- Department of Cardiovascular Diseases, Physiology and Biomedical Engineering, Center of Regenerative Medicine, Mayo Clinic, Scottsdale, AZ, United States
circular RNAs (circRNAs) are a type of non-coding RNAs that are widely present in eukaryotic cells. They have the characteristics of stable structure, high abundance, and cell or tissue specific expression. circRNAs are single-stranded RNAs that are covalently back spliced to form closed circular loops. They may participate in gene expression and regulation through a variety of action modes. circRNAs can encode proteins or function by acting as miRNA sponges for protein translation. Since 2016, a growing number of research studies have shown that circRNAs play important role in the pathogenesis of cardiovascular disease. With the construction of circRNA database, the differential expression of circRNAs in the heart tissue samples from different species and the gradual elucidation of its mode of action in disease may become an ideal diagnosis biomarker and an effective therapeutic target. What can be expected surely has a broader application prospect. In this review, we summarize recent publications on circRNA biogenesis, expression profiles, functions, and the most recent studies of circRNAs in the field of cardiovascular diseases with special emphasis on cardiac regeneration.
Introduction
Circular RNAs (circRNAs) are single-stranded RNAs that, unlike linear RNA, form a covalently closed continuous loop without 5′ end caps or 3′ Poly (A) tails. The concept of “circular RNA” was introduced by Sanger et al. when the team found that viroids are single-stranded covalently closed circRNA molecules (1). The cytoplasmic localization of circular RNA in eukaryotic cells was discovered by Hsu et al. through the electron microscope in 1979 (2). These pilot studies established the foundation of this research field.
Transcription of circRNAs had been a mystery for many years. The circular transcription of the Sry gene was discovered in mice in the early 1990s (3). In 2012, Salzman et al. (4) discovered that circRNA is a transformed transcript produced by reverse splicing of mRNA precursor and found that it is abundantly present in different types of human cells. As the field advances rapidly, a large number of circRNAs were discovered with the utilization of high-throughput sequencing technology, and their biological functions were intensively investigated. In 2016, Hansen et al. (5) found that circular RNA can act as a sponge of microRNA (miRNA) to regulate the growth and development of cells. This study shed new light on the circRNA field. Most recently, Li et al. developed a quickly screening and discovering tool for functional circular RNAs based on the CRISPR-Cas13d system, and discovered a set of functionalities that are important for cell growth and embryonic development (6). This technology provided a new research tool to the circRNA field.
Cardiovascular diseases (CVDs) are the leading cause of mortality worldwide. Several lines of evidence showed that circRNAs play important roles in regulating cardiovascular function. Jakobi et al. were the first group to provide a comprehensive catalog of RNase R-resistant circRNA species for the adult murine heart and explored the circRNA landscape of heart tissue (7). Over the next years, studies had reported that circRNAs are involved in the regulation of the physiology and pathology of the cardiovascular system. In particular, it is noted that circRNAs are involved in the pathogenesis of CVDs, such as myocardial infarction (MI) (8–14), heart failure (15, 16) and coronary artery disease (CAD) (17–23). Some circRNAs served as potential biomarkers for the diagnosis of CVDs (24–26). These findings suggest that circRNAs may be the new target molecules for the diagnosis and treatment of CVDs. In this review, we summarize circRNA classification, biogenesis, properties, functions, and some new research progress in the field of CVDs.
Classification of circRNA
circRNAs can be divided into three types according to the different sources of the sequences: ecRNAs (exonic circRNAs) which are derived from single or multiple exons (4, 27), ciRNAs (circular intronic RNAs) which are derived from introns (28), EIciRNAs (exon-intron circRNAs) which are composed of exons and introns (29) and tricRNAs (tRNA intronic circRNAs) which are formed by splicing tRNA introns (30).
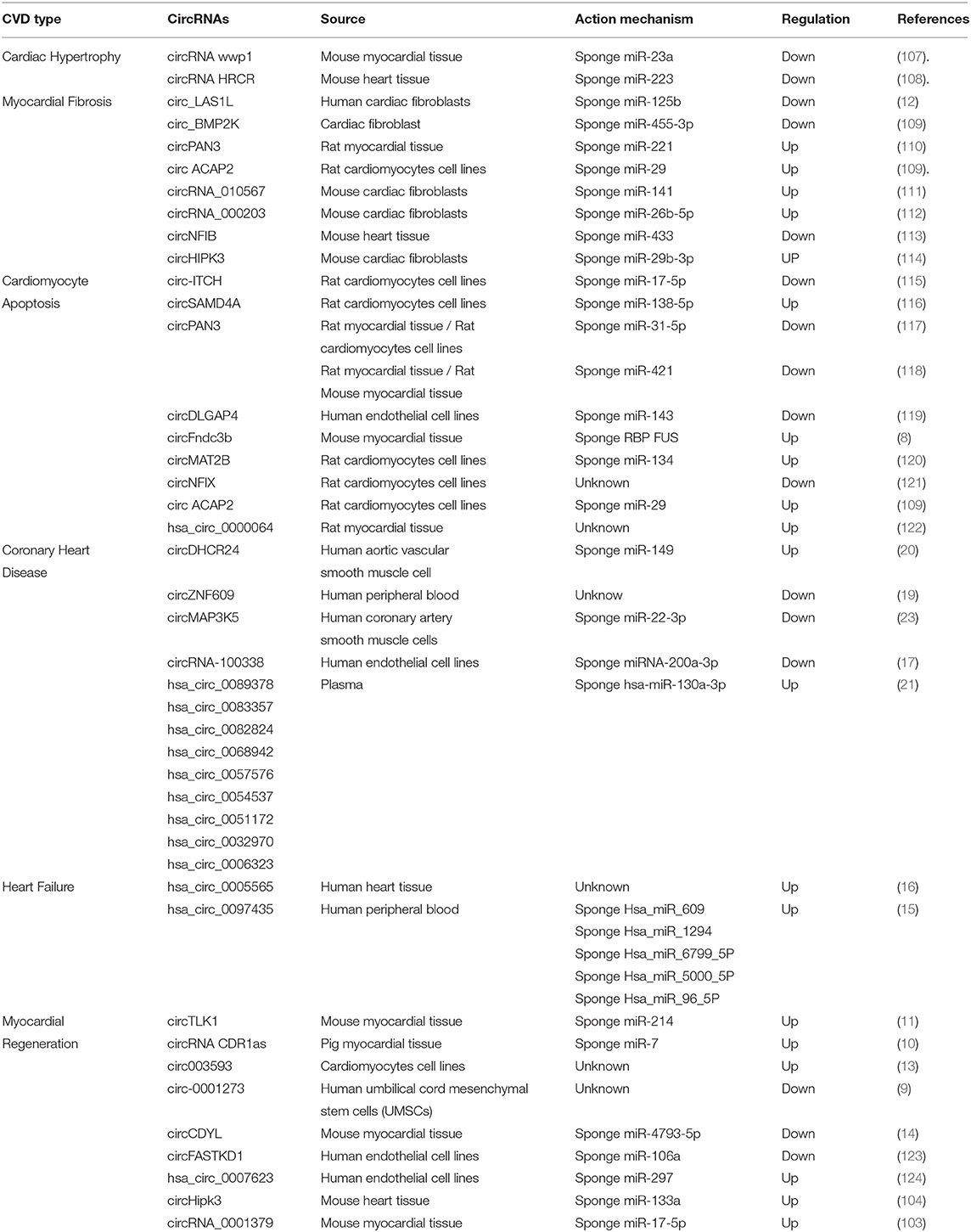
Biogenesis of circRNA
There are four primary models for the formation of circRNA loops from pre-mRNAs (Figure 1), namely lariat-driven circularization (exon skipping), intron-pairing driven circularization (direct back-splicing), circular intronic RNAs, and RNA-binding protein (RBP)-driven circularization.
