can increase following stimulation of proliferation. It has been shown that miR-19, miR-199a and miR-590, can promote proliferation of cardiac cells, while both miR-15 and miR-133 inhibit cardiac cell proliferation (C). This figure adapted from Boon and Dimmeler (76).
CircRNAs are mostly found in eukaryotic cells. Unlike normal linear RNA molecules, the 3-cap and 5-tail of circRNAs are covalently joined to form a closed loop. CircRNAs are characterized by plentiful amounts, tissue specificity, and evolutionary conservation. CircRNAs can act as a competitive endogenous RNA (ceRNA) and a sponge that binds to complementary miRNA sequences to moderate gene expression (77, 78).
MiR-431 has been shown to stimulate the regeneration and differentiation of axons and skeletal muscles by targeting Smad4 (79). Emerging evidence has shown that miR-431 plays an important role in several diseases, including but not limited to hepatocellular carcinoma (80) and colorectal cancer (81). The possible underlying mechanism was suggested to be hypoxia-induced apoptosis (82). Nevertheless, the involvement of miR-431 in cardiac hypoxia/reoxygenation (H/R) is still unclear. It is known that autophagy and cell apoptosis are two important processes involved in H/R and MI (83). Autophagy-related 3 (ATG3) is an E2 enzyme essential for the lipidation of LC3 during autophagy (84). There is evidence that ATG3 is associated with autophagy in several diseases, including breast cancer (85), hepatocellular carcinoma (86), and chondrocyte disturbance (87), however, the role of miR-431 in myocardial damage is not yet understood. Zhou et al. tried to clarify the mechanism of how miR-431 affected apoptosis and autophagy of cardiomyocytes in H/R. They found that H/R was associated with decreased cell viability, increased apoptosis, and lower miR-431 expression in human cardiomyocytes. Inhibition of miR-431 decreased cell viability and promoted cell apoptosis. Additionally, it was shown that miR-431 inhibited ATG3 expression by targeting the 3'-untranslated region of ATG3 mRNA. Exogenous ATG3 overexpression was able to reverse the effects of miR-431 up-regulation on cell survival and apoptosis in H/R-treated human cardiomyocytes. H/R treatment stimulated autophagy in human cardiomyocytes, which was significantly decreased after transfection with a miR-431-mimic. Their findings indicated that miR-431up-regulation could regulate ATG3 expression, and ameliorate H/R-stimulated myocardial injury (88).
Treatment with imatinib mesylate and doxorubicin increased miR-205 expression in the plasma and heart of mice (89). Moreover, miR-205 downregulation significantly decreased myocardial apoptosis in rats with chronic heart failure (90). Cheng et al. sought to understand how Remifentanil (a short-acting synthetic opioid analgesic drug) exerted its protective effects on myocardial I/R damage. The authors found that remifentanil was able to enhance cell survival and reduce apoptosis of H/R-treated cardiomyocytes. Increased miR-205 expression and reduced PINK1 (PTEN induced putative kinase 1) expression was observed following H/R treatment. Preconditioning with remifentanil decreased miR-205 and increased PINK1 expression. Additionally, miR-205 up-regulation decreased the expression of PINK1 and reversed the remifentanil-stimulated increase in cell viability and reduction of apoptosis in H/R-treated cardiomyocytes. The investigators observed that the use of a miR-205 antagomir was associated with a better remifentanil-induced reduction of lactate dehydrogenase (LDH) activity and lower infarct size in rats with I/R injury. Ultimately, miR-205 may be the reason for the protective effect of remifentanil on myocardial I/R damage, making it a possible target for the management of MI (91).
It was reported that miR-147 could inhibit apoptosis in L6 myoblasts subjected to cyclic mechanical stretching (92). MiR-147 downregulation was associated with inhibition of proliferation in gastric cancer cells (93). Nonetheless, the role of miR-147 in MI remains largely unknown. Wu et al. designed an in vivo study to evaluate the function of miR-147 in MI, and the mechanism of action (94). Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was used to measure the expression of Bax mRNA, miR-147, and Bcl-2 mRNA. Inflammatory cytokines including interleukin (IL)-6, IL-β, TNF-α, as well as lactate dehydrogenase (LDH) were measured by enzyme-linked immunosorbent assay (ELISA). Additionally, the MTT [3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium bromide] assay was employed to measure cell viability. Cell apoptosis was measured by terminal dexynucleotidyl transferase (TdT)-mediated dUTP nick end labeling (TUNEL). Additionally, in vivo cardiac function was evaluated by echocardiography. Western blotting was employed to measure the expression of homeodomain interacting protein kinase 2 (HIPK2). Results indicated the lower levels of miR-147 in the rat MI model and also in H9c2 cells treated with H2O2. Moreover, H2O2 treatment was associated with higher levels of inflammatory cytokines and caused apoptosis in H9c2 cells. These effects were abrogated by miR-147 up-regulation. MiR-147 up-regulation reversed the increased Bax expression and reduced the Bcl-2 expression in H9c2 cells induced by H2O2. Furthermore, miR-147 up-regulation was associated with better cardiac function and lower serum LDH values in MI rats. TargetScan analysis demonstrated that HIPK2 could bind to miR-147. Additionally, miR-147 up-regulation was able to inhibit HIPK2 expression (94). The findings of this study provide the postulate that lower levels of miR-147 can be expected in patients with MI, while up-regulation could inhibit myocardial apoptosis and inflammation, and improve cardiac function through regulating HIPK2 (94).
MiR-130 can worsen myocardial injury following MI by targeting peroxisome proliferator-activated receptor gamma (PPAR-γ). In H9c2 cells, miR-130 by targeting PPAR-γ aggravates acute MI-induced injury. In vivo experiments confirmed that miR-130 downregulation promotes PPAR-γ-mediated cardioprotective effects by suppressing NFκB-mediated inflammation and TGF-β1-mediated myocardial fibrosis (85). Pan et al. designed a study to evaluate the diagnostic value of miR-130 in patients with MI. The plasma expression of miR-130 in patients suffering from MI was measured by qRT-PCR. The plasma value of cardiac troponin I (cTnI) and creatine kinase-MB (CK-MB) was examined by ELISA. The authors evaluated the diagnostic value of miR-130 using a receiver operating characteristic (ROC) curve. The plasma values of miR-130, CK-MB, and cTnI in the MI group were significantly higher relative to the control group. The peak of miR-130 expression occurred 6 h after MI onset and the peak was earlier compared to CK-MB and cTnI. A positive correlation was found between miR-130 (6 h after MI onset) and the values of CK-MB and cTnI (12 h after MI onset). The optimal cut-off point was estimated as 1.58 ng/mL with a sensitivity of 82.5% and specificity of 77.5%. The area under the curve (AUC) was measured to be 0.922. These findings suggest that miR-130 could be utilized as a biomarker for MI diagnosis (95).
According to evidence, in patients with HF, the cardiac microRNA-132-3p level is increased in cardiac tissue in response to cardiomyocyte stress (96). It mechanistically drives cardiac remodeling processes leading to heart failure through downregulating the expression of the anti-hypertrophic, pro-autophagic transcription factor Forkhead box O3 (FOXO3) and also inhibiting the expression of genes involved in intracellular calcium handling and contractility (97). As a result, miR-132 appears to be a potentially promising molecular pathophysiological target in the treatment of HF. In this regard, a randomized, placebo-controlled, double-blind, phase 1b dose-escalation study (NCT04045405) was conducted to evaluate the safety, pharmacokinetics, and exploratory pharmacodynamic effects of CDR132L; a synthetic antisense oligonucleotide (ASO) inhibitor, in heart failure patients (98). Based on the results, CDR132L is safe and tolerable, has confirmed linear plasma pharmacokinetics and shows no signs of accumulation, and improves cardiac function (98). Currently, in a randomized, parallel, 3-arm, placebo-controlled phase II clinical trial (NCT05350969) the safety and efficacy of CDR132L in patients with reduced left ventricular ejection fraction (≤45%) after MI is also under investigation (33).
It has been found that miR-204 could affect several physiological and pathological processes, including apoptosis, cell proliferation, and inflammation. Additionally, its capability to affect signaling pathways and their downstream effectors has been demonstrated (99, 100). Therefore, there is a possibility that miR-204 may participate in the process of MI by modulating different pathological responses.
Wang et al. evaluated the effect of miR-204 in rats with MI by targeting the silent information regulator 1 (Sirt1)/p53 signaling pathway. They divided 36 rats into three equal groups, including the sham-operated group, the MI model group, and the miR-204 mimic MI group. The rats in the sham-operation group merely underwent thoracotomy, without any MI injury. MI injuries were created in the MI model group and the miR-204 mimic group and treated with normal saline or miR-204 mimic, respectively. Histological staining revealed a normal morphology in the sham-operated group and severe myocardial tissue damage in the model group. The damage was less in the miR-204 mimic group relative to the MI model group. The authors found a higher Caspase-3 expression in cardiac tissue of the model group and miR-204 mimic group relative to the sham-operated group. The miR-204 level was significantly higher in the model group compared to the miR-204 mimic group. It was shown that Sirt1 (Sirtuin 1) could act as the target gene of miR-204. Higher expression of Sirt1 and p53 was detected in the model group and the miR-204 mimic group compared to the sham-operation group. However, their expression level was notably lower in the miR-204 mimic group compared to the model group. Regarding miR-204 expression, both the miR-204 mimic group and the model group had lower expression; nonetheless, its expression was considerably higher in miR-204 mimic group relative to the model group. A higher apoptosis rate was detected in both the model group and the miR-204 mimic group compared to the sham-operation group. But apoptosis was lower in the miR-204 mimic group compared to the model group. They concluded that miR-204 was able to decrease the apoptosis rate in MI via targeting the Sirt1/p53 signaling pathway (101) (Figure 1, Table 1).
LncRNAs have important regulatory roles in cardiac cell growth, cell survival, and fibrosis in MI. They exert their effects on heart disease through affecting epigenetic gene modifications, transcriptional, and post-translational regulation via direct binding to proteins, miRNAs, and other lncRNAs (138). Although the number of discovered lncRNAs has increased over the years, there is still not much information about the mechanisms of action and the functions performed by these biomolecules. One of the reasons for this delay is that many lncRNAs show unique temporal and spatial expression patterns (139). Furthermore, unlike microRNAs and mRNAs, most lncRNAs have poor sequence conservation among different species, which makes it difficult to obtain functional information and mechanisms of action of the vast majority of lncRNAs and, consequently, translate findings from animal models to humans (139, 140). Besides, the link between basic research and clinical trials in animal models. Therefore, the non-conservation of the nucleotide sequence of lncRNAs among different species is a big and influential challenge, because it makes it difficult to transfer the findings obtained in preclinical studies to humans (141). Hence, clinical trials are limited to working with only those lncRNAs that have human counterparts.
In addition to the issues listed above, the secondary and tertiary structures of lncRNAs must be explored, as they may have structural homologs in other species (142, 143). Finally, drug delivery to the target lncRNA of interest remains a difficulty (144). Even though there are still many challenges to utilizing lncRNAs as a therapeutic approach in CVDs, lncRNAs are promising candidates for the clinic, and due to their specific expression pattern associated with various pathologies, they are recognized as a tool with great application power in personalized medicine (140). In the following, we will discuss some studies about the role of lncRNAs in heart disease and their mechanisms of action.
In an in vitro/in vivo study, Su et al. reported that hypoxia increased the expression of the lncRNA taurine upregulated gene 1 (TUG1), then TUG1 sponged miRNA-132-3p and activated HDAC3, which in turn induced several targets protective gene, stimulated intracellular ROS accumulation, and exacerbated acute MI injury (145).
It was discovered that the lncRNA X inactive specific transcript (XIST) was involved in the inactivation of chromosome X in female cells during embryonic development (146), and it was also overexpressed in several malignancies, such as lung and gastric cancer (147, 148). High levels of XIST were observed in a rat model of acute MI, and its knockdown was shown to suppress cardiomyocyte apoptosis via downregulating miR-449 (149).
Zhou et al. examined the function and underlying mechanism of lncRNA XIST in hypoxia-stimulated injury in H9c2 cells. Cell viability, invasion, migration, and apoptosis were assessed utilizing MTT, transwell, and flow cytometry assays, respectively. Gene expression was measured using Western blotting or qRT-PCR. XIST up-regulation was found in H9c2 cells after hypoxia-stimulated injury, and its knockdown ameliorated the cell damage. XIST overexpression enhanced the expression of B-cell lymphoma 2-associated X (Bax) by binding to miR-150-5p. Thus, it was concluded that XIST was able to protect cardiomyocytes from hypoxia-stimulated damage by modulating the miR-150-5p/Bax axis, suggesting that XIST could a target for MI treatment (150).
The lncRNA called “smooth muscle and endothelial cell-enriched migration/differentiation-associated lncRNA” (SENCR) is located on chromosome 11, and is expressed in human vascular cells (151). There is much evidence in support of the role of SENCR in inhibiting the proliferation and migration of human aortic-vascular smooth muscle cells. SENCR could reduce the formation and progression of atherosclerotic lesions by modulating the miR-4731-5p/FOXO3a signaling pathway (152).
Chen et al. designed a study to evaluate the role and underlying mechanism of SENCR on H/R-stimulated apoptosis in cardiomyocytes. qRT-PCR was employed to quantify the expression of SENCR in the serum of the case group (MI patients) and the control group (non-MI patients with chest pain). Cell viability, cell apoptosis, and inflammatory responses were assessed by MTT, flow cytometry, and ELISA, assays. The luciferase reporter assay was employed to identify the mechanism of SENCR. Downregulation of SENCR was detected in MI patients compared to the control group, and its expression was negatively correlated with cTnI and CK-MB levels in MI patients. H/R-stimulated apoptosis and inflammation were attenuated by SENCR up-regulation. SENCR up-regulation was associated with the downregulation of miR-1 expression. SENCR up-regulation also decreased apoptosis, mitigated inflammatory responses, and increased cell survival in cardiomyocytes. SENCR was shown to be able to reverse H/R myocardial damage via downregulating miR-1. A significant correlation between SENCR expression and the clinicopathological characteristics of MI patients was found (153).
