
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Conserv. Sci., 26 September 2022
Sec. Animal Conservation
Volume 3 - 2022 | https://doi.org/10.3389/fcosc.2022.971280
 Robert M. Zink1*
Robert M. Zink1* Luke B. Klicka2
Luke B. Klicka2More than 170 subspecies are listed as threatened or endangered under the US Endangered Species Act. Most of these subspecies were described decades ago on the basis of geographical variation in morphology using relatively primitive taxonomic methods. The US Fish and Wildlife Service defaults to subspecies descriptions by taxonomists working with specific groups of organisms, but there is no single definition of subspecies across plants and animals. Valid tests today usually entail molecular analyses of variation within and among populations, although there is no reason that behavioral, ecological or molecular characters could not be used, and include tests for significant differences between samples of the putative endangered subspecies and its nearest geographic relatives. We evaluated data gathered since subspecies listed under the ESA were described finding about one-third are valid (distinct evolutionary taxa), one-third are not, and one-third have not been tested. Therefore, it should not be assumed that because a subspecies occurs in a checklist, it is taxonomically valid. If the US Fish and Wildlife Service intends to continue listing subspecies, we suggest that they convene taxonomic experts representing various groups of organisms to provide a minimal set of criteria for a subspecies to be listed under the ESA.
Taxonomic assignments inevitably shape perceptions of biological diversity. Therefore, it is disconcerting that many subspecies and species descriptions trace to very limited information, often gathered in the last century, on the distributions of a small number of (usually morphological) traits with unknown genetic basis. Yet once a Latin binomial or trinomial is in the literature, the group of organisms to which it refers almost automatically assumes an aura of reality that may or may not be commensurate with its true evolutionary distinctiveness. Given the overriding importance of taxonomy on biodiversity recognition and management, increased attention should be devoted to taxonomic assessments (from molecular as well as other data) (Avise 1992).
The legislative basis for much of the conservation effort in the United States is the Endangered Species Act (ESA), passed in 1973 and modified in 1978, 1982, and 1988. Given its title, one might expect it to apply only to species, but in fact it also can be used to list subspecies and distinct population segments (vertebrates only) as threatened or endangered. Listing decisions usually come about when either a U.S. citizen or organization, or the Fish and Wildlife Service itself, determines that the population size of one of these taxonomic entities places it in danger of extinction (endangered) or in almost as much peril (threatened). The species or subspecies is placed on a list of candidate species, and the Fish and Wildlife Service is directed to use the best available scientific or commercial data in making a ruling as to whether the taxon merits listing as endangered or threatened. In this paper we examine the taxonomic category or rank of subspecies. In particular, we determine whether modern tests of subspecies limits have confirmed the validity of listed subspecies, most of which were described more than a half-century ago, as described by National Academy Member John C. Avise in the opening quote.
Systematists, taxonomists, and evolutionary biologists have struggled to define the term species for a century and a half. The biological species concept recognizes a species as a diagnosably distinct population or group of populations that is reproductively isolated from other such populations or groups. Lineage concepts, on the other hand, such as the phylogenetic species concept, recognize diagnosable populations or groups of populations as species irrespective of whether they can hybridize with other such groups (Cracraft, 1983; de Queiroz, 2007). That is, diagnosable subspecies of biological species would more than likely be considered phylogenetic species (Barrowclough et al., 2016). Many other species concepts have been offered, including recent ones that search for congruence among multiple loci or character sets: so-called species-delimitation approaches (Malaney et al., 2017). Below the level of the species, some species concepts recognize subspecies.
A subspecies is a formal taxonomic category that is specified by three Latin names: the genus name, the species name, and the subspecies name. Definitions of subspecies range from whatever a taxonomist says is valid to multi-character genetic and morphological assessments (Zink, 1997). Some favor a rule in which 75% of individuals in a subspecies must be separable from 99% in another subspecies—clearly an arbitrary standard. Taylor et al. (2017) suggest that “a subspecies is a population, or collection of populations, that appears to be a separately evolving lineage with discontinuities resulting from geography, ecological specialization, or other forces that restrict gene flow to the point that the population or collection of populations is diagnosably distinct”. We believe that this definition provides sufficient criteria for recognizing a subspecies as valid and potentially qualified to be listed under the ESA should it become threatened or endangered. Although subspecies are listed under the ESA, the US Fish and Wildlife Service (USFWS) has no set definition of subspecies, instead relying on peer-reviewed literature that reflects the views of taxonomists in different groups. The USFWS maintains a website called the Integrated Taxonomic Information System (ITIS) where subspecies are classified as valid or invalid.1 According to the USFWS, “ITIS taxonomy is based on the latest scientific consensus available and is provided as a general reference source for interested parties” but the evidence for subspecies validity is not given, and the evaluations cannot be verified. It would be beneficial to all involved if the USFWS would at least provide a minimal set of criteria for determining whether a subspecies is listable. For a review of historical views on subspecies, see Supplementary Information.
Rigorous descriptions of subspecies has not historically been the status quo. Consider the Rio Grande subspecies (Meleagris gallopavo intermedia) of the North American wild turkey. The subspecific part of the scientific name, intermedia, was justified by the author (Sennett, 1879) because it was his opinion that the turkey’s appearance was “intermediate” between two other subspecies. Exactly where it starts and stops being intermediate was not noted. To evaluate subspecies, one needs to refer to the specimens on which the original descriptions were based. Although an outlier, the subspecies of white-tailed deer (Odocoileus virginianus leucurus) from the Columbia River area was described on the basis of a single specimen, which was later consumed by the hunter who harvested it, and no remains were deposited as a museum voucher specimen (Gavin and May, 1988).
There are examples in which subspecies correspond to genetically or morphologically defined units that have experienced evolutionarily independent histories and therefore qualify for listing under the ESA. For example, the spotted owl (Strix occidentalis) has three subspecies: the northern spotted owl (S. o. caurina), California spotted owl (S. o. occidentalis), and Mexican spotted owl (S. o. lucida). Barrowclough et al. (2006, 2011) show that each subspecies is genetically distinct, with a narrow hybrid zone between northern and California spotted owls. Vázquez-Miranda et al. (2017) show that subspecies of the LeConte’s thrasher (Toxostoma lecontei) found in the Vizcaino Desert of Baja California, and populations to the north, are genetically distinct and qualify as units worthy of conservation status. Catanach et al. (2021) provide a textbook example of how a subspecies should be tested with modern methods. They examined the status of the hawk Accipter straitus venator from Puerto Rico using ultra-conserved elements (nuclear DNA), mitochondrial DNA (mtDNA), and morphology. Their study shows that the specimens from the island formed a discrete genetic cluster, and in fact they suggest A. s. venator be raised to a full species. These are three examples of subspecies that meet the criteria of Taylor et al. (2017). For a discussion of the views of scientific societies on subspecies see Supplementary Information.
