- Department of Pediatrics, Indiana University School of Medicine, Indianapolis, IN, United States
With the large-scale genome-wide sequencing, long non-coding RNAs (lncRNAs) have been found to compose of a large portion of the human transcriptome. Recent studies demonstrated the multidimensional functions of lncRNAs in heart development and disease. The subcellular localization of lncRNA is considered as a key factor that determines lncRNA function. Cytosolic lncRNAs mainly regulate mRNA stability, mRNA translation, miRNA processing and function, whereas nuclear lncRNAs epigenetically regulate chromatin remodeling, structure, and gene transcription. In this review, we summarize the molecular mechanisms of cytosolic and nuclear lncRNAs in heart development and disease separately, and emphasize the recent progress to dictate the crosstalk of cytosolic and nuclear lncRNAs in orchestrating the same biological process. Given the low evolutionary conservation of most lncRNAs, deeper understanding of human lncRNA will uncover a new layer of human regulatory mechanism underlying heart development and disease, and benefit the future clinical treatment for human heart disease.
Introduction
The heart is a central organ of the circulatory system, which pumps blood and drives oxygen and nutrients throughout the whole body. According to American Heart Association, heart disease is one of the leading causes of death in the United States. Approximately 655,000 Americans die of heart disease each year (1). Although heart disease could be caused by various factors, the most direct and common reason has been recognized as genetic variations in coding genes. During the past decade, accumulated evidence demonstrates that non-coding RNAs (ncRNAs) are also highly relevant to cardiovascular diseases (2). Non-coding RNAs are transcripts without prominent protein coding potential, which include two major groups, short non-coding RNAs (sncRNAs) and long non-coding RNAs (lncRNAs) (3). SncRNAs include transfer RNAs (t-RNAs), ribosomal RNAs (r-RNAs), small nuclear RNAs (snRNAs), microRNAs (miRNAs), small interfering RNAs (siRNAs) and P-element-induced wimpy testis (PIWI) interacting RNAs (piRNAs). SncRNAs are broadly involved in transcriptional and translational regulations (3). LncRNAs are over 200 bp transcripts and lncRNA genes compose a large portion of the human genome. LncRNAs display multidimensional functions at various regulatory levels, such as histone modification, DNA methylation, gene transcription, post-transcription, translation, RNA and protein stability (4). Many lncRNAs have been reported to be involved in cardiovascular development and disease, although their underlying molecular mechanisms in pathological process remain elusive (5). Therefore, understanding the roles of lncRNAs in heart development and disease will reveal the molecular basis of cardiogenesis, and the molecular etiology of human cardiovascular diseases. For example, a conserved lncRNA H19 represses cardiac hypertrophy by preventing and reversing experimental pressure-overload-induced heart failure, and Duchenne and Becker muscular dystrophy associated cardiomyopathy (6, 7). Hence, we summarize the current knowledge of characterized lncRNA mechanisms in heart development and disease (Table 1), and further discuss the clinical potential of lncRNA in heart disease therapy.
Long Non-Coding RNA Functions in Heart Development and Disease
The establishment of in vitro cardiomyocyte (CM) differentiation from mouse and human pluripotent stem cells (hPSCs) allows modeling early events of cardiogenesis in dish. Furthermore, whole transcriptomic profiling and CRISPR/Cas-9 mediated approaches have paved the way toward discovering and functional assessment of crucial lncRNAs in early human cardiac development by using hPSCs (45). Currently, hundreds of lncRNAs have been identified in the human cardiac precursor cells (CPCs), such as cardiac mesoderm enhancer-associated non-coding RNA (Carmen), which promotes cardiac specification and differentiation of CPCs (46). A human-specific lncRNA, Heart Brake LncRNA 1 (HBL1), represses CM differentiation from human hPSCs via counteracting miR-1 function (8). A mouse-specific lncRNA, Braveheart (Bvht), is required for the commitment of nascent mesoderm toward a cardiac fate (33). A heart field related lncRNA, Linc1405, controls cardiac mesoderm specification and cardiogenesis in mESC and in vivo (42). A lateral mesoderm-specific lncRNA Fendrr (FOXF1 Adjacent Non-Coding Developmental Regulatory RNA) plays an essential role in heart and body wall development in vivo (35). In addition to control early cardiac lineage specification, lncRNAs also play important roles in CM maturation and proliferation via various mechanisms, such as regulating the expression ratio of Myh6/Myh7 (47), sarcomere organization (43), cardiac myogenesis (36), metabolic maturation (44, 48) and cardiac conduction (27, 31, 49). LncRNA Mhrt (myosin heavy-chain-associated RNA transcripts) is required for maintaining the ratio of Myh6/Myh7 during mouse heart development and maturation, which is important for CM maturation (47). LncRNA CPR (cardiomyocyte proliferation regulator) induces hypertrophic responses of mature CMs, including increased sarcomere organization and CM surface area (43).
Evidence of the association between deregulation of lncRNAs and heart diseases has been reports for various cardiovascular disease models, such as cardiac hypotrophy (6), muscular dystrophy (7), coronary artery disease (CAD) (50–52), myocardial infarction (32, 53), diabetic cardiomyopathy (54), non-Ischemic cardiomyopathy (NICM) and heart failure (55). Murine and human lncRNA H19 display an anti-hypotrophy function, and CM-restricted H19 gene delivery can suppress the development of cardiac hypertrophy and later on heart failure (6). Recently, Zhang et al. found that H19 inhibits dystrophin degradation, preserves skeletal and cardiac muscle histology, and improves cardiomyocyte strength and heart function in muscular dystrophy cells and murine model (7). H19 also suppresses apoptosis and autophagy of CMs under diabetic condition (12, 56). In myocardial infarction, lncRNA Meg3 is upregulated in infarcted mouse heart and promotes CM death (32). Although a large number of lncRNAs have been found to be associated with heart development and disease (Table 1), the mechanisms of most lncRNAs remain elusive. Particularly, the deeper understating of lncRNA mechanisms will shed light on the clinical potential of lncRNAs, with the findings of novel therapeutic targets or druggable lncRNAs. Interestingly, many lncRNAs show restricted expression patterns in the cytoplasm or nucleus although some lncRNAs express in both, suggesting the differential functions executed by lncRNAs in different subcellular localizations, which are summarized in the following sections.
Mechanisms of Long Non-Coding RNAs in Cytoplasm
The subcellular localization is considered as a key factor determining lncRNA function (57, 58). Although the nucleus is the location for RNA biogenesis and processing, many mature lncRNAs are transported into cytoplasm, showing high cytosolic expressing levels (59). In the cytoplasm, lncRNA-mediated mechanisms have been found to mainly regulate mRNA stability, translation of mRNA, and microRNA (miRNA) related functions (60).