The lncRNA plasmacytoma variant translocation 1 (PVT1), which is located on chromosome 8q24, is a potential oncogene in several malignancies (154). Furthermore, an in vivo study showed that PVT1 was remarkably overexpressed in the hypertrophic mouse heart, and its inhibition was associated with a smaller cardiomyocyte size in Angiotensin II-treated cardiomyocytes, suggesting a crucial role of PVT1 in cardiomyocyte hypertrophy (155). However, its role in other cardiac diseases is not yet clear.
Ouyang et al. established a study to assess the involvement of lncRNA PVT1 in H/R-treated AC16 cardiomyocytes (156). They found out that treatment decreased cell survival and enhanced apoptosis, and its suppression ameliorated the H/R damage and besides, decreased excessive autophagy. Additionally, the authors demonstrated that it may act as a ceRNA to sponge miR-186 in AC16 cells, and rescue research revealed that prohibition of miR-186 was able to suppress the effects of PVT1 suppression in H/R-treated AC16 cells. Thus, PVT1 can be suggested as a valuable target for myocardial I/R injury treatment (156).
Testis-specific transcript Y-linked 15 (TTTY15) is a transcription-mediated chimeric lncRNA with key regulatory functions in some CVDs (157, 158). Bioinformatics and experimental analysis revealed that TTTY15 could promote myocardial cell injury by regulating the miR-98-5p/C-reactive protein (159).
Chen et al. aimed at investigating the role of lncRNA TTTY15 in myocardial I/R damage and its putative interaction mechanisms with miR-374a-5p. In vivo and in vitro experiments revealed enhanced TTTY15 expression in myocardial I/R damage. Besides, TTTY15 inhibited autophagy as well as myocardial I/R damage via modulating the expression of miR-374a-5p. TTTY15 modulated miR-374a-5p expression, thereby regulating FOXO1 expression and autophagy in myocardial I/R damage. Furthermore, TTTY15 inhibition ameliorated in vivo myocardial I/R injury. According to their findings, they concluded that TTTY15 can be taken into account as a precious target for myocardial I/R injury treatment (160).
RNA Component of Mitochondrial RNA Processing Endoribonuclease (RMRP), a recently explored lncRNA consisting 267 nucleotides, has been demonstrated to exhibit carcinogenic effects in a variety of malignancies, including gastric cancer (161), glioma (162), and lung cancer (163). Steinbusch et al. pointed out that RMRP has a pivotal role in chondrocyte hypertrophy and it can contribute to cartilage-hair hypoplasia (164), suggesting the mandatory function of this lncRNA in several diseases. Of note, dysregulation of RMRP has been observed in patients suffering from ischemic heart failure (165). However, there is a scarcity of data regarding whether RMRP is dysregulated in MI and myocardial I/R damage.
Kong et al. sought to clarify the roles of RMRP and its mechanism in myocardial I/R damage (166). To induce myocardial damage, the H9c2 cardiomyocytes were cultured under hypoxia. Moreover, a rat myocardial I/R damage model was established by subjection to 60 min ischemia and 24 h reperfusion. The authors declared that RMRP up-regulation was associated with worsen hypoxia-induced damage in cardiomyocytes. A negative correlation between RMRP and miR-206 was also found and RMRP up-regulation aggravated hypoxia damage via downregulation of miR-206. Additionally, it was shown that miR-206 exerted its effects on hypoxia damage by targeting autophagy related 3 (ATG3). Also, RMRP up-regulation was associated with activation of PI3K/AKT/mTOR signaling pathway in hypoxia-treated H9c2 cells via regulation of miR-206/ATG3. In summary, it can be concluded that RMRP overexpression has the potential to aggravate myocardial injury through the downregulation of miR-206 and consequently overexpression of ATG3. In the setting of myocardial I/R damage, PI3K/Akt/mTOR pathway activation serves as a central downstream mediator of the RMPR/miR-206/ATG3 axis (166).
Clarifying the mechanisms and pathways behind exercise-stimulated adaption will offer new therapeutic targets that can be utilized as treatments for CVDs (140). The preventive and therapeutic effects of exercise on CVDs have been well-established, hence, it can be considered another strategy aimed at regulating the expression of lncRNAs (140). Evidence demonstrated that aerobic exercise can decrease cardiomyocytes' apoptosis and heart fibrosis areas following MI via modulating the expressions of myocardial infarction associated transcript (MIAT), H19, and lncRNA GAS5. In addition, exercise can prohibit the expression of MIAT, known pro-fibrotic lncRNA in the MI settings, which exerts its impacts by sequestering miRNA-24 and provoking the TGF-beta signaling pathway (140).
In neonatal and adult mice, suppression of CAREL (cardiac regeneration-related long non-coding ribonucleic acid) enhanced heart function and was associated with higher cell proliferation and regeneration (167). CRRL (cardiomyocyte regeneration-related lncRNA), like CAREL, is a negative regulator of cardiomyocyte proliferation and cardiac repair. CRRL achieves these effects by binding to miR-199a-3p, inhibiting its activity and boosting levels of its target Hopx (168).
lncRNA ZNFX1 antisense 1 (ZFAS1) is made from a snoRNA host gene. It is one of the lncRNAs involved in cardiac hypertrophy, as its knockdown rescued contractile dysfunction (169, 170). Sarcoplasmic/endoplasmic reticulum Ca2+ ATPase 2a (SERCA2a) is a key protein in the Ca2+ cycle of HF so its dysregulation is a hallmark of HF (171). LncRNA ZFAS1 as a SERCA2a inhibitor binds to SERCA2A protein and disrupts its activity, leading to changed Ca2+ transient and intracellular Ca2+ load in the heart and contractile dysfunction in an animal model of MI (170) (Table 2).
Luo et al. examined circRNA expression throughout the stages of MI progression in an animal model of MI, and in cardiomyocytes with H/R injury in vitro (191). They found high expression of circRNA PVT1 (circPVT1) in both MI tissues and H/R-treated cardiomyocytes. The effect of circPVT1 on cardiac function and cardiomyocyte viability was investigated using loss-of-function assays. Echocardiography was employed to assess cardiac function seven days after MI. Lower circPVT1 expression substantially reduced the infarct size by 60% and avoided the MI-induced decrease in fractional shortening (FS) and ejection fraction (EF). Laboratory findings demonstrated that silencing of circPVT1 was associated with higher cell survival and proliferation, and decreased apoptosis. A significant association between circPVT1 expression and both miR-200a and miR-125b was found. The authors suggested that circPVT1 might sponge miR-200a and miR-125b by acting as a ceRNA (191). Overexpression of miR-125b and miR-200a partly abolished the effects of circPVT1 on cardiomyocyte function. Moreover, the authors reported that the circPVT1/miR-125b/miR-200a axis was able to regulate the SIRT7, p53/TRAF6, PDCD4, and Keap1/Nrf2, signaling pathways. In summary, their findings suggested that circPVT1 exerted a cardioprotective effect against MI and H/R damage by inhibiting apoptosis mediated by miR-200a and miR-125b (191).
CircROBO2 is a recently discovered circRNA, but its role in MI is not fully understood. Chen et al. investigated the pathophysiology of circROBO2 in MI (192). Accordingly, Western blotting and qRT-PCR were employed to assess the expression levels of circROBO2, TRADD, and miR-1184 in MI and sham-operated mouse models at protein and mRNA levels, respectively. Luciferase reporter gene analysis and RNA immunoprecipitation (RIP) were used to investigate the association between miR-1184, circROBO2, and TRADD. Flow cytometry was used to verify the involvement of circROBO2, miR-1184, and TRADD in myocardial cell apoptosis. To investigate the effects of circROBO2 on myocardial damage, researchers used ultrasound echocardiography, serum LDH, serum CK-MB, MI area and measured myocardiocyte apoptosis (192). Compared to the control group, miR-1184 expression levels were significantly lower in the MI group; however, circROBO2 and TRADD expression levels were substantially higher. After overexpression of TRADD, circROBO2 behaved like a sponge for miR-1184. Furthermore, miR-1184 up-regulation increased the protective effect of circROBO2 knockdown by inhibiting TRADD expression. The authors concluded that circROBO2 knockdown lowered cardiomyocyte apoptosis by boosting miR-1184 expression, and reducing TRADD expression in the myocardium after MI (192).
Dysregulation of CircHIPK3 has been detected in several diseases including but not limited to diabetes, malignancies, preeclampsia, and retinal vascular dysfunction (193). It was shown that circHIPK3 suppressed proliferation and induced apoptosis of cardiomyocytes with ischemic-reperfusion injury via binding to miR-124-3p (194). Upon hypoxic injury, cardiomyocyte-derived exosomal circHIPK3 plays a crucial role in maintaining cardiac microvascular endothelial cell function by targeting the MIR29A/IGF-1 pathway and via modulating the miR-29a/IGF-1 axis (16).
Wu et al. examined the underlying ceRNA network involving circHIPK3 in MI (195). A hypoxic model was used to establish MI in vivo, then, the expression levels and association between miR-93-5p, circHIPK3, and Rac1 were evaluated. Gain and loss-of-function experiments were used to assess the ceRNA mechanism (195). CircHIPK3 suppression was associated with decreased myocardial apoptosis, a lower infarct size, myocardial collagen deposition, and improved cardiac function. CircHIPK3 sponged miR-93-5p, while miR-93-5p targeted Rac1. Up-regulation of MiR-93-5p was associated with attenuation of MI-induced cardiomyocyte damage and abrogated the detrimental effect of circHIPK3. CircHIPK3 acted as a sponge for miR-93-5p, thereby enhancing the activation of the Rac1/PI3K/AKT signaling pathway. In conclusion, circHIPK3 suppression was associated with overexpression of miR-93-5p and inhibition of the Rac1/PI3K/Akt signaling pathway, leading to amelioration of MI-induced cardiac dysfunction (195).
The novel circRNA, circMAT2B was shown to play a major role in moderating glucose metabolism in hypoxic conditions (196). Moreover, elevated levels of circMAT2B were found in hepatocellular carcinoma patients, suggesting it could be a target for hepatocellular carcinoma treatment. Nevertheless, its role in MI is still unclear.
Zhu et al. investigated how circMAT2B affects MI. An oxygen-glucose deprivation (OGD)-induced H9c2 cell model was used to simulate MI. H9c2 cells were transfected with ex-circMAT2B plasmid for overexpression, si-circMAT2B for a knockdown, a miR-133 inhibitor, and appropriate controls. The expression levels of miR-133 and circMAT2B were evaluated using qRT-PCR. Apoptosis, reactive oxygen species (ROS) production, cell viability, and secretion of inflammatory cytokines were evaluated using flow cytometry, ROS assay kit, CCK-8 assay, and ELISA, respectively. Furthermore, Western blotting was employed to identify apoptosis and related pathways. It was found that OGD treatment significantly increased circMAT2B expression. Additionally, circMAT2B knockdown dramatically reduced the OGD-stimulated increase in apoptosis, ROS production, and the expression of inflammatory cytokines. SI-circMAT2B increased miR-133 expression. CircMAT2B suppression abrogated OGD-induced H9c2 cell damage and relieved the OGD-stimulated inhibition of the Raf/MEK/ERK and PI3K/AKT signaling pathways via upregulation of miR-133. Collectively, circMAT2B suppression could inhibit inflammation in OGD-induced cardiomyocyte damage in H9c2 cells by upregulating miR-133 (197).
The circRNA Postn (circPostn) is thought to affect cancer development via modulating cell apoptosis and proliferation (198). One in vivo study showed that circPostn is up-regulated in MI (199). Nonetheless, its role in MI-induced myocardial damage and regeneration is not yet clear.
Cheng et al. investigated the effects of circPostn on myocardial damage and remodeling following MI. They found higher plasma levels of circPostn in patients suffering from MI, as well as mice and human cardiomyocytes treated with H/R. CircPostn knockdown considerably reduced MI-induced myocardial damage and infarct size. CircPostn knockdown also increased the left ventricular FS and EF, as well as decreased the left ventricular anterior wall thickness in diastole (LVAWd) and the left ventricular posterior wall thickness in diastole (LVPWd). The expression of collagen 1α1 and collagen 3α1 was increased in MI in vivo but was reduced when circPostn was depleted. Collagen and smooth muscle actin protein expression was increased in MI in vivo and decreased by circPostn knockdown. Moreover, suppression of circPostn decreased the expression of the atrial natriuretic peptide as well as brain natriuretic peptide. Also, the circPostn depletion was able to decrease cardiomyocyte apoptosis in vivo. CircPostn acted mechanically as a miR-96-5p sponge and miR-96-5p targeted BNIP3 in human cardiomyocytes, where circPostn increased BNIP3 level through miR-96-5p targeting. circPostn enhanced H/R-stimulated cardiomyocyte damage via affecting the miR-96-5p/BNIP3 axis. Therefore, the authors infer that circPostn results in MI-stimulated myocardial damage and heart remodeling via moderating the miR-96-5p/BNIP3 axis. The results illustrate a unique understanding of the mechanism behind the regulatory effects of circPostn on MI-stimulated heart dysfunction. miR-96-5p, circPostn, and BNIP3 can be considered as possible therapeutic for the management of MI-stimulated heart damage (200) (Table 3).