Remsen (2005) and Taylor et al. (2017) make it clear that a valid subspecies should be a discrete taxonomic entity with diagnostic boundaries defined by concordant patterns of morphology or genetics. Others (e.g., Haig et al., 2006; Winker, 2010) suggest that subspecies are not discretely differentiated populations but can have “fuzzy” edges owing either to ongoing introgression (gene flow) or to insufficient time having elapsed since the cessation of genetic exchange (the so-called lag effect). Winker (2010) considers subspecies a gold mine of testable hypotheses in evolutionary biology. Indeed, this can be an important function of subspecies, but such subspecies should not be construed as worthy of conservation status under the ESA—only those that are discretely differentiated should be considered worthy of conservation status. The reason is that otherwise, there will be thousands of such arbitrarily defined subspecies that could be accorded taxonomic trinomials and therefore qualify for listing.
Barrowclough (1982) wrote “A named subspecies carries at least the connotation of phenotypic uniformity over an area” and added “a useful subspecies concept will have to have as a goal the same objective as other taxonomic categories-predictiveness”. By predictiveness, Barrowclough (1982) noted that a subspecies must be supported by a “concordance of geographically varying characters that do not simply form clines”. A strong test of a listed subspecies, as envisioned by Taylor et al. (2017), would include the comparison of statistically adequate samples from throughout listed subspecies with samples of other subspecies, preferably those geographically adjacent. Listed subspecies should have at least two geographically spaced samples (if possible), allowing a researcher to test whether each sample is more closely related to the other than to samples from other subspecies. There should be no sampling gaps that would give the illusion of real genetic or morphological discontinuities (see Rising, 2001) owing simply to geographic distance between sampling localities. Evidence of taxonomic distinctiveness could be gathered from several character systems, including morphological, behavioral, molecular or ecological, with preference perhaps given to modern molecular methods. All data must be publicly available, and the analyses must be clearly described. The data should show concordant geographic splits in multiple character systems (Barrowclough, 1982), which would confirm a hypothesis of evolutionary independence. This sets a high bar for taxonomic descriptions of subspecies (Luo et al., 2018).
Molecular methods have revolutionized tests of subspecies and their evolutionary independence (Avise, 1992). The foundation of the ESA rests on the assumption that listed entities are evolutionarily independent. If one examines morphological characters, which are likely under strong selection, one does not expect a single evolutionary history to emerge. The reason is that characters often respond idiosyncratically to opposing environmental dimensions, and therefore picking one morphological character to draw subspecies boundaries ignores alternative patterns in other characters. Only when a suite of morphological or genetic characters all show the same pattern can one safely infer that the pattern reflects the history of population subdivision. It is also the case that some valid, evolutionarily distinct lineages (e.g., subspecies) have experienced morphological stasis and represent cryptic taxa, and only molecular datasets reveal their existence (Moyle and Campbell, 2022). If this is widespread it raises the question of whether all declining populations need to be studied irrespective of degree of morphological distinctiveness using genomics.
Unlike morphological characters, molecular characters used to date are often considered “selectively neutral”—that is, not influenced unduly by natural selection—and hence the only reason for congruent geographic patterns is that they reflect a common underlying evolutionary history. Patten and Remsen (2017) claimed that neutral genetic characters should not be expected to map to subspecies boundaries. However, subspecies described by morphology often are inconsistent with evolutionarily independent groupings because the one or two characters used to in subspecies description do not reflect the historical pattern of population fragmentation. That is, characters responding to selection gradients might not be concordant with the history of population isolation revealed by selectively neutral characters. It is not surprising that many subspecies lack neutral molecular genetic support. However, it is not the genetics that failed; instead, the conflicting patterns among morphological characters result in their failure to reflect true evolutionary patterns, those upon which subspecies should be based.
New molecular methods, often-called next-gen, have resulted in the possibility of surveying thousands to millions of loci, often in the form of single nucleotide polymorphisms. The El Segundo blue butterfly (Euphilotes battoides allyni) is a federally listed subspecies found along the coast of southern California. Dupuis et al. (2020) used a sophisticated molecular analysis of 54,305 SNPs and found that this subspecies is distinct. However, north and south along the coast are six additional, equally distinct genetic clusters. Either there are too few subspecies of E. battoides or the newer techniques will find minor differences of statistical importance irrespective of subspecies boundaries—differences of tenuous biological significance.
Saglam et al. (2017) used nuclear genomics to test subspecies limits in two trout, Oncorhynchus clarkii seleniris and O. c. henshawi, which were ambiguous with mtDNA. Their analyses of 500,000 reads per individual found that both subspecies were highly distinctive. A subspecies of great interest to conservation biologists is the southwestern willow flycatcher (Empidonax traillii extimus). In the only authoritative statement on subspecies in North America, the American Ornithologists’ Union (1957) Checklist, this subspecies was not accepted, although it had been described nine years earlier (Phillips, 1948). Data sets on mtDNA, amplified fragment length polymorphisms, niche modeling, and song vocalizations supported the AOU’s decision to not designate E. t. extimus as a valid subspecies (Zink, 2015; Zink, 2016; see Theimer et al., 2016; Mahoney et al., 2020). Ruegg et al. (2021) analyzed variation in 105,000 single nucleotide polymorphisms (SNPs) from 175 individuals and concluded that the subspecies was valid. However, there are sampling gaps between E. t. extimus and the subspecies to the north (E. t. adastus), there are no samples from the southern extent of the range in Mexico (see below), and there is no assessment of isolation by distance. The samples from California, within the range of E. t. extimus, do not group with those in the eastern part of the subspecies’ range. Thus, Ruegg et al. (2021) found geographic differentiation in genetic variation throughout the range of the species, but whether the data recover the limits of E. t. extimus as described by Phillips (1948), and that which is listed under the ESA, is unclear. Similarly, Vandergast et al. (2022) claimed that the coastal California gnatcatcher (P. c. californica) was distinct, in contrast to a similar next-gen study by Vázquez-Miranda et al. (2022). Vandergast et al. (2022) used a phenogram instead of a phylogenetic analysis, and did not emphasize that their genetic measures showed a cline that was not stepped; additionally, they did not clarify that the limits they suggested do not match the listed subspecies. Therefore, they found some genetic variation but it is not supportive of subspecies limits.
Thus, the next-generation sequencing methods need to be interpreted with caution so as not to confuse sampling and genetic gaps (see below) and so as not to cherry-pick SNPs that favor one hypothesis over another. Given examination of enough SNPs, it would be likely to find some in only one or a few populations, making it seem like support for their distinctiveness. That is, one might exclude characters that suggest a different pattern, whereas overall differentiation should be assessed across all characters (e.g., SNPs). That is, conflicting characters should be a part of the analysis so as not to bias the result to a preconceived conclusion. In addition, next-gen methods do not guarantee similar findings from different labs, as in the case of the California gnatcatcher (Vandergast et al., 2022; Vázquez-Miranda et al., 2022).
Costs of preservation vary widely within and among different groups of organisms (Gordon et al., 2020). At the level of full species, the average cost of preserving a bird species in the United States is $2,571,017, with a wide range of variation. According to Gordon et al. (2020), mammals cost 8–26 times more on average to conserve than plants, and bird species cost 5–30 times more to conserve than plants and 6–14 times more than aquatic invertebrates.
