- 1Department of Cardiology, Center for Translational Medicine, Institute of Precision Medicine, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, China
- 2National Health Commission (NHC) Key Laboratory of Assisted Circulation, Sun Yat-sen University, Guangzhou, China
- 3National-Guangdong Joint Engineering Laboratory for Diagnosis and Treatment of Vascular Diseases, Guangzhou, China
Cardiac remodeling occurs after the heart is exposed to stress, which is manifested by pathological processes such as cardiomyocyte hypertrophy and apoptosis, dendritic cells activation and cytokine secretion, proliferation and activation of fibroblasts, and finally leads to heart failure. Circular RNAs (circRNAs) are recently recognized as a specific type of non-coding RNAs that are expressed in different species, in different stages of development, and in different pathological conditions. Growing evidences have implicated that circRNAs play important regulatory roles in the pathogenesis of a variety of cardiovascular diseases. In this review, we summarize the biological origin, characteristics, functional classification of circRNAs and their regulatory functions in cardiomyocytes, endothelial cells, fibroblasts, immune cells, and exosomes in the pathogenesis of cardiac remodeling.
Introduction
The heart is made up of a variety of cells, including cardiomyocytes and non-cardiomyocytes (fibroblasts, smooth muscle cells, endothelial cells, and immune cells etc.,). These cells communicate with each other in both physiological and pathological conditions through direct cell-cell interaction and paracrine signaling. Fibroblasts, the major component of connective tissue, produce the extracellular matrix (ECM) scaffold that organizes different cellular components of the heart (1). Endothelial cells (ECs) are located on the inner surface of blood vessels and lymphatics, controlling vasomotor tension and regulating angiogenesis (2, 3). Resident and recruited immune cells regulate cardiac microenvironmental homeostasis and inflammation during maladaptive remodeling (4). Pathological conditions, such as hypertension or myocardial infarction (MI), induce maladaptive reactions in cardiomyocytes and non-cardiomyocytes, leading to the deterioration of cardiac function and eventually heart failure. However, molecular and cellular mechanisms of cardiac remodeling have not been fully understood.
Other than linear splicing, the sequence of primary transcripts from gene loci is also found to be processed by back-splicing to generate circular RNAs (circRNAs). CircRNAs can be classified as reverse spliced exons (5–7) or intron-derived RNAs (8, 9). Back-splicing is generally thought as a rare event, although, mammalian circRNAs have been reported decades ago (7). The recent deep sequencing data showed evidences that an unexpectedly large number of circRNAs are in fact expressed (5, 6). And thousands of circRNAs have been identified from different cells and tissues (10–13). More importantly, emerging evidences indicated that circRNAs regulate different cellular behaviors, including proliferation, differentiation, apoptosis, and migration (12, 14).
Early report demonstrated the altered expression of circRNAs in human failing heart (15), indicating the participation of circRNAs in the regulation of the pathogenesis of cardiac diseases. Recent studies have further shown that circRNAs are involved in a variety of cardiovascular diseases, including cardiac remodeling, by regulating the pathophysiology of cardiomyocytes, fibroblasts, endothelial cells, and immune cells (16–19). However, the underlying mechanisms of circRNAs' regulatory functions are not fully understood. A more comprehensive understanding of circRNAs will promote the development of circRNA-based diagnosis and therapeutic interventions in cardiovascular disease. In this review, we will focus on the nature of circRNA and how these circular molecules regulate the pathogenesis of cardiac remodeling.
The Identification, Processing, and Characterization of circRNAs
Identification of circRNA
In 1976, Sanger used the term “circRNA” for the first time to describe viroids, which was his identification of a single stranded, covalently closed RNA molecules that are infectious (20). Early detection of circRNA was rare and its function was poorly understood. They were believed as by-products of linear RNA and thought as “junk RNA” (21). Three decades ago, circRNAs were accidentally discovered in mammals (22, 23). More and more circRNAs have recently been identified by taking the advantage of the breakthrough of high-throughput sequencing, and the biological functions of these emerging molecules have been investigated and uncovered rapidly (6). circRNAs are currently considered as a special type of non-coding RNAs, although, some studies have suggested that circRNAs may have protein-coding capability in vivo.
General Characteristics of circRNAs
Most circRNAs are exonic and have some important characteristics: (I) CircRNAs are expressed in large quantities in many species, from plants to mammals (5). Multiple circRNA isoforms are often processed from a single host gene by selective splicing. Notably, more than 100 circRNA isoforms of the Ryanodine receptor 2 (RyR2) gene are expressed in human hearts (24). (II) CircRNAs are usually expressed in a cell type- and/or developmental stage-specific manner (6, 25, 26). The expression profiles of circRNAs are different in four stages of cardiac differentiation: undifferentiated stage, mesoderm stage, cardiac progenitor cell stage, and final cardiomyocyte stage (26). (III) CircRNAs are not easily degraded by RNA exonuclease because of their covalently closed circular structure. CircRNAs are more stable and have a longer half-lives than linear RNAs (27, 28). These features of circRNAs make these molecules potential candidates for disease diagnosis and prognosis biomarkers, especially the presence of circRNAs in plasma.
Categorization of circRNA
CircRNAs can be divided into three subtypes according to the mode of biogenesis: circRNA, Exon-intron circRNAs (EIciRNA), and ciRNAs. Most circRNAs are derived from exons in linear transcripts, lacking introns, and mainly present in the cytoplasm. In contrast, ciRNAs lack exon sequences, are present in the nucleus and have no obvious enrichment of miRNA binding sites (8, 29). EIciRNA sequences contain exons and introns, which are mainly located in the nucleus and form a protein-RNA complex with U1 snRNP and Polymerase II to regulate the transcription of their parent genes (30, 31). CircRNAs can also be classified as intragenic circRNAs and intergenic circRNAs based on the position of circRNA-originated locus in the genome.
Mechanisms of circRNA Formation
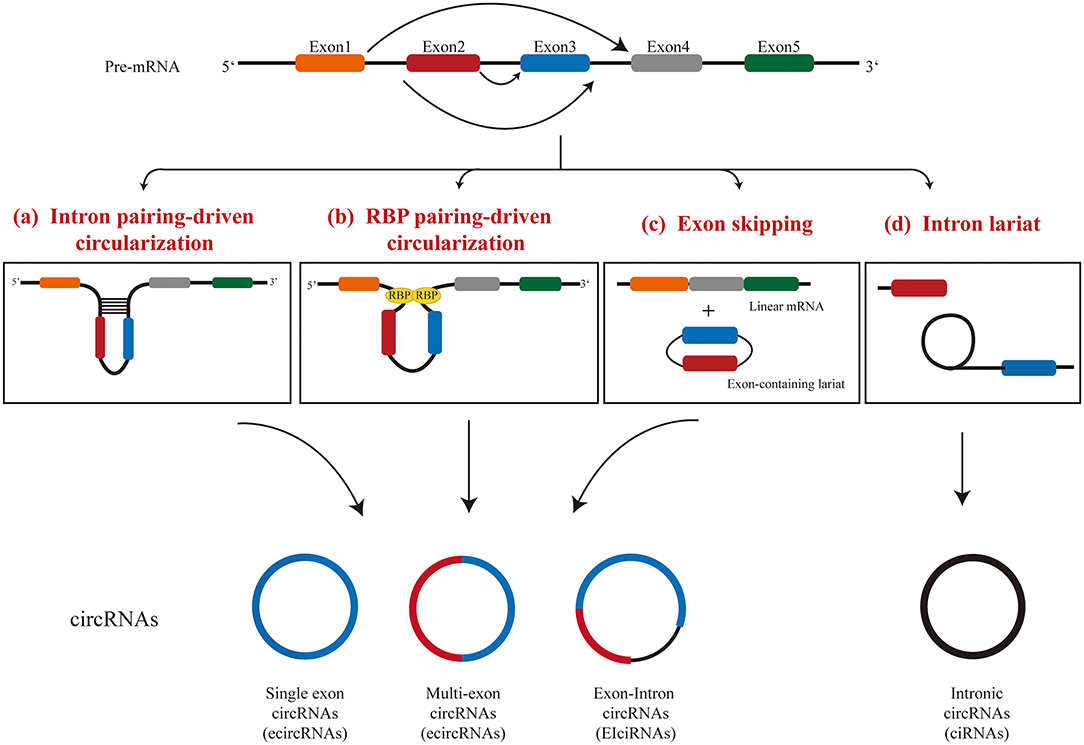
CircRNAs are produced by a unique splicing mechanism called backsplicing (5, 32). Classical splicing events include a typical donor (GU) at the 5 “end of the intron and an receptor (AG) at the 3” end of the intron (33). The circRNAs are formed because the splicing does not finish in the linear manner between intronic donor splicing site next to an exon to the receptor splicing site before the downstream exon, but to the receptor splicing site before the upstream exon. This process produces a covalently closed RNA molecule with or without exons. Three models have been proposed for the formation of circRNAs: (I) intron pairing-driven circularization, (II) RNA binding protein-driven circularization, and (III) lariat-driven circularization (Figure 1).
Intron Pairing-Driven Circularization
Compared with linear splicing, the reverse complementary sequences between introns bracketing circRNAs were significantly more abundant (9). In intron-pairing-driven circularization, cis-acting elements, hairpin structures, or complementary sequences located in the flanking introns of an exon are often used for direct base pairing (5, 9, 34). The minimal intron region required for circRNA circularization has been identified (35): even if the intron is <100 nucleotides and contains a typical splicing site sequence and a short reverse repeat sequence, it is sufficient for exon cyclization. This process appears to be more complicated than canonical base pairing, since not all reverse repeats lead to exon cyclization. It is worth noting that multiple exon cyclization events can occur in one gene locus, and exon cyclization efficiency can be modulated by RNA pairing within flanking introns or by competition for RNA pairing within a single intron (36). Alternative formation of reverse repeats of introns, such as repeated ALU pairs, and the competition between them often result in alternative circularization, which leads to the occurrence of producing multiple circRNA transcripts from a gene locus (36). However, how base pairing between introns affects the assembly of the spliceosome during back-splicing is still not fully understood and warrants further investigation.
RBP-Driven Circularization
The protein-protein interaction between RNA binding proteins (RBPs) makes the splicing sites of pre-mRNA come closer, which further facilitates the spliceosome to participate in the back-splicing reaction. Multiple RBPs have been shown to regulate the generation of circRNAs. Both the RNA-binding motif protein 20 (RBM20) (37) and splicing factor muscleblind (MBL) (38) were shown to increase the generation of circRNAs by binding to specific intron motifs. RBM20 was identified as an important splicing factor in the heart with the function of regulating the formation of circRNAs in TTN gene locus (37).
A further study suggested that the RBP-driven circularization and the intron-pairing-driven circularization may work together to regulate the formation of circRNAs (39). Intronic repeats in flanking introns are believed to provide an opportunity for RBM20 to facilitate the circularization event. After that, a subset of proteins are recruited, which further regulate the formation of circRNA by modulating the activity of spliceosomes. Each gene locus may require a different set of protein factors for the generation of multiple circRNAs.