Exosomes are bilayer-surrounded membrane vesicles measuring from 30 to 100 nm in diameter and are secreted by most cell types. Exosomes can act as carriers to transport biomolecules including, DNA, proteins, RNA, and lipids (225, 226).
Exosomes can display cell molecules characteristic of their source cells on their surface and when they bind to recipient cells, they release their contents into the cytosol of the recipient cells through receptor-ligand interactions, endocytosis, and fusion with the cell membrane, thereby modifying the recipient cell functions (227). A relatively large number of proteins, miRNAs, and lncRNAs can be detected inside exosomes. Emerging evidence suggests that exosomes can act as regulators of cell differentiation, proliferation, and apoptosis in CVDs (228–230). By acting as ceRNA, they can bind to specific miRNAs and act as miRNA sponges in cells. Consequently, they can reduce the activity of miRNAs and thus regulate the expression of their target genes (231).
Exosomal ncRNAs act as regulators of myocardial structure and function in CADs, thus providing new insight into the mechanisms and therapeutic targets for the diagnosis and treatment of these diseases (232). Sirt1 is involved in immune responses and is known as a regulator of inflammation in CVDs (233, 234). It plays a critical role in the regulation of cardiac cell development and CVDs (235, 236). In MI patients, Sirt1 is downregulated, whereas its upregulation could alleviate MI-induced myocardial damage (237). Sirt1 activation could inhibit NLRP3 inflammasome activation and subsequent caspase-1 cleavage and IL-1β secretion, suggesting the protective effect of Sirt1 on vascular endothelial cells (238).
Mao et al. investigated the effects and the underlying mechanism of lncRNA KLF3-AS1 contained in exosomes derived from human mesenchymal stem cells (hMSCs), on the pyroptosis of cardiomyocytes and the treatment of MI (239). Exosomes were transfected with KLF3-AS1 and tested in vitro and in vivo. The effects of exosomal KLF3-AS1 on cell viability, MI area, pyroptosis, and apoptosis were assessed. The dual-luciferase reporter assay was used to determine correlations between KLF3-AS1, miR-138-5p, and Sirt1. Transfection of miR-138-5p and sh-Sirt1 into normal cardiomyocytes was performed to see whether an increase in miR-138-5p or sh-Sirt1 could affect the cardioprotective activity of KLF3-AS1. The delivery of KLF3-AS1 in exosomes resulted in reduced infarct size, less apoptosis, and pyroptosis, and reduced MI progression. KLF3-AS1 can bind to miR-138-5p to modulate the expression of Sirt1. It was found that inhibition of miR-138-5p was associated with decreased pyroptosis and ameliorated the effects of MI. The authors concluded that lncRNA KLF3-AS1 in exosomes released from hMSCs could bind to miR-138-5p to moderate Sirt1 expression to suppress pyroptosis and slow MI progression (239).
Recently, Chen et al. conducted an in vitro study to evaluate the role of exosome-mediated lncRNAs ZEB1-AS1 (Zinc finger e-box-binding homeobox 1 antisense 1) and its underlying mechanisms in atherosclerosis (240). Exosomes were extracted from oxidized low-density lipoprotein (ox-LDL)-treated human umbilical vein endothelial cells (HUVECs). They demonstrated that exo-lncRNA ZEB1-AS1 derived from ox-LDL-induced HUVECs amplified cell injuries by miR-590-5p/ ETS1 (E26 oncogene homolog 1) axis through the TGF-β/Smad pathway, suggesting that stopping ZEB1-AS1 might be an efficient strategy to treat atherosclerosis (240).
It has been shown that serum MMP-9 could be used to differentiate between MI and unstable angina (UA) with a sensitivity and specificity of 80%. MMP-9 levels also predicted unfavorable outcomes in ST-elevation MI (STEMI) patients with a sensitivity of 72.4% and a specificity of 83% (241). Because MI patients show an imbalance between MMPs and their inhibitors (TIMPs), an increase in MMP leading to the degradation of the fibrous cap can be a major cause of plaque instability (242, 243). Increased MMP-9 levels stimulate plaque rupture and result in acute MI. Moreover, high levels of inflammatory cytokines following acute MI promote the additional production and release of MMP-9 (244).
Exosomal miR-221/222 secreted by human aortic smooth muscle cells (HAOSMCs) was found to inhibit HUVECs autophagy in by partially modulating the PTEN/Akt signaling pathway when HUVECs and HAOSMCs were co-cultured (245). According to research on the influence of exosomes on cardiac fibroblasts, hypoxia upregulates the lncRNA AK139128 expression in cardiomyocytes and exosomes, and exo-lncRNA AK139128 derived from hypoxic cardiomyocytes endorses apoptosis, prevents cell proliferation, and modulates fibroblast activity (246). One study showed that the increased expression of miR-125b of mesenchymal stem cells-secreted exosomes enhances myocardial cell survival in rats after I/R by regulating Sirt7, decreasing myocardial cell apoptosis and inflammatory response, and increasing heart function (247). Another investigation found that miR-425 and miR-744 levels in plasma exosome samples from heart failure patients were considerably lower (248). Overexpression of miR-425 or miR-744 in cultured cardiac fibroblasts suppresses TGF1 expression and dramatically decreases angiotensin-induced collagen production and fibrogenesis (248, 249).
Numerous studies have shown that secreted exosomes from myocardial cells contain some microRNA such as miR-125b, miR-126, miR-25-3p, miR-144, and miR-146a which are overexpressed and exert an anti-atherosclerotic role by stopping myocardial apoptosis and facilitating ischemic cardiac repair (250–254). In addition, exosomal miRNAs, miR-25-3p and miR-146a also inhibit the inflammatory response, while exosomal miR-301 prevents myocardial autophagy (254–256). Besides, according to Wang et al., enhanced expression of EXO-MSC-derived miR-21 plays a cardioprotective role by inhibiting apoptosis, promoting angiogenesis, and increasing cell survival through the PTEN/Akt pathway (257).
Chen et al. investigated the association between acute MI and plasma exosomes containing lncRNA NEAT1 (258). They categorized the participants into three groups: the control group, the unstable angina group, and the acute STEMI, overexpression of NEAT1 and MMP-9 was found and they were positively correlated with each other. Furthermore, lower levels of miR-204 were found in STEMI patients and no significant correlation was found between expression levels of NEAT1 or MMP-9 and miR-204. The authors concluded that miR-204, exosomal NEAT1, and MMP-9 could all be considered diagnostic markers for STEMI (258) (Table 4).
We comprehensively review the mechanisms of epigenetics such as DNA methylation, histone modification, and non-coding RNA in various cardiovascular diseases. Moreover, basic and clinical studies on epigenetic therapy for these diseases are summarized.
Existing treatments for CADs have not been equally effective in all patients, suggesting that interindividual variability is an aspect that plays a significant role in personalized treatment. Genetic testing and genomic mapping combined with emerging transcriptional technologies such as microarrays and RNA-Seq, as well as methylation or acetylation patterns and bioinformatics tools can help determine the genetic and epigenetic structures that define an individual's risk profile and identify novel therapeutic epigenetic targets. In this context, the development of small molecule drugs as epigenetic or miRNA modulators (e.g., HDAC inhibitors) has opened up new approaches for the treatment of cardiovascular diseases. Evidence has revealed that ncRNAs can act either as pro-MI or anti-MI molecules by modulating signaling pathways relevant to myocardial cell death or cardiomyocyte regeneration. Up to now, many advances have identified ncRNA-mediated signaling pathways involved in MI, which have contributed to improvement in our understanding of cardiovascular pathogenesis. Abnormal plasma levels of many ncRNAs have been observed in patients with MI, which suggests their potential in the diagnosis and treatment of MI. Importantly, some ncRNA-based therapeutic approaches have shown promising results in animal models of MI, which provides some hope that this approach could be used for the clinical management of MI soon.
The role of exosomal ncRNAs in the progression and development of CADs, such as atherosclerosis, acute coronary syndrome, HF, myocardial I/R injury, and pulmonary hypertension has been the focus of many recent studies. Tissue-specific changes of exosomal ncRNAs could play a role in the initiation and progression of these complications. In terms of biomarkers, exosomal ncRNAs could be superior compared to non-exosomal ncRNAs. Many ncRNAs are harbored inside exosomes, which protects them from degradation and enhances their stability. Moreover, exosomal ncRNAs exist in considerable amounts in different body fluids that can be non-invasively sampled. Hence, exosome ncRNAs are expected to become a new tool for the diagnosis and treatment of CADs.
Considering the epigenetic roles of some RNAs and their capacities to control gene expression, the establishment of novel approaches for modulating the expression of some ncRNAs supported by the development of epigenetic-related drugs with higher specificity, fewer side effects, and lower drug resistance for various types of CADs will be the goal of future development. It is hoped that in the near future, the discovery and continuous research and development of epigenetic regulatory drugs for CADs will have a wider application perspective and will be more beneficial for patients with cardiovascular diseases. To this end, more large-scale basic and clinical trials are still needed to better understand the epigenetic molecular mechanism regulating CADs and to find more strategies to avoid and treat cardiovascular diseases, so as to better guide clinical treatments.
HM was involved in conception, design, statistical analysis, and drafting of the manuscript. SF, AM, MP, MS, MZ, FZ, ST, MR, AS, and MH contributed to data collection and manuscript drafting. All authors approved the final version for submission.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Abi Khalil C. The emerging role of epigenetics in cardiovascular disease. Ther Adv Chronic Dis. (2014) 5:178–87. doi: 10.1177/2040622314529325
2. Momtaz S, Niaz K, Maqbool F, Abdollahi M, Rastrelli L, Nabavi SM. STAT3 targeting by polyphenols: novel therapeutic strategy for melanoma. Biofactors. (2017) 43:347–70. doi: 10.1002/biof.1345
3. Del Re DP, Amgalan D. fundamental mechanisms of regulated cell death and implications for heart disease. Physiol Rev. (2019) 99:1765–817. doi: 10.1152/physrev.00022.2018
4. Mercola M, Ruiz-Lozano P, Schneider MD. Cardiac muscle regeneration: lessons from development. Genes Dev. (2011) 25:299–309. doi: 10.1101/gad.2018411
5. Panzeri I, Rossetti G, Abrignani S, Pagani M. Long intergenic non-coding RNAs: novel drivers of human lymphocyte differentiation. Front Immunol. (2015) 6:175. doi: 10.3389/fimmu.2015.00175
6. Poller W, Dimmeler S, Heymans S, Zeller T, Haas J, Karakas M, et al. Non-coding RNAs in cardiovascular diseases: diagnostic and therapeutic perspectives. Eur Heart J. (2018) 39:2704–16. doi: 10.1093/eurheartj/ehx165
7. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5
8. Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. (2005) 120:623–34. doi: 10.1016/j.cell.2004.12.038
9. Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. (2007) 128:1105–18. doi: 10.1016/j.cell.2007.01.038
10. Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. (2008) 133:128–41. doi: 10.1016/j.cell.2008.02.033
11. Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. (2008) 18:997–1006. doi: 10.1038/cr.2008.282
12. Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. (2008) 141:672–5. doi: 10.1111/j.1365-2141.2008.07077.x
13. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci. (2008) 105:10513–8. doi: 10.1073/pnas.0804549105
14. Ai J, Zhang R, Li Y, Pu J, Lu Y, Jiao J, et al. Circulating microRNA-1 as a potential novel biomarker for acute myocardial infarction. Biochem Biophys Res Commun. (2010) 391:73–7. doi: 10.1016/j.bbrc.2009.11.005
15. Wang Y, Zhao R, Shen C, Liu W, Yuan J, Li C, et al. Exosomal CircHIPK3 released from hypoxia-induced cardiomyocytes regulates cardiac angiogenesis after myocardial infarction. Oxid Med Cell Longevity. (2020) 2020:8418407. doi: 10.1155/2020/8418407
16. Wang Y, Zhao R, Liu W, Wang Z, Rong J, Long X, et al. Exosomal circHIPK3 released from hypoxia-pretreated cardiomyocytes regulates oxidative damage in cardiac microvascular endothelial cells via the miR-29a/IGF-1 pathway. Oxid Med Cell Longevity. (2019) 2019:7954657. doi: 10.1155/2019/7954657
17. Kim JK, Samaranayake M, Pradhan S. Epigenetic mechanisms in mammals. Cell Mol Life Sci. (2009) 66:596–612. doi: 10.1007/s00018-008-8432-4
18. Wang KC, Chang HY. Epigenomics: technologies and applications. Circ Res. (2018) 122:1191–9. doi: 10.1161/CIRCRESAHA.118.310998
19. Yang J, Xu WW, Hu SJ. Heart failure: advanced development in genetics and epigenetics. Biomed Res Int. (2015) 2015:352734. doi: 10.1155/2015/352734
20. Prasher D, Greenway SC, Singh RB. The impact of epigenetics on cardiovascular disease. Biochem Cell Biol. (2020) 98:12–22. doi: 10.1139/bcb-2019-0045
21. Li P, Ge J, Li H. Lysine acetyltransferases and lysine deacetylases as targets for cardiovascular disease. Nat Rev Cardiol. (2020) 17:96–115. doi: 10.1038/s41569-019-0235-9
22. Xie M, Hill JA. HDAC-dependent ventricular remodeling. Trends Cardiovasc Med. (2013) 23:229–35. doi: 10.1016/j.tcm.2012.12.006
23. Eom GH, Kook H. Posttranslational modifications of histone deacetylases: implications for cardiovascular diseases. Pharmacol Ther. (2014) 143:168–80. doi: 10.1016/j.pharmthera.2014.02.012
24. Tang J, Zhuang S. Histone acetylation and DNA methylation in ischemia/reperfusion injury. Clin Sci. (2019) 133:597–609. doi: 10.1042/CS20180465
25. Siebert V, Allencherril J, Ye Y, Wehrens XHT, Birnbaum Y. The role of non-coding RNAs in ischemic myocardial reperfusion injury. Cardiovasc Drugs Ther. (2019) 33:489–98. doi: 10.1007/s10557-019-06893-x
26. Tingare A, Thienpont B, Roderick HL. Epigenetics in the heart: the role of histone modifications in cardiac remodelling. Biochem Soc Trans. (2013) 41:789–96. doi: 10.1042/BST20130012
27. Hyun K, Jeon J, Park K, Kim J. Writing, erasing and reading histone lysine methylations. Exp Mol Med. (2017) 49:e324. doi: 10.1038/emm.2017.11
28. Gillette TG, Hill JA. Readers, writers, and erasers: chromatin as the whiteboard of heart disease. Circ Res. (2015) 116:1245–53. doi: 10.1161/CIRCRESAHA.116.303630
29. Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol. (2014) 15:703–8. doi: 10.1038/nrm3890
30. McKinsey TA. Therapeutic potential for HDAC inhibitors in the heart. Annu Rev Pharmacol Toxicol. (2012) 52:303–19. doi: 10.1146/annurev-pharmtox-010611-134712
31. Yang G, Weng X, Zhao Y, Zhang X, Hu Y, Dai X, et al. The histone H3K9 methyltransferase SUV39H links SIRT1 repression to myocardial infarction. Nat Commun. (2017) 8:1–15. doi: 10.1038/ncomms14941
32. Sung P-H, Luo C-W, Chiang JY, Yip H-K. The combination of G9a histone methyltransferase inhibitors with erythropoietin protects heart against damage from acute myocardial infarction. Am J Transl Res. (2020) 12:3255.
33. Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harbor Perspect Biol. (2014) 6:a018713. doi: 10.1101/cshperspect.a018713
34. Han A, He J, Wu Y, Liu JO, Chen L. Mechanism of recruitment of class II histone deacetylases by myocyte enhancer factor-2. J Mol Biol. (2005) 345:91–102. doi: 10.1016/j.jmb.2004.10.033
35. Zhang M, Yang X, Zimmerman RJ, Wang Q, Ross MA, Granger JM, et al. CaMKII exacerbates heart failure progression by activating class I HDACs. J Mol Cell Cardiol. (2020) 149:73–81. doi: 10.1016/j.yjmcc.2020.09.007
36. Zhang L, Wang H, Zhao Y, Wang J, Dubielecka PM, Zhuang S, et al. Myocyte-specific overexpressing HDAC4 promotes myocardial ischemia/reperfusion injury. Mol Med. (2018) 24:37. doi: 10.1186/s10020-018-0037-2
37. Zhang LX, DeNicola M, Qin X, Du J, Ma J, Tina Zhao Y, et al. Specific inhibition of HDAC4 in cardiac progenitor cells enhances myocardial repairs. Am J Physiol Cell Physiol. (2014) 307:C358–72. doi: 10.1152/ajpcell.00187.2013
38. Dashti F, Mirazimi SMA, Rabiei N, Fathazam R, Rabiei N, Piroozmand H, et al. The role of non-coding RNAs in chemotherapy for gastrointestinal cancers. Mol Ther Nucleic Acids. (2021) 26:892–926. doi: 10.1016/j.omtn.2021.10.004
39. Ferguson BS, Harrison BC, Jeong MY, Reid BG, Wempe MF, Wagner FF, et al. Signal-dependent repression of DUSP5 by class I HDACs controls nuclear ERK activity and cardiomyocyte hypertrophy. Proc Natl Acad Sci. (2013) 110:9806–11. doi: 10.1073/pnas.1301509110
40. Herr DJ, Baarine M, Aune SE, Li X, Ball LE, Lemasters JJ, et al. HDAC1 localizes to the mitochondria of cardiac myocytes and contributes to early cardiac reperfusion injury. J Mol Cell Cardiol. (2018) 114:309–19. doi: 10.1016/j.yjmcc.2017.12.004
41. Eom GH, Cho YK, Ko J-H, Shin S, Choe N, Kim Y, et al. Casein kinase-2α1 induces hypertrophic response by phosphorylation of histone deacetylase 2 S394 and its activation in the heart. Circulation. (2011) 123:2392–403. doi: 10.1161/CIRCULATIONAHA.110.003665
42. Trivedi CM, Lu MM, Wang Q, Epstein JA. Transgenic overexpression of Hdac3 in the heart produces increased postnatal cardiac myocyte proliferation but does not induce hypertrophy. J Biol Chem. (2008) 283:26484–9. doi: 10.1074/jbc.M803686200
43. Cao DJ, Wang ZV, Battiprolu PK, Jiang N, Morales CR, Kong Y, et al. Histone deacetylase (HDAC) inhibitors attenuate cardiac hypertrophy by suppressing autophagy. Proc Natl Acad Sci USA. (2011) 108:4123–8. doi: 10.1073/pnas.1015081108
44. Packer M. Cardioprotective effects of sirtuin-1 and its downstream effectors: potential role in mediating the heart failure benefits of SGLT2 (sodium-glucose cotransporter 2) inhibitors. Circ Heart Fail. (2020) 13:e007197. doi: 10.1161/CIRCHEARTFAILURE.120.007197
45. Bratkowski M, Yang X, Liu X. An evolutionarily conserved structural platform for PRC2 inhibition by a class of Ezh2 inhibitors. Sci Rep. (2018) 8:1–11. doi: 10.1038/s41598-018-27175-w
46. Gan L, Yang Y, Li Q, Feng Y, Liu T, Guo W. Epigenetic regulation of cancer progression by EZH2: from biological insights to therapeutic potential. Biomarker Res. (2018) 6:1–10. doi: 10.1186/s40364-018-0122-2
47. Karantanos T, Christofides A, Bardhan K, Li L, Boussiotis VA. Regulation of T cell differentiation and function by EZH2. Front Immunol. (2016) 7:172. doi: 10.3389/fimmu.2016.00346
48. Wang S, Guo N, Li S, He Y, Zheng D, Li L, et al. EZH2 dynamically associates with non-coding RNAs in mouse hearts after acute angiotensin II treatment. Front Cardiovasc Med. (2021) 8:585691. doi: 10.3389/fcvm.2021.585691
49. Delgado-Olguín P, Huang Y, Li X, Christodoulou D, Seidman CE, Seidman J, et al. Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. Nat Genet. (2012) 44:343–7. doi: 10.1038/ng.1068
50. He A, Ma Q, Cao J, Von Gise A, Zhou P, Xie H, et al. Polycomb repressive complex 2 regulates normal development of the mouse heart. Circ Res. (2012) 110:406–15. doi: 10.1161/CIRCRESAHA.111.252205
51. Zhao L, You T, Lu Y, Lin S, Li F, Xu H. Elevated EZH2 in ischemic heart disease epigenetically mediates suppression of NaV1. 5 expression. J Mol Cell Cardiol. (2021) 153:95–103. doi: 10.1016/j.yjmcc.2020.12.012
52. Jiao W, Hao J, Xie Y, Meng M, Gao W. EZH2 mitigates the cardioprotective effects of mesenchymal stem cell-secreted exosomes against infarction via HMGA2-mediated PI3K/AKT signaling. BMC Cardiovasc Disord. (2022) 22:1–13. doi: 10.1186/s12872-022-02533-9
53. Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. (2007) 447:425–32. doi: 10.1038/nature05918
54. Ehrlich M, Lacey M. DNA methylation and differentiation: silencing, upregulation and modulation of gene expression. Epigenomics. (2013) 5:553–68. doi: 10.2217/epi.13.43
55. Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. (2013) 14:204–20. doi: 10.1038/nrg3354
56. Campanero MR, Armstrong MI, Flemington EK. CpG methylation as a mechanism for the regulation of E2F activity. Proc Natl Acad Sci USA. (2000) 97:6481–6. doi: 10.1073/pnas.100340697
57. Meng H, Cao Y, Qin J, Song X, Zhang Q, Shi Y, et al. DNA methylation, its mediators and genome integrity. Int J Biol Sci. (2015) 11:604. doi: 10.7150/ijbs.11218
58. Gowher H, Jeltsch A. Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: the enzyme modifies DNA in a non-processive manner and also methylates non-CpA sites. J Mol Biol. (2001) 309:1201–8. doi: 10.1006/jmbi.2001.4710
59. Tammen SA, Friso S, Choi S-W. Epigenetics: the link between nature and nurture. Mol Aspects Med. (2013) 34:753–64. doi: 10.1016/j.mam.2012.07.018
60. Neri F, Krepelova A, Incarnato D, Maldotti M, Parlato C, Galvagni F, et al. Dnmt3L antagonizes DNA methylation at bivalent promoters and favors DNA methylation at gene bodies in ESCs. Cell. (2013) 155:121–34. doi: 10.1016/j.cell.2013.08.056
61. Gu T-P, Guo F, Yang H, Wu H-P, Xu G-F, Liu W, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. (2011) 477:606–10. doi: 10.1038/nature10443
62. Iqbal K, Jin S-G, Pfeifer GP, Szabó PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci USA. (2011) 108:3642–7. doi: 10.1073/pnas.1014033108
63. Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. (2011) 146:67–79. doi: 10.1016/j.cell.2011.06.020
64. Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. (2011) 333:1300–3. doi: 10.1126/science.1210597
65. Leoni C, Montagner S, Rinaldi A, Bertoni F, Polletti S, Balestrieri C, et al. Dnmt3a restrains mast cell inflammatory responses. Proc Natl Acad Sci USA. (2017) 114:E1490–9. doi: 10.1073/pnas.1616420114
66. Cull AH, Snetsinger B, Buckstein R, Wells RA, Rauh MJ. Tet2 restrains inflammatory gene expression in macrophages. Exp Hematol. (2017) 55:56–70.e13. doi: 10.1016/j.exphem.2017.08.001
67. Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. (2017) 377:111–21. doi: 10.1056/NEJMoa1701719
68. Xia Y, Brewer A, Bell JT. DNA methylation signatures of incident coronary heart disease: findings from epigenome-wide association studies. Clin Epigenet. (2021) 13:1–16. doi: 10.1186/s13148-021-01175-6
69. Westerman K, Sebastiani P, Jacques P, Liu S, DeMeo D, Ordovás JM. DNA methylation modules associate with incident cardiovascular disease and cumulative risk factor exposure. Clin Epigenet. (2019) 11:1–14. doi: 10.1186/s13148-019-0705-2
70. Sharma P, Kumar J, Garg G, Kumar A, Patowary A, Karthikeyan G, et al. Detection of altered global DNA methylation in coronary artery disease patients. DNA Cell Biol. (2008) 27:357–65. doi: 10.1089/dna.2007.0694
71. Ma SC, Zhang HP, Kong FQ, Zhang H, Yang C, He YY, et al. Integration of gene expression and DNA methylation profiles provides a molecular subtype for risk assessment in atherosclerosis Corrigendum in/10.3892/mmr.2016.5915. Mol Med Rep. (2016) 13:4791–9. doi: 10.3892/mmr.2016.5120
72. Ke J, Dong N, Wang L, Li Y, Dasgupta C, Zhang L, et al. Role of DNA methylation in perinatal nicotine-induced development of heart ischemia-sensitive phenotype in rat offspring. Oncotarget. (2017) 8:76865–80. doi: 10.18632/oncotarget.20172
73. Mattick JS, Taft RJ, Faulkner GJ. A global view of genomic information–moving beyond the gene and the master regulator. Trends Genet. (2010) 26:21–8. doi: 10.1016/j.tig.2009.11.002
74. Sun X, Haider Ali MSS, Moran M. The role of interactions of long non-coding RNAs and heterogeneous nuclear ribonucleoproteins in regulating cellular functions. Biochem J. (2017) 474:2925–35. doi: 10.1042/BCJ20170280
75. Archer K, Broskova Z, Bayoumi AS, Teoh JP, Davila A, Tang Y, et al. Long non-coding RNAs as master regulators in cardiovascular diseases. Int J Mol Sci. (2015) 16:23651–67. doi: 10.3390/ijms161023651
76. Boon RA, Dimmeler SJNRC. MicroRNAs in myocardial infarction. Nat Rev Cardiol. (2015) 12:135–42. doi: 10.1038/nrcardio.2014.207
77. Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. (2015) 365:141–8. doi: 10.1016/j.canlet.2015.06.003
78. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. (2011) 146:353–8. doi: 10.1016/j.cell.2011.07.014
79. Lee KP, Shin YJ, Panda AC, Abdelmohsen K, Kim JY, Lee SM, et al. miR-431 promotes differentiation and regeneration of old skeletal muscle by targeting Smad4. Genes Dev. (2015) 29:1605–17. doi: 10.1101/gad.263574.115
80. Sun K, Zeng T, Huang D, Liu Z, Huang S, Liu J, et al. MicroRNA-431 inhibits migration and invasion of hepatocellular carcinoma cells by targeting the ZEB1-mediated epithelial-mensenchymal transition. FEBS Open Bio. (2015) 5:900–7. doi: 10.1016/j.fob.2015.11.001
81. Su WB, Liu ZY. MiR-431 inhibits colorectal cancer cell invasion via repressing CUL4B. Eur Rev Med Pharmacol Sci. (2018) 22:3047–52.