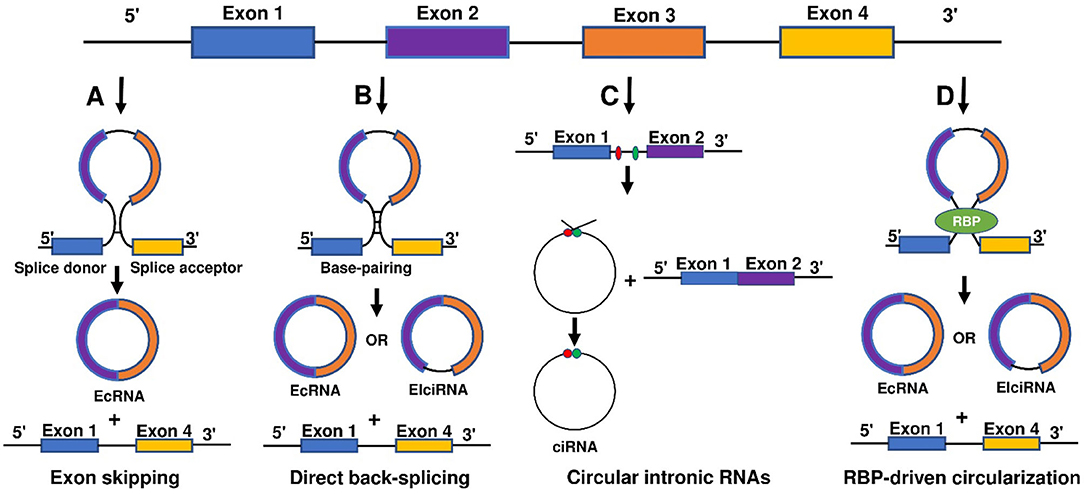
Figure 1. Biogenesis of circ RNAs. There are four main models for the formation of circRNA loops from pre-mRNAs including (A) lariat-driven circularization (exon skipping), (B) intron-pairing driven circularization (direct back-splicing), (C) circular intronic RNAs, and (D) RNA-binding protein (RBP)-driven circularization.
Lariat-driven circularization (exon skipping) is formed by connecting the splice site of 30 nucleotides upstream of the exon to the site of 50 nucleotides downstream (Figure 1A). This connection leads to exon-skipping and the formation of an RNA lariat consisting of several exons and introns. The introns are then removed to generate circRNAs (27, 31).
Intron-pairing driven circularization (direct back-splicing) is formed when pre-mRNA flanking introns contain inverted complementary sequences (Figure 1B). The complementary pairing on both sides of the intron can lead to alternative cyclization and then a generation of various circRNAs, including ecircRNAs and EIciRNAs (27, 32). Furthermore, longer introns can be found in the flanking sequences of circRNAs, and reverse complementary sequences in longer introns can aid the formation of circRNAs (29, 33).
Circular intronic RNAs are produced by eukaryotic spliceosome-mediated splicing (Figure 1C). The lariat intron generated from the splicing reaction evades normal debranching and degradation, and the 3′ “tail” downstream from the branchpoint is trimmed leading to the formation of a stable circRNA. Conserved motifs at both ends, including the 7-nt GU-rich element near the 5′ splice site and the 11-nt C-rich element near the branch point site, are combined to prevent introns form circular branches, which promote the formation of loop structures (28, 34).
Reverse complementary sequences, such as Alu repeats, are located in upstream and downstream introns. RBP-driven circularization is formed when certain transactivator RNA binding proteins that bind to each flanking intron trigger the splicing of the donor and acceptor sites close enough to form circRNA (Figure 1D) (32, 34–38).
Function of circRNA
Despite the rapid growth in the field, the biological functions of circRNAs in eukaryotic cells have not been fully understood. circRNAs share some common characters. First, circRNAs are widely distributed and abundantly expressed in a diverse of cells. circRNAs can be found in a large amount in the cytoplasm of eukaryotic cells derived from animals and plants (27, 39). In humans, more than 30,000 circRNAs have been discovered and are still increasing year by year (40, 41). Second, circRNAs are stable. Due to the covalently closed structure, circRNA is resistant to degradation by ribonuclease (RNase) or exonuclease and is more stable than linear RNA (42). The expression of circRNA differs according to time, tissues, or species (39, 43). circRNA profiles change at different stages of cardiac differentiation or during cardiogenic differentiation of induced pluripotent stem cells (44, 45). Moreover, circRNAs are evolutionarily conserved (43, 46). In 2016, Werfel et al. (47) reported high homology of 1288 circRNAs across humans, mice, and rats. However, many studies have also illustrated that circRNAs are species-specific (48, 49). circRNAs show different expression profiles between normal and diseased tissues (45, 47, 50). Increasing evidence suggests some circRNAs are derived from genomic loci associated with human diseases, and contribute to transcriptional, post-transcriptional, and translational regulations (51). To summarize, there are four main modes for circRNA function (Figure 2).
Figure 2. Mechanisms of circRNA functions. There are five main action mechanisms of circRNAs, including (A) circRNAs can act as miRNAs sponge, (B) circRNAs can function as RBP sponges, (C) circRNAs mediated regulation of the transcription of parental genes, (D) circRNAs can be translated into proteins via some modification, and (E) circRNAs can act as dynamic scaffolding molecules that modulate protein-protein interactions.
1) circRNAs can act as competitive endogenous RNAs (ceRNAs) to regulate gene expression by microRNAs (miRNAs) sponge effects (Figure 2A). miRNAs are important post-transcriptional regulators of gene expression that act by direct base pairing to target sites within untranslated regions of messenger RNAs (mRNAs) (52, 53). circRNAs contain miRNA response elements (MREs) that promote the binding between circRNAs and miRNAs. This binding can decrease the level of functional miRNAs and increase the expression of miRNA targets (53, 54). It has been reported that circRNAs regulate cell function by acting as miRNA sponges. For example, circFOXK2 promotes cell growth, migration, invasion, and apoptosis by binding to multiple sites and functioning as a sponge for miR-942 (55). Similarly, circRNA_100876 regulates the progression of triple-negative breast cancer by functioning as a sponge for miR-136 (56). Other circRNAs, such as circSLC26A4, circRNA_0000253, and circRNA_ANKIB1 can also function as the sponge of miR-1287-5p (57), miRNA-141-5p (58), and miR-195a-5p (59), respectively. circALMS1_6 may participate in the regulation of cardiac remodeling by functioning as a sponge for miR-133 (60).
