- 1Humanitas Clinical and Research Center, Milan, Italy
- 2Department of Molecular and Translational Medicine, University of Brescia, Brescia, Italy
Epigenetics refers to heritable mechanisms able to modulate gene expression that do not involve alteration of the genomic DNA sequence. Classically, mechanisms such as DNA methylation and histone modifications were part of this classification. Today, this field of study has been expanded and includes also the large class of non-coding RNAs (ncRNAs). Indeed, with the extraordinary possibilities introduced by the next-generation sequencing approaches, our knowledge of the mammalian transcriptome has greatly improved. Today, we have identifying thousands of ncRNAs, and unsurprisingly, a direct association between ncRNA dysregulation and development of cardiovascular pathologies has been identified. This class of gene modulators is further divided into short-ncRNAs and long-non-coding RNAs (lncRNAs). Among the short-ncRNA sub-group, the best-characterized players are represented by highly conserved RNAs named microRNAs (miRNAs). miRNAs principally inhibit gene expression, and their involvement in cardiovascular diseases has been largely studied. On the other hand, due to the different roles played by lncRNAs, their involvement in cardiovascular pathology development is still limited, and further studies are needed. For instance, in order to define their roles in the cellular processes associated with the development of diseases, we need to better characterize the details of their mechanisms of action; only then might we be able to develop innovative therapeutic strategies. In this review, we would like to give an overview of the current knowledge on the function of ncRNAs and their involvement in the development of vascular diseases.
Introduction
Originally, biologists assumed that the direction of information within a cell was a linear process going from DNA to RNA to protein and named this concept as the central dogma of molecular biology (1). However, in the last decades, the discovery of additional mechanisms able to modulate DNA transcription has clearly demonstrated that the layers of gene regulation are more complex. Modulation of gene expression might happen by direct methylation of DNA or alteration of DNA accessibility through histone modifications, events refer to as classical epigenetics. On the other hand, recent discoveries originating from the human genome project first, and then from the data generated through the next-generation sequencing approach, suggest that even more layers of regulation characterize this epigenetic landscape. Today, we know that only 2% of our DNA is translated into protein, and that the previously defined “junk RNA” is instead a fundamental part of the machinery involved in gene regulation (Figure 1). Thereby, it is clear that a new genomic era has started, and we can name this as the “non-coding RNA (ncRNA) revolution.” This provides an important new perspective on the centrality of RNA in gene regulation.
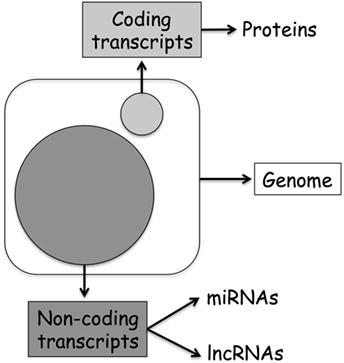
Figure 1. Human genome organization: coding vs. non-coding transcripts. The large circle represents ncDNA, while the small coding DNA.
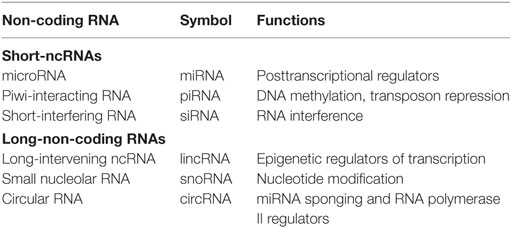
Non-coding RNAs can be divided into two main classes: small-ncRNAs (<200 nucleotides long), which includes microRNAs (miRNAs), piwi-interacting RNAs, and short-interfering RNAs; and long-non-coding RNAs (lncRNAs) (>200 nucleotides long), which include natural antisense transcripts, small nucleolar RNAs, and other types of lncRNAs (2–5) (Table 1).
Even though the biology of ncRNAs is not completely understood, the time to generate therapeutics based on RNA biology is fast approaching. Indeed, recent discoveries on the role of ncRNAs have completely changed our view of cell biology. Until few decades ago, we thought that phenotypes depended only on genetic variants, but the existence of non-Mendelian inheritance indicates that this view is not completely correct. In addition to this, the improvements in RNA stabilization chemistry and the ability to make informed decisions about where a defined molecule should perform its action is allowing progress in the generation of these new types of drugs. In this review, we focus in particular on the role of miRNAs and lncRNAs in vascular biology and disease.
microRNAs
microRNAs are a class of endogenous ncRNA between 16 and 29 nucleotides long. miRNAs are very well conserved through different species, but they do not always regulate the same genes in different organisms. Generally, miRNAs negatively regulate gene expression by binding to the 3′UTR of their target mRNAs, inhibiting transcription or translation (6). Even if rare, they might also block gene expression using alternative methods to 3′UTR binding, such as deadenylation of the polyA tail of the targeted mRNA (7). Furthermore, few studies demonstrated also that miRNAs might bind the promoter of some genes, enhancing RNA polymerase II (PolII) recruitment and thereby increasing gene transcription (8–10).