Lariat-Driven Circularization
Interestingly, a circRNA can also be produced during linear splicing by lariat-driven circularization, in which circRNAs may be generated during exon-skipping events (40) or intron removal in pre-mRNA splicing (8). TTN gene is an example of generating circRNAs through exon-skipping events with more than 80 circRNAs generated through this mechanism in the heart (37). Lariat RNAs are the intermediate product of splicing of pre-mRNA. Under normal circumstances, lariat RNAs released in canonical splicing undergo debranching at the 2′-5′ phosphodiester bond, and are then degraded by exonucleases (41). However, the specific structure of 7 nt GU-rich near the 5′ splice site and 11 nt C-rich near the branching site of lariats prevent the debranching event, and therefore, these RNA molecules remain circular (8, 42). These type of circular RNAs become mature after the 3′ tail of the lariat is degraded up to the branching point (43).
The Regulatory Mechanisms of circRNAs
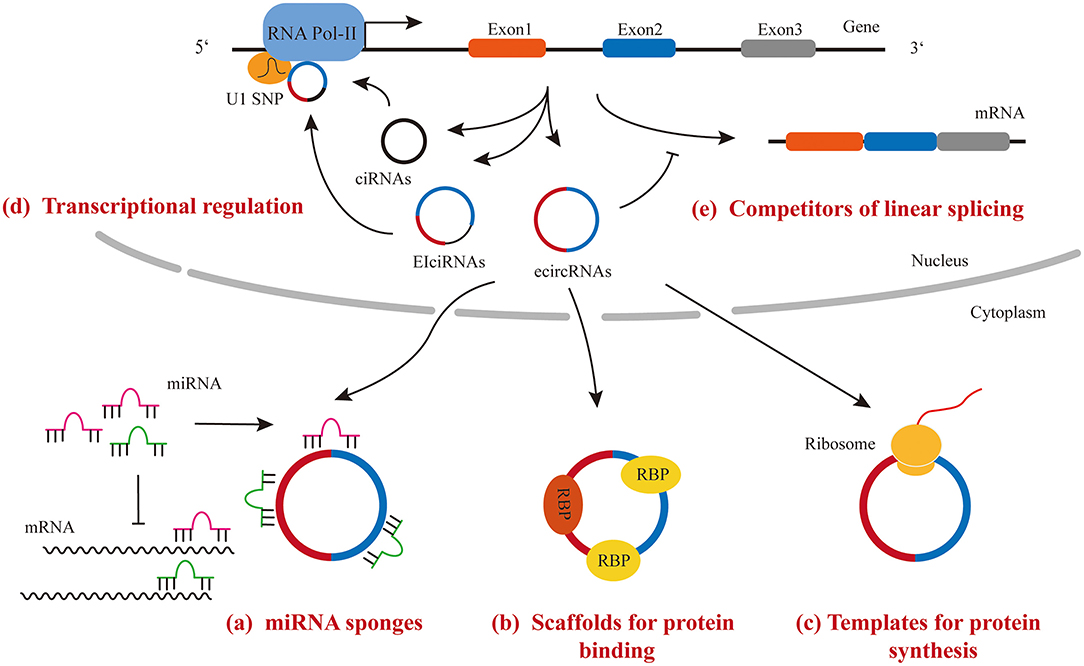
Accumulating data have shown that circRNAs exert their regulatory function through the following mechanisms: (1) functioning as miRNA sponges to sequester miRNAs and de-repress their targets; (2) functioning as scaffolds to bind RNA binding proteins and regulate the activity of downstream signaling; (3) binding to snRNP and polymerase II to regulate transcriptional activity; (4) functioning as competitors for parental gene splicing and expression; (5) functioning as templates for protein synthesis (Figure 2).
MicroRNA Sponge
MiRNAs repress gene expression post-transcriptionally by binding to the 3′ UTR of target mRNAs. CircRNAs have been demonstrated to possess multiple miRNA binding sites by both computational prediction and experimental assays. The interaction between circRNA and miRNA leads to miRNA retention and then lowering their bioactivity, which is referred to the “sponge effect.” For example, a CDR1 locus-derived circRNA, CDR1as, has 63 highly conserved miR-7 binding sites (6, 44). Since no linear transcript of CDR1as were detected, the knockout strategy is simple and the removal of DNA sequence for circRNA from the genome will not affect the expression of any linear transcript from the same DNA locus. The high expression level of CDR1as and the presence of large amount miR-7 binding sites per molecules makes the circRNA a competitive inhibitor of endogenous miR-7.
Scaffold for Protein Interaction
Some circRNAs possess protein binding motifs. Therefore, these circRNAs interact with selected proteins and regulate their activity or localization. For example, the interaction between circPABPN1 and HuR prevents HuR from binding to PABPN1 mRNA and reduces its translation (45). In another example, circMBL, a circRNA derived from MBL gene locus and containing the conserved binding site of MBL, binds to MBL and regulates the splicing of its own pre-mRNA (38). However, computational analysis predicted that the density of RBP binding sites is lower in circRNAs than in 3′ UTR regions of protein-coding genes (46).
Transcriptional Regulation
Most circRNAs are presented in the cytoplasm and act as either miRNA sponges or scaffolds. However, ciRNA and EIciRNA, such as circEIF3J and circPAIP2, remain in the nucleus and interact with U1 snRNA and RNA polymerase II complex to enhance the transcriptional activity of their parent gene (31). However, the underlying mechanism of the regulatory function of EIciRNA remains unclear.
Competitors of Linear Splicing and Gene Expression
The process of circRNA formation also affects the expression of host gene. Some evidences indicated that circRNA formation competes strongly with the linear splicing of pre-mRNA, and therefore, regulates host gene expression (38, 47). For example, an increase in linear splicing efficiency in Drosophila S2 cells led to a decrease in circRNA expression (38). In another example, a decrease in spliceosome components in drosophila cells resulted in an increase in circRNA levels and a decrease in its associated linear mRNA expression (47). Since linear- and back-splicing use the same typical splicing receptor and donor, it is not surprising that the level of circRNAs is negatively associated with the level of their linear mRNA isoforms.
Templates for Protein Synthesis
Although, circRNAs were first identified as non-coding RNAs, some of these circular RNA molecules were found to have the protein/peptide coding capability. Given that circRNAs are mostly localized in the cytoplasm and contain protein-coding exons, people wonder whether they can be loaded into ribosomes and serve as a template for protein/peptide synthesis. Interestingly, studies showed the initiation of translation of circRNAs can occur either at the internal ribosomal entry site (IRES) or at nucleotides with m6A modification in 5′ untranslated region (UTR) (48, 49), although, they lack the cap-dependent translation elements. So far, only a few endogenous circRNAs, such as circFBXW7, circMBL, and circ-ZnF609, have been shown to possess the effective open reading frame for protein/peptide translation (50–55). The function of most circRNA-derived peptides is unknown. It is worthy to note that circRNA-derived peptides were found to be expressed under different stress conditions, such as the translation of circ-ZnF609 in response to heat shock (50, 51). Although, translation of circRNAs does not appear to be a common function of circRNAs, the next important task in this field is to determine the regulatory function of circRNA-derived proteins/peptides.
Participation of circRNAs in the Pathogenesis of Cardiac Remodeling
Due to the limited regenerative capacity of myocardial tissue, the heart undergoes extensive remodeling to compensate the loss of cells or response to stress. During remodeling, hypertrophic growth and limited proliferation occur in cardiomyocytes. In addition, non-cardiomyocytes such as cardiac fibroblasts, endothelial cells, smooth muscle cells and immune cells, are all shown to actively participate in this disease progress. For example, the dying cardiomyocytes secret cytokines to activate the proliferation and differentiation of cardiac fibroblast, and to recruit immune cells to clean up the dead cardiomyocytes; ischemic and oxidative stress also trigger the proliferation of endothelial cells and angiogenesis for re-establishment of blood supply. Different cell types are tightly act together and communicate with each other either through cell-cell junction or cytokines, or even through exosomes during this process.
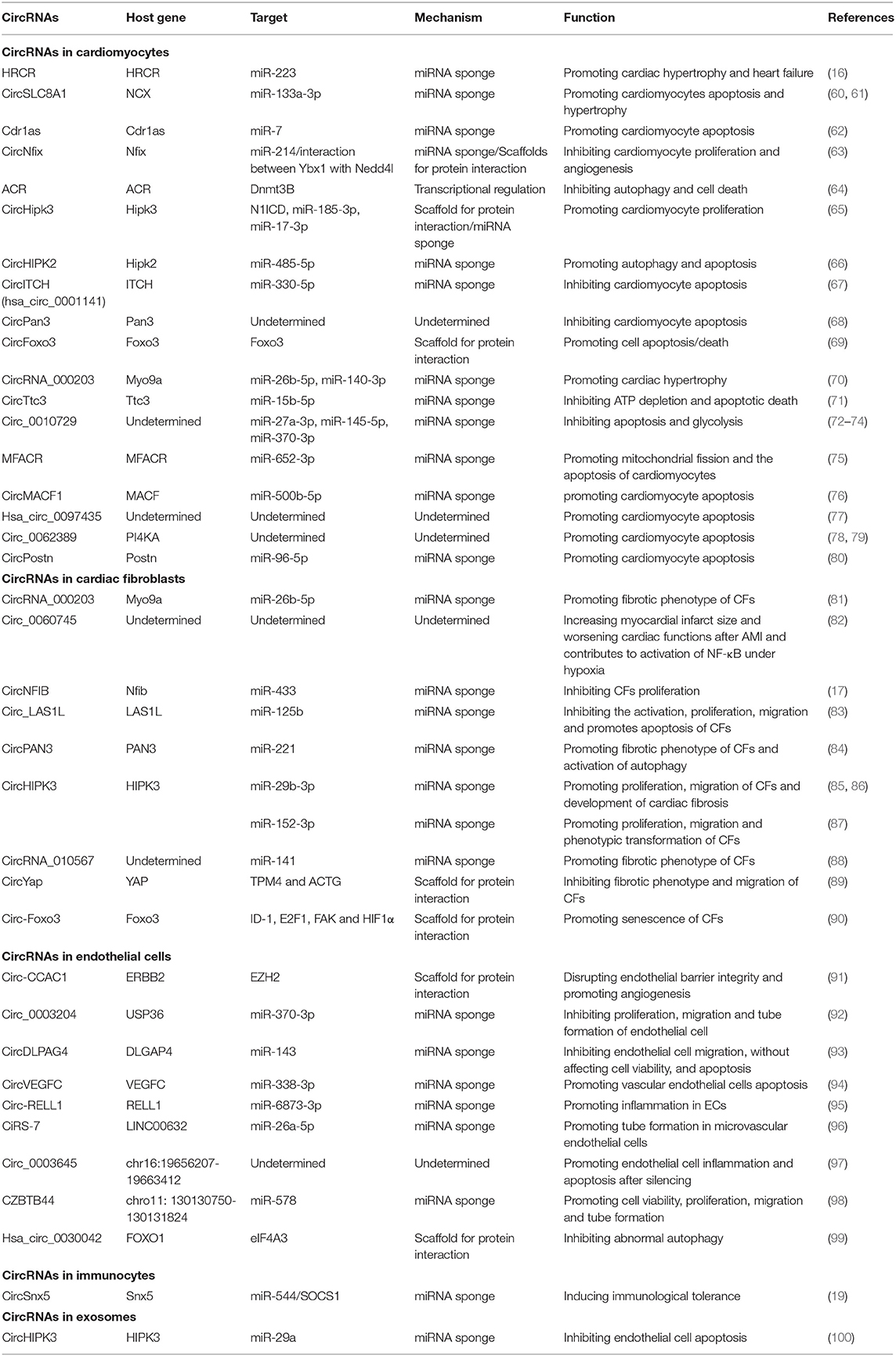
More and more evidences demonstrated that non-coding RNAs constitute a regulatory network in almost all forms of human diseases, including cardiac remodeling and heart failure. These RNA molecules incorporate into the known protein regulatory network to orchestrate a highly complicated gene regulatory network in human diseases. Cracking the “code” of this network will provide us a roadmap to fully understand mechanisms beneath disease phenotype we observed, and eventually lead us to a better and more effective therapy. Previous studies have demonstrated that some non-coding RNAs, such as microRNAs and long non-coding RNAs, have altered expression and play important regulatory roles in cardiac remodeling (14, 56, 57). Due to the different structure of circular RNAs, this type of non-coding RNAs have not been broadly studied until recently (58). Studies showed that circular RNAs are widely presented in different mammalian cell types (5). Growing evidence shows that circRNAs play important roles in cell proliferation, apoptosis, migration, and differentiation (43, 50, 59). Importantly, RNA-sequencing data showed that a subset of circular RNAs are dysregulated in diseased heart (37), supporting the idea of circular RNAs possessing regulatory functions in cardiac remodeling. Here, we systematically review the recent study progress of circRNAs in cardiac remodeling (Table 1 and Figure 3) and discuss the function of some representative circRNAs in cardiomyocytes, fibroblasts, endothelial cells, immune cells, and exosomes in detail.
