- 1Department of Neurology, The Affiliated Hospital of Qingdao University, Qingdao, China
- 2Department of Critical Care Medicine, The Affiliated Hospital of Qingdao University, Qingdao, China
Atherosclerosis is a complex disease closely related to the function of endothelial cells (ECs), monocytes/macrophages, and vascular smooth muscle cells (VSMCs). Despite a good understanding of the pathogenesis of atherosclerosis, the underlying molecular mechanisms are still only poorly understood. Therefore, atherosclerosis continues to be an important clinical issue worthy of further research. Recent evidence has shown that long non-coding RNAs (lncRNAs) and RNA-binding proteins (RBPs) can serve as important regulators of cellular function in atherosclerosis. Besides, several studies have shown that lncRNAs are partly dependent on the specific interaction with RBPs to exert their function. This review summarizes the important contributions of lncRNAs and RBPs in atherosclerosis and provides novel and comprehensible interaction models of lncRNAs and RBPs.
Introduction
Atherosclerosis is the main cause of large-artery atherosclerotic (LAA) stroke (1). While its etiology is complicated and multifactorial; the exact mechanism is still unknown. Generally, when stimulated by dyslipidemia, hypertension, or pro-inflammatory mediators, endothelial cells (ECs) are injured, which enhances the expression of cell adhesion molecules (AMs), causing leukocytes to adhere on their surface (2). Low-density lipoprotein (LDL) penetrates the ECs and the space between the ECs (3). Monocytes migrate and differentiate into tissue macrophages and can form macrophage-derived foam cells by endocytosing the oxidized modified LDL (ox-LDL), leading to intracellular cholesterol accumulation (2). Vascular smooth muscle cells (VSMCs) migrate into the intima and engulf lipids to form muscle-derived foam cells. Once the initial process is completed, the atherosclerotic plaque progresses owing to the persistent accumulation of lipids and foam cells. Atherosclerosis is usually asymptomatic; however, unstable plaques may rupture and provoke thrombosis. Therefore, addressing the molecular mechanism of atherosclerosis is crucial to lay the foundation and highlight the prevention and treatment of stroke.
Recent, research has identified a functional genetic material called long non-coding RNA (lncRNA), which exert significant biological roles in multiple diseases. Although, the function of lncRNA is complex and still controversial, there is sufficient evidence to suggest that many lncRNAs have important cellular functions (4). Significantly, lncRNA regulates plaque development in all stages. They are involved in the process of atherosclerosis such as the regulation of ECs, macrophages, and VSMCs (5). Yan et al. found that lncRNA-RNCR3 was significantly upregulated in ECs and VSMCs cultured in vitro after ox-LDL treatment, and downregulation of RNCR3 accelerated the progress of atherosclerosis, exacerbated hypercholesterolemia and inflammatory factor release, and decreased ECs and VSMCs proliferation (6). Furthermore, studies found that some lncRNAs may be regarded as novel diagnostic biomarkers in LAA (7). Some small-molecule epigenetics drugs have not only been approved by the US Food and Drug Administration but also shown effects in preclinical studies of atherosclerosis (8). However, despite studies have shown that lncRNAs have a variety of important functions, their mechanism of action by which they regulate atherosclerosis is still complex and poorly understood.
Several studies have supported that lncRNAs perform many different functions by directly interacting with RNA-binding proteins (RBPs) (9). In the cytoplasm, for instance, exert their function by interacting with RBP through sequence motifs or by forming unique structural motifs (4). The expression of most RBPs is ubiquitous, usually higher than the average level of cellular protein. With the increase in the number of RBPs in higher eukaryotes, the relative sizes of different RBPs against different RNA targets remain unchanged throughout the phylogeny. However, the non-codingRNA-binding protein family and mRBP family have the lowest conservation rates, only 20% of the ncRNA-binding protein family has a homologous family in yeast (10). Furthermore, it has been found that 98% of RBP homologous families are universally expressed, and their deep evolutionary protection supports their superior basic cell functions. Among them, 20% families have tissue-specific and pervasive analogs, and members of the family are enriched in certain tissues (10). RBPs bound to the same type of RNA usually affect the same tissue and exhibits similar pathology (10). Nevertheless, their molecular function is largely determined by the localization of lncRNA and RBPs (11). Given that lncRNAs represent pivotal regulators in atherosclerosis, it is not unexpected that RBPs play key roles in atherosclerosis. Similarly, it has previously been observed that the RBP human antigen R(HuR) can regulate the progress of atherosclerosis (12). Hence, we also intend to focus on the effect of RBPs for a better understanding of the molecular mechanism in atherosclerosis.
lncRNAs and RBPs are a major area of interest within the field of atherosclerosis. At present, the mechanism of lncRNAs and lncRNAs-RBPs interaction in atherosclerosis remains an area of high research interest. Therefore, it is important to elucidate these molecular interactions to better understand the underlying mechanisms of atherosclerosis. In this review, we first briefly introduce the biology of lncRNA and RBPs. Next, we discuss how lncRNAs and RBPs regulate endothelial cells, macrophages, and vascular smooth muscle cells in atherosclerosis. Finally, we provide three novel and simple interaction models between lncRNAs and RBPs.
The Biology of Long Non-coding RNA and RNA-Binding Protein
lncRNAs are defined as non-coding transcripts of more than 200 nucleotides in length, that do not translate into functional proteins (4), and they have low conservation with species (13). Based on the origin from different genomic locations, we classify lncRNAs are classified into intergenic lncRNAs (also known as lincRNA), intronic lncRNAs, sense lncRNAs, and antisense lncRNAs (14). Elements that determine the extent of lncRNA expression include core promoters, enhancers, and transposable elements (15). Like coding protein genes, most lncRNAs are transcribed by RNA polymerase II, but the promoters of non-coding protein genes have fewer overlapping transcription factor binding motifs and therefore give low levels of lncRNA expression (15). Moreover, many lncRNAs can be produced from enhancers, which are genomic binding sites encoding sequence-specific activator or repressor transcription factors (TFs) regions, and these elements often confer more tissue-specific expression (15). Transposable elements (TEs) are also an important component of lncRNAs biology. Approximately 75% of lncRNAs transcripts contain sequence elements derived from TEs. In addition, 25% of TEs overlap with the transcription start site (TSS) and poly (A) signal (PAS) of lncRNA genes (16). Thus, they are important drivers of lncRNA expression. The localization of lncRNAs within the cell determines its function (17). lncRNAs have been found to exist in the nucleolus, chromatin speckles, and paratopes (18). In addition, some lncRNAs can be transferred into the cytoplasm, further be selectively localized in the mitochondria, ribosomes, extracellular membranes, and exosomes (11).
Accumulated evidence shows that lncRNAs can bind to DNA, RNA, and proteins; change the stability and translation of cytoplasmic mRNAs; and interfere with signaling pathways (4). According to the molecular mechanism of action, lncRNAs can be divided into three subgroups: (1) lncRNA loci containing enhancers that regulate gene expression; (2) lncRNA loci whose transcriptional behavior, rather than the transcript itself, has an important role in regulating neighboring genes; and (3) lncRNA transcripts that achieve their cellular functions by interacting with DNA, other RNAs, and proteins (19, 20). However, the mechanism of lncRNA directly interacting with RNA-binding proteins have attracted our attention.
RNA-binding proteins are proteins that bind RNA through one or more RNA-binding domains (RBDs) and alter the fate or function of the bound RNA, its activity, or the expression of the target gene (21). The structures and mechanisms by which RBP binds and regulates RNA are very diverse (22). Because the structure and function of RBDs provide some insights about the binding preference and target of RBP, it is usually classified according to its specific RBDs (10). Normally, RBPs assemble with RNA to form ribonucleoprotein particles (RNPs) that mature, process, regulate, or transport RNAs (10, 23). The RBDs are the functional unit of protein-bound RNA (22). Most RBPs contain an RNA recognition motif (RRM), a K homology (KH) domain, a DEAD motif, a double-stranded RNA-binding motif (DSRM), or a zinc-finger domain (10). Additionally, some RBPs lacking conventional RNA-binding domains have been discovered (21, 22). As an RBP that needs to typically bind to AU-rich elements (ARE, core sequence 5′-AUUUA), thereby govern the fate of mRNA transcripts from biogenesis, stabilization, translation to RNA decay (24). Based on target-RNA categorization, among the 20,500 protein-coding genes in humans, nearly 39% of RBPs were involved in the non-coding RNA metabolic processes (10).
Similar to lncRNAs, RBP regulatory function in atherosclerosis has also garnered attention. Therefore, understanding the regulatory molecules of lncRNAs and RBPs in atherosclerosis as well as the interaction mode between lncRNA and RBP will lay a foundation for future understanding and prevention and treatment of atherosclerosis from the perspective of molecular biology.