Long Non-coding RNA Counteracts microRNA
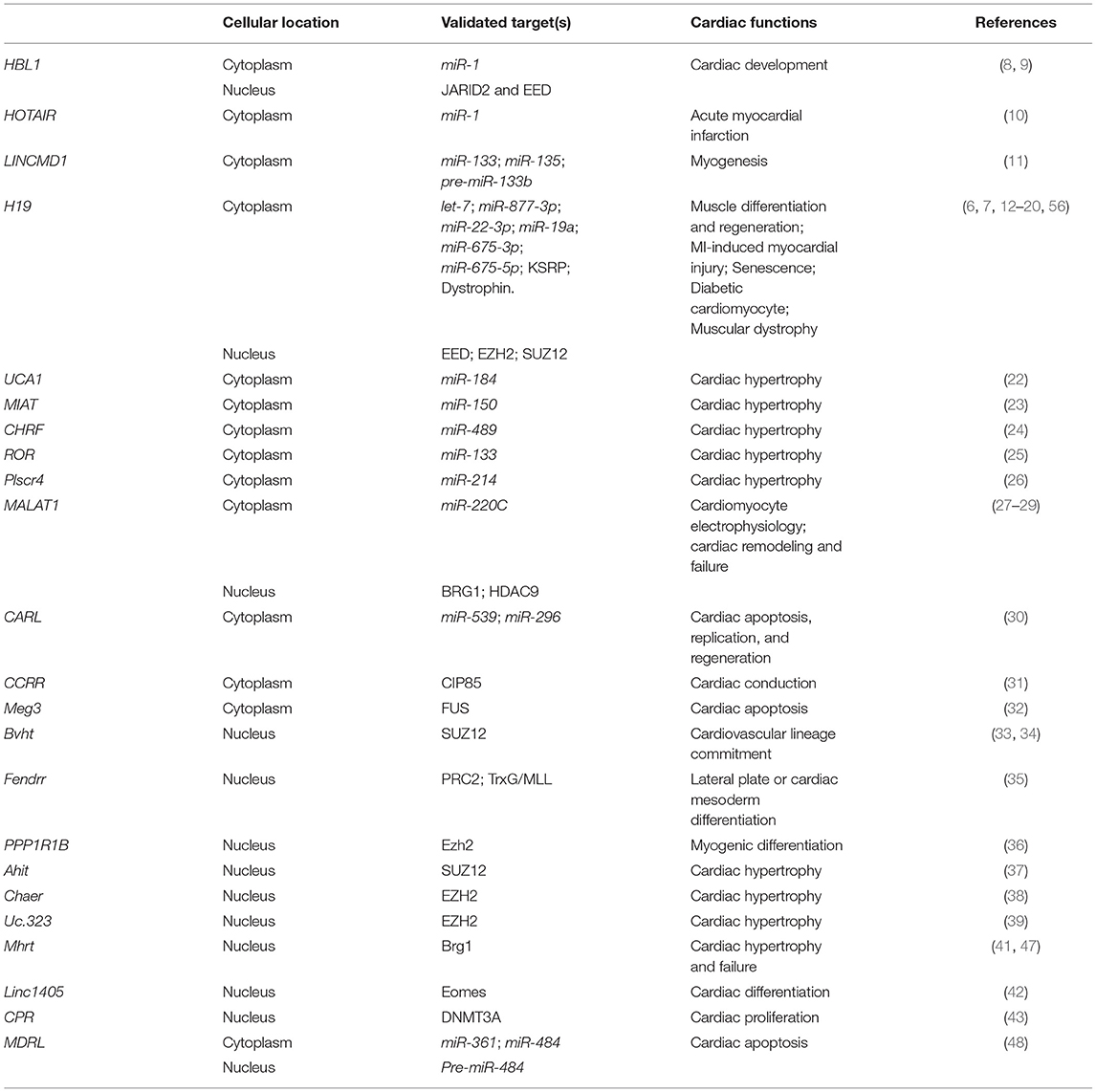
Since the first discovery of competing endogenous RNA (ceRNA), hundreds of lncRNAs have been found to function as miRNA sponge to counteract endogenous miRNAs. The ceRNAs can modulate miRNA activity through sequestration, thereby increasing the expression of miRNA target genes (61). During heart development, several lncRNAs have been identified to counteract miRNAs and regulate expressions of genes essential for stem cells pluripotency or lineage specification. Using hPSCs, HBL1 was identified as a modulator to fine-tune human CM development via sponging miR-1 (8). HBL1 is a human-specific lncRNA highly expressed in hPSCs and gradually diminishes during CM differentiation. Loss of HBL1 increases CM differentiation from hPSCs. HBL1 expresses in both nucleus and cytoplasm of undifferentiated hPSCs. In the cytoplasm, HBL1 binds with miR-1 to fine-tune its activity and further regulate cardiogenic gene expressions (Figure 1). Additionally, lncRNA HOTAIR (HOX antisense intergenic RNA), which was initially described as a regulator of cancer progression, also displays a cardioprotective role in acute myocardial infarction, which is partially through the interaction and negative regulation of miR-1 (10).
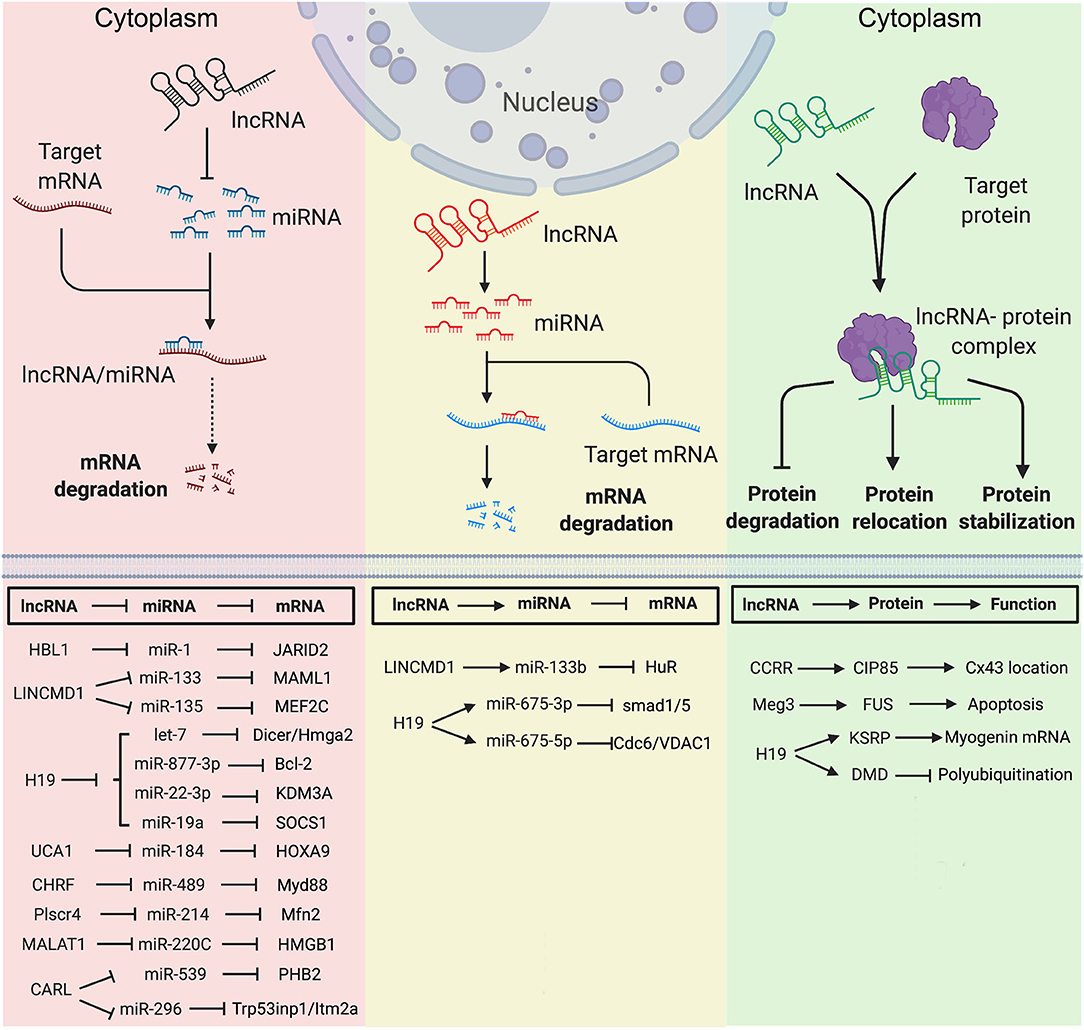
Figure 1. Mechanisms of Long non-coding RNAs in cytoplasm in heart development and diseases. (Left) lncRNA functions as miRNA sponge. (Middle) lncRNA functions as miRNA precursor. (Right) lncRNA functions as protein scaffold. Created with BioRender.com.
LINCMD1 (Long Intergenic Non-protein Coding RNA, Muscle Differentiation 1) is a muscle-specific ceRNA, which is required for muscle differentiation and plays an important role in myogenesis. LINCMD1 acts as ceRNAs for two muscle-specific microRNAs, miR-133 and miR-135, which target the MAML1 (expression of mastermind-like-1) and MEF2C (myocyte-specific enhancer factor 2C) genes, respectively (Figure 1). MAML1 and MEF2C are transcriptional coactivators which positively regulate muscle-specific gene expression. Depletion of LINCMD1 represses the expression of MAML1 and MEF2C, whereas overexpression of LINCMD1 increases MAML1 and MEF2C expression levels and promotes muscle differentiation (11).
It was reported that approximately 378,295 ceRNA interactions appeared in the cardiovascular disease-related ceRNA interactions (62). H19 is a lncRNA with high expression level in embryos (63, 64). H19 is conserved in both human and mouse and has no coding potential. H19 is required for muscle differentiation and regeneration via acting as a natural molecular sponge for the let-7 family of miRNAs (13). Depletion of H19 causes precocious muscle differentiation, which can be repressed by let-7 overexpression (Figure 1) (14). In H2O2-treated CMs and mouse ischemia-reperfusion (I/R) hearts, H19 functions as a ceRNA for miR-877-3p, which targets Bcl-2 to further regulate mitochondria-mediated apoptosis in myocardial I/RI (Figure 1) (15). Additionally, Zhang et al. reported that H19 functions as a ceRNA of miR-22-3p, which directly targets KDM3A gene to ameliorate MI-induced myocardial injury (Figure 1) (16). H19 is also a pro-senescence lncRNA in CMs by counteracting miR-19a to upregulate SOCS1 expression and further activate the p53/p21 pathway to promote CM senescence (Figure 1) (17).