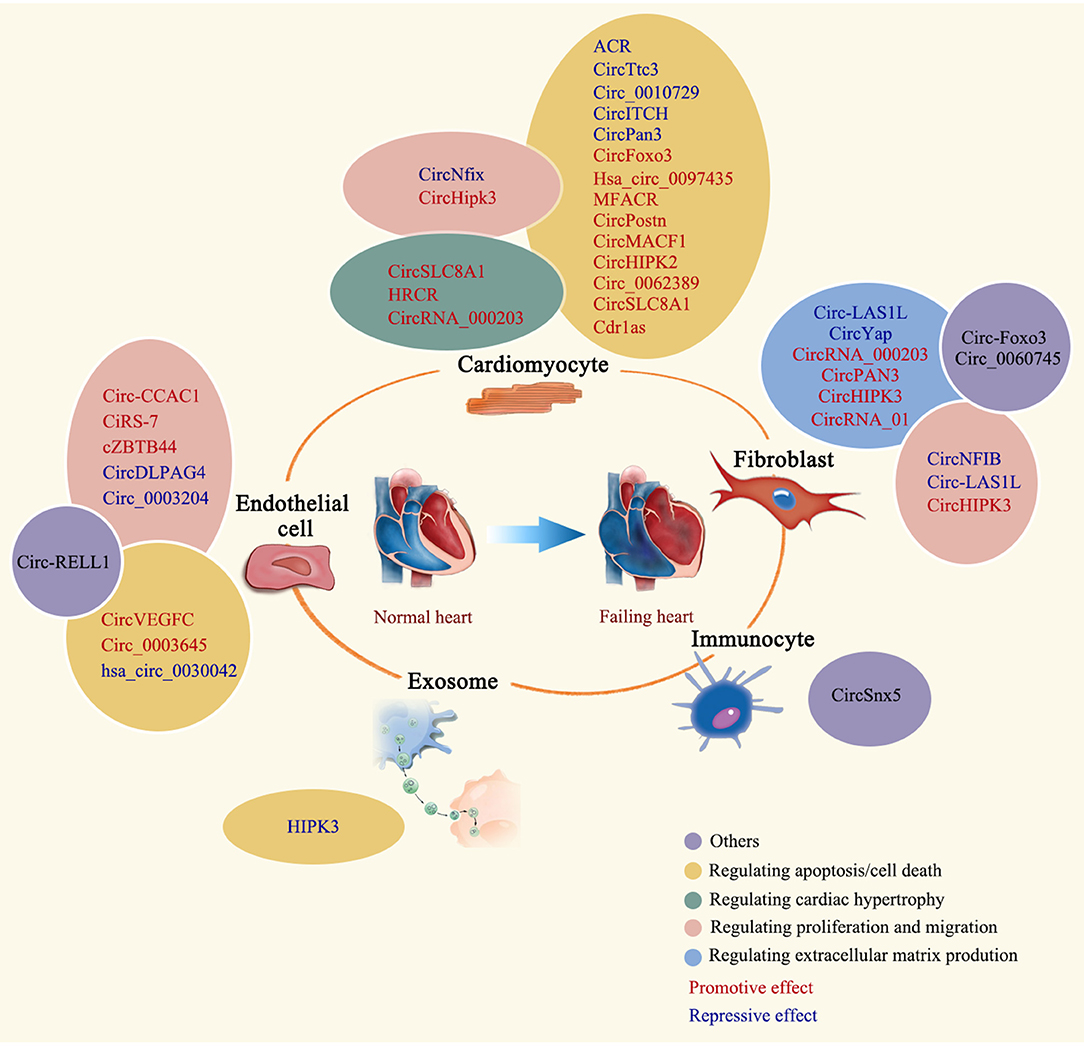
Figure 3. Regulatory roles of circRNAs in cardiac remodeling. The regulatory role of summarized circRNAs is indicated by colored ovals. CircRNAs are labeled in different colors to indicate the promotive or repressive function.
Circular RNAs in Cardiomyocyte
As the main contractile cells in the beating heart, the alteration of cardiomyocytes is the central of cardiac remodeling in various disease status. In adult heart, cardiomyocytes occupied around 75% of left ventricular volume (101). Cardiomyocytes undergo hypertrophic growth, apoptosis/necrosis, and limited proliferation during cardiac remodeling. Non-coding RNAs, such as microRNAs and long non-coding RNAs, have been demonstrated to regulate the pathophysiology of cardiomyocytes in diseased heart (102). Here, we discussed some emerging examples of circRNAs in cardiomyocytes during cardiac remodeling.
HRCR
HRCR was identified as the first circRNA regulating cardiac hypertrophy (16). The expression of circRNA HRCR was shown substantially decreased in mice in response to ISO or TAC treatment. HRCR has a protective function on cardiac hypertrophy and heart failure. Forced expression of HRCR mediated with adenoviral constructs in mouse heart decreases cardiomyocyte hypertrophic growth, interstitial fibrosis and preserves the cardiac function upon ISO treatment. Mechanistically, six target sites for miR-223 were identified in HRCR. HRCR sequesters and decreases the activity of miR-223, and upregulates the expression of target of miR-223, apoptosis repressor with CARD domain (ARC), which is a known regulator of cardiomyocyte hypertrophy and apoptosis (16).
CircSLC8A1
CircSLC8A1 (also named CircNCX1, or named circSlc8a1-1 in mouse), which is enriched in cardiomyocytes, was identified as the most abundant circRNA in human and mouse heart (103), whose host gene encodes the protein of sodium-calcium exchanger (NCX). CircSLC8A1 is generated from the 2nd exon of the host gene SLC8A1. Although, the expression of CircSLC8A1 remains unaltered under some disease conditions such as cardiac hypertrophy in mouse and failing heart in human, it has been confirmed to be involved in regulation of hypertrophic growth of cardiomyocytes as an endogenous sponge for miR-133a (24, 37, 60, 104). Of note, inhibition of circSLC8A1 promotes TAC-induced hypertrophy and HF in mouse (60). Cardiac-specific overexpression of circSlc8a1 in vivo mediated by the AAV9 increases heart weight and results in cardiac dilatation. Different from the expression in hypertrophy, the expression of circSLC8A1 have been demonstrated abnormally increased in dilated cardiomyopathy (105, 106), and upregulated in ischemic rat cardiac cells and mouse heart (61). Through a similar mechanism of acting as miR-133a-3p sponge, the NCX-derived circRNA increased the levels of CDIP1, a target for miR-133a-3p, which promotes cardiomyocyte apoptosis. Therefore, circSLC8A1 could exert different regulatory function in cardiomyocytes depending on the type of stress. Interestingly, circSLC8A1 interacts with the mouse ribosome or rat Argonaute 2 protein, which indicates circSLC8A1 is likely involved in the regulation of mRNA translation (106). Interesting, the expression level of circSLC8A1 is increased and positively correlated with the expression of CK-MB in the pericardial fluid of acute ischemic heart disease patients, which shows the potential of using this circRNA as an auxiliary diagnostic marker for clinical acute coronary syndromes (107).
CircNfix
CircNfix was identified as a super enhancer-associated circRNA by an integrated analysis with RNA-seq data and super enhancer catalogs (63). CircNfix was shown to regulate cardiomyocyte proliferation and angiogenesis. In vivo knockdown of circNfix mediated by cTNT-driven shRNA expression through AAV9 viral delivery system promotes cardiomyocyte proliferation evidenced by the increased the expression of the proliferation markers and the total cardiomyocyte number in infarcted mouse hearts, which leads to an improved cardiac function after MI. Although, CircNfix functions as a miRNA sponge to decrease the promotive function of miR-214 on cardiomyocyte proliferation, unexpectedly, it also promotes Ybx1 degradation through ubiquitination by enhancing the stability of the interaction between Ybx1 with Nedd4l, an E3 ubiquitin ligase. This observation indicates that CircNfix also functions as a scaffold for protein docking. Since transcriptional factor Ybx1 activates the expression of Ccna2 and Ccnb1 (108), CircNfix further decreases the proliferation of cardiomyocyte during myocardial infarction.
ACR
A recent study demonstrated that a circRNA ARC (autophagy-related circular RNA) plays an important role in cardiomyocyte autophagy (64). The expression of circRNA ACR is markedly decreased after the heart subjected to ischemia/reperfusion. ACR attenuates the increased autophagy level upon ischemia/reperfusion injury and plays a protective role in cardiomyocytes in vivo. Mice with overexpression of ACR in the heart had a less cardiomyocyte death in ventricular tissue and a smaller infarction. Mechanistically, instead of a miRNA sponge, ACR acts as a regulator of chromatin modification by binding to Dnmt3B and inhibiting Dnmt3B-mediated DNA methylation of Pink1 promoter. Since Pink1 targets and phosphorylates FAM65B, which was shown to have a regulatory role in autophagy, ARC mediates cardiomyocyte autophagy through a Dnmt3B/Pink1/FAM65B signaling cascade.