lncRNA and RBP as Regulators of Atherosclerosis
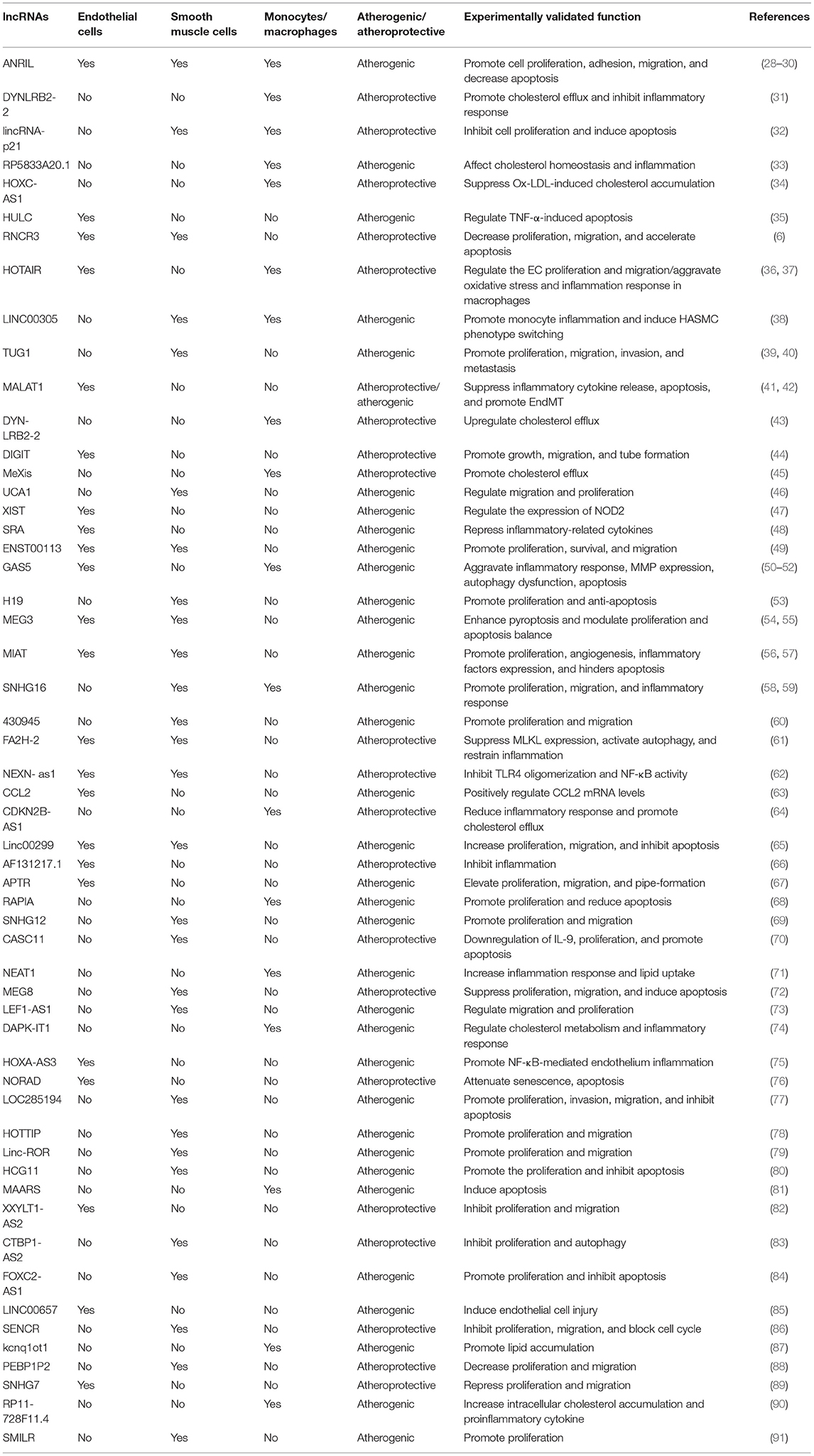
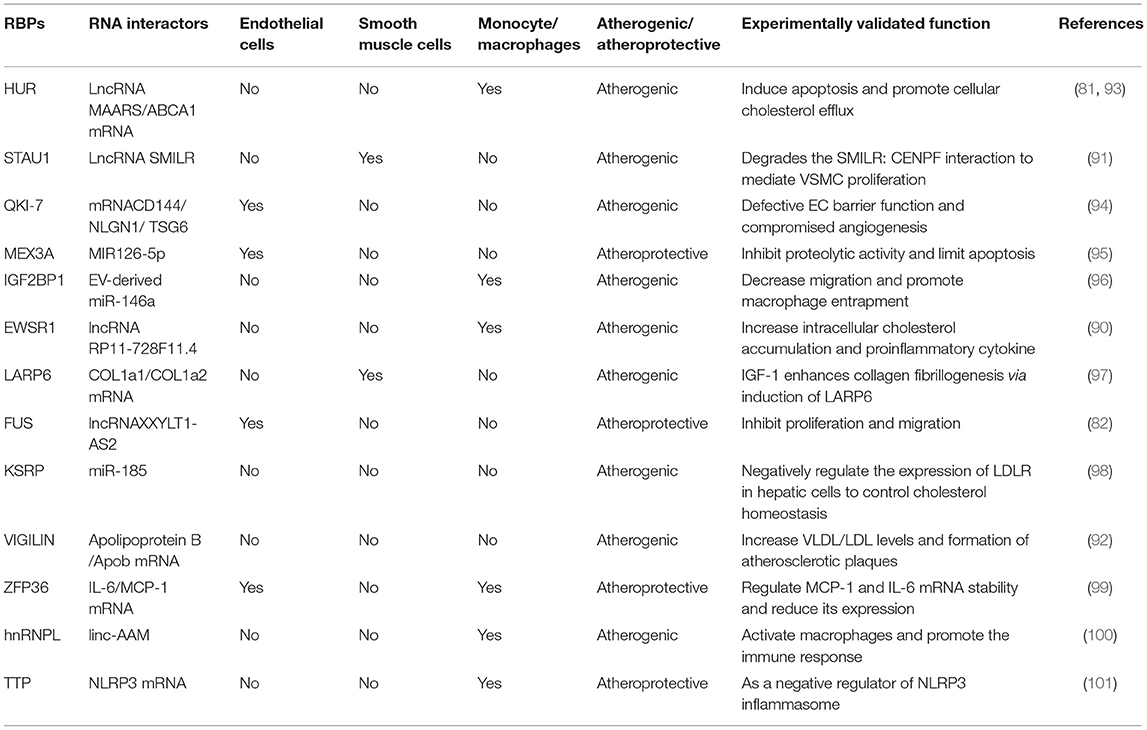
There is increasing evidence that lncRNAs and RBPs can serve as important regulators of cellular function in atherosclerosis. It is reported that lncRNA ANRIL was the first non-coding RNA identified to be associated with atherosclerosis and expressed in endothelial cells, smooth muscle cells, and immune cells (25). Nevertheless, the regulatory mechanisms of non-coding RNA in atherosclerosis were poorly studied at that time. Subsequent researchers verified that ANRIL can promote cell proliferation, migration, and inhibit apoptosis through various mechanisms, such as trans-regulation of target genes or spongy miR-399-5p, and regulation of the RAS/RAF/ERK signaling pathway (26–30). The research on regulatory mechanisms of lncRNAs in atherosclerosis has started a boom. Additionally, through a PubMed search from March 2012 through March 2021, many lncRNAs related to atherosclerosis have been discovered in the past 10 years. We have summarized their regulatory mechanisms in detail in Table 1. However, RBPs have yet to be extensively researched in atherosclerosis. The few studies that have investigated RBPs involved in atherosclerosis suggest an important role in proliferation, migration, and apoptosis. and, a small amount of RBPs have been shown to regulate cholesterol homeostasis. For example, the RBP VIGILIN can regulate the hepatic very low-density lipoprotein (VLDL) secretion, and inhibition of VIGILIN decreases hepatic VLDL secretion and circulating LDL-C levels (92). By retrieving information from PubMed up to January 2021, we summarize RBPs' regulatory mechanisms as follows (Table 2). Given that ECs, monocytes/macrophages, and VSMCs are crucial in the development of atherosclerosis, we have summed up the significance of lncRNAs and RBPs in each of these cells below.
Endothelial Cells
The vascular endothelium is a single layer of ECs that constitute the intima of arteries, veins, and capillaries (102). It is widely acknowledged that EC dysfunction is a key step in atherosclerosis initiation (103). Endothelial stimulation by NF-κB signaling increases the expression of EC adhesion molecules and promotes monocyte recruitment in the vessel wall, which can cause oxidative stress and promote the progression of inflammation (103). A large and growing body of literature has reported that lncRNA plays a role in ECs. Analysis of human umbilical vein endothelial cells after ox-LDL stimulation by lncRNA expression microarray reveals a large number of differentially expressed lncRNAs (104). Clopidogrel, a commonly antiplatelet medication, has been found to inhibit the expression of lncRNA HIF1A-AS1 to reduce EC injury (105).
More recently, several published studies have described that lncRNAs may contribute to their role in inflammation, proliferation, migration, and apoptosis. For example, Hu et al. observed that the expression of NEXN was upregulated by lncRNA NEXN-AS1 (62). Prior studies found that initiation of the TLR4/NF-κB signaling pathway would induce the expression of inflammatory molecules such as MCP1, TNF-α, and IL-6 (62, 106, 107). Theoretically, however, NEXN is a filamentous actin-binding protein (108), which can cause TLR4/NF-κB inactivity, diminish inflammatory molecules, suppresses monocyte recruitment, and prevent atherosclerosis (62). Experimental studies have also found that lncRNAs can regulate EC gene expression; for instance, lncRNA-CCL2 upregulates the levels of its adjacent CCL2 gene, which is a pro-atherosclerotic chemokine gene in the EC lines. Further, the lncRNA APTR can promote the proliferation and migration of ECs (67). HULC can promote the apoptosis of ECs (35). These are the interesting examples used to demonstrate that lncRNA regulates atherosclerotic ECs. Overall, many more studies have confirmed the value of lncRNAs to ECs, even though other mechanisms are not yet clear and need further study in the future.
Similar to lncRNAs, evidence has shown that RBPs play important roles in EC function. For example, the RBP QKI-7 promotes degradation of CD144/NLGN1/TSG6 mRNA, which plays a negative role in EC barrier function and angiogenesis, thus aggravating the progress of atherosclerosis (94). More recently, a variety of RBPs have been shown to affect autophagy, apoptosis, proliferation, and migration of ECs. MEX3A is an RBP that can form a ternary complex with AGO2 on the autophagosome surface and facilitates its nuclear localization to inhibiting proteolytic activity and limiting apoptosis (95). Another interesting study revealed that FUS can also regulate endothelial function. FUS is an atheroprotective factor that inhibits EC proliferation and migration by directly interacting with the lncRNAXXYLT1-AS2. If XXYLT1-AS2/FUS is inhibited, the expression of adhesion molecules (VCAM-1) and chemoattractant proteins (MCP-1) is increased, and monocytes can more easily adhere to ECs (82). The growing evidence reported here draws attention to the contributions of RBPs, particularly in human EC models. However, the significance of these RBPs to whole organism development and function is less well-understood.
Macrophages
During the development of atherosclerosis, macrophages differentiate from monocytes entering the intima and become macrophage-derived foam cells by taking up ox-LDL (109). At the same time, macrophages further secrete cytokines such as inflammatory cytokines (IL-1, IL6, TNF) and chemokines (CCL2, CCL5, CXCL1), and protein hydrolases to exacerbate plaque inflammation by recruiting reinforcements such as monocytes, neutrophils, and T-lymphocytes (110, 111). Macrophage are also defined as an immune cell that can express pattern recognition receptors (PRR) including NOD-like receptors, scavenger receptors, and Toll-like receptors, and are often triggered by “damage” signals from ox-LDL (109, 112). The proliferation of inflammatory macrophages and phenotypic switching are the main factors involved in the progression of atherosclerosis. Macrophages can shift toward a pro-inflammatory phenotype that we call M1 type, or to an anti-inflammatory phenotype that we call M2 type (112).
When macrophages are activated, they trigger an innate immune response. IFN-γ which belongs to Th1 cytokines and lipopolysaccharides can activate M1 macrophages to make pro-inflammatory cytokines. These proinflammatory cytokines contain TNF-α, IL-1β, IL-6, IL-12, and IL-23 (113). M2 macrophages are stimulated by Th2 cytokines that include IL-4 and IL-13, and further produce anti-inflammatory cytokines such as TGF-β and IL-10 (113, 114). Macrophages differentiate into different morphological and functional phenotypes according to changes in the microenvironment (115, 116).
Macrophage polarization is a plastic process. Pathways that have been described in the regulation of macrophage polarization include the PI3K/Akt pathway, Notch pathway, JAK-STAT pathway, TGF-β signal pathway, and Wnt/β-catenin pathway, among others. The PI3K/Akt pathway is activated by TLR4 and other pathogen recognition receptors, cytokines and chemokines, and Fc receptors, which regulate downstream signals that control cytokine production. Akt promotes M2-type macrophage polarization and inhibits M1-type polarization. It is essential for the anti-inflammatory response (117). Signal regulatory protein (SIRP) α is abundantly expressed in macrophages, and plays a critical role in regulating innate immune activation, and can as a novel target of Notch-mediated macrophage polarization (118). SOCS proteins function as feedback inhibitors for cytokines that use the JAK/STAT pathway, given that SOCS3 deficiency, IFN-γ-induced STAT1 and STAT3, and GM-CSF–induced STAT5 will be activated, promote M1 polarization (119). Conversely, IL-4 not merely activates STAT6 but promotes the differentiation of TH2 cells that stimulate M2 macrophage responses (120). Therefore, through the JAK/STAT pathway, cytokines can lead to the activation of transcription factors that dictate M1/M2 polarization and mediate inflammatory responses in macrophages. Likewise, Interleukin-4 (IL-4) regulates macrophages polarization via the TGF-β1/Smad pathway (121). The Wnt/β-catenin pathway activation promotes differentiation of macrophages in the M2 direction, which exhibits anti-inflammatory activity (122).
