- 1College of Medicine, Mohammed Bin Rashid University of Medicine and Health Sciences, Dubai, United Arab Emirates
- 2Meakins-Christie Laboratories, Research Institute of the McGill University Health Center, Montreal, QC, Canada
- 3College of Medicine, Sharjah Institute for Medical Research, University of Sharjah, Sharjah, United Arab Emirates
Background: Coronavirus disease 2019 (COVID-19) is a viral respiratory illness caused by the novel coronavirus SARS-CoV-2. The presence of the pre-existing cardiac disease is associated with an increased likelihood of severe clinical course and mortality in patients with COVID-19. Besides, current evidence indicates that a significant number of patients with COVID-19 also exhibit cardiovascular involvement even in the absence of known cardiac risk factors. Therefore, there is a need to understand the underlying mechanisms and genetic predispositions that explain cardiovascular involvement in COVID-19.
Objectives: In silico analysis of publicly available datasets to decipher the molecular basis, potential pathways, and the role of the endothelium in the pathogenesis of cardiac and vascular injuries in COVID-19.
Materials and Methods: Consistent significant differentially expressed genes (DEGs) shared by endothelium and peripheral immune cells were identified in five microarray transcriptomic profiling datasets in patients with venous thromboembolism “VTE,” acute coronary syndrome, heart failure and/or cardiogenic shock (main cardiovascular injuries related to COVID-19) compared to healthy controls. The identified genes were further examined in the publicly available transcriptomic dataset for cell/tissue specificity in lung tissue, in different ethnicities and in SARS-CoV-2 infected vs. mock-infected lung tissues and cardiomyocytes.
Results: We identified 36 DEGs in blood and endothelium known to play key roles in endothelium and vascular biology, regulation of cellular response to stress as well as endothelial cell migration. Some of these genes were upregulated significantly in SARS-CoV-2 infected lung tissues. On the other hand, some genes with cardioprotective functions were downregulated in SARS-CoV-2 infected cardiomyocytes.
Conclusion: In conclusion, our findings from the analysis of publicly available transcriptomic datasets identified shared core genes pertinent to cardiac and vascular-related injuries and their probable role in genetic susceptibility to cardiovascular injury in patients with COVID-19.
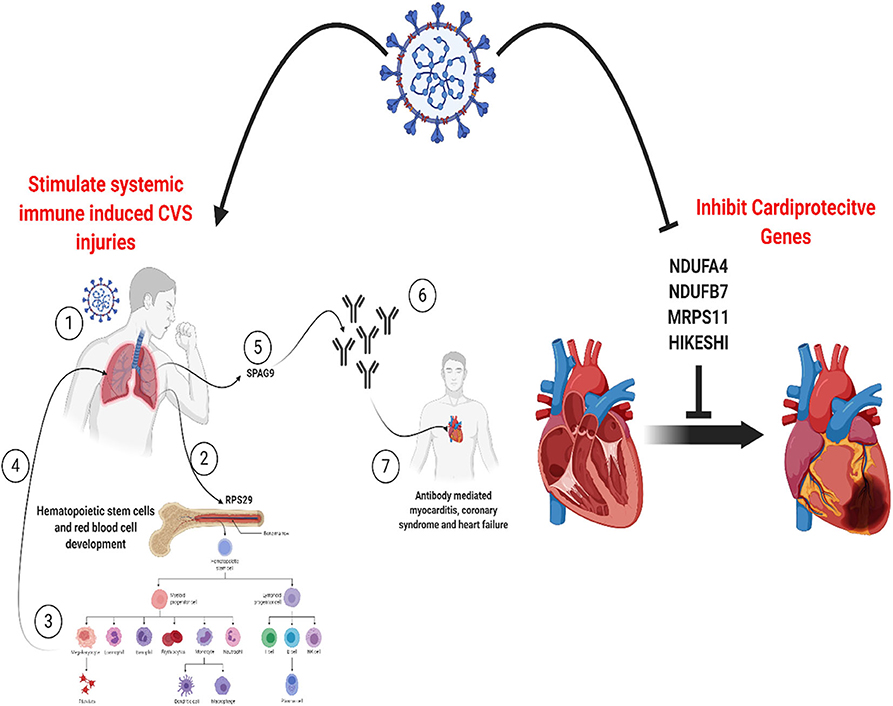
Graphical Abstract. SARS-CoV-2 can induce cardiovascular injures in COVID-19 patients by manipulating a core set of genes specific to endothelium in the lungs, heart, and vessels. This can activate pathways for systemic immune-mediated cardiovascular injuries or increase vulnerability to cardiac injury via inhibition of cardioprotective proteins. Created with BioRender.com.
Introduction
Coronavirus disease 2019 (COVID-19) is a viral respiratory illness caused by the novel coronavirus SARS-CoV-2. To date (1st September 2020), the number of laboratory-confirmed cases of COVID-19 has exceeded 25 million globally, with over 800,000 fatalities (1). The clinical spectrum of COVID-19 ranges from asymptomatic infection to mild to moderate disease in the majority of patients (2, 3). However, some patients exhibit a more severe clinical course characterized by multisystemic and life-threatening manifestations with pneumonia and acute respiratory distress as prominent features (2–4). Patients with pre-existing cardiac disease, hypertension, diabetes, and obesity are more likely to have a severe clinical course with a higher risk of mortality (5–7). In a meta-analysis of 8 studies, including 46,248 patients, cardiovascular disease was the third most common comorbidity in patients with COVID-19 (4). Moreover, there is increasing evidence that a significant number of patients with COVID-19 have cardiovascular involvement, which further increases the likelihood of mortality (5, 6, 8, 9). Notably, even in the absence of known cardiac risk factors, patients with COVID-19 may have an increased risk of cardiovascular injury with a report from China documenting high levels of troponin or cardiac arrest in up to 12% of patients without prior history of cardiovascular disease (6).
The acute cardiovascular syndrome associated with COVID-19 includes a variety of clinical presentations of acute cardiac injury, cardiomyopathy, and hemodynamic instability. Myocardial injury, arrhythmias, cardiac arrests, heart failure, and coagulation abnormality were reported in 7–33% of patients with COVID-19 in China (3, 9). The angiotensin-converting enzyme 2 (ACE-2) receptors used for cellular entry by SARS-CoV-2 are expressed in the lung as well as in various organs, including the heart and endothelial cells (10–12). Direct SARS-CoV-2 infection of the endothelial cells, along with diffuse endothelial inflammation, has been reported (11). The cytokine storm and profound inflammation seen in patients with severe COVID-19 are associated with macrophage and endothelial activation and surges in the levels of interleukin (IL)-1, IL-6, IL-8, and Tumor Necrosis Factor-alpha (TNF-α). Emerging data also indicate a hypercoagulable state in a cluster of patients with COVID-19 with a high incidence of venous thromboembolism (VTE) despite the use of prophylactic anticoagulants (13). Studies have shown that IL-6, one of the significant cytokines described in the cytokine storm, is associated with vascular leakage, activation of the coagulation cascade, and cardiomyopathy (14, 15).
One of the proposed mechanisms of cardiovascular injury in COVID-19 is direct injury to myocardial cells due to viral invasion of the vascular endothelium and myocardium (16). The second postulate is the impact of tissue hypoxia, destabilization of coronary plaque, and micro-thrombogenesis caused by the systematic inflammation associated with cytokine storm (16). In addition, the potential role of genetic susceptibility to COVID-19 related cardiac events has recently been highlighted as a possible contributor to the high mortality among African American patients with COVID-19 (17). As cardiovascular involvement in COVID-19 is now recognized as a predictor of mortality, there is a need to understand the underlying mechanisms and genetic predisposition.
Endothelial cells, like other structural cells, when physiologically activated or during injury like the case of cardiovascular diseases with or without COVID-19, can release increased levels of circulating phospholipid-rich microvesicles that can affect recipient cells locally or via the systemic circulation (18). Such vesicles, called exosomes, may enclose a range of parent cell molecules, including nucleic acids (DNA, mRNA, microRNA, and lncRNA), proteins, and lipids (19). Necrotic or apoptotic processes induced during vascular endothelium damage can lead to the dissemination of such exosomes such that mRNA detected in the circulation can be representative of cells that do not circulate (20). Sampling and molecular analysis of such circulating cells, extracellular vesicles, nucleic acids, which is referred to as liquid biopsy, is emerging as a promising approach for research in cardiovascular injuries (19). Recently endothelial, granulocyte, and platelet-derived exosomes were used to discriminate and map coronary atherosclerotic plaque and calcification in asymptomatic patients (21). In line with this paradigm, we carried out in silico analysis of publicly available datasets derived from different cell sources to decipher the molecular basis, potential pathways, and the role of the endothelium in the pathogenesis of cardiac and vascular injuries in COVID-19.
