
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Catal., 26 July 2023
Sec. Biocatalysis
Volume 3 - 2023 | https://doi.org/10.3389/fctls.2023.1231765
This article is part of the Research TopicExploiting the Power of Biocatalysis: Accessing Optimized Natural Products AnaloguesView all 5 articles
Introduction: The antimicrobial pyrrolnitrin from Pseudomonas strains is formed in four steps from tryptophan and comprises two flavin-dependent halogenases. Both PrnC and PrnA can carry out regioselective chlorination and bromination and are carrier protein-independent. Whilst the tryptophan halogenase PrnA has been studied in detail in the past, this study focuses on the pyrrole halogenating enzyme PrnC.
Methods: The halogenating enzyme PrnC, as well as the essential electron suppliers, the flavin reductases, have been produced soluble in E. coli. Furthermore, a screening of a rational compound library revealed that the pyrrole is essential for substrate recognition; however, the substitution pattern of the benzene ring is not limiting the catalysis.
Results and discussion: This renders PrnC to be a synthetically valuable enzyme for the synthesis of pyrrolnitrin congeners. For its natural substrate monodechloroaminopyrrolnitrin (MDA), the KM value was determined as 14.4 ± 1.2 µM and a kcat of 1.66 ± 0.02 min−1, which is comparable to other halogenases.
Almost 50 years ago, scientists still believed that halogenated compounds did not occur naturally but were artefacts from isolation processes. Twenty years later, the scientific community knew a lot more about the distribution of biosynthetic pathways of secondary metabolites that contain halogens in their scaffolds (Gribble, 2003; Gribble, 2004). Overviews on the plethora of halogenated compounds can be found in various reviews (van Pée, 1996; Büchler et al., 2019; Dachwitz et al., 2020; Menon et al., 2020). In many cases, it is indisputable for both natural compounds and active product ingredients that the impact of halogenations on biological activity can be instrumental or at least strongly modulate the activity (Lu et al., 2012; Latham et al., 2018; Costa et al., 2019; Bradley et al., 2020; Kinner et al., 2022). Furthermore, halogen moieties are vital for many follow-up functionalizations via transition-metal catalysis. These methods supported not only total syntheses but also biocatalytic and semisynthetic synthesis routes within the last 20 years (Schnepel et al., 2016; Baudoin, 2020). In parallel, considerable progress has been made in the investigation of halogenating enzymes and their mode of action. Many reviews showed the catalytic cycles of different classes, pointing out the bottlenecks of the different enzyme classes on the one hand and successful applications that have been established on the other hand (Blasiak and Drennan, 2008; Smith et al., 2013; Weichold et al., 2016; Latham et al., 2018; Büchler et al., 2019; Fejzagić et al., 2019; Minges and Sewald, 2020b; Menon et al., 2020).
With the discovery of flavin-dependent tryptophan-converting halogenases, regioselective halogenation was achieved (Hölzer et al., 2001; Dong et al., 2005; Zehner et al., 2005; Lingkon and Bellizzi, 2020), a feature that is not realized by the class of haloperoxidases which were known before the flavin-dependent halogenases (Fl-Hals) (Hohaus et al., 1997b; a). Today, Fl-Hals are well known in quantity and broaden our understanding of their mechanism (Minges and Sewald, 2020b). In addition to tryptophan, increasing numbers of phenol-converting Fl-Hals are being reported; however, in many cases, they lack strong selectivity, and fewer of them work in a carrier-independent manner as most described tryptophan-halogenases (Wagner et al., 2009; Menon et al., 2017; Mori et al., 2019; Menon et al., 2020).
Fl-Hals have already entered research stages of applicability rather than sheer understanding. Site-directed mutagenesis could vary regioselectivity (Lang et al., 2011; Shepherd et al., 2015; Andorfer and Lewis, 2018), and even upscaling by the use of cross-linked enzyme aggregates (CLEAs) to gram-scale conversions were possible (Frese and Sewald, 2015b; a). Finally, follow-up chemistry has already been carried out such as cross-coupling reactions in crude lysate (Gruß and Sewald, 2020). The first candidate that was found to halogenate tryptophan and its derivatives was PrnA (Hohaus et al., 1997b; a; Keller et al., 2000; Dong et al., 2005), the initial enzyme of the antimicrobial pyrrolnitrin (5) (van Pée et al., 1981; Hill et al., 1994; Hammer et al., 1997; Kirner et al., 1998; Lee and Zhao, 2007; Pawar et al., 2019) biosynthesis cluster from Pseudomonas fluorescens and related species (Scheme 1). It therefore was the basis for understanding the mechanistically relevant motifs, especially in solving the protein structure by X-ray crystallography (Dong et al., 2005).
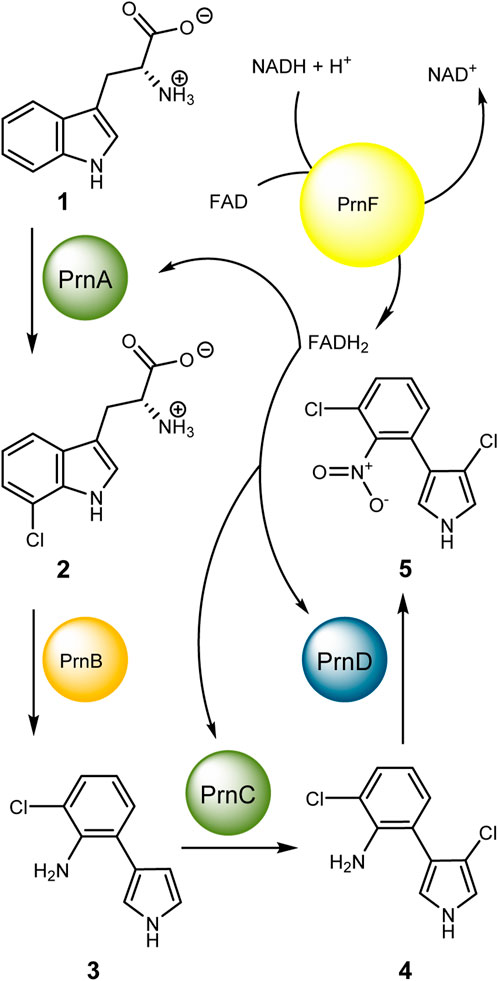
SCHEME 1. Biosynthetic pathway of pyrrolnitrin (PRN) from Pseudomonas fluorescens and related strains. After chlorination of tryptophan in position 7 by the Fl-Hal PrnA, PrnB carries out a ring rearrangement including a decarboxylation reaction to yield the 3-substituted aryl-pyrrole—MDA (3). PrnC, the second Fl-Hal of this pathway successively chlorinates position 4 of the pyrrole and therefore produces the substrate (APRN, 4) for PrnD and PrnF—a two-component system—with PrnD as arylamine N-oxygenase and PrnF as a reductase component. However, PrnF is not specific to PrnD and contributes to the halogenation reactions as well.
