
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CASE REPORT article
Front. Cardiovasc. Med. , 06 August 2019
Sec. Cardiovascular Epidemiology and Prevention
Volume 6 - 2019 | https://doi.org/10.3389/fcvm.2019.00112
 Vassili Choutko1†
Vassili Choutko1† Vladimir Lazarevic2†
Vladimir Lazarevic2† Nadia Gaïa2
Nadia Gaïa2 Myriam Girard2
Myriam Girard2 Gesuele Renzi3,4
Gesuele Renzi3,4 Stefano Leo2
Stefano Leo2 Peter M. Keller5
Peter M. Keller5 Christoph Huber6
Christoph Huber6 Jacques Schrenzel2,3,4*
Jacques Schrenzel2,3,4*The most common causes of infective endocarditis (IE) are Staphylococcus, Streptococcus, Enterococcus, and HACEK-related organisms. In 15–30% of the IE cases, standard blood cultures remain sterile. We aimed at identifying the causative agent of a blood-culture-negative IE by whole metagenome shotgun sequencing (WMGS). A 54-year old woman diagnosed with community-onset pneumonia by a general practitioner, was admitted with dyspnea, cough and fever. The patient's blood cultures were repeatedly negative. The transesophageal echocardiography and transthoracic echocardiography showed an echo density on the left coronary leaflet of the aortic valve and signs suggestive of a ruptured abscess of the mitro-aortic junction. The patient underwent a semi-urgent aortic valve replacement by a mechanical prosthetic valve. We extracted DNA from the surgically-removed fresh valve tissue. The extraction procedure included bacterial/fungal DNA enrichment procedure. Nextera XT library prepared from the valve DNA extract was sequenced (2 × 250) on an Illumina MiSeq instrument. Sequence reads were mapped against bacterial genomic sequences, 16S rRNA genes and clade-specific taxonomic markers. Most of the 103,136 sequencing reads classified as bacterial were assigned to Neisseria meningitidis. In line with these data, mapping of reads against clade-specific and 16S rRNA gene markers revealed N. meningitidis as the most represented species. Assembled metagenomic fragments had the best average nucleotide identity (ANI) with N. meningitidis. Comparison of assembled contigs to reference alleles showed that this strain belongs to the ST-41/44 complex. N. meningitidis is commonly associated with meningitis and/or septicemia but should not be neglected as a causative agent of IE, which became exceedingly rare with the introduction of antibiotics. Our data show that WMGS may be used as a diagnostic procedure to strengthen the diagnosis of IE and to obtain draft genomic sequence of the pathogen and typing information.
Infective endocarditis (IE) remains a deadly disease despite significant progress in the diagnostic procedures and therapies. The most common cases of IE involve staphylococci, streptococci and enterococci as the preponderant microorganisms (>88%) recovered from blood cultures (1). The HACEK group (Haemophilus spp., Aggregatibacter spp., Cardiobacterium spp., Eikenella corrodens, and Klebsiella spp.) of fastidious oropharyngeal bacteria and other gram-negative organisms were reported to cause infection in 0.9–1.6% and 7.4% of IE patients with positive blood culture, respectively (1, 2).
A recent study reported that in 31% of the 918 cases suspected for IE (1), no causative microorganism could be grown using standard blood culture methods. Nevertheless, causative agents can be identified in ~50% of blood culture-negative patients (3) using other methods, including serology, the analysis of the excised valve by microscopy and PCR, and culture-based analyses of samples from the excised valve and other body sites. To increase the probability of pathogen identification, additional diagnostic tests have been proposed such as immunohistology, blood PCR, culture on specific media, prolonged culture period and tissue culture (4, 5). In 70–80% of the patients for whom pathogens remained unidentified by culture and other procedures, antibiotics had been given before the blood was taken for culture (3, 6).
Identifying the causative agent of IE is mandatory to initiate an appropriate antibiotic regimen. To increase the likelihood of identifying the pathogen, it is recommended to take three sets of blood cultures before an antibiotic therapy starts (7). The initial choice of empirical therapy is guided by many factors such as the nature (native or prosthetic) of the infected valve, the exposure to previous antibiotic therapy and the epidemiological type (community or nosocomial) of the infection.
Our case reports an unusual community-acquired blood-culture negative IE due to Neisseria meningitidis and details the next-generation sequencing (NGS) analysis performed to support this rare diagnosis.