Materials and Methods
Identification of Differentially Expressed Genes in Blood Cells Following Cardiovascular Injuries
Datasets
Publicly available transcriptomic datasets were retrieved from Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/). Microarray gene expression datasets with the word “venous thromboembolism, acute coronary syndrome, arrhythmia, viral myocarditis, heart failure, and/or cardiogenic shock” were selected. Then we selected datasets with human patients' samples that were compared with age-matched healthy controls and where the samples studied were either whole blood, peripheral blood cells, or endothelium. No datasets of viral myocarditis or cardiogenic shock fulfilled these inclusion criteria. The five datasets (215 patients and 109 healthy control) that fulfilled the inclusion criteria are shown in Table 1.
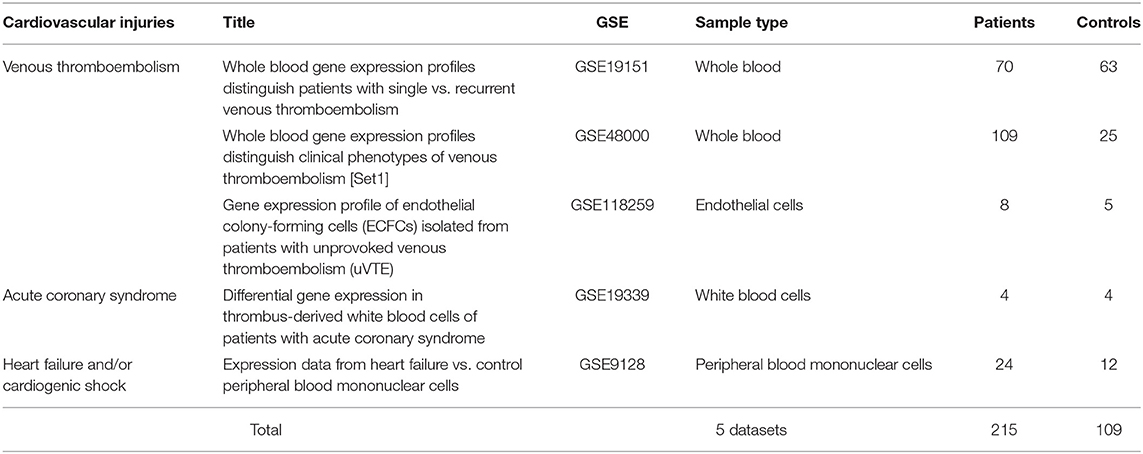
Table 1. List and details of publicly available transcriptomic datasets used in the identification of differentially expressed genes (DEGs) in cardiovascular injuries.
DEGs
We used GEOquery and limma R packages through the GEO2R tool for each dataset (22). We selected the differentially expressed probes, as previously described (23). Briefly, we sorted the genes related to the filtered probes according to the False Discovery Rate (FDR) and selected the top 2,000 differentially expressed probes with FDR <0.05 from each dataset. The annotated genes in each dataset were intersected with DEGs from all other datasets. Enriched Ontology Clustering for the identified genes was performed using the Metascape (http://metascape.org/gp/index.html#/main/step1).
Identification of DEGs in Different Ethnicities
In light of the premise for a potential role for genetic susceptibility to cardiovascular injuries associated with COVID-19, we further explored for the expression of the identified DEGs in the publicly available dataset (GSE17078) of blood outgrowth endothelial cells from 27 healthy subjects of diverse ages and grouped into Caucasian and African Americans.
Virus Perturbations From GEO
In order to explore if the identified genes showed differential expression during viral infections and to identify the viruses that affect their expression, we used the “Gene-virus associations by differential expression of gene following viral infection” database. “https://amp.pharm.mssm.edu/Harmonizome/dataset/GEO+Signatures+of+Differentially+Expressed+Genes+for+Viral+Infections.”
Lung Gene Expression
To identify which lung cells specifically express the genes of interest at a significantly higher level compared to other cells, we explored LungGENS (Lung Gene Expression iN Single-cell), a web-based resource for querying lung single-cell gene expression databases (24).
Identification of DEGs in SARS-CoV-2 Infected Cells and Lungs
The expression of the shortlisted genes was explored in the dataset (GSE147507), where RNA-sequencing of transformed alveolar lung cells (A549) were mock-treated (n = 6) or infected with SARS-CoV-2 (USA-WA1/2020) (n = 6) (25). The same dataset contains uninfected human lung biopsies, one male (age: 72 years), and one female (age: 60 years), which were used as biological replicates and were compared to lung samples derived from a single deceased male patient with COVID-19 (age: 74 years). The retrieved data were used to identify DEGs between infected and uninfected lung samples using BioJupies online tool (https://amp.pharm.mssm.edu/biojupies/). The normalized gene expression was used further to estimate infiltrating immune cells in the lungs.
Estimation of Infiltrating Immune Cells in the Lungs
The normalized gene expression was uploaded to CIBERSORT (https://cibersort.stanford.edu/) to quantify immune cell fractions from bulk lung tissue gene expression profile (26).
Map of Protein Expression Across Human Tissues
Tissue specificity of the identified genes was investigated using The Human Protein Atlas (https://www.proteinatlas.org/) (27). A blood cell-type expression (RNA) option was used to examine the cell specificity of the identified genes. Normalized expressions (NX) for 18 blood cell types and total peripheral blood mononuclear cells (PBMC) were explored.
Identification of Differentially Expressed Genes in SARS-CoV-2 Infected Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes
The expression of the shortlisted genes was explored in the transcriptomic dataset “GSE150392” which is derived from human-induced pluripotent stem cell-derived cardiomyocytes infected in vitro with SARS-CoV-2. The genes which showed significant differential expression between SARS-CoV-2 and mock-infected cells were identified.
Results
Whole Blood and Endothelium Shared DEGs in Patients With Venous Thromboembolism
DEGs in the whole blood of patients with VTE relative to healthy controls (GSE19151 and GSE48000) were intersected with DEGs in endothelial cells of patients with VTE relative to healthy controls (GSE118259), and 36 genes were identified as DEGs common to the three datasets, suggestive of their role in VTE (Figure 1, Table 2).
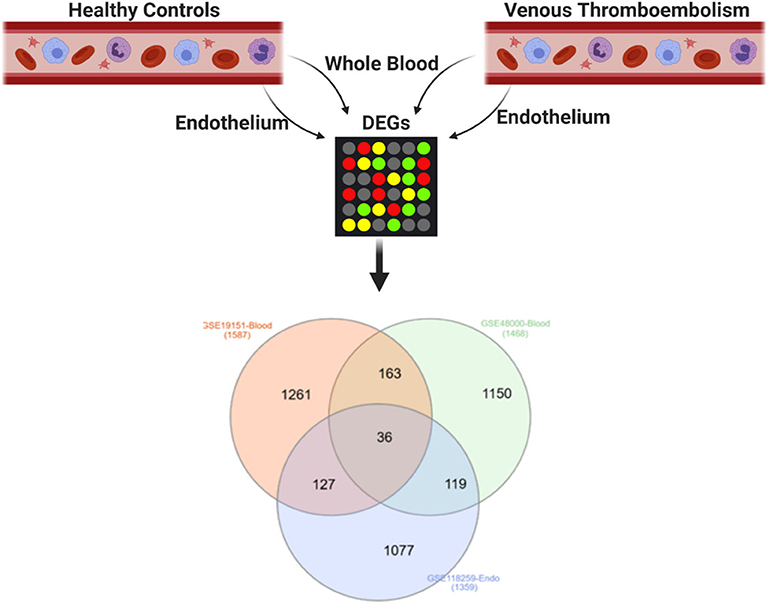
Figure 1. Shared DEGs in the blood and endothelium of patients with venous thromboembolism compared to healthy controls. DEGs in the whole blood of patients with venous thromboembolism (GSE19151 and GSE48000) were intersected with DEGs in endothelial cells of patients with venous thromboembolism (GSE118259). Created with BioRender.com.