microRNAs are regarded as central players in physiology and pathophysiology of the vascular system (11–13). For different reasons, they are interesting targets for therapeutic intervention. A single miRNA can modulate hundreds of genes, including targets in the same pathway; thereby the modulation of their activity might have a major effect on cellular processes. miRNAs are essential regulators of vascular functions, playing a central role in endothelial cell (EC) and smooth muscle cell (SMC) biology (12–14). Finally, the idea to generate RNA therapeutics benefits from previous experience in the field of RNAi (15), that even though includes some specific limitations (16), might hugely impact the generation of a new class of drugs.
Vascular homeostasis is fundamental for the efficiency of the entire cardiovascular system. Pathological alteration of vascular ECs and SMCs leads to the development of different diseases, including atherosclerosis, restenosis, and even diabetes (17, 18). A dysfunctional endothelium contributes to increased stiffness of vessels and impaired distensibility of the arteries and also alters the amount of energy and oxygen transport to cardiac cells, consequentially triggering myocardial damage (19). It has been demonstrated that alteration of EC miRNA biogenesis by knocking down Dicer, a key enzyme involved in miRNA maturation, induces endothelial dysfunction (20, 21). miRNA-126, miRNA-92, and recently the miRNA-143/145 cluster have been shown to play a critical role on EC biology. Through zebrafish and mouse loss-of-function models, miRNA-126 has been shown to regulate the angiogenic ability of ECs. In both models, altered vessel organization during embryogenesis and adulthood was observed (22, 23). A negative effect on vessel angiogenesis is instead played by miRNA-92a: in a mouse model of myocardial infarction, forced modulation of its expression by antagomiRs accelerated vessel growth, and then recovery of cardiac functionality (24). Instead, miRNAs not usually expressed by ECs might be activated by alteration of laminar blood flow, such as the so-called SMC-specific miR-143/145 cluster. miRNA activation is regulated at the transcriptional level, by Kruppel-like factor 2 (KLF2) (25), and posttranscriptionally, by modulation of AMP-activated protein kinase α2, leading to EC-dependent vascular complications (26).
Alteration of the SMC miRNome might lead to vascular complications as well. For instance, neointimal formation, mainly due to SMC dysregulation, is a hallmark of many vascular diseases, including atherosclerosis (27) and pulmonary hypertension (28). The balloon injury model is a common-used model for the study of vascular restenosis, and many miRNAs have been identified by this approach. miRNA-21 has been shown to promote SMC proliferation and migration, inhibiting apoptotic pathways regulated by tropomyosin 1 (29), PTEN, and Bcl-2 (30). Transforming growth factor beta (TGFβ) is a central mediator of vascular remodeling processes (31) following acute injuries; miR-21 has been demonstrated to be regulated by TGFβ at the posttranscriptional level (32), highlighting the potential to generate RNA therapeutics vs. this miRNA. Indeed, through genetic ablation of miR-21 in the mouse, it was possible to reduce neointimal formation in vein graft failure (33) and in-stent restenosis models (34). Similar results were obtained using anti-miRNA technologies (35) (the alternative chemistries to generate therapeutics will be discussed later).
Among the list of the most expressed and fundamental miRNAs for SMCs is also the miRNA-143/145 cluster. It was originally associated with cancer development (36, 37), then more recently it has been demonstrated to play a central role on SMC biology, regulating SMC fate during development (38) and SMC phenotypic switch in adulthood (39). These features of the cluster have been demonstrated by different approaches, including gene therapy (40), but also through the generation of a knock-out mouse model (41–45), in which reduced neointimal formation was observed. Similar results have been obtained also by modulating miRNA-133 expression. Even though its expression in SMCs is not comparable to that of the miRNA-143/145 cluster, it is able to regulate SMC transition from the contractile to the proliferative phenotype. Indeed, its over-expression in rat carotid artery using adenoviral vectors reduced balloon injury-induced neointimal formation (46).