Increasing evidence suggests that lncRNAs play crucial roles during the differentiation of monocyte/macrophage, proliferation, decay, and phenotypic switching of macrophages. However, our insight into the contribution of lncRNAs is still in the early stages. For example, gene overexpression and knockdown experiments show that lncRNA MALAT1 has a positive effect on the pyroptosis of normal macrophages, and downregulation of lncRNA-MALAT1 can abate the pyroptosis of macrophages in rats with diabetic atherosclerosis (123). Additionally, LXR, a ligand-activated nuclear receptor, is also a transcription factor that regulates the expression of genes related to the macrophage's response to cholesterol, including ABCA1, which encodes the plasma membrane transport protein ABCA1, to promote macrophage cholesterol efflux. Sallam et al. found that the LXR-activated lncRNA Mexis increased ABCA1 protein expression levels while enhancing cholesterol efflux (45), which provides novel insights into the prevention and treatment of atherosclerosis. Similarly, the lncRNA DYNLRB2-2 exerts its atheroprotective effect by increasing cholesterol efflux and decreasing inflammatory responses (31). lncRNA HOXC-AS1 can prevent atherosclerosis by decreasing cholesterol accumulation by the mechanism of upregulating HOXC6 expression (34).
Additional studies have revealed that RBPs play a vital role in macrophage migration, apoptosis, and cholesterol regulation and is a crucial driver of atherosclerosis and integration factors of metabolic and inflammatory signals. RBP IGF2BP1 interacts with extracellular vesicle-derived miR-146a to decrease cell migration and promotes macrophage entrapment (96). A recent study identified that HuR not only interacts with ABCA1mRNA to promote cellular cholesterol efflux (93); but also interacts with lncRNA MAARS in the macrophage nucleus, preventing its shuttling to the cytosol and interfering with its RNA-stabilizing function to induce apoptosis (81). The RNA-binding protein tristetraprolin (TTP, encoded by ZFP36) has been found early in eukaryotes (124). This was characterized by the RNA-binding tandem zinc finger (TZF) domain and often be associated with cancer, as well as other inflammatory diseases (101, 125–127). Its function is as a decay signal of RNA by binding to AREs, and adenylate/uridylate-rich RNA motifs (128). As is known, the NLRP3 inflammasome drives the progression of atherosclerosis. Studies have shown that TTP possibly binds to a main ARE and inhibits the expression of NLRP3 in macrophages and other cell types, while, directly affecting TTP expression (101). However, its main test in animal experiments, further research some wonderful strategies to targeted therapy are promising.
Smooth Muscle Cells
It is known that VSMCs are the pivotal cells in the media layer of arteries. Their effects include regulation of arterial contraction, compliance, and production of extracellular matrix (ECM) (129, 130). VSMCs are essential in the stages of atherosclerotic plaque formation and are beneficial and essential for plaque stability (129, 131). Ox-LDL, proinflammatory cytokines, high levels of nitric oxide (NO), and mechanical damage can induce VSMC apoptosis. Furthermore, VSMC apoptosis contributes to plaque inflammation (131). As mentioned before, ox-LDL can stimulate VSMC necrosis and highly oxidized LDL induces necrosis, while mildly oxidized LDL induces ER stress/apoptosis (131). ECs, macrophages, and VSMCs may present autophagic activation in human atherosclerotic plaques, but this needs validation in further studies (131, 132). During atherosclerosis, VSMCs undergo complex structural and functional changes that produce a wide range of phenotypes, including foam cell formation (133).
Published studies have identified many lncRNAs that play a role in controlling VSMCs proliferation, migration, and apoptosis. For example, lncRNA SNHG16 regulates smooth muscle cell proliferation and migration through sponging miR-205 and regulating Smad2. Overexpression of SNHG16 promotes VSMC proliferation and migration, whereas downregulation of SNHG16 inhibits PDGF-bb-stimulated VSMC proliferation and migration (59). Furthermore, the imbalance between proliferation and apoptosis of VSMCs also plays a crucial role during the early stage of atherosclerosis. Li et al. identified lncRNA-MEG3 as a crucial regulator in the balance between proliferation and apoptosis of VSMCs, which could sponge miR-26a as a competing endogenous RNA (55). In addition to lncRNA-MEG3, Chen et al. revealed that targeting LOC285194 can boost cell proliferation and obstruct apoptosis (77). Another novel regulatory factor, lncRNA 430945, significantly suppressed VSMC proliferation and migration via ROR2 (60).
There are fewer RBPs that were found to play a regulatory role in atherosclerotic SMCs than lncRNAs. RNA-sequencing identified smooth muscle–induced lncRNA (SMILR) as a novel intergenic lncRNA activated by VSMCs proliferation. Recent evidence suggests that SMILR interacts with CENPF mRNA and STAU1 in the Cell Cycle Network (91). Because STAU1 is a universally expressed and multifunctional RNA-binding protein, STAU1 participates in mRNA transport and localization to mediate further translation (134).
Taken together, these studies show that lncRNAs are crucially important in the proliferation, migration, apoptosis, and senescence of ECs, VSMCs, and monocytes/macrophages, resulting in two different outcomes of promoting the progression of atherosclerosis or protecting from atherosclerosis. Given the recent curiosity in relating proliferative pathways to plaque development, lncRNAs, and RBPs may be a hopeful direction.
Emerging RBPs Interact With lncRNA in Atherosclerosis
Thus far, few RBPs associated with atherosclerosis have been found to interact with lncRNAs to exert regulatory functions. Although these RBPs are not particularly well-reported, it is worthwhile to further investigate the related aspects of the mechanism, which will be beneficial to provide a basis for future disease treatment through etiology. Collectively, the interaction of lncRNA and RBP in atherosclerosis is shown in Figure 1.
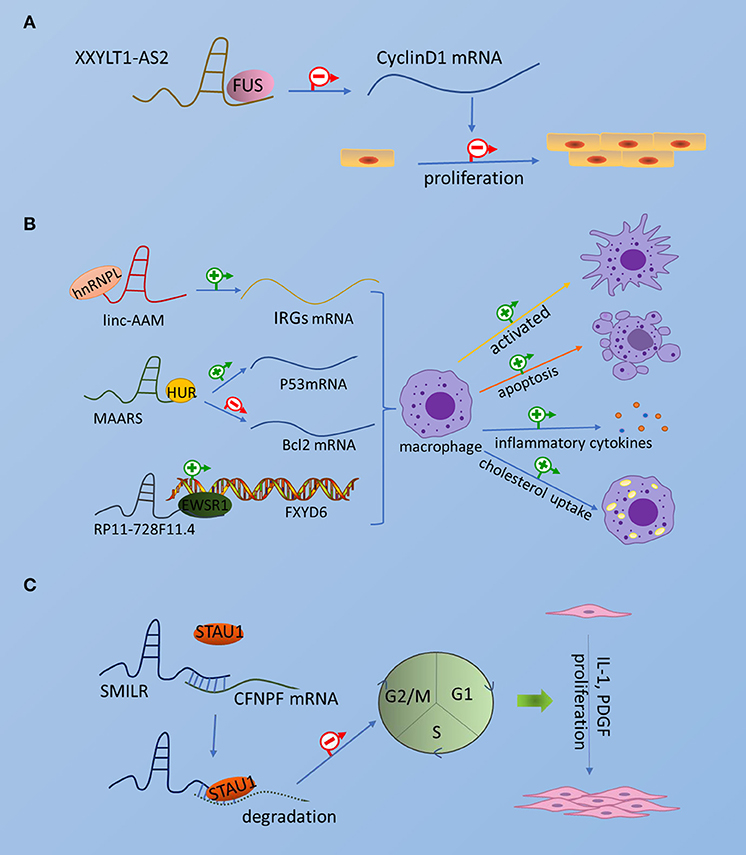
Figure 1. The interaction of lncRNA and RBP in atherosclerosis. The picture schematically represents the interaction of emerging RBPs and lncRNAs in atherosclerotic endothelial cells, smooth muscle cells, and macrophages. (A) Endothelial cells. (B) Smooth muscle cells. (C) Macrophages.
Human Antigen R
HuR (also known as Elavl1) is an RBP that is essential in cellular responses to the immune system and cell cycle (135) and has anti-apoptotic functions (136). Studies have found that HuR is expressed in endothelial cells, VSMCs, and macrophages in the atherosclerotic plaque (137). Nevertheless, the role of HuR in atherosclerosis may be highly dependent on the cell type specificity. According to Feinberg et al. (81), lncRNA MAARS can interact with HuR/ELAVL1, 14 HuR-specific AREs have been identified in the MAARS transcript and the ARE-specific binding of MAARS to HuR was confirmed. The interaction of lncRNA MAARS and HuR can induce macrophage apoptosis and decrease their efferocytosis in advanced plaques by regulating HuR targets such as p53, p27, caspase-9, and Bcl2 to alter HuR cytosolic shuttling. Interestingly, Zhang et al. (138) used smooth muscle-specific HuR knockout mice (HuRSMKO) to investigate the function of HuR in atherosclerosis. The plaque load was increased in the HuRSMKO mouse model of atherosclerosis compared with controls. It was verified that HuR could bind mRNA of adenosine 5-monophosphate-activated protein kinase (AMPK) α1 and AMPKα2, thus improving their stability and translational ability. In contrast, HuR deficiency lead to decreased p-AMPK and LC3II levels and increased p62 levels, resulting in defective autophagy. In turn, AMPK activation induces autophagy and inhibits atherosclerosis in HuRSMKO mice. However, previous studies reported a novel mechanism by which the anti-inflammatory cytokine IL-19 can decrease HuR mRNA expression, leading to decreases in mRNA stability of pro-inflammatory cytokines, to mediated atheroprotective effects. However, the lack of IL-19 leads to increased atherosclerosis (137). In addition, studies have revealed the role of endothelial HuR deficiency in attenuating atherosclerosis, and this effect may be partly due to the decreased expression of proatherogenic molecules and suppressed local inflammation (139). Collectively, HuR as direct or indirect regulators in atherosclerosis is a complex concept and needs to be further explored.
Heterogeneous Nuclear Ribonucleoproteins
The hnRNPs are a type of ribonucleoprotein (RNP) and belong to a large family of RBPs that play an essential role in the cellular nucleic acid metabolism. Their function varies depending on the cellular localization (140, 141). The hnRNPs family has many members such as hnRNPA/B, hnRNPC, hnRNPD, hnRNPE, hnRNPF/H, hnRNPG, hnRNP I (PTBP1)/L, hnRNPK, and hnRNPM/Q, hnRNP P2 (FUS/TLS), and hnRNP R/U and they have different and complex functions (140). The reported functions are as follows: alternative splicing, formation of RNP complex with pre-mRNA, mRNA stabilization, RNA transport, transcriptional and translational regulation, and inhibition of cell differentiation (140, 142). Nuclear lncRNAs interact with heterogeneous nuclear ribonucleoproteins in regulating cellular functions such as glucose and lipid metabolism, immune response, DNA damage response, and others (141).