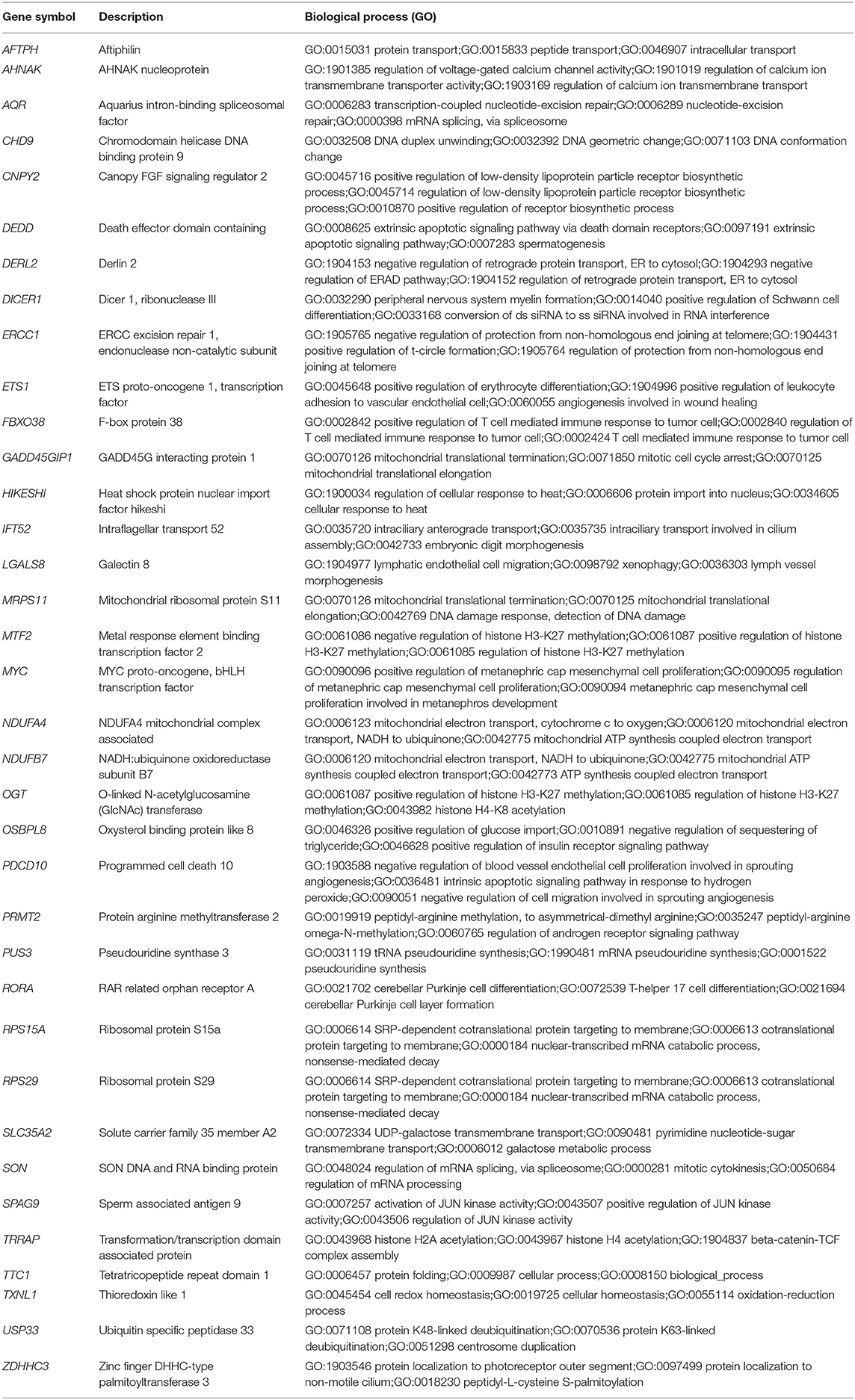
Table 2. List of the 36 shared DEGs in the blood and endothelium of patients with venous thromboembolism compared to healthy controls.
The 36 Shared DEGs Play an Essential Role in Endothelium Biology
To understand the role of the identified 36 genes, we explored their shared biological pathways and found that several of these genes were vital for pathways involved in cell homeostasis, response to stress, and cellular metabolism. These include pathways related to targets of C-MYC transcriptional activation (MYC, TRRAP, PDCD10, OGT, USP33, ZDHHC3, and ETS1), regulation of cellular response to stress (ERCC1, MYC, SPAG9, PDCD10, DERL2, and HIKESHI), and endothelial cell migration (ETS1, LGALS8, and PDCD10). Figure 2 shows the list of biological pathways associated with the DEGs. Four genes MYC, ETS1, OGT, and PDCD10 were shown to be common between the top pathways indicating their significant molecular and biological role:. They are all enriched in the PID MYC ACTIV PATHWAY, suggesting that they are targets of C-MYC transcriptional activation. The proto-oncogene c-Myc is vital for vascular development. Gene expression analysis of c-Myc-deficient endothelial cells showed that the senescent phenotype of c-Myc is needed for the prevention of vascular pro-inflammatory phenotype (28). Global or endothelial and hematopoietic cell-specific loss of c-Myc leads to defects in vasculogenesis and primitive erythropoiesis (29).
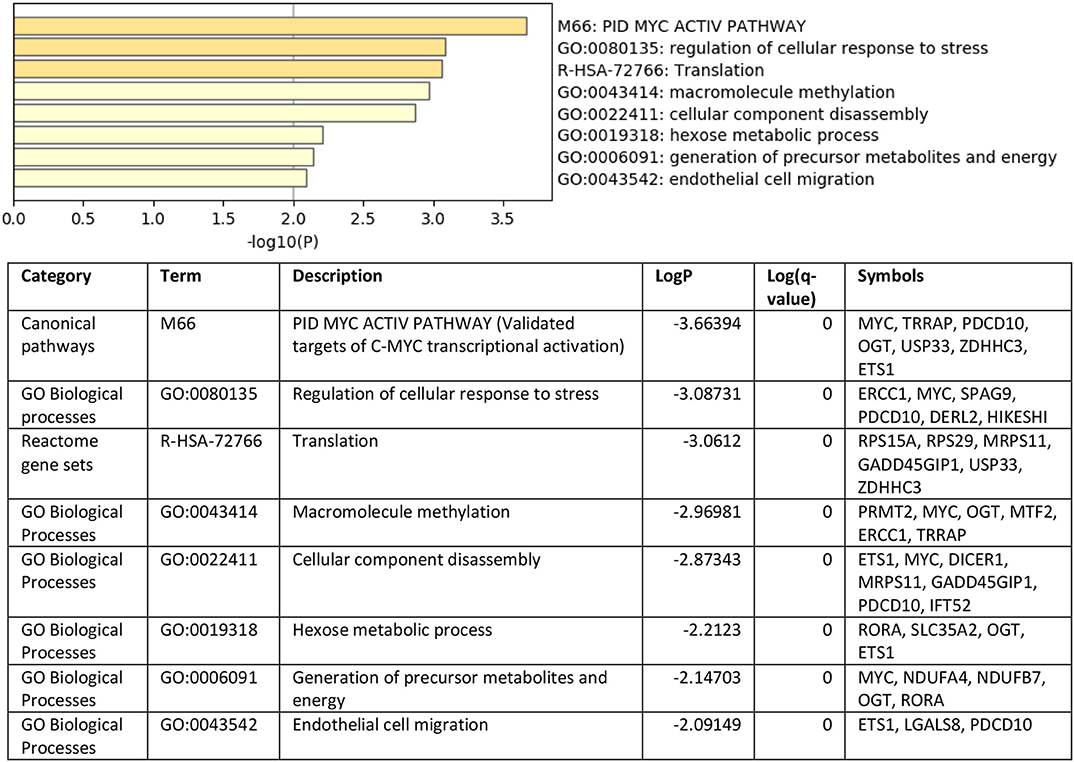
Figure 2. Top pathways enriched with shared DEGs in the blood and endothelium of patients with VTE compared to healthy controls. Created with BioRender.com.
SON, OGT, and RORA Are Differentially Expressed in the Peripheral Blood of Patients With Acute Coronary Syndrome and Heart Failure
The 36 genes identified to be specific to VTE were intersected with DEGs in thrombus-derived white blood cells of patients with acute coronary syndrome vs. controls (GSE19339) and peripheral blood mononuclear cells of patients with heart failure vs. control (GSE9128) (Figure 3). Four genes were shared between VTE and acute coronary syndrome (MTF2, TXNL1, PRMT2, and ERCC2), and ten genes were shared between VTE and heart failure (DICER1, CHD9, MYC, HIKESHI, USP33, AQR, DEDD, DERL2, CNPY2, and PUS3). Only three genes, SON (SON DNA and RNA binding protein), OGT (O-linked N-acetylglucosamine [GlcNAc] transferase), and RORA (RAR related orphan receptor A) were shared by all the three conditions.