Cell-to-cell communication can be mediated by miRNAs transported by different carriers. Recent reports have shown the involvement of proteins, extracellular vesicles, and membrane structures in miRNA-mediated cell-to-cell cross talk. miRNAs, circulating in the blood stream protected by high-density lipoprotein, can be delivered to recipient cells where they inhibit target mRNAs (47); in addition, in plasma, miRNAs can form complexes with Argonaute 2 (48). Extracellular vesicles seem also to be involved in miRNA transport, and known players identified in this category are apoptotic bodies (diameter raging between 800 and 5,000 nm) (49) and exosomes (diameter < 150 nm) (50). Indeed, Zernecke and colleagues demonstrated that EC-derived apoptotic bodies are able to vehicle miRNA-126 to other cells in order to protect injured vessels from atherosclerosis development (51). Similarly, miRNA-126 might be transferred from ECs to SMCs also via exosomes to regulate cell turnover of the recipient cells (52). Actually, the importance of exosomes in cell-to-cell miRNA-mediated communication has been highlighted by many other reports. For instance, a recent work of Dimmeler’s group showed the involvement of these small vesicles in the transfer of the miRNA-143/145 cluster from ECs to SMCs (25). KLF2 induces the secretion of miRNA-143/145-enriched exosomes that are then taken up by SMCs, inducing an atheroprotective SMC phenotype through inhibition of specific target genes. In the opposite direction, miRNA-143 might, by transfer from SMCs to ECs of the pulmonary vasculature, induce a pro-migratory, angiogenic EC phenotype (53). Apart from exosome-mediated transfer, we recently showed that TGFβ triggers miRNA-143/145 cluster transfer from SMCs to ECs. We showed that its transfer is mediated by tiny cytoplasmic structures known as tunneling nanotubes. Once in the receiving cells, the miRNA-143/145 cluster modulates hexokinase 2 and integrin beta-8, negatively modulating EC proliferation and angiogenesis, thereby influencing vessel stabilization (54).
In summary, different miRNAs have been shown to be involved in vascular remodeling processes in vitro and in vivo. There are a lot of opportunities to develop therapeutics, but many facts of miRNA biology need to be elucidated in order to reduce potential long-term side effects.
Long-non-coding RNAs
Accumulating evidence suggests that lncRNAs play an important role in development and diseases through epigenetic control over gene expression (55, 56). LncRNAs are usually transcribed by PolII, spliced, 5′-capped, and 3′-polyadenylated, similarly to protein-coding mRNAs (57). The low level of lncRNA sequence homology between human and other species has been the subject of a long debate, but now conservation is usually evaluated in terms of secondary structure, highlighting a higher conservation level between species (58). Originally, it was thought that lncRNAs did not have any coding potential, while recent pioneer studies have demonstrated that they might actually encode for small peptides (59–61).
One way to classify lncRNAs is through their association with nearby protein-coding genes. The following are the most accepted categories of ncRNAs: (a) sense and (b) antisense, transcribed on the same or opposite strand of a protein-coding gene; (c) intronic and (d) intergenic, arising from an intron of a protein-coding gene or from a region located between two protein-coding genes; (e) enhancer, located in the enhancer region of a protein-coding gene; and (f) circular RNAs (circRNAs), a covalently closed RNA usually deriving from an alternative splicing of a protein-coding gene (Figure 2). LncRNAs play a wide range of roles in the cell, such as transcriptional and posttranscriptional regulation of gene expression. Based on their location, we distinguish between nuclear and cytoplasmic lncRNAs. In the nucleus, they might act as modifiers of chromatin, being involved in the spatial localization of DNA-associated proteins to specific genomic loci. For instance, they might show an enhancer-like activity, inducing transcription (62), or do the exact contrary, inhibiting gene expression by recruiting DNA and histone methyltransferases (63) or histone modifiers, such as the polycomb repressive complex 2 (64) and histone H3 lysine 9 methyltransferases (65). Cytoplasmic lncRNAs might work as activators or repressors of gene expression as well. Positive gene regulation might be obtained when working as miRNA sponges: specific miRNA-target genes are then de-repressed (66–68). On the contrary, protein inhibition might take place by binding to complementary mRNA sequences (69, 70).
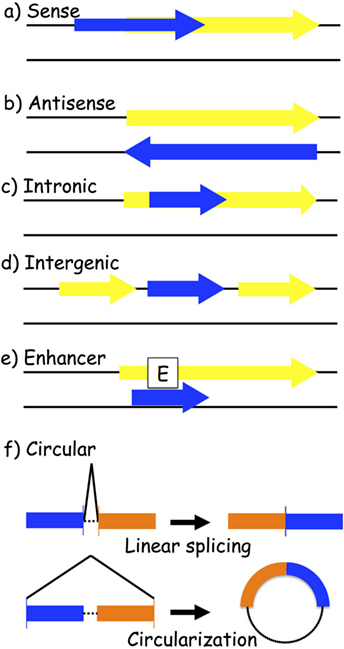
Figure 2. Biogenesis of long-non-coding RNAs (lncRNAs). LncRNA transcripts (blue box) are classified based on their genomic location in relation to the closest gene (yellow box): (a) sense lncRNAs are transcribed on the same strand as an exon; (b) antisense lncRNAs are transcribed on the opposite strand of an exon; (c) intronic lncRNAs; (d) intergenic lncRNAs are located between two different genes; (e) enhancer lncRNAs arise from an enhancer region of a protein-coding gene; and (f) circular RNAs are exonic or intronic sequences that circularize following an alternative splicing of linear transcripts.