Many lncRNAs have been reported to play a ceRNA role in hypertrophic cardiomyopathy. LncRNA UCA1 regulates cardiac hypertrophy via the UCA1/miR-184/HOXA9 axis (Figure 1) (22). MIAT promotes cardiac hypertrophy through targeting miR-150 (23). LncRNA CHRF (cardiac hypertrophy related factor) regulates cardiac hypertrophy via the CHRF/miR-489/Myd88 axis (24). LncRNA ROR mediates cardiac remodeling and promotes cardiac hypertrophy via interacting with miR-133 (25). Plscr4 negatively regulates cardiac hypertrophy in vivo and in vitro via the miR-214/Mfn2 axis (26). MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) reduces transient outward potassium current of CMs by targeting miR-220C and its downstream target gene HMGB1 (Figure 1) (27). CARL (cardiac apoptosis-related lncRNA) significantly increases in CMs since the neonatal stage of mouse (44). CARL can negatively regulate mitochondrial fission and apoptosis through the miR-539/PHB2 axis (44). It can also directly target miR-296 and its downstream genes Trp53inp1 and Itm2a, further regulating CM replication and cardiac regeneration after injury (Figure 1) (30).
To date, hundreds of publications have reported the ceRNA role of lncRNAs under normal and diseased conditions. Given the cascading effects exerted by the gene networks comprising ceRNA-miRNA-coding genes, lncRNA and its downstream gene networks are potential new targets for cardiovascular disease therapy.
Long Non-coding RNA Forms miRNA Precursor
LncRNAs can be transcribed as miRNA precursors, which produce mature miRNAs via further processing. Therefore, lncRNAs could indirectly regulate the expression of miRNA downstream target genes. For example, transcript of LINCMD1 hosts a pre-miR-133b transcript. The RNA-binding protein HuR is a component of LINCMD regulatory circuitry to regulate muscle differentiation (65). During the early stage of muscle differentiation, HuR binds to LINCMD1 and promotes miR-133 biogenesis from the LINCMD1 transcript. HuR/LINCMD1 complex is then targeted by miR-133 in the cytoplasm (Figure 1). Thus, the ceRNA function of LINCMD1 reinforces HuR expression via counteracting miR-133 in a positive feedforward loop (65). In this case, LINCMD1 plays dual roles in fine-tuning the dynamic of muscle differentiation and regeneration.
Interestingly, the exon 1 of H19 hosts transcripts of miR-675-3p and miR-675-5p. MiR-675-3p regulates the bone morphogenetic protein (BMP) signaling pathway by directly targeting Smad1 and Smad5 mRNAs (Figure 1) (18). MiR-675-5p could target DNA replication initiation factor Cdc6 mRNA (18). Therefore, H19 exhibits a pro-differentiation function in primary myoblasts and regenerating skeletal muscles (19). In the rat model of diabetic cardiomyopathy, overexpression of H19 can attenuate apoptosis of diabetic CMs and improve left ventricular function, whereas knockdown of H19 shows opposite functions. Mechanistically, H19 expression is significant downregulated in the hearts of rats with diabetic cardiomyopathy, which leads to a reduced level of miR-675 and an increased level of miR-675 target-gene VDAC1. Enhanced VDAC1 can induce apoptosis of CMs when exposed to high glucose (12).
Long Non-coding RNA Functions as Protein Scaffold
In cytoplasm, lncRNA can regulate protein location and stability by directly binding with target protein(s). As an anti-arrhythmic lncRNA, CCRR (cardiac conduction regulatory RNA) is downregulated in both mouse and human heart failure (31). CCRR knockdown induces arrhythmias, and its overexpression improves cardiac conduction. CCRR is also required for maintaining the proper distribution of connexin43 (CX43) in the intercalated discs (Figure 1). Mechanically, CCRR directly binds with CX43-interacting protein CIP85 and prevents CX43 from backward trafficking and subsequent degradation in the cytoplasm of CMs (31).
Meg3 is upregulated in infarcted mouse hearts and human failing hearts. Meg3 expression is directly regulated by p53 under hypoxic condition. It has been reported that Meg3 has a pro-apoptotic function in rodent CMs (32). Meg3 shRNA delivered by the adeno-associated virus serotype 9 (AAV9) can significantly improve cardiac function. Meg3 functions as protein scaffold to direct bind with RNA-binding protein FUS and regulates apoptotic signaling pathway (Figure 1) (32).
Except for the functions mentioned above, H19 also interacts with proteins in the cytoplasm. In the undifferentiated multipotent mesenchymal C2C12 cells, H19 interacts with a multifunctional RNA binding protein KSRP (K homology-type splicing regulatory protein) (20). To maintain the undifferentiated state of C2C12 cells, cytoplasmic H19 post-transcriptionally modulates gene expression via acting as a protein scaffold of KSRP and promotes its interaction with RNA exosome, which further enhances the KSRP-promoted mRNA decay of myogenic genes (20). Recently, in muscular dystrophy (MD) patients, H19 was found to directly interact with dystrophin and inhibit E3-ligase-dependent polyubiquitination at Lys3584 for protein degradation. Non-silent mutation (C3340Y) of dystrophin results in defective interaction between dystrophin and H19, which causes ubiquitination and degradation of dystrophin (Figure 1) (7). In both Dmd mouse model and human iPSC-derived skeletal muscle cells from patients with Becker MD, simultaneous administration of H19 RNA mimic and nifenazone, an analgesic for rheumatic conditions, could effectively inhibit dystrophin degradation, preserve skeletal and cardiac muscle histology, and improve cardiac strength and heart function. This suggests a protective role of H19 in both Becker and Duchenne muscular dystrophy, providing a potential RNA therapy for MD patients (7).
Long Non-Coding RNA Functions in Nucleus
Compared to cytoplasm, RNAs are processed in nucleus where many lncRNAs reside and execute functions. Nuclear lncRNAs play a variety of crucial roles with complex molecular mechanisms, including regulating chromatin organization, transcription, and different nuclear condensates (66).
Long Non-coding RNA Interacts With the Polycomb-Repressive Complex 2 (PRC2)
Multiple nuclear lncRNAs have been found to regulate lineage differentiation by interacting with PRC2. Histone-modifying complex PRC2 plays a pivotal role in determining the epigenetic state of genes controlling pluripotency, lineage commitment, and cell differentiation (67). A heart-associated lncRNA, Bvht is required for the commitment of nascent mesoderm to a cardiac fate from mouse ESCs (33). In the nucleus, Bvht can activate the core cardiovascular gene network by interacting with SUZ12, a component of PRC2, during CM differentiation (Figure 2). In Bvht-depleted cells, SUZ12 and PRC2 associated chromatin modification H3K27me3 are deposited at promoters of cardiogenic genes, such as Mesp1, which is a master regulator of cardiovascular fate commitment (33). Additionally, deletion of a 5′ asymmetric G-rich internal loop (AGIL) in Bvht can dramatically impair CM differentiation (34). Through AGIL, Bvht can interact with a cellular nucleic acid binding protein CNBP (ZNF9), which is known as a zinc-finger protein to bind with single-stranded G-rich sequences. Together, in the nucleus, Bvht controls cardiovascular lineage commitment by interacting with SUZ12/PRC2 and CNBP through defined RNA motifs (33, 34).