2) circRNAs can function as RBP sponges and RBPs can also participate in back-splicing (Figure 2B) (61–66). RBPs are a group of proteins involved in gene transcription and translation. circRNAs can interact with RBP and inhibit their activities (66–68). circMbl absorbs MBL proteins and regulates the subsequent physiological processes (32). circPABPN1 can bind to HuR to suppress the translation of PABPN1 mRNA (69). circANRIL competitively recruits PES1 to inhibit ribosome biogenesis (70). circFoxo3 interacts with different RBPs to participate in the processes of cardiomyocyte senescence and cell cycle progression (71). circAmotl1 can protect cardiomyocytes and promote cell proliferation and wound healing by binding to PDK1, AKT1, and STAT3 (72, 73). The studies confirm the involvement of circRNAs in post-transcriptional regulation by chelating RBP. In 2020, Okholm et al. (74) conducted an extensive screen of circRNA-RBP interactions and analyzed circRNA-RBP interactions using a large set of eCLIP data with binding sites of 150 RBPs in the ENCODE cell lines HepG2 and K562 with deep-sequenced total RNA samples. Through this study, they confirmed the interactions between circCDYL and RBPs in bladder cancer cells.
3) circRNAs mediated regulation of the transcription of parental genes (Figure 2C). For example, circβ-catenin can produce a novel 370-amino acid β-catenin isoform using the start codon as the linear β-catenin mRNA transcript and terminates translation at a new stop codon created by circularization (75). ci-ankrd52 is a circular intronic RNA that is abundant in the nucleus and has little enrichment at microRNA target sites. ci-ankrd52 can bind to the transcription sites and acts as a positive regulator of Pol II transcription (28). For the circRNAs that interact with RNA polymerase II, exons are typically circularized with introns which are ‘retained' between exons. These circRNAs are termed exon-intron circRNAs or EIciRNAs. They are mainly localized in the nucleus and interact with U1 snRNP to promote transcription of their parental genes (29).
4) circRNAs can be translated into proteins via some modification (Figure 2D). As we know, the translation is performed by ribosomes and involves initiation, elongation, termination and ribosome recycling. Base-modification N6-methyladenosine (m6A) is a common form of base modification in RNAs. It can promote efficient initiation of protein translation from circRNAs in human cells. Legnini et al. revealed that m6A-driven translation of circRNAs is widespread with hundreds of endogenous circRNAs carrying the translation potential (76). circ-ZNF609 is an example of a protein-coding circRNA in eukaryotes. It is related to heavy polysomes and can be translated into a protein in a splicing-dependent and cap-independent manner (77). circRNAs play biological functions through the formation of complexes with proteins; otherwise, a novel protein circFAM188B-103aa encoded by circFAM188B that promotes the proliferation but inhibits the differentiation of chicken SMSCs was identified (78). Moreover, artificial (79) and endogenous circRNAs containing an internal ribosome entry site (IRES) that directly recruits ribosomes (80) can also be translated into protein. Additionally, circRNA with an infinite ORF has hundred-fold higher productivity than linear transcript by rolling circle amplification in an IRES-independent manner (81).
5) circRNAs may bind, store, sort, and sequester proteins to particular subcellular locations, can and act as dynamic scaffolding molecules that modulate protein-protein interactions (Figure 2E). circRNAs can bind to RNAs and can also bind, store, sort or sequester selected proteins such as RBPs to modulate their activity or localization (82). RNA-binding protein 3 (RBM3) dynamically adjusts the proliferation of hepatocellular carcinoma cells by regulating the production of SCD-circRNA2 encoded by the 3′-UTR of the stearoyl-CoA desaturase (SCD) gene (83). Recent studies have shown that RBP quaking could also modify the formation of circRNA through forming RNA-protein complexes (RPCs) (36). In addition to interacting with RBPs, circRNAs can function as protein sponges by adsorbing one or more proteins in binding sites, thereby acting as protein scaffolding by the mediating interaction between proteins. For example, CircFOXO3 could mediate the formation of circFOXO3-p21-CDK2 ternary complex and then serve as scaffolding, affecting the cell cycle progression of cancer (84).
Sequencing of circRNA
RNA-seq emerges as a powerful research tool to study the expression and function of non-coding RNAs including circRNAs (85). The technology of circRNA-seq generally includes library construction, computer sequencing, data analysis and processing, and function prediction (86). Either full transcriptome or circRNA profiling may be used to sequence circRNAs. The full transcriptome profiling is aimed to explore the expression patterns of both coding and non-coding RNA. This approach is suitable for the study of the biological function of circRNA. The circRNA profiling is focused on enriched circRNAs and this approach is appropriate to discover unknown circRNAs. Technically, the main difference between the two approaches is the construction of the sequencing library. The circRNA sequencing library not only requires the removal of most rRNA and poly (A), but also requires the use of ribonuclease RNase R to remove the interference of linear RNA. It has been reported that the abundance of circRNAs decreases after de-linear RNA because some circRNAs are sensitive to RNase R mediated digestion (87, 88). It is worthy to note that the alternative splicing of circRNA requires distinguishing the source of sense and antisense chains in the sequencing results. Therefore, constructing a chain-specific library is ideal as it may improve the accuracy of circRNA sequencing (87). Thus far, more than 100,000 unique human circRNAs have been discovered (89, 90).
After obtaining the circRNA sequencing data, the prediction and identification of circRNAs were carried out based on the identification software such as find circ, CIRCexplorer2, and CIRI (91–93). Real-time fluorescence quantitative PCR (quantitative real-time PCR, qRT-PCR), Northern blot hybridization (Northern blot), in situ hybridization (In situ hybridization, ISH), RPAD (RNase R treatment, polyadenylation, and poly (A) + RNA Depletion) and other techniques are used to validate the data of circRNA sequencing (94–97).
Microarray chip is another efficient tool for circRNA analysis, and it is commonly used in clinics for disease diagnosis. Compared to RNA-seq, microarray chip analysis is different in the following aspects: (2) microarray analysis of circRNA requires a known reference sequence, while RNA-seq can be utilized to analyze unknown circRNAs; (3) microarray chip analysis can be used to quantify circRNA expression when comparing with RNA-seq (98); and (4) microarray chip analysis can efficiently detect reverse splice site sequences and obtain a larger number of circRNAs than RNA-seq (99). However, some limitations of microarray chip analysis include: (2) high total RNA input is required during sample pretreatment; and (3) unlike full transcriptome sequencing, microarray chip does not give the linear RNA data (100). If the reference sequence is unknown, many studies usually use RNA-seq to determine the full transcriptional sequence, then analyze the circRNAs by microarray.
circRNA and CVDs
With the development of deep sequencing technology, we can now understand the types and differential expression of circRNAs and their associated miRNAs in cardiovascular tissues (101–103). Many circRNAs have been reported to be associated with CVDs and their expression pattern are different between healthy and diseased human hearts (15, 19, 104, 105). Quantitative proteomics may be used to discover the regulatory networks of circRNAs in cardiovascular tissues (106). Here, we summarized recent publications on the roles of circRNAs in the development and treatment of CVDs (Table 1).
1) Cardiac hypertrophy. Cardiac hypertrophy is the heart's response to a variety of extrinsic and intrinsic stimuli that impose increased biomechanical stress and can be caused by various cardiovascular diseases. circRNA wwp1 exerts inhibitory roles of cardiac hypertrophy via down-regulation of ANF and miR-23a in isoproterenol hydrochloride-induced cardiac hypertrophy (107). A circRNA HRCR functions as an endogenous miR-223 sponge to sequester and inhibit miR-223 activity, resulting in an increase of ARC expression and protection of the heart from pathological hypertrophy and heart failure (108). Modulation of circRNAs levels may provide a promising therapeutic target for the treatment of cardiac hypertrophy.