A 54-year-old woman presented to the emergency department with dyspnea associated with cough and fever. Her past medical history included allergic asthma and an episode of pulmonary embolism 2 months prior to this hospitalization.
Dyspnea began 2 weeks before her arrival at the hospital. Because of the worsening of symptoms with the development of fever, the patient visited a general practitioner who diagnosed community-onset pneumonia and prescribed co-amoxicillin.
The next morning, she presented at the emergency department of our hospital because of increased dyspnea. Physical examination revealed sinus tachycardia at 110/min, central temperature of 39.2°C and a normal blood pressure of 113/69 mmHg. Cardiac examination showed a systolic murmur maximal at the aortic area and chest auscultation was compatible with left congestive heart failure. She had otherwise no systemic stigmata of infective endocarditis.
Blood tests revealed white blood cell 20.2 × 109/l (neutrophils 15.86 × 109/l, no left shift), hemoglobin 100 g/l, thrombocytes 407 × 109/l, urea 6.4 mmol/l, creatinine 91 μmol/l, C-reactive protein 119 mg/l. The coagulation tests were abnormal but difficult to interpret, since she was taking rivaroxaban for the last 2 months. Two sets (aerobic and anaerobic) of blood cultures were carried out (BD BACTEC™ FX blood culture system) on days 0 and 1.
A chest X-ray was suggestive of a pneumonia at the right pulmonary base but also revealed signs of congestive heart failure. The patient was admitted to the floor and an antibiotic therapy with co-amoxicillin and clarithromycin was started for a severe community acquired pneumonia. An electrocardiogram showed a first-degree heart block without other abnormalities.
Shortly after admission, the patient presented an oppressive chest pain that resolved spontaneously. The electrocardiogram showed flattened T waves in all derivations with the exception of V5-V6 where they were inverted. Cardiac troponin (measured by a high sensitivity assay) was elevated at 112 ng/l. Total creatine kinase was normal with a concentration of 97 U/l, with no significant changes found by repeating the measurement after a 4-h interval.
The patient was admitted to the intensive care unit and intubated. The transesophageal echocardiography (TEE) and transthoracic echocardiography (TTE), performed at day 1, showed an echo density of 14 mm on the left coronary leaflet of the aortic valve and signs suggestive of a ruptured abscess of the mitro-aortic junction that caused a severe aortic regurgitation.
Antibiotic therapy for severe community acquired pneumonia was replaced by ceftriaxone and gentamicin to empirically manage infective endocarditis due to penicillin-sensitive or -resistant streptococci (5). At day 2, after performing a full body and coronary computed tomography scan that did not show secondary lesions nor coronary pathology, the patient underwent a semi-urgent aortic valve replacement by a mechanical prosthetic valve. The cardiac abscess cavity was closed by a pericardial patch.
The cardiac surgeon reported that the valve had an unusual exophytic structure, like a cauliflower. The histological examination of the valve showed a myxoid degeneration of the leaflets with the presence of vegetations and no bacterial or fungal elements evidenced by Gram and periodic acid–Schiff (PAS) staining.
The blood cultures remained sterile 5 days after admission. In the meantime (day 4), the patient received in addition to ceftriaxone and gentamycin, doxycycline for empirical coverage of Coxiella burnetti, Brucella spp., and Bartonella henselae. The serologies for those pathogens revealed negative at day 13. A culture of the valve remained sterile on day 8, after a 6-day incubation. A broad range PCR on the valve tissue showed (day 5) the presence of Neisseria sp., that was confirmed by specific qPCR assays leading to the identification of N. meningitidis serogroup B by targeting ctrA (8), sodC (9), tauE, metA, shlA (10), and siaD markers (11) on day 9. A search for hypocomplementemia showed normal levels of C3 and C4.
The antibiotic regimen was changed (day 13) for ceftriaxone alone for a total duration of 6 weeks.
In this case report, the initial diagnosis of pneumonia led to some delay in diagnosing community-acquired IE. Retrospectively, we can attribute the symptoms of acute dyspnoea and fever to the IE complicated by an acute aortic valve insufficiency.