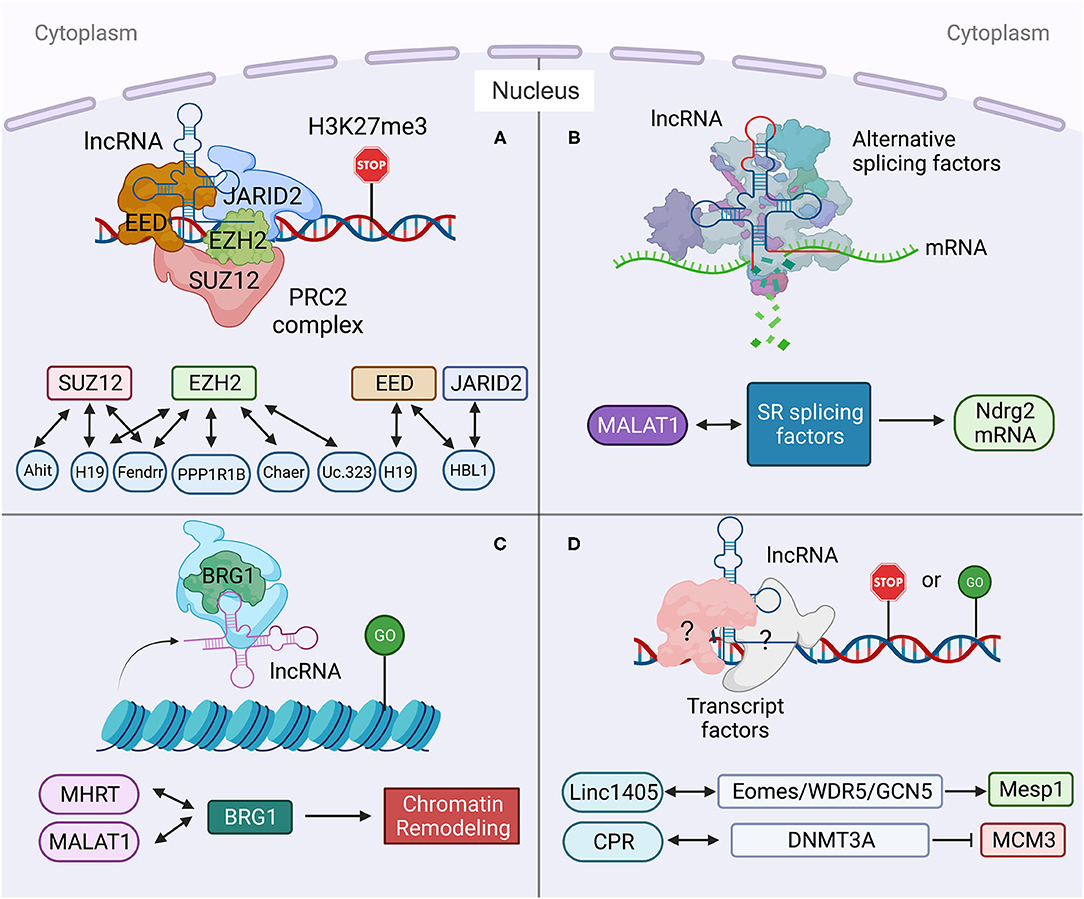
Figure 2. Long non-coding RNA mechanisms in nucleus in heart development and diseases. (A) lncRNA targets to PCR2 complex and regulates its downstream gene transcription. (B) lncRNA regulates pre-mRNA alternative splicing. (C) lncRNA regulates chromatin remodeling. (D) lncRNA interacts with transcription factors to regulate gene transcription. Created with BioRender.com.
In mouse, a lateral mesoderm-specific lncRNA Fendrr is essential for heart development (35). During mouse embryo development, Fendrr binds with both PRC2 via the EZH2 subunit and TrxG/MLL complexes and acts as modulators of PRC2 or TrxG/MLL activity (Figure 2) (35). Fendrr deficient embryos show upregulation of several transcription factors controlling lateral plate or cardiac mesoderm differentiation, accompanied with a drastic reduction of PRC2 occupancy and decreased H3K27 trimethylation and/or increased H3K4 trimethylation at those gene promoters. So, similar to Bvht, Fendrr plays an essential role in controlling cardiac lineage fate commitment via PRC2 (35).
The interaction between lncRNA and PRC2 complex is conserved in human and mouse. LncRNA PPP1R1B was found to bind with EZH2, a key PRC2 subunit (Figure 2) (36, 68). Silencing of PPP1R1B compromises myotube development in both mouse C2C12 and human skeletal myoblasts (36). In hiPSCs-CMs, PPP1R1B deficient also impairs myogenic differentiation (36). PPP1R1B regulates the expression of myogenic transcription factors, such as MyoD, Myogenin, and Tbx5, by interacting with PRC2 at the chromatin interface. PPP1R1B interacts with PRC2 to suppress H3K27me3 histone modification on the MyoD1 and Myogenin promoters. In the nucleus, PPP1R1B modulates PRC2 occupancy on promoters of essential myogenic genes to regulate myogenic differentiation during heart and skeletal muscle development (36).
Our recent study found that nuclear HBL1 interacts with two PRC2 subunits, JARID2 and EED in human pluripotent stem cells (Figure 2) (9). During human cardiogenesis, loss of HBL1 disrupts genome-wide PRC2 occupancy, reduces H3K27me3 chromatin modification on essential cardiogenic genes, and therefore enhances cardiogenic gene transcription in undifferentiated hPSCs and later-on differentiation. At the pluripotency stage, deletions of HBL1 and JARID2 both reduce PRC2 occupancy on 62 overlapped cardiogenic genes. Therefore, HBL1 precisely controls cardiogenic gene transcription via modulating PRC2 occupancy.
H19 plays important functions in both cytoplasm and nucleus. In diabetic cardiomyopathy, cytosolic H19 forms miR-675-3p and miR-675-5p and attenuates apoptosis of CMs (12). Under the same pathological condition, Zhuo et al. reported that H19 directly binds with EZH2, a subunit of PRC2, in CM nucleus to affect the anti-autophagy function (56). Loss of H19 was found to reduce EZH2 and H3K27me3 occupancy on the promoter of DIRAS3, which regulates the formation of autophagosome initiation complex (Figure 2) (21), and causes DIRAS3 downregulation. Consistent with its cytosolic function (12), overexpression of H19 can inhibit cell death of CMs caused by high glucose via this nuclear mechanism. Recently, Viereck et al. reported the interaction between H19 and PRC2 complex subunits EED, EZH2 and SUZ12 in the nuclear lysate of HL-1 CMs (Figure 2) (6). In pressure overload-induced left ventricular hypertrophy mice, H19 ablation aggravates cardiac hypertrophy compared to wild-type mice. Taken together, H19 physically interacts with PRC2 to suppress H3K27me3 modification at the Tescalcin locus, which is an anti-hypertrophic gene, to promote Tescalcin expression and in turn repress the NFAT signaling pathway (6).
Many other lncRNAs also have been found to interact with PRC2 complex under heart disease conditions. Ahit suppresses cardiac hypertrophy through binding with SUZ12 to regulate PRC2 occupancy on the MEF2A (myocyte enhancer factor 2A) promoter (Figure 2) (37). Chaer is required for the development of cardiac hypertrophy through direct binding with PRC2 subunit EZH2 to further regulate expressions of Anf, Myh7 and Acta1 genes (Figure 2) (38). Uc.323 protects CMs against cardiac hypertrophy by binding with EZH2 to regulate CPT1b gene expression (Figure 2) (39).
Taken together, lncRNAs play important roles in cardiac development and diseases by interacting with PRC2 complex to affect PRC2-related epigenetic modifications.
Long Non-coding RNA Regulates Alternative Splicing of Pre-mRNA
Alternative splicing (AS) of pre-mRNA enhances diversities of transcriptome and proteomic of the genome in higher eukaryotes (69). During tissue- or cell-type specification, the serine/arginine (SR) splicing factors regulate AS in a concentration or phosphorylation dependent manner (70, 71). During human cardiovascular differentiation, stage-specific RNA alternative splicing and lineage-enriched lncRNAs were identified by whole RNA-seq (45). As a long nuclear-retained regulatory RNA (nrRNA), MALAT1 interacts with SR splicing factors in the nuclear speckle domains (Figure 2) (28). MALAT1 regulates mRNA alternative splicing by modulating the levels of phosphorylated SR proteins (28). During pressure overload-induced cardiac remodeling and failure, Malat1 was found to be an alternative splicing regulator of Ndrg2, which shows skipped exon 3 in hypertrophic mouse hearts (72, 73).