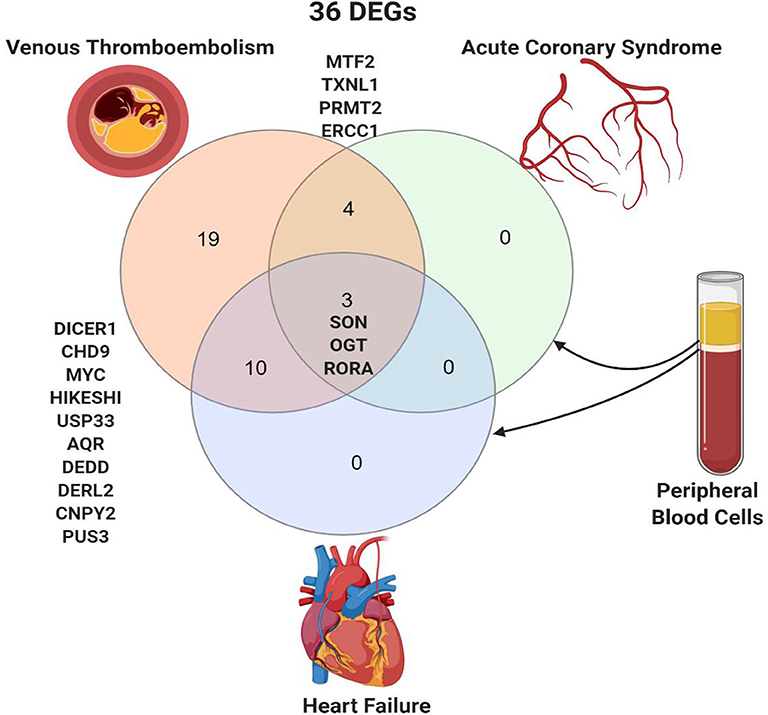
Figure 3. Shared peripheral blood DEGs in patients with venous thromboembolism, acute coronary syndrome, and heart failure. Created with BioRender.com.
SON, OGT, and RORA Expression in Healthy Endothelium of African Americans
To explore the premise of genetic susceptibility for COVID-19 related cardiac events, we explored the gene expression of the three shared DEGs (SON, OGT, and RORA) in the publicly available dataset (GSE17078) of blood outgrowth endothelial cells from 27 healthy Caucasian and African American subjects. The findings show that SON, OGT, and RORA are significantly downregulated in the healthy endothelium of African Americans compared to Caucasians (Figure 4).
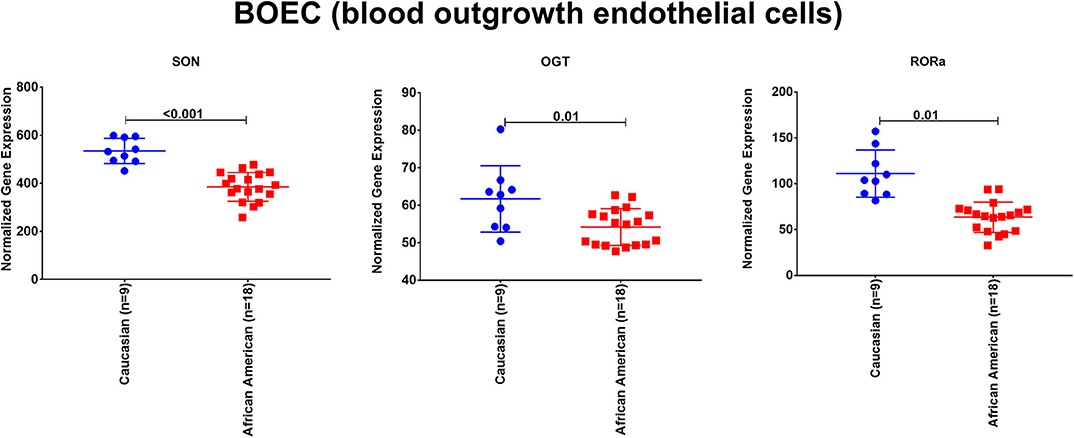
Figure 4. The mRNA expression of SON, OGT, and RORA in the publicly available dataset (GSE17078) of blood outgrowth endothelial cells from 27 healthy subjects of diverse ages and grouped into Caucasian and African Americans. Created with BioRender.com.
Expression of the DEGs in Viral Infections
All 36 DEGs showed differential expression during viral infections as per the “Gene-virus associations by differential expression of gene following viral infection” database. The most frequently identified viruses affecting most of the genes were SARS-CoV strains from infected lung epithelial cells (SARS-CoV, SARS-dORF6, or SARS-BatSRBD “GSE47960, GSE47961 and GSE50000”; icSARS-CoV or the icSARS-dORF6 mutant “GSE37827”) and a SARS CoV MA15 infection in C57Bl/6 mouse model “GSE33266.” The genes which showed consistent differential expression in different datasets in response to SARS-CoV infections were SON, OGT, PRMT2 (protein arginine methyltransferase 2), TXNL1 (thioredoxin like 1), CNPY2 (canopy FGF signaling regulator 2), MRPS11 (mitochondrial ribosomal protein S11), SPAG9 (sperm associated antigen 9), MTF2 (metal response element-binding transcription factor 2), CHD9 (chromodomain helicase DNA binding protein 9), and RPS29 (ribosomal protein S29).
Lung Single-Cell Expression of DEGs
The cellular composition of the lung is 40–50% endothelial cells, which differentiate in parallel with epithelial cells to form gas exchange units which are in contact with the external environment and thus need to ensure a rapid immune response (30). In lung diseases, including infections, the transcriptomes of endothelial cells, pericyte/smooth muscle cells, fibroblasts, and macrophage clusters showed that endothelial cells had the most differentially expressed gene profile compared to other cell types (31). We speculated that if we found common differentially expressed genes shared between the two cell types and which could be affected by SARS-CoV-2 infection, then we may be able further to understand the link between COVID-19 and associated endothelium injuries. Querying lung single-cell gene expression databases showed that expression of some of the 36 DEGs was significantly higher in lung endothelial cells (PRMT2, OGT, MTF2, and CHD9), lung fibroblasts (PRMT2, OGT, MTF2, CHD9, TXNL1, CNPY2, SPAG9, MRPS11, SON, and RPS29) and epithelial cells (TXNL1, CNPY2, and SPAG9). The only gene whose expression was found to also be related to myeloid/immune cells was RPS29. Figure 5 shows the DEGs and their expression in different cells. Details of peak expressions for each DEG in different cells is provided in Supplementary Table.
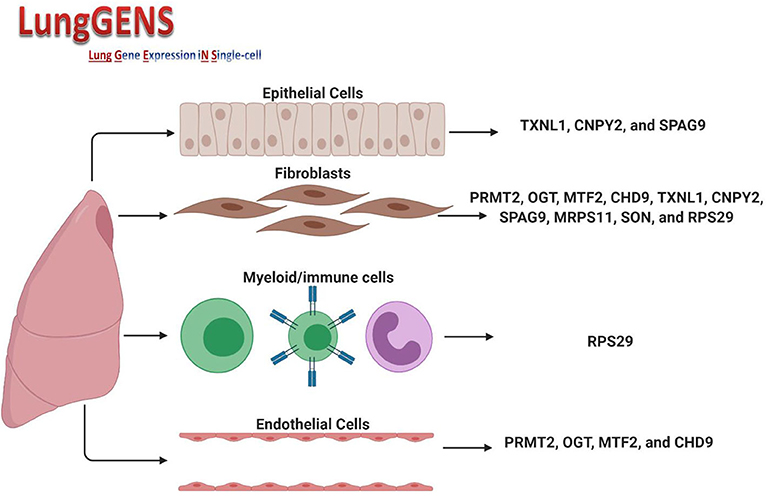
Figure 5. Expression of DEGs in different lung cell types. Created with BioRender.com.
SNPs in the Identified DEGs With Significant Association to COVID-19
We searched for the COVID-19 GWAS (https://grasp.nhlbi.nih.gov/Covid19GWASResults.aspx) looking for Annotated top results (only variants with P <1E-5) in 1,723 positive cases vs. 11,409 negative controls and found that none of the 36 genes identified carry SNPs with significant association to COVID-19, indicating that these genes are differentially expressed during infection or disease as a dynamic response to stimuli or condition.
RPS29 and SPAG9 in SARS-CoV-2 Infected Lung Epithelial Cells
The expression of the 36 DEGs was examined in mock vs. SARS-CoV-2 infected lung epithelial cells. Although most of the genes were upregulated by the virus infection, only RPS29 and SPAG9 showed significant upregulation, as shown in Figure 6.