CircHipk3
Similar to ACR, some of circRNAs have been demonstrated to serve as a signaling regulator by binding with protein in cardiomyocytes. A recent study found an increased expression level of circHipk3 in the fetal or neonatal mouse heart (65). AAV9-mediated overexpression of circHipk3 attenuated cardiac dysfunction and fibrosis in a mouse model of myocardial infarction. CircHipk3 was shown to interact with N1ICD protein to increase N1ICD acetylation level and stability, which was partially responsible for the beneficial effect of circHipk3 in cardiomyocytes. On the contrary, silencing circHIPK3 has a protective effect in a variety of heart diseases. The hypertrophic growth of cardiomyocytes was markedly inhibited by the knockdown of circHIPK3 in a TAC-induced cardiac hypertrophy model (109). Since CircHIPK3 is a sponge of miR-185-3p, decreased level of CircHIPK3 in the knockdown increases the inhibitory effect of miR-185-3p on CaSR. In addition, knockdown of circHIPK3 benefits the heart after myocardial infarction through a circ-HIPK3/miR-17-3p/ADCY6 signaling cascade in cardiomyocytes (110). While in a model of LPS-induced myocarditis, knockdown of circHIPK3 significantly represses cardiomyocyte apoptosis and alleviates oxidative stress and inflammation in cardiac tissue (111). Interestingly, it was reported that circHIPK2, originated from the second exon of another HIPK family member HIPK2, facilitated autophagy in H2O2-caused myocardial injury via sponging miR-485-5p and de-repressing miR-485-5p target, ATG101 (66, 112).
Other circRNAs in Cardiomyocytes
Doxorubicin is widely used in tumor chemotherapy, however, with the dose-dependent cardiotoxicity. Doxorubicin-induced cardiotoxicity involves many molecular mechanisms, including induction of reactive oxygen species (113), inhibition of the activity of topoisomerase II (114), interruption of calcium homeostasis, induction of mitochondrial dysfunction, and destruction of sarcomere function (115). Recent studies showed that circRNAs are involved in doxorubicin-induced cardiomyopathy. A previous study identified 356 differentially expressed circRNAs in doxorubicin-treated human hearts (67). CircITCH, a circRNA highly conserved between human and mice, was significantly downregulated in DOX-induced cardiomyopathy. AAV9-mediated overexpression of circITCH ameliorates oxidative stress and DNA damage, cell death, contractile dysfunction, and calcium handling defects in DOX-induced cardiomyopathy in a mouse model by acting as a miR-330-5p sponge to de-repress the expression of SIRT6, BIRC5, and ATP2A2. In another recent study, overexpression of circPan3 was shown to attenuate DOX-induced cardiomyocyte apoptosis with unknown mechanism (68). In another example, circ-Foxo3 interacts with the anti-senescence proteins ID1 and E2F1, and anti-stress proteins FAK and HIF1α to prevent their nuclear translocation for transcription (90). Therefore, silencing of circ-Foxo3 relieved the cardiac injury induced by doxorubicin. In addition, circFoxo3 levels were significantly higher in I/R injury resulted from 24 h of cold storage and reperfusion in heart transplantation (69). In vivo and in vitro experiments demonstrates that knockdown of circFoxo3 improves heart graft function and reduced cell apoptosis/death and mitochondrial damage.
Circular RNAs in the Activation of Cardiac Fibroblasts
Cardiac fibrosis, generally referred to an aberrant accumulation of extracellular matrix (ECM) proteins in the interstitial space of heart tissue, is closely associated with pathological cardiac remodeling. It manifests as deposition of scar, increasing stiffness, decreasing contraction and impaired heart function, which ultimately resulting in heart failure (116). Although, cardiac fibrosis is a complex process and involves many types of cells in the heart, such as cardiomyocytes, fibroblasts, lymphocytes, and pericytes (117–119), extensive studies have proved that cardiac fibroblasts (CFs) play a pivotal role in this process (120). When suffered cardiac injury, the proliferation and migration of CFs are increased. Moreover, cytokines, especially the transforming growth factor-β (TGF-β), and growth factors are also secreted, which contributes to fibroblast activation and ultimately transform CFs into myofibroblasts (121). In this situation, myofibroblasts begin to express α-smooth muscle actin (α-SMA) and enhance the secretion of ECM proteins, such as collagen type I and collagen type III, which results in the formation of scar and eventually leads to cardiac fibrosis (122). Recent studies have shown circRNAs actively participate in the pathogenesis of cardiac fibrosis, especially in the activation of cardiac fibroblasts.
CircRNA_000203
CircRNA_000203, derived from Myo9a, is upregulated in the myocardium of diabetic mouse and Ang-II-treated cardiac fibroblasts. Overexpression of circRNA_000203 in mouse CFs in vitro increased the expression of Col1a2, Col3a1, and α-SMA, indicating the activation of CFs and the accumulation of ECM. Moreover, RNA pull-down confirmed that circRNA_000203 is a sponge of miR-26b-5p. The targeting of CTGF and Col1a2 by miR-26b was experimentally confirmed, which was interfered by the overexpression of circRNA_000203 (81). Interestingly, circRNA_000203 is also upregulated in Ang-II-treated cardiomyocytes. Forced expression of circRNA_000203 promotes the hypertrophic growth of cardiomyocytes and transgenic mice with cardiac-specific overexpression of circRNA_000203 have an advanced phenotype in a model of Ang-II-induced cardiac hypertrophy (70).
CircNFIB
CircNFIB (mmu_circ_0011794), generated from the exon regions of Nfib, was identified as a candidate circRNA to sponge miR-433, a miRNA promoting cardiac fibrosis (123). CircNFIB has a decreased expression in both 3-week post-MI mice hearts and TGF-β-treated CFs. The proliferation of CFs, induced by TGF-β treatment, was significantly inhibited by the overexpression of circNFIB in vitro (17). Mechanistically, circNFIB de-represses AZIN1 and JNK1, which are targeted by miR-433. Overexpression of circNFIB increased the expression of AZIN1 and JNK1 and impaired the activation p38/ERK/Smad3, thus confirming the pivotal role of circNFIB as a competing endogenous RNA (ceRNA) in cardiac fibrosis. However, the role of circNFIB in vivo in cardiac remodeling is still unclear and requires for further investigation.
Circ_LAS1L
CircRNAs have been found to be involved in acute myocardial infarction (AMI) in recent years (124). Bie et al. (125) demonstrated the crucial function of miR-125b/SFRP5 axis in CF growth and activation in previous report. Further, analysis showed that miR-125b targets circ_LAS1L with two binding sites, which was confirmed by RIP and RNA Pull-down. The expression level of circ_LAS1L is significantly downregulated while miR-125b expression is increased in AMI patients. Forced expression of circ_LAS1L upregulates the expression of SFRP5 and downregulates the expression of α-SMA, collagen I, and collagen III in CFs. Interestingly, gain function of miR-125b together with overexpression of circ_LAS1L appears not to modulate CFs proliferation, apoptosis, and migration. However, SFRP5 siRNA, instead of miR-125b mimics, bypassed the counter effect of circ_LAS1L on CF proliferation and migration, indicating that circ_LAS1L functions as a sponge of miR-125b to modulate CFs proliferation and migration in vitro (83). Whether circ_LAS1L has a repressive function on cardiac fibrosis upon cardiac injury in vivo warrant investigation in the future.
CircRNA_PAN3
CircPAN3, a circRNA generated from the PAN3 locus, has been found to maintain the self-renewal of intestinal stem cells (126), to modulate drug resistance in acute myeloid leukemia (AML) (127, 128), and to recede myocardial ischaemia/reperfusion injury (129). Recently, Li et al. reported circPAN3 as a new profibrotic factor in cardiac fibrosis (84). The expression of circPAN3 increases significantly in fibrotic regions of rat heart induced by myocardial infarction (MI). Silencing of circPAN3 in MI hearts reduces the level of fibrosis, including decreased expression of fibrotic markers, and inhibits cardiac myocyte apoptosis and autophagy. Consistently, knockdown of circPAN3 represses TGF-β induced proliferation, migration and autophagy of CFs in vitro. In the molecular level, circPAN3 was demonstrated to interact with miR-221 and sequester miR-221 from regulate its targets, FoxO3, as a sponge. Gain function of miR-221 decreases the expression of FoxO3 and ATG7, two known targets for miR-221 and have been proved crucial in autophagy in previous studies (130). These data indicate that circPAN3 promotes fibrosis via miR-221/FoxO3/ATG7 cascade-mediated autophagy.
Circ_0060745
A recent study reported that circ_0060745 expression level in CFs is increased dramatically in the myocardium of AMI mice (82). Knockdown and overexpression of circ_0060745 improved and deteriorated the cardiac function, respectively. In addition, the silencing of circ_0060745 leads to less cell apoptosis in the infarcted areas while circ_0060745 overexpression had the opposite effect. Further, analysis found that the expression of inflammatory cytokines, including IL-6, IL-12, IL-1β, and TNF-α, are decreased upon knockdown of circ_0060745, which could suppress peritoneal macrophage migration. The downregulation of the inflammatory cytokines is induced by the inhibition of NF-κB activation in the circ_0060745 knockdown.
CircHIPK3
CircHIPK3 (mmu_circ_0001052), originated from exon 2 of HIPK3, has been reported to play an important role in cancers (131). Recent studies suggested it also regulates cardiac fibrosis via different signaling cascades. Ni et al. found circHIPK3 promoted CF proliferation, migration and activation by modulating the activity of a known fibrosis-related microRNA, miR-29b-3p, through sponging (85). Similarly, another study showed circHIPK3 induced cardiac fibrosis through a circHIPK3/miR-29b-3p/Col1a1/Col3a1 signaling cascade in mouse diabetic cardiomyopathy model (86). Interestingly, circHIPK3 modulates CFs function in a hypoxia condition in a similar sponge manner, but through a different signaling cascade, the circHIPK3/miR-152-3p/TGF-β2 axis (87). Therefore, circHIPK3 is likely to be an important upstream node of the miRNA-mediated posttranscriptional gene regulation network in cardiac fibrosis.
CircYap
CircYap hsa_circ_0002320, generated from exons 5 and exon 6 of YAP pre-mRNA, is the highest expressed isoforms derived from Yap gene locus in human hearts. The expression level of circYap decreases significantly in hypertrophic patient hearts of patients and in pressure-overloaded mouse hearts. The forced expression of circYap alleviates the declined heart function and increased cardiac fibrosis in a TAC-induced mouse cardiac hypertrophy model. Overexpression of circYap in cardiac fibroblasts (MCF) in vitro suppresses the expression of fibrotic markers and migration of cardiac fibroblasts. Mechanistically, circYap interacts with both tropomyosin-4 (TMP4) and gamma-actin (ACTG), and enhances the interaction between TMP4 and ACTG, which subsequently inhibits the actin polymerization and cardiac fibrosis (89).