Long Non-coding RNA and Chromatin Remodeling
In addition to interactions with splicing factors and epigenetic factors, lncRNAs have also been shown to interact with chromatin remodeling complexes (74). Mhrt (myosin heavy-chain-associated RNA transcripts) is a cardiac-specific lncRNA located in the murine myosin heavy chain 7 locus and is suppressed by the BRG1-HDAC-PARP chromatin repressor complex in cardiomyopathy (75). Overexpression of Mhrt protects mouse heart from hypertrophy and failure (47). Mhrt directly binds with BRG1, which is a chromatin-remodeling factor and the ATPase subunit of the SWI/SNF complex (Figure 2) (40), to remove SWI/SNF from its occupied genomic regions on target genes, thus regulating chromatin remodeling and gene transcription. Mhrt binds with the helicase domain of BRG1, which is crucial for tethering BRG1to its targets. In turn, BRG1 represses Mhrt in stress-induced cardiac hypertrophy and failure (41). This MHRT-BRG1 feedback circuit is also conserved in the human heart (47). MALAT1 can also form RNA-protein complex with chromatin-remodeling enzyme BRG1 and histone deacetylase HDAC9 in vascular smooth muscle cells (Figure 2). This HDAC9-MALAT1-BRG1 complex represses expression of contractile protein genes in association with gain of H3K27me3 histone modification (29).
Long Non-coding RNA Interacts With Transcription Factors
Besides chromatin-remodeling factors and epigenetic factors, transcription factors have also been found to interact with lncRNAs in heart development and disease. Linc1405 is highly expressed in heart during mouse embryo development and critical for proper cardiac differentiation (42). Linc1405 interdependently interacts with Eomes, which physically mediates Eomes/WDR5/GCN5 complex binding at the enhancer region of Mesp1 gene to activate its expression (Figure 2) (42). Mesp1 is one of the earliest key regulators of cardiac lineage specification (76). Disruption of Mesp1 in mice results in embryonic lethality due to a cardiac mesoderm deficiency (77). Therefore, linc1405 guides Eomes/WDR5/GCN5 complex to directly target Mesp1 and affect expression of Mesp1 downstream genes to control cardiac differentiation (42).
Recently, lncRNA CPR (cardiomyocyte proliferation regulator) was found to play an important role in the regulation of CM proliferation (43). Deletion of CPR in CMs increases CM proliferation, reduces scar formation, and improves heart function after myocardial injury. Mechanically, CPR represses CM proliferation by suppressing the transcription of MCM3, which regulates initiation of eukaryotic genome replication and cell cycle (78) by direct binding with DNMT3A. Further, DNMT3A promotes CpG methylation of MCM3 promoter and represses transcription of MCM3 (Figure 2) (43).
Crosstalk of Cytosolic and Nuclear Portions of the Same Long Non-coding RNA
Many lncRNAs, such as HBL1 (8, 9), H19 (6, 7, 13, 14, 19, 20), MDRL (48) and LncMyoD (79), express in both cytoplasm and nucleus to display different functional mechanisms. However, how the cytosolic and nuclear mechanisms mediated by the same lncRNA could crosstalk with each other has been rarely studied. Recently, we reported the function of nuclear HBL1 in human cardiogenesis (9), following our previous characterization of cytosolic HBL1 role during human CM differentiating (8). We also defined the mechanism by which cytosolic and nuclear HBL1 crosstalk to control cardiogenic gene transcription (9). HBL1 functions as a miR-1 sponge in cytoplasm and governs PRC2 occupancy on cardiogenic genes in nucleus (Figure 3). In the meanwhile, miR-1 was found to bind with 3'UTR of JARID2 mRNA to repress its expression, and JARID2 deficiency reduces PRC2 occupancy on cardiogenic genes. This conserved miR-1-JARID2 axis thus allows precise regulation of nuclear PRC2 occupancy on cardiogenic genes through miR-1 activity in cytosol (Figure 3). In the cytoplasm, HBL1 counteracts miR-1, which further determines mRNA and protein level of JARID2. After JARID2 protein entering nucleus, nuclear HBL1 binds with JARID2 and EED to determine PRC2 occupancy on cardiogenic genes (Figure 3). Together, this HBL1/miR-1-HBL1/JARID2/PRC2 mechanism coordinates to fine-tune the chromatin state of essential cardiogenic genes in human cardiogenesis (8, 9).
Figure 3. Long non-coding RNA mechanisms cohere with cytoplasm and nucleus in heart development and diseases. (Left) the functions of HBL1 in both cytoplasm and nucleus. (Middle) H19 has multiple functions in both cytoplasm and nucleus. (Right) the functions of MDRL in both cytoplasm and nucleus. Created with BioRender.com.
H19 has been well-studied in heart development and various heart diseases, including diabetic cardiomyopathy (12, 56), myocardial infarction (15), cardiac hypertrophy (6), muscular dystrophies (7) and heart failure (17). H19 displays multiple functions in both cytoplasm and nucleus. Under cardiac hypertrophy, H19 acts as a miR-675 precursor to regulate the expression of miR-675 downstream gene VDAC1 and CM apoptosis in cytoplasm (Figure 3) (12); In nucleus, H19 regulates PRC2 occupancy on the promoters of DIRAS3 and Tescalcin to repress cardiac hypotrophy (Figure 3) (6, 56). Consistently, all those studies reported that overexpression of H19 in CMs can mitigate cardiac hypertrophy (6, 12, 56). These observations raise a question whether the cytosolic and nuclear functions of H19 could coordinate to regulate cardiac hypertrophy, which remains to be further investigated.
MDRL (mitochondrial dynamic related lncRNA) is another well-studied lncRNA with defined mechanisms in both nucleus and cytoplasm. MDRL functions as a ceRNA of miR-361, which directly affects miR-484 expression in mouse CMs (Figure 3) (48). MDRL inhibits mitochondrial fission and apoptosis through two miRNAs, miR-361 and miR-484. In nucleus, MDRL affects the processing of pre-miR-484 by targeting miR-361. In cytoplasm, MDRL regulates the mitochondrial network through both miR-361 and miR-484 (Figure 3). This work defined the complex functions of MDRL in both miRNA processing and downstream gene expression (48). All these findings suggest that clinical application of lncRNA should rely on deeper mechanistic studies, especially the differential roles of the same lncRNA in nucleus and cytoplasm.
Conclusions
We summarized the biological functions and molecular mechanisms of lncRNAs in heart development and disease. In heart development, lncRNAs Carmen, HBL1, Bvht, Fendrr, Bvht and CRP regulate cardiac fate commitment, lineage differentiation, CM maturation/proliferation, and sarcomere organization etc. via both nuclear and cytoplasmic mechanisms. In heart diseases, lncRNAs are involved in the pathogenesis of cardiac hypotrophy, muscular dystrophy, myocardial infarction, diabetic cardiomyopathy, non-Ischemic cardiomyopathy (NICM) and heart failure and so on. With current progresses of genome-wide sequencing and functional screening studies, more functional lncRNAs have been identified in organogenesis and diseases, although the detailed molecular mechanisms of most lncRNAs have not been clearly defined. For example, lncRNAs ALIEN is expressed in undifferentiated pluripotent stem cells and impairs cardiovascular differentiation from pluripotent stem cells with molecular mechanism to be further studied (80). LncRNA GASL1 is downregulated in chronic heart failure and can inhibit CM apoptosis through TGF-β1 signaling pathway, but how it regulates TGF-β1 is unclear (81). A group of lncRNAs are enriched in peripheral blood under different heart disease conditions (82, 83). For example, lncRNA Heat2 expression is increased in the blood of heart failure patients (84); lncRNA MT-LIPCAR, transcribed from mitochondrial DNA, is positively associated with left ventricular diastolic dysfunction (54, 85). Although these lncRNAs might be utilized as disease markers or possess therapeutic penitential, their molecular mechanisms still require further characterizations.
The subcellular location of lncRNA is critical for its function, particularly for those lncRNAs highly expressed in both nucleus and cytoplasm (86). Cytosolic lncRNAs mainly function as regulators of mRNA stability, mRNA translation, miRNA processing and function, whereas nuclear lncRNAs can epigenetically regulate chromatin remodeling, structure, and gene transcription. Therefore, the balanced doses and transportation of lncRNA between cytoplasm and nucleus are expected to be a new research topic in the lncRNA field. During the last two decades, the translational potential of non-coding RNAs in heart disease therapy has gradually emerged. Nowadays, accumulated evidence indicates that lncRNAs provide a new layer of regulatory mechanism on top of coding genes. Since many lncRNAs have low evolutionary conservation (87), studies of lncRNAs might also reveal unique molecular mechanisms of heart development and disease in the human. Given the complex mechanisms, it is expected lncRNAs could offer new preventive and treatment approaches for human diseases including cardiovascular disease. Although, currently, there is no lncRNA therapeutic approach has progressed into preclinical or clinical trial, H19 has been tested as a potential clinical therapeutic target in the Yucatan mini-pig (88). The expression changes of lncRNAs under different setting of heart diseases make it difficult for clinical applications. For example, in cardiac hypertrophy, Mhrt is downregulated (47), while Chaer and Chrf are upregulated (24, 38). MALAT1 and Whispr expressions are upregulated in cardiac fibrosis, whereas Meg3 and GAS5 expressions are downregulated (89–92). Nevertheless, upregulated lncRNAs can be repressed by using shRNA, locked nucleic acids (LNAs) or GapmeR, and downregulated lncRNAs can be enhanced by using virus such as adenovirus, adeno-associated virus (AAV), and lentivirus (93). Although no clinical trial exists for lncRNA therapy in heart disease, the success of non-coding RNA miR-132 based clinical trial paved the way. Recently, phase 1b clinical study to assess safety, pharmacokinetics and pharmacodynamics parameters of CDR132L, a miR-132 inhibitor, has been completed (94). CDR132L is safe and well tolerated. Importantly, it improves cardiac function of heart failure patients. Therefore, the clinical applications of lncRNAs have a bright future, with fully and clearly characterized molecular mechanisms.