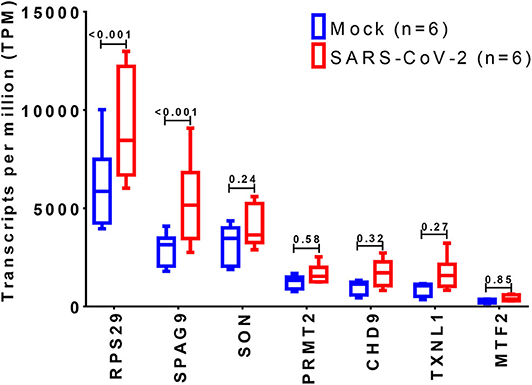
Figure 6. Identification of differentially expressed genes in SARS-CoV-2 infected cells. The shortlisted genes expression was explored in the dataset (GSE147507), where RNA-Sequencing of transformed alveolar lung cells (A549) were mock-treated (n = 6) or infected with SARS-CoV-2 (USA-WA1/2020) (n = 6). Created with BioRender.com.
SPAG9 and RPS29 in Immune Cells
We sought to identify which immune cell expresses the highest level of RPS29 and SPAG9. Our findings indicate that RPS29 showed low cell type specificity but was higher in T cells while SPAG9 was enriched specifically in neutrophils and basophils (Figure 7).
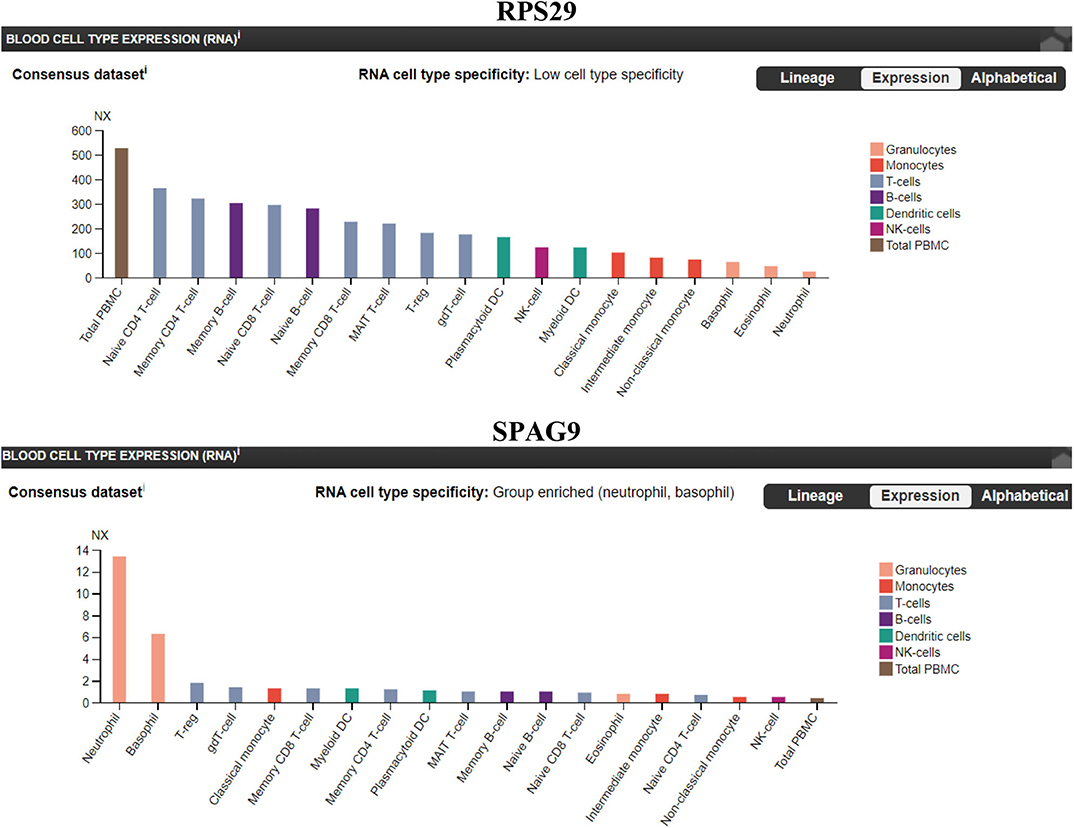
Figure 7. Immune cells specificity of the identified genes (RPS29 and SPAG9) using The Human Protein Atlas. A blood cell-type expression (RNA) option was used to examine the cell specificity of the identified genes. Normalized eXpression (NX) for 18 blood cell types and total peripheral blood mononuclear cells (PBMC) were explored. Created with BioRender.com.
Cardiac Protective Genes Are Downregulated in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes Infected With SARS-CoV-2
We explored the novel transcriptomic dataset “GSE150392” which is derived from in vitro work in which human-induced pluripotent stem cell-derived cardiomyocytes were infected with SARS-CoV-2. Nine of the 36 DEGs showed significant differential expression between SARS-CoV-2 and mock-infected cells (Table 3). Four genes (NDUFA4, NDUFB7, MRPS11, and HIKESHI) were downregulated by SARS-CoV-2 while the remaining five genes (CHD9, MTF2, RORA, MYC, and ETS1) were upregulated.
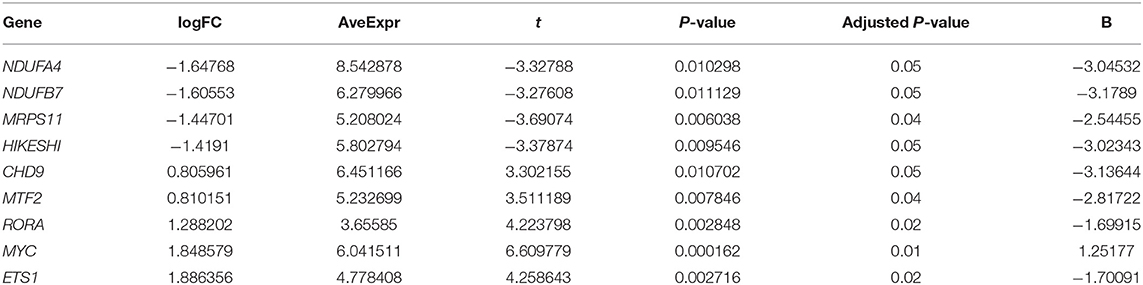
Table 3. List of genes significantly altered in human-induced pluripotent stem cell-derived cardiomyocytes infected with SARS-CoV-2 in vitro extracted from (GSE150392) dataset.
Discussion
Although respiratory failure has been the primary concern in COVID-19 infection, cardiac injury manifested by a rise in high-sensitivity troponin has gained considerable attention due to its reported association with mortality (3, 5). A higher incidence of acute onset heart failure, myocardial infarction, myocarditis, and cardiac arrest in COVID-19 patients is documented in the literature (9). On the basis of this, we hypothesized that a common molecular pathway shared between these common cardiovascular diseases might be activated in SARS-CoV-2 infection and thus provide an explanation for the high rate of cardiovascular complications seen in COVID-19 patients. To achieve that, we started by comparing cases to control in each of these diseases to find their shared DEGs, and then determine if these genes were also triggered specifically in COVID-19. The dataset we used for validation was Lung cells infected with SARS-CoV-2 (which is one of the few datasets available). As these identified genes were found to be expressed in lung cells, we postulate that they might represent the core machinery genes and the link between COVID-19, which is, in essence, lung infection and cardiovascular injuries, which are systemic consequences. While it would be ideal to utilize datasets derived from COVID-19 patients with cardiovascular outcomes for such comparative analysis, these are currently not available. Nevertheless, the findings from this study provide important new information that expands our current understanding of cardiovascular injuries in COVID-19.