The coastal California gnatcatcher (Polioptila californica californica) is listed as threatened. Its range includes the densely populated area of southern California from Palos Verde Peninsula south to the border with Baja California (and farther south to the end of Baja California Sur, where it becomes relatively common). The validity of the subspecies has been challenged (Zink et al., 2000; Zink et al., 2013; Zink et al., 2016) and defended (McCormack and Maley, 2015), and recent genomics data show that it is not evolutionarily distinct and hence not a valid subspecies (Vázquez-Miranda et al., 2022). The USFWS has suggested that excluding this habitat has come at a cost of at least $1 billion (Gordon, 2018). Fortunately, much of the land occupied by the coastal California gnatcatcher is currently preserved by habitat conservation plans (Winchell and Doherty, 2018), and gnatcatcher populations are apparently genetically connected (Vandergast et al., 2019).
Most ESA-listed subspecies were described before 1950 (137 of 175), and 150 (86%) were described before 1966 (see Figure 1), using methods that involved assessments of morphological variation. We note that in fishes, more emphasis is placed on protecting Evolutionary Significant Units, which are not afforded names in the Linnean System; that is, relatively few subspecies of fish are listed under the ESA. The ESA also allows protection of Distinct Population Segments (US Fish and Wildlife Service, 1996), although we do not discuss this category in this paper. It was not until 1966 that the first molecular methods appeared that could be used to test subspecies limits.
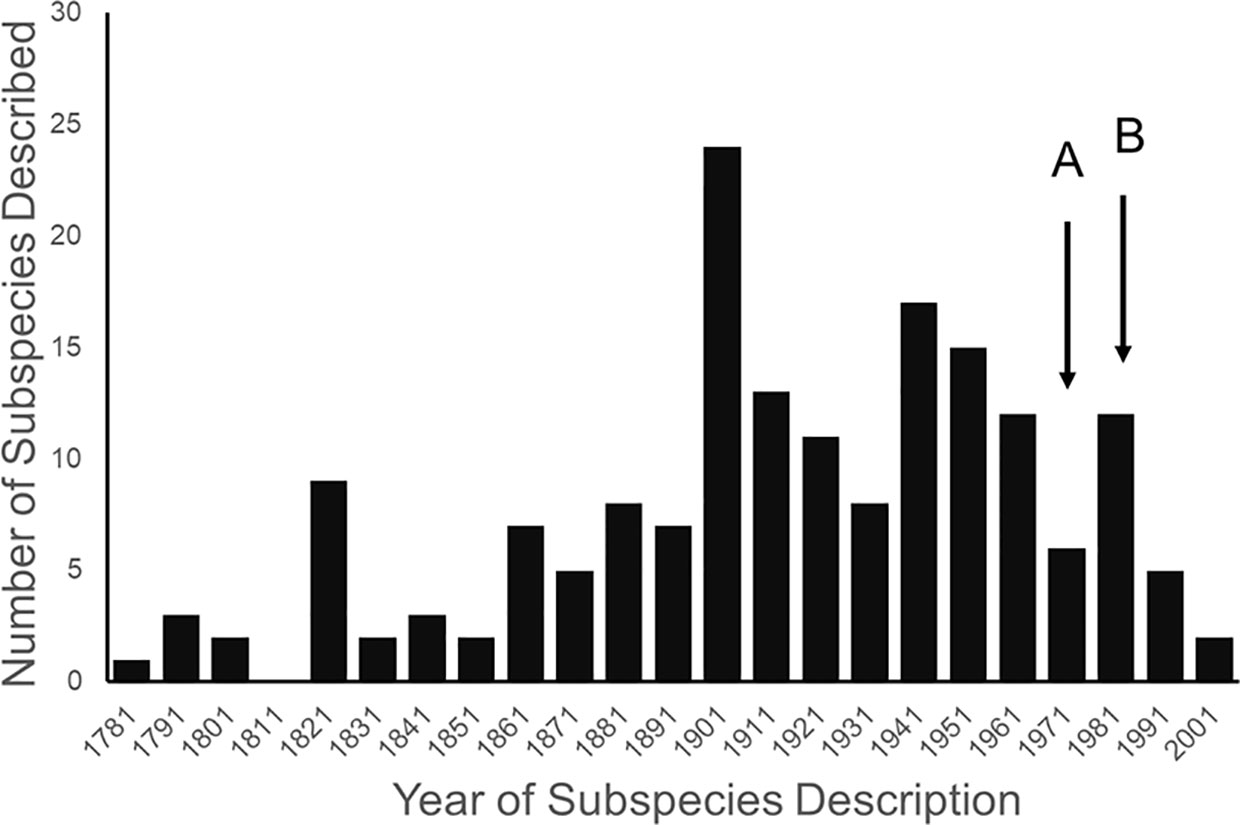
Figure 1 Distribution of years in which subspecies listed under the endangered species act were described. Arrow (A) marks the beginning of the use of allozymes in studying genetic variation in natural populations, and arrow (B) marks the beginning of the use of mitochondrial DNA.
The molecular methods used evolved from relatively crude assessment of distinguishing alleles at protein-coding loci (allozyme electrophoresis) to studies involving thousands of base-pairs at the DNA level. Most molecular examinations (n = 92) of subspecies limits used mtDNA (n = 67), and some were combinations of mtDNA and microsatellites (n = 19) or mtDNA and nuclear DNA (n = 14). Evaluations of listed subspecies vary widely in their sampling size, from a single individual to over 100 samples. Given the variation in the areal extent of listed subspecies’ distributions, a diversity in sampling size is not surprising; however, the relative percentage of the distribution covered by sampling also varies widely. In several cases researchers were able to include only a single population represented by one individual, thus making inferences of population distinctiveness difficult.
We examined 165 listed subspecies to determine how many were supported by modern analyses (see Table 1: 11 have been removed the USFWS in ITIS; https://www.itis.gov/). As noted earlier, a valid test would include multiple samples within the subspecies and comparisons with adjacent samples. Seven subspecies have been removed from the list because, according to the ITIS website, they have been elevated to species or there was a data error in the description. We found that the bulk of the remaining subspecies are distributed relatively equally among three categories: supported, not supported, and not tested (Figure 2). This summary suggests that a listed subspecies has a fifty-fifty chance of being consistent with the criteria listed by Taylor et al., 2017).