The fact that blood cultures remained sterile may be ascribed to the administration of one dose of co-amoxicillin prior to the patient's hospitalization and possibly the fastidious nature of the microorganism. The atypical clinical presentation associated with a pneumonia, made us suspect an infection due to Chlamydia pneumoniae, Mycoplasma pneumoniae, or Legionella pneumophila in addition to common IE-causing gram-positive cocci and bacteria from the HACEK group.
We tested the suitability of whole metagenomic shotgun sequencing, as a method that does not target specific species, for use in diagnosing bacterial infections (Supplementary Material). We extracted DNA from a 30-mg heart valve specimen using a procedure that enables enrichment of microbial DNA and therefore better assessment of bacterial community by NGS. The concentration of bacterial DNA in the sample extract (4.4 pg/μL) was roughly 10-fold lower than human DNA concentration (45.69 pg/μL), and ~20-fold higher than bacterial DNA concentration in the negative control (NEC) (0.23 pg/μL), as assessed by qPCR experiments.
Illumina sequencing followed by quality control checks, yielded 1,709,287 and 166,806 reads pairs for the sample and NEC metagenomic libraries, respectively. Most of the 103,136 reads classified by CLARK as bacterial in the sample data set were assigned to Neisseria (98.81%) at the genus level and, at the species level, to N. meningitidis (51.91%) (Figure 1). The most abundant organism from the genera other than Neisseria was Cutibacterium (Propionibacterium) acnes, a known reagent contaminant (12). Mapping of reads against clade-specific markers using MetaPhlAn2 revealed only two species: highly dominant N. meningitidis (99.38%) and unclassified Propionibacteriaceae (0.62%). In line with these analyses, the majority of reads mapping to 16S rRNA genes were classified to genus Neisseria with a k-mer based (Wang) approach and EzBioCloud reference 16S database. Taxonomic assignments of metagenomic 16S rRNA gene fragments, based on UBLAST-alignments, showed that both unique and multiple top hits were by far most frequently assigned to N. meningitidis.
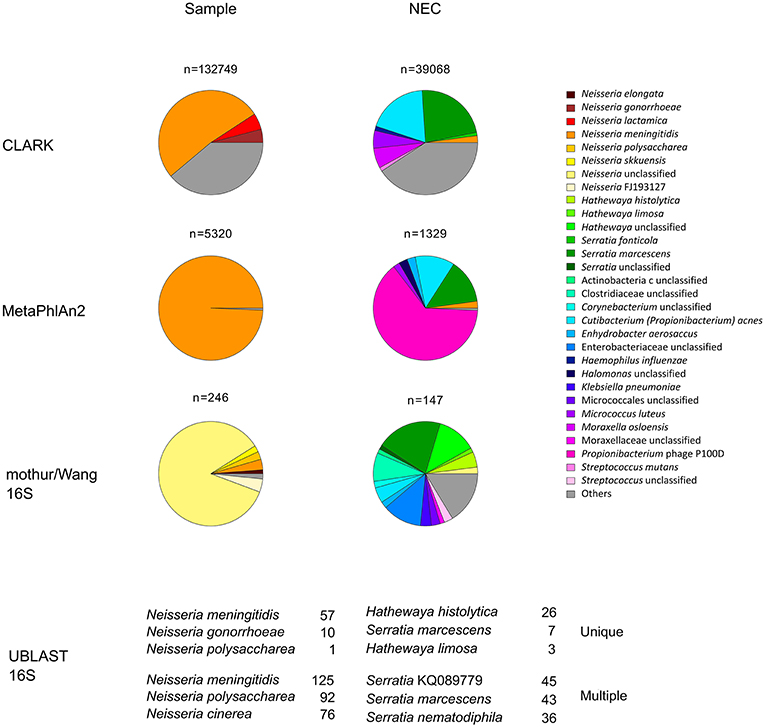
Figure 1. Relative abundance of prokaryotic taxa in metagenomic dataset. Proportions were determined by classifying at the species level sequences of genome fragments (CLARK), read mapping to clade markers (MetaPhlAn2) or by classifying 16S rRNA gene fragment sequences (mothur/Wang 16S and UBLAST 16S). All taxa with relative abundance <1% were summed up and represented as “others.” For simplicity, in the mothur/Wang 16S approach, only the results for the forward sequence reads are presented. They were further analyzed by UBLAST-alignments to the EzBioCloud reference 16S database (UBLAST 16S) and the top three most frequently identified species based on unique or multiple UBLAST hits are indicated. n, number of hits obtained for each sample and method used; NEC, negative extraction control.