From our comprehensive in silico approach, we identified 36 DEGs in the blood and endothelium of patients with VTE. Among these were genes known to play key roles in endothelium and vascular biology, with several being vital for pathways for C-MYC transcriptional activation, regulation of cellular response to stress as well as endothelial cell migration. In addition, some of the genes involved in endothelial cell migration (ETS1, LGALS8, and PDCD10) are also known to be associated with perturbations during viral infection (32–37). Notably, of the 36 DEGs identified, three genes, namely SON, OGT, and RORA, were also expressed in the peripheral blood of patients with acute coronary syndrome and heart failure. These findings implicate SON, OGT, and RORA as shared core genes in cardiac and vascular-related injuries. As these DEGs were also shared with mesenchymal cells of the lung, we speculate that they may represent the missing link between lung damage and related cardiovascular injuries reported in patients with COVID-19 patients. SON gene encodes an RNA-binding protein that promotes the splicing of many cell-cycle and DNA-repair transcripts and maintains accurate splicing for a subset of Human pre-mRNAs (38). SON is involved in pathways regulating virus infection like influenza virus infection as its deletion can lead to reduced influenza viral RNA levels and decreased viral infection suggesting that SON is needed for influenza virus replication (39). In human-induced pluripotent stem cell-derived multipotent cardiac progenitor cells, knockdown of SON reduced proliferation and differentiation of cardiomyocytes, while increasing fibroblasts (40). OGT is an O-GlcNAc transferase that catalyzes the addition of the O-GlcNAc post-translational modification to proteins, which is essential in regulating the stress response, differentiation, nutrient sensing, and autophagy (41). O-GlcNAc level is increased during ischemia-reperfusion or hemorrhagic shock with a cardioprotective effect making augmentation of O-GlcNAc levels a potential new therapeutic option for cardiovascular dysfunction or ischemia/reperfusion (42). RORA is a nuclear receptor retinoic acid-related orphan receptor-α that has been recently identified in the heart to inhibit ANG II-induced pathological hypertrophy and cardiomyocyte death, repress IL-6 transcription, and its level is reduced in failing mouse and human hearts (43). RORA deficient staggered mice subjected to myocardial ischemia/reperfusion injury show significantly increased myocardial infarct size, myocardial apoptosis, and exacerbated contractile dysfunction compared to wild-type mice (44). Moreover, mice with cardiomyocyte-specific RORA overexpression were less vulnerable to injury (44). RORA has been described as a transcription factor which ties metabolic and inflammatory signaling pathways. In fact, macrophages from staggerer mice (which have a deletion in RORA) overexpress Il1b following LPS stimulation suggesting an anti-inflammatory role for RORA (45). One mechanism that has been postulated involves the role of RORA in inducing IκBα, which negatively regulated the NFκB signaling pathway (46). However, it has been suggested that RORA may play a dual role in tissue and cell-dependent manner. For example, in adipose tissue RORA may play a pro-inflammatory role by driving endoplasmic reticulum stress (47). Interestingly in human-induced pluripotent stem, cell-derived cardiomyocytes infected in vitro with SARS-CoV-2, the expression of RORA was upregulated, and we speculate that this might be a cardioprotective response to direct viral invasion. Furthermore, SON, OGT, and RORA regulate the maintenance and differentiation of stem cells, including endothelial progenitor cells (EPCs) (6). They are preferentially expressed in undifferentiated stem cells but downregulated during stem cell differentiation (7–9). The ability of vascular endothelial cells to repair relies on the EPCs (11). As the occurrence of cardiovascular events during COVID-19 suggests that targeting the endothelium is part of the viral infection course, we surmise COVID-19 patients who have a pre-existing genetic propensity for low SON, OGT, and RORA expression may therefore be more susceptible to cardiac damage. Our findings of significant downregulation of SON, OGT, and RORA in healthy endothelium of African Americans is consistent with this hypothesis. This may explain the increased risk of cardiovascular injury among African American patients with COVID-19.
All the 36 DEGs showed differential expression during viral infections, and the most frequently identified viruses were SARS-CoV strains. Specifically, in SARS-CoV-2 infected lung epithelial cells, RPS29 and SPAG9 genes were significantly upregulated. RPS29, which was the only DEG found to be specific to myeloid/immune Cells (S1.21) with TPM of 1,205.92 and intermediate fibroblast 2 (S2.5) with TPM of 1,098.99 in the lung, encodes for a ribosomal protein with an established role in hematopoietic stem cells and red blood cell development (48). RPS29 is a component of the small 40S ribosomal subunit and needed for rRNA processing and ribosome biogenesis (49). Germ-line mutation in RPS29 cause Diamond-Blackfan anemia, which is an inherited bone marrow failure syndrome (49). RNA-seq analysis of acute myocardial infarction samples has shown that RPS29 was one of the top upregulated genes (50). Interestingly, RPS29 has been reported to be upregulated in A549 cells infected with the novel H3N2 Swine Influenza virus and the 2009 H1N1 pandemic Influenza virus (51). It was also upregulated in inflammatory conditions like periodontitis and associated with raised IFN-α (52). It is likely that the upregulation of RPS20 in viral infection provides a mechanism for stimulation of hematopoietic stem cells and red blood cell development for increased production of immune cells like neutrophils for recruitment to the site of infection. SPAG9 is known to induce an immune response and to regulate JNK and mitogen-activated protein kinases (MAPKs) signaling pathways, cell cycle progression, and matrix metalloproteinases (53). SPAG9 is involved in the trafficking of endocytic vesicles within the intercellular bridge (54). SPAG9 antibody in serum appears to be related to the type of lung cancer, indicating its specificity to lung-related tissues (55). It is one of the cardiac cytoskeleton and sarcomere assembly and function genes which are enhanced in mice with the deleted muscleblind-like family of splice regulators involved in cardiac dysfunction (56). The virus-induced upregulation of SPAG9 might induce antibodies against it that might cross-react with the heart cytoskeleton and cause cardiac damage in the form of myocarditis and cardiac dysfunction. In Figure 8, we illustrate the pathway for the postulated role of RPS29 and SPAG9 genes in SARS-COV-2 related cardiovascular injuries.
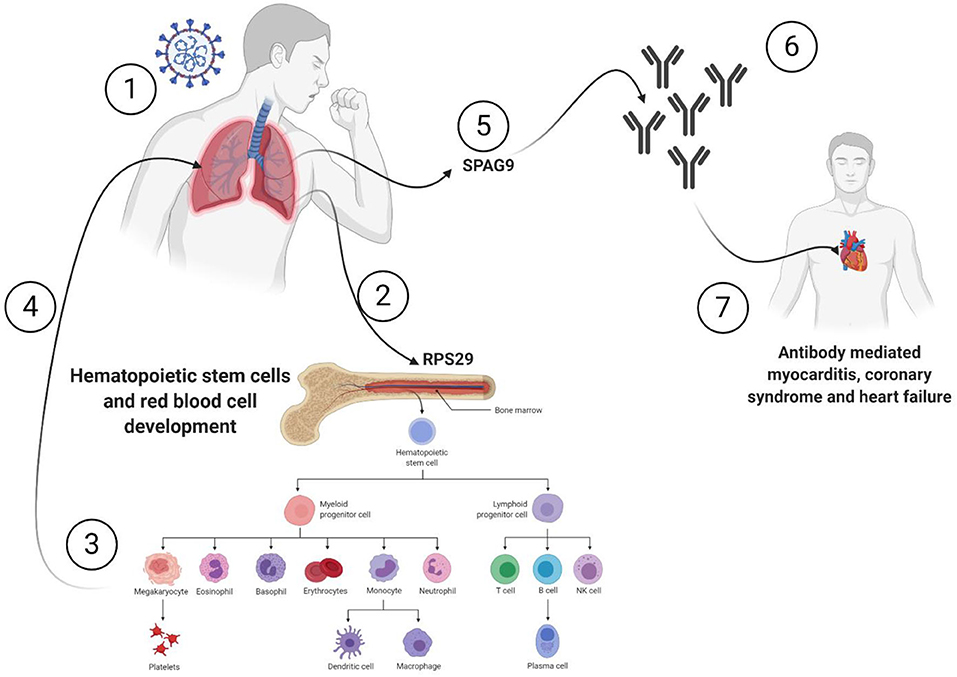
Figure 8. Role of RPS29 and SPAG9 genes in SARS-COV-2 related cardiovascular injuries. (1) Lung viral infection (2) upregulates RPS29 that can (3) stimulate hematopoietic stem cells and red blood cell development to provide immune cells like neutrophils (4) to reach the lung, (5) the virus-induced upregulation of SPAG9 might (6) induce antibodies against it that might cross-(7) react with the heart cytoskeleton and cause cardiac damage in the form of myocarditis and cardiac dysfunction. Created with BioRender.com.
Our analysis of the SARS-CoV-2 infected cardiomyocyte derived dataset showed that of the 36 DEGs identified in this study, four genes (NDUFA4L2; NDUFB7; MRPS11; HIKESHI) which are known to be cardioprotective were downregulated. NDUFA4L2 plays a role in protecting cardiomyocytes from apoptosis and mitochondrial dysfunction during ischemia/reperfusion event, while NDUFB7 has been linked with mitochondrial dysfunction and cardiomyocyte senescence (57, 58). MRPS11 is a mitochondrial gene involved in sex-specific cardiac structure and function alterations (59). Heat shock proteins are involved in protecting the heart against heart failure by facilitating the removal of misfolded and degraded proteins (60), and HIKESHI plays a role in heat-shock stress response regulation to protect cells from heat shock damages. This finding suggests that in addition to the proposed RPS29 and SPAG9 induced cardiac damage pathway alluded to earlier, SARS-CoV-2 also employs a mechanism of downregulation of cardioprotective genes to promote cardiac injury.
