- 1University of Oklahoma Health Sciences Center, Oklahoma City, OK, United States
- 2Oklahoma Medical Research Foundation, Oklahoma City, OK, United States
- 3University of Tennessee Health Science Center, Memphis, TN, United States
- 4Tongji Medical College of Huazhong University of Science and Technology, Wuhan, China
Background: Tetraspanins and integrins are integral membrane proteins. Tetraspanins interact with integrins to modulate the dynamics of adhesion, migration, proliferation, and signaling in the form of membrane domains called tetraspanin-enriched microdomains (TEMs). TEMs also contain other cell adhesion proteins like immunoglobulin superfamily (IgSF) proteins and claudins. Cardiovascular functions of these TEM proteins have emerged and remain to be further revealed.
Objectives: The aims of this study are to explore the roles of these TEM proteins in the cardiovascular system using bioinformatics tools and databases and to highlight the TEM proteins that may functionally associate with cardiovascular physiology and pathology.
Methods: For human samples, three databases—GTEx, NCBI-dbGaP, and NCBI-GEO—were used for the analyses. The dbGaP database was used for GWAS analysis to determine the association between target genes and human phenotypes. GEO is an NCBI public repository that archives genomics data. GTEx was used for the analyses of tissue-specific mRNA expression levels and eQTL. For murine samples, GeneNetwork was used to find gene–phenotype correlations and gene–gene correlations of expression levels in mice. The analysis of cardiovascular data was the focus of this study.
Results: Some integrins and tetraspanins, such as ITGA8 and Cd151, are highly expressed in the human cardiovascular system. TEM components are associated with multiple cardiovascular pathophysiological events in humans. GWAS and GEO analyses showed that human Cd82 and ITGA9 are associated with blood pressure. Data from mice also suggest that various cardiovascular phenotypes are correlated with integrins and tetraspanins. For instance, Cd82 and ITGA9, again, have correlations with blood pressure in mice.
Conclusion: ITGA9 is related to blood pressure in both species. KEGG analysis also linked ITGA9 to metabolism and MAPK signaling pathway. This work provides an example of using integrated bioinformatics approaches across different species to identify the connections of structurally and/or functionally related molecules to certain categories of diseases.
Introduction
Tetraspanin-Enriched Microdomains
Tetraspanin-enriched microdomains (TEMs) are ubiquitously present in a variety of cells such as endothelial cells (ECs), vascular smooth muscle cells (VSMCs), leukocytes, and platelets. Mainly formed by tetraspanins, TEMs also contain other membrane proteins such as integrins, immunoglobulin superfamily (IgSF) proteins, and claudins (1) (Figure 1).
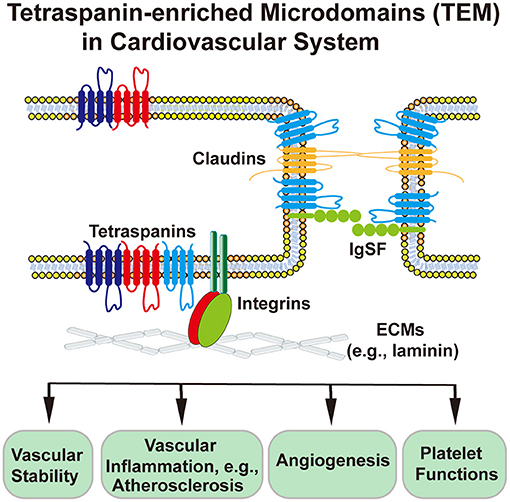
Figure 1. Schematic presentation of tetraspanin-enriched microdomains (TEMs) at the cell–ECM and cell–cell interfaces and TEM-involved or TEM-regulated cardiovascular functions. Endothelial cell is used as an example of cardiovascular cells. In endothelial cells, tetraspanins associate with each other and with integrins, claudins, and IgSF proteins, to regulate physiological and pathological activities of heat, vessel, and blood.
Besides four membrane-spanning segments, each tetraspanin has a highly conserved CCG motif in large extracellular loop and a few polar residues in transmembrane domains (2). The functions of the 33 human tetraspanins are quite diverse, with unknown underlying general mechanism. Integrins are heterodimeric receptors mainly for extracellular matrices (ECMs), conducting both inside-out and outside-in signaling for various cellular functions (3) such as cell adhesion, migration, proliferation, and survival (4, 5).
Recent Progress in Vascular Pathophysiology of TEMs
Tetraspanins
Studies on tetraspanins in blood vessels were largely limited to CD9, CD63, CD151, and Tspan12 (6, 7). A recent study revealed that CD82 restrains pathological angiogenesis by facilitating cell adhesion protein endocytosis and then confining EC movement (8). The miR-K6-5p of Kaposi's sarcoma-associated herpesvirus represses CD82 expression by targeting its coding sequence and promotes EC invasion and angiogenesis by activating c-Met signaling (9). Postnatal angiogenesis becomes severely impaired in tetraspanin Tspan8-null and Cd151-null; Tspan8-null mice, together with reduced EC–ECM adhesion and EC migration upon Cd151 and Tspan8 ablations (10), resembling the vascular impacts of Cd151-null mice reported earlier. Tspan8 appears to support the lymphangiogenesis induced by pancreatic cancer. CD151-integrin α6β1 complexes drive embryonic stem cells on Laminin 111 to differentiate to ECs, but CD151–integrin α3β1 complexes do not (11). Tetraspanin Tspan18 is expressed in developing vasculature and primary ECs, and its silencing via morpholino in zebrafish leads to vascular defects in angiogenesis, vessel stability, and arterial-venous specification through vascular endothelial growth factor (VEGF)/VEGFR and Notch signaling (12). VEGF-induced expression of tm4sf18, a tetraspanin-related four-transmembrane protein, in ECs serves as a positive feedback mechanism to amplify VEGF signaling and therefore enhance angiogenesis, in contrast to the well-established negative feedback mechanism of Notch in VEGF-dependent angiogenesis (13).
A recent study showed that CD9 antibody or Cd9 ablation attenuates the atherosclerotic plaque progression in mouse as CD9 promotes senescence of ECs (14). Consistently, CD9 expression becomes increased in human arteries upon aging and in atherosclerotic arteries in human (15). CD151 sustains EC–EC and EC–ECM adhesions and confines cytoskeletal tension within ECs, to support vascular stability (16). Differential expressions of Tspan7 in ischemic and non-ischemic muscles between C57BL/6 and BALB/c mouse strains, which display differential susceptibility to hindlimb ischemia and were used to identify the genes associated with vascular inflammatory responses to hindlimb ischemia, suggest a regulatory role of Tspan7 in peripheral arterial disease (17). Given that tetraspanins serve not only as markers but also as regulators of exosomes, direct participation of the exosomes from VSMCs in artery calcification (13) suggests tetraspanin involvement in this vascular event. Tspan2 may be associated with VSMC differentiation as its selective expression in VSMCs is dramatically upregulated by TGFβ or myocardin, two activators of VSMC differentiation, but becomes downregulated in mouse carotid arteries after ligation injury and in human arteriovenous fistula samples after occlusion by dedifferentiated neointimal VSMCs (18). Human-induced pluripotent stem cell (hiPSC)-derived cardiomyocyte progenitors that express CD82 almost exclusively differentiate into cardiomyocytes both in vitro and in vivo, and CD82 is involved in fate commitment to cardiomyocytes through Wnt signaling inhibition (19). Hence, CD82 may serve as a marker for prospectively isolating cardiomyocyte progenitors.
Mutations in TSPAN12 cause familial exudative vitreoretinopathy (FEVR) (20, 21). As a co-receptor of Norrin, Tspan12 interacts with Norrin and its receptor Frizzled-4 via its large extracellular loop and enhances the selectivity of Norrin binding to Frizzled-4, while FEVR-linked mutations in TSPAN12 disrupt the co-receptor function of Tspan12 (22). Besides its importance in retinal vasculature development, Tspan12 in endothelia is also needed for the maintenance of the blood–retina barrier (23). As an exciting advancement, Tspan12 antibody displayed therapeutic benefits for vasoproliferative retinopathies in mouse models by limiting β-catenin signaling, and a combination of the antibody and VEGF antagonist markedly improved the efficacy of VEGF antagonist (24). Similarly, norrin restores diabetes-caused or VEGF-induced disruption of the blood–retina barrier by enhancing tight junction and through β-catenin (25). Intriguingly, VEGF facilitates norrin signaling by elevating the presence of Tspan12 at the EC surface in a MAPK signaling-dependent manner. Tspan12 also supports norrin signaling for the maintenance of the blood–brain barrier (26).
In contrast to increased bleeding upon murine Cd151 ablation, Cd82 ablation reduces bleed time in mouse by enhancing integrin αIIbβ3 expression on platelets and clot retraction (27). Both CD151 and tetraspanin TSSC6 are physically associated with ADP purinergic receptor P2Y12 and functionally complement P2Y12 roles in platelet aggregation and thrombus stabilization (28, 29). Moreover, Tspan18 is required for the proper formation of thrombi in response to inflammatory stimuli and deposition of platelets in response to vascular injury by promoting Orai1/Ca2+ signaling in and von Willebrand factor release from ECs (30). CD63 is required for stabilizing P-selectin at the EC surface to recruit leukocytes (31), and annexin A8 facilitates trafficking of CD63 from late endosome to Weibel–Palade body and EC surface and stabilizes the presence of CD63 and P-selectin on the EC surface for leukocyte recruitment (32).