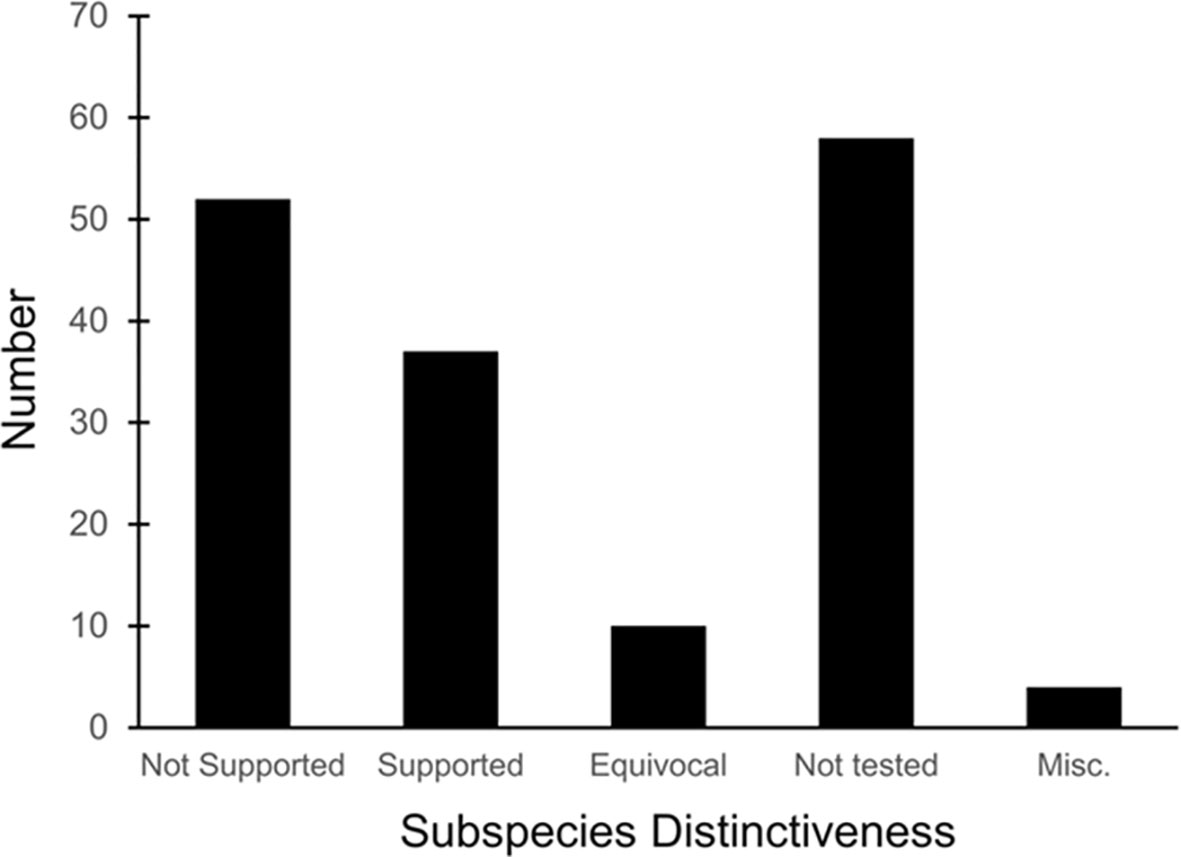
Figure 2 Distribution of results of evaluation of listed vertebrate subspecies. “Not tested” means samples from listed subspecies were not compared with samples from adjacent subspecies, there were too few samples, or samples were not examined.
We found that the ITIS classification departs from our summary. More than 40 of the 51 subspecies (78%) that were not supported by our evaluation were considered valid on the ITIS website.
The molecular methods used to test subspecies have evolved greatly over the past few decades, owing to a large increase in resolving power. With the new potential to describe genomes of individuals, some issues should be recognized. First, if sampling is not evenly spaced, sampling gaps will give the illusion of discrete taxonomic boundaries (Figure S1). Also, if a gap in the range is caused by anthropogenic elimination of intermediate areas, the populations might appear distinct, albeit not from natural evolutionary processes. Subspecies limits cannot be tested without a clear and rigorous sampling protocol.
It is possible for a subspecies that is not evolutionary distinct (a category that includes many subspecies) to be ecologically important—perhaps important enough to merit listing. Examples might include keystone species such as large carnivores: the Florida panther, for instance. However, providing quantitative data of ecological importance might be as large a task as documenting taxonomic distinctiveness. The lack of consistency among subspecies definitions used in ESA listings is a major failing of taxonomists. To further the use of taxonomic work in conservation decisions, this failing ought to be addressed.
We understand that some will have the view that all subspecies proposed for listing should be accepted at face value because they might be valid but not protected because of no current tests of their validity, hence, their loss would be lamentable (a Pascal’s Wager argument). We argue here that given the high cost of subspecies preservation and the fact that roughly 50% of subspecies tested are supported by modern methods, it should be unacceptable to list a subspecies under the ESA without modern analyses confirming Taylor et al. (2017) criteria. It should be incumbent upon the USFWS to seek consensus among taxonomists who work on different groups or organisms to agree on a list of minimal criteria for a subspecies to be listed under the ESA so that listing decisions are transparent.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
RZ designed project, conducted analyses, wrote the draft. LK gathered raw data and edited the manuscript. Both authors contributed to the article and approved the submitted version.
We thank M. Cronin for valuable suggestions on the manuscript. The project received support from the Center for Growth and Opportunity at Utah State University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcosc.2022.971280/full#supplementary-material
American Ornithologists’ Union (1957). The check-list of North American birds. fifth ed (Baltimore, Maryland: Amer. Ornith. Union).
Avise J. C. (1992). Molecular population structure and the biogeographic history of a regional fauna: a case history with lessons for conservation biology. Oikos, 62–76. doi: 10.2307/3545516
Avise J. C., Nelson W. S. (1989). Molecular genetic relationships of the extinct dusky seaside sparrow. Science 243 (4891), 646–648. doi: 10.1126/science.243.4891.646.
Bangs M. R., Douglas M. R., Chafin T. K., Douglas M. E. (2020). Gene flow and species delimitation in fishes of western North America: flannelmouth (Catostomus latipinnis) and bluehead sucker (C. pantosteus discobolus). Ecol. Evol. 10 (13), 6477–6493. doi: 10.1002/ece3.6384
Barrowclough G. F., Cracraft J., Klicka J., Zink R. M. (2016). How many kinds of birds are there and why does it matter? PloS One 11 (11), e0166307. doi: 10.1371/journal.pone.0166307
Barrowclough G. F., Groth J. G., Mertz L. A., Gutiérrez R. J. (2006). Genetic structure of Mexican spotted owl (Strix occidentalis lucida) populations in a fragmented landscape. Auk 123 (4), 1090–1102. doi: 10.1093/auk/123.4.1090
Barrowclough G. F., Gutiérrez R. J., Groth J. G., Lai J. E., Rock D. F. (2011). The hybrid zone between northern and California spotted owls in the Cascade–Sierran suture zone. Condor 113 (3), 581–589. doi: 10.1525/cond.2011.100203
Benedict B. D., Castellanos A. A., Light J. E. (2019). Phylogeographic assessment of the Heermann’s kangaroo rat (Dipodomys heermanni). J. Mammalogy 100 (1), 72–91. doi: 10.1093/jmammal/gyy166
Branch L. C., Clark A. M., Moler P. E., Bowen B. W. (2003). Fragmented landscapes, habitat specificity, and conservation genetics of three lizards in Florida scrub. Conserv. Genet. 4 (2), 199–212. doi: 10.1023/A:1023398908793
Brown V. A., Brooke A., Fordyce J. A., McCracken G. F. (2011). Genetic analysis of populations of the threatened bat Pteropus mariannus. Conserv. Genet. 12 (4), 933–941. doi: 10.1007/s10592-011-0196-y
Buchalski M. R., Sacks B. N., Gille D. A., Penedo M. C. T., Ernest H. B., Morrison S. A., et al. (2016). Phylogeographic and population genetic structure of bighorn sheep (Ovis canadensis) in North American deserts. J. Mammalogy 97 (3), 823–838. doi: 10.1093/jmammal/gyw011
Buehler D. M., Baker A. J., Piersma T. (2006). Reconstructing palaeoflyways of the late Pleistocene and early Holocene red knot Calidris canutus. Ardea -Wageningen- 94 (3), 485–498.