Little effort has been made to investigate the second halogenase from the pyrrolnitrin pathway. PrnC halogenates the pyrrole moiety of monodechloroaminopyrrolnitrin (3) (Hohaus et al., 1997b; a). Similar to flavin-dependent monooxygenases (Huijbers et al., 2014; Sucharitakul et al., 2014), Fl-Hals are not self-sufficient and, therefore, require flavin reductases (Fl-R) to provide NAD(P)H as redox equivalent for the oxidation of chloride to electrophilic “chloronium” species (Dong et al., 2005; Shepherd et al., 2015). The pyrrolnitrin cluster comprises PrnF as NADH:FAD-oxidoreductase (Lee and Zhao, 2007; Tiwari et al., 2012). With the detection of the four most important genes involved in pyrrolnitrin production—prnA to prnD (Kirner et al., 1998)—a heterologous expression of this cluster in Escherichia coli has been carried out (Hammer et al., 1997).
Investigations of the pathway with prnA substitutions revealed that PrnC does not convert MDA (3) analogs derived from 6-chloro-tryptophan rather than the natural intermediate 7-chloro-tryptophan (2) (Seibold et al., 2006). However, the 5- and 7-positions do not seem to be a sterical problem as methyl groups are accepted as well (Hamill et al., 1970). Similar results were found in feeding experiments when the 6-position of the former indole ring system was substituted by a moiety larger than fluorine or hydrogen, e.g., trifluoromethyl group (Chang et al., 1981; Kirner et al., 1998). Finally, the fermentation processes of Pseudomonas aureofaciens with sodium bromide yielded brominated derivatives of PRN (5) not only for the tryptophan halogenation but also for the pyrrole halogenation step (van Pée et al., 1983; Pawar et al., 2019).
Therefore, the objective of our study was to investigate PrnC as a purified enzyme concerning its substrate specificity and kinetic parameters. From our point of view, PrnC shows potential not only because of its strong regioselectivity in contrast to other pyrrole halogenases like HalB (Wynands and van Pee, 2004; Wynands, 2007) but also because of its carrier-independency unlike many other pyrrole-converting halogenases, e.g., PltA (Dorrestein et al., 2005) or Arm21 (Qiao et al., 2019; Menon et al., 2020). The combination of both properties seems to be unique for this pyrrole halogenase. To realize the application of PrnC, it is also necessary to establish the required electron supply chain and, ideally, a cofactor regeneration system.
Furthermore, the production of pyrrolnitrin (5), its intermediates, and functionalized aryl pyrroles generally allows for longer synthetic routes with (de)protection, halogen exchange, and cross-coupling reactions (Morrison et al., 2009). While aryl pyrroles can be addressed in shorter routes (Smith et al., 2002), late-stage halogenations by chemical reagents presumably suffer from lack of selectivity and, therefore, are part of this study too (Rodriguez et al., 2014; Song et al., 2019). The use of the halogenase PrnC may provide concise synthesis routes and the ability for late-stage decorations.
Gene amplification of the Pseudomonas protegens Pf-5 genome resulted in a construct, pET28a (+)-prnC-His9 (Supporting Material Supporting Material Section 1.2), suitable for gene expression in E. coli production strains. In accordance with literature for other Fl-Hals, especially with regard to expression of thal (Seibold et al., 2006) or prnA (Shepherd et al., 2015), E. coli ArcticExpress (DE3) was chosen for protein production to avoid inclusion bodies. Additional cold shock via temperature and induction by supplementation with 4% (v/v) ethanol (Salotra et al., 1995; Jones et al., 1996) yielded 6 g/L cell dry weight. Protein isolation via immobilized metal affinity chromatography (IMAC, see Supporting Material Section 1.4) and buffer exchange enabled the concentration of protein samples to approximately 4.3 mg/mL (65 µM) as maximum, whereas typical protein yields have been estimated as −1 mg/g cells [−15 mg/L culture], which translates to 8 mg (2 mg/mL; 30 µM). After verifying protein by MALDI-TOF analysis (see Supplementary Figure S2), the aim was to identify an optimal buffer system. The storage buffer was 25 mM phosphate buffer and 300 mM NaCl; pH 7.4 with 10% glycerol in accordance with literature information (Gao and Ellis, 2005; Lee and Zhao, 2007; Shepherd et al., 2015).
For initial experiments, flavin reduction was carried out by E. coli native reductase SsuE (see Supplementary Sections S1.3.2, 1.5.1). Problems of solubility and proper folding (Gao and Ellis, 2005) were encountered at the beginning, and the reductase was used as cleared lysates with an volumetric activity of 1.10 ± 0.06 U/mL (Gao and Ellis, 2005; Menon et al., 2016). However, to exclude limitations in kinetic studies, the pyrrolnitrin pathway reductase PrnF was produced in E. coli and yielded 7.7 mg/g cells and ca. 39 mg in total (2.76 mg/mL). The activity was determined to be 197 ± 21 U/mL [71 ± 8 U/mg; 25 s-1 (Tiwari et al., 2012)]. The latter was used for Michaelis–Menten kinetics and substrate scope investigation.
The natural substrate MDA has been synthesized and used for biocatalytic conversions. The optimization has been carried out using a factorial design. Initially, three different pH values 6, 7, and 8 have been tested for stability (see Supplementary Section S1.5). Considering the tendency of pyrroles to polymerize under acidic conditions (Lu et al., 2006; Joule and Mills, 2010) and the highest activity over time for pH = 7, the pH was tested ranging from neutral to slightly basic conditions.
