- 1Department of Crop Science, University of Ghana, Legon, Accra, Ghana
- 2Department of Entomology and Plant Pathology, University of Tennessee, Knoxville, TN, United States
Cryptolepis sanguinolenta is an important medicinal plant used in the treatment of malaria in Ghana. Overharvesting, destruction of entire plant populations and poor seed viability have resulted in a substantial decrease in wild populations thereby threatening its long-term potential and survivability. In this study, fifteen polymorphic microsatellite loci were used to evaluate the genetic diversity and population structure of 179 C. sanguinolenta individuals among eight subpopulations in Ghana. The subpopulations were separated by a distance of 8.3 – 233.3 km. Our results indicated relatively high levels of genetic diversity (Ho= 0.41; He=0.61) and high gene flow (Nm=7.06), an indication of greater stability and adaptability within the ecosystem, limited genetic differentiation (mean FST=0.05; highest FST=0.1), which suggested insignificant differentiation among the subpopulations. The high levels of gene flow resulting from the wind-dispersed seeds might have contributed to the limited genetic differentiation among the subpopulations. The Bayesian cluster analysis revealed the presence of a population structure (K=2). A lack of isolation by distance (r=0.012; P=0.34) indicated an increase in the genetic similarity among the subpopulations as the geographic distance between them decreased. This study described the genetic diversity and population structure in the current C. sanguinolenta accessions and laid a foundation for future breeding efforts.
1 Introduction
According to a World Health Organization (WHO) 2021 report, 241 million cases of malaria were reported in 2020 (WHO, 2021). Most of these cases occurred in the African region (228 million), followed by the Eastern Mediterranean (5.75 million), and the South-East Asian region (5 million). Globally, malaria deaths reached 627,000, out of which, 96% are accounted for in sub-Saharan Africa (WHO, 2021). In addition, children under five accounted for 80% of the malaria deaths in Africa (WHO, 2021). Although efforts have been made towards reducing the global burden of malaria, drug resistance, accessibility, and the affordability of antimalarial medications continue to threaten this effort. To mitigate these issues, alternative approaches to using traditional medicine could provide more sustainable and cost-effective treatment options.
An estimated 80% of the world’s population almost exclusively depends on local medicinal plants for the prevention and treatment of various diseases because they are readily accessible and affordable (Kirby, 1996; WHO, 2002). Several efficacious medicines such as aspirin (an analgesic) derived from Salix alba L., Digoxin (cadiotonic) from Digitalis lantana Ehrh., Artemisinin (anti-malarial) from Artemisia annua L., and Quinine from Cinchona ledgeriana Moens ex Trimen. are used for the of malaria treatment (Luo et al., 1998) and originate from plants (Licciardi and Underwood, 2011; Veeresham, 2012). In the fight against malaria, medicinal plants with a history of use as potential antimalarials in resource-poor communities should be considered in the development of intervention approaches (Abebe and Garedew, 2019). In that capacity, steps to conserve and promote sustainable, resilient, and climate-smart agro-ecosystems must be encouraged while documenting and preserving traditional medicinal plants knowledge limited to local healers and elders in these communities. Abebe and Garedew (2019) identified 25 plant species from 22 families that had been used in traditional treatments of malaria in Ethiopia with Cyperus spp., Allium sativum L., Lepidium sativum L., and Echinops kebericho Mesfin. are utilized most extensively.
With an increase in anthropogenic activities, there is an urgent need to prioritize the protection of wild populations and the cultivation of useful medicinal plants to ensure future availability for healthcare needs. Cryptolepis sanguinolenta, locally known in Ghana as ‘Nibima’ in Twi, ‘Gangnamau’ in Hausa, or ‘Kadze’ in Ewe, is a perennial vine belonging to the Apocynaceae family with a history of use in Africa (Luo et al., 1998; Osafo et al., 2017). It grows in rainforests, deciduous forest belts, and secondary forest clearings along the west coast of Africa from Nigeria through Ghana to Senegal (Dokosi, 1998). In Ghana, however, it is located along the slopes of the Akwapim and Kwahu mountain ranges (Addae-Kyereme, 2004) with a long-standing history of use in the treatment of malaria.
Roots, the portion of the plant of commercial and medicinal value, contain the highest concentration of indoloquinoline and cryptolepine, which have the most potent antiplasmodial activity (Dwuma-Badu et al., 1978; Tackie et al., 1991; Cimanga et al., 1997).
Minor alkaloids including hydroquinone, quindoline, cryptoquindoline, cryptoheptine and 11-hydroxy derivatives of cryptolepine with biological/pharmacological activities have also been isolated (Dokosi, 1998). The combined presence of these alkaloids accounts for the biological and/or pharmacological activity of C. sanguinolenta. The effectiveness of its extracts is comparable to chloroquine when taken orally (Boye, 1989). Apart from its antimalarial properties (Ansah et al., 2005; Tempesta, 2010; Osei-Djarbeng et al., 2015), it is recognized for its anticancer (Ansah and Mensah, 2013), antihyperglycemic (WHO, 2002), antimicrobial (Sawer et al., 2005) and antimycrobacterial (Gibbons et al., 2003) potential. Preparations from C. sanguinolenta were approved in February 2021 for clinical trials for the treatment of COVID 19 by the Food and Drugs Authority of Ghana (FDA, 2021) and has been shown to inhibit hepatitis B virus replication (Domfeh et al., 2021).
In Ghana, C. sanguinolenta harvesting is done by smallholder farmers (plant collectors) who use the roots to make antimalarial decoctions. “Nibima” is one of the popular decoctions sold in local markets and is produced by the Centre for Plant Medicine Research (CPMR) (https://www.cpmr.org.gh/product-category/decoctions). Overtime, the plants have been overharvested and these plant collectors must travel further into the forest, often with overnight stays, to gather C. sanguinolenta (Amissah, personal communication 2021). Destructive harvesting of the entire plant, along with its root system, is not sustainable in the long term and has already resulted in a substantial decrease in wild populations (Jansen and Schmelzer, 2010; Amissah et al., 2016a).
Over collection and habitat loss poses a significant threat to the conservation efforts of endangered plant species, especially those used for medicinal purposes (Kala, 2000; Reed and Frankham, 2003). Human activities, such as illegal poaching or disruptive harvesting of endemic, medicinal plants, can negatively impact biodiversity at the gene, species, community and ecosystem levels (Meffe and Carroll, 1997; Templeton et al., 2001). This can result in habitat fragmentation, erosion of natural and adaptive genetic diversity, reduction in effective population size, lower evolutionary potential and ultimately contribute to species extinction (Templeton et al., 2001; Soares et al., 2019). Understanding the population genetics of a plant species provides insight into the effects of natural selection, genetic drift, mutation and gene flow that can be used to develop suitable conservation methods and assist with future breeding programmes for plant species on the verge of extinction. Population genetics studies have played a significant role in the conservation of several species. For example, genetic diversity and inbreeding were significantly influenced by the size of the population of Chersophilus duponti, a threatened lark in the Alaudidae (Meffe and Carroll, 1997) and Hypericum cumulicola, a rare species of flowering plant found in small and isolated populations (Oakley, 2015).
As is the practice with several important medicinal plants, there is very little effort devoted to the conservation of C. sanguinolenta. There is an urgent need for establishing a conservation strategy for medicinal plant species in general and specifically for C. sanguinolenta. Currently, there is very limited knowledge of the population dynamics of C. sanguinolenta. As part of research efforts geared at conserving this species, our group has developed protocols for the cultivation of C. sanguinolenta (Jansen and Schmelzer, 2010), in addition, we have developed microsatellite loci for future genetic diversity studies (Amissah et al., 2016b). To address these knowledge gaps, this study seeks to: i) measure the genetic diversity of C. sanguinolenta across eight subpopulations from natural habitats within three regions of Ghana; and ii) describe the spatial structure of these subpopulations.
2 Materials and methods
2.1 Sampling localities, DNA extraction, isolation and PCR amplification
One hundred and eighty-six plants of C. sanguinolenta were collected and assigned to eight subpopulations of which each was represented by 17 to 29 individuals located within the Ashanti (Asuafo-Sekyere West), Eastern (Hwehwee-Ata n’ Ata, Hwehwee-Yaw Gyeni, Kwahu Abene, Kwahu Pepease, OgenyaKrobo/Krobo Gyekiti), and Brong Ahafo (Dromankese Nkoranza North) region in the period between April and December 2015 (Figure 1) .