IgSF Proteins and Claudins
IgSF proteins EWI-2 and EWI-F associate physically with tetraspanins CD9, CD81, and CD82 (1, 33) (Figure 1). EWI-2 connects tetraspanins to actin cytoskeleton, interacts with phospholipids with its cytoplasmic domain (34, 35), and associates indirectly with some integrins via tetraspanins (36, 37). Claudins 1, 7, and 11 are associated with tetraspanins CD9, CD81, Tspan8, and Tspan3 (Figure 1). As claudins are essential constituents of tight junction, TEMs to which claudins partition in ECs are predicted to regulate the function of endothelial barriers such as the blood–brain barrier and the blood–retina barrier (38).
Integrins
Besides being successful therapeutic targets of thrombosis, integrins are emerging targets for other cardiovascular diseases, especially atherosclerosis (39), due to their crucial functions in platelets and ECs (40, 41). Integrin ανβ3 is used as an angiogenesis marker for early-stage atherosclerosis imaging (42), in addition to (i) its supporting role in tumor and other pathological angiogenesis such as Matrigel plug and aortic ring angiogenesis and (ii) its potential as a therapeutic target against tumor angiogenesis (43–45). Interestingly, exosomal integrin αvβ6 released by prostate cancer cells promotes vascular morphogenesis, probably by confining STAT1 phosphorylation (46). Integrin β3 was also recently shown to regulate VSMCs during atherogenesis (47). Other integrins were found to be important in blood vessel maturation (48) and stability (49). For example, heterozygous ablation of endothelial integrin β1 or inhibition of integrin β1 by its function-blocking antibody reduced acute vascular leakage caused by bacterial endotoxin (50). Integrins in VSMCs regulate the contractile function and affect arterial stiffening (51), while blockades of integrin α5β1–fibronectin and α5β1–fibrin interactions enhance the calcification of valvular interstitial cell of cardiac valves (52). In addition to ECs and VSMCs, integrins in leukocytes, mainly β1 and β2 integrins, are well-documented for their roles in vascular inflammatory response (53). Recent advances reveal that sharp upregulation of integrin α3β1 in neutrophils during sepsis is associated with Toll-like receptor-induced vascular inflammatory responses and cytokine productions (54) and that neutrophils aid oxidation-dependent modification of extracellular matrix during vascular inflammation and then the oxidation promotes integrins αMβ2- and αDβ2-mediated vascular infiltration of macrophages (55). Apparently, there is still much to learn about the roles of integrins in the pathogenesis of cardiovascular diseases (56, 57), for the purpose of disease treatment.
To expand our knowledge about TEMs (Figure 1), we combine multiple bioinformatics tools and databases to investigate putative linkages of each member of the tetraspanin and integrin superfamilies to cardiovascular events systematically. Bioinformatics proves beneficial when it comes to predicting the disease risk factors and targets.
Methods
Databases With Information From Human Samples
The Genotype-Tissue Expression Database
The Genotype-Tissue Expression (GTEx) database provides human gene expression levels and their regulatory relationships to genetic variations, stressing the concept of expression quantitative trait loci (eQTL). This database was used to gather the mRNA levels of all integrins and tetraspanins. Version 8 of GTEx, which includes 17,382 samples from 948 human donors, was downloaded on January 2021 at https://gtexportal.org/.
The Database of Genotypes and Phenotypes
The Database of Genotypes and Phenotypes (dbGaP) is a National Center for Biotechnology Information (NCBI)-affiliated database investigating the associations between human genotypes and phenotypes (58). The results from genome-wide association studies (GWAS) housed in National Human Genome Research Institute were merged with the other genetic databases from NCBI, such as Gene and dbSNP, to suggest potential genotype–phenotype correlations that are more general than the typical mutation–disease linkage. We used the public data interface Phenotype–Genotype Integrator (PheGenI) (59) to detect every gene belonging to the two functionally related families for their associations with cardiovascular phenotypes. Phenotypes such as platelet function and cholesterol were also included for their relevance to coronary heart disease and thrombosis. All deposited datasets (1,232 in total) in this database were included in this study. The data was downloaded in November 2018 at https://www.ncbi.nlm.nih.gov/gap/. Linear regression or logistic regression was used for statistical analysis.
Gene Expression Omnibus for Literature Mining and Transcriptional Correlation Network Construction
Gene Expression Omnibus (GEO) is an NCBI database that deposits a wide range of high-throughput genomics data across species and under numerous normal or diseased conditions. We utilized all human datasets to predict the correlated genes of our molecules of interest.
The literature-mining approach IRIDESCENT (60–62) was used to map relations between genes, diseases, and phenotypes. Briefly, it uses a thesaurus of terms, including spelling and abbreviation variants, to identify when they co-occur within PubMed abstracts. The network of relationships can then be interrogated for commonalities. The latest version was run on MEDLINE on January 2021, which included 31,907,626 total records.
For GAMMA analysis, data for the microarray platform GPL570 was downloaded in August 2018 from NCBI's GEO repository (https://www.ncbi.nlm.nih.gov/geo/), consisting of 80,600 samples from 2,698 experiments. Sample data (GSM files) was quantile normalized prior to calculating a matrix of gene–gene correlations using Pearson's correlation coefficient (r) (ALGLIB, Sergey Bochkanov, 2007, https://www.alglib.net/). Using the K-nearest neighbors (KNN) approach (K = 40), a set of the most highly correlated genes was obtained for each gene as described previously (63). Using a “guilt by association” approach, literature-based commonalities were identified for the most highly correlated genes with IRIDESCENT (62). The result is a list of literature-based enrichments for the correlated genes (63) that are, in turn, predicted functions for individual genes. The categories of commonalities identified are diseases, phenotypes, genes, chemicals, drugs, and Gene Ontology categories. GAMMA identifies the top 40 most correlated genes for each input gene, and then IRIDESCENT provides an analysis of what those genes have in common in MEDLINE in terms of (1) the name of the connected concept or commonality (e.g., diseases, genes, etc), (2) the number of genes (out of 40) connected to the concept in MEDLINE or shared relationships, and (3) an observed to expected value that reflects how many connections were observed relative to the number expected by chance, given the number of connections of each concept in the analysis. The “Score” reflects a balance between how unusual the observed number of connections to a concept is, given how connected each object is in the subnetwork being analyzed, and how many connections were made. If only the observed to expected (Obs/Exp) value is used to score connections between concepts, the highest values tend to be between relatively rare concepts. These could be potentially interesting, but at the same time, the association is based upon less evidence. Conversely, the most connected concepts are almost always the most abundant concepts in general. Thus, the score provides a way to balance the search for a statistically unusual number of links between concepts with the amount of evidence available.
Databases With Information From Mouse Samples
GeneNetwork
The GeneNetwork website (http://genenetwork.org/) (64) provides genomic data from various mouse strains and different murine tissue samples. The main purpose of this database is to investigate genome-to-phenome relationships in mice (65). We used this database to (i) associate integrins and tetraspanins with mouse cardiovascular phenotypes; (ii) find correlations between expression levels of different genes, specifically in mouse aorta samples; and (iii) predict tissue-specific signaling pathways of a protein of interest by the gene–gene associations. The mouse aorta transcriptome data [UCLA BXD Aorta Affy M430 2.0 (Jan16) RMA] and cardiovascular phenotype data were downloaded on December 2019 from GeneNetwork and used for Pearson correlation coefficient analysis to identify gene–phenotype and gene–gene associations, with the resulted p < 0.05 as the significance threshold. The analysis was carried out using R package v4.03 (66), and the aorta transcriptome data were deposited to and can be accessed on GeneNetwork and GEO under the accession numbers of GN819 and GSE66569, respectively. A total of 38 mouse lines, consisting of 36 BXD strains and two C57BL/6J and DBA2/J parental strains, were included in the aorta transcriptome data analysis. Gene set enrichment analysis was performed at the Webgestalt website (http://www.webgestalt.org) (67), to investigate the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. The p value generated from the test was automatically adjusted to account for multiple comparisons using the Benjamini and Hochberg correction (68). A minimum overlap of five genes and an adjusted p < 0.05 were required to determine the genes significantly overrepresented in those categories.
Results
Some Integrins and Tetraspanins, Such as ITGA8 and Cd151, Are Highly Distributed in the Human Cardiovascular System
The expression level of each gene of tetraspanins and integrins in the human cardiovascular system was determined by GTEx. According to the data, as shown in the heatmaps (Figure 2A), the integrin and tetraspanin superfamilies have contrast compositions in the cardiovascular system, suggesting a possible discrepancy or divergence in their functions. The mRNA levels of integrins α3, α5, α7, α8, αV, β1, and β5 and tetraspanins TSPAN3, TSPAN4, TSPAN9, CD9, CD63, CD81, and CD151 range from moderate to high. In mouse aortas, tetraspanins CD151, CD81, TSPAN3, and CD9; integrins β5, β1, and αV; and integrin-associated protein Fermt2/Kindlin2 are relatively highly expressed (Table 1), although this study/dataset did not dissect the distributions of these expressions in EC and/or VSMC compartments of the aortas. Given that VSMCs constitute the predominant cell mass in aorta, these expressions more likely represent the expressions from the VSMCs of aorta.