2) Cardiac fibrosis. Activation and phenotypical transition of cardiac fibroblasts contribute to cardiac fibrosis. It was reported that circ_BMP2K enhances the regulatory effects of miR-455-3p on its target gene SUMO1 which leads to the inhibition of TGF-β1 or Ang II to induce the activation of cardiac fibroblasts (109). circRNA_010567, circRNA_000203, and circHIPK3 were upregulated in Angiotensin-II (Ang-II)-induced activation of cardiac fibroblasts (111, 112, 114). circNFIB was downregulated in TGF-β induced activation of primary adult cardiac fibroblasts (113). Some studies showed that targeting circRNAs improve myocardial ischemic and reperfusion injuries by attenuating myocardial fibrosis. For example, circ_LAS1L is down-regulated in patients with acute myocardial infarction and regulates cardiac fibroblast activation, growth, and migration by inhibiting miR-125b/SFRP5 pathway (12). circPAN3 knockdown attenuated autophagy-mediated cardiac fibrosis after myocardial infarction via miR-221/FoxO3/ATG7 axis (110). The roles of circRNAs in cardiac fibrosis has been summarized in a recent review article (54).
3) Cardiomyocyte apoptosis. It was reported that some circRNAs may be involved in injury-induced cardiomyocyte apoptosis. For example, circRNA ITCH mediates H2O2-induced myocardial cell apoptosis by upregulating miR-17-5p via wnt/β-catenin signaling pathway (115). circSAMD4A aggravates hypoxia/reoxygenation (H/R)-induced cardiomyocyte apoptosis and inflammatory response by sponging miR-138-5p (116). MicroRNA-31-5p acts as a negative regulator of circPAN3 by directly suppressing QKI in doxorubicin-induced apoptosis of cardiomyocytes (117). circPAN3 also ameliorates myocardial ischemia and reperfusion injury by regulating miR-421/Pink1 axis-mediated suppression of autophagy (118). HECTD1 overexpression increases cell viability and decreases cell apoptosis and migration, and circDLPAG4/HECTD1 mediates ischemia/reperfusion injury in endothelial cells via ER stress (119). Down-regulation of circFndc3b was observed in mice with myocardial infarction, and overexpression of circFndc3b increases angiogenic activity and reduces cell apoptosis in cardiac endothelial cells and cardiomyocytes which led to improved left ventricular functions (8). Other studies showed that miR-133 was regulated by circMAT2B. CircMAT2B knockdown attenuates oxygen-glucose deprivation-induced injury through up-regulating miR-133 in H9c2 cells (120). circNFIX can serve as a pro-apoptosis factor in cardiomyocytes (121). The expression of circ ACAP2 is induced by myocardial infarction which leads to increased cardiomyocyte apoptosis by sponging miR-29 (109). Salidroside inhibits apoptosis and autophagy of cardiomyocytes by regulation of circular RNA hsa_circ_0000064 in cardiac ischemia-reperfusion injury (122). These data suggest that circRNAs may be the new targets for designing cardioprotective treatments against cardiomyocyte death.
4) Coronary heart disease. circRNAs are involved in the pathogenesis of atherosclerosis and coronary heart disease. It has been reported that suppression of circDHCR24 alleviates aortic smooth muscle cell proliferation and migration by targeting miR-149-5p/MMP9 axis in human aortic vascular smooth muscle cells after PDGF-BB treatment (20). Recent studies showed that the expression level of circZNF609 in peripheral blood leukocytes of patients with coronary artery disease was significantly decreased, and circZNF609 regulates the release of inflammatory cytokines such as IL-6, IL-10, and TNF-α by serving as sponges to different miRNAs that control the expression of these cytokines (19). circMAP3K5 was downregulated in patients with coronary heart disease and acted as a microRNA-22-3p sponge to promote resolution of intimal hyperplasia via TET2-mediated smooth muscle cell differentiation (23). circRNA-100338 may induce angiogenesis after myocardial ischemia-reperfusion injury by sponging miR-200a-3p in human coronary endothelial cells (17). The level of hsa_circ_0001445 in plasma was associated with the severity of coronary atherosclerosis. In vitro, hsa_circ_0001445 was downregulated in extracellular vesicles secreted by human coronary smooth muscle cells upon exposure to atherogenic conditions (22). The 3 circRNAs (hsa_circ_0016868, hsa_circ_0001364, hsa_circ_0006731) have been verified by the coronary artery segments Sanger sequencing obtained from an 81-year-old male patient with the sudden death of myocardial infarction (18). In addition, a recent study on the differential expression of circRNAs in plasma samples from patients with coronary heart disease identified 9 circRNAs that promote the expression of transient receptor potential cation channel subfamily M member 3 by inhibiting hsa-miR-130a-3p (21). These data demonstrate that circRNAs play important roles in the pathogenesis of coronary heart disease and may serve as diagnostic or therapeutic targets for coronary heart disease.
5) Heart failure. Despite the detailed roles of circRNAs in the progression of human heart failure remains elusive, recent high-throughput sequencing studies identified many circRNAs with changed expression in patients with heart failure. The top highly expressed EAT circRNAs corresponded to genes involved in cell proliferation and inflammatory responses. A recent study on circRNA expression profile in epicardial adipose tissue in patients with heart failure showed that EAT circRNAs may contribute to the pathogenesis of metabolic disorders (16). Another study showed that the upregulation of Hsa_circ_0097435 contributes to the pathogenesis of heart failure via sponging multiple microRNAs (15).
6) Cardiac regeneration and repair. Recent studies indicated that modulating circRNA activity may have great therapeutic potential for myocardial regeneration and repair. Up- or down-regulation of circRNAs and miRNAs and circRNA-miRNA coexpression had been shown to change the expression of the genes associated with myocardial ischemia and reperfusion injuries (101, 103). It has been reported that circRNAs can regulate inflammatory factors to improve myocardial ischemia and reperfusion injury. The circTLK1 exacerbates myocardial ischemia and reperfusion injury via targeting miR-214/RIPK1 through TNF signaling pathway (11). circ003593 has also been shown to confer cardioprotection through NLRP3 inflammasome myocardial infarct rats (13). circRNA CDR1as was identified in pig hearts. Elevated circRNA CDR1as in the infarction region of the pig heart is negatively associated with infarct size and positively associated with improved heart function (10). circ-0001273 can remarkably inhibit myocardial cell apoptosis and promote repair in myocardial infarction hearts (9). circCDYL was downregulated in myocardial tissues and hypoxia myocardial cells after acute myocardial infarction. circCDYL overexpression and downregulation can promote and inhibit the proliferation of cardiomyocytes in vitro, respectively. Additionally, circCDYL can promote the proliferation of cardiomyocytes through the miR-4793-5p/APP pathway (14). The downregulation of circFASTKD1 induces angiogenesis and improves cardiac function and repair after myocardial infarction (123). Hsa_circ_0007623 can bind to miR-297 and acts as a sponge of microRNA-297 which promotes cardiac repair after acute myocardial ischemia and protects cardiac function (124). circHipk3 was overexpressed in the fetal or neonatal heart of mice and functioned to promote the proliferation of cardiomyocyte and endothelial cells which leads to angiogenesis. Further study showed that circHipk3 regulates cardiac regeneration in mice post myocardial infarction by interacting with Notch1 and miR-133a (104). These findings highlight the physiological role of circRNAs in cardiac repair and indicate that modulation of circRNA may represent a potential strategy to promote cardiac function and remodeling after myocardial injuries.