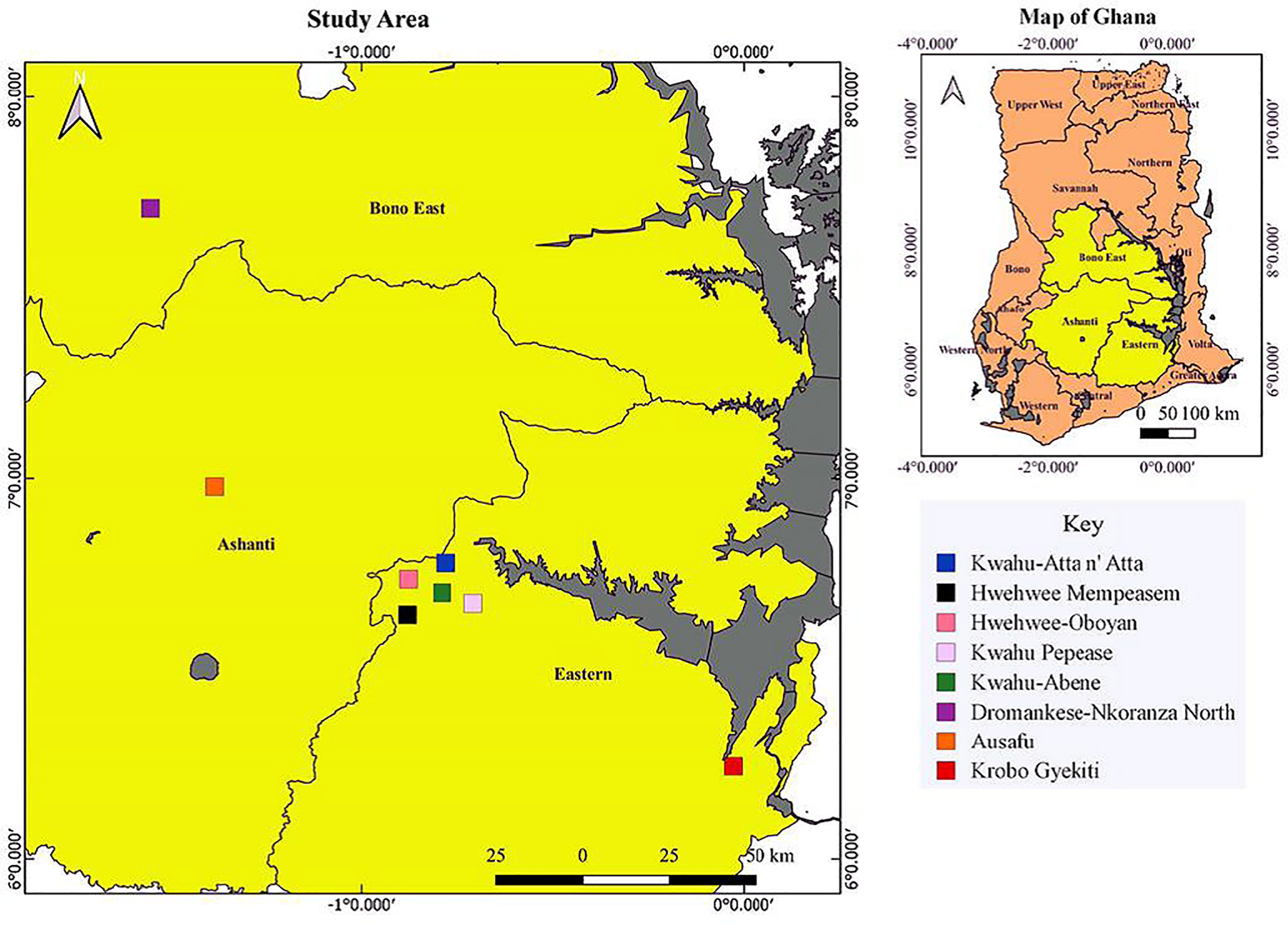
Figure 1 Distribution map of Cryptolepis sanguinolenta subpopulations across three different regions in Ghana.
The identification of plants collected from each location was based on morphological characteristics of the species and verified using herbarium samples from an earlier study. Samples were maintained and grown at the Department of Crop Science of the University of Ghana under field conditions. The site falls within the Coastal Savannah agroecological zone of Ghana with a total annual rainfall of approximately 748 mm and a temperature of between 24 - 32°C. The soil at the site is sandy loam and classified as the Adenta series by the Soil Survey Staff (1999).
2.2 Genotyping
Genomic DNA was isolated from fresh C. sanguinolenta leaf samples using a CTAB protocol (Porebski et al., 1997). The concentration and purity of DNA samples were measured with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE, USA) before analysis. Polymerase chain reaction (PCR) amplification was performed in 10 µl reaction mixtures containing 1 µl DNA template (4 ng/µl), 1 µl of GeneAmp 10x PCR Buffer II (Applied Biosystems, Branchburg, NJ, USA), 1 µl of 25 mM MgCl2, 1 µl of each 2 mM of dNTPs, 1 µl of 2.5 µM specific polymorphic microsatellite primers, 0.08 µl of 5 U AmpliTaq Gold® DNA polymerase (Applied Biosystems), and 4.92 µl of sterile nanopure water. Nine polymorphic microsatellite primer pairs, developed from a previous study (Amissah et al., 2016b), as well as six additional polymorphic microsatellite primer pairs (Table 1) were selected and used for the population studies.
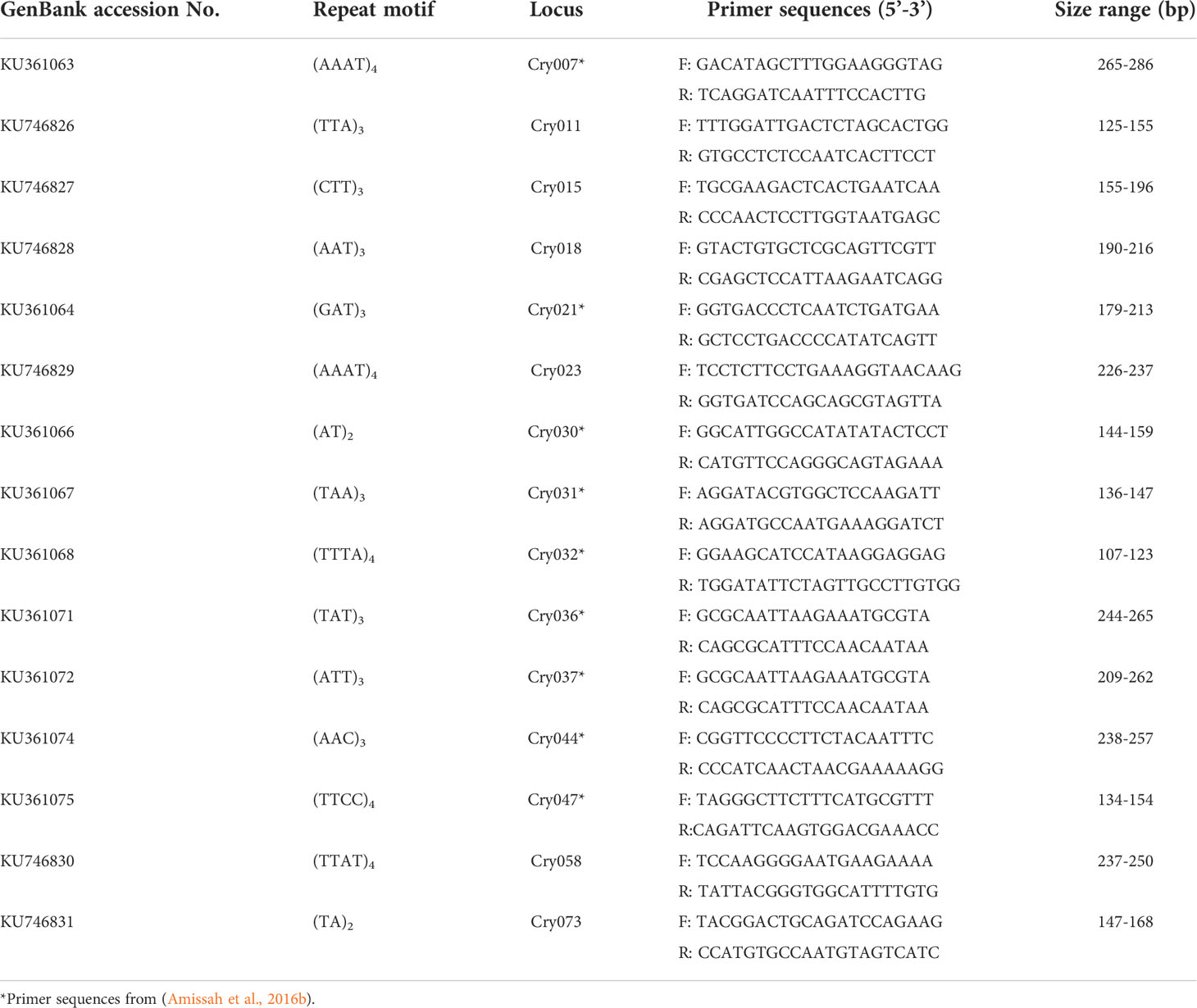
Table 1 Microsatellite loci used to analyze 179 Cryptolepis sanguinolenta samples collected from eight locations in Ghana.
Amplification reactions were temperature cycled in 96-well plates using an Eppendorf Mastecycler Thermocycler (Eppendorf AG, Hamburg, Germany) with the following program: 94°C for 3 min, followed by 35 cycles of 94°C for 40 seconds, 58°C for 40 seconds, 72°C for 30 seconds, and a final extension at 72°C for 4 min. PCR products were analyzed with the QIAxcelCapillary Electrophoresis System (Qiagen, Valencia, CA, USA) using an internal 25-bp DNA size ladder. Data were automatically recorded and exported using QIAxcel ScreenGel Software,which provided both a gel view and an electropherogram of the separated PCR products (alleles).
2.3 Data analyses
2.3.1 Loci polymorphism and population diversity
The 186 individuals were analyzed using 15 species-specific polymorphic microsatellite primers. DNA of seven individual samples failed to amplify across several loci and therefore were excluded from further analyses resulting in 179 individuals in the study. The program FLEXIBIN (Amos et al., 2007) was used to assign alleles into appropriate classes. To achieve the best allelic class fit, the program uses a simple algorithm through a series of steps and phases to find the best binning combinations.
Standard measures of genetic diversity among 179 individuals of C. sanguinolenta were determined using Nei’s gene diversity (Amos et al., 2007) in FSTAT version 2.9.3.2 (Goudet, 2001; Goudet, 2005) and program R version 4.1.2 using packages poppr (Kamvar et al., 2014; Kamvar et al., 2015) and hierfstat (Goudet, 2005). Analyses included the number of alleles observed at each locus (N) and across subpopulations (Na), number of effective alleles (Ne), and allelic richness (AR), a measure of the number of alleles independent of sample size. Allelic richness, calculated using a rarefication method based on a minimum sample size of 15 diploid individuals or 30 genes, is used as an estimate of the long-term evolutionary potential of given individual(s) to adapt and persist in a population (El Mousadik and Petit, 1996). We also estimated observed and expected heterozygosity (gene diversity) under Hardy Weinberg equilibrium (Ho and He, respectively), linkage disequilibrium (rbard) (non-random association of alleles between loci), and Shannon-Weiner diversity index (H), which considers both allele richness and evenness of the allelic distribution. Further analyses included the number of unique private alleles across loci and subpopulations. F-statistics estimates included inbreeding coefficient (FIS) and fixation index (FST). In addition, gene flow (Nm) estimates were calculated using GenAlEx version 6.502 (Peakall and Smouse, 2006; Peakall and Smouse, 2012).