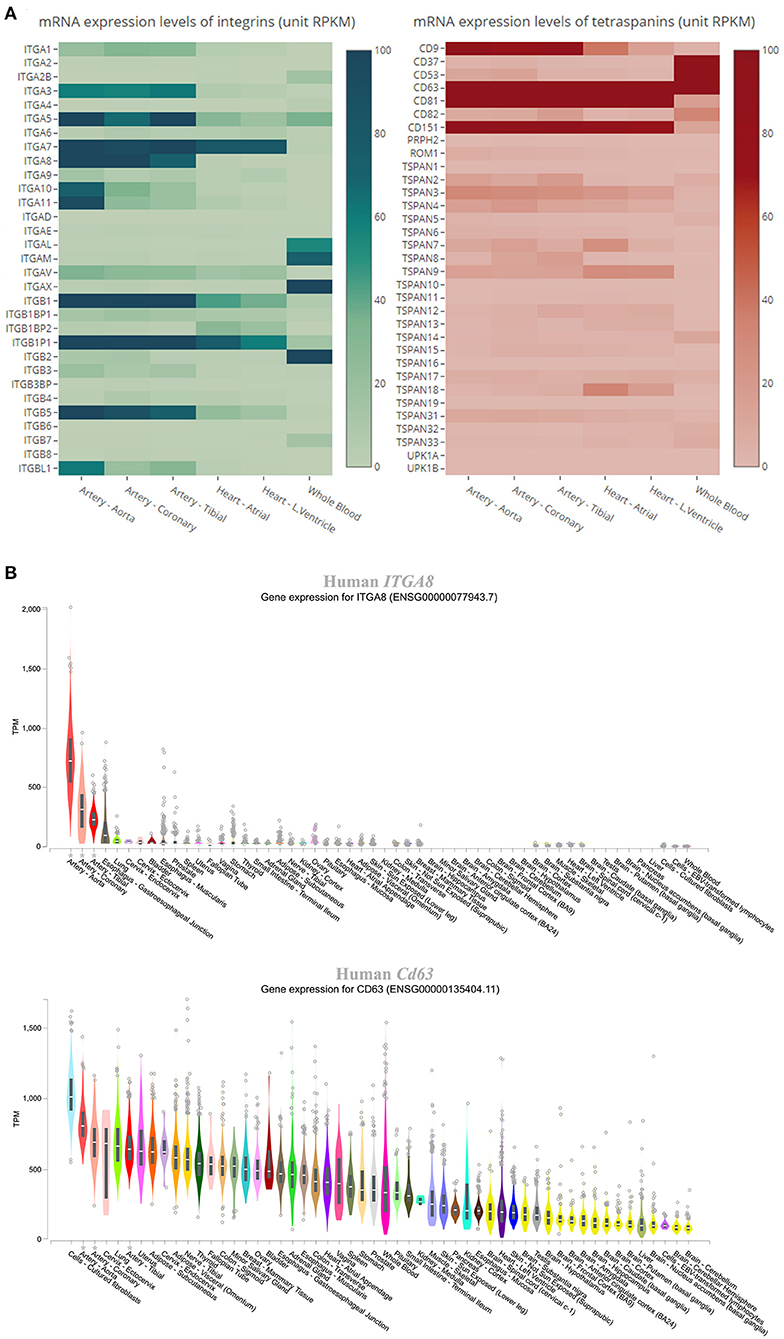
Figure 2. Expression levels of integrins and tetraspanins in human cardiovascular tissues. (A) Heatmaps showing the mRNA levels in the units of RPKM (reads per kilobase of transcript, per million mapped reads) of the integrins and tetraspanins in human aorta, coronary artery, tibial artery, heart atrium, left ventricle, and whole blood. (B) Box plot showing the expression levels, presented as TPM (transcripts per kilobase million), of ITGA8 and Cd63 across various human tissues. All data were extracted from GTEx.
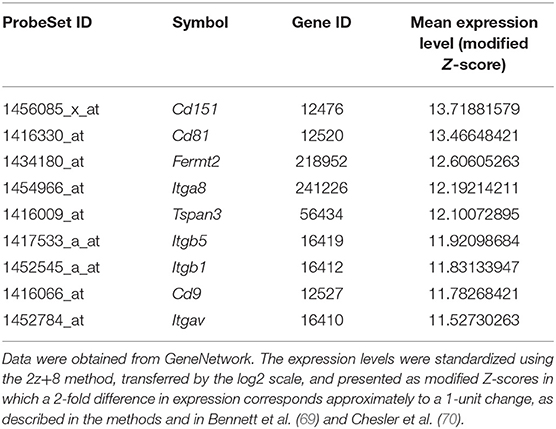
Table 1. Mouse integrins, tetraspanins, and integrin-associated proteins with the gene expression levels that are significantly higher than the median expression level of all genes in mouse aortas.
The genes that are abundant in both human and mouse cardiovascular systems were chosen for comparisons of their expression levels across various human tissues, using data from GTEx. All of the genes are highly expressed at the mRNA level in arteries, including the aorta, coronary arteries, and tibial arteries. Figure 2B shows genes ITGA8 and Cd63, which are highly expressed in the arteries, as examples for integrin and tetraspanin families, respectively. We must acknowledge that, as human tissue specimen examined in GTEx largely came from healthy subjects, the low expressions for some of these tetraspanin and integrin genes at physiological condition do not dismiss the possibility of upregulation of their expressions under pathological conditions. For other organs such as the liver and lung, their expressions in the vasculature within the organs are difficult to be determined.
GWAS Shows Associations Between Integrins or Tetraspanins and Cardiovascular Events
In order to find associations between the genes of our interest and cardiovascular phenotypes, we used the database interface Phenotype–Genotype Integrator of NCBI-dbGaP for the investigation. The results of GWAS analysis provided insights into the correlations of integrins and tetraspanins and human diseases on a genome scale (Figure 3A). Integrin gene ITGA8 has missense single-nucleotide polymorphism (SNP) variant (denoted red in Figure 3A) rs7895372 associated with cholesterol in an Oceania population (p = 6.000 × 10−6). Other associations involving missense variations are as follows: ITGA9 with arterial pressure through rs146867977 in African Americans (p = 7.254 × 10−6), ITGAE with platelet function tests through rs2976230 in African Americans (p = 1.523 × 10−5), and ITGAM with platelet function tests through two missense variants, rs1143678 (p = 7.013 × 10−7) and rs1143683 (p = 9.980 × 10−7), in Europeans. In addition, ITGA2 has two coding sequence synonymous (cds-syn) variants, rs1062535 (p = 1.096 × 10−5) and rs1126643 (p = 1.723 × 10−5), that are also associated with platelet function tests in Europeans (denoted in red in Figure 3A).