Temperature (20°C and 40°C), the concentration of NADPH (50 μM and 100 µM), sodium chloride (10 mM and 100 mM), and FAD (10 μM and 500 µM), pH (7 and 8), and the activity of the flavin reductase SsuE (7 mU/mL and 34 mU/mL) (Gao and Ellis, 2005) (Table 1) can influence the overall conversion. To figure out both the importance of these factors and putative interactions, a factorial design has been used; the empirical levels are given in the aforementioned listing and in Table 1. The results are shown in detail in Supplementary Table S4. The influence of each factorial is shown in Figure 1. Overall, the conversion ranged from 0.16% to 100%. As proposed in the book “Novel Specific Halogenating Enzymes from Bacteria” by van Peé et al., our data underlined the findings that the halide source has an inhibitory effect on enzyme activity, and a pH value close to 7 is beneficial (Altmann et al., 1997). An increase in the substrate NADPH and temperature resulted in an increase in the conversion; in contrast, a lower concentration of FAD was beneficial. The same observation has been made for Thal previously (Moritzer and Niemann, 2019). The best results (−100%) were reproducibly obtained for the reaction condition 40°C, 7 mU/mL SsuE, pH 7, 10 µM FAD, 1,000 µM NADPH, and 10 mM sodium chloride. Even though full conversion could be obtained, the supply of the reductase SsuE is tedious, and this reductase could only be used as a crude extract due to solubility issues. The next optimization step was to change the electron supply from SsuE (E. coli) to the flavin reductase PrnF (Lee and Zhao, 2007; Tiwari et al., 2012), the native reductase from the biosynthetic cluster of pyrrolnitrin (5, P. protegens Pf-5). In accordance with literature, a soluble expression of ssuE (Gao and Ellis, 2005) was rather difficult in homologous vector-based overexpression. In contrast, prnF yielded soluble protein expressed in E. coli BL21 (DE3), enabling full conversion within minutes for higher substrate concentrations.
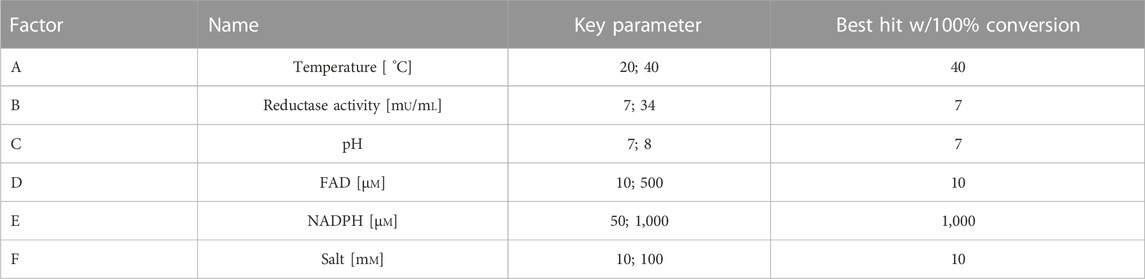
TABLE 1. Parameters of the factorial design of experiment (DoE) approach. Each parameter was tested with two values in combination with the others.
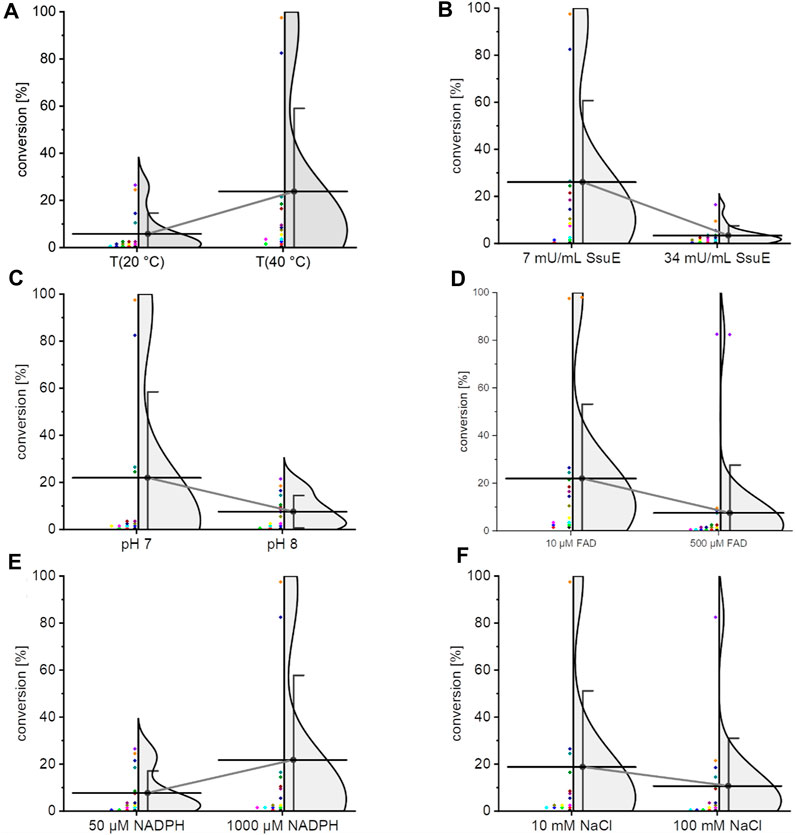
FIGURE 1. Statistical overview of the factorial design for the optimization of MDA conversion. Only the variation of one parameter is shown; the full composition is shown in Supplementary Table S4. Diamonds (♦) represent the data as conversion of MDA. The mean value ± standard deviation is indicated by a black circle (whiskers). The density fit (gray) is a Kernel density estimation. The individual experiments have been color-coded. To illustrate the respective tendency, the mean values were connected. The factorials are temperature (A), SsuE activity (B), pH (C), FAD concentration (D), NADPH concentration, (E) and sodium chloride concentration (F).
Under these conditions, steady-state kinetics have been recorded employing GC-MS (see Figure 2). The initial velocities of the formation of the halogenation product APRN (4) fit with the Michaelis–Menten equation (see Supplementary Section S1.5.3). PrnC offers a kcat of 1.66 ± 0.02 min-1 and a KM of 14.4 ± 1.2 µM, resulting in a catalytic efficiency of 1.9 ± 0.18 s-1mM-1 (Figure 3). PrnC showed a moderate tolerance for higher substrate concentrations. Even though the reaction rate was higher at 40°C, PrnC was deactivated faster. As the compounds MDA (3) and APRN (4) showed less stability under these conditions as well, we chose 30°C as an optimum temperature for these experiments (Supplementary Sections S1.4.4, 1.5.3). The stability of these compounds has not been quantified as this is a qualitative observation.