In an atherosclerosis model, the level of hnRNPK protein was elevated in SMCs, and its subcellular localization was related to the cell cycle. Early in the cell cycle, a slight increase in cytoplasmic hnRNPK may be associated with increased neogenesis, but at the end of the cell cycle, hnRNPK accumulated in the cytoplasm and decreased in the nucleus, indicating translocation of nuclear hnRNPK to the cytoplasm and suggesting that hnRNPK regulates vascular smooth muscle proliferation (143). Chen et al. found that linC-AAM can interact with the RBP hnRNPL through the CACACA motif to activate macrophages and promote immune response; In their experiments, they localized the lncRNA in the nucleus by RNA fluorescence in situ hybridization (FISH) and detected hnRNPL by RNA pull-down assay and confirmed its activation of macrophages and promotion of immune response gene expression by linc-AAM silencing or knockout (KO) and overexpression of lncRNA (100). Studies of hnRNPC expression in atherosclerosis are particularly scarce. A previous study found that hnRNP is mainly expressed in ECs and that hnPRNC is involved in vascular cell signaling pathways activated by low physiological levels of hydrogen peroxide, which can regulate vascular cell proliferation (144). The hnRNP P2 as the RBP fused in sarcoma/translocated in liposarcoma (FUS/TLS), is also called FUS, which mutants often be reported to relate to cancer and neurodegeneration in humans (140, 145). Recently, a study found that lncRNA that interacts with FUS can regulate proliferation and migration of ECs in atherosclerosis (82).
Ewing Sarcoma Breakpoint Region 1/EWS RNA Binding Protein 1
EWSR1 plays an important role in neurodegeneration, epigenetic alteration, and cellular functions such as autophagy and mitochondrial activity (146). A growing body of evidence indicates that lncRNA is an important player in atherosclerosis. For example, researchers revealed that lncRNA RP11-728F11.4 interaction with the RNA-binding protein upregulated the cognate gene FXYD6 in atherosclerotic plaques. Knockdown or overexpression of RP11-728F11.4 affected cholesterol uptake, inflammatory molecule production, levels of lipids in monocytes-derived macrophage (90). Hence, EWSR1 is also a key regulator in atherosclerosis.
Staufen 1
STAU1 is a double-stranded (ds) RNA-binding protein known to be involved in mRNA decay. It binds dsRNA structures that are formed not only by intramolecular base-pairing of 3′UTR sequences but also by intermolecular base-pairing of 3'UTR sequences with a lncRNA via partially complementary Alu elements (134). The lncRNA SMILR is a novel intergenic lncRNA activated by VSMC proliferation and is related to atherosclerosis. It was shown to interact with centromere protein F (CENPF) mRNA to promote VSMC proliferation. Interestingly, using RNA pull-down and mass spectrometric analysis, STAU1 was found likely to bind to SMILR within the first half of its sequence, which is the predicted interaction site with CENPF. Knockdown of STAU1 upregulates the expression of SMILR and CENPF mRNA (91).
The structures and mechanisms via which RNA-binding proteins interact with transcripts are varied and complex. Not to mention the binding to lncRNAs, because lncRNAs are involved in a variety of biological functions.
The Interaction ModeL of lncRNA and RBP
RBPs Regulate the Expression of lncRNAs
Degradation, repression and overexpression of lncRNAs are important for the regulation of biological messaging; however, in addition to physiological factors (infection, tumor) (147, 148), physical factors (temperature) (149), that has been found to affect lncRNA normal expression. A few known lncRNAs have been reported to be affected by their binding protein. For example, Bachand et al. identified a class of poly(A)-binding protein nuclear 1(PABPN1) sensitive lncRNAs, PABPN1 promotes post-transcriptional regulation of sensitive lncRNAs through polyadenylation (150). Similarly, the serine/arginine-rich splicing factor 1 (SRSF1) plays a positive role on the regulation of lncRNA NEAT1 in gliomas (151). Based on RNA-Binding Protein Immunoprecipitation (RIP) analysis, the RBP SRSF1 directly interacts with NEAT1, if knock down the SRSF1, the NEAT1 would fast degradation (151). There are also reports that HuR plays a similar role in lncRNA NEAT1 (152). In short, RBPs can regulate the stability of lncRNA. Furthermore, the study found that PTB-associated splicing factor (PSF) a protein that has both RNA-binding domains and DNA-binding domains, binds to and represses the lncRNA CTBP1-AS promoter. CTBP1 expression is generally upregulated in prostate cancer, and they could be promising targets for therapeutic options of prostate cancer (153). Furthermore, Tian et al. found that TTP regulates lncRNA HOTAIR expression by a posttranscriptional mechanism. HOTAIR is a downstream target of TTP, and according to the AUUUA consensus sequence, the researcher predicted four TTP-binding sites of HOTAIR. They also used RIP to investigate the role of TTP in the regulation of HOTAIR expression (154).
Taken together, the expression of RBPs effect the level of lncRNAs, but the mechanism is yet unclear and needs further investigation, the same RBPs can regulate different lncRNAs, by contrast, the same lncRNA can be regulated by different RBPs in a different environment.
lncRNAs Regulate the Target Gene Expression by Interacting With RBPs
In addition to binding to lncRNAs to affect the expression of lncRNAs, RBPs can also affect the expression of lncRNA target genes. For example, Chen et al. found that exosomal lncRNA LNMAT2 recruited hnRNPA2B1 to the PROX1 promoter to upregulate PROX1 expression by directly interacting with hnRNPA2B1, leading to lymphangiogenesis and lymphatic metastasis in bladder cancer (155). The exosome-mediated lncRNA AFAP1-AS1 can bind to AUF1 and activate ERBB2 translation to regulate the resistance of trastuzumab (156). The lncRNA RP11-728F11.4 was shown to interact with the RBP EWSR1 and upregulate the expression of the homologous gene FXYD6, which encodes a Na+/K+- ATPase regulator and Na+-ATPase. The increased activity of Na+-ATPase, intracellular cholesterol accumulation, and pro-inflammatory cytokine production increased atherosclerotic lesions (90).
The molecular mechanisms by which lncRNAs bind to RBP to affect the expression of lncRNA target genes are complex and much is still unknown. We know that lncRNA can act as a scaffold, specifically, lncRNA acts as a structural component for nucleic acid-protein (also name ribonucleoprotein) complexes formed with proteins to connect multiple proteins to regulate the expression of their target genes. For instance, hnRNPK and YBX1 are lncRNA SCAT7-interacting proteins, and recruitment of the FGFR2 and FGFR3 promoter regions SCAT7-hnRNPK-YBX1 RNP complexes promotes transcriptional activation of the FGF/FGFR pathway, resulting in sustained cell proliferation via the PI3K/AKT and Ras/MAPK pathways (157). Further, hnRNPK has been reported to bind to lincRNA-p21 to repress the target p53 transcription (158).
lncRNAs Regulate the Activity of Their Specific Binding Proteins
The binding of lncRNA to RBP can regulate the expression of lncRNA and affect the expression of the target gene, and it can also affect the activity of the binding proteins; the outcomes are similar, in that all of them will affect the corresponding signaling pathway and thus change the regulatory outcome of the molecule. For example, the interaction between lncRNA and UPFI can regulate the expression of other mRNA but also decrease the stability of the lncRNAs. In some cases, the binding of lncRNA and UPF1 will affect the expression of UPF1, though the specific mechanisms involved need to be further investigated (159). Additionally, the lncRNA MALAT1 recruits SRSF2 and binds tightly to it, thus making the AKT2 (serine/threonine kinase2) effectively phosphorylate SRSF2 and form a stable combination of PKCδ pre-mRNA, promoting selective splicing of PKCdII in HT22 cells (160). Apart from phosphorylation modification to alter protein activity, ubiquitination is also a common modality. Studies have found that lncRNA mamRNA supports antagonistic RBPs Mmi1 and Mei2 to ensure their mutual inhibition, allowing Mmi1 to target Mei2 for ubiquitin-mediated downregulation, and in turn, allowing Mei2 accumulation to impede Mmi1 activity and fine-tune mitotic growth during meiotic mRNA degradation (161). The regulation mode between the above lncRNA and RBP is shown in Figure 2.
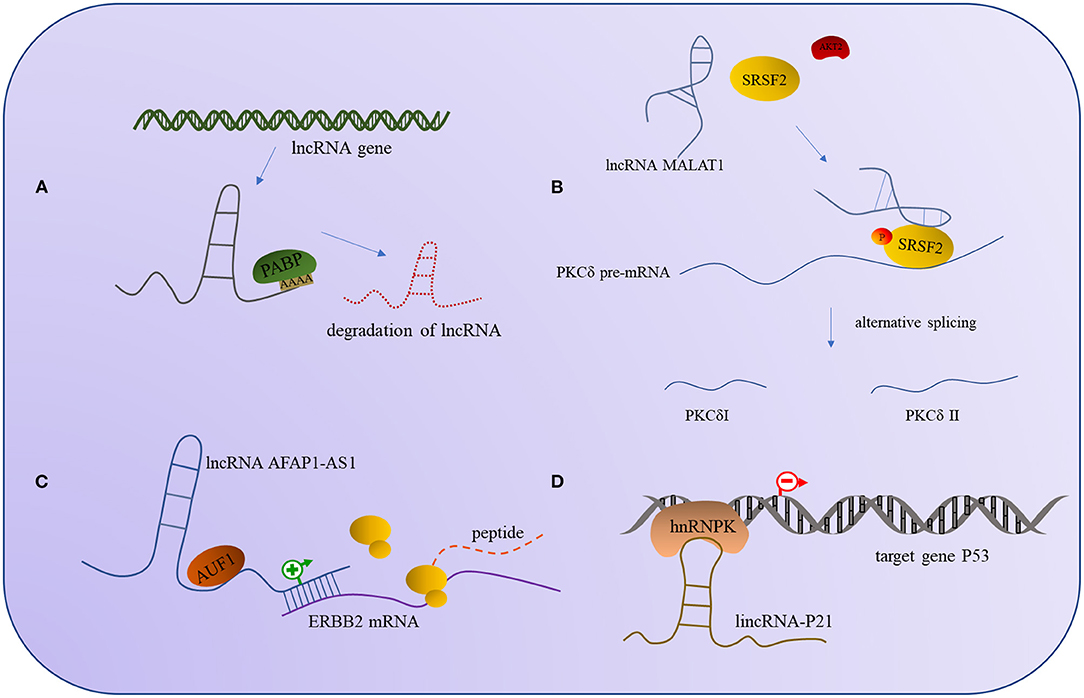
Figure 2. The interaction models between lncRNA and RNA-binding protein. (A) PABP interacts with lncRNA induces the degradation of lncRNA. (B) The interaction of lncRNA AFAP1-AS1 and RBP AUF1 can promote the expression of ERBB2 mRNA. (C) The combination of lncRNA MALAT1 and SRSF2 can promote the phosphorylation of SRSF2 by AKT2 to alternative splicing PKCδ. (D) The interaction of hnRNPK and lincRNA-P21 can repress the expression of the target gene P53.
The limitation is that there were few extensive studies so far on the mechanism of lncRNA and RBP interaction in the field of atherosclerosis; most of the model construction in this review is based on studies on tumor-specific conditions.