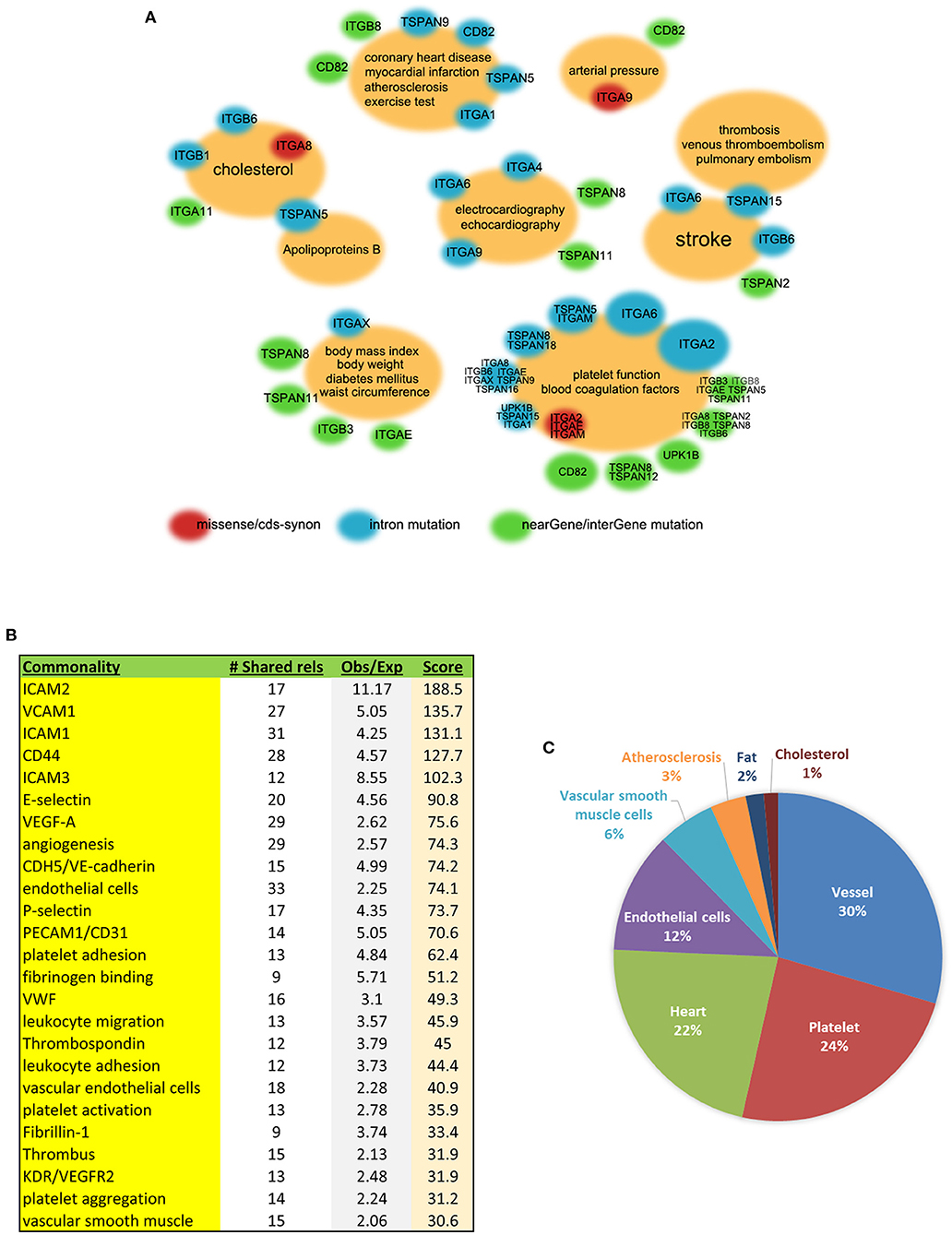
Figure 3. Integrin and/or tetraspanin genes associated with multiple cardiovascular phenotypes in humans. (A) Association clouds with GWAS phenotypes at the centers and related genes at the surrounding. Small red clouds with gene names denote an association between a missense and/or cds-syn mutation(s) of the genes and the phenotype(s) in the center; blue denotes intron mutation; green denotes the 3′ or 5′ untranslated region, near-gene, or intergene SNPs. The size of the surrounding clouds represents the number of associations. Data from dbGaP. (B) The 66 members of the tetraspanin and integrin gene families were analyzed for the commonalities that they share within the published literature. The “# Shared rels” is how many of these 66 genes shared the commonality; “Obs/Exp” is the ratio of observed to expected connections, reflecting enrichment of the association; and “Score” is the product of the two (see methods). (C) GAMMA prediction charts based on GEO co-expression datasets displaying various predicted gene-function associations in the human cardiovascular system.
TSPAN5, TSPAN15, Cd82, ITGA2, and ITGA6 are worth mentioning for their multiple non-coding SNP associations within the introns (denoted in blue in Figure 3A). TSPAN5 has a dozen of SNPs associated with cholesterol, LDL-cholesterol, and apolipoprotein B in Europeans. TSPAN15 is correlated with a few different diseases involving thrombosis—venous thromboembolism (71), pulmonary embolism, and stroke in three independent GWAS studies conducted among European or African American populations. CD82 has one intron SNP rs730129 that is associated with atherosclerosis and other near-gene or intergenic SNPs that are associated with myocardial infarction, blood pressure, and platelet function tests. In addition to the cds-syn variants mentioned above, ITGA2 has multiple intron SNPs that are associated with platelet function tests in Europeans, with results coming from the same study (72). ITGA6 has a potentially versatile role in the cardiovascular system due to containing intron SNPs that correlate with VLDL, echocardiography, platelet function tests, and stroke. There are other near-gene or intergene SNP associations, as shown in green in Figure 3A.
We also investigated EWI-2 and EWI-F, and neither has any association with cardiovascular diseases in human from the analysis. This is probably due to differential expressions between them and their associated tetraspanins in the cardiovascular system. At least in human and mouse large arteries, the expression levels of EWI-2 and EWI-F are much lower than those of Cd81 and Cd9. For example, based on the GTEx database, the numbers of Cd81 transcripts in human aorta, coronary artery, and tibial artery are ~8.2, 8.8, and 5.5 times as high as the numbers of EWI-2 transcripts, respectively (data not shown). The association of claudins with cardiovascular diseases did not emerge from the analysis, probably due to less presence of tissues such as the brain and retina, in which tight junction is crucial for vascular integrity, in the datasets.
GEO Analysis Reveals That Cd9, Cd82, Cd151, TSPAN18, UPK1B, and Most Integrins Are Co-expressed With the Genes Related to Cardiovascular Functions
All human datasets from GEO were processed with specific data mining to extract the co-expressed genes of tetraspanins and integrins in the cardiovascular system. The software IRIDESCENT was used to identify functional commonality between tetraspanins and integrins by grouping their co-expressed genes with terms such as molecules, events, and phenotypes. The 40 most correlated genes with an entire list of tetraspanin and integrin family members were first identified, and then, for each one, common terms associated with the cardiovascular system were identified using literature mining, as described in the methods. Strong commonalities of the integrin and tetraspanin family members highlight their roles in the interactions between ECs and blood cells (e.g., platelets, leukocytes) and between vascular cells and ECM (Figure 3B), further supporting functional connections of tetraspanins to cell adhesion proteins.
We also manually selected those that are closely related to the cardiovascular system. Associations were found between Cd9, Cd82, Cd151, TSPAN18, UPK1B, most of the integrins, and various cardiovascular functions (Figure 3C). The most prominent involvements were vessel-related events including microvessels, neovascularization, and vascular development and remodeling, which account for 30% of all associations. Other associations included platelets, heart, ECs, VSMCs, atherosclerosis, fat, and cholesterol. Notably, Cd9, Cd151, ITGAV, and ITGBL1 were associated with two distinct, well-recognized smooth muscle cell markers, SM22alpha (gene symbol: TAGLN) and smooth muscle alpha-actin (ACTA2), suggesting their potential roles in the regulation of VSMCs.
Genome-Wide Studies Reveal That ITGAE, Eight Other Integrins, TSPAN5, and Six Other Tetraspanins Are Relevant to Cardiovascular Events in Mice
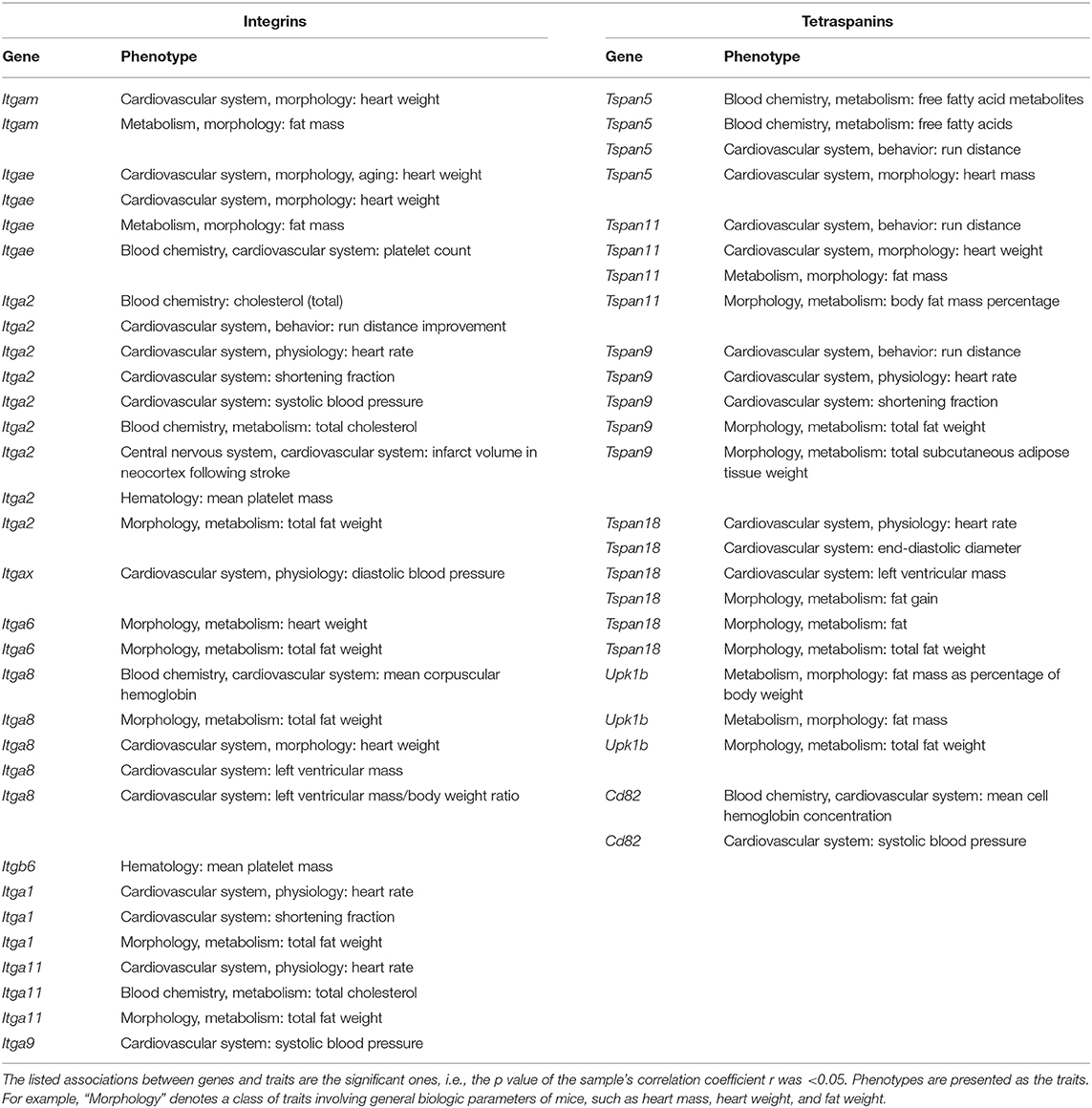
Table 1 shows the expression levels of relatively highly expressed genes. Results from a variety of genome-wide studies focusing on murine cardiovascular system were deposited in GeneNetwork. Similar to human GWAS, we extracted the data that fell within the spectrum of the genes and phenotypes in which we were interested (Table 2). Among integrins, ITGA2, ITGA9, and ITGAX were associated with blood pressure, ITGA2 and ITGA11 with cholesterol, ITGA2 and ITGB6 with platelet mass, and ITGAE with platelet count. ITGA8 and ITGAE were associated with heart weight, whereas ITGA1, ITGA2, and ITGA11 were correlated with heart rate. ITGA1 and ITGA2 were associated with shortening fraction in echocardiography.