In the last decade, our understanding on the roles of lncRNAs in pathologies such as cancer has grown exponentially, whereas knowledge on their role in vascular diseases remains limited.
Interaction between SMCs and ECs is critical for vessel physiology (71), thereby the identification of lncRNAs playing a role on both cell types is of great interest. For instance, the vascular-enriched lncRNA SENCR has been shown to regulate the differentiation status of SMCs, downregulating myocardin, and several contractile proteins. Moreover, SMCs with a decreased expression of SENCR acquire a pro-migratory phenotype, through the upregulation of midkine and pleiotrophin (72). On the other hand, the same lncRNA regulates proliferative, migratory, and angiogenic capabilities of ECs (73). Importantly, the same studies showed that expression of SENCR is altered in samples from patients with vascular diseases, including limb ischemia and coronary disease.
Evidence shows that lncRNA-p21 is downregulated in the coronary vessels of patients with heart disease (74). In vitro studies confirmed the ability of lncRNA-p21 to inhibit SMC proliferation and to induce cellular apoptosis. Similarly, lncRNA-p21 reduces EC proliferation and increases cellular apoptosis in in vitro models, mainly through the regulation of miRNA-130 (75). Moreover, Wu and colleagues have downregulated lncRNA-p21 in vivo, observing increased neointimal hyperplasia formation in injured carotids (74).
Smooth muscle cell-enriched or EC-enriched lncRNAs have been identified as well. Recently, Baker’s group, through RNA-sequencing analysis, identified an SMC-specific lncRNA named SMILR, which regulates SMC proliferation. The authors showed the clinical association between the level of SMILR in plasma and the stability of atherosclerotic plaques (76). Through a similar approach (RNA sequencing), several lncRNAs modulated by angiotensin II have been identified in SMCs. In particular, lnc-Ang362, a host transcript for miRNA-221 and miRNA-222, regulates the expression of these two miRNAs, resulting in a decrease of SMC proliferation (77).
Hypoxia regulates the metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) in ECs. Inhibition of MALAT1 in vitro activates migration of EC tip cells, while proliferation of EC stalk cells is disturbed, resulting in impaired angiogenesis, as observed also in retina vascularization and collateral vessel formation in ischemic limbs (78).
Very recently, a new class of lncRNAs named circRNAs has been identified. They can be either exonic or intronic in origin. CircRNAs derive from alternative splicing of linear genes, and they might play different functions in relation of their origin (79). Exonic circRNAs are predominately located in cytoplasm, and they have been shown to bind miRNAs, modulating their direct effects on specific target genes (80). For instance, circRNA-7 contains more than 70 binding sites for miRNA-7, acting as its natural cellular sponge (67, 68). On the other hand, intronic circRNAs are mainly located in the nucleus and are poorly enriched in miRNA-binding sites (79). A recent report by Zhang and colleagues showed an ability of circ-ankrd52 to regulate its parental gene by stabilizing the PolII machinery (81). Moreover, other reports showed the enrichment of circRNAs in exosomes; their detection the plasma of patients suggests a potential role as clinical biomarkers (82, 83).
However, as of today, only one study has been published on the role of circRNAs in vascular cells, with particular attention to regulation by hypoxia in ECs (84). This calls for a more detailed analysis of their role in vascular development and disease.
Therapeutic Prospective for ncRNAs
The need to translate ncRNA findings from bench to bedside is a direct consequence of the need to identify new therapeutic approaches to treat vascular diseases. For this purpose, different technologies are currently available. In particular, miRNA therapeutics have already entered phase I clinical studies.
In vivo modulation of miRNA expression might be reached in different ways. For instance, lentiviral or adenoviral vectors carrying specific miRNAs have been utilized in animal models of vascular diseases. Over-expression of miRNA-145 by this approach reduced neointimal formation and atherosclerosis development in rodents (40, 42, 85). However, even if this approach might be optimized for specific local delivery using stent technologies or similar, concerns have been raised on the use of this method in humans. Thereby, the most developed methodology for miRNA modulation is based on chemically modified oligonucleotides. In order to increase miRNA levels, mimic oligonucleotides might be used. In a model of pulmonary hypertension in mice, Montgomery and colleagues systemically delivered miRNA-29 conjugated with cholesterol, observing a reduction of pulmonary fibrosis (86).