Conclusion and Perspective
Taken together, the studies discussed in this review show that lncRNAs and RBPs play biological functions in atherosclerotic ECs, SMCs, and macrophages; simultaneously, this review provided novel and understandable interaction models of lncRNAs and RBPs. Further extensive research is needed in the future to understand the functions and mechanisms of lncRNAs and RBPs, and provide novel and effective methods for the diagnosis, prevention and treatment of atherosclerosis.
Author Contributions
YD drafted the manuscript. RY, SZ, and QX provided insightful discussions and comments on the manuscript. HZ, XP, and XZ discussed and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81771259) and the Natural Science Foundation of Shandong Province (ZR2020MH138).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Banerjee C, Chimowitz MI. Stroke caused by atherosclerosis of the major intracranial arteries. Circul Res. (2017) 120:502–13. doi: 10.1161/CIRCRESAHA.116.308441
2. Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. (2011) 473:317–25. doi: 10.1038/nature10146
3. Falk E. Pathogenesis of atherosclerosis. J Am Coll Cardiol. (2006) 47(8 Suppl):C7–12. doi: 10.1016/j.jacc.2005.09.068
4. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. (2021) 22:96–118. doi: 10.1038/s41580-020-00315-9
5. Fasolo F, Di Gregoli K, Maegdefessel L, Johnson JL. Non-coding RNAs in cardiovascular cell biology and atherosclerosis. Cardiovasc Res. (2019) 115:1732–56. doi: 10.1093/cvr/cvz203
6. Shan K, Jiang Q, Wang XQ, Wang YN, Yang H, Yao MD, et al. Role of long non-coding RNA-RNCR3 in atherosclerosis-related vascular dysfunction. Cell Death Dis. (2016) 7:e2248. doi: 10.1038/cddis.2016.145
7. Zhang S, Wang J, Qu MJ, Wang K, Ma AJ, Pan XD, et al. Novel insights into the potential diagnostic value of circulating exosomal IncRNA-related networks in large artery atherosclerotic stroke. Front Mol Biosci. (2021) 8:682769. doi: 10.3389/fmolb.2021.682769
8. Xu S, Kamato D, Little PJ, Nakagawa S, Pelisek J, Jin ZG. Targeting epigenetics and non-coding RNAs in atherosclerosis: from mechanisms to therapeutics. Pharmacol Ther. (2019) 196:15–43. doi: 10.1016/j.pharmthera.2018.11.003
9. Ferrè F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinformatics. (2016) 17:106–16. doi: 10.1093/bib/bbv031
10. Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet. (2014) 15:829–45. doi: 10.1038/nrg3813
11. Carlevaro-Fita J, Johnson R. Global positioning system: understanding long noncoding RNAs through subcellular localization. Mol Cell. (2019) 73:869–83. doi: 10.1016/j.molcel.2019.02.008
12. Barton M, Meyer MR. HuR-ry up: how hydrogen sulfide protects against atherosclerosis. Circulation. (2019) 139:115–8. doi: 10.1161/CIRCULATIONAHA.118.036854
13. Qian X, Zhao J, Yeung PY, Zhang QC, Kwok CK. Revealing lncRNA structures and interactions by sequencing-based approaches. Trends Biochem Sci. (2019) 44:33–52. doi: 10.1016/j.tibs.2018.09.012
14. Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. (2013) 10:925–33. doi: 10.4161/rna.24604
15. Ali T, Grote P. Beyond the RNA-dependent function of LncRNA genes. Elife. (2020) 9:e60583. doi: 10.7554/eLife.60583
16. Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, et al. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet. (2013) 9:e1003470. doi: 10.1371/journal.pgen.1003470
17. Cuevas-Diaz Duran R, Wei H, Kim DH, Wu JQ. Invited review: long non-coding RNAs: important regulators in the development, function and disorders of the central nervous system. Neuropathol Appl Neurobiol. (2019) 45:538–56. doi: 10.1111/nan.12541
18. Guo CJ, Xu G, Chen LL. Mechanisms of long noncoding RNA nuclear retention. Trends Biochem Sci. (2020) 45:947–60. doi: 10.1016/j.tibs.2020.07.001
19. Constanty F, Shkumatava A. lncRNAs in development and differentiation: from sequence motifs to functional characterization. Development. (2021) 148:182741. doi: 10.1242/dev.182741
20. Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. (2017) 18:206. doi: 10.1186/s13059-017-1348-2
21. Hentze MW, Castello A, Schwarzl T, Preiss T. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol. (2018) 19:327–41. doi: 10.1038/nrm.2017.130
22. Corley M, Burns MC, Yeo GW. How RNA-binding proteins interact with RNA: molecules and mechanisms. Mol Cell. (2020) 78:9–29. doi: 10.1016/j.molcel.2020.03.011
23. Gebauer F, Schwarzl T, Valcárcel J, Hentze MW. RNA-binding proteins in human genetic disease. Nat Rev Genet. (2021) 22:185–98. doi: 10.1038/s41576-020-00302-y
24. Siang DTC, Lim YC, Kyaw AMM, Win KN, Chia SY, Degirmenci U, et al. The RNA-binding protein HuR is a negative regulator in adipogenesis. Nat Commun. (2020) 11:213. doi: 10.1038/s41467-019-14001-8
25. Broadbent HM, Peden JF, Lorkowski S, Goel A, Ongen H, Green F, et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. (2008) 17:806–14. doi: 10.1093/hmg/ddm352
26. Congrains A, Kamide K, Katsuya T, Yasuda O, Oguro R, Yamamoto K, et al. CVD-associated non-coding RNA, ANRIL, modulates expression of atherogenic pathways in VSMC. Biochem Biophys Res Commun. (2012) 419:612–6. doi: 10.1016/j.bbrc.2012.02.050
27. Congrains A, Kamide K, Oguro R, Yasuda O, Miyata K, Yamamoto E, et al. Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis. (2012) 220:449–55. doi: 10.1016/j.atherosclerosis.2011.11.017
28. Motterle A, Pu X, Wood H, Xiao Q, Gor S, Ng FL, et al. Functional analyses of coronary artery disease associated variation on chromosome 9p21 in vascular smooth muscle cells. Hum Mol Genet. (2012) 21:4021–9. doi: 10.1093/hmg/dds224
29. Holdt LM, Hoffmann S, Sass K, Langenberger D, Scholz M, Krohn K, et al. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. (2013) 9:e1003588. doi: 10.1371/journal.pgen.1003588
30. Huang T, Zhao HY, Zhang XB, Gao XL, Peng WP, Zhou Y, et al. LncRNA ANRIL regulates cell proliferation and migration via sponging miR-339-5p and regulating FRS2 expression in atherosclerosis. Eur Rev Med Pharmacol Sci. (2020) 24:1956–69. doi: 10.26355/eurrev_202002_20373
31. Hu YW, Yang JY, Ma X, Chen ZP, Hu YR, Zhao JY, et al. A lincRNA-DYNLRB2-2/GPR119/GLP-1R/ABCA1-dependent signal transduction pathway is essential for the regulation of cholesterol homeostasis. J Lipid Res. (2014) 55:681–97. doi: 10.1194/jlr.M044669
32. Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen C, et al. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation. (2014) 130:1452–65. doi: 10.1161/CIRCULATIONAHA.114.011675
33. Hu YW, Zhao JY, Li SF, Huang JL, Qiu YR, Ma X, et al. RP5-833A20.1/miR-382-5p/NFIA-dependent signal transduction pathway contributes to the regulation of cholesterol homeostasis and inflammatory reaction. Arterioscler Thromb Vasc Biol. (2015) 35:87–101. doi: 10.1161/ATVBAHA.114.304296
34. Huang C, Hu YW, Zhao JJ, Ma X, Zhang Y, Guo FX, et al. Long noncoding RNA HOXC-AS1 suppresses Ox-LDL-induced cholesterol accumulation through promoting HOXC6 expression in THP-1 macrophages. DNA Cell Biol. (2016) 35:722–9. doi: 10.1089/dna.2016.3422
35. Ma Y, Huang D, Yang F, Tian M, Wang Y, Shen D, et al. Long noncoding RNA highly upregulated in liver cancer regulates the tumor necrosis factor-α-induced apoptosis in human vascular endothelial cells. DNA Cell Biol. (2016) 35:296–300. doi: 10.1089/dna.2015.3203
36. Peng Y, Meng K, Jiang L, Zhong Y, Yang Y, Lan Y, et al. Thymic stromal lymphopoietin-induced HOTAIR activation promotes endothelial cell proliferation and migration in atherosclerosis. Biosci Rep. (2017) 37:BSR20170351. doi: 10.1042/BSR20170351
37. Liu J, Huang GQ, Ke ZP. Silence of long intergenic noncoding RNA HOTAIR ameliorates oxidative stress and inflammation response in ox-LDL-treated human macrophages by upregulating miR-330-5p. J Cell Physiol. (2019) 234:5134–42. doi: 10.1002/jcp.27317
38. Zhang DD, Wang WT, Xiong J, Xie XM, Cui SS, Zhao ZG, et al. Long noncoding RNA LINC00305 promotes inflammation by activating the AHRR-NF-κB pathway in human monocytes. Sci Rep. (2017) 7:46204. doi: 10.1038/srep46204
39. Li FP, Lin DQ, Gao LY. LncRNA TUG1 promotes proliferation of vascular smooth muscle cell and atherosclerosis through regulating miRNA-21/PTEN axis. Eur Rev Med Pharmacol Sci. (2018) 22:7439–47. doi: 10.26355/eurrev_201811_16284
40. Tang Y, Hu J, Zhong Z, Liu Y, Wang Y. Long noncoding RNA TUG1 promotes the function in ox-LDL-Treated HA-VSMCs via miR-141-3p/ROR2 Axis. Cardiovasc Ther. (2020) 2020:6758934. doi: 10.1155/2020/6758934
41. Li S, Sun Y, Zhong L, Xiao Z, Yang M, Chen M, et al. The suppression of ox-LDL-induced inflammatory cytokine release and apoptosis of HCAECs by long non-coding RNA-MALAT1 via regulating microRNA-155/SOCS1 pathway. Nutr Metab Cardiovasc Dis. (2018) 28:1175–87. doi: 10.1016/j.numecd.2018.06.017
42. Li H, Zhao Q, Chang L, Wei C, Bei H, Yin Y, et al. LncRNA MALAT1 modulates ox-LDL induced EndMT through the Wnt/beta-catenin signaling pathway. Lipids Health Dis. (2019) 18:62. doi: 10.1186/s12944-019-1006-7