In conclusion, our findings from the analysis of publicly available transcriptomic datasets identified three shared core genes pertinent to cardiac and vascular-related injuries. The possibility for their role in genetic susceptibility to cardiovascular injury in patients with COVID-19 was highlighted. In addition, it is likely that a combination of RPS29 and SPAG9 genes induced pathways, as well as downregulation of cardioprotective genes, contribute to cardiac and vascular events in patients with COVID-19.
Given that our analysis is in silico, experimental validation of our findings suggesting the potential role in genetic susceptibility such as in vitro experiments on endothelial cells exposed to SARS-CoV-2 antigens are needed to enable a better understanding of cardiovascular events associated with SARS-CoV-2 infection. The main limitation here is that the study is performed on the premise that venous thromboembolism, acute coronary syndrome, and heart failure might be common during COVID-19 infection. However, in silico analysis of studies with patients with COVID−19 infection vs. those without COVID-19 and a similar CVR outcome will be useful in pinpointing specific genes.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2020.582399/full#supplementary-material
References
1. Hopkins J. Coronavirus resource center John Hopkins University of medicine. In: COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). ArcGIS; Johns Hopkins University (2020).
2. Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. (2020) 395:507–13. doi: 10.1016/S0140-6736(20)30211-7
3. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. (2020) 395:497–506. doi: 10.1016/S0140-6736(20)30183-5
4. Yang J, Zheng Y, Gou X, Pu K, Chen Z, Guo Q, et al. Prevalence of comorbidities and its effects in patients infected with SARS-CoV-2: a systematic review and meta-analysis. Int J Infect Dis. (2020) 94:91–5. doi: 10.1016/j.ijid.2020.03.017
5. Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019. Novel coronavirus-infected pneumonia in Wuhan, China. JAMA. (2020) 323:1061–9. doi: 10.1001/jama.2020.1585
6. Zheng YY, Ma YT, Zhang JY, Xie X. COVID-19 and the cardiovascular system. Nat Rev Cardiol. (2020) 17:259–60. doi: 10.1038/s41569-020-0360-5
7. Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. (2020) 395:1054–62. doi: 10.1016/S0140-6736(20)30566-3
8. Kwenandar F, Japar KV, Damay V, Hariyanto TI, Tanaka M, Lugito NPH, et al. Coronavirus disease 2019 and cardiovascular system: a narrative review. Int J Cardiol Heart Vasc. (2020) 29:100557. doi: 10.1016/j.ijcha.2020.100557
9. Shi S, Qin M, Shen B, Cai Y, Liu T, Yang F, et al. Association of cardiac injury with mortality in hospitalized patients with COVID-19 in Wuhan, China. JAMA Cardiol. (2020) 5:802–10. doi: 10.1001/jamacardio.2020.0950
10. Ferrario CM, Jessup J, Chappell MC, Averill DB, Brosnihan KB, Tallant EA, et al. Effect of angiotensin-converting enzyme inhibition and angiotensin II receptor blockers on cardiac angiotensin-converting enzyme 2. Circulation. (2005) 111:2605–10. doi: 10.1161/CIRCULATIONAHA.104.510461
11. Varga Z, Flammer AJ, Steiger P, Haberecker M, Andermatt R, Zinkernagel AS, et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet. (2020) 395:1417–8. doi: 10.1016/S0140-6736(20)30937-5
12. Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. (2004) 203:631–7. doi: 10.1002/path.1570
13. Klok FA, Kruip M, van der Meer NJM, Arbous MS, Gommers D, Kant KM, et al. Incidence of thrombotic complications in critically ill ICU patients with COVID-19. Thromb Res. (2020) 191:145–7. doi: 10.1016/j.thromres.2020.04.013
14. Kanda T, Takahashi T. Interleukin-6 and cardiovascular diseases. Japan Heart J. (2004) 45:183–93. doi: 10.1536/jhj.45.183
15. Levi M, Keller TT, van Gorp E, ten Cate H. Infection and inflammation and the coagulation system. Cardiovasc Res. (2003) 60:26–39. doi: 10.1016/S0008-6363(02)00857-X
16. Clerkin KJ, Fried JA, Raikhelkar J, Sayer G, Griffin JM, Masoumi A, et al. COVID-19 and cardiovascular disease. Circulation. (2020) 141:1648–55. doi: 10.1161/CIRCULATIONAHA.120.046941
17. Giudicessi JR, Roden DM, Wilde AAM, Ackerman MJ. Genetic susceptibility for COVID-19-associated sudden cardiac death in African Americans. Heart rhythm. (2020) 17:1487–92. doi: 10.1016/j.hrthm.2020.04.045
18. Badimon L, Suades R, Vilella-Figuerola A, Crespo J, Vilahur G, Escate R, et al. Liquid biopsies: microvesicles in cardiovascular disease. Antioxid Redox Signal. (2019) 33:645–62. doi: 10.1089/ars.2019.7922
19. Zhou B, Xu K, Zheng X, Chen T, Wang J, Song Y, et al. Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct Target Ther. (2020) 5:144. doi: 10.1038/s41392-020-00258-9
20. Carrió I, Flotats A. Liquid biopsies and molecular imaging: friends or foes? Clin Transl Imaging. (2020) 8:47–50. doi: 10.1007/s40336-019-00350-3
21. Chiva-Blanch G, Padró T, Alonso R, Crespo J, Perez de Isla L, Mata P, et al. Liquid biopsy of extracellular microvesicles maps coronary calcification and atherosclerotic plaque in asymptomatic patients with familial hypercholesterolemia. Arterioscler Thromb Vasc Biol. (2019) 39:945–55. doi: 10.1161/ATVBAHA.118.312414
22. Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. (2012) 41:D991–5. doi: 10.1093/nar/gks1193
23. Hachim MY, Al Heialy S, Hachim IY, Halwani R, Senok AC, Maghazachi AA, et al. Interferon-Induced Transmembrane Protein (IFITM3) is upregulated explicitly in sars-cov-2 infected lung epithelial cells. Front Immunol. (2020) 11:1372. doi: 10.3389/fimmu.2020.01372
24. Du Y, Guo M, Whitsett JA, Xu Y. 'LungGENS': a web-based tool for mapping single-cell gene expression in the developing lung. Thorax. (2015) 70:1092–4. doi: 10.1136/thoraxjnl-2015-207035
25. Blanco-Melo D, Nilsson-Payant BE, Liu W-C, Møller R, Panis M, Sachs D, et al. SARS-CoV-2 launches a unique transcriptional signature from in vitro, ex vivo, and in vivo systems. bioRxiv [Preprint]. (2020). doi: 10.1101/2020.03.24.004655
26. Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol. (2018) 1711:243–59. doi: 10.1007/978-1-4939-7493-1_12
27. Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Tissue-based map of the human proteome. Science. (2015) 347:1260419. doi: 10.1126/science.1260419
28. Florea V, Bhagavatula N, Simovic G, Macedo FY, Fock RA, Rodrigues CO. c-Myc is essential to prevent endothelial pro-inflammatory senescent phenotype. PLoS ONE. (2013) 8:e73146. doi: 10.1371/journal.pone.0073146
29. Testini C, Smith RO, Jin Y, Martinsson P, Sun Y, Hedlund M, et al. Myc-dependent endothelial proliferation is controlled by phosphotyrosine 1212 in VEGF receptor-2. EMBO Rep. (2019) 20:e47845. doi: 10.15252/embr.201947845
30. Jambusaria A, Hong Z, Zhang L, Srivastava S, Jana A, Toth PT, et al. Endothelial heterogeneity across distinct vascular beds during homeostasis and inflammation. Elife. (2020) 9:e51413. doi: 10.7554/eLife.51413
31. Saygin D, Tabib T, Bittar HET, Valenzi E, Sembrat J, Chan SY, et al. Transcriptional profiling of lung cell populations in idiopathic pulmonary arterial hypertension. Pulm Circ. (2020) 10:1–15. doi: 10.1177/2045894020908782