As for tetraspanins, TSPAN5 was associated with free fatty acids in the blood, while TSPAN11 was associated with heart weight, TSPAN9 with end-diastolic diameter in echocardiography, and TSPAN18 with heart rate. Cd82 was associated with blood pressure.
Other loose associations (i.e., the ones with relatively larger p values) with cardiovascular phenotypes include the following: ITGA2 with the infarct size in neocortex after experimental stroke; ITGA2, TSPAN5, TSPAN9, and TSPAN11 with run distance performance; and UPK1B with fat, fat mass, or fat weight.
Based on the Data From Both Species, TSPAN5 and ITGA8 Are Associated With Lipid Metabolism, Cd82 and ITGA9 With Blood Pressure, and ITGA2, ITGAE, and ITGB6 With Platelet Functions
We choose ITGA9 for further analysis. At the protein level, more than 90% of sequences identity between human and mouse ITGA9 make them highly comparable. The KEGG pathway-based gene set enrichment analysis using the co-expression data from mouse aorta suggested that ITGA9 is involved in metabolism, MAPK signaling, actin cytoskeleton, focal adhesion, and Wnt signaling. Figure 4A shows the top 10 enriched pathways according to the adjusted p value.
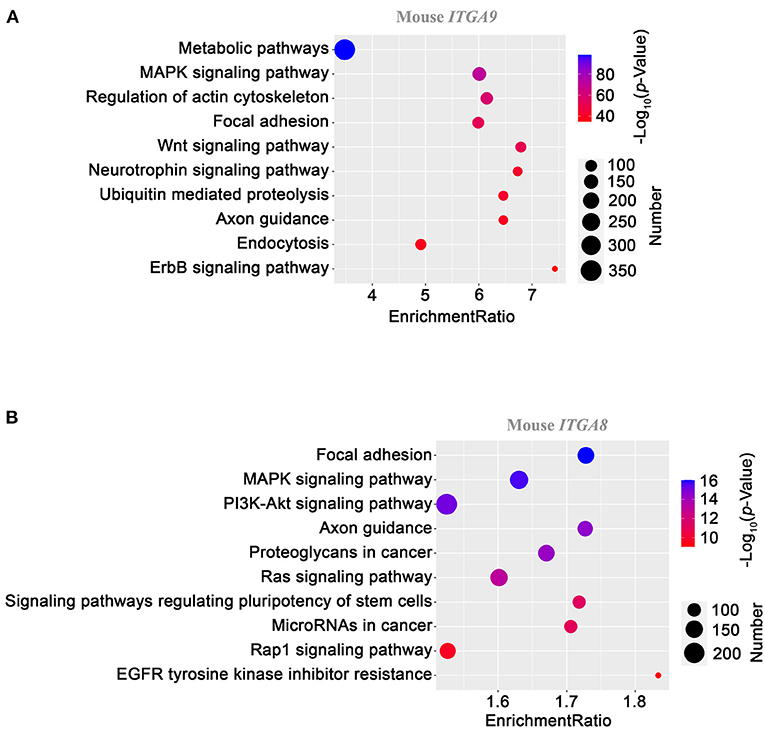
Figure 4. KEGG pathway enrichment analysis on ITGA9 (A) and ITGA8 (B) using gene correlation data from mouse aortas. Gene set enrichment analysis was performed as described in the Methods section. The dot size presents the number of enriched genes with the indicated pathway, and the dot color codes the level of –log10 of the p value. The enrichment ratio is defined as the number of observed genes divided by the number of expected genes in a KEGG category.
A similar analysis was applied to ITGA8, given its potential involvement in cholesterol metabolism. Focal adhesion, MAPK signaling, and PI3K-Akt signaling are the top three pathways (Figure 4B), reminiscent of the readouts from ITGA9 analysis.
Discussion
High-throughput data, such as the data of genome, transcriptome, proteome, metabolome, and phenome, not only convey massive amounts of insightful information but also contain considerable noise (73). The noise can be caused by, for example, factors such as biological variation or statistical variance. Here, we have integrated the analyses of human and mouse data to determine possible significant “signals” by matching the readouts with the area of our research interest. Such a comprehensive approach in principle likely provides better conclusion for establishing a close relationship between a molecular change and a pathophysiological event.
Both integrins and tetraspanins exhibit markedly diverse expression levels across their family spectra in the cardiovascular system, so do the expression levels of a given member of the families across different human tissues. Some of them such as Cd151 and ITGA8 are highly expressed in arteries, probably to some extent reflecting their functional connection, although tissue specificity of a gene does not necessarily correlate with the gene expression level in the tissue. CD151, as stated earlier, is a modulator of vascular functions, and integrin α8 is a crucial regulator of smooth muscle cells (74). As shown in the expression heatmap, ITGA9 has a moderate expression level in human aorta compared with other integrins, but it exhibits an overall low-expression profile across all other tissues. A previous study showed that the abundance of a protein in a certain state is the result of optimization of the cost vs. benefit (75). Also, the expression of mRNA is not always correlated with the expression of protein (76). Hence, ITGA9 may affect vascular function like the regulation of blood pressure without having a high mRNA expression profile in the cardiovascular system.
Moreover, we found many intronic variants in the human GWAS database. TSPAN5, for example, has various intronic SNPs correlated with cholesterol or “cholesterol, LDL.” A previous study (77) and a literature review (78) led to our belief that these introns may contain transcriptional regulatory elements, such as DNA methylation loci, and therefore may alter the quantity or feature of the transcripts of this gene. For example, the intronic SNPs could be transcriptional regulatory elements to alter the RNA splicing of TSPAN5. Similarly, the intergenic variants may influence the phenotype through transcriptional modifications. Previous studies showed that cds-syn variants may cause the instability of the mRNA or even the alteration of protein conformation (79, 80). The associations of missense variants are easier to understand, since the changes in protein sequences could directly lead to functional loss or gain (81, 82). The statistical significance of a genome association was generally determined by a p value that is <5 × 10−8 (83). None of the exon variants that we found were significant following this standard, but we still considered the association to be not random and be suggestive if the same association was found in mice. Some of the variants located in introns meet the criteria: for instance, an association between an SNP within TSPAN15 (rs78707713) and venous thromboembolism has a p value of 2 × 10−16. Thus, it is likely that this gene is involved in the pathogenesis of venous thromboembolism, either as a cause or a result.
The data organization and processing in GeneNetwork are similar to those in the human databases that we used, and complement the results from the human databases. ITGA9 has associations with the same phenotype, i.e., blood pressure, in both species. In humans, the relevant variant, rs146867977, is located within the exon of ITGA9 and causes a missense alteration. In mice, blood pressure as a trait was found to have a maximum association “peak” that falls within the ITGA9 gene sequence. ITGA9 has a very long genomic DNA sequence in both humans and mice (~360,000 and ~290,000 bp, respectively). The chance of having a random association, therefore, increases. A previous study found that a different SNP of ITGA9 is associated with hypertension (rs7640747, p = 4.8 × 10−7) (84), consistent with our prediction. Interestingly, the pathway analysis reflected what has already been reported, assuring us of the reliability of our analyses. For instance, MAP kinase and downstream molecules are indeed activated by integrins (85, 86) and so are actin cytoskeleton (87), Wnt signaling (88), and focal adhesion (89). Of course, in vivo experiments are needed to determine whether integrin α9 actually regulates blood pressure.
Furthermore, we have combined transcriptional correlations derived from GEO expression datasets with a literature-mining tool, IRIDESCENT, to predict gene-function associations using a “guilt by association” approach (63). The ability of the approach to predict a number of already-established literature commonalities lends additional plausibility to the unestablished ones. Predicted associations with blood vessels, for example, were shown by earlier studies to rely on integrins and tetraspanins in biological processes such as angiogenesis (49, 90).