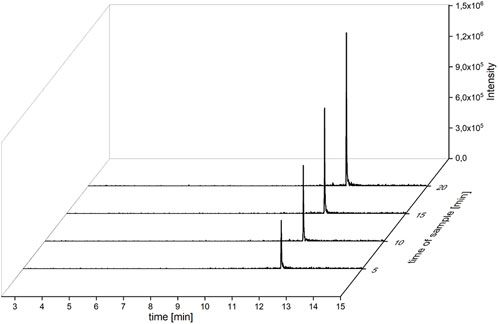
FIGURE 2. GC traces of APRN (4) after 5 to 20 min reaction course. This exemplary dataset for the determination of Michealis–Menten kinetics was taken at the concentration of 50 µm MDA (3). Depicted are the extracted ion chromatograms (EICs).
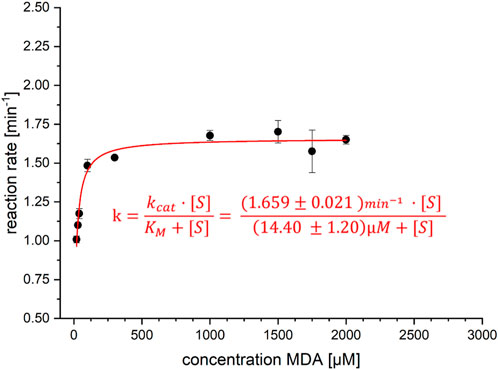
FIGURE 3. Michaelis–Menten kinetics for PrnC. Reactions were carried out with a total volume of 1 mL and stopped by adding 500 µL of ethyl acetate containing the internal standard (IS) m-chlorotoluene. Reaction rates were determined by an increase in the product area (APRN, 4) over time, normalized to the area of IS. By division of these normalized areas with the equation of the calibration y = 0.0019x + 0.0014 [area APRN/area IS · µM], activities in [µM/min] could be calculated. The reaction rates are calculated at the enzyme concentration of 1 mg/mL, and the molecular weight of 66,241 g/mol and kcat can be calculated. Analysis was based on GC-MS. The corrected R2 value is given as 0.941.
As it was shown in literature that the halogenation of tryptophan is not necessary for the conversion in the pyrrolnitrin (5) pathway (Chang et al., 1981; Kirner et al., 1998), we wanted to find out how substrate-specific PrnC halogenates with respect to the naturally occurring moieties (13, 14, and 15, see Scheme 2). Additionally, we tested one heterocycle as an aryl moiety (16) attached to the pyrrole, exchanged the pyrrole with a less reactive furan (20), and finally tested the aryl lacking pyrrole-2-carbaldehyde (19, Figure 4). As some of the compounds in these experiments are possibly less reactive, temperature was increased to 35°C (see Supplementary Section S1.6). Based on the conversion, the aryl moiety seems to be essential as no chlorination product could be detected for the pyrrole-2-carbaldehyde (19). The same is true for the furan (20) derivative of MDA (3). As this compound is less reactive, at least under these conditions, no halogenation was possible. However, it could also be anticipated that furan lacks the hydrogen-bridge donor, and, thus, there might be no activation within the active site. The conversion of the surrogate substrate with the pyridyl moiety (16) achieved 30% after 60 min. All other compounds showed full conversion in the given time, as no substrate was detectable anymore. However, it must be mentioned that for the 3-phenylpyrrole (15), no single but only double chlorination was found. This leads us to the hypothesis that in this case, no specific orientation of the substrate is possible in the active site.
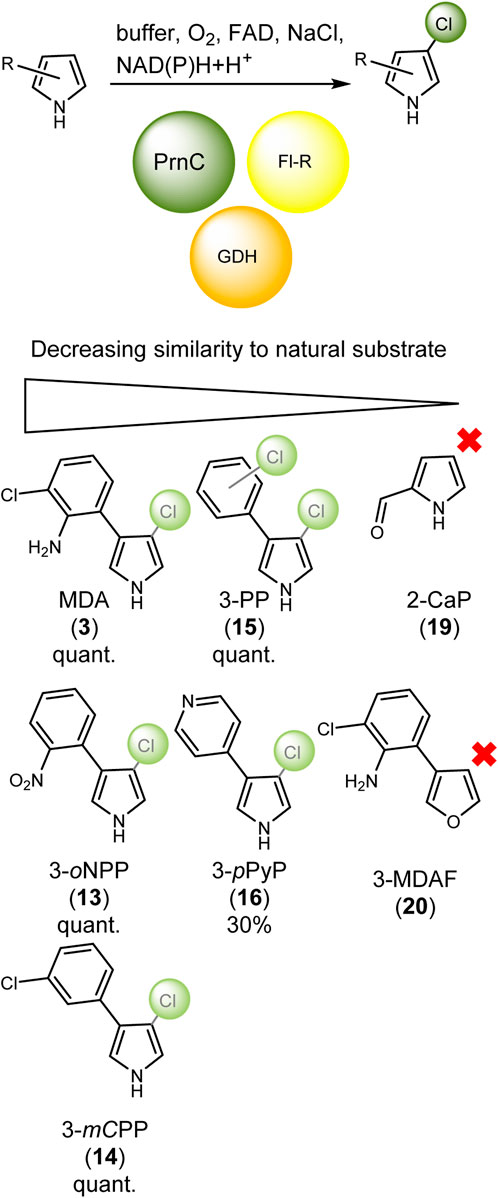
SCHEME 2. Simplified catalytic reaction for the chlorination of MDA via PrnC to produce APRN (4) and derivatives. PrnC is represented by the Fl-Hal in green, the flavin-reductase as Fl-R in yellow, and the cofactor recycling system for the glucose dehydrogenase (GDH) in orange. Surrogates are sorted by similarity to the natural substrate. The chlorination site is indicated in pale green (cf. Figure 4). Those substrates that were not converted are indicated with a red cross. Conversions are given as quantitative or for 3-mCPP as 30%.