Future Perspectives
The studies we discussed in the paper highlight the significance of circRNAs in the pathogenesis of CVDs. circRNAs are stable and abundantly present in the circulatory system which enables them to serve as biomarkers for the diagnosis and treatment of CVDs; however, there are some critical issues to be addressed. Firstly, there is no reliable methodology for the detection of circRNAs. In terms of circRNA detection, newer, simpler, and more reliable methods will be expected to appear. This will provide convenience for us to study circRNAs, facilitate the faster output of research results, and obtain more target circRNAs with diagnostic and therapeutic significance. Secondly, some circRNA biomarkers come from small samples and populations. This makes us question the reliability and representativeness of the research results. Thirdly, the mechanism underlying circRNA functions in the cardiovascular system remain largely elusive. For the function and mechanism studies, the current research methods are limited and difficult to operate. New research protocols need to be further explored. Rapid development can be achieved only by breaking through the technological bottleneck in the field of circRNA research. Lastly, there is a lack of efficient approaches for modulating circRNA expression in the cardiovascular system. It is supposed to be that in the future there will be new and more diverse methods in modulating the overexpression and inhibition of circRNAs. This will promote the development of the mechanism underlying circRNA functions. Then the research results can be quickly used for clinical diagnosis and treatment in the field of vascular diseases.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work is supported by NIH R01 grant HL142627 and American Heart Association Grant 20TPA35490001.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Sanger HL Riesner D and Gross HJ Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. (1976) 73:3852–6. doi: 10.1073/pnas.73.11.3852
2. Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. (1979) 280:339–40. doi: 10.1038/280339a0
3. Capel B, Nicolis AS, Hacker S, Walter M, Koopman P, Goodfellow P. Lovell-Badge R. Cell-Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. (1993) 73:1019–30. doi: 10.1016/0092-8674(93)90279-Y
4. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. (2012) 7:e30733. doi: 10.1371/journal.pone.0030733
5. Hansen TB, Veno MT, Damgaard CK, Kjems J. Comparison of circular RNA prediction tools. Nucleic Acids Res. (2016) 44:e58. doi: 10.1093/nar/gkv1458
6. Li S, Li X, Xue W, Zhang L, Yang LZ, Cao SM, et al. Screening for functional circular RNAs using the CRISPR-Cas13 system. Nat Methods. (2021) 18:51–59. doi: 10.1038/s41592-020-01011-4
7. Jakobi T, Czaja-Hasse LF, Reinhardt R, Dieterich C. Profiling and validation of the circular RNA repertoire in adult murine hearts. Gen Prot Bioinform. (2016) 14:216–23. doi: 10.1016/j.gpb.2016.02.003
8. Garikipati VNS, Verma SK, Cheng Z, Liang D, Truongcao MM, Cimini M, et al. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat Commun. (2019) 10:4317. doi: 10.1038/s41467-019-11777-7
9. Lei D, Wang Y, Zhang L, Wang Z. Circ_0010729 regulates hypoxia-induced cardiomyocyte injuries by activating TRAF5 via sponging miR-27a-3p. Life Sci. (2020) 262:118511. doi: 10.1016/j.lfs.2020.118511
10. Mester-Tonczar J, Winkler J, Einzinger P, Hasimbegovic E, Kastner N, Lukovic D, et al. Association between circular RNA CDR1as and post-infarction cardiac function in pig ischemic heart failure: influence of the anti-fibrotic natural compounds bufalin and lycorine. Biomolecules. (2020) 10:1180. doi: 10.3390/biom10081180
11. Song YF, Zhao L, Wang BC, Sun JJ, Hu JL, Zhu XL, et al. The circular RNA TLK1 exacerbates myocardial ischemia/reperfusion injury via targeting miR-214/RIPK1 through TNF signaling pathway. Free Radic Biol Med. (2020) 155:69–80. doi: 10.1016/j.freeradbiomed.2020.05.013
12. Sun LY, Zhao JC, Ge XM, Zhang H, Wang CM, Bie ZD. Circ_LAS1L regulates cardiac fibroblast activation, growth, and migration through miR-125b/SFRP5 pathway. Cell Biochem Funct. (2020) 38:443–50. doi: 10.1002/cbf.3486
13. Xiao Y, Oumarou DB, Wang S, Liu Y. Circular RNA involved in the protective effect of Malva sylvestris L. on myocardial ischemic/re-perfused Injury Front Pharmacol. (2020) 11:520486. doi: 10.3389/fphar.2020.520486
14. Zhang M, Wang Z, Cheng Q, Wang Z, Lv X, Wang Z, et al. Circular RNA (circRNA) CDYL induces myocardial regeneration by ceRNA amyocardial infarction. Med Sci Monit. (2020) 26:e923188. doi: 10.12659/MSM.923188
15. Han J, Zhang L, Hu L, Yu H, Xu F, Yang B, et al. Circular RNA-expression profiling reveals a potential role of Hsa_circ_0097435 in heart failure via sponging multiple MicroRNAs. Front Genet. (2020) 11:212. doi: 10.3389/fgene.2020.00212
16. Zheng ML, Du XP, Zhao L, Yang XC. Expression profile of circular RNAs in epicardial adipose tissue in heart failure. Chin Med J. (2020) 133:2565–72. doi: 10.1097/CM9.0000000000001056
17. Chang H, Wu ZB, Zhang L. Circ-100338 induces angiogenesis after myocardial ischemia-reperfusion injury by sponging miR-200a-3p. Eur Rev Med Pharmacol Sci. (2020) 24:6323–32.doi: 10.26355/eurrev_202006_21530
18. Hou C, Gu L, Guo Y, Zhou Y, Hua L, Chen J, et al. Association between circular RNA expression content and severity of coronary atherosclerosis in human coronary artery. J Clin Lab Anal. (2020) 34:e23552. doi: 10.1002/jcla.23552
19. Liang B, Li M, Deng Q, Wang C, Rong J, He S, et al. CircRNA ZNF609 in peripheral blood leukocytes acts as a protective factor and a potential biomarker for coronary artery disease. Ann Transl Med. (2020) 8:741. doi: 10.21037/atm-19-4728
20. Peng W, Li T, Pi S, Huang L, Liu Y. Suppression of circular RNA circDHCR24 alleviates aortic smooth muscle cell proliferation and migration by targeting miR-149-5p/MMP9 axis. Biochem Biophys Res Commun. (2020) 529:753–9. doi: 10.1016/j.bbrc.2020.06.067
21. Pan RY, Liu P, Zhou HT, Sun WX, Song J, Shu J, et al. Circular RNAs promote TRPM3 expression by inhibiting hsa-miR-130a-3p in coronary artery disease patients. Oncotarget. (2020) 8:60280–90. doi: 10.18632/oncotarget.19941
22. Vilades D, Martinez-Camblor P, Ferrero-Gregori A, Bar C, Lu D, Xiao K, et al. Plasma circular RNA hsa_circ_0001445 and coronary artery disease: performance as a biomarker. FASEB J. (2020) 34:4403–14. doi: 10.1096/fj.201902507R