Circular RNAs in Endothelial Cells
Endothelial cells (ECs) play a central role in cardiac remodeling, regeneration, as well as angiogenesis in the treatment of cardiovascular diseases (132, 133). Therefore, it is of great significance to identify factors that promoting and inhibiting angiogenesis and their underlying molecular mechanisms. To date, many studies have confirmed that circRNAs are involved in regulating the proliferation, migration, apoptosis, and tubule formation of ECs, which further mediates the dynamics of ECs and regulates angiogenesis.
Circ-ZnF609
The level of circ-ZNF609 in peripheral blood leukocytes of coronary artery disease patients is significantly decreased (134). In another study of circRNAs in retinal vascular dysfunction, Circ-ZnF609 was found significantly upregulated under conditions of high glucose and hypoxia stress in vivo and in vitro (18). Silence of circ-ZnF609 increased endothelial cell migration and tube formation, and protected endothelial cells against oxidative stress and hypoxia stress in vitro. Circ-ZnF609 acts as an endogenous miR-615-5p sponge to sequester miR-615-5p and inhibit its function, leading to the increased expression of MEF2A. Overexpression of MEF2A rescues the endothelial cell migration, tube formation, and apoptosis mediated by the silence of circ-ZnF609, which further demonstrates the regulatory mechanism of circ-ZnF609 on ECs via circ-ZnF609/miR-615-5p/MEF-2A signaling cascade.
Interestingly, circ-ZnF609 was also identified as a circRNA regulating muscle differentiation in mice and humans, and its expression is altered in Duchenne muscular dystrophy (DMD) myoblasts (50). Circ-ZnF609 specifically controls the proliferation of myoblasts. It was demonstrated that circ-ZnF609 is associated with heavy polysomes and translated into a protein in a splicing-dependent and cap-independent manner, providing an example of protein encoding circRNA in eukaryotes. Whether circ-ZnF609 functions as a miRNA sponge or protein-coding circRNA, or both, or it has a preferable manner depended on cell type, needs to be studied in the future. Furthermore, the role of endothelial circ-ZnF609 in the heart upon ischemia/reperfusion in vivo warrants investigation.
CircFndc3b
CircFndc3b is differentially expressed in the mouse hearts after myocardial infarction (MI) and in the heart tissues of patients with ischemic cardiomyopathy (135). Overexpression of circFndc3b in cardiac endothelial cells increases the expression of vascular endothelial growth factor-A, enhances angiogenesis, and reduces the apoptosis of cardiomyocytes and endothelial cells. In post-MI hearts, adeno-associated virus-mediated overexpression of circFndc3b reduces myocardial apoptosis, enhances neovascularization, and improves left ventricular function. CircFndc3b interacts with the RNA-binding protein FUS in sarcoma to regulate VEGF expression and signal transduction. These findings highlight the physiological role of circRNA in heart repair and suggest that regulating the expression of CircFndc3b is a potential therapeutic strategy for protecting the heart from myocardial infarction.
CZNF292
Hypoxia condition is introduced in the ischemic region of the diseased heart. To identify hypoxia-related circRNAs, endothelial circRNAs were screened from human umbilical vein endothelial cells cultured under normal or hypoxia conditions (136). CZNF292 is one of candidates identified in the screen. In vitro, target-specific depletion of CZNF292 with siRNAs inhibits angiogenic germination and spherical germination of endothelial cells, suggesting that CZNF292 has pro-angiogenic function. The overexpression of CZNF292 further confirms its pro-proliferation effect. Interestingly, the circRNA appears not associate with Argonaute, indicating it unlikely functions as a microRNA sponge. These data suggest that endothelial circRNAs could mediate angiogenesis under hypoxia condition with undetermined mechanism. Although, the in vivo function of CZNF292 in the heart is still unknown, it is interesting to investigate whetherCZNF292 promotes angiogenesis in vivo and benefit the heart after ischemia/reperfusion injury.
Circular RNAs in the Dynamics of Cardiac Immunocytes
Inflammation and fibrosis are two key factors in cardiac remodeling. Inflammatory response is generally caused by acute cell death, for instance, the sudden loss of cardiomyocytes after myocardial infarction. Necrotic cardiomyocytes crack and release cellular contents, which activates the inflammation reaction for cleaning dead cells and matrix debris (137, 138). During the cardiac inflammatory process, immunocytes accumulate in myocardium, infiltrate surrounding tissue and further regulate inflammatory reaction (139, 140). Among immunocytes, dendritic cells (DCs), derived from bone marrow, are antigen-presenting cells and crucial in immune response. Moreover, cardiac DCs has been reported to have heart-protective effects in acute myocardial infarction (AMI), such as the deletion of DCs in mice deteriorates the cardiac remodeling (141). In addition, cardiac specific tDCs (tolerogenic dendritic cells) can evoke the generation of Tregs, which can promote a macrophage-specific repair program after AMI (142).
Recently, Yu et al. reported a novel DC-expressed circRNA, named circSnx5, has a vital function in maintaining cardiac immune homeostasis. CircSnx5, generated from the snx5 gene locus, represses the maturation of DCs when its expression is upregulated in DCs. Knockdown of circSnx5 results in an inflammatory phenotype of dendritic cells. Mechanistically, circSnx5 sponges miR-544 and de-represses the downstream target of miR-544, the suppressor of cytokine signaling 1 (SOCS1). On the other hand, circSnx5 directly influences the nuclear translocation of PU.1 to regulate the expression of downstream MHC class II, which is critical to DC's function. In addition, the injury and inflammation of cardiac tissue is decreased, and the cardiac function is improved after introducing circSnx5-overexpressing DCs into experimental autoimmune myocarditis (EAM) mice. Thus, all these results confirm that circSnx5 has a protective effect on AMI (19).
Circular RNA Messenger in Exosomes
It is well-known that exosomes are involved in the intercellular communication. Correct communication between cells has been shown critical in preserving body homeostasis and health (102). Cell-cell communication via exosomes is involved in the pathological processes of some chronic diseases such as cancer and heart diseases (143, 144), but the disease progression regulated by circRNAs from the shuttling exosomes in the heart were less studied (145).
A recent study has provided evidence supporting the role of exosomal circRNAs in multiple physiological processes including the regulation of the heart function. Wang et al. found that exosomal circHIPK3 is highly expressed in hypoxic exosomes secreted from cardiomyocytes. Silencing of circHIPK3 is associated with increased levels of apoptosis, ROS, MDA, and proapoptotic proteins in cardiac microvascular endothelial cells (CMVECs) (100). The upregulated circHIPK3 sponges miR-29a, an apoptosis-suppressing miRNA, to de-repress the expression of IGF-1, and subsequently regulates the oxidative damage in CMVECs.
Conclusion
In conclusion, circRNAs are identified as new players to participate in the process of human diseases. Although, the biogenesis and molecular mechanism of circRNAs are still not fully understood, emerging evidences have demonstrated that these circular RNA molecules are broadly present in mammalian cells with different regulatory functions. The heart is composed of a variety of cell types that interact with each other through direct contact or paracrine signaling. More and more studies have proved that circRNAs are involved in the process of cardiac remodeling and have important regulatory functions in cardiomyocytes, endothelial cells, fibroblasts and immune cells during this disease process. The regulation of circRNAs in different cell types of the heart adds a new layer of regulation to the known gene regulation network of cardiovascular disease. Currently, we are still facing challenges in the study of circRNAs. For example, most of circRNAs can not be knocked out for the loss-of-function study since targeting circRNAs using CRISPR–Cas9 or DNA recombination strategy is likely to affect the splicing or expression of linear host genes. Although, the expression of circRNAs could be knocked down by specific siRNAs, the narrow junction of back-splicing limits the design of siRNA for a portion of circRNAs. Therefore, approaches for circRNA study are urged to be improved, which will lead us to better understand the function of circRNAs in human diseases.
Overall, roles of circRNAs in the pathogenesis of cardiovascular disease still remains largely unknown. Unlike linear RNA molecules, the stability of circRNAs grants the advantage of these circular RNA molecules in therapeutic applications, such as disease diagnosis and transgene delivery in gene therapy. Given that extracellular vesicles or exosomes contain circRNAs, capturing tissue-specific and disease-specific vesicles or exosomes for circRNAs profiling could be a good strategy to identify biomarkers for disease diagnosis. Therefore, deeper and systematic studies of circRNAs in the content of different diseases, such as cardiac hypertrophy and heart failure, is the prerequisite of moving the knowledge of circRNAs into the therapeutic applications against the deadly cardiovascular disease.
Author Contributions
TY, TL, TD, and Z-PH prepared the manuscript. TY, TL, and TD wrote the main parts of the article and produced graphics. YD and YC reviewed and edited the manuscript. Z-PH drafted the final version of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81873463 to Z-PH), the Guangdong Basic and Applied Basic Research Foundation (2019B151502003 to Z-PH and 2018A030313448 to YC), the Guangdong Science and Technology Department (2018A050506026 to Z-PH), the Fundamental Research Funds for the Central Universities (20ykzd06 to Z-PH), the Traditional Chinese Medicine Bureau of Guangdong Province (20181056), and the donation for scientific research from the Terry Fox Foundation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank members of the Huang laboratory for advice and technical support.