On the other hand, normalization of an abnormally upregulated miRNA might be obtained utilizing antisense molecules. Different chemistries are available for this purpose: the first used technology was named “antagomiR,” in which the modified miRNA antisense oligo is conjugated to 2′-O-methyl-cholesterol, while the entire molecule is stabilized by phosphorothioate bonds (87). More recently, pharmaceutical companies have started to produce different oligonucleotides to improve miRNA targeting. For instance, Santaris patented a chemistry called “locked nucleic acids” (LNAs) in which the oligonucleotides are shorter than the target miRNA (8- or 15-base antisense molecules), thereby making them easier to deliver and without the need for cholesterol conjugation. Similarly, lncRNAs might be inhibited using molecules names GapmeRs: these are antisense oligonucleotides in which DNA monomers are flanked by LNA-modified sequences. The LNA stretches increase the stability of the oligonucleotide, while DNA gaps trigger the activity of RNase H due to the presence of DNA:RNA duplexes (88).
In term of clinical applications, there are only three ongoing clinical trials. The most advanced is a phase IIa clinical trial on anti-miRNA-122 by Santaris (http://ClinicalTrials.gov identifier: NCT01200420): the anti-miRNA-122 LNA inhibits hepatitis C replication through miRNA-122 reduction in liver (89–91). Mirna Therapeutics is instead running a phase I study in which miRNA-34a is delivered through liposomal injections to treat solid cancers in patients with liver involvement or inoperable primary liver cancer (http://ClinicalTrials.gov identifier: NCT01829971) (92). Finally, Regulus Therapeutics very recently started a phase I clinical study on an anti-miRNA-21 molecule to treat Alport syndrome, a kidney disease that alters the glomerular basement membrane (http://ClinicalTrials.gov identifier: NCT02855268) (93).
Several preclinical studies have confirmed the potentiality to develop miRNA-based therapeutics, and this has been confirmed by the above-described clinical trials. However, to take full advantage of this approach, many questions remain open: for example, what is the role of the miRNA passenger strand, since it has been demonstrated that in some cases it has a biological function. On the other hand, our knowledge on lncRNAs is still limited, in particular related to vascular diseases. Consequently, clinical application of lncRNA modifiers is much more distant and more studies are needed.
Author Contributions
LE and MQ equally contributed to the preparation of the submitted mini review.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer, CP, and handling Editor declared their shared affiliation and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
The authors would like to thank Michael V. G. Latronico and Gianluigi Condorelli for critically reviewing the manuscript.
Funding
This work was supported by grants from the Italian Ministry of Health (# GR2010_2302354) and Fondazione Veronesi to LE; grants from the Italian Ministry of Health (# GR2011_02349075) and CARIPLO Foundation (# 2014-1108) to MQ.
References
2. Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science (2008) 319:1787–9. doi: 10.1126/science.1155472
3. Collins LJ, Chen XS. Ancestral RNA: the RNA biology of the eukaryotic ancestor. RNA Biol (2009) 6:495–502. doi:10.4161/rna.6.5.9551
4. Collins LJ, Penny D. The RNA infrastructure: dark matter of the eukaryotic cell? Trends Genet (2009) 25:120–8. doi:10.1016/j.tig.2008.12.003
5. Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet (2009) 5:e1000459. doi:10.1371/journal.pgen.1000459
6. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell (2004) 116:281–97. doi:10.1016/S0092-8674(04)00045-5
7. Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA (2009) 15:21–32. doi:10.1261/rna.1399509
8. Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A (2008) 105:1608–13. doi:10.1073/pnas.0707594105
9. Qu H, Zheng L, Pu J, Mei H, Xiang X, Zhao X, et al. miRNA-558 promotes tumorigenesis and aggressiveness of neuroblastoma cells through activating the transcription of heparanase. Hum Mol Genet (2015) 24:2539–51. doi:10.1093/hmg/ddv018
10. Zhang Y, Fan M, Zhang X, Huang F, Wu K, Zhang J, et al. Cellular microRNAs up-regulate transcription via interaction with promoter TATA-box motifs. RNA (2014) 20:1878–89. doi:10.1261/rna.045633.114