Bulgin N. L., Gibbs H. L., Vickery P., Baker A. J. (2003). Ancestral polymorphisms in genetic markers obscure detection of evolutionarily distinct populations in the endangered Florida grasshopper sparrow (Ammodramus savannarum floridanus). Mol. Ecol. 12 (4), 831–844. doi: 10.1046/j.1365-294X.2003.01774.x
Byerly P. A. (2021). Ecology and conservation genomics of roseate terns (North America: Doctoral dissertation, University of Louisiana at Lafayette).
Caballero I. C., Ashley M. V. (2011). Genetic analysis of the endemic island loggerhead shrike, Lanius ludovicianus anthonyi. Conserv. Genet. 12 (6), 1485–1493. doi: 10.1007/s10592-011-0247-4
Campbell D. C., Piller K. R. (2017). Let’s jump in: a phylogenetic study of the Great Basin springfishes and poolfishes, Crenichthys and Empetrichthys (Cyprinodontiformes: Goodeidae). PloS One 12 (10), e0185425. doi: 10.1371/journal.pone.0185425
Catanach T. A., Halley M. R., Allen J. M., Johnson J. A., Thorstrom R., Palhano S., et al. (2021). Systematics and conservation of an endemic radiation of Accipiter hawks in the Caribbean islands. Auk 138 (3), ukab041. doi: 10.1093/ornithology/ukab041
Colgan D. J., Soheili S. (2008). Evolutionary lineages in Emballonura and Mosia bats (Mammalia: Microchiroptera) from the southwestern Pacific. Pacific Sci. 62 (2), 219–232. doi: 10.2984/1534-6188(2008)62[219:ELIEAM]2.0.CO;2
Cracraft J. (1983). “Species concepts and speciation analysis,” in Current ornithology (New York, NY: Springer), 159–187.
Cronin M. A., Cánovas A., Bannasch D. L., Oberbauer A. M., Medrano J. F. (2015). Wolf subspecies: reply to Weckworth et al. and Fredrickson et al. J. Heredity 106 (4), 417–419. doi: 10.1093/jhered/esv029
Degner J. F., Stout I. J., Roth J. D., Parkinson C. L. (2007). Population genetics and conservation of the threatened southeastern beach mouse (Peromyscus polionotus niveiventris): subspecies and evolutionary units. Conserv. Genet. 8 (6), 1441–1452. doi: 10.1007/s10592-007-9295-1
De Moya R. S., Savage W. K., Tenney C., Bao X., Wahlberg N., Hill R. I. (2017). Interrelationships and diversification of Argynnis Fabricius and Speyeria Scudder butterflies. Systematic Entomology 42 (4), 635–649. doi: 10.1111/syen.12236
de Queiroz K. (2007). Species concepts and species delimitation. Systematic Biol. 56 (6), 879–886. doi: 10.1080/10635150701701083
Draheim H. M., Miller M. P., Baird P., Haig S. M. (2010). Subspecific status and population genetic structure of least terns (Sternula antillarum) inferred by mitochondrial DNA control-region sequences and microsatellite DNA. Auk 127 (4), 807–819. doi: 10.1525/auk.2010.09222
Drovetski S. V., Pearson S. F., Rohwer S. (2005). Streaked horned lark Eremophila alpestris strigata has distinct mitochondrial DNA. Conserv. Genet. 6 (6), 875–883. doi: 10.1007/s10592-005-9074-9.
Dupuis J. R., Geib S. M., Osborne K. H., Rubinoff D. (2020). Genomics confirms surprising ecological divergence and isolation in an endangered butterfly. Biodiversity Conserv. 29 (6), 1897–1921. doi: 10.1007/s10531-020-01950-6
Dupuis J. R., Oliver J. C., Brunet B. M., Longcore T., Johnson J. J., Sperling F. A. (2018). Genomic data indicate ubiquitous evolutionary distinctiveness among populations of California metalmark butterflies. Conserv. Genet. 19 (5), 1097–1108. doi: 10.1007/s10592-018-1081-8
Fitak R. R., Koprowski J. L., Culver M. (2013). Severe reduction in genetic variation in a montane isolate: the endangered Mount Graham red squirrel (Tamiasciurus hudsonicus grahamensis). Conserv. Genet. 14 (6), 1233–1241. doi: 10.1007/s10592-013-0511-x
Fleischer R. C., Slikas B., Beadell J., Atkins C., Mcintosh C. E., Conant S. (2007). Genetic variability and taxonomic status of the Nihoa and Laysan millerbirds. Condor 109 (4), 954–962. doi: 10.1093/condor/109.4.954
Fredrickson R. J., Hedrick P. W., Wayne R. K., vonHoldt B. M., Phillips M. K. (2015). Mexican Wolves are a valid subspecies and an appropriate conservation target. J. Heredity 106 (4), 415–416. doi: 10.1093/jhered/esv028
Gavin T. A., May B. (1988). Taxonomic status and genetic purity of Columbian white-tailed deer. J. Wildlife Manage., 1–10. doi: 10.2307/3801048
Girard P., Takekawa J. Y., Beissinger S. R. (2010). Uncloaking a cryptic, threatened rail with molecular markers: origins, connectivity and demography of a recently-discovered population. Conserv. Genet. 11 (6), 2409–2418. doi: 10.1007/s10592-010-0126-4
Gompert Z., Nice C. C., Fordyce J. A., Forister M. L., Shapiro A. M. (2006). Identifying units for conservation using molecular systematics: the cautionary tale of the Karner blue butterfly. Mol. Ecol. 15 (7), 1759–1768. doi: 10.1111/j.1365-294X.2006.02905.x
Gordon R. (2018) Whatever the cost of the endangered species act, it’s huge. competitive enterprise institute, no. 247. Available at: https://cei.org/sites/default/files/Robert_Gordon_-_%E2%80%9CWhatever_the_Cost%E2%80%9D_of_the_Endangered_Species_Act%2C_It%E2%80%99s_Huge.pdf.