2.3.2 Population structure and historical demography
Genetic structure analysis of C. sanguinolenta was performed using Bayesian cluster analysis (Peakall and Smouse, 2012) in program STRUCTURE version 2.3.4 (Pritchard and Donnelly, 2001) with a series of complementary approaches including analysis of molecular variance (AMOVA) (Excoffier and Lischer, 2010), and Mantel test for isolation by distance. The program STRUCTURE was used with 20 independent runs for each k value between one and ten at 500,000 MCMC (Markov Chain Monte Carlo) repetitions and a burn-in period of 500,000 iterations using an admixture model with correlated allele frequencies (without prior assumption of population origin). The admixture model assumes that individuals with admixed ancestry can inherit a portion of their genome from ancestors in population k (Porras-Hurtado et al., 2013). STRUCTURE HARVESTER (Earl and VonHoldt, 2012), a web-based program for obtaining K values and summarizing output data from STRUCTURE, was used to estimate the optimum value of genetic clusters using Evanno’s method (Evanno et al., 2005). Data was visualized using program web-based program POPHELPER (Francis, 2017). Because the STRUCTURE-like approach assumes panmictic populations and unlinked markers (Pritchard et al., 2000), alternatively, model-free approaches that utilize multivariate K-means clustering such as Discriminant Analysis of Principal Components (DAPC) are often used. Here, we used DAPC to further investigate the underlying structure among C. sanguinolenta populations using R package adegenet (Jombart, 2008; Jombart and Ahmed, 2011). The initial selection includes a principal component analysis (PCA) identification, followed by the selection of appropriate PCA vectors that can explain the majority of variance while minimizing over-fit of the DAPC, and finally using a selected number of PCAs that could reveal differences between groups and minimize within-group variations (Ony et al., 2021). We also performed pairwise FST analysis for all pairs of subpopulations using 10,000 permutations in GenAlEx. Significant FST values were calculated after sequential Bonferonni corrections (Rice, 1989; Bergamaschi and Lama, 2015).
The hierarchical distribution of genetic variation of the C. sanguinolenta subpopulations was measured using Arlequin version 3.5.1.2 (Excoffier and Lischer, 2010). AMOVA was used to partition the total variance into hierarchical components (among subpopulations and within subpopulations). Although AMOVA findings can complement STRUCTURE results, these outcomes should not be taken as independent confirmation of implied hierarchical structure. AMOVA analyses for C. sanguinolenta subpopulations were partitioned based on all sampling localities clustered as one hierarchal group and clusters identified by STRUCTURE. For STRUCTURE partitioning, each individual was clustered into one of the two groups as determined by the program.
To determine whether or not geographical structure is best explained by hierarchical clustering, the correlation between pairwise genetic and geographic distance was estimated using mass package (Venables and Ripley, 2002) in program R. Statistical significance was evaluated using the Mantel test using 9,999 permutations. R program was also utilized for the construction of UPGMA (unweighted pair group method of arithmetic averages) dendrogram based on Bruvo’s distance with 1,000 bootstrap replicates and threshold set at 50% using package poppr (Kamvar et al., 2014; Kamvar et al., 2015). Lastly, C. sanguinolenta populations were tested for evidence of bottlenecks using the software BOTTLENECK version 1.2 (Piry et al., 1999). The program evaluates the presence of population bottleneck as an excess (recent population bottleneck) or deficit (recent population expansion) of genetic diversity, relative to the number of alleles present in the C. sanguinolenta.
3 Results
3.1 Population diversity
One hundred and one alleles were identified using 15 polymorphic microsatellite loci across the three regions in Ghana where C. sanguinolenta plants are found (Table 2).
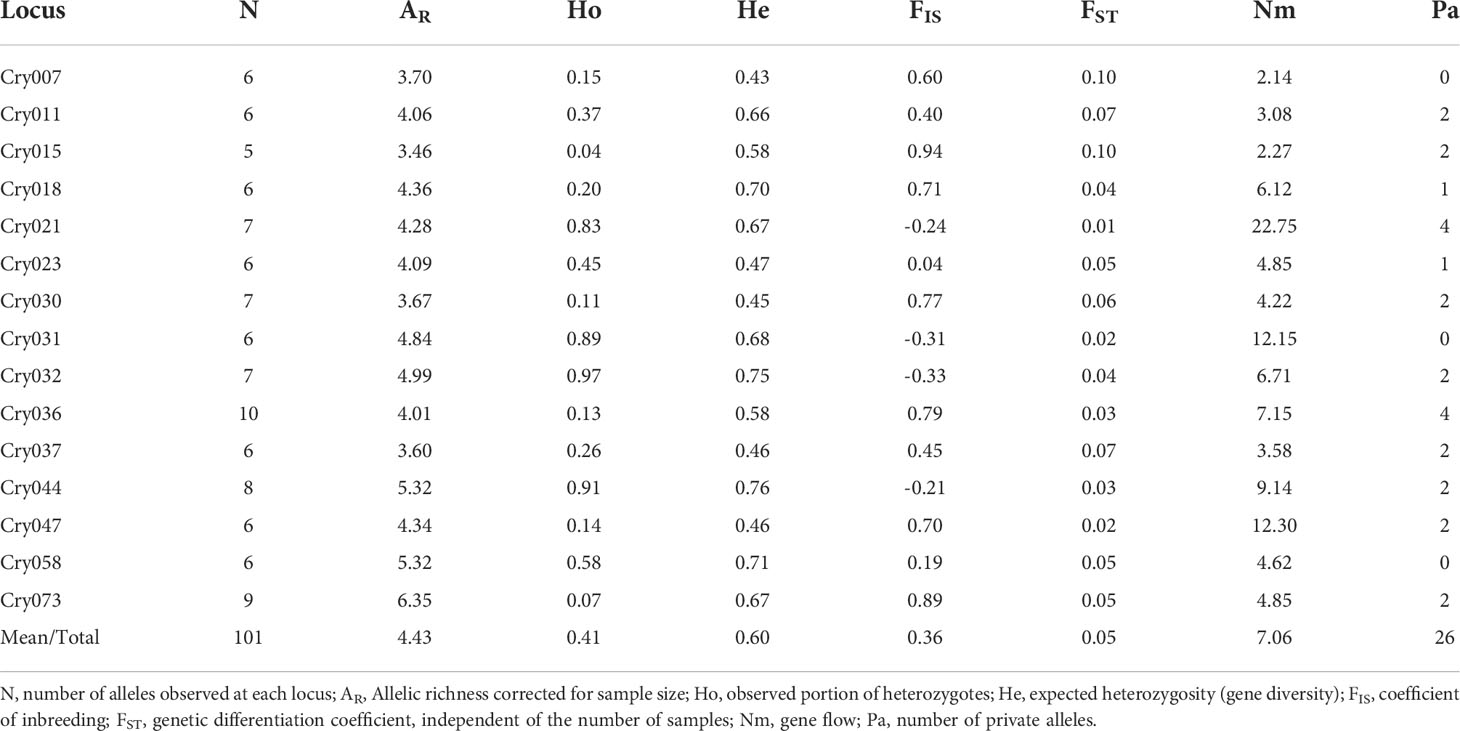
Table 2 Diversity indices and gene flow estimates for fifteen microsatellite loci over Cryptolepis sanguinolenta populations.
The mean allelic richness (AR), as assessed with 15 microsatellites was 4.43 and the number of alleles varied from 5 (Cry015) to 10 (Cry036) with a total number of 101 alleles (Table 2). Overall, subpopulations differentiation was low (FST=0.05) and indicated little genetic differentiation, high gene flow (Nm=7.06), significant inbreeding (FIS=0.36) and structure among the C. sanguinolenta subpopulations (Ho<He) (Table 2). Expected heterozygosity across all loci ranged from 0.43 to 0.76 with a mean of 0.60. Twelve of the fifteen tested microsatellite loci yielded private alleles and resulted in the discovery of 26 unique alleles (Table 2).
The effective number of alleles (Ne) ranged from 2.49 to 2.72 with an average of 2.60 across all subpopulations. Shannon-Weiner diversity index (H) and allelic richness (AR) values were high across all subpopulations (H=3.10, AR=4.15) but generally higher for subpopulations from the Eastern region (H=3.12, AR=4.21 for six Eastern subpopulations) when compared to Ashanti (H=3.09, AR=3.89) and Brong-Ahafo (H=3.0, AR=4.02) regions (Table 3).
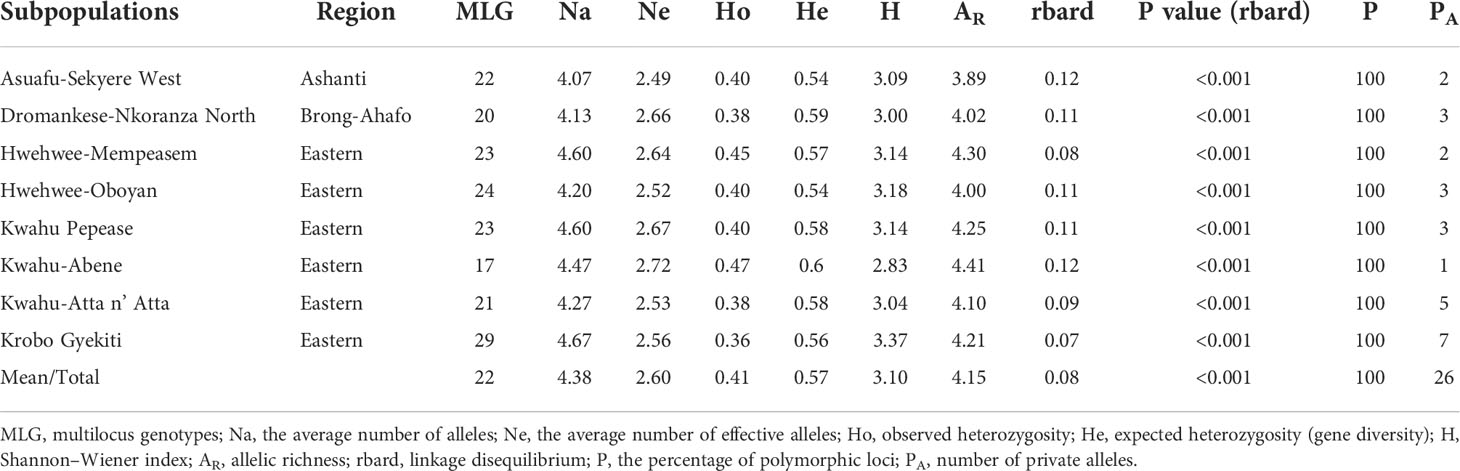
Table 3 Genetic diversity estimates for sampled Cryptolepis sanguinolenta individuals averaged across eight subpopulations and 15 microsatellite loci.