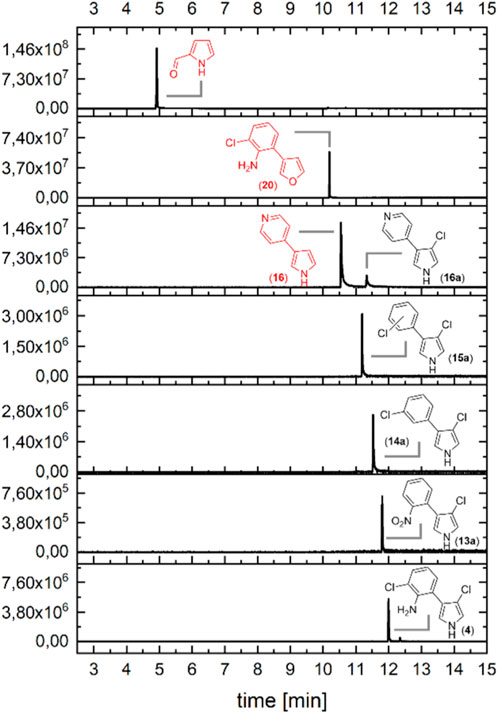
FIGURE 4. Conversion of MDA and derivatives after incubation with PrnC for 60 min at 35°C and 700 rpm in 0.5 mL. Extracted ion chromatograms (EICs) of the GC-MS analysis after extraction with 250 µL ethyl acetate containing the internal standard are shown. The substrates that were not completely or entirely converted are shown in red. Biocatalytic products of synthesized starting materials are labeled with an additional “a.” APRN (4), and the key structure was labeled according to Scheme 1.
To compare the biocatalytic reaction of PrnC to common chemical chlorination reactions, three typical reagents were used (see Supplementary Section S1.7). In the first approach, MDA (3) should be halogenated under similar conditions as biocatalysis, namely, under buffered conditions. Therefore, the enzyme solution has been replaced by hypochlorous acid (HOCl), N-chlorosuccinimide (NCS), and Palau’Chlor® (Supplementary Material). As these reactions did not yield any halogenation product, more typical conditions for halogenations were chosen. Therefore, we compared the reaction of MDA (3) in chloroform at 50°C with NCS and Palau’Chlor® as chlorine sources. In accordance with literature (Rodriguez et al., 2014), we chose elevated temperatures for the activation of the pyrrole ring in the first approach. However, this led to a mixture of four to five products (see Figure 5), and only approximately 1%–3% of APRN (4) was formed. The main product in both cases was the compound with a retention time of 11.53 min (66%–69%). To decrease side reactions and therefore the number of chlorination products, temperature was dropped to room temperature, and MDA was used in protected (3a) and unprotected (3) forms. It was possible to reduce the number of products to two and three. In the case of MDA (3), no APRN (4) was found, but almost selective chlorination (92%) of an unidentified product with a retention time of 11.84 min occurred. The reaction for protected MDA (3a) showed APRN (4) formation with 2%. Interestingly, the ratio of the side products changed to 24% (11.84 min) and 73% (12.3 min).
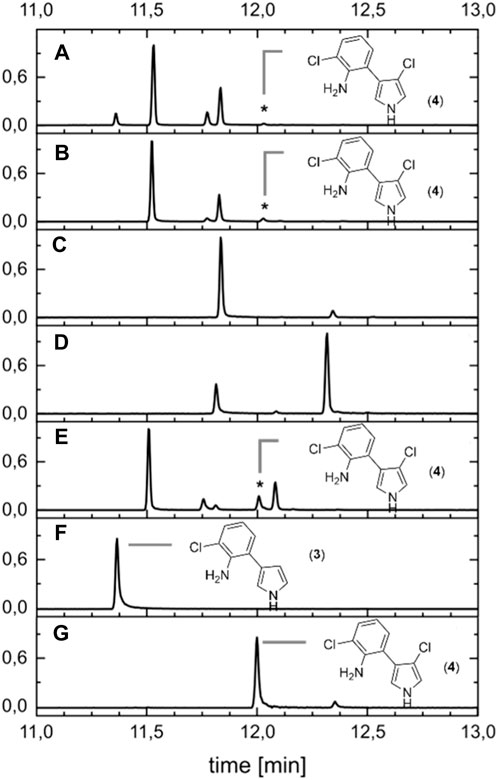
FIGURE 5. Chemical chlorination of protected and unprotected MDA with NCS and Palau’Chlor®. Chemical reaction did not yield any or only yielded a low percentage of APRN (*). (A) Chlorination of MDA at 50 °C with NCS. (B) Chlorination of MDA at 50 °C with Palau’Chlor®. (C) Reaction of MDA at ambient temperature. (D) Chlorination of N-TIPS-MDA with Palau’Chlor® and consecutive de-protection. (E) Chlorination of N-TIPS-MDA with NCS at 0°C and consecutive de-protection. (F) MDA as reference without reaction. (G) Biocatalytic conversion of MDA to APRN.
After tackling the problem of insolubility with the E. coli ArcticExpress (DE3) strain, we were able to detect initial activity for PrnC in reaction mixtures containing flavin reductase SsuE. However, the conversion rates at the beginning were very low, so we started optimization of the reaction conditions beginning with the pH value. To increase activity even further and consider interactions of different reaction compounds and parameters, we applied the factorial design of experiment approach. Even though the difference between the worst and best conditions was approximately 600-fold, we were not satisfied with the reaction rate. After replacing flavin reductase SsuE lysates with purified PrnF, full conversion of 60 µM MDA (3) was possible in less than 1 hour. The kinetic parameters were determined for PrnC with KM = 14.40 ± 1.20 µM and kcat = 1.66 ± 0.02 min-1 (kcat/KM = 0.115 ± 0.011 μM-1 min-1) due to synthesis of the natural product (4) and calibration (see Supplementary Section S1.5.5). These parameters were in range with those of PrnA (Lang et al., 2011; Minges and Sewald, 2020a), RebH (Dachwitz et al., 2020), and RadH (Menon et al., 2017), ranging from 0.0006 μM-1 min-1 to 12 μM-1 min-1 for various substrates, but pyrroles have not been reported. As the synthesis route of APRN (4) has a few bottlenecks and needs relatively high effort, we also wanted to compare the chemical late-stage halogenation reactions to the biocatalysis.
After finding a compromise between substrate and product instability at high temperatures and low reaction rates at low temperatures, we screened for surrogate substrates to figure out the substrate specificity of PrnC. As already stated in the literature using biosynthesis and knockout approaches, we wanted to verify and add insights about the substrate scope. This showed that substrates similar to natural MDA (3) were converted at approximately the same reaction rates. Thus, this enzyme is promiscuous enough to accept substrate derivatives, and this opens the possibility of using this enzyme for synthetic applications. In particular, the extraordinary regioselectivity is shown in comparison with chemical halogenation techniques, which yielded many by-products and were hardly able to form the natural product APRN (4). Here, we could show that even with screening for different parameters, including chlorination reagents, temperature, and protecting groups, it is either not possible to yield APRN (4) at all or if yielded, only in low amounts of a maximum of approximately 10%. To the best of our knowledge and referring to literature for immobilized reactions of tryptophan halogenases in a gram scale, PrnC is the only flavin-dependent halogenase that is able to address the position of aryl pyrroles selectively, and does not additionally require carrier proteins, thus indicating the need for continued investigation.