Organisms other than N. meningitidis dominated the negative control. Small percentage of Neisseria reads identified in negative controls may be ascribed to cross-contamination during DNA extraction or sequencing library preparation, or to assignment of sequence reads to the wrong index during demultiplexing. By combining the results of bacterial DNA quantification from qPCR experiments with the relative abundance of bacterial taxa obtained by NGS (13), we estimated that the Neisseria DNA load in NEC was about 430 and 890 times lower than that of the valve specimen extract, for CLARK and MetaPhlAn2-based analyses, respectively.
The reads from the sample dataset assigned by CLARK to genus Neisseria were assembled into 362 contigs. The evaluation of assembled contigs with QUAST revealed that they covered 88.1% of the reference NZ-05/33 genome. The metagenomic fragments that mapped to the reference genome sequence (Figure 2) were more represented in the origin-half of the chromosome. Similar observations was made for Brucella melitensis DNA fragments found in a necrotic hepatic lesion of a patient with a chronic brucelloma (15). Such a pattern suggests that the infective agent was actively replicating (16, 17).
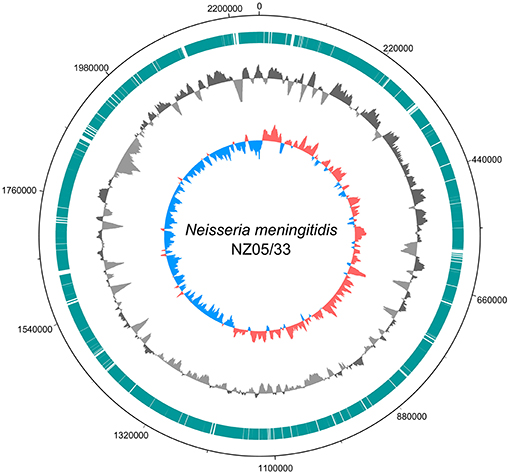
Figure 2. Ring diagram showing the genome of the reference N. meningitidis strain NZ-05/33 chromosome sequence (GenBank accession number NC_017518.1) and homologous metagenome sequences from this study. Circles depict (from outside to inside): chromosomal position; chromosomal regions to which mapped selected (classified by CLARK as belonging to genus Neisseria) metagenomic reads (turquoise bars); sequencing depth of the chromosomal regions obtained by mapping selected metagenomic reads (dark gray for above-average and light gray for below-average) using a 10,000 nt window in steps of 200 nt; GC skew with a 10,000 nt window in steps of 200 nt (red for positive, blue for negative). The figure was generated using DNAPlotter (14).
Assembled sequence contigs had average nucleotide identity (ANI) values of 97.03–99.47% with 1,368 N. meningitidis, 94.76–95.02% with 473 N. gonorrhoeae, and ≤94.52% with other Neisseria species isolates whose genomic sequences were available in the NCBI database. Comparison of assembled contigs to reference alleles from the Neisseria PubMLST database showed that the Neisseria strain in question perfectly matched (100% identity and 100% alignment) the seven MLST alleles (abcZ_3, adk_6, aroE_34, fumC_5, gdh_22, pdhC_6, pgm_9) of the ST-41/44 complex. This clonal complex is by far most frequently associated with serogroup B strains (18, 19).
Mapping of contigs to the ResFinder database did not reveal any acquired antimicrobial resistance gene but identified an aspartate at position 121 of the porin porB gene, that in N. gonorrhoeae contributes to a partial resistance to penicillins, tetracyclines, and cephalosporins (20).
The most common pathogen among Neisseria species causing IE remain Neisseria gonorrhoeae, with 71 cases reported since 1939, involving predominantly left-sided native valves in young males (21, 22), and Neisseria elongata with 18–24 cases (23, 24) described in literature. N. meningitidis can cause a broad spectrum of clinical manifestations and is largely known for worldwide diseases, such as purulent meningitis and/or septicemia associated with high morbidity and mortality. IE caused by N. meningitidis was more common before the antibiotic era with a trend of affecting the left side of the heart. Since 1960s, meningococcal IE has become a rare disease with only 13 reported cases (25). Nevertheless, N. meningitidis should not be neglected as a causative agent of IE. Our data show that whole metagenome shotgun sequencing (WMGS) may be used as a diagnostic procedure to strengthen the diagnosis of IE, especially in culture-negative cases, and to obtain draft genomic sequence of the pathogen and typing information.