Gordon E. R., Butt N., Rosner-Katz H., Binley A. D., Bennett J. R. (2020). Relative costs of conserving threatened species across taxonomic groups. Conserv. Biol. 34 (1), 276–281. doi: 10.1111/cobi.13382
Haas S. E., Kimball R. T. (2009). Genetic divergence among snail kite subspecies: implications for the conservation of the endangered Florida snail kite (Rostrhamus sociabilis). Ibis 151, 181–185. doi: 10.1111/j.1474-919X.2008.00872.x
Haig S. M., Beever E. A., Chambers S. M., Draheim H. M., Dugger B. D., Dunham S., et al. (2006). Taxonomic considerations in listing subspecies under the US endangered species act. Conserv. Biol. 20 (6), 1584–1594. doi: 10.1111/j.1523-1739.2006.00530.x
Hamm C. A., Rademacher V., Landis D. A., Williams B. L. (2014). Conservation genetics and the implication for recovery of the endangered Mitchell’s satyr butterfly, Neonympha mitchellii mitchellii. J. Heredity 105 (1), 19–27. doi: 10.1093/jhered/est073
Hendricks S., Navarro A. Y., Wang T., Wilder A., Ryder O. A., Shier D. M. (2020). Patterns of genetic partitioning and gene flow in the endangered San Bernardino kangaroo rat (Dipodomys merriami parvus) and implications for conservation management. Conserv. Genet. 21 (5), 819–833. doi: 10.1007/s10592-020-01289-z
Hofman C. A., Rick T. C., Hawkins M. T. R., Funk W. C., Ralls K., Boser C. L., et al. (2015). Mitochondrial genomes suggest rapid evolution of dwarf California channel islands foxes (Urocyon littoralis). PLoS One 10 (2), e0118240. doi: 10.1371/journal.pone.0118240
Holycross A. T., Douglas M. E. (2007). Geographic isolation, genetic divergence, and ecological non-exchangeability define ESUs in a threatened sky-island rattlesnake. Biol. Conserv. 134 (1), 142–154. doi: 10.1016/j.biocon.2006.07.020
Jackson J. D. U., Bruford M. W., Székely T., DaCosta J. M., Sorenson M. D., Russo I. R. M., et al. (2020). Population differentiation and historical demography of the threatened snowy plover Charadrius nivosus (Cassi). Conserv. Genet. 21 (3), 387–404. doi: 10.1007/s10592-020-01256-8
Jackson D. J., Cook J. A. (2020). A precarious future for distinctive peripheral populations of meadow voles (Microtus pennsylvanicus). J. Mammalogy 101 (1), 36–51. doi: 10.1093/jmammal/gyz196
Janzen F. J., Krenz J. G., Haselkorn T. S., Brodie E. D. Jr., Brodie E. D. III (2002). Molecular phylogeography of common garter snakes (Thamnophis sirtalis) in western North America: implications for regional historical forces. Mol. Ecol. 11 (9), 1739–1751. doi: 10.1046/j.1365-294X.2002.01571.x
Jordan S., Simon C., Polhemus D. (2003). Molecular systematics and adaptive radiation of hawaii’s endemic damselfly genus Megalagrion (Odonata: Coenagrionidae). Systematic Biol. 52 (1), 89–109. doi: 10.1080/10635150390132803
Karin B. R., Cicero C., Koo M. S., Bowie R. C. (2018). The role of history and ecology as drivers of song divergence in Bell’s and sagebrush sparrows (Artemisiospiza, Aves: Passerellidae). Biol. J. Linn. Soc. 125 (2), 421–440. doi: 10.1093/biolinnean/bly090
Klicka L. B., Kus B. E., Title P. O., Burns K. J. (2016). Conservation genomics reveals multiple evolutionary units within Bell’s vireo (Vireo bellii). Conserv. Genet. 17 (2), 455–471. doi: 10.1007/s10592-015-0796-z
Klimova A., Munguia-Vega A., Hoffman J. I., Culver M. (2014). Genetic diversity and demography of two endangered captive pronghorn subspecies from the Sonoran desert. J. Mammalogy 95 (6), 1263–1277. doi: 10.1644/13-MAMM-A-321
Klütsch C. F., Manseau M., Wilson P. J. (2012). Phylogeographical analysis of mtDNA data indicates [sic] postglacial expansion from multiple glacial refugia in woodland caribou (Rangifer tarandus caribou). PLoS One 7 (12), e52661. doi: 10.1371/journal.pone.0052661
Larson S., Gagne R. B., Bodkin J., Murray M. J., Ralls K., Bowen L., et al. (2021). Translocations maintain genetic diversity and increase connectivity in sea otters, Enhydra lutris. Marine Mammal Sci. 37: 1475–1497.
Luo A., Ling C., Ho S. Y., Zhu C. D. (2018). Comparison of methods for molecular species delimitation across a range of speciation scenarios. Systematic Biol. 67 (5), 830–846. doi: 10.1093/sysbio/syy011
Mahoney S. M., Reudink M. W., Pasch B., Theimer T. C. (2020). Song but not plumage varies geographically among willow flycatcher Empidonax traillii subspecies. J. Avian Biol. 51 (12). doi: 10.1111/jav.02621.
Malaney J. L., Cook J. A. (2013). Using biogeographical history to inform conservation: the case of Preble’s meadow jumping mouse. Mol. Ecol. 22 (24), 6000–6017. doi: 10.1111/mec.12476
Malaney J. L., Demboski J. R., Cook J. A. (2017). Integrative species delimitation of the widespread North American jumping mice (Zapodinae). Mol. Phylogenet. Evol. 114, 137–152. doi: 10.1016/j.ympev.2017.06.001
Malaney J. L., Frey J. K., Cook J. A. (2012). The biogeographic legacy of an imperilled taxon provides a foundation for assessing lineage diversification, demography and conservation genetics. Diversity Distributions 18 (7), 689–703. doi: 10.1111/j.1472-4642.2011.00866.x
Maldonado J. E., Vilà C., Wayne R. K. (2001). Tripartite genetic subdivisions in the ornate shrew (Sorex ornatus). Mol. Ecol. 10 (1), 127–147. doi: 10.1046/j.1365-294X.2001.01178.x
Maley J. M., Brumfield R. T. (2013). Mitochondrial and next-generation sequence data used to infer phylogenetic relationships and species limits in the Clapper/King rail complex R.longirostris/R. elegans. Condor 115 (2), 316–329. doi: 10.1525/cond.2013.110138
Martin A. P. (2010). The conservation genetics of Ash Meadows pupfish populations. I. The Warm Springs pupfish Cyprinodon nevadensis pectoralis. Conserv. Genet. 11 (5), 1847–1857. doi: 10.1007/s10592-010-0077-9.