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
JG conducted the experiments and wrote the initial manuscript. TC and JP designed the scientific project, helped analyze the data, and revised the manuscript. All authors contributed to the article and approved the submitted version.
This project was realized within the Bioeconomy Science Center. The scientific activities of the Bioeconomy Science Center were financially supported by the Ministry of Culture and Science within the framework of the NRW Strategieprojekt BioSC.
The authors would like to thank Birgit Henßen, Bea Paschold, and Alexander Veljko Fejzagić for their support and precious discussions enabling this research.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fctls.2023.1231765/full#supplementary-material
Altmann, A., Burd, W., Hammer, P. E., Hohaus, K., Hill, D. S., Ligon, J. M., et al. (1997). “Novel specific halogenating enzymes from bacteria,” in Mechanisms of biohalogenation and dehalogenation. Editors D. B. Janssen, K. Soda, and R. Wever (Amsterdam, Netherlands: North-Holland Publishing Co: Verhandelingen Natuurkunde), 43–54.
Andorfer, M. C., and Lewis, J. C. (2018). Understanding and improving the activity of flavin-dependent halogenases via random and targeted mutagenesis. Annu. Rev. Biochem. 87, 159–185. doi:10.1146/annurev-biochem-062917-012042
Baudoin, O. (2020). Multiple catalytic C−H bond functionalization for natural product synthesis. Angew. Chem. Int. Ed. 59 (41), 17798–17809. doi:10.1002/anie.202001224
Blasiak, L. C., and Drennan, C. L. (2008). Structural perspective on enzymatic halogenation. Accounts Chem. Res. 42 (1), 147–155. doi:10.1021/ar800088r
Bradley, S. A., Zhang, J., and Jensen, M. K. (2020). Deploying microbial synthesis for halogenating and diversifying medicinal alkaloid scaffolds. Front. Bioeng. Biotechnol. 8, 594126. doi:10.3389/fbioe.2020.594126
Büchler, J., Papadopoulou, A., and Buller, R. (2019). Recent advances in flavin-dependent halogenase biocatalysis: Sourcing, engineering, and application. Catalysts 9 (12), 1030. doi:10.3390/catal9121030
Chang, C., Floss, H., Hook, D., Mabe, J., Manni, P., Martin, L., et al. (1981). The biosynthesis of the antibiotic pyrrolnitrin by Pseudomonas aureofaciens. J. Antibiotics 34 (5), 555–566. doi:10.7164/antibiotics.34.555
Costa, P. J., Nunes, R., and Vila-Viçosa, D. (2019). Halogen bonding in halocarbon-protein complexes and computational tools for rational drug design. Expert Opin. Drug Discov. 14 (8), 805–820. doi:10.1080/17460441.2019.1619692
Dachwitz, S., Widmann, C., Frese, M., Niemann, H. H., and Sewald, N. (2020). Enzymatic halogenation: Enzyme mining, mechanisms, and implementation in reaction cascades. London, United Kingdom: Royal Society of Chemistry.
Dong, C., Flecks, S., Unversucht, S., Haupt, C., Van Pee, K.-H., and Naismith, J. H. (2005). Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination. Science 309 (5744), 2216–2219. doi:10.1126/science.1116510
Dorrestein, P. C., Yeh, E., Garneau-Tsodikova, S., Kelleher, N. L., and Walsh, C. T. (2005). Dichlorination of a pyrrolyl-S-carrier protein by FADH2-dependent halogenase PltA during pyoluteorin biosynthesis. Proc. Natl. Acad. Sci. 102 (39), 13843–13848. doi:10.1073/pnas.0506964102
Fejzagić, A. V., Gebauer, J., Huwa, N., and Classen, T. (2019). Halogenating enzymes for active agent synthesis: First steps are done and many have to follow. Molecules 24 (21), 4008. doi:10.3390/molecules24214008
Frese, M., and Sewald, N. (2015a). Enzymatic halogenation of tryptophan on a gram scale. Angew. Chem. Int. Ed. 54 (1), 298–301. doi:10.1002/anie.201408561
Frese, M., and Sewald, N. (2015b). Enzymatische Halogenierung von Tryptophan im Gramm-Maßstab. Angew. Chem. 127 (1), 302–305. doi:10.1002/ange.201408561
Gao, B., and Ellis, H. R. (2005). Altered mechanism of the alkanesulfonate FMN reductase with the monooxygenase enzyme. Biochem. Biophysical Res. Commun. 331 (4), 1137–1145. doi:10.1016/j.bbrc.2005.04.033
Gribble, G. W. (2004). Natural organohalogens: A new frontier for medicinal agents? J. Chem. Educ. 81 (10), 1441. doi:10.1021/ed081p1441
Gribble, G. W. (2003). The diversity of naturally produced organohalogens. Chemosphere 52 (2), 289–297. doi:10.1016/s0045-6535(03)00207-8
Gruß, H., and Sewald, N. (2020). Late-stage diversification of tryptophan-derived biomolecules. Chemistry–A Eur. J. 26, 5328–5340. doi:10.1002/chem.201903756
Hamill, R., Elander, R., Mabe, J., and Gorman, M. (1970). Metabolism of tryptophan by Pseudomonas aureofaciens: III. Production of substituted pyrrolnitrins from tryptophan analogues. Appl. Microbiol. 19 (5), 721–725. doi:10.1128/am.19.5.721-725.1970
Hammer, P. E., Hill, D. S., Lam, S. T., Van Pée, K.-H., and Ligon, J. M. (1997). Four genes from Pseudomonas fluorescens that encode the biosynthesis of pyrrolnitrin. Appl. Environ. Microbiol. 63 (6), 2147–2154. doi:10.1128/Aem.63.6.2147-2154.1997
Hill, D., Stein, J., Torkewitz, N., Morse, A., Howell, C., Pachlatko, J., et al. (1994). Cloning of genes involved in the synthesis of pyrrolnitrin from Pseudomonas fluorescens and role of pyrrolnitrin synthesis in biological control of plant disease. Appl. Environ. Microbiol. 60 (1), 78–85. doi:10.1128/aem.60.1.78-85.1994
Hohaus, K., Altmann, A., Burd, W., Fischer, I., Hammer, P. E., Hill, D. S., et al. (1997a). NADH-abhängige Halogenasen sind wahrscheinlich eher an der Biosynthese von Halogenmetaboliten beteiligt als Haloperoxidasen. Angew. Chem. 109 (18), 2102–2104. doi:10.1002/ange.19971091829
Hohaus, K., Altmann, A., Burd, W., Fischer, I., Hammer, P. E., Hill, D. S., et al. (1997b). NADH-Dependent halogenases are more likely to Be involved in halometaolite biosynthesis than haloperoxidases. Angewandte Chemie Int. Ed. Engl. 36 (18), 2012–2013. doi:10.1002/anie.199720121
Hölzer, M., Burd, W., Reißig, H. U., and Pée, K. H. v. (2001). Substrate specificity and regioselectivity of tryptophan 7-halogenase from Pseudomonas fluorescens BL915. Adv. Synthesis Catal. 343 (67), 591–595. doi:10.1002/1615-4169(200108)343:6/7<591:aid-adsc591>3.0.co;2-e
Huijbers, M. M., Montersino, S., Westphal, A. H., Tischler, D., and van Berkel, W. J. (2014). Flavin dependent monooxygenases. Archives Biochem. Biophysics 544, 2–17. doi:10.1016/j.abb.2013.12.005
Jones, P. G., Mitta, M., Kim, Y., Jiang, W., and Inouye, M. (1996). Cold shock induces a major ribosomal-associated protein that unwinds double-stranded RNA in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 93 (1), 76–80. doi:10.1073/pnas.93.1.76
Joule, J. A., and Mills, K. (2010). Heterocyclic chemistry. England, UK: School of Chemistr The University of Manchester.