82. Zhao Y, Ponnusamy M, Dong Y, Zhang L, Wang K, Li P. Effects of miRNAs on myocardial apoptosis by modulating mitochondria related proteins. Clin Exp Pharmacol Physiol. (2017) 44:431–40. doi: 10.1111/1440-1681.12720
83. Dong Y, Undyala VV, Gottlieb RA, Mentzer RM Jr, Przyklenk K. Autophagy: definition, molecular machinery, and potential role in myocardial ischemia-reperfusion injury. J Cardiovasc Pharmacol Ther. (2010) 15:220–30. doi: 10.1177/1074248410370327
84. Radoshevich L, Murrow L, Chen N, Fernandez E, Roy S, Fung C, et al. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell. (2010) 142:590–600. doi: 10.1016/j.cell.2010.07.018
85. Gu J, Wang Y, Wang X, Zhou D, Wang X, Zhou M, et al. Effect of the LncRNA GAS5-MiR-23a-ATG3 axis in regulating autophagy in patients with breast cancer. Cell Physiol Biochem. (2018) 48:194–207. doi: 10.1159/000491718
86. Yang L, Peng X, Jin H, Liu J. Long non-coding RNA PVT1 promotes autophagy as ceRNA to target ATG3 by sponging microRNA-365 in hepatocellular carcinoma. Gene. (2019) 697:94–102. doi: 10.1016/j.gene.2019.02.036
87. D'Adamo S, Alvarez-Garcia O, Muramatsu Y, Flamigni F, Lotz MK. MicroRNA-155 suppresses autophagy in chondrocytes by modulating expression of autophagy proteins. Osteoarthritis Cartilage. (2016) 24:1082–91. doi: 10.1016/j.joca.2016.01.005
88. Zhou K, Xu Y, Wang Q, Dong L. Overexpression of miR-431 attenuates hypoxia/reoxygenation-induced myocardial damage via autophagy-related 3. Acta Biochim Biophys Sin. (2021) 53:140–8. doi: 10.1093/abbs/gmaa154
89. Hanousková B, Skála M, Brynychová V, Zárybnický T, Skarková V, Kazimírová P, et al. Imatinib-induced changes in the expression profile of microRNA in the plasma and heart of mice-A comparison with doxorubicin. Biomed Pharmacother. (2019) 115:108883. doi: 10.1016/j.biopha.2019.108883
90. Xuan Y, Liu S, Li Y, Dong J, Luo J, Liu T, et al. Short-term vagus nerve stimulation reduces myocardial apoptosis by downregulating microRNA-205 in rats with chronic heart failure. Mol Med Rep. (2017) 16:5847–54. doi: 10.3892/mmr.2017.7344
91. Cheng L, Wu Y, Tang J, Zhang C, Cheng H, Jiang Q, et al. Remifentanil protects against myocardial ischemia/reperfusion injury via miR-205-mediated regulation of PINK1. J Toxicol Sci. (2021) 46:263–71. doi: 10.2131/jts.46.263
92. Du Y, Yang F, Lv D, Zhang Q, Yuan X. MiR-147 inhibits cyclic mechanical stretch-induced apoptosis in L6 myoblasts via ameliorating endoplasmic reticulum stress by targeting BRMS1. Cell Stress Chaperones. (2019) 24:1151–61. doi: 10.1007/s12192-019-01037-4
93. Shen J, Niu W, Zhang H, Jun M, Zhang H. Downregulation of MicroRNA-147 inhibits cell proliferation and increases the chemosensitivity of gastric cancer cells to 5-fluorouracil by directly targeting PTEN. Oncol Res. (2018) 26:901–11. doi: 10.3727/096504017X15061902533715
94. Wu CG, Huang C. MicroRNA-147 inhibits myocardial inflammation and apoptosis following myocardial infarction via targeting HIPK2. Eur Rev Med Pharmacol Sci. (2020) 24:6279–87.
95. Pan X, He Y, Chen Z, Yan G, Ma G. Circulating miR-130 is a potential bio signature for early prognosis of acute myocardial infarction. J Thor Dis. (2020) 12:7320–5. doi: 10.21037/jtd-20-3207
96. Foinquinos A, Batkai S, Genschel C, Viereck J, Rump S, Gyöngyösi M, et al. Preclinical development of a miR-132 inhibitor for heart failure treatment. Nat Communic. (2020) 11:1–10. doi: 10.1038/s41467-020-14349-2
97. Ponikowski P, Voors AA, Anker SD, Bueno H, Cleland JG, Coats AJ, et al. 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure. Kardiologia Polska. (2016) 74:1037–147. doi: 10.5603/KP.2016.0141
98. Täubel J, Hauke W, Rump S, Viereck J, Batkai S, Poetzsch J, et al. Novel antisense therapy targeting microRNA-132 in patients with heart failure: results of a first-in-human Phase 1b randomized, double-blind, placebo-controlled study. Eur Heart J. (2021) 42:178–88. doi: 10.1093/eurheartj/ehaa898
99. Yuan H, Zhang J, Li F, Li W, Wang H. Sinomenine exerts antitumour effect in gastric cancer cells via enhancement of miR-204 expression. Basic Clin Pharmacol Toxicol. (2019) 125:450–9. doi: 10.1111/bcpt.13285
100. Zhou LY, Zhai M, Huang Y, Xu S, An T, Wang YH, et al. The circular RNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the Pink1/ FAM65B pathway. Cell Death Differ. (2019) 26:1299–315. doi: 10.1038/s41418-018-0206-4
101. Wang L, Xi J, Liu T, Zhang Z, Zhang P. MiR-204 reduces apoptosis in rats with myocardial infarction by targeting SIRT1/p53 signaling pathway. Eur Rev Med Pharmacol Sci. (2020) 24:12306–14.
102. Xiao J, Zhu X, He B, Zhang Y, Kang B, Wang Z, et al. MiR-204 regulates cardiomyocyte autophagy induced by ischemia-reperfusion through LC3-II. J Biomed Sci. (2011) 18:35. doi: 10.1186/1423-0127-18-35
103. D'Alessandra Y, Devanna P, Limana F, Straino S, Di Carlo A, Brambilla PG, et al. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur Heart J. (2010) 31:2765–73. doi: 10.1093/eurheartj/ehq167
104. Wang F, Long G, Zhao C, Li H, Chaugai S, Wang Y, et al. Atherosclerosis-related circulating miRNAs as novel and sensitive predictors for acute myocardial infarction. PLoS ONE. (2014) 9:e105734. doi: 10.1371/journal.pone.0105734
105. Tang Y, Wang Y, Park KM, Hu Q, Teoh JP, Broskova Z, et al. MicroRNA-150 protects the mouse heart from ischaemic injury by regulating cell death. Cardiovasc Res. (2015) 106:387–97. doi: 10.1093/cvr/cvv121
106. Li H, Zhang P, Li F, Yuan G, Wang X, Zhang A, et al. Plasma miR-22–5p, miR-132–5p, and miR-150–3p are associated with acute myocardial infarction. Biomed Res Int. (2019) 2019:5012648. doi: 10.1155/2019/5012648
107. Chen L, Bai J, Liu J, Lu H, Zheng K. A Four-MicroRNA panel in peripheral blood identified as an early biomarker to diagnose acute myocardial infarction. Front Physiol. (2021) 12:669590. doi: 10.3389/fphys.2021.669590
108. Parahuleva MS, Euler G, Mardini A, Parviz B, Schieffer B, Schulz R, et al. Identification of microRNAs as potential cellular monocytic biomarkers in the early phase of myocardial infarction: a pilot study. Sci Rep. (2017) 7:15974. doi: 10.1038/s41598-017-16263-y
109. Wu Q, Zhou X, Han X, Zhuo Y, Zhu S, Zhao Y, et al. Genome-wide identification and functional analysis of long noncoding RNAs involved in the response to graphene oxide. Biomaterials. (2016) 102:277–91. doi: 10.1016/j.biomaterials.2016.06.041
110. Yan Y, Dang H, Zhang X, Wang X, Liu X. The protective role of MiR-206 in regulating cardiomyocytes apoptosis induced by ischemic injury by targeting PTP1B. Biosci Rep. (2020) 40:BSR20191000. doi: 10.1042/BSR20191000
111. Fang J, Song XW, Tian J, Chen HY, Li DF, Wang JF, et al. Overexpression of microRNA-378 attenuates ischemia-induced apoptosis by inhibiting caspase-3 expression in cardiac myocytes. Apoptosis. (2012) 17:410–23. doi: 10.1007/s10495-011-0683-0
112. Vogel B, Keller A, Frese KS, Kloos W, Kayvanpour E, Sedaghat-Hamedani F, et al. Refining diagnostic microRNA signatures by whole-miRNome kinetic analysis in acute myocardial infarction. Clin Chem. (2013) 59:410–8. doi: 10.1373/clinchem.2011.181370
113. Hsu A, Chen SJ, Chang YS, Chen HC, Chu PH. Systemic approach to identify serum microRNAs as potential biomarkers for acute myocardial infarction. Biomed Res Int. (2014) 2014:418628. doi: 10.1155/2014/418628
114. Huang S, Chen M, Li L, He M, Hu D, Zhang X, et al. Circulating MicroRNAs and the occurrence of acute myocardial infarction in Chinese populations. Circ Cardiovasc Genet. (2014) 7:189–98. doi: 10.1161/CIRCGENETICS.113.000294
115. Kim EN, Kim CJ, Kim SR, Song JA, Choe H, Kim KB, et al. High serum CRP influences myocardial miRNA profiles in ischemia-reperfusion injury of rat heart. PLoS ONE. (2019) 14:e0216610. doi: 10.1371/journal.pone.0216610
116. Eryilmaz U, Akgüllü Ç, Beşer N, Yildiz Ö, Kurt Ömürlü I, Bozdogan B. Circulating microRNAs in patients with ST-elevation myocardial infarction. Anatol J Cardiol. (2016) 16:392–6. doi: 10.5152/AnatolJCardiol.2015.6603
117. Garikipati VN, Krishnamurthy P, Verma SK, Khan M, Abramova T, Mackie AR, et al. Negative regulation of miR-375 by interleukin-10 enhances bone marrow-derived progenitor cell-mediated myocardial repair and function after myocardial infarction. Stem Cells. (2015) 33:3519–29. doi: 10.1002/stem.2121
118. Feng G, Yan Z, Li C, Hou Y. microRNA-208a in an early stage myocardial infarction rat model and the effect on cAMP-PKA signaling pathway. Mol Med Rep. (2016) 14:1631–5. doi: 10.3892/mmr.2016.5402
119. He Y, Zhong J, Huang S, Shui X, Kong D, Chen C, et al. Elevated circulating miR-126–3p expression in patients with acute myocardial infarction: its diagnostic value. Int J Clin Exp Pathol. (2017) 10:11051–6.
120. Wang KJ, Zhao X, Liu YZ, Zeng QT, Mao XB, Li SN, et al. Circulating MiR-19b-3p, MiR-134–5p and MiR-186–5p are promising novel biomarkers for early diagnosis of acute myocardial infarction. Cell Physiol Biochem. (2016) 38:1015–29. doi: 10.1159/000443053
121. Xue S, Liu D, Zhu W, Su Z, Zhang L, Zhou C, et al. Circulating MiR-17–5p, MiR-126–5p and MiR-145–3p are novel biomarkers for diagnosis of acute myocardial infarction. Front Physiol. (2019) 10:123. doi: 10.3389/fphys.2019.00123
122. Yao XL, Lu XL, Yan CY, Wan QL, Cheng GC, Li YM. Circulating miR-122–5p as a potential novel biomarker for diagnosis of acute myocardial infarction. Int J Clin Exp Pathol. (2015) 8:16014–9.
123. Yuan J, Chen H, Ge D, Xu Y, Xu H, Yang Y, et al. Mir-21 promotes cardiac fibrosis after myocardial infarction via targeting Smad7. Cell Physiol Biochem. (2017) 42:2207–19. doi: 10.1159/000479995
124. Liu J, Sun F, Wang Y, Yang W, Xiao H, Zhang Y, et al. Suppression of microRNA-16 protects against acute myocardial infarction by reversing beta2-adrenergic receptor down-regulation in rats. Oncotarget. (2017) 8:20122–32. doi: 10.18632/oncotarget.15391
125. Jia K, Shi P, Han X, Chen T, Tang H, Wang J. Diagnostic value of miR-30d-5p and miR-125b-5p in acute myocardial infarction. Mol Med Rep. (2016) 14:184–94. doi: 10.3892/mmr.2016.5246
126. Guo ML, Guo LL, Weng YQ. Implication of peripheral blood miRNA-124 in predicting acute myocardial infarction. Eur Rev Med Pharmacol Sci. (2017) 21:1054–9. doi: 10.1016/j.jacc.2017.07.071
127. Yuan X, Pan J, Wen L, Gong B, Li J, Gao H, et al. MiR-144–3p enhances cardiac fibrosis after myocardial infarction by targeting PTEN. Front Cell Dev Biol. (2019) 7:249. doi: 10.3389/fcell.2019.00249
128. Pedretti S, Brulhart-Meynet MC, Montecucco F, Lecour S, James RW, Frias MA. HDL protects against myocardial ischemia reperfusion injury via miR-34b and miR-337 expression which requires STAT3. PLoS ONE. (2019) 14:e0218432. doi: 10.1371/journal.pone.0218432
129. Bye A, Røsjø H, Nauman J, Silva GJ, Follestad T, Omland T, et al. Circulating microRNAs predict future fatal myocardial infarction in healthy individuals - the HUNT study. J Mol Cell Cardiol. (2016) 97:162–8. doi: 10.1016/j.yjmcc.2016.05.009
130. Zhang R, Lan C, Pei H, Duan G, Huang L, Li L. Expression of circulating miR-486 and miR-150 in patients with acute myocardial infarction. BMC Cardiovasc Disord. (2015) 15:51. doi: 10.1186/s12872-015-0042-0
131. Jakob P, Kacprowski T, Briand-Schumacher S, Heg D, Klingenberg R, Stähli BE, et al. Profiling and validation of circulating microRNAs for cardiovascular events in patients presenting with ST-segment elevation myocardial infarction. Eur Heart J. (2017) 38:511–5. doi: 10.1093/eurheartj/ehw563
132. Schulte C, Molz S, Appelbaum S, Karakas M, Ojeda F, Lau DM, et al. miRNA-197 and miRNA-223 predict cardiovascular death in a cohort of patients with symptomatic coronary artery disease. PLoS ONE. (2015) 10:e0145930. doi: 10.1371/journal.pone.0145930
133. Karakas M, Schulte C, Appelbaum S, Ojeda F, Lackner KJ, Münzel T, et al. Circulating microRNAs strongly predict cardiovascular death in patients with coronary artery disease-results from the large AtheroGene study. Eur Heart J. (2017) 38:516–23. doi: 10.1093/eurheartj/ehw250
134. Zhu J, Yao K, Wang Q, Guo J, Shi H, Ma L, et al. Circulating miR-181a as a potential novel biomarker for diagnosis of acute myocardial infarction. Cell Physiol Biochem. (2016) 40:1591–602. doi: 10.1159/000453209
135. Ward JA, Esa N, Pidikiti R, Freedman JE, Keaney JF, Tanriverdi K, et al. Circulating cell and plasma microRNA profiles differ between non-ST-segment and ST-segment-elevation myocardial infarction. Fam Med Med Sci Res. (2013) 2:108.