Author Contributions
LH and LY summarized the references and prepared the manuscript. LH drafted the illustrations. LY supervised the project. All authors contributed to the article and approved the submitted version.
Funding
This was supported by NIH (RO1 HL 147871, R21 HD095049) and AHA (20EIA35260114, 19TPA34850038) to LY, and 2019 AHA postdoc fellowship Award (19POST34380871) to LH.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
lncRNA, long non-coding RNA; ceRNA, competing endogenous RNA; miRNAs microRNAs, ; iPSCs, induced pluripotent stem cells; CPCs, cardiac precursor cells; CM, cardiomyocyte; CAD, coronary artery disease; NICM, non-Ischemic cardiomyopathy; PRC2, polycomb-repressive complex 2; AS, alternative splicing; JARID2, jumonji and AT-rich interaction domain containing 2; EED, embryonic ectoderm development; EZH2, enhancer of zeste homolog 2.
References
1. Virani SS, Alonso A, Aparicio HJ, Benjamin EJ, Bittencourt MS, Callaway CW, et al. Heart disease and stroke statistics-2021 update: a report from the american heart association. Circulation. (2021) 143:e254–e743. doi: 10.1161/CIR.0000000000000950
2. Das S, Shah R, Dimmeler S, Freedman JE, Holley C, Lee JM, et al. Noncoding RNAs in cardiovascular disease: current knowledge, tools and technologies for investigation, and future directions: a scientific statement from the american heart association. Circ Genom Precis Med. (2020) 13:e000062. doi: 10.1161/HCG.0000000000000062
3. Zhang P, Wu W, Chen Q, Chen M. Non-coding RNAs and their integrated networks. J Integr Bioinform. (2019) 16:20190027. doi: 10.1515/jib-2019-0027
4. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. (2021) 22:96–118. doi: 10.1038/s41580-020-00315-9
5. Gomes CPC, Spencer H, Ford KL, Michel LYM, Baker AH, Emanueli C, et al. The function and therapeutic potential of long non-coding RNAs in cardiovascular development and disease. Mol Ther Nucleic Acids. (2017) 8:494–507. doi: 10.1016/j.omtn.2017.07.014
6. Viereck J, Buhrke A, Foinquinos A, Chatterjee S, Kleeberger JA, Xiao K, et al. Targeting muscle-enriched long non-coding RNA H19 reverses pathological cardiac hypertrophy. Eur Heart J. (2020) 41:3462–74. doi: 10.1093/eurheartj/ehaa519
7. Zhang Y, Li Y, Hu Q, Xi Y, Xing Z, Zhang Z, et al. The lncRNA H19 alleviates muscular dystrophy by stabilizing dystrophin. Nat Cell Biol. (2020) 22:1332–45. doi: 10.1038/s41556-020-00595-5
8. Liu J, Li Y, Lin B, Sheng Y, Yang L. HBL1 is a human long noncoding RNA that modulates cardiomyocyte development from pluripotent stem cells by counteracting MIR1. Dev Cell. (2017) 42:333–348 e5. doi: 10.1016/j.devcel.2017.07.023
9. Liu J, Liu S, Han L, Sheng Y, Zhang Y, Kim IM, et al. LncRNA HBL1 is required for genome-wide PRC2 occupancy and function in cardiogenesis from human pluripotent stem cells. Development. (2021) 148:dev199628. doi: 10.1242/dev.199628
10. Gao L, Liu Y, Guo S, Yao R, Wu L, Xiao L, et al. Circulating long noncoding RNA HOTAIR is an essential mediator of acute myocardial infarction. Cell Physiol Biochem. (2017) 44:1497–508. doi: 10.1159/000485588
11. Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous. RNCell A. (2011) 147:358–69. doi: 10.1016/j.cell.2011.09.028
12. Li X, Wang H, Yao B, Xu W, Chen J, Zhou X. lncRNA H19/miR-675 axis regulates cardiomyocyte apoptosis by targeting VDAC1 in diabetic cardiomyopathy. Sci Rep. (2016) 6:36340. doi: 10.1038/srep36340
13. Poirier F, Chan C, Timmons P, Robertson E, Evans M, Rigby P. The murine H19 gene is activated during embryonic stem cell differentiation in vitro and at the time of implantation in the developing embryo. Development. (1991) 113:1105–14. doi: 10.1242/dev.113.4.1105
14. Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. (2013) 52:101–12. doi: 10.1016/j.molcel.2013.08.027
15. Li X, Luo S, Zhang J, Yuan Y, Jiang W, Zhu H, et al. lncRNA H19 alleviated myocardial I/RI via suppressing miR-877-3p/Bcl-2-mediated mitochondrial apoptosis. Mol Ther Nucleic Acids. (2019) 17:297–309. doi: 10.1016/j.omtn.2019.05.031
16. Zhang BF, Jiang H, Chen J, Hu Q, Yang S, Liu XP, et al. LncRNA H19 ameliorates myocardial infarction-induced myocardial injury and maladaptive cardiac remodelling by regulating KDM3A. J Cell Mol Med. (2020) 24:1099–115. doi: 10.1111/jcmm.14846
17. Zhuang Y, Li T, Xiao H, Wu J, Su S, Dong X, et al. LncRNA-H19 drives cardiomyocyte senescence by targeting miR-19a/socs1/p53 axis. Front Pharmacol. (2021) 12:631835. doi: 10.3389/fphar.2021.631835
18. Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. (2014) 28:491–501. doi: 10.1101/gad.234419.113
19. Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. (2007) 13:313–6. doi: 10.1261/rna.351707
20. Giovarelli M, Bucci G, Ramos A, Bordo D, Wilusz CJ, Chen CY, et al. H19 long noncoding RNA controls the mRNA decay promoting function of KSRP. Proc Natl Acad Sci U S A. (2014) 111:E5023–8. doi: 10.1073/pnas.1415098111
21. Lu Z, Baquero MT, Yang H, Yang M, Reger AS, Kim C, et al. DIRAS3 regulates the autophagosome initiation complex in dormant ovarian cancer cells. Autophagy. (2014) 10:1071–92. doi: 10.4161/auto.28577
22. Zhou G, Li C, Feng J, Zhang J, Fang Y. lncRNA UCA1 Is a Novel regulator in cardiomyocyte hypertrophy through targeting the miR-184/HOXA9 axis. Cardiorenal Med. (2018) 8:130–9. doi: 10.1159/000487204
23. Zhu X, Yuan Y, Rao S, Wang P. LncRNA MIAT enhances cardiac hypertrophy partly through sponging miR-150. Eur Rev Med Pharmacol Sci. (2016) 20:3653.