References
1. Porter KE, Turner NA. Cardiac fibroblasts: at the heart of myocardial remodeling. Pharmacol Ther. (2009) 123:255–78. doi: 10.1016/j.pharmthera.2009.05.002
2. Aird WC. Phenotypic heterogeneity of the endothelium: I. Structure, function, and mechanisms. Circ Res. (2007) 100:158–73. doi: 10.1161/01.RES.0000255691.76142.4a
3. Colliva A, Braga L, Giacca M, Zacchigna S. Endothelial cell-cardiomyocyte crosstalk in heart development and disease. J Physiol. (2020) 598:2923–39. doi: 10.1113/JP276758
4. Frieler RA, Mortensen RM. Immune cell and other non-cardiomyocyte regulation of cardiac hypertrophy and remodeling. Circulation. (2015) 131:1019–30. doi: 10.1161/CIRCULATIONAHA.114.008788
5. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. (2013) 19:141–57. doi: 10.1261/rna.035667.112
6. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. (2013) 495:333–8. doi: 10.1038/nature11928
7. Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, et al. Scrambled exons. Cell. (1991) 64:607–13. doi: 10.1016/0092-8674(91)90244-S
8. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long non-coding RNAs. Mol Cell. (2013) 51:792–806. doi: 10.1016/j.molcel.2013.08.017
9. Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. (2015) 10:170–7. doi: 10.1016/j.celrep.2014.12.019
10. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. (2013) 9:e1003777. doi: 10.1371/journal.pgen.1003777
11. Maass PG, GlaŽar P, Memczak S, Dittmar G, Hollfinger I, Schreyer L, et al. A map of human circular RNAs in clinically relevant tissues. J Mol Med. (2017) 95:1179–89. doi: 10.1007/s00109-017-1582-9
12. Fasolo F, Di Gregoli K, Maegdefessel L, Johnson JL. Non-coding RNAs in cardiovascular cell biology and atherosclerosis. Cardiovasc Res. (2019) 115:1732–56. doi: 10.1093/cvr/cvz203
13. Schiano C, Benincasa G, Franzese M, Della Mura N, Pane K, Salvatore, et al. Epigenetic-sensitive pathways in personalized therapy of major cardiovascular diseases. Pharmacol Ther. (2020) 210:107514. doi: 10.1016/j.pharmthera.2020.107514
14. Kumarswamy R, Thum T. Non-coding RNAs in cardiac remodeling and heart failure. Circ Res. (2013) 113:676–89. doi: 10.1161/CIRCRESAHA.113.300226
15. Devaux Y, Creemers EE, Boon RA, Werfel S, Thum T, Engelhardt S, et al. Circular RNAs in heart failure. Eur J Heart Fail. (2017) 19:701–9. doi: 10.1002/ejhf.801
16. Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. (2016) 37:2602–11. doi: 10.1093/eurheartj/ehv713
17. Zhu Y, Pan W, Yang T, Meng X, Jiang Z, Tao L, et al. Upregulation of circular RNA CircNFIB attenuates cardiac fibrosis by sponging miR-433. Front Genet. (2019) 10:564. doi: 10.3389/fgene.2019.00564
18. Liu C, Yao MD, Li CP, Shan K, Yang H, Wang JJ, et al. Silencing Of circular RNA-ZNF609 ameliorates vascular endothelial dysfunction. Theranostics. (2017) 7:2863–77. doi: 10.7150/thno.19353
19. Chen Q, Mang G, Wu J, Sun P, Li T, Zhang H, et al. Circular RNA circSnx5 controls immunogenicity of dendritic cells through the miR-544/SOCS1 axis and PU.1 activity regulation. Mol Ther. (2020) 28:2503–18. doi: 10.1016/j.ymthe.2020.07.001
20. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. (1976) 73:3852–6. doi: 10.1073/pnas.73.11.3852
21. Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules. Faseb J. (1993) 7:155–60. doi: 10.1096/fasebj.7.1.7678559
22. Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. (1993) 73:1019–30. doi: 10.1016/0092-8674(93)90279-Y
23. Cocquerelle C, Daubersies P, Majérus MA, Kerckaert JP, Bailleul B. Splicing with inverted order of exons occurs proximal to large introns. Embo J. (1992) 11:1095–8. doi: 10.1002/j.1460-2075.1992.tb05148.x
24. Werfel S, Nothjunge S, Schwarzmayr T, Strom TM, Meitinger T, Engelhardt S. Characterization of circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol. (2016) 98:103–7. doi: 10.1016/j.yjmcc.2016.07.007
25. Jakobi T, Czaja-Hasse LF, Reinhardt R, Dieterich C. Profiling and validation of the circular RNA repertoire in adult murine hearts. Genomics Proteomics Bioinformatics. (2016) 14:216–23. doi: 10.1016/j.gpb.2016.02.003
26. Li Y, Zhang J, Huo C, Ding N, Li J, Xiao J, et al. Dynamic organization of lncRNA and circular RNA regulators collectively controlled cardiac differentiation in humans. EBioMedicine. (2017) 24:137–46. doi: 10.1016/j.ebiom.2017.09.015
27. Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular RNA: a new star of non-coding RNAs. Cancer Lett. (2015) 365:141–8. doi: 10.1016/j.canlet.2015.06.003
28. Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. (2006) 34:e63. doi: 10.1093/nar/gkl151
29. Wu TY, Zhang TH, Qu LM, Feng JP, Tian LL, Zhang BH, et al. MiR-19a is correlated with prognosis and apoptosis of laryngeal squamous cell carcinoma by regulating TIMP-2 expression. Int J Clin Exp Pathol. (2014) 7:56–63.
30. Chen L, Huang C, Wang X, Shan G. Circular RNAs in eukaryotic cells. Curr Genomics. (2015) 16:312–8. doi: 10.2174/1389202916666150707161554
31. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. (2015) 22:256–64. doi: 10.1038/nsmb.2959
32. Eger N, Schoppe L, Schuster S, Laufs U, Boeckel JN. Circular RNA splicing. Adv Exp Med Biol. (2018) 1087:41–52. doi: 10.1007/978-981-13-1426-1_4
33. Burset M, Seledtsov IA, Solovyev VV. Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res. (2000) 28:4364–75. doi: 10.1093/nar/28.21.4364
34. Dubin RA, Kazmi MA, Ostrer H. Inverted repeats are necessary for circularization of the mouse testis Sry transcript. Gene. (1995) 167:245–8. doi: 10.1016/0378-1119(95)00639-7
35. Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. (2014) 28:2233–47. doi: 10.1101/gad.251926.114
36. Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. (2014) 159:134–47. doi: 10.1016/j.cell.2014.09.001
37. Khan MA, Reckman YJ, Aufiero S, van den Hoogenhof MM, van der Made I, Beqqali A, et al. RBM20 regulates circular rna production from the titin gene. Circ Res. (2016) 119:996–1003. doi: 10.1161/CIRCRESAHA.116.309568
38. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. (2014) 56:55–66. doi: 10.1016/j.molcel.2014.08.019
39. Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, Cherry, et al. Combinatorial control of drosophila circular RNA expression by intronic repeats, hnRNPs, SR proteins. Genes Dev. (2015) 29:2168–82. doi: 10.1101/gad.270421.115
40. Zaphiropoulos PG. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci U S A. (1996) 93:6536–41. doi: 10.1073/pnas.93.13.6536
41. Wilkinson ME, Charenton C, Nagai K. RNA splicing by the spliceosome. Annu Rev Biochem. (2020) 89:359–88. doi: 10.1146/annurev-biochem-091719-064225
42. Shen T, Han M, Wei G, Ni T. An intriguing RNA species–perspectives of circularized RNA. Protein Cell. (2015) 6:871–80. doi: 10.1007/s13238-015-0202-0
43. Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. (2014) 20:1829–42. doi: 10.1261/rna.047126.114
44. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. (2013) 495:384–8. doi: 10.1038/nature11993
45. Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. (2017) 14:361–9. doi: 10.1080/15476286.2017.1279788
46. You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. (2015) 18:603–10. doi: 10.1038/nn.3975
47. Liang D, Tatomer DC, Luo Z, Wu H, Yang L, Chen LL, et al. The output of protein-coding genes shifts to circular RNAs When the pre-mRNA processing machinery is limiting. Mol Cell. (2017) 68:940–54.e943. doi: 10.1016/j.molcel.2017.10.034
48. Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, et al. 5' UTR m(6)A promotes cap-independent translation. Cell. (2015) 163:999–1010. doi: 10.1016/j.cell.2015.10.012
49. Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. (2015) 526:591–4. doi: 10.1038/nature15377
50. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. (2017) 6:22–37.e29. doi: 10.1016/j.molcel.2017.02.017
51. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of circRNAs. Mol Cell. (2017) 66:9–21.e27. doi: 10.1016/j.molcel.2017.02.021
52. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. (2017) 27:626–41. doi: 10.1038/cr.2017.31
53. Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. (2018) 110:304–15. doi: 10.1093/jnci/djx166
54. Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. (2018) 37:1805–14. doi: 10.1038/s41388-017-0019-9
55. Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun. (2018) 9:4475. doi: 10.1038/s41467-018-06862-2
56. Nakamura M, Sadoshima J. Mechanisms of physiological and pathological cardiac hypertrophy. Nat Rev Cardiol. (2018) 15:387–407. doi: 10.1038/s41569-018-0007-y
57. Poller W, Dimmeler S, Heymans S, Zeller T, Haas J, Karakas M, et al. Non-coding RNAs in cardiovascular diseases: diagnostic and therapeutic perspectives. Eur Heart J. (2018) 39:2704–16. doi: 10.1093/eurheartj/ehx165
58. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. (2019) 20:675–91. doi: 10.1038/s41576-019-0158-7
59. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. (2016) 44:2846–58. doi: 10.1093/nar/gkw027
60. Lim TB, Aliwarga E, Luu TDA, Li YP, Ng SL, Annadoray L, et al. Targeting the highly abundant circular RNA circSlc8a1 in cardiomyocytes attenuates pressure overload induced hypertrophy. Cardiovasc Res. (2019) 115:1998–2007. doi: 10.1093/cvr/cvz130
61. Li M, Ding W, Tariq MA, Chang W, Zhang X, Xu W, et al. A circular transcript of ncx1 gene mediates ischemic myocardial injury by targeting miR-133a-3p. Theranostics. (2018) 8:5855–69. doi: 10.7150/thno.27285
62. Geng HH, Li R, Su YM, Xiao J, Pan M, Cai XX, et al. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS ONE. (2016) 11:e0151753. doi: 10.1371/journal.pone.0151753
63. Huang S, Li X, Zheng H, Si X, Li B, Wei G, et al. Loss of super-enhancer-regulated circRNA nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation. (2019) 139:2857–76. doi: 10.1161/CIRCULATIONAHA.118.038361
64. Zhou LY, Zhai M, Huang Y, Xu S, An T, Wang YH, et al. The circular RNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the Pink1/FAM65B pathway. Cell Death Differ. (2019) 26:1299–315. doi: 10.1038/s41418-018-0206-4
65. Si X, Zheng H, Wei G, Li M, Li W, Wang H, et al. circRNA Hipk3 induces cardiac regeneration after myocardial infarction in mice by binding to notch1 and miR-133a. Mol Ther Nucleic Acids. (2020) 21:636–55. doi: 10.1016/j.omtn.2020.06.024
66. Zhou J, Li L, Hu H, Wu J, Chen H, Feng, et al. Circ-HIPK2 accelerates cell apoptosis and autophagy in myocardial oxidative injury by sponging miR-485-5p and targeting ATG101. J Cardiovasc Pharmacol. (2020) 76:427–36. doi: 10.1097/FJC.0000000000000879
67. Han D, Wang Y, Wang Y, Dai X, Zhou T, Chen J, et al. The tumor-suppressive human circular RNA CircITCH sponges miR-330-5p to ameliorate doxorubicin-induced cardiotoxicity through upregulating SIRT6, survivin, and SERCA2a. Circ Res. (2020) 127:e108–25. doi: 10.1161/CIRCRESAHA.119.316061
68. Ji X, Ding W, Xu T, Zheng X, Zhang J, Liu M, et al. MicroRNA-31-5p attenuates doxorubicin-induced cardiotoxicity via quaking and circular RNA Pan3. J Mol Cell Cardiol. (2020) 140:56–67. doi: 10.1016/j.yjmcc.2020.02.009
69. Su Y, Zhu C, Wang B, Zheng H, McAlister V, Lacefield JC, et al. Circular RNA Foxo3 in cardiac ischemia-reperfusion injury in heart transplantation: a new regulator and target. Am J Transplant. (2020). doi: 10.1111/ajt.16475. [Epub ahead of print].