136. Wang Y, Pan X, Fan Y, Hu X, Liu X, Xiang M, et al. Dysregulated expression of microRNAs and mRNAs in myocardial infarction. Am J Transl Res. (2015) 7:2291–304.
137. Wei T, Folkersen L, Ehrenborg E, Gabrielsen A. MicroRNA 486–3P as a stability marker in acute coronary syndrome. Biosci Rep. (2016) 36:e00351. doi: 10.1042/BSR20160023
138. Hobuß L, Bär C, Thum TJFip. Long non-coding RNAs: at the heart of cardiac dysfunction? Front Physiol. (2019) 10:30. doi: 10.3389/fphys.2019.00030
139. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. (2012) 22:1775–89. doi: 10.1101/gr.132159.111
140. Correia CCM, Rodrigues LF, de Avila Pelozin BR, Oliveira EM, Fernandes T. Long non-coding RNAs in cardiovascular diseases: potential function as biomarkers and therapeutic targets of exercise training. Non Coding RNA. (2021) 7:65. doi: 10.3390/ncrna7040065
141. Diederichs S. The four dimensions of noncoding RNA conservation. Trends Genet. (2014) 30:121–3. doi: 10.1016/j.tig.2014.01.004
142. Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. (2014) 505:696–700. doi: 10.1038/nature12756
143. Spitale RC, Crisalli P, Flynn RA, Torre EA, Kool ET, Chang HY. RNA SHAPE analysis in living cells. Nat Chem Biol. (2013) 9:18–20. doi: 10.1038/nchembio.1131
144. Das S, Shah R, Dimmeler S, Freedman JE, Holley C, Lee J-M, et al. Noncoding RNAs in cardiovascular disease: current knowledge, tools and technologies for investigation, and future directions: a scientific statement from the american heart association. Circulation. (2020) 13:e000062. doi: 10.1161/HCG.0000000000000062
145. Su Q, Liu Y, Lv X-W, Dai R-X, Yang X-H, Kong B-H. LncRNA TUG1 mediates ischemic myocardial injury by targeting miR-132–3p/HDAC3 axis. Am J Physiol Heart Circul Physiol. (2020) 318:H332–44. doi: 10.1152/ajpheart.00444.2019
146. Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. (2013) 341:1237973. doi: 10.1126/science.1237973
147. Chen DL, Ju HQ, Lu YX, Chen LZ, Zeng ZL, Zhang DS, et al. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin Cancer Res. (2016) 35:142. doi: 10.1186/s13046-016-0420-1
148. Tang Y, He R, An J, Deng P, Huang L, Yang W. lncRNA XIST interacts with miR-140 to modulate lung cancer growth by targeting iASPP. Oncol Rep. (2017) 38:941–8. doi: 10.3892/or.2017.5751
149. Zhang M, Liu HY, Han YL, Wang L, Zhai DD, Ma T, et al. Silence of lncRNA XIST represses myocardial cell apoptosis in rats with acute myocardial infarction through regulating miR-449. Eur Rev Med Pharmacol Sci. (2019) 23:8566–72.
150. Zhou J, Li D, Yang BP, Cui WJ. LncRNA XIST inhibits hypoxia-induced cardiomyocyte apoptosis via mediating miR-150–5p/Bax in acute myocardial infarction. Eur Rev Med Pharmacol Sci. (2020) 24:1357–66.
151. Bell RD, Long X, Lin M, Bergmann JH, Nanda V, Cowan SL, et al. Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arterioscler Thromb Vasc Biol. (2014) 34:1249–59. doi: 10.1161/ATVBAHA.114.303240
152. Ye F, Zhang J, Zhang Q, Zhang J, Chen C. Preliminary study on the mechanism of long noncoding RNA SENCR regulating the proliferation and migration of vascular smooth muscle cells. J Cell Physiol. (2020) 235:9635–43. doi: 10.1002/jcp.29775
153. Chen M, Guo Y, Sun Z, Meng X. Long non-coding RNA SENCR alleviates hypoxia/reoxygenation-induced cardiomyocyte apoptosis and inflammatory response by sponging miR-1. Cardiovasc Diagn Ther. (2021) 11:707–15. doi: 10.21037/cdt-20-1037
154. Pan X, Zheng G, Gao C. LncRNA PVT1: a novel therapeutic target for cancers. Clin Lab. (2018) 64:655–62. doi: 10.7754/Clin.Lab.2018.171216
155. Yu YH, Hu ZY, Li MH, Li B, Wang ZM, Chen SL. Cardiac hypertrophy is positively regulated by long non-coding RNA PVT1. Int J Clin Exp Pathol. (2015) 8:2582–9.
156. Ouyang M, Lu J, Ding Q, Qin T, Peng C, Guo Q. Knockdown of long non-coding RNA PVT1 protects human AC16 cardiomyocytes from hypoxia/reoxygenation-induced apoptosis and autophagy by regulating miR-186/Beclin-1 axis. Gene. (2020) 754:144775. doi: 10.1016/j.gene.2020.144775
157. Zheng J, Zhuo YY, Zhang C, Tang GY, Gu XY, Wang F. LncRNA TTTY15 regulates hypoxia-induced vascular endothelial cell injury via targeting miR-186–5p in cardiovascular disease. Eur Rev Med Pharmacol Sci. (2020) 24:3293–301.
158. Huang S, Tao W, Guo Z, Cao J, Huang X. Suppression of long noncoding RNA TTTY15 attenuates hypoxia-induced cardiomyocytes injury by targeting miR-455–5p. Gene. (2019) 701:1–8. doi: 10.1016/j.gene.2019.02.098
159. Ma R, Gao L, Liu Y, Du P, Chen X, Li G. LncRNA TTTY15 knockdown alleviates H2O2-stimulated myocardial cell injury by regulating the miR-98–5p/CRP pathway. Mol Cell Bioch. (2021) 476:81–92. doi: 10.1007/s11010-020-03887-4
160. Chen YQ, Yang X, Xu W, Yan Y, Chen XM, Huang ZQ. Knockdown of lncRNA TTTY15 alleviates myocardial ischemia-reperfusion injury through the miR-374a-5p/FOXO1 axis. IUBMB Life. (2021) 73:273–85. doi: 10.1002/iub.2428
161. Song H, Sun W, Ye G, Ding X, Liu Z, Zhang S, et al. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med. (2013) 11:225. doi: 10.1186/1479-5876-11-225
162. Feng W, Li L, Xu X, Jiao Y, Du W. Up-regulation of the long non-coding RNA RMRP contributes to glioma progression and promotes glioma cell proliferation and invasion. Arch Med Sci. (2017) 13:1315–21. doi: 10.5114/aoms.2017.66747
163. Meng Q, Ren M, Li Y, Song X. LncRNA-RMRP acts as an oncogene in lung cancer. PLoS ONE. (2016) 11:e0164845. doi: 10.1371/journal.pone.0164845
164. Steinbusch MMF, Caron MMJ, Surtel DAM, Friedrich F, Lausch E, Pruijn GJM, et al. Expression of RMRP RNA is regulated in chondrocyte hypertrophy and determines chondrogenic differentiation. Sci Rep. (2017) 7:6440. doi: 10.1038/s41598-017-06809-5
165. Greco S, Zaccagnini G, Perfetti A, Fuschi P, Valaperta R, Voellenkle C, et al. Long noncoding RNA dysregulation in ischemic heart failure. J Transl Med. (2016) 14:183. doi: 10.1186/s12967-016-0926-5
166. Kong F, Jin J, Lv X, Han Y, Liang X, Gao Y, et al. Long noncoding RNA RMRP upregulation aggravates myocardial ischemia-reperfusion injury by sponging miR-206 to target ATG3 expression. Biomed Pharmacother. (2019) 109:716–25. doi: 10.1016/j.biopha.2018.10.079
167. Cai B, Ma W, Ding F, Zhang L, Huang Q, Wang X, et al. The long noncoding RNA CAREL controls cardiac regeneration. J Am Coll Cardiol. (2018) 72:534–50. doi: 10.1016/j.jacc.2018.04.085
168. Chen G, Li H, Li X, Li B, Zhong L, Huang S, et al. Loss of long non-coding RNA CRRL promotes cardiomyocyte regeneration and improves cardiac repair by functioning as a competing endogenous RNA. J Mol Cell Cardiol. (2018) 122:152–64. doi: 10.1016/j.yjmcc.2018.08.013
169. Huang P, Yang D, Yu L, Shi Y. Downregulation of lncRNA ZFAS1 protects H9c2 cardiomyocytes from ischemia/reperfusion-induced apoptosis via the miR-590-3p/NF-κB signaling pathway. Mol Med Rep. (2020) 22:2300–6. doi: 10.3892/mmr.2020.11340
170. Zhang Y, Jiao L, Sun L, Li Y, Gao Y, Xu C, et al. LncRNA ZFAS1 as a SERCA2a inhibitor to cause intracellular Ca2+ overload and contractile dysfunction in a mouse model of myocardial infarction. Circ Res. (2018) 122:1354–68. doi: 10.1161/CIRCRESAHA.117.312117
171. Zhihao L, Jingyu N, Lan L, Michael S, Rui G, Xiyun B, et al. SERCA2a: a key protein in the Ca2+ cycle of the heart failure. Heart Fail Rev. (2020) 25:523–35. doi: 10.1007/s10741-019-09873-3
172. Trembinski DJ, Bink DI, Theodorou K, Sommer J, Fischer A, van Bergen A, et al. Aging-regulated anti-apoptotic long non-coding RNA Sarrah augments recovery from acute myocardial infarction. Nat Commun. (2020) 11:2039. doi: 10.1038/s41467-020-15995-2
173. Wang K, Long B, Zhou LY, Liu F, Zhou QY, Liu CY, et al. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun. (2014) 5:3596. doi: 10.1038/ncomms4596
174. Qu X, Song X, Yuan W, Shu Y, Wang Y, Zhao X, et al. Expression signature of lncRNAs and their potential roles in cardiac fibrosis of post-infarct mice. Biosci Rep. (2016) 36:e00337. doi: 10.1042/BSR20150278
175. Liu CY, Zhang YH, Li RB, Zhou LY, An T, Zhang RC, et al. LncRNA CAIF inhibits autophagy and attenuates myocardial infarction by blocking p53-mediated myocardin transcription. Nat Commun. (2018) 9:29. doi: 10.1038/s41467-017-02280-y
176. Li J, Xie J, Wang YZ, Gan YR, Wei L, Ding GW, et al. Overexpression of lncRNA Dancr inhibits apoptosis and enhances autophagy to protect cardiomyocytes from endoplasmic reticulum stress injury via sponging microRNA-6324. Mol Med Rep. (2021) 23:116. doi: 10.3892/mmr.2020.11755
177. Chen Z, Liu R, Niu Q, Wang H, Yang Z, Bao Y. Morphine Postconditioning alleviates autophage in ischemia-reperfusion induced cardiac injury through up-regulating lncRNA UCA1. Biomed Pharmacother. (2018) 108:1357–64. doi: 10.1016/j.biopha.2018.09.119
178. Shi HJ, Wang MW, Sun JT, Wang H, Li YF, Chen BR, et al. A novel long noncoding RNA FAF inhibits apoptosis via upregulating FGF9 through PI3K/AKT signaling pathway in ischemia-hypoxia cardiomyocytes. J Cell Physiol. (2019) 234:21973–87. doi: 10.1002/jcp.28760
179. Du L, Chen J, Wu Y, Xia G, Chen M, Zhao P, et al. Long Non-coding RNA N1LR protects against myocardial ischemic/reperfusion injury through regulating the TGF-β signaling pathway. Front Cardiovasc Med. (2021) 8:654969. doi: 10.3389/fcvm.2021.654969
180. Song N, Li XM, Luo JY, Zhai H, Zhao Q, Zhou XR, et al. Construction and analysis for differentially expressed long non-coding RNAs and mRNAs in acute myocardial infarction. Sci Rep. (2020) 10:6989. doi: 10.1038/s41598-020-63840-9
181. Zangrando J, Zhang L, Vausort M, Maskali F, Marie PY, Wagner DR, et al. Identification of candidate long non-coding RNAs in response to myocardial infarction. BMC Genomics. (2014) 15:460. doi: 10.1186/1471-2164-15-460
182. Su Q, Liu Y, Lv XW, Ye ZL, Sun YH, Kong BH, et al. Inhibition of lncRNA TUG1 upregulates miR-142–3p to ameliorate myocardial injury during ischemia and reperfusion via targeting HMGB1- and Rac1-induced autophagy. J Mol Cell Cardiol. (2019) 133:12–25. doi: 10.1016/j.yjmcc.2019.05.021
183. Zhao J, Chen F, Ma W, Zhang P. Suppression of long noncoding RNA NEAT1 attenuates hypoxia-induced cardiomyocytes injury by targeting miR-378a-3p. Gene. (2020) 731:144324. doi: 10.1016/j.gene.2019.144324
184. Huang Z, Ye B, Wang Z, Han J, Lin L, Shan P, et al. Inhibition of LncRNA-HRIM increases cell viability by regulating autophagy levels during hypoxia/reoxygenation in myocytes. Cell Physiol Biochem. (2018) 46:1341–51. doi: 10.1159/000489149
185. Wang JJ, Bie ZD, Sun CF. Long noncoding RNA AK088388 regulates autophagy through miR-30a to affect cardiomyocyte injury. J Cell Biochem. (2019) 120:10155–63. doi: 10.1002/jcb.28300
186. Tong G, Wang Y, Xu C, Xu Y, Ye X, Zhou L, et al. Long non-coding RNA FOXD3-AS1 aggravates ischemia/reperfusion injury of cardiomyocytes through promoting autophagy. Am J Transl Res. (2019) 11:5634–44.