43. Li Y, Shen S, Ding S, Wang L. LincRNA DYN-LRB2-2 upregulates cholesterol efflux by decreasing TLR2 expression in macrophages. J Cell Biochem. (2018) 119:1911–21. doi: 10.1002/jcb.26352
44. Miao C, Cao H, Zhang Y, Guo X, Wang Z, Wang J. LncRNA DIGIT accelerates tube formation of vascular endothelial cells by sponging miR-134. Int Heart J. (2018) 59:1086–95. doi: 10.1536/ihj.17-290
45. Sallam T, Jones M, Thomas BJ, Wu X, Gilliland T, Qian K, et al. Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA. Nat Med. (2018) 24: 304–12. doi: 10.1038/nm.4479
46. Tian S, Yuan Y, Li Z, Gao M, Lu Y, Gao H. LncRNA UCA1 sponges miR-26a to regulate the migration and proliferation of vascular smooth muscle cells. Gene. (2018) 673:159–66. doi: 10.1016/j.gene.2018.06.031
47. Xu X, Ma C, Liu C, Duan Z, Zhang L. Knockdown of long noncoding RNA XIST alleviates oxidative low-density lipoprotein-mediated endothelial cells injury through modulation of miR-320/NOD2 axis. Biochem Biophys Res Commun. (2018) 503:586–92. doi: 10.1016/j.bbrc.2018.06.042
48. Yang S, Sun J. LncRNA SRA deregulation contributes to the development of atherosclerosis by causing dysfunction of endothelial cells through repressing the expression of adipose triglyceride lipase. Mol Med Rep. (2018) 18:5207–14. doi: 10.3892/mmr.2018.9497
49. Yao X, Yan C, Zhang L, Li Y, Wan Q. LncRNA ENST00113 promotes proliferation, survival, and migration by activating PI3K/Akt/mTOR signaling pathway in atherosclerosis. Medicine. (2018) 97:e0473. doi: 10.1097/MD.0000000000010473
50. Ye J, Wang C, Wang D, Yuan H. LncRBA GSA5, up-regulated by ox-LDL, aggravates inflammatory response and MMP expression in THP-1 macrophages by acting like a sponge for miR-221. Exp Cell Res. (2018) 369:348–55. doi: 10.1016/j.yexcr.2018.05.039
51. Liang W, Fan T, Liu L, Zhang L. Knockdown of growth-arrest specific transcript 5 restores oxidized low-density lipoprotein-induced impaired autophagy flux via upregulating miR-26a in human endothelial cells. Eur J Pharmacol. (2019) 843:154–61. doi: 10.1016/j.ejphar.2018.11.005
52. Chen L, Yang W, Guo Y, Chen W, Zheng P, Zeng J, et al. Exosomal lncRNA GAS5 regulates the apoptosis of macrophages and vascular endothelial cells in atherosclerosis. PLoS ONE. (2017) 12:e0185406. doi: 10.1371/journal.pone.0185406
53. Zhang L, Cheng H, Yue Y, Li S, Zhang D, He R. H19 knockdown suppresses proliferation and induces apoptosis by regulating miR-148b/WNT/beta-catenin in ox-LDL -stimulated vascular smooth muscle cells. J Biomed Sci. (2018) 25:11. doi: 10.1186/s12929-018-0418-4
54. Zhang Y, Liu X, Bai X, Lin Y, Li Z, Fu J, et al. Melatonin prevents endothelial cell pyroptosis via regulation of long noncoding RNA MEG3/miR-223/NLRP3 axis. J Pineal Res. (2018) 64:e12449. doi: 10.1111/jpi.12449
55. Bai Y, Zhang Q, Su Y, Pu Z, Li K. Modulation of the proliferation/apoptosis balance of vascular smooth muscle cells in atherosclerosis by lncRNA-MEG3 via regulation of miR-26a/Smad1 Axis. Int Heart J. (2019) 60:444–50. doi: 10.1536/ihj.18-195
56. Zhong X, Ma X, Zhang L, Li Y, Li Y, He R. MIAT promotes proliferation and hinders apoptosis by modulating miR-181b/STAT3 axis in ox-LDL-induced atherosclerosis cell models. Biomed Pharmacother. (2018) 97:1078–85. doi: 10.1016/j.biopha.2017.11.052
57. Sun G, Li Y, Ji Z. Up-regulation of MIAT aggravates the atherosclerotic damage in atherosclerosis mice through the activation of PI3K/Akt signaling pathway. Drug Deliv. (2019) 26:641–9. doi: 10.1080/10717544.2019.1628116
58. An JH, Chen ZY, Ma QL, Wang HJ, Zhang JQ, Shi FW. LncRNA SNHG16 promoted proliferation and inflammatory response of macrophages through miR-17-5p/NF-kappaB signaling pathway in patients with atherosclerosis. Eur Rev Med Pharmacol Sci. (2019) 23:8665–77. doi: 10.26355/eurrev_201910_19184
59. Lin Y, Tian G, Zhang H, Yuan W, Xie Y, Yang Y, et al. Long non-coding RNA SNHG16 regulates human aortic smooth muscle cell proliferation and migration via sponging miR-205 and modulating Smad2. J Cell Mol Med. (2019) 23:6919–29. doi: 10.1111/jcmm.14576
60. Cui C, Wang X, Shang XM, Li L, Ma Y, Zhao GY, et al. lncRNA 430945 promotes the proliferation and migration of vascular smooth muscle cells via the ROR2/RhoA signaling pathway in atherosclerosis. Mol Med Rep. (2019) 19:4663–72. doi: 10.3892/mmr.2019.10137
61. Guo FX, Wu Q, Li P, Zheng L, Ye S, Dai XY, et al. The role of the LncRNA-FA2H-2-MLKL pathway in atherosclerosis by regulation of autophagy flux and inflammation through mTOR-dependent signaling. Cell Death Differ. (2019) 26:1670–87. doi: 10.1038/s41418-018-0235-z
62. Hu YW, Guo FX, Xu YJ, Li P, Lu ZF, McVey DG, et al. Long noncoding RNA NEXN-AS1 mitigates atherosclerosis by regulating the actin-binding protein NEXN. J Clin Invest. (2019) 129:1115–28. doi: 10.1172/JCI98230
63. Khyzha N, Khor M, DiStefano PV, Wang L, Matic L, Hedin U, et al. Regulation of CCL2 expression in human vascular endothelial cells by a neighboring divergently transcribed long noncoding RNA. Proc Natl Acad Sci USA. (2019) 116:16410–9. doi: 10.1073/pnas.1904108116
64. Li H, Han S, Sun Q, Yao Y, Li S, Yuan C, et al. Long non-coding RNA CDKN2B-AS1 reduces inflammatory response and promotes cholesterol efflux in atherosclerosis by inhibiting ADAM10 expression. Aging. (2019) 11:1695–715. doi: 10.18632/aging.101863
65. Liu Y, Chen Y, Tan L, Zhao H, Xiao N. Linc00299/miR-490-3p/AURKA axis regulates cell growth and migration in atherosclerosis. Heart Vessels. (2019) 34:1370–80. doi: 10.1007/s00380-019-01356-7
66. Lu Q, Meng Q, Qi M, Li F, Liu B. Shear-sensitive lncRNA AF131217.1 Inhibits Inflammation in HUVECs via regulation of KLF4. Hypertension. (2019) 73:e25–34. doi: 10.1161/HYPERTENSIONAHA.118.12476
67. Song A, Feng R, Gao J, Yang C. Long noncoding RNA Alu-mediated p21 transcriptional regulator promotes proliferation, migration, and pipe-formation of human microvascular endothelial cells by sponging miR-126. J Cell Biochem. (2019) 120:19858–67. doi: 10.1002/jcb.29291
68. Sun C, Fu Y, Gu X, Xi X, Peng X, Wang C, et al. Macrophage-enriched lncRNA RAPIA: a novel therapeutic target for atherosclerosis. Arterioscler Thromb Vasc Biol. (2020) 40:1464–78. doi: 10.1161/ATVBAHA.119.313749
69. Sun Y, Zhao JT, Chi BJ, Wang KF. Long noncoding RNA SNHG12 promotes vascular smooth muscle cell proliferation and migration via regulating miR-199a-5p/HIF-1alpha. Cell Biol Int. (2020) 44:1714–26. doi: 10.1002/cbin.11365
70. Tao K, Hu Z, Zhang Y, Jiang D, Cheng H. LncRNA CASC11 improves atherosclerosis by downregulating IL-9 and regulating vascular smooth muscle cell apoptosis and proliferation. Biosci Biotechnol Biochem. (2019) 83:1284–8. doi: 10.1080/09168451.2019.1597621
71. Wang L, Xia JW, Ke ZP, Zhang BH. Blockade of NEAT1 represses inflammation response and lipid uptake via modulating miR-342-3p in human macrophages THP-1 cells. J Cell Physiol. (2019) 234:5319–26. doi: 10.1002/jcp.27340
72. Zhang B, Dong Y, Zhao Z. LncRNA MEG8 regulates vascular smooth muscle cell proliferation, migration and apoptosis by targeting PPARalpha. Biochem Biophys Res Commun. (2019) 510:171–6. doi: 10.1016/j.bbrc.2019.01.074
73. Zhang L, Zhou C, Qin Q, Liu Z, Li P. LncRNA LEF1-AS1 regulates the migration and proliferation of vascular smooth muscle cells by targeting miR-544a/PTEN axis. J Cell Biochem. (2019) 120:14670–8. doi: 10.1002/jcb.28728
74. Zhen Z, Ren S, Ji H, Ding X, Zou P, Lu J. The lncRNA DAPK-IT1 regulates cholesterol metabolism and inflammatory response in macrophages and promotes atherogenesis. Biochem Biophys Res Commun. (2019) 516 1234–41. doi: 10.1016/j.bbrc.2019.06.113
75. Zhu X, Chen D, Liu Y, Yu J, Qiao L, Lin S, et al. Long noncoding RNA HOXA-AS3 integrates NF-kappaB signaling to regulate endothelium inflammation. Mol Cell Biol. (2019) 39:e00139–19. doi: 10.1128/MCB.00139-19
76. Bian W, Jing X, Yang Z, Shi Z, Chen R, Xu A, et al. Downregulation of LncRNA NORAD promotes Ox-LDL-induced vascular endothelial cell injury and atherosclerosis. Aging. (2020) 12:6385–400. doi: 10.18632/aging.103034
77. Cheng Q, Zhang M, Zhang M, Ning L, Chen D. Long non-coding RNA LOC285194 regulates vascular smooth muscle cell apoptosis in atherosclerosis. Bioengineered. (2020) 11:53–60. doi: 10.1080/21655979.2019.1705054
78. Guo X, Liu Y, Zheng X, Han Y, Cheng J. HOTTIP knockdown inhibits cell proliferation and migration via regulating miR-490-3p/HMGB1 axis and PI3K-AKT signaling pathway in ox-LDL-induced VSMCs. Life Sci. (2020) 248:117445. doi: 10.1016/j.lfs.2020.117445
79. Ji Z, Chi J, Sun H, Ru A, Ni T, Zhang J, et al. Linc-ROR targets FGF2 to regulate HASMC proliferation and migration via sponging miR-195-5p. Gene. (2020) 725:144143. doi: 10.1016/j.gene.2019.144143
80. Liu Y, Cui X, Wang C, Zhao S. LncRNA HCG11 regulates proliferation and apoptosis of vascular smooth muscle cell through targeting miR-144-3p/FOXF1 axis in atherosclerosis. Biol Res. (2020) 53:44. doi: 10.1186/s40659-020-00306-2
81. Simion V, Zhou H, Haemmig S, Pierce JB, Mendes S, Tesmenitsky Y, et al. A macrophage-specific lncRNA regulates apoptosis and atherosclerosis by tethering HuR in the nucleus. Nat Commun. (2020) 11:6135. doi: 10.1038/s41467-020-19664-2