70. Li H, Xu JD, Fang XH, Zhu JN, Yang J, Pan R, et al. Circular RNA circRNA_000203 aggravates cardiac hypertrophy via suppressing miR-26b-5p and miR-140-3p binding to Gata4. Cardiovasc Res. (2020) 116:1323–34. doi: 10.1093/cvr/cvz215
71. Cai L, Qi B, Wu X, Peng S, Zhou G, Wei Y, et al. Circular RNA Ttc3 regulates cardiac function after myocardial infarction by sponging miR-15b. J Mol Cell Cardiol. (2019) 130:10–22. doi: 10.1016/j.yjmcc.2019.03.007
72. Lei D, Wang Y, Zhang L, Wang Z. Circ_0010729 regulates hypoxia-induced cardiomyocyte injuries by activating TRAF5 via sponging miR-27a-3p. Life Sci. (2020) 262:118511. doi: 10.1016/j.lfs.2020.118511
73. Jin Q, Chen Y. Silencing circular RNA circ_0010729 protects human cardiomyocytes from oxygen-glucose deprivation-induced injury by up-regulating microRNA-145-5p. Mol Cell Biochem. (2019) 462:185–94. doi: 10.1007/s11010-019-03621-9
74. Zhang J, Gao C, Zhang J, Ye F. Circ_0010729 knockdown protects cardiomyocytes against hypoxic dysfunction via miR-370-3p/TRAF6 axis. EXCLI journal. (2020) 19:1520–32. doi: 10.17179/excli2020-2809
75. Wang K, Gan TY, Li N, Liu CY, Zhou LY, Gao JN, et al. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. (2017) 24:1111–20. doi: 10.1038/cdd.2017.61
76. Zhao B, Li G, Peng J, Ren L, Lei L, Ye H, et al. CircMACF1 attenuates acute myocardial infarction through miR-500b-5p-EMP1 axis. J Cardiovasc Transl Res. (2021) 14:161–72. doi: 10.1007/s12265-020-09976-5
77. Han J, Zhang L, Hu L, Yu H, Xu F, Yang B, et al. Circular RNA-expression profiling reveals a potential role of Hsa_circ_0097435 in heart failure via sponging multiple MicroRNAs. Front Genet. (2020) 11:212. doi: 10.3389/fgene.2020.00212
78. Zhang Y, Chen B. Silencing circ_0062389 alleviates cardiomyocyte apoptosis in heart failure rats via modulating TGF-β1/Smad3 signaling pathway. Gene. (2021) 766:145154. doi: 10.1016/j.gene.2020.145154
79. She Y, Han Y, Zhou G, Jia F, Yang T, Shen Z. hsa_circ_0062389 promotes the progression of non-small cell lung cancer by sponging miR-103a-3p to mediate CCNE1 expression. Cancer Genet. (2020) 241:12–9. doi: 10.1016/j.cancergen.2019.12.004
80. Cheng N, Wang MY, Wu YB, Cui HM, Wei SX, Liu B, et al. Circular RNA POSTN promotes myocardial infarction-induced myocardial injury and cardiac remodeling by regulating miR-96-5p/BNIP3 axis. Front Cell Dev Biol. (2020) 8:618574. doi: 10.3389/fcell.2020.618574
81. Tang CM, Zhang M, Huang L, Hu ZQ, Zhu JN, Xiao Z, et al. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci Rep. (2017) 7:40342. doi: 10.1038/srep40342
82. Zhai C, Qian G, Wu H, Pan H, Xie S, Sun Z, et al. Knockdown of circ_0060745 alleviates acute myocardial infarction by suppressing NF-kappaB activation. J Cell Mol Med. (2020) 24:12401–10. doi: 10.1111/jcmm.15748
83. Sun LY, Zhao JC, Ge XM, Zhang H, Wang CM, Bie ZD. Circ_LAS1L regulates cardiac fibroblast activation, growth, and migration through miR-125b/SFRP5 pathway. Cell Biochem Funct. (2020) 38:443–50. doi: 10.1002/cbf.3486
84. Li F, Long TY, Bi SS, Sheikh SA, Zhang CL. CircPAN3 exerts a profibrotic role via sponging miR-221 through FoxO3/ATG7-activated autophagy in a rat model of myocardial infarction. Life Sci. (2020) 257:118015. doi: 10.1016/j.lfs.2020.118015
85. Ni H, Li W, Zhuge Y, Xu S, Wang Y, Chen Y, et al. Inhibition of circHIPK3 prevents angiotensin II-induced cardiac fibrosis by sponging miR-29b-3p. Int J Cardiol. (2019) 292:188–96. doi: 10.1016/j.ijcard.2019.04.006
86. Wang W, Zhang S, Xu L, Feng Y, Wu X, Zhang M, et al. Involvement of circHIPK3 in the pathogenesis of diabetic cardiomyopathy in mice. Diabetologia. (2021) 64:681–92. doi: 10.1007/s00125-020-05353-8
87. Liu W, Wang Y, Qiu Z, Zhao R, Liu Z, Chen W, et al. CircHIPK3 regulates cardiac fibroblast proliferation, migration and phenotypic switching through the miR-152-3p/TGF-β2 axis under hypoxia. PeerJ. (2020) 8:e9796. doi: 10.7717/peerj.9796
88. Zhou B, Yu JW. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-β1. Biochem Biophys Res Commun. (2017) 487:769–75. doi: 10.1016/j.bbrc.2017.04.044
89. Wu N, Xu J, Du WW, Li X, Awan FM, Li F, et al. YAP circular RNA, circYap, attenuates cardiac fibrosis via binding with tropomyosin-4 and gamma-actin decreasing actin polymerization. Mol Ther. (2021) 29:1138–50. doi: 10.1016/j.ymthe.2020.12.004
90. Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. (2017) 38:1402–12. doi: 10.1093/eurheartj/ehw001
91. Xu Y, Leng K, Yao Y, Kang P, Liao G, Han Y, et al. A circular RNA, cholangiocarcinoma-associated circular RNA 1, contributes to cholangiocarcinoma progression, induces angiogenesis, and disrupts vascular endothelial barriers. Hepatology. (2021) 73:1419–35. doi: 10.1002/hep.31493
92. Zhang S, Song G, Yuan J, Qiao S, Xu S, Si Z, et al. Circular RNA circ_0003204 inhibits proliferation, migration and tube formation of endothelial cell in atherosclerosis via miR-370-3p/TGFβR2/phosph-SMAD3 axis. J Biomed Sci. (2020) 27:11. doi: 10.1186/s12929-019-0595-9
93. Chen L, Luo W, Zhang W, Chu H, Wang J, Dai X, et al. circDLPAG4/HECTD1 mediates ischaemia/reperfusion injury in endothelial cells via ER stress. RNA Biol. (2020) 17:240–53. doi: 10.1080/15476286.2019.1676114
94. Wei H, Cao C, Wei X, Meng M, Wu B, Meng L, et al. Circular RNA circVEGFC accelerates high glucose-induced vascular endothelial cells apoptosis through miR-338-3p/HIF-1α/VEGFA axis. Aging. (2020) 12:14365–75. doi: 10.18632/aging.103478
95. Huang HS, Huang XY, Yu HZ, Xue Y, Zhu PL. Circular RNA circ-RELL1 regulates inflammatory response by miR-6873-3p/MyD88/NF-κB axis in endothelial cells. Biochem Biophys Res Commun. (2020) 525:512–9. doi: 10.1016/j.bbrc.2020.02.109
96. Cui M, Shen W, Qin W, Wang X, Li Y, Xu F, et al. Circular RNA ciRS-7 promotes tube formation in microvascular endothelial cells through downregulation of miR-26a-5p. J Biochem Mol Toxicol. (2020) 34:e22468. doi: 10.1002/jbt.22468
97. Qin M, Wang W, Zhou H, Wang X, Wang F, Wang H. Circular RNA circ_0003645 silencing alleviates inflammation and apoptosis via the NF-κB pathway in endothelial cells induced by oxLDL. Gene. (2020) 755:144900. doi: 10.1016/j.gene.2020.144900
98. Zhou RM, Shi LJ, Shan K, Sun YN, Wang SS, Zhang SJ, et al. Circular RNA-ZBTB44 regulates the development of choroidal neovascularization. Theranostics. (2020) 10:3293–307. doi: 10.7150/thno.39488
99. Yu F, Zhang Y, Wang Z, Gong W, Zhang C. hsa_circ_0030042 regulates abnormal autophagy and protects atherosclerotic plaque stability by targeting eIF4A3. Theranostics. (2021) 11:5404–17. doi: 10.7150/thno.48389
100. Wang Y, Zhao R, Liu W, Wang Z, Rong J, Long X, et al. Exosomal circHIPK3 released from hypoxia-pretreated cardiomyocytes regulates oxidative damage in cardiac microvascular endothelial cells via the miR-29a/IGF-1 pathway. Oxid Med Cell Longev. (2019) 2019:1–28. doi: 10.1155/2019/7954657
101. Vliegen HW, van der Laarse A, Cornelisse CJ, Eulderink F. Myocardial changes in pressure overload-induced left ventricular hypertrophy. A study on tissue composition, polyploidization and multinucleation. Eur Heart J. (1991) 12:488–94. doi: 10.1093/oxfordjournals.eurheartj.a059928
102. Abbas N, Perbellini F, Thum T. Non-coding RNAs: emerging players in cardiomyocyte proliferation and cardiac regeneration. Basic Res Cardiol. (2020) 115:52. doi: 10.1007/s00395-020-0816-0
103. Tan WLW, Lim BTS, Anene-Nzelu CGO, Ackers-Johnson M, Dashi A, See K, et al. A landscape of circular RNA expression in the human heart. Cardiovasc Res. (2017) 113:298–309. doi: 10.1093/cvr/cvw250
104. Wu HJ, Zhang CY, Zhang S, Chang M, Wang HY. Microarray expression profile of circular RNAs in heart tissue of mice with myocardial infarction-induced heart failure. Cell Physiol Biochem. (2016) 39:205–16. doi: 10.1159/000445617
105. Lei W, Feng T, Fang X, Yu Y, Yang J, Zhao ZA, et al. Signature of circular RNAs in human induced pluripotent stem cells and derived cardiomyocytes. Stem Cell Res Ther. (2018) 9:56. doi: 10.1186/s13287-018-0793-5
106. Siede D, Rapti K, Gorska AA, Katus HA, Altmüller J, Boeckel JN, et al. Identification of circular RNAs with host gene-independent expression in human model systems for cardiac differentiation and disease. J Mol Cell Cardiol. (2017) 109:48–56. doi: 10.1016/j.yjmcc.2017.06.015
107. Tian M, Xue J, Dai C, Jiang E, Zhu B, Pang H. CircSLC8A1 and circNFIX can be used as auxiliary diagnostic markers for sudden cardiac death caused by acute ischemic heart disease. Sci Rep. (2021) 11:4695. doi: 10.1038/s41598-021-84056-5