23. Zeng Z, Xia L, Fan S, Zheng J, Qin J, Fan X, et al. Circular RNA CircMAP3K5 acts as a MicroRNA-22-3p sponge to promote resolution of intimal hyperplasia via TET2-mediated smooth muscle cell differentiation. Circulation. (2021) 143:354–71. doi: 10.1161/CIRCULATIONAHA.120.049715
24. Vausort M, Salgado-Somoza A, Zhang L, Leszek P, Scholz M, Teren A, et al. Myocardial infarction-associated circular RNA predicting left ventricular dysfunction. J Am Coll Cardiol. (2016) 68:1247–8. doi: 10.1016/j.jacc.2016.06.040
25. Zhao Z, Li X, Gao C, Jian D, Hao P, Rao L, et al. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci Rep. (2017) 7:39918. doi: 10.1038/srep39918
26. Zhang J, Xu Y, Xu S, Liu Y, Yu L, Li Z, et al. Plasma circular RNAs, Hsa_circRNA_025016, predict postoperative atrial fibrillation after isolated off-pump coronary artery bypass grafting. J Am Heart Assoc. (2018) 7:6642. doi: 10.1161/JAHA.117.006642
27. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. (2013) 19:141–157. doi: 10.1261/rna.035667.112
28. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell. (2013) 51:792–806. doi: 10.1016/j.molcel.2013.08.017
29. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. (2015) 22:256–64. doi: 10.1038/nsmb.2959
30. Schmidt CA, Matera AG. tRNA introns: presence, processing, and purpose. Wiley Interdiscip Rev RNA. (2020) 11:e1583. doi: 10.1002/wrna.1583
31. Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. (2014) 159:134–47. doi: 10.1016/j.cell.2014.09.001
32. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. (2014) 56:55–66. doi: 10.1016/j.molcel.2014.08.019
33. Petkovic S, Muller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. (2015) 43:2454–65. doi: 10.1093/nar/gkv045
34. Shi Y, He R, Yang Y, He Y, Shao K, Zhan L, et al. Circular RNAs: Novel biomarkers for cervical, ovarian and endometrial cancer. Oncol Rep. (2020) 44:1787–98. doi: 10.3892/or.2020.7780
35. Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, et al. Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. (2015) 5:8057. doi: 10.1038/srep08057
36. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. (2015) 160:1125–1134. doi: 10.1016/j.cell.2015.02.014
37. Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, Cherry S, et al. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, SR proteins. Genes Dev. (2015) 29:2168–82. doi: 10.1101/gad.270421.115
38. Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, et al. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol Cell. (2017) 67:214–27 e217. doi: 10.1016/j.molcel.2017.05.023
39. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. (2013) 9:e1003777. doi: 10.1371/annotation/f782282b-eefa-4c8d-985c-b1484e845855
40. Xu T, Wu J, Han P, Zhao Z, Song X. Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. BMC Genomics. (2017) 18:680. doi: 10.1186/s12864-017-4029-3
41. Zeng X, Lin W, Guo M, Zou Q. A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput Biol. (2017) 13:e1005420. doi: 10.1371/journal.pcbi.1005420
42. Li XF, Lytton J. A circularized sodium-calcium exchanger exon 2 transcript. J Biol Chem. (1999) 274:8153–60. doi: 10.1074/jbc.274.12.8153
43. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. (2013) 495:333–8. doi: 10.1038/nature11928
44. Li Y, Zhang J, Huo C, Ding N, Li J, Xiao J, et al. Dynamic Organization of lncRNA and circular RNA regulators collectively controlled cardiac differentiation in humans. EBioMedicine. (2017) 24:137–46. doi: 10.1016/j.ebiom.2017.09.015
45. Siede D, Rapti K, Gorska AA, Katus HA, Altmuller J, Boeckel JN, et al. Identification of circular RNAs with host gene-independent expression in human model systems for cardiac differentiation and disease. J Mol Cell Cardiol. (2017) 109:48–56. doi: 10.1016/j.yjmcc.2017.06.015
46. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. (2014) 15:409. doi: 10.1186/s13059-014-0409-z
47. Werfel S, Nothjunge S, Schwarzmayr T, Strom TM, Meitinger T, Engelhardt S. Characterization of circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol. (2016) 98:103–7. doi: 10.1016/j.yjmcc.2016.07.007
48. Aufiero S, Reckman YJ, Pinto YM, Creemers EE. Circular RNAs open a new chapter in cardiovascular biology. Nat Rev Cardiol. (2019) 16:503–14. doi: 10.1038/s41569-019-0185-2
49. Lim TB, Lavenniah A, Foo RS. Circles in the heart and cardiovascular system. Cardiovasc Res. (2020) 116:269–78. doi: 10.1093/cvr/cvz227
50. Gupta SK, Garg A, Bar C, Chatterjee S, Foinquinos A, Milting H, et al. Quaking inhibits doxorubicin-mediated cardiotoxicity through regulation of cardiac circular RNA expression. Circ Res. (2018) 122:246–54. doi: 10.1161/C.I.R.C.R.E.S.A.H.A.117.311335
51. Dong R, Zhang XO, Zhang Y, Ma XK, Chen LL, Yang L. CircRNA-derived pseudogenes. Cell Res. (2016) 26:747–50. doi: 10.1038/cr.2016.42
52. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. (2009) 136:215–33. doi: 10.1016/j.cell.2009.01.002
53. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. (2013) 495:384–8. doi: 10.1038/nature11993
54. Zhang L, Zhang Y, Wang Y, Zhao Y, Ding H, Li P. Circular RNAs: functions and clinical significance in cardiovascular disease. Front Cell Dev Biol. (2020) 8:584051. doi: 10.3389/fcell.2020.584051
55. Wong CH, Lou UK, Li Y, Chan SL, Tong JH, To KF, et al. CircFOXK2 promotes growth and metastasis of pancreatic ductal adenocarcinoma by complexing with RNA-binding proteins and sponging MiR-942. Cancer Res. (2020) 80:2138–49. doi: 10.1158/0008-5472.CAN-19-3268
56. Yu X, Xiao W, Song H, Jin Y, Xu J, Liu X. CircRNA_100876 sponges miR-136 to promote proliferation and metastasis of gastric cancer by upregulating MIEN1 expression. Gene. (2020) 748:144678. doi: 10.1016/j.gene.2020.144678
57. Ji F, Du R, Chen T, Zhang M, Zhu Y, Luo X, et al. Circular RNA circSLC26A4 accelerates cervical cancer progression via miR-1287-5p/HOXA7 axis. Mol Ther Nucleic Acids. (2020) 19:413–20. doi: 10.1016/j.omtn.2019.11.032
58. Song J, Chen ZH, Zheng CJ, Song KH, Xu GY, Xu S, et al. Exosome-Transported circRNA_0000253 competitively adsorbs MicroRNA-141-5p and increases IDD. Mol Ther Nucleic Acids. (2020) 21:1087–99. doi: 10.1016/j.omtn.2020.07.039
59. Li C, Mu J, Shi Y, Xin H. LncRNA CCDC26 interacts with CELF2 protein to enhance myeloid leukemia cell proliferation and invasion via the circRNA_ANKIB1/miR-195-5p/PRR11 axis. Cell Transplant. (2021) 30: 80. doi: 10.1177/0963689720986080