The datasets generated for this study can be found in European Nucleotide Archive (ENA), PRJEB24753.
According to hospital protocol, no formal ethics approval was required. The patient agreed and provided written informed consent for publication of this case report.
VC, VL, CH, and JS analyzed and interpreted patient data. MG, NG, SL, GR, and PK performed the experiments. NG, SL, and VL analyzed the metagenomics data. VC, VL, and JS wrote the manuscript. All authors read and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2019.00112/full#supplementary-material
1. Fournier P-E, Gouriet F, Casalta J-P, Lepidi H, Chaudet H, Thuny F, et al. Blood culture-negative endocarditis: improving the diagnostic yield using new diagnostic tools. Medicine. (2017) 96:e8392. doi: 10.1097/md.0000000000008392
2. Chambers ST, Murdoch D, Morris A, Holland D, Pappas P, Almela M, et al. HACEK infective endocarditis: characteristics and outcomes from a large, multi-national cohort. PLoS ONE. (2013) 8:e63181. doi: 10.1371/journal.pone.0063181
3. Lamas CC, Eykyn SJ. Blood culture negative endocarditis: analysis of 63 cases presenting over 25 years. Heart. (2003) 89:258–62. doi: 10.1136/heart.89.3.258
4. Brouqui P, Raoult D. Endocarditis due to rare and fastidious bacteria. Clin Microbiol Rev. (2001) 14:177–207. doi: 10.1128/cmr.14.1.177-207.2001
5. Habib G, Lancellotti P, Antunes MJ, Bongiorni MG, Casalta J-P, Del Zotti F, et al. 2015 ESC Guidelines for the management of infective endocarditisThe Task Force for the Management of Infective Endocarditis of the European Society of Cardiology (ESC) endorsed by: European Association for Cardio-Thoracic Surgery (EACTS), the European Association of Nuclear Medicine (EANM). Eur Heart J. (2015) 36:3075–128. doi: 10.1093/eurheartj/ehv319
6. Houpikian P, Raoult D. Blood culture-negative endocarditis in a reference center: etiologic diagnosis of 348 cases. Medicine. (2005) 84:162–73. doi: 10.1097/01.md.0000165658.82869.17
7. Auckenthaler RW. Laboratory diagnosis of infective endocarditis. Eur Heart J. (1984) 5(Suppl. C):49–51. doi: 10.1093/eurheartj/5.suppl_c.49
8. Corless CE, Guiver M, Borrow R, Edwards-Jones V, Fox AJ, Kaczmarski EB. Simultaneous detection of Neisseria meningitidis, Haemophilus influenzae, and Streptococcus pneumoniae in suspected cases of meningitis and septicemia using real-time PCR. J Clin Microbiol. (2001) 39:1553–8. doi: 10.1128/jcm.39.4.1553-1558.2001
9. Dolan Thomas J, Hatcher CP, Satterfield DA, Theodore MJ, Bach MC, Linscott KB, et al. sodC-based real-time PCR for detection of Neisseria meningitidis. PLoS ONE. (2011) 6:e19361. doi: 10.1371/journal.pone.0019361
10. Diene SM, Bertelli C, Pillonel T, Jacquier N, Croxatto A, Jaton K, et al. Comparative genomics of Neisseria meningitidis strains: new targets for molecular diagnostics. Clin Microbiol Infect. (2016) 22:568.e561–8.e567. doi: 10.1016/j.cmi.2016.03.022
11. Mölling P, Jacobsson S, Bäckman A, Olcén P. Direct and rapid identification and genogrouping of meningococci and porA amplification by LightCycler PCR. J Clin Microbiol. (2002) 40:4531–5. doi: 10.1128/jcm.40.12.4531-4535.2002.