Mean observed (Ho) and expected heterozygosity, or gene diversity (He), were 0.41 and 0.57, respectively, indicating the presence of population structure (Table 3). Weak but significant linkage disequilibrium was detected across all eight sub-populations (rbard = 0.08, P<0.01) (Table 3).
Private alleles (total PA=26) were identified from all eight subpopulations of C. sanguinolenta. (Table 3). Krobo Gyekiti (PA=7), Kwahu-Atta n’ Atta (PA=5), Kwahu Pepease (PA=3), and Hwehwee-Oboyan (PA=3) subpopulations from the eastern region had the highest number of private alleles per subpopulations (Table 3). The highest average number of alleles was found in the eastern region (Hwehwee-Mempeasem, Kwahu Pepease and Krobo Gyekiti subpopulations) compared to Ashanti and Brong-Ahafo regions (Asuafu-Sekyere and Dromankese-Nkoranza North subpopulations) (Table 3).
3.2 Population structure and historical demography
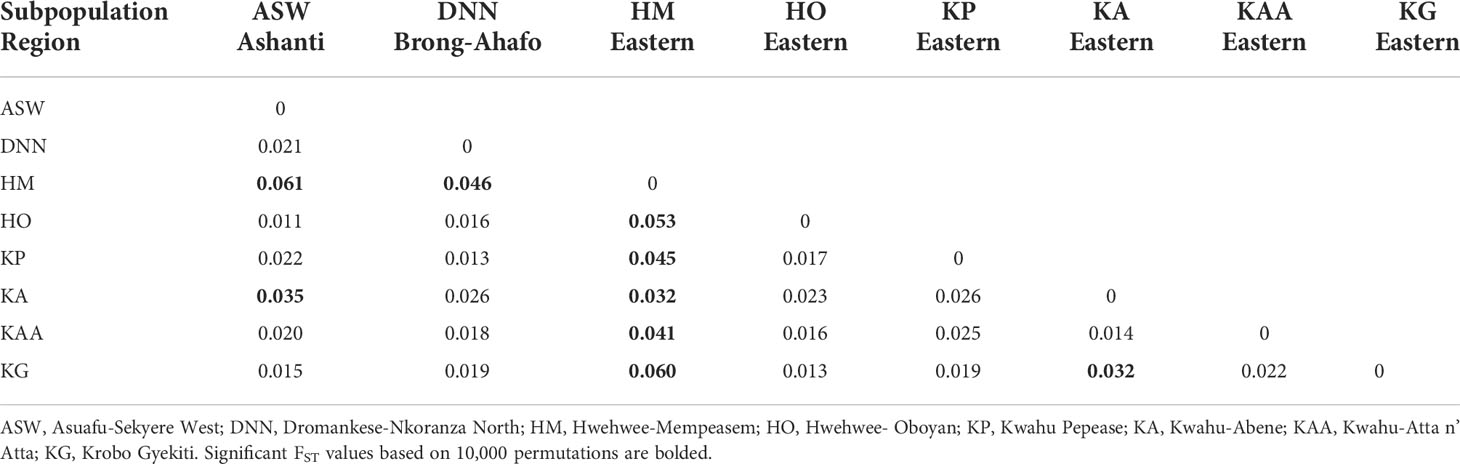
Subpopulations of C. sanguinolenta demonstrated very low levels of population differentiation (FST=0.05) (Table 2) despite the distribution of the plant across three different regions in Ghana. Comparisons among Asuafu-Sekyere West and Dromankese-Nkoranza North subpopulations resulted in non-significant pairwise FST values (Table 4). However, significant values were obtained for the Hwehwee-Mempeasem subpopulation from the east when compared to the other seven C. sanguinolenta subpopulations in Ghana (Table 4). Analyses of all other subpopulations resulted in non-significant FST values (Table 4) except for Kwahu-Abene and Krobo Gyekiti subpopulation from the eastern region.
The greatest pairwise genetic differentiation was among Asuafu-Sekyere West and Hwehwee-Mempeasem (FST=0.061; P<0.001) and Hwehwee-Mempeasem and Krobo Gyekiti subpopulations (FST=0.060; P<0.001) (Table 4), which could be biased by differences in sample size among the regions (Table 3; Table 4). Population structuring is best explained by the presence of two genetic clusters (K=2), even though we examined delta k for the presence of three and four genetic clusters (Figure 2).
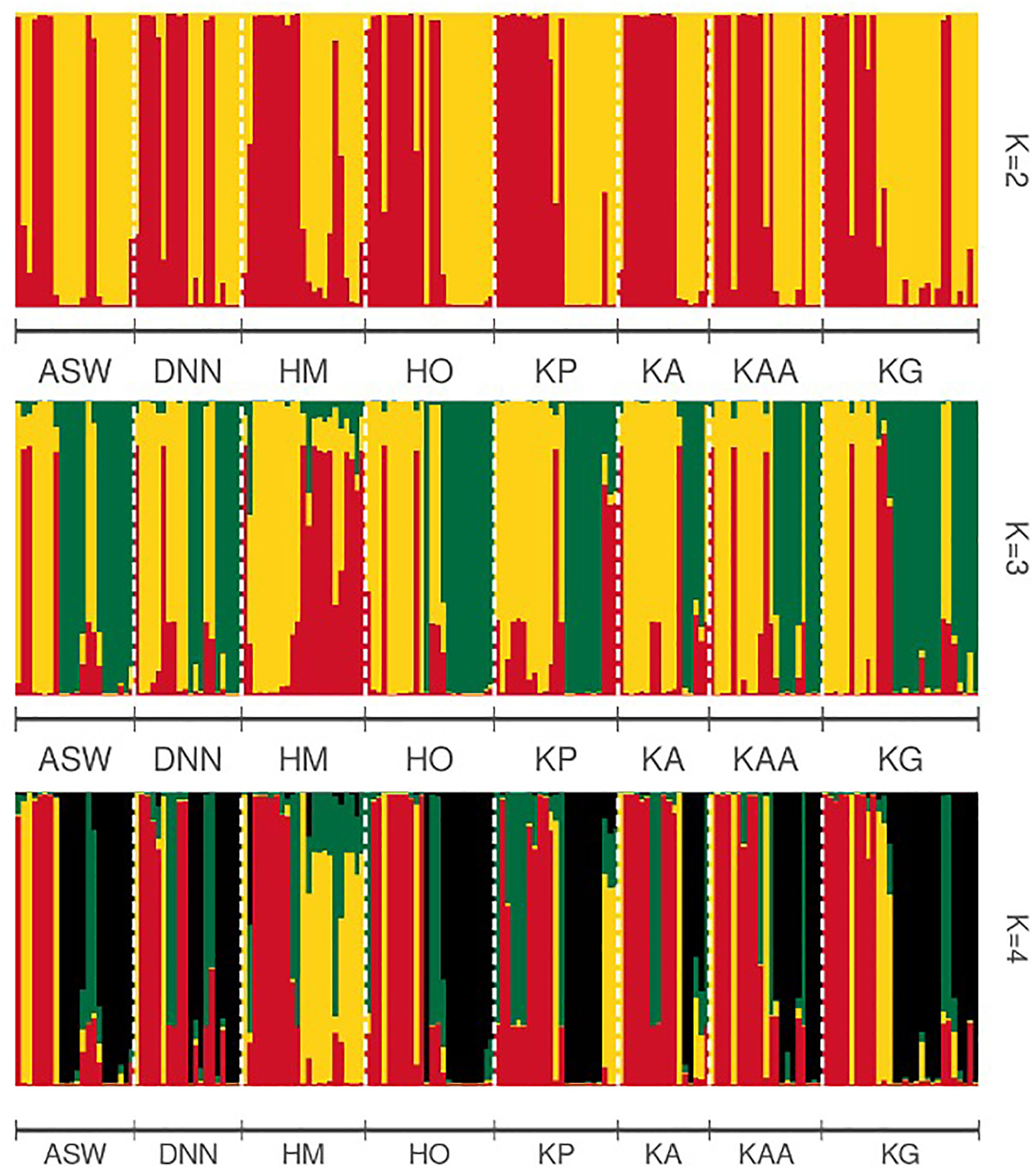
Figure 2 STRUCTURE bar plot for eight Cryptolepis sanguinolenta subpopulations showing results for (K=2-4). Each bar indicates the individual assignment probability of that individual belonging to one of the clusters indicated by different colors, based on twenty iterations. Evanno’s method indicated that two genetic clusters (K = 2) best explain the population structure in this species. Population designations are ASW, Asuafu-Sekyere West; DNN, Dromankese-Nkoranza North; HM, Hwehwee-Mempeasem; HO, Hwehwee-Oboyan; KP, Kwahu Pepease; KA, Kwahu-Abene; KAA, Kwahu-Atta n’ Atta; KG, Krobo Gyekiti.
All cluster groups had representatives of individuals from the eight subpopulations present, with high levels of admixture among the individuals in each subpopulation, and no evidence of geographical structuring (Figure 2). Consistent with the lack of isolation by distance (P=0.34; r=0.012; Figure 3), our findings are further supported by the Discriminant Analysis of Principal Components (DAPC) plot that indicated the presence of two distinct groups with mixed individuals from different subpopulation (Figure 4).