Keller, S., Wage, T., Hohaus, K., Hölzer, M., Eichhorn, E., and van Pée, K. H. (2000). Purification and partial characterization of tryptophan 7-halogenase (PrnA) from Pseudomonas fluorescens. Angew. Chem. 112 (13), 2300–2302. doi:10.1002/1521-3773(20000703)39:13<2300:aid-anie2300>3.0.co;2-i
Kinner, A., Nerke, P., Siedentop, R., Steinmetz, T., Classen, T., Rosenthal, K., et al. (2022). Recent advances in biocatalysis for drug synthesis. Biomedicines 10 (5), 964. doi:10.3390/biomedicines10050964
Kirner, S., Hammer, P. E., Hill, D. S., Altmann, A., Fischer, I., Weislo, L. J., et al. (1998). Functions encoded by pyrrolnitrin biosynthetic genes from Pseudomonas fluorescens. J. Bacteriol. 180 (7), 1939–1943. doi:10.1128/JB.180.7.1939-1943.1998
Lang, A., Polnick, S., Nicke, T., William, P., Patallo, E. P., Naismith, J. H., et al. (2011). Changing the regioselectivity of the tryptophan 7-halogenase PrnA by site-directed mutagenesis. Angewandte Chemie, Int. Ed. Engl. 50 (13), 2951–2953. doi:10.1002/anie.201007896
Latham, J., Brandenburger, E., Shepherd, S. A., Menon, B. R. K., and Micklefield, J. (2018). Development of halogenase enzymes for use in synthesis. Chem. Rev. 118 (1), 232–269. doi:10.1021/acs.chemrev.7b00032
Lee, J. K., and Zhao, H. (2007). Identification and characterization of the flavin:NADH reductase (PrnF) involved in a novel two-component arylamine oxygenase. J. Bacteriol. 189 (23), 8556–8563. doi:10.1128/JB.01050-07
Lingkon, K., and Bellizzi, J. J. (2020). Structure and activity of the thermophilic tryptophan-6 halogenase BorH. Chembiochem 21 (8), 1121–1128. doi:10.1002/cbic.201900667
Lu, G., Li, C., and Shi, G. (2006). Polypyrrole micro-and nanowires synthesized by electrochemical polymerization of pyrrole in the aqueous solutions of pyrenesulfonic acid. Polymer 47 (6), 1778–1784. doi:10.1016/j.polymer.2006.01.081
Lu, Y., Liu, Y., Xu, Z., Li, H., Liu, H., and Zhu, W. (2012). Halogen bonding for rational drug design and new drug discovery. Expert Opin. Drug Discov. 7 (5), 375–383. doi:10.1517/17460441.2012.678829
Menon, B. R. K., Brandenburger, E., Sharif, H. H., Klemstein, U., Shepherd, S. A., Greaney, M. F., et al. (2017). RadH: A versatile halogenase for integration into synthetic pathways. Angew. Chem. Int. Ed. 56 (39), 11841–11845. doi:10.1002/anie.201706342
Menon, B. R., Latham, J., Dunstan, M. S., Brandenburger, E., Klemstein, U., Leys, D., et al. (2016). Structure and biocatalytic scope of thermophilic flavin-dependent halogenase and flavin reductase enzymes. Org. Biomol. Chem. 14 (39), 9354–9361. doi:10.1039/c6ob01861k
Menon, B. R., Richmond, D., and Menon, N. (2020). Halogenases for biosynthetic pathway engineering: Toward new routes to naturals and non-naturals. Catal. Rev. 64, 533–591. doi:10.1080/01614940.2020.1823788
Minges, H., and Sewald, N. (2020a). Recent advances in synthetic application and engineering of halogenases. Chemcatchem 12 (18), 4450–4470. doi:10.1002/cctc.202000531
Minges, H., and Sewald, N. (2020b). Recent advances in synthetic application and engineering of halogenases. ChemCatChem 12, 4450–4470. doi:10.1002/cctc.202000531
Mori, S., Pang, A. H., Chandrika, N. T., Garneau-Tsodikova, S., and Tsodikov, O. V. (2019). Publisher Correction: Unusual substrate and halide versatility of phenolic halogenase PltM. Nat. Commun. 10 (1), 2053. doi:10.1038/s41467-019-09731-8
Moritzer, A.-C., and Niemann, H. H. (2019). Binding of FAD and tryptophan to the tryptophan 6-halogenase Thal is negatively coupled. Protein Sci. 28 (12), 2112–2118. doi:10.1002/pro.3739
Morrison, M. D., Hanthorn, J. J., and Pratt, D. A. (2009). Synthesis of pyrrolnitrin and related halogenated phenylpyrroles. Org. Lett. 11 (5), 1051–1054. doi:10.1021/ol8026957
Pawar, S., Chaudhari, A., Prabha, R., Shukla, R., and Singh, D. P. (2019). Microbial pyrrolnitrin: Natural metabolite with immense practical utility. Biomolecules 9 (9), 443. doi:10.3390/biom9090443
Qiao, Y., Yan, J., Jia, J., Xue, J., Qu, X., Hu, Y., et al. (2019). Characterization of the biosynthetic gene cluster for the antibiotic armeniaspirols in streptomyces armeniacus. J. Nat. Prod. 82, 318–323. doi:10.1021/acs.jnatprod.8b00753
Rodriguez, R. A., Pan, C.-M., Yabe, Y., Kawamata, Y., Eastgate, M. D., and Baran, P. S. (2014). Palau’chlor: A practical and reactive chlorinating reagent. J. Am. Chem. Soc. 136 (19), 6908–6911. doi:10.1021/ja5031744