108. Jurchott K, Bergmann S, Stein U, Walther W, Janz M, Manni I, et al. YB-1 as a cell cycle-regulated transcription factor facilitating cyclin A and cyclin B1 gene expression. J Biol Chem. (2003) 278:27988–96. doi: 10.1074/jbc.M212966200
109. Xu X, Wang J, Wang X. Silencing of circhipk3 inhibits pressure overload-induced cardiac hypertrophy and dysfunction by sponging miR-185-3p. Drug Des Devel Ther. (2020) 14:5699–710. doi: 10.2147/DDDT.S245199
110. Deng Y, Wang J, Xie G, Zeng X, Li H. Circ-HIPK3 strengthens the effects of adrenaline in heart failure by MiR-17-3p - ADCY6 axis. Int J Biol Sci. (2019) 15:2484–96. doi: 10.7150/ijbs.36149
111. Fan S, Hu K, Zhang D, Liu F. Interference of circRNA HIPK3 alleviates cardiac dysfunction in lipopolysaccharide-induced mice models and apoptosis in H9C2 cardiomyocytes. Ann Transl Med. (2020) 8:1147. doi: 10.21037/atm-20-5306
112. Li S, Ma Y, Tan Y, Ma X, Zhao M, Chen B, et al. Profiling and functional analysis of circular RNAs in acute promyelocytic leukemia and their dynamic regulation during all-trans retinoic acid treatment. Cell Death Dis. (2018) 9:651. doi: 10.1038/s41419-018-0699-2
113. Wei S, Ma W, Li X, Jiang C, Sun T, Li Y, et al. Involvement of ROS/NLRP3 inflammasome signaling pathway in doxorubicin-induced cardiotoxicity. Cardiovasc Toxicol. (2020) 20:507–19. doi: 10.1007/s12012-020-09576-4
114. Hao E, Mukhopadhyay P, Cao Z, Erdélyi K, Holovac E, Liaudet L, et al. Cannabidiol protects against doxorubicin-induced cardiomyopathy by modulating mitochondrial function and biogenesis. Mol Med. (2015) 21:38–45. doi: 10.2119/molmed.2014.00261
115. Renu K, VG A, PB TP, Arunachalam S. Molecular mechanism of doxorubicin-induced cardiomyopathy - an update. Eur J Pharmacol. (2018) 818:241–53. doi: 10.1016/j.ejphar.2017.10.043
116. Ma ZG, Yuan YP, Wu HM, Zhang X, Tang QZ. Cardiac fibrosis: new insights into the pathogenesis. Int J Biol Sci. (2018) 14:1645–57. doi: 10.7150/ijbs.28103
117. Hu S, Liu H, Hu Z, Li L, Yang Y. Follistatin-like 1: a dual regulator that promotes cardiomyocyte proliferation and fibrosis. J Cell Physiol. (2020) 235:5893–902. doi: 10.1002/jcp.29588
118. Zarak-Crnkovic M, Kania G, Jazwa-Kusior A, Czepiel M, Wijnen WJ, Czyz J, et al. Heart non-specific effector CD4(+). T cells protect from postinflammatory fibrosis and cardiac dysfunction in experimental autoimmune myocarditis. Basic Res Cardiol. (2019) 115:6. doi: 10.1007/s00395-019-0766-6
119. Su H, Zeng H, Liu B, Chen JX. Sirtuin 3 is essential for hypertension-induced cardiac fibrosis via mediating pericyte transition. J Cell Mol Med. (2020) 24:8057–68. doi: 10.1111/jcmm.15437
120. Travers JG, Kamal FA, Robbins J, Yutzey KE, Blaxall BC. Cardiac fibrosis: the fibroblast awakens. Circ Res. (2016) 118:1021–40. doi: 10.1161/CIRCRESAHA.115.306565
121. Souders CA, Bowers SL, Baudino TA. Cardiac fibroblast: the renaissance cell. Circ Res. (2009) 105:1164–76. doi: 10.1161/CIRCRESAHA.109.209809
122. Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, Brown RA. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat Rev Mol Cell Biol. (2002) 3:349–63. doi: 10.1038/nrm809
123. Tao L, Bei Y, Chen P, Lei Z, Fu S, Zhang H, et al. Crucial role of miR-433 in regulating cardiac fibrosis. Theranostics. (2016) 6:2068–83. doi: 10.7150/thno.15007
124. Guo Y, Luo F, Liu Q, Xu D. Regulatory non-coding RNAs in acute myocardial infarction. J Cell Mol Med. (2017) 21:1013–23. doi: 10.1111/jcmm.13032
125. Bie ZD, Sun LY, Geng CL, Meng QG, Lin XJ, Wang YF, et al. MiR-125b regulates SFRP5 expression to promote growth and activation of cardiac fibroblasts. Cell Biol Int. (2016) 40:1224–34. doi: 10.1002/cbin.10677
126. Zhu P, Zhu X, Wu J, He L, Lu T, Wang Y, et al. IL-13 secreted by ILC2s promotes the self-renewal of intestinal stem cells through circular RNA circPan3. Nat Immunol. (2019) 20:183–94. doi: 10.1038/s41590-018-0297-6
127. Shang J, Chen WM, Liu S, Wang ZH, Wei TN, Chen, et al. CircPAN3 contributes to drug resistance in acute myeloid leukemia through regulation of autophagy. Leuk Res. (2019) 85:106198. doi: 10.1016/j.leukres.2019.106198
128. Shang J, Chen WM, Wang ZH, Wei TN, Chen ZZ, Wu WB. CircPAN3 mediates drug resistance in acute myeloid leukemia through the miR-153-5p/miR-183-5p-XIAP axis. Exp Hematol. (2019) 70:42–54.e43. doi: 10.1016/j.exphem.2018.10.011
129. Zhang CL, Long TY, Bi SS, Sheikh SA, Li F. CircPAN3 ameliorates myocardial ischaemia/reperfusion injury by targeting miR-421/Pink1 axis-mediated autophagy suppression. Lab Invest. (2021) 101:89–103. doi: 10.1038/s41374-020-00483-4
130. Cai J, Li R, Xu X, Zhang L, Lian R, Fang L, et al. CK1alpha suppresses lung tumour growth by stabilizing PTEN and inducing autophagy. Nat Cell Biol. (2018) 20:465–78. doi: 10.1038/s41556-018-0065-8
131. Wen J, Liao J, Liang J, Chen XP, Zhang, Chu B. Circular RNA HIPK3: a key circular RNA in a variety of human cancers. Front Oncol. (2020) 10:773. doi: 10.3389/fonc.2020.00773
132. Pinto AR, Ilinykh A, Ivey MJ, Kuwabara JT, D'Antoni ML, Debuque R, et al. Revisiting cardiac cellular composition. Circ Res. (2016) 118:400–9. doi: 10.1161/CIRCRESAHA.115.307778
133. Talman V, Kivelä R. Cardiomyocyte-Endothelial cell interactions in cardiac remodeling and regeneration. Front Cardiovasc Med. (2018) 5:101. doi: 10.3389/fcvm.2018.00101
134. Liang B, Li M, Deng Q, Wang C, Rong J, He S, et al. CircRNA ZNF609 in peripheral blood leukocytes acts as a protective factor and a potential biomarker for coronary artery disease. Ann Transl Med. (2020) 8:741. doi: 10.21037/atm-19-4728
135. Garikipati VNS, Verma SK, Cheng Z, Liang D, Truongcao MM, Cimini M, et al. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat Commun. (2019) 10:4317. doi: 10.1038/s41467-019-11777-7
136. Boeckel JN, Jaé N, Heumüller AW, Chen W, Boon RA, Stellos K, et al. Identification and characterization of hypoxia-regulated endothelial circular RNA. Circ Res. (2015) 117:884–90. doi: 10.1161/CIRCRESAHA.115.306319
137. Frangogiannis NG. Regulation of the inflammatory response in cardiac repair. Circ Res. (2012) 110:159–73. doi: 10.1161/CIRCRESAHA.111.243162
138. Prabhu SD, Frangogiannis N.G. The biological basis for cardiac repair after myocardial infarction: from inflammation to fibrosis. Circ Res. (2016) 119:91–112. doi: 10.1161/CIRCRESAHA.116.303577
139. Bacmeister L, Schwarzl M, Warnke S, Stoffers B, Blankenberg S, Westermann D, et al. Inflammation and fibrosis in murine models of heart failure. Basic Res Cardiol. (2019) 114:19. doi: 10.1007/s00395-019-0722-5
140. Andreadou I, Cabrera-Fuentes HA, Devaux Y, Frangogiannis NG, Frantz S, Guzik T, et al. Immune cells as targets for cardioprotection: new players and novel therapeutic opportunities. Cardiovasc Res. (2019) 115:1117–30. doi: 10.1093/cvr/cvz050
141. Anzai A, Anzai T, Nagai S, Maekawa Y, Naito K, Kaneko H, et al. Regulatory role of dendritic cells in postinfarction healing and left ventricular remodeling. Circulation. (2012) 125:1234–45. doi: 10.1161/CIRCULATIONAHA.111.052126
142. Choo EH, Lee JH, Park EH, Park HE, Jung NC, Kim TH, et al. Infarcted myocardium-primed dendritic cells improve remodeling and cardiac function after myocardial infarction by modulating the regulatory T cell and macrophage polarization. Circulation. (2017) 135:1444–57. doi: 10.1161/CIRCULATIONAHA.116.023106
143. Zhang Y, Hu YW, Zheng L, Wang Q. Characteristics and roles of exosomes in cardiovascular disease. DNA Cell Biol. (2017) 36:202–11. doi: 10.1089/dna.2016.3496
144. Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. (2016) 126:1208–15. doi: 10.1172/JCI81135
Keywords: cardiac remodeling, non-coding RNA, circular RNA, heart disease, gene regulation
Citation: Yang T, Long T, Du T, Chen Y, Dong Y and Huang Z-P (2021) Circle the Cardiac Remodeling With circRNAs. Front. Cardiovasc. Med. 8:702586. doi: 10.3389/fcvm.2021.702586
Received: 29 April 2021; Accepted: 02 June 2021;
Published: 25 June 2021.
Edited by:
Jinwei Tian, The Second Affiliated Hospital of Harbin Medical University, ChinaReviewed by:
Michinari Nakamura, Rutgers University, Newark, United StatesMurugavel Ponnusamy, Qingdao University, China
Copyright © 2021 Yang, Long, Du, Chen, Dong and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhan-Peng Huang, aHVhbmd6aHAyN0BtYWlsLnN5c3UuZWR1LmNu; Yugang Dong, ZG9uZ3hnQG1haWwuc3lzdS5lZHUuY24=; Yili Chen, Y2hlbnlpbDdAbWFpbC5zeXN1LmVkdS5jbg==
†These authors have contributed equally to this work
 Tiqun Yang
Tiqun Yang Tianxin Long
Tianxin Long Tailai Du
Tailai Du Yili Chen1,2*
Yili Chen1,2* Yugang Dong
Yugang Dong Zhan-Peng Huang
Zhan-Peng Huang