24. Wang K, Liu F, Zhou LY, Long B, Yuan SM, Wang Y, et al. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res. (2014) 114:1377–88. doi: 10.1161/CIRCRESAHA.114.302476
25. Jiang F, Zhou X, Huang J. Long non-coding RNA-ROR mediates the reprogramming in cardiac hypertrophy. PLoS ONE. (2016) 11:e0152767. doi: 10.1371/journal.pone.0152767
26. Lv L, Li T, Li X, Xu C, Liu Q, Jiang H, et al. The lncRNA Plscr4 controls cardiac hypertrophy by regulating miR-214. Mol Ther Nucleic Acids. (2018) 10:387–97. doi: 10.1016/j.omtn.2017.12.018
27. Zhu P, Yang M, Ren H, Shen G, Chen J, Zhang J, et al. Long noncoding RNA MALAT1 downregulates cardiac transient outward potassium current by regulating miR-200c/HMGB1 pathway. J Cell Biochem. (2018) 119:10239–49. doi: 10.1002/jcb.27366
28. Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. (2010) 39:925–38. doi: 10.1016/j.molcel.2010.08.011
29. C.L. Lino Cardenas, Kessinger CW, Cheng Y, MacDonald C, MacGillivray T, Ghoshhajra B, et al. An HDAC9-MALAT1-BRG1 complex mediates smooth muscle dysfunction in thoracic aortic aneurysm. Nat Commun. (2018) 9:1009. doi: 10.1038/s41467-018-03394-7
30. Cai B, Ma W, Ding F, Zhang L, Huang Q, Wang X, et al. The long noncoding RNA CAREL controls cardiac regeneration. J Am Coll Cardiol. (2018) 72:534–50. doi: 10.1016/j.jacc.2018.04.085
31. Zhang Y, Sun L, Xuan L, Pan Z, Hu X, Liu H, et al. Long non-coding RNA CCRR controls cardiac conduction via regulating intercellular coupling. Nat Commun. (2018) 9:4176. doi: 10.1038/s41467-018-06637-9
32. Wu H, Zhao ZA, Liu J, Hao K, Yu Y, Han X, et al. Long noncoding RNA Meg3 regulates cardiomyocyte apoptosis in myocardial infarction. Gene Ther. (2018) 25:511–23. doi: 10.1038/s41434-018-0045-4
33. Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. (2013) 152:570–83. doi: 10.1016/j.cell.2013.01.003
34. Xue Z, Hennelly S, Doyle B, Gulati AA, Novikova IV, Sanbonmatsu KY, et al. A G-rich motif in the lncRNA braveheart interacts with a zinc-finger transcription factor to specify the cardiovascular lineage. Mol Cell. (2016) 64:37–50. doi: 10.1016/j.molcel.2016.08.010
35. Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. (2013) 24:206–14. doi: 10.1016/j.devcel.2012.12.012
36. Kang X, Zhao Y, Van Arsdell G, Nelson SF, Touma M. Ppp1r1b-lncRNA inhibits PRC2 at myogenic regulatory genes to promote cardiac and skeletal muscle development in mouse and human. RNA. (2020) 26:481–91. doi: 10.1261/rna.073692.119
37. Yu J, Yang Y, Xu Z, Lan C, Chen C, Li C, et al. Long noncoding RNA ahit protects against cardiac hypertrophy through SUZ12 (Suppressor of Zeste 12 Protein Homolog)-mediated downregulation of MEF2A (Myocyte Enhancer Factor 2A). Circ Heart Fail. (2020) 13:e006525. doi: 10.1161/CIRCHEARTFAILURE.119.006525
38. Wang Z, Zhang XJ Ji YX, Zhang P, Deng KQ, Gong J, et al. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat Med. (2016) 22:1131–9. doi: 10.1038/nm.4179
39. Sun Y, Fan W, Xue R, Dong B, Liang Z, Chen C, et al. Transcribed ultraconserved regions, Uc323, ameliorates cardiac hypertrophy by regulating the transcription of CPT1b (Carnitine Palmitoyl transferase 1b). Hypertension. (2020) 75:79–90. doi: 10.1161/HYPERTENSIONAHA.119.13173
40. Alver BH, Kim KH, Lu P, Wang X, Manchester HE, Wang W, et al. The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat Commun. (2017) 8:14648. doi: 10.1038/ncomms14648
41. Han P, Chang CP. Long non-coding RNA and chromatin remodeling. RNA Biol. (2015) 12:1094–8. doi: 10.1080/15476286.2015.1063770
42. Guo X, Xu Y, Wang Z, Wu Y, Chen J, Wang G, et al. A Linc1405/Eomes complex promotes cardiac mesoderm specification and cardiogenesis. Cell Stem Cell. (2018) 22:893–908 e6. doi: 10.1016/j.stem.2018.04.013
43. Ponnusamy M, Liu F, Zhang YH Li RB, Zhai M, Liu F, et al. Long noncoding RNA CPR (Cardiomyocyte Proliferation Regulator) regulates cardiomyocyte proliferation and cardiac repair. Circulation. (2019) 139:2668–84. doi: 10.1161/CIRCULATIONAHA.118.035832
44. Wang K, Long B, Zhou LY, Liu F, Zhou QY, Liu CY, et al. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun. (2014) 5:3596. doi: 10.1038/ncomms4596
45. Li Y, Lin B, Yang L. Comparative transcriptomic analysis of multiple cardiovascular fates from embryonic stem cells predicts novel regulators in human cardiogenesis. Sci Rep. (2015) 5:9758. doi: 10.1038/srep09758
46. Ounzain S, Micheletti R, Arnan C, Plaisance I, Cecchi D, Schroen B, et al. CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J Mol Cell Cardiol. (2015) 89:98–112. doi: 10.1016/j.yjmcc.2015.09.016
47. Han P, Li W, Lin CH, Yang J, Shang C, Nuernberg ST, et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. (2014) 514:102–6. doi: 10.1038/nature13596
48. Wang K, Sun T, Li N, Wang Y, Wang JX, Zhou LY, et al. MDRL lncRNA regulates the processing of miR-484 primary transcript by targeting miR-361. PLoS Genet. (2014) 10:e1004467. doi: 10.1371/journal.pgen.1004467
49. Long QQ, Wang H, Gao W, Fan Y, Li YF, Ma Y, et al. Long noncoding RNA Kcna2 antisense RNA contributes to ventricular arrhythmias via silencing Kcna2 in rats with congestive heart failure. J Am Heart Assoc. (2017) 6:e005965. doi: 10.1161/JAHA.117.005965
50. Zhang YH, Pan X, Zeng T, Chen L, Huang T, Cai YD. Identifying the RNA signatures of coronary artery disease from combined lncRNA and mRNA expression profiles. Genomics. (2020) 112:4945–58. doi: 10.1016/j.ygeno.2020.09.016
51. Holdt LM, Teupser D Long Noncoding RNA ANRIL. Lnc-ing genetic variation at the chromosome 9p21 locus to molecular mechanisms of atherosclerosis. Front Cardiovasc Med. (2018) 5:145. doi: 10.3389/fcvm.2018.00145
52. Yari M, Bitarafan S, Broumand MA, Fazeli Z, Rahimi M, Ghaderian SMH, et al. Association between long noncoding RNA ANRIL expression variants and susceptibility to coronary artery disease. Int J Mol Cell Med. (2018) 7:1–7. doi: 10.22088/IJMCM.BUMS.7.1.1
53. Zhang J, Yu L, Xu Y, Liu Y, Li Z, Xue X, et al. Long noncoding RNA upregulated in hypothermia treated cardiomyocytes protects against myocardial infarction through improving mitochondrial function. Int J Cardiol. (2018) 266:213–7. doi: 10.1016/j.ijcard.2017.12.097
54. de Gonzalo-Calvo D, Kenneweg F, Bang C, Toro R, van der Meer RW, Rijzewijk LJ, et al. Circulating long-non coding RNAs as biomarkers of left ventricular diastolic function and remodelling in patients with well-controlled type 2 diabetes. Sci Rep. (2016) 6:37354. doi: 10.1038/srep37354
55. Yang KC, Yamada KA, Patel AY, Topkara VK, George I, Cheema FH, et al. Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation. (2014) 129:1009–21. doi: 10.1161/CIRCULATIONAHA.113.003863
56. Zhuo C, Jiang CR, Lin X, Shao M. LncRNA H19 inhibits autophagy by epigenetically silencing of DIRAS3 in diabetic cardiomyopathy. Oncotarget. (2017) 8:1429–37. doi: 10.18632/oncotarget.13637
57. Buxbaum AR, Haimovich G, Singer RH. In the right place at the right time: visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol. (2015) 16:95–109. doi: 10.1038/nrm3918
58. Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol. (2021) 220:e202009045. doi: 10.1083/jcb.202009045
59. Rashid F, Shah A, Shan G. Long Non-coding RNAs in the cytoplasm. Gen Proteom Bioinform. (2016) 14:73–80. doi: 10.1016/j.gpb.2016.03.005