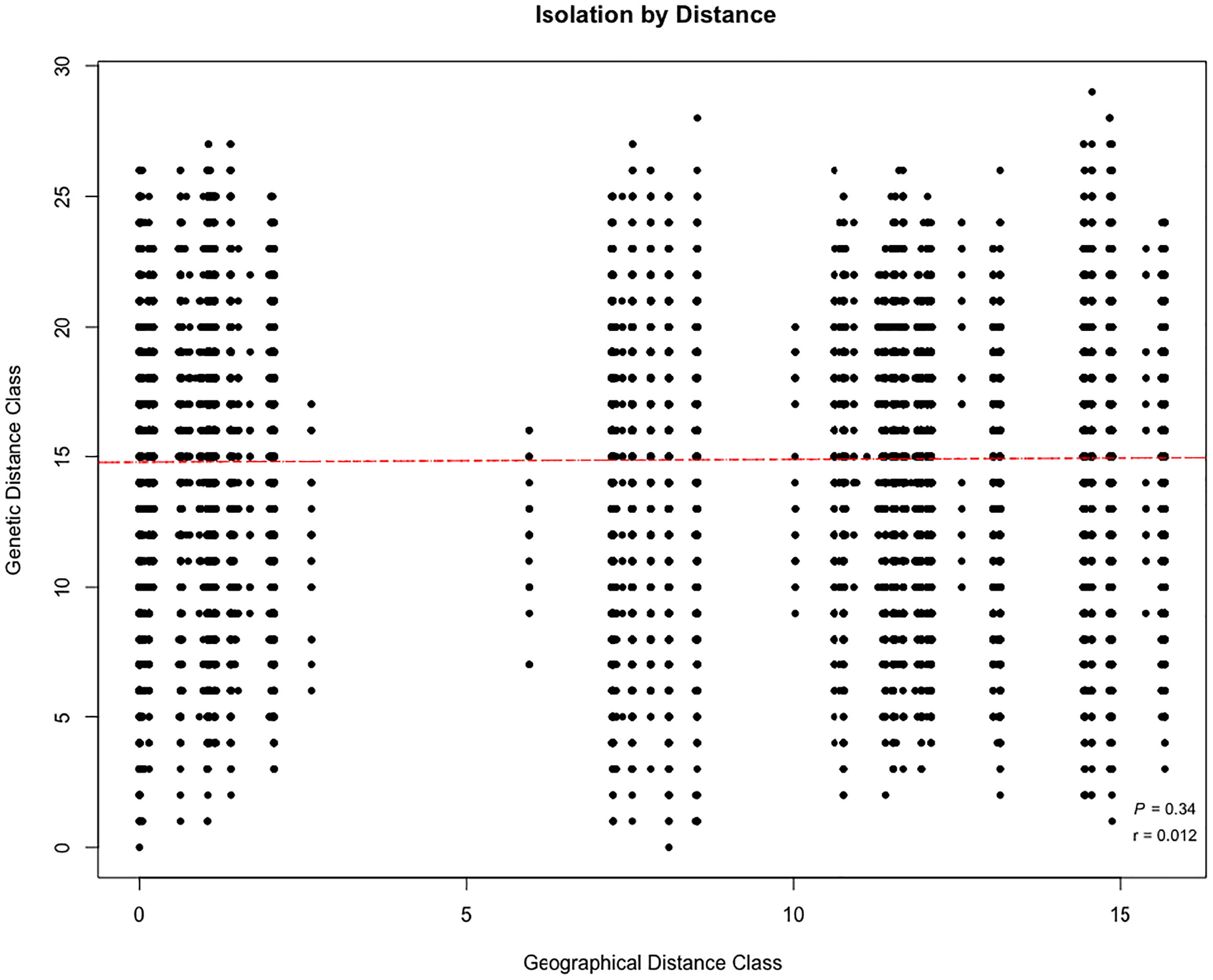
Figure 3 Pairwise genetic and geographical distances of 179 Cryptolepis sanguinolenta samples collected from 8 subpopulations in Ghana displayed as a scatterplot.
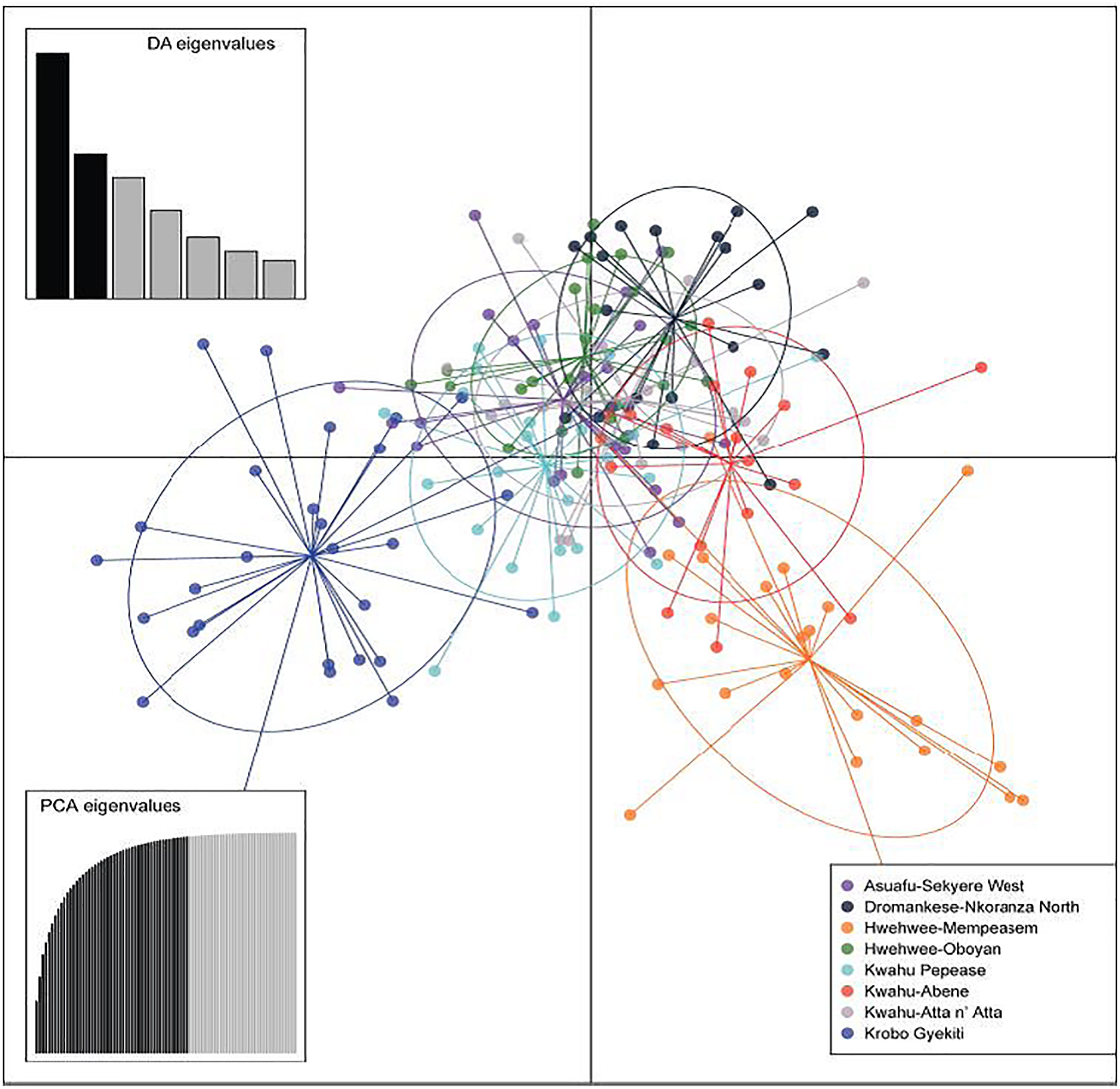
Figure 4 Discriminant Analysis of Principal Components (DAPC) plot of Cryptolepis sanguinolenta individuals across eight subpopulations in Ghana. Data was constructed using 1,000 permutations. Discriminant Analysis eigenvalues are also presented.
To test whether Isolation by Distance (IBD) occurs on a smaller scale, C. sanguinolenta individuals were clustered based on geographical subpopulation assignment and analyzed separately. Three subpopulations from the eastern region indicated significant evidence of isolation by distance - Kwahu Pepease, Kwahu-Atta n’ Atta and Krobo Gyekiti (P<0.05). When C. sanguinolenta individuals were grouped based on STRUCTURE findings, the two clusters did not have significant correlation between genetic and geographic distance (P=0.06). The UPGMA dendrogram revealed a similar pattern of two groups and a lack of geographical structuring among the examined subpopulations (Figure 5).
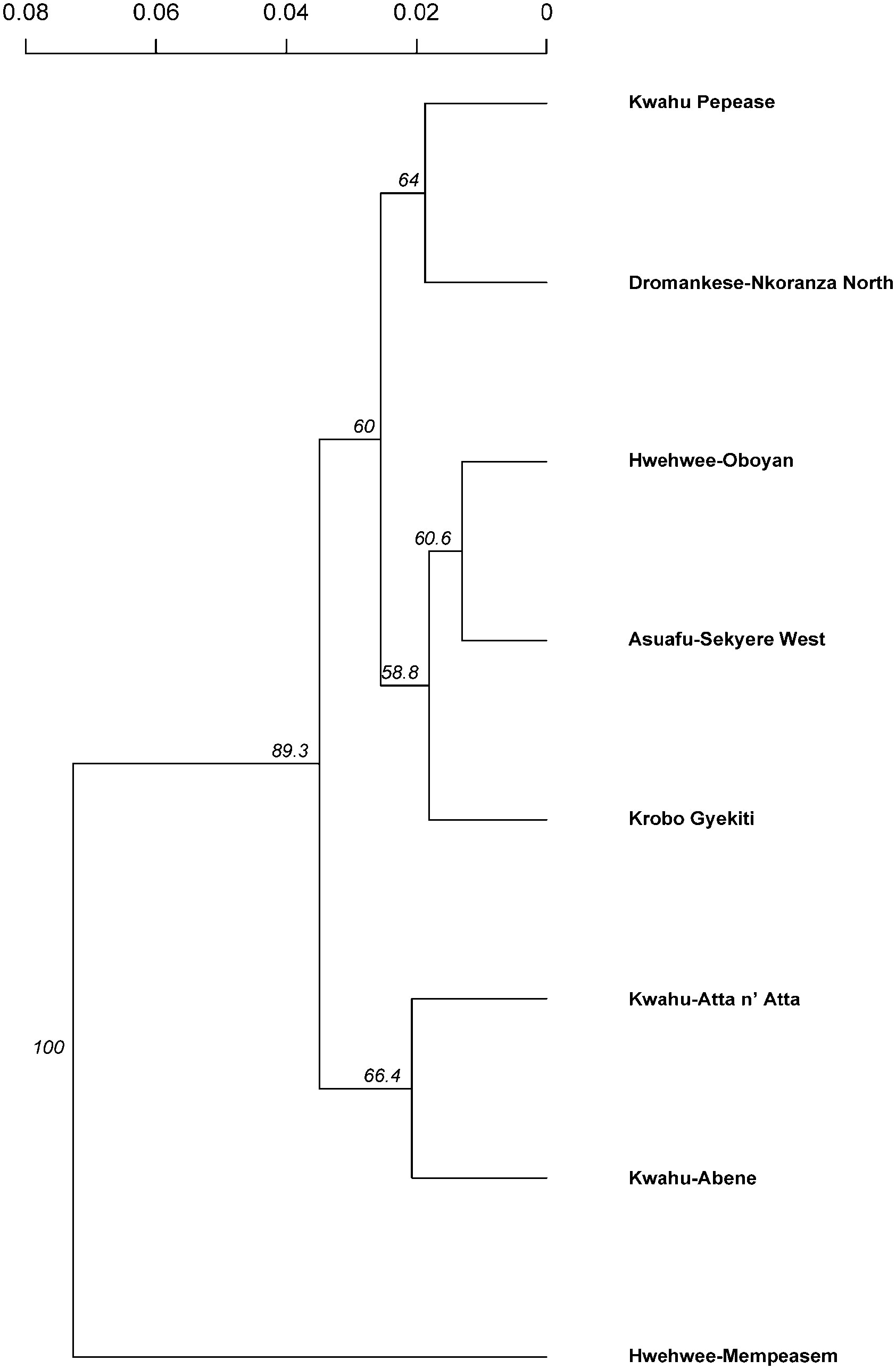
Figure 5 An unweighted pair group method with arithmetic mean (UPGMA) dendrogram of Cryptolepis sanguinolenta populations using 15 polymorphic microsatellite primers based on Bruvo’s genetic distance. Numbers indicate the percentage of bootstrap support using 1,000 replications (threshold set at 50%).