12. Lusk RW. Diverse and widespread contamination evident in the unmapped depths of high throughput sequencing data. PLoS ONE. (2014) 9:e110808. doi: 10.1371/journal.pone.0110808
13. Lazarevic V, Gaïa N, Girard M, Schrenzel J. Decontamination of 16S rRNA gene amplicon sequence datasets based on bacterial load assessment by qPCR. BMC Microbiol. (2016) 16:73. doi: 10.1186/s12866-016-0689-4
14. Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. (2009) 25:119–20. doi: 10.1093/bioinformatics/btn578
15. Lazarevic V, Gaïa N, Girard M, Leo S, Cherkaoui A, Renzi G, et al. When bacterial culture fails, metagenomics can help: a case of chronic hepatic brucelloma assessed by next-generation sequencing. Front Microbiol. (2018) 9:1566. doi: 10.3389/fmicb.2018.01566
16. Chandler MG, Pritchard RH. The effect of gene concentration and relative gene dosage on gene output in Escherichia coli. Mol Gen Genet. (1975) 138:127–41. doi: 10.1007/bf02428117
17. Korem T, Zeevi D, Suez J, Weinberger A, Avnit-Sagi T, Pompan-Lotan M, et al. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science. (2015) 349:1101–6. doi: 10.1126/science.aac4812
18. Yazdankhah SP, Kriz P, Tzanakaki G, Kremastinou J, Kalmusova J, Musilek M, et al. Distribution of serogroups and genotypes among disease-associated and carried isolates of Neisseria meningitidis from the Czech Republic, Greece, and Norway. J Clin Microbiol. (2004) 42:5146–53. doi: 10.1128/jcm.42.11.5146-5153.2004
19. Jounio U, Saukkoriipi A, Bratcher HB, Bloigu A, Juvonen R, Silvennoinen-Kassinen S, et al. Genotypic and phenotypic characterization of carriage and invasive disease isolates of Neisseria meningitidis in Finland. J Clin Microbiol. (2012) 50:264–73. doi: 10.1128/jcm.05385-11
20. Olesky M, Hobbs M, Nicholas RA. Identification and analysis of amino acid mutations in porin IB that mediate intermediate-level resistance to penicillin and tetracycline in Neisseria gonorrhoeae. Antimicrob Agents Chemother. (2002) 46:2811–20. doi: 10.1128/aac.46.9.2811-2820.2002
21. de Campos FPF, Kawabata VS, Bittencourt MS, Lovisolo SM, Felipe-Silva A, de Lemos APS. Gonococcal endocarditis: an ever-present threat. Autopsy Case Rep. (2016) 6:19–25. doi: 10.4322/acr.2016.037
22. Olayemi G, Oferczak M, Elagizi A, El-Abbassi I, Eschete M, Crowe J. Gonococcal endocarditis: the gift that stops giving! An uncommon presentation of a common disease. J La State Med Soc. (2017) 169:47.
23. Rossella P, Patrizia C, Fabio Oreste T, Renato T, Emanuele G, Vincenzo A, et al. Native mitral valve endocarditis caused by Neisseria elongata subsp. nitroreducens in a patient with marfan syndrome: first case in Italy and review of the literature. Case Rep Infect Dis. (2016) 2016:4956205. doi: 10.1155/2016/4956205
24. Samannodi M, Vakkalanka S, Zhao A, Hocko M. Neisseria elongata endocarditis of a native aortic valve. BMJ Case Rep. (2016) 2016:bcr2015213311. doi: 10.1136/bcr-2015-213311
Keywords: next-generation sequencing, cardiac valve, endocarditis, culture-negative infection, clinical metagenomics, Neisseria meningitidis
Citation: Choutko V, Lazarevic V, Gaïa N, Girard M, Renzi G, Leo S, Keller PM, Huber C and Schrenzel J (2019) Rare Case of Community-Acquired Endocarditis Caused by Neisseria meningitidis Assessed by Clinical Metagenomics. Front. Cardiovasc. Med. 6:112. doi: 10.3389/fcvm.2019.00112
Received: 23 April 2019; Accepted: 23 July 2019;
Published: 06 August 2019.
Edited by:
Ailin Barseghian, University of California, Irvine, United StatesReviewed by:
Christoph Sinning, Universitäres Herzzentrum Hamburg GmbH (UHZ), GermanyCopyright © 2019 Choutko, Lazarevic, Gaïa, Girard, Renzi, Leo, Keller, Huber and Schrenzel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jacques Schrenzel, amFjcXVlcy5zY2hyZW56ZWxAaGN1Z2UuY2g=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.