11. Latronico MV, Elia L, Condorelli G, Catalucci D. Heart failure: targeting transcriptional and post-transcriptional control mechanisms of hypertrophy for treatment. Int J Biochem Cell Biol (2008) 40:1643–8. doi:10.1016/j.biocel.2008.03.002
12. Quintavalle M, Condorelli G, Elia L. Arterial remodeling and atherosclerosis: miRNAs involvement. Vascul Pharmacol (2011) 55:106–10. doi:10.1016/j.vph.2011.08.216
13. Elia L, Condorelli G. RNA (Epi)genetics in cardiovascular diseases. J Mol Cell Cardiol (2015) 89:11–6. doi:10.1016/j.yjmcc.2015.07.012
14. Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res (2008) 79:581–8. doi:10.1093/cvr/cvn156
15. Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nat Mater (2013) 12:967–77. doi:10.1038/nmat3765
16. Aagaard L, Rossi JJ. RNAi therapeutics: principles, prospects and challenges. Adv Drug Deliv Rev (2007) 59:75–86. doi:10.1016/j.addr.2007.03.005
17. Deanfield JE, Halcox JP, Rabelink TJ. Endothelial function and dysfunction: testing and clinical relevance. Circulation (2007) 115:1285–95. doi:10.1161/CIRCULATIONAHA.106.652859
18. Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev (2004) 84:767–801. doi:10.1152/physrev.00041.2003
19. Marti CN, Gheorghiade M, Kalogeropoulos AP, Georgiopoulou VV, Quyyumi AA, Butler J. Endothelial dysfunction, arterial stiffness, and heart failure. J Am Coll Cardiol (2012) 60:1455–69. doi:10.1016/j.jacc.2011.11.082
20. Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res (2007) 100:1164–73. doi:10.1161/01.RES.0000265065.26744.17
21. Suarez Y, Fernandez-Hernando C, Yu J, Gerber SA, Harrison KD, Pober JS, et al. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci U S A (2008) 105:14082–7. doi:10.1073/pnas.0804597105
22. Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell (2008) 15:272–84. doi:10.1016/j.devcel.2008.07.008
23. Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell (2008) 15:261–71. doi:10.1016/j.devcel.2008.07.002
24. Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science (2009) 324:1710–3. doi:10.1126/science.1174381
25. Hergenreider E, Heydt S, Treguer K, Boettger T, Horrevoets AJ, Zeiher AM, et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol (2012) 14:249–56. doi:10.1038/ncb2441
26. Kohlstedt K, Trouvain C, Boettger T, Shi L, Fisslthaler B, Fleming I. AMP-activated protein kinase regulates endothelial cell angiotensin-converting enzyme expression via p53 and the post-transcriptional regulation of microRNA-143/145. Circ Res (2013) 112:1150–8. doi:10.1161/CIRCRESAHA.113.301282
27. Hoglund VJ, Dong XR, Majesky MW. Neointima formation: a local affair. Arterioscler Thromb Vasc Biol (2010) 30:1877–9. doi:10.1161/ATVBAHA.110.211433
28. Jonigk D, Golpon H, Bockmeyer CL, Maegel L, Hoeper MM, Gottlieb J, et al. Plexiform lesions in pulmonary arterial hypertension composition, architecture, and microenvironment. Am J Pathol (2011) 179:167–79. doi:10.1016/j.ajpath.2011.03.040
29. Wang M, Li W, Chang GQ, Ye CS, Ou JS, Li XX, et al. MicroRNA-21 regulates vascular smooth muscle cell function via targeting tropomyosin 1 in arteriosclerosis obliterans of lower extremities. Arterioscler Thromb Vasc Biol (2011) 31:2044–53. doi:10.1161/ATVBAHA.111.229559
30. Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, et al. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res (2007) 100:1579–88. doi:10.1161/CIRCRESAHA.106.141986
31. Goumans MJ, Liu Z, ten Dijke P. TGF-beta signaling in vascular biology and dysfunction. Cell Res (2009) 19:116–27. doi:10.1038/cr.2008.326
32. Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature (2008) 454:56–61. doi:10.1038/nature07086
33. McDonald RA, White KM, Wu J, Cooley BC, Robertson KE, Halliday CA, et al. miRNA-21 is dysregulated in response to vein grafting in multiple models and genetic ablation in mice attenuates neointima formation. Eur Heart J (2013) 34:1636–43. doi:10.1093/eurheartj/eht105
34. McDonald RA, Halliday CA, Miller AM, Diver LA, Dakin RS, Montgomery J, et al. Reducing in-stent restenosis: therapeutic manipulation of miRNA in vascular remodeling and inflammation. J Am Coll Cardiol (2015) 65:2314–27. doi:10.1016/j.jacc.2015.03.549
35. Wang D, Deuse T, Stubbendorff M, Chernogubova E, Erben RG, Eken SM, et al. Local MicroRNA modulation using a novel anti-miR-21-eluting stent effectively prevents experimental in-stent restenosis. Arterioscler Thromb Vasc Biol (2015) 35:1945–53. doi:10.1161/ATVBAHA.115.305597
36. Akao Y, Nakagawa Y, Kitade Y, Kinoshita T, Naoe T. Downregulation of microRNAs-143 and -145 in B-cell malignancies. Cancer Sci (2007) 98:1914–20. doi:10.1111/j.1349-7006.2007.00618.x