This bioinformatics study has several limitations. First, we rely upon deposited public data, which is often incompletely annotated and of varying quality both on the individual experiment level (e.g., some studies may be statistically underpowered) and the aggregate level (e.g., most studies are small scale, but some are very large, creating the possibility of batch effects). Thus, genes identified with the highest confidence will tend to be those with larger transcriptional changes, and the relevance of a gene to cardiovascular biology on a different level (e.g., translation, protein–protein interactions, etc) will not be detectable with these methods. Another limitation of data mining for cardiovascular diseases is that, unlike cancer-related databases, data sources and analysis tools are limited currently. Thus, we cannot make comparisons across platforms. In addition, as the human data was not stratified by age, gender, or ethnicity, we may not understand the data in great depth or may miss new associations. Moreover, GWAS-based phenotypes may be indirect or non-specific. For example, a mutation affecting energy production or waste disposal would likely affect many things, including cardiovascular function. In part, we attempt to compensate for this limited perspective with the transcriptional network analysis, which will tend to weight specific associations (e.g., cardiomyocytes) higher than general ones. Given very limited availability of mouse GWAS data (one dataset from mouse vs. hundreds of datasets from human), our observation of the little overlap in gene–phenotype associations between humans and mice could reflect an incomplete view. Nonetheless, the databases used here are major repositories with high-quality uniform datasets that combine numerous large-scale studies. New techniques such as single-cell RNAseq, when widely applied to the cardiovascular research field, will bring more details into the association studies, adding a dimension of marker-specific subpopulations and subgroups for more thorough analysis (91). Apparently, the conclusions from all bioinformatics analyses need to be confirmed by experimental approaches, so do the observations made by this bioinformatics study, in order to establish causal relationships between TEM genes and cardiovascular pathophysiology.
Conclusion
On the basis of combined analyses on human and mouse data, our study identified and predicted that ITGA9 is highly relevant to cardiovascular diseases among integrins and tetraspanins and promising for further experimental studies, although this integrin was already found to support lymphatic and venous valve formation and thrombosis (92–94). Other highlighted associations are TSPAN5 and ITGA8 with lipid metabolism, Cd82 with blood pressure, and ITGA2, ITGAE, and ITGB6 with platelet functions. This work provides an example of using integrated bioinformatics approaches to predict the links of structurally and functionally related genes and proteins to given diseases in a comprehensive and systematic manner.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
GS, JC, JW, and FX performed analysis and wrote the manuscript. YD wrote the manuscript. LL, YW, and D-wW supervised the study. XZ conceived, designed, and supervised the study and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
XZ was supported by NIH grants HL132553 and HL137819 and research grants from OCASCR, a program of TSET. JW was supported by NIH grant GM104938.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We also thank Ms. Kathy Kyler for English editing.
Abbreviations
cds-syn, coding sequence synonymous; ECs, endothelial cells; ECM, extracellular matrix; eQTL, expression quantitative trait loci; FEVR, familial exudative vitreoretinopathy; GEO, Gene Expression Omnibus; GWAS, genome-wide association studies; GTEx, Genotype-Tissue Expression; KEGG, Kyoto Encyclopedia of Genes and Genomes; SNP, single-nucleotide polymorphism; TEM, tetraspanin-enriched microdomain; VSMC, vascular smooth muscle cell.
References
1. Yanez-Mo M, Barreiro O, Gordon-Alonso M, Sala-Valdes M, Sanchez-Madrid F. Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol. (2009) 19:434–46. doi: 10.1016/j.tcb.2009.06.004
2. Bassani S, Cingolani LA. Tetraspanins: Interactions and interplay with integrins. Int J Biochem Cell Biol. (2012) 44:703–8. doi: 10.1016/j.biocel.2012.01.020
3. Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. (2002) 110:673–87. doi: 10.1016/s0092-8674(02)00971-6
4. Bridgewater RE, Norman JC, Caswell PT. Integrin trafficking at a glance. J Cell Sci. (2012) 125:3695–701. doi: 10.1242/jcs.095810
5. Ross TD, Coon BG, Yun S, Baeyens N, Tanaka K, Ouyang M, et al. Integrins in mechanotransduction. Curr Opin Cell Biol. (2013) 25:613–8. doi: 10.1016/j.ceb.2013.05.006
6. Zhang F, Kotha J, Jennings LK, Zhang XA. Tetraspanins and vascular functions. Cardiovascular Res. (2009) 83:7–15. doi: 10.1093/cvr/cvp080
7. Bailey RL, Herbert JM, Khan K, Heath VL, Bicknell R, Tomlinson MG. The emerging role of tetraspanin microdomains on endothelial cells. Biochem. Soc. Transact. (2011) 39:1667–73. doi: 10.1042/BST20110745
8. Wei Q, Zhang F, Richardson MM, Roy NH, Rodgers W, Liu Y, et al. CD82 restrains pathological angiogenesis by altering lipid raft clustering and CD44 trafficking in endothelial cells. Circulation. (2014) 130:1493–504. doi: 10.1161/CIRCULATIONAHA.114.011096
9. Li W, Hu M, Wang C, Lu H, Chen F, Xu J, et al. A viral microRNA downregulates metastasis suppressor CD82 and induces cell invasion and angiogenesis by activating the c-Met signaling. Oncogene. (2017) 36:5407–20. doi: 10.1038/onc.2017.139
10. Zhao K, Erb U, Hackert T, Zoller M, Yue S. Distorted leukocyte migration, angiogenesis, wound repair and metastasis in Tspan8 and Tspan8/CD151 double knockout mice indicate complementary activities of Tspan8 and CD51. Biochim Biophys Acta. (2018) 1865:379–91. doi: 10.1016/j.bbamcr.2017.11.007
11. Toya SP, Wary KK, Mittal M, Li F, Toth PT, Park C, et al. Integrin alpha6beta1 expressed in ESCs instructs the differentiation to endothelial cells. Stem Cells. (2015) 33:1719–29. doi: 10.1002/stem.1974
12. Li GX, Zhang S, Liu R, Singh B, Singh S, Quinn DI, et al. Tetraspanin18 regulates angiogenesis through VEGFR2 and Notch pathways. Biol. Open. (2020) 10:bio050096. doi: 10.1242/bio.050096
13. Kapustin AN, Chatrou ML, Drozdov I, Zheng Y, Davidson SM, Soong D, et al. Vascular smooth muscle cell calcification is mediated by regulated exosome secretion. Circulat Res. (2015) 116:1312–23. doi: 10.1161/CIRCRESAHA.116.305012
14. Cho JH, Kim EC, Son Y, Lee DW, Park YS, Choi JH, et al. CD9 induces cellular senescence and aggravates atherosclerotic plaque formation. Cell Death Different. (2020) 27:2681–96. doi: 10.1038/s41418-020-0537-9
15. Kim JR, Choi JH. CD9 expression in vascular aging and atherosclerosis. Histol. Histopathol. (2020) 18268:1449–53. doi: 10.14670/HH-18-268
16. Zhang F, Michaelson JE, Moshiach S, Sachs N, Zhao W, Sun Y, et al. Tetraspanin CD151 maintains vascular stability by balancing the forces of cell adhesion and cytoskeletal tension. Blood. (2011) 118:4274–84. doi: 10.1182/blood-2011-03-339531
17. Chu LH, Vijay CG, Annex BH, Bader JS, Popel AS. PADPIN: protein-protein interaction networks of angiogenesis, arteriogenesis, and inflammation in peripheral arterial disease. Physiol Genom. (2015) 47:331–43. doi: 10.1152/physiolgenomics.00125.2014
18. Zhao J, Wu W, Zhang W, Lu YW, Tou E, Ye J, et al. Selective expression of TSPAN2 in vascular smooth muscle is independently regulated by TGF-beta1/SMAD and myocardin/serum response factor. FASEB J. (2017) 31:2576–91. doi: 10.1096/fj.201601021R
19. Takeda M, Kanki Y, Masumoto H, Funakoshi S, Hatani T, Fukushima H, et al. Identification of cardiomyocyte-fated progenitors from human-induced pluripotent stem cells marked with CD82. Cell Rep. (2018) 22:546–56. doi: 10.1016/j.celrep.2017.12.057
20. Poulter JA, Ali M, Gilmour DF, Rice A, Kondo H, Hayashi K, et al. Mutations in TSPAN12 cause autosomal-dominant familial exudative vitreoretinopathy. Am J Human Genet. (2010) 86:248–53. doi: 10.1016/j.ajhg.2010.01.012
21. Nikopoulos K, Gilissen C, Hoischen A, van Nouhuys CE, Boonstra FN, Blokland EA, et al. Next-generation sequencing of a 40 Mb linkage interval reveals TSPAN12 mutations in patients with familial exudative vitreoretinopathy. Am J Human Genet. (2010) 86:240–7. doi: 10.1016/j.ajhg.2009.12.016
22. Lai MB, Zhang C, Shi J, Johnson V, Khandan L, McVey J, et al. TSPAN12 is a norrin Co-receptor that Amplifies Frizzled4 ligand selectivity and signaling. Cell Rep. (2017) 19:2809–22. doi: 10.1016/j.celrep.2017.06.004
23. Zhang C, Lai MB, Pedler MG, Johnson V, Adams RH, Petrash JM, et al. Endothelial cell-specific inactivation of TSPAN12 (Tetraspanin 12) reveals pathological consequences of barrier defects in an otherwise intact vasculature. Arterioscler Thrombosis Vascular Biol. (2018) 38:2691–705. doi: 10.1161/ATVBAHA.118.311689
24. Bucher F, Zhang D, Aguilar E, Sakimoto S, Diaz-Aguilar S, Rosenfeld M, et al. Antibody-mediated inhibition of Tspan12 ameliorates vasoproliferative retinopathy through suppression of beta-catenin signaling. Circulation. (2017) 136:180–95. doi: 10.1161/CIRCULATIONAHA.116.025604