187. Yu SY, Dong B, Fang ZF, Hu XQ, Tang L, Zhou SH. Knockdown of lncRNA AK139328 alleviates myocardial ischaemia/reperfusion injury in diabetic mice via modulating miR-204–3p and inhibiting autophagy. J Cell Mol Med. (2018) 22:4886–98. doi: 10.1111/jcmm.13754
188. Zhang J, Gao C, Meng M, Tang H. Long Noncoding RNA MHRT protects cardiomyocytes against H2O2-induced apoptosis. Biomol Ther. (2016) 24:19–24. doi: 10.4062/biomolther.2015.066
189. Xia J, Jiang N, Li Y, Wei Y, Zhang X. The long noncoding RNA THRIL knockdown protects hypoxia-induced injuries of H9C2 cells through regulating miR-99a. Cardiol J. (2019) 26:564–74. doi: 10.5603/CJ.a2018.0054
190. Hu C, Li J, Du Y, Li J, Yang Y, Jia Y, et al. Impact of chronic intermittent hypoxia on the long non-coding RNA and mRNA expression profiles in myocardial infarction. J Cell Mol Med. (2021) 25:421–33. doi: 10.1111/jcmm.16097
191. Luo C, Ling GX, Lei BF, Feng X, Xie XY, Fang C, et al. Circular RNA PVT1 silencing prevents ischemia-reperfusion injury in rat by targeting microRNA-125b and microRNA-200a. J Mol Cell Cardiol. (2021) 159:80–90. doi: 10.1016/j.yjmcc.2021.05.019
192. Chen TP, Zhang NJ, Wang HJ, Hu SG, Geng X. Knockdown of circROBO2 attenuates acute myocardial infarction through regulating the miR-1184/TRADD axis. Mol Med. (2021) 27:21. doi: 10.1186/s10020-021-00275-6
193. Wen J, Liao J, Liang J, Chen XP, Zhang B, Chu L. Circular RNA HIPK3: a key circular RNA in a variety of human cancers. Front Oncol. (2020) 10:773. doi: 10.3389/fonc.2020.00773
194. Bai M, Pan CL, Jiang GX, Zhang YM, Zhang Z. CircHIPK3 aggravates myocardial ischemia-reperfusion injury by binding to miRNA-124–3p. Eur Rev Med Pharmacol Sci. (2019) 23:10107–14.
195. Wu Y, Wu M, Yang J, Li Y, Peng W, Wu M, et al. Silencing CircHIPK3 sponges miR-93–5p to inhibit the activation of Rac1/PI3K/AKT pathway and improves myocardial infarction-induced cardiac dysfunction. Front Cardiovasc Med. (2021) 8:645378. doi: 10.3389/fcvm.2021.645378
196. Liu YY, Zhang LY, Du WZ. Circular RNA circ-PVT1 contributes to paclitaxel resistance of gastric cancer cells through the regulation of ZEB1 expression by sponging miR-124–3p. Biosci Rep. (2019) 39:BSR20193045. doi: 10.1042/BSR20193045
197. Zhu Y, Zou C, Jia Y, Zhang H, Ma X, Zhang J. Knockdown of circular RNA circMAT2B reduces oxygen-glucose deprivation-induced inflammatory injury in H9c2 cells through up-regulating miR-133. Cell Cycle. (2020) 19:2622–30. doi: 10.1080/15384101.2020.1814025
198. Long N, Chu L, Jia J, Peng S, Gao Y, Yang H, et al. CircPOSTN/miR-361–5p/TPX2 axis regulates cell growth, apoptosis and aerobic glycolysis in glioma cells. Cancer Cell Int. (2020) 20:374. doi: 10.1186/s12935-020-01454-x
199. Garikipati VNS, Verma SK, Cheng Z, Liang D, Truongcao MM, Cimini M, et al. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat Commun. (2019) 10:4317. doi: 10.1038/s41467-019-11777-7
200. Cheng N, Wang MY, Wu YB, Cui HM, Wei SX, Liu B, et al. Circular RNA POSTN promotes myocardial infarction-induced myocardial injury and cardiac remodeling by regulating miR-96–5p/BNIP3 axis. Front Cell Dev Biol. (2020) 8:618574. doi: 10.3389/fcell.2020.618574
201. Zhao B, Li G, Peng J, Ren L, Lei L, Ye H, et al. CircMACF1 Attenuates acute myocardial infarction through miR-500b-5p-EMP1 axis. J Cardiovasc Transl Res. (2021) 14:161–72. doi: 10.1007/s12265-020-09976-5
202. Zhu Y, Zhao P, Sun L, Lu Y, Zhu W, Zhang J, et al. Overexpression of circRNA SNRK targets miR-103–3p to reduce apoptosis and promote cardiac repair through GSK3β/β-catenin pathway in rats with myocardial infarction. Cell Death Discov. (2021) 7:84. doi: 10.1038/s41420-021-00467-3
203. Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li X, et al. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics. (2017) 7:3842–55. doi: 10.7150/thno.19764
204. Jin P, Li LH, Shi Y, Hu NB. Salidroside inhibits apoptosis and autophagy of cardiomyocyte by regulation of circular RNA hsa_circ_0000064 in cardiac ischemia-reperfusion injury. Gene. (2021) 767:145075. doi: 10.1016/j.gene.2020.145075
205. Zhang M, Wang Z, Cheng Q, Wang Z, Lv X, Wang Z, et al. Circular RNA (circRNA) CDYL induces myocardial regeneration by ceRNA after myocardial infarction. Med Sci Monit. (2020) 26:e923188. doi: 10.12659/MSM.923188
206. Mester-Tonczar J, Einzinger P, Winkler J, Kastner N, Spannbauer A, Zlabinger K, et al. Novel identified circular transcript of RCAN2, circ-RCAN2, shows deviated expression pattern in pig reperfused infarcted myocardium and hypoxic porcine cardiac progenitor cells in vitro. Int J Mol Sci. (2021) 22:1390. doi: 10.3390/ijms22031390
207. Salgado-Somoza A, Zhang L, Vausort M, Devaux Y. The circular RNA MICRA for risk stratification after myocardial infarction. Int J Cardiol Heart Vasc. (2017) 17:33–6. doi: 10.1016/j.ijcha.2017.11.001
208. Sun LY, Zhao JC, Ge XM, Zhang H, Wang CM, Bie ZD. Circ_LAS1L regulates cardiac fibroblast activation, growth, and migration through miR-125b/SFRP5 pathway. Cell Biochem Funct. (2020) 38:443–50. doi: 10.1002/cbf.3486
209. Zhu Y, Pan W, Yang T, Meng X, Jiang Z, Tao L, et al. Upregulation of circular RNA CircNFIB attenuates cardiac fibrosis by sponging miR-433. Front Genet. (2019) 10:564. doi: 10.3389/fgene.2019.00564
210. Zhang C-L, Long T-Y, Bi S-S, Sheikh S-A, Li F. CircPAN3 ameliorates myocardial ischaemia/reperfusion injury by targeting miR-421/Pink1 axis-mediated autophagy suppression. Lab Invest. (2021) 101:89–103. doi: 10.1038/s41374-020-00483-4
211. Wang S, Cheng Z, Chen X, Lu G, Zhu X, Xu G. CircUBXN7 mitigates H/R-induced cell apoptosis and inflammatory response through the miR-622-MCL1 axis. Am J Transl Res. (2021) 13:8711–27.
212. Cai X, Li B, Wang Y, Zhu H, Zhang P, Jiang P, et al. CircJARID2 regulates hypoxia-induced injury in H9c2 cells by affecting miR-9–5p-mediated BNIP3. J Cardiovasc Pharmacol. (2021) 78:e77–85. doi: 10.1097/FJC.0000000000001033
213. Tan J, Pan W, Chen H, Du Y, Jiang P, Zeng D, et al. Circ_0124644 Serves as a ceRNA for miR-590–3p to promote hypoxia-induced cardiomyocytes injury via regulating SOX4. Front Genet. (2021) 12:667724. doi: 10.3389/fgene.2021.667724
214. Cai L, Qi B, Wu X, Peng S, Zhou G, Wei Y, et al. Circular RNA Ttc3 regulates cardiac function after myocardial infarction by sponging miR-15b. J Mol Cell Cardiol. (2019) 130:10–22. doi: 10.1016/j.yjmcc.2019.03.007
215. Wang X, Cheng Z, Xu J, Feng M, Zhang H, Zhang L, et al. Circular RNA Arhgap12 modulates doxorubicin-induced cardiotoxicity by sponging miR-135a-5p. Life Sci. (2021) 265:118788. doi: 10.1016/j.lfs.2020.118788
216. Ren Q, Li H, Wang X. The circular RNA ZNF292 alleviates OGD-induced injury in H9c2 cells via targeting BNIP3. Cell Cycle. (2019) 18:3365–77. doi: 10.1080/15384101.2019.1676585
217. Bian Y, Pang P, Li X, Yu S, Wang X, Liu K, et al. CircHelz activates NLRP3 inflammasome to promote myocardial injury by sponging miR-133a-3p in mouse ischemic heart. J Mol Cell Cardiol. (2021) 158:128–39. doi: 10.1016/j.yjmcc.2021.05.010
218. Tian M, Xue J, Dai C, Jiang E, Zhu B, Pang H. CircSLC8A1 and circNFIX can be used as auxiliary diagnostic markers for sudden cardiac death caused by acute ischemic heart disease. Sci Rep. (2021) 11:4695. doi: 10.1038/s41598-021-84056-5
219. Gao WQ, Hu XM, Zhang Q, Yang L, Lv XZ, Chen S, et al. Downregulation of circFASTKD1 ameliorates myocardial infarction by promoting angiogenesis. Aging. (2020) 13:3588–604. doi: 10.18632/aging.202305
220. Li F, Long T-Y, Bi S-S, Sheikh SA, Zhang C-L. circPAN3 exerts a profibrotic role via sponging miR-221 through FoxO3/ATG7-activated autophagy in a rat model of myocardial infarction. Life Sci. (2020) 257:118015. doi: 10.1016/j.lfs.2020.118015
221. Tang CM, Zhang M, Huang L, Hu ZQ, Zhu JN, Xiao Z, et al. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci Rep. (2017) 7:40342. doi: 10.1038/srep40342
222. Wang Y, Li C, Zhao R, Qiu Z, Shen C, Wang Z, et al. CircUbe3a from M2 macrophage-derived small extracellular vesicles mediates myocardial fibrosis after acute myocardial infarction. Theranostics. (2021) 11:6315. doi: 10.7150/thno.52843
223. Zhao C, Liu J, Ge W, Li Z, Lv M, Feng Y, et al. Identification of regulatory circRNAs involved in the pathogenesis of acute myocardial infarction. Front Genet. (2020) 11:626492. doi: 10.3389/fgene.2020.626492
224. Ren K, Li B, Jiang L, Liu Z, Wu F, Zhang Y, et al. circ_0023461 silencing protects cardiomyocytes from hypoxia-induced dysfunction through targeting miR-370–3p/PDE4D signaling. Oxid Med Cell Longev. (2021) 2021:8379962. doi: 10.1155/2021/8379962
225. Ibrahim A, Marbán E. Exosomes: fundamental biology and roles in cardiovascular physiology. Annu Rev Physiol. (2016) 78:67–83. doi: 10.1146/annurev-physiol-021115-104929
226. Jafari A, Babajani A, Abdollahpour-Alitappeh M, Ahmadi N, Rezaei-Tavirani M. Exosomes and cancer: from molecular mechanisms to clinical applications. Med Oncol. (2021) 38:1–17. doi: 10.1007/s12032-021-01491-0
227. Tkach M, Théry C. Communication by extracellular vesicles: where we are and where we need to go. Cell. (2016) 164:1226–32. doi: 10.1016/j.cell.2016.01.043
228. Li X, Zhou J, Huang K. Inhibition of the lncRNA Mirt1 attenuates acute myocardial infarction by suppressing NF-κB activation. Cell Physiol Biochem. (2017) 42:1153–64. doi: 10.1159/000478870
229. Gao L, Liu Y, Guo S, Yao R, Wu L, Xiao L, et al. Circulating long noncoding RNA HOTAIR is an essential mediator of acute myocardial infarction. Cell Physiol Biochem. (2017) 44:1497–508. doi: 10.1159/000485588
230. Yuan Z, Huang W. New developments in exosomal lncRNAs in cardiovascular diseases. Front Cardiovasc Med. (2021) 709169. doi: 10.3389/fcvm.2021.709169
231. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. (2014) 505:344–52. doi: 10.1038/nature12986
232. Li C, Ni Y-Q, Xu H, Xiang Q-Y, Zhao Y, Zhan J-K, et al. Roles and mechanisms of exosomal non-coding RNAs in human health and diseases. Signal Transd Targeted Ther. (2021) 6:1–31. doi: 10.1038/s41392-021-00779-x
233. Winnik S, Auwerx J, Sinclair DA, Matter CM. Protective effects of sirtuins in cardiovascular diseases: from bench to bedside. Eur Heart J. (2015) 36:3404–12. doi: 10.1093/eurheartj/ehv290