37. Michael MZ, O’ Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res (2003) 1:882–91.
38. Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature (2009) 460:705–10. doi:10.1038/nature08195
39. Quintavalle M, Elia L, Condorelli G, Courtneidge SA. MicroRNA control of podosome formation in vascular smooth muscle cells in vivo and in vitro. J Cell Biol (2010) 189:13–22. doi:10.1083/jcb.200912096
40. Cheng Y, Liu X, Yang J, Lin Y, Xu DZ, Lu Q, et al. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ Res (2009) 105:158–66. doi:10.1161/CIRCRESAHA.109.197517
41. Boettger T, Beetz N, Kostin S, Schneider J, Kruger M, Hein L, et al. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J Clin Invest (2009) 119:2634–47. doi:10.1172/JCI38864
42. Elia L, Quintavalle M, Zhang J, Contu R, Cossu L, Latronico MV, et al. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ (2009) 16:1590–8. doi:10.1038/cdd.2009.153
43. Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF, et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev (2009) 23:2166–78. doi:10.1101/gad.1842409
44. Norata GD, Pinna C, Zappella F, Elia L, Sala A, Condorelli G, et al. MicroRNA 143-145 deficiency impairs vascular function. Int J Immunopathol Pharmacol (2012) 25:467–74. doi:10.1177/039463201202500216
45. Sala F, Aranda JF, Rotllan N, Ramirez CM, Aryal B, Elia L, et al. MiR-143/145 deficiency protects against progression of atherosclerosis in Ldlr-/-mice. Thromb Haemost (2014) 112:796–802. doi:10.1160/TH13-11-0905
46. Torella D, Iaconetti C, Catalucci D, Ellison GM, Leone A, Waring CD, et al. MicroRNA-133 controls vascular smooth muscle cell phenotypic switch in vitro and vascular remodeling in vivo. Circ Res (2011) 109:880–93. doi:10.1161/CIRCRESAHA.111.240150
47. Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol (2011) 13:423–33. doi:10.1038/ncb2210
48. Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A (2011) 108:5003–8. doi:10.1073/pnas.1019055108
49. Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol (2007) 35:495–516. doi:10.1080/01926230701320337
50. Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A (2016) 113:E968–77. doi:10.1073/pnas.1521230113
51. Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal (2009) 2:ra81. doi:10.1126/scisignal.2000610
52. Zhou J, Li YS, Nguyen P, Wang KC, Weiss A, Kuo YC, et al. Regulation of vascular smooth muscle cell turnover by endothelial cell-secreted microRNA-126: role of shear stress. Circ Res (2013) 113:40–51. doi:10.1161/CIRCRESAHA.113.280883
53. Deng L, Blanco FJ, Stevens H, Lu R, Caudrillier A, McBride M, et al. MicroRNA-143 activation regulates smooth muscle and endothelial cell crosstalk in pulmonary arterial hypertension. Circ Res (2015) 117:870–83. doi:10.1161/CIRCRESAHA.115.306806
54. Climent M, Quintavalle M, Miragoli M, Chen J, Condorelli G, Elia L. TGFbeta triggers miR-143/145 transfer from smooth muscle cells to endothelial cells, thereby modulating vessel stabilization. Circ Res (2015) 116:1753–64. doi:10.1161/CIRCRESAHA.116.305178
55. Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature (2009) 458:223–7. doi:10.1038/nature07672
56. Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A (2009) 106:11667–72. doi:10.1073/pnas.0904715106
57. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet (2016) 17:47–62. doi:10.1038/nrg.2015.10
58. Diederichs S. The four dimensions of noncoding RNA conservation. Trends Genet (2014) 30:121–3. doi:10.1016/j.tig.2014.01.004
59. Nelson BR, Makarewich CA, Anderson DM, Winders BR, Troupes CD, Wu F, et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science (2016) 351:271–5. doi:10.1126/science.aad4076
60. Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell (2015) 160:595–606. doi:10.1016/j.cell.2015.01.009
61. Magny EG, Pueyo JI, Pearl FM, Cespedes MA, Niven JE, Bishop SA, et al. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science (2013) 341:1116–20. doi:10.1126/science.1238802
62. Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell (2010) 143:46–58. doi:10.1016/j.cell.2010.09.001
63. Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J (2012) 31:522–33. doi:10.1038/emboj.2011.459
64. Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science (2008) 322:750–6. doi:10.1126/science.1163045
65. Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, et al. The air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science (2008) 322:1717–20. doi:10.1126/science.1163802
66. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell (2011) 146:353–8. doi:10.1016/j.cell.2011.07.014
67. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature (2013) 495:384–8. doi:10.1038/nature11993
68. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature (2013) 495:333–8. doi:10.1038/nature11928
69. Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature (2012) 491:454–7. doi:10.1038/nature11508
70. Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell (2012) 47:648–55. doi:10.1016/j.molcel.2012.06.027
71. Jain RK. Molecular regulation of vessel maturation. Nat Med (2003) 9:685–93. doi:10.1038/nm0603-685
72. Bell RD, Long X, Lin M, Bergmann JH, Nanda V, Cowan SL, et al. Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arterioscler Thromb Vasc Biol (2014) 34:1249–59. doi:10.1161/ATVBAHA.114.303240
73. Boulberdaa M, Scott E, Ballantyne M, Garcia R, Descamps B, Angelini GD, et al. A role for the long noncoding RNA SENCR in commitment and function of endothelial cells. Mol Ther (2016) 24:978–90. doi:10.1038/mt.2016.41
74. Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen C, et al. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation (2014) 130:1452–65. doi:10.1161/CIRCULATIONAHA.114.011675
75. He C, Ding JW, Li S, Wu H, Jiang YR, Yang W, et al. The role of long intergenic noncoding RNA p21 in vascular endothelial cells. DNA Cell Biol (2015) 34:677–83. doi:10.1089/dna.2015.2966
76. Ballantyne MD, Pinel K, Dakin R, Vesey AT, Diver L, Mackenzie R, et al. Smooth muscle enriched long noncoding RNA (SMILR) regulates cell proliferation. Circulation (2016) 133:2050–65. doi:10.1161/CIRCULATIONAHA.115.021019
77. Leung A, Trac C, Jin W, Lanting L, Akbany A, Saetrom P, et al. Novel long noncoding RNAs are regulated by angiotensin II in vascular smooth muscle cells. Circ Res (2013) 113:266–78. doi:10.1161/CIRCRESAHA.112.300849
78. Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res (2014) 114:1389–97. doi:10.1161/CIRCRESAHA.114.303265
79. Vicens Q, Westhof E. Biogenesis of circular RNAs. Cell (2014) 159:13–4. doi:10.1016/j.cell.2014.09.005
80. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell (2014) 56:55–66. doi:10.1016/j.molcel.2014.08.019
81. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell (2013) 51:792–806. doi:10.1016/j.molcel.2013.08.017
82. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res (2015) 25:981–4. doi:10.1038/cr.2015.82
83. Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One (2015) 10:e0141214. doi:10.1371/journal.pone.0141214
84. Boeckel JN, Jae N, Heumuller AW, Chen W, Boon RA, Stellos K, et al. Identification and characterization of hypoxia-regulated endothelial circular RNA. Circ Res (2015) 117:884–90. doi:10.1161/CIRCRESAHA.115.306319
85. Lovren F, Pan Y, Quan A, Singh KK, Shukla PC, Gupta N, et al. MicroRNA-145 targeted therapy reduces atherosclerosis. Circulation (2012) 126:S81–90. doi:10.1161/CIRCULATIONAHA.111.084186
86. Montgomery RL, Yu G, Latimer PA, Stack C, Robinson K, Dalby CM, et al. MicroRNA mimicry blocks pulmonary fibrosis. EMBO Mol Med (2014) 6:1347–56. doi:10.15252/emmm.201303604
87. Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature (2005) 438:685–9. doi:10.1038/nature04303
88. Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature (2012) 488:111–5. doi:10.1038/nature11362
89. Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science (2010) 327:198–201. doi:10.1126/science.1178178
90. Janssen HL, Reesink HW, Zeuzem S, Lawitz E, Rodriguez-Torres M, Chen A, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med (2013) 368:1685–94. doi:10.1056/NEJMoa1209026
91. Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med (2013) 368:1685–94. doi:10.1056/NEJMoa1209026
92. Bouchie A. First microRNA mimic enters clinic. Nat Biotechnol (2013) 31:577. doi:10.1038/nbt0713-577
Keywords: microRNAs, long-non-coding RNAs, vascular diseases, RNA therapeutics, gene expression
Citation: Elia L and Quintavalle M (2017) Epigenetics and Vascular Diseases: Influence of Non-coding RNAs and Their Clinical Implications. Front. Cardiovasc. Med. 4:26. doi: 10.3389/fcvm.2017.00026
Received: 16 January 2017; Accepted: 12 April 2017;
Published: 27 April 2017
Edited by:
Giovanni Esposito, University of Naples Federico II, ItalyReviewed by:
Alexander N. Kapustin, King’s College London, UKCinzia Perrino, University of Naples Federico II, Italy
Copyright: © 2017 Elia and Quintavalle. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leonardo Elia, bGVvbmFyZG8uZWxpYSYjeDAwMDQwO3VuaWJzLml0;
Manuela Quintavalle, bWFudWVsYS5xdWludGF2YWxsZSYjeDAwMDQwO2h1bWFuaXRhc3Jlc2VhcmNoLml0
 Leonardo Elia
Leonardo Elia Manuela Quintavalle
Manuela Quintavalle