Matocq M. D., Kelly P. A., Phillips S. E., Maldonado J. E. (2012). Reconstructing the evolutionary history of an endangered subspecies across the changing landscape of the great central valley of California. Mol. Ecol. 21 (24), 5918–5933. doi: 10.1111/mec.12079
McCormack J. E., Maley J. M. (2015). Interpreting negative results with taxonomic and conservation implications: Another look at the distinctness of coastal California gnatcatchers. Auk 132, 380–388. doi: 10.1642/AUK-14-184.1
McHugh A., Bierzychudek P., Greever C., Marzulla T., Van Buskirk R., Binford G. (2013). A molecular phylogenetic analysis of Speyeria and its implications for the management of the threatened Speyeria zerene hippolyta. J. Insect Conserv. 17 (6), 1237–1253. doi: 10.1007/s10841-013-9605-5
Metcalf J. L., Stowell S. L., Kennedy C. M., Rogers K. B., McDonald D., Epp J., et al. (2012). Historical stocking data and 19th century DNA reveal human-induced changes to native diversity and distribution of cutthroat trout. Mol. Ecol. 21 (21), 5194–5207. doi: 10.1111/mec.12028
Miller M. P., Mullins T. D., Haig S. M. (2016). Genetic diversity and population structure in the threatened Oregon silverspot butterfly (Speyeria zerene hippolyta) in western Oregon and northwestern California–implications for future translocations and the establishment of new populations (U.S. Geological Survey), 23. doi: 10.3133/ofr20161162
Miller M. P., Mullins T. D., Haig S. M., Takano L., Garcia K. (2015). Genetic structure, diversity, and interisland dispersal in the endangered Mariana common moorhen (Gallinula chloropus guami). Condor: Ornithological Appl. 117 (4), 660–669. doi: 10.1650/CONDOR-15-42.1
Miller C. R., Waits L. P., Joyce P. (2006). Phylogeography and mitochondrial diversity of extirpated brown bear (Ursus arctos) populations in the contiguous united states and Mexico. Mol. Ecol. 15 (14), 4477–4485. doi: 10.1111/j.1365-294X.2006.03097.x
Moyle P. B., Campbell M. A. (2022). Cryptic species of freshwater sculpin (Cottidae, Cottus) in California, USA. Zootaxa 5154 (5), 501–507. doi: 10.11646/zootaxa.5154.5.1
Nagarajan R. P., Goodbla A., Graves E., Baerwald M., Holyoak M., Schreier A. (2020). Non-invasive genetic monitoring for the threatened valley elderberry longhorn beetle. PloS One 15 (1), e0227333. doi: 10.1371/journal.pone.0227333
Neuwald J. L. (2010). Population isolation exacerbates conservation genetic concerns in the endangered Amargosa vole, Microtus californicus scirpensis. Biol. Conserv. 143 (9), 2028–2038. doi: 10.1016/j.biocon.2010.05.007
Nikolakis Z. L., Orton R. W., Crother B. I. (2022). Fine-scale population structure within an Eastern Nearctic snake complex (Pituophis melanoleucus). Zoologica Scripta 51 (2), 133–146. doi: 10.1111/zsc.12522
Patten M. A., Remsen J. V. Jr. (2017). Complementary roles of phenotype and genotype in subspecies delimitation. J. Heredity 108 (4), 462–464. doi: 10.1093/jhered/esx013
Patton J. L., Williams D. F., Kelly P. A., Cypher B. L., Phillips S. E. (2019). Geographic variation and evolutionary history of Dipodomys nitratoides (Rodentia: Heteromyidae), a species in severe decline. J. Mammalogy 100 (5), 1546–1563. doi: 10.1093/jmammal/gyz128
Pertoldi C., Tokarska M., Wójcik J. M., Kawałko A., Randi E., Kristensen T. N., et al. (2010). Phylogenetic relationships among the European and American bison and seven cattle breeds reconstructed using the BovineSNP50 illumina genotyping BeadChip. Acta Theriologica 55 (2), 97–108. doi: 10.4098/j.at.0001-7051.002.2010
Phillips A. R. (1948). Geographic variation in Empidonax traillii. Auk 65, 507–514. doi: 10.2307/4080601
Piaggio A. J., Jeffers J. (2013). On the edge: A genetic assessment of Aplodontia rufa from the edge of their distribution. Western North Am. Nat. 73 (4), 485–496. doi: 10.3398/064.073.0413
Piaggio A. J., Navo K. W., Stihler C. W. (2009). Intraspecific comparison of population structure, genetic diversity, and dispersal among three subspecies of townsend’s big-eared bats, Corynorhinus townsendii townsendii, C. t. pallescens, and the endangered C. t. virginianus. Conserv. Genet. 10 (1), 143–159. doi: 10.3398/064.073.0413.
Piaggio A. J., Perkins S. L. (2005). Molecular phylogeny of north American long-eared bats (Vespertilionidae: Corynorhinus): inter- and intraspecific relationships inferred from mitochondrial and nuclear DNA sequences. Mol. Phylogenet. Evol. 37 (3), 762–775. doi: 10.1016/j.ympev.2005.03.029
Proshek B., Dupuis J. R., Engberg A., Davenport K., Opler P. A., Powell J. A., et al. (2015). Genetic evaluation of the evolutionary distinctness of a federally endangered butterfly, Lange’s metalmark. BMC Evolutionary Biol. 15 (1), 1–15. doi: 10.1186/s12862-015-0354-9.
Ramey R. R., Liu H. P., Epps C. W., Carpenter L. M., Wehausen J. D. (2005). Genetic relatedness of the Preble’s meadow jumping mouse (Zapus hudsonius preblei) to nearby subspecies of Z. hudsonius as inferred from variation in cranial morphology, mitochondrial DNA and microsatellite DNA: implications for taxonomy and conservation. Anim. Conserv. Forum 8 (3), 329–346. doi: 10.1017/S1367943005002313.
Remsen J. V. Jr. (2005). Pattern, process, and rigor meet classification. Auk 122 (2), 403–413. doi: 10.1093/auk/122.2.403
Richmond J. Q., Wood D. A., Swaim K. E., Fisher R. N., Vandergast A. G. (2016). Historical habitat barriers prevent ring-like genetic continuity throughout the distribution of threatened Alameda striped racers (Coluber lateralis euryxanthus). Herpetologica 72 (3), 202–213. doi: 10.1655/Herpetologica-D-15-00046.1
Rippert J. S. (2017). Population genetics and functional connectivity of the riparian brush rabbit (Sylvilagus bachmani riparius): Implications for the conservation of an endangered lagomorph (Doctoral dissertation, University of Nevada, Reno).
Rising J. D. (2001). Geographic variation in size and shape of savannah sparrows (Passerculus sandwichensis) (Cooper Ornithological Society).
Rogers S. O., Watson B. T., Neves R. J. (2001). Life history and population biology of the endangered tan riffleshell (Epioblasma florentina walkeri) (Bivalvia: Unionidae). J. North Am. Benthological Soc. 20 (4), 582–594. doi: 10.2307/1468089
Ruegg K., Anderson E. C., Somveille M., Bay R. A., Whitfield M., Paxton E. H., et al. (2021). Linking climate niches across seasons to assess population vulnerability in a migratory bird. Global Change Biol. 27 (15), 3519–3531. doi: 10.1111/gcb.15639
Ruiz-García M., Pinedo-Castro M. (2013). “Population genetics and phylogeographic analyses of the jaguarundi (Puma yagouaroundi) by means of three mitochondrial markers: the first molecular population study of this species,” in Molecular population genetics, phylogenetics, evolutionary biology and conservation of the Neotropical carnivores, 245–288.
Saglam I. K., Prince D. J., Meek M., Ali O. A., Miller M. R., Peacock M., et al. (2017). Genomic analysis reveals genetic distinctiveness of the Paiute cutthroat trout Oncorhynchus clarkii seleniris. Trans. Am. Fisheries Soc. 146 (6), 1291–1302. doi: 10.1080/00028487.2017.1356373
Saremi N. F., Supple M. A., Byrne A., Cahill J. A., Coutinho L. L., Dalén L., et al. (2019). Puma genomes from north and south America provide insights into the genomic consequences of inbreeding. Nat. Commun. 10 (1), 1–10. doi: 10.1038/s41467-019-12741-1.
Sennett G. B. (1879). Further notes on the ornithology of the Lower Rio Grande of Texas, from observations made during the spring of 1878. Bull. United States Geographical Survey 5, 371–440.
Shiraiwa K., Cong Q., Grishin N. V. (2014). A new heraclides swallowtail (Lepidoptera, Papilionidae) from North America is recognized by the pattern on its neck. Zookeys 468), 85. doi: 10.3897/zookeys.468.8565.