60. Dong K, He X, Su H, Fulton DJR, Zhou J. Genomic analysis of circular RNAs in heart. BMC Med Genomics. (2020) 13:167. doi: 10.1186/s12920-020-00817-7
61. Pan X, Shen HB. RNA-protein binding motifs mining with a new hybrid deep learning based cross-domain knowledge integration approach. BMC Bioinform. (2017) 18:136. doi: 10.1186/s12859-017-1561-8
62. Pan X, Rijnbeek P, Yan J, Shen HB. Prediction of RNA-protein sequence and structure binding preferences using deep convolutional and recurrent neural networks. BMC Gen. (2018) 19:511. doi: 10.1186/s12864-018-4889-1
63. Pan X, Shen HB. Predicting RNA-protein binding sites and motifs through combining local and global deep convolutional neural networks. Bioinformatics. (2018) 34:3427–36. doi: 10.1093/bioinformatics/bty364
64. Wang Z, Lei X, Wu FX. Identifying cancer-specific circRNA-RBP binding sites based on deep learning. Molecules. (2019) 24:35. doi: 10.3390/molecules24224035
65. Bhuyan R, Bagchi A. Prediction of the differentially expressed circRNAs to decipher their roles in the onset of human colorectal cancers. Gene. (2020) 762:145035. doi: 10.1016/j.gene.2020.145035
66. Wawrzyniak O, Zarebska Z, Kuczynski K, Gotz-Wieckowska A, Rolle K. Protein-related circular RNAs in human pathologies. Cells. (2020) 9:1841. doi: 10.3390/cells9081841
67. Tang Q, Hann SS. Biological roles and mechanisms of circular RNA in human cancers. Onco Targets Ther. (2020) 13:2067–92. doi: 10.2147/OTT.S233672
68. Zang J, Lu D, Xu A. The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. J Neurosci Res. (2020) 98:87–97. doi: 10.1002/jnr.24356
69. Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. (2017) 14:361–9. doi: 10.1080/15476286.2017.1279788
70. Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. (2016) 7:12429. doi: 10.1038/ncomms12429
71. Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. (2017) 38:1402–12. doi: 10.1093/eurheartj/ehw001
72. Yang ZG, Awan FM, Du WW, Zeng Y, Lyu J, Wu .upta S, et al. The circular RNA interacts with STAT3, increasing its nuclear translocation and wound repair by modulating Dnmt3a and miR-17 function. Mol Ther. (2017) 25:2062–74. doi: 10.1016/j.ymthe.2017.05.022
73. Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li X, et al. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics. (2017) 7:3842–55. doi: 10.7150/thno.19764
74. Okholm TLH, Sathe S, Park SS, Kamstrup AB, Rasmussen AM, Shankar A, et al. Transcriptome-wide profiles of circular RNA and RNA-binding protein interactions reveal effects on circular RNA biogenesis and cancer pathway expression. Genome Med. (2020) 12:112. doi: 10.1186/s13073-020-00812-8
75. Kalyana-Sundaram S, Kumar-Sinha C, Shankar S, Robinson DR, Wu YM, Cao X, et al. Expressed pseudogenes in the transcriptional landscape of human cancers. Cell. (2012) 149:1622–34. doi: 10.1016/j.cell.2012.04.041
76. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. (2017) 27:626–41. doi: 10.1038/cr.2017.31
77. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. (2017) 66:22–37 e29. doi: 10.1016/j.molcel.2017.02.017
78. Yin H, Shen X, Zhao J, Cao X, He H, Han S, et al. Circular RNA circfam188b encodes a protein that regulates proliferation and differentiation of chicken skeletal muscle satellite cells. Front Cell Dev Biol. (2020) 8:522588. doi: 10.3389/fcell.2020.522588
79. Cy C, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. (1995) 268:415–7. doi: 10.1126/science.7536344
80. Macejak GD, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. (1991) 353:90–4. doi: 10.1038/353090a0
81. Abe N, Hiroshima M, Maruyama H, Nakashima Y, Nakano Y, Matsuda A, et al. Rolling circle amplification in a prokaryotic translation system using small circular RNA. Angew Chem Int Ed Engl. (2013) 52:7004–8. doi: 10.1002/anie.201302044
82. Hentze MW, Preiss T. Circular RNAs: splicing's enigma variations. EMBO J. (2013) 32:923–5. doi: 10.1038/emboj.2013.53
83. Dong W, Dai ZH, Liu FC, Guo XG, Ge CM, Ding J, et al. The RNA-binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD-circRNA 2 production. EBioMed. (2019) 45:155–67. doi: 10.1016/j.ebiom.2019.06.030
84. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. (2016) 44:2846–58. doi: 10.1093/nar/gkw027
85. Van Dijk EL, Auger H, Jaszczyszyn Y, Thermes C. Ten years of next-generation sequencing technology. Trends Genet. (2014) 30:418–26. doi: 10.1016/j.tig.2014.07.001
86. Wang J, Ren Q, Hua L, Chen J, Zhang J, Bai H, et al. Comprehensive analysis of differentially expressed mRNA, lncRNA and circRNA and their ceRNA networks in the longissimus dorsi muscle of two different pig breeds. Int J Mol Sci. (2019) 20:1107. doi: 10.3390/ijms20051107
87. Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. (2016) 26:1277–87. doi: 10.1101/gr.202895.115
88. Dahl M, Daugaard I, Andersen MS, Hansen TB, Gronbaek K, Kjems J, et al. Enzyme-free digital counting of endogenous circular RNA molecules in B-cell malignancies. Lab Invest. (2018) 98:1657–69. doi: 10.1038/s41374-018-0108-6
89. Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. (2014) 20:1666–70. doi: 10.1261/rna.043687.113
90. Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. (2019) 176:869–81 e813. doi: 10.1016/j.cell.2018.12.021
91. Sekar S, Cuyugan L, Adkins J, Geiger P, Liang WS. Circular RNA expression and regulatory network prediction in posterior cingulate astrocytes in elderly subjects. BMC Genomics. (2018) 19:340. doi: 10.1186/s12864-018-4670-5
92. Sekar S, Geiger P, Cuyugan L, Boyle A, Serrano G, Beach TG, et al. Identification of circular RNAs using RNA sequencing. J Vis Exp. (2019). doi: 10.3791/59981
93. Zhang QL, Ji XY, Li HW, Guo J, Wang F, Deng XY, et al. Identification of circular RNAs and their altered expression under poly(I:C) challenge in key antiviral immune pathways in amphioxus. Fish Shellfish Immunol. (2019) 86:1053–7. doi: 10.1016/j.fsi.2018.12.061
94. Panda AC, De S, Grammatikakis I, Munk R, Yang X, Piao Y, et al. High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res. (2017) 45:e116. doi: 10.1093/nar/gkx297
95. Chuang TJ, Chen YJ, Chen CY, Mai TL, Wang YD, Yeh CS, et al. Integrative transcriptome sequencing reveals extensive alternative trans-splicing and cis-backsplicing in human cells. Nucleic Acids Res. (2018) 46:3671–91. doi: 10.1093/nar/gky032
96. Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. (2018) 110:166. doi: 10.1093/jnci/djx166