25. Diaz-Coranguez M, Lin CM, Liebner S, Antonetti DA. Norrin restores blood-retinal barrier properties after vascular endothelial growth factor-induced permeability. J Biol Chem. (2020) 295:4647–60. doi: 10.1074/jbc.RA119.011273
26. Wang Y, Cho C, Williams J, Smallwood PM, Zhang C, Junge HJ, et al. Interplay of the norrin and Wnt7a/Wnt7b signaling systems in blood-brain barrier and blood-retina barrier development and maintenance. Proc. Natl. Acad. Sci. USA. (2018) 115:E11827–36. doi: 10.1073/pnas.1813217115
27. Uchtmann K, Park ER, Bergsma A, Segula J, Edick MJ, Miranti CK. Homozygous loss of mouse tetraspanin CD82 enhances integrin alphaIIbbeta3 expression and clot retraction in platelets. Exp Cell Res. (2015) 339:261–9. doi: 10.1016/j.yexcr.2015.11.006
28. Makkawi M, Moheimani F, Alserihi R, Howells D, Wright M, Ashman L, et al. A complementary role for tetraspanin superfamily member CD151 and ADP purinergic P2Y12 receptor in platelets. Thrombosis Haemostasis. (2015) 114:1004–19. doi: 10.1160/TH14-11-0967
29. Makkawi M, Howells D, Wright MD, Jackson DE. A complementary role for tetraspanin superfamily member TSSC6 and ADP purinergic P2Y12 receptor in platelets. Thrombosis Res. (2018) 161:12–21. doi: 10.1016/j.thromres.2017.11.009
30. Noy PJ, Gavin RL, Colombo D, Haining EJ, Reyat JS, Payne H, et al. Tspan18 is a novel regulator of the Ca(2+) channel Orai1 and von Willebrand factor release in endothelial cells. Haematologica. (2019) 104:1892–905. doi: 10.3324/haematol.2018.194241
31. Doyle EL, Ridger V, Ferraro F, Turmaine M, Saftig P, Cutler DF. CD63 is an essential cofactor to leukocyte recruitment by endothelial P-selectin. Blood. (2011) 118:4265–73. doi: 10.1182/blood-2010-11-321489
32. Poeter M, Brandherm I, Rossaint J, Rosso G, Shahin V, Skryabin BV, et al. Annexin A8 controls leukocyte recruitment to activated endothelial cells via cell surface delivery of CD63. Nat Commun. (2014) 5:3738. doi: 10.1038/ncomms4738
33. Zhang XA, Lane WS, Charrin S, Rubinstein E, Liu L. EWI2/PGRL associates with the metastasis suppressor KAI1/CD82 and inhibits the migration of prostate cancer cells. Cancer Res. (2003) 63:2665–74.
34. Sala-Valdes M, Ursa A, Charrin S, Rubinstein E, Hemler ME, Sanchez-Madrid F, et al. EWI-2 and EWI-F link the tetraspanin web to the actin cytoskeleton through their direct association with ezrin-radixin-moesin proteins. J Biol Chem. (2006) 281:19665–75. doi: 10.1074/jbc.M602116200
35. He B, Zhang YH, Richardson MM, Zhang JS, Rubinstein E, Zhang XA. Differential functions of phospholipid binding and palmitoylation of tumour suppressor EWI2/PGRL. Biochem J. (2011) 437:399–411. doi: 10.1042/BJ20101381
36. Stipp CS, Kolesnikova TV, Hemler ME. EWI-2 regulates alpha3beta1 integrin-dependent cell functions on laminin-5. J Cell Biol. (2003) 163:1167–77. doi: 10.1083/jcb.200309113
37. Kolesnikova TV, Stipp CS, Rao RM, Lane WS, Luscinskas FW, Hemler ME. EWI-2 modulates lymphocyte integrin alpha4beta1 functions. Blood. (2004) 103:3013–9. doi: 10.1182/blood-2003-07-2201
38. Cong X, Kong W. Endothelial tight junctions and their regulatory signaling pathways in vascular homeostasis and disease. Cell Signal. (2020) 66:109485. doi: 10.1016/j.cellsig.2019.109485
39. Finney AC, Stokes KY, Pattillo CB, Orr AW. Integrin signaling in atherosclerosis. Cell Mol Life Sci. (2017) 74:2263–82. doi: 10.1007/s00018-017-2490-4
40. Hillis GS, Flapan AD. Cell adhesion molecules in cardiovascular disease: a clinical perspective. Heart. (1998) 79:429–31. doi: 10.1136/hrt.79.5.429
41. Clemetson KJ, Clemetson JM. Integrins and cardiovascular disease. Cell Mol Life Sci. (1998) 54:502–13. doi: 10.1007/s000180050179
42. Winter PM, Morawski AM, Caruthers SD, Fuhrhop RW, Zhang H, Williams TA, et al. Molecular imaging of angiogenesis in early-stage atherosclerosis with alpha(v)beta3-integrin-targeted nanoparticles. Circulation. (2003) 108:2270–4. doi: 10.1161/01.CIR.0000093185.16083.95
43. Demircioglu F, Hodivala-Dilke K. alphavbeta3 Integrin and tumour blood vessels-learning from the past to shape the future. Curr Opin Cell Biol. (2016) 42:121–7. doi: 10.1016/j.ceb.2016.07.008
44. Mahabeleshwar GH, Feng W, Phillips DR, Byzova TV. Integrin signaling is critical for pathological angiogenesis. J Exp Med. (2006) 203:2495–507. doi: 10.1084/jem.20060807
45. Steri V, Ellison TS, Gontarczyk AM, Weilbaecher K, Schneider JG, Edwards D, et al. Acute depletion of endothelial beta3-integrin transiently inhibits tumor growth and angiogenesis in mice. Circulat Res. (2014) 114:79–91. doi: 10.1161/CIRCRESAHA.114.301591
46. Krishn SR, Salem I, Quaglia F, Naranjo NM, Agarwal E, Liu Q, et al. The alphavbeta6 integrin in cancer cell-derived small extracellular vesicles enhances angiogenesis. J Extracell Vesicles. (2020) 9:1763594. doi: 10.1080/20013078.2020.1763594
47. Misra A, Feng Z, Chandran RR, Kabir I, Rotllan N, Aryal B, et al. Integrin beta3 regulates clonality and fate of smooth muscle-derived atherosclerotic plaque cells. Nat Commun. (2018) 9:2073. doi: 10.1038/s41467-018-04447-7
48. Garmy-Susini B, Jin H, Zhu Y, Sung RJ, Hwang R, Varner J. Integrin alpha4beta1-VCAM-1-mediated adhesion between endothelial and mural cells is required for blood vessel maturation. J Clin Invest. (2005) 115:1542–51. doi: 10.1172/JCI23445
49. Yamamoto H, Ehling M, Kato K, Kanai K, van Lessen M, Frye M, et al. Integrin beta1 controls VE-cadherin localization and blood vessel stability. Nat Commun. (2015) 6:6429. doi: 10.1038/ncomms7429
50. Hakanpaa L, Kiss EA, Jacquemet G, Miinalainen I, Lerche M, Guzman C, et al. Targeting beta1-integrin inhibits vascular leakage in endotoxemia. Proc Natl Acad Sci USA. (2018) 115:E6467–76. doi: 10.1073/pnas.1722317115
51. Lacolley P, Regnault V, Segers P, Laurent S. Vascular smooth muscle cells and arterial stiffening: relevance in development, aging, and disease. Physiol Rev. (2017) 97:1555–617. doi: 10.1152/physrev.00003.2017
52. Gu X, Masters KS. Regulation of valvular interstitial cell calcification by adhesive peptide sequences. J Biomed Mater Res. Part A. (2010) 93:1620–30. doi: 10.1002/jbm.a.32660
53. Ou Z, Dolmatova EV, Lassegue B, Griendling KK. β1 and β2 integrins: central players in regulating vascular permeability and leukocyte recruitment during acute inflammation. Am J Physiol. (2020) 320:H734–9. doi: 10.1152/ajpheart.00518.2020
54. Lerman YV, Lim K, Hyun YM, Falkner KL, Yang H, Pietropaoli AP, et al. Sepsis lethality via exacerbated tissue infiltration and TLR-induced cytokine production by neutrophils is integrin alpha3beta1-dependent. Blood. (2014) 124:3515–23. doi: 10.1182/blood-2014-01-552943
55. Yakubenko VP, Cui K, Ardell CL, Brown KE, West XZ, Gao D, et al. Oxidative modifications of extracellular matrix promote the second wave of inflammation via beta2 integrins. Blood. (2018) 132:78–88. doi: 10.1182/blood-2017-10-810176
56. Mortality GBD, Causes of Death C. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. (2015) 385:117–71. doi: 10.1016/S0140-673661682-2
57. Avraamides CJ, Garmy-Susini B, Varner JA. Integrins in angiogenesis and lymphangiogenesis. Nat Rev Cancer. (2008) 8:604–17. doi: 10.1038/nrc2353
58. Tryka KA, Hao L, Sturcke A, Jin Y, Wang ZY, Ziyabari L, et al. NCBI's database of genotypes and phenotypes: dbGaP. Nucleic Acids Res. (2014) 42:D975–9. doi: 10.1093/nar/gkt1211
59. Ramos EM, Hoffman D, Junkins HA, Maglott D, Phan L, Sherry ST, et al. Phenotype-Genotype Integrator (PheGenI): synthesizing genome-wide association study (GWAS) data with existing genomic resources. Eur J Human Genet. (2014) 22:144–7. doi: 10.1038/ejhg.2013.96
60. Wren JD. Extending the mutual information measure to rank inferred literature relationships. BMC Bioinformat. (2004) 5:145. doi: 10.1186/1471-2105-5-145
61. Wren JD, Bekeredjian R, Stewart JA, Shohet RV, Garner HR. Knowledge discovery by automated identification and ranking of implicit relationships. Bioinformatics. (2004) 20:389–8. doi: 10.1093/bioinformatics/btg421
62. Wren JD, Garner HR. Shared relationship analysis: ranking set cohesion and commonalities within a literature-derived relationship network. Bioinformatics. (2004) 20:191–8. doi: 10.1093/bioinformatics/btg390
63. Wren JD. A global meta-analysis of microarray expression data to predict unknown gene functions and estimate the literature-data divide. Bioinformatics. (2009) 25:1694–701. doi: 10.1093/bioinformatics/btp290
64. Mulligan MK, Mozhui K, Prins P, Williams RW. GeneNetwork: a toolbox for systems genetics. Methods Mol Biol. (2017) 1488:75–120. doi: 10.1007/978-1-4939-6427-7_4
65. Wang X, Pandey AK, Mulligan MK, Williams EG, Mozhui K, Li Z, et al. Joint mouse-human phenome-wide association to test gene function and disease risk. Nat Commun. (2016) 7:10464. doi: 10.1038/ncomms10464
66. R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna (2020). Available online at: https://www.R-project.org/
67. Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. (2019) 47:W199–205. doi: 10.1093/nar/gkz401
68. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc Seri B. (1995) 57:289–300.