82. Wang Q, Yang Y, Fu X, Wang Z, Liu Y, Li M, et al. Long noncoding RNA XXYLT1-AS2 regulates proliferation and adhesion by targeting the RNA binding protein FUS in HUVEC. Atherosclerosis. (2020) 298:58–69. doi: 10.1016/j.atherosclerosis.2020.02.018
83. Wang Y, Zhang CX, Ge SL, Gong WH. CTBP1AS2 inhibits proliferation and induces autophagy in oxLDLstimulated vascular smooth muscle cells by regulating miR1955p/ATG14. Int J Mol Med. (2020) 46:839–48. doi: 10.3892/ijmm.2020.4624
84. Wang YQ, Xu ZM, Wang XL, Zheng JK, Du Q, Yang JX, et al. LncRNA FOXC2-AS1 regulated proliferation and apoptosis of vascular smooth muscle cell through targeting miR-1253/FOXF1 axis in atherosclerosis. Eur Rev Med Pharmacol Sci. (2020) 24:3302–14. doi: 10.26355/eurrev_202003_20698
85. Wu H, Liu T, Hou H. Knockdown of LINC00657 inhibits ox-LDL-induced endothelial cell injury by regulating miR-30c-5p/Wnt7b/beta-catenin. Mol Cell Biochem. (2020) 472:145–55. doi: 10.1007/s11010-020-03793-9
86. Ye F, Zhang J, Zhang Q, Zhang J, Chen C. Preliminary study on the mechanism of long noncoding RNA SENCR regulating the proliferation and migration of vascular smooth muscle cells. J Cell Physiol. (2020) 235:9635–43. doi: 10.1002/jcp.29775
87. Yu XH, Deng WY, Chen JJ, Xu XD, Liu XX, Chen L, et al. LncRNA kcnq1ot1 promotes lipid accumulation and accelerates atherosclerosis via functioning as a ceRNA through the miR-452-3p/HDAC3/ABCA1 axis. Cell Death Dis. (2020) 11:1043. doi: 10.1038/s41419-020-03263-6
88. He X, Lian Z, Yang Y, Wang Z, Fu X, Liu Y, et al. Long Non-coding RNA PEBP1P2 suppresses proliferative VSMCs phenotypic switching and proliferation in atherosclerosis. Mol Ther Nucleic Acids. (2020) 22:84–98. doi: 10.1016/j.omtn.2020.08.013
89. Zhang S, Zhu X, Li G. E2F1/SNHG7/miR-186-5p/MMP2 axis modulates the proliferation and migration of vascular endothelial cell in atherosclerosis. Life Sci. (2020) 257:118013. doi: 10.1016/j.lfs.2020.118013
90. Dong XH, Lu ZF, Kang CM, Li XH, Haworth KE, Ma X, et al. The long noncoding RNA RP11-728F11.4 promotes atherosclerosis. Arterioscler Thromb Vasc Biol. (2021) 41:1191–204. doi: 10.1161/ATVBAHA.120.315114
91. Mahmoud AD, Ballantyne MD, Miscianinov V, Pinel K, Hung J, Scanlon JP, et al. The human-specific and smooth muscle cell-enriched LncRNA SMILR promotes proliferation by regulating mitotic CENPF mRNA and drives cell-cycle progression which can be targeted to limit vascular remodeling. Circul Res. (2019) 125:535–51. doi: 10.1161/CIRCRESAHA.119.314876
92. Mobin MB, Gerstberger S, Teupser D, Campana B, Charisse K, Heim MH, et al. The RNA-binding protein vigilin regulates VLDL secretion through modulation of Apob mRNA translation. Nat Commun. (2016) 7:12848. doi: 10.1038/ncomms12848
93. Ramírez CM, Lin CS, Abdelmohsen K, Goedeke L, Yoon JH, Madrigal-Matute J, et al. RNA binding protein HuR regulates the expression of ABCA1. J Lipid Res. (2014) 55:1066–76. doi: 10.1194/jlr.M044925
94. Yang C, Eleftheriadou M, Kelaini S, Morrison T, González MV, Caines R, et al. Targeting QKI-7 in vivo restores endothelial cell function in diabetes. Nat Commun. (2020) 11:3812. doi: 10.1038/s41467-020-17468-y
95. Santovito D, Egea V, Bidzhekov K, Natarelli L, Mourão A, Blanchet X, et al. Autophagy unleashes noncanonical microRNA functions. Autophagy. (2020) 16:2294–6. doi: 10.1080/15548627.2020.1830523
96. Nguyen MA, Karunakaran D, Geoffrion M, Cheng HS, Tandoc K, Perisic Matic L, et al. Extracellular vesicles secreted by atherogenic macrophages transfer microRNA to inhibit cell migration. Arterioscler Thromb Vasc Biol. (2018) 38:49–63. doi: 10.1161/ATVBAHA.117.309795
97. Blackstock CD, Higashi Y, Sukhanov S, Shai SY, Stefanovic B, Tabony AM, et al. Insulin-like growth factor-1 increases synthesis of collagen type I via induction of the mRNA-binding protein LARP6 expression and binding to the 5' stem-loop of COL1a1 and COL1a2 mRNA. J Biol Chem. (2014) 289:7264–74. doi: 10.1074/jbc.M113.518951
98. Jiang H, Zhang J, Du Y, Jia X, Yang F, Si S, et al. microRNA-185 modulates low density lipoprotein receptor expression as a key posttranscriptional regulator. Atherosclerosis. (2015) 243:523–32. doi: 10.1016/j.atherosclerosis.2015.10.026
99. Zhang H, Taylor WR, Joseph G, Caracciolo V, Gonzales DM, Sidell N, et al. mRNA-binding protein ZFP36 is expressed in atherosclerotic lesions and reduces inflammation in aortic endothelial cells. Arterioscler Thromb Vasc Biol. (2013) 33:1212–20. doi: 10.1161/ATVBAHA.113.301496
100. Chen X, He Y, Zhu Y, Du J, Sun H. linc-AAM facilitates gene expression contributing to macrophage activation and adaptive immune responses. Cell Rep. (2021) 34:108584. doi: 10.1016/j.celrep.2020.108584
101. Haneklaus M, O'Neil JD, Clark AR, Masters SL, O'Neill LAJ. The RNA-binding protein Tristetraprolin (TTP) is a critical negative regulator of the NLRP3 inflammasome. J Biol Chem. (2017) 292:6869–81. doi: 10.1074/jbc.M116.772947
102. Kruger-Genge A, Blocki A, Franke RP, Jung F. Vascular endothelial cell biology: an update. Int J Mol Sci. (2019) 20:4411. doi: 10.3390/ijms20184411
103. Eelen G, de Zeeuw P, Treps L, Harjes U, Wong BW, Carmeliet P. Endothelial cell metabolism. Physiol Rev. (2018) 98:3–58. doi: 10.1152/physrev.00001.2017
104. Singh KK, Matkar PN, Pan Y, Quan A, Gupta V, Teoh H, et al. Endothelial long non-coding RNAs regulated by oxidized LDL. Mol Cell Biochem. (2017) 431:139–49. doi: 10.1007/s11010-017-2984-2
105. Wang J, Chen L, Li H, Yang J, Gong Z, Wang B, et al. Clopidogrel reduces apoptosis and promotes proliferation of human vascular endothelial cells induced by palmitic acid via suppression of the long non-coding RNA HIF1A-AS1 in vitro. Mol Cell Biochem. (2015) 404:203–10. doi: 10.1007/s11010-015-2379-1
106. Hennessy EJ, Parker AE, O'Neill LA. Targeting toll-like receptors: emerging therapeutics? Nat Rev Drug Discov. (2010) 9:293–307. doi: 10.1038/nrd3203
107. Michelsen KS, Wong MH, Shah PK, Zhang W, Yano J, Doherty TM, et al. Lack of Toll-like receptor 4 or myeloid differentiation factor 88 reduces atherosclerosis and alters plaque phenotype in mice deficient in apolipoprotein E. Proc Natl Acad Sci USA. (2004) 101:10679–84. doi: 10.1073/pnas.0403249101
108. Ohtsuka T, Nakanishi H, Ikeda W, Satoh A, Momose Y, Nishioka H, et al. Nexilin: a novel actin filament-binding protein localized at cell-matrix adherens junction. J Cell Biol. (1998) 143:1227–38. doi: 10.1083/jcb.143.5.1227
109. Moore KJ, Sheedy FJ, Fisher EA. Macrophages in atherosclerosis: a dynamic balance. Nat Rev Immunol. (2013) 13:709–21. doi: 10.1038/nri3520
110. Tong Y, Cai L, Yang S, Liu S, Wang Z, Cheng J. The research progress of vascular macrophages and atherosclerosis. Oxid Med Cell Longev. (2020) 2020:7308736. doi: 10.1155/2020/7308736
111. Tabas I, Lichtman AH. Monocyte-macrophages and T Cells in atherosclerosis. Immunity. (2017) 47:621–34. doi: 10.1016/j.immuni.2017.09.008
112. Schultze JL, Schmieder A, Goerdt S. Macrophage activation in human diseases. Semin Immunol. (2015) 27:249–56. doi: 10.1016/j.smim.2015.07.003
113. Shapouri-Moghaddam A, Mohammadian S, Vazini H, Taghadosi M, Esmaeili SA, Mardani F, et al. Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol. (2018) 233:6425–40. doi: 10.1002/jcp.26429
114. Jinnouchi H, Guo L, Sakamoto A, Torii S, Sato Y, Cornelissen A, et al. Diversity of macrophage phenotypes and responses in atherosclerosis. Cell Mol Life Sci. (2020) 77:1919–32. doi: 10.1007/s00018-019-03371-3
115. Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. (2005) 5:953–64. doi: 10.1038/nri1733
116. Guilliams M, Mildner A, Yona S. Developmental and functional heterogeneity of monocytes. Immunity. (2018) 49:595–613. doi: 10.1016/j.immuni.2018.10.005
117. Vergadi E, Ieronymaki E, Lyroni K, Vaporidi K, Tsatsanis C. Akt signaling pathway in macrophage activation and M1/M2 polarization. J Immunol. (2017) 198:1006–14. doi: 10.4049/jimmunol.1601515
118. Lin Y, Zhao JL, Zheng QJ, Jiang X, Tian J, Liang SQ, et al. Notch signaling modulates macrophage polarization and phagocytosis through direct suppression of signal regulatory protein α expression. Front Immunol. (2018) 9:1744. doi: 10.3389/fimmu.2018.01744
119. Qin H, Holdbrooks AT, Liu Y, Reynolds SL, Yanagisawa LL, Benveniste EN. SOCS3 deficiency promotes M1 macrophage polarization and inflammation. J Immunol. (2012) 189:3439–48. doi: 10.4049/jimmunol.1201168
120. O'Shea JJ, Pesu M, Borie DC, Changelian PS. A new modality for immunosuppression: targeting the JAK/STAT pathway. Nat Rev Drug Discov. (2004) 3:555–64. doi: 10.1038/nrd1441
121. Zhang J, Shi H, Zhang N, Hu L, Jing W, Pan J. Interleukin-4-loaded hydrogel scaffold regulates macrophages polarization to promote bone mesenchymal stem cells osteogenic differentiation via TGF-β1/Smad pathway for repair of bone defect. Cell Prolif. (2020) 53:e12907. doi: 10.1111/cpr.12907