Slikas B., Jones I. B., Derrickson S. R., Fleischer R. C. (2000). Phylogenetic relationships of Micronesian white-eyes based on mitochondrial sequence data. Auk 117 (2), 355–365. doi: 10.1093/auk/117.2.355
Statham M. J., Rich A. R., Lisius S. K., Sacks B. N. (2012). Discovery of a remnant population of Sierra Nevada red fox (Vulpes vulpes necator). Northwest Sci. 86, 122–132. doi: 10.3955/046.086.0204
Storfer A., Mech S. G., Reudink M. W., Lew K. (2014). Inbreeding and strong population subdivision in an endangered salamander. Conserv. Genet. 15 (1), 137–151. doi: 10.1007/s10592-013-0526-3
Swei A., Brylski P. V., Spencer W. D., Dodd S. C., Patton J. L. (2003). Hierarchical genetic structure in fragmented populations of the little pocket mouse (Perognathus longimembris) in southern California. Conserv. Genet. 4 (4), 501–514. doi: 10.1023/A:1024768831808
Taylor B. L., Perrin W. F., Reeves R. R., Rosel P. E., Wang J. Y., Cipriano F., et al. (2017). Why we should develop guidelines and quantitative standards for using genetic data to delimit subspecies for data-poor organisms like cetaceans. Mar. Mammal Sci. 33 (S1), 12–26. doi: 10.1111/mms.12413
Theimer T. C., Smith A. D., Mahoney S. M., Ironside K. E. (2016). Available data support protection of the southwestern willow flycatcher under the endangered species act. Condor 118, 289–299. doi: 10.1650/CONDOR-15-71.1
Tonione M., Johnson J. R., Routman E. J. (2011). Microsatellite analysis supports mitochondrial phylogeography of the hellbender (Cryptobranchus alleganiensis). Genetica 139 (2), 209–219. doi: 10.1007/s10709-010-9538-9
Tursi R. M., Hughes P. T., Hoffman E. A. (2013). Taxonomy versus phylogeny: evolutionary history of marsh rabbits without hopping to conclusions. Diversity Distributions 19 (2), 120–133. doi: 10.1111/j.1472-4642.2012.00915.x
US Fish and Wildlife Service (1996). Policy regarding the recognition of distinct vertebrate population segments under the endangered species act Vol. 61 (Federal Register), 4722.
Vandergast A. G., Kus B. E., Preston K. L., Barr K. R. (2019). Distinguishing recent dispersal from historical genetic connectivity in the coastal California gnatcatcher. Sci. Rep. 9 (1), 1–12. doi: 10.1038/s41598-018-37712-2
Vandergast A. G., Kus B. E., Wood D. A., Milano E. R., Preston K. L. (2022). Subspecies differentiation and range-wide genetic structure are driven by climate in the California gnatcatcher, a flagship species for coastal sage scrub conservation. Evolutionary Appl. 00, 1–17. doi: 10.1111/eva.13429.
Vázquez-Miranda H., Griffin J. A., Sheppard J. M., Herman J. M., Rojas-Soto O., Zink R. M. (2017). Morphological and molecular evolution and their consequences for conservation and taxonomy in the Le Conte’s Thrasher Toxostoma lecontei. J. Avian Biol. 48 (7), 941–954. doi: 10.1111/jav.01057
Vázquez-Miranda H., Zink R. M., Pinto B. J. (2022). Comparative phylogenomic patterns in the Baja California avifauna, their conservation implications, and the stages in lineage divergence. Mol. Phylogenet. Evol. 171, 107466. doi: 10.1016/j.ympev.2022.107466
Vogler A. P., De Salle R. (1994). “Mitochondrial DNA evolution and the application of the phylogenetic species concept in the Cicindela dorsalis complex (Coleoptera: Cicindelidae),” in Carabid beetles: Ecology and evolution, 79–85.
Weyandt S. E., van den Bussche R., Hamilton M. J., Leslie D. M. (2005). Unraveling the effects of sex and dispersal: conservation genetics of the endangered Ozark big-eared bat (Corynorhinus townsendii ingens). J. Mammalogy 85, 140–148. doi: 10.1644/04-MAMM-F-067R1.1
Williford D., Deyoung R. W., Honeycutt R. L., Brennan L. A., Hernández F., Wehland E. M., et al. (2014). Contemporary genetic structure of the northern bobwhite west of the Mississippi river. J. Wildlife Manage. 78 (5), 914–929. doi: 10.1002/jwmg.733
Winchell C. S., Doherty P. F. Jr. (2018). Restoring habitat for coastal California gnatcatchers (Polioptila californica californica). Condor: Ornithological Appl. 120 (3), 581–595. doi: 10.1650/CONDOR-17-221.1
Winker K. (2010). Subspecies represent geographically partitioned variation, a gold mine of evolutionary biology, and a challenge for conservation. Ornithological Monogr. 67 (1), 6–23. doi: 10.1525/om.2010.67.1.6
Wood D. A., Emmons I. D., Nowak E. M., Christman B. L., Holycross A. T., Vandergast A. G. (2018). Conservation genomics of the mogollon narrow-headed gartersnake (Thamnophis rufipunctatus) and northern Mexican gartersnake (Thamnophis eques megalops) (No. 2018-1141) (US Geological Survey).
Young D. L., Allard M. W. (1997). Conservation genetics of the plain pigeon Columba inornata in Puerto Rico and the Dominican republic. Mol. Ecol. 6 (9), 877–879. doi: 10.1111/j.1365-294X.1997.tb00142.x
Zink R. M. (1997). “Species concepts,” in Bulletin of the British ornithologists' club, vol. 117. , 97–109.
Zink R. M. (2015). Genetics, morphology, and ecological niche modeling do not support the subspecies status of the endangered southwestern willow flycatcher (Empidonax traillii extimus). Condor: Ornithological Appl. 117 (1), 76–86. doi: 10.1650/CONDOR-14-27.1
Zink R. M. (2016). Current topics in avian conservation genetics with special reference to the southwestern willow flycatcher. Open Ornithology J. 9, 60–69. doi: 10.2174/1874453201609010060
Zink R. M., Barrowclough G. F., Atwood J. L., Blackwell R. C. (2000). Genetics, taxonomy and conservation of the threatened California gnatcatcher. Conserv. Biol. 14, 1394–1405. doi: 10.1046/j.1523-1739.2000.99082.x
Zink R. M., Groth J. G., Vázquez-Miranda H., Barrowclough G. F. (2013). Phylogeography of the California gnatcatcher (Polioptila californica) using multilocus DNA sequences and ecological niche modeling: implications for conservation. Auk 130, 449–458. doi: 10.1525/auk.2013.12241
Zink R. M., Groth J. G., Vázquez-Miranda H., Barrowclough G. F. (2016). Geographic variation, null hypotheses and subspecies limits in the California gnatcatcher: a response to McCormack and Maley. Auk 133, 59–68. doi: 10.1642/AUK-15-63.1
Keywords: endangered species act, subspecies, taxonomic status, listing criteria, taxonomic methods
Citation: Zink RM and Klicka LB (2022) The taxonomic basis of subspecies listed as threatened and endangered under the endangered species act. Front. Conserv. Sci. 3:971280. doi: 10.3389/fcosc.2022.971280
Received: 16 June 2022; Accepted: 23 August 2022;
Published: 26 September 2022.
Edited by:
William J. McShea, Smithsonian Conservation Biology Institute (SI), United StatesReviewed by:
Andrew Hope, Kansas State University, United StatesCopyright © 2022 Zink and Klicka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert M. Zink, cnppbmsyQHVubC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.