69. Bennett BJ, Davis RC, Civelek M, Orozco L, Wu J, Qi H, et al. Genetic architecture of atherosclerosis in mice: a systems genetics analysis of common inbred strains. PLoS Genet. (2015) 11:e1005711. doi: 10.1371/journal.pgen.1005711
70. Chesler EJ, Lu L, Shou S, Qu Y, Gu J, Wang J, et al. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat Genet. (2005) 37:233–42. doi: 10.1038/ng1518
71. Germain M, Chasman DI, de Haan H, Tang W, Lindstrom S, Weng LC, et al. Meta-analysis of 65:734 individuals identifies TSPAN15 and SLC44A2 as two susceptibility loci for venous thromboembolism. Am J human Genet. (2015) 96:532–42. doi: 10.1016/j.ajhg.2015.01.019
72. Qayyum R, Snively BM, Ziv E, Nalls MA, Liu Y, Tang W, et al. A meta-analysis and genome-wide association study of platelet count and mean platelet volume in african americans. PLoS Genet. (2012) 8:e1002491. doi: 10.1371/journal.pgen.1002491
73. He S, Chen H, Zhu Z, Ward DG, Cooper HJ, Viant MR, et al. Robust twin boosting for feature selection from high-dimensional omics data with label noise. Informat Sci. (2015) 291:1–18. doi: 10.1016/j.ins.2014.08.048
74. Zargham R, Thibault G. Alpha 8 integrin expression is required for maintenance of the smooth muscle cell differentiated phenotype. Cardiovascular Res. (2006) 71:170–8. doi: 10.1016/j.cardiores.2006.03.003
75. Dekel E, Alon U. Optimality and evolutionary tuning of the expression level of a protein. Nature. (2005) 436, 588–592. doi: 10.1038/nature03842
76. Wu L, Candille SI, Choi Y, Xie D, Jiang L, Li-Pook-Than J, et al. Variation and genetic control of protein abundance in humans. Nature 499, 79–82 (2013). doi: 10.1038/nature12223
77. Tokuhiro S, Yamada R, Chang X, Suzuki A, Kochi Y, Sawada T, et al. An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis. Nature genetics 35:341–8 (2003). doi: 10.1038/ng1267
78. Zhang F, Lupski JR. Non-coding genetic variants in human disease. Human Mol Genet. (2015) 24:R102–10. doi: 10.1093/hmg/ddv259
79. Friedrich U, Datta S, Schubert T, Plossl K, Schneider M, Grassmann F, et al. Synonymous variants in HTRA1 implicated in AMD susceptibility impair its capacity to regulate TGF-beta signaling. Human Mol Genet. 24:6361–73 (2015). doi: 10.1093/hmg/ddv346
80. Chamary JV, Parmley JL, Hurst LD. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat Rev Genet. (2006) 7:98–108. doi: 10.1038/nrg1770
81. Walerych D, Lisek K, Sommaggio R, Piazza S, Ciani Y, Dalla E, et al. Proteasome machinery is instrumental in a common gain-of-function program of the p53 missense mutants in cancer. Nat Cell Biol. (2016) 18:897–909. doi: 10.1038/ncb3380
82. MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. (2012) 335:823–8. doi: 10.1126/science.1215040
83. Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. (2016) 48:481–7. doi: 10.1038/ng.3538
84. Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. (2009) 41:677–87. doi: 10.1038/ng.384
85. Aikawa R, Nagai T, Kudoh S, Zou Y, Tanaka M, Tamura M, et al. Integrins play a critical role in mechanical stress-induced p38 MAPK activation. Hypertension. (2002) 39:233–8. doi: 10.1161/hy0202.102699
86. Yee KL, Weaver VM, Hammer DA. Integrin-mediated signalling through the MAP-kinase pathway. IET Syst Biol. (2008) 2:8–15. doi: 10.1049/iet-syb:20060058
87. Delon I, Brown NH. Integrins and the actin cytoskeleton. Curr Opin Cell Biol. (2007) 19:43–50. doi: 10.1016/j.ceb.2006.12.013
88. Du J, Zu Y, Li J, Du S, Xu Y, Zhang L, et al. Extracellular matrix stiffness dictates Wnt expression through integrin pathway. Sci Rep. (2016) 6:20395. doi: 10.1038/srep20395
89. Cavalcanti-Adam EA, Volberg T, Micoulet A, Kessler H, Geiger B, Spatz JP. Cell spreading and focal adhesion dynamics are regulated by spacing of integrin ligands. Biophys J. (2007) 92:2964–74. doi: 10.1529/biophysj.106.089730
90. Nikolopoulos SN, Blaikie P, Yoshioka T, Guo W, Giancotti FG. Integrin beta4 signaling promotes tumor angiogenesis. Cancer Cell. (2004) 6:471–83. doi: 10.1016/j.ccr.2004.09.029
91. Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet. (2013) 14:618–30. doi: 10.1038/nrg3542
92. Hoye AM, Couchman JR, Wewer UM, Fukami K, Yoneda A. The newcomer in the integrin family: integrin alpha9 in biology and cancer. Adv Biol Regulat. (2012) 52:326–39. doi: 10.1016/j.jbior.2012.03.004
93. Dhanesha N, Jain M, Tripathi AK, Doddapattar P, Chorawala M, Bathla G, et al. Targeting myeloid-specific integrin alpha9beta1 improves short- and long-term stroke outcomes in murine models with preexisting comorbidities by limiting thrombosis and inflammation. Circulat Res. (2020) 126:1779–94. doi: 10.1161/CIRCRESAHA.120.316659
Keywords: angiogenesis, atherosclerosis, blood pressure, integrin, platelet
Citation: Sun G, Chen J, Ding Y, Wren JD, Xu F, Lu L, Wang Y, Wang D-w and Zhang XA (2021) A Bioinformatics Perspective on the Links Between Tetraspanin-Enriched Microdomains and Cardiovascular Pathophysiology. Front. Cardiovasc. Med. 8:630471. doi: 10.3389/fcvm.2021.630471
Received: 17 November 2020; Accepted: 15 February 2021;
Published: 30 March 2021.
Edited by:
Filip Swirski, Massachusetts General Hospital and Harvard Medical School, United StatesReviewed by:
Anthony Wayne Orr, Louisiana State University Health Shreveport, United StatesBlanca Camoretti-Mercado, University of South Florida, United States
Copyright © 2021 Sun, Chen, Ding, Wren, Xu, Lu, Wang, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin A. Zhang, eGluLXpoYW5nLTEmI3gwMDA0MDtvdWhzYy5lZHU=
†These authors have contributed equally to this work
 Ge Sun1†
Ge Sun1† Junxiong Chen
Junxiong Chen Jonathan D. Wren
Jonathan D. Wren Fuyi Xu
Fuyi Xu Lu Lu
Lu Lu Dao-wen Wang
Dao-wen Wang Xin A. Zhang
Xin A. Zhang