122. Abaricia JO, Shah AH, Chaubal M, Hotchkiss KM, Olivares-Navarrete R. Wnt signaling modulates macrophage polarization and is regulated by biomaterial surface properties. Biomaterials. (2020) 243:119920. doi: 10.1016/j.biomaterials.2020.119920
123. Han Y, Qiu H, Pei X, Fan Y, Tian H, Geng J. Low-dose sinapic acid abates the pyroptosis of macrophages by downregulation of lncRNA-MALAT1 in rats with diabetic atherosclerosis. J Cardiovasc Pharmacol. (2018) 71:104–12. doi: 10.1097/FJC.0000000000000550
124. Wells ML, Perera L, Blackshear PJ. An ancient family of RNA-binding proteins: still important! Trends Biochem Sci. (2017) 42:285–96. doi: 10.1016/j.tibs.2016.12.003
125. Patial S, Blackshear PJ. Tristetraprolin as a therapeutic target in inflammatory disease. Trends Pharmacol Sci. (2016) 37:811–21. doi: 10.1016/j.tips.2016.07.002
126. Lai WS, Wells ML, Perera L, Blackshear PJ. The tandem zinc finger RNA binding domain of members of the tristetraprolin protein family. Wiley Interdiscip Rev RNA. (2019) 10:e1531. doi: 10.1002/wrna.1531
127. Saini Y, Chen J, Patial S. The tristetraprolin family of RNA-binding proteins in cancer: progress and future prospects. Cancers. (2020) 12:1539. doi: 10.3390/cancers12061539
128. Jonas K, Calin GA, Pichler M. RNA-Binding proteins as important regulators of long non-coding RNAs in cancer. Int J Mol Sci. (2020) 21:2969. doi: 10.3390/ijms21082969
129. Basatemur GL, Jørgensen HF, Clarke MCH, Bennett MR, Mallat Z. Vascular smooth muscle cells in atherosclerosis. Nat Rev Cardiol. (2019) 16:727–44. doi: 10.1038/s41569-019-0227-9
130. Hu D, Yin C, Luo S, Habenicht AJR, Mohanta SK. Vascular smooth muscle cells contribute to atherosclerosis immunity. Front Immunol. (2019) 10:1101. doi: 10.3389/fimmu.2019.01101
131. Grootaert MOJ, Moulis M, Roth L, Martinet W, Vindis C, Bennett MR, et al. Vascular smooth muscle cell death, autophagy and senescence in atherosclerosis. Cardiovasc Res. (2018) 114:622–34. doi: 10.1093/cvr/cvy007
132. Perrotta I. The use of electron microscopy for the detection of autophagy in human atherosclerosis. Micron. (2013) 50:7–13. doi: 10.1016/j.micron.2013.03.007
133. Allahverdian S, Chaabane C, Boukais K, Francis GA, Bochaton-Piallat ML. Smooth muscle cell fate and plasticity in atherosclerosis. Cardiovasc Res. (2018) 114:540–50. doi: 10.1093/cvr/cvy022
134. Park E, Maquat LE. Staufen-mediated mRNA decay. Wiley Interdiscip Rev RNA. (2013) 4:423–35. doi: 10.1002/wrna.1168
135. Sun Q, Tripathi V, Yoon JH, Singh DK, Hao Q, Min KW, et al. MIR100 host gene-encoded lncRNAs regulate cell cycle by modulating the interaction between HuR and its target mRNAs. Nucleic Acids Res. (2018) 46:10405–16. doi: 10.1093/nar/gky696
136. Abdelmohsen K, Lal A, Kim HH, Gorospe M. Posttranscriptional orchestration of an anti-apoptotic program by HuR. Cell Cycle. (2007) 6:1288–92. doi: 10.4161/cc.6.11.4299
137. Ray M, Gabunia K, Vrakas CN, Herman AB, Kako F, Kelemen SE, et al. Genetic deletion of IL-19 (Interleukin-19) exacerbates atherogenesis in Il19(-/-) × Ldlr(-/-) double knockout mice by dysregulation of mRNA stability protein HuR (Human Antigen R). Arterioscler Thromb Vasc Biol. (2018) 38:1297–308. doi: 10.1161/ATVBAHA.118.310929
138. Liu S, Jiang X, Cui X, Wang J, Liu S, Li H, et al. Smooth muscle-specific HuR knockout induces defective autophagy and atherosclerosis. Cell Death Dis. (2021) 12:385. doi: 10.1038/s41419-021-03671-2
139. Fu X, Zhai S, Yuan J. Endothelial HuR deletion reduces the expression of proatherogenic molecules and attenuates atherosclerosis. Int Immunopharmacol. (2018) 65:248–55. doi: 10.1016/j.intimp.2018.09.023
140. Geuens T, Bouhy D, Timmerman V. The hnRNP family: insights into their role in health and disease. Hum Genet. (2016) 135:851–67. doi: 10.1007/s00439-016-1683-5
141. Sun X, Haider Ali MSS, Moran M. The role of interactions of long non-coding RNAs and heterogeneous nuclear ribonucleoproteins in regulating cellular functions. Biochem J. (2017) 474:2925–35. doi: 10.1042/BCJ20170280
142. Fabbiano F, Corsi J, Gurrieri E, Trevisan C, Notarangelo M, D'Agostino VG. RNA packaging into extracellular vesicles: an orchestra of RNA-binding proteins? J Extracell Vesicles. (2020) 10:e12043. doi: 10.1002/jev2.12043
143. Laury-Kleintop LD, Tresini M, Hammond O. Compartmentalization of hnRNP-K during cell cycle progression and its interaction with calponin in the cytoplasm. J Cell Biochem. (2005) 95:1042–56. doi: 10.1002/jcb.20486
144. Panchenko MP, Silva N, Stone JR. Up-regulation of a hydrogen peroxide-responsive pre-mRNA binding protein in atherosclerosis and intimal hyperplasia. Cardiovasc Pathol. (2009) 18:167–72. doi: 10.1016/j.carpath.2008.03.008
145. Brooke GN, Culley RL, Dart DA, Mann DJ, Gaughan L, McCracken SR, et al. FUS/TLS is a novel mediator of androgen-dependent cell-cycle progression and prostate cancer growth. Cancer Res. (2011) 71:914–24. doi: 10.1158/0008-5472.CAN-10-0874
146. Lee J, Nguyen PT, Shim HS, Hyeon SJ, Im H, Choi MH, et al. EWSR1, a multifunctional protein, regulates cellular function and aging via genetic and epigenetic pathways. Biochim Biophys Acta Mol Basis Dis. (2019) 1865:1938–45. doi: 10.1016/j.bbadis.2018.10.042
147. Rodrigues AC, Adamoski D, Genelhould G, Zhen F, Yamaguto GE, Araujo-Souza PS, et al. NEAT1 and MALAT1 are highly expressed in saliva and nasopharyngeal swab samples of COVID-19 patients. Mol Oral Microbiol. (2021). doi: 10.1111/omi.12351. [Epub ahead of print].
148. Marin-Bejar O, Mas AM, Gonzalez J, Martinez D, Athie A, Morales X, et al. The human lncRNA LINC-PINT inhibits tumor cell invasion through a highly conserved sequence element. Genome Biol. (2017) 18:202. doi: 10.1186/s13059-017-1331-y
149. Chen HS, Tong HS, Zhao Y, Hong CY, Bin JP, Su L. Differential expression pattern of exosome long non-coding RNAs (lncRNAs) and MicroRNAs (miRNAs) in vascular endothelial cells under heat stroke. Med Sci Monit. (2018) 24:7965–74. doi: 10.12659/MSM.909983
150. Beaulieu YB, Kleinman CL, Landry-Voyer AM, Majewski J, Bachand F. Polyadenylation-dependent control of long noncoding RNA expression by the poly(A)-binding protein nuclear 1. PLoS Genet. (2012) 8:e1003078. doi: 10.1371/journal.pgen.1003078
151. Zhou X, Li X, Yu L, Wang R, Hua D, Shi C, et al. The RNA-binding protein SRSF1 is a key cell cycle regulator via stabilizing NEAT1 in glioma. Int J Biochem Cell Biol. (2019) 113:75–86. doi: 10.1016/j.biocel.2019.06.003
152. Chai Y, Liu J, Zhang Z, Liu L. HuR-regulated lncRNA NEAT1 stability in tumorigenesis and progression of ovarian cancer. Cancer Med. (2016) 5:1588–98. doi: 10.1002/cam4.710
153. Takayama K, Horie-Inoue K, Katayama S, Suzuki T, Tsutsumi S, Ikeda K, et al. Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J. (2013) 32:1665–80. doi: 10.1038/emboj.2013.99
154. Tian FJ, He XY, Wang J, Li X, Ma XL, Wu F, et al. Elevated tristetraprolin impairs trophoblast invasion in women with recurrent miscarriage by destabilization of HOTAIR. Mol Ther Nucleic Acids. (2018) 12:600–9. doi: 10.1016/j.omtn.2018.07.001
155. Chen C, Luo Y, He W, Zhao Y, Kong Y, Liu H, et al. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J Clin Invest. (2020) 130:404–21. doi: 10.1172/JCI130892
156. Han M, Gu Y, Lu P, Li J, Cao H, Li X, et al. Exosome-mediated lncRNA AFAP1-AS1 promotes trastuzumab resistance through binding with AUF1 and activating ERBB2 translation. Mol Cancer. (2020) 19:26. doi: 10.1186/s12943-020-1145-5
157. Ali MM, Akhade VS, Kosalai ST, Subhash S, Statello L, Meryet-Figuiere M, et al. PAN-cancer analysis of S-phase enriched lncRNAs identifies oncogenic drivers and biomarkers. Nat Commun. (2018) 9:883. doi: 10.1038/s41467-018-03265-1
158. Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. (2010) 142:409–19. doi: 10.1016/j.cell.2010.06.040
159. He J, Ma X. Interaction between LncRNA and UPF1 in tumors. Front Genet. (2021) 12:624905. doi: 10.3389/fgene.2021.624905
160. El Bassit G, Patel RS, Carter G, Shibu V, Patel AA, Song S, et al. MALAT1 in human adipose stem cells modulates survival and alternative splicing of PKCδII in HT22 cells. Endocrinology. (2017) 158:183–95. doi: 10.1210/en.2016-1819
Keywords: long non-coding RNA, RNA-binding protein, atherosclerosis, endothelial cells, macrophages, smooth muscle cells
Citation: Ding Y, Yin R, Zhang S, Xiao Q, Zhao H, Pan X and Zhu X (2021) The Combined Regulation of Long Non-coding RNA and RNA-Binding Proteins in Atherosclerosis. Front. Cardiovasc. Med. 8:731958. doi: 10.3389/fcvm.2021.731958
Received: 30 June 2021; Accepted: 07 October 2021;
Published: 02 November 2021.
Edited by:
Lingfang Zeng, King's College London, United KingdomReviewed by:
Andriana Margariti, Queen's University Belfast, United KingdomViorel Simion, Dewpoint Therapeutics, United States
Copyright © 2021 Ding, Yin, Zhang, Xiao, Zhao, Pan and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongqin Zhao, emhhb2hvbmdxaW55eUAxNjMuY29t; Xudong Pan, ZHJwYW4wMjJAMTYzLmNvbQ==; Xiaoyan Zhu, enh5c2RqbUAxNjMuY29t; emh1eGlhb3lhbkBxZHUuZWR1LmNu
 Yuanyuan Ding1
Yuanyuan Ding1 Shuai Zhang
Shuai Zhang Xiaoyan Zhu
Xiaoyan Zhu