60. Noh JH, Kim KM, McClusky WG, Abdelmohsen K, Gorospe M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip Rev RNA. (2018) 9:e1471. doi: 10.1002/wrna.1471
61. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. (2016) 17:272–83. doi: 10.1038/nrg.2016.20
62. Song C, Zhang J, Qi H, Feng C, Chen Y, Cao Y, et al. The global view of mRNA-related ceRNA cross-talks across cardiovascular diseases. Sci Rep. (2017) 7:10185. doi: 10.1038/s41598-017-10547-z
63. Bhat SA, Ahmad SM, Mumtaz PT, Malik AA, Dar MA, Urwat U, et al. Long non-coding RNAs: mechanism of action and functional utility. Noncoding RNA Res. (2016) 1:43–50. doi: 10.1016/j.ncrna.2016.11.002
64. White NM, Cabanski CR, Silva-Fisher JM, Dang HX, Govindan R, Maher CA. Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol. (2014) 15:429. doi: 10.1186/s13059-014-0429-8
65. Legnini I, Morlando M, Mangiavacchi A, Fatica A, Bozzoni I. A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Molecular cell. (2014) 53:506–514. doi: 10.1016/j.molcel.2013.12.012
66. Guo CJ, Xu G, Chen LL. Mechanisms of long noncoding RNA nuclear retention. Trends Biochem Sci. (2020) 45:947–60. doi: 10.1016/j.tibs.2020.07.001
67. Deevy O, Bracken AP. PRC2 functions in development and congenital disorders. Development. (2019) 146:dev181354. doi: 10.1242/dev.181354
68. Gan L, Yang Y, Li Q, Feng Y, Liu T, Guo W. Epigenetic regulation of cancer progression by EZH2: from biological insights to therapeutic potential. Biomark Res. (2018) 6:10. doi: 10.1186/s40364-018-0122-2
69. Bonnal SC, Lopez-Oreja I, Valcarcel J. Roles and mechanisms of alternative splicing in cancer - implications for care. Nat Rev Clin Oncol. (2020) 17:457–74. doi: 10.1038/s41571-020-0350-x
70. Naro C, Sette C. Phosphorylation-mediated regulation of alternative splicing in cancer. Int J Cell Biol. (2013) 2013:151839. doi: 10.1155/2013/151839
71. Wang Y, Liu J, Huang BO, Xu YM Li J, Huang LF, et al. Mechanism of alternative splicing and its regulation. Biomed Rep. (2015) 3:152–8. doi: 10.3892/br.2014.407
72. Song HK, Hong SE, Kim T, Kim DH. Deep RNA sequencing reveals novel cardiac transcriptomic signatures for physiological and pathological hypertrophy. PLoS ONE. (2012) 7:e35552. doi: 10.1371/journal.pone.0035552
73. Peters T, Hermans-Beijnsberger S, Beqqali A, Bitsch N, Nakagawa S, Prasanth KV, et al. Long Non-Coding RNA Malat-1 Is Dispensable during Pressure Overload-Induced Cardiac Remodeling and Failure in Mice. PLoS ONE. (2016) 11:e0150236. doi: 10.1371/journal.pone.0150236
74. Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. (2016) 73:2491–509. doi: 10.1007/s00018-016-2174-5
75. Hang CT, Yang J, Han P, Cheng HL, Shang C, Ashley E, et al. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. (2010) 466:62–7. doi: 10.1038/nature09130
76. Saga Y, Kitajima S, Miyagawa-Tomita S. Mesp1 expression is the earliest sign of cardiovascular development. Trends Cardiovasc Med. (2000) 10:345–52. doi: 10.1016/S1050-1738(01)00069-X
77. Saga Y, Hata N, Kobayashi S, Magnuson T, Seldin MF, Taketo MM. MesP1: a novel basic helix-loop-helix protein expressed in the nascent mesodermal cells during mouse gastrulation. Development. (1996) 122:2769–78. doi: 10.1242/dev.122.9.2769
78. Sclafani RA, Holzen TM. Cell cycle regulation of DNA replication. Annu Rev Genet. (2007) 41:237–80. doi: 10.1146/annurev.genet.41.110306.130308
79. C. Gong, Li Z, Ramanujan K, Clay I, Zhang Y, Lemire-Brachat S, et al. A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Dev Cell. (2015) 34:181–91. doi: 10.1016/j.devcel.2015.05.009
80. Kurian L, Aguirre A, Sancho-Martinez I, Benner C, Hishida T, Nguyen TB, et al. Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. Circulation. (2015) 131:1278–90. doi: 10.1161/CIRCULATIONAHA.114.013303
81. Deng H, Ouyang W, Zhang L, Xiao X, Huang Z, Zhu W. LncRNA GASL1 is downregulated in chronic heart failure and regulates cardiomyocyte apoptosis. Cell Mol Biol Lett. (2019) 24:41. doi: 10.1186/s11658-019-0165-x
82. Zhu YJ, Mao D, Gao W, Hu H. Peripheral whole blood lncRNA expression analysis in patients with eosinophilic asthma. Medicine (Baltimore). (2018) 97:e9817. doi: 10.1097/MD.0000000000009817
83. Hermans-Beijnsberger S, van Bilsen M, Schroen B. Long non-coding RNAs in the failing heart and vasculature. Noncoding RNA Res. (2018) 3:118–130. doi: 10.1016/j.ncrna.2018.04.002
84. Boeckel JN, Perret MF, Glaser SF, Seeger T, Heumuller AW, Chen W, et al. Identification and regulation of the long non-coding RNA Heat2 in heart failure. J Mol Cell Cardiol. (2019) 126:13–22. doi: 10.1016/j.yjmcc.2018.11.004
85. Dorn GW, LIPCAR. a mitochondrial lnc in the noncoding RNA chain? Circ Res. (2014) 114:1548–50. doi: 10.1161/CIRCRESAHA.114.304028
86. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. (2012) 22:1775–89. doi: 10.1101/gr.132159.111
87. Johnsson P, Lipovich L, Grander D, Morris KV. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta. (2014) 1840:1063–71. doi: 10.1016/j.bbagen.2013.10.035
88. Li DY, Busch A, Jin H, Chernogubova E, Pelisek J, Karlsson J, et al. H19 induces abdominal aortic aneurysm development and progression. Circulation. (2018) 138:1551–68. doi: 10.1161/CIRCULATIONAHA.117.032184
89. Huang S, Zhang L, Song J, Wang Z, Huang X, Guo Z, et al. Long noncoding RNA MALAT1 mediates cardiac fibrosis in experimental postinfarct myocardium mice model. J Cell Physiol. (2019) 234:2997–3006. doi: 10.1002/jcp.27117
90. Micheletti R, Plaisance I, Abraham BJ, Sarre A, Ting CC, Alexanian M, et al. The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Sci Transl Med. (2017) 9:eaai9118. doi: 10.1126/scitranslmed.aai9118
91. Tao H, Zhang JG, Qin RH Dai C, Shi P, Yang JJ, et al. LncRNA GAS5 controls cardiac fibroblast activation and fibrosis by targeting miR-21 via PTEN/MMP-2 signaling pathway. Toxicology. (2017) 386:11–8. doi: 10.1016/j.tox.2017.05.007
92. Piccoli MT, Gupta SK, Viereck J, Foinquinos A, Samolovac S, Kramer FL, et al. Inhibition of the cardiac fibroblast-enriched lncRNA Meg3 prevents cardiac fibrosis and diastolic dysfunction. Circ Res. (2017) 121:575–83. doi: 10.1161/CIRCRESAHA.117.310624
93. Huang CK, Kafert-Kasting S, Thum T. Preclinical and clinical development of noncoding RNA therapeutics for cardiovascular disease. Circ Res. (2020) 126:663–78. doi: 10.1161/CIRCRESAHA.119.315856
94. Taubel J, Hauke W, Rump S, Viereck J, Batkai S, Poetzsch J, et al. Novel antisense therapy targeting microRNA-132 in patients with heart failure: results of a first-in-human Phase 1b randomized, double-blind, placebo-controlled study. Eur Heart J. (2021) 42:178–88. doi: 10.1093/eurheartj/ehaa898
Keywords: long non-coding RNA, microRNA, heart development, heart disease, ceRNA, PRC2 complex
Citation: Han L and Yang L (2021) Multidimensional Mechanistic Spectrum of Long Non-coding RNAs in Heart Development and Disease. Front. Cardiovasc. Med. 8:728746. doi: 10.3389/fcvm.2021.728746
Received: 22 June 2021; Accepted: 16 August 2021;
Published: 16 September 2021.
Edited by:
Jiandong Liu, University of North Carolina at Chapel Hill, United StatesCopyright © 2021 Han and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Yang, bHlhbmc3QGl1LmVkdQ==
 Lei Han
Lei Han Lei Yang
Lei Yang