234. Li Y, Wang P, Yang X, Wang W, Zhang J, He Y, et al. SIRT1 inhibits inflammatory response partly through regulation of NLRP3 inflammasome in vascular endothelial cells. Mol Immunol. (2016) 77:148–56. doi: 10.1016/j.molimm.2016.07.018
235. Chen Z, Peng IC, Cui X, Li YS, Chien S, Shyy JY. Shear stress, SIRT1, and vascular homeostasis. Proc Natl Acad Sci USA. (2010) 107:10268–73. doi: 10.1073/pnas.1003833107
236. Pillai VB, Sundaresan NR, Gupta MP. Regulation of Akt signaling by sirtuins: its implication in cardiac hypertrophy and aging. Circ Res. (2014) 114:368–78. doi: 10.1161/CIRCRESAHA.113.300536
237. Mori J, Patel VB, Abo Alrob O, Basu R, Altamimi T, Desaulniers J, et al. Angiotensin 1–7 ameliorates diabetic cardiomyopathy and diastolic dysfunction in db/db mice by reducing lipotoxicity and inflammation. Circ Heart Fail. (2014) 7:327–39. doi: 10.1161/CIRCHEARTFAILURE.113.000672
238. Li Y, Yang X, He Y, Wang W, Zhang J, Zhang W, et al. Negative regulation of NLRP3 inflammasome by SIRT1 in vascular endothelial cells. Immunobiology. (2017) 222:552–61. doi: 10.1016/j.imbio.2016.11.002
239. Mao Q, Liang XL, Zhang CL, Pang YH, Lu YX. LncRNA KLF3-AS1 in human mesenchymal stem cell-derived exosomes ameliorates pyroptosis of cardiomyocytes and myocardial infarction through miR-138–5p/Sirt1 axis. Stem Cell Res Ther. (2019) 10:393. doi: 10.1186/s13287-019-1522-4
240. Chen D, Wang K, Zheng Y, Wang G, Jiang M. Exosomes-mediated LncRNA ZEB1-AS1 facilitates cell injuries by miR-590–5p/ETS1 axis through the TGF-β/Smad pathway in oxidized low-density lipoprotein-induced human umbilical vein endothelial cells. J Cardiovasc Pharmacol. (2021) 77:480–90. doi: 10.1097/FJC.0000000000000974
241. Hamed GM, Fattah MF. Clinical Relevance of matrix metalloproteinase 9 in patients with acute coronary syndrome. Clin Appl Thromb Hemost. (2015) 21:705–11. doi: 10.1177/1076029614567309
242. Sakakura K, Nakano M, Otsuka F, Ladich E, Kolodgie FD, Virmani R. Pathophysiology of atherosclerosis plaque progression. Heart Lung Circ. (2013) 22:399–411. doi: 10.1016/j.hlc.2013.03.001
243. Johnson JL. Metalloproteinases in atherosclerosis. Eur J Pharmacol. (2017) 816:93–106. doi: 10.1016/j.ejphar.2017.09.007
244. Lahdentausta L, Leskelä J, Winkelmann A, Tervahartiala T, Sorsa T, Pesonen E, et al. Serum MMP-9 diagnostics, prognostics, and activation in acute coronary syndrome and its recurrence. J Cardiovasc Transl Res. (2018) 11:210–20. doi: 10.1007/s12265-018-9789-x
245. Li L, Wang Z, Hu X, Wan T, Wu H, Jiang W, et al. Human aortic smooth muscle cell-derived exosomal miR-221/222 inhibits autophagy via a PTEN/Akt signaling pathway in human umbilical vein endothelial cells. Biochem Biophys Res Commun. (2016) 479:343–50. doi: 10.1016/j.bbrc.2016.09.078
246. Wang L, Zhang J. Exosomal lncRNA AK139128 derived from hypoxic cardiomyocytes promotes apoptosis and inhibits cell proliferation in cardiac fibroblasts. Int J Nanomed. (2020) 15:3363. doi: 10.2147/IJN.S240660
247. Chen Q, Liu Y, Ding X, Li Q, Qiu F, Wang M, et al. Bone marrow mesenchymal stem cell-secreted exosomes carrying microRNA-125b protect against myocardial ischemia reperfusion injury via targeting SIRT7. Mol Cell Biochem. (2020) 465:103–14. doi: 10.1007/s11010-019-03671-z
248. Wang L, Liu J, Xu B, Liu Y-L, Liu Z. Reduced exosome miR-425 and miR-744 in the plasma represents the progression of fibrosis and heart failure. Kaohsiung J Med Sci. (2018) 34:626–33. doi: 10.1016/j.kjms.2018.05.008
249. Liu J, Wu J, Li L, Li T, Wang J. The role of exosomal non-coding RNAs in coronary artery disease. Front Pharmacol. (2020) 11:603104. doi: 10.3389/fphar.2020.603104
250. Zhu L-P, Tian T, Wang J-Y, He J-N, Chen T, Pan M, et al. Hypoxia-elicited mesenchymal stem cell-derived exosomes facilitates cardiac repair through miR-125b-mediated prevention of cell death in myocardial infarction. Theranostics. (2018) 8:6163. doi: 10.7150/thno.28021
251. Wang W, Zheng Y, Wang M, Yan M, Jiang J, Li Z. Exosomes derived miR-126 attenuates oxidative stress and apoptosis from ischemia and reperfusion injury by targeting ERRFI1. Gene. (2019) 690:75–80. doi: 10.1016/j.gene.2018.12.044
252. Peng Y, Zhao J-L, Peng Z-Y, Xu W-F, Yu G-L. Exosomal miR-25–3p from mesenchymal stem cells alleviates myocardial infarction by targeting pro-apoptotic proteins and EZH2. Cell Death Dis. (2020) 11:1–15. doi: 10.1038/s41419-020-2545-6
253. Wen Z, Mai Z, Zhu X, Wu T, Chen Y, Geng D, et al. Mesenchymal stem cell-derived exosomes ameliorate cardiomyocyte apoptosis in hypoxic conditions through microRNA144 by targeting the PTEN/AKT pathway. Stem Cell Res Ther. (2020) 11:1–17. doi: 10.1186/s13287-020-1563-8
254. Pan J, Alimujiang M, Chen Q, Shi H, Luo X. Exosomes derived from miR-146a-modified adipose-derived stem cells attenuate acute myocardial infarction– induced myocardial damage via downregulation of early growth response factor 1. J Cell Biochem. (2019) 120:4433–43. doi: 10.1002/jcb.27731
255. Peng L, Chun-guang Q, Bei-fang L, Xue-zhi D, Zi-hao W, Yun-fu L, et al. Clinical impact of circulating miR-133, miR-1291 and miR-663b in plasma of patients with acute myocardial infarction. Diagn Pathol. (2014) 9:89. doi: 10.1186/1746-1596-9-89
256. Li Y, Yang R, Guo B, Zhang H, Zhang H, Liu S, et al. Exosomal miR-301 derived from mesenchymal stem cells protects myocardial infarction by inhibiting myocardial autophagy. Biochem Biophys Res Commun. (2019) 514:323–8. doi: 10.1016/j.bbrc.2019.04.138
257. Wang K, Jiang Z, Webster KA, Chen J, Hu H, Zhou Y, et al. Enhanced cardioprotection by human endometrium mesenchymal stem cells driven by exosomal microRNA-21. Stem Cells Transl Med. (2017) 6:209–22. doi: 10.5966/sctm.2015-0386
258. Chen Z, Yan Y, Wu J, Qi C, Liu J, Wang J. Expression level and diagnostic value of exosomal NEAT1/miR-204/MMP-9 in acute ST-segment elevation myocardial infarction. IUBMB Life. (2020) 72:2499–507. doi: 10.1002/iub.2376
259. Su J, Li J, Yu Q, Wang J, Li X, Yang J, et al. Exosomal miRNAs as potential biomarkers for acute myocardial infarction. IUBMB Life. (2020) 72:384–400. doi: 10.1002/iub.2189
260. Liang C, Zhang L, Lian X, Zhu T, Zhang Y, Gu N. Circulating exosomal SOCS2-AS1 acts as a novel biomarker in predicting the diagnosis of coronary artery disease. Biomed Res Int. (2020) 2020:9182091. doi: 10.1155/2020/9182091
261. Zhao X, Jia Y, Chen H, Yao H, Guo W. Plasma-derived exosomal miR-183 associates with protein kinase activity and may serve as a novel predictive biomarker of myocardial ischemic injury. Exp Ther Med. (2019) 18:179–87. doi: 10.3892/etm.2019.7555
262. Shyu KG, Wang BW, Fang WJ, Pan CM, Lin CM. Hyperbaric oxygen-induced long non-coding RNA MALAT1 exosomes suppress MicroRNA-92a expression in a rat model of acute myocardial infarction. J Cell Mol Med. (2020) 24:12945–54. doi: 10.1111/jcmm.15889
263. Huang P, Wang L, Li Q, Tian X, Xu J, Xu J, et al. Atorvastatin enhances the therapeutic efficacy of mesenchymal stem cells-derived exosomes in acute myocardial infarction via up-regulating long non-coding RNA H19. Cardiovasc Res. (2020) 116:353–67. doi: 10.1093/cvr/cvz139
264. Sun L, Zhu W, Zhao P, Wang Q, Fan B, Zhu Y, et al. Long noncoding RNA UCA1 from hypoxia-conditioned hMSC-derived exosomes: a novel molecular target for cardioprotection through miR-873–5p/XIAP axis. Cell Death Dis. (2020) 11:696. doi: 10.1038/s41419-020-02783-5
265. Zheng ML, Liu XY, Han RJ, Yuan W, Sun K, Zhong JC, et al. Circulating exosomal long non-coding RNAs in patients with acute myocardial infarction. J Cell Mol Med. (2020) 24:9388–96. doi: 10.1111/jcmm.15589
266. Li C, Song J, Li X, Zhang T, Li Z. Circular RNA 0001273 in exosomes derived from human umbilical cord mesenchymal stem cells (UMSCs) in myocardial infarction. Eur Rev Med Pharmacol Sci. (2020) 24:10086–95.
267. Shan K, Jiang Q, Wang X-Q, Wang Y, Yang H, Yao M-D, et al. Role of long non-coding RNA-RNCR3 in atherosclerosis-related vascular dysfunction. Cell Death Dis. (2016) 7:e2248–e. doi: 10.1038/cddis.2016.145
268. Chen H, Xia W, Hou M. LncRNA-NEAT1 from the competing endogenous RNA network promotes cardioprotective efficacy of mesenchymal stem cell-derived exosomes induced by macrophage migration inhibitory factor via the miR-142–3p/FOXO1 signaling pathway. Stem Cell Res Ther. (2020) 11:1–15. doi: 10.1186/s13287-020-1556-7
269. Liu Y, Zhang W, Gu J, Sun Y, Cui H, Bu J, et al. Exosome-mediated miR-106a-3p derived from ox-LDL exposed macrophages accelerated cell proliferation and repressed cell apoptosis of human vascular smooth muscle cells. Eur Rev Med Pharmacol Sci. (2020) 24:7039–50.
270. Sun L, Zhu W, Zhao P, Zhang J, Lu Y, Zhu Y, et al. Down-regulated exosomal MicroRNA-221–3p derived from senescent mesenchymal stem cells impairs heart repair. Front Cell Dev Biol. (2020) 8:263. doi: 10.3389/fcell.2020.00263
271. Sun X-H, Wang X, Zhang Y, Hui J. Exosomes of bone-marrow stromal cells inhibit cardiomyocyte apoptosis under ischemic and hypoxic conditions via miR-486–5p targeting the PTEN/PI3K/AKT signaling pathway. Thromb Res. (2019) 177:23–32. doi: 10.1016/j.thromres.2019.02.002
Keywords: myocardial infarction, non-coding RNAs, microRNAs, long non-coding RNAs, circular RNAs, exosome
Citation: Fadaei S, Zarepour F, Parvaresh M, Motamedzadeh A, Tamehri Zadeh SS, Sheida A, Shabani M, Hamblin MR, Rezaee M, Zarei M and Mirzaei H (2022) Epigenetic regulation in myocardial infarction: Non-coding RNAs and exosomal non-coding RNAs. Front. Cardiovasc. Med. 9:1014961. doi: 10.3389/fcvm.2022.1014961
Received: 09 August 2022; Accepted: 17 October 2022;
Published: 10 November 2022.
Edited by:
Dongtak Jeong, Hanyang Universiy, Erica, South KoreaReviewed by:
Changwon Kho, Pusan National University, South KoreaCopyright © 2022 Fadaei, Zarepour, Parvaresh, Motamedzadeh, Tamehri Zadeh, Sheida, Shabani, Hamblin, Rezaee, Zarei and Mirzaei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mehdi Rezaee, bS5yZXphZWlAYWJ6dW1zLmFjLmly; Maryam Zarei, ZHIubWFyeWFtemFyZWlAZ21haWwuY29t; Hamed Mirzaei, bWlyemFlaS1oQGthdW1zLmFjLmly; aC5taXJ6YWVpMjAwMkBnbWFpbC5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.