97. Wang L, Long H, Zheng Q, Bo X, Xiao X, Li B. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol Cancer. (2019) 18:119. doi: 10.1186/s12943-019-1046-7
98. Labaj PP, Leparc GG, Linggi BE, Markillie LM, Wiley HS, Kreil DP. Characterization and improvement of RNA-Seq precision in quantitative transcript expression profiling. Bioinformatics. (2011) 27:i383–91. doi: 10.1093/bioinformatics/btr247
99. Li S, Teng S, Xu J, Su G, Zhang Y, Zhao J, et al. Microarray is an efficient tool for circRNA profiling. Brief Bioinform. (2019) 20:1420–33. doi: 10.1093/bib/bby006
100. López-Jiménez E, Ana R, Andrés-León E. RNA sequencing and prediction tools for circular RNAs analysis. Adv Exp Med Biol. (2018) 1087:17–33. doi: 10.1007/978-981-13-1426-1_2
101. Lin F, Yang Y, Guo Q, Xie M, Sun S, Wang X, et al. Analysis of the molecular mechanism of acute coronary syndrome based on circRNA-miRNA network regulation. Evid Based Complement Alternat Med. (2020) 2020:1584052. doi: 10.1155/2020/1584052
102. Liu T, Zhang G, Wang Y, Rao M, Zhang Y, Guo A, et al. Identification of Circular RNA-MicroRNA-Messenger RNA regulatory network in atrial fibrillation by integrated analysis. Biomed Res Int. (2020) 2020:8037273. doi: 10.1155/2020/8037273
103. Sun Z, Yu T, Jiao Y, He D, Wu J, Duan W, et al. Expression profiles and ontology analysis of circular RNAs in a mouse model of myocardial ischemia/reperfusion injury. Biomed Res Int. (2020) 2020:2346369. doi: 10.1155/2020/2346369
104. Si X, Zheng H, Wei G, Li M, Li W, Wang H, et al. circRNA Hipk3 induces cardiac regeneration after myocardial infarction in mice by binding to Notch1 and miR-133a. Mol Ther Nucleic Acids. (2020) 21:636–55. doi: 10.1016/j.omtn.2020.06.024
105. Sun JY, Shi Y, Cai XY, Liu J. Potential diagnostic and therapeutic value of circular RNAs in cardiovascular diseases. Cell Signal. (2020) 71:109604. doi: 10.1016/j.cellsig.2020.109604
106. Chen JX, Hua L, Zhao CH, Jia QW, Zhang J, Yuan JX, et al. Quantitative proteomics reveals the regulatory networks of circular RNA BTBD7_hsa_circ_0000563 in human coronary artery. J Clin Lab Anal. (2020) 34:e23495. doi: 10.1002/jcla.23495
107. Ming-Hui Yang HW, Sheng-Na Han, Xin J.ia, Si hang, Fei-Fei D., ai Meng-Jiao Z, hou Zhongnan, et al. Circular RNA expression in isoproterenol hydrochloride-induced cardiac hypertrophy. Aging. (2020) 12:2530–44. doi: 10.18632/aging.102761
108. Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. (2016) 37:2602–11. doi: 10.1093/eurheartj/ehv713
109. Liu X, Wang M, Li Q, Liu W, Jiang H. CircRNA ACAP2 Induces Myocardial Apoptosis After Myocardial Infarction by Sponging miR-29. Minerva Medica. (2020).
110. Li F, Long TY, Bi SS, Sheikh SA, Zhang CL. circPAN3 exerts a profibrotic role via sponging miR-221 through FoxO3/ATG7-activated autophagy in a rat model of myocardial infarction. Life Sci. (2020) 257:118015. doi: 10.1016/j.lfs.2020.118015
111. Zhou B, Yu JW. A novel identified circular RNA. circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-beta1. Biochem Biophys Res Commun. (2017) 487:769–75. doi: 10.1016/j.bbrc.2017.04.044
112. Tang CM, Zhang M, Huang L, Hu ZQ, Zhu JN, Xiao Z, et al. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci Rep. (2017) 7:40342. doi: 10.1038/srep40342
113. Zhu Y, Pan W, Yang T, Meng X, Jiang Z, Tao L, et al. Upregulation of circular RNA CircNFIB attenuates cardiac fibrosis by sponging miR-433. Front Genet. (2019) 10:564. doi: 10.3389/fgene.2019.00564
114. Ni H, Li W, Zhuge Y, Xu S, Wang Y, Chen Y, et al. Inhibition of circHIPK3 prevents angiotensin II-induced cardiac fibrosis by sponging miR-29b-3p. Int J Cardiol. (2019) 292:188–96. doi: 10.1016/j.ijcard.2019.04.006
115. Zhang N, Wang X. Circular RNA ITCH mediates H2O2-induced myocardial cell apoptosis by targeting miR-17-5p via wnt/beta-catenin signalling pathway. Int J Exp Pathol. (2020) 102:22–31. doi: 10.1111/iep.12367
116. Hu X, Ma R, Cao J, Du X, Cai X, Fan Y. CircSAMD4A aggravates H/R-induced cardiomyocyte apoptosis and inflammatory response by sponging miR-138-5p. J Cell Mol Med. (2020). doi: 10.1111/jcmm.16093
117. Ji X, Ding W, Xu T, Zheng X, Zhang J, Liu M, et al. MicroRNA-31-5p attenuates doxorubicin-induced cardiotoxicity via quaking and circular RNA Pan3. J Mol Cell Cardiol. (2020) 140:56–67. doi: 10.1016/j.yjmcc.2020.02.009
118. Zhang CL, Long TY, Bi SS, Sheikh SA, Li F. CircPAN3 ameliorates myocardial ischaemia/reperfusion injury by targeting miR-421/Pink1 axis-mediated autophagy suppression. Lab Invest. (2021) 101:89–103. doi: 10.1038/s41374-020-00483-4
119. Chen L, Luo W, Zhang W, Chu H, Wang J, Dai X, et al. circDLPAG4/HECTD1 mediates ischaemia/reperfusion injury in endothelial cells via ER stress. RNA Biol. (2020) 17:240–53. doi: 10.1080/15476286.2019.1676114
120. Zhu Y, Zou C, Jia Y, Zhang H, Ma X, Zhang J. Knockdown of circular RNA circMAT2B reduces oxygen-glucose deprivation-induced inflammatory injury in H9c2 cells through up-regulating miR-133. Cell Cycle. (2020) 19:2622–30. doi: 10.1080/15384101.2020.1814025
121. Cui X, Dong Y, Li M, Wang X, Jiang M, Yang W, et al. A circular RNA from NFIX facilitates oxidative stress-induced H9c2 cells apoptosis. In Vitro Cell Dev Biol Anim. (2020) 56:715–22. doi: 10.1007/s11626-020-00476-z
122. Jin P, Li LH, Shi Y, Hu NB. Salidroside inhibits apoptosis and autophagy of cardiomyocyte by regulation of circular RNA hsa_circ_0000064 in cardiac ischemia-reperfusion injury. Gene. (2021) 767:145075. doi: 10.1016/j.gene.2020.145075
123. Gao WQ, Hu XM, Zhang Q, Yang L, Lv XZ, Chen S, et al. (2021). Downregulation of circFASTKD1 ameliorates myocardial infarction by promoting angiogenesis. Aging. 13:3588–604. doi: 10.18632/aging.202305
Keywords: circular RNA, cardiovascular disease, pathogenesis, cardiomyocyte, regeneration
Citation: Tang L, Li P, Jang M and Zhu W (2021) Circular RNAs and Cardiovascular Regeneration. Front. Cardiovasc. Med. 8:672600. doi: 10.3389/fcvm.2021.672600
Received: 26 February 2021; Accepted: 22 March 2021;
Published: 13 April 2021.
Edited by:
Junjie Yang, University of Alabama at Birmingham, United StatesReviewed by:
Jiacheng Sun, University of Alabama at Birmingham, United StatesShiyue Xu, The First Affiliated Hospital of Sun Yat-sen University, China
Copyright © 2021 Tang, Li, Jang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wuqiang Zhu, emh1Lnd1cWlhbmdAbWF5by5lZHU=
 Ling Tang
Ling Tang Pengsheng Li
Pengsheng Li Michelle Jang
Michelle Jang Wuqiang Zhu
Wuqiang Zhu