32. Ghosh S, MarElia-Bennett CB, Hildreth BE, Lefler JE, Sharma SM, Ostrowski MC. Redundant function of Ets1 and Ets2 in regulating M-phase progression in post-natal angiogenesis. bioRxiv [Preprint]. (2020). doi: 10.1101/2020.02.19.956417
33. Harris TA, Yamakuchi M, Kondo M, Oettgen P, Lowenstein CJ. Ets-1 and Ets-2 regulate the expression of MicroRNA-126 in endothelial cells. Arterioscler Thromb Vasc Biol. (2010) 30:1990–7. doi: 10.1161/ATVBAHA.110.211706
34. Gutierrez KD, Morris VA, Wu D, Barcy S, Lagunoff M. Ets-1 is required for the activation of VEGFR3 during latent kaposi's sarcoma-associated herpesvirus infection of endothelial cells. J Virol. (2013) 87:6758–68. doi: 10.1128/JVI.03241-12
35. Herkt CE, Caffrey BE, Surmann K, Blankenburg S, Gesell Salazar M, Jung AL, et al. A MicroRNA network controls legionella pneumophila replication in human macrophages via LGALS8 and MX1. mBio. (2020) 11:e03155–19. doi: 10.1128/mBio.03155-19
36. Cattaneo V, Tribulatti MV, Carabelli J, Carestia A, Schattner M, Campetella O. Galectin-8 elicits pro-inflammatory activities in the endothelium. Glycobiology. (2014) 24:966–73. doi: 10.1093/glycob/cwu060
37. Sarmento L, Afonso CL, Estevez C, Wasilenko J, Pantin-Jackwood M. Differential host gene expression in cells infected with highly pathogenic H5N1 avian influenza viruses. Vet Immunol Immunopathol. (2008) 125:291–302. doi: 10.1016/j.vetimm.2008.05.021
38. Sharma A, Markey M, Torres-Muñoz K, Varia S, Kadakia M, Bubulya A, et al. Son maintains accurate splicing for a subset of human pre-mRNAs. J Cell Sci. (2011) 124:4286–98. doi: 10.1242/jcs.092239
39. Lu X, Ng HH, Bubulya PA. The role of SON in splicing, development, and disease. Wiley Interdiscip Rev RNA. (2014) 5:637–46. doi: 10.1002/wrna.1235
40. Schroeder AM, Allahyari M, Vogler G, Missinato MA, Nielsen T, Yu MS, et al. Model system identification of novel congenital heart disease gene candidates: focus on RPL13. Hum Mol Genet. (2019) 28:3954–69. doi: 10.1093/hmg/ddz213
41. Pravata VM, Muha V, Gundogdu M, Ferenbach AT, Kakade PS, Vandadi V, et al. Catalytic deficiency of O-GlcNAc transferase leads to X-linked intellectual disability. Proc Natl Acad Sci USA. (2019) 116:14961–70. doi: 10.1073/pnas.1900065116
42. Ferron M, Denis M, Persello A, Rathagirishnan R, Lauzier B. Protein O-GlcNAcylation in cardiac pathologies: past, present, future. Front Endocrinol. (2019) 9:819. doi: 10.3389/fendo.2018.00819
43. Beak JY, Kang HS, Huang W, Myers PH, Bowles DE, Jetten AM, et al. The nuclear receptor RORα protects against angiotensin II-induced cardiac hypertrophy and heart failure. Am J Physiol Heart Circ Physiol. (2019) 316:H186–200. doi: 10.1152/ajpheart.00531.2018
44. He B, Zhao Y, Xu L, Gao L, Su Y, Lin N, et al. The nuclear melatonin receptor RORα is a novel endogenous defender against myocardial ischemia/reperfusion injury. J Pineal Res. (2016) 60:313–26. doi: 10.1111/jpi.12312
45. Nejati Moharrami N, Bjørkøy Tande E, Ryan L, Espevik T, Boyartchuk V. RORα controls inflammatory state of human macrophages. PLoS ONE. (2018) 13:e0207374. doi: 10.1371/journal.pone.0207374
46. Delerive P, Monté D, Dubois G, Trottein F, Fruchart-Najib J, Mariani J, et al. The orphan nuclear receptor ROR alpha is a negative regulator of the inflammatory response. EMBO Rep. (2001) 2:42–8. doi: 10.1093/embo-reports/kve007
47. Liu Y, Chen Y, Zhang J, Liu Y, Zhang Y, Su Z. Retinoic acid receptor-related orphan receptor α stimulates adipose tissue inflammation by modulating endoplasmic reticulum stress. J Biol Chem. (2017) 292:13959–69. doi: 10.1074/jbc.M117.782391
48. Taylor AM, Humphries JM, White RM, Murphey RD, Burns CE, Zon LI. Hematopoietic defects in rps29 mutant zebrafish depend upon p53 activation. Exp Hematol. (2012) 40:228–37.e5. doi: 10.1016/j.exphem.2011.11.007
49. Mirabello L, Macari ER, Jessop L, Ellis SR, Myers T, Giri N, et al. Whole-exome sequencing and functional studies identify RPS29 as a novel gene mutated in multicase Diamond-Blackfan anemia families. Blood. (2014) 124:24–32. doi: 10.1182/blood-2013-11-540278
50. Zhuo L-A, Wen Y-T, Wang Y, Liang Z-F, Wu G, Nong M-D, et al. LncRNA SNHG8 is identified as a key regulator of acute myocardial infarction by RNA-seq analysis. Lipids Health Dis. (2019) 18:201. doi: 10.1186/s12944-019-1142-0
51. Gao L, Gao J, Liang Y, Li R, Xiao Q, Zhang Z, et al. Integration analysis of a miRNA-mRNA expression in A549 cells infected with a novel H3N2 swine influenza virus and the 2009. H1N1 pandemic influenza virus. Infect Genet Evol. (2019) 74:103922. doi: 10.1016/j.meegid.2019.103922
52. Wright H, Matthews J, Chapple I, Ling-Mountford N, Cooper P. Periodontitis associates with a type 1 IFN signature in peripheral blood neutrophils. J Immunol. (2008) 181:5775–84. doi: 10.4049/jimmunol.181.8.5775
53. Pan J, Yu H, Guo Z, Liu Q, Ding M, Xu K, et al. Emerging role of sperm-associated antigen 9 in tumorigenesis. Biomed Pharmacother. (2018) 103:1212–6. doi: 10.1016/j.biopha.2018.04.168
54. Montagnac G, Sibarita J-B, Loubéry S, Daviet L, Romao M, Raposo G, et al. ARF6 interacts with JIP4 to Control a motor switch mechanism regulating endosome traffic in Cytokinesis. Curr Biol. (2009) 19:184–95. doi: 10.1016/j.cub.2008.12.043
55. Ren B, Wei X, Zou G, He J, Xu G, Xu F, et al. Cancer testis antigen SPAG9 is a promising marker for the diagnosis and treatment of lung cancer. Oncol Rep. (2016) 35:2599–605. doi: 10.3892/or.2016.4645
56. Dixon DM, Choi J, El-Ghazali A, Park SY, Roos KP, Jordan MC, et al. Loss of muscleblind-like 1 results in cardiac pathology and persistence of embryonic splice isoforms. Sci Rep. (2015) 5:9042. doi: 10.1038/srep09042
57. Jones DP, True HD, Patel J. Leukocyte trafficking in cardiovascular disease: insights from experimental models. Mediators Inflamm. (2017) 2017:9746169. doi: 10.1155/2017/9746169
58. Anderson R, Lagnado A, Maggiorani D, Walaszczyk A, Dookun E, Chapman J, et al. Length-independent telomere damage drives post-mitotic cardiomyocyte senescence. EMBO J. (2019) 38:e100492. doi: 10.15252/embj.2018100492
59. Wang H, Sun X, Chou J, Lin M, Ferrario CM, Zapata-Sudo G, et al. Inflammatory and mitochondrial gene expression data in GPER-deficient cardiomyocytes from male and female mice. Data Brief. (2016) 10:465–73. doi: 10.1016/j.dib.2016.11.057
Keywords: COVID-19, SARS-CoV-2, cardiac and vascular injuries, transcriptomic, DEG (differentially expressed gene) analysis
Citation: Hachim MY, Al Heialy S, Senok A, Hamid Q and Alsheikh-Ali A (2020) Molecular Basis of Cardiac and Vascular Injuries Associated With COVID-19. Front. Cardiovasc. Med. 7:582399. doi: 10.3389/fcvm.2020.582399
Received: 11 July 2020; Accepted: 18 September 2020;
Published: 03 November 2020.
Edited by:
Mireille Ouimet, University of Ottawa, CanadaReviewed by:
Giuseppe Danilo Norata, University of Milan, ItalyWai Ho Tang, Guangzhou Medical University, China
Copyright © 2020 Hachim, Al Heialy, Senok, Hamid and Alsheikh-Ali. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abiola Senok, QWJpb2xhLnNlbm9rQG1icnUuYWMuYWU=; Alawi Alsheikh-Ali, QWxhd2kuQWxzaGVpa2hhbGlAbWJydS5hYy5hZQ==
 Mahmood Yaseen Hachim
Mahmood Yaseen Hachim Saba Al Heialy
Saba Al Heialy Abiola Senok
Abiola Senok Qutayba Hamid2,3
Qutayba Hamid2,3 Alawi Alsheikh-Ali
Alawi Alsheikh-Ali