Salotra, P., Singh, D. K., Seal, K. P., Krishna, N., Jaffe, H., and Bhatnagar, R. (1995). Expression of DnaK and GroEL homologs in Leuconostoc esenteroides in response to heat shock, cold shock or chemical stress. FEMS Microbiol. Lett. 131 (1), 57–62. doi:10.1111/j.1574-6968.1995.tb07754.x
Schnepel, C., Minges, H., Frese, M., and Sewald, N. (2016). A high-throughput fluorescence assay to determine the activity of tryptophan halogenases. Angew. Chem. Int. Ed. 55 (45), 14159–14163. doi:10.1002/anie.201605635
Seibold, C., Schnerr, H., Rumpf, J., Kunzendorf, A., Hatscher, C., Wage, T., et al. (2006). A flavin-dependent tryptophan 6-halogenase and its use in modification of pyrrolnitrin biosynthesis. Biocatal. Biotransformation 24 (6), 401–408. doi:10.1080/10242420601033738
Shepherd, S. A., Karthikeyan, C., Latham, J., Struck, A.-W., Thompson, M. L., Menon, B. R., et al. (2015). Extending the biocatalytic scope of regiocomplementary flavin-dependent halogenase enzymes. Chem. Sci. 6 (6), 3454–3460. doi:10.1039/c5sc00913h
Smith, D. R., Grüschow, S., and Goss, R. J. (2013). Scope and potential of halogenases in biosynthetic applications. Curr. Opin. Chem. Biol. 17 (2), 276–283. doi:10.1016/j.cbpa.2013.01.018
Smith, N. D., Huang, D., and Cosford, N. D. (2002). One-step synthesis of 3-aryl-and 3, 4-diaryl-(1 H)-pyrroles using tosylmethyl Isocyanide (TosMIC). Org. Lett. 4 (20), 3537–3539. doi:10.1021/ol0267073
Song, S., Li, X., Wei, J., Wang, W., Zhang, Y., Ai, L., et al. (2019). DMSO-catalysed late-stage chlorination of (hetero) arenes. Nat. Catal. 3, 107–115. doi:10.1038/s41929-019-0398-0
Sucharitakul, J., Tinikul, R., and Chaiyen, P. (2014). Mechanisms of reduced flavin transfer in the two-component flavin-dependent monooxygenases. Archives Biochem. Biophysics 555-556, 33–46. doi:10.1016/j.abb.2014.05.009
Tiwari, M. K., Singh, R. K., Lee, J.-K., and Zhao, H. (2012). Mechanistic studies on the flavin: NADH reductase (PrnF) from Pseudomonas fluorescens involved in arylamine oxygenation. Bioorg. Med. Chem. Lett. 22 (3), 1344–1347. doi:10.1016/j.bmcl.2011.12.078
van Pée, K. H., Salcher, O., and Lingens, F. (1981). Synthese von 7-Chlor-l-und 7-Chlor-D-tryptophan; Biosynthese von Pyrrolnitrin. Liebigs Ann. Chem. 1981 (2), 233–239. doi:10.1002/jlac.198119810207
van Pée, K.-H. (1996). Biosynthesis of halogenated metabolites by bacteria. Annu. Rev. ;icrobiology 50 (1), 375–399. doi:10.1146/annurev.micro.50.1.375
van Pée, K.-H., Salcher, O., Fischer, P., Bokel, M., and Lingens, F. (1983). The biosynthesis of brominated pyrrolnitrin derivatives by Pseudomonas aureofaciens. J. Antibiotics 36 (12), 1735–1742. doi:10.7164/antibiotics.36.1735
Wagner, C., El Omari, M., and Konig, G. M. (2009). Biohalogenation: nature’s way to synthesize halogenated metabolites. J. Nat. Prod. 72 (3), 540–553. doi:10.1021/np800651m
Weichold, V., Milbredt, D., and van Pee, K. H. (2016). Specific enzymatic halogenation-from the discovery of halogenated enzymes to their applications in vitro and in vivo. Angew. Chem. Int. Ed. 55 (22), 6374–6389. doi:10.1002/anie.201509573
Wynands, I. (2007), Detektion und Überexpression von Genen FADH2-abhängiger Halogenasen aus Actinoplanes sp. ATCC 33002. Dresden, Germany: Technische Universität Dresden.
Wynands, I., and van Pee, K. H. (2004). A novel halogenase gene from the pentachloropseudilin producer sp. ATCC 33002 and detection of in vitro halogenase activity. FEMS Microbiol. Lett. 237 (2), 363–367. doi:10.1016/j.femsle.2004.06.053
Keywords: flavin-dependent halogenase, halogenation, natural compound, biocatalysis, pyrrolnitrin
Citation: Gebauer J, Pietruszka J and Classen T (2023) Expression and characterization of PrnC—a flavin-dependent halogenase from the pyrrolnitrin biosynthetic pathway of Pseudomonas protegens Pf-5. Front. Catal. 3:1231765. doi: 10.3389/fctls.2023.1231765
Received: 30 May 2023; Accepted: 07 July 2023;
Published: 26 July 2023.
Edited by:
Mohammed Nabil Ahmed Khalil, Cairo University, EgyptReviewed by:
Max Julian Cryle, Monash University, AustraliaCopyright © 2023 Gebauer, Pietruszka and Classen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas Classen, dC5jbGFzc2VuQGZ6LWp1ZWxpY2guZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.