Two different analyses of molecular variance (AMOVA) were conducted. The results of the first AMOVA (one hierarchical grouping), showed that the variation within subpopulations was higher (96.95%), than among subpopulations (3.05%) (Table 5). A similar trend of high individual variation (74.56%) and relatively lower variation among subpopulations (25.44%) was obtained after data was partitioned based on the STRUCTURE results (Table 5).
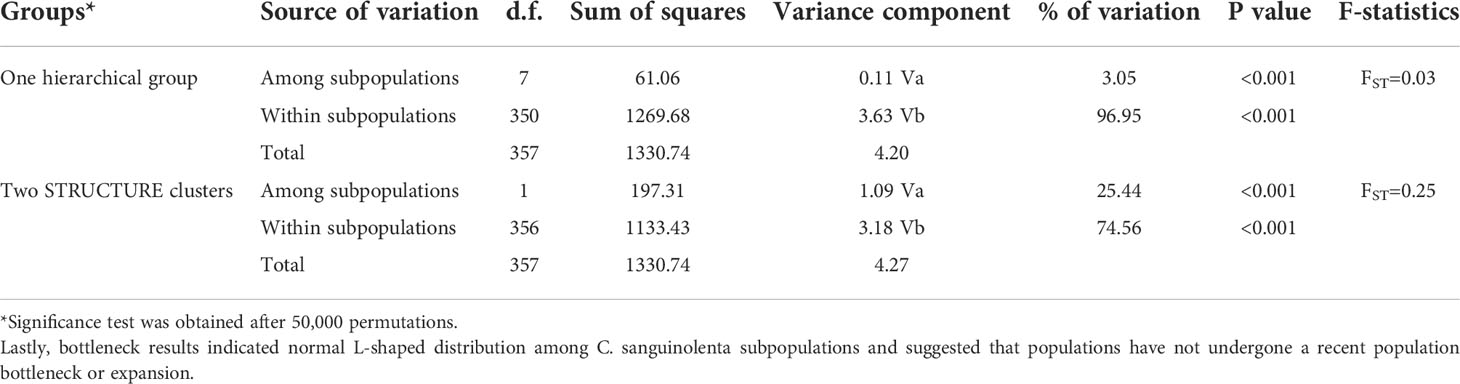
Table 5 Analysis of molecular variance (AMOVA) for Cryptolepis sanguinolenta subpopulations using 15 microsatellite loci.
4 Discussion
The large-scale use of artemisinin since its initial discovery in the early 1970s, has resulted in worldwide drug resistance prompting a switch in strategy to artemisinin-based combination therapy (ACT) in 2005 (Tu, 2011; WHO, 2020). The problem is further compounded by the increased resistance of malaria vectors to many insecticides currently available (Balkew et al., 2012; Massebo et al., 2013), which are cost-prohibitive on a large-scale and unsustainable in the long term. According to reports from WHO (WHO, 2015), artemisinin-based combination therapy (ACTs) had been adopted as a first-line treatment policy in several countries but may not be readily available in rural, remote and poverty-stricken communities of Western Africa, including Ghana. As a result, in Ghana, Mali, Zambia and Nigeria, traditional medicine approach is practiced with 60% of children who reported high fever resulting from malaria infection (Abdullahi, 2011).
People living in developing countries have limited access to modern medicine and sophisticated treatments (Reed and Frankham, 2003), thus often relying on medicinal plants that offer an alternative, cost-effective, and freely available traditional approach for malaria control (Turkson et al., 2019). In some areas, the traditional medicine approach is the only source of medical care available (Romero-Daza, 2002; Abdullahi, 2011). Based on African Health Monitor Report in 2003, there is one healer for every 500 individuals vs. one medical doctor for every 40,000 patients in Africa; for Ghana, there is one healer for every 200 individuals vs. one medical doctor for every 20,000 patients (Chatora, 2003; Abdullahi, 2011; WHO, 2014). These staggering numbers, combined with the fact that the majority of doctors reside in urban areas, stress the importance of traditional healers and indigenous plants in treatments of communities in rural Africa. The healers, also known as traditional doctors, approach the patients holistically, treating not only the given condition but restoring the social, emotional, and spiritual equilibrium of their patients based on community history and customs (Hillenbrand, 2006; Abdullahi, 2011).
Traditional African knowledge of indigenous plants have been utilized for healing and treating various ailments, including malaria, for centuries. Preservation and conservation of these nearly lost species is imperative, and documentation of alternative medicine applications and approaches used by traditional healers as an ancient and culture-bound method of healing for centuries should be prioritized (Abdullahi, 2011; Veeresham, 2012). These plants are largely understudied, and very limited financial and technological resources are available for research related to drug development in these countries (Reed and Frankham, 2003). Indigenous C. sanguinolenta contains not only antimalarial, anti-cancer, antihyperglycemic, antimicrobial, and antimycobacterial properties, but can be utilized to promote sustainable and climate-smart agro-ecosystems across West Africa. This, in turn, would allow sustainable commercialization of this plant as a cash crop across its native distribution, thus providing farmers with additional income while preventing destructive collection.
Despite overharvesting and near extinction of these indigenous plants, our findings provided evidence of high genetic diversity among C. sanguinolenta subpopulations across Ghana. In declining and isolated populations unable to respond to a variety of selection pressures, genetic variation can be reduced rapidly as a result of the limited number of individuals and inbreeding within the populations that are often driven by random genetic drift (Ellstrand and Elam, 1993; Hawley et al., 2006; Luan et al., 2006). This can further decrease species fitness, prospects for adaptive change, ultimately causing species extinction (Fischer and Matthies, 1998; Severns, 2003; Hawley et al., 2006; Hadziabdic, 2010). However, this is the first comprehensive study focusing on the population genetic and spatial structure of this medicinal plant, thus we can expect these individuals to be somewhat resistant to apparent population bottleneck and genetic drift effects, at least in the short term. Long term monitoring, better propagation systems, and selective breeding processes are necessary for better informed conservation efforts.
Our results indicated the presence of a weak population structure low genetic differentiation, and high gene flow among wild C. sanguinolenta subpopulations. Findings from this study can be attributed to the specialized wind dispersal mechanism of C. sanguinolenta seeds over long distances, as the main factor affecting variation in gene flow and population structure similarly reported in Cecropia obtusifolia (Epperson and Alvarez-Buylla, 1997), and some woody plant species (Hamrick et al., 1992), and thus providing long-term potential for the conservation of C. sanguinolenta. Gene flow is a directional transfer of genetic material across populations as a result of seed dispersal over a period of time and in some tree and shrub species, it is positively correlated with the direction of wind flows and rates (Kling and Ackerly, 2021). The signal of a weak population structure could be due to high gene flow in the species as reported by Waples (1998), since gene flow is known to be the predominant evolutionary force for the genetic structure (García-Verdugo et al., 2010). The unlimited gene flow and low magnitude of genetic differentiation observedcould be attributed to the relatively larger genetic neighbourhood and sampling size. According to Franceschinelli and Kesseli (1999), the relative magnitude of genetic differentiation is directly influenced by the size of the sampling area such that genetic differentiation increased with decreased sample plots in Helicteres brevispira.
In a large genetic neighbourhood, proposed by Wright (Wright, 1943; Wright, 1946), uniform selection pressures may limit differentiation among individuals. Alternatively, local differences and a relatively slow dispersal process of new alleles in these smaller geographical areas may increase differentiation between the groups (Wright, 1943; Wright, 1946). The presence of a contiguous genetic neighbourhood in C. sanguinolenta subpopulations in the present study is supported by STRUCTURE findings and indicated the presence of two distinct genetic clusters and extensive admixture as a result of gene flow. The high gene flow found in our study indicates that the effective population size of current C. sanguinolenta wild plants may be sufficient to offset the impact of population decline and the effect it has on genetic diversity. Similar to findings by Ratnaningrum et al. (2017), larger gene flow was found to be the main emphasis for natural genetical processes in Santalum album population.
The Mantel test revealed no correlation between Bruvo’s distance and geographic distance suggesting that genetic differentiation in the 8 subpopulations is not caused by geographic isolation due to distance. A possible explanation for the absence of the influence of isolation distance on genetic diversity could be due to high gene flow, sexual reproduction, and wind dispersal of seeds in C. sanguinolenta. The results of the Mantel test are similar to findings in Miscanthus, M. sinensis (Nie et al., 2014; Zhao et al., 2017).
These findings have practical implications for the conservation of C. sanguinolenta. First, the absence of geographic distance on genetic differentiation in C. sanguinolenta subpopulations, suggests that sampling from one geographic area could help cover a significant proportion of the genetic diversity in C. sanguinolenta. We recommend a protection strategy as follows: 1) protection of the plants in the Kwahu-Abene area, and 2) scaling-up the sampling to other C. sanguinolenta populations with high genetic diversity such as Hwehwee-Mempeasem, Hwehwee-Oboyan, Kwahu Pepease, and Asuafu-Sekyere West. In addition, ex situ conservation by encouraging the cultivation of C. sanguinolenta among smallholder farmers should be explored to ensure its availability in the needed quantities, thus contributing to a decrease in the harvest volumes from the wild, as a way of conserving the species in situ.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author contributions
Conceptualization JNA, RNT, and DH; methodology JNA and SLB; validation JNA and SLB; formal analysis JNA and DH; investigation, JNA; resources JNA, DH, and RNT; data curation JNA and DH; writing—original draft preparation JNA and DH; writing—review and editing all authors; visualization JNA; supervision JNA; project administration JNA; funding acquisition JNA. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by USDA NACA, grant number 58-6062-006, and the Volkswagen Foundation of Germany, grant number (85456). Funding from the Fulbright Scholar Fellowship supported DH’s visit to Ghana to draft portions of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdullahi A. A. (2011). Trends and challenges of traditional medicine in Africa. Afr. J. Traditional Complementary Altern. Medicines 8(5S):115–123. doi: 10.4314/ajtcam.v8i5S.5
Abebe D., Garedew B. (2019). Ethnobotanical survey of plants traditionally used for malaria prevention and treatment in indigenous villages of tepi town south West Ethiopia. J. Pharmacognosy Phytother 11(1):9–16. doi: 10.5897/JPP2018.0532
Amissah J., Osei-Safo D., Asare C., Missah-Assihene B., Danquah E., Addae-Mensah I. (2016a). Influence of age and staking on the growth and cryptolepine concentration in cultivated roots of Cryptolepis sanguinolenta (Lindl.) schlt. J. Med Plants Res. 10(9):113–121. doi: 10.5897/JMPR2015.5793
Amissah N., Wadl P., Hadziabdic D., Boggess S., Trigiano R. (2016b). Characterization of thirteen microsatellite loci from the ghanian antimalarial plant Cryptolepis sanguinolenta. J. Med Plants Res. 10(14):183–187. doi: 10.5897/JMPR2016.6047
Amos W., Hoffman J., Frodsham A., Zhang L., Best S., Hill A. (2007). Automated binning of microsatellite alleles: Problems and solutions. Mol. Ecol. Notes 7(1):10–14. doi: 10.1111/j.1471-8286.2006.01560.x
Ansah C., Khan A., Gooderham N. J. (2005). In vitro genotoxicity of the West African anti-malarial herbal Cryptolepis sanguinolenta and its major alkaloid cryptolepine. Toxicology 208(1):141–147. doi: 10.1016/j.tox.2004.11.026
Ansah C., Mensah K. B. (2013). A review of the anticancer potential of the antimalarial herbal Cryptolepis sanguinolenta and its major alkaloid cryptolepine. Ghana Med. J. 47(3):137–147.
Balkew M., Getachew A., Chibsa S., Olana D., Reithinger R., Brogdon ,. W. (2012). Insecticide resistance: A challenge to malaria vector control in Ethiopia. Malaria J. 11(1):1–2. doi: 10.1186/1475-2875-11-S1-P139
Bergamaschi A. C., Lama M. A.D. (2015). Gene variation and genetic differentiation among populations of the solitary mud dauber wasp Trypoxylon (Trypargilum) albitarse Fabricius 1804 (Hymenoptera, Crabronidae). Genetics and Molecular Biology 38, 519–526. doi: 10.1590/S1415-475738420150097
Boye G. L. (1989). “Studies on antimalarial action of cryptolepis sanguinolenta extract,” in International symposium on East-West medicine (Seoul, Korea), 242–251.
Chatora R. (2003). An overview of the traditional medicine situation in the African region. Afr. Health Monitor 4(1):4–7.
Cimanga K., De Bruyne T., Pieters L., Vlietinck A. J., Turger C. A. (1997). In vitro and in vivo antiplasmodial activity of cryptolepine and related alkaloids from Cryptolepis sanguinolenta. J. Natural Products 60(7):688–691. doi: 10.1021/np9605246
Domfeh S. A., Narkwa P. W., Quaye O., Kusi K. A., Rivera O., Danaah M. M., et al. (2021). Cryptolepine and nibima inhibit hepatitis b virus replication. Sci. Afr. 13, e00942. doi: 10.1016/j.sciaf.2021.e00942
Dwuma-Badu D., Ayim J. S., Fiagbe N. I., Knapp J. E., Schiff P. L. Jr., Slatkin D. J. (1978). Constituents of West African medicinal plants XX: Quindoline from Cryptolepis sanguinolenta. J. Pharm. Sci. 67(3):433–434. doi: 10.1002/jps.2600670350
Earl D. A., VonHoldt B. M. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the evanno method. Conserv. Genet. Resour. 4(2):359–361. doi: 10.1007/s12686-011-9548-7
Ellstrand N. C., Elam D. R. (1993). Population genetic consequences of small population size: implications for plant conservation. Annu. Rev. Ecol. Systematics 24(1):217–242. doi: 10.1146/annurev.es.24.110193.001245
El Mousadik A., Petit R. (1996). High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.) skeels] endemic to Morocco. Theor. Appl. Genet. 92(7):832–839. doi: 10.1007/BF00221895
Epperson B. K., Alvarez-Buylla E. R. (1997). Limited seed dispersal and genetic structure in life stages of cecropia obtusifolia. Evolution 51(1):275–282. doi: 10.1111/j.1558-5646.1997.tb02409.x
Evanno G., Regnaut S., Goudet J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 14(8):2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Excoffier L., Lischer H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and windows. Mol. Ecol. Resour. 10(3):564–567. doi: 10.1111/j.1755-0998.2010.02847.x
FDA (2021). Food and drugs authority approves first herbal medicine for clinical trial on covid-19 treatment, 1st 2021 ed. Food and Drugs Authority, Ghana
Fischer M., Matthies D. (1998). Effects of population size on performance in the rare plant Gentianella germanica. J. Ecol. 86(2):195–204. doi: 10.1046/j.1365-2745.1998.00246.x
Franceschinelli E. V., Kesseli R. (1999). Population structure and gene flow of the Brazilian shrub helicteres brevispira. Heredity 82(4):355–363. doi: 10.1038/sj.hdy.6884790
Francis R. M. (2017). Pophelper: an r package and web app to analyse and visualize population structure. Mol. Ecol. Resour. 17(1):27–32. doi: 10.1111/1755-0998.12509
García-Verdugo C., Forrest A. D., Fay M. F., Vargas P. (2010). The relevance of gene flow in metapopulation dynamics of an oceanic island endemic, olea europaea subsp. guanchica. Evolution: Int. J. Organic Evol. 64(12):3525–3536. doi: 10.1111/j.1558-5646.2010.01091.x
Gibbons S., Fallah F., Wright C. W. (2003). Cryptolepine hydrochloride: a potent antimycobacterial alkaloid derived from Cryptolepis sanguinolenta. Phytother Research: Int. J. Devoted to Pharmacol. Toxicological Eval. Natural Product Derivatives 17(4):434–436. doi: 10.1002/ptr.1284
Goudet J. (2001) FSTAT, a program to estimate and test gene diversities and fixation indices, version 2.9. 3. Available at: http://www2.unil.ch/popgen/softwares/fstat.htm.
Goudet J. (2005). Hierfstat, a package for r to compute and test hierarchical f-statistics. Mol. Ecol. Notes 5(1):184–186. doi: 10.1111/j.1471-8286.2004.00828.x
Hadziabdic D. (2010). Evaluation of genetic diversity of flowering dogwood (Cornus florida l.) in the eastern united states using microsatellites. PhD diss., University of Tennessee, 2010. Available at: https://trace.tennessee.edu/utk_graddiss/694.
Hamrick J. L., Godt M. J. W., Sherman-Broyles S. L. (1992). “Factors influencing levels of genetic diversity in woody plant species,” in Population genetics of forest trees (Springer Dordrecht).
Hawley D. M., Hanley D., Dhondt A. A., Lovette I. J. (2006). Molecular evidence for a founder effect in invasive house finch (Carpodacus mexicanus) populations experiencing an emergent disease epidemic. Mol. Ecol. 15(1):263–275. doi: 10.1111/j.1365-294X.2005.02767.x
Hillenbrand E. (2006). Improving traditional-conventional medicine collaboration: Perspectives from cameroonian traditional practitioners. Nordic J. Afr. Stud. 15(1):1–15. doi: 10.53228/njas.v15i1.7
Jansen P., Schmelzer G. (2010). Cryptolepis sanguinolenta (Lindl.) schltr. In: Schmelzer G.H., Gurib-Fakim A. (Eds.), Prota 11(2). Medicinal plants/Plantes médicinales 2. [CD-Rom]. PROTA, Wageningen, Netherlands
Jombart T. (2008). Adegenet: a r package for the multivariate analysis of genetic markers. Bioinformatics 24(11):1403–1405. doi: 10.1093/bioinformatics/btn129
Jombart T., Ahmed I. (2011). Adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27(21):3070–3071. doi: 10.1093/bioinformatics/btr521
Kala C. P. (2000). Status and conservation of rare and endangered medicinal plants in the Indian trans-himalaya. Biol. Conserv. 93(3):371–379. doi: 10.1016/S0006-3207(99)00128-7
Kamvar Z. N., Brooks J. C., Grünwald N. J. (2015). Novel r tools for analysis of genome-wide population genetic data with emphasis on clonality. Front. Genet. 208. doi: 10.3389/fgene.2015.00208
Kamvar Z. N., Tabima J. F., Grünwald N. J. (2014). Poppr: an r package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2, e281. doi: 10.7717/peerj.281
Kirby G. C. (1996). Medicinal plants and the control of protozoal disease, with particular reference to malaria. Trans. R. Soc. Trop. Med. Hygiene 90(6):605–609. doi: 10.1016/S0035-9203(96)90404-6
Kling M. M., Ackerly D. D. (2021). Global wind patterns shape genetic differentiation, asymmetric gene flow, and genetic diversity in trees. Proc. Natl. Acad. Sci. 118(17), e2017317118. doi: 10.1073/pnas.2017317118
Licciardi P. V., Underwood J. R. (2011). Plant-derived medicines: a novel class of immunological adjuvants. Int. Immunopharmacol. 11(3):390–398. doi: 10.1016/j.intimp.2010.10.014
Luan S., Chiang T.-Y., Gong X. (2006). High genetic diversity vs. low genetic differentiation in Nouelia insignis (Asteraceae), a narrowly distributed and endemic species in China, revealed by ISSR fingerprinting. Ann. Bot. 98(3):583–589. doi: 10.1093/aob/mcl129
Luo J., Fort D., Carlson T., Noamesi B., Nii-Amon-Kotei D., King S., et al. (1998). Cryptolepis sanguinolenta: An ethnobotanical approach to drug discovery and the isolation of a potentially useful new antihyperglycaemic agent. Diabetic Med. 15(5):367–374. doi: 10.1002/(SICI)1096-9136(199805)15:5<367::AID-DIA576>3.0.CO;2-G
Massebo F., Balkew M., Gebre-Michael T., Lindtjørn B. (2013). Blood meal origins and insecticide susceptibility of Anopheles arabiensis from chano in south-West Ethiopia. Parasites Vectors 6(1):1–10. doi: 10.1186/1756-3305-6-44
Meffe G., Carroll C., contributors (1997). “Conservation management case studies,” in Principles of conservation biology, 2nd ed.Eds. Meffe G. K., Carroll C. R. (Sunderland, Mass: Sinauer Associates), 419–478.
Nie G., Zhang X.-Q., Huang L.-K., Xu W.-Z., Wang J.-P., Zhang Y.-W., et al. (2014). Genetic variability and population structure of the potential bioenergy crop Miscanthus sinensis (Poaceae) in southwest China based on SRAP markers. Molecules 19(8):12881–12897. doi: 10.3390/molecules190812881
Oakley C. G. (2015). The influence of natural variation in population size on ecological and quantitative genetics of the endangered endemic plant Hypericum cumulicola. Int. J. Plant Sci. 176(1):11–19. doi: 10.1086/67794611
Ony M., Klingeman W. E., Zobel J., Trigiano R. N., Ginzel M., Nowicki M., et al. (2021). Genetic diversity in north American Cercis canadensis reveals an ancient population bottleneck that originated after the last glacial maximum. Sci. Rep. 11(1):1–16. doi: 10.1038/s41598-021-01020-z
Osafo N., Mensah K. B., Yeboah O. K. (2017). Phytochemical and pharmacological review of cryptolepis sanguinolenta (Lindl.) schlechter. Adv. Pharmacol. Sci. 2017, 1–13. doi: 10.1155/2017/3026370
Osei-Djarbeng S. N., Agyekum-Attobra E., Nkansah R., Solaga D., Osei-Asante S., Owusu-Dapaah G. (2015). Medicinal plants constituting antimalarial herbal preparations in the ghanaian market. Br. J. Pharm. Res. 5(3):153. doi: 10.9734/BJPR/2015/14896
Peakall R., Smouse P. E. (2006). GENALEX 6: genetic analysis in excel. population genetic software for teaching and research. Mol. Ecol. Notes 6(1):288–295. doi: 10.1111/j.1471-8286.2005.01155.x
Peakall R., Smouse P. E. (2012). GENALEX 6.5: genetic analysis in excel. Population Genet. software Teach. Res. an update. Bioinf. 28(1):2537–2539. doi: 10.1093/bioinformatics/bts460
Piry S., Luikart G., Cornuet J. M. (1999). Computer note. BOTTLENECK: a computer program for detecting recent reductions in the effective size using allele frequency data. J. Heredity 90(4):502–503. doi: 10.1093/jhered/90.4.502
Porebski S., Bailey L. G., Baum B. R. (1997). Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 15(1):8–15. doi: 10.1007/BF02772108
Porras-Hurtado L., Ruiz Y., Santos C., Phillips C., Carracedo Á., Lareu M. (2013). An overview of STRUCTURE: Applications, parameter settings, and supporting software. Front. Genet. 4(98). doi: 10.3389/fgene.2013.00098
Pritchard J. K., Donnelly P. (2001). Case–control studies of association in structured or admixed populations. Theor. Population Biol. 60(3):227–237. doi: 10.1006/tpbi.2001.1543
Pritchard J. K., Stephens M., Donnelly P. (2000). Inference of population structure using multilocus genotype data. Genetics 155(2):945–959. doi: 10.1093/genetics/155.2.945
Ratnaningrum Y. W., Indrioko S., Faridah E., Syahbudin A. (2017). Gene flow and selection evidence of sandalwood (Santalum album) under various population structures in gunung sewu (Java, Indonesia), and its effects on genetic differentiation. Biodiversitas J. Biol. Diversity 18(4):1493–1505. doi: 10.13057/biodiv/d180427
Reed D. H., Frankham R. (2003). Correlation between fitness and genetic diversity. Conserv. Biol. 17(1):230–237. doi: 10.1046/j.1523-1739.2003.01236.x
Rice W. R. (1989). Analyzing tables of statistical tests. Evolution 43(1):223–225. doi: 10.2307/2409177
Romero-Daza N. (2002). Traditional medicine in Africa. Ann. Am. Acad. Political Soc. Sci. 583(1):173–176. doi: 10.1177/000271620258300111
Sawer I., Berry M., Ford J. (2005). The killing effect of cryptolepine on Staphylococcus aureus. Lett. Appl. Microbiol. 40(1):24–29. doi: 10.1111/j.1472-765X.2004.01625.x
Severns P. (2003). Inbreeding and small population size reduce seed set in a threatened and fragmented plant species, lupinus sulphureus ssp. kincaidii (Fabaceae). Biol. Conserv. 110(2):221–229. doi: 10.1016/S0006-3207(02)00191-X
Soares L. A. S. S., Cazetta E., Santos L. R., Franca D., Gaiotto F. A. (2019). Anthropogenic disturbances eroding the genetic diversity of a threatened palm tree: A multiscale approach. Front. Genet. 1090. doi: 10.3389/fgene.2019.01090
Soil Survey Staff (1999). Soil taxonomy: A basic system of soil classification for making and interpreting soil surveys 2nd edition (Natural Resources Conservation Service. U.S.: Department of Agriculture Handbook). 436
Tackie A. N., Sharaf M. H., Schiff P. L. Jr., Boye G. L., Crouch R. C., Martin G. E. (1991). Assignment of the proton and carbon NMR spectra of the indoloquinoline alkaloid cryptolepine. J. Heterocyclic Chem. 28(5):1429–1435. doi: 10.1002/jhet.5570280540
Tempesta M. S. (2010). The clinical efficacy of Cryptolepis sanguinolenta in the treatment of malaria. Ghana Med. J. 44(1):1.
Templeton A. R., Robertson R. J., Brisson J., Strasburg J. (2001). Disrupting evolutionary processes: The effect of habitat fragmentation on collared lizards in the Missouri ozarks. Proc. Natl. Acad. Sci. 98(10):5426–5432. doi: 10.1073/pnas.091093098
Tu Y. (2011). The discovery of artemisinin (qinghaosu) and gifts from Chinese medicine. Nat. Med. 17(10):1217–1220. doi: 10.1038/nm.2471
Turkson B. K., Mensah M., Sarpong K. (2019). Clinical evaluation of efficacy of a herbal formulation used in the treatment of malaria. Eur. J. Med Plants 27, (1–7). doi: 10.9734/ejmp/2019/v27i330117
Veeresham C. (2012). Natural products derived from plants as a source of drugs. J. Advanced Pharm. Technol. Res. 3(4):200. doi: 10.4103/2231-4040.104709
Venables W., Ripley B. D. (2002). Statistics complements to modern applied statistics, 4th ed. Springer, New York
Waples R. S. (1998). Separating the wheat from the chaff: patterns of genetic differentiation in high gene flow species. J. Heredity 89(5):438–450. doi: 10.1093/jhered/89.5.438
WHO (2002). “Traditional medicine-growing needs and potential,” in WHO policy perspectives med, vol. 2. , 6. Geneva
WHO (2014). Global status report on noncommunicable diseases 2014 (Switzerland: World Health Organization).
WHO (2020). The state of food security and nutrition in the world 2020: transforming food systems for affordable healthy diets (Rome: Food & Agriculture Org).
Wright S. (1946). Isolation by distance under diverse systems of mating. Genetics 31(1):39. doi: 10.1093/genetics/31.1.39
Keywords: anti-malarial, indigenous plant, genetic diversity, gene flow, medicinal plants
Citation: Amissah JN, Hadziabdic D, Boggess SL and Trigiano RN (2022) Genetic diversity and population structure of the antimalarial plant Cryptolepis sanguinolenta in Ghana. Front. Conserv. Sci. 3:1020981. doi: 10.3389/fcosc.2022.1020981
Received: 16 August 2022; Accepted: 15 September 2022;
Published: 04 October 2022.
Edited by:
Richard T. Corlett, Xishuangbanna Tropical Botanical Garden (CAS), ChinaReviewed by:
Pe Rajasekharan, Indian Institute of Horticultural Research (ICAR), IndiaEnwei Tian, Southern Medical University, China
Copyright © 2022 Amissah, Hadziabdic, Boggess and Trigiano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jacqueline Naalamle Amissah, am5hbWlzc2FoQHVnLmVkdS5naA==; Denita Hadziabdic, ZGhhZHppYWJAdXRrLmVkdQ==
 Jacqueline Naalamle Amissah
Jacqueline Naalamle Amissah Denita Hadziabdic
Denita Hadziabdic Sarah L. Boggess
Sarah L. Boggess Robert N. Trigiano
Robert N. Trigiano